Abstract
During the development of multicellular organisms, transcriptional regulation plays an important role in the control of cell growth, differentiation and morphogenesis. SUMOylation is a reversible post-translational process involved in transcriptional regulation through the modification of transcription factors and through chromatin remodelling (either modifying chromatin remodelers or acting as a ‘molecular glue’ by promoting recruitment of chromatin regulators). SUMO modification results in changes in the activity, stability, interactions or localization of its substrates, which affects cellular processes such as cell cycle progression, DNA maintenance and repair or nucleocytoplasmic transport. This review focuses on the role of SUMO machinery and the modification of target proteins during embryonic development and organogenesis of animals, from invertebrates to mammals.
Keywords: development, SUMO, ubiquitin
Introduction
SUMO belongs to the Ubiquitin-like modifier (UbL) family of proteins and attaches covalently to target proteins in a transient and reversible process termed SUMOylation. SUMO proteins are highly conserved in eukaryotes, but the number of paralogues varies among species. A single SUMO gene has been identified in S. cerevisiae (smt3), C. elegans (smo-1) and the insect Drosophila melanogaster (smt3), whereas three SUMO paralogues are found in mammals and eight in plants. There are three SUMO genes in the human genome, SUMO1 to 3. Human SUMO2 and SUMO3 share 97% identity at amino acid level (referred as SUMO2/3), and they share 47% sequence identity with SUMO1. SUMO4 shares 87% identity with SUMO2, and its expression is limited to some tissues [1].
SUMO proteins are synthesized as precursors that need to be matured by SUMO isopeptidases to expose the C-terminal di-glycine motif. The matured SUMO is activated by the heterodimeric E1 enzyme, comprised by SUMO-activating enzyme subunit 1 (SEA1, Aos1) and 2 (SAE2, Uba2). E1 forms a thioester bond between its catalytic cysteine and the SUMO C-terminal glycine. Once activated, SUMO is passed to the catalytic cysteine of the only E2 conjugating enzyme UBC9 (Ubiquitin Conjugating Enzyme E2 I). Finally, SUMO is transferred to the substrate either directly or through a SUMO E3 ligase (Figure 1 and Table 1). The transfer through the E3 ligases ensures a higher conjugation rate and the use of a particular E3 confers substrate specificity. The SUMO E3 ligases best characterized are Protein inhibitor of activated STAT 1 to 4 (PIAS1 to 4) and the Ran-binding protein 2 (RanBP2) Recently, the zinc finger protein ZNF451 was shown to have SUMO E3 ligase activity and to assemble efficiently SUMO2/3 chains (Table 1) [2,3]. The proteases involved in maturation and in the reverse de-conjugation are the ubiquitin-like protein-specific proteases (Ulps) in yeast and invertebrates and sentrin-specific proteases (SENPs) in mammals (SENP1–3 and SENP5–7). Moreover, two additional SUMO isopeptidases have been described in humans, deSUMOylating isopeptidase (DeSI), and the ubiquitin-specific protease-like 1 (USPL1) (Table 1) [4–6].
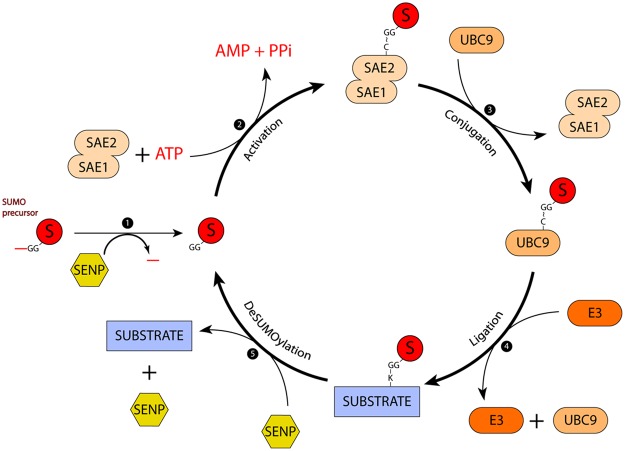
Figure 1. The SUMOylation/deSUMOylation cycle.
(1) First, SENPs process newly synthesized SUMO precursor into mature SUMO. (2) Then, SUMO's exposed di-glycine forms a thioester bond with the SAE2's catalytic cysteine in an ATP-dependent manner. (3) SUMO is then passed from the SAE1/SAE2 E1 activating heterodimer to the E2 conjugating enzyme UBC9, which also forms a thioester bond. (4) Substrates can be directly modified by E2-SUMO, but E3s might enhance conjugation rates by binding either E2-SUMO or substrates. (5) SENPs cleave the isopeptide bond and SUMO as well as the substrate are recycled. S: SUMO. ∼: thioester bond.
Table 1. E3 SUMO ligases and deSUMOylases.
| Organism | Name | Tissue expression/Subcellular localization | Mammalian Ortholog | Biological/Cellular process | References |
|---|---|---|---|---|---|
| E3 Ligases | |||||
| S. cerevisiae | Ull1/Siz1 | - | PIAS4 | Sumoylation of septins and histone H3, Mitosis | [99,100] |
| Nfi1/Siz2 | - | PIAS4 | Septin regulation | [101,102] | |
| Cst9/Zip3 | - | RNF212 | Synaptonemal complex formation, Meiosis | [103] | |
| Mms21 | - | NSE2 | DNA replication and repair | [104,105] | |
| C. elegans | GEI-17 | Germ cells, embryo, pharinx, neurons | PIAS2-4 | Meiosis, Telomere positioning, DNA damage | [32,106–108] |
| Drosophila | tonalli | Salivary gland, Ring gland, imaginal discs, other tissues | ZMIZ1, ZMIZ2 | Chromatin modification | [109–112] |
| Suppressor of variegation 2-10 | Nervous system, reproductive system, other tissues | PIAS1-4 | Chromatin modification, JAK-STAT signaling | [113–115] | |
| Mammals | RNF212 | Ubiquitous, Germ cells | - | Meiotic recombination | [23] |
| PIAS1 | Ubiquitous, Germ cells | - | Embryogenesis, Neuronal differentiation, Cardiac development | [50,58,81] | |
| PIAS2 | Testis, pancreas, others/ PML body | - | Post-Synaptic dendritic differentiation | [82] | |
| PIAS3 | Ubiquitous/Nucleus | - | Neuronal differentiation, Steroidogenic tissue, Retinal differentiation | [66,79,87] | |
| PIAS4 | Ubiquitous, enhanced in testis/ PML body | - | Early embryogenesis stage | [51] | |
| RanBP2 | Ubiquitous/Nuclear membrane and vesicles | - | Macromolecular transport | [116] | |
| NSE2 | Ubiquitous/Nucleus | - | Myogenic differentiation, DNA damage repair | [117] | |
| ZNF451 | Ubiquitous/Nucleoplasm | - | SUMO chain formation | [2] | |
| Pc2/CBX4 | Ubiquitous/Nucleoplasm and nuclear bodies | - | Heart development | [54] | |
| TOPORS | Ubiquitous/Nucleoplasm | - | Chromatin modification | [118] | |
| SLX4 | Ubiquitous/Nucleoplasm, cytosol and cell junctions | - | Genome maintenance | [119] | |
| hDREF | Ubiquitous | - | Nucleosome remodeling and cell proliferation | [120] | |
| MAPL | Nucleoplasm, mitochondria and cytosol | - | Mitochondrial fission | [121] | |
| Krox20 | Ubiquitous | - | Hindbrain development | [78] | |
| ZMIZ1 | Ovary, prostate, spleen and testis/Nucleoplasm | - | Embryonic development, vascular development | [111,122,123] | |
| ZMIZ2 | Gallbladder, testis and germ cells/Nuceloplasm and mitochondria | - | Embryonic development of neural tissue | [111,124] | |
| TRIM 1–19–22—27–28–32–39 | Subcellular proteinaceous bodies | - | Senescence, Apoptosis, Innate immunity, Antiviral defense, Gene silencing, Autophagy, Genomic stability | [125–130] | |
| DeSUMOylases | |||||
| S. cerevisiae | Ulp1 | Nuclear pore | SENP1, SENP2 | Cell cycle progression, Telomeric silencing, DNA damage | [131,132] |
| Ulp2 | Nucleoplasm | SENP6 | Cell cycle, DNA damage, DNA replication | [133,134] | |
| C. elegans | ulp-1 | Neurons, intestine, germ line, body wall muscle cell | SENP1 | Meiosis, embryonic development | [135] |
| ulp-2 | Hypodermis and neuroblasts/Citosol and nucleus | SENP7 | Embryonic development | [136] | |
| ulp-4 | Body wall muculature, hypodermis/Mitochondrial matrix | SENP7 | Cholesterol metabolism cell cycle, Unfolded protein response | [137–139] | |
| ulp-5 | n.s. | Predicted deSUMOylase | [138] | ||
| Drosophila | Ulp1 | Early embryo, embryonic CNS, adult germ line, other tissues | SENP1, SENP2 | Central nervous system projection neuron axonogenesis, negative regulation of Toll signaling pathway and inmflamatory response | [140–142] |
| CG12717 | n.s. | SENP6, SENP7 | Predicted deSUMOylase | [141] | |
| Veloren | Early embryo, embryonic CNS | SENP7, SENP6 | Axon targeting, negative regulation of cell death | [141] | |
| Mammals | SENP1 | Ubiquitous, enhanced in testis/Nuclear pore and nuclear foci | - | Embryogenesis, Mitotic progression, Senescence, Hematopoiesis | [47,143–146] |
| SENP2 | Ubiquitous/Nuclear pore, nuclear foci, cytoplasm | - | Trophoblast development, cardiac development, Myogenesis | [48,54,147,148] | |
| SENP3 | Ubiquitous/Nucleolus | - | Osteogenic differentiation, Sarcomere organization, Oocyte meiosis, Ribosome biogenesis | [29,60,149,150] | |
| SENP5 | Nucleolus and mitochondria | - | Ribosome biogenesis, RNAPI-mediated transcription 47S rRNA, tmitochondrial fragmentation during mitosis | [151,152] | |
| SENP6 | Ubiquitous/Nucleoplasm | - | Osteochondro-progenitor homeostasis, Hematopoiesis | [153,154] | |
| SENP7 | Ubiquitous/Nucleoplasm | - | Neuronal differentiation,Chromatin remodeling | [76,155] | |
| DeSI1 | Ubiquitous, enhanced in gastrontestinal tract, pancreas and muscle tissues/Cytoplasm and nucleus | - | Modulation transcriptional repressor activity | [5] | |
| DeSI2 | Cytoplasm | - | Modulation transcriptional repressor activity | [5] | |
| USPL1 | Ubiquitous, enhanced in gastrointestinal tract and kidney/Cajal bodies | - | RNAPII-mediated snRNA transcription | [4,156] | |
The organism, tissue expression, subcellular localization, orthologs and biological and cellular processes of the E3 SUMO ligases and deSUMOylases are shown.
n.s.: not specified.
SUMOylation modulates the function of target proteins by changing their subcellular localization, modifying their DNA-binding or chromatin association ability, recruiting histone-deacetylases and other corepressors or interfering with other post-translational modifications. This review is focused on the role of SUMO during embryonic development highlighting the most recent studies related to organogenesis (Table 2). In each section, we first review the expression and/or roles of SUMOs, E1, E2, ligases or proteases and then, we provide examples of SUMO target proteins and the effect of SUMOylation in their function during development.
Table 2. SUMOylation components in developmental processes.
| Organ/Process | SUMO/Ubc9 | E3 Ligase | De SUMOylase | Target | Organism | Pathway/Function | Reference |
|---|---|---|---|---|---|---|---|
| Germ cells | Ubc9 | Yeast | Chromosome synapsis during meiosis | [7] | |||
| Smt3 | Yeast | Chromosome synapsis during meiosis | [8] | ||||
| Lwr | Drosophila | Defects in meiotic chromosome segregation | [10] | ||||
| SMO-1 | C. elegans | Sterility | [12] | ||||
| UBC9 | C. elegans | Sterility | [12] | ||||
| Spermatogenesis | RNF212 | Mouse | Formation of axis-associated SUMO conjugates | [22] | |||
| CDK1 | Mouse | n.s. | [25] | ||||
| RNAPII | Mouse | n.s. | [25] | ||||
| CDC5 | Mouse | n.s. | [25] | ||||
| PIWIL2 or MILI | Mouse | n.s. | [25] | ||||
| DDX4 | Mouse | n.s. | [25] | ||||
| TARDBP or TDP-43 | Mouse | n.s. | [25] | ||||
| STK31 | Mouse | n.s. | [25] | ||||
| Oogenesis | SUMO1 | PLK1 | Mouse | microtubule and spindle pole organization | [28] | ||
| SUMO2/3 | PLK2 | Mouse | kinetochore | [28] | |||
| SENP2 | Mouse | metaphase II spindle organization | [27] | ||||
| SENP3 | Mouse | G2-M transition and spindle assembly | [29] | ||||
| SENP7 | Mouse | meiosis and egg maturation | [30] | ||||
| GEI-17 | KLP-19 | C. elegans | Recruitment to the Ring Complex | [31] | |||
| BUB-1 | C. elegans | Localization between segregating chromosomes during early anaphase I | [32] | ||||
| CLS-2 | C. elegans | Localization to central spindle | [32] | ||||
| SUMO1 | Septin2 | Mouse | Chromosome congression and meiosis progression | [33] | |||
| Embryogenesis and ZGA | Ubc9 | Mouse | nuclear organization and chromosome segregation at postimplantation stage | [36] | |||
| Smt3, Lwr | Drosophila | early embryogenesis | [37–39] | ||||
| UBC9 | C. elegans | embryogenesis | [40] | ||||
| ubc9 | Zebrafish | embryogenesis | [41] | ||||
| SUMO2 | Mouse | Embryogenesis, stage E10.5, growth, cell proliferation, cell survival | [42] | ||||
| SENP1 | HIF1alpha, GATA1 | Mouse | Mid-gestational embryogenesis; Erythropoiesis; placental development | [45–47] | |||
| SENP2 | p53/Mdm2 | Mouse | Cell cycle progression during mouse trophoblast development, endoreduplication | [48] | |||
| PIAS1 | Mouse | Embryogenesis, stages E10.5 and E12.5, red blood cells, angiogenesis, capillary plexus and blood vessel formation, heart development | [50] | ||||
| PIAS4 | DPPA2 | Mouse | zygotic genome activation; chromosome segregation; heterochromoatine state | [51,52] | |||
| Heart | SENP2 | cyclin and cyclin-dependent kinase inhibitors | Transgenic mice overexpressing human SENP2 Mouse | cardiomyocyte proliferation | [54,55] | ||
| SENP5 | Mouse | cell death | [56] | ||||
| SUMO1 | SENP2 | Pc2/CBX4 | Mouse | SUMO1-conjugated CBX4 decreases cardiomyocyte proliferation/suppressing the expression of Gata4 and Gata6/regulation of chromatin remodelling complexes | [54] | ||
| SUMO1 | PIAS1 | GATA4 | Mouse | enhanced transcriptional activity | [57,58] | ||
| SUMO1 | PIAS1 | Nkx2.5 | Mouse | enhanced transcriptional activity | [53,59] | ||
| SRF | Mouse | enhanced transcriptional activity/ synergy with Nkx2.5 in activation of cardiac target genes | [59,157,158] | ||||
| MEF2 | Mouse | reduced transcriptional activity | [159–162] | ||||
| SUMO1 | PIAS1 | myocardin | Mouse | enhanced transcriptional activity | [163] | ||
| Prox1 | Human | downregulation corepressor activity, increased transcriptional activity | [164,165] | ||||
| Tbx2/Tbx5 | Human, mouse, C. elegans | pharyngeal muscle development | [108,166] | ||||
| Osteogenic differentiation | SENP3 | RbBP5 | Human | activation of HOX gene DLX3 | [60] | ||
| Adrenal Gland | SUMO1 | PIAS1, PIAS3 | SF1 | Human, mouse | Attenuation of its transcriptional capacity | [64–66] | |
| Smt3 | Ftz-f1 | Drosophila | Sterol uptake. Attenuation of its transcriptional capacity | [69,70] | |||
| SMO-1 | NHR-25 | C. elegans | cell fate of reproductive organs | [71] | |||
| Neuronal development and differentiation | SENP2 | Drp1 | mouse | neurodegeneration through the modulation of mitochondrial morphogenesis | [75] | ||
| SENP7 | neuronal differentiation | [76] | |||||
| Braf35 | Mouse | repression of neuronal specific genes and inhibition of neuronal differentiation | [77] | ||||
| Krox20 | Nab | negative regulation | [78] | ||||
| SUMO1, 2, 3 | PIAS1, 3 | SENP2 | FOXP2 | Human | modulates transcriptional activity on downstream target genes (DISC1, SRPX2, and MiR200c); Purkinje cell development, cerebellar motor function and vocal communication | [79–81] | |
| PIAS2 | MEF2A | Human, rat | represses transcriptional activity; postsynaptic dendritic differentiation | [82] | |||
| Retinal proliferation and differentiation | ubc9 | Sp1 | Xenopus laevis | promotes retinal progenitor proliferation by repressing the cell cycle exit; suppresses p27Xic1 expression | [84] | ||
| Smt3, Aos1/Uba2, Lwr | Drosophila | proliferating cells in the developing eye | [85,86] | ||||
| PIAS3 | Nr2e3 | Mouse | specification of the rod subtype in the retina while preventing cone-like characteristics | [87] |
Proteins modified by SUMO in Germ Cells, embryogenesis, ZGA, Heart, osteogenic differentiation, adrenal gland, Neuronal development and differentiation and retinal proliferation and differentiation. The organism, SUMO ligases, DeSUMOylases, their targets, and the pathway and function are shown.
n.s.: not specified.
ZGA: zygotic genome activation.
SUMO in the germ cells
Primordial germ cells are a specialized population of cells that undergo meiosis to generate gametes, oocytes and spermatozoa. This involves several tightly coordinated processes such as pairing of homologous chromosomes, formation of the synaptonemal complex (SC) and the completion of meiotic recombination that leads to physical attachments between homologous chromosomes.
In yeast, both Ubc9 and Smt3 localize to synapsed regions of meiotic chromosomes. An ubc9-t mutant exhibited inefficient synapsis [7] and a meiotic smt3 reduction-of-function strain displayed abnormal levels of crossover recombination and diminished SC assembly [8]. These studies show that SUMOylation regulates chromosome synapsis during meiosis in budding yeast [9]. In Drosophila, mutations of the Drosophila UBC9 homologue lesswright (lwr), associated with either insertions in the 5′unstranslate region (lwr5486) or with a point mutation (G-to-A) in the coding region that leads to substitution of Arg104 by His (lwr5), show defects in meiotic chromosome segregation [10]. In C. elegans, the homolog of SUMO1, SMO-1 and the E2 conjugation enzyme UBC9 localize to germline nuclei throughout prophase I [11]. Both, the smo-1(ok359) null mutant and ubc9(tm2610) mutant, with deletion of the sequences that encoded for the catalytic domain, were sterile. Although the germ cells enter the meiotic prophase, they have defects in meiotic progression and failed to form normal sperm and oocytes [12]. All these studies show the important roles for SUMOylation during meiosis that include the maintenance of meiotic centromeric heterochromatin, meiotic DNA double-strand break repair and homologous recombination, centromeric coupling and the assembly of the SC [13].
Spermatogenesis
The roles that SUMO plays during spermatogenesis include meiotic sex chromosome inactivation, centromeric heterochromatin organization, XY body formation, microtubule nucleation and nuclear restructuring [14–18]. In mouse prophase I of meiosis, SUMO1 is localized to the XY body in spermatocytes, whereas only SUMO2/3 are detected near centromeres in metaphase I spermatocytes [14,15,19]. During human meiotic prophase, SUMO1 is associated with XY chromosome axes and also found in centromeric and pericentromeric heterochromatin [17,20].
The proteasome is involved in ensuring that homologous chromosomes pair each other during meiosis [21]. SUMO acts in coordination with ubiquitin-proteasome to regulate major transitions of meiotic recombination. Interestingly, in mouse, a SUMO-ubiquitin relay recruits proteasomes to the axes between homologous chromosomes to mediate chromosome pairing and recombination between homologs. The Ring Finger Protein 212 (RNF212), involved in SUMO conjugation, mediates the formation of axis-associated SUMO conjugates, while the ubiquitin ligase Cyclin B1 Interacting Protein 1 (CCNB1IP1 or HEI10) antagonizes RNF212 by promoting its turnover from synapses chromosomes [22,23]. Recently, novel proteins modified by SUMO during spermatogenesis have been identified in human and mouse: Cyclin Dependent Kinase 1 (CDK1), RNA polymerase II (RNAP II), Cell Division Cycle 5 Like (CDC5), Piwi Like RNA-Mediated Gene Silencing 2 (PIWIL2 or MILI), DEAD-Box Helicase 4 (DDX4), TAR DNA Binding Protein (TARDBP or TDP-43) and Serine/Threonine Kinase 31 (STK31); but the functional role of SUMOylation of these factors in spermatogenesis has not been reported [24,25].
Oogenesis
During mouse oocyte maturation and growth, different expression patterns and protein localizations have been described for SUMO1 and SUMO2/3 [18]. In transcriptionally active oocytes, both SUMO1 and SUMO2/3 are localized to the nucleoplasm and chromatin. In transcriptionally quiescent oocytes, SUMO1 is weakly detected with chromatin, while SUMO2/3 is localized throughout the nucleoplasm and on chromatin [26]. During oocyte maturation, SUMO1 is localized to the spindle poles in prometaphase I, metaphase I and II stages and around the separating homologues in anaphase I and telophase I stages of first meiosis. SUMO 2/3 is mainly concentrated near centromeres [27]. Interestingly, the SUMOylation of the Polo-like kinase 1 (PLK1) by different SUMO paralogues correlates with its different functions and localizations: PLK1 modification by SUMO1 is related to its function in microtubule and spindle pole organization, whereas modification by SUMO2/3 regulates its function at the kinetochore [28].
Studies in mouse show the important roles of deSUMOylases during oogenesis. The overexpression of Senp2 led to defects in metaphase II spindle organization in mature eggs [27]. Other examples are the regulation of G2-M transition and spindle assembly by SENP3 [29] and the meiotic arrest and decrease of mature eggs in SENP7 deficient oocytes [30].
SUMO modification plays also a role in chromosome congression in oocyte meiosis in C. elegans by regulating the multi-protein ring complex (RC) assembly [31]. There, the SUMO E3 ligase GEI-17 modifies and recruits the kinesin KLP-19 to the RC. Recently, the same group showed that SUMO regulates the dynamic localization of the central spindle proteins Mitotic Checkpoint Serine/Threonine Kinase (BUB-1) and CLS-2 during female meiosis [32]. Few SUMO modified proteins have been identified in mouse oocyte. As an example, the GTP binding protein Septin2 is modified by SUMO1 and its inhibition showed that it plays an essential role in regulating chromosome congression and meiosis progression [33]. Thus, SUMO1 plays crucial roles during meiotic oocyte maturation by regulating spindle organization, chromosome congression and chromosome segregation [34]. In addition to the role in oogenesis, SUMOylation is also required for the communication of the oocyte with the ovarian somatic cells [35].
SUMO in embryogenesis and zygotic genome activation (ZGA)
SUMO plays important roles in embryogenesis, as revealed by embryonic lethality when the conjugating enzyme UBC9 is deleted or knocked-down. Ubc9 deficient mouse embryos show severe defects in nuclear organization and chromosome segregation, and die at early post implantation stage [36]. In Drosophila embryogenesis, Smt3 coordinates multiple regulatory pathways and loss of function mutations in Ubc9 results in impaired embryogenesis [37–39]. Embryonic arrest is also observed in Ubc9 knockdown in C. elegans and zebrafish [40,41].
In mouse, the SUMOs orthologues have non-overlapping roles during embryonic development. SUMO2 is expressed at higher levels than SUMO3 in early embryonic stages and is indispensable for embryonic development, as shown by the phenotype of the null mutant mice. Sumo2−/− embryos die at stage E10.5 and exhibit severe growth retardation with reduced cell proliferation and increased cell death. However, embryos deficient in SUMO3 are viable [42]. Similarly, SUMO1 deficient mice are viable, likely because its function can be compensated by SUMO2 or SUMO3 [43,44].
Consistently with a central role for SUMOylation in embryonic development, knockout mice of the SUMO proteases SENP1 or SENP2 are mid-gestational embryonic lethal [45,46]. Mutations of SENP1 in mouse produce defects in erythropoiesis by impairing the physiological deSUMOylation of the hematopoietic factors Hypoxia Inducible Factor 1 Subunit Alpha (HIF1α) and GATA Binding Protein 1 (GATA1) [45,47]. SENP2 mutations cause deficiencies in cell cycle progression during mouse trophoblast development: SENP2 ablation disturbs the p53/Mdm2 pathway, affecting the expansion of trophoblasts progenitors and their maturation [48]. Moreover, deficiency of SUMO E3 ligases, such as PIAS1/PIASy double-knockout mice, impairs embryonic development between E10.5 and E12.5 in mouse [49,50].
While early phases of embryonic development are driven by maternal determinants, development comes under the control of the zygotic genome activation (ZGA) after fertilization. Two recent studies have analyzed the function of SUMOylation in regulating the maternal to zygotic transition and ZGA. Overexpression of the E3 ligase PIAS4 in mouse zygotes inhibited ZGA and impaired early embryo development. PIAS4 effect is partially caused by the SUMOylation of Developmental Pluripotency Associated 2 (DPPA2), which converts this transcriptional activator to a potent inhibitor of zygotic transcriptional program [51]. In agreement, another study shows that overexpression of PIAS4 after fertilization led to a failure of chromosome segregation and impaired ZGA, due to the enhanced SUMO ligase activity. Overexpressed PIAS4 disturbed the demethylation of histone H3 lysine 9 trimethylation (H3K9me3), affecting the heterochromatin state [52].
SUMO in developing tissues and organs
To illustrate the role of SUMO in different developmental pathways, we selected several examples in which SUMO components and SUMOylation of transcription factors play fundamental roles in organ development.
Heart development
The enrichment of SUMO1 and SUMO2 mRNAs in cardiac chamber regions undergoing proliferation and differentiation suggests a central role for SUMOylation in heart development [53]. A critical issue to achieve correct cardiac development is the balance between SUMOylation/deSUMOylation. Deletion of the deSUMOylating enzyme SENP2 in mice caused defects in cardiac development due to decreased cardiomyocyte proliferation: knockout of SENP2 lead to accumulation of SUMO1-conjugated Chromobox 4 or Polycomb 2 Homolog (Pc2/CBX4), a subunit of the polycomb repressive complex 1 (PRC1). SUMOylation of Pc2/CBX4 facilitated its binding to H3K27me3, suppressing the expression of the cardiac transcription factors encoding genes Gata4 and 6 [54], revealing a role for SUMOylation in the regulation of chromatin remodelling complexes during cardiogenesis. Moreover, SENP2 overexpression produced abnormal cardiomyocyte proliferation, with dysregulation of cyclin and cyclin-dependent kinase inhibitors, leading to cardiac defects [55]. Likewise, overexpression of SENP5 in mouse cardiomyocytes increased cell death and led to cardiomyopathy. Indeed, dysregulated levels of SENP5 and SUMO conjugation are observed in human failing hearts [56]. A role for PIAS1 has also been described for erythropoiesis and angiogenesis in the yolk sac. PIAS1 regulates proliferation in cells from the endoderm and mesoderm and its inactivation reduces the myocardium muscle mass, impairing cardiac development [50].
SUMO modifies a multitude of transcription factors that are important for normal cardiac development. These factors include NK2 homeobox 5 (Nkx2.5), GATA4 and 6, Serum Response Factor (SRF), myocyte enhancer factor-2 (MEF2), myocardin, T-box transcription factors-2 and -5 (TBX2 and 5) and prospero-related homeobox (Prox1) [57]. The zinc finger-containing transcription factor GATA4 is modified by SUMO1 in its transactivation domain, which results in enhanced transcriptional activity. The E3 ligase PIAS1 enhances the GATA4 SUMOylation efficiency via its RING finger domain [58]. Similarly, SUMO1 modification of the homeodomain transcription factor Nkx2.5 by PIAS1 increased its transcriptional activity by enhancing the physical association with its binding partners [53,59].
Osteogenic differentiation
Mesenchymal stem cells have the ability to differentiate into multiple cell types including adipocytes, chondrocytes and osteocytes. SUMOylation is required for the epigenetic control of gene expression during osteogenic differentiation of human stem cells. Notably, some studies show that SUMO affects the expression of HOX genes, which are evolutionary conserved master regulators that determine body plan in vertebrate development. For instance, the SUMO isopeptidase SENP3 associates with the Lysine Methyltransferase 2A and 2D (KMT2A/KMT2D or MLL1/MLL2) histone methyltransferase complexes and catalyzes the deSUMOylation of RB Binding Protein 5 (RbBP5), which is required for activation of HOX genes such as Distal-Less Homeobox (DLX3) [60]. A recent study by this group further showed that flightless-I-homolog (FLII), member of the gelsolin family of actin-remodelling proteins, determines the SENP3 recruitment and MILL1/2 complex assembly on the DLX3 gene [61].
Adrenal gland development and hormone synthesis
SUMO function is necessary in cell fate determination during adrenal gland development. SUMOylation components are expressed in human adrenal cortex and SUMO modification of transcription factors Steroidogenic Factor 1 (SF-1 or NR5A1), Wilms Tumor Protein 1 (WT1), GLI Family Zinc Finger 3 (GLI3), Spalt Like Transcription Factor 1 (SALL1) and Nuclear Receptor Subfamily 0 Group B Member 1 (NR0B1 or DAX1) have been described [62,63]. SF-1, member of the NR5A subfamily of nuclear receptors, is crucial for the development of the adrenal gland and for the expression of steroidogenic genes. SF-1 interacts with UBC9, PIAS1 and PIAS3 and is modified by SUMO, which results in attenuation of its transcriptional capacity [64–66]. Interestingly, a knock-in mouse model expressing a non-SUMOylatable form of SF-1 exhibits endocrine abnormalities and changes in cell fate, due in part to the inappropriate activation of the Hedgehog signalling [67]. The resulting mutant adrenal glands in this model exhibit a persistent foetal tissue, suggesting that SUMOylation interferes as well with normal maturation. Indeed, another recent study shows that foetal adrenal cortex regression is controlled by the synergistic interaction between SF-1 SUMOylation and DAX1, a nuclear receptor corepressor that interacts with SF-1 and inhibits genes involved in adrenal development and steroidogenesis [68]. In Drosophila, SUMO is as well required in steroidogenic tissues for the synthesis of steroid hormones [69]. Drosophila SF-1 homolog Fushi Tarazu Transcription Factor 1 (Ftz-f1) is modified by SUMO and is involved in sterol uptake, in part through the scavenger receptor member Snmp1 [70]. In addition, SUMOylation of the C. elegans homologue NHR-25 regulates it activity and maintains proper cell fate during development of the reproductive organs [71].
Neuronal development and differentiation
SUMOylation exerts a central role during embryonic brain development. Several studies have analyzed the spatiotemporal distribution of the SUMO moieties, UBC9, SAE1, SENP1 and SENP6 in the developing mouse and rat brains [72–74]. Total conjugation by SUMO1 and SUMO2/3 peaked at E12, whereas the highest levels of UBC9 expression were detected between E15 and E18.
Similarly to the heart, a controlled SUMOylation and deSUMOylation balance is important in the developing brain. A mouse model deficient for SENP2 in neural progenitors shows increased neuronal SUMOylation levels and produces neurodegeneration through the modulation of mitochondrial morphogenesis. This degeneration is a consequence of the hyper-SUMOylation of Dynamin-related protein 1 (Drp1), which promotes its association with mitochondria and neuronal apoptosis [75]. Recent studies showed that SENP7 is involved as well in proper neuronal differentiation [76].
SUMO regulates the function of several transcription factors during neuronal differentiation, including PHD Finger Protein 21A (PHF21A or Braf35), Early Growth Response 2 (EGR2 or Krox20), Myocyte Enhancer Factor 2A (MEF2A) and Forkhead Box P2 (Foxp2). In mouse developing brain, Braf35, a subunit of the LSD1-CoREST histone demethylase complex, is expressed in immature neurons. SUMOylation of Braf35 is required for the repression of neuronal specific genes and for the inhibition of neuronal differentiation [77]. An interesting case of cross-regulation is exemplified by the zinc finger transcription factor Krox20, which has essential roles in vertebrate hindbrain segmentation. Krox20 was described as a SUMO ligase for its coregulators, the NGFI-A Binding (Nab) proteins. As a consequence, the SUMOylation of Nab by Krox20 negatively modulates Krox20 activity and the extension of Krox20-positive territories [78]. During neuronal differentiation in the cerebellum, SUMOylation of the transcription factor FOXP2 increases, as a result of the function of PIAS1 and PIAS3 SUMO ligases and isopeptidase SENP2 [79,80]. This modification is required for the regulation of cerebellar motor function and vocal communication [81]. SUMOylation is also involved in the postsynaptic dendritic differentiation in the cerebellar cortex. The E3 ligase PIAS2 induces SUMO modification of the transcription factor Myocyte Enhancer Factor 2A (MEF2A), repressing the MEF2-dependent transcription in neurons [82]. Furthermore, the SUMOylation machinery participates in the synapsis plasticity and is associated with neurodegenerative diseases [83].
Retinal proliferation and differentiation
During embryonic development, the vertebrate retina originates from the central nervous system. In Xenopus laevis, ubc9 controls retinal progenitor proliferation by repressing the cell cycle exit in an high mobility group box 3 (hmgb3)-dependent manner. This function is partially mediated by the SUMOylation of the transcription factor Sp1, which suppressed p27Xic1 expression leading to the promotion of retinal progenitor proliferation [84]. In Drosophila, knockdown of Smt3 or E1 and E2 enzymes disrupts the proliferating cells in the developing eye, as well as in other imaginal tissues [85,86].
Two retina photoreceptors, rods and cones, arise from a common progenitor. Interestingly, SUMOylation promotes the specification of the rod subtype in the retina while preventing cone-like characteristics. This function involves the E3 SUMO ligase PIAS3 that SUMOylates the transcription factor Nuclear Receptor Subfamily 2 Group E Member 3 (Nr2e3) and converts it into a repressor of cone-specific gene expression [87]. In addition, PIAS3 has been involved in establishing dorsoventral patterning and visual response of cone photoreceptors in the mouse retina [88].
Conclusions
The development of organisms requires a fine-tune regulation of diverse signalling pathways. Several findings during the last years have unravelled the crucial role of SUMOylation for developmental and differentiation processes through the modification of relevant transcription factors. To these previous examples, we could add the role of SUMOylation during limb development. For instance, wing formation in Drosophila depends on the regulation of the SALL transcription factors by SUMO [89,90], or the role of SUMOylation in hedgehog signalling, a pathway that is relevant in limb formation [91]. In addition, SUMOylation of epigenetic regulators modifies their transcriptional activity, localization or stability. Additional complexity is driven by the interplay of SUMOylation with other post-translational modifications such as acetylation, phosphorylation or ubiquitylation. By the modification of transcription factor and chromatin remodelers, SUMO is involved in the regulation of cell division, cell lineage commitment, specification, and differentiation during the developmental processes.
A fine balance of SUMOylation/deSUMOylation is required during embryonic development and for normal cardiac and neuronal development. The disruption of the SUMO homeostasis led to inhibition of cell cycle progression, changes in gene expression through chromatin remodelling and to apoptosis. Thus, in normal physiological conditions, the isopeptidase activity is essential to maintain a stable fraction of SUMO modified proteins. Interestingly, changes in SENPs levels that disrupts SUMO equilibrium are observed also in several carcinomas. For example, in hepatocellular carcinoma, the complex between Cbx4 and PIASy mediates hypoxia-induced angiogenesis through enhancing HIF-1α sumoylation and increasing the transcriptional activity of HIF-1 [92].
A crucial step to understand the diverse cellular functions of this modification is the detection of targets of SUMOylation in vivo, due to the low abundance of the SUMOylated forms for any given target. For this reason, the technology to identify SUMO targets is crucial. Emergence of new techniques for the analysis of protein SUMOylation and characterization of the SUMO pathway across species and organs have been described [93–96]. Remarkably, as dysregulation of SUMO conjugation is associated to different human diseases it represents a potential therapeutic target. The study of in vivo role of SUMOylation in higher eukaryotes and also in more simple organisms with powerful genetic tools such as yeasts and invertebrates [97,98] will allow to elucidate more functions of SUMO targets during development.
Perspectives
SUMOylation has been proven crucial in the regulation of developmental processes.
Fine-tuning of key transcription factors function and signalling pathways components by SUMOylation contributes to modulate developmental processes.
Generation of new technologies to identify SUMOylation targets in vivo in model systems and in an organ specific manner will be crucial to achieve a more complete knowledge of the role of SUMO during development.
Acknowledgements
We apologize to those whose related publication could not be cited due to space limitations. We are grateful to all members of Barrio's Lab for comments and suggestions. R.B. acknowledges grants BFU2017-84653-P (MINECO/AEI/FEDER/EU), SEV-2016-0644 (Severo Ochoa Excellence Program, MINECO/AEI), 765445-EU (UbiCODE Program, EU), SAF2017-90900-REDT (UBIRed Program, MINECO/AEI).
Abbreviations
- CDC5
cell division cycle 5
- CDK1
cyclin dependent kinase 1
- DDX4
DEAD-box helicase 4
- GATA1
GATA binding protein 1
- MEF2
myocyte enhancer factor-2
- PLK1
Polo-like kinase 1
- RC
ring complex
- RNF212
ring finger protein 212
- RanBP2
Ran-binding protein 2
- RbBP5
RB Binding Protein 5
- SC
synaptonemal complex
- SENPs
sentrin-specific proteases
- SRF
serum response factor
- STK31
Serine/Threonine Kinase 31
- USPL1
ubiquitin-specific protease-like 1
- ZGA
zygotic genome activation
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Flotho A. and Melchior F. (2013) Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385 10.1146/annurev-biochem-061909-093311 [DOI] [PubMed] [Google Scholar]
- 2.Koidl S., Eisenhardt N., Fatouros C., Droescher M., Chaugule V.K. and Pichler A. (2016) The SUMO2/3 specific E3 ligase ZNF451-1 regulates PML stability. Int. J. Biochem. Cell. Biol. 79, 478–487 10.1016/j.biocel.2016.06.011 [DOI] [PubMed] [Google Scholar]
- 3.Pichler A., Fatouros C., Lee H. and Eisenhardt N. (2017) SUMO conjugation: a mechanistic view. Biomol. Concepts 8, 13–36 10.1515/bmc-2016-0030 [DOI] [PubMed] [Google Scholar]
- 4.Schulz S., Chachami G., Kozaczkiewicz L., Winter U., Stankovic-Valentin N., Haas P. et al. (2012) Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 13, 930–938 10.1038/embor.2012.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin E.J., Shin H.M., Nam E., Kim W.S., Kim J.H., Oh B.H. et al. (2012) DeSUMOylating isopeptidase: a second class of SUMO protease. EMBO Rep. 13, 339–346 10.1038/embor.2012.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suh H.Y., Kim J.H., Woo J.S., Ku B., Shin E.J., Yun Y. et al. (2012) Crystal structure of DeSI-1, a novel deSUMOylase belonging to a putative isopeptidase superfamily. Proteins 80, 2099–2104 10.1002/prot.24093 [DOI] [PubMed] [Google Scholar]
- 7.Hooker G.W. and Roeder G.S. (2006) A role for SUMO in meiotic chromosome synapsis. Curr. Biol. 16, 1238–1243 10.1016/j.cub.2006.04.045 [DOI] [PubMed] [Google Scholar]
- 8.Voelkel-Meiman K., Taylor L.F., Mukherjee P., Humphryes N., Tsubouchi H. and Macqueen A.J. (2013) SUMO localizes to the central element of synaptonemal complex and is required for the full synapsis of meiotic chromosomes in budding yeast. PLoS Genet. 9, e1003837 10.1371/journal.pgen.1003837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cahoon C.K. and Hawley R.S. (2016) Regulating the construction and demolition of the synaptonemal complex. Nat. Struct. Mol. Biol. 23, 369–377 10.1038/nsmb.3208 [DOI] [PubMed] [Google Scholar]
- 10.Apionishev S., Malhotra D., Raghavachari S., Tanda S. and Rasooly R.S. (2001) The Drosophila UBC9 homologue lesswright mediates the disjunction of homologues in meiosis I. Genes Cells 6, 215–224 10.1046/j.1365-2443.2001.00413.x [DOI] [PubMed] [Google Scholar]
- 11.Reichman R., Shi Z., Malone R. and Smolikove S. (2018) Mitotic and meiotic functions for the SUMOylation pathway in the Caenorhabditis elegans germline. Genetics 208, 1421–1441 10.1534/genetics.118.300787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broday L., Kolotuev I., Didier C., Bhoumik A., Gupta B.P., Sternberg P.W. et al. (2004) The small ubiquitin-like modifier (SUMO) is required for gonadal and uterine-vulval morphogenesis in Caenorhabditis elegans. Genes Dev. 18, 2380–2391 10.1101/gad.1227104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nottke A.C., Kim H.M. and Colaiacovo M.P. (2017) Wrestling with chromosomes: the roles of SUMO during meiosis. Adv. Exp. Med. Biol. 963, 185–196 10.1007/978-3-319-50044-7_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers R.S., Inselman A., Handel M.A. and Matunis M.J. (2004) SUMO modified proteins localize to the XY body of pachytene spermatocytes. Chromosoma 113, 233–243 10.1007/s00412-004-0311-7 [DOI] [PubMed] [Google Scholar]
- 15.Vigodner M. and Morris P.L. (2005) Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation, and nuclear reshaping. Dev. Biol. 282, 480–492 10.1016/j.ydbio.2005.03.034 [DOI] [PubMed] [Google Scholar]
- 16.Brown P.W., Hwang K., Schlegel P.N. and Morris P.L. (2008) Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and SUMOylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Hum. Reprod. 23, 2850–2857 10.1093/humrep/den300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metzler-Guillemain C., Depetris D., Luciani J.J., Mignon-Ravix C., Mitchell M.J. and Mattei M.G. (2008) In human pachytene spermatocytes, SUMO protein is restricted to the constitutive heterochromatin. Chromosome Res. 16, 761–782 10.1007/s10577-008-1225-7 [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez A. and Pangas S.A. (2016) Regulation of germ cell function by SUMOylation. Cell Tissue Res. 363, 47–55 10.1007/s00441-015-2286-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.La Salle S., Sun F., Zhang X.D., Matunis M.J. and Handel M.A. (2008) Developmental control of sumoylation pathway proteins in mouse male germ cells. Dev. Biol. 321, 227–237 10.1016/j.ydbio.2008.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vigodner M., Ishikawa T., Schlegel P.N. and Morris P.L. (2006) SUMO-1, human male germ cell development, and the androgen receptor in the testis of men with normal and abnormal spermatogenesis. Am. J. Physiol. Endocrinol. Metab. 290, E1022–E1033 10.1152/ajpendo.00527.2005 [DOI] [PubMed] [Google Scholar]
- 21.Ahuja J.S., Sandhu R., Mainpal R., Lawson C., Henley H., Hunt P.A. et al. (2017) Control of meiotic pairing and recombination by chromosomally tethered 26S proteasome. Science 355, 408–411 10.1126/science.aaf4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiao H., Prasada Rao H.B., Yang Y., Fong J.H., Cloutier J.M., Deacon D.C. et al. (2014) Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat. Genet. 46, 194–199 10.1038/ng.2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao H.B., Qiao H., Bhatt S.K., Bailey L.R., Tran H.D., Bourne S.L. et al. (2017) A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355, 403–407 10.1126/science.aaf6407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchiani S., Tamburrino L., Ricci B., Nosi D., Cambi M., Piomboni P. et al. (2014) SUMO1 in human sperm: new targets, role in motility and morphology and relationship with DNA damage. Reproduction 148, 453–467 10.1530/REP-14-0173 [DOI] [PubMed] [Google Scholar]
- 25.Xiao Y., Pollack D., Andrusier M., Levy A., Callaway M., Nieves E. et al. (2016) Identification of cell-specific targets of sumoylation during mouse spermatogenesis. Reproduction 151, 149–166 10.1530/REP-15-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ihara M., Stein P. and Schultz R.M. (2008) UBE2I (UBC9), a SUMO-conjugating enzyme, localizes to nuclear speckles and stimulates transcription in mouse oocytes. Biol. Reprod. 79, 906–913 10.1095/biolreprod.108.070474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z.B., Ou X.H., Tong J.S., Li S., Wei L., Ouyang Y.C. et al. (2010) The SUMO pathway functions in mouse oocyte maturation. Cell Cycle 9, 2640–2646 10.4161/cc.9.13.12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feitosa W.B., Hwang K.S. and Morris P.L. (2018) Temporal and SUMO-specific SUMOylation contribute to the dynamics of Polo-like kinase 1 (PLK1) and spindle integrity during mouse oocyte meiosis. Dev. Biol. 434, 278–291 10.1016/j.ydbio.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang C.J., Wu D., Khan F.A. and Huo L.J. (2015) The SUMO protease SENP3 orchestrates G2-M transition and spindle assembly in mouse oocytes. Sci. Rep. 5, 15600 10.1038/srep15600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang C.J.J., Wu D., Jiao X.F.F., Khan F.A., Xiong C.L.L., Liu X.M.M. et al. (2017) Maternal SENP7 programs meiosis architecture and embryo survival in mouse. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1195–1206 10.1016/j.bbamcr.2017.03.005 [DOI] [PubMed] [Google Scholar]
- 31.Pelisch F., Tammsalu T., Wang B., Jaffray E.G.. Gartner A. and Hay R.T. (2017) A SUMO-dependent protein network regulates chromosome congression during oocyte meiosis. Mol. Cell. 65, 66–77 10.1016/j.molcel.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelisch F., Bel Borja L., Jaffray E.G. and Hay R.T. (2019) Sumoylation regulates protein dynamics during meiotic chromosome segregation in C. elegans oocytes. J. Cell Sci. 132, jcs232330 10.1242/jcs.232330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu J.L., Lin S.L., Li M., Ouyang Y.C., Hou Y., Schatten H. et al. (2010) Septin2 is modified by SUMOylation and required for chromosome congression in mouse oocytes. Cell Cycle 9, 1607–1616 10.4161/cc.9.8.11463 [DOI] [PubMed] [Google Scholar]
- 34.Yuan Y.F., Zhai R., Liu X.M., Khan H.A., Zhen Y.H. and Huo L.J. (2014) SUMO-1 plays crucial roles for spindle organization, chromosome congression, and chromosome segregation during mouse oocyte meiotic maturation. Mol. Reprod. Dev. 81, 712–724 10.1002/mrd.22339 [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez A., Briley S.M., Patton B.K., Tripurani S.K., Rajapakshe K., Coarfa C. et al. (2019) Loss of the E2 SUMO-conjugating enzyme Ube2i in oocytes during ovarian folliculogenesis causes infertility in mice. Development 146, dev176701 10.1242/dev.176701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C. et al. (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9, 769–779 10.1016/j.devcel.2005.10.007 [DOI] [PubMed] [Google Scholar]
- 37.Epps J.L. and Tanda S. (1998) The Drosophila semushi mutation blocks nuclear import of bicoid during embryogenesis. Curr. Biol. 8, 1277–1280 10.1016/S0960-9822(07)00538-6 [DOI] [PubMed] [Google Scholar]
- 38.Huang L., Ohsako S. and Tanda S. (2005) The lesswright mutation activates Rel-related proteins, leading to overproduction of larval hemocytes in Drosophila melanogaster. Dev. Biol. 28, 407–420 10.1016/j.ydbio.2005.02.006 [DOI] [PubMed] [Google Scholar]
- 39.Nie M., Xie Y., Loo J.A. and Courey A.J. (2009) Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS ONE 4, e5905 10.1371/journal.pone.0005905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones D., Crowe E., Stevens T.A. and Candido E.P.M. (2002) Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 3, RESEARCH0002 10.1186/gb-2001-3-1-research0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nowak M. and Hammerschmidt M. (2006) Ubc9 regulates mitosis and cell survival during zebrafish development. Mol. Biol. Cell 17, 5324–5336 10.1091/mbc.e06-05-0413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L., Wansleeben C., Zhao S., Miao P., Paschen W. and Yang W. (2014) SUMO 2 is essential while SUMO 3 is dispensable for mouse embryonic development. EMBO Rep. 15, 878–885 10.15252/embr.201438534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evdokimov E., Sharma P., Loskett S.J., Lualdi M. and Kuehn M.R. (2008) Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J. Cell Sci. 121, 4106–4113 10.1242/jcs.038570 [DOI] [PubMed] [Google Scholar]
- 44.Zhang F.-P., Mikkonen L., Toppari J., Palvimo J.J., Thesleff I. and Janne O.A. (2008) Sumo-1 function is dispensable in normal mouse development. Mol. Cell. Biol. 28, 5381–5390 10.1128/MCB.00651-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng J., Kang X., Zhang S. and Yeh E.T.H. (2007) SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 131, 584–595 10.1016/j.cell.2007.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaguchi T., Sharma P., Athanasiou M., Kumar A., Yamada S. and Kuehn M.R. (2005) Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol. Cell. Biol. 25, 5171–5182 10.1128/MCB.25.12.5171-5182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu L., Ji W., Zhang H., Renda M.J., He Y., Lin S. et al. (2010) SENP1-mediated GATA1 deSUMOylation is critical for definitive erythropoiesis. J. Cell Biol. 207, 1183–1195 10.1084/jem.20092215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiu S.Y., Asai N., Costantini F. and Hsu W. (2008) SUMO-specific protease 2 is essential for modulating p53-mdm2 in development of trophoblast stem cell niches and lineages. PLoS Biol. 6, e310 10.1371/journal.pbio.0060310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tahk S., Liu B., Chernishof V., Wong K.A., Wu H. and Shuai K. (2007) Control of specificity and magnitude of NF-kappa B and STAT1-mediated gene activation through PIASy and PIAS1 cooperation. Proc. Natl Acad. Sci. U.S.A. 104, 11643–8 10.1073/pnas.0701877104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Constanzo J.D., Deng M., Rindhe S., Kj T., Cc Z. and Scaglioni P.P. (2016) Pias1 is essential for erythroid and vascular development in the mouse embryo. Dev. Biol. 415, 98–110 10.1016/j.ydbio.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan Y.L., Zhang C., Hao J., Wang X.L., Ming J., Mi L. et al. (2019) DPPA2/4 and SUMO E3 ligase PIAS4 opposingly regulate zygotic transcriptional program. PLoS Biol. 17, 1–31 10.1371/journal.pbio.3000324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higuchi C., Yamamoto M., Shin S.W., Miyamoto K. and Matsumoto K. (2019) Perturbation of maternal PIASy abundance disrupts zygotic genome activation and embryonic development via SUMOylation pathway. Biol. Open. 8, bio048652 10.1242/bio.048652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Costa M.W., Lee S., Furtado M.B., Xin L., Sparrow D.B., Martinez C.G. et al. (2011) Complex SUMO-1 regulation of cardiac transcription factor NKX2-5. PLoS ONE 6, e24812 10.1371/journal.pone.0024812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang X., Qi Y., Zuo Y., Wang Q., Zou Y., Schwartz R.J. et al. (2010) SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell 38, 191–201 10.1016/j.molcel.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim E.Y., Chen L., Ma Y., Yu W., Chang J., Moskowitz I.P. et al. (2012) Enhanced desumoylation in murine hearts by overexpressed SENP2 leads to congenital heart defects and cardiac dysfunction. J. Mol. Cell Cardiol. 52, 638–649 10.1016/j.yjmcc.2011.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim E.Y., Zhang Y., Beketaev I., Segura A.M., Yu W., Xi Y. et al. (2015) SENP5, a SUMO isopeptidase, induces apoptosis and cardiomyopathy. J. Mol. Cell. Cardiol. 78, 154–164 10.1016/j.yjmcc.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 57.Wang J. and Schwartz R.J. (2010) Sumoylation and regulation of cardiac gene expression. Circ. Res. 107, 19–29 10.1161/CIRCRESAHA.110.220491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J., Feng X.H. and Schwartz R.J. (2004) SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. J. Biol. Chem. 279, 49091–8 10.1074/jbc.M407494200 [DOI] [PubMed] [Google Scholar]
- 59.Wang J., Zhang H., Iyer D., Feng X.H. and Schwartz R.J. (2008) Regulation of cardiac specific nkx2.5 gene activity by small ubiquitin-like modifier. J. Biol. Chem. 283, 23235–23243 10.1074/jbc.M709748200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nayak A., Viale-Bouroncle S., Morsczeck C. and Muller S. (2014) The SUMO-specific isopeptidase SENP3 regulates MLL1/MLL2 methyltransferase complexes and controls osteogenic differentiation. Mol. Cell 55, 47–58 10.1016/j.molcel.2014.05.011 [DOI] [PubMed] [Google Scholar]
- 61.Nayak A., Reck A., Morsczeck C. and Müller S. (2017) Flightless-I governs cell fate by recruiting the SUMO isopeptidase SENP3 to distinct HOX genes. Epigenetics Chromatin 10, 15 10.1186/s13072-017-0122-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dumontet T., Sahut-Barnola I., Dufour D., Lefrançois-Martinez A.M., Berthon A., Montanier N. et al. (2019) Hormonal and spatial control of SUMOylation in the human and mouse adrenal cortex. FASEB J. 33, 10218–10230 10.1096/fj.201900557R [DOI] [PubMed] [Google Scholar]
- 63.Talamillo A., Martin D., Hjerpe R., Sanchez J. and Barrio R. (2010) SUMO and ubiquitin modifications during steroid hormone synthesis and function. Biochem. Soc. Trans. 38, 54–59 10.1042/BST0380054 [DOI] [PubMed] [Google Scholar]
- 64.Campbell L.A., Faivre E.J., Show M.D., Ingraham J.G., Flinders J., Gross J.D. et al. (2008) Decreased recognition of SUMO-sensitive target genes following modification of SF-1 (NR5A1). Mol. Cell. Biol. 28, 7476–7486 10.1128/MCB.00103-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen W.Y., Lee W.C., Hsu N.C., Huang F. and Chung B.C. (2004) SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1). J. Biol. Chem. 279, 38730–5 10.1074/jbc.M405006200 [DOI] [PubMed] [Google Scholar]
- 66.Komatsu T., Mizusaki H., Mukai T., Ogawa H., Baba D., Shirakawa M. et al. (2004) Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4 binding protein/steroidogenic factor 1 (Ad4BP/SF-1) regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol. Endocrinol. 18, 2451–2462 10.1210/me.2004-0173 [DOI] [PubMed] [Google Scholar]
- 67.Lee F.Y., Faivre E.J., Suzawa M., Lontok E., Ebert D., Cai F. et al. (2011) Eliminating SF-1 (NR5A1) sumoylation in vivo results in ectopic hedgehog signaling and disruption of endocrine development. Dev. Cell 21, 315–327 10.1016/j.devcel.2011.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xing Y., Morohashi K.I., Ingraham H.A. and Hammer G.D. (2017) Timing of adrenal regression controlled by synergistic interaction between Sf1 SUMOylation and Dax1. Development 144, 3798–3807 10.1242/dev.150516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Talamillo A., Sánchez J., Cantera R., Pérez C., Martín D., Caminero E. et al. (2008) Smt3 is required for Drosophila melanogaster metamorphosis. Development 135, 1659–1668 10.1242/dev.020685 [DOI] [PubMed] [Google Scholar]
- 70.Talamillo A., Herboso L., Pirone L., Pérez C., González M., Sánchez J. et al. (2013) Scavenger receptors mediate the role of SUMO and Ftz-f1 in Drosophila steroidogenesis. PLoS Genet. 9, e1003473 10.1371/journal.pgen.1003473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ward J.D., Yamamoto K.R. and Asahina M. (2014) SUMO as a nuclear hormone receptor effector. Worm 3, e29317 10.4161/worm.29317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hasegawa Y., Yoshida D., Nakamura Y. and Sakakibara S.I. (2014) Spatiotemporal distribution of SUMOylation components during mouse brain development. J. Comp. Neurol. 522, 3020–3036 10.1002/cne.23563 [DOI] [PubMed] [Google Scholar]
- 73.Loriol C., Khayachi A., Poupon G., Gwizdek C. and Martin S. (2013) Activity-dependent regulation of the sumoylation machinery in rat hippocampal neurons. Biol. Cell 105, 30–45 10.1111/boc.201200016 [DOI] [PubMed] [Google Scholar]
- 74.Watanabe M., Takahashi K., Tomizawa K., Mizusawa H. and Takahashi H. (2008) Developmental regulation of Ubc9 in the rat nervous system. Acta Biochim. Pol. 55, 681–686 10.18388/abp.2008_3027 [DOI] [PubMed] [Google Scholar]
- 75.Fu J., Yu H.M.I., Chiu S.Y., Mirando A.J., Maruyama E.O., Cheng J.G. et al. (2014) Disruption of SUMO-specific protease 2 induces mitochondria mediated neurodegeneration. PLoS Genet. 10, e1004579 10.1371/journal.pgen.1004579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Juarez-Vicente, F., Luna-Pelaez, N. and Garcia-Dominguez, M. The sumo protease Senp7 is required for proper neuronal differentiation. Biochim. Biophys. Acta Mol. Cell Res. 2016;1863:1490-1498. 10.1016/j.bbamcr.2016.03.028 [DOI] [PubMed] [Google Scholar]
- 77.Ceballos-Chávez M., Rivero S., García-Gutiérrez P., Rodríguez-Paredes M., García-Domínguez M., Bhattacharya S. et al. (2012) Control of neuronal differentiation by sumoylation of BRAF35, a subunit of the LSD1-CoREST histone demethylase complex. Proc. Natl Acad. Sci. U.S.A. 109, 8085–8090 10.1073/pnas.1121522109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garcia-Gutiérrez P., Juárez-Vicente F., Gallardo-Chamizo F., Charnay P. and García-Domínguez M. (2011) The transcription factor Krox20 is an E3 ligase that sumoylates its Nab coregulators. EMBO Rep. 12, 1018–1023 10.1038/embor.2011.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Estruch S.B., Graham S.A., Deriziotis P. and Fisher S.E. (2016) The language-related transcription factor FOXP2 is post-translationally modified with small ubiquitin-like modifiers. Sci. Rep. 6, 20911 10.1038/srep20911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meredith L.J., Wang C.M., Nascimento L., Liu R., Wang L. and Yang W.H. (2016) The key regulator for language and speech development, FOXP2, is a novel substrate for SUMOylation. J. Cell. Biochem. 117, 426–438 10.1002/jcb.25288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Usui N., Co M., Harper M., Rieger M.A., Dougherty J.D. and Konopka G. (2017) Sumoylation of FOXP2 regulates motor function and vocal communication through Purkinje cell development. Biol. Psychiatry 81, 220–230 10.1016/j.biopsych.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shalizi A., Bilimoria P.M., Stegmüller J., Gaudillière B., Yang Y., Shuai K. et al. (2007) PIASx is a MEF2 SUMO E3 ligase that promotes postsynaptic dendritic morphogenesis. J. Neurosci. 27, 10037–10046 10.1523/JNEUROSCI.0361-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Henley J.M., Craig T.J. and Wilkinson K.A. (2014) Neuronal SUMOylation: mechanisms, physiology, and roles in neuronal dysfunction. Physiol. Rev. 94, 1249–1285 10.1152/physrev.00008.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Terada K. and Furukawa T. (2010) Sumoylation controls retinal progenitor proliferation by repressing cell cycle exit in Xenopus laevis. Dev. Biol. 347, 180–194 10.1016/j.ydbio.2010.08.023 [DOI] [PubMed] [Google Scholar]
- 85.Kanakousaki K. and Gibson M.C. (2012) A differential requirement for SUMOylation in proliferating and non-proliferating cells during Drosophila development. Development 139, 2751–2762 10.1242/dev.082974 [DOI] [PubMed] [Google Scholar]
- 86.Takanaka Y. and Courey A.J. (2005) SUMO enhances vestigial function during wing morphogenesis. Mech. Dev. 122, 1130–1137 10.1016/j.mod.2005.05.004 [DOI] [PubMed] [Google Scholar]
- 87.Onishi A., Peng G.H., Hsu C., Alexis U., Chen S. and Blackshaw S. (2009) Pias3-dependent SUMOylation directs rod photoreceptor development. Neuron 61, 234–246 10.1016/j.neuron.2008.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Campla C.K., Breit H., Dong L., Gumerson J.D., Roger J.E. and Swaroop A. (2017) Pias3 is necessary for dorso-ventral patterning and visual response of retinal cones but is not required for rod photoreceptor differentiation. Biol. Open 6, 881–890 10.1242/bio.024679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sánchez J., Talamillo A., González M., Sánchez-Pulido L., Jiménez S., Pirone L. et al. (2011) Drosophila Sal and Salr are transcriptional repressors. Biochem. J. 438, 437–445 10.1042/BJ20110229 [DOI] [PubMed] [Google Scholar]
- 90.Sánchez J., Talamillo A., Lopitz-Otsoa F., Pérez C., Hjerpe R., Sutherland J.D. et al. (2010) Sumoylation modulates the activity of spalt-like proteins during wing development in Drosophila. J. Biol. Chem. 285, 25841–9 10.1074/jbc.M110.124024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu A. (2019) Proteostasis in the Hedgehog signaling pathway. Semin. Cell Dev. Biol. 93, 153–163 10.1016/j.semcdb.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li J., Xu Y., Long X.D., Wang W., Jiao H.K., Mei Z. et al. (2014) Cbx4 governs HIF-1alpha to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell 25, 118–131 10.1016/j.ccr.2013.12.008 [DOI] [PubMed] [Google Scholar]
- 93.Hendriks I.A., Lyon D., Su D., Skotte N.H., Daniel J.A., Jensen L.J. et al. (2018) Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun. 9, 2456 10.1038/s41467-018-04957-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lang V., Da Silva-Ferrada E., Barrio R., Sutherland J.D. and Rodriguez M.S. (2016) Using biotinylated SUMO-traps to analyze SUMOylated proteins. Methods Mol. Biol. 1475, 109–121 10.1007/978-1-4939-6358-4_8 [DOI] [PubMed] [Google Scholar]
- 95.Pirone L., Xolalpa W., Sigursson J.O., Ramirez J., Pérez C., González M. et al. (2017) A comprehensive platform for the analysis of ubiquitin-like protein modifications using in vivo biotinylation. Sci. Rep. 7, 40756 10.1038/srep40756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sheng Z., Wang X., Ma Y., Zhang D., Yang Y., Zhang P. et al. (2019) MS-based strategies for identification of protein SUMOylation modification. Electrophoresis 40, 2877–2887 10.1002/elps.201900100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abed M., Bitman-Lotan E. and Orian A. (2018) The biology of SUMO-targeted ubiquitin ligases in Drosophila development, immunity, and cancer. J. Dev. Biol. 6, E2 10.3390/jdb6010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Broday L. (2017) The SUMO system in Caenorhabditis elegans development. Int. J. Dev. Biol. 61, 159–164 10.1387/ijdb.160388LB [DOI] [PubMed] [Google Scholar]
- 99.Takahashi Y., Kahyo T., Toh E.A., Yasuda H. and Kikuchi Y. (2001) Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276, 48973–7 10.1074/jbc.M109295200 [DOI] [PubMed] [Google Scholar]
- 100.Takahashi Y., Yong-Gonzalez V., Kikuchi Y. and Strunnikov A. (2006) SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics 172, 783–794 10.1534/genetics.105.047167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Johnson E.S. and Gupta A.A. (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106, 735–744 10.1016/S0092-8674(01)00491-3 [DOI] [PubMed] [Google Scholar]
- 102.Takahashi Y., Toh E.A. and Kikuchi Y. (2003) Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J. Biochem. 133, 415–422 10.1093/jb/mvg054 [DOI] [PubMed] [Google Scholar]
- 103.Serrentino M.E., Chaplais E., Sommermeyer V. and Borde V. (2013) Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet. 9, e1003416 10.1371/journal.pgen.1003416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mahendrawada L., Rai R., Kothiwal D. and Laloraya S. (2017) Interplay between Top1 and Mms21/Nse2 mediated sumoylation in stable maintenance of long chromosomes. Curr. Genet. 63, 627–645 10.1007/s00294-016-0665-4 [DOI] [PubMed] [Google Scholar]
- 105.Zhao X. and Blobel G. (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl Acad. Sci. U.S.A. 102, 4777–4782 10.1073/pnas.0500537102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ferreira H.C., Towbin B.D., Jegou T. and Gasser S.M. (2013) The shelterin protein POT-1 anchors Caenorhabditis elegans telomeres through SUN-1 at the nuclear periphery. J. Cell Biol. 203, 727–735 10.1083/jcb.201307181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pelisch F., Sonneville R., Pourkarimi E., Agostinho A., Blow J.J., Gartner A. et al. (2015) Erratum: Dynamic SUMO modification regulates mitotic chromosome assembly and cell cycle progression in Caenorhabditis elegans. Nat. Commun. 6, 6352 10.1038/ncomms7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roy Chowdhuri S., Crum T., Woollard A., Aslam S. and Okkema P.G. (2006) The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans. Dev. Biol. 295, 664–677 10.1016/j.ydbio.2006.04.001 [DOI] [PubMed] [Google Scholar]
- 109.Gutierrez L., Zurita M., Kennison J.A. and Vazquez M. (2003) The Drosophila trithorax group gene tonalli (tna) interacts genetically with the Brahma remodeling complex and encodes an SP-RING finger protein. Development 130, 343–354 10.1242/dev.00222 [DOI] [PubMed] [Google Scholar]
- 110.Monribot-Villanueva J., Juarez-Uribe R.A., Palomera-Sanchez Z., Gutierrez-Aguiar L., Zurita M., Kennison J.A. et al. (2013) Tnaa, an SP-RING protein, interacts with Osa, a subunit of the chromatin remodeling complex BRAHMA and with the SUMOylation pathway in Drosophila melanogaster. PLoS ONE 8, e62251 10.1371/journal.pone.0062251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rodriguez-Magadan H., Merino E., Schnabel D., Ramirez L. and Lomeli H. (2008) Spatial and temporal expression of Zimp7 and Zimp10 PIAS-like proteins in the developing mouse embryo. Gene Expr. Patterns 8, 206–213 10.1016/j.modgep.2007.10.005 [DOI] [PubMed] [Google Scholar]
- 112.Rosales-Vega M., Hernandez-Becerril A., Murillo-Maldonado J.M., Zurita M. and Vazquez M. (2018) The role of the trithorax group TnaA isoforms in Hox gene expression, and in Drosophila late development. PLoS ONE 13, e0206587 10.1371/journal.pone.0206587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hari K.L., Cook K.R. and Karpen G.H. (2001) The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 15, 1334–1348 10.1101/gad.877901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Le H.D., Donaldson K.M., Cook K.R. and Karpen G.H. (2004) A high proportion of genes involved in position effect variegation also affect chromosome inheritance. Chromosoma 112, 269–276 10.1007/s00412-003-0272-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mohr S.E. and Boswell R.E. (1999) Zimp encodes a homologue of mouse Miz1 and PIAS3 and is an essential gene in Drosophila melanogaster. Gene 229, 109–116 10.1016/S0378-1119(99)00033-5 [DOI] [PubMed] [Google Scholar]
- 116.Hamada M., Haeger A., Jeganathan K.B., van Ree J.H., Malureanu L., Walde S. et al. (2011) Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. J Cell Biol. 194, 597–612 10.1083/jcb.201102018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Berkholz J., Michalick L. and Munz B. (2014) The E3 SUMO ligase Nse2 regulates sumoylation and nuclear-to-cytoplasmic translocation of skNAC-Smyd1 in myogenesis. J. Cell Sci. 127, 3794–3804 10.1242/jcs.150334 [DOI] [PubMed] [Google Scholar]
- 118.Pungaliya P., Kulkarni D., Park H.J., Marshall H., Zheng H., Lackland H. et al. (2007) TOPORS functions as a SUMO-1 E3 ligase for chromatin-modifying proteins. J. Proteome Res. 6, 3918–3923 10.1021/pr0703674 [DOI] [PubMed] [Google Scholar]
- 119.Guervilly J.H., Takedachi A., Naim V., Scaglione S., Chawhan C., Lovera Y. et al. (2015) The SLX4 complex is a SUMO E3 ligase that impacts on replication stress outcome and genome stability. Mol. Cell 57, 123–137 10.1016/j.molcel.2014.11.014 [DOI] [PubMed] [Google Scholar]
- 120.Yamashita D., Moriuchi T., Osumi T. and Hirose F. (2016) Transcription factor hDREF is a novel SUMO E3 ligase of Mi2alpha. J. Biol. Chem. 291, 11619–11634 10.1074/jbc.M115.713370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Braschi E., Zunino R. and McBride H.M. (2009) MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 10, 748–754 10.1038/embor.2009.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sharma M., Li X., Wang Y., Zarnegar M., Huang C.Y., Palvimo J.J. et al. (2003) Hzimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. EMBO J. 22, 6101–6114 10.1093/emboj/cdg585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Beliakoff J., Lee J., Ueno H., Aiyer A., Weissman I.L., Barsh G.S. et al. (2008) The PIAS-like protein Zimp10 is essential for embryonic viability and proper vascular development. Mol. Cell. Biol. 28, 282–292 10.1128/MCB.00771-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rodriguez-Magadan H., Ramirez L., Schnabel D., Vazquez M. and Lomeli H. (2010) Sexually dimorphic gene expression of the Zimp7 and Zimp10 genes in embryonic gonads. Gene Expr. Patterns 10, 16–23 10.1016/j.gep.2009.11.004 [DOI] [PubMed] [Google Scholar]
- 125.Chu Y. and Yang X. (2011) SUMO e3 ligase activity of TRIM proteins. Oncogene 30, 1108–1116 10.1038/onc.2010.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ivanov A.V., Peng H., Yurchenko V., Yap K.L., Negorev D.G., Schultz D.C. et al. (2007) PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823–837 10.1016/j.molcel.2007.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liang Q., Deng H., Li X., Wu X., Tang Q., Chang T.H. et al. (2011) Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J. Immunol. 187, 4754–4763 10.4049/jimmunol.1101704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Neo S.H., Itahana Y., Alagu J., Kitagawa M., Guo A.K., Lee S.H. et al. (2015) TRIM28 is an E3 ligase for ARF-mediated NPM1/B23 SUMOylation that represses centrosome amplification. Mol. Cell. Biol. 35, 2851–2863 10.1128/MCB.01064-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang Y., Fiskus W., Yong B., Atadja P., Takahashi Y., Pandita T.K. et al. (2013) Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc. Natl Acad. Sci. U.S.A. 110, 6841–6846 10.1073/pnas.1217692110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hannoun Z., Maarifi G. and Chelbi-Alix M.K. (2016) The implication of SUMO in intrinsic and innate immunity. Cytokine Growth Factor Rev. 29, 3–16 10.1016/j.cytogfr.2016.04.003 [DOI] [PubMed] [Google Scholar]
- 131.Li S.J. and Hochstrasser M. (1999) A new protease required for cell-cycle progression in yeast. Nature 398, 246–251 10.1038/18457 [DOI] [PubMed] [Google Scholar]
- 132.Nie M. and Boddy M.N. (2015) Pli1(PIAS1) SUMO ligase protected by the nuclear pore-associated SUMO protease Ulp1SENP1/2. J. Biol. Chem. 290, 22678–22685 10.1074/jbc.M115.673038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kroetz M.B., Su D. and Hochstrasser M. (2009) Essential role of nuclear localization for yeast Ulp2 SUMO protease function. Mol. Biol. Cell 20, 2196–2206 10.1091/mbc.e08-10-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li S.J. and Hochstrasser M. (2000) The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20, 2367–2377 10.1128/MCB.20.7.2367-2377.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Davis-Roca A.C., Divekar N.S., Ng R.K. and Wignall S.M. (2018) Dynamic SUMO remodeling drives a series of critical events during the meiotic divisions in Caenorhabditis elegans. PLoS Genet. 14, e1007626 10.1371/journal.pgen.1007626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tsur A., Bening Abu-Shach U. and Broday L. (2015) ULP-2 SUMO protease regulates E-Cadherin recruitment to adherens junctions. Dev. Cell 35, 63–77 10.1016/j.devcel.2015.08.019 [DOI] [PubMed] [Google Scholar]
- 137.Gao K., Li Y., Hu S. and Liu Y. (2019) SUMO peptidase ULP-4 regulates mitochondrial UPR-mediated innate immunity and lifespan extension. eLife 8, e41792 10.7554/eLife.41792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pelisch F., Sonneville R., Pourkarimi E., Agostinho A., Blow J.J., Gartner A. et al. (2014) Dynamic SUMO modification regulates mitotic chromosome assembly and cell cycle progression in Caenorhabditis elegans. Nat. Commun. 5, 5485 10.1038/ncomms6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sapir A., Tsur A., Koorman T., Ching K., Mishra P., Bardenheier A. et al. (2014) Controlled sumoylation of the mevalonate pathway enzyme HMGS-1 regulates metabolism during aging. Proc. Natl Acad. Sci. U.S.A. 111, E3880–E3889 10.1073/pnas.1414748111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Anjum S.G., Xu W., Nikkholgh N., Basu S., Nie Y., Thomas M. et al. (2013) Regulation of Toll signaling and inflammation by beta-arrestin and the SUMO protease Ulp1. Genetics 195, 1307–1317 10.1534/genetics.113.157859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Berdnik D., Favaloro V. and Luo L. (2012) The SUMO protease Verloren regulates dendrite and axon targeting in olfactory projection neurons. J. Neurosci. 32, 8331–8340 10.1523/JNEUROSCI.6574-10.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hashiyama K., Shigenobu S. and Kobayashi S. (2009) Expression of genes involved in sumoylation in the Drosophila germline. Gene Expr. Patterns 9, 50–53 10.1016/j.gep.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 143.Cubenas-Potts C. and Matunis M.J. (2013) SUMO: a multifaceted modifier of chromatin structure and function. Dev. Cell 24, 1–12 10.1016/j.devcel.2012.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sharma P., Yamada S., Lualdi M., Dasso M. and Kuehn M.R. (2013) Senp1 is essential for desumoylating Sumo1-modified proteins but dispensable for Sumo2 and Sumo3 deconjugation in the mouse embryo. Cell Rep. 3, 1640–1650 10.1016/j.celrep.2013.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Van Nguyen T., Angkasekwinai P., Dou H., Lin F.M., Lu L.S., Cheng J. et al. (2012) SUMO-specific protease 1 is critical for early lymphoid development through regulation of STAT5 activation. Mol. Cell 45, 210–221 10.1016/j.molcel.2011.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yates K.E., Korbel G.A., Shtutman M., Roninson I.B. and DiMaio D. (2008) Repression of the SUMO-specific protease Senp1 induces p53-dependent premature senescence in normal human fibroblasts. Aging Cell 7, 609–621 10.1111/j.1474-9726.2008.00411.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Maruyama E.O., Lin H., Chiu S.Y., Yu H.M., Porter G.A. and Hsu W. (2016) Extraembryonic but not embryonic SUMO-specific protease 2 is required for heart development. Sci. Rep. 6, 20999 10.1038/srep20999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Qi Y., Zuo Y., Yeh E.T. and Cheng J. (2014) An essential role of small ubiquitin-like modifier (SUMO)-specific Protease 2 in myostatin expression and myogenesis. J. Biol. Chem. 289, 3288–3293 10.1074/jbc.M113.518282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Haindl M., Harasim T., Eick D. and Muller S. (2008) The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 9, 273–279 10.1038/embor.2008.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nayak A., Lopez-Davila A.J., Kefalakes E., Holler T., Kraft T. and Amrute-Nayak M. (2019) Regulation of SETD7 methyltransferase by SENP3 is crucial for sarcomere organization and cachexia. Cell Rep. 27, 2725–36 e4 10.1016/j.celrep.2019.04.107 [DOI] [PubMed] [Google Scholar]
- 151.Yun C., Wang Y., Mukhopadhyay D., Backlund P., Kolli N., Yergey A. et al. (2008) Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J. Cell Biol. 183, 589–595 10.1083/jcb.200807185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zunino R., Schauss A., Rippstein P., Andrade-Navarro M. and McBride H.M. (2007) The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J. Cell Sci. 120, 1178–1188 10.1242/jcs.03418 [DOI] [PubMed] [Google Scholar]
- 153.Li J., Lu D., Dou H., Liu H., Weaver K., Wang W. et al. (2018) Desumoylase SENP6 maintains osteochondroprogenitor homeostasis by suppressing the p53 pathway. Nat. Commun. 9, 143 10.1038/s41467-017-02413-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen D., Wang P., Lewis R.L., Daigh C.A., Ho C., Chen X. et al. (2007) A microarray analysis of the emergence of embryonic definitive hematopoiesis. Exp. Hematol. 35, 1344–1357 10.1016/j.exphem.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 155.Garvin A.J., Densham R.M., Blair-Reid S.A., Pratt K.M., Stone H.R., Weekes D. et al. (2013) The deSUMOylase SENP7 promotes chromatin relaxation for homologous recombination DNA repair. EMBO Rep. 14, 975–983 10.1038/embor.2013.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hutten S., Chachami G., Winter U., Melchior F. and Lamond A.I. (2014) A role for the Cajal-body-associated SUMO isopeptidase USPL1 in snRNA transcription mediated by RNA polymerase II. J. Cell Sci. 127, 1065–1078 10.1242/jcs.141788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Matsuzaki K., Minami T., Tojo M., Honda Y., Uchimura Y., Saitoh H. et al. (2003) Serum response factor is modulated by the SUMO-1 conjugation system. Biochem. Biophys. Res. Commun. 306, 32–38 10.1016/S0006-291X(03)00910-0 [DOI] [PubMed] [Google Scholar]
- 158.Wang Y., Morishima M., Zheng M., Uchino T., Mannen K., Takahashi A. et al. (2007) Transcription factors Csx/Nkx2.5 and GATA4 distinctly regulate expression of Ca2+ channels in neonatal rat heart. J. Mol. Cell. Cardiol. 42, 1045–1053 10.1016/j.yjmcc.2007.03.905 [DOI] [PubMed] [Google Scholar]
- 159.Kang J., Gocke C.B. and Yu H. (2006) Phosphorylation-facilitated sumoylation of MEF2C negatively regulates its transcriptional activity. BMC Biochem. 7, 5 10.1186/1471-2091-7-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gocke C.B., Yu H. and Kang J. (2005) Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J. Biol. Chem. 280, 5004–5012 10.1074/jbc.M411718200 [DOI] [PubMed] [Google Scholar]
- 161.Riquelme C., Barthel K.K. and Liu X. (2006) SUMO-1 modification of MEF2A regulates its transcriptional activity. J. Cell. Mol. Med. 10, 132–144 10.1111/j.1582-4934.2006.tb00295.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gregoire S. and Yang X.J. (2005) Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell. Biol. 25, 2273–2287 10.1128/MCB.25.6.2273-2287.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang J., Li A., Wang Z., Feng X., Olson E.N. and Schwartz R.J. (2007) Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol. Cell. Biol. 27, 622–632 10.1128/MCB.01160-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pan M.R., Chang T.M., Chang H.C., Su J.L., Wang H.W. and Hung W.C. (2009) Sumoylation of Prox1 controls its ability to induce VEGFR3 expression and lymphatic phenotypes in endothelial cells. J. Cell Sci. 122, 3358–3364 10.1242/jcs.050005 [DOI] [PubMed] [Google Scholar]
- 165.Shan S.F., Wang L.F., Zhai J.W., Qin Y., Ouyang H.F., Kong Y.Y. et al. (2008) Modulation of transcriptional corepressor activity of prospero-related homeobox protein (Prox1) by SUMO modification. FEBS Lett. 582, 3723–3728 10.1016/j.febslet.2008.09.057 [DOI] [PubMed] [Google Scholar]
- 166.Huber P., Crum T., Clary L.M., Ronan T., Packard A.V. and Okkema P.G. (2013) Function of the C. elegans T-box factor TBX-2 depends on SUMOylation. Cell. Mol. Life Sci. 70, 4157–4168 10.1007/s00018-013-1336-y [DOI] [PMC free article] [PubMed] [Google Scholar]