Abstract
Introduction
The secretory carrier-associated membrane protein 3 (SCAMP3) is a component of post-Golgi membranes, functions as a protein carrier and is critical for subcellular protein transportation. Limited studies revealed an elevated expression of SCAMP3 in breast cancer and hepatocellular carcinoma; however, its role in glioma remains unknown. The aim of our study is to investigate the expression pattern and functional mechanisms of SCAMP3 in glioma.
Methods
mRNA and protein levels of SCAMP3 were examined in glioma tissues together with nontumorous brain tissues by using quantitative real-time-PCR and immunohistochemistry staining. The prognostic role of SCAMP3 in glioma was evaluated through univariate and multivariate analyses. In vitro and in vivo assays were conducted to explore the underlying mechanisms of SCAMP3-induced glioma progression.
Results
The expression level of SCAMP3 was higher in glioma tissues than that in normal brain tissues. High protein level of SCAMP3 was correlated with larger tumor size and advanced WHO grade. Glioma patients with high-SCAMP3 level had worse overall survival. In addition, SCAMP3 was defined as an independent risk factor of glioma prognosis. Cellular and xenograft studies revealed that SCAMP3 promotes glioma proliferation possibly through enhancing EGFR and mTORC1 signaling.
Discussion
Our studies revealed that high-SCAMP3 expression level was closely related to the unfavorable clinical features and poor prognosis of glioma patients. SCAMP3 may serve as an invaluable prognostic indicator and novel therapeutic target for glioma treatment.
Keywords: glioma, SCAMP3, proliferation, prognosis, EGFR
Introduction
Glioma is the most common primary malignancies in the central nervous system of adults, which is characterized with high recurrence incidence and poor prognosis.1 The median overall survival for glioma is approximately 15 months due to its rapid proliferation and metastasis.2 Despite more and more attentions had been paid on glioma mechanisms, the therapeutic effect of radiation and chemotherapy following surgery remains unsatisfied. Glioma is highly heterogeneous which can be induced by various molecule aberrations.3,4 Therefore, identifying dysregulated molecular and elucidating their signaling pathways are invaluable for understanding its progression mechanisms as well as developing novel therapeutic targets.5,6
Besides signaling proteins, intensive studies had explored the nonnegligible role of carrier proteins that modulating protein subcellular trafficking and secretion. For example, the secretory carrier-associated membrane protein (SCAMP) family, which is widely distributed as a component of post-Golgi membranes, functions as protein carriers and are critical for membrane protein trafficking and recycling.7 Interestingly, accumulating evidence are revealing the tumor-related role of SCAMPs. For example, the SCAMP1 was reported to prevent invasion of breast cancer via enhancing protein trafficking of MTSS1 (metastasis suppressor protein 1), consequently promoted tumor cell apoptosis.8 In contrast with SCAMP1, SCAMP3 was identified to be upregulated in hepatocellular carcinoma and seemed like a pro-oncogenic protein,9 although both SCAMP1 and SCAMP3 participate in EGFR signaling.10 Nevertheless, whether SCAMP3 plays roles in glioma have not been determined.
The present study initially investigated the expression pattern of SCAMP3 in clinical glioma tissues from both mRNA and protein levels. Our data demonstrated that SCAMP3 is remarkably upregulated in gliomas compared with normal brain tissues. Furthermore, univariate and multivariate analyses revealed SCAMP3 as an independent risk factor for glioma prognosis. Cellular assays implicated a significant effect of SCAMP3 on enhancing glioma cell proliferation. Finally, we explored the potential underlying mechanisms of SCAMP3-induced glioma proliferation via cellular and xenograft strategies.
Methods
Patients and Samples
This study was approved by the Ethics Committee of the Yantai Affiliated Hospital of Binzhou Medical University. Written informed consents were obtained from all patients or direct relatives. Formalin-fixed paraffin-embedded tissue samples were collected from those underwent surgical resection during 2010–2017 (n=107). All the enrolled patients satisfied the following criteria: 1) did not receive adjunctive therapy before surgery; 2) underwent gross total resection (GTR) or subtotal resection (STR) surgical treatment; 3) survive for at least 6 months since the date of surgery to exclude surgery-related death; and 4) follow-up information was intact. Another 14 fresh-frozen glioma samples were also collected in our Hospital. Parallelly, we purchased normal brain tissue samples from Cureline (South San Francisco, USA; n=14), a global human biospecimens biotech company with local IRBs and ethical committees' approval.
Antibodies and Materials
Mouse EGF and rapamycin were purchased from Invitrogen (Carlsbad, CA, USA) and used at a working concentration of 100ng/mL and 20nM, respectively. Gefitinib was purchased from VWR (Philadelphia, PA, USA) and used at a working concentration of 1μM. The pcDNA3.0-SCAMP3 plasmid construct was generated as described by others.11 The sequence of SCAMP3-siRNA oligonucleotide was 5′-CCUAAGAACUAUGGCUCAUTT-3′.12 The antibodies used in this study include anti-SCAMP3 (Proteintech, 26888-1-AP), anti-Ki67 (Proteintech, 27309-1-AP), anti-EGFR, anti-p70-S6K (Cell Signaling, cat# 2708), anti-phospho-p70-S6K (Cell Signaling, cat# 9234), anti-4E-BP1 (Cell Signaling, cat# 9644), anti-phospho-4E-BP1 (Cell Signaling, cat# 2855), anti-GAPDH (Cell Signaling, cat# 5174).
RNA Isolation and Quantitative Real-Time PCR
Quantitative real-time PCR was performed on RNAs extracted from gliomas or purchased normal brain tissues. Total RNA was isolated according to the manufacturer’s protocol.13 The concentration and purity of RNA samples were tested using the A260–A280 nm absorption ratio. The quantitative real-time PCR was conducted using followed primers obtained from Origene (Rockville, MD, USA):
SCAMP3 -Forward: 5′- GGATGTGCTCTTTGTCCTCCAG-3′
SCAMP3-Reverse: 5′- GCACAGCAATGCCAGTGAAGAG-3′
GAPDH-Forward: 5′- CGAGATCCCTCCAAAATCAA −3′
GAPDH-Reverse: 5′- TTCACACCCATGACGAACAT −3′
Immunohistochemistry (IHC) and IHC Analysis
The IHC staining was performed for all the 107 glioma FFPE tissues using the standard method described by others.14 The expression of SCAMP3 was evaluated independently by two pathologists based on both the staining intensity and positive percentage. Briefly, the staining intensity was scored as 1 (weak staining), 2 (moderate staining), 3 (strong staining). The percentage of positive stained cells was scored as 1 (0–25% positive), 2 (26–50% positive), 3 (50–75% positive), and 4 (76–100% positive). The expression level of SCAMP3 was finally evaluated by multiplying the two scores above (range 1–12). Similarly, the Ki-67 levels in xenografts were evaluated by IHC based on the percentage of positive stained cells.
Cell Culture and Transfection
Human glioma cell line U251 was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). U251 cells were cultured in DMEM supplemented with 10% fetal bovine serum (Gibco, Gaithersburg, MD, USA) and 1% penicillin/streptomycin under 5% CO2 in a 37-degree incubator. U251 cells were transiently transfected with pcDNA3.0-SCAMP3 plasmid or SCAMP3-siRNA, which was achieved by using Lipofectamine 2000 (Invitrogen, Pittsburgh, PA, USA) according to the manufacturer’s instructions. The transfection reagent was used as negative control. U251 cells stably transfected with pcDNA3.0-SCAMP3 were selected by using G418 antibiotics, while U251 cells with stably SCAMP3-knockdown was achieved by using SCAMP3-shRNA (Santa Cruz Biotechnology, cat #sc-41294-SH) transfection.
Cell Proliferation Assay
The proliferation capacity of U251 cells was assessed using (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (MTS reagent) (Abcam, USA). In this assay, 103 transfected cells were seeded into a 96-well plate in 100 μL DMEM and measurements were performed 24, 48, 72 and 96 h after seeding according to the manufacturer’s protocol. In brief, 20 μL MTS reagent was added to each well 3 hours before the designated time point. The absorbance was measured at 490 nm using a microplate reader. Each experiment was performed in triplicate and repeated three times.
Colony Formation Assay
Briefly, 103 transfected cells were seeded into a well of a six-well plate and cultured in a 37-degree incubator for 2 weeks. Then, the cells were fixed with formalin for 30 min and stained with 0.1% crystal violet for 15 min. The number of formed colonies was calculated. Each experiment was performed in triplicate and repeated three times.
Migration and Invasion Assays
Cell migration experiments were performed using 24-well plates with 8.0-μm-pore transwell chambers (Corning, NY, USA). Approximately 2×104 transfected cells were seeded into the upper chambers, and 600 μL complete DMEM was added into the lower chambers. After 24 hours of incubation, the cells attached to the bottom of chambers were fixed with methanol, stained with 0.5% crystal violet and counted in 5 random microscopy fields. The invasion assay was conducted similarly except that the chambers were pre-coated with 2.5 mg/mL Matrigel.
Western Blot
Total protein in cultured cells was extracted with RIPA lysis buffer containing protease inhibitors and phosphatase inhibitors. Approximately 20 μg of total protein was subjected to Western blot by separating them in 10% SDS-PAGE and electro-transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). Membranes were blocked with 5% FBS for 1 hour and then incubated with the primary antibodies as described above overnight. Then, the membranes were incubated with corresponding secondary antibodies and the enhanced chemiluminescence (ECL) was added to detect specific bands using films.
Xenograft Model of Glioma in Nude Mice
Male BALB/c nu/nu mice (5 weeks old) were purchased from Shanghai Institute of Materia Medical, Chinese Academy of Sciences, and housed under specific pathogen-free conditions. The experimental protocol was conducted under the supervision of Binzhou Medical University Animal Center. Mice were randomly assigned to experimental group and control group, and 5×106 stable-transfected cells were subcutaneously implanted into mice. Tumor volumes were measured every 5 days to obtain the growth curve. After 20 days, all mice were sacrificed, and the xenografts were isolated and weighed.
Statistical Analysis
Statistical analysis was performed with SPSS 20.0 (SPSS Inc., Chicago, IL, USA). Overall survival time was expressed as the mean ± SD (standard deviation) or median (interquartile range). The correlations between SCAMP3 expression and patients’ clinicopathological characteristics were tested by using the χ2 test. Univariate survival analysis was performed using the Kaplan–Meier method, and the significance of the difference between the groups was analyzed using the Log-rank test. The independent prognostic significance of each variable was assessed using Cox proportional-hazards regression model. Data from cellular and xenograft experiments were compared by Student’s t-test. All statistical tests were two-tailed and P<0.05 was considered as statistically significant.
Results
Patients’ Information
Of the 107 enrolled glioma patients, there were 42 females (39.3%) and 65 males (60.7%) with the age ranged from 22 to 67 years old at the time of diagnosis. There were 67 cases (59.8%) with the largest tumor diameter less than 5.0 cm, while the other 43 cases (40.2%) with a tumor size less than 5.0 cm. Among all the patients, only 21 cases (19.6%) were classified as WHO grade II, 34 cases (31.8%) with WHO grade III, and the other 52 cases (48.6%) with WHO grade IV. We also assessed the disease severity by calculating Karnofsky Performance Score (KPS),15 which indicated that 65 patients (60.7%) with KPS score less than 90, and 42 cases (39.3%) with KPS score larger than 90. The surgery pattern of our cohort was also retrieved, showing that 84 patients (78.5%) underwent gross total resection, and the other 23 cases (21.5%) underwent subtotal resection. Patients’ clinicopathological information was summarized in Table 1. The follow-up time was ranged from 6 to 72 months.
Table 1.
Clinicopathological Characteristics of Glioma Patients
| Variables | Cases | Percentage |
|---|---|---|
| Sex | ||
| Female | 42 | 39.3 |
| Male | 65 | 60.7 |
| Age (Years) | ||
| ≤50 | 50 | 46.7 |
| >50 | 57 | 53.3 |
| Tumor Size | ||
| ≤5 cm | 64 | 59.8 |
| >5 cm | 43 | 40.2 |
| WHO Grade | ||
| II | 21 | 19.6 |
| III | 34 | 31.8 |
| IV | 52 | 48.6 |
| KPS Score | ||
| ≤90 | 65 | 60.7 |
| >90 | 42 | 39.3% |
| Resection Pattern | ||
| GTR | 84 | 78.5 |
| STR | 23 | 21.5 |
| SCAMP3 Level | ||
| Low | 63 | 58.9 |
| High | 44 | 41.1 |
Abbreviations: SCAMP3, secretory carrier-associated membrane protein 3; KPS, Karnofsky performance score; GTR, gross total resection; STR, subtotal resection.
SCAMP3 Expression Is Upregulated in Gliomas and Correlated with Tumor Size
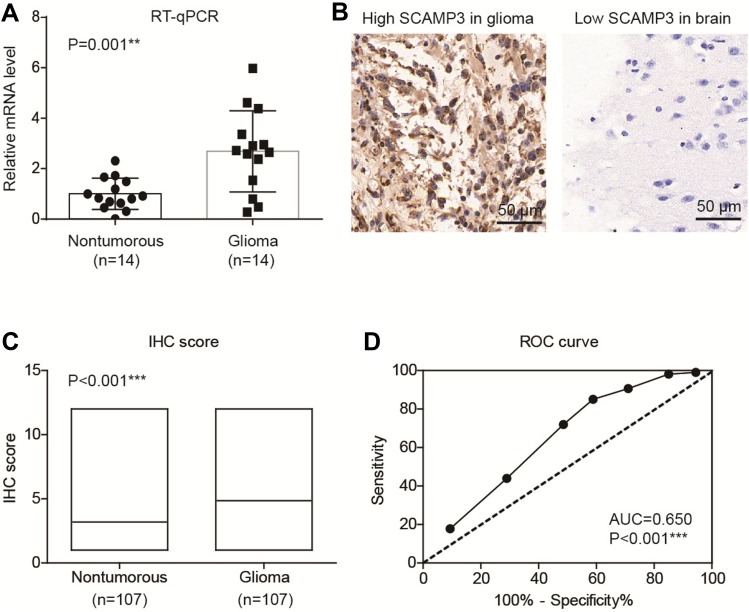
By performing RT-qPCR targeting SCAMP3 in fresh-frozen glioma and normal brain tissues, we found that SCAMP3-mRNA was approximately 2.7 folds higher in gliomas than that in normal brains (Figure 1A, P=0.001). Therefore, we tested the protein expression of SCAMP3 in the FFPE tissues by the IHC method, which demonstrated its cytoplasm and membrane localization (Figure 1B). Consistent with RT-qPCR data, the mean IHC score of gliomas is remarkably higher than that of nontumorous tissues (Figure 1C, P<0.001). By generating the receiver operating characteristic (ROC) curve based on the IHC scores (Figure 1D), we classified patients into low-SCAMP3 group (IHC score <6) and high-SCAMP3 group (IHC score ≥6). Accordingly, 63 patients were sub-grouped as low-SCAMP3 expression and the other 44 patients as high-SCAMP3 expression.
Figure 1.
SCAMP3 is upregulated in glioma tissues. (A) RT-qPCR results demonstrated that SCAMP3-mRNA is higher in gliomas compared with normal brain tissues. (B) Representative IHC images of SCAMP3 staining in gliomas and brain tissues. Magnification: 400×. (C) The IHC score of SCAMP3 was significantly higher in gliomas than that in nontumorous tissues. (D) ROC curve was plotted to identify the cutoff value to divide patients into high-SCAMP3 group and low-SCAMP3 group based on the IHC scores. The area under curve is 0.650 and IHC score=5.0 was identified as the cutoff score. **P<0.01 by Student’s t-test, ***P<0.001 by Student’s t-test.
We tested the correlations between SCAMP3 expression and clinicopathological features of all the patients (Table 2). A significant correlation between tumor size and SCAMP3 was observed, showing that patients with higher SCAMP3 are more likely to possess larger tumor size (P=0.011). Similarly, higher SCAMP3 is positively correlated with advanced WHO grade (P=0.010). The high-SCAMP3 percentage is only 23.8% and 29.4% in WHO grade II and grade III patients, respectively. However, up to 55.8% WHO grade III patients showed high-SCAMP3 level, indicating the pro-oncogenic effect of SCAMP3 in glioma.
Table 2.
Correlation Between SCAMP3 and Clinicopathological Characteristics of Glioma Patients
| Variables | Cases | SCAMP3 Expression | P value | |
|---|---|---|---|---|
| (n = 107) | Low (n = 63) | High (n = 44) | ||
| Sex | 0.769 | |||
| Female | 42 | 24 | 18 (42.9%) | |
| Male | 65 | 39 | 26 (40.0%) | |
| Age (years) | 0.176 | |||
| ≤50 | 50 | 26 | 24 (48.0%) | |
| >50 | 57 | 37 | 20 (35.1%) | |
| Tumor Size | 0.011* | |||
| ≤5 cm | 64 | 44 | 20 (31.3%) | |
| >5 cm | 43 | 19 | 24 (55.8%) | |
| WHO Grade | 0.010* | |||
| II | 21 | 16 | 5 (23.8%) | |
| III | 34 | 24 | 10 (29.4%) | |
| IV | 52 | 23 | 29 (55.8%) | |
| KPS Score | 0.609 | |||
| ≤90 | 65 | 37 | 28 (43.1%) | |
| >90 | 42 | 26 | 16 (38.1%) | |
| Resection Pattern | 0.461 | |||
| GTR | 84 | 51 | 33 (39.3%) | |
| STR | 23 | 12 | 11 (47.8%) | |
Note: *P<0.05 by Chi-square test.
Abbreviations: SCAMP3, secretory carrier-associated membrane protein 3; KPS, Karnofsky performance score; GTR, gross total resection; STR, subtotal resection.
Higher SCAMP3 Indicates Unfavorable Overall Survival of Glioma Patients
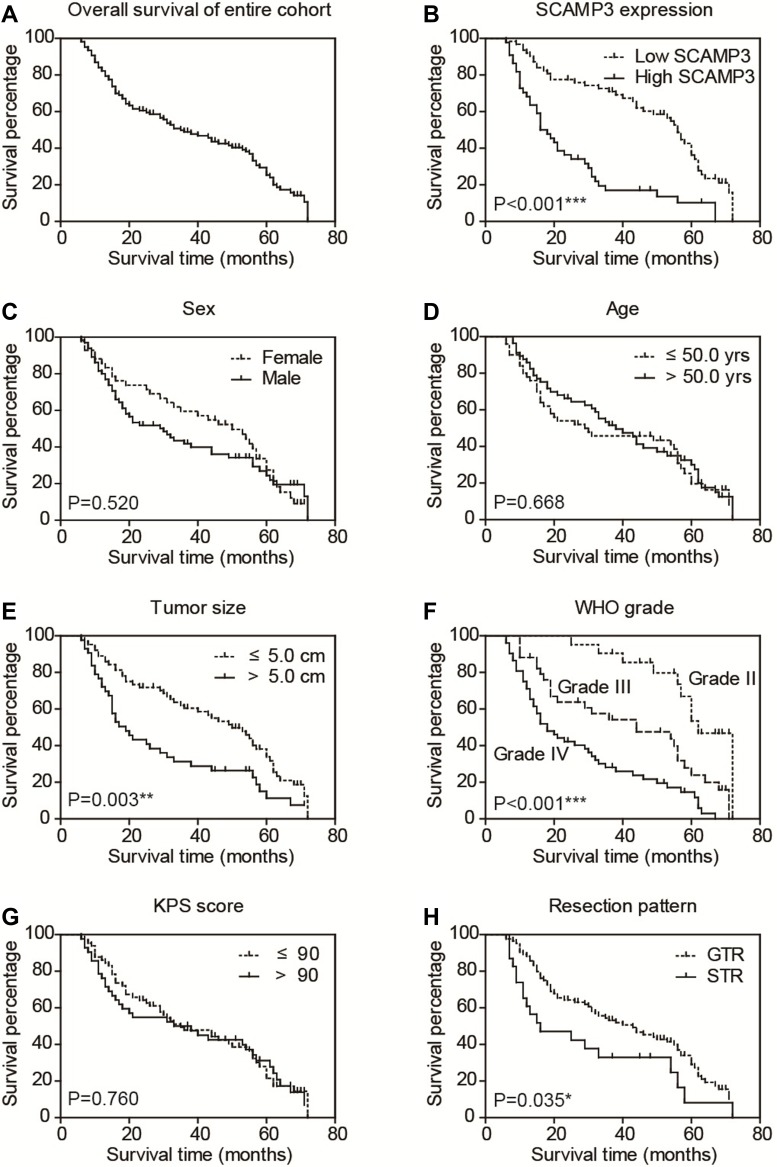
To illustrate the clinical relevance of SCAMP3 expression in glioma, overall survival data were assessed using the Kaplan–Meier method. The overall survival time of our entire cohort was 35 months and the 3-year overall survival rate was 48.84% (Figure 2A). Of note, survival analysis suggested that patients with high-SCAMP3 expression in glioma tissue had shorter overall survival time than those with low expression (Figure 2B, P<0.001). The 3-year overall survival rate was 71.0% for low-SCAMP3 patients while 17.0% for high-SCAMP3 patients (Table 3). The prognostic effects of other related variables were also analyzed (Figure 2C–H), which indicated significant effects of tumor size (P=0.003), WHO grade (P<0.001), and surgery pattern (P=0.035). Briefly, patients with larger tumor size, with advanced WHO grade, or who underwent subtotal resection instead of gross total resection showed poorer clinical outcomes. In contrast, no statistically significant effect was observed regarding patients’ sex, age, or KPS score.
Figure 2.
Overall survival of glioma patients. The overall survival curve of our entire cohort was plotted by Kaplan–Meier analysis (A). Besides, the prognostic effect of each variable was assessed by Log-rank test, including SCAMP3 level (B), patients’ sex (C), age (D), tumor size (E), WHO grade (F), KPS score (G) and resection pattern (H), respectively. *P<0.05 by Log-rank test, **P<0.01 by Log-rank test, ***P<0.001 by Log-rank test.
Table 3.
Overall Survival of Glioma Patients by Univariate Analysis
| Variables | Patients | Overall Survival Time (Months) | P value | |
|---|---|---|---|---|
| (n, %) | Mean ± SD | 3-Year OS | ||
| Sex | 0.520 | |||
| Female | 42 (39.3%) | 42.6 ± 3.4 | 59.5% | |
| Male | 65 (60.7%) | 35.8 ± 3.1 | 48.5% | |
| Age (Years) | 0.668 | |||
| ≤50 | 50 (46.7%) | 36.8 ± 3.5 | 45.8% | |
| >50 | 57 (53.3%) | 39.8 ± 3.1 | 51.6% | |
| Tumor Size | 0.003** | |||
| ≤5 cm | 64 (59.8%) | 44.5 ± 2.9 | 60.4% | |
| >5 cm | 43 (40.2%) | 29.3 ± 3.4 | 31.3% | |
| WHO Grade | <0.001*** | |||
| II | 21 (19.6%) | 60.7 ± 3.3 | 90.5% | |
| III | 34 (31.8%) | 41.4 ± 4.0 | 54.3% | |
| IV | 52 (48.6%) | 27.4 ± 2.8 | 28.2% | |
| KPS Score | 0.760 | |||
| ≤90 | 65 (60.7%) | 39.2 ± 2.9 | 47.8% | |
| >90 | 42 (39.3%) | 37.3 ± 3.9 | 50.0% | |
| Resection Pattern | 0.035* | |||
| GTR | 84 (78.5%) | 40.8 ± 2.5 | 53.2% | |
| STR | 23 (21.5%) | 29.4 ± 5.0 | 33.0% | |
| SCAMP3 Expression | <0.001*** | |||
| Low | 63 (58.9%) | 48.0 ± 2.8 | 71.0% | |
| High | 44 (41.1%) | 24.7 ± 2.9 | 17.0% | |
Notes: *P<0.05, **P<0.01, ***P<0.001 by Log-rank test.
Multivariate analysis was further conducted to identify independent prognostic factors by subjecting those significant variables of Kaplan–Meier analysis into a Cox regression model (Table 4). Accordingly, high-SCAMP3 level was confirmed as an independent risk factor (HR=1.996, 95% CI 1.184–3.366, P=0.010). Besides, advanced WHO grade (HR=1.827, 95% CI 1.411–2.365, P<0.001) and subtotal surgical resection (HR=2.655, 95% CI 1.463–4.817, P=0.001) were also independently associated with poorer prognosis.
Table 4.
Multivariate Analysis of the OS of Glioma Patients
| Variables | Hazard Ratio | 95% Confidence Interval | P value |
|---|---|---|---|
| Tumor size (>5 cm vs ≤5 cm) |
1.474 | 0.900–2.416 | 0.123 |
| WHO grade (IV vs II–III) |
1.827 | 1.411–2.365 | <0.001*** |
| Resection pattern (STR vs ≤GTR) |
2.655 | 1.463–4.817 | 0.001** |
| SCAMP3 expression (High vs Low) |
1.996 | 1.184–3.366 | 0.010* |
Notes: *P<0.05, **P<0.01, ***P<0.001 by Cox regression test.
Abbreviations: SCAMP3, secretory carrier-associated membrane protein 3; GTR, gross total resection; STR, subtotal resection.
SCAMP3 Facilitates Glioma Cell Proliferation Through EGFR and mTORC1 Signaling Pathways
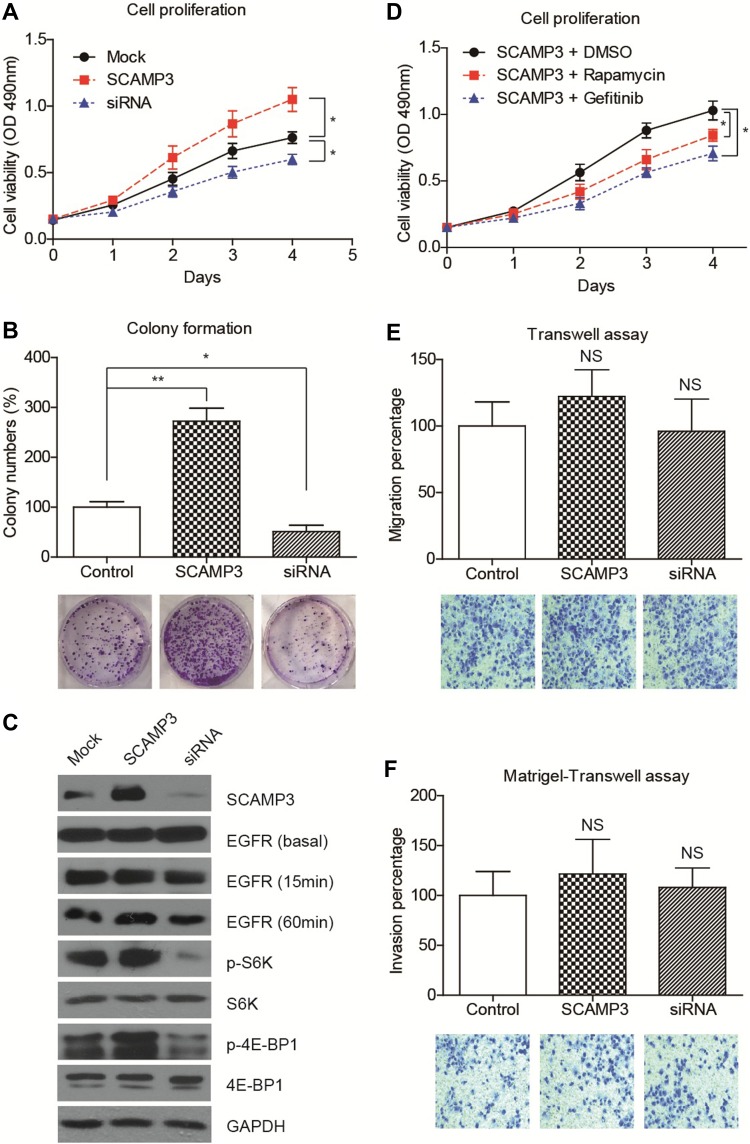
Considering that SCAMP3 expression is associated with patient survival in human glioma, we sought to further understand the potential role of SCAMP3 in the regulation of glioma cells’ biological phonotype. Firstly, an MTS assay was performed to evaluate the potential effect of SCAMP3-overexpression or SCAMP3-silencing on the proliferation of glioma cell line U251 (Figure 3A). Our results showed that overexpression of SCAMP3 promoted the proliferation of U251 cells. In contrast, knockdown of SCAMP3 induced an inhibited proliferation curve compared with control group. Consistent with the proliferation curve, colony formation assays also revealed that the ability of colony formation was enhanced in SCAMP3-overexpressing cells while decreased in SCAMP3-silencing cells (Figure 3B). The transfection efficiency of overexpression and knockdown was validated by Western blot (Figure 3C). Since EGFR is a critical protein that participates in glioma progression and previous studies showed that SCAMP3 regulates EGFR recycling in HEK293 cells,11 we were interested to test whether the EGFR signaling was affected by SCAMP3 in glioma U251 cells. There was no difference regarding the basal EGFR level nor its level after 15 min EGF stimulation. However, after stimulated with EGF for 60 min, the EGFR protein level was significantly lower in SCAMP3-siRNA cells than that in the SCAMP3-overexpression cells (Figure 3C), showing the possibility that SCAMP3 may promote EGFR recycling and escape from degradation after EGF stimulation. Besides EGFR, SCAMP3 was recently reported to be involved in mTORC1 signaling in osteosarcoma Saos-2 cells.16 Therefore, we also tested the alterations of mTORC1 downstream signaling after silencing or overexpressing SCAMP3. Although no difference was observed on total p70-S6K or total 4E-BP1 protein level, their phosphorylation status was significantly upregulated upon overexpressing SCAMP3. As expected, SCAMP3-siRNA transfection showed opposite effects (Figure 3C). Moreover, we conducted rescue experiments by treating SCAMP3-overexpressing cells with rapamycin (mTOR inhibitor) and gefitinib (EGFR inhibitor), respectively. Either inhibiting mTOR or EGFR significantly attenuated the proliferation-promoting effect of SCAMP3 on glioma cells (Figure 3D). It has been well acknowledged that both EGFR and mTORC1 are major contributors for malignancies including glioma, our data provided evidence that SCAMP3 promotes glioma progression at least partially by enhancing EGFR and mTORC1 signaling pathways. In contrast with the significant role on promoting glioma proliferation, SCAMP3 showed no statistically significant effect on the migration or invasion of glioma cells (Figure 3E and 3F).
Figure 3.
SCAMP3 promotes glioma cell proliferation. (A) The proliferation curves of transfected U251 cells were plotted based on MTS data, which indicated that overexpressing SCAMP3 enhanced while silencing SCAMP3 inhibited cell proliferation. (B) Colony formation assays were conducted for transfected U251 cells. Consistent with the proliferation results, SCAMP3 transfection resulted in more colonies while SCAMP3-knockdown attenuated colony formation. (C) Immunoblotting results confirmed the transfection efficiencies of SCAMP3 overexpression and knockdown. Besides, silencing SCAMP3 leads to impaired EGFR recycling, as well as the activation of S6K and 4E-BP1 proteins. (D) U251 cells transfected with SCAMP3 plasmids were further treated with rapamycin or gefitinib and then subjected to MTS assay. Either inhibiting mTOR or EGFR can significantly attenuate the proliferation-promoting effect of SCAMP3 on glioma cells. (E) The alteration of migration capacity by overexpressing or silencing SCAMP3 was tested and revealed no statistical significance. (F) Matrigel-transwell assay demonstrated that SCAMP3 has no significant effect on U251 cell invasion. Data were analyzed by Student’s t-test compared with control group. *P<0.05, **P<0.01. NS, not significant.
SCAMP3 Enhances Glioma Growth by in vivo Assay
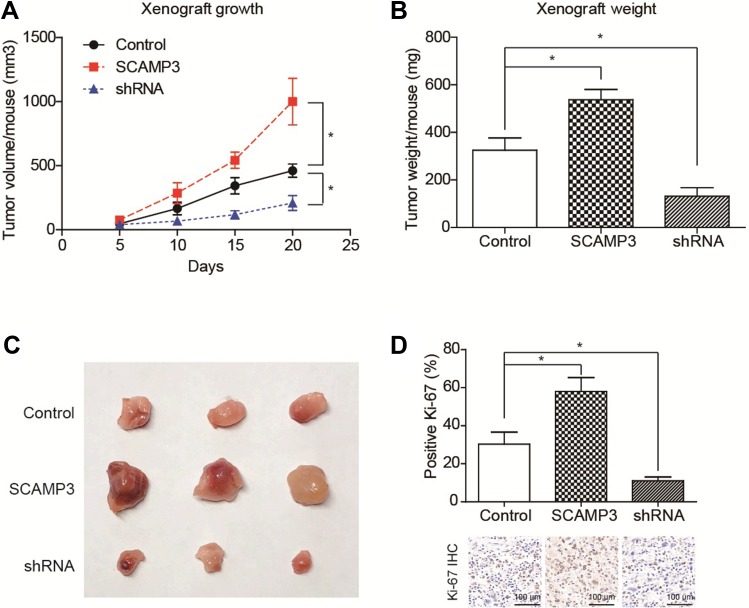
We then established a subcutaneous xenograft tumor model to verify the effects of SCAMP3 on glioma progression in vivo. The tumor growth curve was plotted and showed an enhanced growth rate in the SCAMP3-overexpression group (Figure 4A). Consistently, 20 days after tumor incubation, the tumor volume in the SCAMP3-overexpression group was significantly larger than that of the control group, and the tumor volume in SCAMP3-shRNA group was significantly smaller (Figure 4B and 4C). We further investigated the proliferation ability of glioma cells in these tumor samples by IHC examination targeting Ki-67. The results revealed that Ki-67 expression was markedly enhanced in gliomas with SCAMP3-overexpression. In contrast, the Ki-67 level was decreased in SCAMP3-shRNA group compared with control group (Figure 4D). Taken together, our data indicated that SCAMP3 promoted glioma proliferation both in vitro and in vivo.
Figure 4.
SCAMP3 promotes glioma growth in vivo. (A) The growth curve of implanted xenografts was plotted, which showed that U251-SCAMP3 cells possessed the highest growth rate. In contrast, U251 cells with SCAMP3-shRNA transfection showed impaired growth capacity compared with control cells. (B) The final tumor weight was tested after xenograft isolation and revealed consistent results with growth curves. (C) The photographed images of isolated xenografts. (D) The proliferation index was determined using the percentage of Ki-67 positive cells in isolated xenografts by IHC staining. As a result, SCAMP3-overexpression groups showed highest proliferation index while SCAMP3-shRNA groups showed lowest proliferation index. *P<0.05 by Student’s t-test compared with control group.
Discussions
Rapid proliferation and metastasis are the predominant obstacles troubling the clinical technicians and limiting the outcome of glioma patients. Therefore, identifying the molecule associated with glioma progression will be invaluable for risk evaluation and individual therapy development. Here we showed that SCAMP3 is upregulated in glioma tissues for the first time. The possible tumor-related role of SCAMP3 was initially reported by Skawran B et.al, which showed that SCAMP3 gene transcription was enriched in hepatocellular carcinoma samples by microarray assay.17 Similarly, another study by Naboulsi et al screened the protein alterations of hepatocellular carcinoma by a quantitative proteomic method and demonstrated that SCAMP3 was upregulated compared to nontumorous liver tissues.18 The following IHC experiments also verified a higher level of SCAMP3 in hepatocellular carcinoma, further implying the tumor-related role of this endocytosis-associated protein. The later study by Suárez-Arroyo et al reported that SCAMP3 showed higher levels in the invasive ductal carcinoma as well as inflammatory breast cancer, compared with normal breast tissues by proteomic and IHC methods.19 Although the above data showed that SCAMP3 may be a candidate oncogene in different malignancies, no further validation test was conducted regarding its clinical significance nor oncogenic mechanism. Our data emphasized the critical role of SCAMP3 not only by confirming its upregulation in glioma tissues, but also by analyzing its positive correlation with tumor size and WHO grade.
Of note, a previous study by Ghosh et al demonstrated that SCAMP3 expression was elevated in the cancer stem cells of glioblastoma, in comparison with its expression in healthy neural stem cells.20 Taking into consideration that cancer stem cells are characterized by high proliferative capability, which is responsible for oncogenesis and progression of cancers, the elevated SCAMP3 in glioblastoma cancer stem cells may be involved in its tumorigenesis process. Therefore, we next verified its pro-oncogenic effect by ectopic overexpression of SCAMP3 or silencing SCAMP3 in U251 human glioma cells. As a result, its proliferation-related role was validated although no significant effect was observed on metastasis. Consistent with our data in glioma cell line, another group recently reported that SCAMP3 knockdown significantly suppressed cell growth but not cell motility abilities of melanoma.21 Additionally, our in vivo results further confirmed the significant effect of SCAMP3 on promoting glioma growth in xenografts.
To date, no molecular mechanism underlying the role of SCAMP3 in tumors was reported. However, the relationship between SCAMP3 with EGFR has been demonstrated in HEK293 cells, on that SCAMP3 facilitates EGFR escaping from proteome-dependent degradation, consequently enhances EGFR recycling.11 Therefore, we tested whether there exists crosstalk between EGFR and SCAMP3 in glioma cells. Our data showed that silencing SCAMP3 has no effect on basal EGFR, but indeed decreased the EGFR level after long time stimulation by EGF, revealing its possible role in modulating EGFR recycling and signaling. Besides EGFR, the activation of mTORC1 was recently reported to be correlated with SCAMP3 in osteosarcoma cells,16 thus promoted us to test alteration of mTORC1 downstream signaling. In accordance with the data in osteosarcoma cells, overexpressing SCAMP3 remarkably resulted in a hyperphosphorylated status of both S6K and 4E-BP1 proteins, the two most well-known mTORC1 downstream effectors. Moreover, either EGFR or mTOR inhibitor can attenuate the SCAMP3-induced cell proliferation. Taken together, our cellular data implied that SCAMP3 may exert oncogenic effect by facilitating EGFR and mTORC1 signaling pathways. These findings also provide evidence for combination therapy development considering the intensive attentions drawn by EGFR inhibitors in glioma treatment.22,23
Our study has several limitations. Firstly, the survival data were obtained from a single medical center and may lead to regional bias. More clinicopathological information and a larger cohort from multiple hospitals will be helpful for better validating our conclusion. Secondly, whether SCAMP3 enhances cell cycle or attenuates cell apoptosis needs additional investigation. Thirdly, the detailed and individual effect of EGFR and mTORC1 in SCAMP3-induced glioma proliferation remains unknown. Further cellular experiments will be necessary to fully illustrate the underlying mechanisms of SCAMP3 in promoting glioma progression. Nevertheless, here we provided the initial evidence about the potential roles of SCAMP3 in glioma progression from clinical, cellular, and animal aspects.
Conclusions
In conclusion, our results established an oncogenic role of SCAMP3 in the progression of glioma and verified that SCAMP3 is a novel predictor for overall survival of glioma patients. Mechanism studies indicated that SCAMP3 can enhance the proliferation of glioma by upregulating EGFR and mTORC1 signaling both in vitro and in vivo. Our data will not only be helpful for glioma prognosis prediction but also provide evidence for its potential as a novel therapeutic target.
Acknowledgment
This study was supported by the grant of Medical Health Technology Development Plan of Shandong Province (2018WS536, China).
Data Sharing Statement
Data will be available upon request.
Ethics and Consent Statement
This research was conducted ethically in accordance with the World Medical Association Declaration of Helsinki for human sample analyses. Subjects have given their written informed consent and the study protocol was approved by the Ethics Committee of the Yantai Affiliated Hospital of Binzhou Medical University. The animal experiments were conducted according to “Guide for the Care and Use of Laboratory Animals, 8th edition” under the supervision of Binzhou Medical University Animal Center.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Weller M, Wick W, Aldape K, et al. Glioma. Nat Rev Dis Primers. 2015;1(1):15017. doi: 10.1038/nrdp.2015.17 [DOI] [PubMed] [Google Scholar]
- 2.Kline C, Felton E, Allen IE, Tahir P, Mueller S. Survival outcomes in pediatric recurrent high-grade glioma: results of a 20-year systematic review and meta-analysis. J Neurooncol. 2018;137(1):103–110. doi: 10.1007/s11060-017-2701-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molinaro AM, Taylor JW, Wiencke JK, Wrensch MR. Genetic and molecular epidemiology of adult diffuse glioma. Nat Rev Neurol. 2019;15(7):405–417. doi: 10.1038/s41582-019-0220-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melin BS, Barnholtz-Sloan JS, Wrensch MR, et al. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49(5):789. doi: 10.1038/ng.3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gessler F. Molecular Regulation of Glioma Cell Migration: Assessment and Therapeutic Implications. University of Cambridge; 2019. [Google Scholar]
- 6.Duan J, Huang W, Shi H. Positive expression of KIF20A indicates poor prognosis of glioma patients. Onco Targets Ther. 2016;9:6741–6749. doi: 10.2147/ott.s115974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernández-Chacón R, Achiriloaie M, Janz R, Albanesi JP, Südhof TC. SCAMP1 function in endocytosis. J Biol Chem. 2000;275(17):12752–12756. doi: 10.1074/jbc.275.17.12752 [DOI] [PubMed] [Google Scholar]
- 8.Vadakekolathu J, Al-Juboori SIK, Johnson C, et al. MTSS1 and SCAMP1 cooperate to prevent invasion in breast cancer. Cell Death Dis. 2018;9(3):344. doi: 10.1038/s41419-018-0364-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Sheng J, Zhang Y, et al. Overexpression of SCAMP3 is an indicator of poor prognosis in hepatocellular carcinoma. Oncotarget. 2017;8(65):109247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu TT, Castle JD. Tyrosine phosphorylation of selected secretory carrier membrane proteins, SCAMP1 and SCAMP3, and association with the EGF receptor. Mol Biol Cell. 1998;9(7):1661–1674. doi: 10.1091/mbc.9.7.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoh QL, Castle AM, Hubbard CH, Katsumata O, Castle JD. SCAMP3 negatively regulates epidermal growth factor receptor degradation and promotes receptor recycling. Mol Biol Cell. 2009;20(6):1816–1832. doi: 10.1091/mbc.e08-09-0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tseng H-W, Li S-C, Tsai K-W. Metformin treatment suppresses melanoma cell growth and motility through modulation of microRNA expression. Cancers. 2019;11(2):209. doi: 10.3390/cancers11020209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, Fan H, Liu H, et al. WNT5B exerts oncogenic effects and is negatively regulated by miR-5587-3p in lung adenocarcinoma progression. Oncogene. 2019. doi: 10.1038/s41388-019-1071-4 [DOI] [PubMed] [Google Scholar]
- 14.Zhang Q, Fan H, Zou Q, et al. TEAD4 exerts pro-metastatic effects and is negatively regulated by miR6839-3p in lung adenocarcinoma progression. J Cell Mol Med. 2018;22(7):3560–3571. doi: 10.1111/jcmm.13634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karnofsky DA, Abelmann WH, Craver LF, Burchenal JH. The use of the nitrogen mustards in the palliative treatment of carcinoma. With particular reference to bronchogenic carcinoma. Cancer. 1948;1(4):634–656. doi: [DOI] [Google Scholar]
- 16.Beaumatin F, O’Prey J, Barthet VJ, et al. mTORC1 activation requires DRAM-1 by facilitating lysosomal amino acid efflux. Mol Cell. 2019;76(1):163–76. e8. doi: 10.1016/j.molcel.2019.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skawran B, Steinemann D, Weigmann A, et al. Gene expression profiling in hepatocellular carcinoma: upregulation of genes in amplified chromosome regions. Mod Pathol. 2008;21(5):505–516. doi: 10.1038/modpathol.3800998 [DOI] [PubMed] [Google Scholar]
- 18.Naboulsi W, Bracht T, Megger DA, et al. Quantitative proteome analysis reveals the correlation between endocytosis-associated proteins and hepatocellular carcinoma dedifferentiation. Biochim Biophys Acta. 2016;1864(11):1579–1585. doi: 10.1016/j.bbapap.2016.08.005 [DOI] [PubMed] [Google Scholar]
- 19.Suarez-Arroyo IJ, Feliz-Mosquea YR, Perez-Laspiur J, et al. The proteome signature of the inflammatory breast cancer plasma membrane identifies novel molecular markers of disease. Am J Cancer Res. 2016;6(8):1720–1740. [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh D, Ulasov IV, Chen L, et al. TGFbeta-responsive HMOX1 expression is associated with stemness and invasion in glioblastoma multiforme. Stem Cells. 2016;34(9):2276–2289. doi: 10.1002/stem.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tseng HW, Li SC. Metformin treatment suppresses melanoma cell growth and motility through modulation of microRNA expression. Cancers. 2019;11(2):209. doi: 10.3390/cancers11020209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neyns B, Sadones J, Joosens E, et al. Stratified Phase II trial of cetuximab in patients with recurrent high-grade glioma. Ann Oncol. 2009;20(9):1596–1603. doi: 10.1093/annonc/mdp032 [DOI] [PubMed] [Google Scholar]
- 23.Freeman AC, Platt SR, Holmes S, et al. Convection-enhanced delivery of cetuximab conjugated iron-oxide nanoparticles for treatment of spontaneous canine intracranial gliomas. J Neurooncol. 2018;137(3):653–663. doi: 10.1007/s11060-018-2764-1 [DOI] [PubMed] [Google Scholar]