Highlights
-
•
Software packages were applied to mitigate the effects of artifacts and to produce robust tensor estimation.
-
•
Opposite phase-encoding directions were used in DTI acquisition to improve correction for EPI distortions.
-
•
Advanced tensor-based registration of DTI images was obtained using a population-specific template.
Keywords: Diffusion tensor imaging (DTI), Nonlinear registration, Tract-based spatial statistics (TBSS), Pediatric
Abstract
Analysis of scalar maps obtained by diffusion tensor imaging (DTI) produce valuable information about the microstructure of the brain white matter. The DTI scanning of child populations, compared with adult groups, requires specifically designed data acquisition protocols that take into consideration the trade-off between the scanning time, diffusion strength, number of diffusion directions, and the applied analysis techniques. Furthermore, inadequate normalization of DTI images and non-robust tensor reconstruction have profound effects on data analyses and may produce biased statistical results. Here, we present an acquisition sequence that was specifically designed for pediatric populations, and describe the analysis steps of the DTI data collected from extremely preterm-born young school-aged children and their age- and gender-matched controls. The protocol utilizes multiple software packages to address the effects of artifacts and to produce robust tensor estimation. The computation of a population-specific template and the nonlinear registration of tensorial images with this template were implemented to improve alignment of brain images from the children.
Graphical abstract

Specifications table
| Subject Area | Neuroscience |
| More specific subject area | Neuroimaging/ Brain imaging |
| Protocol name | DTI data analysis protocol for children |
| Reagents/tools | The scanning was performed using a 3 T MAGNETOM Skyra scanner (Siemens Healthcare, Erlangen, Germany) and a 30-channel head coil. |
| Experimental design | Diffusion-weighted images were acquired using a spin-echo-based single shot EPI sequence with full k-space coverage and GRAPPA parallel acquisition option (TR 9000 ms, TE 80 ms, FOV 240 mm, matrix size 96 × 96, slice thickness 2.5 mm, 70 contiguous axial slices, acceleration factor 2). The DTI dataset (45 vol: 30 uniformly distributed diffusion gradient directions at b = 1000 s/mm2, 6 directions at b = 500 s/mm2, 6 directions at b = 300 s/mm2, and 3 non diffusion-weighted images at b = 0 s/mm2) was collected twice with reversed phase-encoding directions (anterior-posterior and posterior-anterior). |
| Trial registration | |
| Ethics | Ethical approval for the study was obtained from the Ethics Committee I of the Hospital District of Helsinki and Uusimaa. All children gave assent and caregivers provided informed written consent prior to participation in accordance with the Declaration of Helsinki. |
| Value of the Protocol |
|
Background
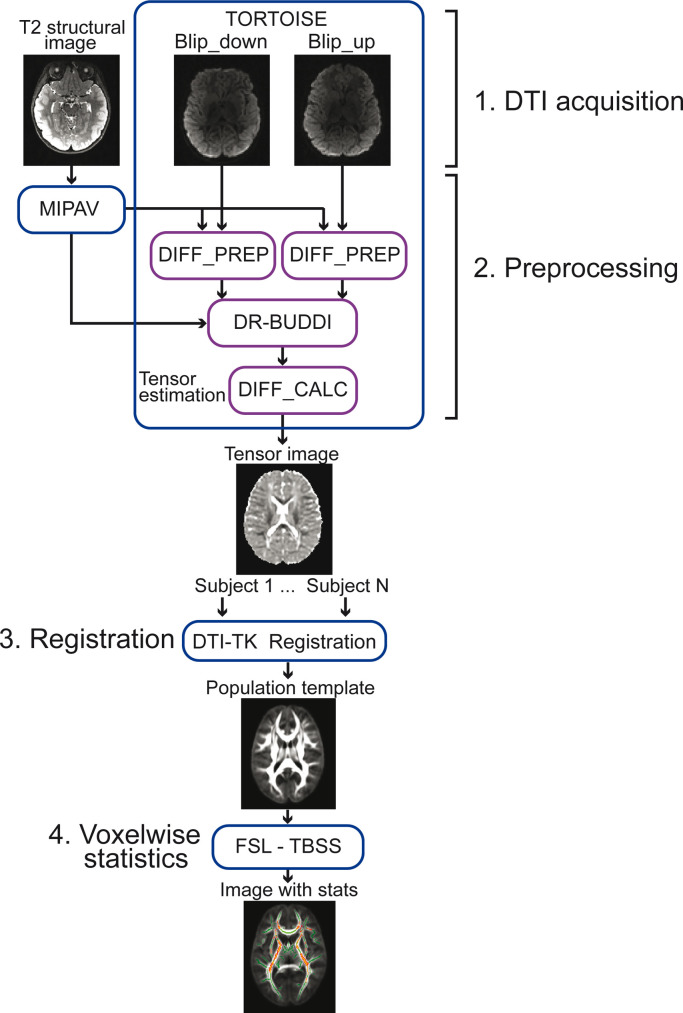
The results of statistical analyses on brain imaging data depend partially on the quality of the collected data and on the preprocessing steps of the data analysis. Moreover, the collection and analysis of brain images from children is associated with multiple obstacles including subject compliance during long scanning sessions [1], larger amount of motion in children compared to adults [2], and the lack of available templates for specific age ranges [3]. The common sources of noise that affect the diffusion tensor imaging (DTI) data include eddy currents induced by rapid switching of gradient coils, geometric distortions resulting from the strong external magnetic field, and subject motion in the scanner [4]. Developmental and pathologic processes of the brain may influence the microstructural composition and architecture of the affected tissues and alter the diffusion of water within the tissues [5]. With the protocol described here, we investigated the white matter microstructure of extremely preterm-born (birth < 28 weeks of gestation) young school-aged children from a large cohort that also included age-matched term-born children [6]. Earlier studies applying diffusion weighted magnetic resonance imaging (MRI) on preterm-born children have yielded variable results, some reporting lower fractional anisotropy (FA) values in preterm-born children compared with controls [7], [8], [9], [10], some higher FA values [11]. Multiple reasons may underlie variability in the results including the extent and cause of prematurity, the age of children at brain imaging, and the applied imaging parameters and preprocessing methods of the data. The current protocol (Fig. 1) was designed for the analysis of DTI data collected from young, school-aged children. It includes four major parts: 1) the acquisition of diffusion weighted images (DWIs), 2) preprocessing of DWIs, 3) nonlinear registration of the images to a population-specific template, and 4) voxelwise statistical analysis. The protocol utilizes the preprocessing methods that have been tested in earlier studies on pediatric DTI datasets and showed the best performance compared with other tools [12,13].
Fig. 1.
Illustration of the protocol that was designed for the analysis of diffusion tensor imaging (DTI) data collected from young school-aged children. The protocol has four major parts: (1) The acquisition of diffusion weighted images (DWIs), (2) preprocessing of DWIs, (3) nonlinear registration of the images to a population-specific template, and (4) voxelwise statistical analysis. MIPAV = Medical Image Processing, Analysis, and Visualization; TBSS = tract-based spatial statistics.
We applied an acquisition sequence that was based on the advanced protocol from the NIH MRI project [14]. This project resulted in a publicly-available pediatric DTI database, and developed special preprocessing algorithms and tools for DTI data analyses. In addition to images with high and low b-values, the protocol also included images with intermediate b-values in order to account for the higher water content in immature brain structures of young subjects [15]. The intermediate DWIs have higher signal-to-noise ratio and they are more similar in contrast intensity with the T2-weighted images that are used as targets for registration. An acquisition of several intermediate b-values facilitates the registration of the DWIs with the highest b-value by sequentially computing a new reference target based on the lower b-values [16]. The acquisition sequence resulted in two datasets that were collected with opposite phase encoding directions which allowed the application of advanced correction techniques for the echo-planar imaging (EPI) distortions.
To preprocess the data we used the TORTOISE toolbox (http://www.tortoisedti.org) [17] which allows to mitigate the effect of motion artifacts in the presence of EPI distortions. A previous study compared TORTOISE with other motion correction techniques and showed that TORTOISE outperforms other DTI preprocessing tools due to its improved estimation of motion parameters based on normalized mutual information cost function [12]. Considering that the prevalence of motion in child populations during the collection of neuroimaging data is high [18], this tool is especially well suited for pediatric studies. Another source of variability that impacts the statistics of voxel-wise analyses corresponds to the template selection and registration of DWIs to this template [19]. To minimize the impact of misregistration we used an advanced DTI-TK nonlinear registration method (http://dti-tk.sourceforge.net) [20] which, unlike standard methods that are based on scalar maps, makes use of the directional information from tensorial datasets. Finally, the tract-based spatial statistics (TBSS) approach was applied to analyze the white matter microstructure. The TBSS is a suitable technique that evaluates the whole-brain white matter and does not require a priori selection of specific regions of interest.
We applied the protocol to the analysis of DTI data collected from 15 extremely preterm-born young school-aged children [8 males, mean (standard deviation, SD) gestational age at birth 26.7 (1.0) weeks, mean (SD) age at scanning 7.6 y (0.1)], and from 16 age- and gender-matched term-born controls [9 males, mean (SD) gestational age at birth 40 (0.9) weeks, mean (SD) age at scanning 7.6 y (0.2)] [6]. The children were from a large cohort that was recruited for a multimodal follow-up study (KeKeKe, Extremely Preterm Birth and Development of the Central Nervous System [21]). Based on a neuropsychological assessment at 6 years, the cognitive performance of the preterm-born children was within normal range, and they had not been diagnosed with neurological or neurosensory impairments.
DTI acquisition sequence
The acquisition sequence included two repetitions of 45 images (6 images with, b = 300 s/mm2, 6 images with b = 500 s/mm2, 30 images with b = 1000 s/mm2 and 3 non-weighted b = 0 s/mm2 images). Both repetitions were collected with the same acquisition parameters except for the phase encoding direction which was reversed for the second repetition from anterior-posterior into posterior-anterior (blip-down and blip-up, respectively). The DWIs were collected using a spin-echo-based single shot EPI sequence with full k-space coverage and GRAPPA parallel acquisition option (TR 9000 ms, TE 80 ms, FOV 240 mm, matrix size 96 × 96, slice thickness 2.5 mm, 70 contiguous axial slices, acceleration factor 2). For each subject, a 3D T2-weighted image set was collected using SPACE sequence (TR 3200 ms, TE 411 ms, FOV 256 mm, matrix size 256 × 256, 176 sagittal slices, 1.0 mm isotropic voxels). The DTI imaging session lasted around 25 min during which the children watched video movies/cartoons.
DTI data preprocessing and registration
The DTI data were preprocessed using TORTOISE 2.5.2b software (http://www.tortoisedti.org) [17]. First, the individual T2-weighted images from each subject were midsagittally aligned and then put into AC (anterior commissure) -PC (posterior commissure) alignment using the MIPAV (Medical Image Processing, Analysis, and Visualization) 7.0.1 toolbox in order to standardize initial data orientation. These T2 images were used to provide a reference frame for the registration of the DWIs. The diffusion gradient direction files were created using the dcm2nii package of MRIcron that converts DICOM images to NIfTI format. For each dataset phase-encoding direction, the corresponding raw DICOM images from the scanner (45 vol: 30 gradient directions at b = 1000 s/mm2, 6 directions at b = 500 s/mm2, 6 directions at b = 300 s/mm2, and 3 non-diffusion-weighted images at b = 0 s/mm2) were imported into the TORTOISE DIFF_PREP module. The main registration settings of the DIFF_PREP were kept as default except that the final resolution of the DWIs was set to 1.5 × 1.5 × 1.5 mm isotropic voxels and the “keep intermediate data” flag was selected to save the upsampled datasets and transformation parameters. The DTI datasets were corrected for motion and eddy currents distortion, and the b-matrix was reoriented to preserve the orientation information [22]. For each subject, the upsampled datasets with opposite phase-encoding directions were combined within the TORTOISE DR-BUDDI module [23] to correct for susceptibility artifacts induced by EPI distortions. The tensors were estimated using the nonlinear RESTORE algorithm [24] from the DIFF_CALC module which generated the 4D tensor datasets containing the 6 tensor elements. This algorithm produces robust results even in the presence of outliers induced by cardiac pulsation and head motion, and thus, it can be an optimal choice for studies of non-sedated subjects.
After the correction, all tensorial datasets were fed into the DTI-TK (http://dti-tk.sourceforge.net) [20] nonlinear registration pipeline. This whole-tensor method showed the best performance compared with seven other algorithms in the registration of fiber tracts in a study of white matter pathologies in the developing brain [13]. Therefore, considering the impact of extreme prematurity on white matter structure and volume especially around the periventricular regions, we decided to adapt this method for the registration of our data to the template [6]. The datasets from all children were bootstrapped and resampled into 1.5 × 1.75 × 2.25 mm resolution (suggested by DTI-TK tutorial) in order to obtain the initial population template. Following this, the tensorial images underwent rigid and affine alignment with the initial template using Euclidean Distance Squared similarity metric. Finally, the deformable registration was applied to align the tensorial datasets with the template which was iteratively optimized for 6 times.
Voxelwise analysis of the white matter tracts
The voxelwise analysis of white matter tracts was performed using TBSS [25] from the FSL (FMRIB (Functional Magnetic Resonance Imaging of the Brain) Software Library, https://fsl.fmrib.ox.ac.uk/fsl) 5.0 toolbox. The mean FA image was created and thinned to produce the white matter skeleton (thresholded at FA > 0.2). The TVtool command from DTI-TK toolbox generated individual maps of FA, mean diffusivity (MD), axial diffusivity (AD) and radial diffusivity (RD) for each child. For each analysis, the individual maps (FA, MD, RD and AD) were projected onto the FA skeleton. The group differences in the white matter measures were estimated using an unpaired t-test with nonparametric permutation method (5000 permutations). Statistical maps were thresholded at P < 0.05 using a threshold-free cluster enhancement (TFCE) method with family-wise error (FWE) correction for multiple comparisons.
Quantitative analysis of DTI-based metrics obtained from major tracts
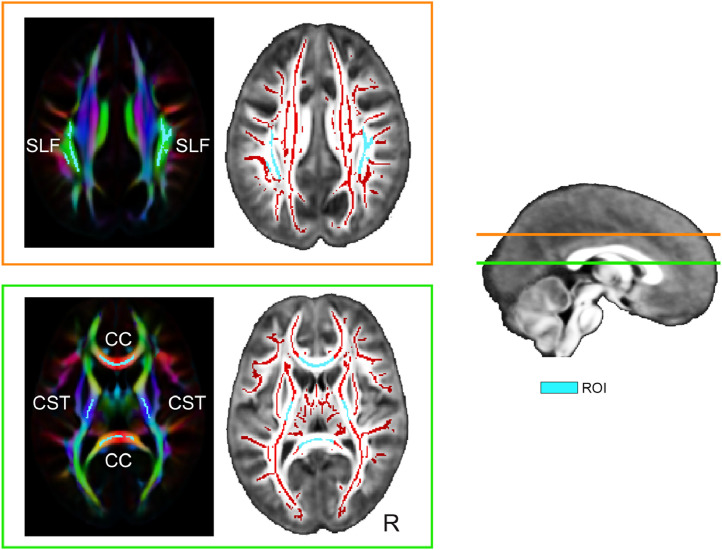
Table 1 reports the mean and SD values of the FA, MD, AD, and RD for both the preterm-born and control children of five masks representing major commissural, projection and association tracts. The masks were drawn on the mean FA skeleton map obtained from the TBSS analysis. In addition, the directionally encoded color (DEC) map of the mean FA template was used as a guide to validate the belonging of the portions of the FA skeleton to the white matter tracts of interest. The masks were drawn to the corpus callosum (CC) (3223 voxels) that included both the genu and splenium of the CC, to regions of the left (386 voxels) and right (394 voxels) corticospinal tract (CST), and the left (588 voxels) and right (504 voxels) superior longitudinal fasciculus (SLF). Fig. 2 shows the five masks on the mean FA skeleton and DEC maps.
Table 1.
Summary of diffusion tensor imaging metrics. Mean (SD) fractional anisotropy (FA), and mean (MD), axial (AD), and radial (RD) diffusivity values for the extremely preterm-born (n = 15) and term-born (n = 16) children.
| Child group | Tract | FA | MD | AD | RD |
|---|---|---|---|---|---|
| Preterm-born | CC | 0.82 (0.02) | 0.87 (0.04) | 2.00 (0.08) | 0.31 (0.03) |
| CST_L | 0.75 (0.02) | 0.78 (0.03) | 1.65 (0.07) | 0.35 (0.03) | |
| CST_R | 0.75 (0.02) | 0.78 (0.03) | 1.65 (0.07) | 0.35 (0.02) | |
| SLF_L | 0.55 (0.04) | 0.78 (0.04) | 1.30 (0.05) | 0.52 (0.05) | |
| SLF_R | 0.58 (0.04) | 0.78 (0.04) | 1.34 (0.05) | 0.49 (0.05) | |
| Term-born | CC | 0.83 (0.02) | 0.88 (0.05) | 2.04 (0.08) | 0.31 (0.04) |
| CST_L | 0.73 (0.03) | 0.82 (0.06) | 1.69 (0.09) | 0.39 (0.05) | |
| CST_R | 0.74 (0.03) | 0.82 (0.05) | 1.69 (0.08) | 0.38 (0.05) | |
| SLF_L | 0.51 (0.03) | 0.81 (0.06) | 1.29 (0.09) | 0.57 (0.06) | |
| SLF_R | 0.53 (0.03) | 0.81 (0.05) | 1.33 (0.08) | 0.55 (0.05) |
SD = standard deviation; CC = corpus callosum; CST = corticospinal tract; SLF = superior longitudinal fasciculus; L = left; R = right.
Fig. 2.
The DTI masks (cyan) are shown on directionally encoded color (DEC) map of the mean FA template and on the mean FA skeleton (red) obtained from TBSS analysis. The DEC map shows the preferred orientation of fibers using the red (left-right), green (anterior-posterior) and blue (superior-inferior) color scheme. The horizontal lines in the sagittal brain slice of the population template indicate the level of axial slice planes that are presented in the frames with the corresponding colors. CC = corpus callosum; CST = corticospinal tract; SLF = superior longitudinal tract; ROI = region of interest; R = right.
Testing and applying the protocol
We tested the protocol and the outcome of the TBSS analysis by performing a between-groups analysis on the normally developing 7.5-year-old control children, as suggested by Bach et al. (2014) [26]. In our original study, where the current protocol was applied, we performed a split-sample analysis by using the TBSS approach after the DTI-TK registration to test the within group consistency of the control children (please see supplements in [6]). The results showed no significant differences between the subsamples (P < 0.05, TFCE corrected) indicating that there were no unexpected differences between the subjects within the groups. The purpose of the split-sample analysis performed here was to evaluate the impact of the template selection on the statistical results. For this purpose, we repeated the split-sample comparison of the control children described in [6], but instead of the DTI-TK registration, we used the FSL registration method before the TBSS analysis. The DTI data from the control children were divided into two equal-sized subsamples (8 vs. 8), and the FA values of the subsample groups were compared using the FSL registration and the TBSS approach. Compared with the DTI-TK registration that works with tensorial datasets, the FSL registration utilizes only scalar FA maps. We obtained the FA images generated by TORTOISE during tensor estimation and forwarded them into the FSL TBSS registration pipeline. The FA image of each subject was non-linearly aligned with the FA images of the other subjects. The FA image with the smallest average warp score obtained from the registration with all other subjects was selected as the target template and aligned into the 1 × 1 × 1 mm MNI152 standard space using affine registration. Following this, the FA images were aligned into the FA target template using non-linear transformation, and then into the MNI152 space using affine transformation. The FA map was thinned to produce the FA skeleton (thresholded at default FA > 0.2), and each subject's FA image was projected onto this skeleton. The group differences in FA values were estimated using an unpaired t-test with nonparametric permutation method with 5000 permutations. Statistical maps were thresholded at P < 0.05 using the TFCE method with FWE correction for multiple comparisons. As expected, and in line with the result obtained with the DTI-TK registration, the analysis showed no significant differences between the subsamples.
In our previous study where the current protocol was applied, we found significant correlations between the FA and the response times in the 1- and 2-back tasks in the control children [6]. Here we investigated whether the template selection before the TBSS analysis affects the correlation results. We conducted a whole brain analysis of correlations between the FA and the response times of the 1-back task by performing the TBSS analyses after both the DTI-TK and FSL registrations. These analyses were done on a subsample of 15 control children, as the response times from one control child were not available. Fig. 3A shows the results of the TBSS analysis after the DTI-TK registration to the population template, and Fig. 3B after the FSL registration. In the case of the FSL registration, there were several significant associations between response times and FA e.g. in the CC and CST, and in the cerebellar white matter. In this analysis, the FA maps of all children were registered to the most representative child's FA map from the sample (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/TBSS/UserGuide#tbss_2_reg). The TBSS after the DTI-TK registration also showed significant correlations between the response times and FA in several tracts, but there were also some notable differences compared with the results obtained using the FSL registration. As shown in Fig. 3A, the TBSS after the DTI-TK registration showed no significant correlations in cerebellar regions. One possible explanation for these differences is that the registration of scalar maps is prone to misalignment effects that may produce false positive findings in the TBSS when FSL registration is used [19]. Another source of discrepancies between the results may relate to the projection of voxels to the population skeleton. If the white matter region is centered between two branches of the skeleton, the values of this region can contribute to the statistics of two anatomical locations and produce biased statistical results [27]. The FSL uses the subject with the most typical FA from the sample as a template, whereas the DTI-TK creates the template using the information about the whole tensor of all subjects and this type of information produces the best performance in registration of subjects to the template [28].
Fig. 3.
Results of correlation analyses between the fractional anisotropy (FA) and response times of 1-back tasks in normally developing, young school-aged control children (n = 15) born at term-age. The images illustrate significant correlations (P < 0.05, shown in red-yellow) between the FA and response times when the tract-based spatial statistical (TBSS) analysis (P < 0.05, TFCE-corrected) was performed (A) using the DTI-TK nonlinear registration to the population template, and (B) when TBSS analysis was performed using the FSL registration. The horizontal lines in the sagittal brain slices of the population template (A), and the most representative subject's template (B), indicate the level of the selected axial slice planes that are presented in the frames with the corresponding colors. CC = corpus callosum; CST = corticospinal tract; MCP = middle cerebellar peduncle; R = right.
Advantages and limitations of the protocol
The main advantage of this protocol is that the collection of DTI datasets with opposite phase encoding directions allows the application of special correction techniques to reduce the effect of artefacts in the presence of EPI-induced distortions. Another advantage is related to the application of DTI-TK nonlinear registration that improves the alignment of subjects to the population template. The main limitation of the protocol is the relatively long computation time that is required to obtain the population template and to perform the registration of all subjects to this template.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
We wish to thank Dr. Carlo Pierpaoli for his suggestions and help in designing the protocols and handling of the DTI data, and Marita Kattelus for her invaluable help in brain imaging. This work was supported by grants from Päivikki and Sakari Sohlberg Foundation (S.C.), Aalto Brain Centre (S.C.), Foundation for Pediatric Research (M.M., P.L., S.A.), Hospital District of Helsinki and Uusimaa Medical Imaging Centre (J.P.), Finnish Cultural Foundation (M.T.), Instrumentarium Science Foundation (M.T.), Ella and Georg Ehnrooth Foundation (M.T.), Arvo and Lea Ylppö Foundation (A.L., P.L.), The Finnish Medical Foundation (P.L.), Finska Läkaresällskapet (S.A.), Governmental Subsidy for Clinical Research (S.A.), Helsinki University Central Hospital Research Funds (TYH 2014104 to A.L., and TYH 2016202 and TYH 2017205 to V.V.) and Yrjö Jahnsson Foundation (V.V).
References
- 1.Vanderwal T., Kelly C., Eilbott J., Mayes L.C., Castellanos F.X. Inscapes: a movie paradigm to improve compliance in functional magnetic resonance imaging. Neuroimage. 2015;122:222–232. doi: 10.1016/j.neuroimage.2015.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poldrack R.A., Pare-Blagoev E.J., Grant P.E. Pediatric functional magnetic resonance imaging: progress and challenges. Top. Magn. Reson. Imaging. 2002;13:61–70. doi: 10.1097/00002142-200202000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Tamnes C.K., Roalf D.R., Goddings A.L., Lebel C. Diffusion MRI of white matter microstructure development in childhood and adolescence: methods, challenges and progress. Dev. Cogn. Neurosci. 2018;33:161–175. doi: 10.1016/j.dcn.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Bihan D., Poupon C., Amadon A., Lethimonnier F. Artifacts and pitfalls in diffusion MRI. J. Magn. Reson. Imaging. 2006;24(3):478–488. doi: 10.1002/jmri.20683. [DOI] [PubMed] [Google Scholar]
- 5.Alexander A.L., Lee J.E., Lazar M., Field A.S. Diffusion tensor imaging of the brain. Neurotherapeutics. 2007;4(3):316–329. doi: 10.1016/j.nurt.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tokariev M., Vuontela V., Lönnberg P., Lano A., Perkola J., Wolford E., Andersson S., Metsäranta M., Carlson S. Altered working memory-related brain responses and white matter microstructure in extremely preterm-born children at school age. Brain Cogn. 2019;136(November) doi: 10.1016/j.bandc.2019.103615. Epub 2019 Sep25. [DOI] [PubMed] [Google Scholar]
- 7.Li K., Sun Z., Han Y., Gao L., Yuan L., Zeng D. Fractional anisotropy alterations in individuals born preterm: a diffusion tensor imaging meta-analysis. Dev. Med. Child Neurol. 2015;57:328–338. doi: 10.1111/dmcn.12618. [DOI] [PubMed] [Google Scholar]
- 8.Skranes J., Vangberg T.R., Kulseng S., Indredavik M.S., Evensen K.A., Martinussen M., Dale A.M., Haraldseth O., Brubakk A.-.M. Clinical findings and white matter abnormalities seen on diffusion tensor imaging in adolescents with very low birth weight. Brain. 2007;130(Pt 3):654–666. doi: 10.1093/brain/awm001. [DOI] [PubMed] [Google Scholar]
- 9.Vangberg T.R., Skranes J., Dale A.M., Martinussen M., Brubakk A.M., Haraldseth O. Changes in white matter diffusion anisotropy in adolescents born prematurely. Neuroimage. 2006;32(4):1538–1548. doi: 10.1016/j.neuroimage.2006.04.230. [DOI] [PubMed] [Google Scholar]
- 10.Vollmer B., Lundequist A., Mårtensson G., Nagy Z., Lagercrantz H., Smedler A.C., Forssberg H. Correlation between white matter microstructure and executive functions suggests early developmental influence on long fibre tracts in preterm born adolescents. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0178893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman H.M., Lee E.S., Loe I.M., Yeom K.W., Grill-Spector K., Luna B. White matter microstructure on diffusion tensor imaging is associated with conventional magnetic resonance imaging findings and cognitive function in adolescents born preterm. Dev. Med. Child Neurol. 2012;54(9):809–814. doi: 10.1111/j.1469-8749.2012.04378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor P.A., Alhamud A., van der Kouwe A., Saleh M.G., Laughton B., Meintjes E. Assessing the performance of different DTI motion correction strategies in the presence of EPI distortion correction. Hum. Brain Mapp. 2016;37(12):4405–4424. doi: 10.1002/hbm.23318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Gupta A., Liu Z., Zhang H., Escolar M.L., Gilmore J.H., Gouttard S., Fillard P., Maltbie E., Gerig G., Styner M. DTI registration in atlas based fiber analysis of infantile Krabbe disease. Neuroimage. 2011;55(4):1577–1586. doi: 10.1016/j.neuroimage.2011.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker L., Chang L.C., Nayak A., Irfanoglu M.O., Botteron K.N., McCracken J., McKinstry R.C., Rivkin M.J., Wang D.J., Rumsey J., Pierpaoli C., Brain Development Cooperative Group The diffusion tensor imaging (DTI) component of the NIH MRI study of normal brain development (PedsDTI) Neuroimage. 2016;124(Pt B):1125–1130. doi: 10.1016/j.neuroimage.2015.05.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sundgren P.C., Dong Q., Gómez-Hassan D., Mukherji S.K., Maly P., Welsh R. Diffusion tensor imaging of the brain: review of clinical applications. Neuroradiology. 2004;46(5):339–350. doi: 10.1007/s00234-003-1114-x. [DOI] [PubMed] [Google Scholar]
- 16.Rohde G.K., Barnett A.S., Basser P.J., Marenco S., Pierpaoli C. Comprehensive approach for correction of motion and distortion in diffusion-weighted MRI. Magn. Reson. Med. 2004;51:103–114. doi: 10.1002/mrm.10677. [DOI] [PubMed] [Google Scholar]
- 17.Pierpaoli C., Walker L., Irfanoglu M.O., Barnett A., Basser P., Chang L.-C., Koay C., Pajevic S., Rohde G., Sarlls J., Wu M. TORTOISE: an integrated software package for processing of diffusion MRI data. ISMRM 18th annual meeting 2010; Stockholm, Sweden; 2010. #1597. [Google Scholar]
- 18.Power J.D., Mitra A., Laumann T.O., Snyder A.Z., Schlaggar B.L., Petersen S.E. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keihaninejad S., Ryan N.S., Malone I.B., Modat M., Cash D., Ridgway G.R., Zhang H., Fox N.C., Ourselin S. The importance of group-wise registration in tract based spatial statistics study of neurodegeneration: a simulation study in Alzheimer's disease. PLoS ONE. 2012;7(11):e45996. doi: 10.1371/journal.pone.0045996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H., Avants B.B., Yushkevich P.A., Woo J.H., Wang S., McCluskey L.H., Elman L.B., Melhem E.R., Gee J.C. High-dimensional spatial normalization of diffusion tensor images improves the detection of white matter differences: an example study using amyotrophic lateral sclerosis. IEEE Trans. Med. Imaging. 2007;26(11):1585–1597. doi: 10.1109/TMI.2007.906784. [DOI] [PubMed] [Google Scholar]
- 21.Rahkonen P., Nevalainen P., Lauronen L., Pihko E., Lano A., Vanhatalo S., Pesonen A.-K., Heinonen K., Räikkönen K., Valanne L., Autti T., Andersson S., Metsäranta M. Cortical somatosensory processing measured by magnetoencephalography predicts neurodevelopment in extremely low-gestational-age infants. Ped. Res. 2013;73:763–771. doi: 10.1038/pr.2013.46. [DOI] [PubMed] [Google Scholar]
- 22.Leemans A., Jones D.K. The B-matrix must be rotated when correcting for subject motion in DTI data. Magn. Reson. Med. 2009;61(6):1336–1349. doi: 10.1002/mrm.21890. [DOI] [PubMed] [Google Scholar]
- 23.Irfanoglu M.O., Modi P., Nayak A., Hutchinson E.B., Sarlls J., Pierpaoli C. DR-BUDDI (Diffeomorphic registration for blip-up blip-Down diffusion imaging) method for correcting echo planar imaging distortions. Neuroimage. 2015;106:284–299. doi: 10.1016/j.neuroimage.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang L.C., Jones D.K., Pierpaoli C. RESTORE: robust estimation of tensors by outlier rejection. Magn. Reson. Med. 2005;53(5):1088–1095. doi: 10.1002/mrm.20426. [DOI] [PubMed] [Google Scholar]
- 25.Smith S.M., Jenkinson M., Johansen-Berg H., Rueckert D., Nichols T.E., Mackay C.E., Watkins K.E., Ciccarelli O., Cader M.Z., Matthews P.M., Behrens T.E. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31(4):1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 26.Bach M., Laun F.B., Leemans A., Tax C.M., Biessels G.J., Stieltjes B., Maier-Hein K.H. Methodological considerations on tract-based spatial statistics (TBSS) Neuroimage. 2014;100:358–369. doi: 10.1016/j.neuroimage.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 27.Zalesky A. Moderating registration misalignment in voxelwise comparisons of DTI data: a performance evaluation of skeleton projection. Magn. Reson. Imaging. 2011;29(1):111–125. doi: 10.1016/j.mri.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Shen Y., Liu D., Li G., Guo Z., Fan Y., Niu Y. Evaluations of diffusion tensor image registration based on fiber tractography. Biomed. Eng. Online. 2017;16(1):9. doi: 10.1186/s12938-016-0299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]