Abstract
Radiotherapy with or without concurrent chemotherapy is the standard treatment for nasopharyngeal carcinoma (NPC) patients, whose efficacy is limited partly by intrinsic and acquired radioresistance. DNA methyltransferase 3B (DNMT3B) has been reported to participate in tumorigenesis via DNA methylation, but its role in mediating progression and radioresistance of NPC remains unclear. Therefore, we conducted the following studies to explore the relationship between DNMT3B and NPC. Here, we found that DNMT3B was elevated in NPC tissues and predicted the poor prognosis of NPC patients. We demonstrated for the first time that ionizing radiation could induce DNMT3B, which might be one of the reasons for radioresistance. Silencing of DNMT3B inhibited migration and invasion via suppressing epithelial-mesenchymal transition (EMT) in NPC cells. Furthermore, silencing DNMT3B restored and activated p53 and p21 via DNA demethylation, which led to cell cycle arrest and apoptosis, resulting in increased radiosensitivity of NPC both in vitro and in vivo. DNMT3B functions as a novel oncogene in the radioresistance of NPC through regulating EMT, cell cycle, and apoptosis. Therefore, DNMT3B could be a potential target for NPC treatment.
Keywords: DNMT3B, p53, p21, DNA methylation, nasopharyngeal carcinoma, radioresistance
Introduction
Nasopharyngeal carcinoma (NPC) is a head and neck cancer with a specific geographic distribution. It affected about 130,000 patients worldwide in 2018, with the highest incidence occurring in Southeast Asia, especially in southern China.1,2 Radiotherapy (RT) with or without concurrent chemotherapy is the standard treatment for NPC patients. Overall, the prognosis of NPC has dramatically improved in the past few decades owing to better radiotherapy technology and effective application of chemotherapy.3 However, NPC radioresistance and metastasis are still major hurdles. Therefore, there is a need for continuous studies into the mechanism of refractory NPC to expand the clinical benefit to a more extensive population and improve patient prognosis.
DNA methylation provides an important epigenetic mechanism for initiation and progression of cancers. Its main function is to enhance the transcriptional silencing of genes, especially tumor suppressor genes. Aberrant methylation of tumor suppressor genes has been found in most human cancers, which in turns leads to gene silencing and causes carcinogenesis and development of cancers.4 DNA methyltransferases (DNMTs) are involved in transcriptional silencing and play an important role in the DNA methylation of malignant cancers. There are five DNMTs encoded by human genome: DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L. DNMT1, DNMT3A, and DNMT3B are canonical cytosine-5 DNMTs that catalyze the addition of methylation marks to genomic DNA. In contrast, DNMT2 and DNMT3L are non-canonical family members, since they do not possess the catalytic activity of DNMT. DNMT3A and DNMT3B encode de novo methyltransferases, while DNMT1 encodes a maintenance methyltransferase. They are essential for mammalian development and involved in the development of cancers.5 Specific DNMT inhibitors such as 5-azacytidine and 5-aza-2′-deoxycytidine have been extensively studied in various cancers,6, 7, 8 including NPC,9,10 for decades. However, these drugs had dose-limiting toxicity in vivo, and their anti-tumor effect varied in different solid tumors. Therefore, it is necessary to continue in-depth study with DNMTs.
DNMT3B has been explored in recent years because of its role in tumor initiation, progression, and metastasis via DNA methylation.11, 12, 13 Overexpression of DNMT3B, but not DNMT1 or DNMT3A, significantly contributed to the elevated DNMT activity in breast cancer cells, eventually resulting in a hypermethylator phenotype.14 DNMT3B also played a vital role in CpG island methylator phenotype of colorectal cancer.15 A high expression level of DNMT3B was found to be closely related to poor prognosis of several types of cancers.16, 17, 18 Moreover, it has been reported that knockdown of DNMT3B could sensitize esophageal and prostate cancer cells to radiation.19,20 However, few studies were carried out to explore the relationship between DNMT3B and NPC.
p53 is an important tumor suppressor gene involved in cell cycle arrest, cellular senescence, and apoptosis.21 It is also a key molecule to determine tissue-specific cellular responses to ionizing radiation (IR). Radiosensitive tissues have a high basal mRNA expression level of p53, and a decline in radiation sensitivity is accompanied by a reduction in p53 mRNA expression.22 However, more than 50% of human tumors harbor mutations in the p53 gene, and most tumors had dysfunctional p53 signaling.23 The dysfunction and inactivation of p53 was also closely correlated with NPC initiation, development, and radioresistance.24 Therefore, the restoration of p53 function is potentially of great value in NPC patients. p21 (cyclin-dependent kinase inhibitor 1A, CDKN1A) is a universal cell-cycle inhibitor directly controlled by p53 and p53-independent pathways. It can block the activities of cyclin-dependent kinases (CDKs) and contribute to cellular senescence and tumor inhibition.25,26
In the present study, we demonstrated for the first time that DNMT3B was upregulated in NPC compared to normal tissue and predicted the poor prognosis of NPC patients. Silencing of DNMT3B induced cell cycle arrest and apoptosis through restoration of p53 and p21 function via DNA demethylation, eventually promoting radiosensitivity in NPC. Thus, DNMT3B could be a potential target in clinical practice of NPC.
Results
DNMT3B Expression Is Upregulated in NPC Patients and Predicts Poor Prognosis
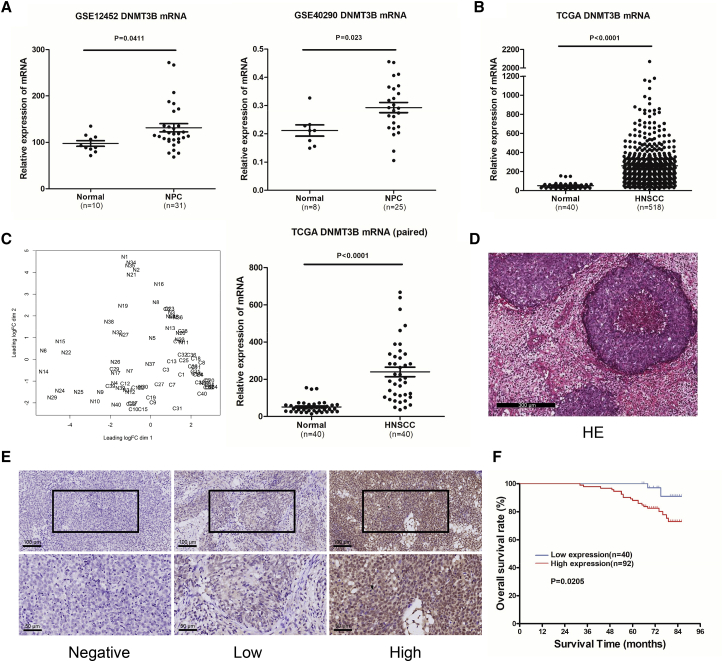
To determine the role of DNMT3B in NPC, we first analyzed the available human NPC datasets from Gene Expression Omnibus (GEO) databases (GEO: GSE12452 and GSE40290) and found that DNMT3B was upregulated in NPC samples compared with normal nasopharyngeal epithelial samples (Figure 1A). Considering the limitation of small sample size in GEO databases and the lack of specific NPC samples in The Cancer Genome Atlas (TCGA) databases, we analyzed the DNMT3B expression in head and neck squamous cell carcinoma (HNSCC) with TCGA. The HNSCC samples expressed higher DNMT3B levels compared to normal tissues (Figure 1B). We further isolated 40 paired samples, tumor versus adjacent non-tumor tissues, from HNSCC with detailed patient information (Table S1) and got similar results (Figure 1C). To corroborate the above results, we conducted a tissue microarray to investigate DNMT3B in NPC. Figure 1D shows a representative image of H&E staining for NPC. Then, we performed immunohistochemistry (IHC) staining for DNMT3B in the NPC tissue microarray. DNMT3B was mainly located in the nuclei of NPC cells (Figure 1E). High expression of DNMT3B was observed in 92/132 (69.7%) NPC samples and low DNMT3B expression was observed in 40/132 (30.3%) (Table 1). DNMT3B expression was not statistically associated with age, histologic classification, or N category, but it was significantly related to sex (p = 0.029), distant metastasis (p = 0.004), and clinical stage (p = 0.006). Moreover, Kaplan-Meier analysis showed that the expression of DNMT3B was significantly correlated with overall survival in NPC patients. As shown in Figure 1F, NPC patients with high DNMT3B expression had shorter overall survival than did those with low DNMT3B levels (p = 0.0205), suggesting that higher levels of DNMT3B predicted poor prognosis in NPC patients. However, DNMT3B expression is not related to disease-free survival in NPC patients (Figure S1). When compared with other factors, including clinical stage and DNMT3B expression level, metastasis status was the independent factor for overall survival (OS) through the Cox regression approach (Table S5). From TCGA data, we found that there was no significant relationship between DNMT3B and OS in HNSCC patients. However, for 5-year OS, patients with a low level of DNMT3B had a higher survival rate than did patients with a high level of DNMT3B (51.9% versus 42.6%) (Figure S2).
Figure 1.
DNMT3B Expression Is Upregulated and Associated with Poor Prognosis in NPC Patients
(A) Expression of DNMT3B in 31 NPC tissue and 10 normal tissue samples from the ArrayExpress microarray database (GEO: GSE12452 and GSE40290). (B) Expression of DNMT3B in head and neck squamous cell carcinoma (HNSCC) and normal tissue samples from The Cancer Genome Atlas (TCGA). (C) 40 paired samples isolated from TCGA, HNSCC versus adjacent non-tumor tissues; expression level comparison of DNMT3B in the 40 paired samples. (D) Representative image of H&E staining of NPC samples. (E) Representative images of IHC staining showing negative (left), low (middle), and high (right) DNMT3B expression of NPC tissues from tissue microarray. Upper, original magnification, ×200; lower, original magnification, ×400. (F) Kaplan-Meier overall survival curves for all 132 patients with NPC stratified by high (n = 92) versus low (n = 40) DNMT3B expression. The p value was calculated using the log rank test.
Table 1.
Association between DNMT3B Expression and the Clinicopathological Features of NPC Patients
| Feature | Total | DNMT3B |
p Value | |
|---|---|---|---|---|
| Low Expression | High Expression | |||
| Age (years) | ||||
| ≥45 | 77 | 23 | 54 | 0.898 |
| <45 | 55 | 17 | 38 | |
| Sex | ||||
| Male | 101 | 36 | 65 | 0.029 |
| Female | 31 | 4 | 27 | |
| Histologic Classification | ||||
| U | 116 | 36 | 80 | 0.840 |
| D | 16 | 4 | 12 | |
| N Category | ||||
| N0 | 36 | 11 | 25 | 0.969 |
| N1–3 | 96 | 29 | 67 | |
| Metastasis | ||||
| No | 111 | 39 | 72 | 0.004 |
| Yes | 21 | 1 | 20 | |
| Clinical Stage | ||||
| I+II | 72 | 29 | 43 | 0.006 |
| III+IV | 60 | 11 | 49 | |
NPC, nasopharyngeal carcinoma; U, undifferentiated non-keratinized carcinoma; D, differentiated non-keratinized carcinoma.
DNMT3B Is Upregulated after Exposure to IR and Involved in Radioresistance of NPC
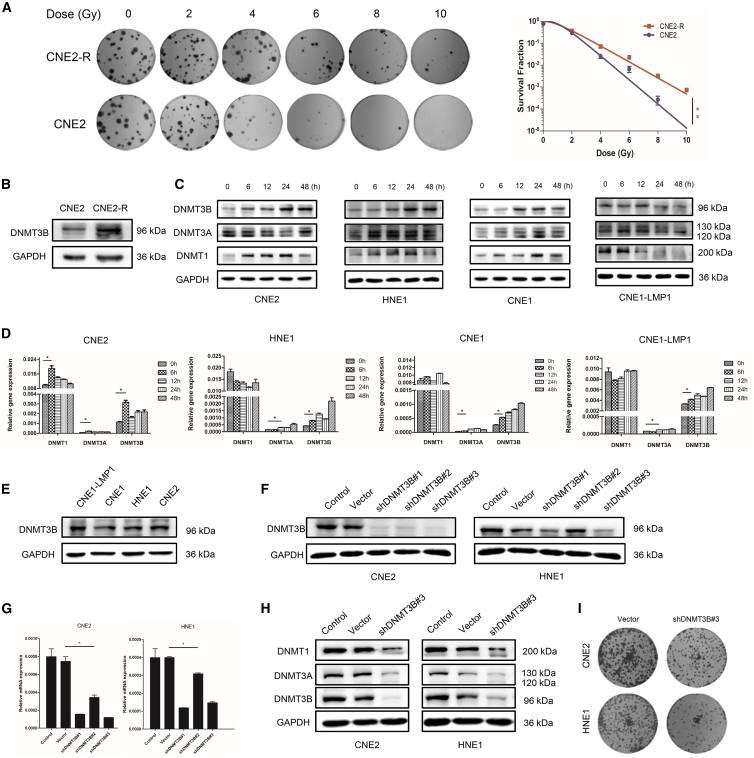
Radiotherapy with or without concurrent chemotherapy is the standard treatment for NPC patients. We found that DNMT3B was highly expressed in NPC patients and predicted poor prognosis, after which we assumed that there was a correlation between DNMT3B and radiation. Consequently, we conducted the following experiments to verify our conjecture. The CNE2-R cell line was constructed from the CNE2 cell line according to the published methods.27 The significantly higher survival fraction in the CNE2-R cell line than that in the CNE2 cell line demonstrated the successful construction of CNE2-R (Figure 2A). Immunoblotting showed that DNMT3B expression was upregulated in CNE2-R (Figure 2B). The expression levels of DNMTs were then assessed by immunoblotting and qRT-PCR in the irradiated NPC cells at different time points (Figures 2C and 2D). The results showed that DNMT3B expression increased gradually after irradiation, both in mRNA and protein. However, DNMT1 and DNMT3A did not respond to IR consistently in mRNA and protein. The results above supported the concept that DNMT3B could be induced by radiation in NPC and might be related to radioresistance. To further explore the role of DNMT3B in radioresistance of NPC, we detected the basic expression of DNMT3B in four NPC cell lines. We found that the expression of DNMT3B was higher in CNE2, HNE1, and CNE1-LMP1 when compared to CNE1 (Figure 2E). The result was consistent with previous studies,28,29 which showed that the Epstein-Barr virus (EBV) oncogene product latent membrane protein 1 (LMP1) induced and upregulated DNMT3B expression in NPC. Therefore, we chose CNE2 and HNE1 to investigate the role of DNMT3B in NPC. We established the DNMT3B-silenced CNE2 and HNE1 cell lines. As shown in Figures 2F and 2G, DNMT3B was significantly reduced by DNMT-specific short hairpin RNA (shRNA) (shDNMT)3B#3. Thus, cells transfected with shDNMT3B#3 were chosen for further experiments. In a clone formation assay, silencing of DNMT3B inhibited the proliferation of NPC cell lines in vitro (Figure 2I). Also, note that DNMT1 and DNMT3A were also downregulated after silencing DNMT3B in CNE2 and HNE1 cell lines (Figure 2H), which indicated that DNMT3B contributed to the main DNMT activity in NPC. In breast cancer cells, overexpression of DNMT3B, not DNMT1 or DNMT3A, also significantly contributed to elevated DNMT activity.14
Figure 2.
DNMT3B Is Upregulated after Exposure to Ionizing Radiation and Involved in Radioresistance of NPC
(A) Representative images from colony formation assay, showing survival fraction of CNE2-R cells and CNE2 cells with radiation. (B) DNMT3B expression of CNE2-R and CNE2 cells detected by immunoblotting. (C) The proteins of DNMT family members from whole-cell lysates were analyzed using immunoblotting in different NPC cell lines after irradiation. (D) Quantitative analysis of mRNA of DNMT family members in different NPC cell lines after irradiation. (E) DNMT3B protein levels were analyzed by immunoblotting in different NPC cell lines. (F and G) Selection of the most appropriate NPC cell lines and verification of the silencing efficiency of shDNMT3Bs through immunoblotting (F) and RT-PCR (G). (H) Expression levels of DNMT1, DNMT3A, and DNMT3B after silencing DNMT3B in CNE2 and HNE1 cell lines. (I) Representative images from colony formation assay showing cell viability after silencing DNMT3B. ∗p < 0.05, ∗∗p < 0.01.
Silencing of DNMT3B Inhibits Migration and Invasion via Suppressing the Epithelial-Mesenchymal Transition in NPC Cells
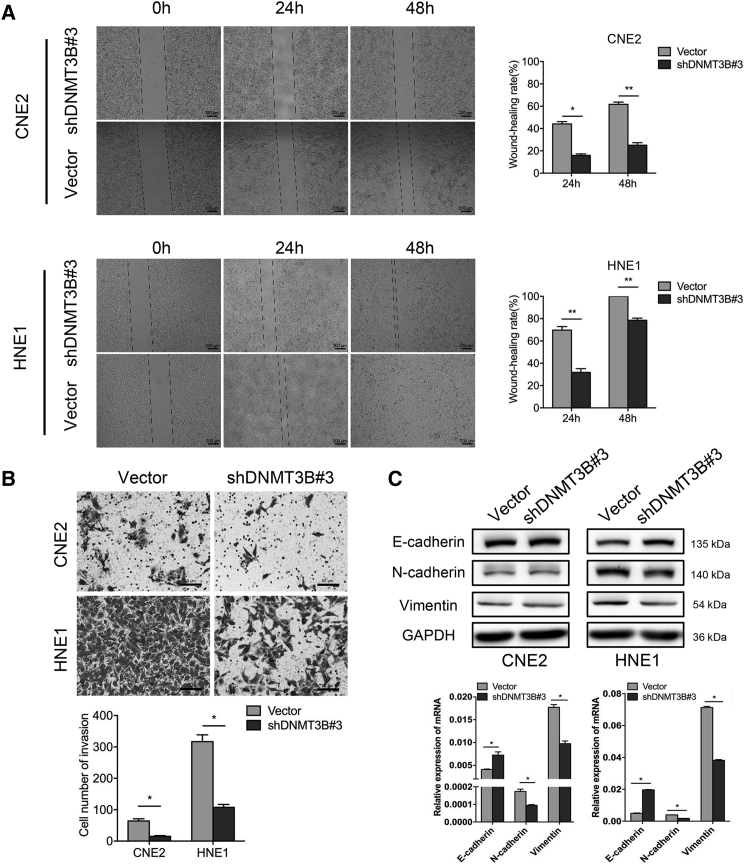
A wound-healing migration assay and a Transwell invasion assay were performed to explore the migration and invasion ability of transfected cells. The wound healing rate of CNE2-shDNMT3B#3 was significantly decreased compared to CNE2-vector (15.98% ± 1.81% versus 44.16% ± 2.90%, 24 h, p < 0.05; 25.08% ± 3.15% versus 61.74% ± 2.78%, 48 h, p < 0.01). There were similar results with HNE1 (31.88% ± 4.75% versus 69.72% ± 4.50%, 24 h, p < 0.001; 78.52% ± 2.69% versus 100.00% ± 0%, 48 h, p < 0.001) (Figure 3A). We also found that silencing DNMT3B inhibited the invasion ability of NPC cells. The numbers of CNE2-shDNMT3B#3 and HNE1-shDNMT3B#3 cells passing through the extracellular matrix (ECM) gel and polycarbonate membrane were 15 ± 4 and 107 ± 14, respectively, compared with CNE2-vector (64 ± 10) and HNE1-vector (317 ± 31) cells (Figure 3B). The difference was statistically significant (p < 0.05). We then detected epithelial-mesenchymal transition (EMT) marker proteins to explore whether the effect of DNMT3B on cell migration and invasion was a consequence of EMT. We found that epithelial marker protein E-cadherin was increased, whereas mesenchymal marker proteins N-cadherin and vimentin were decreased in cells transfected with shDNMT3B#3, especially in HNE1 (Figure 3C). The gene levels of EMT markers showed the same tendency. The results indicated that silencing DNMT3B inhibited EMT in NPC cells.
Figure 3.
Silencing of DNMT3B Inhibits Migration and Invasion through Reversing the Epithelial-Mesenchymal Transition
(A) Wound-healing assay was applied to test the migration ability in transfected NPC cells (original magnification, ×50), and wound-healing rates at the indicated times (0, 24, and 48 h) were quantitatively analyzed. (B) Representative images of Transwell assays (original magnification, ×200) and quantitative assessment of the number of cells invading to the lower chamber. (C) EMT-related protein and mRNA levels were analyzed by immunoblotting and RT-PCR after silencing of DNMT3B. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Silencing DNMT3B Causes G1 Phase Arrest and Promotes Apoptosis through Restoring the p53/p21 Signaling Pathway in NPC Cells
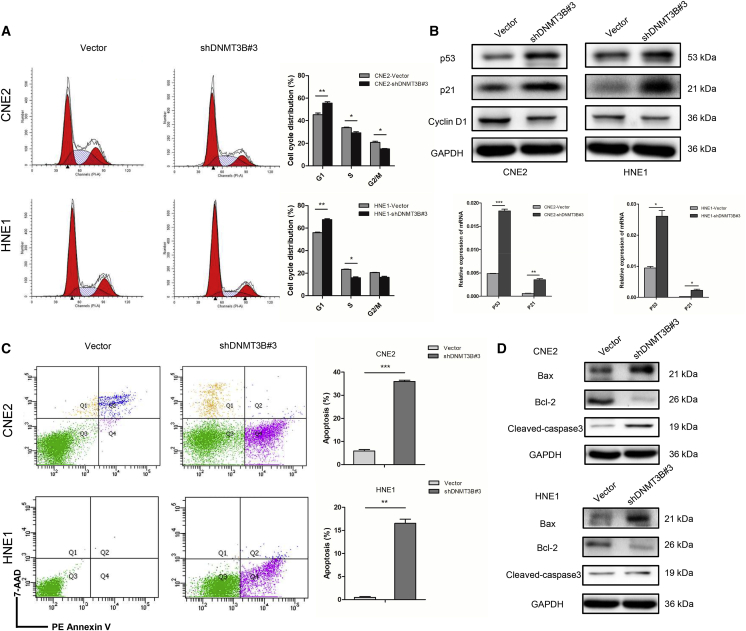
We found that silencing DNMT3B could inhibit the proliferation of NPC cell lines (Figure 2I). Therefore, a series of experiments were conducted to investigate whether DNMT3B would affect cell cycle distribution and apoptosis. The results indicated that silencing of DNMT3B caused G1 phase arrest and decreased the transition to the S phase (Figure 4A). Consistent with this, we found that p53 and p21 were activated by silencing of DNMT3B, and G1 cell cycle promoter cyclin D1 was downregulated. The mRNA levels of p53 and p21 showed the same tendency (Figure 4B; Figure S3). In addition, silencing DNMT3B promoted apoptosis in NPC cells (Figure 4C). Apoptosis-related protein Bax was increased in shDNMT3B#3 groups, whereas Bcl-2 was decreased. Cleaved caspase-3 was another standard apoptotic protein marker, also upregulated when silencing DNMT3B (Figure 4D). The results indicated that silencing of DNMT3B caused G1 phase arrest and promoted apoptosis through restoring the function of p53 and p21.
Figure 4.
Silencing of DNMT3B Causes G1 Arrest and Promotes Apoptosis by Restoring p53 and p21
(A) Cell cycle distribution measured by flow cytometry in CNE2 and HNE1 cells transfected with DNMT3B-specific (shDNMT3B#3) or control shRNA. (B) Protein expression of p53, p21, and cyclin D1, and mRNA expression of p53 and p21 in CNE2 and HNE1 cells transfected with DNMT3B-specific or control shRNA. (C) Cell apoptosis measured by flow cytometry in DNMT3B-knockdown CNE2 and HNE1 cells. (D) Bax, Bcl-2, and cleaved caspase-3 were detected by immunoblotting in CNE2 and HNE1 cells transfected with DNMT3B-specific or control shRNA. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Restoration and Activation of p53 and p21 Results from DNA Demethylation through Silencing DNMT3B in NPC Cells
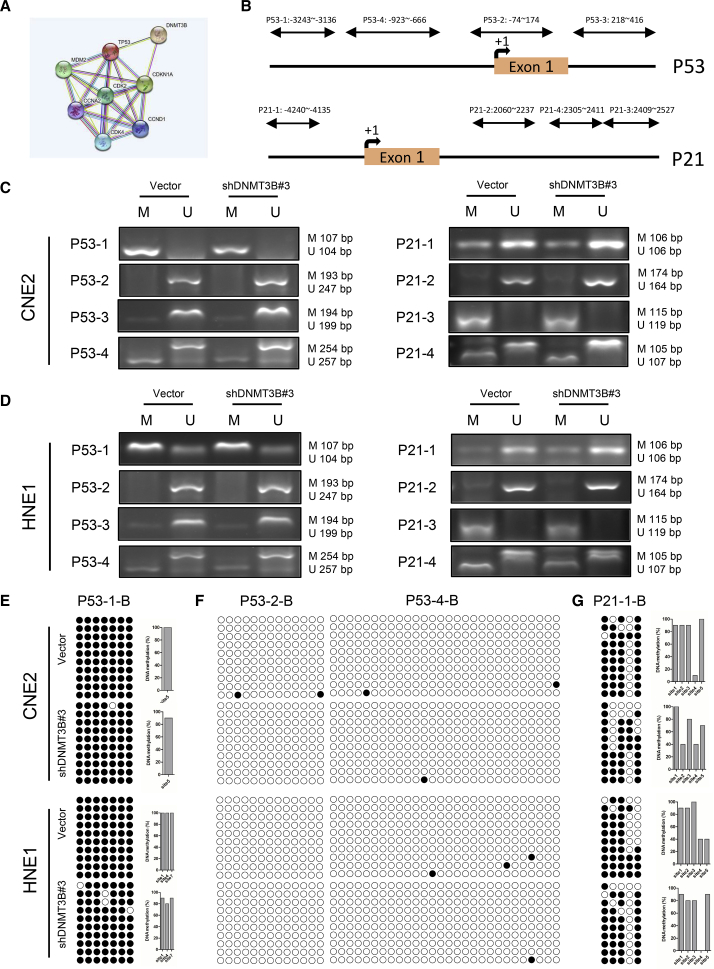
The results above demonstrated that silencing DNMT3B activated the p53/p21 signaling pathway. We used the STRING database to further explore the relationship between DNMT3B and p53/p21. STRING is a database of known and predicted protein-protein interactions. The interactions include direct (physical) and indirect (functional) associations. We found that DNMT3B was predicted to regulate p53 and p21 via epigenetic modification (Figure 5A). We then designed multiple primers (Table S2) for methylation-specific PCR (MSP) of p53 and p21 to investigate how DNMT3B regulated p53 and p21 (Figure 5B). MSPs of p53 and p21 were performed and revealed that silencing of DNMT3B decreased methylation of p53 and p21, respectively (Figures 5C and 5D). We further conducted bisulfite sequencing PCR (BSP) to explore the demethylation of p53 and p21 by silencing of DNMT3B. BSP with p53-1-B, p53-2-B, p53-4-B, and p21-1-B primers (Table S3) showed demethylation at different sites (Figures 5E–5G), the same as BSP with p53-3-B, p21-2-B, and p21-3-B primers (Figure S4). These data were consistent with upregulation of p53 and p21 in mRNA and protein levels after DNMT3B was knocked down, suggesting that silencing of DNMT3B restored and activated p53 and p21 function via DNA demethylation.
Figure 5.
Restoration and Activation of p53 and p21 Results from DNA Demethylation through Silencing DNMT3B
(A) DNMT3B was predicted to regulate p53 and p21 in the STRING database through epigenetic modification. (B) Sites of primers designed for methylation-specific PCR of p53 and p21. (C and D) DNA from CNE2 (C) and HNE1 (D) cells transfected with DNMT3B-specific or control shRNA was treated with bisulfite. Representative results obtained by methylation-specific PCR analysis of p53 (left) and p21 (right) promoters are shown. “M” represents methylated alleles and “U” represents nonmethylated alleles. (E–G) DNA from CNE2 and HNE1 cells transfected with DNMT3B-specific or control shRNA was treated with bisulfite. The results obtained by BSP analysis of p53 and p21 promoters with P53-1-B (E), P53-2-B, P53-4-B (F), and P21-1-B (G) primers are shown. ●, methylation at CpG dinucleotides; ○, demethylation at CpG dinucleotides.
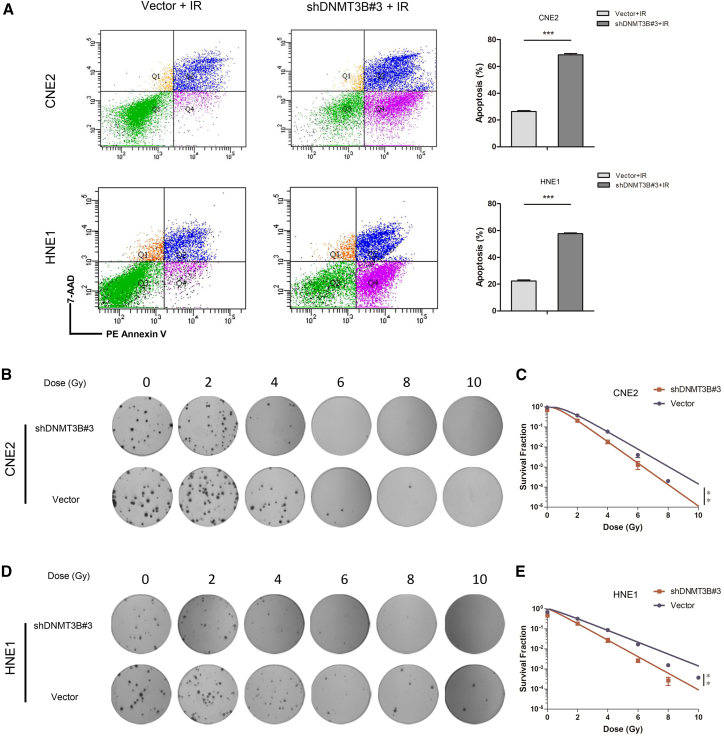
Silencing DNMT3B Enhances Radiosensitivity via Promoting Superior Apoptosis in NPC Cells after Exposure to IR
The p53/p21 pathway was involved in regulation of radiosensitivity of cancers. Silencing of DNMT3B restored and activated p53/p21 signaling pathway via DNA demethylation. On the one hand, it induced significant apoptosis; on the other hand, it caused G1 phase arrest and decreased the S phase. It is well known that S-phase cells appear to be resistant to irradiation, accounting for radioresistance.30 Reducing the S-phase ratio predicted radiosensitivity of NPC. As shown in Figure 6A, the proportion of apoptotic cells significantly increased in DNMT3B-silenced cells compared to control groups with radiation (CNE2, p < 0.001; HNE1, p < 0.001). Additionally, colony formation showed that CNE2-shDNMT3B#3 and HNE1-shDNMT3B#3 cells had statistically lower colony survival rates compared to vector groups after irradiation (Figures 6B–6E) (p < 0.01). These results suggested that silencing of DNMT3B enhanced radiosensitivity of NPC cells through the p53/p21 signaling pathway in vitro.
Figure 6.
Silencing of DNMT3B Enhances Radiosensitivity via Promoting Superior Apoptosis in NPC Cells
(A) Apoptotic rates determined by flow cytometry. Compared to control groups, the apoptotic rate significantly increased in the DNMT3B-knockdown group after irradiation. (B and D) Representative images from colony formation assays in transfected NPC cells after irradiation (B, CNE2 cells; D, HNE1 cells). (C and E) Survival fraction curves of clone formation assays in transfected NPC cells with radiation (C, CNE2 cells; E, HNE1 cells). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
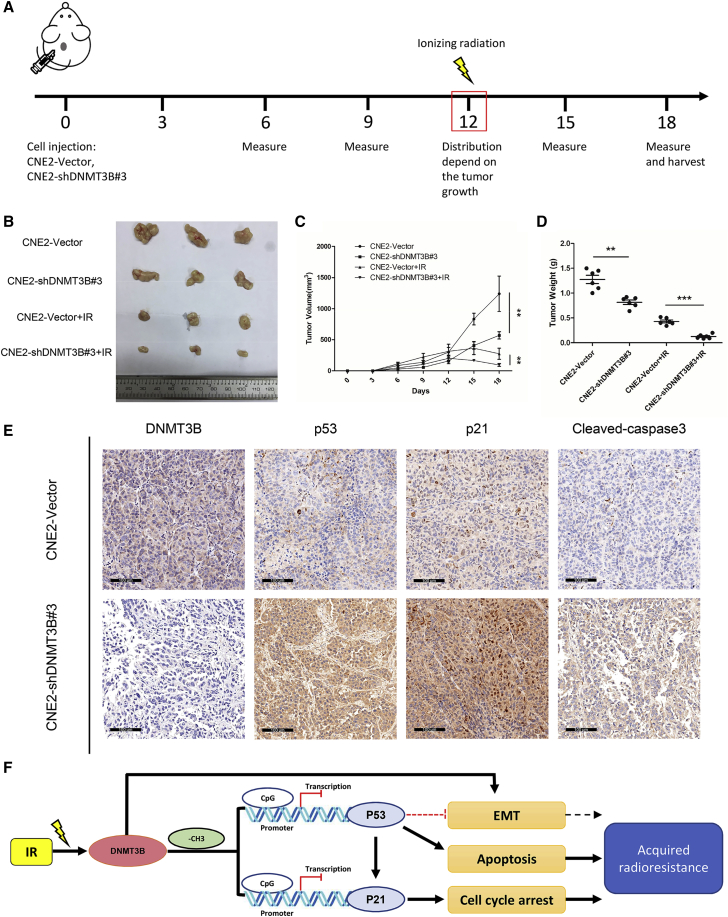
Silencing DNMT3B Promotes NPC Radiosensitivity In Vivo
A xenograft mouse model was applied to further verify whether DNMT3B participated in tumor radiosensitivity in vivo. The overall scheme of animal experiment is shown in Figure 7A. The tumor sizes were smaller and growth rates were lower in CNE2-shDNMT3B#3 compared to CNE2-vector groups, the same as for tumor weights. Irradiated groups had a significantly lower rate of tumor growth than did non-irradiated groups after 8 Gy irradiation. Also, note that the tumor sizes in CNE2-shDNMT3B#3 groups with irradiation were significantly smaller than those in irradiated CNE2-vector groups (Figures 7B–7D). Tumor tissue IHC showed that silencing of DNMT3B increased the expression of p53, p21, and cleaved caspase-3, which was consistent with the results in vitro (Figure 7E). The flowchart of our experiments is shown in Figure 7F. Activation of DNMT3B after IR results in downregulation and inactivation of p53 and p21 via DNA methylation and then leads to inhibition of apoptosis and promotion of cell growth, eventually causing acquired radioresistance of NPC. This process could be reversed by silencing DNMT3B. Thus, DNMT3B could be a potential target for NPC treatment.
Figure 7.
Silencing of DNMT3B Promotes Tumor Radiosensitivity In Vivo
(A) Overall scheme of animal experiments. (B) Representative tumor samples from each group are shown. (C) Tumor volumes from each group were tracked for 18 days. (D) Tumor weights in each group. (E) Representative IHC staining of DNMT3B, p53, p21 and cleaved caspase-3 from tumor samples in each non-irradiated group (original magnification, ×200). (F) Conceptual diagram of our study. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
The latest clinical trial results from the Sun Yat-sen University Cancer Center showed that induction chemotherapy added to chemoradiotherapy significantly improved recurrence-free survival and overall survival, as compared with chemoradiotherapy alone, among patients with locoregionally advanced NPC.3 However, radioresistance and distant metastasis remain major obstacles to improve the prognosis of patients with NPC.
NPC is a complex multifactorial disease that possesses aberrant gene function driven by genetic and epigenetic alterations. The DNMT family consists of a conserved set of DNA-modifying enzymes that play a central role in epigenetic regulation of malignant cancers. For instance, DNMT1 was upregulated by the interleukin-6 (IL-6)/Janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) pathway to enhance lung cancer initiation through downregulation of p53 and p21 resulting from DNA hypermethylation.31 Downregulation of DNMT3A inhibited lung cancer stem cell-like phenotypes by repressing Wnt/β-catenin signaling.32 DNMT3B was also reported to contribute to tumorigenesis via transcriptional silencing, and its expression was correlated with poor prognosis in various cancer patients.11,16 In addition, DNMT3B was found to interact with DNMT1 and DNMT3A, contributing to the main DNMT activity in various cancers, including breast cancer and colorectal cancer.
In this study, we found that DNMT3B expression was higher in NPC tissues than that in normal tissues from GEO databases. In the tissue microarray, the expression level of DNMT3B was significantly associated with distant metastasis and clinical stage of NPC patients, which indicated that DNMT3B might participate in the progression and metastasis of NPC. In addition, NPC patients with a high level of DNMT3B had shorter overall survival. These results suggested that DNMT3B might be an available prognostic marker for NPC patients.
We further investigated the relationship between DNMT3B and NPC under the effect of IR. We found that DNMT3B expression increased gradually after radiation over time. It was upregulated significantly in CNE2-R. Hu et al.20 demonstrated that DNMT3B could also be induced by IR in prostate cancer cells, and knockdown of DNMT3B sensitized prostate cancer cells to radiation. Thus, we supposed that DNMT3B could be induced by radiation in NPC and related to radioresistance.
A series of experiments with radiation were then performed to explore the role of DNMT3B in radiotherapy of NPC. We found that downregulation of DNMT3B dramatically caused G1 phase arrest and promoted apoptosis through restoring and activating the p53/p21 signaling pathway in vitro and in vivo, resulting in improvement of radiosensitivity of NPC. The dysfunction and suppression of p53 was closely correlated with radioresistance of NPC. Huang et al.33 found that EBV-encoded LMP1 contributed to radioresistance in NPC by suppressing the p53-mediated apoptosis pathway. Wild-type p53 activity has been shown to be essential for the efficacy of radiation and chemotherapy, which has led to development of strategies to restore normal p53 function in tumors. The injection of the exogenous p53 gene into NPC patients significantly improved radiosensitivity.34 Our study suggested that the function of p53 was restored after DNMT3B silencing and then activated the p53/p21 signaling pathway, downregulated cyclin D1, and led to cell cycle arrest. At the same time, p53 activated apoptosis-related protein Bax and got rid of the combination of Bcl-2, having finally induced apoptosis, which was consistent with a previous study.35 We also found that the combination of DNMT3B silencing and radiation significantly promoted superior apoptosis and improved the radiosensitivity of NPC cell lines. Li et al.36 recently found a steady increase in p53 and p21 protein levels in the NPC CNE2 cell line over time when exposed to IR, leading to cancer cell apoptosis and senescence. We supposed that silencing DNMT3B enhanced the activation of p53 and p21 responding to IR, then promoted NPC radiosensitivity.
The STRING database was analyzed to further study the mechanism responsible for DNMT3B-mediated regulation of p53 and p21. We found that DNMT3B was predicted to regulate p53 p21 through epigenetic modification. The MSP and BSP results showed that p53 and p21 were restored and reactivated via DNA demethylation after silencing DNMT3B. Peng et al.37 also found that epigenetic silencing of p53 was induced by repeated exposure to PM2.5 through the reactive oxygen species (ROS)-Akt-DNMT3B pathway-mediated promoter hypermethylation. Jiang et al.38 demonstrated that p53 DNA methylation was mediated by miR-125b-targeted DNMT3B, involving vascular smooth muscle cell proliferation induced by homocysteine. We are the first to propose that DNMT3B mediated methylation of p21. This may be due to direct epigenetic regulation of p21 by DNMT3B, or indirect regulation through DNMT1.31,39 Furthermore, note that silencing of DNMT3B decreased methylation of p53 and p21, respectively. This indicated that p21 also functioned as a cell-cycle inhibitor in a p53-independent cell cycle arrest pathway in our study.
Consistent with results of the tissue microarray, we found that silencing of DNMT3B inhibited the migration and invasion of NPC cells through reversing EMT. This may result from the inhibition and reversal of methylation of E-cadherin promoter following DNMT3B silencing.13 Recently, it has been shown that wild-type p53 can reverse EMT back to epithelial characteristics.40,41 Restoration of p53 function by silencing DNMT3B may also account for the reversal of EMT. Since EMT has been implicated in increased resistance to radiotherapy in NPC,36,42 inhibition of EMT after silencing DNMT3B may reverse the radioresistance of NPC.
In conclusion, our results indicated that radiation-induced upregulation of DNMT3B might be one of the reasons for NPC radioresistance. Silencing of DNMT3B inhibited the migration and invasion of NPC cells through reversing EMT. Furthermore, silencing DNMT3B restored and activated p53 and p21 via DNA demethylation, which could lead to cell cycle arrest and apoptosis, resulting in increased radiosensitivity. Therefore, DNMT3B could be a potential target for radiosensitization in the treatment of NPC.
Materials and Methods
Cell Lines and Culture
Human NPC cell lines CNE2, HNE1, CNE1, and CNE1-LMP1 were obtained from the Cancer Research Institute of Central South University (Changsha, China). The CNE2-radioresistance cell line (CNE2-R) was constructed from the CNE2 cell line as described in a previous publication.27 HNE1 and CNE2 were derived from a poorly differentiated NPC and lost the EBV genome during multiple cell passages. CNE1 is an EBV-negative, well-differentiated cell line, and CNE1-LMP1 was established by introducing LMP1 cDNA into CNE1 cells. The two cell lines, CNE2 and HNE1, mainly used in the study were authenticated using short tandem repeat profiling according to the suggestions from ATCC and authentication of NPC tumor lines,43 the same as our previous study.42 All cell lines were tested for mycoplasma contamination and cultured for less than 2 months in RPMI 1640 media (Promoter Biotech, China) with 10% fetal bovine serum (Gibco, South America) at 37°C in a humidified atmosphere with 5% CO2.
Construction of Lentivirus Vectors and Cell Transfection
To establish stable transfected cell lines, the shDNMT3B lentivirus particle and empty-vector lentivirus particle were purchased from GeneChem (GV493, Shanghai, China) and used to transfect CNE2 and HNE1 cells following the manufacturer’s instructions. Cells were then subcultured in selective medium with 3 μg/mL puromycin (Sigma, USA) for at least 2 weeks to select stable cell lines. Immunoblotting and real-time PCR were applied to detect the efficiency of transfection. The target sequences of DNMT3B and empty vector shRNA are as follows: shRNA 1, 5′-ATGCAACGATCTCTCAAAT-3′; shRNA 2, 5′-TGTCATTGTTTGATGGCAT-3′; shRNA 3, 5′-CTCAAGACAAATTGCTATA-3′; empty vector shRNA, 5′-TTCTCCGAACGTGTCACGT-3′.
X-Ray Irradiation
An RS2000 X-ray biological research irradiator (3-mm copper filter, 160 kV, 25 mA; Rad Source Technologies, Suwanee, GA, USA) was used to perform X-ray irradiation. The dose rate was 1.151 Gy/min. Single irradiation doses were 2–10 Gy.
Immunoblotting
Immunoblotting was performed to assess the expression of proteins semiquantitatively. Total protein was extracted using radioimmunoprecipitation assay (RIPA) buffer (P0013B, Beyotime Biotechnology, China). Equal amounts of protein samples, measured by microplate reader (Synergy H1, BioTek, USA) and Beyotime protein assay reagent, were separated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking, each membrane was incubated with the indicated primary antibodies at 4°C overnight. Then, it was incubated with secondary antibodies at room temperature for 1.5 h. The blots were visualized by West Dura extended duration substrate (34076, Thermo Fisher Scientific, USA). Anti-DNMT3B (67259), anti-E-cadherin (3195), anti-N-cadherin (14215), and anti-cleaved caspase-3 (9664) were purchased from Cell Signaling Technology (USA). Anti-p53 (10442), anti-p21 (10355), anti-cyclin D1 (60186), anti-DNMT1 (24206), anti-DNMT3A (20954), anti-Bax (50599), and anti-Bcl-2 (12789) were from Proteintech (Wuhan, China). Anti-vimentin (ab92547) and anti-GAPDH (ab8245) were obtained from Abcam (USA).
RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted using TRIzol following the manufacturer’s instructions. Reverse transcription was performed using the PrimeScript RT reagent kit (Perfect Real Time) (RR037A, Takara, Japan). A fast real-time PCR system (7900HT, Applied Biosystems, USA) was utilized to run real-time PCR. The PCR was performed with SYBR Premix Ex Taq (RR420A, Takara, Japan), in accord with the the manufacturer’s protocol. Primers for PCR were designed by Primer Premier 5.0 software (Table S4).
MSP
Genomic DNA of cells was isolated using a TIANamp genomic DNA kit (DP304, Tiangen, China). Then, DNA was treated with a DNA bisulfite conversion kit (DP215, Tiangen, China) in accord with the manufacturer’s instructions. The sodium bisulfite-modified DNA was amplified using an MSP kit (EM101, Tiangen, China). The PCR products were separated on 2.5% agarose gels and visualized using ethidium bromide staining. Primer sequences designed for MSP are listed in Table S2.
BSP
Genomic DNA of cells was isolated using the TIANamp genomic DNA kit (DP304, Tiangen, China). Then, DNA was treated with a DNA bisulfite conversion kit (DP215, Tiangen, China) in accord with the the manufacturer’s instructions. Aliquots of the bisulfite-treated DNA were subjected to 45 cycles of PCR amplification. PCR primers in the same regions of MSP were designed using the CpG island search engine MethPrimer, which are shown in Table S3. The final PCR products were cloned using the pMD19-T vector cloning kit (Takara) and clones (at least 10 for each promoter region for each gene) were sequenced using an automated ABI Prism 3730XL DNA sequencer (Applied Biosystems, USA).
Wound-Healing Migration Assay
A wound-healing migration assay was used to assess the migration ability of NPC cells. Cells were seeded in 24-well plates and grown to a confluent monolayer overnight with 10% fetal bovine serum. A 10-μL pipette tip was used to draw a straight line on the bottom of the 24-well plates, then cells were incubated in serum-free PRMI 1640 medium. Migration was visualized at the indicated times (0, 24, and 48 h after scratch) under a microscope. The migration distances were measured by ImageJ analysis software (National Institutes of Health, Bethesda, MD, USA). Each experiment was repeated at least three times.
Transwell Invasion Assay
Transwell 24-well plates with 8μm diameter filter (Corning Life Sciences, NY) were used to detect the invasion ability of cells. Filters were precoated with 40 μL of diluted ECM gel overnight. In the upper chamber, approximately 1 × 105 cells in 200 μL of serum-free RPMI 1640 medium were seeded. The lower chamber contained 500 μL of RPMI 1640 medium with 10% fetal bovine serum. After incubation for 20–24 h, the lower chamber was fixed in 4% paraformaldehyde for 20 min, stained with crystal violet, and counted under an upright microscopy. Each experiment was repeated at least three times.
Colony Formation Assay
A colony formation assay was used to detect the survival fraction of NPC cells after radiation. Cells were seeded in six-well plates and incubated at 37°C for 24 h for attachment. Plates were irradiated with 0, 2, 4, 6, 8, or 10 Gy separately the following day. After 10–14 days of incubation, the cells were fixed with methanol for 15 min and then stained with 0.1% crystal violet for 15 min. The plates were imaged, and the colonies were counted by three different investigators. The experiment was carried out in triplicate for each group. At least 50 cells per clone indicated a valid clone. The survival fraction (%) = [number of valid clones/(number of inoculated cells × 0 Gy colony formation rate)] × 100%. The survival fraction curves of the clone formation assays were fitted in a “single-hit multi-target” model as described previously.42 All data were analyzed with GraphPad Prism 5 software.
Cell Cycle Analysis
Cell cycle analysis was applied to detect cell cycle distribution of NPC cells after silencing DNMT3B. Cells were prepared and digested with RNaseA at 37°C for 30 min and then stained with propidium iodide (PI) for 30 min at 4°C according to the manufacturer’s protocol (Promoter, Wuhan, China). The cells were analyzed by flow cytometry in a FACSCanto II flow cytometer (BD Biosciences, USA).
Apoptosis Analysis
Apoptosis analysis was applied to detect the apoptotic proportion of cells. Cells were irradiated with 6 Gy and harvested after 72 h of incubation. A phycoerythrin (PE)-annexin V apoptosis detection kit I (559763, BD Biosciences, USA) was used to perform apoptosis analysis following the manufacturer’s instructions. A FACSCanto II flow cytometer (BD Biosciences, USA) was applied to detect apoptotic cells. Each experiment was repeated at least three times.
In Vivo Xenograft Assay
A xenograft mouse model was conducted to further validate the results. Five- to 6-week-old female BALB/c nude mice (Animal Care Center of Hunan SJA Laboratory Animal, Changsha, China) were housed in a specific pathogen-free, temperature and humidity-controlled environment with food and water in their cages. This study was reviewed and approved by the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. All animal experiments were conducted strictly in accordance with Animal Study Guidelines of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology Animal Care Facility and National Institutes of Health guidelines. In brief, cells (1.0 × 107/200 μL) were injected subcutaneously into the right armpit of mice. The mice were then randomly divided into four groups, and each group had six mice: CNE2-vector, CNE2-shDNMT3B#3, CNE2-vector+IR, and CNE2-shDNMT3B#3+IR. The animals received IR (8 Gy) on day 12 after injection. Tumor size was measured twice a week with a caliper, and tumor volume was calculated using the following formula: (long length × short length2)/2. The experiment was terminated after 3 weeks of tracking.
STRING Database
The interaction proteins network of DNMT3B was constructed using the STRING database (https://string-db.org/), which is an online database of predicted functional associations between proteins. An interaction with a combined score >0.4 was considered statistically significant.
GEO and TCGA Data Analysis
The GEO database was applied and selected to analyze DNMT3B expression level in NPC samples and normal nasopharyngeal epithelial samples. After adjusting, mRNA microarray data from GEO: GSE12452 and GSE40290 were background subtracted, normalized, and analyzed using BRB-ArrayTools (http://brb.nci.nih.gov/BRB-ArrayTools/). TCGA database (https://www.cancer.gov/tcga/) was used and downloaded to analyze DNMT3B in HNSCC. Raw data were normalized by TMM (trimmed mean of M-values) and then observed by BCV (biological coefficient of variation) to conduct quality control.
Tissue Microarray and IHC Staining
A human NPC tissue microarray was purchased from Outdo Biotech (Shanghai, China). A total of 132 NPC samples without adjacent normal nasopharynx tissues were obtained with patient information, including age, sex, histologic classification, lymph node status, metastasis status, and clinical stage. Before the biopsy, no patient received radiotherapy or chemotherapy. The human NPC tissue microarray was stained with anti-DNMT3B antibody (1:100, ab2851, Abcam, USA) following the standard protocols. Two pathologists reviewed and scored IHC staining for each sample independently. DNMT3B expression was scored according to the staining scope and intensity. Specifically, staining scope was scored as 1 (0%–25%), 2 (25%–50%), 3 (50%–75%), and 4 (75%–100%), and staining intensity was scored as 0 (negative), 1 (weakly positive; light yellow), 2 (moderately positive; yellow-brown), and 3 (strongly positive; dark brown). DNMT3B expression score = staining scope × intensity. An expression score ≥6 was defined as high expression, and those <6 were defined as low expression. Additionally, tissue IHC was employed to detect DNMT3B, p53, p21, and cleaved caspase-3 expression of xenograft mouse tissue with the indicated antibodies, including anti-DNMT3B (1:100, ab2851, Abcam, USA), anti-p53 (2527, Cell Signaling Technology, USA), anti-p21 (10355, Proteintech, China), and anti-cleaved caspase-3 (9664, Cell Signaling Technology, USA).
Statistical Analysis
SPSS software (version 19.0; SPSS, Chicago, IL, USA) was used for statistical analysis. Data are shown as mean ± standard deviation (SD). A Student’s t test and ANOVA test were applied to test statistical significance. The chi-square test or Fisher’s exact test was used to analyze the relationship between DNMT3B expression and clinicopathological features. Kaplan-Meier analysis and a log rank test were used to compare survival. p < 0.05 was considered significant (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Author Contributions
Guoqing Hu, Guangyuan Hu, G.L., and Q.M. conceived the idea and designed the study. C.W. performed most of the experiments. E.G., J.M., and L.S. performed the animal experiments. C.W., S.P., X.N., and M.L. analyzed the GEO and TCGA database and made the figures. C.W., S.P., W.S., L.Z., and D.L. wrote the manuscript. All authors approved the final version of the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81572960 and 81773231).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.04.007.
Supplemental Information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y.P., Chan A.T.C., Le Q.T., Blanchard P., Sun Y., Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. doi: 10.1016/S0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y., Chen L., Hu G.Q., Zhang N., Zhu X.D., Yang K.Y., Jin F., Shi M., Chen Y.P., Hu W.H. Gemcitabine and cisplatin induction chemotherapy in nasopharyngeal carcinoma. N. Engl. J. Med. 2019;381:1124–1135. doi: 10.1056/NEJMoa1905287. [DOI] [PubMed] [Google Scholar]
- 4.Mohammad H.P., Barbash O., Creasy C.L. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat. Med. 2019;25:403–418. doi: 10.1038/s41591-019-0376-8. [DOI] [PubMed] [Google Scholar]
- 5.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018;19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 6.Zhu W.G., Hileman T., Ke Y., Wang P., Lu S., Duan W., Dai Z., Tong T., Villalona-Calero M.A., Plass C., Otterson G.A. 5-Aza-2′-deoxycytidine activates the p53/p21Waf1/Cip1 pathway to inhibit cell proliferation. J. Biol. Chem. 2004;279:15161–15166. doi: 10.1074/jbc.M311703200. [DOI] [PubMed] [Google Scholar]
- 7.Montalban-Bravo G., Garcia-Manero G. Myelodysplastic syndromes: 2018 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2018;93:129–147. doi: 10.1002/ajh.24930. [DOI] [PubMed] [Google Scholar]
- 8.Agrawal K., Das V., Vyas P., Hajdúch M. Nucleosidic DNA demethylating epigenetic drugs—a comprehensive review from discovery to clinic. Pharmacol. Ther. 2018;188:45–79. doi: 10.1016/j.pharmthera.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Jiang W., Li Y.Q., Liu N., Sun Y., He Q.M., Jiang N., Xu Y.F., Chen L., Ma J. 5-Azacytidine enhances the radiosensitivity of CNE2 and SUNE1 cells in vitro and in vivo possibly by altering DNA methylation. PLoS ONE. 2014;9:e93273. doi: 10.1371/journal.pone.0093273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S., Zhang R., Claret F.X., Yang H. Involvement of microRNA-24 and DNA methylation in resistance of nasopharyngeal carcinoma to ionizing radiation. Mol. Cancer Ther. 2014;13:3163–3174. doi: 10.1158/1535-7163.MCT-14-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linhart H.G., Lin H., Yamada Y., Moran E., Steine E.J., Gokhale S., Lo G., Cantu E., Ehrich M., He T. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y.C., Tang Y.A., Shieh J.M., Lin R.K., Hsu H.S., Wang Y.C. DNMT3B overexpression by deregulation of FOXO3a-mediated transcription repression and MDM2 overexpression in lung cancer. J Thorac Oncol. 2014;9:1305–1315. doi: 10.1097/JTO.0000000000000240. [DOI] [PubMed] [Google Scholar]
- 13.Chen L.H., Hsu W.L., Tseng Y.J., Liu D.W., Weng C.F. Involvement of DNMT 3B promotes epithelial-mesenchymal transition and gene expression profile of invasive head and neck squamous cell carcinomas cell lines. BMC Cancer. 2016;16:431. doi: 10.1186/s12885-016-2468-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roll J.D., Rivenbark A.G., Jones W.D., Coleman W.B. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol. Cancer. 2008;7:15. doi: 10.1186/1476-4598-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nosho K., Shima K., Irahara N., Kure S., Baba Y., Kirkner G.J., Chen L., Gokhale S., Hazra A., Spiegelman D. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clin Cancer Res. 2009;15:3663–3671. doi: 10.1158/1078-0432.CCR-08-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girault I., Tozlu S., Lidereau R., Bieche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415–4422. [PubMed] [Google Scholar]
- 17.Amara K., Ziadi S., Hachana M., Soltani N., Korbi S., Trimeche M. DNA methyltransferase DNMT3b protein overexpression as a prognostic factor in patients with diffuse large B-cell lymphomas. Cancer Sci. 2010;101:1722–1730. doi: 10.1111/j.1349-7006.2010.01569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayette S., Thomas X., Jallades L., Chabane K., Charlot C., Tigaud I., Gazzo S., Morisset S., Cornillet-Lefebvre P., Plesa A. High DNA methyltransferase DNMT3B levels: a poor prognostic marker in acute myeloid leukemia. PLoS ONE. 2012;7:e51527. doi: 10.1371/journal.pone.0051527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen M.F., Lu M.S., Lin P.Y., Chen P.T., Chen W.C., Lee K.D. The role of DNA methyltransferase 3b in esophageal squamous cell carcinoma. Cancer. 2012;118:4074–4089. doi: 10.1002/cncr.26736. [DOI] [PubMed] [Google Scholar]
- 20.Xue G., Ren Z., Chen Y., Zhu J., Du Y., Pan D., Li X., Hu B. A feedback regulation between miR-145 and DNA methyltransferase 3b in prostate cancer cell and their responses to irradiation. Cancer Lett. 2015;361:121–127. doi: 10.1016/j.canlet.2015.02.046. [DOI] [PubMed] [Google Scholar]
- 21.Bieging K.T., Mello S.S., Attardi L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudkov A.V., Komarova E.A. The role of p53 in determining sensitivity to radiotherapy. Nat. Rev. Cancer. 2003;3:117–129. doi: 10.1038/nrc992. [DOI] [PubMed] [Google Scholar]
- 23.Bykov V.J.N., Eriksson S.E., Bianchi J., Wiman K.G. Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer. 2018;18:89–102. doi: 10.1038/nrc.2017.109. [DOI] [PubMed] [Google Scholar]
- 24.Liu F.F. Novel gene therapy approach for nasopharyngeal carcinoma. Semin. Cancer Biol. 2002;12:505–515. doi: 10.1016/s1044579x02000937. [DOI] [PubMed] [Google Scholar]
- 25.El-Deiry W.S. p21(WAF1) mediates cell-cycle inhibition, relevant to cancer suppression and therapy. Cancer Res. 2016;76:5189–5191. doi: 10.1158/0008-5472.CAN-16-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abbas T., Dutta A. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nie X., Guo E., Wu C., Liu D., Sun W., Zhang L., Long G., Mei Q., Wu K., Xiong H., Hu G. SALL4 induces radioresistance in nasopharyngeal carcinoma via the ATM/Chk2/p53 pathway. Cancer Med. 2019;8:1779–1792. doi: 10.1002/cam4.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seo S.Y., Kim E.O., Jang K.L. Epstein-Barr virus latent membrane protein 1 suppresses the growth-inhibitory effect of retinoic acid by inhibiting retinoic acid receptor-β2 expression via DNA methylation. Cancer Lett. 2008;270:66–76. doi: 10.1016/j.canlet.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 29.Peng H., Chen Y., Gong P., Cai L., Lyu X., Jiang Q., Wang J., Lu J., Yao K., Liu K. Higher methylation intensity induced by EBV LMP1 via NF-κB/DNMT3b signaling contributes to silencing of PTEN gene. Oncotarget. 2016;7:40025–40037. doi: 10.18632/oncotarget.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pawlik T.M., Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2004;59:928–942. doi: 10.1016/j.ijrobp.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Liu C.C., Lin J.H., Hsu T.W., Su K., Li A.F., Hsu H.S., Hung S.C. IL-6 enriched lung cancer stem-like cell population by inhibition of cell cycle regulators via DNMT1 upregulation. Int. J. Cancer. 2015;136:547–559. doi: 10.1002/ijc.29033. [DOI] [PubMed] [Google Scholar]
- 32.Liu T., Wu X., Chen T., Luo Z., Hu X. Downregulation of DNMT3A by miR-708-5p inhibits lung cancer stem cell-like phenotypes through repressing Wnt/β-catenin signaling. Clin Cancer Res. 2018;24:1748–1760. doi: 10.1158/1078-0432.CCR-17-1169. [DOI] [PubMed] [Google Scholar]
- 33.Yang C.F., Peng L.X., Huang T.J., Yang G.D., Chu Q.Q., Liang Y.Y., Cao X., Xie P., Zheng L.S., Huang H.B. Cancer stem-like cell characteristics induced by EB virus-encoded LMP1 contribute to radioresistance in nasopharyngeal carcinoma by suppressing the p53-mediated apoptosis pathway. Cancer Lett. 2014;344:260–271. doi: 10.1016/j.canlet.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Pan J.J., Zhang S.W., Chen C.B., Xiao S.W., Sun Y., Liu C.Q., Su X., Li D.M., Xu G., Xu B., Lu Y.Y. Effect of recombinant adenovirus-p53 combined with radiotherapy on long-term prognosis of advanced nasopharyngeal carcinoma. J. Clin. Oncol. 2009;27:799–804. doi: 10.1200/JCO.2008.18.9670. [DOI] [PubMed] [Google Scholar]
- 35.Mello S.S., Attardi L.D. Deciphering p53 signaling in tumor suppression. Curr. Opin. Cell Biol. 2018;51:65–72. doi: 10.1016/j.ceb.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X., Liu Y., Yin L., Peng Y., Peng Y., Gao Y., Yuan B., Zhu Q., Cao T., Xie B. Radiation-promoted CDC6 protein stability contributes to radioresistance by regulating senescence and epithelial to mesenchymal transition. Oncogene. 2019;38:549–563. doi: 10.1038/s41388-018-0460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou W., Tian D., He J., Wang Y., Zhang L., Cui L., Jia L., Zhang L., Li L., Shu Y. Repeated PM2.5 exposure inhibits BEAS-2B cell P53 expression through ROS-Akt-DNMT3B pathway-mediated promoter hypermethylation. Oncotarget. 2016;7:20691–20703. doi: 10.18632/oncotarget.7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao C., Zhang H., Zhao L., Zhou L., Zhang M., Xu H., Han X., Li G., Yang X., Jiang Y. miR-125b targets DNMT3b and mediates p53 DNA methylation involving in the vascular smooth muscle cells proliferation induced by homocysteine. Exp. Cell Res. 2016;347:95–104. doi: 10.1016/j.yexcr.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Zheng Q.H., Ma L.W., Zhu W.G., Zhang Z.Y., Tong T.J. p21Waf1/Cip1 plays a critical role in modulating senescence through changes of DNA methylation. J. Cell. Biochem. 2006;98:1230–1248. doi: 10.1002/jcb.20838. [DOI] [PubMed] [Google Scholar]
- 40.Powell E., Piwnica-Worms D., Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discov. 2014;4:405–414. doi: 10.1158/2159-8290.CD-13-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang C.J., Chao C.H., Xia W., Yang J.Y., Xiong Y., Li C.W., Yu W.H., Rehman S.K., Hsu J.L., Lee H.H. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo M., Wu C., Guo E., Peng S., Zhang L., Sun W., Liu D., Hu G., Hu G. FOXO3a knockdown promotes radioresistance in nasopharyngeal carcinoma by inducing epithelial-mesenchymal transition and the Wnt/β-catenin signaling pathway. Cancer Lett. 2019;455:26–35. doi: 10.1016/j.canlet.2019.04.019. [DOI] [PubMed] [Google Scholar]
- 43.Chan S.Y., Choy K.W., Tsao S.W., Tao Q., Tang T., Chung G.T., Lo K.W. Authentication of nasopharyngeal carcinoma tumor lines. Int. J. Cancer. 2008;122:2169–2171. doi: 10.1002/ijc.23374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.