Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has emerged in Chinese people in December 2019 and has currently spread worldwide causing the COVID-19 pandemic with more than 150,000 deaths. In order for a SARS-CoV like virus circulating in wild life for a very long time to infect the index case-patient, a number of conditions must be met, foremost among which is the encounter with humans and the presence in homo sapiens of a cellular receptor allowing the virus to bind. Recently it was shown that the SARS-CoV-2 spike protein, binds to the human angiotensin I converting enzyme 2 (ACE2). This molecule is a peptidase expressed at the surface of lung epithelial cells and other tissues, that regulates the renin-angiotensin-aldosterone system. Humans are not equal with respect to the expression levels of the cellular ACE2. Moreover, ACE2 polymorphisms were recently described in human populations. Here we review the most recent evidence that ACE2 expression and/or polymorphism could influence both the susceptibility of people to SARS-CoV-2 infection and the outcome of the COVID-19 disease. Further exploration of the relationship between the virus, the peptidase function of ACE2 and the levels of angiotensin II in SARS-CoV-2 infected patients should help to better understand the pathophysiology of the disease and the multi-organ failures observed in severe COVID-19 cases, particularly heart failure.
Keywords: COVID-19, SARS-CoV-2, Hypertension, Cardiac failure, ACE2
Introduction
Over the past 20 years, seven coronaviruses responsible for more or less severe respiratory diseases have emerged in humans. Several of them, including SARS-CoV-2 (a Betacoronavirus lineage b/Sarbecovirus), can cause patients lung injury and sometimes multi-organ failure with adverse myocardial remodeling, myocardial stress, and cardiomyopathy.1 , 2 Recently, SARS-CoV-2 was reported to be a human angiotensin I converting enzyme 2 (ACE2)-tropic virus3 , 4 able to bind the alveolar pneumocytes which express ACE2 at their surface.5 , 6 Yet, in humans the ACE2 mRNAs were found expressed in virtually all organs including the heart, blood vessels, kidney and testis, opening the possibility for this virus to infect other tissues beside lung.7 , 8 ACE2 is a known peptidase that regulates the renin-angioten-aldosterone system (RAAS), thus controlling blood pressure. Therefore, it is not surprising that initials reports suggested that hypertension, diabetes and cardiovascular diseases were the most frequent comorbidity in COVID-19 disease.9
The human coronaviruses
Coronaviruses (CoV) circulate in bats and generally pass over an intermediate animal host before crossing species barrier to infect humans.10 Different species of bats in China carry genetically diverse coronaviruses, some of which are direct ancestors of SARS-CoV.11, 12, 13 Indeed, the first SARS-CoV that caused a human outbreak derived from SARS-like CoV circulating in Chinese horseshoe Rhinolophus bats which apparently adapted to wild Himalayan palm-civet before spreading in humans.14 The MERS-CoV originated from a Pipistrellus bat CoV and was probably transmitted to humans through contact with infected camels.15, 16, 17 Soon after the first outbreak of SARS-CoV-2 in humans, it was reported that this new virus was related to a bat-borne coronavirus (BatCoV RaTG13) present in the Rhinolophus affinis bat species.18 The identification of an intermediate animal hosts has been the subject of intense research and it was claimed that a pangolin (Manis javanica) was the intermediate host for SARS-CoV-2.19 The SARS-CoV-2 receptor ACE2 from bat and pangolin and several other species, were found to resemble that of human.20
Before 2003, although human coronavirus 229E (HCoV-229E) (Alphacoronavirus) and HCoV-OC43 (Betacoronavirus lineage a) described in the 1960s were known to be agents of respiratory infections, they lent little attention. In the early 2000s, two other coronaviruses responsible for similar diseases were identified, the HCoV-NL63 (Alphacoronavirus) and HCoV-HKU1 (Betacoronavirus lineage a). Even if the health authorities pay little attention to these viruses, sometimes they can cause deaths in people with fragile health. A study in Switzerland reported that among 279 subjects who had bronchoalveolar lavage for investigation of respiratory symptoms, 29 were tested positive for HCoV (detection rate: 10.4%).21 A large-scale polymerase chain reaction (PCR) screening of 11,661 nasal samples from European patients with respiratory disease, found 35 HCoV-229E (0.30%), 61 HCoV-HKU1 (0.52%), 75 HCoV-NL63 (0.64%), and 111 HCoV-OC43 (0.85%).22 A similar study in Africa on 5573 nasal samples from child hospitalized for pneumonia found 114 HCoV-229E (2.05%), 163 HCoV-NL63 (2.93%), and 111 HCoV-OC43 (1.99%).23 Two Chinese studies involving almost 25,000 throat and nasal swab samples from patients with acute respiratory tract infections revealed 114 HCoV-229E (0.37%–0.57%), 61 HCoV-HKU1 (0.18%–0.33%), 104 HCoV-NL63 (0.33%–0.52%), and 523 HCoV-OC43 (1.36%–3.04%), respectively.24 , 25 The fatality rate of the coronaviruses causing the common winter cold was estimated 0.5%–1.5%.26
Coronaviruses strongly gained in notoriety when SARS-CoV (Betacoronavirus lineage b) emerged in China in March 2003 and was proven responsible for the severe acute respiratory syndrome (SARS) outbreak in humans.27 The SARS-CoV adapted to humans and became able to spread from person-to-person leading to a fatality rate of 9.6% in infected patients, causing global concern. The Middle East Respiratory Syndrome (MERS) caused by the MERS-CoV (Betacoronavirus lineage 2c), was reported in Saudi Arabia in 2012. This epidemic which has been one of the least deadly in absolute number of deaths, was the one which has created the most fears in health authorities and the most important panic in the populations due to its high fatality rate (case fatality rate of 34.7%).28 The SARS-CoV-2 that emerged in China at the end of 2019, is responsible for respiratory infections including pneumonia with a mortality rate estimated about 1%–2.5%,2 increasing with age and the existence of underlying diseases. Under chest computerized tomography (CT) scans, the majority of patients show bilateral ground glass-like opacities and subsegmental areas of consolidation indicative of SARS-CoV-2 induced pneumonia.
The MERS-CoV, SARS-CoV, SARS-CoV-2 and their cellular receptors
Already for SARS-CoV, it was demonstrated that this virus used the angiotensin I converting enzyme 2 (ACE2) to enter human cells.29 The novel Betacoronavirus SARS-CoV-2 (formerly 2019-nCoV), that cause COVID-19 disease, has 79.5% nucleotide identity with SARS-CoV.1 It is worth noting that HCoV-NL63, SARS-CoV and SARS-CoV-2 spike proteins bind ACE230 expressed at high levels in type I and II alveolar cells in the lung, whereas MERS-CoV bind the dipeptidyl peptidase 4 (DPP4)/CD26), a multifunctional serine peptidase known involved in T cell activation.31 The analysis of SARS-CoV-2 spike (S) protein and ACE2 three-dimensional (3-D) structures allowed identification of regions in the peptidase domain of ACE2 required for viral spike binding.3 Three very elegant papers published in the recent weeks characterized SARS-CoV-2 entry in target cells through interactions with ACE2 and serine protease TMPRSS2 priming as well as the 3-D structures involved in these interactions.3 , 32 , 33
The human monocarboxypeptidase ACE-2 was originally cloned from human heart failure and lymphoma cDNA libraries.7 Although the ACE2 gene is usually considered silent in immune cells, the expression of ACE2 mRNAs was reported in a subset of CD14+ CD16-human monocytes.34 ACE2 is also expressed by enterocytes of the small intestine and expected to regulate the expression of the gut antimicrobial peptides.35 Moreover, this peptidase is also present on the arterial and venous endothelial cells, and arterial smooth muscle.36 In normal human lung, the ACE2 protein is found on type I and II alveolar epithelial lung cells.37 High expression of ACE2 was also reported on the epithelial cells of oral mucosa.38 Single-cell RNA-seq analysis indicated that Asian men have a higher ACE2 mRNA expression in lung than women and that Asian people express higher amount of ACE2 than Caucasian and African American populations,39 but this observation remains controversial.40 Until recently, the genetic basis of ACE2 expression in different populations remained largely unknown.41
ACE2 structure and function
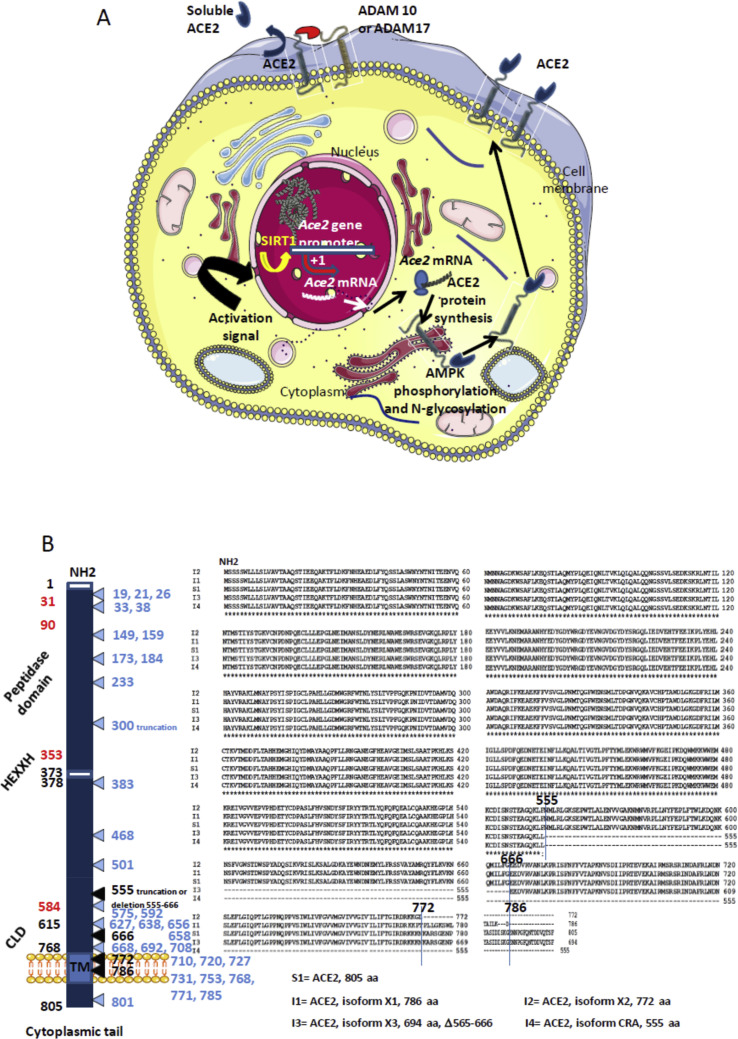
The ACE2 gene span 39.98 kb of genomic DNA and contains 18 exons. It maps to chromosome X at position Xp22.8 It encodes a type I cell-surface glycoprotein of about 100 kDa, composed by 805 amino acids and characterized by a N-terminal signal peptide of 17 amino acid residues, a peptidase domain (PD) (residues 19–615) with its HEXXH zinc binding metalloprotease motif, a C-terminal Collectrin (a regulator of renal amino acid transport and insulin)-like domain (CLD) (residues 616–768) that includes a ferredoxin-like fold “Neck” domain (615–726), that end with an hydrophobic transmembrane hydrophobic helix region of 22 amino acid residues followed by an intracellular segment of 43 amino acid residues.7 , 42 The histidine motif HEXXH identified as an important component in a wide variety of zinc-dependent metalloproteases consists of five residues, the first histidine followed by glutamic acid being conserved, then the two variable amino acids and a final histidine.43 Crystal structure analysis have suggested the presence of several hinge regions and N-glycosylations.44
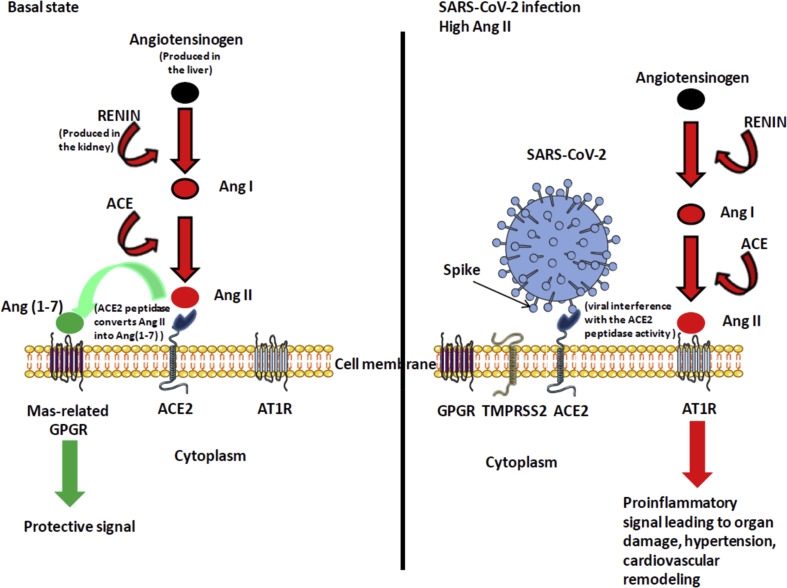
ACE2 belongs to the family of ACE members which have a wider tissues distribution. The juxtamembrane, transmembrane and cytoplasmic tail of ACE2 do not resemble ACE but these two proteins share the CLD region, a 220 amino-acid domain. Angiotensin converting enzymes (ACE) are zinc metallopeptidases. ACE, is a widely distributed protein of 170 kDa encoded by a 21 kb gene located on chromosome 17 (17q23),45 , 46 that converts the inactive decapeptide, angiotensin (Ang) I to an active vasoconstrictor octapeptide Ang II [Asp-Arg-Val-Tyr-Ile-His-Pro-Phe] that controls the blood pressure,47, 48, 49, 50 and through inactivation of bradykinin vasodilatator.51 AngII also triggers the release of aldosterone that regulates the capacity of kidney to absorb sodium and water.52 Moreover, Ang II stimulates DPP4 activity likely via the seven-transmembrane receptor (7TM) angiotensin II type A receptor (AT1R)-mediated transactivation of epidermal growth factor receptor53 and DPP4 inhibitors are described as a new class of anti-diabetic treatments the cardiovascular safety of which has been confirmed whereas their impact on hypertension is under evaluation.54 Ang II also mediates cell proliferation by stimulating various cytokines.55 ACE2, known for its diverse biological functions, including regulation of blood pressure through the renin-angiotensin-aldosterone system (RAAS), converts the octapeptide AngII to the heptapeptide Ang (1–7) by hydrolysis of the C-terminal residue. Ang (1–7) is expect to exert its action through the MAS-related (MAS1) G protein-coupled receptor (GPGR).56 , 57 In the pancreas ACE2 play an important glycemia-protective role.58 Low ACE2 expression in the kidney is also associated with progressive renal diseases including diabetic nephropathy.59 A soluble form of the catalytic ACE2 ectodomain can be released in the circulation following cleavage between amino acids 716 and 741 by sheddase ADAM10 and ADAM17.60, 61, 62. The transcriptional regulation of ACE2 is under the control of DNA-binding protein such as Sirtuin 1 (SIRT1).63 (Fig. 1 A).
Figure 1.
A. Schematic representation of the regulation of ACE2. The transcription of the Ace2 gene is under control of the SIRT1 DNA-binding protein that binds the Ace2 gene promotor. Post-transcriptional regulation by miRNA (miRNA143, miRNA421) could occur (not shown). Following translation the newly synthesized ACE2 proteins are likely target of post-transcriptional modifications such as phosporylation of Ser680 by AMPK that enhances the stability of ACE2, and N-glycosylations. Once expressed at the cell membrane the ACE2 protein can be regulated by sheddases (ADAM10, ADAM17) that cleave the ACE2 extracellular domain and release a circulating soluble form sACE2 capable to interact with integrins (ITGB1). B. Schematic representation (left) of the ACE2 molecule and its major domains. The amino acids position is in black. Some of the amino acids important for viral tropism are in red (previous studies showed that residues 31, 41, and regions 82–84 and 353–357 are important for viral spike binding). Clustal Omega multiple sequence alignment (EMBL-EBI bioinformatic tool; Copyright © EMBL 2020) of human ACE2 and its different isoforms (right). The comparison of the reference Homo sapiens ACE2 protein sequence (S1=Genbank: BAB40370.1) with 9 others ACE2 sequences from the NCBI reference sequence database (S2=UniProtKB Q9BYF1.2; S3=NCBI NP_001358344.1; S4=NCBI NP_068576.1; S5= GenBank EAW98892.1; S6= GenBank AAH48094.2; S7= GenBank AAH39902.1; S8= GenBank AAO25651.1; S9= GenBank BAD99267.1; GenBank AAF99721.1), showed 100% amino acids identity (not shown). The Clustal MSA was also used for the comparison of the human ACE2 S1 sequence and available sequences of ACE2 isoforms: the isoform X1 (I1) = NCBI XP_011543851.1; isoform X2 (I2) = NCBI XP_011543853.1; isoform X3(I3) = NCBI XP_011543854.1; isoform CRA (I4) = GenBank EAW98891.1. The figure illustrates that these isoforms correspond to deletions in the CLD domain, or truncations in the transmembrane domain. A very elegant work by Cao and colleagues42 has recently analyzed 1700 ACE2 variants in search of ACE2 protein polymorphism. The mutations and truncations found by this team are shown in light blue.
ACE2 polymorphism and diseases
ACE2 limits the adverse vasoconstrictor and profibrotic effects of AngII. The hydrolysis of AngII into Ang (1–7) reduces the oxidative stress of AngII on endothelial cerebral arteries.64 Ang (1–7) was reported to have vasodilatory and antifibrotic actions.65 Disruption of ACE2 results in increased AngII levels and impaired cardiac function.66 Reduced levels of cardiac ACE2 have been reported in hypertension (HT) and diabetic heart disease.67 , 68 Low expression of ACE2 mRNA was associated to HT, dyslipidemia and/or heart failure.69
A polymorphism of ACE2 gene was first documented in the Chinese population with three ACE2 variants (rs4240157, rs4646155, and rs4830542) associated with HT,70, 71, 72, 73, 74 in a Nicotine Dependence in Teens Canadian cohort rs2074192, rs233575, and rs2158083 mutations were significantly associated with pathological variations of blood pressure.75 ACE2 rs21068809 mutation (C > T) has been reported associated with clinical manifestations of HT.76 In Indian the study of 246 HT patients and 274 normotensive people indicated an association of HT with ACE2 rs21068809 mutation.77 In Brazilian patients, the combination of ACE I/D and ACE2 G8790A polymorphisms revels susceptibility to HT.78 The RAAS pathway can also be regulated by a polymorphism in ACE. In African-American with hypertension an ACE polymorphism was reported.79
Very recently, Cao and colleagues reported the results of a large investigation (1700 variants) of coding sequences variants in ACE2 and the allele frequency differences between populations in ACE2 gene from the China Metabolic Analytics Project and 1000 Genome Project database and other large scale genome databases.41 They found one variant with a truncation Gln300 in China. In addition, they reported 32 variants among which seven hotspot variants in different populations.
Viral ACE2 receptor polymorphism and coronaviruses infection
It remains possible that ACE2 gene polymorphism, human ACE2 mRNA expression and human ACE2 protein polymorphism influence SARS-CoV-2 susceptibility and COVID-19 disease outcome.
For more than two decades, in the field of the human immunodeficiency virus (HIV), a retrovirus transmitted by sexual intercourse, it was demonstrated that the binding of the gp120 viral envelope glycoprotein to the CD4 receptor,80 , 81 to CXCR482 , 83 or CCR5 coreceptor,84 triggers cell signaling. These molecules play a crucial role in the permanent molecular crosstalk between the cell and its environment. In this viral model, the study of the CCR5 co-receptor polymorphism clearly showed that a natural Δ32 deletion prevented the infection by HIV of homozygous people carrying this genotype.85 , 86 For the MERS-CoV, attachment of the spike (S) glycoprotein to human cells require the host cell typeII transmembrane protein dipeptidyl peptidase 4 (DPP4/CD26).87 , 88 Following interaction with DPP4, the S protein of MERS-CoV undergoes proteolytic activation through the cellular serine protease TMPRSS2 and cysteine protease cathepsin L once inside endosomes.89 Soluble forms of DPP4 can be released in the blood circulation after cleavage by the kallikrein-related peptidase 5 (KLK5).90 It was recently reported that among fourteen characterized mutants forms of DPP4, four polymorphisms (K267E, K267N, A291P and Δ346-348) strongly reduce the binding and penetration of MERS-CoV into target cells and the viral replication.91
Regarding SARS-CoV, the S1 domain of the spike protein mediates ACE2 receptor binding whereas the S2 domain is a membrane-associated portion that likely undergoes post-binding transconformational modifications allowing membrane fusion. The viral receptor binding domain (RBD) located in S1 has been narrowed down to amino acid residues 318 to 510.92 A co–crystal structure of ACE2 to the RBD revealed that residues 424 to 494 are involved in direct contact with the first α-helix and Lys353 and proximal residues at the N-terminus of β-sheet 5 of ACE2.93 By altering the His353 amino acid in rat ACE2 and modifying a glycosylation site (Asp 90) that may alter the conformation of the α-helix 1 of ACE2, Li and colleagues93 converted the rat ACE2 into an efficient receptor for SARS-CoV. A point mutation Leu584Ala in ACE2, markedly attenuated the shedding of the enzyme and facilitated SARS-CoV entry into target cells.61 A soluble form of ACE2 lacking the cytoplasmic and transmembrane domain of the molecule was reported capable of blocking binding of SARS-CoV spike protein to ACE2.94 Expression of ACE2 was found down regulated in cells infected by SARS-CoV.95 A recombinant SARS-CoV spike protein was found to down regulated ACE2 expression through release of sACE2 and thereby promotes lung injury.96 Among other antiviral effect of Chloroquine on SARS-CoV in vitro one could be attributable to a deficit in the glycosylation of the ACE2 virus cell surface receptor.97 , 98 Regarding the HCoV-NL63 that also employ ACE2 for cell entry a recombinant SARS-CoV/HCoV-NL63 spike protein trigger shedding of sACE2.99
Very recently, investigation of SARS-CoV-2 cell entry through ACE2 binding showed important commonalities between SARS-CoV and SARS-CoV-2 infection, including similar choice of entry receptors.32 SARS-CoV and SARS-CoV-2 share about 76% amino acids identity and most amino acid residues essential for ACE2 binding were conserved in the SARS-CoV-2 spike S1 domain. Another recent paper published reported the structural basis of SARS-CoV2 interaction with ACE2.3 The trimeric SARS-CoV-2 S1 spike binds the PD domain of ACE2 and the cleavage of ACE2 C-terminal segment (residues 697 to 716) by the transmembrane protease serine 2 (TMPRSS2) enhances the S-protein-driven viral entry. By comparing the 805 amino acid residues of the 10 human ACE2 proteins and the 4 different ACE2 isoforms available through GeneBank using Clustal Omega multiple sequence alignment, a 100% identity among the complete ACE2 sequences was observed and the isoforms corresponded to a deletion in the CLD domain, or truncation in the transmembrane domain. The role of these isoforms in SARS-CoV-2 infection and COVID-19 outcome, remains speculative. According to the recent work by Cao and colleagues,41 32 variants of ACE2 where characterized among which seven hotspot variants (Lys26Arg, Ile486Val, Ala627Val, Asn638Ser, Ser692Pro, Asn720Asp, and Leu731Ile/Phe) in different populations (Fig. 1B). This open the possibility that some people could be less susceptible to SARS-CoV-2 infection than others.
Discussion
ACE2 protein at the surface of lung alveolar epithelial cells allows infection of the respiratory tract with SARS-CoV-2. It can be hypothesized that the ACE2 levels correlate with susceptibility to SARS-CoV-2 infection. Apparently, men have a higher ACE2 expression in lung than women and Asian people express ACE2 higher than Caucasian and African American populations.37 This is in agreement with the finding that conversion of Ang II to Ang (1–7) by ACE2 was higher in males than female,100 suggesting an over-expression of ACE2 in men. Because ACE2 is encoded by a gene located on the X chromosome and men express more ACE2 than women it could be speculated that depending the allele expressed by women, they could be considered of lower sensitivity against the most severe adverse effects of the infection.99 , 101 All clinical reports published to date indicate that men represent between 66% and 75% of the most severe cases of COVID-19. During early SARS-CoV-2 infection and viral spread within body tissues, the ACE2 function is likely impaired either by steric hindrance of the peptidase domain of ACE2 following virus binding or by down regulation of ACE2 mRNA expression and ACE2 protein. In severe COVID-19 disease, the presence of the viral receptor on other tissues than lung may explain the multi-organ failure sometimes observed in clinic. We therefore suggest that quantification of ACE2 and AngII be added to the COVID-19 patients biological monitoring.
It is known that ACE2 can shift the RAAS balance by conversion of Ang II to Ang (1–7). Consequently, HT and COVID-19 recently become a question of concern for international professional societies of cardiology regarding: i) the susceptibility of patients with HT to get COVID-19; ii) the severity of the disease; and, iii) the use of ACE inhibitors (ACEi) and AngII receptor blockers (ARBs, that targets the AT1R). It is known that HT inhibitors increase the cell-surface expression of ACE2. It was demonstrated that ACEi can increase intestinal ACE2 mRNA expression.102 Although data are lacking regarding the effects of such drugs on ACE2 mRNA expression in lung epithelial cells, there is a concern that patients taking those treatments can favor virus capture. In patients with HT who received long-term olmesartan (ARB) treatment, urinary ACE2 levels were higher than among untreated control patients.103 In contrast to HT, in patients suffering from idiopathic pulmonary fibrosis, the expression levels of ACE2 are markedly decreased.30 ACE2 is a major actor toward resolution of inflammation and fibrosis.104 In an animal model of bleomycin-induced pulmonary fibrosis, treatment with intraperitoneal injection of recombinant human ACE2 improved the lung function and decreased lung inflammation and fibrosis.105 Moreover, impaired phosphorylation of ACE2 Ser680 by AMP-activated protein kinase in pulmonary endothelium leads to a labile ACE2 and hence pulmonary HT.106 We must also paid attention to molecules such as xanthenone (XNT) and dimiazene aceturate (DIZE, an anti-trypanosomal drug) described as ACE2 activators.107 In a rat model of ischemic heart disease, the subcutaneous infusion of DIZE significantly increased cardiac ACE2 mRNA expression and ACE2 protein catalytic activity, reduced ACE mRNA expression, and improved cardiac remodeling.108 The possible beneficial properties of other molecules such as exenatide (a glucagon-like peptide-1 agonist) which induces an increase in vasodilatory and a decrease in vasoconstrictive mediators must also be investigated.109 In addition, it was recently reported that heparin (anticoagulant) treatment is associated with decreased mortality in severe COVID-19 patients with coagulopathy.110
In a Chinese cohort of 1099 patients with COVID-19, 165 (13%) individuals were patients with HT, among which 24% suffered from severe COVID-19, a percentage of 3.7%, slightly higher to that of the general population of COVID-19 patients.111 In a smaller cohort of 191 patients with COVID-19, 58 (30%) were patients with hypertension and 48% of them died, which is surprisingly high percentage (14.6%).112 These results suggest that the prevalence of patients with HT was higher in patients who developed severe COVID-19 disease than those who do not. By mid march 2020, the international professional societies of cardiology recommended continuing patients' treatment.113 Indeed, when the SARS-CoV-2 spike binds its ACE2 receptor in the α helix 1 (Lys31, Tyr 41) and β5 region (Lys353) it likely reduces the catalytic properties of ACE2 that is usually associated with reduced inflammation. The lack of Ang (1–7) generation may increase lung injury114 , 115 and cardiovascular risks, Ang II acting like an inflammatory cytokine.116 In a murine model it was observed that lung inflammation aggravates AngII-induced induced abdominal aortic aneurysms.117
Mutations might modify the expression level of ACE2 protein as shown in a murine model.118 The deletion of ACE2 in mice model was associated with increased circulation and tissue AngII levels and led to cardiovascular damage.119 , 120 It remain possible that i) mutations affecting the human ACE2 gene; ii) transcriptional variation in ACE2 mRNA expression; iii) post-transcriptional modifications that act on the ACE2 viral receptor (such as N-glycosylation), and; iv) putative ACE2 protein mutations, may influence the outcome of COVID-19 by acting on blood pressure through the RAAS and possible increasing of lung and heart damages through the oxidative stress triggered by Ang II. Recently an high rate fatality of SARS-CoV-2 was reported in Iran,121 without satisfactory explanation. If underreporting of the number of infected people can be excluded, it could be hypothesized: i) a more aggressive variant clade of SARS-CoV-2122; ii) a variation in ACE2; or, iii) a variation in genes like those encoding Toll-like receptors. Since it is known that Ang (1–7) prevents inflammation by inhibiting the resistin/Toll-like receptor 4 (TLR4)/MAPK/NF-kB pathway123 and that there is a high variability of the TLR4 gene in different ethnic groups in Iran,124 it remains possible that SARS-CoV-2 triggers increased inflammation in Iranian patients by suppressing the ACE2-mediated metabolism of AngII to Ang (1–7). This could be related to the observation that mice deficient in the TLR3/TLR4 adaptor TRIF are highly susceptible to SARS-CoV infection including severe inflammatory induction.125
The mechanism of acute myocardial injury caused by SARS-CoV-2 during severe COVD-19 disease might be related to the inhibition of ACE2 catalytic activity.126 (Fig. 2 ). Interestingly, a recent study posted as a pre-print paper127 pointed out a list of 97 approved drugs that may have a therapeutic potential against COVID-19 including anti-diabetics (metaformin), statins (simvastatin) and ARBs (sartans). Medical records of patients currently treated with these compounds may help to identify whether those drugs have a beneficial or adverse effect on COVID-19 patients. Metaformin was also identified as a potential drug-repurposing against SARS-CoV-2 in another study.128 It should be remembered that a large number of data suggest that there is a mild or severe cytokine storm in severe COVID-19 patients which is an important cause of death. To reduce the pro-inflammatory effect of AngII and the cytokine storm observed in severe cases of COVID-19, it might make sense to continue treating patients with ACE inhibitors and ARBs, a conclusion shared by recent recommendations of the international societies of cardiology.129 However, Fang and colleagues115 recently reported that the most distinctive comorbidities in patients who died from COVID-19 are HT, coronary heart diseases, cerebrovascular diseases and diabetes, and among them several were treated by ACE inhibitors. How should clinicians navigate this uncertainty for patients who are taking ACE inhibitors and ARBs and become infected with SARS-CoV-2? Do these molecules have a harmful effect in the outcome of the disease or is the link that is made highlights only a confounding factor which confirms that HT is a major factor of comorbidity? In agreement with others,130, 131, 132, 133 we consider that it is of special importance to rapidly evaluate whether these drugs are more beneficial than harmful in severe COVID-19 patients.
Figure 2.
Simplified diagram of the renin-angiotensin system in normal and pathologic conditions. The left panel indicates that ACE2 converts Ang II to Ang (1–7) leading to protective signal. The right panel illustrates the possible dysfunction of signals when SARS-CoV-2 is attached to its ACE2 receptor. Under this condition Ang (1–7) is no longer synthetized, Ang II accumulates and binds AT1R, leading to proinflammatory signals that trigger both tissues damage (in particular lung and heart) and hypertension.
Funding
This work was supported by the French Government under the « Investissements d'avenir » (Investments for the Future) program managed by the Agence Nationale de la Recherche (French ANR: National Agency for Research), (reference: Méditerranée Infection 10-IAHU-03).
Ethical approval
Not required.
Declaration of Competing Interest
CD declare a link of interest with the Sanofi and Merck pharmaceutical companies. JMR and DR declare that they have no competing interests.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020 doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu Y., Zhao Y.B., Wang Q., Li J.Y., Zhou Z.J., Liao C.H. Predicting the angiotensin converting enzyme 2 (ACE2) utilizing capability as the receptor of SARS-CoV-2. Microb Inf. 2020 doi: 10.1016/j.micinf.2020.03.003. Pre-proof. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. bioRxiv. 2020 doi: 10.1101/2020.01.26.919985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang P.H., Cheng Y. Increasing host cellular receptor—angiotensinconverting enzyme 2 (ACE2) expression by coronavirus may facilitate 2019-nCoV infection. bioRxiv. 2020 doi: 10.1101/2020.02.24.963348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N. A novel angiotensin-converting enzyme-related carboxypeptidase. (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 8.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 9.Bavishi C., Maddox T.M., Messerli F.H. Coronavirus disease 2019 (COVID-19) infection and renin angiotensin system blockers. JAMA Cardiol. 2020 doi: 10.1001/jamacardio.2020.1282. Published online April 3, 2020. [DOI] [PubMed] [Google Scholar]
- 10.Afelt A., Devaux C.A., Serra-Cobo J., Frutos R. Bats, bat-borne viruses, and environmental changes. IntechOpen. 2018:113–131. [chapter 8] [Google Scholar]
- 11.Hu B., Ge X., Wang L.F., Shi Z. Bat origin of human coronaviruses. Virol J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25(1):35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afelt A., Frutos R., Devaux C. Bats, coronaviruses, and deforestation: toward the emergence of novel infectious diseases? Front Microbiol. 2018;9:702. doi: 10.3389/fmicb.2018.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci USA. 2005;102(7):2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q., Qi J., Yuan Y., Xuan Y., Han P., Wan Y. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16(3):328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabir J.S.M., Lam T.T.Y., Ahmed M.M.M., Li L., Shen Y., Abo-Aba S.E.M. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351(6268):81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 17.Anthony S.J., Gilardi K., Menachery V.D., Goldstein T., Ssebide B., Mbabazi R. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. MBio. 2017;8(2) doi: 10.1128/mBio.00373-17. e00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu P., Chen W., Chen J.P. Viral metagenomics revealed Sendai virus and coronavirus infection of Malayan pangolins (Manis javanica) Viruses. 2019;11(11):979. doi: 10.3390/v11110979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luan J., Lu Y., Jin X., Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem Biophys Res Commun. 2020 doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garbino J., Crespo S., Aubert J.D., Rochat T., Ninet B., Deffernez C. A prospective hospital-based study of the clinical impact of non–severe acute respiratory syndrome (Non-SARS)–Related human coronavirus infection. Clin Inf Dis. 2006;43:1009–1015. doi: 10.1086/507898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaunt E.R., Hardie A., Claas E.C.J., Simmonds P., Templeton K.E. (2010). Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48(8):2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiyula P.K., Agoti C.N., Munywoki P.K., Njeru R., Bett A., Otieno J.R. Human coronavirus NL63 molecular epidemiology and evolutionary patterns in rural coastal Kenya. J Inf Dis. 2018;217(11):1728–1739. doi: 10.1093/infdis/jiy098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng Z.Q., Chen D.H., Tan W.P., Qiu S.Y., Xu D., Liang H.X. Epidemiology and clinical characteristics of human coronaviruses OC43, 229E, NL63, and HKU1: a study of hospitalized children with acute respiratory tract infection in Guangzhou, China. Eur J Clin Microbiol Inf Dis. 2018;37:363–369. doi: 10.1007/s10096-017-3144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S.F., Tuo J.L., Huang X.B., Zhu X., Zhang D.M., Zhou K. Epidemiology characteristics of human coronaviruses in patients with respiratory infection symptoms and phylogenetic analysis of HCoV-OC43 during 2010-2015 in Guangzhou. PLoS One. 2018;13(1):e0191789. doi: 10.1371/journal.pone.0191789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi W.I. Comparison of the clinical characteristics and mortality of adults infected with human coronaviruses 229E and OC43. Res Square. 2019 doi: 10.21203/rs.2.15496/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 28.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 29.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X., Molina-Molina M., Abdul-Hafez A., Uhal V., Xaubet A., Uhal B.D. Angiotensin converting enzyme-2 is protective but downregulated in human and experimental lung fibrosis. Am J Physiol Lung Cell Mol Physiol. 2008;295:L178–L185. doi: 10.1152/ajplung.00009.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner L., Klemann C., Stephan M., von Hörsten S. Unravelling the immunological roles of dipeptidyl peptidase 4 (DPP4) activity and/or structure homologue (DASH) proteins. Clin Exp Immunol. 2016;184(3):265–283. doi: 10.1111/cei.12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:1–10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutkowska-Zapala M., Suski M., Szatanek R., Lenart M., Weglarczyk K., Olszanecki R. Human monocyte subsets exhibit divergent angiotensin I-converting activity. Clin Exp Immunol. 2015;181:126–132. doi: 10.1111/cei.12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashimoto T., Perlot T., Rehman A., Trichereau J., Ishiguro H., Paolino M. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Zurich Open Rep Arch (Univ Zurich) 2012 doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamming I., Timens W., Bulthuis M., Lely T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS Coronavirus. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun P., Lu X., Xu C., Sun W., Pan B. Understanding of COVID-19 based on current evidence. J Med Virol. 2020 doi: 10.1002/jmv.25722. Published online February 25, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of orla mucosa. Int J Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. bioRxiv. 2020 doi: 10.1101/2020.01.26.919985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai G. 2020. Tobacco-use disparity in gene expression of ACE2, the receptor of 2019-nCov. Preprint at. [DOI] [Google Scholar]
- 41.Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Disc. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H., Wada J., Hida K., Tsuchiyama Y., Hiragushi K., Shikata K. Collectrin, a collecting duct-specific transmembrane glycoprotein, is a novel homolog of ACE2 and is developmentally regulated in embryonic kidneys. J Biol Chem. 2001;276(20):17132–17139. doi: 10.1074/jbc.M006723200. [DOI] [PubMed] [Google Scholar]
- 43.Jongeneel C.V., Bouvier J., Bairoch A. A unique signature identifies a family sequence, which could have better zinc-binding activity than of zinc-dependent metallopeptidases. FEBS Letters. 1989;242:211–214. doi: 10.1016/0014-5793(89)80471-5. [DOI] [PubMed] [Google Scholar]
- 44.Watermeyer J.M., Sewell B.T., Scwager S.L., Natesh R., Corradi H.R., Acharya K.R. Structure of testis ACE glycosylation mutants and evidence for conserved domain movement. Biochemistry. 2006;45(42):12655–12663. doi: 10.1021/bi061146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howard T.E., Shai S.Y., Langford K.G., Martin B.M., Bernstein K.E. Transcription of testicular angiotensin-converting enzyme (ACE) is initiated within the 12th intron of the somatic ACE gene. Mol Cell Biol. 1990;10:4294–4302. doi: 10.1128/mcb.10.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sayed-Tabatabaei F.A., Oostra B.A., Isaacs A., van Duijn C.M., Witteman J.C.M. ACE polymorphism. Circ Res. 2006;98:1123–1133. doi: 10.1161/01.RES.0000223145.74217.e7. [DOI] [PubMed] [Google Scholar]
- 47.Erdos E.G., Skidgel R.A. The angiotensin I-converting enzyme. Lab Invest. 1987;56:345–348. [PubMed] [Google Scholar]
- 48.Ferrario C.M., Chappell M.C., Tallant E.A., Brosnihan K.B., Diz D.I. Counterregulatory actions of angiotensin-(1–7) Hypertension. 1997;30:535–541. doi: 10.1161/01.hyp.30.3.535. [DOI] [PubMed] [Google Scholar]
- 49.Oudit G.Y., Crackower M.A., Backx P.H., Penninger J.M. The role of ACE2 in cardiovascular physiology. Trends Cardiovasc Med. 2003;13:93–101. doi: 10.1016/s1050-1738(02)00233-5. [DOI] [PubMed] [Google Scholar]
- 50.Turner A.J. Exploring the structure and function of zinc metallopeptidases: old enzymes and new discoveries. Biochem Soc Trans. 2003;31:723–727. doi: 10.1042/bst0310723. [DOI] [PubMed] [Google Scholar]
- 51.Jaspard E., Wei I., Alhenc-Gelas F. Differences in the properties and enzymatic specifities of the two active sites of angiotensin I-converting enzyme (kininase II). Studies with bradykinin and other natural peptides. J Biol Chem. 1993;268:9496–9503. [PubMed] [Google Scholar]
- 52.Brewster U.C., Perazella M.A. The renin-angiotensin-aldosterone system and the kidney disease. Am J Med. 2004;116:263–272. doi: 10.1016/j.amjmed.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 53.Aroor A., Zuberek M., Duta C., Meuth A., Sowers J.R., Whaley-Connell A. Angiotensin II stimulation of DPP4 activity regulates megalin in the proximal tubules. Int J Mol Sci. 2016;17(5):780. doi: 10.3390/ijms17050780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawase H., Bando Y.K., Nishimura K., Aoyama M., Monji A., Murohara T. A dipeptidyl peptidase-4 inhibitor ameliorates hypertensive cardiac remodeling via angiotensin-II/sodium-proton pump exchanger-1 axis. J Mol Cell Cardiol. 2016;98:37–47. doi: 10.1016/j.yjmcc.2016.06.066. [DOI] [PubMed] [Google Scholar]
- 55.Carluccio M., Soccio M., De Caterina R. Aspects of gene polymorphisms in cardiovascular disease : the renin-angiotensin system. Eur J Clin Invest. 2001;31:476–488. doi: 10.1046/j.1365-2362.2001.00839.x. [DOI] [PubMed] [Google Scholar]
- 56.Santos R.A., Simoes e Silva A.C., Maric C., Silva D.M.R., Machado R.P., de Buhr I. Angiotensin-(1-7) is an endogenous ligand for the G protein-coupled receptor Mas. Proc Natl Acad Sci U S A. 2003;100:8258–8263. doi: 10.1073/pnas.1432869100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karnik S.S., Singh K.D., Tirupula K., Unal H. Significance of angiotensin 1-7 coupling with MAS1 receptor and other GPGRs to the renin-angiotensin system: IUPHAR Review 22. Br J Pharmacol. 2017;174(9):737–753. doi: 10.1111/bph.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pedersen K.B., Chhabra K.H., Nguyen V.K., Xia H., Lazartigues E. The transcription factor HNF1α induces expression of angiotensin-converting enzyme 2 (ACE2) in pancreatic islets from evolutionarily conserved promoter motifs. Biochim Biophys Acta. 2013;(11):1829. doi: 10.1016/j.bbagrm.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reich H.N., Oudit G.Y., Penninger J.M., Scholey J.W., Herzenberg A.M. Decreased glomerular and tubular expression of ACE2 in patients with type 2 diabetes and kidney disease. Kidney Int. 2008;74(12):1610–1616. doi: 10.1038/ki.2008.497. [DOI] [PubMed] [Google Scholar]
- 60.Jia H.P., Look D.C., Tan P., Shi L., Hickey M., Gakhar L. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am J Physiol Lung Cell Mol Physiol. 2009;297(1):L84–L96. doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao F., Zimpelmann J., Agaybi S., Gurley S.B., Puente L., Burns K.D. Characterization of angiotensin-converting enzyme 2 ectodomain shedding from mouse proximal tubular cells. PLoS One. 2014;9(1):e85958. doi: 10.1371/journal.pone.0085958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Devaux C.A., Mezouar S., Mege J.L. The E-cadherin cleavage associated to pathogenic bacteria infections can favor bacterial invasion and transmigration, dysregulation of the immune response and cancer induction in humans. Front Microbiol. 2019;10:2598. doi: 10.3389/fmicb.2019.02598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patel V.B., Zhong J.C., Grant M.B., Oudit G.Y. Role of the ACE2/angiotensin 1–7 axis of the renin-angiotensin system in heart failure. Circ Res. 2016;118(8):1313–1326. doi: 10.1161/CIRCRESAHA.116.307708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pena Silva R.A., Chu Y., Miller J.D., Mitchell I.J., Penninger J.M., Faraci F.M. Impact of ACE2 deficiency and oxidative stress on cerebrovascular function with aging. Stroke. 2012;43(12):3358–3363. doi: 10.1161/STROKEAHA.112.667063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tallant E.A., Clark M.A. Molecular mechanisms of inhibition of vascular growth by angiotensin-(1-7) Hypertension. 2003;42:574–579. doi: 10.1161/01.HYP.0000090322.55782.30. [DOI] [PubMed] [Google Scholar]
- 66.Crackower M.A., Sarao R., Oudit G.Y., Yagil C., Kozieradzki I., Scanga S.E. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 67.Diez-Freire C., Vazquez J., Correa de Adjounian M.F., Ferrari M.F.R., Yuan L., Silver X. ACE2 gene transfer attenuates hypertension-linked pathophysiological changes in the SHR. Physiol Genom. 2006;27:12–19. doi: 10.1152/physiolgenomics.00312.2005. [DOI] [PubMed] [Google Scholar]
- 68.Tikellis C., Pickering R., Tsorotes D., Du X.J., Kiriazis H., Nguyen-Huu T.P. Interaction of diabetes and ACE2 in the pathogenesis of cardiovascular disease in experimental diabetes. Clin Sci (Lond) 2012;123:519–529. doi: 10.1042/CS20110668. [DOI] [PubMed] [Google Scholar]
- 69.Velkoska E., Patel S.K., Burrell L.M. Angiotensin converting enzyme 2 and diminazene: role in cardiovascular and blood pressure regulation. Curr Opin Nephrol Hypertens. 2016;25:384–395. doi: 10.1097/MNH.0000000000000254. [DOI] [PubMed] [Google Scholar]
- 70.Yi L., Gu Y.H., Wang X.L., An L.Z., Xie X.D., Shao W. Association of ACE, ACE2 and UTS2 polymorphisms with essential hypertension in Han and Dongxiang populations from North-Western China. J Int Med Res. 2006;34:272–283. doi: 10.1177/147323000603400306. [DOI] [PubMed] [Google Scholar]
- 71.Niu W., Qi Y., Hou S., Zhou W., Qiu C. Correlation of angiotensin-converting enzyme 2 gene polymorphisms with stage 2 hypertension in Han Chinese. Transl Res. 2007;150:374–380. doi: 10.1016/j.trsl.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 72.Fan X.H., Wang Y.B., Wang H., Sun K., Zhang W.L., Song X.D. Polymorphisms of angiotensin-converting enzyme (ACE) and ACE2 are not associated with orthostatic blood pressure dysregulation in hypertensive patients. Acta Pharmacol Sin. 2009;30:1237–1244. doi: 10.1038/aps.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Y.Y., Liu D., Zhang P., Zhong J.C. Impact of ACE2 gene polymorphism on antihypertensive efficacy of ACE inhibitors. J Hum Hypertens. 2016;30:766–771. doi: 10.1038/jhh.2016.24. [DOI] [PubMed] [Google Scholar]
- 74.Luo Y., Liu C., Guan T., Li Y., Lai Y., Li F. Association of ACE2 genetic polymorphisms with hypertension-related target organ damages in south Xinjiang. Hypertens Res. 2019;42(5):681–689. doi: 10.1038/s41440-018-0166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malard L., Kakinami L., O'Loughlin J., Roy-Gagnon M.H., Labbe A., Pilote L. The association between the angiotensin-converting enzyme-2 gene and blood pressure in a cohort study of adolescents. BMC Med Genet. 2013;14:117. doi: 10.1186/1471-2350-14-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Q., Tang X., Yu C.Q., Chen D.F., Tian J., Cao Y. Correlation of angiotensin-converting enzyme 2 gene polymorphism with antihypertensive effects of benazepril. Beijing Da Xue Xue Bao. 2010;42:293–298. [PubMed] [Google Scholar]
- 77.Patnaik M., Pati P., Swain S.N., Mohaprata M.K., Dwibedi B., Kar S.K. Association of angiotensin-converting enzyme and angiotensin-converting enzyme-2 gene polymorphisms with essential hypertension in the population of Odisha, India. Ann Hum Biol. 2014;41:143–150. doi: 10.3109/03014460.2013.837195. [DOI] [PubMed] [Google Scholar]
- 78.Pinheiro D.S., Santos R.S., Veiga Jardim P.C.B., Silva E.G., Reis A.A.S., Pedrino G.R. The combination of ACE I/D and ACE2 G8790A polymorphisms revels susceptibility to hypertension: a genetic association study in Brazilian patients. PLoS One. 2019;14(8):e0221248. doi: 10.1371/journal.pone.0221248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duru K., Farrow S., Wang J.M., Lockette W., Kurtz T. Frequency of a deletion polymorphism in the gene for angiotensin converting enzyme is increased in african-Americans with hypertension. Am J Hypertens. 1994;7(8):759–762. doi: 10.1093/ajh/7.8.759. [DOI] [PubMed] [Google Scholar]
- 80.Benkirane M., Jeang K.T., Devaux C. The cytoplasmic domain of CD4 plays a critical role during the early stages of HIV infection in T-cells. EMBO J. 1994;13(23):5559–5569. doi: 10.1002/j.1460-2075.1994.tb06893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Briant L., Coudronnière N., Robert-Hebmann V., Benkirane M., Devaux C. Binding of HIV-1 virions or gp120-anti-gp120 immune complexes to HIV-1-infected quiescent peripheral blood mononuclear cells reveals latent infection. J Immunol. 1996;156(10):3994–4004. [PubMed] [Google Scholar]
- 82.Biard-Piechaczyk M., Robert-Hebmann V., Richard V., Roland J., Hipskind R.A., Devaux C. Caspase-dependent apoptosis of cells expressing the chemokine receptor CXCR4 is induced by cell membrane-associated human immunodeficiency virus type 1 envelope glycoprotein. Virology. 2000;268(2):329–344. doi: 10.1006/viro.1999.0151. [DOI] [PubMed] [Google Scholar]
- 83.Roland J., Murphy B., Robert-Hebmann V., Ahr B., Delauzun V., Nye K. Role of the intracytoplasmic domains of CXCR4 in SDF-1 mediated signaling. Blood. 2003;101:399–406. doi: 10.1182/blood-2002-03-0978. [DOI] [PubMed] [Google Scholar]
- 84.Alkhatib G., Locati M., Kennedy P.E., Murphy P.M., Berger E.A. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology. 1997;234:340–348. doi: 10.1006/viro.1997.8673. [DOI] [PubMed] [Google Scholar]
- 85.Liu R., Paxton W.A., Choe S., Ceradini D., Martin S.R., Horuk R. Homozygous defect in HIV-1 corecptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 86.Samson M., Libert F., Doranz B.J., Rucker J., Liesnard C., Farber C.M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 87.Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23(8):986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shirato K., Kawase M., Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol. 2013;87(23):12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nargis T., Kumar K., Ghosh A.R., Sharma A., Rudra D., Sen D. KLK5 induces shedding of DPP4 from circulatory Th17 cells in type 2 diabetes. Mol Metabolism. 2017;6(2017):1529–1539. doi: 10.1016/j.molmet.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kleine-Weber H., Schroeder S., Krüger N., Prokscha A., Naim H.Y., Müller M.A. Polymorphisms in dipeptidyl peptidase 4 reduce host cell entry of Middle East respiratory syndrome coronavirus. Emerg Microb Infect. 2020;9(1):155–168. doi: 10.1080/22221751.2020.1713705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Babcock G.J., Esshaki D.J., Thomas W.D., Jr., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lambert D., Yarski M., Warner F.J., Thornhill P., Parkin E.T., Smith A.I. Tumor necrosis factor- convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J Biol Chem. 2005;280(34):30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuba K., Imai Y., Rao S., Goa H., Guo F., Guan B. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Glowacka I., Bertram S., Herzog P., Pfefferle S., Steffen I., Muench M.O. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J Virol. 2010;84(2):1198–1205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Burrell L.M., Harrap S.B., Velkoska E., Patel S.K. The ACE2 gene: its potential as a functional candidate for cardiovascular disease. Clin Sci (Lond) 2013;124(2):65–76. doi: 10.1042/CS20120269. [DOI] [PubMed] [Google Scholar]
- 100.Gwathmey T.M., Shaltout H.A., Nixon P.A., O'Shea T.M., Rose J.C., Washburn L.K. Gender differences in urinary ACE and ACE2 activities in adolescents. FASEB J. 2008;22(1):940. [Google Scholar]
- 101.White M.C., Fleeman, Arnold A.C. Sex differences in the metabolic effects of the renin-angiotensin system. Biol Sex Differ. 2019;10:31. doi: 10.1186/s13293-019-0247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vuille-dit-Bille R.N., Camargo S.M., Emmenegger L., Sasse T., Kummer E., Jando J. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids. 2015;47:693–705. doi: 10.1007/s00726-014-1889-6. [DOI] [PubMed] [Google Scholar]
- 103.Furuhashi M., Moniwa N., Mita T., Fuseya T., Ishimura S., Ohno K. Urinary angiotensinconverting enzyme 2 in hypertensive patients may be increased by olmesartan, an angiotensin II receptor blocker. Am J Hypertens. 2015;28:15–21. doi: 10.1093/ajh/hpu086. [DOI] [PubMed] [Google Scholar]
- 104.Marshall R.P., Gohlke P., Chambers R.C., Howell D.C., Bottoms S.E., Unger T. Angiotensin II and the fibroproliferative response to acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2004;286:L156–L164. doi: 10.1152/ajplung.00313.2002. [DOI] [PubMed] [Google Scholar]
- 105.Rey-Parra G.J., Vadivel A., Coltan L., Hall A., Eaton F., Schuster M. Angiotensin converting enzyme 2 abrogates bleomycin-induced lung injury. J Mol Med (Berl) 2012;90(6):637–647. doi: 10.1007/s00109-012-0859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang J., Dong J., Martin M., He M., Gongol B., Marin T.L. AMP-activated protein kinase phosphorylation of angiotensin-converting enzyme 2 in endothelium mitigates pulmonary hypertension. Am J Respir Crit Care Med. 2018;198(4):509–520. doi: 10.1164/rccm.201712-2570OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Velkoska E., Patel S.K., Burrell L.M. Angiotensin converting enzyme 2 and diminazene: role in cardiovascular and blood pressure regulation. Curr Opinion. 2016;25(5):384–395. doi: 10.1097/MNH.0000000000000254. [DOI] [PubMed] [Google Scholar]
- 108.Qi Y., Zhang J., Cole-Jeffrey C.T., Shenoy V., Espejo A., Hanna M. Diminazene aceturate enhances angiotensin-converting enzyme 2 activity and attenuates ischemia-induced cardiac pathophysiology. Hypertension. 2013;62:746–752. doi: 10.1161/HYPERTENSIONAHA.113.01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chaudhuri A., Ghanim H., Makdissi A., Green K., Abuaysheh S., Batra M. Exenatide induces an increase in vasodilatory and a decrease in vasoconstrictive mediators. Diabetes Obes Metabol. 2017;19:729–733. doi: 10.1111/dom.12835. [DOI] [PubMed] [Google Scholar]
- 110.Tang N., Bai H., Chen X., Gong J., Li D., Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J Thromb Haemost. 2020 Mar 27 doi: 10.1111/jth.14817. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guan W., Ni Z., Hu Y., Liang W.H., Ou C.Q., He J.X. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult in patients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30566-3. Published March 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sparks M., Hiremath S. 2020. The Coronavirus Conundrum: ACE2 and hypertension edition. NephJC.http://www.nephjc.com/news/covidace2 Up-date march 17. [Google Scholar]
- 114.Forrester S.J., Booz G.W., Sigmund C.D., Coffman T.M., Kawai T., Rizzo V. Angiotensin II signal transduction: an update on mechanisms of physiology and pathophysiology. Physiol Rev. 2018;98(3):1627–1738. doi: 10.1152/physrev.00038.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet. 2020 doi: 10.1016/S2213-2600(20)30116-8. Published online March 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brasier A.R., Recinos A., III, Eledrisi M.S. Vascular inflammation and the renin-angiotensin system. Arterioscler Thromb Vasc Biol. 2002;22:1257–1266. doi: 10.1161/01.atv.0000021412.56621.a2. [DOI] [PubMed] [Google Scholar]
- 117.Liu C.L., Wang Y., Liao M., Wemmelund H., Ren J., Fernandes C. Allergic lung inflammation aggravates angiotensin II-induced abdominal aortic aneurysms in mice. Arterioscler Thromb Vasc Biol. 2016;36(1):69–77. doi: 10.1161/ATVBAHA.115.305911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wysocki J., Ye M., Soler M.J., Gurley S.B., Xiao H.D., Bernstein K.E. ACE and ACE2 activity in diabetic mice. Diabetes. 2006;55:2132–2139. doi: 10.2337/db06-0033. [DOI] [PubMed] [Google Scholar]
- 119.Yamamoto K., Ohishi M., Katsuya T., Ito N., Ikushima M., Kaibe M. Deletion of angiotensin-converting enzyme 2 accelerates pressure overload-induced cardiac dysfunction by increasing local angiotensin II. Hypertension. 2006;47:718–726. doi: 10.1161/01.HYP.0000205833.89478.5b. [DOI] [PubMed] [Google Scholar]
- 120.Rabelo L.A., Todiras M., Nunes-Souza V., Qadri F., Szijarto I.A., Gollasch M. Genetic deletion of ACE2 induces vascular dysfunction in C57BL/6 mice: role of nitric oxide imbalance and oxidative stress. PLoS One. 2016;11:e0150255. doi: 10.1371/journal.pone.0150255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Johns Hopkins University Coronavirus resource center. https://coronavirus.jhu.edu/map.html
- 122.Eden J.S., Rockett R., Carter I., Rahman H., de Ligt J., Hadfield J. An emergent clade of SARS-CoV-2 linked to returned travellers from Iran. bioRxiv. 2020 doi: 10.1101/2020.03.15.992818. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sousa Santos S.H., Oliveira Andrade J.M., Rodrigues Fernandes L., Sinisterra R.D.M., Sousa F.B., Feltenberger J.D. Oral Angiotensin-(1–7) prevented obesity and hepatic inflammation by inhibition of resistin/TLR4/MAPK/NF-κB in rats fed with high-fat diet. Peptides. 2013;46:47–52. doi: 10.1016/j.peptides.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 124.Ioana M., Ferwerda B., Farjadian S., Ioana L., Ghaderi A., Oosting M. High variability of TLR4 gene in different ethnic groups in Iran. Innate Immun. 2012;18(3):492–502. doi: 10.1177/1753425911423043. [DOI] [PubMed] [Google Scholar]
- 125.Totura A.L., Whitmore A., Agnihothram S., Schaefer A., Katze M.G., Heise M.T. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio. 2015;6(3) doi: 10.1128/mBio.00638-15. e00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zheng Y.Y., Ma Y.T., Zhang J.Y., Xie X. COVID-19 and the cardiovascular system. Nat Rev Cardiol. 2020 doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nabirotchkin S., Peluffo A.E., Bouaziz J., Cohen D. 2020. Focusing the unfolded protein response and autophagy related pathways to reposition common approved drugs against COVID-19. Preprints, 2020030302. [DOI] [Google Scholar]
- 128.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug repurposing. bioRxiv. 2020 doi: 10.1101/2020.03.22.002386. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.COVID-19 Resource center Cardiology societies recommend patients taking ACE inhibitors, ARBs who contract COVID-19 should continue treatment. https://www.healio.com/cardiology/vascular-medicine/news/online/%7Bfe7f0842-aecb-417b-9ecf-3fe7e0ddd991%7D/cardiology-societies-recommend-patients-taking-ace-inhibitors-arbs-who-contract-covid-19-should-continue-treatment
- 130.Fang L., Karakiulakis G., Roth M. Antihypertensive drugs and risk of COVID-19 ? Lancet Resp Med. 2020 doi: 10.1016/S2213-2600(20)30159-4. Published Online March 26, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Patel A.B., Verma A. COVID-19 and angiotensin-converting enzyme inhibitors and angiotensin receptor blockers: what is the evidence? JAMA. 2020 doi: 10.1001/jama.2020.4812. Published Online Mar 24; [DOI] [PubMed] [Google Scholar]
- 132.Vaduganathan M., Vardeny O., Michel T., McMurray J.J.V., Pfeffer M.A., Solomon S.D. Renin-angiotensin-aldosterone system inhibitors in patients with covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.South A.M., Tomlinson L., Edmonston D., Hiremath S., Sparks M.A. Controversies of renin-angiotensin system inhibition during the COVID-19 pandemic. Nat Rev Nephrol. 2020 doi: 10.1038/s41581-020-0279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]