Abstract
Eukaryotic life depends upon the interplay between vast networks of signaling pathways composed of upwards of 109–1010 proteins per cell. The integrity and normal operation of the cell requires that these proteins act in a precise spatial and temporal manner. The ubiquitin system is absolutely central to this process and perturbation of its function contributes directly to the onset and progression of a wide variety of diseases, including cancer, metabolic syndromes, neurodegenerative diseases, autoimmunity, inflammatory disorders, infectious diseases, and muscle dystrophies. Whilst the individual components and the overall architecture of the ubiquitin system have been delineated in some detail, how ubiquitination might be successfully targeted, or harnessed, to develop novel therapeutic approaches to the treatment of disease, currently remains relatively poorly understood. In this review, we will provide an overview of the current status of selected small molecule ubiquitin system inhibitors. We will further discuss the unique challenges of targeting this ubiquitous and highly complex machinery, and explore and highlight potential ways in which these challenges might be met.
Keywords: chemical probe, drug development, inhibitor, post-translational modification, PROTAC, ubiquitin
The ubiquitin system: a brief introduction
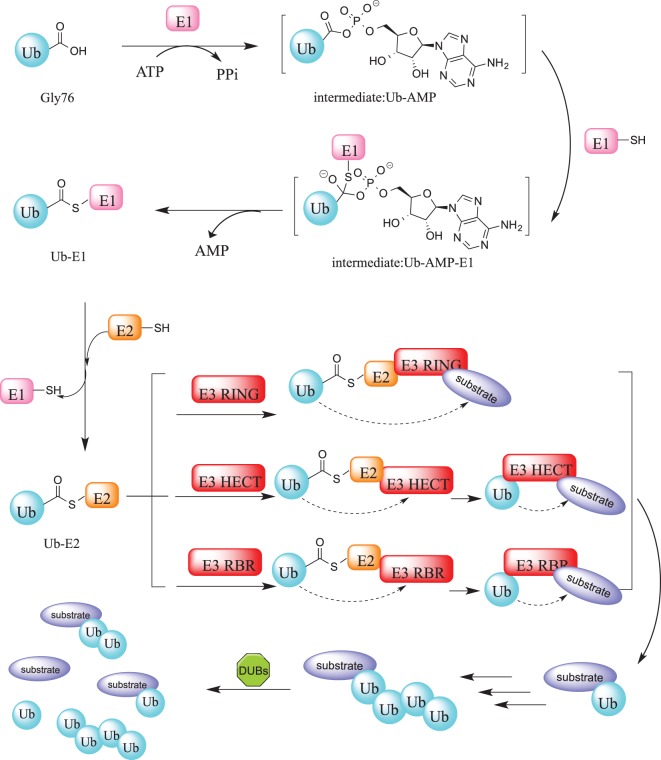
Ubiquitination is indispensable for the survival of all known eukaryotic cells and is vital for most if not all cellular processes [1,2]. The system is composed of a cascade of enzymes that reversibly catalyze the covalent attachment of ubiquitin, an 8.5 KDa protein module, at a defined position on specific protein substrates. Although it is best known for triggering protein degradation via the ubiquitin–proteasome system (UPS) [3], it also plays fundamental roles in a broad range of phenomena such as the DNA damage response, protein localization, intracellular signaling, autophagy, and endocytosis [4–9]. Ubiquitination is carried out sequentially by three specialized enzyme classes: E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligating enzyme). Deubiquitination is mediated by deubiquitinating enzymes (DUBs) that cleave ubiquitin from the substrate protein or trim ubiquitin moieties from a polyubiquitin chain (Figure 1) [10]. The balance between ubiquitination and deubiquitination is tightly coupled and is a critical determinant of protein levels and activity.
Figure 1. The ubiquitination and deubiquitination cascade.
Protein ubiquitination is initiated by the E1 enzyme that catalyzes the adenylation of the C-terminal glycine residue of Ub, forming Ub-AMP. This phosphoester bond subsequently undergoes a nucleophilic substitution involving the sulfhydryl group of the E1 active site cysteine residue. Following this, Ub is transferred to an E2 enzyme active site cysteine residue through a trans thioesterification reaction that yields an activated thioester-linked E2-Ub complex. Finally, an E3 ligase catalyzes the transfer of Ub from the E2 enzyme to a target substrates creating an isopeptide bond between the C-terminal glycine of ubiquitin and the substrate lysine. Depending on the particular class of E3 ligase, this reaction is stepwise (HECT or RBR E3s) or direct without the formation of intermediates (RING E3s). The ubiquitination process is reversed by DUBs through cleavage of ubiquitin from the substrate.
A definitive link between aberrant ubiquitin signaling and disease
Several lines of evidence support the notion that dysregulation of ubiquitination is closely associated with multiple human diseases, including numerous cancer types [11–13], cardiovascular disease [14], viral diseases [15–18], neurodegenerative disorders [19], and congestive heart failure [20]. Disruption of normal (de-)ubiquitination can result via many distinct, though not mutually exclusive, mechanisms. Firstly, components of the ubiquitin machinery can directly suffer missense mutations or small deletions, exemplified by PARKIN E3 ligase mutations, which cause a familial form of Parkinson's disease [21]; BRCA1-associated protein 1 (BAP1, a UCH family member) mutations that are found in many cancer types including melanoma, mesothelioma and renal cell carcinoma [22]; mutations in USP7, USP8, USP9X that are associated with neurological disorders [23], Cushing disease [24,25], and developmental disorders [26], respectively; mutations of STAMBP, which encodes a JAMM family DUB, can lead to microcephaly–capillary malformation syndrome [27]. Secondly, perturbation of the ubiquitin machinery can also be caused by gene rearrangements, illustrated by the TRE17/USP6 translocation that is causally linked to aneurysmal bone cysts [28]. Thirdly, the expression of DUBs and E3 ligases have been found to be up-regulated or lost in a wide variety of tumors: gene amplification of the RING-type E3s MDM2 or MDMX, exerts oncogenic effects in primary colorectal cancers [29] and sarcomagenesis [30]; ubiquitin-dependent degradation of AMP-activated protein kinase (AMPK), which controls cellular energy status, is mediated by the cancer-specific MAGE-A3/6-TRIM28 ubiquitin ligase [31]; overexpression of USP28 stabilizes the MYC proto-oncogene in colon and breast carcinomas [32].
The vast majority of cellular proteins are subject to ubiquitination and the net effect of corrupting ubiquitin signaling is that the levels and the activity of target proteins are either augmented (gain of function) or suppressed (loss of function). It is not surprising that these changes can play a major role in disease. In this light, the ubiquitin system potentially offers an inroad to the development of novel targeted treatments. In the case of cancer, for example, the past decade has witnessed a shift away from traditional chemotherapies towards precision treatments that specifically target tumor cells and thus result in more limited cytotoxicity in healthy tissues. This approach has largely focused on the design of kinase inhibitors and whilst there have been improvements in patient outcomes, the promised revolution has thus far failed to completely materialize. The two principal reasons for this are the acquisition of resistance and the fact that this class of inhibitors is limited in range since they were designed only for so-called ‘druggable’ targets. In the ensuing sections, we will discuss how either inhibiting parts of the ubiquitin system or, by contrast, actually harnessing the destructive power of the ubiquitin machinery, could each be a powerful solution to circumvent these problems and potentially hasten a new dawn for targeted therapies.
Therapeutic targeting of the ubiquitin machinery
A first question to consider is, what represents an optimal target? The overall architecture of the ubiquitin apparatus can be conveniently viewed as being composed of the following: the proteasome, E1, E2, and E3 enzymes that catalyze ubiquitination, the DUBS that mediate deubiquitination, and the ubiquitin molecule itself. Related to this, are two further questions, specifically, what is the optimal means for achieving inhibition, e.g. small molecules, peptides, antibodies, RNA-based reagents? And, what particular biochemical phenomenon is targeted: inhibition of activity by direct binding to and disruption of a specific regulatory domain/function; blocking regulatory protein–protein interactions? The therapeutic potential of this pathway is now being exploited and many UPS modulators have now entered the clinic (Table 1). There are many peptide-based inhibitors of the proteasome, such as Carfilzomib (see Table 1). Recently, microRNAs have been shown to modulate the UPS. By example, miR-137 has been reported to target Mib1 (an ubiquitin ligase), which controls neuronal maturation [53]. Both the messenger RNA and protein levels of USP25 have been shown to be regulated by direct binding of miR-200c to the 3′-untranslated region of USP25 [54]. This raises the possibility that RNA-based regulation of the UPS could serve as a potential future therapeutic method. However, to date, the majority of UPS inhibitors are cell permeable small molecules (see Table 1).
Table 1. Proteasome, E1, E3, or DUB modulators in clinic use or in (pre)-clinical development.
| Target | Compound ID | Structure | Highest clinical stage and status1 | Reference |
|---|---|---|---|---|
| Proteasome inhibitors | ||||
| proteasome | Bortezomib | 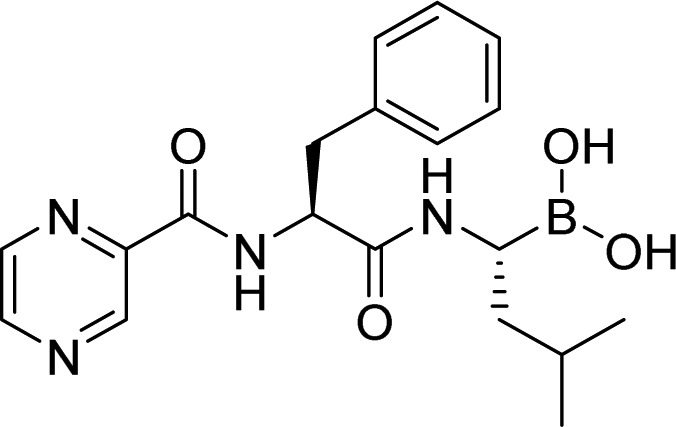 |
FDA approved | Teicher et al. [33] |
| Carfilzomib | 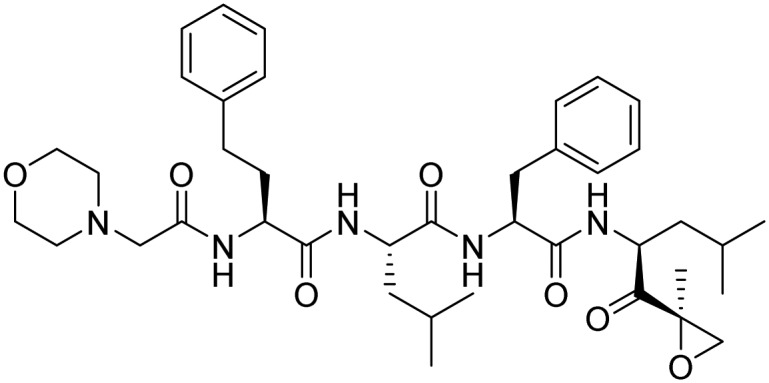 |
FDA approved | Kim and Crews [34] | |
| Ixazomib | 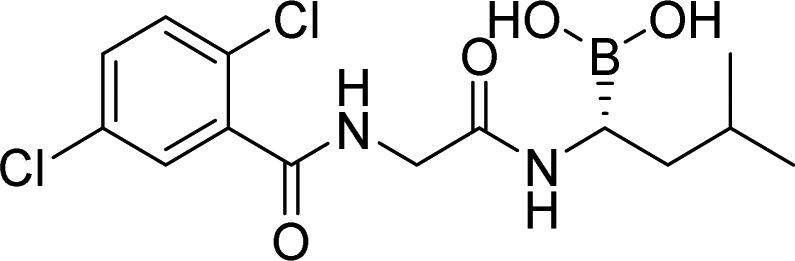 |
FDA approved | Kupperman et al. [35] | |
| Oprozomib | 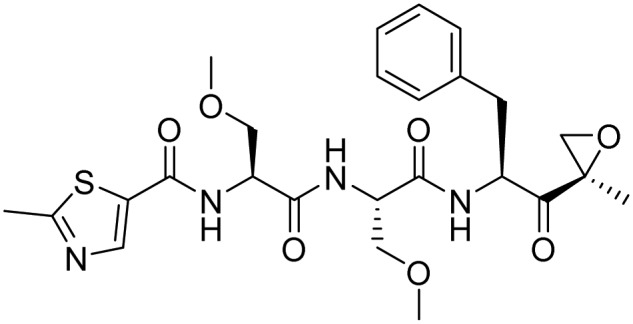 |
Phase 1/Phase 2, completed | Chauhan et al. [36] | |
| Delanzomib | 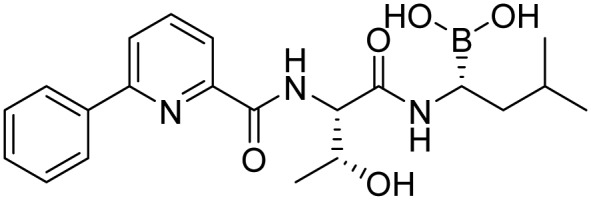 |
Phase 1/Phase 2, terminated | Piva et al. [37] | |
| Marizomib | 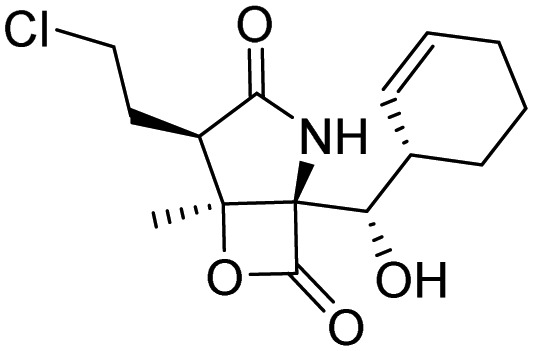 |
Phase 3, recruiting | Potts et al. [38] | |
| E1 modulator | ||||
| UAE (Uba1) | TAK-243 | 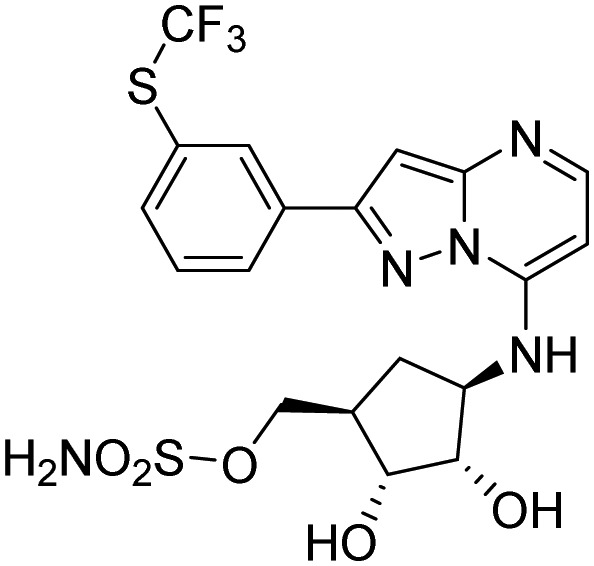 |
Phase 1, completed | Milhollen et al. [39] Hyer et al. [40] |
| E3 modulators | ||||
| CRBN | Thalidomide | 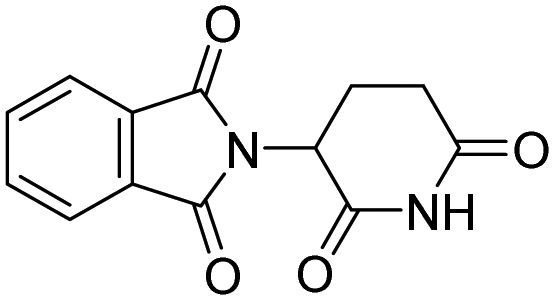 |
FDA approved | Ito et al. [41] |
| Lenalidomide |  |
FDA approved | Krönke, et al. [42, 43] | |
| Pomalidomide | 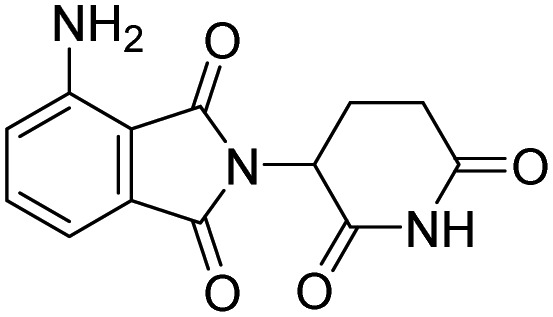 |
FDA approved | Fischer et al. [44] | |
| Avadomide | 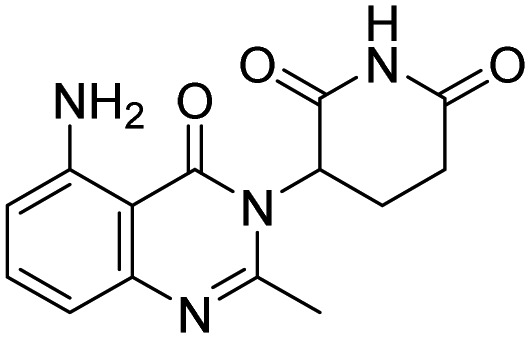 |
Phase 1/Phase 2, active | Rasco et al. [45] | |
| Iberdomide | 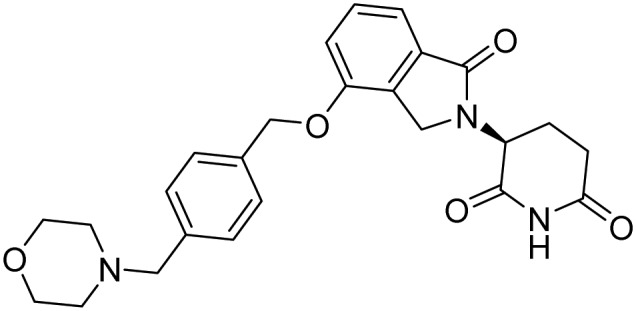 |
Phase 1/Phase 2, recruiting | Bjorklund et al. [46] | |
| MDM2 | APG-115 | 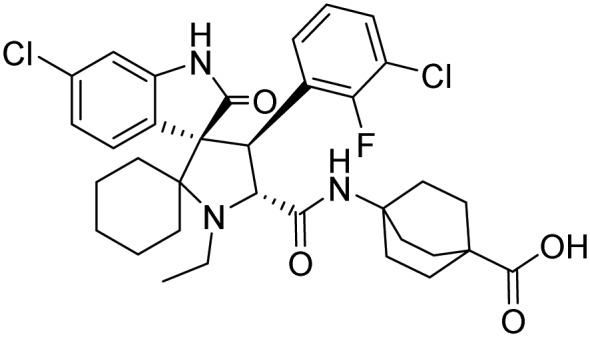 |
Phase 1/Phase 2, recruiting | Rasco et al. [47] |
| CGM097 | 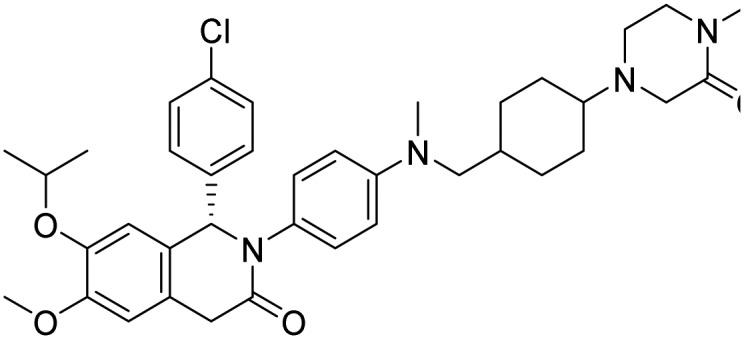 |
Phase 1, active | Holzer et al. [48] | |
| Cul4-DCAF15 | Tasisulam | 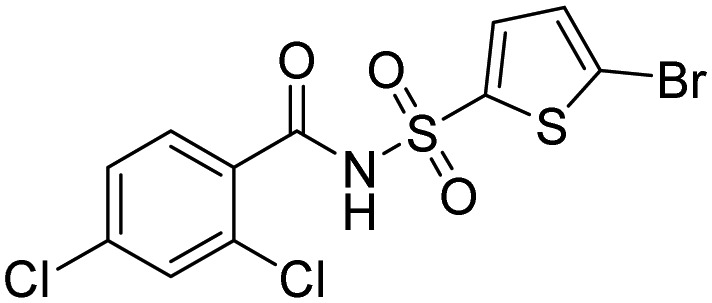 |
Phase 3, terminated | Han et al. [49] |
| Indisulam | 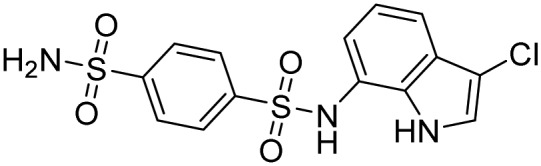 |
Phase 2, completed | ||
| NSC-339004 | 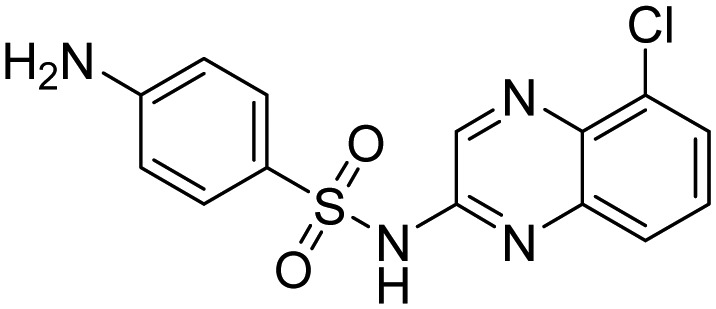 |
Phase 2, Completed | ||
| Keap1 | Bardoxolone methyl | 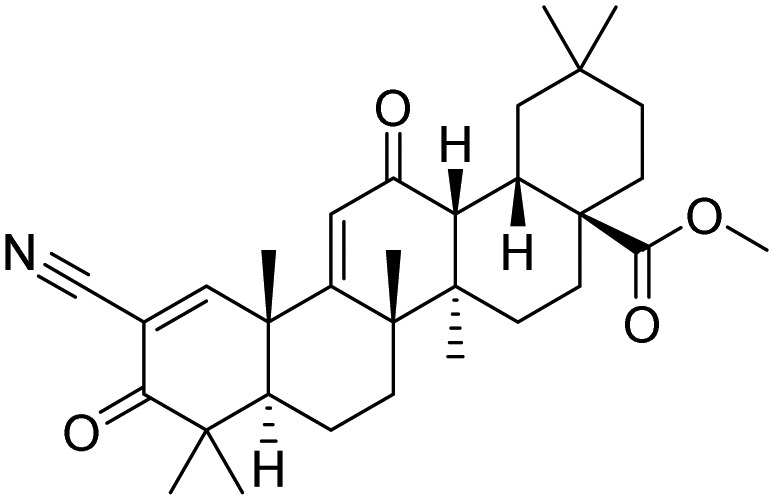 |
Phase 3, recruiting | Gross et al. [50] |
| Omaveloxolone | 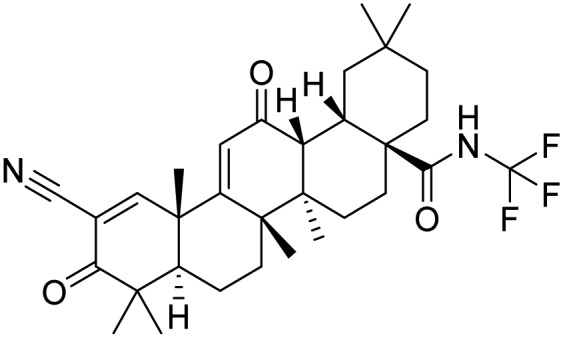 |
Phase 2, completed | Lynch et al. [51] | |
| Deubiquitinase inhibitors | ||||
| USP14 > UCH37; many other DUBs inhibited | VLX1570 | 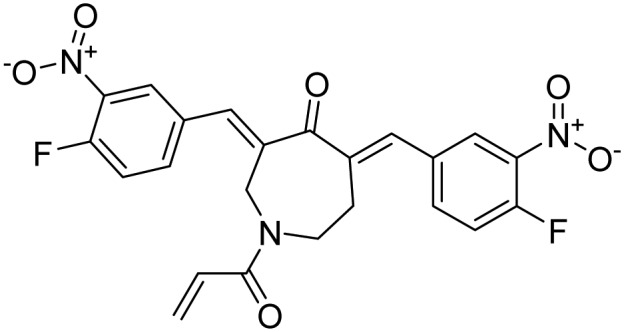 |
Phase 1/Phase 2, prematurely ended | Paulus et al. [52] |
Data from ClinicalTrials.gov and EU Clinical Trials Register.
Whilst inhibitors of the proteasome and of the E1 enzyme have shown efficacy, an obvious drawback of such compounds is that they influence generically a large number of proteins/cellular networks which renders them toxic. To address this problem, in recent years efforts have focussed on identifying inhibitors of specific E3 ligases or DUBs. Here, we would like to update the very recent development of selected small molecule inhibitors against components of the ubiquitin system. A comprehensive description of earlier compounds has been provided elsewhere [1,55–58].
Targeting E3 ligases
The human proteome encodes more than 600 E3 ligases that fall into three main groups: the really interesting new gene (RING) class comprising ∼600 members, the homologous to E6AP C terminus (HECT) class which includes at least 28 members and the RING between RING (RBR) class encompassing 14 members [59–61]. A detailed ubiquitin taxonomy, specifically, a precise mapping of the enzyme to substrate, is currently lacking, however, the sheer number of ligases points strongly a high degree of specificity. This suggests that selective blocking of E3 ligase function would not result in the levels of toxicity associated with inhibition of the proteasome, ubiquitin or the broader spectrum E1- or E2-enzymes. Below we discuss recent advances in the development of such inhibitors.
RING-type E3 ligase inhibitors
TRAF6 is an E3 ligase known for its critical role in many immune signaling pathways. The molecule C25-140 (1, Table 2 was identified in a high-throughput screen designed to identify compounds that disrupt the TRAF6–Ubc13 protein–protein interaction [62]. Although C25-140 was also shown to inhibit cIAP1 [62], the relatively greater selectivity of this compound was revealed by its inability to block multiple other E3 ligases including RING-type (e.g. cIAP1 and MDM2) and HECT-type (e.g. ITCH and E6AP) enzymes. Indeed, it has been argued that the identified off-target activity of C25-140 could prove to be beneficial for the treatment of autoimmune and inflammatory diseases since illicit TNFα signaling has been shown to underlie the development of these diseases [62]. RNF4 is an E3 ubiquitin ligase that promotes ubiquitination and subsequent proteasomal degradation of SUMOylated proteins [69,70]. TRH 1-23 (2, Table 2) is a promising RNF4 inhibitor that was recently identified in an activity-based protein profiling (ABPP) screen. An analog of TRH 1-23, named CCW-16 (3), was found to be a more a potent RNF4 inhibitor, exhibiting an IC50 of 1.8 μM [63].
Table 2. Recently reported structures of small molecule inhibitors of E3 ligases.
| Target | Compound ID | Structure | Reference |
|---|---|---|---|
| TRAF6 | 1, C25-140 | 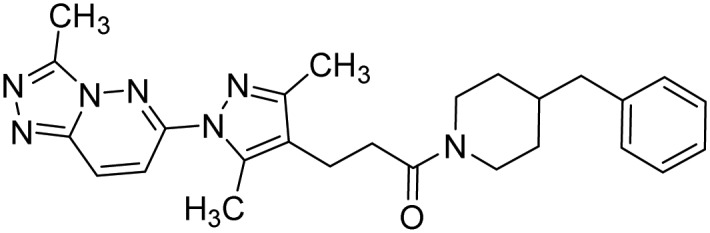 |
Brenke et al. [62] |
| RNF4 | 2, TRH 1-23 | 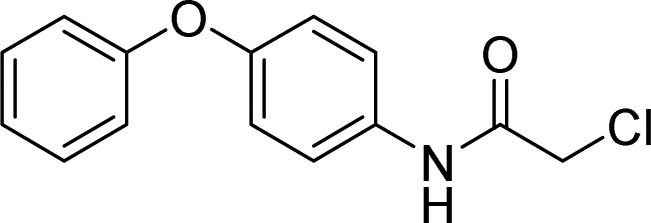 |
Ward et al. [63] |
| 3, CCW16 | 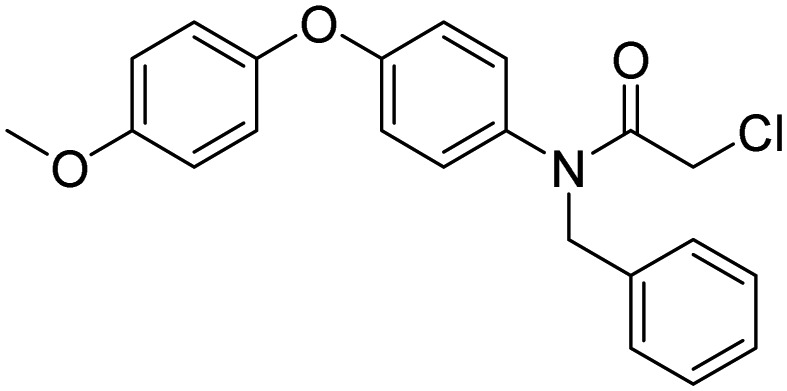 |
||
| VHL |
4a: X = F, Z = H 4b: X = H, Z = F |
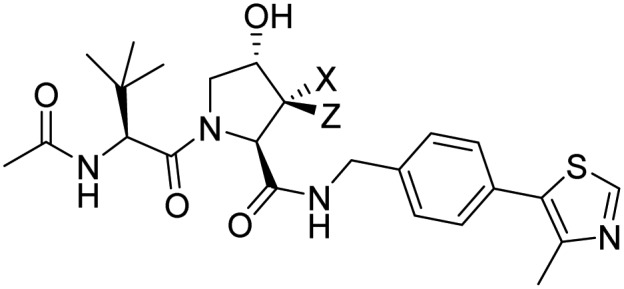 |
Testa et al. [64] |
| WWP2 | 5 | 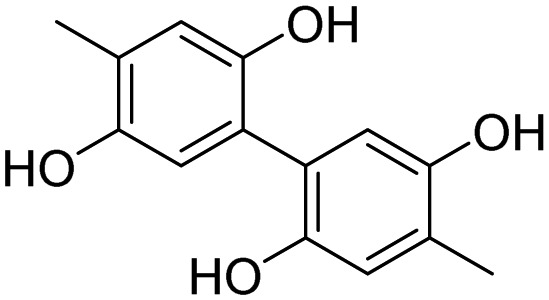 |
Watt et al. [65] |
| 6 | 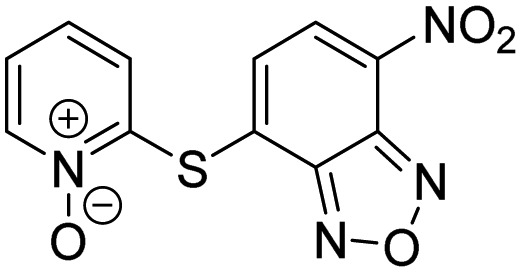 |
||
| 7 | 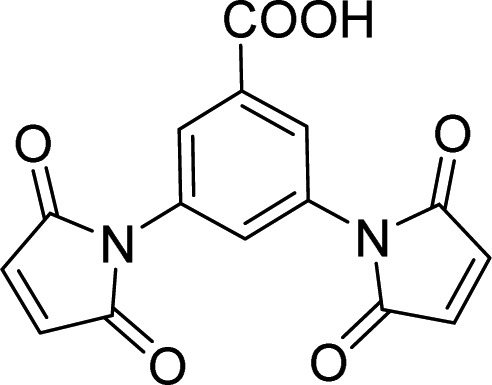 |
||
| 8 | 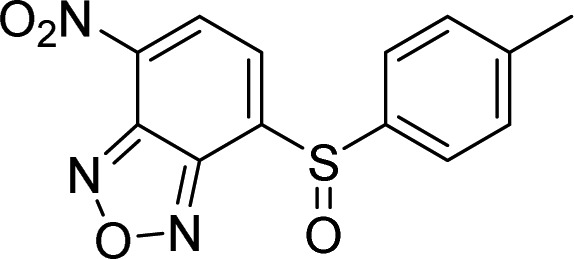 |
||
| 9 | 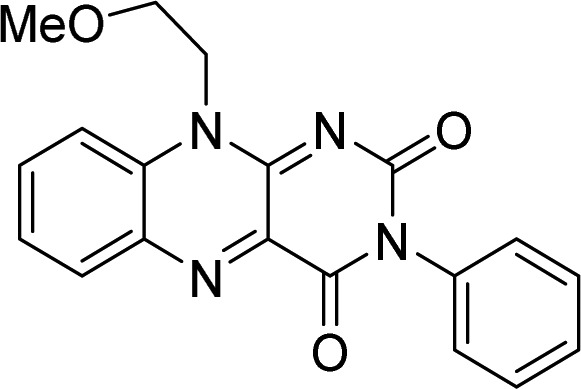 |
||
| SMURF1 | 10, HS-152 | 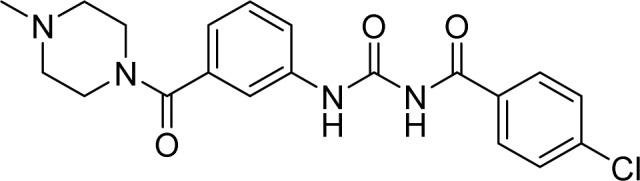 |
Tian et al. [66] |
| HOIP | 11, Bendamustine | 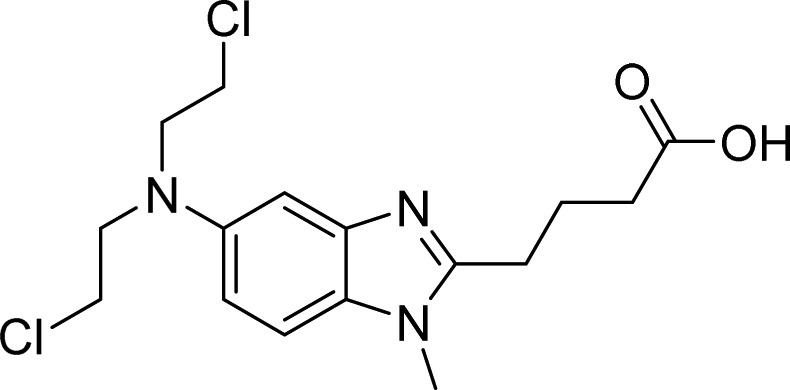 |
De Cesare et al. [67] |
| 12 | 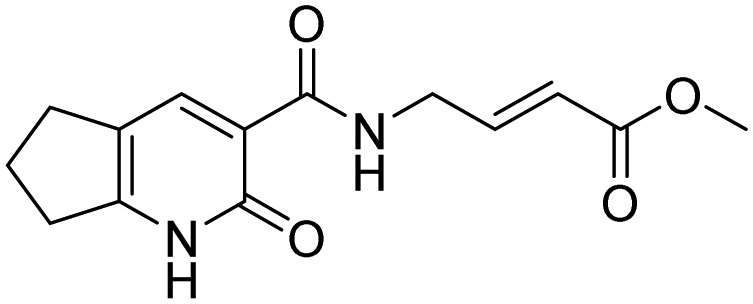 |
Johansson et al. [68] | |
| 13 | 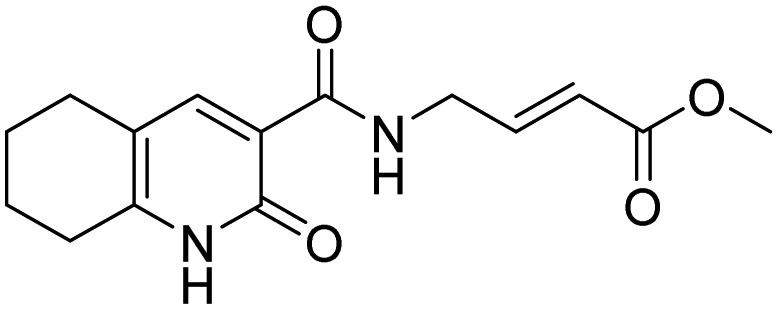 |
||
| 14 | 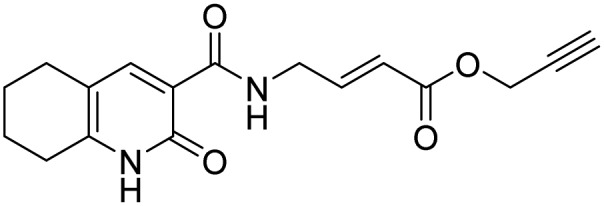 |
Von Hippel–Lindau (VHL) E3 Ubiquitin Ligase is one of the few E3 ligases, including cereblon, VHL, MDM2, and cIAP, that have been successfully exploited in targeted protein degradation strategies [71,72]. Recognition of hydroxyproline (Hyp) by VHL is highly stereoselective and conformation-dependent. Fluoro-hydroxyproline (F-Hyp) containing compounds (4a, 4b) have been reported to be VHL Ligands with higher binding affinity and improved metabolica stability compared with the parent Hyp-containing ligand. The ligand 4a has been employed successfully in novel VHL-based PROTAC reagents [64] (see section targeted protein degradation).
HECT-type E3 ligase inhibitors
High-throughput screening of small molecule libraries [65] has yielded many compounds (see Table 2, structures 5–9) that show promise as potentially therapeutically relevant WWP2 E3 ligase inhibitors (IC50 values are in the low µM range). The sites of interaction between inhibitor and target have been determined by NMR, which may facilitate the process of refining these WWP2 inhibitors. A potent SMURF1 small molecule inhibitor named HS-152 (10) was discovered using a cell-based high-throughput screening campaign [66]. This compound blocks the catalytic activity of SMURF HECT domains thereby abrogating SMURF1-mediated SMAD1 degradation as well as SMURF1-mediated RHOA/RHOB degradation.
RBR-type E3 ligase inhibitors
A label-free MALDI-TOF mass spectrometry approach was used to test 1430 FDA approved drugs for their ability to specifically inhibit E3 ligases representative of the three different enzyme classes. By this means, bendamustine (11, Table 2), a chemotherapeutic reagent commonly used in the treatment of chronic lymphocytic leukemia and lymphomas, was found to inhibit HOIP with an IC50 of 6.4 µM, while it did not significantly affect the activity of either MDM2 or ITCH [67]. A fragment-based covalent ligand screen, with the aim of identifying inhibitors that covalently target the active site cysteine residue of HOIP, has highlighted three selective small molecules (see Table 2, compounds 12–14) as promising HOIP inhibitors, which displayed limited off-target activity against other RBR or HECT E3 ligases [68].
Targeted protein degradation
Whilst selective inhibition of the activity of specific E3 ligases could prove to be of significant therapeutic value, a potentially revolutionary innovation currently gaining widespread attention takes an entirely different approach. Rather than inhibiting E3 ligases, targeted protein degradation, including proteolysis targeting chimeras (PROTACs) [73], immunomodulatory drugs (IMiDs) [74], and specific and nongenetic IAP-dependent protein erasers (SNIPER) [75,76], harnesses the destructive power of the host cell ubiquitin machinery to eliminate unwanted disease-causing proteins in a targeted manner. Since a detailed account, this technology is beyond the scope of this review, we refer readers to many recent reviews that comprehensively summarize recent progress in this field [77–84].
Targeting deubiquitination
The human proteome contains ∼100 DUBs belonging to two distinct enzyme classes: the cysteine proteases and the metalloproteases. The cysteine protease class comprises six different enzyme families: ubiquitin-specific proteases (USPs), ubiquitin carboxyl-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Machado-Joseph (Josephin) domain-containing proteases (MJDs), motif interacting with Ub-containing novel DUB family (MINDY) [85], and zinc finger containing ubiquitin peptidase 1 (ZUFSP/ZUP1) [86–89]. The metalloprotease enzyme class consists of a single family of enzymes: JAB1/MPN/MOV34 (JAMMs). DUBs strip protein substrates of ubiquitin thus rescuing them from degradation via the proteasome. Accordingly, inhibiting DUBs, in common with the PROTAC technology, can selectively eliminate targets by rendering them susceptible to proteasomal degradation. Numerous small molecule DUB inhibitors have been reported, however, until recently, only few of them showed selectivity [90].
USP7
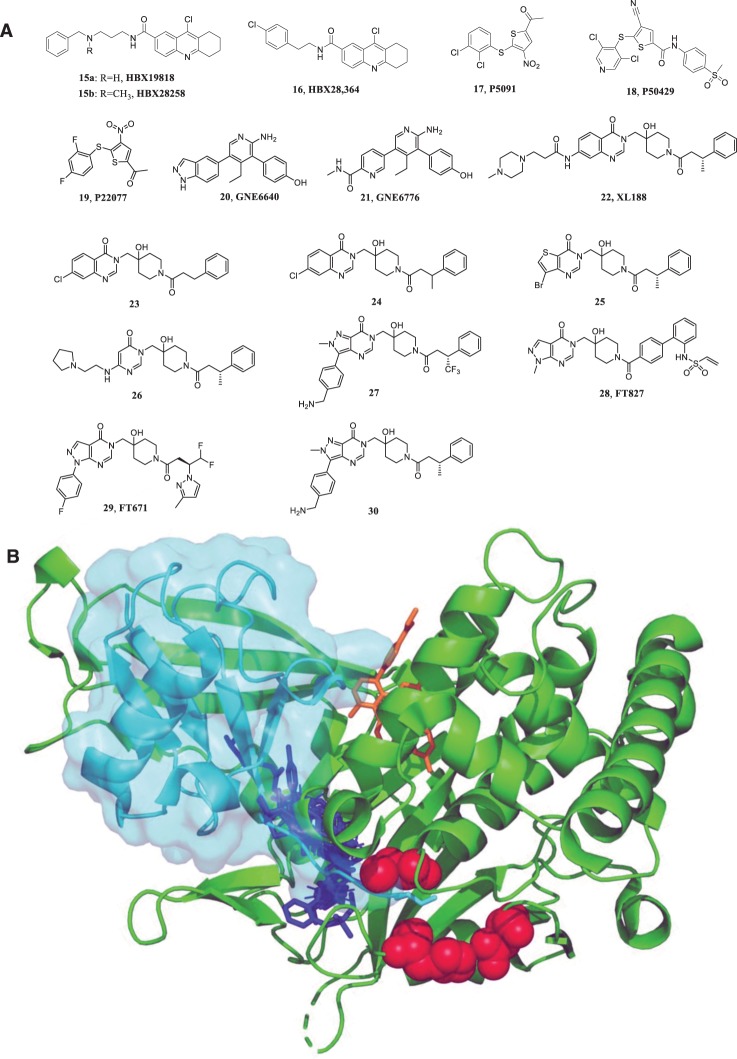
The ground-breaking work on ubiquitin-specific protease (USP) 7 inhibitors represents a paradigm for the discovery and characterization of highly selective small molecule inhibitors of DUBs. USP7 or HAUSP (herpesvirus associated ubiquitin-specific protease) is best known for its role as a regulator of the tumor suppressor p53 [91,92] although it has many other reported targets. USP7 deubiquitinates murine double minute-2 (MDM2), an E3 ligase of p53, resulting in p53 degradation. Consequently, it would be expected that a USP7 inhibitor would promote stabilization of p53 leading to suppression of cell growth and/or apoptosis that may be beneficial for the treatment of certain cancers. The 9-chloro derivatives of amidotetrahydroacridine, HBX19818 (15a) and HBX28259 (15b) (see Figure 2), have been reported to be irreversible inhibitors of USP7 that function via nucleophilic attack of catalytic cysteine residues mediated by the reactive chloride moiety and resulting in covalent linkage to the protein [93]. HBX 19818 was found to specifically inhibit USP7 but not a panel of different DUBs under physiological conditions. This compound was shown to inhibit HCT116 proliferation and to induce apoptosis in a dose-dependent manner. Interestingly, HBX 28364 (16), which lacks the basic alkyl amine side chain of HBX 19818, does not show any inhibitory activity. Crystallographic analysis of USP7-inhibitor complexes has enabled structure-guided optimization of potent and highly specific USP7 inhibitors, and mainly three binding regions have been unmasked to date (red, blue, and orange in Figure 2B) [94]. P5091 (17, Figure 2) was initially discovered as a dual USP7/USP47 inhibitor [95], which causes apoptosis of multiple myeloma cells and prolonged survival of transplanted mice (using xenograft tumor models) [96]. Further optimization of P5091 led to the discovery of P50429 (18) and P22077 (19) that inhibit the proliferation of HCT116 cells [95] and neuroblastoma growth [97], respectively. A combination of solution NMR and mass spectrometry studies, revealed that P22077 and P50429 bind irreversibly to the catalytic cysteine C223 of USP7 (red in figure 2B). This interaction induced a conformational switch in the enzyme associated with an active site rearrangement [98]. Medicinal chemistry optimization of a fragment yielded the small molecules GNE6640 (20) and GNE6776 (21) shown in Figure 2. This class of allosteric inhibitors has been demonstrated to bind to the ‘palm’ region of the USP7 catalytic domain thereby impeding ubiquitin binding (orange, Figure 2B) [99,100]. NMR-based saturation transfer difference (STD) experiments showed that these compounds bind to a novel functional site 12 Å away from the catalytic cysteine residue [100]. These molecules potently inhibited the activity of USP7 (IC50 values of 1.34 μM and 0.75 μM, respectively) and did not inhibit the activity of a panel of 36 DUBs at concentrations of 100 μM, which is convincing evidence of high target selectivity [99]. Cocrystal structures have further shown that GNE6640 and GNE6776 interact with acidic residues that commonly promote hydrogen-bond interactions with Lys48 of ubiquitin. A group of pyrimidinone analogs (22–30) [101–104], reported by several laboratories independently, have been shown to bind to the same catalytic region that is normally occupied by the C-terminal tail of ubiquitin (blue structure, Figure 2B). One of these compounds (30, Figure 2) strongly inhibited the proliferation of several cancer cell lines with equal or greater efficacy compared with known MDM2 antagonists (IC50 = 6 nM). All of these highly selective inhibitors bind non-covalently to USP7, with the exception of FT827, which harbors a vinylsulfonamide moiety that extends to the catalytic triad and covalently binds to the active cysteine [103].
Figure 2. Small molecule inhibitors of USP7 and their representative binding domains.
(A) Structures of small molecule inhibitors targeting USP7. (B) The binding characteristics of current inhibitors of USP7. The catalytic triad of the catalytic domain is shown as red spheres. Compounds P50429 and P22077 both bind to this region. The allosteric inhibitors (GNE6640 and GNE6776, shown in orange) occupy a functional site that is 12 Å away from the catalytic cysteine. The pyrimidinone analogs (shown in blue) bind to the catalytic region that is normally occupied by the C terminus of ubiquitin (shown in cyan).
Other DUBs
Ubiquitin-specific protease 14 (USP14), a proteasome associated DUB, has been reported to be involved in tumorigenesis in many cancer types. It also plays an important role in neurodegenerative disorders. IU1 (31, see Table 3) was first reported in 2010 as a selective small molecule inhibitor of USP14, which could enhance the degradation of several proteasome substrates such as tau and TDP-43 [105]. Chemical modification of IU1 resulted in IU1-47 (32), which is 10-fold more potent than IU1 but retained specificity for USP14. Following IU1-47 treatment, both endogenous wild-type tau and pathological mutantforms of tau (in murine primary neurons and in Human iPSC-derived neurons) could be significantly degraded, and this effect was dependent on Lys174 in tau [106]. An analog of IU1 termed 1B10 (33) was found recently to be more potent than IU1, and also had a better membrane permeability [107]. UCHL1, belonging to the Ubiquitin C-terminal Hydrolase subfamily, is considered to be a promising therapeutic target in neurodegenerative diseases, cancer, and liver/lung fibrosis. Very recently, several cyanopyrrolidine-based UCHL1 inhibitors, that covalently bind to the active cysteine, have been reported. Compound 34, for example, was first reported as a UCHL1 inhibitor in a patent application by Mission Therapeutics [108]. But it was found to inhibit both UCHL1 and UCHL3 with IC50 values of 0.67 ± 1.0 μM and 6.4 ± 1.1 μM, respectively [109]. A more selective and potent UCHL1 inhibitor (35a) [118], as well as a structurally related activity-based probe (35b) [110], were reported to label UCHL1 in living cells at low micromolar concentrations, and block pro-fibrotic responses in IPF cellular models without substantial associated cytotoxicity. The cell permeable small molecules 6RK73 (36) and 8RK64 (37) were found to target UCHL1 but no other DUBs [111]. However, they did inhibit the non-DUB protein Park7 as an off-target. After chemically decorating fluorophores, such as BodipyFL-alkyne, BodipyTMR-alkyne, and Rhodamine110-alkyne, the first examples of a UCHL1-selective probe were generated and the Bodipy-labeled reagent was shown to react with UCHL1 in zebrafish embryos. OTUB2 is an ovarian tumor domain (OTU) family member that preferentially cleaves Lys63-linked polyubiquitination chains and to a lesser degree, Lys11-linked and Lys48-linked chains. Recently, the first highly selective OTUB2 covalent inhibitor named OTUB2-COV-1 (38, Table 3) was identified by combining a new high-throughput thiol-reactivity assay with high-throughput crystallography [112].
Table 3. Selected structures of currently reported small molecule inhibitors targeting DUBs.
| Target | Compound ID | Structure | Reference |
|---|---|---|---|
| USP14 | 31, IU1 | 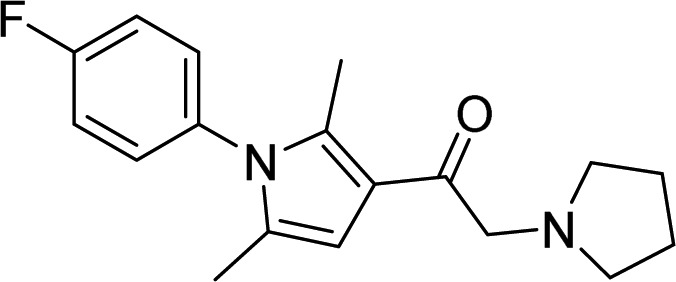 |
Lee et al. [105] |
| 32, IU1-47 | 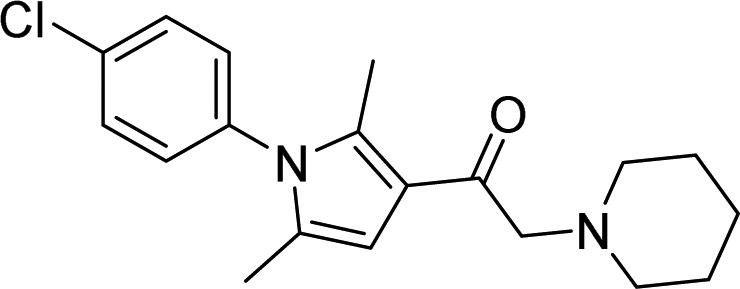 |
Boselli et al. [106] | |
| 33, 1B10 | 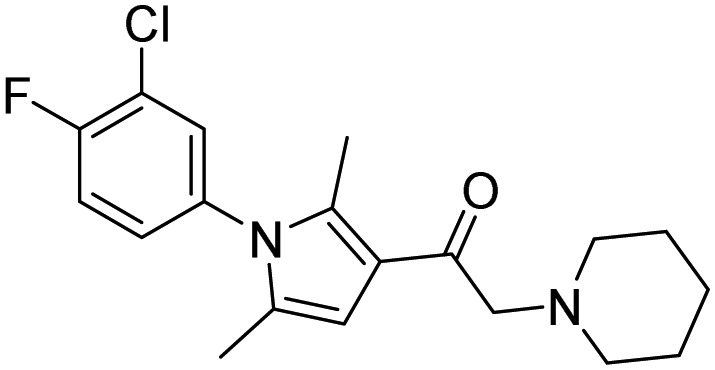 |
Palmer et al. [107] | |
| UCHL1 | 34 | 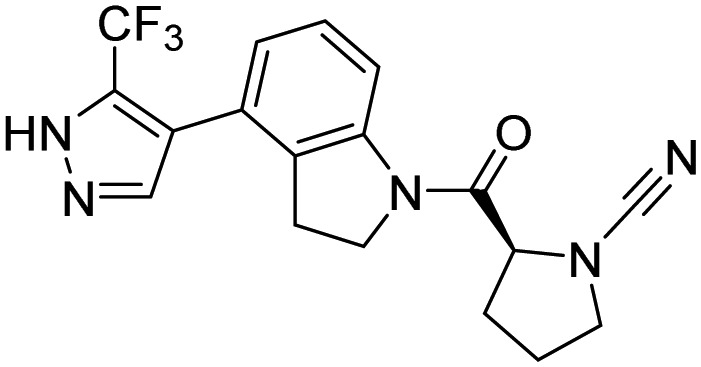 |
Krabill et al. [108] Kemp et al. [109] |
|
35a:R = H 35b:R = CCH |
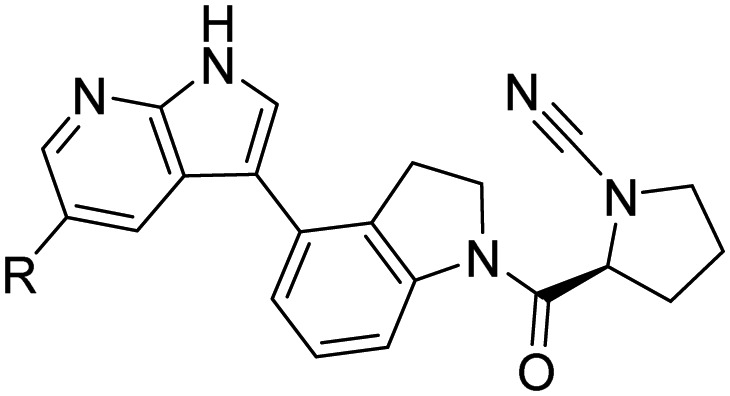 |
Nattawadee et al. [110] | |
| 36, 6RK73 | 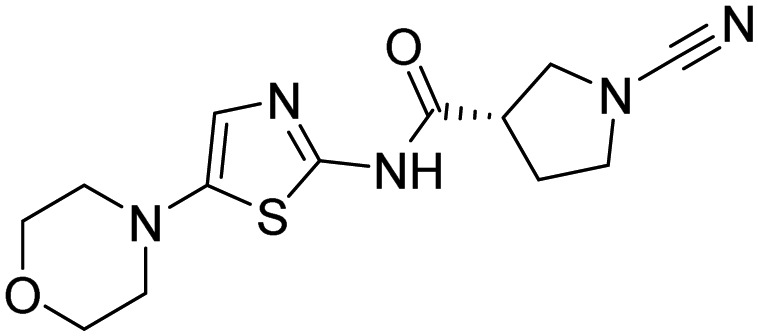 |
Geurink et al. [111] | |
| 37, 8RK64 | 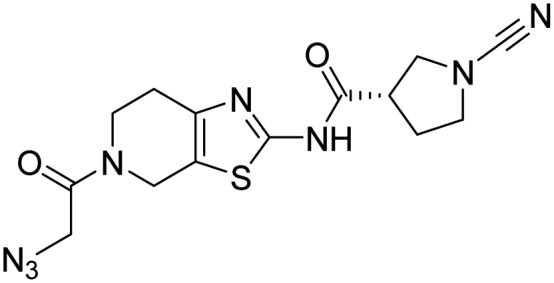 |
||
| OTUB2 | 38, OTUB2-COV-1 | 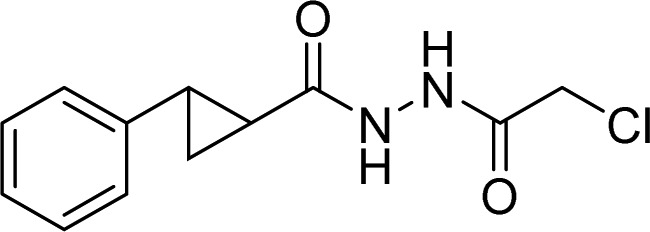 |
Resnick et al. [112] |
| Rpn11 | 39, 8TQ | 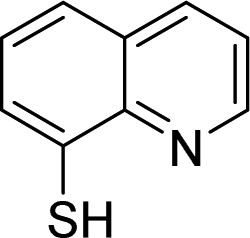 |
Perez et al. [113] |
| 40, Capzimin | 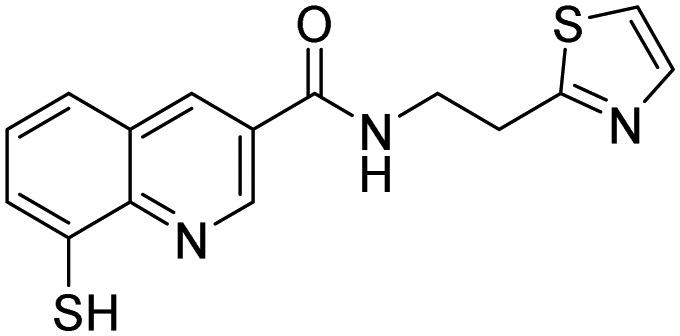 |
Li et al. [114] | |
| Rpn11, also Csn5 and AMSH | 41, SOP11 | 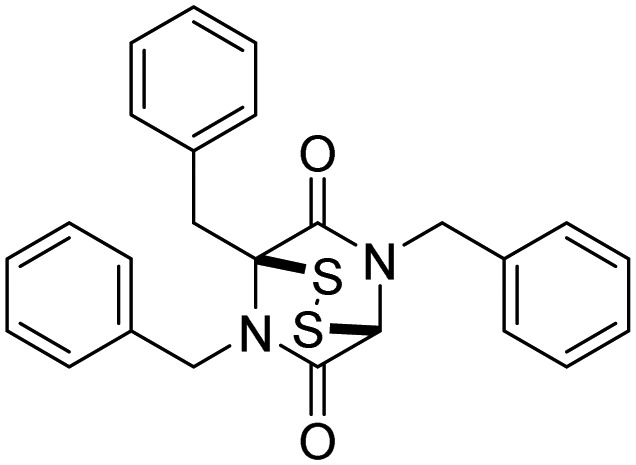 |
Li et al. [115] |
| RPN11 and other JAMM metalloproteases | 42, Thilolutin | 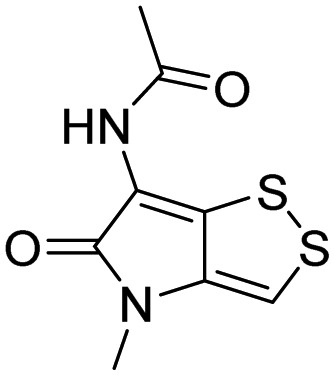 |
Lauinger et al. [116] |
| STAMBP | 43, BC-1471 | 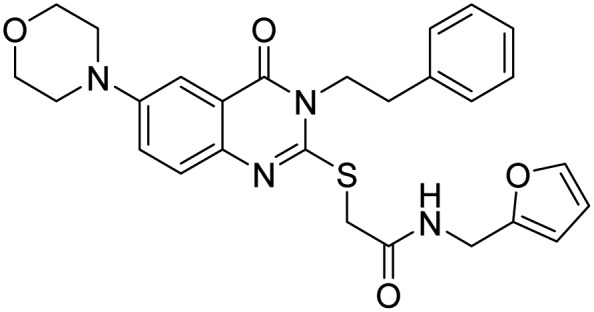 |
Bednash et al. [117] |
Rpn11 (POH1/PSMD14), contains Jab1/MPN metalloenzyme (JAMM) motif, is an essential catalytic subunit of the 19S regulatory particle of the proteasome that cleaves polyubiquitin chains from substrates prior to their proteasomal degradation [119]. Several small molecule inhibitors targeting RPN11 have recently been identified. Quinoline-8-thiol (39, 8TQ, see Table 3) and its derivatives were discovered by screening a library of metal-binding pharmacophores [113]. Further molecular refinement of the 8TQ scaffold yielded several compounds capable of inhibiting RPN11 at submicromolar concentrations. The 8TQ derivative Capzimin (40) was found to be a potent Rpn11 inhibitor (IC50 = 2.4 μM) [114]. Unlike classical proteasome inhibitors that target the 20S core particle, Capzimin stabilizes proteasome substrates and inhibits tumor cell proliferation including the proliferation of cells that are resistant to bortezomib. Through the use of a novel assay for monitoring proteasome-mediated protein degradation, Li et al. [115] also identified epidithiodiketopiperazines as being a new class of RPN11 inhibitors. The most promising candidate, SOP11 (41), quenched RPN11 protease activity by chelating the active site Zn2+ ion. This inhibition stabilized the levels of many substrates that would otherwise have been degraded by the proteasome in cells. However, SOP11 also inhibits the activity of other JAMM family members in vitro, including Csn5 and AMSH. Thiolutin (42), a disulfide-containing antibiotic and anti-angiogenic compound produced by Streptomyces, was reported to function as a zinc chelator that inhibits RPN11 and other JAMM metalloproteases [116]. STAMBP (or AMSH), is a K63-specific JAMM-type DUB that protects endosome cargo proteins from lysosomal degradation and also controls the levels of ubiquitination of ESCRT proteins. BC-1471 (43) was discovered as a specific STAMBP inhibitor (IC50 = 0.33 μM) that selectively blocked deubiquitination of Ub-NALP7 by recombinant STAMBP [117] but did not significantly inhibit the activity of a panel of 38 different DUBs at the concentration tested.
Targeting ubiquitin: a new mechanism
Targeting ubiquitin itself could also prove to be a valid approach to perturb ubiquitin signaling. Although not drug-like, molecules called ubistatins have been shown to bind to ubiquitin in a chemical genetic screen [120,121], although these molecules have features also found in ‘frequent hitters’. An additional advance in a similar direction came through the discovery of a cyclic peptide ubiquitin binder, by using the RaPID system [122]. These peptides have been shown to efficiently trigger apoptosis in cancer cells by binding tightly and with a high degree of specificity to K48-linked Ub chains, thus disrupting their interaction with the proteasome and their subsequent degradation. This unique mode of action could open new opportunities for therapeutic intervention.
Future challenges
The last two decades, most strikingly in the area of oncology, have witnessed a shift away from broad-spectrum medicines such as traditional chemotherapies towards precision treatments that specifically target diseased cells and thus result in more limited off-target cytotoxicity. Whilst there have been some improvements in patient outcomes, the promised revolution has thus far largely failed to materialize. The UPS is integral to virtually all known eukaryotic cellular processes and in recent years it has become clear that specifically targeting (or harnessing in the case of the targeted protein degradation technology) this system could open a new front in the treatment of numerous diseases. Towards this aim, steady progress is being made, evidenced by the number of unique drugs entering the clinic. However, in order to maximize the potential of manipulating the UPS, many challenges will have to be overcome and new opportunities will need to be explored while unique dimensions of this complex pathway are being elucidated.
Exploiting E3 ligase and DUB structural diversity
In this review, we have highlighted the potential of targeting E3 ligases and DUBs. Because each enzyme class interacts with a defined set of substrates, inhibitors of these enzymes should in principle exhibit greater specificity and reduced on-target toxicity compared with proteasome inhibitors, for example. There are, however, possible pitfalls. An obvious potential limitation relates to the fact that RING-type E3 domains and HECT domains share a high degree of structural similarity, as do the catalytic pockets of DUBs. These sites seem to be the most suited to drug development [123], but this similarity increases the probability of obtaining inhibitors that block the activity of multiple enzymes. This might not necessarily be a bad thing if the treatment is effective and unwanted toxicity is significantly lower than currently available options. One way to obtain selective inhibitors would be to target the non-catalytic regions rather than catalytic pockets. For instance, USP7 contains a central binding cleft in the N-terminal TRAF-like domain that is specifically recognized by [PA]-xx-S amino acid motifs found in p53, Hdm2, and Epstein–Barr nuclear antigen 1 [91]; the rhodanese domain of USP8 includes a ligase recognition site [124]; a non-catalytic ubiquitin-like (UBL) region of USP11 has been shown to interact with a helical motif but this site is absent from paralogs USP4 and USP15 [125]. However, current knowledge of such binding sites is still lacking for most DUBs and E3 ligases. Moreover, DUBs are flexible enzymes, that often can allosterically alternate between active and inactive conformations to regulate their activity [126,127].
Complexity of the ubiquitin code
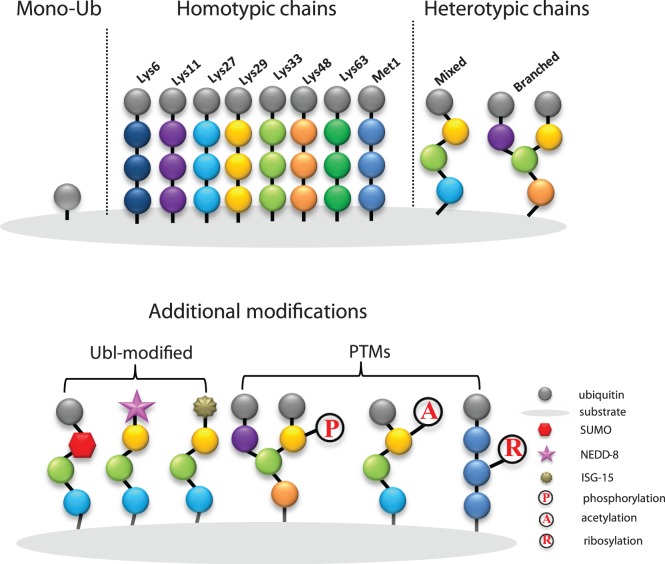
The versatile function of ubiquitination requires the ubiquitin system to be remarkably complex. An essential feature of this complexity which is referred to as the ‘ubiquitin code’, consists of the diverse architecture of Ub chains, Ub chain length and post-translational modifications (PTMs) at multiple sites (Figure 3). The first layer of this complexity results from the fact that the carboxy terminus of Ub can be conjugated to any of seven lysine residues (Lys6, Lys 11, Lys 27, Lys 29, Lys 33, Lys 48, Lys 63) or the N-terminal methionine residue (Met1) on the proximal Ub moiety yielding eight homotypic polyubiquitin chains [128]. The diversity of chain length and topology determines the fate of the substrate, while K48-polyubiquitin is considered to be a signal for degradation by the proteasome [129]. Heterotypic chains and branched chains form the second layer of ubiquitination complexity. In this case, several different linkage types can be contained in one ubiquitin chain or multiple lysine sites on one ubiquitin molecule can be modified to form branched structures [127]. The huge number of distinct chain architectures, resulting from these different linkages, carry important signals in cellular signal transduction pathways [130]. It has been reported that branched chains can enhance substrate recognition by the proteasome and ultimately drive turnover of cell-cycle regulators during mitosis [131]. However, the function of heterotypic/branched chains are still poorly understood because technologies to study them in vivo are limited. The complexity of the ubiquitin code is further expanded through the cross-communication between ubiquitin and other PTMs. Phosphorylation [132–134], acetylation [133, 135], and more recently ribosylation [136–139] are all found on ubiquitin chains, and ubiquitin can be connected to UBL modifiers, such as small ubiquitin-related modifier (SUMO) [140], neuronal precursor cell-expressed developmentally down-regulated protein 8 (NEDD8) [141], and interferon-stimulated gene 15 (ISG15) [142]. In sum, whilst the ubiquitin code is evidently more intricate than is currently known, future approaches to manipulate the code could produce selective inhibitors of specific proteins/biological phenomenon.
Figure 3. The diversity of ubiquitin modifications.
Monoubiquitin is the simplest modification. Eight distinct homotypic polyubiquitin chains are formed by each ubiquitin molecule linking to another via a Lys or Met1 at the same position. Heterotypic chains consist of more than one linkage type in linear or branched mode. Modifications of ubiquitin and UBL modifiers, such as SUMO, NEDD-8 or ISG-15, as well as with other PTMs such as phosphorylation (P), acetylation (A) and ribosylation generates additional levels of complexity.
Functional redundancy
Functional redundancy, that is, the tendency of one protein to compensate for the loss of function of a different protein, is a common biological phenomenon and is one the major causes of resistance to targeted treatments, particularly in oncology. Despite the very large numbers of E3 ligases and DUBs, the UPS exhibits a significant degree of functional redundancy. How can this problem be surmounted to produce clinically robust therapies? To date, a detailed ubiquitin taxonomy is absent such that there is an imprecise mapping of enzymes to the substrates they target. Producing a more comprehensive map would go some way to solving this problem by helping to define suitable combination therapies that are less susceptible to redundancy.
Conclusion
One ultimate goal for a biomedical researcher is to design therapies that effectively treat the disease, do not cause off-target toxicity and that are not susceptible to resistance. During the past decade, we have witnessed dramatic progress in ubiquitin system chemistry and biochemical research into the pathway, resulting in some knowledge of the ‘ubiquitin code’, and UPS enzyme function and their mechanisms of regulation. Parallel to these discoveries has been the development of an increasing number of inhibitors targeting this system, which could prove to be an efficacious and selective way to treat diseases such as cancer.
Perspectives
We are evidently still far away from having a complete picture of ubiquitin biology. In the coming years, fully deciphering the nature of the Ub code will become a priority as little is known about the biological relevance of most ubiquitin chain linkage types (such as K27-, K29-, and K33-linked polyUb chains), or additional layers of complexity of the ubiquitin code (branched and hybrid chains, mixed PTMs). In this respect, methods for unraveling the secrets of the Ub code, such as ubiquitin chain restriction analysis (UbiCRest) [143,144] and Ub-clipping technology [145], will be important. To optimize the prospects of developing E3 or DUB inhibitors for clinical use, mapping the E3-substrate and DUB-substrate relationships are urgently needed as well as structural insight into how specific substrates are recognized and how their ubiquitination is regulated in time and space and under different cellular conditions. This represents an important and at the same time very challenging task. Furthermore, developing novel screening technologies for inhibitor discovery is crucial as the high concentrations of reducing agents used in assays result in very high false-positive rates [146] and as a result reported Ub system inhibitors can be unreliable. With advances in bioinformatics and novel technologies for high-throughput screening and other tools (such as activity-based probes, high-throughput crystallography, and the use of mass spectrometry), the development of specific E3 and DUB inhibitors may become within reach. In addition to blocking the UPS, targeted protein degradation technology could prove to be an essential part of modern medicines armory to treat disease.
Acknowledgement
The authors thank Dr. Robbert Kim for help with Figure 2.
Abbreviations
- BAP1
BRCA1-associated protein 1
- DUBs
deubiquitinating enzymes
- HECT
homologous to E6AP C terminus
- MDM2
murine double minute-2
- OTUs
ovarian tumor proteases
- PROTACs
proteolysis targeting chimeras
- PTMs
post-translational modifications
- RBR
RING between RING
- RING
really interesting new gene
- SUMO
small ubiquitin-related modifier
- UBL
ubiquitin-like
- UCHs
ubiquitin carboxyl-terminal hydrolases
- UPS
ubiquitin–proteasome system
- USP14
ubiquitin-specific protease 14
- USPs
ubiquitin-specific proteases
- VHL
Von Hippel–Lindau
- ZUFSP/ZUP1
zinc finger containing ubiquitin peptidase 1
Competing Interests
H.O. holds shares in Ubiq Bio B.V.
Funding
H. Wu received funding from the European Union Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie grant agreement no. 707404. This work was further supported by a VICI grant from the Netherlands Foundation for Scientific Research (N.W.O.) to H.O.
References
- 1.Wertz I.E. and Wang X. (2018) From discovery to bedside: targeting the ubiquitin system. Cell Chem. Biol. 26, 156–177 10.1016/j.chembiol.2018.10.022 [DOI] [PubMed] [Google Scholar]
- 2.Ovaa H. and Vertegaal A.C. (2018) Probing ubiquitin and SUMO conjugation and deconjugation. Biochem. Soc. Trans. 46, 423–436 10.1042/BST20170086 [DOI] [PubMed] [Google Scholar]
- 3.Ciechanover A. (2005) Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6, 79–87 10.1038/nrm1552 [DOI] [PubMed] [Google Scholar]
- 4.Villa-Hernández S., Bueno A. and Bermejo R. (2017) The multiple roles of ubiquitylation in regulating challenged DNA replication. Adv. Exp. Med. Biol. 1042, 395–419 10.1007/978-981-10-6955-0_18 [DOI] [PubMed] [Google Scholar]
- 5.Schwertman P., Bekker-Jensen S. and Mailand N. (2016) Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 17, 379–394 10.1038/nrm.2016.58 [DOI] [PubMed] [Google Scholar]
- 6.Grumati P. and Dikic I. (2018) Ubiquitin signaling and autophagy. J. Bio. Chem. 293, 5404–5413 10.1074/jbc.TM117.000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacomin A.-C., Taillebourg E. and Fauvarque M.-O. (2018) Deubiquitinating enzymes related to autophagy: new therapeutic opportunities? Cells 7, 112 10.3390/cells7080112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leznicki P. and Kulathu Y. (2017) Mechanisms of regulation and diversification of deubiquitylating enzyme function. J. Cell Sci. 130, 1997–2006 10.1242/jcs.201855 [DOI] [PubMed] [Google Scholar]
- 9.Ebner P., Versteeg G.A. and Ikeda F. (2017) Ubiquitin enzymes in the regulation of immune responses. Crit. Rev. Biochem. Mol. Biol. 52, 425–460 10.1080/10409238.2017.1325829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clague M.J., Urbé S. and Komander D. (2019) Breaking the chains: deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 20, 338–352 10.1038/s41580-019-0099-1 [DOI] [PubMed] [Google Scholar]
- 11.Senft D., Qi J. and Ronai Z.A. (2018) Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 18, 69–88 10.1038/nrc.2017.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Besse A., Besse L., Kraus M., Mendez-Lopez M., Bader J., Xin B.T. et al. (2019) Proteasome inhibition in multiple myeloma: head-to-head comparison of currently available proteasome inhibitors. Cell Chem. Biol. 26, 340–351 10.1016/j.chembiol.2018.11.007 [DOI] [PubMed] [Google Scholar]
- 13.Hoeller D. and Dikic I.J.N. (2009) Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 10.1038/nature07960 [DOI] [PubMed] [Google Scholar]
- 14.Shukla S.K. and Rafiq K.J.T.R. (2019) Proteasome biology and therapeutics in cardiac diseases. Transl. Res. 205, 64–76 10.1016/j.trsl.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Shi F., Hu J., Xie L., Bode A.M. and Cao Y. (2019) The role of deubiquitinases in oncovirus and host interactions. J. Oncol. 2019, 2128410 10.1155/2019/2128410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perng Y.C. and Lenschow D.J.J.N.R.M. (2018) ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 16, 423–439 10.1038/s41579-018-0020-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seissler T., Marquet R. and Paillart J.C.J.V. (2017) Hijacking of the ubiquitin/proteasome pathway by the HIV auxiliary proteins. Viruses 9, 322 10.3390/v9110322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gan J., Qiao N., Strahan R., Zhu C., Liu L., Verma S.C. et al. (2016) Manipulation of ubiquitin/SUMO pathways in human herpesviruses infection. Rev. Med. Virol. 26, 435–445 10.1002/rmv.1900 [DOI] [PubMed] [Google Scholar]
- 19.Boland B., Yu W.H., Corti O., Mollereau B., Henriques A., Bezard E. et al. (2018) Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 17, 660–688 10.1038/nrd.2018.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popovic D., Vucic D. and Dikic I. (2014) Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 10.1038/nm.3739 [DOI] [PubMed] [Google Scholar]
- 21.Pickrell A.M. and Youle R.J. (2015) The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron 85, 257–273 10.1016/j.neuron.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murali R., Wiesner T. and Scolyer R.A. (2013) Tumours associated with BAP1 mutations. Pathology 45, 116–126 10.1097/PAT.0b013e32835d0efb [DOI] [PubMed] [Google Scholar]
- 23.Hao Y.H., Fountain Jr M.D., Tacer K.F., Xia F., Bi W., Kang S.-H.L. et al. (2015) USP7 acts as a molecular rheostat to promote WASH-dependent endosomal protein recycling and is mutated in a human neurodevelopmental disorder. Mol. Cell 59, 956–969 10.1016/j.molcel.2015.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Z.Y., Song Z.J., Chen J.H., Wang Y.F., Li S.Q., Zhou L.F. et al. (2015) Recurrent gain-of-function USP8 mutations in cushing's disease. Cell Res. 25, 306–317 10.1038/cr.2015.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reincke M., Sbiera S., Hayakawa A., Theodoropoulou M., Osswald A., Beuschlein F. et al. (2015) Mutations in the deubiquitinase gene USP8 cause cushing's disease. Nat. Genet. 47, 31–38 10.1038/ng.3166 [DOI] [PubMed] [Google Scholar]
- 26.Homan C.C., Kumar R., Nguyen L.S., Haan E., Raymond F.L., Abidi F. et al. (2014) Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am. J. Hum. Genet. 94, 470–478 10.1016/j.ajhg.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonell L.M., Mirzaa G.M., Alcantara D., Schwartzentruber J., Carter M.T., Lee L.J. et al. (2013) Mutations in STAMBP, encoding a deubiquitinating enzyme, cause microcephaly–capillary malformation syndrome. Nat. Genet. 45, 556–562 10.1038/ng.2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira A.M. and Chou M.M. (2014) USP6-induced neoplasms: the biologic spectrum of aneurysmal bone cyst and nodular fasciitis. Hum. Pathol. 45, 1–11 10.1016/j.humpath.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 29.Forslund A., Zeng Z., Qin L.-X., Rosenberg S., Ndubuisi M., Pincas H. et al. (2008) MDM2 gene amplification is correlated to tumor progression but not to the presence of SNP309 or TP53 mutational status in primary colorectal cancers. Mol. Cancer Res. 6, 205–211 10.1158/1541-7786.MCR-07-0239 [DOI] [PubMed] [Google Scholar]
- 30.Ito M., Barys L., O'Reilly T., Young S., Gorbatcheva B., Monahan J. et al. (2011) Comprehensive mapping of p53 pathway alterations reveals an apparent role for both SNP309 and MDM2 amplification in sarcomagenesis. Clin. Cancer Res. 17, 416–426 10.1158/1078-0432.CCR-10-2050 [DOI] [PubMed] [Google Scholar]
- 31.Pineda C.T., Ramanathan S., Tacer K.F., Weon J.L., Potts M.B., Ou Y.H. et al. (2015) Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell 160, 715–728 10.1016/j.cell.2015.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popov N., Wanzel M., Madiredjo M., Zhang D., Beijersbergen R., Bernards R. et al. (2007) The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 9, 765–774 10.1038/ncb1601 [DOI] [PubMed] [Google Scholar]
- 33.Teicher B.A., Ara G., Herbst R., Palombella V.J. and Adams J.J. (1999) The proteasome inhibitor PS-341 in cancer therapy. Clin. Cancer Res. 5, 2638–2645 PMID: [PubMed] [Google Scholar]
- 34.Kim K.B. and Crews C.M. (2013) From epoxomicin to carfilzomib: chemistry, biology, and medical outcomes. Nat. Prod. Rep. 30, 600–604 10.1039/c3np20126k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kupperman E., Lee E.C., Cao Y., Bannerman B., Fitzgerald M., Berger A. et al. (2010) Evaluation of the proteasome inhibitor MLN9708 in preclinical models of human cancer. Cancer Res. 70, 1970–1980 10.1158/0008-5472.CAN-09-2766 [DOI] [PubMed] [Google Scholar]
- 36.Chauhan D., Catley L., Li G., Podar K., Hideshima T., Velankar M. et al. (2005) A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from bortezomib. Cancer Cell 8, 407–419 10.1016/j.ccr.2005.10.013 [DOI] [PubMed] [Google Scholar]
- 37.Piva R., Ruggeri B., Williams M., Costa G., Tamagno I., Ferrero D. et al. (2008) CEP-18770: a novel, orally active proteasome inhibitor with a tumor-selective pharmacologic profile competitive with bortezomib. Blood 111, 2765–2775 10.1182/blood-2007-07-100651 [DOI] [PubMed] [Google Scholar]
- 38.Potts B., Albitar M., Anderson K., Baritaki S., Berkers C., Bonavida B. et al. (2011) Marizomib, a proteasome inhibitor for all seasons: preclinical profile and a framework for clinical trials. Curr. Cancer Drug Targets 11, 254–284 10.2174/156800911794519716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milhollen M., Sappal D., Duffy J., Hoar K., Huck J., Sha P. et al. (2014) 577 characterization of the cellular mechanism of action of the first in class investigational inhibitor of the Ubiquitin Activating Enzyme, MLN7243. . Euro. J. Cancer 50, 186 10.1016/S0959-8049(14)70703-8 [DOI] [Google Scholar]
- 40.Hyer M.L., Milhollen M.A., Ciavarri J., Fleming P., Traore T., Sappal D. et al. (2018) A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med. 24, 186–193 10.1038/nm.4474 [DOI] [PubMed] [Google Scholar]
- 41.Ito T., Ando H., Suzuki T., Ogura T., Hotta K., Imamura Y. et al. (2010) Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 10.1126/science.1177319 [DOI] [PubMed] [Google Scholar]
- 42.Krönke J., Udeshi N.D., Narla A., Grauman P., Hurst S.N., McConkey M. et al. (2014) Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 10.1126/science.1244851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krönke J., Fink E.C., Hollenbach P.W., MacBeth K.J., Hurst S.N., Udeshi N.D. et al. (2015) Lenalidomide induces ubiquitination and degradation of CK1α in del (5q) MDS. Nature 523, 183–188 10.1038/nature14610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer E.S., Böhm K., Lydeard J.R., Yang H., Stadler M.B., Cavadini S. et al. (2014) Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 10.1038/nature13527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasco D.W., Papadopoulos K.P., Pourdehnad M., Gandhi A.K., Hagner P.R., Li Y. et al. (2019) A first-in-human study of novel cereblon modulator avadomide (CC-122) in advanced malignancies. Clin. Cancer Res. 25, 90–98 10.1158/1078-0432.CCR-18-1203 [DOI] [PubMed] [Google Scholar]
- 46.Bjorklund C.C., Kang J., Amatangelo M., Polonskaia A., Katz M., Chiu H. et al. (2019) Iberdomide (CC-220) is a potent cereblon E3 ligase modulator with antitumor and immunostimulatory activities in lenalidomide-and pomalidomide-resistant multiple myeloma cells with dysregulated CRBN. Leukemia 10.1038/s41375-019-0620-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rasco D.W., Lakhani N.J., Li Y., Men L., Wang H., Ji J. et al. (2019) A phase I study of a novel MDM2 antagonist APG-115 in patients with advanced solid tumors. J. Clin. oncol. 37, 3126–3126 10.1200/JCO.2019.37.15_suppl.3126 [DOI] [Google Scholar]
- 48.Holzer P., Masuya K., Furet P., Kallen J., Valat-Stachyra T., Ferretti S. et al. (2015) Discovery of a dihydroisoquinolinone derivative (NVP-CGM097): a highly potent and selective MDM2 inhibitor undergoing phase 1 clinical trials in p53wt tumors. J. Med. Chem. 58, 6348–6358 10.1021/acs.jmedchem.5b00810 [DOI] [PubMed] [Google Scholar]
- 49.Han T., Goralski M., Gaskill N., Capota E., Kim J., Ting T.C. et al. (2017) Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356, eaal3755 10.1126/science.aal3755 [DOI] [PubMed] [Google Scholar]
- 50.Gross O., Appel G., Block G., Chin M., Goldsberry A., Inker L. et al. (2018) SP121 a phase 2/3 study of the efficacy and safety of bardoxolone methyl in patients with alport syndrome. Nephrol. Dial. Transplant. 33, i384–i385 10.1093/ndt/gfy104.SP121 [DOI] [Google Scholar]
- 51.Lynch D.R., Farmer J., Hauser L., Blair I.A., Wang Q.Q., Mesaros C. et al. (2019) Safety, pharmacodynamics, and potential benefit of omaveloxolone in Friedreich ataxia. Ann. Clin. Transl. Neurol. 6, 15–26 10.1002/acn3.660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X., D'Arcy P., Caulfield T.R., Paulus A., Chitta K., Mohanty C. et al. (2015) Synthesis and evaluation of derivatives of the proteasome deubiquitinase inhibitor b-AP15. Chem. Biol. Drug Des. 86, 1036–1048 10.1111/cbdd.12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smrt R.D., Szulwach K.E., Pfeiffer R.L., Li X., Guo W., Pathania M. et al. (2010) MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 28, 1060–1070 10.1002/stem.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J., Tan Q., Yan M., Liu L., Lin H., Zhao F. et al. (2014) miRNA-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Mol. Cancer 13, 166 10.1186/1476-4598-13-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang X. and Dixit V.M. (2016) Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 26, 484–498 10.1038/cr.2016.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel K., Ahmed Z.S., Huang X., Yang Q., Ekinci E., Neslund-Dudas C.M. et al. (2018) Discovering proteasomal deubiquitinating enzyme inhibitors for cancer therapy: lessons from rational design, nature and old drug reposition. Future Med. Chem. 10, 2087–2108 10.4155/fmc-2018-0091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D'Arcy P., Wang X. and Linder S. (2015) Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol. Ther. 147, 32–54 10.1016/j.pharmthera.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 58.Schauer N., Magin R.S., Liu X., Doherty L. and Buhrlage S.J. (2019) Advances in discovering deubiquitinating enzyme (DUB) inhibitors. J. Med. Chem. 10.1021/acs.jmedchem.9b01138 [DOI] [PubMed] [Google Scholar]
- 59.Berndsen C.E. and Wolberger C. (2014) New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21, 301–307 10.1038/nsmb.2780 [DOI] [PubMed] [Google Scholar]
- 60.Dove K.K. and Klevit R.E. (2017) RING-between-RING E3 ligases: emerging themes amid the variations. J. Mol. Biol. 429, 3363–3375 10.1016/j.jmb.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber J., Polo S. and Maspero E. (2019) HECT e3 ligases: a tale with multiple facets. Front. Physiol. 10, 370 10.3389/fphys.2019.00370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brenke J.K., Popowicz G.M., Schorpp K., Rothenaigner I., Roesner M., Meininger I. et al. (2018) Targeting TRAF6 E3 ligase activity with a small-molecule inhibitor combats autoimmunity. J. Biol. Chem. 293, 13191–13203 10.1074/jbc.RA118.002649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward C.C., Kleinman J.I., Brittain S.M., Lee P.S., Chung C.Y.S., Kim K. et al. (2019) Covalent ligand screening uncovers a RNF4 E3 ligase recruiter for targeted protein degradation applications. ACS. Chem. Biol. 14, 2430–2440 10.1021/acschembio.8b01083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Testa A., Lucas X., Castro G.V., Chan K.-H., Wright J.E., Runcie A.C. et al. (2018) 3-Fluoro-4-hydroxyprolines: synthesis, conformational analysis, and stereoselective recognition by the VHL E3 ubiquitin ligase for targeted protein degradation. J. Am. Chem. Soc. 140, 9299–9313 10.1021/jacs.8b05807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watt J.E., Hughes G.R., Walpole S., Monaco S., Stephenson G.R., Bulman Page P.C. et al. (2018) Discovery of small molecule WWP2 ubiquitin ligase inhibitors. Chemistry 24, 17677–17680 10.1002/chem.201804169 [DOI] [PubMed] [Google Scholar]
- 66.Tian M., Zeng T., Liu M., Han S., Lin H., Lin Q. et al. (2019) A cell-based high-throughput screening method based on a ubiquitin-reference technique for identifying modulators of E3 ligases. J. Biol. Chem. 294, 2880–2891 10.1074/jbc.RA118.003822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Cesare V., Johnson C., Barlow V., Hastie J., Knebel A. and Trost M.J. (2018) The MALDI-TOF E2/E3 ligase assay as universal tool for drug discovery in the ubiquitin pathway. Cell Chem. Biol. 5, 1117–1127 10.1016/j.chembiol.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johansson H., Isabella Tsai Y.C., Fantom K., Chung C.-W., Kümper S., Martino L. et al. (2019) Fragment-based covalent ligand screening enables rapid discovery of inhibitors for the RBR E3 ubiquitin ligase HOIP. J. Am. Chem. Soc. 141, 2703–2712 10.1021/jacs.8b13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staudinger J.L. (2017) The molecular interface between the SUMO and ubiquitin systems, in SUMO regulation of cellular processes. Adv. Exp. Med. Biol. 963, 99–110 10.1007/978-3-319-50044-7_6 [DOI] [PubMed] [Google Scholar]
- 70.Plechanovová A., Jaffray E.G.. McMahon S.A., Johnson K.A., Navrátilová I., Naismith J.H. et al. (2011) Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat. Struct. Mol. Biol. 18, 1052–1059 10.1038/nsmb.2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lai A.C. and Crews C.M. (2017) Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 16, 101–114 10.1038/nrd.2016.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rape M. (2018) Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 19, 59–70 10.1038/nrm.2017.83 [DOI] [PubMed] [Google Scholar]
- 73.Moon S. and Lee B.-H. (2018) Chemically induced cellular proteolysis: an emerging therapeutic strategy for undruggable targets. Mol. Cells 41, 933–942 10.14348/molcells.2018.0372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang S., Han L., Han J., Li P., Ding Q., Zhang Q.-J. et al. (2019) Uncoupling of PARP1 trapping and inhibition using selective PARP1 degradation. Nat. Chem. Biol. 15, 1223–1231 10.1038/s41589-019-0379-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Okuhira K., Ohoka N., Sai K., Nishimaki-Mogami T., Itoh Y., Ishikawa M. et al. (2011) Specific degradation of CRABP-II via cIAP1-mediated ubiquitylation induced by hybrid molecules that crosslink cIAP1 and the target protein. FEBS. Lett. 585, 1147–1152 10.1016/j.febslet.2011.03.019 [DOI] [PubMed] [Google Scholar]
- 76.Ohoka N., Okuhira K., Ito M., Nagai K., Shibata N., Hattori T. et al. (2017) In vivo knockdown of pathogenic proteins via specific and nongenetic inhibitor of apoptosis protein (IAP)-dependent protein erasers (SNIPERs). J. Biol. Chem. 292, 4556–4570 10.1074/jbc.M116.768853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bondeson D.P. and Crews C.M. (2017) Targeted protein degradation by small molecules. Annu. Rev. Pharmacol. Toxicol. 57, 107–123 10.1146/annurev-pharmtox-010715-103507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lucas X. and Ciulli A. (2017) Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies. Curr. Opin. Struct. Biol. 44, 101–110 10.1016/j.sbi.2016.12.015 [DOI] [PubMed] [Google Scholar]
- 79.Schapira M., Calabrese M.F., Bullock A.N. and Crews C.M. (2019) Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discov. 18, 949–963 10.1038/s41573-019-0047-y [DOI] [PubMed] [Google Scholar]
- 80.Watt G.F., Scott-Stevens P. and Gaohua L. (2019) Targeted protein degradation in vivo with proteolysis targeting chimeras: current status and future considerations. Drug Discov. Today Technol. 31, 69–80 10.1016/j.ddtec.2019.02.005 [DOI] [PubMed] [Google Scholar]
- 81.Pettersson M. and Crews C.M. (2019) PROteolysis TArgeting Chimeras (PROTACs)-past, present and future. Drug Discov. Today Technol. 31, 15–27 10.1016/j.ddtec.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ohoka N., Ujikawa O., Shimokawa K., Sameshima T., Shibata N., Hattori T. et al. (2018) Different degradation mechanisms of inhibitor of apoptosis proteins (IAPs) by the specific and nongenetic IAP-dependent protein eraser (SNIPER). Chem. Pharm. Bull (Tokyo) 67, 203–209 10.1248/cpb.c18-00567 [DOI] [PubMed] [Google Scholar]
- 83.Bochicchio A., Jordaan S., Losasso V., Chetty S., Perera R.C., Ippoliti E. et al. (2017) Designing the sniper: improving targeted human cytolytic fusion proteins for anti-cancer therapy via molecular simulation. Biomedicines 5, E9 10.3390/biomedicines5010009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Naito M., Ohoka N. and Shibata N. (2019) SNIPERs-Hijacking IAP activity to induce protein degradation. Drug Discov. Today Technol. 31, 35–42 10.1016/j.ddtec.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 85.Rehman S.A.A., Kristariyanto Y.A., Choi S.-Y., Nkosi P.J., Weidlich S., Labib K. et al. (2016) MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell 63, 146–155 10.1016/j.molcel.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kwasna D., Rehman S.A.A., Natarajan J., Matthews S., Madden R., De Cesare V. et al. (2018) Discovery and characterization of ZUFSP/ZUP1, a distinct deubiquitinase class important for genome stability. Mol. Cell 70, 150–164 10.1016/j.molcel.2018.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haahr P., Borgermann N., Guo X., Typas D., Achuthankutty D., Hoffmann S. et al. (2018) ZUFSP deubiquitylates K63-linked polyubiquitin chains to promote genome stability. Mol. Cell 70, 165–174 10.1016/j.molcel.2018.02.024 [DOI] [PubMed] [Google Scholar]
- 88.Hewings D.S., Heideker J., Ma T.P., AhYoung A.P., El Oualid F., Amore A. et al. (2018) Reactive-site-centric chemoproteomics identifies a distinct class of deubiquitinase enzymes. Nat. Commun. 9, 1162–1179 10.1038/s41467-018-03511-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hermanns T., Pichlo C., Woiwode I., Klopffleisch K., Witting K.F., Ovaa H. et al. (2018) A family of unconventional deubiquitinases with modular chain specificity determinants. Nat Commun. 9, 799–812 10.1038/s41467-018-03148-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ritorto M.S., Ewan R., Perez-Oliva A.B., Knebel A., Buhrlage S.J., Wightman M. et al. (2014) Screening of DUB activity and specificity by MALDI-TOF mass spectrometry. Nat. Commun. 5, 4763 10.1038/ncomms5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sheng Y., Saridakis V., Sarkari F., Duan S., Wu T., Arrowsmith C.H. et al. (2006) Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat. Struct. Mol. Biol. 13, 285–291 10.1038/nsmb1067 [DOI] [PubMed] [Google Scholar]
- 92.Colland F., Formstecher E., Jacq X., Reverdy C., Planquette C., Conrath S. et al. (2009) Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 8, 2286–2295 10.1158/1535-7163.MCT-09-0097 [DOI] [PubMed] [Google Scholar]
- 93.Reverdy C., Conrath S., Lopez R., Planquette C., Atmanene C., Collura V. et al. (2012) Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem. Biol. 19, 467–477 10.1016/j.chembiol.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 94.Pozhidaeva A. and Bezsonova I. (2019) USP7: structure, substrate specificity, and inhibition. DNA Repair (Amst) 76, 30–39 10.1016/j.dnarep.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weinstock J., Wu J., Cao P., Kingsbury W.D., McDermott J.L., Kodrasov M.P. et al. (2012) Selective dual inhibitors of the cancer-related deubiquitylating proteases USP7 and USP47. ACS. Med. Chem. Lett. 3, 789–792 10.1021/ml200276j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chauhan D., Tian Z., Nicholson B., Kumar K.S., Zhou B., Carrasco R. et al. (2012) A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell 22, 345–358 10.1016/j.ccr.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fan Y., Cheng J., Vasudevan S., Dou J., Zhang H., Patel R. et al. (2013) USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 4, e867 10.1038/cddis.2013.400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pozhidaeva A., Valles G., Wang F., Wu J., Sterner D.E., Nguyen P. et al. (2017) USP7-specific inhibitors target and modify the enzyme's active site via distinct chemical mechanisms. Cell Chem. Biol. 24, 1501–1512 10.1016/j.chembiol.2017.09.004 [DOI] [PubMed] [Google Scholar]
- 99.Kategaya L., Di Lello P., Rougé L., Pastor R., Clark K.R., Drummond J. et al. (2017) USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 550, 534–538 10.1038/nature24006 [DOI] [PubMed] [Google Scholar]
- 100.Di Lello P., Pastor R., Murray J.M., Blake R.A., Cohen F., Crawford T.D. et al. (2017) Discovery of small-molecule inhibitors of ubiquitin specific protease 7 (USP7) using integrated NMR and in silico techniques. J. Med. Chem. 60, 10056–10070 10.1021/acs.jmedchem.7b01293 [DOI] [PubMed] [Google Scholar]
- 101.Gavory G., O'Dowd C.R., Helm M.D., Flasz J., Arkoudis E., Dossang A. et al. (2018) Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nat. Chem. Biol. 14, 118–125 10.1038/nchembio.2528 [DOI] [PubMed] [Google Scholar]
- 102.Lamberto I., Liu X., Seo H.-S., Schauer N.J., Iacob R.E., Hu W. et al. (2017) Structure-guided development of a potent and selective non-covalent active-site inhibitor of USP7. Cell Chem. Biol. 24, 1490–1500 10.1016/j.chembiol.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Turnbull A.P., Ioannidis S., Krajewski W.W., Pinto-Fernandez A., Heride C., Martin A.C. et al. (2017) Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 550, 481–486 10.1038/nature24451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.O'Dowd C.R., Helm M.D., Rountree J.S., Flasz J.T., Arkoudis E., Miel H. et al. (2018) Identification and structure-guided development of pyrimidinone based USP7 inhibitors. ACS. Med. Chem. Lett. 9, 238–243 10.1021/acsmedchemlett.7b00512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee B., Lee M.J., Park S., Oh D.C., Elsasser S., Chen P.C. et al. (2010) Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 10.1038/nature09299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boselli M., Lee B.H., Robert J., Prado M.A., Min S.W., Cheng C. et al. (2017) An inhibitor of the proteasomal deubiquitinating enzyme USP14 induces tau elimination in cultured neurons. J. Biol. Chem. 292, 19209–19225 10.1074/jbc.M117.815126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Palmer A.L., de Jong A., Leestemaker Y., Geurink P.P., Wijdeven R.H., Ovaa H. et al. (2018) Inhibition of the deubiquitinase Usp14 diminishes direct MHC class I antigen presentation. J. Immunol. 200, 928–936 10.4049/jimmunol.1700273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kemp M. I. and Woodrow M. D. (2018), Cyanopyrrolidine derivatives as inhibitors for dubs. US20180362460A1
- 109.Krabill A.D., Chen H., Hussain S., Feng C., Abdullah A., Das C. et al. (2019) Biochemical and cellular characterization of a cyanopyrrolidine covalent ubiquitin C-terminal hydrolase L1 inhibitor. Chembiochem 21, 712–722 10.1002/cbic.201900434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nattawadee P., Aurélien G., Thomas L.-H., Sofía L.-O., Edward J., Christelle W. (2019) Discovery of a potent and selective covalent inhibitor and activity-based probe for the deubiquitylating enzyme UCHL1, with anti-fibrotic activity. ChemRxiv 10.26434/chemrxiv.10058429.v1 [DOI] [Google Scholar]
- 111.Geurink P.P., Kooij R., Sapmaz A., Liu S., Xin B.-T., Janssen G.M.C. et al. (2019) A small-molecule activity-based probe for monitoring ubiquitin C-terminal hydrolase L1 (UCHL1) activity in live cells and zebrafish embryos. BioRxiv 10.1101/827642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Resnick E., Bradley A., Gan J., Douangamath A., Krojer T., Sethi R. et al. (2019) Rapid covalent-probe discovery by electrophile-fragment screening. J. Am. Chem. Soc. 141, 8951–8968 10.1021/jacs.9b02822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Perez C., Li J., Parlati F., Rouffet M., Ma Y., Mackinnon A.L. et al. (2017) Discovery of an inhibitor of the proteasome subunit Rpn11. J. Med. Chem. 60, 1343–1361 10.1021/acs.jmedchem.6b01379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li J., Yakushi T., Parlati F., Mackinnon A.L., Perez C., Ma Y. et al. (2017) Capzimin is a potent and specific inhibitor of proteasome isopeptidase Rpn11. Nat. Chem. Biol. 13, 486–493 10.1038/nchembio.2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li J., Zhang Y., Dos Santos B.D.S.S., Wang F., Ma Y., Perez C. et al. (2018) Epidithiodiketopiperazines inhibit protein degradation by targeting proteasome deubiquitinase Rpn11. Cell Chem. Biol. 25, 1350–1358 10.1016/j.chembiol.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lauinger L., Li J., Shostak A., Cemel I.A., Ha N., Zhang Y. et al. (2017) Thiolutin is a zinc chelator that inhibits the Rpn11 and other JAMM metalloproteases. Nat. Chem. Biol. 13, 709–714 10.1038/nchembio.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bednash J.S., Weathington N., Londino J., Rojas M., Gulick D.L., Fort R. et al. (2017) Targeting the deubiquitinase STAMBP inhibits NALP7 inflammasome activity. Nat. Commun. 8, 15203 10.1038/ncomms15203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kemp M., Stockley M. and Jones A. (2018), Cyanopyrrolidines as dub inhibitors for the treatment of cancer. WO2017009650A1
- 119.Verma R., Aravind L., Oania R., McDonald W.H., Yates J.R., Koonin E.V. et al. (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615 10.1126/science.1075898 [DOI] [PubMed] [Google Scholar]
- 120.Verma R., Peters N.R., D'Onofrio M., Tochtrop G.P., Sakamoto K.M., Varadan R. et al. (2004) Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science 306, 117–120 10.1126/science.1100946 [DOI] [PubMed] [Google Scholar]
- 121.Nakasone M.A., Lewis T.A., Walker O., Thakur A., Mansour W., Castañeda C.A. et al. (2017) Structural basis for the inhibitory effects of ubistatins in the ubiquitin-proteasome pathway. Structure 25, 1839–1855 10.1016/j.str.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nawatha M., Rogers J.M., Bonn S.M., Livneh I., Lemma B., Mali S.M. et al. (2019) De novo macrocyclic peptides that specifically modulate Lys48-linked ubiquitin chains. Nat. Chem. 11, 644–652 10.1038/s41557-019-0278-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Weissman A.M. (2013) Ubiquitin and drug discovery: challenges and opportunities. Cell Biochem. Biophys. 67, 1–2 10.1007/s12013-013-9620-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Avvakumov G.V., Walker J.R., Xue S., Finerty P.J., Mackenzie F., Newman E.M. et al. (2006) Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8). J. Biol. Chem. 281, 38061–38070 10.1074/jbc.M606704200 [DOI] [PubMed] [Google Scholar]
- 125.Spiliotopoulos A., Ferreras L.B., Densham R.M., Caulton S.G., Maddison B.C., Morris J.R. et al. (2019) Discovery of peptide ligands targeting a specific ubiquitin-like domain-binding site in the deubiquitinase USP11. J. Biol. Chem. 294, 424–436 10.1074/jbc.RA118.004469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sahtoe D.D. and Sixma T.K. (2015) Layers of DUB regulation. Trends Biochem. Sci. 40, 456–467 10.1016/j.tibs.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 127.Mevissen T.E. and Komander D. (2017) Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192 10.1146/annurev-biochem-061516-044916 [DOI] [PubMed] [Google Scholar]
- 128.Swatek K.N. and Komander D. (2016) Ubiquitin modifications. Cell Res. 26, 399–422 10.1038/cr.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mattern M., Sutherland J., Kadimisetty K., Barrio R. and Rodriguez M.S. (2019) Using ubiquitin binders to decipher the ubiquitin code. Trends Biochem. Sci. 44, 599–615 10.1016/j.tibs.2019.01.011 [DOI] [PubMed] [Google Scholar]
- 130.Ohtake F. and Tsuchiya H. (2016) The emerging complexity of ubiquitin architecture. J. Biochem. 161, 125–133 10.1093/jb/mvw088 [DOI] [PubMed] [Google Scholar]
- 131.Meyer H.J. and Rape M.J.C. (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157, 910–921 10.1016/j.cell.2014.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Swaney D.L., Beltrao P., Starita L., Guo A., Rush J., Fields S. et al. (2013) Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 10, 676–682 10.1038/nmeth.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ohtake F., Saeki Y., Sakamoto K., Ohtake K., Nishikawa H., Tsuchiya H. et al. (2015) Ubiquitin acetylation inhibits polyubiquitin chain elongation. EMBO. Rep. 16, 192–201 10.15252/embr.201439152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huguenin-Dezot N., De Cesare V., Peltier J., Knebel A., Kristaryianto Y.A., Rogerson D.T. et al. (2016) Synthesis of isomeric phosphoubiquitin chains reveals that phosphorylation controls deubiquitinase activity and specificity. Cell Rep. 16, 1180–1193 10.1016/j.celrep.2016.06.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee S.Y., Choi Y.S., Kim E.H., Cheong H.K., Lee Y.J., Lee J.G. et al. (2018) Nonenzymatic acetylation of ubiquitin Lys side chains is modulated by their neighboring residues. FEBS. J. 285, 1277–1289 10.1111/febs.14404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang C.S., Jividen K., Spencer A., Dworak N., Ni L., Oostdyk L.T. et al. (2017) Ubiquitin modification by the E3 ligase/ADP-ribosyltransferase Dtx3L/Parp9. Mol. Cell 66, 503–516 10.1016/j.molcel.2017.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Qiu J., Sheedlo M.J., Yu K., Tan Y., Nakayasu E.S., Das C. et al. (2016) Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533, 120–124 10.1038/nature17657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kalayil S., Bhogaraju S., Bonn F., Shin D., Liu Y., Gan N. et al. (2018) Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination. Nature 557, 734–738 10.1038/s41586-018-0145-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bhogaraju S., Kalayil S., Liu Y., Bonn F., Colby T., Matic I. et al. (2016) Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell 167, 1636–1649 10.1016/j.cell.2016.11.019 [DOI] [PubMed] [Google Scholar]
- 140.Hendriks I.A., Lyon D., Su D., Skotte N.H., Daniel J.A., Jensen L.J. et al. (2018) Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun. 9, 2456 10.1038/s41467-018-04957-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Maghames C.M., Lobato-Gil S., Perrin A., Trauchessec H., Rodriguez M.S., Urbach S. et al. (2018) NEDDylation promotes nuclear protein aggregation and protects the ubiquitin proteasome system upon proteotoxic stress. Nat. Commun. 9, 4376 10.1038/s41467-018-06365-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fan J.B., Arimoto K.-L., Motamedchaboki K., Yan M., Wolf D.A. and Zhang D.E. (2015) Identification and characterization of a novel ISG15-ubiquitin mixed chain and its role in regulating protein homeostasis. Sci. Rep. 5, 12704 10.1038/srep12704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hospenthal M.K., Mevissen T.E. and Komander D.J. (2015) Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest). Nat. Protoc. 10, 349–361 10.1038/nprot.2015.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mevissen T.E., Hospenthal M.K., Geurink P.P., Elliott P.R., Akutsu M., Arnaudo N. et al. (2013) OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154, 169–184 10.1016/j.cell.2013.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Swatek K.N., Usher J.L., Kueck A.F., Gladkova C., Mevissen T.E., Pruneda J.N. et al. (2019) Insights into ubiquitin chain architecture using Ub-clipping. Nature 572, 533–537 10.1038/s41586-019-1482-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Harrigan J.A., Jacq X., Martin N.M. and Jackson S.P. (2018) Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–78 10.1038/nrd.2017.152 [DOI] [PMC free article] [PubMed] [Google Scholar]