Abstract
Purpose
Exome sequencing (ES) is increasingly used for the diagnosis of rare genetic disease. However, some pathogenic sequence variants within the exome go undetected due to the technical difficulty of identifying them. Mobile element insertions (MEIs) are a known cause of genetic disease in humans but have been historically difficult to detect via ES and similar targeted sequencing methods.
Methods
We developed and applied a novel MEI detection method prospectively to samples received for clinical ES beginning in November 2017. Positive MEI findings were confirmed by an orthogonal method and reported back to the ordering provider. In this study, we examined 89,874 samples from 38,871 cases.
Results
Diagnostic MEIs were present in 0.03% (95% binomial test confidence interval: 0.02–0.06%) of all cases and account for 0.15% (95% binomial test confidence interval: 0.08–0.25%) of cases with a molecular diagnosis. One diagnostic MEI was a novel founder event. Most patients with pathogenic MEIs had prior genetic testing, three of whom had previous negative DNA sequencing analysis of the diagnostic gene.
Conclusion
MEI detection from ES is a valuable diagnostic tool, reveals molecular findings that may be undetected by other sequencing assays, and increases diagnostic yield by 0.15%.
Keywords: diagnostics, exome sequencing, rare disease, Mendelian disease, mobile elements
INTRODUCTION
Mobile elements are discrete segments of genomic DNA that can insert new copies elsewhere in the genome through an RNA intermediate. In humans, the vast majority of mobile elements no longer retain the ability to create new insertions.1 A minority of mobile elements, primarily from the L1, Alu, and SVA families, remain active and are capable of producing new insertions.2–4 It is estimated that 1 in 12–14 live human births has a de novo MEI.5 As such, MEIs are an endogenous and ongoing source of variation in human genomes.
MEIs cause disease by directly disrupting coding sequence or otherwise altering messenger RNA (mRNA). For example, the first disease-causing MEI variant identified in humans was a hemizygous variant in F8, causing hemophilia through loss of function.6 A recent review created a comprehensive catalog of 124 disease-causing MEIs, and a subsequent study identified an additional 34 disease-causing MEIs in a number of genes and conditions.7,8
MEIs are not routinely detected in clinical diagnostic testing except for some known founder events with moderate population frequencies.9–11 Specialized variant-calling algorithms are needed to detect MEIs from next-generation sequencing (NGS) data, and the paucity of known disease-causing MEIs is thus likely due to ascertainment bias.
Recently, diagnostic laboratories have been evaluating hereditary cancer genes for MEIs.8,12 To date, there has been no systematic analysis of the role MEIs play more broadly in rare disease; however, it is estimated that MEIs are responsible for disease in 0.04% to 0.1% of individuals with suspicion of genetic disease.5,13 We applied MEI discovery to a prospective cohort of clinical exome samples. In doing so, we determined the overall burden of diagnostic MEIs in a referral population for exome sequencing.
MATERIALS AND METHODS
Patients
We analyzed 89,874 clinical exome sequencing (ES) samples from 6 November 2017 to 31 August 2019 (Table S1, Fig. S1). Samples were sequenced by Illumina HiSeq or NovaSeq 2 × 100 or 2 × 150 reads after hybridization capture using either Agilent Clinical Research Exome or IDT xGen Exome v1.0 baits as previously described.14 The study was conducted in accordance with all guidelines set forth by the Western Institutional Review Board, Puyallup, Washington (WIRB 20162523). Informed consent for genetic testing was obtained from all individuals undergoing testing, and WIRB waived authorization for use of de-identified aggregate data for these purposes. All positive findings were confirmed by Sanger sequencing and reported back to the ordering provider (Table S2). Patients for whom clinical data, including photos, are reported provided consent for their information to be presented. All reported variants were submitted to ClinVar and SCV IDs are pending. The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page (http://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/).
MEI detection
MEI detection tools that rely on discordant read pairs have reduced sensitivity on NGS capture data relative to selection-free genome sequencing or to larger insert libraries where there would be higher rates of discordant read pairs. MEIs are underrepresented in targeted sequence fragments, and smaller insert-size libraries used to increase on-target percentage lead to fewer discordant read pairs spanning MEIs.
Our clinical exomes have low rates of discordant read pairs (mean 1.4%, Fig. S2). Even so, MEIs occurring within a targeted capture region produce reads that partially map to the reference genome and partially map to MEI sequence (i.e., clipped reads). We therefore developed a custom MEI detection tool called SCRAMble (Soft Clipped Read Alignment Mapper) for application to targeted capture sequencing.
In brief, SCRAMble identifies clusters of soft clipped reads in a BAM file, builds consensus sequences, aligns to representative L1Ta, AluYa5, and SVA-E sequences, and outputs MEI calls (Fig. S3, Table S3). We estimate a technical sensitivity of 85.0–91.5%. and a precision of 93.8–99.9% (Supplementary Methods, Table S4, Fig. S4). All reported MEI calls were confirmed by Sanger sequencing.
Comparison with other MEI callers
We compared runtimes and MEI calls for MELT,15 Mobster,16 and SCRAMble for 1075 sequential ES samples from January 2019 (Fig. S5). We also resequenced 12 of the 14 MEI positive samples from our cohort, ran all three MEI callers, and examined recall of the pathogenic MEI (Supplementary Methods). SCRAMble had the highest overall recall rate of known MEI polymorphisms in targeted regions (Table S5). SCRAMble also had the highest recall rate of pathogenic MEIs in resequenced positive samples (Table S5). For targeted sequencing, it may be valuable to apply MEI detection that does not rely upon discordant read pairs.
Statistical analysis
All statistical analyses, unless otherwise noted, were performed in the R computing environment.
Code availability
SCRAMble source code is available on GitHub for noncommercial use (https://github.com/GeneDx/scramble).
RESULTS
In November 2017, we began prospectively detecting MEIs in samples referred to our laboratory for clinical ES. In this study, we examined 89,874 samples from 38,871 cases. Among these individuals were 21,806 complete child–parent trios. The probands were referred for genetic testing for a variety of clinical indications with neurodevelopmental delay being the most common (Fig. S1). SCRAMble does not distinguish between heterozygous and homozygous genotypes, however, we use postprocessing steps in our pipeline to estimate genotype based on the SCRAMble call and local sequence coverage. Herein, we refer to MEIs detected in a given sample as “calls” and refer to unique MEI sites as “variants.” There were 1,101,790 calls of 23,014 MEI variants (12,380 Alus, 8531 L1s, and 2103 SVAs) for an average of 12.2 MEI calls per person (Fig. S6). The MEIs in this study show hallmarks of L1-mediated target primed reverse transcription including a median target site duplication of 14 bp, variable 5′ truncation, and similarity to the 5′TT/AAAA3′ endonuclease recognition sequence at the insertion site (unpublished data).
In this study, 8753/38,871 cases (22.5%) had a positive molecular diagnosis. Fourteen MEIs were classified as pathogenic or likely pathogenic by applying American College of Medical Genetics and Genomics (ACMG) criteria as previously described14 and confirmed by Sanger sequencing (Table 1). Of these, 13 were sufficient to explain the patients’ phenotypes, and one was identified as a secondary finding in BRCA2. Thus, diagnostic MEIs accounted for 13/8753 (0.15%, binomial test 95% confidence interval [CI] 0.08–0.25%) of the positive cases and 13/38,871 (0.03%, binomial test 95% CI 0.02–0.06%) of all cases. An Alu insertion identified in MAK in one patient (Table 1) is a known founder event in Ashkenazi Jews.11 The secondary finding MEI in another patient (Table 1) was of the same MEI family, in the same orientation, and in the same position as a pathogenic variant that was previously reported;8 however, Qian et al. reported a full length Alu while the Alu observed here was truncated at the 5’ end by 220 bp (Table 1). The remaining pathogenic MEIs have not been previously described to the best of our knowledge.
Table 1.
Reported pathogenic mobile element insertions (MEIs).
| Gene | Classification | Transcript | Annotation | Inheritance | Orientation | TSD | 5’ truncation |
|---|---|---|---|---|---|---|---|
| ATM | LPATH | NM_139312 | c.169–4insL1 | AR | − | 21 bp | 8 bp |
| BRCA2a | PATH | NM_000059.3 | c.3407_3408insAlu | AD | + | 17 bp | 220 bp |
| CHD8 | LPATH | NM_001170629.1 | c.5419_5420insAlu | AD | + | 16 bp | None |
| DEPDC5 | PATH | NM_001242896 | c.2958_2959insAlu | AD | − | 15 bp | 211 bp |
| EFTUD2 | PATH | NM_001258353 | c.1277_1278insAlu | AD | − | 16 bp | 2 bp |
| ETFB | PATH | NM_001985 | c.426_427insAlu | AR | + | 16 bp | None |
| MAK | PATH | NM_001242385 | c.1297_1298insAlu | AR | − | 11 bp | None |
| NSD1 | PATH | NM_022455.4 | c.1926_1927insAlu | AD | − | 17 bp | None |
| OCRL | PATH | NM_001587 | c.2557_2558insL1 | XL | − | 17 bp | 5657 bp |
| OFD1 | PATH | NM_001330209 | c.416_417insAlu | XL | − | 15 bp | None |
| RPL11 | PATH | NM_000975 | c.375_376insSVA | AD | − | 16 bp | 716 bp |
| SLC26A3 | LPATH | NM_000111 | c.131+3insAlu | AR | − | 13 bp | 1 bp |
| USH2A | PATH | NM_206933.2 | c.8932_8933insAlu | AR | − | 14 bp | 1 bp |
| ZEB2 | PATH | NM_014795 | c.1600_1601insAlu | AD | − | 9 bp | None |
Orientation is relative to hg19.
AD autosomal dominant, AR autosomal recessive, Ins insertion, LPATH likely pathogenic, PATH pathogenic, TSD target site duplication, XL X-linked.
aSecondary finding. All other MEIs are considered diagnostic.
Twelve of the 13 probands with a diagnostic MEI had prior genetic tests that were negative, 8 of whom had sequencing-based tests. For three of the patients, the gene that contained the diagnostic MEI had been previously sequenced, but no positive results had been reported. Among the 13 diagnosed patients were those with multiple congenital anomalies, neurodevelopmental delay, and abnormalities of the eye, musculature, and metabolism. These are too few diagnostic MEIs from which to draw broader conclusions; however, we expect that the yield of diagnostic MEIs for various disease areas mirrors the referral population for ES and that pathogenic MEIs are not restricted by any particular disease.
In one case, prenatal ES identified a novel homozygous Alu insertion in exon 4 of ETFB, in a fetus with bilaterally enlarged microcystic kidneys, echogenic bowel, unilateral postaxial polydactyly, and an abnormal placenta. Biallelic loss of function variants in ETFB cause glutaric acidemia IIB.17 The parents were each confirmed to be heterozygous for the MEI. Consanguinity was denied and confirmed by kinship analysis from ES data. In addition, a 574-kb run of homozygosity was found in the proband covering the MEI site suggesting a possible Eastern European founder event approximately 35 generations ago (Fig. S7).
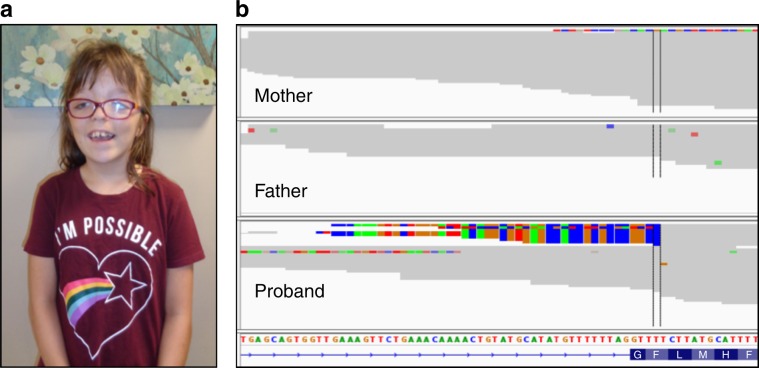
In another case, a female with global developmental delay, macrocephaly, agenesis of the corpus callosum, gray matter dysplasia, interhemispheric cysts, natal teeth, cleft tongue, lingual cyst, dysmorphic facial features, and digital anomalies was referred for ES with a differential diagnosis of oral–facial–digital (OFD) syndrome (Fig. 1). The family had previously pursued genetic testing by Sanger sequencing of OFD1 with negative results, but ES with MEI detection revealed a de novo Alu insertion in exon 6 (Fig. 1).
Fig. 1. Case report: de novo Alu insertion in OFD1 causes oral–facial–digital syndrome.
a Patient with a clinical diagnosis of oral–facial–digital syndrome for whom a diagnostic Alu was identified in exon 6 of the OFD1 gene. b Clipped read evidence of a minus strand Alu can be seen in proband, but not in either parent indicating a de novo event.
A 40-year-old male patient with intellectual disability, dilated cardiomyopathy, macrocephaly, kidney disease, scoliosis, and 2–3 toe syndactyly was referred for ES. This patient previously had microarray, fragile X, and Marfan-like connective tissue NGS panel testing, all with negative results. By applying MEI detection, an Alu insertion was detected in exon 10 of NSD1 that was absent from the mother and presumed de novo. No sample from the father was submitted for testing. Heterozygous, loss of function variants in NSD1 have been identified in patients with Sotos syndrome, which has substantial phenotypic overlap with the patient.
DISCUSSION
Mobile elements are a source of pathogenic variants in a referral patient population with suspected genetic etiology. By applying mobile element detection to our clinical diagnostic pipeline, we are able to improve diagnostic yield and provide clinicians and patients with diagnoses that are otherwise not apparent. MEIs account for positive molecular diagnosis in more than 1 in 700 diagnosed cases, a similar rate to one previously described.18 Similarly, Gardner et al. describe an analogous study using nearly 10,000 exome sequenced trios for probands with neurodevelopmental delay (NDD).5 We observe similar diagnostic rates (4/9738, 0.04% in Gardner et al., and 13/38,871, 0.03% in this study, two-sided Fisher p value 0.76). Thus, despite using different MEI detection methods (MELT15 vs. SCRAMble), different sequencing methods,14,19 and having independent cohorts, our study and Gardner et al., largely recapitulate each other.
We identified 13 diagnostic, pathogenic MEIs that were sufficient to explain patient phenotypes in individuals with rare disease and one pathogenic MEI as a secondary finding. Of note, all but one of these MEIs were novel and would not have been detected by genotyping of known founder events. The pathogenic MEIs described here, although few, were of substantial consequence to the patients and families involved. For the case of the 11-year-old girl with a clinical diagnosis of oral–facial–digital syndrome (Fig. 1), OFD1 had already been sequenced before the diagnostic MEI was found. Her first sequencing test was by Sanger sequencing. We suspect that the Alu insertion caused preferential polymerase chain reaction (PCR) amplification of the shorter, wild-type allele causing allele dropout and blinding the lab to the MEI. Before having a confirmed molecular diagnosis, there was the potential of unanticipated comorbidities of a condition of unknown molecular etiology. With a definitive molecular diagnosis, the patient’s condition can be confidently managed. Likewise, a de novo diagnostic MEI was found in NSD1 that led to a diagnosis of Sotos syndrome in an adult male. A confirmed diagnosis allows the proband to be appropriately screened and managed for potential comorbidities of Sotos syndrome such as cardiac and renal anomalies.
A limitation of our current diagnostic pipeline for MEIs is our inability to interpret noncoding MEIs on a case-by-case basis. There are now multiple examples of noncoding MEIs that cause Mendelian disease.7 It is possible we are detecting more disease-causing MEIs, but are unable to classify them as pathogenic since they are in noncoding regions.
Altogether, diagnostic MEIs accounted for a small proportion of positive cases (0.15%). We consider this the lower limit of the true diagnostic rate since MEI-containing sequence fragments are not captured as efficiently as wild-type alleles in library preparation and are thus underrepresented in capture-based NGS data. We expect the rate of diagnostic MEIs to climb as (1) improvements to bioinformatics tools and wet lab methods, including the transition to PCR-free clinical genome sequencing, increase the sensitivity for detecting MEIs, and (2) RNA and functional studies are performed to better understand the impact of regulatory and intronic MEIs. In the meantime, MEI discovery and genotyping are providing much needed answers for patients and families, including those who have already had genetic testing performed.
Supplementary information
Acknowledgements
The authors kindly thank the patients, families, and medical providers who participated in this study. The authors also thank Erin Ryan, Claire Teigen, Amanda Singleton, Omer Gokcumen, Kirsty McWalter, and Vlad Gainullin.
Disclosure
R.I.T., K.G., S.L., K.A., C.B., J.S., Z.Z., B.F., H.S., J.J. and K.R. are employed by GeneDx. J.J. and K.R. are shareholders of OPKO. The other authors declare no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41436-020-0749-x) contains supplementary material, which is available to authorized users.
References
- 1.Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Bennett EA, Keller H, Mills RE, et al. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brouha B, Schustak J, Badge RM, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Xing J, Grover D, et al. SVA elements: a hominid-specific retroposon family. J Mol Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 5.Gardner EJ, Prigmore E, Gallone G, et al. Contribution of retrotransposition to developmental disorders. Nat Commun. 2019;10:4630. doi: 10.1038/s41467-019-12520-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kazazian HH, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 7.Hancks DC, Kazazian HH., Jr Roles for retrotransposon insertions in human disease. Mob DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian Y, Mancini-DiNardo D, Judkins T, et al. Identification of pathogenic retrotransposon insertions in cancer predisposition genes. Cancer Genet. 2017;216–7:159–169. doi: 10.1016/j.cancergen.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe M, Kobayashi K, Jin F, et al. Founder SVA retrotransposal insertion in Fukuyama-type congenital muscular dystrophy and its origin in Japanese and Northeast Asian populations. Am J Med Genet. 2005;138:344–348. doi: 10.1002/ajmg.a.30978. [DOI] [PubMed] [Google Scholar]
- 10.Teugels E, De Brakeleer S, Goelen G, Lissens W, Sermijn E, De Grève J. De novo Alu element insertions targeted to a sequence common to the BRCA1 and BRCA2 genes. Hum Mutat. 2005;26:284–284. doi: 10.1002/humu.9366. [DOI] [PubMed] [Google Scholar]
- 11.Tucker BA, Scheetz TE, Mullins RF, et al. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci USA. 2011;108:E569–76. doi: 10.1073/pnas.1108918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vysotskaia VS, Hogan GJ, Gould GM, et al. Development and validation of a 36-gene sequencing assay for hereditary cancer risk assessment. PeerJ. 2017;5:e3046. doi: 10.7717/peerj.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J-M, Chuzhanova N, Stenson PD, Férec C, Cooper DN. Meta-analysis of gross insertions causing human genetic disease: novel mutational mechanisms and the role of replication slippage. Hum Mutat. 2005;25:207–221. doi: 10.1002/humu.20133. [DOI] [PubMed] [Google Scholar]
- 14.Retterer K, Juusola J, Cho MT, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 15.Gardner EJ, Lam VK, Harris DN, et al. The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res. 2017;27:1916–1929. doi: 10.1101/gr.218032.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thung DT, de Ligt J, Vissers LE, et al. Mobster: accurate detection of mobile element insertions in next generation sequencing data. Genome Biol. 2014;15:488. doi: 10.1186/s13059-014-0488-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colombo I, Finocchiaro G, Garavaglia B, et al. Mutations and polymorphisms of the gene encoding the beta-subunit of the electron transfer flavoprotein in three patients with glutaric acidemia type II. Hum Mol Genet. 1994;3:429–435. doi: 10.1093/hmg/3.3.429. [DOI] [PubMed] [Google Scholar]
- 18.Wimmer K, Callens T, Wernstedt A, Messiaen L, Starink T. The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion. PLoS Genet. 2011;7:e1002371.. doi: 10.1371/journal.pgen.1002371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.