Abstract
Ribosome biogenesis is the fine-tuned, essential process that generates mature ribosomal subunits and ultimately enables all protein synthesis within a cell. Novel regulators of ribosome biogenesis continue to be discovered in higher eukaryotes. While many known regulatory factors are proteins or small nucleolar ribonucleoproteins, microRNAs (miRNAs), and long non-coding RNAs (lncRNAs) are emerging as a novel modulatory layer controlling ribosome production. Here, we summarize work uncovering non-coding RNAs (ncRNAs) as novel regulators of ribosome biogenesis and highlight their links to diseases of defective ribosome biogenesis. It is still unclear how many miRNAs or lncRNAs are involved in phenotypic or pathological disease outcomes caused by impaired ribosome production, as in the ribosomopathies, or by increased ribosome production, as in cancer. In time, we hypothesize that many more ncRNA regulators of ribosome biogenesis will be discovered, which will be followed by an effort to establish connections between disease pathologies and the molecular mechanisms of this additional layer of ribosome biogenesis control.
Keywords: cancer, gene regulation, non-coding RNA, nucleolus, ribosomopathies
Introduction
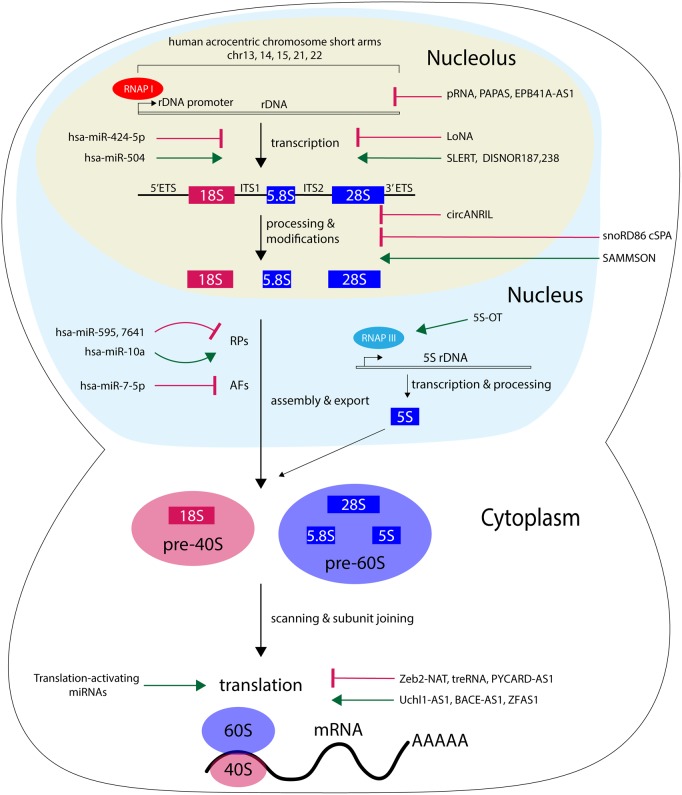
Ribosome biogenesis is the essential, complex, and tightly regulated process in which large and small ribosomal subunits are assembled. This process accounts for the majority of energy usage in a eukaryotic cell (reviewed in [1,2]). Throughout this process, the co-ordinated effort of all three RNA polymerases (RNAPI, II, III), ∼80 ribosomal proteins (RPs), and over 200 assembly factors (AFs) is required (Figure 1). Ribosome biogenesis starts in the nucleolus when RNAPI engages tandemly repeated ribosomal DNA (rDNA) arrays to transcribe the polycistronic precursor ribosomal RNA (rRNA). In humans, this 47S pre-rRNA is processed to generate the mature 18S, 5.8S, and 28S rRNAs, which are modified by C/D and H/ACA box small nucleolar ribonucleoproteins (snoRNPs) to undergo 2′-O-methylation and pseudouridylation, respectively. The mature rRNAs are assembled with the RNAPIII-transcribed 5S rRNA and RPs to form the 40S (18S and small RPs) and 60S ribosomal subunits (28S, 5.8S, 5S, and large RPs) that translate messenger RNAs (mRNAs) into polypeptides in the cytoplasm. More in-depth regulation of ribosome biogenesis continues to be discovered, including regulation by micro and long non-coding RNAs (Figure 1; Tables 1 and 2).
Figure 1. Ribosome biogenesis is regulated at multiple steps by microRNAs (miRNAs) and long non-coding RNAs (lncRNAs).
The pre-rRNA is transcribed by RNAPI (red) and the 5S rRNA is transcribed by RNAPIII (blue). Pre-rRNA is processed in a series of steps to remove the external and internal transcribed spacer sequences (5′ ETS and 3′ ETS; ITS1 and ITS2). The rRNAs are assembled with ribosomal proteins (RPs) with assistance from assembly factors (AFs) to make the mature 40S ribosomal subunit (in red; 18S rRNA) and 60S ribosomal subunit (in blue; 28S, 5.8S and 5S rRNAs) to translate mRNAs in the cytoplasm. Control by the indicated miRNAs (left) and lncRNAs (right) can activate (green pointed arrow) or inhibit (red bar-headed arrow) specific steps in ribosome biogenesis.
Table 1. MicroRNAs and microRNA machineries involved in ribosome biogenesis.
| MicroRNA | Ribosome biogenesis steps controlled | Regulation of ribosome biogenesis | mRNA targets | Molecular function | Associated phenotypes or diseases | Ref. |
|---|---|---|---|---|---|---|
| miR-504 | pre-rRNA transcription | Negative | TP53 | miR-504 is a mirtron of FGF13 that targets TP53, derepressing transcription of FGF13 and dampening pre-rRNA synthesis. TP53 protein represses transcription of miR-504 and FGF13. FGF13 represses pre-rRNA transcription in the nucleolus, which may mitigate oncogene-associated proteotoxic stress. | Cancer | (Figures 1 and 3A) [25] |
| miR-24, miR-130a, miR-145 | pre-rRNA transcription, RP gene transcription (indirectly via MYC down-regulation) | Negative | MYC | RPL5, RPL11, and RPS14 facilitate miRNA-mediated translational repression of MYC mRNA. | Cancer | (Figure 3B) [29-32] |
| Let-7 family | pre-rRNA transcription, RP gene transcription (indirectly via MYC down-regulation) | Negative | MYC, HRAS | RPL22 knockdown up-regulates LIN28B and decreases mature levels of Let-7 miRNA paralogs, unsilencing MYC and HRAS oncogenes. | Cancer, Diamond–Blackfan anemia | [33] |
| miR-7641 | RP gene transcription | Negative | RPS16, TNFS10; other RPs indirectly | Inhibition of miR-7641 sensitizes cancer cells, improving doxorubicin apoptotic response. miR-7641 mimic reduces levels of RPS16 directly and 8 other RPs indirectly. | Cancer | (Figure 1) [36] |
| miR-7 (Chinese hamster ovary cells) | pre-rRNA transcription and processing (indirectly via Akt pathway) | Negative | Possible direct targets: STRN3, CALU, CNN3, BMS1, among others | miR-7 depletion in Chinese hamster ovary (CHO) cells unleashes cell proliferation and antibody production via derepression of the Akt pathway and ribosome biogenesis. | miR-7 depletion increases proliferation and antibody production | (Figure 1) [42] |
| miR-424-5p | pre-rRNA transcription | Negative | POLR1A, UBTF, RRN3, SMAD7, CDC25A | Ribosome-related miR-424-5p targets including POLR1A and UBTF were found by Ago2 pulldown. miR-424-5p overexpression reduces muscle size in mice. | COPD, sarcopenia, muscle loss in ICU and aortic surgery patients | (Figure 1) [49] |
| miR-595 | 60S assembly; induces nucleolar stress response | Negative | RPL27A | miR-595 reduces RPL27A levels, inducing TP53 activation, nucleolar structural disruption, and blockade of erythroid proliferation and differentiation. Conversely, RPL27A overexpression leads to enhanced proliferation. | Myelodysplasia | (Figure 1) [47] |
| miR-145, miR-146a | Effects on ribosome biogenesis unclear; miRNAs associated with ribosomopathy | Unclear | TIRAP, TRAF6 | Codepletion of miR-145 and miR-146a activates the innate immune response via IL-6 and phenocopies 5q- syndrome at the cellular level, leading to thrombocytosis and megakaryopoesis. | 5q- syndrome ribosomopathy (myelodysplasia) | [46] |
| Zebrafish miRs (dre-miR-125c, dre-miR-140*, dre-miR-2191, dre-miR-30b, dre-miR-459*) |
Effects on ribosome biogenesis unclear; miRNAs associated with ribosomopathy | Unclear | Unidentified | miRs associated with a RPL5-deficient zebrafish model of Diamond–Blackfan anemia were identified bioinformatically. | Zebrafish model of Diamond–Blackfan anemia | [48] |
| miR-369-3p, Let-7, miRcxcr-4 (synthetic) | Translational efficiency | Positive | TNF, HMGA2, CX synthetic transcript | MicroRNAs can induce translation of targets by binding 3′ UTR AU-rich elements (AREs). Requisite FXR1 interacts with AGO2 to mediate increased translation efficiency. The translation-activating FXR1a-associated miRNP binds the 3′UTR of targets with shortened poly(A) tails. | Activation of translation of ARE-containing mRNAs | (Figure 1, translation-activating miRNAs) [52,54] |
| miR-10a | Translational efficiency | Positive | RPS: 2, 6, 16, 18, 20 RPL: 9, 13A, 15, 23 |
miR-10a directly binds RP mRNA downstream of 5′TOP motif in 5′ UTR. Overexpressing miR-10a increases mature rRNA levels, protein synthesis, and 3T3 cell proliferation, while inhibiting miR-10a has an opposite effect. | Increased RP levels and global translation | (Figures 1 and 3C) [55] |
| Dicer and Drosha | Translational efficiency | Positive | Senescent cells have lower translation, though mRNA levels do not change significantly. Dicer and Drosha expression is lower in senescent cells, and Dicer/Drosha knockdown recapitulates translation down-regulation and mRNA changes observed in senescent cells. | Cellular senescence | [102] |
Table 2. lncRNAs involved in ribosome biogenesis.
| lncRNA(s) | Ribosome biogenesis steps controlled (cellular localization) | Regulation of ribosome biogenesis | Molecular function | Associated phenotypes or diseases | Ref. |
|---|---|---|---|---|---|
| Long nucleolar RNA (LoNA) | pre-rRNA transcription, processing/modifications, translation (nucleolar) | Negative | Two 5′-end nucleolin binding sites and two 3′-end C/D box snoRNA sequences sequester nucleolin and inhibit functional snoRNP formation respectively. | Neurodegeneration (Alzheimer's disease) | (Figures 1 and 5A) [59,61] |
| Promoter associated RNA (pRNA) | pre-rRNA transcription (nucleolar) | Negative | TIP5 interacting stem loop recruits NoRC, DNA:RNA triplex formation recruits DNMT3b. | Embryonic stem cell exit from pluripotency | (Figures 1 and 5B) [66,67,69] |
| Promoter and pre-rRNA antisense (PAPAS) | pre-rRNA transcription (nucleolar) | Negative | Chromatin modifications by recruiting either suv4-20h2 or CHD4/NuRD complexes to the rDNA under various stress conditions. | Quiescence and proliferation | (Figure 1) [67,71–74] |
| snoRNA-ended lncRNA that enhances pre-rRNA transcription (SLERT) | pre-rRNA transcription (nucleolar) | Positive | H/ACA snoRNA ended for nucleolar localization, interacts with DDX21 to inhibit its repressive function. | Cancer (oncogenic) | (Figure 1) [75] |
| Circular antisense lncRNA in INK4 locus (circANRIL) | pre-rRNA processing (nucleolar) | Negative | Binds LSU processing factor PES1 to inhibit ITS2 cleavage. | Atherosclerosis, Kawasaki disease | (Figures 1 and 5C) [78,79] |
| alu-containing RNAs (aluRNA) | nucleolar structure, pre-rRNA transcription (nucleolar) | Positive | alu elements from spliced introns are enriched in nucleoli and necessary to maintain nucleolar structure and pre-rRNA transcription through interactions with nucleolin and nucleophosmin. | N/A | [81] |
| Intergenic spacer RNAs (IGS) | nucleolar stress response, nucleolar structure (nucleolar detention center) | N/A | Expressed under various stress conditions and sequester proteins within nucleolar foci for inactivation. | Amyloidgenesis | [83,103–105] |
| 5S rRNA overlapped transcripts (5S-OT) | 5S rRNA transcription (nuclear) | Positive | Associates with 5S rDNA gene clusters to increase 5S transcription. | Cell differentiation | (Figure 1) [84] |
| Erythrocyte membrane protein band 4.1 like 4A antisense 1 (EPB41L4A-AS1) | pre-rRNA transcription (nucleolar) | Negative | Interacts with HDAC2 and NPM1 to increase their nucleolar localization. | Cancer (tumor suppressor) | (Figure 1) [97] |
| snoRNA 86 cytoplasmic 5′ snoRNA capped 3′ polyadenylated RNA (snoRD86 cSPA) | pre-rRNA processing (cytoplasmic) | Negative | Product of alternative splicing of NOP56 pre-mRNA that undergoes nonsense mediated decay. | N/A | (Figure 1) [85] |
| Survival associated mitochondrial melanoma specific oncogenic non-coding RNA (SAMMSON) | pre-rRNA processing (cytoplasmic) | Positive | Interacts with P32 to increase P32's mitochondrial localization and CARF to increase XRN2 nucleolar localization. | Melanoma (oncogenic) | (Figure 1) [88,92] |
| Distal junction transcripts (DISNOR 187, DISNOR 238) | pre-rRNA transcription (nucleolar) | Positive | Distal junction transcripts lead to nucleolar stress response and decreased pre-rRNA transcription upon depletion. | N/A | (Figure 1) [98] |
| Antisense/translation reprogramming lncRNAs | translation (cytoplasmic) | Both | Inhibit or enhance translation of their sense and other transcripts under various stress inductions. | N/A | Examples (Figure 1): BACE1-AS [106] PYCARD-AS1 [107] treRNA [108] Uchl1-AS1 [109] Zeb2-NAT [110] ZFAS1 [111] |
miRNAs and ribosome biogenesis
Canonical microRNA biogenesis and target regulation
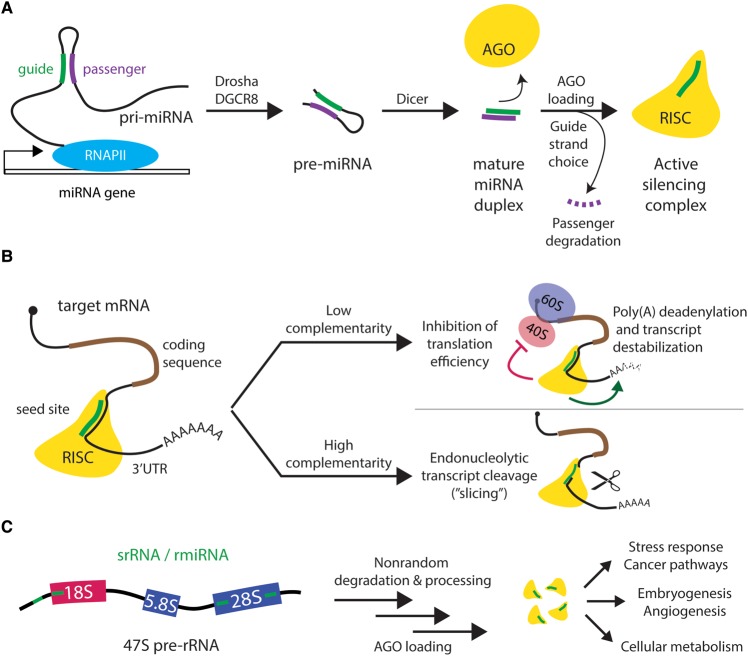
MicroRNAs (miRNAs or miRs) are a class of short (∼21 nucleotide, nt) non-coding RNAs (ncRNAs) that post-transcriptionally regulate mRNA stability and translatability. Canonical miRNA biogenesis is well-studied; typically, a primary miRNA (pri-miRNA) transcript is synthesized by RNAPII from a miRNA gene and then undergoes two processing steps before being loaded into the Argonaute protein (AGO) to form an active RNA-induced silencing complex (RISC) (Figure 2A). The active silencing complex binds mRNA transcripts at a site usually within the 3′UTR of the targets. Based on the degree of miRNA:mRNA complementarity, RISC can induce transcript destabilization or translational repression (low complementarity) or transcript cleavage (high complementarity) (Figure 2B), thereby down-regulating target expression post-transcriptionally [3,4].
Figure 2. Biogenesis and function of microRNAs (miRNAs).
(A) Canonical generation and activation of cellular miRNAs. In most cases, a primary miRNA transcript (pri-miRNA) containing the mature miRNA sequence is synthesized by RNAPII from a miRNA gene. Nuclear processing of the pri-miRNA by a microprocessor complex containing Drosha and DGCR8 results in a trimmed intermediate precursor hairpin (pre-miRNA), which is exported to the cytoplasm for secondary processing by Dicer endonuclease to generate the mature miRNA duplex. Argonaute (AGO) protein binds the miRNA duplex to form an active RNA-induced silencing complex (RISC), retaining the ‘guide' strand and expelling the ‘passenger' strand (miRNA*) for degradation. Guide strand choice depends on conformational energetics of loading the mature duplex into Argonaute. (B) miRNA-mediated post-transcriptional mRNA regulation depends on target complementarity. The active silencing complex is targeted to mRNA transcripts via complementary hybridization with the 6-base seed region of the loaded miRNA guide, usually at a site within the 3′UTR of the targets downstream of the coding sequence (brown). Based on the degree of miRNA:mRNA complementarity, RISC can induce translational repression, transcript destabilization, or transcript cleavage, thereby down-regulating target expression post-transcriptionally. Low complementarity causes mRNA poly(A) deadenylation and reduces translation efficiency, while high complementarity can trigger endonucleolytic target degradation (slicing). (C) rRNA-hosted miRNAs are stably generated and control diverse cellular processes and outcomes. Small rDNA-derived RNAs or rRNA-hosted miRNA analogs (srRNAs or rmiRNAs) are produced from functional (18S and 28S) and nonfunctional (5′ ETS) regions of the 47S pre-rRNA transcript. The mechanisms for rmiRNA production are poorly understood, but are not likely to be due to random degradation. Mature rmiRNAs have been observed to control diverse developmental, metabolic, and stress pathways.
Noncanonical miRNA sources
While most miRNAs originate from independently transcribed miRNA genes, many noncanonical miRNA sources have been discovered. Mature miRNAs can be processed out of endogenously transcribed intron lariats (mirtrons), short hairpin RNAs (shRNAs), transfer RNAs (tRNAs), and small nuclear and nucleolar RNAs (sn- and snoRNAs) [3]. Notably, miRNA-like molecules interchangeably called small rDNA-derived RNAs or rRNA-hosted miRNA analogs (srRNAs or rmiRNAs) derive from precursor and mature rRNAs through poorly understood mechanisms (Figure 2C) [5,6]. srRNA/rmiRNA generation is likely not due to random degradation but rather controlled processing that creates stable srRNA products, and may be Drosha-dependent or independent [6–8]. Functionally, rmiRNAs have been shown to bind AGO proteins and are implicated in the regulation of various metabolic and developmental pathways. srRNAs in fly and human cells associate with AGO complexes [7]. Differential hepatic srRNA expression was observed in diabetic mice, and specific srRNAs were found to modulate transcription of regulators of gluconeogenesis in mouse hepatoma cells [7]. Several human rmiRNAs were predicted to target stress- and cancer-related pathways [6], and differential rmiRNA expression has been observed upon heat stress in rice [9] and wheat [10]. In zebrafish, rmiRNAs mapping to the 18S and 28S rRNAs were found to be critical for early embryogenesis [8] and blood vessel formation [11], respectively. Additionally, differential spatiotemporal transcription of rDNA sequence variants may also alter rmiRNA expression [8,12]. Novel rRNA-derived miRNAs exemplify the emerging connection between miRNAs and ribosome biogenesis, underscoring how diverse cellular processes can be modulated by miRNAs that interact with ribosome production.
Nucleolar miRNAs
Dozens of miRNAs are stably enriched in the nucleolus in a variety of human and mammalian cell lines, and their localization is resistant to RNAPI transcription inhibition or other cellular and nucleolar stresses [13–15]. These nucleolar miRNAs may have diverse noncanonical biological roles including direct regulation of ribosomal subunit formation [16] or rRNA synthesis [17], the protected formation of pre-silenced miRNA:mRNA pairs away from the crowded and competitive cytoplasm [18], mediation of a defensive response to exogenous genetic material [14], or even targeting for deactivation by nucleolar deaminase editing [13]. These initial observations warrant additional follow-up to better define the identities and roles of nucleolar miRNAs. For a more extensive discussion of miRNAs localized to the nucleolus, we refer the reader to a brief review by Catalanotto et al. [19].
Direct down-regulation of miRNA targets in ribosome biogenesis
A growing cadre of disease-associated miRNAs have been shown to control components, subprocesses, or regulators of ribosome biogenesis (Figure 1, Table 1). Dysregulation of miRNA expression correlates with the progression of many types of cancer [20,21], and the link between cancer and ribosome biogenesis is well-established [22]. We review recent publications that consider (1) direct interplay among cancer, ribosome biogenesis and miRNAs holistically and (2) miRNAs regulators of ribosome biogenesis in ribosomopathies and in chronic obstructive pulmonary disease (COPD).
Upstream control of ribosome biogenesis via miRNA-mediated control of TP53, MYC, and oncogenesis
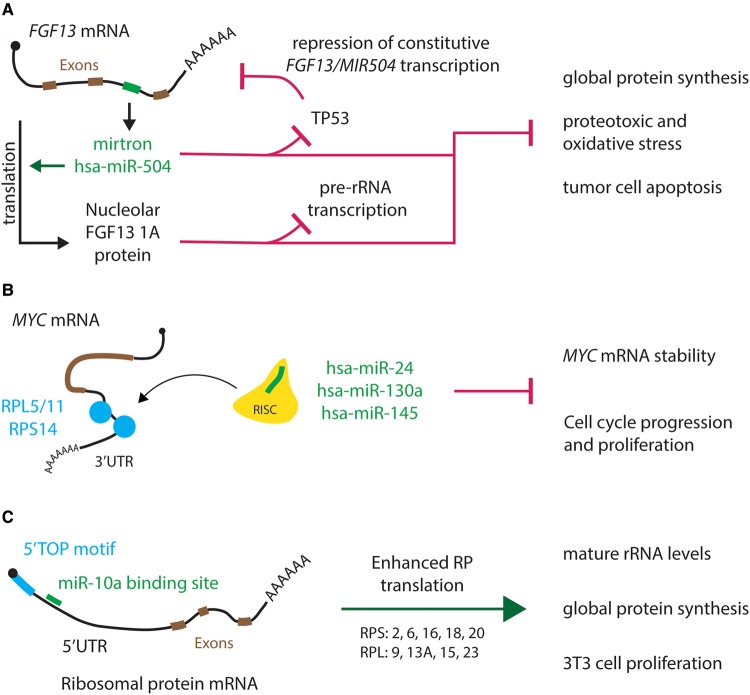
The well-studied cancer switches TP53 and MYC sit as regulatory endpoints of several miRNAs [23] and are intimately connected with ribosome biogenesis [22,24]. A genetic circuit encompassing hsa-miR-504, the nucleolar protein isoform FGF13 1A, and genome guardian TP53 attenuates ribosome biogenesis in a manner promoting cell survival in models of oncogenic escape (Figures 1 and 3A) [25]. miR-504 is an FGF13 mirtron that targets TP53, although constitutive transcription of the FGF13/MIR504 locus itself is negatively regulated by TP53 via understudied mechanisms. Concerted up-regulation of FGF13 1A and miR-504 represses pre-rRNA transcription and TP53 translation, in turn attenuating global protein synthesis, oncogenic proteotoxic and oxidative stress, and tumor cell apoptosis.
Figure 3. Examples of miRNA-mediated control of ribosome biogenesis.
(A) hsa-miR-504 regulates TP53 levels and pre-rRNA transcription. miR-504 is generated from an FGF13 intron (exons in brown, miR-504 mirtron in green) and targets TP53 transcripts. Through an uninvestigated mechanism, TP53 protein dampens constitutive transcription of the FGF13/MIR504 locus. FGF13 up-regulation increases levels of miR-504 and the nucleolar protein isoform FGF13 1A, repressing TP53 translation and pre-rRNA transcription. This results in attenuation of global translation, reduction in oncogenic proteotoxic and oxidative stress, and decreased tumor cell apoptosis. (B) RPL5, RPL11, and RPS14 enable miRNA silencing of MYC. RPL5, RPL11, and RPS14 (blue) can bind the 3′UTR of MYC transcripts, and can guide active RISC complexes (yellow) loaded with miRNAs targeting MYC (green) to the mRNA. This RP-guided, miRNA-mediated MYC repression modulates cell cycle progression and proliferation, and attenuates ribosome biogenesis indirectly. (C) hsa-miR-10a enhances RP translation efficiency by binding 5′TOP mRNAs. miR-10a (green) was found to bind the 5′UTR of at least five small and four large RP mRNAs containing a 5′TOP motif (blue), increasing their translation efficiency. Augmented RP production enhances the cellular capacity for ribosome biogenesis and proliferation.
Several RPs are necessary for miRNA-mediated regulation of the MYC oncogene, which itself controls transcription of rDNA, RPs, AFs, and translation initiation factors [24]. uL5 (RPL11), uL18 (RPL5), and uS11 (RPS14), which stabilize TP53 in the nucleolar stress response [26–28], have been shown to escort the armed RISC complex to MYC transcripts for silencing by hsa-miR-24 [29,30] or hsa-miR-145 (Figure 3B) [31]. UV irradiation also induces uL5-guided MYC repression by hsa-miR-130a [32]. Via LIN28B, eL22 (RPL22) indirectly controls maturation of hsa-miR-let-7 family paralogs [33] that repress MYC, KRAS, HMGA1 and HMGA2, and other oncogenes [34,35].
Additionally, miRNAs affecting cancer treatment outcomes or oncogene expression have links with ribosome biogenesis. hsa-miR-7641 has been shown to repress uS9 (RPS16) directly and eight other RPs indirectly, and miR-7641 depletion sensitized breast and colon cancer cells to doxorubicin-induced apoptosis (Figure 1) [36]. The tumor suppressor hsa-miR-7-5p, which has been reported to target oncogenic EGFR, BCL2 [37], RELA (p65) [38], among others [39–41], may also play a role in down-regulating ribosome biogenesis. Depletion of the miR-7 homolog in Chinese hamster ovary (CHO) cells unleashed cell proliferation and antibody production, correlating with derepression of ribosomal assembly factor BMS1 and Akt pathway activators STRN3 and Ezrin (Figure 1) [42]. Overall, miRNAs can fine-tune the balance between proliferation and oncogenesis by modulating upstream regulators of ribosome biogenesis such as TP53 and MYC, as well as other oncogenes, RPs, and AFs downstream of pre-rRNA transcription.
MicroRNA dysregulation in ribosomopathies
Ribosomopathies are diseases of impaired ribosome biogenesis that result in tissue-specific defects in affected patients. Interruption of particular ribosome biogenesis subprocesses have been shown to cause a wide range of clinical presentations including anemia, leukemia, neutropenia, craniofacial defects, cancer, and other diseases of major organs [43–45]. miRNAs are involved in two classical hematological ribosomopathies, myelodysplastic syndrome (MDS) and Diamond–Blackfan anemia (DBA). As part of the 5q- MDS deletion, hsa-miR-145 and hsa-miR-146a [46] are lost, while deletion of hsa-miR-595 occurs in –7/7q- MDS [47]. In murine hematopoietic progenitor cells, codepletion of miR-145 and miR-146a homologs relieved repression of TIRAP and TRAF6, activating the innate immune response via IL-6 and leading to 5q- syndrome-like thrombocytosis and megakaryopoiesis [46]; these features were mirrored in similar experiments on patient bone marrow. miR-595 silences RPL27A resulting in TP53 activation, disruption of nucleolar architecture independent of TP53, reduction of mature 60S subunits, and blockade of erythroid proliferation and differentiation (Figure 1) [47]. RPL27A derepression enhanced proliferation in cell lines and was observed in –7/7q- MDS patients, though the authors cite the need for further translational studies [47]. Finally, tens of miRNAs were found to be differentially expressed in RP-depleted zebrafish models of the ribosomopathy DBA [48]. Potential targets of the up-regulated miRNAs were enriched for functions in transcriptional regulation and neuronal and cellular development. It will be of interest to reexamine other ribosomopathies for miRNAs important for pathogenesis.
miRNA regulation of ribosome biogenesis in chronic obstructive pulmonary disease
Other disease-associated miRNAs have been connected to ribosome biogenesis. COPD and intensive care unit patients with muscle wasting exhibit elevated expression of hsa-miR-424-5p, which targets the RNAPI pre-initiation complex factors POLR1A and UBTF (Figure 1) [49]. miR-424-5p was found to down-regulate mature rRNA levels in myoblasts and to cause muscle atrophy in mice [49]. We hypothesize that yet-undiscovered miRNA regulators of ribosome biogenesis may also play central roles in diseases arising from defects in growth-sensitive biological processes, such as development and angiogenesis, that are already known to be partially controlled by miRNAs [50,51].
Direct up-regulation of miRNA targets in ribosome biogenesis
Although miRNAs generally down-regulate the expression of their target genes, select cases have unveiled miRNA-mediated enhancement of transcript-specific or global translation (Table 1). Both endogenous and synthetic miRNAs can increase translation efficiency of target transcripts by a specialized microRNA ribonucleoprotein (miRNP) complex that lacks the normally present repressor protein GW182/TNRC6 but contains the RNA binder FXR1a [52–54]. hsa-miR-10a binds the 5′UTR of RP transcripts harboring a 5′TOP motif, up-regulating their translation, augmenting ribosome biogenesis, and enhancing proliferation (Figures 1 and 3C) [55]. Such noncanonical translational activation is also implicated in cellular quiescence and senescence, two paused growth states instigated by mTOR deactivation that feature diminished ribosome biogenesis [56,57]. Translation-activating miRNAs were originally discovered in quiescent human cells [52,53], and down-regulation of miRNA processing machinery initiates global translation reduction and induction of senescence (Figure 1) [55]. In summary, miRNAs can also enhance ribosome biogenesis and global protein synthesis via targeted RP up-regulation or mechanisms implicated in cellular aging.
lncRNAs and ribosome biogenesis
Long non-coding RNAs biogenesis and function
Besides the essential non-coding rRNAs, other long non-coding RNAs (lncRNAs) have been increasingly found to possess significant roles in eukaryotic ribosome biogenesis, with some even dysregulated in disease and proposed as therapeutic targets or biomarkers. lncRNAs are RNAs over 200 nt and without protein-coding potential. lncRNAs are transcribed in a variety of genomic contexts: intergenic, antisense, intronic, and overlapping orientations with respect to protein-coding genes [58]. This leads to their cis functions, where they regulate the expression of nearby genes. In addition, lncRNAs have diverse trans functions to regulate distant genes. Recent work has established many diverse cellular roles for these non-coding portions of the genome.
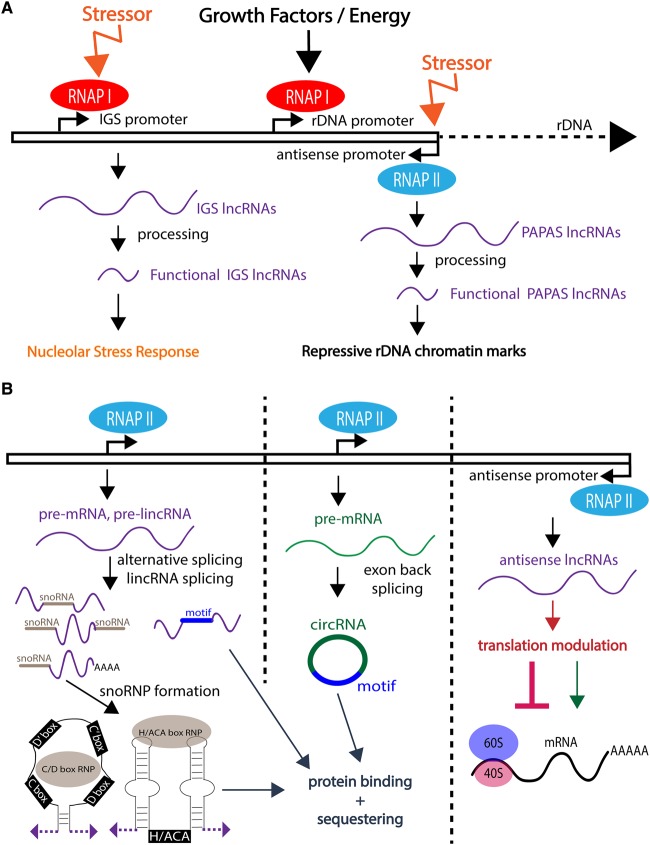
lncRNAs utilize different biogenesis pathways and sequence properties to perform roles in ribosome biogenesis. Some transcripts are transcribed by RNAPI from the rDNA gene locus (Figure 4A), while others are independently transcribed by RNAPII (lincRNAs) or produced by alternative splicing of RNAPII transcripts (Figure 4B). Ribosome biogenesis-specific lncRNA roles may be direct, modulatory, or stress response-specific. These molecular mechanisms include protein binding or sequestration, rDNA chromatin modifications, snoRNP formation, and transcript-specific translation modulations (Table 2). Here, we discuss examples of lncRNAs that function within the nucleolus to modulate rDNA transcription, pre-rRNA processing, and nucleolar structure as well as lncRNAs that alter ribosome biogenesis levels and translation located outside the nucleolus.
Figure 4. Biogenesis of diverse long non-coding RNAs (lncRNAs) involved in ribosome biogenesis.
(A) lncRNAs involved in ribosome biogenesis transcribed from ribosomal DNA (rDNA) gene loci. (Left) Intergenic spacer (IGS) RNAs are transcribed by RNA polymerase I (RNAPI) under stress conditions and processed into functional RNAs. (Middle) pre-rRNA transcription is controlled by growth factors and available energy regulated by several signaling pathways. (Right) Promoter and pre-rRNA antisense (PAPAS) RNAs are transcribed in an antisense orientation relative to the rDNA and rDNA promoter sequences and processed into functional RNAs. (B) lncRNAs involved in ribosome biogenesis transcribed by RNAPII in a variety of genomic contexts. (Left) lncRNAs are transcribed by RNAPII either independently long intergenic noncoding RNAs (lincRNAs) or via alternative splicing of other RNAPII transcripts. These RNAs can include small nucleolar RNA (snoRNA) sequences (gray) that form mature noncanonical C/D or H/ACA box small nucleolar ribonucleoproteins (snoRNPs) or binding motifs (blue). (Middle) RNAPII transcribes pre-mRNAs that can undergo back-splicing of exons to produce circular RNAs (circRNAs) (green). These RNAs can then contain motifs (blue) to bind and sequester proteins (blue-gray). (Right) Antisense lncRNAs are transcribed by RNAPII in the opposite direct of their sense transcript partners to regulate translation of specific, usually sense, transcript mRNAs (red).
Nucleolar lncRNAs
Long nucleolar RNA (LoNA): 1 RNA, 2 functions
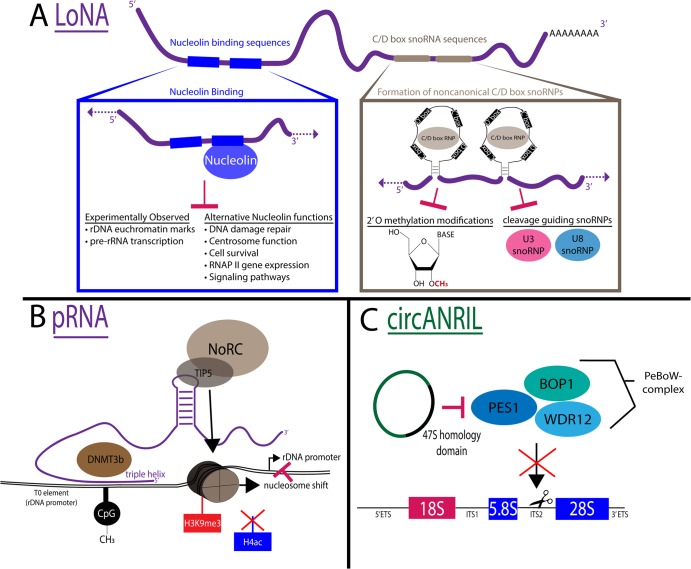
LoNA is a 1.5 kb polyadenylated lncRNA that performs dual independent functions to regulate RNAPI transcription, pre-rRNA processing, and modification (Figures 1 and 5A). LoNA was discovered by Li et al. [59] as the 4th most abundant lncRNA present in mouse neuroblastoma cell nucleoli. Overexpression of LoNA reduces pre-rRNA levels, mature rRNA levels, and global translation; conversely, knockdown enhances ribosome production. Two 5′-end nucleolin binding motifs sequester nucleolin from establishing rDNA euchromatin modifications and enhancing RNAPI transcription (Figure 5A) (nucleolin functions reviewed in [60]). At the same time, LoNA reduces pre-rRNA processing and rRNA modification by competition binding using its two 3′-end C/D box snoRNA sequences that form noncanonical C/D box snoRNPs. Additionally, LoNA levels are increased in Alzheimer's disease model mice, connecting a lncRNA to the pathogenesis of a neurodegenerative disease known to have associations with nucleolar stress and reduced rRNA production [59,61,62] (reviewed in [63]).
Figure 5. Examples of molecular mechanisms of long non-coding RNAs (lncRNAs) involved in ribosome biogenesis.
(A) Long nucleolar RNA (LoNA) is a multifunctional polyadenylated lncRNA that regulates pre-rRNA transcription, modification, and processing. (Left) It contains two nucleolin binding sequences (blue), one of which is functional to bind and inhibit nucleolin (blue) function. (Right) It contains two C/D box small nucleolar RNA (snoRNA) sequences that are able to form noncanonical C/D box small nucleolar ribonucleoproteins (snoRNPs) (gray) to inhibit modification and processing of the pre-rRNA. (B) pRNA is a multifunctional lncRNA that regulates ribosomal DNA (rDNA) chromatin modifications. (Left) It forms a DNA:RNA triple helix with the T0 element of the rDNA promoter to recruit the chromatin modifier DNMT3b (brown) to methylate CpG (black). (Right) It contains a stem-loop structure that interacts with TIP5 (gray) the large subunit of the nucleolar remodeling complex (NoRC) (brown) to promote H3K9me3 histone methylations (red), remove H4ac acetylation modifications (blue), and cause a nucleosome shift to block rDNA promoter access. (C) circANRIL (green) binds PeBoW complex (PES1, BOP1, WDR12,) (green/blue) to inhibit pre-rRNA processing of internal transcribed spacer 2 (ITS2) through 47S homology domain (black).
Other lncRNAs that modulate pre-rRNA transcription
Other lncRNAs have been implicated in the control of pre-rRNA transcription through direct roles in rDNA chromatin modifications. Although pre-rRNA is the most highly transcribed RNA, rDNA genes can adopt inactive, silent, or active chromatin states. Here, we focus on rDNA chromatin activity dynamics regulating rDNA transcription; however, it is important to note these dynamics also play an important role in genome stability and organization (reviewed in [64,65]).
Promoter associated RNA (pRNA), is transcribed by RNAPI from the intergenic spacer (IGS) region of the rDNA and degraded by the exosome to produce 150–300 nt molecules (Figure 4A) [66]. pRNA is required for rDNA silencing through the recruitment of the nucleolar remodeling complex (NoRC) via direct interaction (Figures 1 and 5B) [67]. Additionally, a 5′ portion of pRNA forms a DNA:RNA triplex with the T0 regulatory element of the rDNA promoter. While triplex formation has only been observed in vitro, it suggests that pRNA recruits the DNA methyltransferase DNMT3b to further establish repressive chromatin marks by CpG methylation at this element (Figure 5B) [68]. pRNA's function is also necessary for exit from pluripotency in embryonic stem cells [69] and was speculated to be important for maintaining genomic stability [70].
There are many IGS transcripts that regulate rDNA chromatin modifications, including promoter and pre-rRNA antisense transcripts (PAPAS) (Figures 1 and 4A). These transcripts inactivate pre-rRNA transcription upon a variety of stressors, including density arrest and serum deprivation that lead to H4K20me3 modifications at the rDNA promoter [71]. Additionally, hypo-osmotic, hypotonic, and heat shock stressors promote nucleosome remodeling and deacetylase (NuRD) complex recruitment and H4ac inhibition by a similarly proposed DNA:RNA triplex formation as that as pRNA at the rDNA promoter [72–74].
Pre-rRNA transcription modulation also occurs beyond the chromatin level by a snoRNA-ended lncRNA that enhances pre-rRNA transcription (SLERT) (Figure 1). Both of its ends bear H/ACA snoRNA sequences that are essential for its nucleolar localization [75]. Additionally, there is a 143-nt internal sequence that interacts with DDX21, an RNA helicase that forms rings around multiple RNAPI complexes to inhibit pre-rRNA transcription. This interaction relieves suppression and increases pre-rRNA transcription [75]. DDX21 is also important for pre-rRNA processing steps [76], but SLERT's role in pre-rRNA processing has not yet been explored.
Circular antisense non-coding RNA in the INK4 locus (circANRIL), a long non-coding circular RNA that inhibits pre-rRNA processing
Only a few lncRNAs have been reported to modulate pre-rRNA processing, but similarly to LoNA and SLERT, these lncRNAs modulate pre-rRNA processing via sequestration mechanisms. circANRIL binds and sequesters PES1, a member of the PES1, BOP1, WDR12 (PeBoW) complex that is essential for large subunit processing through internal transcribed spacer 2 (ITS2) cleavage [77]. PES1 binds circANRIL at a 47S pre-rRNA homology domain bearing sequence identities across the pre-rRNA primary transcript (Figure 5C) [78]. This interaction most likely inhibits PeBoW complex formation and PES1 interaction with the pre-rRNA, leading to nucleolar stress (Figure 1). Identification of other lncRNAs with pre-rRNA homology could provide competition regulation like circANRIL, which is proposed to be present in sufficient amounts in cells to modulate ribosome production. This anti-proliferative function is supported by observations of lower circANRIL abundance in childhood systemic vasculitis (Kawasaki disease) patients [79] and association with a higher risk of atherosclerosis in a human cohort [78].
lncRNAs in nucleolar structure maintenance
RNAPII transcription inhibition disrupts nucleolar structure, indicating a possible function of RNAPII transcripts within the nucleolus [80]. Caudron-Herger et al. [81] recently identified RNAPII alu-repeat transcripts that are partially responsible for maintaining nucleolar structure. These 100–300 nt RNAs are spliced from introns and enriched in the nucleolus. They interact with nucleolin and nucleophosmin to enhance RNAPI transcription and support nucleolar structure [81]. These RNAs provide a possible mechanism for the coordination of RNAPI and RNAPII transcription, particularly since alu elements represent a large portion (∼10%) of the human genome [82].
Some nucleolar lncRNAs are not directly involved in ribosome biogenesis, but arise under different stress conditions and are associated with the nucleolar stress response. This stress response elicits outputs to inhibit ribosome synthesis and induce apoptosis. These lncRNAs include multiple IGS RNAs that are transcribed by RNAPI (Figure 4A) and form foci within the nucleolus under stress conditions, termed ‘nucleolar detention centers.' These lncRNAs sequester and inactivate proteins within these detention centers. Establishment of this remodeled nucleolus is thought to be important for stress-induced transcriptional inactivation [83] (reviewed in [65]).
Non-nucleolar lncRNAs
5S rRNA transcription regulation by a lncRNA
While transcription of the polycistronic 47S pre-rRNA by RNAPI occurs within the nucleolus, the 5S rRNA is transcribed in the nucleoplasm by RNAPIII. 5S overlapping transcript (5S-OT) regulates this important extra-nucleolar transcription step (Figure 1). 5S-OT is transcribed by RNAPII and is complementary to the 5S rRNA. Its depletion leads to decreases in the production of 5S rRNAs through undiscovered cis-acting mechanisms [84].
lncRNAs that sequester processing factors outside the nucleolus
snoRNA sequences in lncRNAs can be utilized in other unique ways to modulate ribosome biogenesis levels. This includes an orphan C/D box snoRNA, snoRD86 within an intron of the NOP56 pre-mRNA. Upon increased levels of NOP56 and snoRNPs, this intronic snoRNA is able to bind and sequester C/D box snoRNPs to inhibit splicing of the NOP56 pre-mRNA. This ultimately ends in nonsense-mediated decay of the unspliced transcript, containing snoRD86, termed snoRD86 cytoplasmic 5′ snoRNA capped 3′ polyadenylated RNA (cSPA) (Figure 1) [85]. It will be of interest to determine the extent to which splicing feedback regulation occurs for intronic snoRNAs in other ribosome biogenesis factor pre-mRNAs.
While ribosome biogenesis is generally increased in cancer (reviewed in [86]), synchronization of this process with mitochondrial ribosome production is also necessary for cancer cell fitness [87,88]. Survival associated mitochondrial melanoma specific oncogenic non-coding RNA (SAMMSON) is another example of a lncRNA that binds ribosome biogenesis factors to fine-tune ribosome biogenesis in melanoma cells for synchronization of mitochondrial and nucleolar ribosome production (Figure 1). SAMMSON binds p32, a protein essential for mitoribosome formation [89], and increases its mitochondrial localization [88]. Furthermore, SAMMSON increases the nucleolar localization of XRN2, an exonuclease necessary for pre-rRNA processing [90]. It performs this latter task by binding CARF, a negative regulator of XRN2 [91], that normally increases its nucleoplasmic localization [92]. SAMMSON's presence increases nucleolar and mitochondrial ribosome synthesis, while its depletion leads to aberrant pre-rRNA processing intermediates. SAMMSON expression has only been reported in cancer cells; however, it is possibly expressed during embryonic development, since p32 is essential for development [89].
snoRD86 cSPA and SAMMSON utilize binding sequences to sequester RNA binding proteins from performing their functions in pre-rRNA processing. While it is not yet clear how these lncRNAs function in normal cellular metabolism, their study has revealed a mechanism for how ribosome biogenesis can be controlled from outside the nucleolus. Nevertheless, the presence of snoRNA sequences within lncRNAs might not always lead to direct roles in ribosome biogenesis modulation. Prader–Willi Syndrome deletion region (15q11-q13) transcribes several SPAs and snolncRNAs that have no current identified function in ribosome biogenesis (reviewed in [93]).
Translation modulation by lncRNAs
Although lncRNAs have been increasingly observed to be translated into micro peptides (reviewed in [94]), there is a lncRNA subcellular localization system (lncSLdb) that identified 300 lncRNAs associated with mature ribosomes in the cytoplasm through extensive literature mining [95]. This indicates possible lncRNA regulation of final ribosome maturation steps or translation efficiency if not translated into micro peptides. As of now, there are cases of the latter being studied (reviewed in [96]) (see examples in Figure 1 and Table 2).
Future developments of miRNAs and lncRNAs in ribosome biogenesis
Studies continue to emerge and implicate novel nucleolar miRNAs and lncRNAs in ribosome biogenesis regulation. Although canonical miRNA-mediated translational repression occurs in the cytoplasm, new roles have been revealed for miRNAs localized to the nucleolus including regulation of RNAPI and subunit assembly, defense against exogenous nucleic acids, and precise control of miRNA stability and target recognition. The lncRNA erythrocyte membrane protein band 4.1 Like 4A antisense 1 (EPB41L4A-AS1) was identified as a possible regulator of glycolysis and glutaminolysis in cancer. Yet, it interacts with HDAC2 and NPM1 to increase their nucleolar localization [97], raising the question: does EPB41L4A-AS1 expression down-regulate RNAPI transcription if HDAC2 is able to establish heterochromatin marks on the rDNA (Figure 1)? Additionally, lncRNAs expressed from the distal flanking junction of the rDNA (DISNOR 187 and 238) have been recently discovered. Depletion of these leads to decreased pre-rRNA transcription [98], suggesting a functional role of these lncRNAs in the transcription of the rDNA (Figure 1). Further study of nucleolar miRNAs and lncRNAs will illuminate how ncRNAs other than rRNA shape the dynamic landscape of the nucleolus.
Both miRNAs and lncRNAs play important roles in cancer and development, but to date only miRNAs have been associated with ribosomopathies, such as myelodysplasia and DBA. However, one study identified candidate miRNAs and lncRNAs that have altered expression in a DBA zebrafish model compared with wildtype [48]. Continued work will help elucidate new ncRNAs that play a role in ribosomopathy pathogenesis and disease-related nucleolar stress responses.
Published bioinformatic databases may contain more miRNAs and lncRNAs that are candidates for ribosome biogenesis regulation based on their localization and links to disease. RNALocate notes 79 nucleolar human miRNAs, plus 448 nucleolar and 190 ribosome-associated human lncRNAs on top of the lncRNAs in lncSLdb [99]. lncATLAS identified 28 lncRNAs that were enriched in the nucleolus compared with the nucleoplasm by subcellular fractionation followed by RNA-seq in K652 cells [100]. In addition to LoNA, Li et al. [59] also identified several other nucleolar localized lncRNAs by a similar RNA-seq method. It would be of interest to extend these experiments to look for conserved nucleolar miRNAs and lncRNAs across cell types and species. The mammal ncRNA-disease repository (MNDR) database, which catalogs ncRNAs linked to diseases, may provide clues about novel miRNA and lncRNA regulators of ribosome biogenesis based on their involvement in ribosomopathies or otherwise growth-sensitive diseases [101]. Together, bioinformatic databases bearing localization and disease linkage data about miRNAs and lncRNAs can help reveal novel ncRNA regulators of ribosome biogenesis.
Perspectives
miRNAs and lncRNAs play diverse roles in the regulation of ribosome biogenesis, forming an additional dense layer of control over cellular growth and translational output. In diseases where ribosome biogenesis is implicated, such as cancer and various ribosomopathies, these ncRNAs comprise another conduit by which biochemical defects become the basis of medical pathogenesis.
miRNAs and lncRNAs modulate core ribosome biogenesis processes including RNAPI pre-rRNA transcription, pre-rRNA processing, and ribosome assembly as well as nucleolar structural maintenance and global translation.
Future discovery of novel ncRNA regulators of ribosome biogenesis and of expanded regulatory roles for known ncRNAs will be aided by (1) experimental inspection of specific steps in ribosome biogenesis and (2) bioinformatic databases containing localization and disease linkage data.
Acknowledgements
Thank you to the members of the laboratory of S.J.B. for insightful information, questions, and comments throughout the manuscript writing process.
Abbreviations
- AFs
assembly factors
- AGO
Argonaute protein
- COPD
chronic obstructive pulmonary disease
- cSPA
capped 3′ polyadenylated RNA
- DBA
Diamond–Blackfan anemia
- IGS
intergenic spacer
- lncRNAs
long non-coding RNAs
- LoNA
long nucleolar RNA
- MDS
myelodysplastic syndrome
- miRNAs
microRNAs
- mRNA
messenger RNA
- ncRNAs
non-coding RNAs
- pRNA
promoter associated RNA
- rDNA
ribosomal DNA
- RISC
RNA-induced silencing complex
- RPs
ribosomal proteins
- rRNA
ribosomal RNA
- snoRNPs
small nucleolar ribonucleoproteins
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
S.J.B. acknowledges 1R35GM131687 from the National Institutes of Health (NIH), and the Breast Cancer Alliance for support of her laboratory. M.A.M. and C.J.B. acknowledge T32GM007223 from the NIH for funding.
Author Contributions
C.J.B. wrote the miRNA portion and M.A.M. wrote the lncRNA portion. All authors read, commented on, discussed, edited, and approved the final manuscript.
References
- 1.Woolford J.L. Jr and Baserga S.J. (2013) Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henras A.K., Plisson-Chastang C., O'Donohue M.F., Chakraborty A. and Gleizes P.E. (2015) An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 6, 225–242 10.1002/wrna.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel D.P. (2018) Metazoan microRNAs. Cell 173, 20–51 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gebert L.F.R. and MacRae I.J. (2019) Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 20, 21–37 10.1038/s41580-018-0045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert M., Benmoussa A. and Provost P. (2019) Small non-Coding RNAs derived from eukaryotic ribosomal RNA. Noncoding RNA 5, E16 10.3390/ncrna5010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshikawa M. and Fujii Y.R. (2016) Human ribosomal RNA-derived resident microRNAs as the transmitter of information upon the cytoplasmic cancer stress. Biomed. Res. Int. 2016, 7562085 10.1155/2016/7562085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei H., Zhou B., Zhang F., Tu Y., Hu Y., Zhang B. et al. (2013) Profiling and identification of small rDNA-derived RNAs and their potential biological functions. PLoS One 8, e56842 10.1371/journal.pone.0056842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locati M.D., Pagano J.F.B., Abdullah F., Ensink W.A., van Olst M., van Leeuwen S. et al. (2018) Identifying small RNAs derived from maternal- and somatic-type rRNAs in zebrafish development. Genome 61, 371–378 10.1139/gen-2017-0202 [DOI] [PubMed] [Google Scholar]
- 9.Mangrauthia S.K., Sailaja B., Pusuluri M., Jena B., Prasanth V.V., Agarwal S. et al. (2018) Deep sequencing of small RNAs reveals ribosomal origin of microRNAs in Oryza sativa and their regulatory role in high temperature. Gene Rep. 11, 270–278 10.1016/j.genrep.2018.05.002 [DOI] [Google Scholar]
- 10.Wang Y., Li H., Sun Q. and Yao Y. (2016) Characterization of small RNAs derived from tRNAs, rRNAs and snoRNAs and their response to heat stress in wheat seedlings. PLoS One 11, e0150933 10.1371/journal.pone.0150933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y., Duan X., Xu G., Wang X., Wei G., Dong S. et al. (2019) A ribosomal DNA-hosted microRNA regulates zebrafish embryonic angiogenesis. Angiogenesis 22, 211–221 10.1007/s10456-019-09663-3 [DOI] [PubMed] [Google Scholar]
- 12.Locati M.D., Pagano J.F.B., Girard G., Ensink W.A., van Olst M., van Leeuwen S. et al. (2017) Expression of distinct maternal and somatic 5.8S, 18S, and 28S rRNA types during zebrafish development. RNA 23, 1188–1199 10.1261/rna.061515.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Politz J.C., Hogan E.M. and Pederson T. (2009) MicroRNAs with a nucleolar location. RNA 15, 1705–1715 10.1261/rna.1470409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z.F., Liang Y.M., Lau P.N., Shen W., Wang D.K., Cheung W.T. et al. (2013) Dynamic localisation of mature microRNAs in human nucleoli is influenced by exogenous genetic materials. PLoS One 8, e70869 10.1371/journal.pone.0070869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai B., Liu H. and Laiho M. (2014) Small RNA expression and deep sequencing analyses of the nucleolus reveal the presence of nucleolus-associated microRNAs. FEBS Open Bio. 4, 441–449 10.1016/j.fob.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atwood B.L., Woolnough J.L., Lefevre G.M., Saint Just Ribeiro M., Felsenfeld G. and Giles K.E. (2016) Human Argonaute 2 is tethered to ribosomal RNA through microRNA interactions. J. Biol. Chem. 291, 17919–17928 10.1074/jbc.M116.725051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Politz J.C., Zhang F. and Pederson T. (2006) MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl. Acad. Sci. U.S.A. 103, 18957–18962 10.1073/pnas.0609466103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyes-Gutierrez P., Ritland Politz J.C. and Pederson T. (2014) A mRNA and cognate microRNAs localize in the nucleolus. Nucleus 5, 636–642 10.4161/19491034.2014.990864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catalanotto C., Cogoni C. and Zardo G. (2016) MicroRNA in control of gene expression: an overview of nuclear functions. Int. J. Mol. Sci. 17, E1712 10.3390/ijms17101712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin S. and Gregory R.I. (2015) MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 15, 321–333 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Acunzo M., Romano G., Wernicke D. and Croce C.M. (2015) MicroRNA and cancer–a brief overview. Adv. Biol. Regul. 57, 1–9 10.1016/j.jbior.2014.09.013 [DOI] [PubMed] [Google Scholar]
- 22.Derenzini M., Montanaro L. and Trere D. (2017) Ribosome biogenesis and cancer. Acta Histochem. 119, 190–197 10.1016/j.acthis.2017.01.009 [DOI] [PubMed] [Google Scholar]
- 23.Mei Y. and Wu M. (2016) Noncoding RNAs regulating p53 and c-Myc signaling. Adv. Exp. Med. Biol. 927, 337–365 10.1007/978-981-10-1498-7_13 [DOI] [PubMed] [Google Scholar]
- 24.van Riggelen J., Yetil A. and Felsher D.W. (2010) MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 10, 301–309 10.1038/nrc2819 [DOI] [PubMed] [Google Scholar]
- 25.Bublik D.R., Bursac S., Sheffer M., Orsolic I., Shalit T., Tarcic O. et al. (2017) Regulatory module involving FGF13, miR-504, and p53 regulates ribosomal biogenesis and supports cancer cell survival. Proc. Natl. Acad. Sci. U.S.A. 114, E496–E505 10.1073/pnas.1614876114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lessard F., Brakier-Gingras L. and Ferbeyre G. (2019) Ribosomal proteins control tumor suppressor pathways in response to nucleolar stress. Bioessays 41, e1800183 10.1002/bies.201800183 [DOI] [PubMed] [Google Scholar]
- 27.Nicolas E., Parisot P., Pinto-Monteiro C., de Walque R., De Vleeschouwer C. and Lafontaine D.L. (2016) Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat. Commun. 7, 11390 10.1038/ncomms11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X., Hao Q., Liao J., Zhang Q. and Lu H. (2013) Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene 32, 388–396 10.1038/onc.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Challagundla K.B., Sun X.X., Zhang X., DeVine T., Zhang Q., Sears R.C. et al. (2011) Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol. Cell. Biol. 31, 4007–4021 10.1128/MCB.05810-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao J.M., Zhou X., Gatignol A. and Lu H. (2014) Ribosomal proteins L5 and L11 co-operatively inactivate c-Myc via RNA-induced silencing complex. Oncogene 33, 4916–4923 10.1038/onc.2013.430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X., Hao Q., Liao J.M., Liao P. and Lu H. (2013) Ribosomal protein S14 negatively regulates c-Myc activity. J. Biol. Chem. 288, 21793–21801 10.1074/jbc.M112.445122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Challagundla K.B., Sun X.X., Zhang Q. and Dai M.S. (2015) MicroRNA-130a associates with ribosomal protein L11 to suppress c-Myc expression in response to UV irradiation. Oncotarget 6, 1101–1114 10.18632/oncotarget.2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao S., Lee S.Y., Gutierrez A., Perrigoue J., Thapa R.J., Tu Z. et al. (2012) Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood 120, 3764–3773 10.1182/blood-2012-03-415349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balzeau J., Menezes M.R., Cao S. and Hagan J.P. (2017) The LIN28/let-7 pathway in cancer. Front. Genet. 8, 31 10.3389/fgene.2017.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Cao L., Wang Y., Wang X., Liu N. and You Y. (2012) Regulation of let-7 and its target oncogenes (Review). Oncol Lett. 3, 955–960 10.3892/ol.2012.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reza A., Choi Y.J., Yuan Y.G., Das J., Yasuda H. and Kim J.H. (2017) MicroRNA-7641 is a regulator of ribosomal proteins and a promising targeting factor to improve the efficacy of cancer therapy. Sci. Rep. 2017, 8365 10.1038/s41598-017-08737-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J., Tao Y., Zhou Y., Qin N., Chen C., Tian D. et al. (2015) MicroRNA-7: a promising new target in cancer therapy. Cancer Cell Int. 15, 103 10.1186/s12935-015-0259-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giles K.M., Brown R.A., Ganda C., Podgorny M.J., Candy P.A., Wintle L.C. et al. (2016) microRNA-7-5p inhibits melanoma cell proliferation and metastasis by suppressing RelA/NF-κB. Oncotarget 7, 31663–31680 10.18632/oncotarget.9421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Y., Luo X., Li P., Tan J., Wang X., Xiang T. et al. (2015) miR-7-5p suppresses cell proliferation and induces apoptosis of breast cancer cells mainly by targeting REGgamma. Cancer Lett. 358, 27–36 10.1016/j.canlet.2014.12.014 [DOI] [PubMed] [Google Scholar]
- 40.Yin C.Y., Kong W., Jiang J., Xu H. and Zhao W. (2019) miR-7-5p inhibits cell migration and invasion in glioblastoma through targeting SATB1. Oncol Lett. 17, 1819–1825 10.3892/ol.2018.9777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu W., Wang Y., Zhang D., Yu X. and Leng X. (2018) MiR-7-5p functions as a tumor suppressor by targeting SOX18 in pancreatic ductal adenocarcinoma. Biochem. Biophys. Res. Commun. 497, 963–970 10.1016/j.bbrc.2018.02.005 [DOI] [PubMed] [Google Scholar]
- 42.Coleman O., Suda S., Meiller J., Henry M., Riedl M., Barron N. et al. (2019) Increased growth rate and productivity following stable depletion of miR-7 in a mAb producing CHO cell line causes an increase in proteins associated with the Akt pathway and ribosome biogenesis. J. Proteomics 195, 23–32 10.1016/j.jprot.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 43.Farley K.I. and Baserga S.J. (2016) Probing the mechanisms underlying human diseases in making ribosomes. Biochem. Soc. Trans. 44, 1035–1044 10.1042/BST20160064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sulima S.O., Kampen K.R. and De Keersmaecker K. (2019) Cancer biogenesis in ribosomopathies. Cells 8, E229 10.3390/cells8030229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aspesi A. and Ellis S.R. (2019) Rare ribosomopathies: insights into mechanisms of cancer. Nat. Rev. Cancer 19, 228–238 10.1038/s41568-019-0105-0 [DOI] [PubMed] [Google Scholar]
- 46.Starczynowski D.T., Kuchenbauer F., Argiropoulos B., Sung S., Morin R., Muranyi A. et al. (2010) Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat. Med. 16, 49–58 10.1038/nm.2054 [DOI] [PubMed] [Google Scholar]
- 47.Alkhatabi H.A., McLornan D.P., Kulasekararaj A.G., Malik F., Seidl T., Darling D. et al. (2016) RPL27A is a target of miR-595 and may contribute to the myelodysplastic phenotype through ribosomal dysgenesis. Oncotarget 7, 47875–47890 10.18632/oncotarget.10293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wan Y., Zhang Q., Zhang Z.J., Song B.F., Wang X.M., Zhang Y.C. et al. (2016) Transcriptome analysis reveals a ribosome constituents disorder involved in the RPL5 downregulated zebrafish model of Diamond–Blackfan anemia. BMC Med. Genomics 9, 13 10.1186/s12920-016-0174-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Connolly M., Paul R., Farre-Garros R., Natanek S.A., Bloch S., Lee J. et al. (2018) miR-424-5p reduces ribosomal RNA and protein synthesis in muscle wasting. J. Cachexia Sarcopenia Muscle 9, 400–416 10.1002/jcsm.12266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ambros V. (2011) MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 21, 511–517 10.1016/j.gde.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J. and Wang D.Z. (2012) microRNAs in cardiovascular development. J. Mol. Cell. Cardiol. 52, 949–957 10.1016/j.yjmcc.2012.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vasudevan S., Tong Y. and Steitz J.A. (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- 53.Vasudevan S., Tong Y. and Steitz J.A. (2008) Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle 7, 1545–1549 10.4161/cc.7.11.6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bukhari S.I.A., Truesdell S.S., Lee S., Kollu S., Classon A., Boukhali M. et al. (2016) A specialized mechanism of translation mediated by FXR1a-associated microRNP in cellular quiescence. Mol. Cell 61, 760–773 10.1016/j.molcel.2016.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orom U.A., Nielsen F.C. and Lund A.H. (2008) MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30, 460–471 10.1016/j.molcel.2008.05.001 [DOI] [PubMed] [Google Scholar]
- 56.Hein N., Sanij E., Quin J., Hannan K.M., Ganley A. and Hannan R.D. (2012) The nucleolus and ribosomal genes in aging and senescence In Senescence (Nagata, T. ed.), pp. 171–208, IntechOpen [Google Scholar]
- 57.Polymenis M. and Kennedy B.K. (2017) Unbalanced growth, senescence and aging. Adv. Exp. Med. Biol. 1002, 189–208 10.1007/978-3-319-57127-0_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H. et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li D., Zhang J., Wang M., Li X., Gong H., Tang H. et al. (2018) Activity dependent LoNA regulates translation by coordinating rRNA transcription and methylation. Nat. Commun. 9, 1726 10.1038/s41467-018-04072-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ugrinova I., Petrova M., Chalabi-Dchar M. and Bouvet P. (2018) Multifaceted nucleolin protein and its molecular partners in oncogenesis. Adv. Protein Chem. Struct. Biol. 111, 133–164 10.1016/bs.apcsb.2017.08.001 [DOI] [PubMed] [Google Scholar]
- 61.Pietrzak M., Rempala G., Nelson P.T., Zheng J.J. and Hetman M. (2011) Epigenetic silencing of nucleolar rRNA genes in Alzheimer's disease. PLoS One 6, e22585 10.1371/journal.pone.0022585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Payao S.L., Smith M.A., Winter L.M. and Bertolucci P.H. (1998) Ribosomal RNA in Alzheimer's disease and aging. Mech. Ageing Dev. 105, 265–272 10.1016/S0047-6374(98)00095-5 [DOI] [PubMed] [Google Scholar]
- 63.Sia P.I., Wood J.P., Chidlow G., Sharma S., Craig J. and Casson R.J. (2016) Role of the nucleolus in neurodegenerative diseases with particular reference to the retina: a review. Clin. Exp. Ophthalmol. 44, 188–195 10.1111/ceo.12661 [DOI] [PubMed] [Google Scholar]
- 64.Bersaglieri C. and Santoro R. (2019) Genome organization in and around the nucleolus. Cells 8, E579 10.3390/cells8060579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nemeth A. and Grummt I. (2018) Dynamic regulation of nucleolar architecture. Curr. Opin. Cell Biol. 52, 105–111 10.1016/j.ceb.2018.02.013 [DOI] [PubMed] [Google Scholar]
- 66.Mayer C., Schmitz K.M., Li J., Grummt I. and Santoro R. (2006) Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell 22, 351–361 10.1016/j.molcel.2006.03.028 [DOI] [PubMed] [Google Scholar]
- 67.Bierhoff H., Schmitz K., Maass F., Ye J. and Grummt I. (2010) Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb. Symp. Quant Biol. 75, 357–364 10.1101/sqb.2010.75.060 [DOI] [PubMed] [Google Scholar]
- 68.Schmitz K.M., Mayer C., Postepska A. and Grummt I. (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24, 2264–2269 10.1101/gad.590910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Savic N., Bar D., Leone S., Frommel S.C., Weber F.A., Vollenweider E. et al. (2014) lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 15, 720–734 10.1016/j.stem.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 70.Postepska-Igielska A., Krunic D., Schmitt N., Greulich-Bode K.M., Boukamp P. and Grummt I. (2013) The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO Rep. 14, 704–710 10.1038/embor.2013.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bierhoff H., Dammert M.A., Brocks D., Dambacher S., Schotta G. and Grummt I. (2014) Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol. Cell 54, 675–682 10.1016/j.molcel.2014.03.032 [DOI] [PubMed] [Google Scholar]
- 72.Zhao Z., Senturk N., Song C. and Grummt I. (2018) lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev. 32, 836–848 10.1101/gad.311688.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao Z., Dammert M.A., Grummt I. and Bierhoff H. (2016) lncRNA-Induced nucleosome repositioning reinforces transcriptional repression of rRNA genes upon hypotonic stress. Cell Rep. 14, 1876–1882 10.1016/j.celrep.2016.01.073 [DOI] [PubMed] [Google Scholar]
- 74.Zhao Z., Dammert M.A., Hoppe S., Bierhoff H. and Grummt I. (2016) Heat shock represses rRNA synthesis by inactivation of TIF-IA and lncRNA-dependent changes in nucleosome positioning. Nucleic Acids Res. 44, 8144–8152 10.1093/nar/gkw496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xing Y.H., Yao R.W., Zhang Y., Guo C.J., Jiang S., Xu G. et al. (2017) SLERT regulates DDX21 rings associated with Pol I transcription. Cell 169, 664–78.e16 10.1016/j.cell.2017.04.011 [DOI] [PubMed] [Google Scholar]
- 76.Calo E., Flynn R.A., Martin L., Spitale R.C., Chang H.Y. and Wysocka J. (2015) RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518, 249–253 10.1038/nature13923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holzel M., Rohrmoser M., Schlee M., Grimm T., Harasim T., Malamoussi A. et al. (2005) Mammalian WDR12 is a novel member of the Pes1–Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 170, 367–378 10.1083/jcb.200501141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W. et al. (2016) Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7, 12429 10.1038/ncomms12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu J., Zhou Q., Niu Y., Chen J., Zhu Y., Ye S. et al. (2019) Aberrant expression of serum circANRIL and hsa_circ_0123996 in children with Kawasaki disease. J. Clin. Lab. Anal. 33, e22874 10.1002/jcla.22874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weinmann R., Raskas H.J. and Roeder R.G. (1974) Role of DNA-dependent RNA polymerases II and III in transcription of the adenovirus genome late in productive infection. Proc. Natl. Acad. Sci. U.S.A. 71, 3426–3439 10.1073/pnas.71.9.3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caudron-Herger M., Pankert T., Seiler J., Németh A., Voit R., Grummt I. et al. (2015) Alu element-containing RNAs maintain nucleolar structure and function. EMBO J. 34, 2758–2774 10.15252/embj.201591458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Batzer M.A. and Deininger P.L. (2002) Alu repeats and human genomic diversity. Nat. Rev. Genet. 3, 370–379 10.1038/nrg798 [DOI] [PubMed] [Google Scholar]
- 83.Jacob M.D., Audas T.E., Uniacke J., Trinkle-Mulcahy L. and Lee S. (2013) Environmental cues induce a long noncoding RNA-dependent remodeling of the nucleolus. Mol. Biol. Cell 24, 2943–2953 10.1091/mbc.e13-04-0223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu S., Wang X. and Shan G. (2016) Insertion of an Alu element in a lncRNA leads to primate-specific modulation of alternative splicing. Nat. Struct. Mol. Biol. 23, 1011–1019 10.1038/nsmb.3302 [DOI] [PubMed] [Google Scholar]
- 85.Lykke-Andersen S., Ardal B.K., Hollensen A.K., Damgaard C.K. and Jensen T.H. (2018) Box C/D snoRNP autoregulation by a cis-Acting snoRNA in the NOP56 Pre-mRNA. Mol. Cell 72, 99–111.e5 10.1016/j.molcel.2018.08.017 [DOI] [PubMed] [Google Scholar]
- 86.Penzo M., Montanaro L., Trere D. and Derenzini M. (2019) The ribosome biogenesis-cancer connection. Cells 8, E55 10.3390/cells8010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Battersby B.J. and Richter U. (2013) Why translation counts for mitochondria - retrograde signalling links mitochondrial protein synthesis to mitochondrial biogenesis and cell proliferation. J. Cell Sci. 126(Pt, 4331–4338 10.1242/jcs.131888 [DOI] [PubMed] [Google Scholar]
- 88.Leucci E., Vendramin R., Spinazzi M., Laurette P., Fiers M., Wouters J. et al. (2016) Melanoma addiction to the long non-coding RNA SAMMSON. Nature 531, 518–522 10.1038/nature17161 [DOI] [PubMed] [Google Scholar]
- 89.Yagi M., Uchiumi T., Takazaki S., Okuno B., Nomura M., Yoshida S. et al. (2012) P32/gC1qR is indispensable for fetal development and mitochondrial translation: importance of its RNA-binding ability. Nucleic Acids Res. 40, 9717–9737 10.1093/nar/gks774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang M. and Pestov D.G. (2011) 5′-end surveillance by Xrn2 acts as a shared mechanism for mammalian pre-rRNA maturation and decay. Nucleic Acids Res. 39, 1811–1822 10.1093/nar/gkq1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sato S., Ishikawa H., Yoshikawa H., Izumikawa K., Simpson R.J. and Takahashi N. (2015) Collaborator of alternative reading frame protein (CARF) regulates early processing of pre-ribosomal RNA by retaining XRN2 (5′–3′ exoribonuclease) in the nucleoplasm. Nucleic Acids Res. 43, 10397–10410 10.1093/nar/gkv1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vendramin R., Verheyden Y., Ishikawa H., Goedert L., Nicolas E., Saraf K. et al. (2018) SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat. Struct. Mol. Biol. 25, 1035–1046 10.1038/s41594-018-0143-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu H., Yin Q.F., Luo Z., Yao R.W., Zheng C.C., Zhang J. et al. (2016) Unusual processing generates SPA lncRNAs that sequester multiple RNA binding proteins. Mol. Cell 64, 534–548 10.1016/j.molcel.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 94.Hartford C.C.R. and Lal A. (2020) When long non-coding becomes protein-coding. Mol. Cell. Biol. 40, e00528-19 10.1128/MCB.00528-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wen X., Gao L., Guo X., Li X., Huang X., Wang Y. et al. (2018) lncSLdb: a resource for long non-coding RNA subcellular localization. Database (Oxford) 2018, 1–6 10.1093/database/bay085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Verheyden Y., Goedert L. and Leucci E. (2018) Control of nucleolar stress and translational reprogramming by lncRNAs. Cell Stress 3, 19–26 10.15698/cst2019.01.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liao M., Liao W., Xu N., Li B., Liu F., Zhang S. et al. (2019) LncRNA EPB41L4A-AS1 regulates glycolysis and glutaminolysis by mediating nucleolar translocation of HDAC2. EBioMedicine 41, 200–213 10.1016/j.ebiom.2019.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Sluis M., Gailín M., McCarter J.G.W., Mangan H., Grob A. and McStay B. (2019) Human NORs, comprising rDNA arrays and functionally conserved distal elements, are located within dynamic chromosomal regions. Genes Dev. 33, 1688–1701 10.1101/gad.331892.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang T., Tan P., Wang L., Jin N., Li Y., Zhang L. et al. (2017) RNALocate: a resource for RNA subcellular localizations. Nucleic Acids Res. 45, D135–D1D8 10.1093/nar/gkx533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mas-Ponte D., Carlevaro-Fita J., Palumbo E., Hermoso Pulido T., Guigo R. and Johnson R. (2017) LncATLAS database for subcellular localization of long noncoding RNAs. RNA. 23, 1080–1087 10.1261/rna.060814.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cui T., Zhang L., Huang Y., Yi Y., Tan P., Zhao Y. et al. (2018) MNDR v2.0: an updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res. 46, D371–D3D4 10.1093/nar/gkx1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Srikantan S., Marasa B.S., Becker K.G., Gorospe M. and Abdelmohsen K. (2011) Paradoxical microRNAs: individual gene repressors, global translation enhancers. Cell Cycle 10, 751–759 10.4161/cc.10.5.14825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Audas T.E., Audas D.E., Jacob M.D., Ho J.J., Khacho M., Wang M. et al. (2016) Adaptation to stressors by systemic protein amyloidogenesis. Dev. Cell 39, 155–168 10.1016/j.devcel.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Audas T.E., Jacob M.D. and Lee S. (2012) Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell 45, 147–157 10.1016/j.molcel.2011.12.012 [DOI] [PubMed] [Google Scholar]
- 105.Wang M., Tao X., Jacob M.D., Bennett C.A., Ho J.J.D., Gonzalgo M.L. et al. (2018) Stress-induced low complexity RNA activates physiological amyloidogenesis. Cell Rep. 24, 1713-1721.e4 10.1016/j.celrep.2018.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E. et al. (2008) Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14, 723–730 10.1038/nm1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Miao H., Wang L., Zhan H., Dai J., Chang Y., Wu F. et al. (2019) A long noncoding RNA distributed in both nucleus and cytoplasm operates in the PYCARD-regulated apoptosis by coordinating the epigenetic and translational regulation. PLoS Genet. 15, e1008144 10.1371/journal.pgen.1008144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gumireddy K., Li A., Yan J., Setoyama T., Johannes G.J., Orom U.A. et al. (2013) Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 32, 2672–2684 10.1038/emboj.2013.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S. et al. (2012) Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457 10.1038/nature11508 [DOI] [PubMed] [Google Scholar]
- 110.Beltran M., Puig I., Peña C., García J.M., Alvarez A.B., Peña R. et al. (2008) A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 22, 756–769 10.1101/gad.455708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hansji H., Leung E.Y., Baguley B.C., Finlay G.J., Cameron-Smith D., Figueiredo V.C. et al. (2016) ZFAS1: a long noncoding RNA associated with ribosomes in breast cancer cells. Biol. Direct. 11, 62 10.1186/s13062-016-0165-y [DOI] [PMC free article] [PubMed] [Google Scholar]