Abstract
Transmission of viruses from one species to another is not unusual in nature. Despite this, evolutionarily successful transmissions are rare. Such events can cause pandemics and are followed by host–virus coevolution procedures that can increase the fitness potential of viruses. In this perspective article, I recognize eight main types of trans-species viral transmission. I consider two of them as evolutionarily successful, explaining why coronavirus SARS-CoV-2 could be one of them.
Keywords: COVID-19, Mutation, Epidemic, Gene, Infection, Natural selection, Molecular evolution
Transmission of viruses from one species to another is not unusual in nature. Focusing on humans, many people all over the world are in close proximity to animals due to local customs or due to their way of life. One can suppose that single transmission events of animal viruses to humans (viral exposure in human bodies/cells by direct contact) are happening daily, on the order of millions of times. The vast majority of those events are not successful in causing infection. It takes 100s or 1000s of years for a virus to “gain” the right combination of mutations in order to insert in human cells and survive further through spreading in multiple individuals (Longdon et al. 2014). Alternatively, it is just a matter of poor lack, the right virus at the right time to transit to a new host. Newly emerged viruses need an appropriate host cell receptor, ability to proliferate in the cytoplasm, and a way to escape host immunity (Llorente García and Marsh 2020). Thinking evolutionarily, natural selection favors: viruses that successfully infect host’s cells, viruses that are minimally lethal, viruses that are highly contagious (Plauzolles et al. 2015). Maybe the most known examples of such cases in humans are the viruses causing the seasonal common colds (Greenberg 2016). On the other hand, emergence of successful viruses is a rare event. Rare but it happens, since millions of mutations and infections (or transmissions) are happening every day. Pandemic phenomena could be the first presentation of trans-species infections. Through pandemic events, viruses can increase their fitness and become minimally harmful (see below for explanation). SARS-CoV-2 (Huang et al. 2020) may be an example of an evolutionary successful transition. Perhaps, we are all now witnesses of the first manifestations of this virus–host coevolution.
Let’s now see what types of animal-to-human viral transmissions are possible. This is my personal view, under the concept of fitness and evolution. I recognize eight main possibilities, all are represented in Fig. 1: (1A) most animal–human contacts (millions or billions per day) do not result in any viral transmission. The most common reason is the absence of an appropriate receptor in human cells; thus, the virus will be destroyed by the human immune system; (1B) some animal viruses can occasionally infect humans, but symptoms are mild or none. Human-to-human transmission probability is limited or zero. Due to the characteristics of these infections, most of them are unknown to scientists; (1C) some animal viruses can occasionally infect humans, but symptoms are severe, even lethal. Human-to-human transmission probability is limited or zero (e.g., Rabies virus) (Brunker et al. 2018); (1D) these viral transitions are characterized by mild infections and low probability for human-to-human transmission (e.g., cowpox virus) (Pastoret et al. 2000); (1E) this type is similar with 1D, but the difference is that a percentage of the infections are lethal. The fatality percentage varies, depending on the viral species or strain (eg. MERS virus) (Zumla et al. 2015); (1F) this type is characterized by high probability for human-to-human transmission and mild infections (e.g., corona viruses causing common colds in humans, see below); (1G) this type is characterized by high probability for human-to-human transmission and high fatality rate (e.g., Ebola virus) (Malvy et al. 2019); and (1H) this type is characterized by high probability for human-to-human transmission and low fatality rate (e.g., SARS-CoV-2).
Fig. 1.
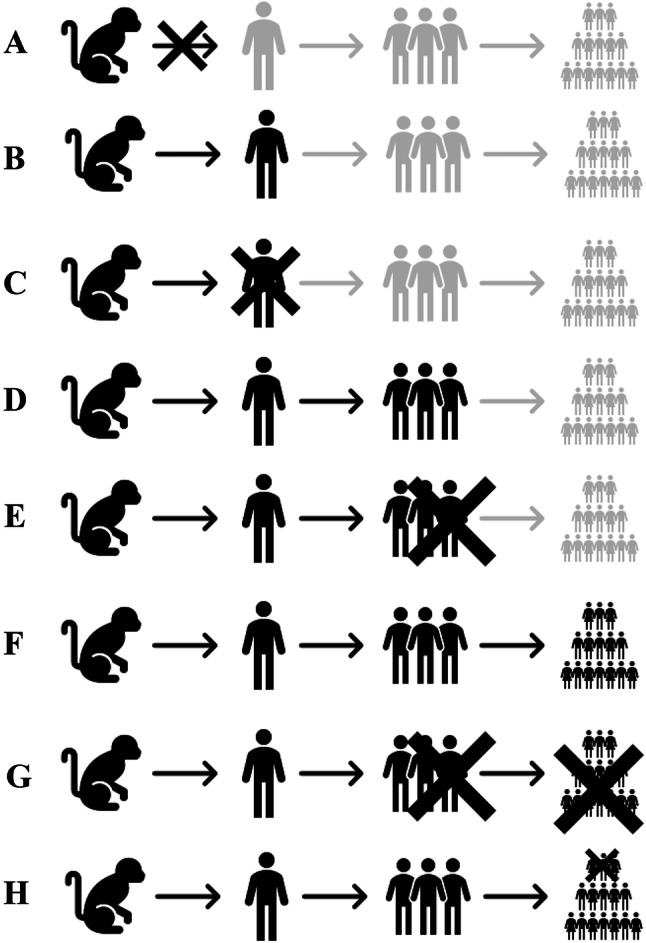
Animal to human viral transition types. See details in text
According to this classification, we could categorize SARS-CoV-2 as type 1H. In fact, we can consider that 1F and 1H are the most evolutionary successful viruses. SARS-CoV-2 is extremely contagious but fatality rate is low, around 3% (World Health Organization, March 3, 2020). If humans fail to eliminate this virus through vaccinations, then its fitness could be increased through years, its fatality rate will probably be reduced further, and we will finally have a type 1F virus. This could happen since many people that are genetically predisposed for severe infections will die and the genetic pool of human populations will be modified accordingly. This is the usual way that viruses increase their fitness (Fumagalli et al. 2010).
Successful animal-to-human transitions are rare events. Focusing on corona viruses, there are four known corona viruses that cause mild infections in humans (originated through animal-to-human transition events): HCoV-229E, HCoV-OC43, HCoV-HKU1, and HCoV-NL63 (Ye et al. 2020; Vijaykrishna et al. 2007). These four viruses are not older than 150 years (Ye et al. 2020; Vijaykrishna et al. 2007). SARS-CoV-2 may become the fifth one, if finally, it will be established in the human populations. Corona viruses seem to transit relatively easily from animals to humans, but they also disappear easily through evolutionary time. We can assume that from the fact that modern humans have been evolved the last 120–150 thousand years (Poznik et al. 2013). It is not clear why evolutionary successful viruses disappear from host populations. Two possible explanations are genetic decay of viruses’ genomes though mutation accumulation or some large-scale extinctions of host populations. We presently know only five corona viruses (including SARS-CoV-2) that have been successfully spread in human populations for the last 150 years. Obviously, coronaviruses like MERS (Zumla et al. 2015) and SARS-CoV-1 (Peiris et al. 2003) did not ever reach a fitness threshold in order to be established in human populations. Of course, human population and population density are increasing in Earth, so probabilities for successful transitions/transmissions are increasing even more.
The outbreak of SARS-CoV-2 reminded scientists of the need to learn more about the evolution of zoonotic diseases. The current pandemic can be a great opportunity to watch in real time the ongoing evolution of a probably evolutionarily successful animal-to-human viral transition. This is a valuable experience and a great source of knowledge for evolutionary biologists.
Acknowledgements
I thank my colleague Dr. Eirini Christaki for critical reading of the first draft of this paper.
Funding
Not applicable.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- Brunker K, Nadin-Davis S, Biek R. Genomic sequencing, evolution and molecular epidemiology of rabies virus. Rev Sci Tech. 2018;37:401–408. doi: 10.20506/rst.37.2.2810. [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Pozzoli U, Cagliani R, et al. Genome-wide identification of susceptibility alleles for viral infections through a population genetics approach. PLoS Genet. 2010 doi: 10.1371/journal.pgen.1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg SB. Update on human rhinovirus and coronavirus infections. Semin Respir Crit Care Med. 2016;37:555–571. doi: 10.1055/s-0036-1584797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente García I, Marsh M. A biophysical perspective on receptor-mediated virus entry with a focus on HIV. Biochim Biophys Acta-Biomembr. 2020 doi: 10.1016/j.bbamem.2019.183158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B, Brockhurst MA, Russell CA, et al. The evolution and genetics of virus host shifts. PLoS Pathog. 2014 doi: 10.1371/journal.ppat.1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvy D, McElroy AK, de Clerck H, et al. Ebola virus disease. Lancet. 2019;393:936–948. doi: 10.1016/S0140-6736(18)33132-5. [DOI] [PubMed] [Google Scholar]
- Pastoret PP, Bennett M, Brochier B, Akakpo AJ. Animals, public health and the example of cowpox. OIE Rev Sci Tech. 2000;12:23–32. doi: 10.20506/rst.19.1.1213. [DOI] [PubMed] [Google Scholar]
- Peiris JSM, Lai ST, Poon LLM, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plauzolles A, Lucas M, Gaudieri S. Influence of host resistance on viral adaptation: hepatitis C virus as a case study. Infect Drug Resist. 2015;8:63–74. doi: 10.2147/IDR.S49891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poznik GD, Henn BM, Yee MC, et al. Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females. Science. 2013;341:562–565. doi: 10.1126/science.1237619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D, Smith GJD, Zhang JX, et al. Evolutionary insights into the ecology of coronaviruses. J Virol. 2007;81:4012–4020. doi: 10.1128/JVI.02605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z-W, Yuan S, Yuen K-S, et al. Zoonotic origins of human coronaviruses. Int J Biol Sci. 2020;16:1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A, Hui DS, Perlman S. Middle east respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]


