Abstract
Most viruses inhibit the innate immune system and/or the RNA degradation processes of host cells to construct an advantageous intracellular environment for their survival. Characteristic RNA sequences within RNA virus genomes or RNAs transcribed from DNA virus genomes contribute toward this inhibition. In this study, we developed a method called “Fate-seq” to comprehensively identify the RNA sequences derived from RNA and DNA viruses, contributing RNA stability in the cells. We examined the stabilization activity of 5,924 RNA fragments derived from 26 different viruses (16 RNA viruses and 10 DNA viruses) using next-generation sequencing of these RNAs fused 3′ downstream of GFP reporter RNA. With the Fate-seq approach, we detected multiple virus-derived RNA sequences that stabilized GFP reporter RNA, including sequences derived from severe acute respiratory syndrome-related coronavirus (SARS-CoV). Comparative genomic analysis revealed that these RNA sequences and their predicted secondary structures are highly conserved between SARS-CoV and the novel coronavirus, SARS-CoV-2, which is responsible for the global outbreak of the coronavirus-associated disease that emerged in December 2019 (COVID-19). These sequences have the potential to enhance the stability of viral RNA genomes, thereby augmenting viral replication efficiency and virulence.
Keywords: Virus, RNA stability, Functional sequence, SARS-CoV, SARS-CoV-2, COVID-19
Highlights
-
•
RNA stability is an important regulator for viral life-cycle.
-
•
Fate-seq enables measurement of stabilization activity of RNAs derived from viruses.
-
•
Conserved sequences among coronaviruses including SARS-CoV-2 have stabilizing activity.
1. Introduction
Viral particles carry either a double-stranded DNA genome or a single- or double-stranded RNA genome. Viral genomes are replicated in the host cells through the expression of viral genes, a process that is a key factor in viral virulence. Host cells attempt to restrict viral amplification by activating innate immune signaling pathways such as RIG-I-mediated response [1] and/or RNA degradation enzymes such as XRN1 and RNase L [2]. To cope with host responses, certain viruses inhibit the innate immune system and RNA degradation through the suppressor activity of characteristic viral RNA sequences. For example, high-dimensional RNA sequences such as those containing stem-loops or the pseudoknots typical of the flavivirus [termed subgenomic flavivirus RNA (sfRNA)] interrupt the progress of XRN1 along the viral genomic RNA, thereby inhibiting RNA degradation and resulting in the accumulation of degradation intermediates [3,4]. These partially degraded viral genomic RNAs inhibit TRIM25, an ubiquitin ligase recognizing RIG-I, which then inhibits the host immune response. The Group C enterovirus contains phylogenetically conserved RNA structures that competitively inhibit RNase L, the antiviral endoribonuclease in host cells [5]. Thus, several viruses have evolved to carry RNA fragments with characteristic high-dimensional structures within their genomic RNA or in RNAs transcribed from their genomic DNA to regulate the fate of viral replication in host cells through the regulation of RNA degradation. However, a systematic study of the sequences regulating viral RNA stability has not yet been performed.
Here, we developed a system called “Fate-seq” to comprehensively identify the RNA sequences derived from the genomes of RNA and DNA viruses that contribute toward the stability of viral RNA in the cells. The Fate-seq system incorporates the following procedures: (i) DNA sequences derived from genomes of RNA viruses and DNA viruses are fragmented in silico (viral sequences), (ii) a vector library is constructed by inserting these viral sequences downstream of the coding sequence (CDS) of a reporter gene in an expression vector (vector library), (iii) the resulting chimeric mRNAs are transcribed in vitro using the vector library as the template (mRNA library), (iv) the mRNA library is transfected into cells and later retrieved at a set time point, and (v) the remaining chimeric mRNAs in the cells are measured with the use of next-generation sequencing (NGS) to assess how the viral sequences influenced the mRNA stability.
Fate-seq revealed that multiple viral sequences, including 21 viral sequences derived from the genome of severe acute respiratory syndrome-related coronavirus (SARS-CoV), were relatively stable. Furthermore, the sequences of the identified viral sequences derived from the SARS-CoV genome were highly conserved within the Coronaviridae family, and some of the conserved sequences have a potential to form notable secondary structures. Additionally, the secondary structures are conserved in the novel coronavirus related to the ongoing coronavirus-associated disease outbreak that first emerged in December 2019 (COVID-19). Thus, the viral sequences identified by Fate-seq may enhance the genomic stability of SARS-CoV and SARS-CoV-2 within their hosts, where they have potential to inhibit the host’s cellular RNA degradation machinery and to contribute toward viral replication efficiency and virulence.
2. Results
2.1. Stability analysis of viral RNA fragments
We developed “Fate-seq” to identify RNA sequences within RNA viral genome and the RNA fragments derived from viral DNA genome to assess their stabilizing activity in human cells. Fate-seq consists of the following four steps: (i) in silico design of viral sequences based on the genomic sequences of DNA and RNA viruses, (ii) construction of a vector library harboring viral sequences, (iii) an mRNA library from the vector library, and (iv) transfection of the mRNA library into cells and quantification of the remaining mRNA after a set incubation period by NGS.
2.1.1. Design of the viral sequences
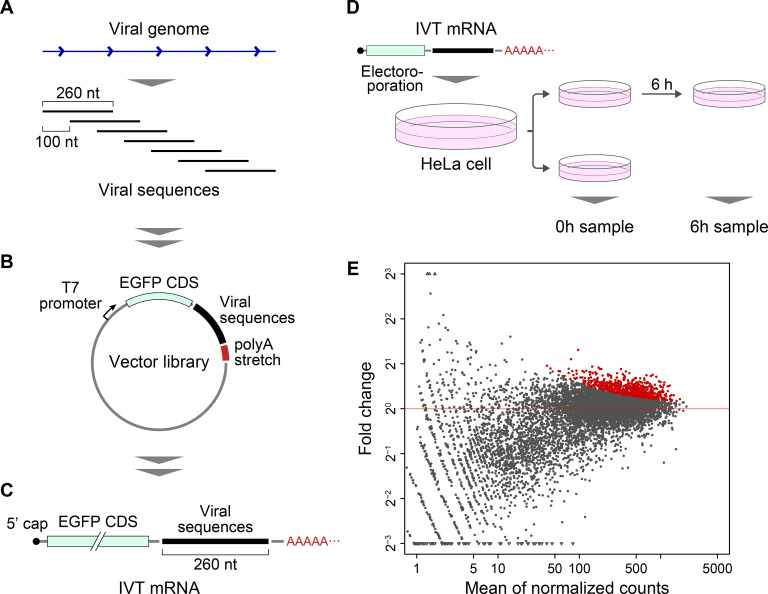
We selected 26 different viruses (10 DNA viruses and 16 RNA viruses) on the basis of Baltimore classification [6] as seeds for identifying viral fragments of interest (Table S1). The genome sequences of these viruses were obtained from the RefSeq database (ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/viral/) and RNA fragments of 260 base in length were generated based on these viral genomic sequences. The fragments were designed by sliding the 260-nt capture region at 100-nt intervals along the viral genomes (Fig. 1 A). Finally, we obtained a total of 5,924 fragments, termed as “viral sequences”, for further characterization.
Fig. 1.
Schematic illustration of Fate-seq. (A) Design of the viral sequences. The viral sequences of 260 nt in length were generated by sliding the capture region at 100-nt intervals along each viral genome. A total of 26 different viruses were used to generate the viral sequences. (B) Structure of the vector library. The viral sequences were inserted immediately downstream of the EGFP coding sequence (CDS) in a reporter vector harboring the T7 promoter, the EGFP CDS, and a polyA stretch (A60). (C) The mRNA library. In vitro transcription (IVT) of the vector library was used to produce the mRNA library, in which each mRNA contained the EGFP CDS, a viral sequence, as well as a polyA stretch. (D) Transfection and retrieval of the mRNA library. The mRNA library was transfected into HeLa cells using electroporation, and the transfected cells were then divided into two wells. The mRNA retrieved from the cells immediately after the transfection was termed the “0 h sample”. The mRNA retrieved from the cells 6 h after the transfection was termed the “6 h sample”. (E) The MA plot comparing read counts between 0 h and 6 h samples. The x-axis average of log2 read counts in 0 h and 6 h. The y-axis represents log2 ratio of 6 h–0 h. The red and black dots represent viral sequences showing significant increases in 6 h compared to 0 h (adjusted p-value<0.05) or not, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.1.2. Construction of the vector library
We constructed a vector library by inserting the designed viral sequences into the NsiI site located in the downstream of the enhanced green fluorescent protein (EGFP) CDS (Fig. 1B).
2.1.3. Construction of the mRNA library
We generated the mRNA library through in vitro transcription (IVT) of the vector library after NsiI digestion (Fig. 1C). Since mRNAs incorporating modified nucleosides such as pseudouridine and methylcytidine may make these RNAs less immunogenic [7], pseudouridine instead of uridine and methylcytidine instead of cytidine were used as substrates for IVT. We also added an anti-reverse cap analog (ARCA) into the substrate mix to act as a modified nucleoside for incorporation as the 5′ cap of the IVT mRNA, to avoid inhibition of translation [8]. Since recognition of the phosphate group at the 5′ end of mRNAs by RIG-I triggers the immune system [9], we dephosphorylated the 5′ ends of the IVT mRNA.
2.1.4. Transfection of the mRNA library into cells and quantification of the retrieved mRNA using NGS analysis
We transfected the mRNA library into a HeLa cells by electroporation and retrieved the total RNA from these cells at 0 or 6 h after transfection (Fig. 1D). The RNA samples were reverse-transcribed into cDNA, and the level of the remaining viral sequences in each sample was quantified using NGS. For reverse transcription into cDNA, we used primers recognizing the primer-binding sites added onto the 5′ and 3′ ends of the viral sequences during design of the viral sequences to allow selective amplification of the inserted viral sequences. These primers were connected with index for multiplex sequencing and sequencing adapters (P7 and P5) (Table S2), to make these sequences be added to the 3′ and 5′ ends of the amplification product during PCR.
2.2. Identification of regions contributing toward the stability of viral genomes
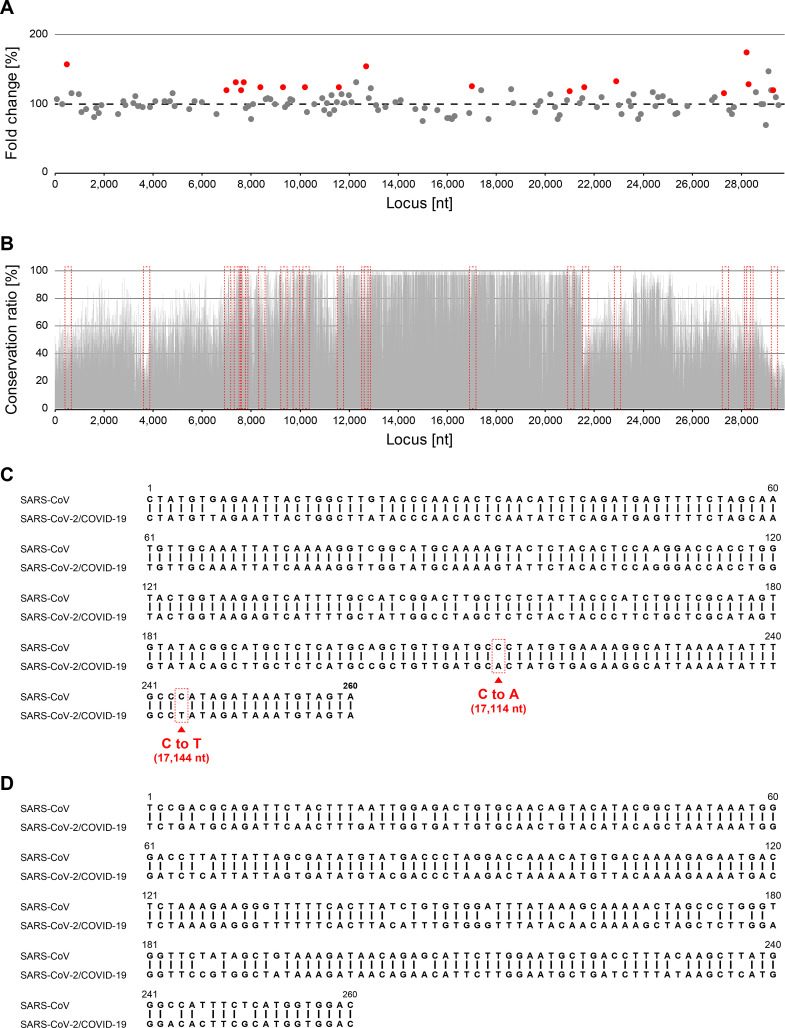
The NGS reads were aligned to the viral genomic sequences used to design the viral sequences. More than 95% of the reads could be aligned, of which approximately 55% and 45% mapped to the forward and reverse strand, respectively. We then quantified the number of reads that aligned on the region of the viral sequences. Note that, since the total abundance of the introduced mRNA library decreased in a time-dependent manner, the counts in each sample were not absolute but were rather indicative of relative abundance among the mRNAs introduced into the cells. To identify viral sequences that confers RNA stability, we searched for the viral sequences with statistically significant increase in 6 h compared to 0 h samples. From the 11,848 viral sequences (corresponding to 5,924 fragments from forward and reverse strands), we filtered lowly-expressed sequences, resulting in the remaining 7,442 sequences. Among these, 625 viral sequences (377 from DNA viruses and 248 from RNA viruses) were significantly increased in 6 h compared to 0 h (adjusted p-value < 0.05) (Fig. 1E; Table S3; Table S4). These included viral sequences derived from various RNA viruses such as SARS-CoV. Since the 1,087 viral sequences derived from RNA viruses were derived directly from the parental viral genomes, these were considered more likely to function in a similar manner in vivo in host cells. Therefore, we focused on the viral sequences derived from RNA viruses, especially SARS-CoV, which is the RNA virus species with the highest number of increase viral sequences (Table S3), for further analysis. The SARS-CoV genome used to design the viral sequences consists of 29,751 bases, generating 296 viral sequences from the forward strand. Among these 296 viral sequences, 21 viral sequences significantly increased in the 6 h samples (Fig. 2 A; Table S5). We termed these sequences “RNA stabilizing regions”.
Fig. 2.
Identification of genomic sequences contributing to the stability of the SARS-CoV genome. (A) Fold changes in the CPM values of individual viral sequences. The x-axis indicates the location of individual viral sequences along the SARS-CoV genome from which these viral sequences were derived. The y-axis indicates the fold change (%) in the counts per million mapped read (CPM) values between the RNA samples extracted at 0 h and 6 h after transfection into HeLa cells. Gray dots indicate viral sequences with a CPM >5 and coefficient of variation (CV) < 10% at both 0 h and 6 h after transfection. Red dots indicate viral sequences that displayed significantly increased abundance in the 6 h sample compared with the 0 h sample (adjusted p-value<0.05). (B) Conservation rate of each base along the SARS-CoV genome. The x-axis indicates the locations of individual bases, while the y-axis indicates the conservation rate of each base compared with other viruses within the Coronaviridae. (C and D) Sequence alignment between 16,901 and 17,160 nt (COV001) (C) and 20,901–21,160 nt (COV002) (D) of the SARS-CoV genome with corresponding regions along the SARS-CoV-2 genome. Vertical lines indicate base homology. Red squares indicate mutations critical for the formation of the stem-loops specific to SARS-CoV-2 (further described in Fig. 3). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.3. Examination of genomic conservation within the Coronaviridae
To examine whether the identified RNA stabilizing regions are evolutionarily conserved, we compared the genomes of 37 viruses belonging to the Coronaviridae family (Table S6), including human and bat coronaviruses from the RefSeq database (ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/viral/) and SARS-CoV-2 from the NCBI Nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore/MN908947). The obtained genome sequences were multiply aligned and the conservation rate of each base was calculated relative to the SARS-CoV genome. The data revealed that while the conservation rate was relatively low upstream and downstream along the genome, it was high near the central region (7,000–21,500 nt; Fig. 2B). Moreover, among the 21 viral sequences found to have significantly increased abundance 6 h after transfection, we found two with sequences exhibiting relatively high rates (>55%) of conservation (16,901–17,160 nt and 20,901–21,160 nt in the SARS-CoV genome; Table S5). In addition, to examine whether these regions play a role in the genomic stability of SARS-CoV-2, we compared the sequences of these regions between SARS-CoV and SARS-CoV-2. This comparison revealed that 92.7% and 84.6% of the nucleotides in the 16,901–17,160 nt and 20,901–21,160 nt regions in the SARS-CoV genome, respectively, were conserved in the corresponding regions of the SARS-CoV-2 genome (Fig. 2C and D). We named these two regions COV001 and COV002, respectively.
2.4. Secondary structure of the stabilizing elements
Some type of high-dimensional structures such as secondary structure regulates RNA stability [10,11]. To predict the potential for secondary structures in the two RNA stabilizing regions identified, we applied the CentroidFold tool [12].
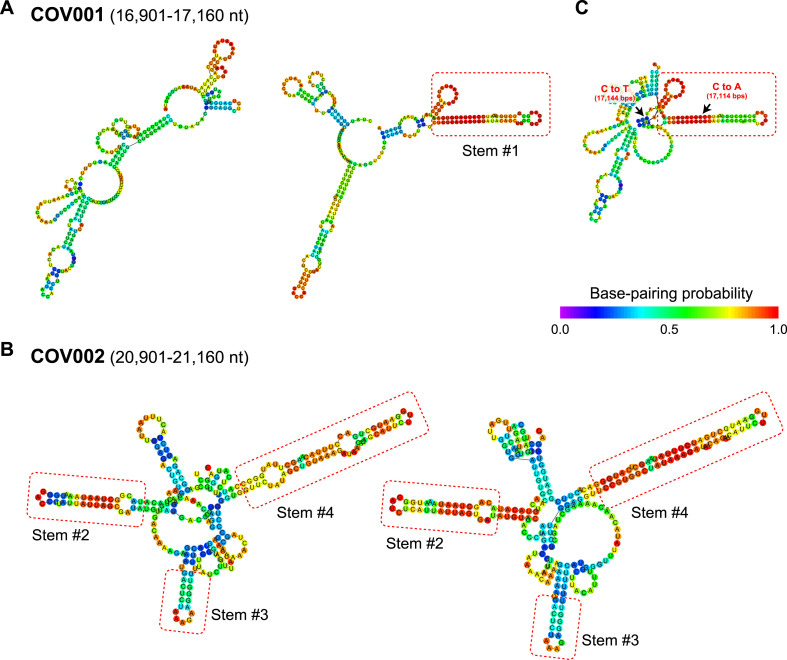
The COV001 region of SARS-CoV did not contain any probable stem-loops, whereas the corresponding region in SARS-CoV-2 did contain one such structure (Stem #1) (Fig. 3 A). In contrast, the three stem-loops predicted in COV002 of SARS-CoV (Stem #2–4) were also inferred in the corresponding genomic region of SARS-CoV-2 (Fig. 3B). Since Stem #1 is specific for SARS-CoV-2 and is predicted to be a long and stable stem-loop, it may contribute toward the specific characteristics of SARS-CoV-2. In silico mutations of COV001 in SARS-CoV of C to A at 17,114 nt and C to T at 17,144 nt formed a putative stem-loop similar in structure to that of Stem #1 (Fig. 3C). Note that the SARS-CoV-2 genomic regions corresponding to COV001 and COV002 were almost completely conserved among the 1,116 SARS-CoV-2 genomic sequences sampled from human hosts (Materials and Methods): 1,107 genomes (99.2%) for COV001 and 1,109 genomes (99.4%) for COV001 were identical (Fig. S1), indicating that their secondary structures are also highly conserved among SARS-CoV-2 strains. The co-occurrence of these mutations in SARS-CoV-2 may enhance the genomic stability of this virus within host cells through inhibition of the host’s RNA degradation machinery, thereby contributing toward enhanced viral replication and virulence.
Fig. 3.
Predicted secondary structure of viral sequences highly conserved between SARS-CoV and SARS-CoV-2. (A) Secondary structure of the COV002 region of SARS-CoV (left) and the corresponding region of the SARS-CoV-2 genome (right). (B) Secondary structure of the COV001 region of SARS-CoV (left) and the corresponding region of the SARS-CoV-2 genome (right). (C) The secondary structure of the COV001 region of SARS-CoV was mutated in silico. The putative stem-loop structure caused by in silico mutation at the positions indicated by the black arrows suggest that these mutations are critical for the formation of a stem-loop similar to that of Stem #1 observed for the corresponding region of SARS-CoV-2. Inferred stem-loops are indicated with red boxes (Stem #1–4), and base coloring was used to indicate the base-pairing probability. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3. Discussion
In this study, we developed “Fate-seq” for identification of RNA sequences contributing RNA stabilization. Using 26 viral genomes as seeds, we evaluated the stability of individual regions of these viral genomes in parallel using genome-derived RNA fragments, and we detected stabilizing elements within multiple viral genomes including that of SARS-CoV. Moreover, comparative genomic analysis of the Coronaviridae indicated that two of these stabilizing regions (COV001 and COV002) were highly conserved in the Coronaviridae, suggesting that these sequences may play an important role in viral function in the host cell. Comparison of the SARS-CoV and SARS-CoV-2 genomes revealed that the sequences of COV001 and COV002 are highly conserved to the corresponding regions of the SARS-CoV-2 genome. Inference of the secondary structure of these sequences indicated that the region in SARS-CoV-2 corresponding to COV001 forms a stem-loop that was not inferred for SARS-CoV. In contrast, both COV002 and the corresponding region in SARS-CoV-2 form multiple stem-loops. This potential difference in the secondary structure may contribute toward the virulence of SARS-CoV-2. The identified sequences, which might affect viral RNA stability and RNA-binding proteins binding to these sequences, may offer possible targets for the treatment of viral infectious diseases including COVID-19.
4. Materials and Methods
4.1. Design and synthesis of viral sequences
The genomic sequences of 26 different viral genomes across all groups of the Baltimore classification [6,13] (Table S1) were acquired from the RefSeq database (ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/viral/). The 5,924 fragments of 260 nt in length were generated in silico by sliding the capture region along 100-nt intervals (see Fig. 1A). To the 5′ and 3′ ends of these fragments, 20-nt sequences including primer binding sites for PCR amplification and recognition sites for homologous recombination were added, which extended the fragments to 300 nt in length. These fragments were then synthesized as a mixture of single-stranded oligonucleotides by Twist Bioscience (San Francisco, CA, USA). The synthesized oligonucleotides were then amplified by 24 cycles of PCR using the KAPA HiFi HotStart ReadyMix PCR Kit (Roche, Basel, Switzerland).
4.2. Construction of the vector library
The vectors, which harbored the T7 promoter, the EGFP CDS, and a NsiI restriction enzyme recognition site followed by a polyA stretch (A60), were linearized and then amplified by PCR using the PrimeSTAR GXL DNA Pol (Takara Bio, Inc., Japan). To generate the vector library, the linearized vector and viral sequences were mixed with 5× In-Fusion HD Enzyme Premix (Takara Bio, Inc.), according to manufacture’s instruction. The vector library was transformed into DH5alpha competent cells, and the vector library was retrieved with the PureYield Plasmid Midiprep (Promega, WI, USA).
4.3. Construction of the mRNA library
The vector library was treated with NsiI restriction enzyme to linearize the vectors through cleavage at the recognition site located immediately upstream of the polyA stretch. The MEGAscript T7 Transcription Kit (Thermo Fisher Scientific, MA, USA) was used for IVT with 300 mM ATP, 300 mM metCTP, 300 mM pseudoUTP, 60 mM GTP, and 240 mM ARCA, in accordance with the manufacturer’s instructions. After IVT, the template vectors were eliminated with 1.0 μL of TURBO DNase (2.0 unit/μL) at 37 °C for 1 h. The mRNA library was purified with the NucleoSpin TriPrep kit (Takara Bio, Inc.). To dephosphorylate the 5′ ends of the synthesized mRNA, 100 units of Antarctic Phosphatase (NEB) were added to the mRNA followed by incubation at 37 °C for 1 h.
4.4. Transfection of mRNA library into the cells by electroporation
HeLa cells (1.0 × 106) were treated with 10 μg IVT mRNAand 100 μL of Nucleofector SF (Lonza, Basel, Switzerland) in a 100-μL cuvette with the “program CN-114” protocol according to the manufacturer’s instructions.
4.5. NGS analysis
The total RNAs were reverse-transcribed with the PrimeScript RT Master Mix (Takara Bio, Inc.). The cDNA was mixed with 30 μL of 1× Tris/EDTA (TE) buffer and 40 μL of Agencourt AMPure XP beads (QIAGEN, Hilden, Germany) for purification. The purified cDNA was eluted with 34.5 μL of 1× TE buffer and amplified with the KAPA HiFi HotStart ReadyMix PCR Kit (Roche) over 18 cycles. For the amplification, we used PCR primers connected with the index for multiplex sequencing and sequencing adapter (P7 and P5), to make these sequences be added to 3′ and 5′ ends of the amplification product. The cDNAs were sequenced by the Hiseq2500 system (single-end, 50 base pair reads).
4.6. Quantification of remaining mRNA after transfection
The Bowtie2 tool [14] was used to map the NGS reads using default parameters. The genome indexes used for read alignment were built using Bowtie2 (bowtie2-build) based on the genomic sequences used to design the viral sequences. The number of reads that aligned exactly to location of the designed viral sequences was counted, and these counts were corrected based on the total number of mapped reads to generate the CPM values.
4.7. Statistical analysis of the increased abundance of viral sequences
From the read count table, viral sequences whose sum of counts across samples were >6 reads were filtered. Then, the DESeq2 [15] package (version 1.24.0) of the R language (version 3.6.1) was used with the default parameters to search for differentially expressed viral sequences between the 0 h and 6 h samples (adjusted p-value<0.05).
4.8. Identification of the conserved regions within Coronaviridae genomes
Genome sequences of viruses from the Coronaviridae family were obtained from the RefSeq database (ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/viral/). The genome sequence of SARS-CoV-2 was obtained from the NCBI Nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore/MN908947). The genomic sequences of these Coronaviridae were multiply aligned using the ClustalW2 tool [16,17] with the Slow/Accurate alignment parameters (all other parameters were set to default). The conservation rate was calculated as the percentage of viruses analyzed that had the same base as that at each position along the SARS-CoV genome.
The LAST web service (http://lastweb.cbrc.jp/) was used to extract COV001 and COV002-corresponding regions in SARS-CoV-2. The genomic sequences of SARS-CoV-2 sampled from human hosts were downloaded from the GISAID database [18]. Only genome sequences that had no ‘N’ nucleotides and were incorporated into the Nextstrain website (https://nextstrain.org/ncov/global; last access: 2020/0) were selected, resulting in 1,116 SARS-CoV-2 genomes (Table S2). MAFFT (v7.450) [19] was used with the parameters ‘--auto --clustalout --reorder’ for multiple alignment of the COV001- and COV002-correspanding regions in MN908947 and 1,116 SARS-CoV-2 whole genome sequences.
4.9. Estimation of secondary structure
The sequences comprising 16,901–17,160 nt (COV001) and 20,901–21,160 nt (COV002) of the SARS-CoV genome and the corresponding regions of the SARS-CoV-2 genome were extracted as described above. The secondary structures in these two regions were inferred with the CentroidFold tool (http://rtools.cbrc.jp/centroidfold/). For in silico validation of the importance of the two observed SARS-CoV-2 mutations, C at 17,114 nt and C at 17,144 nt in the COV001 region of the SARS-CoV genome were replaced with A and T, respectively. The secondary structure of the modified sequence was also inferred using the CentroidFold tool.
Author contributions
H.W., K.K., and N.A. conceived the project. H.W., Y.Y., and N.A. designed and performed the experiments. K.K. E.H., T.T., and, H.O. analyzed the data. H.W., K.K., E.H., T.T., H.O., and N.A. wrote the manuscript. All authors approved the submitted manuscript.
Funding sources
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant numbers 17KK0163, 18H02570, 18KT0016, and 16H06279). K.K. received funding from JSPS KAKENHI (Grant Number: 19K16635) and the Takeda Life Science Foundation. T.T. received funding from JSPS KAKENHI (Grant Number: 19K24361 and 20K19915). H.O. received funding from JSPS KAKENHI (Grant Number: 19K20394).
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
We thank to Ms. Taguchi for cooperation of sample preparation of NGS measurements. The NGS measurements were performed by Prof. Hiroyuki Aburatani at Research Center for Advanced Science and Technology, The University of Tokyo. The computational analysis in this study was performed on the National Institute of Genetics (NIG) supercomputer system at the Research Organization of Information and Systems (ROIS) in Tokyo, Japan, and Human Genome Center (HGC) supercomputer system at the Institute of Medical Science, the University of Tokyo.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2020.05.008.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Sequence conservation among 1,116 SARS-CoV-2 genomes. (A) Number of SARS-CoV-2 genomes whose sequences in the COV001- and COV002-corresponding regions were identical to those of MN908947. (B) Conservation rate of each base along the COV001-corresponding region among the SARS-CoV-2 genomes. (C) Conservation rate of each base along the COV002-corresponding region among the SARS-CoV-2 genomes.
Viruses used for design of viral sequences.
Primers used for PCR amplification.
Viral sequences exhibiting significant increase in read counts.
Quantification of the relative abundance of individual viral sequences.
Significantly increased viral sequences from the SARS-CoV genome.
Coronaviridae viruses used for genome alignments against SARS-CoV.
GISAID accession IDs for 1,116 SARS-CoV-2 genome.
References
- 1.Pichlmair A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Weber F., Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 2.Nogimori T., Nishiura K., Kawashima S., Nagai T., Oishi Y., Hosoda N., Imataka H., Kitamura Y., Kitade Y., Hoshino S.-I. Dom34 mediates targeting of exogenous RNA in the antiviral OAS/RNase L pathway. Nucleic Acids Res. 2019;47:432–449. doi: 10.1093/nar/gky1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moon S.L., Anderson J.R., Kumagai Y., Wilusz C.J., Akira S., Khromykh A.A., Wilusz J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012;18:2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manokaran G., Finol E., Wang C., Gunaratne J., Bahl J., Ong E.Z., Tan H.C., Sessions O.M., Ward A.M., Gubler D.J., Harris E., Garcia-Blanco M.A., Ooi E.E. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Townsend H.L., Jha B.K., Han J.-Q., Maluf N.K., Silverman R.H., Barton D.J. A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L. RNA. 2008;14:1026–1036. doi: 10.1261/rna.958908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baltimore D. Expression of animal virus genomes. Bacteriol. Rev. 1971;35:235–241. doi: 10.1128/mmbr.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durbin A.F., Wang C., Marcotrigiano J., Gehrke L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio. 2016;7 doi: 10.1128/mBio.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng Z.-H., Sharma V., Singleton S.F., Gershon P.D. Synthesis and application of a mRNA cap analog. Org. Lett. 2002;4:161–164. doi: 10.1021/ol0167715. [DOI] [PubMed] [Google Scholar]
- 9.Pichlmair A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Weber F., Reis E Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 10.Brown J.A., Steitz J.A. Intronless β-globin reporter: a tool for studying nuclear RNA stability elements. Methods Mol. Biol. 2016;1428:77–92. doi: 10.1007/978-1-4939-3625-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilusz J.E., JnBaptiste C.K., Lu L.Y., Kuhn C.-D., Joshua-Tor L., Sharp P.A. A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato K., Hamada M., Asai K., Mituyama T. CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 2009;37:W277–W280. doi: 10.1093/nar/gkp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbalenya A.E. Increasing the number of available ranks in virus taxonomy from five to ten and adopting the Baltimore classes as taxa at the basal rank. Arch. Virol. 2018;163:2933–2936. doi: 10.1007/s00705-018-3915-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson J.D., Higgins D.G., Gibson T.J., Clustal W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larkin M.A., Blackshields G., Brown N.P., Chenna R., Mcgettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 18.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence conservation among 1,116 SARS-CoV-2 genomes. (A) Number of SARS-CoV-2 genomes whose sequences in the COV001- and COV002-corresponding regions were identical to those of MN908947. (B) Conservation rate of each base along the COV001-corresponding region among the SARS-CoV-2 genomes. (C) Conservation rate of each base along the COV002-corresponding region among the SARS-CoV-2 genomes.
Viruses used for design of viral sequences.
Primers used for PCR amplification.
Viral sequences exhibiting significant increase in read counts.
Quantification of the relative abundance of individual viral sequences.
Significantly increased viral sequences from the SARS-CoV genome.
Coronaviridae viruses used for genome alignments against SARS-CoV.
GISAID accession IDs for 1,116 SARS-CoV-2 genome.