Abstract
The objective of this study was to quantify the emissions of enteric CH4 from growing Hereford steers raised under feedlot conditions based on contrasting levels of residual feed intake (RFI). A repeated measurements experiment was conducted over 20 d to determine CH4 production from two groups of nine Hereford steers, with contrasting RFI values (mean ± SD): low RFI (LRFI group; −0.78 ± 0.22 kg DMI/d) vs. high RFI (HRFI group; 0.83 ± 0.34 kg DMI/d). Steers were selected from a larger contemporary population in which the RFI was evaluated. Steers were maintained under confined conditions with ad libitum access to water and feed, comprising a total mixed ration of 55% sorghum silage, 21% barley silage, 21% corn grain, and 3% protein–mineral–vitamin–premix, provided twice a day. Before the beginning of CH4 measurements, the live weight of both groups of animals was determined, which on average (±SEM) was 357.0 ± 5.11 and 334.0 ± 10.17 kg in the LRFI and HRFI groups, respectively. Methane emission (g/d) was measured on each animal with the sulfur hexafluoride (SF6) tracer technique, during two consecutive periods of 5 d. Individual daily intake and feeding behavior characteristics were measured using a GrowSafe automated feeding system (Model 6000, GrowSafe Systems Ltd, Airdrie, Alberta, Canada). Methanogens in the ruminal content were quantified using quantitative polymerase chain reaction with primers targeting the mcrA gene. Methane emission was near 27% lower in animals with LRFI when expressed in absolute terms (g/d; 26.8%; P = 0.009), by unit of dry matter intake (g CH4/kg; 27.9%, P = 0.021), or as % of gross energy intake (26.7%; P = 0.027). These differences could not be explained by differences in amount of total of methanogens (average = 9.82 log10 units; P = 0.857). However, there were some differences in animal feeding behavior that could explain these differences (e.g., LRFI animals tended to spend less time in feeders). Our results suggest that, in Hereford steers, the selection by RFI values is a promising mitigation strategy for the reduction of the emission of enteric CH4.
Keywords: enteric methane, feeding behavior, methanogens, residual feed intake, steers
INTRODUCTION
Emissions from global livestock represent 14.5% of anthropogenic greenhouse gases emissions and 44% of livestock emissions are in the form of methane (CH4) (FAO, 2013). There are several alternatives to reduce CH4 emissions such as improving feed quality, using CH4 inhibitors, and breeding for lower CH4.
Residual feed intake (RFI) is calculated as the difference between observed and predicted animal intake in relation to performance. Negative RFI values indicate high efficiency of converting feed to products. According to Arthur and Herd (2005), RFI has been described as an animal characteristic of medium heritability, which has been associated with CH4 emissions (Cassandro et al., 2013). Animals with low RFI (LRFI) are reported to be more efficient (consume less feed than expected at equal body weight and gain) and produce less emissions compared with high RFI (HRFI) animals (Basarab et al., 2013). Consequently, animal selection for LRFI has been proposed as an alternative to mitigate CH4 emissions (Hegarty et al., 2007; Basarab et al., 2013; Pickering et al., 2015; de Haas et al., 2017) although some studies on grazing conditions did not find differences between divergent RFI animals (Velazco et al., 2017). Up to now, research on this topic is limited. Although it is recognized that RFI selection will contribute for reducing CH4 emission intensity (emissions per unit of product; Waghorn and Hegarty, 2011), some authors reported no differences between LRFI and HRFI animals in emission per kg of DM ingested (Hegarty et al., 2007). Inconsistencies among results may have been caused by different and sometimes limited RFI ranges used in the experiments. Other animal characteristics, such as feeding behavior or microbiota, have not been reported quantitatively as secondary variables for the interpretation of CH4 emission related to RFI.
Most information available regarding this topic has been developed on dairy cattle or Angus beef cattle. Hereford is one of the few beef cattle breeds publishing estimated breeding values for RFI (i.e., Uruguay and Canada, Ravagnolo et al., 2018). It would be of interest the study of the association of this new trait with CH4 emission on the breed.
The goal of this study was to quantify the emissions of enteric CH4 from growing Hereford steers under confined conditions in relation to contrasting levels of RFI.
MATERIALS AND METHODS
The study was carried out in December 2014 at Kiyú Test Station of the Hereford Breeders Association located in San José, Uruguay (GPS Coordinates: S Latitude 34° 35.797′, W length 56° 42.302′).
Experimental Design, Treatments, Animals, and Management
The study was conducted with 112 Hereford steers that were part of a 3-yr project, with the goal of building a training population of 1,000 animals for genomic selection for RFI in the Uruguayan Hereford breed (Navajas et al., 2014). One hundred twelve animals corresponded to one of the three RFI tests of the first year of the project and were originally obtained from five commercial farms.
The estimate of RFI was based on measurements of individual feed intake using the GrowSafe automated feeding system (Model 6000, GrowSafe Systems Ltd, Airdrie, Alberta, Canada), in two pens with eight feeders each, with ad libitum access to water and food. Individual feed intake data used for RFI calculation were recorded during a conventional 70-d test, after 28 d of adaptation to diet and feeding system. Animals were fed twice a day with a fully mixed ration (total mixed ration [TMR]) of sorghum silage, barley silage, corn grain, and protein–mineral–vitamin–premix (Table 1).
Table 1.
Ingredients and chemical composition of TMR
| Diet ingredient, % (as-fed basis) | |
|---|---|
| Sorghum silage | 55 |
| Barley silage | 21 |
| Corn grain | 21 |
| Protein–mineral–vitamin premix1 | 3 |
| Chemical composition | |
| DM, % | 44.95 |
| CP, % | 12.57 |
| NDF, % | 47.59 |
| ADF, % | 30.87 |
| TDN, %2 | 64.11 |
| A, % | 7.94 |
| ADL, % | 9.30 |
| DMD, % | 65.02 |
| GE, Mcal/kg DM | 3.93 |
| ME, Mcal/kg DM3 | 2.31 |
1Soybean meal 77%, Mycosorb 0.9%, Rumensin 0.3%, Urea 8.1%, CaCO3 7.3%, NaCl 5.5%, Rovimix Feedlot 0.9%.
2TDN = Total digestible nutrients = 96.03 − (1.034 × ADF, %) (Alemu et al., 2017).
3ME, MJ/kg DM = [(TDN, %/100) × 4.4 Mcal/kg TDN] × 4.184 MJ DE/Mcal × 0.82 MJ ME/MJ DE (Alemu et al., 2017). Values are means.
After completing the test, 112 steers were ranked based on their RFI values that were computed based on the following model as proposed by Basarab et al. (2003), based on Koch et al. (1963):
| (1) |
where DMI was the dry matter intake (DMI) (kg); ADG was average daily gain (kg/d), MLWt was the metabolic weight defined as mid test LW0.75 (kg); Bfat was the subcutaneous fat depth measured at the end of test by ultrasound (mm); b0 is the intercept; and b1, b2, and b3 were the partial regression coefficients for each trait on DMI. The residual (e) is taken to represent RFI. The effect of pen was not included because preliminary analysis indicated that it was not significant (P > 0.01). The R2 of the multiple regression used for RFI estimation was 0.80.
DMI was calculated as the average of 68 valid daily records adjusted by the dry matter percentage. Live weight (LW) measurements were performed every 14 d, early in the morning and without fasting. Two consecutive days of LW measurements were used for the initial, middle, and final weight and one for intermediate measurements. The ADG was calculated by the regression of all LW during the test, considering only those with R2 ≥ 0.95. Subcutaneous dorsal fat depth was measured by ultrasound by certified technicians using the Aloka SSD 500 unit, equipped with a linear matrix transducer of 3.5 MHz and 17.2 cm (Aloka Co. Ltd, Tokyo, Japan). The ultrasound images were collected in the field and interpreted later with the off-line interpretation software Biosoft Toolbox (version 2.1 of Biotronics Inc.).
Based on the RFI ranking, two groups of nine steers (18 animals) with extreme RFI values were selected for the present study (mean ± SD: −0.78 ± 0.22 kg DMI/d vs. 0.83 ± 0.34 kg DMI/d). Both groups were confined for 20 more days and fed the same diet with same feeding regime used during the RFI test. DMI and feeding behavior characteristics were measured using a GrowSafe-automated feeding system. Each feeding event was registered for all the animals according to Basarab et al. (2003), and individual data of duration feeding events, head down times, and meal size were used to calculate meal duration time (s), average meal size (kg), meals per day, head down duration (s), head down duration per meal (s), and feed rate (g/s).
A statistical experimental design of repeated measurements over time, including two treatments and two 5-d measurement periods, was used. The treatments consisted of the two contrasting levels of RFI of the animals: LRFI and HRFI.
Determination of CH4 Emissions
The sulfur hexafluoride (SF6) tracer technique (Johnson et al., 1994), as modified by Gere and Gratton (2010), was used to quantify daily methane (CH4) emissions. Eight days before beginning the CH4 measurements, eight animals from each RFI group (a total of 16 animals) were given an oral permeation tube filled with sulfur hexafluoride (SF6) using a plastic dosing applicator. The SF6 in the tube was used as a marker for gas emissions. Background concentrations of CH4 and SF6 were measured during the same period. Two animals (one of each RFI group) were used as control of SF6 background, so the SF6 permeation tubes were not administered. For CH4 background (environmental baseline), a collection container was placed inside the pen. Daily permeation rates (PRs) of SF6 from the tubes averaged (mean ± SEM) 6.0 ± 0.55 and 5.9 ± 0.58 mg/d in the LRFI and HRFI groups, respectively. The emission of enteric CH4 was measured for the 18 animals during two consecutive 5-d periods following the procedure performed by Dini et al. (2017). The first 8 d of the study were used for the adaptation of the animals to the use of the CH4 collection containers and for the stabilization of rumen SF6 levels. During the study, the LW of the animals was recorded at the beginning and at the end of each measurement period. The average LWs were (mean ± SEM) 357.0 ± 5.11 and 334.0 ± 10.17 kg in the LRFI and HRFI groups, respectively.
The collection of exhaled and eructated gas was performed using two 0.5-liter stainless steel containers per animal. At the beginning of each period, these containers were evacuated, cleaned with N2, and placed on each side of the animal’s head. At the end of each period, the containers were removed from the animals and the post-sampling pressure was measured. Containers with pressure values of 400 to 600 mb were considered valid according to Gere and Gratton (2010) and Gere (2012), as this manipulation ensures good quality samples. Less than 10% of the containers with pressure values <400 mb were considered atypical, and therefore removed from the experiment. Five subsamples were extracted from each container, stored in 12 mL vacutainers (Exetainer; Labco Ltd, Lampeter, Ceredigion, UK), and analyzed using a gas chromatograph (Agilent 7890A, Santa Clara, CA) with a flame ionization detector (FID) and an electron capture detector (ECD) for determining CH4 and SF6 concentrations, respectively. After obtaining a chromatographic analysis of samples, CH4 emissions per animal were calculated using the PR of each SF6 capsule and the concentrations above the background of CH4 and SF6 (in ppm and ppt, respectively) using the following equation:
| (2) |
Determination of Amount of Total Methanogens
After completing the second CH4 measurement period, the 18 steers were reincorporated to the original herd, and maintained on grazing conditions for 6 to 10 mo until slaughter. During the summer, they grazed on sorghum pasture (Sorghum vulgare) in a vegetative stage, supplemented with sorghum silage, and followed by oat pasture (Avena sativa) also in vegetative stage with corn grain supplementation of 6 kg/steer until slaughter. Steers were slaughtered when they reached 500 kg LW (607 ± 11.6-d average age of the slaughter). At the time of slaughter, ruminal content was sampled and stored at −80°C until use. Deoxyribonucleic acid (DNA) was extracted and quantification of the number of copies of the methyl coenzyme-M reductase gene (mcrA) was determined by the quantitative polymerase chain reaction (qPCR). This gene was used as a functional marker to enumerate methanogens (Luton et al., 2002). Reactions (25 μL) were performed in a BioRad CFX 96 thermocycler using Power SYBR Green PCR Master Mix (Applied Biosystems) and primers qmrcA-F and qmrcA-R; reaction conditions were the same as in Denman et al. (2007). Standard curves for absolute quantification and efficiency estimation were performed according to Fraga et al. (2015). Three replicates of each DNA sample (20 ng) were used. A nontemplate (sterile distilled water) negative control was loaded on each plate run.
Chemical Analysis
Samples were taken daily from the TMR and were weighed and dried at 60°C for 48 h. They were ground to pass through a 1-mm screen and analyzed to determine chemical composition. DM, ash (A), and total nitrogen (CP = N × 6.25) content were analyzed according to AOAC (1990) (methods ID 934.01, ID 942.05, and ID 955.04, respectively). The NDF was analyzed with heat stable amylase and sodium sulfite. ADF and ADL were determined using the methods of Van Soest et al. (1991), including residual ash. The in vitro digestibility was determined according to Tilley and Terry (1963), and gross energy (GE) was determined with an adiabatic bomb calorimeter (Autobomb Gallenkamp; Loughborough, Leics, United Kingdom).
Statistical Analysis
Data were analyzed using version 9.0 of SAS software (SAS Institute, Inc., Cary, NC). Intake and CH4 emissions data were analyzed as repeated measures, with the steers as the subject of the repeated measurements, using the PROC MIXED procedure according to the following model:
| (3) |
which included the fixed effect of the treatment (Ti = LRFI and HRFI), the fixed effect of the period (Pj = 1 and 2), their interaction [(TxP)ij], and the residual error (eijk).
Daily weight gain, feeding behavior variables, and the number of copies per mg of the mcrA gene obtained from the ruminal content were analyzed using the PROC MIXED procedure of SAS (SAS Institute Inc., Cary, NC), with the animal as the experimental unit according to the following model:
| (4) |
which included the fixed effect of the treatment (Ti = LRFI and HRFI).
Means were compared with a Tukey–Kramer test. Normality test was applied to all variables (Shapiro and Wilk, 1965). Data of copies of mrcA/mg, average meal size, meal duration time, and feed rate had heterogeneous variances and therefore were analyzed after logarithmic transformation. These data were analyzed by PROC MIXED as described previously. The average values were considered different when P ≤ 0.05 and tended to differ if 0.05 ≤ P ≤ 0.10.
RESULTS
Less efficient animals (HRFI) had a higher DMI and produced more CH4 than the most efficient animals (LRFI), but there were no differences in the ADG (mean = 0.82 kg/d; P = 0.923; Table 2) between the groups evaluated. The most efficient animals spent less time eating (P < 0.001), remained less time with the head down (P = 0.029), and their feed rate tended to be higher (P = 0.062) with respect to HRFI ones (Table 2). However, no differences were found in average meal size (mean = 0.90 kg, P = 0.549), in the number of meals (mean = 13.2, P = 0.627), or in the time they remained with their heads down at each meal (mean = 347.8 s, P = 0.141).
Table 2.
Intake, daily gain, and feeding behavior characteristics for two contrasting levels of RFI of the animals: lower RFI (LRFI, −0.78 ± 0.22 kg DMI/d) and higher RFI (HRFI, 0.83 ± 0.34 kg DMI/d) steers
| LRFI | HRFI | SEM | P value | |
|---|---|---|---|---|
| Intake, kg DM/d | 9.33 | 10.6 | 0.33 | 0.014 |
| GEI, Mcal/d | 37.1 | 42.3 | 1.32 | 0.014 |
| ADG, kg/d | 0.83 | 0.80 | 0.190 | 0.923 |
| Feed rate, g/s | 4.05 | 2.75 | – | – |
| Feed rate, log 10 | 1.32 | 0.98 | 0.120 | 0.062 |
| Meal duration time, s | 11802 | 15404 | 397.7 | <0.0001 |
| Average Meal Size, kg | 0.81 | 0.98 | – | – |
| Average Meal Size, log 10 | −0.23 | −0.12 | 0.121 | 0.549 |
| Meals per day | 12.8 | 13.6 | 1.14 | 0.627 |
| Head down duration/meal, s | 265 | 431 | – | – |
| Head down duration/meal, log 10 | 5.47 | 5.89 | 0.190 | 0.141 |
| Head down duration, s | 3024 | 4519 | 440.9 | 0.029 |
GEI = gross energy intake; ADG = average daily gain. Values are average per treatment (n = 8/treatment).
The most efficient animals (LRFI) exhibited a 26.8% lower CH4 emission (g/d) and a lower CH4 yield when expressed as g/kg DMI (27.9%) or as percent of GEI (26.7%) compared with HRFI animals (Table 3).
Table 3.
Emission of CH4 for two contrasting levels of RFI of the animals: low RFI (LRFI, −0.78 ± 0.22 kg DMI/d) and high RFI (HRFI, 0.83 ± 0.34 kg DMI/d) steers
| Treatment | P value | |||||
|---|---|---|---|---|---|---|
| LRFI | HRFI | SEM | Treatment | Period | Treat*Per | |
| Emissions, g/d | 194 | 265 | 15.9 | 0.009 | 0.423 | 0.911 |
| CH4/kg DMI, g/kg | 20.3 | 28.1 | 1.76 | 0.021 | 0.107 | 0.390 |
| Ym, % | 6.72 | 9.17 | 0.580 | 0.027 | 0.102 | 0.391 |
| CH4/kg NDFI, g/kg | 43.0 | 59.2 | 3.73 | 0.024 | 0.103 | 0.396 |
| CH4/kg ADFI, g/kg | 65.4 | 92.0 | 5.69 | 0.015 | 0.106 | 0.398 |
Treat*Per = interaction between treatment and period; Emissions = daily CH4 emissions; CH4/kg DMI = CH4 emission per kilogram of dry matter intake; Ym = methane yield; CH4/kg NDFI = CH4 emission per kilogram of neutral detergent fiber intake; CH4/kg ADFI = CH4 emission per kilogram of acid detergent fiber. Values are average per treatment (n = 8/treatment).
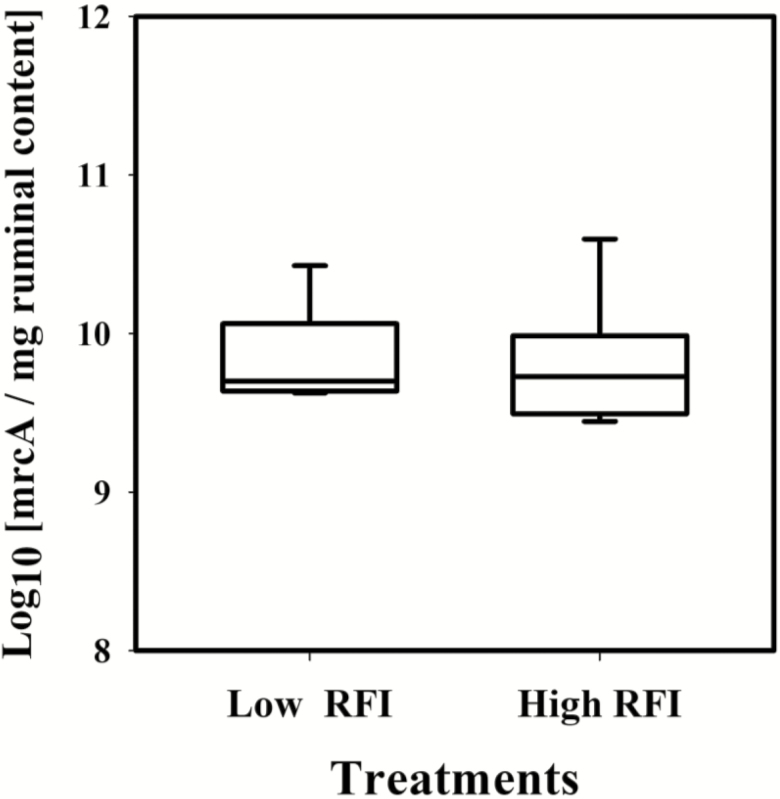
There were no differences in the amount of total methanogens between treatments, as the average number of copies of the mcrA gene was similar (mean = 9.82 log10 units; P = 0.86; Figure 1). The amplification efficiency of the qPCR reaction calculated from the standard curve was 96.6% with R2= 0.99, ensuring accuracy of the method.
Figure 1.
Number of copies of the mrcA gene for two contrasting levels of RFI of the animals: low RFI (LRFI, −0.78 ± 0.22 kg DMI/d) and high RFI (HRFI, 0.83 ± 0.34 kg DMI/d) steers. The line inside each box represents treatment median and the ends of the whiskers represent the minimum and maximum values of all of the data for each treatment.
DISCUSSION
The current study confirmed that high conversion efficient animals with low values of RFI consume a lower quantity of feed and dedicate less time to meals (12% and 23% less in this case, respectively). The lower time spent eating has been proposed by Basarab et al. (2013) as one of the mechanisms involved in the higher efficiency, associated with lower energy spent in feeding activities.
Average methane emissions (194 and 265 g/d for LRFI and HRFI, respectively), were comparable with those observed by Manafiazar et al. (2016) and Alemu et al. (2017) for crossbred beef replacement heifers using the GreenFeed system (205 and 202 to 222 g/d, respectively). As expected, LRFI animals, with a RFI average of −0.78 kg DMI/d, emitted up to 27% less CH4 than the HRFI animals, with a RFI average of 0.83 kg DMI/d. The less time dedicated to meal led to a strong tendency for a higher ingestion rate of the more efficient animals. In this sense, it is demonstrated that animals with higher intake rates also have faster passage rates of particles in the rumen, which is not necessarily associated with lower digestibility (Pérez-Ruchel et al., 2013), at least for high-quality diets. In our study, a higher rate of passage of the rumen particles could explain the lower CH4 emission in LRFI animals (Nkrumah et al., 2006). However, different associations between RFI and methane emissions have been previously reported. A recent study by Alemu et al. (2017) used both GreenFeed system and respiration chambers to evaluate CH4 emissions of crossbreed beef heifers of HRFI and LRFI. These authors reported that LRFI and HRFI animals emitted similar CH4 per day and per kg DMI when measured in respiration chambers, but there were differences in daily CH4 emissions when the GreenFeed system was used. Meanwhile Velazco et al. (2017) found higher predicted DMI and higher CH4 emissions in lower RFI Angus yearling steers and heifers under grazing conditions. Although these results were attributed to diet quality, it would be necessary to take into account other variables affecting RFI, including feeding behavior characteristics (not reported in the aforementioned study) and feeding type and conditions (grazing vs. confined).
In this study, differences in CH4 emissions could not be directly associated with differences in mcrA quantification, as also observed in previous studies (Zhou et al., 2011; Danielsson et al., 2012; Rira et al., 2016), indicating that animals with low values of RFI do not necessarily present less Archaea populations. However, it is necessary to point out that in our study rumen content was obtained at slaughter after 6 to 9 mo of finishing under grazing conditions, which started when the CH4 measurements were completed. Wallace et al. (2014) studied microbiome and methane emissions on beef cattle consuming different diets and observed that the rates archaea/bacteria were similar in rumen samples collected in vivo and postmortem, as well as the correlation between archaea/bacteria and methane emission, with independence of the diet. Since the RFI is an intrinsic condition of the animal, differences between LRFI and HRFI groups on CH4 emmisions and microbiome characteristics should persist along the life with independence of the diet consumed. However, some authors have suggested that the relationship between RFI and methane emission depends on the diet (Velazco et al., 2017). According to Jones et al. (2011), lower RFI cows produced less CH4 than those of HRFI only when they were fed a high-quality pasture. Based on the scarce existing information about the relationship between the RFI, methane emission, and microbiome, it is necessary to consider that the different diet consumed could have weakened the association between Archaea populations and methane emissions in lower and higher RFI groups. Additionally, methanogen quantification could not represent actual methanogenic activity. In future studies, quantification of mcrA mRNAs should shed light on understanding methanogens activity.
There are recent studies that question the use of RFI as a strategy to mitigate enteric CH4 emissions (Jones et al., 2011; Alemu et al., 2017; Velazco et al., 2017). However, it should be noted that in these prior studies, the populations used did not present a strong divergence in RFI, which may have affected the results in relation to the emission of CH4. Jones et al. (2011) reported that the evaluated populations had average RFI values of −0.69 vs. 0.68 kg/d, whereas Alemu et al. (2017) reported values of −0.25 vs. 0.29. In our study, the evaluated animals presented a greater contrast in the values of RFI (−0.78 vs. 0.83 kg/d). Furthermore, Nkrumah et al. (2006) found differences in CH4 emissions with animal populations displaying a HRFI contrast (−1.18 vs. 1.25 kg/d).
The results of this study show that animals with lower RFI emit less CH4, indicating that selection by the level of RFI is a promising mitigation strategy, which can be used synergistically with the management of dietary components. Future research should investigate the association between the RFI and the quantity and activity of methanogens, as well as between these and the emission of CH4. This will provide a more comprehensive understanding of the potential and scope of RFI on the reduction of CH4 emission.
ACKNOWLEDGMENTS
We thank the Uruguayan Hereford Breeders Association, Ministry of Livestock, Agriculture and Fishery, National Meat Institute, Clemente Estable Biological Research Institute, and Rural Association of Uruguay. Financial support was partially provided by the National Agency for Research and Innovation (grant RTS_1_2012_1_3489 and POS_NAC_2013_1_11_718). We are also grateful to our collaborators, Julieta Mariotta, Ana Rabaza, Juan de La Fuente, and Diego Romaso.
LITERATURE CITED
- Alemu A. W., Vyas D., Manafiazar G., Basarab J. A., and Beauchemin K. A.. 2017. Enteric methane emissions from low- and high-residual feed intake beef heifers measured using greenfeed and respiration chamber techniques. J. Anim. Sci. 95:3727–3737. doi:10.2527/jas.2017.1501 [DOI] [PubMed] [Google Scholar]
- AOAC 1990. Official methods of analysis. 15th ed Arlington, VA: Association of Official Analytical Chemists. [Google Scholar]
- Arthur P. F., and Herd R. M.. 2005. Efficiency of feed utilisation by livestock – implications and benefits of genetic improvement. Can. J. Anim. Sci. 85, 281–290. doi:10.4141/A04-062 [Google Scholar]
- Basarab J. A., Beauchemin K. A., Baron V. S., Ominski K. H., Guan L. L., Miller S. P., and Crowley J. J.. 2013. Reducing GHG emissions through genetic improvement for feed efficiency: effects on economically important traits and enteric methane production. Animal 7(Suppl 2):303–315. doi:10.1017/S1751731113000888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basarab J. A., Prince M. A., Aalhus J. L., Okine E. K., Snelling W. M., and Lyle K. L.. 2003. Residual feed intake and body composition in young growing cattle. Can J. Anim. Sci. 83:189–204. doi:10.4141/A02-065 [Google Scholar]
- Cassandro M., Mele M., and Stefanon B.. 2013. Genetic aspects of enteric methane emission in livestock ruminants. Ital. J. Anim. Sci. 12:450–458. doi:10.4081/ijas.2013.e73 [Google Scholar]
- Danielsson R., Schnürer A., Arthurson V., and Bertilsson J.. 2012. Methanogenic population and CH₄ production in Swedish dairy cows fed different levels of forage. Appl. Environ. Microbiol. 78:6172–6179. doi:10.1128/AEM.00675-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denman S. E., Tomkins N. W., and McSweeney C. S.. 2007. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol. Ecol. 62:313–322. doi:10.1111/j.1574-6941.2007.00394.x [DOI] [PubMed] [Google Scholar]
- Dini Y., Gere J. I., Cajarville C., and Ciganda V. S.. 2017. Using highly nutritious pastures to mitigate enteric methane emissions from cattle grazing systems in South America. Animal Production Science. (“in press”). doi:10.1071/AN16803 [Google Scholar]
- Food and Agriculture Organization of the United Nations. 2013. Key facts and findings. [accessed 18 May 2018] http://www.fao.org/newa/story/en/item/197623/icode/.
- Fraga M., Fernández S., Cajarville C., Martínez M., Abin-Carriquiry J. A., and Zunino P.. 2015. In vitro modulation of rumen microbiota and fermentation by native microorganisms isolated from the rumen of a fed exclusively on pasture bovine. Ann. Microbiol. 65:2355–2362. doi:10.1007/s13213-015-1077-2 [Google Scholar]
- Gerber P. J., Hristov A. N., Henderson B., Makkar H., Oh J., Lee C., Meinen R., Montes F., Ott T., Firkins J.,. et al. 2013. Technical options for the mitigation of direct methane and nitrous oxide emissions from livestock: a review. Animal 7(Suppl 2):220–234. doi:10.1017/S1751731113000876 [DOI] [PubMed] [Google Scholar]
- Gere J. I. 2012. La técnica de trazado por SF6 para medir emisiones de metano de rumiantes en pastoreo : desarrollos metodológicos y algunas aplicaciones. [PhD Thesis], Tandil, Argentina: Faculty of Exact Sciences. [Google Scholar]
- Gere J. I., and Gratton R.. 2010. Simple, low cost flow controllers for time averaged atmospheric sampling and other applications. Lat. Am. Appl. Res. 40:377–381. [Google Scholar]
- de Haas Y., Pszczola M., Soyeurt H., Wall E., and Lassen J.. 2017. Invited review: phenotypes to genetically reduce greenhouse gas emissions in dairying. J. Dairy Sci. 100:855–870. doi:10.3168/jds.2016-11246 [DOI] [PubMed] [Google Scholar]
- Hegarty R. S., Goopy J. P., Herd R. M., and McCorkell B.. 2007. Cattle selected for lower residual feed intake have reduced daily methane production. J. Anim. Sci. 85:1479–1486. doi:10.2527/jas.2006-236 [DOI] [PubMed] [Google Scholar]
- Johnson K., Huyler M., Westberg H., Lamb B., and Zimmerman P.. 1994. Measurement of methane emissions from ruminant livestock using a sulfur hexafluoride tracer technique. Environ. Sci. Technol. 28:359–362. doi:10.1021/es00051a025 [DOI] [PubMed] [Google Scholar]
- Jones F. M., Phillips F. A., Naylor T., and Mercer N. B.. 2011. Methane emissions from grazing Angus beef cows selected for divergent residual feed intake. Anim. Feed Sci. Technol. 166–167: 302–307. doi:10.1016/j.anifeedsci.2011.04.020. [Google Scholar]
- Koch R.M., Swiger L.A., Chambers D., and Gregory K. E.. 1963. Efficiency of feed use in beef cattle. J. Anim. Sci. 22:486–494. [Google Scholar]
- Luton P. E., Wayne J. M., Sharp R. J., and Riley P. W.. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148(Pt 11):3521–3530. doi:10.1099/00221287-148-11-3521 [DOI] [PubMed] [Google Scholar]
- Manafiazar G., Zimmerman S., and Basarab J.. 2016. Repeatability and variability of short-term spot measurement of methane and carbon dioxide emissions from beef cattle using GreenFeed emissions monitoring system. Can. J. Anim. Sci. 126:CJAS-2015-0190. doi:10.1139/CJAS-2015-0190 [Google Scholar]
- Navajas E., Pravia M. I., Lema M., Clariget J., Aguilar I., Brito G., Peraza P., Dalla Rizza M., and Montossi F.. 2014. Genetic improvement of feed efficiency and carcass and meat quality of Hereford cattle by genomics. Proceedings of the 60th International Congress Meat Science Technology, ICoMST Punta del Este, Uruguay; p. 17–20. [Google Scholar]
- Nkrumah J. D., Okine E. K., Mathison G. W., Schmid K., Li C., Basarab J. A., Price M. A., Wang Z., and Moore S. S.. 2006. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J. Anim. Sci. 84:145–153. [DOI] [PubMed] [Google Scholar]
- Pérez-Ruchel A., Repetto J. L., and Cajarville C.. 2013. Suitability of live yeast addition to alleviate the adverse effects due to the restriction of the time of access to feed in sheep fed only pasture. J. Anim. Physiol. Anim. Nutr. (Berl). 97:1043–1050. doi:10.1111/jpn.12008 [DOI] [PubMed] [Google Scholar]
- Pickering N. K., Oddy V. H., Basarab J., Cammack K., Hayes B., Hegarty R. S., Lassen J., McEwan J. C., Miller S., Pinares-Patiño C. S.,. et al. 2015. Animal board invited review: genetic possibilities to reduce enteric methane emissions from ruminants. Animal 9:1431–1440. doi:10.1017/S1751731115000968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravagnolo O., Aguilar I., Crowley J.J., Pravia M.I., Lema M., Macedo F.L., Scott S., and Navajas E.A.. 2018. Accuracy of genomic predictions of residual feed intake in Hereford with Uruguayan and Canadian training populations. Proceedings of the World Congress on Genetics Applied to Livestock Production, Volume Electronic Poster Session – Species – Bovine (beef); Auckland, New Zealand 1, p. 723. [Google Scholar]
- Rira M., Morgavi D. P., Popova M., Marie-Magdeleine C., Silou-Etienne T., Archimède H., and Doreau M.. 2016. Ruminal methanogens and bacteria populations in sheep are modified by a tropical environment. Anim. Feed Sci. Technol. 220:226–236. doi: 10.1016/j.anifeedsci.2016.08.010 [Google Scholar]
- Shapiro S., and Wilk M.. 1965. An analysis of variance test for normality (complete samples). Biometrika. 52:591–611. doi:10.1093/biomet/52.3-4.591 [Google Scholar]
- Tilley J. M. A., and Terry R. A.. 1963. A two-stage technique for the in vitro digestion of forage crops. J. Br. Grassl. Soc. 18:104–111. [Google Scholar]
- Van Soest P. J., Robertson J. B., and Lewis B. A.. 1991. Methods for dietary fiber, neutral detergent fiber, and non-starch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74:3583–3597. doi:10.3168/50jds.S0022-0302(91)78551-2 [DOI] [PubMed] [Google Scholar]
- Velazco J. I., Herd R. M., Cottle D. J., and Hegarty R. S.. 2017. Daily methane emissions and emission intensity of grazing beef cattle genetically divergent for residual feed intake. Anim. Prod. Sci. 57:627–635. doi:10.1071/AN15111 [Google Scholar]
- Waghorn G. C., and Hegarty R. S.. 2011. Lowering ruminant methane emissions through improved feed conversion efficiency. Anim. Feed Sci. Technol. 166–167: 291–301. doi: 10.1016/j.anifeedsci.2011.04.019 [Google Scholar]
- Wallace R. J., Rooke J. A., Duthie C. A., Hyslop J. J., Ross D. W., McKain N., de Souza S. M., Snelling T. J., Waterhouse A., and Roehe R.. 2014. Archaeal abundance in post-mortem ruminal digesta may help predict methane emissions from beef cattle. Sci. Rep. 4:5892. doi:10.1038/srep05892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Chung Y. H., Beauchemin K. A., Holtshausen L., Oba M., McAllister T. A., and Guan L. L.. 2011. Relationship between rumen methanogens and methane production in dairy cows fed diets supplemented with a feed enzyme additive. J. Appl. Microbiol. 111:1148–1158. doi:10.1111/j.1365-2672.2011.05126.x [DOI] [PubMed] [Google Scholar]