Abstract
At the end of the year 2019, the novel coronavirus (2019-nCoV) was spreading in Wuhan, China, and the outbreak process has a high speed. It was recognized as a pandemic by the World Health Organization (WHO) on 11 March 2020. Coronaviruses are enveloped and single-stranded RNA that have several families including Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS). The pathogenesis mechanism and disease outcomes of SARS and MERS are now clear to some extent, but little information is available for 2019-nCoV. This newly identified corona virus infection represents flu-like symptoms, but usually the first symptoms are fever and dry cough. There has been no specific treatment against 2019-nCoV up to now, and physicians only apply supportive therapy. In the present article, we made an attempt to review the behavior of the virus around the world, epidemiology, a pathway for influx into the host cells, clinical presentation, as well as the treatments currently in use and future approaches; nitazoxanide may be our dream drug. We hope that this review has a positive impact on public knowledge for helping to deal with the 2019-nCoV and move one step forward toward its treatment in the near future.
Key Words: Novel coronavirus, 2019-nCoV, Treatment approach, Infection, Respiratory problem
Introduction
In late 2019, the local health authorities in Wuhan, China, declared a high load of individuals that had pneumonia with unknown etiology (1). Subsequently, in early January 2020, numerous medical diagnostic laboratories in China reported that a new coronavirus was the cause of occult pneumonia. The World Health Organization (WHO) named this invasive virus briefly as 2019 novel coronavirus (2019-nCoV) (2). Coronaviruses are enveloped, single stranded RNA, and belong to the subfamily Coronavirinae. It has previously been reported that CoVs which causes human diseases have six types, and is categorized into two subgroups including gently and extremely pathogenic CoVs. The 229E, HKU1, OC43 and NL63 are gently pathogenic CoVs, leading to 10–30% of upper respiratory tract infections (3,4). Instead, the extremely pathogenic CoVs, containing Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS), mostly contaminate lower airways and lead to pneumonia (5). Given that the clinical signs of2019-nCoV, SARS-CoV, and MERS-CoV have some resemblances, it can range from asymptomatic infections to severe respiratory illnesses (6). The rapid prevalence of the novel type of CoV reminds us that it is a major global health threat. Chen Y, et al. think that severe prevalence of novel CoV in the future is inevitable as climate is changing and human exposure to animals is increasing; therefore, the need for effective treatment modalities and training for CoVs is necessary (2).
In the present article, we attempted to review the behavior of the virus around the world, epidemiology, a pathway for influx into the host cells, clinical presentation, as well as the treatments currently in use. We hope that this review could have a positive impact on our knowledge of the new virus and help establish effective ways to combat the novel coronavirus 2019 in near future.
Taxonomy
Coronaviruses (CoVs) form a large and important family of viruses found in nature. CoVs have a positive-sense single-stranded RNA (+ssRNA) with a GC content of 32–43% that belongs to the family Coronaviridae and the order Nidovirales. They contain the largest genome among the recognized RNA viruses (genome length is about 26–32 kb) (7). Based on the genomic structure and phylogenetic analysis, the family Coronaviridae is currently classified into two subfamilies, including Torovirinae, and Coronavirinae. The Coronavirinae includes four genera called Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (8). Two genera, Alphacoronavirus and Betacoronavirus, only cause infection in mammals. They account for respiratory infection in humans and enteritis in animals. The members of the genus Betacronavirus are categorized into five subgenera, in which the two genera Hibecovirus and Nobecovirus do not infect humans (9,10). The members of the other three genera, Merbecovirus, Sarbecovirus, and Embecovirus, are the cause of the disease in humans. Merbecovirus containing MERS-CoV and Sarbecovirus containing SARS-CoV are two major zoonotic pathogenic coronaviruses (Table 1 ). Anew pandemic Coronavirus, so called 2019-nCoV, has been recently introduced in Wuhan, China (11). 2019-nCoV is still considered as unclassified Betacoronavirus, but according to early phylogenetic studies, this virus is related to SARS and is over 85% matched with the bat SARS-like CoV (9). Accordingly, the International Committee on Taxonomy of Viruses named it severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Table 1.
Current taxonomy of the human relevant coronaviruses. According the NCBI Taxonomy, National Institutes of Health, USA (https://www.ncbi.nlm.nih.gov/taxonomy) and ICTV Taxonomy
| Family | Subfamily | Genus | Subgenus | Species |
|---|---|---|---|---|
| Coronaviridae | Torovirinae | |||
| Coronavirinae | Alphacoronavirus | Duvinacovirus | Human coronavirus 229E | |
| Setracovirus | Human coronavirus NL63 | |||
| Betacoronavirus | Embecovirus | Betacoronavirus 1 | ||
| Hibecovirus | Human coronavirus HKU1 | |||
| Nobecovirus | ||||
| Sarbecovirus | SARS-CoV | |||
| Merbecovirus | MERS-CoV | |||
| Unclassified Betacoronavirus | 2019-nCoV | |||
| Gammacoronavirus | ||||
| Deltacoronavirus |
2019-nCoV, 2019-novel coronavirus; SARS, Severe Acute Respiratory Syndrome; MERS, Middle East Respiratory Syndrome.
Morphology and Genome Structure
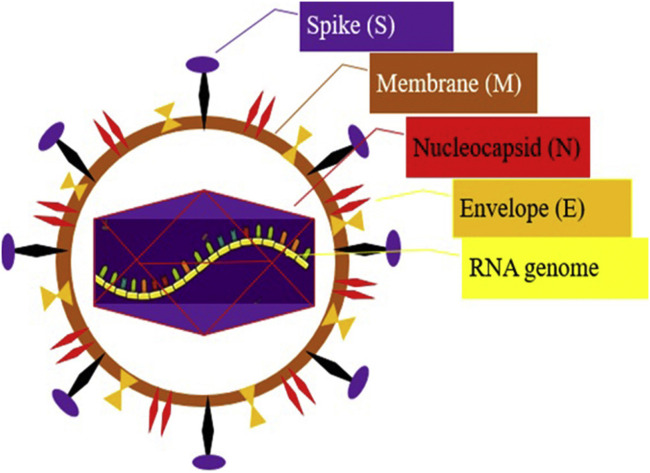
The 2019-nCoV is an enveloped, spherical, and relatively large (about 120 nm in diameter) particle; the envelope of this virus in electron micrographs emerges as a separated pair of electron-dense shells. The viral envelope contains a lipid bilayer by which the membrane (M), envelope (E) and spike (S) structural proteins are harbored (Figure 1 ) (12,13). The genome of CoVs is Monopartite, linear, single-stranded positive-sense RNA (+ssRNA) (∼30 kb in size), Capped at the 5′ end, and poly adenylated at the 3′ end with at least six open reading frames (ORFs) and other subsidiary genes (14). The RNA genome includes 29,891 nucleotides (Gene Bank accession number MN908947), encoding 9860 amino acids. The 5′ terminal two-thirds of the genome contains two ORFs, ORF1 and ORF2 that encodes pp1a and pp1ab polyproteins, which is further split into 11 and 16 proteins, respectively, which encode non-structural proteins (nsps) (13,15). These proteins are processed into the viral polymerase (RdRp) and other non-structural proteins involved in RNA synthesis (16). The structural proteins including spike (S), envelope (E), membrane (M), and nucleocapsid (N) are arranged in the order of one-third 3′ terminal of the genome (15). One of the most important genes that can help researchers to produce vaccine in the future is S gene because this gene product has an effective role in receptor binding and host specificity (17).
Figure 1.
The structure of 2019-CoV.
Proposed a Pathway for 2019-nCoV Influx into the Host Cells
Recently reported that between the SARS-CoV genome sequence and the novel coronavirus exist 82% similarity, thus, named 2019-nCoV by WHO (13). This theory may be indicating that 2019-nCoV uses the same SARS-CoV mechanism i.e. through angiotensin-converting enzyme2 (ACE2) receptor and the TMPRSS2 protease to infect the human cells. Coronavirus spike (S) protein facilitates virus entry into target cells. The binding of SARS to the receptor and its entry into the cell depends on a cellular protease (18). Sequence analysis has shown that some of the 2019-nCoV clusters and bat-associated SARS76 CoV viruses (SARSr-CoV) can use the ACE2 receptor to enter the host cell. Analysis of the receptor-binding motif (RBM) and a part of the receptor-binding domain (RBD) that interacts with ACE2, revealed that most of the essential amino acid residues were retained at 2019-nCoV for ACE2 binding, but in the SARSr-CoV S proteins were not observed. It was found that it does not use ACE2 for entry into the cells and need priming the S protein for binding to ACE2 (19). It has been reported that entry of SARS-CoV-2 to host cells may block via a clinical antagonist of the TMPRSS2, a cellular serine protease, that is recruited via SARS-CoV-2 to S protein priming. These new findings have significant implications for our knowledge about the transmission and pathogenesis of SARS-CoV-2 and treatment approaches (20). Shieh W-J, et al., reported that SARS-CoV might goal a full spectrum of similar cells. The SARS-CoV in the respiratory system leads to infect mostly pneumocytes and macrophages (21). In addition to lung, other tissues can expression of ACE2. Thus, SARS-CoV may extent to extrapulmonary organs. While the affinity of SARS-S and SARS-2-S to ACE2 receptor has been comparable, therefore we still expect this for SARS-CoV-2. Previous studies showed that in the upper airways a mild expression of ACE2 (22,23) may be able to reduce SARS-CoV spread, given that the high dispersion ability of SARS91 CoV-2 against SARS-CoV (24). Definitively, it is important to note that the high-level expression of ACE2 can prevent lung damage. However, its downregulated through SARS-S, which lead to stimulate SARS. To enter the virus into host cells and covers S protein cleavage at the S1/S2 and the S2′ sites, the need to specific proteases in the host cells for priming of coronavirus S proteins (25). The cutting site of SARS-2-S, several arginine residues (multibasic) that shows high sensitivity for cutting. Actually, into the host cells, SARS-2-S was proficiently cleaved, fallowing by cleaved S protein was incorporated into vesicular stomatitis virus (VSV) particles. Notably, the area in which the incision has occurred can define the zoonotic potential of coronaviruses (26,27); the SARS-CoV100 2 and coronavirus have the most closely associated. SARS-CoV can use endosomal cysteine protease to priming S-protein in TMPRSS2 negative cells. Nevertheless, priming of protein S via TMPRSS2 protease is required for virus entry into target cells, and the spread of the virus in infected host cells (28).
Epidemiology
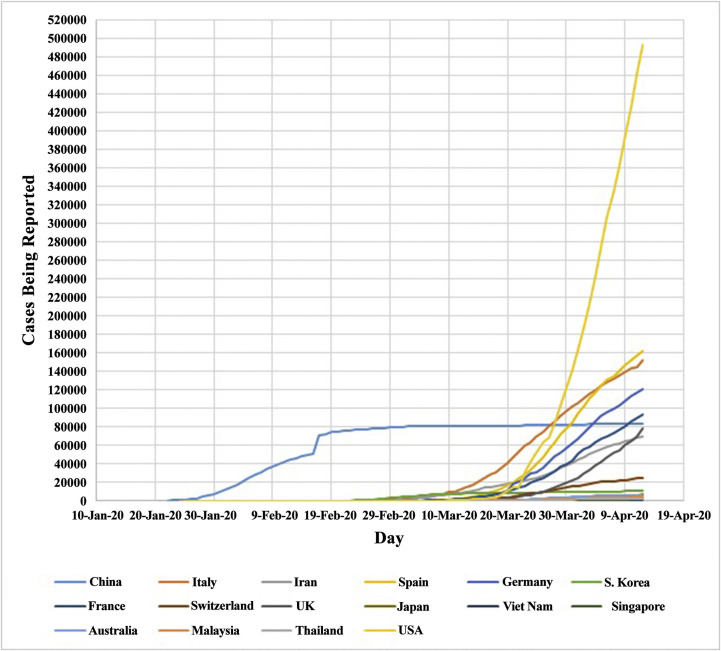
After the first reports of 2019-nCoV outbreak in Wuhan, Hubei, China, in December 2019, the virus quickly spread throughout China during several weeks, and then on the other Asian countries, the Middle East, Africa, the Americas, Oceania, and Europe. In accordance with the daily information of the WHO, the pandemic of 2019-nCoV till now caused 1,861,672 positive cases and 114,980 deaths in the worldwide by 13th April 2020 (29), that lead to a main global health concern (Table 2 ) and (Figure 2 ) (9,30, 31, 32). This information is updated daily at the Situation Report site of the WHO (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports). Another source of information is Worldometer (https://www.worldometers.info/coronavirus/), which is managed by an international team of researchers, developers, and volunteers with the purpose of providing world statistics available in a thought-provoking and time suitable format to all of the audience around the world.
Table 2.
Ten countries with the most reported laboratory-confirmed COVID-19 cases and deaths. Data updated as of 13 April 2020
| Country | Cases | Deaths | Death (%) | Recovered |
|---|---|---|---|---|
| USA | 560,433 | 22,115 | 3.94 | 32,634 |
| Spain | 169,496 | 17,489 | 10.31 | 64,727 |
| Italy | 156,363 | 19,899 | 12.72 | 34,211 |
| France | 132,591 | 14,393 | 10.85 | 27,186 |
| Germany | 127,854 | 3,022 | 2.36 | 64,300 |
| UK | 84,279 | 10,612 | 12.59 | N/A |
| China | 82,160 | 3,341 | 4.06 | 77,663 |
| Iran | 71,686 | 4,474 | 6.24 | 43,894 |
| Turkey | 56,956 | 1,198 | 2.10 | 3,446 |
| Belgium | 30,589 | 3,903 | 12.75 | 6,707 |
USA, United States of America; S. Korea, Southern Korea; UK, United Kingdom.
Figure 2.
Countries with reported laboratory confirmed COVID-19 cases. Data from January 21st 2020 to April 12th 2020. USA, United States of America; S. Korea, Southern Korea; UK, United Kingdom.
Clinical Features and Transmission
2019-nCoV infections can be hidden for 2–14 d without any symptoms generally, but can display from 3–7 d mostly (33). After the incubation period of the disease, the presentations are mostly fever, exhaustion and cough, which are probably accompanied by nasal congestion, runny nose, expectoration, and rarely diarrhea and headache. Typically, fever can be low to moderate, and even with no fever (34). After a week and with progress of displaying the disease, other sings such as dyspnea, cyanosis, and eventually systemic fatal symptoms appear, including malaise or restlessness, poor feeding, bad appetite, less activity, and respiratory failure. The late signs that could occur in severe cases may be accompanied by septic shock, metabolic acidosis, irreversible bleeding and coagulation dysfunction (33,35). The clinical presentations and reports from several investigations are summarized in Table 3 (32,36, 37, 38, 39, 40). The new virus is diffused through droplets of the respiratory system when patients with infection cough, sneeze, or even during the conversation. Furthermore, other ways for transmission of the virus are embracing and close contact (e.g., contact with the mouth, nose or eyes conjunctiva via polluted hands). It has been not recognized so far that spread can happen by mother to infant or breast feeding (33).
Table 3.
Clinical presentations of patients with 2019-nCoV infection
| Blood tests |
Clinical presentations |
CT scan features |
|||
|---|---|---|---|---|---|
| Variables | Changes | Variables | Changes | Variables | Changes |
| WBC | Normal Or Increase | Mean age (year) | 41 | Pneumonia (%) | 77 |
| Lymphocyte | Decrease Or Intense decrease | Mean incubation time (d) | 3 | GGO (%) | 50 |
| CRP | Normal Or Increase | Fever (%) | 88 | Bilateral patched shadowing (%) | 46 |
| LFT | Intense increase | Fatigue (%) | 70 | Interlobular septal thickening (%) | 73 |
| CK | Increase | Dry cough (%) | 68 | GGO with consolidation (%) | 60 |
| Myoglobin | Increase | Myalgia (%) | 35 | ||
| D-dimer | Increase | Headache (%) | 9 | ||
| Serum creatinine | Increase Or Intense decrease | Diarrhea (%) | 5 | ||
| Blood urea nitrogen | Increase Or Intense decrease | ||||
| Total bilirubin | Increase | ||||
| Glucose | Increase | ||||
WBC, White Blood Cell; CRP, C-reactive protein; LFT, Liver Function Tests; CK, Creating Kinase; GGO, Ground-glass opacity.
Lung Imaging and Features
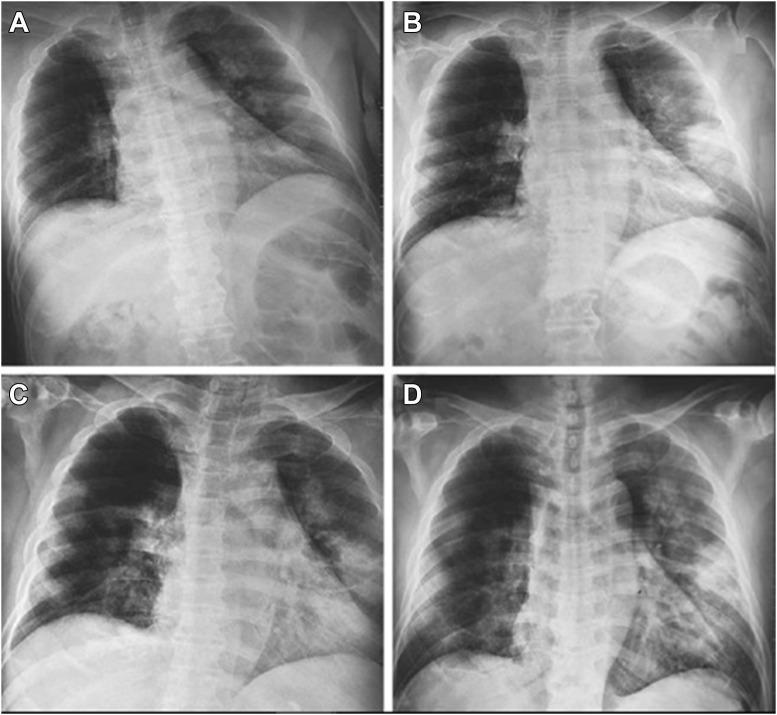
2019-nCoV leads to an acute resolved infection, which could be lethal with a mortality rate of about 2%. In the case of severe infection, it might lead to expiration of the patients due to severe alveolar injury and advanced respiratory failure (41,42). However, post-mortem biopsy might be taken from the liver, heart, and lung tissues. In the lung histological studies, bilateral diffuse alveolar damage with cellular fibromyxoid exudate might be seen. Patent desquamation of the pneumocytes and hyaline membrane construction in the right lung has also been seen to demonstrate acute respiratory failure (43). On the other hand, biopsy assay in the left lung exhibited pulmonary oedema with hyaline membrane construction. In the whole lung tissues, interstitial mononuclear inflammatory infiltrates occur mostly by lymphocytes. Furthermore, syncytial cells with abnormal puffy pneumocytes characterized via bulky nuclei, amphophilic granular cytoplasm, and noticeable nucleoli in the intra-alveolar spaces were recognized. These features display viral cytopathic-like variations (44). Pneumonia patients with initial stage lung X-ray analysis demonstrate several small patchy shadows and interstitial variations significant in the lung periphery (6). In patients with severe disease, more progress to bilateral multiple ground-glass opacity, infiltrating shadows, and pulmonary merging, with rare pleural effusion, can be seen. In both lungs of the youngsters with severe disease, multiple lobar lesions may be observed (45). Moreover, biopsy of the liver indicated modest microvascular steatosis and slight lobular activity; nevertheless, there was no definite proof to support 2019-nCoV contamination or drug-induced liver damage as a result. Subsequently, biopsy observations showed that no clear histological variations were seen in the heart tissue, proposing that 2019-nCoV contamination might not directly damage the heart (46). A radiograph of an infected person's chest is shown in Figure 3 , as an example.
Figure 3.
Chest radiographs of the person with CoV-19 infection. - Chest radiographs obtained at admission (A), on day 3 (B), day 5 (C), and day 6 (D) after admission (32).
Treatment Approaches
Presently, there are no FDA-approved treatments or vaccines for 2019-nCoV. The National Institute of Allergy and Infectious Diseases (NIAID) within the National Institutes of Health (NIH) is leading the funding of federal research and response to 2019-nCoV, while certain stakeholders in the US are choosing to supply their own 2019-nCoV research. Internationally, the UK Medicines and Healthcare Products Regulatory Agency (MHRA) and European Medicines Agency (EMA) have called for targeted efforts to progress treatments against 2019-nCoV. Some companies are looking to repurpose approved drugs that have worked against similar coronaviruses in the past or are hypothesized to attack or immobilize 2019-nCoV based on the mechanism of action. Plasma and stem cells from patients who have recovered from 2019-nCoV also are being investigated (Table 4 ). In addition, different companies are working hard for produce vaccine; experts estimate it could take between 12–18 months to develop a vaccine ready for market. Supplementary Table 1 showing the major vaccine candidates in development for prevention of 2019-nCoV. This information is updated weekly at the Situation Report site of the Regulatory Focus™ (https://www.raps.org/news-and-articles/about-regulatory-focus). However, some approaches have been suggested to prevent further spread of the infection and controlling 2019-nCoV. In the first step, suspected persons should be isolated and kept separately and the severe and acute patients should be detected as soon as possible. During therapeutic measurements, patients need more attention and their vital symptoms, pO2, pCO2, arterial pH, etc. should be regularly checked. Then, the general treatments are started which include resting and supportive therapeutics; adequate calorie and water consumption; keeping water electrolyte balance; and homeostasis. Some treatments are antiviral such as, interferon-α2b nebulization 100,000–400,00 IU/kg) in addition to lopinavir/ritonavir (200 mg/50 mg) (46, 47, 48). Furthermore, antibiotics are also used, but illogical usage of antibiotics should be prevented, and rarely the usage of immunomodulation drugs such as corticosteroids should be applied carefully in common types of infection. In certain conditions, rapid lung radiography and confirmation of the incidence of ARDS, observation of clear toxic signs such as encephalitis or encephalopathy, hemophagocytic syndrome and other severe problems, observation of septic shock, and eventually clear wheezing indications are recommended. Certain drugs such as methylprednisolone intravenously (1–2 mg/kg/d) are suggested for 3–5 d, but not for long-term consumption (46,49, 50, 51, 52). In severe patients, immunoglobulin can also be recommended intravenously. The suggested dose is 1.0 g/kg/d for 2 d, or 400 mg/kg/d for 5 d (50,53,54). In patients with blood circulation problems, drugs that are vasoactive can be used for recovering microcirculation on the basis of sufficient liquid intake (46). Cases who had acute renal damage should be given kidney tests function. In children, if intracranial hypertension and convulsion happens, there is a need to decrease the intracranial pressure and control convulsion to be suitable (53,54). There is speculation that patients with 2019-nCoV who are receiving ACE inhibitors or angiotensin receptor blockers (ARBs) may be at increased risk for adverse outcomes (55,56). The ACE2 is a receptor for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and renin-angiotensin aldosterone system inhibitors can increase the ACE2 levels (57). Finally, in the patients who had respiratory complication despite nasal catheter or mask oxygenation, heated humidified high-flow nasal cannula (HHHFNC), non-invasive ventilation such as continuous positive airway pressure (CPAP), or non-invasive high-frequency ventilation can be used. If improvement is not possible, mechanical ventilation with endotracheal intubation and a protective lung ventilation strategy should be adopted (45). It is worth noting that J.-F. Rossignol at 2016 presented nitazoxanide as a broad-spectrum antiviral agent for Flu and other viral respiratory infections; this drug is undergoing clinical progress. In vitro studies on nitazoxanide showed its efficient function against MERS-CoV and other coronaviruses by inhibiting expression of the viral N protein. He has also declared that the clinical trials and post-marketing experience approve nitazoxanide as an attractive drug candidate for treatment of MERS-CoV (58). There is a hope that nitazoxanide may be effective for treatment of 2019-nCoV through similar mechanisms.
Table 4.
Candidate drugs under testing for 2019-nCoV treatment; updated 10 April 2020
| Drug/Therapy | Medication class | Developer | Original use | Status | |
|---|---|---|---|---|---|
| Investigational candidates for 2019-nCoV | Remdesivir | Antiviral | Gilead Sciences | Treatment for Ebola and Marburg virus infections | Phase 3 clinical trial; (NCT04252664), (NCT04257656), (NCT04292730), (NCT04292899) |
| Mavrilimumab | Monoclonal antibody | Kiniksa Pharmaceuticals | Treatment of rheumatoid arthritis | Phase 2/3 clinical trial | |
| Convalescent plasma | Immunoglobulin | Treatment for Ebola virus infection | FDA approved | ||
| CD24Fc | Recombinant fusion protein | OncoImmune | Treatment of graft-versus-host disease | Phase 3 clinical trial (NCT04317040) | |
| Lenzilumab | Anti-human GM-CSF monoclonal antibody | Humanigen | Against cytokine release syndrome | Phase 3 clinical trial | |
| Leronlimab | Humanized IgG4 monoclonal antibody | CytoDyn | Breast cancer/HIV | ||
| EIDD-2801 | Oral broad-spectrum antiviral | Drug Innovation Ventures at Emory | against infections such as influenza, chikungunya, Ebola and equine encephalitis | launching a clinical trial | |
| Therapeutics Approved for Other Indications | Hydroxychloroquine (Plaquenil) and chloroquine (Aralen) | Quinoline | Sanofi; Mylan, Teva, Novartis, Bayer, Rising Pharmaceuticals | Anti-malaria | Ongoing, (NCT04303507) |
| Lopinavir-ritonavir (Kaletra) | HIV protease inhibitor | AbbVie | Treatment of HIV-1 | Phase 4 randomized controlled trial, (NCT04255017), (NCT04307693) | |
| Tocilizumab | IL-6 receptor antagonist | Roche | Treatment of rheumatoid arthritis | Phase 3 randomized controlled trial, (COVACTA) | |
| Sarilumab | IL-6 receptor antagonist | Sanofi and Regeneron | Treatment of rheumatoid arthritis | Phase 2/3 clinical trial | |
| 2019-nCoV Therapeutics Approved Outside the US | Favilavir | Antiviral agent | Fujifilm Toyama Chemical and Zhejiang Hisun Pharmaceutical | Against many RNA viruses | phase 3 clinical trial |
FDA, Food and Drug Administration; IL, Interleukin.
Conclusion
The high prevalence of the 2019-nCoV has become a global clinical problem these days. However, we do not know much about precision mechanism and function of this new virus at present. Various pharmaceutical companies around the world are working hard to find effective drug and vaccine against this virus. Until then, we must carefully follow WHO health measures to control infection and prevent further transmission of the virus. On the other hand, it is expected that different countries in this current crisis abandon political disputes, share information, and work unitedly against 2019-nCoV all over the world to combat this life-threatening disease of mankind.
Acknowledgments
The authors gratefully acknowledge Department of Biochemistry and Clinical Laboratories, Faculty of Medicine, Tabriz University of Medical Sciences, and professor Nasser Samadi, DCLS, PhD.
(ARCMED_2020_487)
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.arcmed.2020.04.022.
Contributor Information
Mohsen Hemmati-Dinarvand, Email: Hematym@sums.ac.ir.
Atefeh Seghatoleslam, Email: Seghatolea@sums.ac.ir.
Funding
None.
Competing interests
None declared.
Ethical Approval
Not required.
Supplementary Data
References
- 1.Lu H., Stratton C.W., Tang Y.W. Outbreak of Pneumonia of Unknown Etiology in Wuhan China: the Mystery and the Miracle. J Med Virol. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han Q., Lin Q., Jin S. Recent insights into 2019-nCoV: a brief but comprehensive review. J Infect. 2020:80. doi: 10.1016/j.jinf.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G., Fan Y., Lai Y. Coronavirus infections and immune responses. J Med Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorbalenya A.E., Baker S.C., Baric R.S. Severe acute respiratory syndrome-related coronavirus–The species and its viruses, a statement of the Coronavirus Study Group. BioRxiv. 2020 doi: 10.1101/2020.02.07.937862. [Preprint] [DOI] [Google Scholar]
- 8.Adams M.J., Lefkowitz E.J., King A.M. 50 years of the International Committee on Taxonomy of Viruses: progress and prospects. Arch Virol. 2017;162:1441–1446. doi: 10.1007/s00705-016-3215-y. [DOI] [PubMed] [Google Scholar]
- 9.Millán-Oñate J., Rodriguez-Morales A.J., Camacho-Moreno G. A new emerging zoonotic virus of concern: the 2019 novel Coronavirus (COVID-19) Infectio. 2020;24:187–192. [Google Scholar]
- 10.de Groot R.J., Baker S.C., Baric R. Family coronaviridae. Virus taxonomy. 2012:806–828. [Google Scholar]
- 11.Wang Y., Liu D., Shi W. A47 Origin and possible genetic recombination of the middle east respiratory syndrome coronavirus from the first imported case in china: phylogenetics and coalescence analysis. Virus Evol. 2017;3(Suppl_1) doi: 10.1093/ve/vew036.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snijder E.J., Van Der Meer Y., Zevenhoven-Dobbe J. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan J.F., Kok K.H., Zhu Z. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masters P.S. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik Y.S., Sircar S., Bhat S. Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Quart. 2020;40:68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W., Moore M.J., Vasilieva N. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F., Li W., Farzan M. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Scieas. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 20.Zhou P., Yang X.L., Wang X.G. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. BioRxiv. 2020 [Google Scholar]
- 21.Shieh W.J., Hsiao C.H., Paddock C.D. Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum Pathol. 2005;36:303–309. doi: 10.1016/j.humpath.2004.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamming I., Timens W., Bulthuis M.L. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertram S., Heurich A., Lavender H. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PloS one. 2012;7 doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park Y.J., Walls A.C., Wang Z. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat Struct Mol Biol. 2019;26:1151–1157. doi: 10.1038/s41594-019-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haga S., Yamamoto N., Nakai-Murakami C. Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α production and facilitates viral entry. Natl Acad Sci. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menachery V.D., Dinnon K.H., Yount B.L. Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J Virol. 2020;94 doi: 10.1128/JVI.01774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y., Liu C., Du L. Two mutations were critical for bat-to-human transmission of Middle East respiratory syndrome coronavirus. J Virol. 2015;89:9119–9123. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwata-Yoshikawa N., Okamura T., Shimizu Y. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93:e01815–e01818. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., Zheng X., Tong Q. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J Med Virol. 2020 doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shanmugaraj B., Malla A., Phoolcharoen W. Emergence of novel coronavirus 2019-nCoV: need for rapid vaccine and biologics development. Pathogens. 2020;9:148. doi: 10.3390/pathogens9020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya S., Sharma P., Mathur H. Recent apprise on coronavirus and its terrible insinuations. Virus Disease. 2020;18:1. doi: 10.1007/s13337-020-00582-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 33.Chen Z.M., Fu J.F., Shu Q. Diagnosis and treatment recommendations for pediatric respiratory infection caused by the 2019 novel coronavirus. World J Pediatr. 2020 doi: 10.1007/s12519-020-00345-5. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu P., Zhu J., Zhang Z. A familial cluster of infection associated with the 2019 novel coronavirus indicating possible person-to-person transmission during the incubation period. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel A., Jernigan D.B. Initial public health response and interim clinical guidance for the 2019 novel coronavirus outbreak—United States, December 31, 2019–February 4, 2020. Morb Mortal Wkly Rep. 2020;69:140. doi: 10.15585/mmwr.mm6905e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bajema K.L., Oster A.M., McGovern O.L. Persons evaluated for 2019 novel coronavirus—United States, January 2020. Morb Mortal Wkly Rep. 2020;69:166. doi: 10.15585/mmwr.mm6906e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang D., Lin M., Wei L. Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan, China. Jama. 2020;323:1092–1093. doi: 10.1001/jama.2020.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H., Guo J., Wang C. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395:809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Z., Shi L., Wang Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan J.F., Yuan S., Kok K.H. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu F., Zhao S., Yu B. A new coronavirus associated with human respiratory disease in China. Nat. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phan L.T., Nguyen T.V., Luong Q.C. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382:872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y.C., Liao C.H., Chang C.F. A locally transmitted case of SARS-CoV-2 infection in Taiwan. N Engl J Med. 2020;382:1070–1072. doi: 10.1056/NEJMc2001573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.National Recommendations for Diagnosis and Treatment of pneumonia caused by 2019-nCoV (the 4th edition). National Health Commission and National Administrative Office of Chinese Tradition Medicine. https://www.nhc.gov.cn/xcs/zheng cwj/20200 1/42945 63ed3 5b4320 9b31 739bd 0785e 67/files/7a930 91112 67475 a99d4 30696 2c8bf 78.pdf
- 47.Chu C.M., Cheng V.C., Hung I.F. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arabi Y.M., Alothman A., Balkhy H.H. Treatment of Middle East Respiratory Syndrome with a combination of lopinavir-ritonavir and interferon-β1b (MIRACLE trial): study protocol for a randomized controlled trial. Trials. 2018;19:81. doi: 10.1186/s13063-017-2427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perlman S. Another decade, another coronavirus. N Engl J Med. 2020;382:760–762. doi: 10.1056/NEJMe2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu Q., Wang X.F., Qiang Y. Expert consensus on the diagnosis and treatment of viral pneumonia in children (2019) Zhong guo Shi Yong Er Ke Za Zhi. 2019;34:801–807. [Google Scholar]
- 51.Arabi Y.M., Mandourah Y., Al-Hameed F. Corticosteroid therapy for critically ill patients with Middle East respiratory syndrome. Am J Respir Crit Care Med. 2018;197:757–767. doi: 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- 52.WHO Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected. 2020. https://www.who.int/docs/defau lt-sourc e/coron aviru se/clini cal-manag ement -of-novel -cov.pdf?sfvrs n=bc7da 517_2
- 53.The Subspecialty Group of Respiratory Diseases. The Editorial Board, Chinese Journal of Pediatrics Diagnosis and treatment guideline of community acquired pneumonia in children (2013 the First part) Zhong hua Er Ke Za Zhi. 2013;51:745–752. [PubMed] [Google Scholar]
- 54.Wang X.F., Deng L., Liu J.R. Guidelines for the diagnosis and treatment of adenovirus pneumonia in children (2019 edition) Chin J Cli Infect Dis. 2019;12:161–1666. [Google Scholar]
- 55.Zheng Y.Y., Ma Y.T., Zhang J.Y. COVID-19 and the cardiovascular system. Nat Rev Cardiol. 2020;17:259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med. 2020;8:e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wan Y., Shang J., Graham R. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossignol J.F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J Infect Public Health. 2016;9:227–230. doi: 10.1016/j.jiph.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.