INTRODUCTION
Dehorning, a standard cattle management practice to protect animals and humans from injury, is unpleasant, costly, and subject to public scrutiny (Stafford and Mellor, 2011). In the United States, 94% of dairy cattle producers report routine dehorning (USDA, 2009). Horns are recessively inherited; an alternative is to breed for polled (Long and Gregory, 1978).
The frequency of the POLLED allele is very low in U.S dairy cattle (<0.01). Therefore, adding its economic value to the lifetime net merit index (NM$) does not effectively increase POLLED (Cole, 2015). Gene editing to produce high-genetic merit, polled bulls has been proposed as an efficient method to eliminate dehorning (Carlson et al., 2016).
The use of gene editing to eliminate dehorning in Holsteins was previously simulated (Mueller et al., 2018). Dehorning is also an issue in Jerseys, the second largest U.S. dairy breed. The HORNED allele frequency is 1.5% lower in Jerseys compared with Holsteins (Null, 2015), and a higher proportion of polled sires are available (Spurlock et al., 2014).
This simulation tested the hypothesis that gene editing is more efficient than conventional breeding for eliminating HORNED from the Jersey population. The objective was to model the incorporation of POLLED into the U.S. Jersey population using either conventional breeding or gene editing for three polled-mating schemes and quantify changes in HORNED frequency, inbreeding, and rates of genetic gain.
MATERIALS AND METHODS
Simulation
Geneedit.py (Cole, 2017) simulated introgression of POLLED into the Jersey breed via conventional breeding or gene editing. The base population was 35,000 cows distributed over 200 herds and 350 bulls. True breeding values for NM$ were determined by randomly sampling from a normal distribution, with a mean of $0 for cows and $300 for bulls and SD of $200 for both. The proportion of polled bulls was set to 5.4% heterozygous (Pp) and 1.5% homozygous (PP). These bulls averaged 0.5 and 1.3 SD lower NM$, respectively, than horned bulls (NAAB, 2018). The frequency of HORNED was set to 0.978 (Null, 2015). Horned status was determined by randomly selecting one allele each from the sire and dam. The population was limited to 500 bulls and 100,000 cows total over 20 yr, with overlapping generations (Cole, 2015; Mueller et al., 2018).
Mate Allocation
Four mating schemes were modeled (A–D, Table 1). To establish a baseline, NM$ was the only sire selection criterion in scheme D. Polled sires were preferentially selected in mating schemes A–C. Both PP and Pp bulls were used in scheme A. Only PP bulls were used in schemes B and C. In schemes A and B, bulls were limited to 5,000 matings per year; if too few polled bulls were available, horned bulls were used. Matings per bull were not limited in scheme C, so no horned sires were used. All schemes used the modified Pryce scheme to allocate bulls to cows, which penalizes the parent average NM$ for inbreeding (Pryce et al., 2012) and the economic costs of horned and carrier calves (Cole, 2015; Mueller et al., 2018). The average cost of dehorning is $22.50 per animal (Thompson et al., 2017). To account for breeder preferences and marketing opportunities, horned and carrier calves were penalized $40 and $20, respectively (Cole, 2015; Mueller et al., 2018).
Table 1.
Parameters and results of each scenario
| Mating scheme | Scenario | Gene editing | 1° bull-selection criterion | Sire genotype(s) | Matings per year limit | HORNED allele frequency* | Inbreeding %* | NM$*,† |
|---|---|---|---|---|---|---|---|---|
| A | A1 | No | Polled | PP, Pp (pp‡) | Yes | 0.22a,b | 6.0a,b | 2,820a,b |
| A2 | Yes | 0.09a,b | 5.5a,b | 3,184a,b | ||||
| B | B1 | No | Polled | PP (pp‡) | Yes | 0.59a,b | 5.2a,b | 2,936a,b |
| B2 | Yes | 0.01a,b | 5.7a | 3,337a,b | ||||
| C | C1 | No | Polled | PP | No | 0.01b | 14.4a,b | 2,636a,b |
| C2 | Yes | 0.01b | 7.7a | 3,216a,b | ||||
| D | D | No | NM$ | n/a|| | Yes | 0.99 | 7.2 | 3,446 |
*Average values at year 20 of simulation.
†Lifetime NM$.
‡If not enough polled sires available for mating scheme, horned sires were used.
||In scenario D, both PP and Pp sires may have been available, but genotype was not included as a selection criterion.
aSignificant difference (P ≤ 0.01) between scenarios within a mating scheme.
bSignificant difference (P ≤ 0.01) between scenario and baseline (D).
Gene Editing
Gene editing was modeled as an added step to the Kasinathan (2015) production system, which uses advanced reproductive technologies and somatic cell nuclear transfer cloning. In gene-editing schemes, designated with a number 2 in the “scenario” column of Table 1, the live bulls were sorted on NM$ in descending order, and the top 1% of Pp and pp bulls were cloned and edited. All edited bulls were assumed to be PP. Schemes designated with a number 1 in the “scenario” column of Table 1, and the baseline scenario (D), used only conventional breeding.
Analysis
Ten replicates of each scenario were compared using the Student t-test. P values of ≤0.01 were considered significantly different.
RESULTS AND DISCUSSION
HORNED Allele Frequency
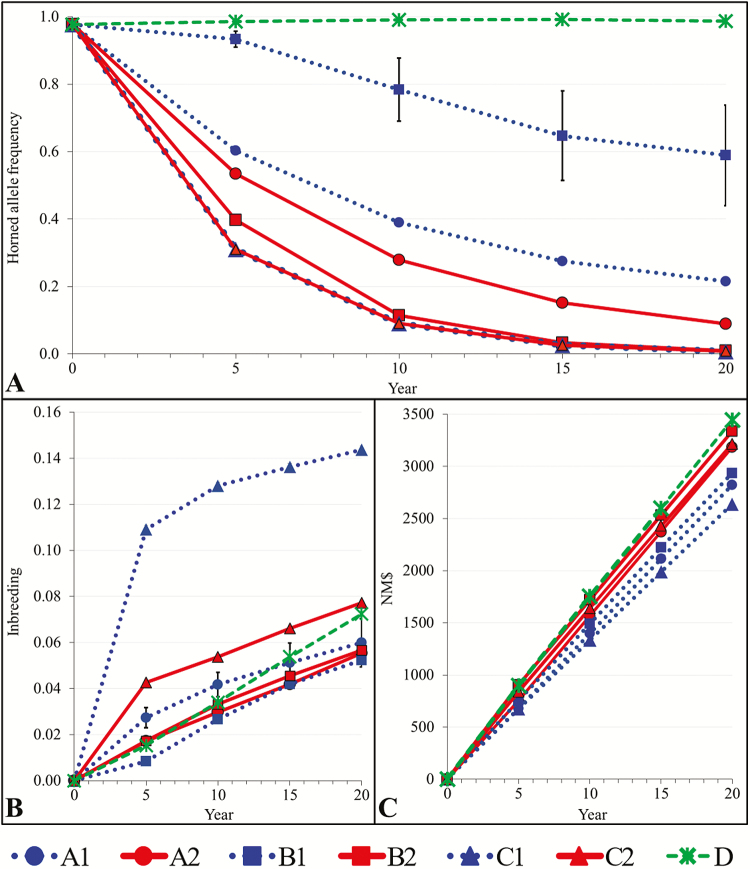
Baseline results (D) support findings by Cole (2015) and Mueller et al., (2018) that including a horned economic penalty ($40) in the selection index is not effective at decreasing the frequency of HORNED (Figure 1A).
Figure 1.
Effect of each mating scenario on (A) HORNED allele frequency, (B) inbreeding, and (C) NM$. Conventional breeding is a dashed green line, conventional breeding for polled scenarios are dotted blue lines, and gene editing for polled scenarios are solid red lines. Bars are used to represent the SEM.
Similar to Holstein, gene editing decreased HORNED frequency as fast, or faster, than conventional breeding in each scheme. Scenarios B1 and B2 exhibited the largest difference (0.58) in HORNED frequency within a mating scheme after 20 yr. Only a small number (n = 8) of PP sires were available in the base population. Since there was also a mating limit, several horned sires were used in B1. Therefore, HORNED frequency did not decrease as quickly as in other scenarios. In contrast, using gene editing in B2 rapidly increased the number of high NM$ PP sires, so fewer horned sires were used each year. As a result, HORNED frequency in B2 decreased significantly faster (P ≤ 0.01) than B1. Since only PP sires were used in scheme C, both conventional breeding and gene editing resulted in the same (P = 0.16) rapid change in HORNED frequency. Results for scheme A were intermediate to B and C.
Inbreeding
In scenario D, inbreeding reached 7% after 20 yr. Polled schemes A and B showed lower inbreeding than D (A1, A2, B1 (P ≤ 0.01); B2 (P = 0.28)) (Figure 1B). In contrast, selecting for polled in Holstein resulted in higher inbreeding vs. D in all but B1 (Mueller et al., 2018). The polled-mating schemes used an additional selection criterion in the breeding objective, so herds used a wider variety of sires, resulting in lower inbreeding levels than D. However, this simulation assumed that all base population animals were initially unrelated, which is unlikely in a production population. Since a mixture of sire genotypes were used in schemes A and B, inbreeding reached 6% regardless of the introgression method after 20 yr. When herds were forced to use only PP sires, inbreeding was significantly higher (14%, P ≤ 0.01) than D for the conventional breeding scheme (C1) but was not significantly different (8%, P = 0.09) for the gene-editing scenario (C2).
Genetic Gain
Polled-mating schemes resulted in significantly slower (P ≤ 0.01) rates of genetic gain (NM$) vs. D (Figure 1C), which is consistent with previous findings (Spurlock et al., 2014; Mueller et al., 2018). In all polled schemes, gene editing resulted in significantly greater (P ≤ 0.01) genetic gain than conventional breeding. The greatest difference in NM$ within a mating scheme after 20 yr, $580, was observed between C1 and C2. Although only PP sires were also used in C2, gene editing allowed for the top NM$ bulls to be PP in just a few years. The greatest average rate of genetic gain per year, $167, was achieved with gene editing in B2, when herds preferentially used PP sires, but could also use pp sires.
Consistent with our hypothesis, these results show that gene editing was more efficient at rapidly reducing the frequency of HORNED, while keeping inbreeding at acceptable levels and maintaining rates of genetic gain in Jersey. Scenario B2, which used both PP and pp sires in combination with gene editing, was the optimal scenario (Figure 1). This is consistent with the results observed in Holstein (Mueller et al., 2018). Scheme C models a case where consumer and market expectations force the dairy industry to eliminate dehorning immediately, thereby requiring the exclusive use of PP sires.
IMPLICATIONS
Our simulations show, given the current NM$ of dairy sires, conventional breeding to decrease HORNED frequency will increase inbreeding and slow the rate of genetic gain (NM$). Resulting economic considerations hinder the dairy industry’s ability to address this animal welfare concern through currently available approaches. Although long-term progress can be made through conventional breeding, the negative impact on inbreeding and NM$ is greater than if gene editing was used. If consumers demand an immediate end to dehorning, producers may have limited time to change their practices. In this case, gene editing will be necessary to avoid long-term detrimental effects to the U.S. dairy industry. This study demonstrates how gene editing to produce high-genetic merit-polled sires could relieve dairy producers’ economic concerns while also alleviating consumers’ animal welfare concerns.
Footnotes
The authors acknowledge funding support from USDA NIFA Grant no. 2015-67015-23316. Cole was supported by appropriated USDA ARS project 8042-31000-002-00.
LITERATURE CITED
- Carlson D. F., Lancto C. A., Zang B., Kim E. S., Walton M., Oldeschulte D., Seabury C., Sonstegard T. S., and Fahrenkrug S. C.. 2016. Production of hornless dairy cattle from genome-edited cell lines. Nat. Biotechnol. 34: 479–481. doi:10.1038/nbt.3560 [DOI] [PubMed] [Google Scholar]
- Cole J. B. 2015. A simple strategy for managing many recessive disorders in a dairy cattle breeding program. Genet. Sel. Evol. 47:94. doi:10.1186/s12711-015-0174-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. B. 2017. Python programs and notebooks used for simulation of gene editing in dairy cattle breeding programs [accessed March 1, 2018]. https://github.com/wintermind/gene-editing.
- Kasinathan P., Wei H., Xiang T., Molina J. A., Metzger J., Broek D., Kasinathan S., Faber D. C., and Allan M. F.. 2015. Acceleration of genetic gain in cattle by reduction of generation interval. Sci. Rep. 5:8674. doi:10.1038/srep08674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C. R., and Gregory K. E.. 1978. Inheritance of the horned, scurred, polled condition in cattle. J. Hered. 69:395–400. [Google Scholar]
- Mueller M. L., Cole J. B., Sonstegard T. S., and Van Eenennaam A. L.. 2018. Simulation of introgression of the polled allele into the Holstein breed via conventional breeding versus gene editing. Proceedings of the 11th World Congress of Genetics Applied to Livestock Production, vol. Technologies – Gene Editing, p. 755; Auckland, New Zealand. http://www.wcgalp.org/; http://www.wcgalp.org/proceedings/2018/simulation-introgression-polled-allele-holstein-breed-conventional-breeding-versus.
- NAAB 2018. Complete list of all active, foreign, genomic and limited status sires [accessed March 1, 2018]. http://www.naab-css.org/db/.
- Null D. J., Cole J. B., Sun C., and Van Raden P. M.. 2015. Assignment of polled status using single nucleotide polymorphism genotypes and predicted gene content. J. Dairy Sci. 98(Suppl. 2):800. (Abstr. 647) [Google Scholar]
- Pryce J. E., Hayes B. J., and Goddard M. E.. 2012. Novel strategies to minimize progeny inbreeding while maximizing genetic gain using genomic information. J. Dairy Sci. 95:377–388. doi:10.3168/jds.2011–4254 [DOI] [PubMed] [Google Scholar]
- Spurlock D. M., Stock M. L., and Coetzee J. F.. 2014. The impact of 3 strategies for incorporating polled genetics into a dairy cattle breeding program on the overall herd genetic merit. J. Dairy Sci. 97:5265–5274. doi:10.3168/jds.2013-7746 [DOI] [PubMed] [Google Scholar]
- Stafford K. J., and Mellor D. J.. 2011. Addressing the pain associated with disbudding and dehorning in cattle. Appl. Anim. Behav. Sci. 135:226–231. doi:10.1016/j.applanim.2011.10.018 [Google Scholar]
- Thompson N. M., Widmar N. O., Schutz M. M., Cole J. B., and Wolf C. A.. 2017. Economic considerations of breeding for polled dairy cows versus dehorning in the United States. J.Dairy Sci. 100:4941–4952. doi:10.3168/jds.2016-12099 [DOI] [PubMed] [Google Scholar]
- USDA 2009. Reference of dairy cattle health and management practices in the U.S [Accessed March 1, 2018]. https://www.aphis.usda.gov/animal_health/nahms/dairy/downloads/dairy07/Dairy07_dr_PartIV.pdf.