Abstract
Weaning of pigs can lead to low-feed intake resulting in a lag in growth performance, reduced gut health, and diarrheal diseases. Epidermal growth factor (EGF), the most abundant growth factor in milk, increased weaned pig BW gain and feed efficiency in our previous work. It is believed that intestinal microbiota plays an important role in gut health and pig growth, but limited data are available on the impact of feed additives, such as EGF, on the microbial communities of the intestines. The objective of the study was to investigate if the positive influence of EGF supplementation on weight gain and gut health was related to differences in intestinal microbiota. To examine the efficacy of EGF, a 21-d animal trial was performed using 72 pigs (two equal blocks of 36 pigs with three barrows and three gilts/pen). Pigs were assigned to one of two dietary treatments at weaning (20 ± 2 d of age; n = 6 pens/treatment) balancing across treatment for litter, gender, and initial BW. Recombinant yeast supernatant containing EGF at 120 μg/kg BW/d and without EGF (control) was added to the feed for 21 d, followed by a common diet for 7 d. Pig performance was measured weekly and ileal digesta was collected at day 21 from six pigs/treatment for microbiome analysis. Pigs fed diets containing EGF fermentation supernatant had greater (P = 0.01) daily gain in week 3 and overall resulting in heavier (P = 0.029) BW at day 28, which was consistent to our previous finding. No difference in alpha-diversity (Chao1, Shanon, and Simpson indices) and beta-diversity (weighted and unweighted UniFrac distances) of ileal digesta microbiota between EGF supplemented and control pigs were observed. The relative abundances of bacterial taxa did not differ among treatment groups at the phylum level. The relative abundances of Corynebacterium (0.0 vs. 0.9%), Blautia (0.003 vs. 0.26%), and Coprococcus (0.0 vs. 0.05%) genera, and Rumminococcaceae family (0.001 vs. 0.08%) were decreased (P < 0.05) in EGF group compared to control and were negatively correlated (P < 0.05, r > 0.60) with growth performance. Pathways related to detoxification and carbohydrate metabolism were differentially represented in the luminal bacterial populations. The improved growth of pigs supplemented with EGF supernatant produced by Pichia pastoris may be related to changes in functional capacity of the gut microbial populations. However, the impact on mucosa-associated or large intestinal communities is still unknown.
Keywords: epidermal growth factor, growth performance, microbiota, weaned pig
INTRODUCTION
Concern over antibiotic resistance is not a new topic. The European Union placed a ban on antibiotics in the mid 2000s (European Union, 2005), the United States transitioned away from antibiotic use under the veterinary feed directive as of 1 January 2017 (US FDA, 2013), and only prescription use of medically important antimicrobials used for livestock will be allowed as of 1 December 2018 in Canada (Government of Canada, 2018). It is estimated that 90% of all weaned pigs have had exposure to antimicrobials at some point during production (USDA, 2007; Deckert et al., 2010) and the weaning phase is the period of most likely exposure due to reduced disease resistance (Cromwell, 2002). As a result, much emphasis is being placed on management and feeding strategies for weaned pigs in order to ensure optimal performance without the use of antibiotics (Nyachoti and Heo, 2013). Nyachoti and Heo (2013) suggested that a three-pronged approach that includes management, nutrition, and feed additives will be necessary to maintain performance at levels achieved prior to the removal of antibiotics.
Epidermal growth factor (EGF) is the most abundant growth factor in milk (Odle et al., 1996) with receptors on the basolateral and luminal intestinal epithelial cells (Kelly et al., 1992) suggesting EGF is a suitable target feed additive to improve piglet health. We have previously demonstrated that a porcine EGF-secreting Lactococcus lactis increased weaned pig BW gain and feed efficiency and improved gut health through enhanced jejunal structure and reduced inflammatory index (Bedford et al. 2014, 2015). However, it is difficult to determine whether the degree of change in the intestinal structure is sufficient to account for the improvement in performance and thus another mechanism may also play a role in the observed response. It is generally acknowledged that intestinal microbiota play an important role in gut health and pig growth. Consequently, there has been a greater research emphasis placed on understanding the interaction between nutritional strategies, disease, overall animal health and the intestinal microbiota.
To further the application of EGF to production settings, we have recently generated a recombinant yeast that can produce porcine EGF at a concentration that is close to 10 times higher than that of L. lactis as a means to improve EGF production efficiency and reduce fermentation costs. The objectives of the study were to test the efficiency of the EGF-containing Pichia pastoris fermentation supernatant on weaned pig performance and to determine whether the positive influence on weight gain, efficiency, and gut health previously observed was related to differences in composition and functional capacity of the intestinal microbiota.
MATERIALS AND METHOD
The use of animals and all experimental procedures were approved by the University of Guelph’s Animal Care Committee in accordance with the Canadian Council on Animal Care guidelines (CCAC, 2009).
Production of EGF Supernatant
The expression vector designed to express porcine EGF (pJ912-EGF) was codon-optimized for optimal expression in P. pastoris and synthesized by DNA2.0 (Menlo Park, CA). Plasmid DNA was linearized with SwaI (New England Biolabs; Beverly, MA) and purified by phenol-chloroform extraction and ethanol precipitation. Purified linearized plasmid DNA was dissolved in 10 µL of nuclease-free H2O. Electrocompetent P. pastoris X-33 (Invitrogen; Carlsbad, CA) was prepared according to the manufacturer’s instructions. Competent cells (100 µL) were mixed with the dissolved purified DNA in a 0.4 cm gap width electroporation cuvette (BTX, San Diego, CA). Electroporation was carried out in an Electro Cell Manipulator 600 (BTX) at 1.5 kV. Transformed cells were plated on YPD agar (1% yeast extract (Difco, Detroit, MI), 2% peptone (Difco), 2% glucose (Fisher Scientific, Pittsburgh, PA, USA), 2% agar (Difco) supplemented with 1 M sorbitol and zeocin (100 µg/mL).
Positive transformant colonies were inoculated into a 500 mL flask containing 50 mL BMGY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate [pH 6.0], 1.4% yeast nitrogen base (Difco), 0.00004% biotin [Difco), 1% glycerol (Fisher Scientific) at 30 °C with shaking at 250 rpm. For high cell density fermentation, P. pastoris cultivation was carried out in BioFlo/CelliGen 115 benchtop bioreactor (New Brunswick Scientific, Edison, NJ). Supernatant samples were harvested at 48 h of methanol induction of EGF expression. To quantify the EGF concentration, both indirect ELISA and western blot methods were employed and the EGF titer was 180 ± 30 mg/L (Figure 1).
Figure 1.
Quantification of EGF secreted by Pichia pastoris. (A) Known concentrations of recombinant pig EGF were used to generate a standard curve to quantify EGF via indirect ELISA. (B) Western blot quantification of EGF in yeast supernatant (SN) after 28 h MeOH induction.
Western Blot and ELISA
Proteins in the fermentation supernatant were separated by 12% SDS-PAGE and then transferred to polyvinylidene difluoride (PVDF) membranes. After blocking in 5% skim milk at room temperature for 1 h, it was incubated with polyclonal rabbit primary antibody against pig EGF (1:1000 dilution; Biomatik, Cambridge, Ontario, Canada) at room temperature for 1 h. Membranes were then incubated in goat anti-rabbit IgG horseradish peroxidase-linked antibody (Cell Signaling Technology, Beverly, MA) at room temperature for 1 h, ECL Plus Western Blotting Detection System kit (Amersham, Piscataway, NJ) was used to detect EGF. The EGF level was also measured using Express ELISA kit for Rabbit Primary Antibodies (GenScript, Piscataway, NJ) according to manufacturer’s instructions. Recombinant human EGF (Sigma Chemical Co., St. Louis, MO) was used as the standard and measured at 450 nm.
Animals, Diets, and Treatments
A total of 72 pigs (two equal blocks of 36 pigs with three barrows and three gilts/pen) at 6.64 ± 0.29 kg BW were assigned to one of two dietary treatments at weaning (20 ± 2 d of age; n = 6 pens/treatment) balancing across treatment for litter, gender, and initial BW. Dietary treatments consisted of P. pastoris fermentation supernatant with EGF (EGF-PP) and P. pastoris fermentation supernatant without EGF (control). Supplementation of fermentation supernatant with or without EGF were included in phases I and II of a three-phase feeding program (Table 1), where phase I was fed for 1 wk and phase II for 2 wk. Based on previous work (Bedford et al., 2014, 2015), EGF supplementation is expected to stimulate intestinal development, thus a common phase III diet was provided to all pigs for one additional week to determine if the expected benefit lasted after EGF was withdrawn. Diets were formulated to meet or exceed NRC (2012) nutrient requirements for weaned pigs. Pigs in the EGF-PP group were given volumes of the fermentation product to achieve 120 µg EGF-containing supernatant/kg BW per day, based on analyzed EGF concentrations in the supernatant. The EGF calculation was based on the initial BW, BW-week 1, and BW-week 2 for entire wk 1, wk 2, and wk 3, respectively. The EGF concentration required in the supernatant per pen was prepared in a total volume of 50 mL and mixed with the feed before being fed. Control pigs were fed matching volumes of fermentation supernatant of P. pastoris without EGF. The fermentation products were mixed in with fresh feed at 9:00 a.m. daily and feed was replenished throughout the day. Feed refusals were removed weekly and weighed for determination of pen feed disappearance. Pigs had ad libitum access to feed and water.
Table 1.
Ingredient composition (as-fed) and nutrient analysis of a three-phase feeding program for newly weaned pigs supplemented with EGF*
| Item | Phase I | Phase II | Phase III |
|---|---|---|---|
| Ingredients (%) | |||
| Wheat | 20.25 | 20.20 | 15.00 |
| Corn | 14.52 | 26.23 | 48.85 |
| Oat groats | 7.50 | 5.00 | — |
| Fish meal | 7.50 | 5.00 | — |
| Soybean meal, dehulled (48% CP) | 20.00 | 25.00 | 30.00 |
| Soy protein concentrate | 2.50 | — | — |
| Whey | 20.80 | 12.85 | — |
| Lys-HCl | 0.28 | 0.34 | 0.35 |
| DL-methionine | 0.17 | 0.18 | 0.12 |
| Threonine | 0.14 | 0.16 | 0.14 |
| Tryptophan | 0.01 | 0.01 | — |
| Limestone | 0.75 | 0.90 | 1.17 |
| Monocalcium phosphate | 0.48 | 0.88 | 0.97 |
| Salt | 0.10 | 0.25 | 0.40 |
| AV fat blend/tallow | 4.50 | 2.50 | 2.50 |
| Vitamin and mineral mix† | 0.50 | 0.50 | 0.50 |
| Calculated nutrient content | |||
| DE (kcal/kg) | 3,654.34 | 3,534.92 | 3,530.14 |
| CP, % | 24.39 | 23.20 | 21.04 |
| Total Lys, % | 1.67 | 1.57 | 1.35 |
| SID Lys, % | 1.51 | 1.42 | 1.22 |
| Ca, % | 0.78 | 0.79 | 0.72 |
| P, % | 0.69 | 0.71 | 0.61 |
| Dig. P, % | 0.38 | 0.38 | 0.28 |
| Analyzed nutrient content | |||
| DM, % | 87.56 | 86.63 | 86.96 |
| CP, % | 22.58 | 21.79 | 20.36 |
| Ca, % | 0.85 | 0.83 | 0.73 |
| P, % | 0.74 | 0.72 | 0.63 |
*Phases I and II diets were fed for 7 and 14 d post-weaning, respectively, and supplemented with either Pichia pastoris fermentation supernatant containing EGF or P. pastoris fermentation supernatant without EGF. All pigs received a common Phase III diet for 7 d.
†Supplied per kg of complete diet: vitamin A, 10,000 IU in the form of retinyl acetate (2.5 mg) and retinyl palmitate (1.7 mg); vitamin D, 1,000 IU in the form of cholecalciferol; vitamin E, 56 IU in the form of DL-α-tocopherol acetate (44 mg);vitamin K, 2.5 mg in the form of menadione; choline, 500 mg; pantothenic acid,15 mg; riboflavin, 5 mg; folic acid, 2 mg; niacin, 25 mg; thiamine, 1.5 mg; vitamin B-6, 1.5 mg; biotin, 0.2 mg; vitamin B-12, 0.025 mg; Se, 0.3 mg in the form of Na2SeO3; Cu, 15 mg in the form of CuSO45H2O; Zn, 104 mg in the form of ZnO; Fe, 100 mg in the form of FeSO4; Mn, 19 mg in the form of MnO2; and I, 0.3 mg in the form of KI 2.
Animal Performance and Sample Collection
Pigs were weighed individually at the end of each week to determine ADG and weekly feed disappearance was determined to calculated ADFI. Feed efficiency as G:F was calculated from daily gain over daily intake. Previous studies indicated a positive response to EGF in the small intestine (Bedford et al., 2014; 2015); because the ileum has greater concentrations of microbial populations than more proximal small intestine sections (Gaskins, 2001), thus microbial populations in the ileum were evaluated. On day 21 post-weaning, one pig from each pen with a BW closest to the pen average was euthanized using an intracardial injection of pentobarbital sodium and phenytoin sodium (Euthansol, 0.3 mL/kg BW; Intervet Canada Corp., Kirkland, Quebec, Canada) for collection of ileal digesta and subsequent microbial composition analyses. The visceral cavity was opened for removal of the small intestine; the ileum was clamped at each end prior to removal to avoid spilling of digesta. The ileal digesta contents were collected in sterile tubes and kept on ice until being stored at −80 °C until further use.
Microbial Assessment
DNA extraction.
Approximately 200 mg of each digesta sample (n = 6 per treatment group) was used for genomic DNA extraction using ZR Fecal DNA MiniPrep (ZYMO Research, Irvine, CA), which included a bead-beating step for the mechanical lysis of the microbial cells. DNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA). DNA samples were normalized to 20 ng/µL, and quality checked by PCR amplification of the 16S rRNA gene using universal primers 27F (5′-GAAGAGTTTGATCATGGCTCAG-3′) and 342R (5′-CTGCTGCCTCCCGTAG-3′) as described (Khafipour et al., 2009). Amplicons were verified by agarose gel electrophoresis.
Library construction and illumina sequencing.
The V4 region of 16S rRNA gene was targeted for PCR amplification using a modified F338 and barcoded R806 primers (Caporaso et al., 2012) as described previously (Derakhshani et al., 2016). PCR reactions were performed in duplicate for each sample and contained 1.0 µL of prenormalized DNA, 1.0 µL of each forward and reverse primers (10 µM), 12 µL HPLC grade water (Fisher Scientific, Ottawa, Ontario, Canada), and 10 µL 5 Prime Hot MasterMix (5 Prime, Inc., Gaithersburg, MD). Reactions consisted of an initial denaturing step at 94 °C for 3 min followed by 30 amplification cycles at 94 °C for 45 s, 62 °C for 60 s, and 72 °C for 90 s; finalized by an extension step at 72 °C for 10 min in an Eppendorf Mastercycler pro (Eppendorf, Hamburg, Germany). PCR products were then purified using a ZR-96 DNA Clean-up Kit (ZYMO Research) to remove primers, deoxy-nucleotide-tri phosphates (dNTPs) and reaction components. The V4 library was then generated by pooling 200 ng of each sample, quantified by PicoGreen dsDNA (Invitrogen, Burlington, Ontario, Canada) and diluted to a final concentration of 5 pM, measured by Qubit 2.0 Fluorometer (Life technologies, Burlington, Ontario, Canada). To improve the unbalanced and biased base composition of the generated 16S rRNA gene libraries, 15% of PhiX control library was spiked into each amplicon pool. Customized sequencing primers for read 1 (5′-TATGGTAATTGTGTG CCAGCMGCCGCGGTAA-3′), read 2 (5′-AGTCAGTCAGCCGGAC TACHVGGG TWTCTAAT-3′), and index read (5′-ATTAGAWACCCBDGTA GTCCGGCTGA CTGACT-3′) were synthesized and purified by polyacrylamide gel electrophoresis (Integrated DNA Technologies, Coralville, IA) and added to the MiSeq Reagent Kit v3 (600-cycle; Illumina, San Diego, CA). The 300 paired-end sequencing reaction was performed on a MiSeq platform (Illumina) at the Gut Microbiome and Large Animal Biosecurity Laboratories, Department of Animal Science, University of Manitoba, Canada. All sequencing data are uploaded into the Sequence Read Archive of NCBI and are available through accession numbers SRR6491189-98.
Bioinformatic analysis.
The FLASH assembler (Magoč and Salzberg, 2011) was used to merge overlapping paired-end Illumina FASTQ files. Sequences with mismatches or ambiguous calls in the overlapping region were discarded. The output FASTQ file was then analyzed by downstream computational pipelines of the open source software package QIIME version 1.9.1 (Caporaso et al., 2010b). Assembled reads were demultiplexed according to the barcode sequences and exposed to additional quality-filters so that reads with ambiguous calls and those with phred quality scores (Q-scores) below 20 were discarded. Chimeric reads were filtered using UCHIME (Edgar et al., 2011) and sequences were assigned to operational taxonomic units (OTU) using the QIIME implementation of UCLUST (Edgar, 2010) at 97% pairwise identity threshold. Taxonomies were assigned to the representative sequence of each OTU using RDP classifier (Wang et al., 2007) and aligned with the Greengenes Core reference database (DeSantis et al., 2006) using PyNAST algorithms (Caporaso et al., 2010a).The phylogenetic tree was built with FastTree 2.1.3. (Price et al., 2010) for further comparisons between microbial communities.
Within-community diversity (alpha-diversity) was calculated by different indices of species richness and evenness including Shannon, Simpson, Chao1, observed number of species, goods coverage, and PD whole tree using the open source software QIIME. An even depth of 15,140 sequences per sample was used to calculate the richness and diversity indices for the ileal digesta, respectively. Beta-diversity was measured by calculating the weighted and unweighted UniFrac distances (Lozupone and Knight, 2005) using QIIME default scripts. Principal coordinate analysis was applied on resulting distance matrices to generated two-dimensional plots using PRIMER 7 software (Warwick and Clarke, 2006).
The open source software, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt, version 1.0.0) was used to predict metagenome function based on 16S rRNA gene sequences (Langille et al., 2013). For the PICRUSt analysis, closed-reference OTUs were picked at 97% similarity against the Greengenes database. The OTUs were normalized for predicted 16S rRNA copy number before predicting gene family abundance for each metagenome based on Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology groups (KOs) using Langille et al. (2013). The correlation plot was generated by R (3.3.1 version) with the corrplot package (Wei et al., 2016).
Statistical Analysis
Statistical analyses of the BW, daily gain, daily intake, and G:F data were performed using PROC MIXED and repeated measures analysis in SAS version 9.4 (SAS Inst. Inc., Cary, NC) with pen as the experimental unit and the main effect of diet, week, and diet by week interaction. Differences in alpha-diversity indices among treatment groups were calculated using GLIMMIX procedure (Tukey studentized range adjustment) with EGF-PP supplementation as fixed-effect and individual animal as random the variable. Permutational multivariate analysis of variance (PERMANOVA, Primer 7; Anderson, 2005) was used to calculate P values of UniFrac distances between EGF and control groups. Multivariate Association with Linear Models (MaAsLin) combined with GLIMMIX model analysis was used to analyze the difference in relative abundance of bacterial taxa at the phylum and genus levels between EGF-PP and control. Welch’s t-test was applied on gene function from PICRUSt outputs, percentage of the predicted metagenome made by a given KEGG functional module, using STAMP 2.0 to compare differences in functional capacity of microbiome between treatment groups. Correlations between bacterial taxa and growth performances were calculated by nonparametric Spearman’s rank correlation analysis using Prism 7. The differences between treatment groups were considered significant at P < 0.05.
RESULTS AND DISCUSSION
Growth Performance
Pigs fed diets containing EGF-PP were heavier (P < 0.04) at days 21 and 28 post wean, due to improved (P = 0.0002) daily gain and G:F in week 3 and no difference in feed disappearance (Table 2). BW, daily gain, and feed intake increased (P < 0.0001) with week post-wean (Table 2). There was an interaction between diet and week for daily gain. In control-fed pigs, daily gain increased (P < 0.001) on a weekly basis up to week 3 followed by a plateau in week 4. Alternatively, in EGF-PP fed pigs, gain increased (P < 0.0001) weekly up to week 3 followed by a decrease (P = 0.0005) in gain in week 4. Overall G:F was greater (P = 0.033) in pigs fed diets containing EGF-PP (0.695 vs. 0.639 ± 0.018). Despite the slight decrease in daily gain from weeks 3 to 4 in pigs fed EGF-PP, daily gain was equivalent in week 4 between groups, such that pigs fed EGF-PP remained 1 kg heavier at day 28. Feed additives targeted for improvement of gut health beyond the stomach must be retained through to the site of desired action, such as the small intestine in this study, in order to have an impact. Cheung et al. (2009) confirmed the presence of EGF in intestinal digesta following a period of EGF feeding. The improvement in growth performance is similar to that previously reported using EGF supernatant produced using L. lactis (Bedford et al., 2015) indicating retained efficacy of the recombinant EGF using a yeast-based production system. In addition, the control group similarly received P. pastoris fermentation broth; thus, the improved performance is due to EGF rather than potential active compounds supplied by the fermentation supernatant.
Table 2.
Growth performance of weaned pigs fed diets with or without EGF supernatant*
| Item | Treatment | SEM† | P value† | |||
|---|---|---|---|---|---|---|
| Control | EGF | Treatment | Week | Treatment * week | ||
| BW, kg | 0.26 | 0.029 | <0.0001 | 0.067 | ||
| Weaning | 6.68 | 6.59 | ||||
| Day 7 | 7.43 | 7.33 | ||||
| Day 14 | 9.39 | 9.48 | ||||
| Day 21 | 13.44a | 14.27b | ||||
| Day 28 | 17.30a | 18.38b | ||||
| Daily gain, g/d | 17 | 0.002 | <0.0001 | 0.044 | ||
| Week 1 | 103 | 105 | ||||
| Week 2 | 282 | 307 | ||||
| Week 3 | 578a | 676b | ||||
| Week 4 | 552 | 585 | ||||
| Daily feed intake, g/d | 33 | 0.449 | <0.0001 | 0.604 | ||
| Week 1 | 185 | 185 | ||||
| Week 2 | 351 | 348 | ||||
| Week 3 | 766 | 768 | ||||
| Week 4 | 1,292 | 1,366 | ||||
| G:F, g/g | 0.04 | 0.033 | <0.0001 | 0.319 | ||
| Week 1 | 0.56 | 0.58 | ||||
| Week 2 | 0.81 | 0.88 | ||||
| Week 3 | 0.75a | 0.88b | ||||
| Week 4 | 0.43 | 0.43 | ||||
*Phases I and II diets were fed for 7 and 14 d post-weaning, respectively, and supplemented with either Pichia pastoris fermentation supernatant containing EGF or P. pastoris fermentation supernatant without EGF (control). All pigs received a common phase III diet for 7 d.
†Data were analyzed as repeated measures. SEM represents SE for treatment * week lsmeans. Within a row, values with different superscript a,b differ P < 0.05.
Microbiota Analysis
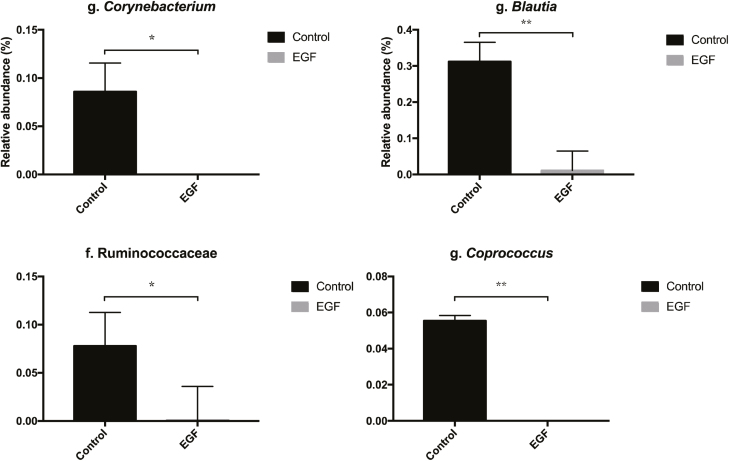
A total of 11 ileum digesta samples were bioinformatically analyzed. One of the control samples was eliminated from bioinformatics analyses due to low sequence reads. In total, 393,964 sequences were generated with an average of 35,815 sequences per sample. There was no difference in alpha-diversity of digesta samples between pigs fed diets containing EGF-PP or control (Table 3). Similarly, there was no separation among microbiota based on weighted and unweighted beta-diversity (Figure 2). There was no effect of EGF supernatant on relative abundances of bacteria phyla in the ileal digesta (Table 4). Firmicutes made up the dominant phyla in both groups representing 99.9 and 99.1 ± 0.8% of the community in pigs fed EGF-PP and control diets, respectively. Lactobacillus (88.3 and 83.8 ± 15.2%), Streptococcus (0.33 and 12.8 ± 10.3%), Clostridium (1.3 and 0.4 ± 0.6%), Sarcina (4.0 and 0.27 ± 3.3%) were the primary genera within the EGF-PP and control-fed pigs. The remaining 6% and 2% consisted of genera representing <1% relative abundance of the community (Table 4). There were four bacterial taxa whose relative abundances were higher (P < 0.05) in control-fed pigs compared to the EGF group (Corynebacterium, Blautia, and Coprococcus genus and Ruminococcaceae family) with each representing 0.10, 0.30, 0.06, and 0.05 %, respectively, of the community (Figure 3). In a longitudinal analysis of fecal bacterial populations in commercial swine herds, Blautia and Coprococcus were identified as part of the common intestinal bacterial community, present at abundances <5% declining in abundance with age (Kim et al., 2011). Species within the Corynebacterium (i.e. C. diptherium) have been associated with intestinal disease in humans and pigs (Percy et al., 1966; Bernard, 2012). Grow-finish pigs fed diets containing resistant starch had greater relative proportions of Coprococcus and Blautia in the cecum and colon that were positively correlated with carbohydrate metabolism (Sun et al., 2016). Digesta samples in the current study were collected at the end of phase II which represents proportional changes in diet ingredients that have been previously associated with impacting gut microbiota (Levesque et al., 2014). Thus, changes in microbial populations due to diet ingredients, which would be expected to be consistent across treatments due to the same base diet being fed, may have masked the impact of EGF supplementation which may be acting through direct contact with luminal bacteria (although less likely) or indirectly through improvement in overall intestinal health.
Table 3.
Alpha-diversity indices of ileal digesta from weaned pigs fed diets supplemented Pichia pastoris fermentation supernatant with or without EGF*
| Item | EGF | Control | Pooled SEM | P value |
|---|---|---|---|---|
| Shannon | 3.35 | 3.37 | 0.28 | 0.95 |
| Simpson | 0.76 | 0.76 | 0.05 | 0.96 |
| Observed species | 421.2 | 429.4 | 44.8 | 0.90 |
| Chao 1 | 1151.6 | 1050.8 | 99.84 | 0.49 |
| Goods coverage | 0.98 | 0.98 | 0.001 | 0.84 |
| PD whole tree | 7.15 | 11.30 | 2.45 | 0.26 |
*Diets were supplemented with the respective P. pastoris fermentation supernatant in phase I (days 0 to 7) and phase II (days 8 to 21). All pigs received a common diet in phase III (days 22 to 28). Ileal digesta was collected on day 28 post-wean.
Figure 2.
Principal coordinates analysis (PCoA) plots of unweighted (A) and weighted (B) UniFrac distances of microbiota of ileal digesta samples from pigs fed diets supplemented with either Pichia pastoris fermentation supernatant containing EGF or P. pastoris fermentation supernatant without EGF (control). For each comparison, the P value was obtained from PERMANOVA and considered significant for P <0.05.
Table 4.
Relative abundance of bacterial taxa in ileal digesta from weaned pigs fed diets supplemented Pichia pastoris fermentation supernatant with or without EGF*
| Taxa | EGF | Control | Pooled SEM | P value |
|---|---|---|---|---|
| p. Actinobacteria | 0.042 | 0.736 | 0.43 | 0.29 |
| g. Actinomyces | 0.010 | 0.222 | 0.09 | 0.24 |
| f. Actinosynnemataceae unclassified | 0.003 | 0.028 | 0.12 | 0.56 |
| g. Corynebacterium | 0.000a | 0.086b | 0.03 | 0.02 |
| f. Micrococcaceae unclassified | 0.004 | 0.178 | 0.05 | 0.22 |
| g. Saccharopolyspora | 0.001 | 0.024 | 0.02 | 0.44 |
| g. Streptomyces | 0.003 | 0.098 | 0.04 | 0.35 |
| g. Bifidobacterium | 0.018 | 0.045 | 0.03 | 0.71 |
| p. Bacteroidetes | 0.007 | 0.044 | 0.03 | 0.32 |
| g. Prevotella | 0.004 | 0.034 | 0.03 | 0.99 |
| p. Cyanobacteria | 0.001 | 0.041 | 0.04 | 0.31 |
| o. Streptophyta unclassified | 0.001 | 0.038 | 0.03 | 0.31 |
| p. Firmicutes | 99.94 | 99.08 | 0.77 | 0.29 |
| o. Bacillales unclassified | 0.001 | 0.151 | 0.04 | 0.44 |
| f. Gemellaceae unclassified | 0.001 | 0.037 | 0.03 | 0.11 |
| g. Lactobacillus | 88.30 | 83.80 | 15.21 | 0.77 |
| g. Weissella | 0.000 | 0.025 | 0.02 | 0.30 |
| g. Lactococcus | 0.012 | 0.638 | 0.57 | 0.30 |
| g. Streptococcus | 0.331 | 12.77 | 10.27 | 0.26 |
| o. Clostridiales unclassified | 0.000 | 0.025 | 0.02 | 0.27 |
| f. Clostridiaceae unclassified 1 | 5.771 | 0.423 | 4.39 | 0.26 |
| g. Clostridium | 1.278 | 0.036 | 0.06 | 0.92 |
| g. Sarcina | 4.048 | 0.272 | 3.28 | 0.28 |
| f. Clostridiaceae unclassified 2 | 0.129 | 0.009 | 0.10 | 0.25 |
| f. Lachnospiraceae unclassified | 0.011 | 0.114 | 0.06 | 0.26 |
| g. [Ruminococcus] | 0.001 | 0.046 | 0.02 | 0.17 |
| g. Blautia | 0.003a | 0.262b | 0.03 | 0.003 |
| g. Coprococcus | 0.000a | 0.055b | 0.02 | 0.003 |
| g. Dorea | 0.001 | 0.028 | 0.02 | 0.17 |
| g. Peptostreptococcus | 0.001 | 0.040 | 0.02 | 0.44 |
| f. Ruminococcaceae unclassified | 0.001a | 0.078b | 0.03 | 0.02 |
| g. Megasphaera | 0.009 | 0.030 | 0.03 | 0.44 |
| g. Veillonella | 0.010 | 0.017 | 0.01 | 0.46 |
| g. [Eubacterium] | 0.003 | 0.034 | 0.02 | 0.20 |
| g. Catenibacterium | 0.001 | 0.024 | 0.02 | 0.44 |
*The table shows the relative abundance of bacterial taxa at genus level (above 0.01%); some taxa could only be classified to family (f), order (o) or class (c) level. Within a row, values with different superscript a,b differ P < 0.05.
Figure 3.
Relative abundance of bacterial taxa in ileal digesta that significantly differed from pigs fed diets supplemented with either Pichia pastoris fermentation supernatant containing EGF or P. pastoris fermentation supernatant without EGF (control). Values are presented as least square means with SEM. *P < 0.05; **P < 0.01.
In the current study, presence of these four taxa were generally negatively associated with performance (Figure 4). Specifically, Coprococcus, Corynebacterium, and Blautia were negatively associated with BW in week 4 (P < 0.05). Coprococcus and Corynebacterium were also negatively associated with BW in week 3 (P < 0.05) and Ruminococcaceae was negatively correlated with feed disappearance (P < 0.05). While there did appear to be a relationship between minor changes in gut bacterial populations and performance, interpretation of correlation data, particularly given the very low abundance of these bacteria, should be made cautiously.
Figure 4.
Correlation between pig growth performances and relative abundance of the four ileal bacterial taxa including Corynebacterium, Blautia, Coprococcus, and Ruminococcaceae. The scale colors (Spearman’s ρ from −1 to +1) indicate whether the correlation is positive (blue colored circle) or negative (red colored circle) between the taxa and growth performances. The * symbol indicates a statistically significant correlation at P <0.05. iBW, BW.weeks 1, 2, 3, and 4 represent BW on days 0, 7, 14 and 28; daily gain (DG); weeks 1, 2, 3, 4 and overall represent ADG in weeks 1, 2, 3, 4, and all 4 weeks. Similarly, daily feed disappearance (DFD) weeks 1, 2, 3, 4, and overall and GF.weeks 1, 2, 3, 4, and overall represent average daily feed disappearance and G:F in weeks 1, 2, 3, 4, and all 4 weeks.
Prediction of Functional and Metabolic Capacity of Ileal Microbiota
A total of 328 level 3 KEGG pathways were predicted in the microbiome of the ileum digesta samples (Supplementary Table 2). Of these, four metabolic pathways were different (P < 0.05) between treatment groups (Figure 5). In pigs fed EGF-PP containing diets, the inositol phosphate metabolism, retinol metabolism, and drug and xenobiotics metabolism related to cytochrome P450 expression were underrepresented compared to the control. The role of these four pathways in explaining the observed growth performance is unclear. Inositol phosphates play a role in regulating cell proliferation and migration. Wahl et al. (1998) reported EGF-stimulated inositol triphosphate formation in cells overexpressing the EGF-receptor which appears opposite of the under-representation of inositol phosphate metabolic pathway in pigs fed EGF-containing diets in the current study. Alternatively, cytochromes P450 are involved in elimination of foreign chemicals from the body, as well as, the metabolism and inactivation of drugs. They are also required for metabolic activation of derivatives that can cause toxicity and cell death (Gonzalez, 2005). The lower predicted representation of drug and xenobiotic metabolism by cytochrome P450 may represent a positive gut metabolic shift. In an in vitro analysis, Ching et al. (1996) reported suppressed expression of constitutive cytochrome P450 2C11 mRNA in rat hepatocytes incubated with EGF.
Figure 5.
Fold change in the number of 16S rRNA sequences annotated to hierarchy level 3 KEGG pathways representing the predicted metabolic capacity of the ileum microbiome with significant differences (P < 0.05) between EGF and control. Weaned pigs were fed diets supplemented with either Pichia pastoris fermentation supernatant containing EGF or P. pastoris fermentation supernatant without EGF (control).
Production of EGF using a recombinant yeast improved weaned pig performance. The effects of EGF on intestinal health (and thus pig growth) may be loosely associated with the metabolic functional capacity of the gut microbiota rather than dramatic changes in the bacterial populations themselves. However, the impact on mucosa-associated bacteria or large intestinal bacterial populations is unknown.
Supplementary Material
Footnotes
The authors would like to acknowledge D. C. Wey and the research station staff for husbandry and assistance with experimental procedures. Research supported by funds from Ontario Ministry of Agriculture, Food and Rural Affairs (OMAFRA; Guelph, Ontario, Canada), AB Vista, The Natural Sciences and Engineering Research Council of Canada (Ottawa, Ontario, Canada), Ontario Pork (Guelph, Ontario, Canada), Ajinimoto Heartland Inc. (Chicago, IL), Swine Innovation Porc Canada, and Royal De Heus (Ede, The Netherlands)
LITERATURE CITED
- Anderson M. 2005. PERMANOVA: a FORTRAN computer program for permutational multivariate analysis of variance. New Zealand: Department of Statistics, University of Auckland; p. 24. [Google Scholar]
- Bedford A., Chen T., Huynh E., Zhu C., Medeiros S., Wey D., de Lange C., and Li J.. 2015. Epidermal growth factor containing supernatant enhances intestine development in early-weaned pigs in vivo: potential mechanism involved. J. Biotechnol. 174:9–19. doi:10.1016/j.biotec.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Bedford A., Huynh E., Fu M., Zhu C., Wey D., de Lange C., and Li J.. 2014. Growth performance of early-weaned pigs is enhanced by feeding epidermal growth factor-expressing Lactococcuslactis fermentation product. J. Biotechnol. 173:47–52. doi:10.1016/j.biotec.2014.01.012 [DOI] [PubMed] [Google Scholar]
- Bernard K. 2012. The genus Corynebacterium and other medically-relevant, coryneform bacteria. J. Clin. Microbiol. 50:3152–8. doi:10.1128/JCM.00796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canadian Council on Animal Care.. 2009. CCAC guidelines on: the care and use of farm animals in research, teaching and testing. Ottawa (Canada): Canadian Council on Animal Care. [Google Scholar]
- Caporaso J. G., Bittinger K., Bushman F. D., DeSantis T. Z., Andersen G. L., and Knight R.. 2010a. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi:10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Pena A. G., Goodrich J. K., Gordon J. I., et al. 2010b. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. doi:10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., Owens S. M., Betley J., Fraser L., Bauer M., et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6:1621–1624. doi:10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung Q. C. K., Yuan Z., Dyce P. W., Wu D., DeLange K., and Li J.. 2009. Generation of epidermal growth factor-expressing Lactococcus lactis and its enhancement on intestinal development and growth of early-weaned mice. Am. J. Clin. Nutr. 89:871–9. doi: 10.3945/ajcn.2008.27073. [DOI] [PubMed] [Google Scholar]
- Ching K. Z., Tenney K. A., Chen J., and Morgan E. T.. 1996. Suppression of constitutive cytochrome P450 gene expression y epidermal growth factor receptor ligands in cultured rate hepatocytes. Drug Metab. Dispos. 24:542–6. [PubMed] [Google Scholar]
- Cromwell G. 2002. Why and how antibiotics are used in swine production. Anim. Biotechnol. 13:7–27. doi:10.1081/ABIO-120005767. PMID:12212945 [DOI] [PubMed] [Google Scholar]
- Deckert A., Gow S., Rosengren L., Leger D., Avery B., Daignault D., Dutil L., Reid-Smith R., and Irwin R.. 2010. Canadian integrated program for antimicrobial resistance surveillance (CIPARS) farm program: results from finsiher surveillance. Zoon. Pub. Health. 57:71–84. doi: 10.111/j31863-2378.2010.01356.x.PMID:21083820 [DOI] [PubMed] [Google Scholar]
- Derakhshani H., Tun H. M., and Khafipour E.. 2016. An extended single-index multiplesed 16S rRNA sequencing for microbial community analysis on MiSeq illumina platform. J. Basic Micro. 56:321–6. doi: 10.1002/jobm.201500420. [DOI] [PubMed] [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K. T., Huber D. Dalevi P. Hu, and Andersen G. L.. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. doi:10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi:10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., and Knight R.. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi:10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Union.. 2005. Ban on antibiotics as growth promoters in animal feed enters into effect (183//2003/EC) [Online] [accessed 26 November 2016] http://europa.eu/rapid/press-release_IP-05-1687_en.htm.
- Gaskins H. R. 2001. Intestinal bacteri and their influence on swine growth. In: A. J. Lewis and L. L. Southern, editors, Swine nutrition. Boca Raton (FL): CRC Press; p. 585–608. [Google Scholar]
- Gonzalez F. J. 2005. Role of cytochromes P450 in chemical toxicity and oxidative stress: studies with CYP2E1. Mut. Res. 569:101–10. doi: 10.1016/j.mrfmmm.2004.04.021 [DOI] [PubMed] [Google Scholar]
- Government of Canada.. 2018. Responsible use of medically important antimicrobials in animals. [accessed 9 March 2018] https://www.canada.ca/en/public-health/services/antibiotic-antimicrobial-resistance/animals/actions/responsible-use-antimicrobials.html.
- Kelly D., McFayden M., King T. P., and Morgan P. J.. 1992. Characterization of autoradiographic localization of the epidermal growth factor receptor in the jejunum of neonatal weaned pigs. Reprod. Fertil. Dev. 4:183–191. doi:10.1071/RD9920183 [DOI] [PubMed] [Google Scholar]
- Khafipour E., Li S., Plaizier J. C., and Krause D. O.. 2009. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl. Environ. Microbiol. 75:7115–24. doi: 10.1128/AEM.00739-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. B., Boewicz K., White B. A., Singer R. S., Sreevatsan S., Tu A. J., and Isaacson R. E.. 2011. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet. Micro. 153:124–133. doi:10.1016/j.vetmic.2011.05.021 [DOI] [PubMed] [Google Scholar]
- Langille M. G., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., Clemente J. C., Burkepile D. E., Vega Thurber R. L., Knight R., et al. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31:814–821. doi:10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque C. L., Hooda S., Swanson K., and deLange C. F. M.. 2014. Alterations in ileal mucosa bacteria related to diet complexity and growth performance in young pigs. PLoS One 9:e108472. doi:10.1371/journal.pone.0108472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., and Knight R.. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. doi:10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T., and Salzberg S. L.. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi:10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan X. C., Tickle T. L., Sokol H., Gevers D., Devaney K. L., Ward D. V., Reves J. A., Shah S. A., LeLeiko N., Snapper S. B., et al. 2012. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13:R79. doi:10.1186/gb-2012-13-9-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyachoti M., and Heo J. M.. 2013. Feed additives and feeding strategies to replace antibiotics. Adv. Pork Prod. 24:123–128. [Google Scholar]
- NRC 2012. Nutrient Requirements of Swine. 11th rev. ed Natl. Acad. Press, Washington, DC. [Google Scholar]
- Odle J., Zijlstra R. T., and Donovan S. M.. 1996. Intestinal effects of milkborne growth factors in neonates of agricultural importance. J. Anim. Sci. 74:2509–2522. doi:10.2527/1996.74102509x [DOI] [PubMed] [Google Scholar]
- Percy D. H., Ruhnke H. L., and Soltys M. A.. 1966. A case of infectuous cystitis pyelonephritis of swine caused by Corynebacterium suis. Can. Vet. J. 7:291–291. [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., and Arkin A. P.. 2010. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi:10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Su Y., and Zhu W.. 2016. Microbiome-metabolome responses in the cecum and colon of pig to a high resistant starch diet. Front. Microbiol. 7:779. doi:10.3389/fmicb.2016.00779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- US FDA United States Food and Drug Administration 2013. Guidance for industry #209. The judicious use of medically important antimicrobial drugs in food-producing animals [accessed 26 November 2016] http://www.fda.gov/downloads/AnimalVeterinary/GuidanceComplianceEnforcement/GuidanceforIndustry/UCM216936.pdf.
- USDA United States Department of Agriculture.. 2007. Swine 2006. Part II: reference of swine health and health management practices in the United States, 2006 [accessed 26 November 2016] https://www.aphis.usda.gov/animal_health/nahms/swine/downloads/swine2006/Swine2006_dr_PartII.pdf.
- Wahl M. I., Nishibe S., Suh P. G., Rhee S. G., and Carpenter G.. 1998. Epidermal growth factor stimulates tyrosine phosphorylation of phospholipase C-II independently of receptor internalization and extracellular calcium. Proc. Nat. Acad. Sci. 86:1568–73. doi: 10.1073/pnas.86.5.1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick R., and Clarke K.. 2006. Primer 6. PRIMER-E Ltd, Plymouth, UK. [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., and Cole J. R.. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. doi:10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Simko V., and Wei M. T.. 2016. Package ‘corrplot’[J]. Statistician 56:316–324. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.