Abstract
Background
Leprosy has been treated with multidrug therapy, which has been distributed for free across the globe and regarded as highly efficient. However, the impossibility of growing Mycobacterium leprae in axenic media has historically impaired assessments of M. leprae resistance, a parameter only recently detectable through molecular methods.
Methods
A systematic, population-based search for M. leprae resistance in suspected leprosy relapse cases and contacts was performed in Prata Village, an isolated, hyperendemic, former leprosy colony located in the Brazilian Amazon. Results led to an extended active search involving the entire Prata population. Confirmed leprosy cases were investigated for bacterial resistance using a combination of in vivo testing and direct sequencing of resistance genes folP1, rpoB, and gyrA. A molecular epidemiology analysis was performed using data from 17 variable number tandem repeats (VNTR).
Results
Mycobacterium leprae was obtained from biopsies of 37 leprosy cases (18 relapses and 19 new cases): 16 (43.24%) displayed drug-resistance variants. Multidrug resistance to rifampicin and dapsone was observed in 8 relapses and 4 new cases. Single resistance to rifampicin was detected in 1 new case. Resistance to dapsone was present in 2 relapses and 1 new case. Combined molecular resistance and VNTR data revealed evidence of intra-familial primary transmission of resistant M. leprae.
Conclusions
A comprehensive, population-based systematic approach to investigate M. leprae resistance in a unique population revealed an alarming scenario of the emergence and transmission of resistant strains. These findings may be used for the development of new strategies for surveillance of drug resistance in other populations.
Keywords: leprosy, M. leprae, multidrug resistance, primary resistance, transmission
Our study, in a unique population, revealed the highest proportion of resistant Mycobacterium leprae isolates to single and multiple drugs ever reported and demonstrated intra-familial transmission of resistant M. leprae: immediate, strong attention to drug resistance in leprosy is suggested.
Over the past 20 years, the global number of new cases of leprosy has remained stable, irrespective of available effective treatment, suggesting that better prophylactic and diagnostic tools are necessary to improve disease control and achieve reduced incidence rates; in this scenario, maintaining high therapeutic efficacy is of critical importance [1].
The first cases of Mycobacterium leprae secondary and primary drug resistance (DR) to dapsone (DDS) were reported in 1964 [2] and 1977 [3], respectively; the first case of resistance to rifampicin (RIF) was described in 1976 [4]. In 1981, the World Health Organization (WHO) recommend multidrug therapy (MDT) against leprosy, composed by DDS, RIF, and clofazimine [5]; in 1996, the first case of primary multidrug resistance (MDR) was reported [6], followed by the description of the first case of MDR to DDS, RIF, and ofloxacin [7]. Clofazimine resistance has hardly ever been detected in M. leprae, possibly due to the absence of a homolog for the Rv0678 efflux pump found in M. tuberculosis [6, 8, 9]
The inability of M. leprae to grow in artificial media has been a major limitation for the detection of DR/MDR in leprosy: the classic Shepard’s method, based on bacterial growth in the mouse footpad, is a labor-intensive and time-consuming procedure [10]. In the 1990s, molecular tools became available for sequencing drug resistance–determining regions (DRDR) of M. leprae genes folP1, rpoB, and gyrA, associated with resistance to DDS, RIF, and ofloxacin, respectively [11]. Consequently, there has been an increase in the number of sporadic case reports of DR and MDR isolates of M. leprae [11–14]. In 2009, the WHO launched a drug resistance surveillance program restricted to references and sentinel centers, which focused on pretreated individuals rediagnosed with leprosy [15].
To date, reports of M. leprae resistance rates range from 2.05% (of 243 isolates in Colombia) [16] to 16% (of 24 isolates in Guinea-Conakry) [17]. An Indian study of 239 relapses and 11 new cases found 21.6% of cases to be DR and 6.8% to be MDR [18]. Finally, a recent, large study of 1932 M. leprae strains obtained between 2009 and 2015 in sentinel centers of 19 countries determined that 8.0% (154) of the isolates presented some degree of resistance. Primary and secondary resistance to RIF were 2.0% (16/789) and 5.2% (58/1143), respectively; 20 cases (1.0%) were resistant to both DDS and RIF [19, 20].
Here, we present a population-based study on M. leprae resistance to MDT, performed in a hyperendemic population of a former leprosy colony located in the Brazilian Amazon. We propose that DR/MDR contributes to the maintenance of the endemicity in the village through reactivation/relapse and primary drug–resistant disease.
SUBJECTS AND METHODS
Ethics Statement
The ethics committees for human research of all involved institutions approved all methods and procedures used in this study (protocol number PUCPR 274.776) and written informed consent was obtained from all participants. The animal experiments were conducted in accordance with the guidelines of the Brazilian Committee for Care and Use of Laboratory Animals (protocol number ILSL 07/1).
Target Population
This study involved the entire population of the Vila do Santo Antonio do Prata (Prata village), located in the Amazonic state of Pará, Brazil. In the 1920s, the village became a venue for compulsory isolation of leprosy patients who were diagnosed across the northern and northeastern states of Brazil. Isolation was mandatory until 1962; however, the strong social stigma associated with leprosy has been very present and, to date, emigration of affected individuals from the village and immigration of nonaffected individuals to the village has been limited. As a result, the Prata Village remains mostly socially and geographically isolated and, despite efforts towards disease control, leprosy is still highly prevalent and homogenously distributed across the entire community [21].
At the time of enrollment, the village had an estimated population of around 3000 individuals, mainly composed of descendants of founding, leprosy-affected individuals. A previous, population-based genetic epidemiology study of the Prata village revealed a cumulative prevalence of leprosy of 12.8%, among the highest ever reported [21]. The same study revealed the presence of a strong genetic component controlling susceptibility to leprosy in the community, a finding compatible with the hypothesis that, due to the history of the village, genetic risk factors for leprosy are enriched within this population [21]. All inhabitants share the same limited environment: the village has only 1 active church, 2 elementary schools, 1 social club, and 1 large central square, used by the population for leisure activities such as soccer games and outdoor celebrations. In this context, it is reasonable to assume that all individuals are constantly, equally, and heavily exposed to M. leprae. Combined, these peculiar characteristics present the Prata village as a unique model for epidemiological, population-based leprosy studies.
Recruitment Strategy and Procedures
Recruitment followed a 2-stage strategy. In the first stage, in 2009, following WHO recommendations, we investigated M. leprae resistance, focusing on suspected cases of relapse [15]—as defined by individuals with new skin lesions and/or enlarged tender nerves who had been released from treatment at least 5 years before—and household contacts. The resulting high number of cases of M. leprae with MDR led to the second stage, in 2013, of active case-finding, expanding the survey to the entire population (Table 1). Results of the DR/MDR analysis for all samples/patients were immediately released to the local health officials in order to inform therapeutic decisions. In 2017, individuals identified as cases or relapses in the first and/or second stage of recruitment were reached for follow-up. Detailed descriptions of the recruitment strategy and procedures are available in the Supplementary Methods.
Table 1.
Epidemiological Description of the Enrolled Patients
| 2009 | 2013 | |||
|---|---|---|---|---|
| Relapse | Contacts | Individuals | ||
| Number of suspected cases examined | 117 | 85 | 611 | |
| … | Relapse | New case | Relapse | New case |
| Leprosy cases | 12 | 10 | 11 | 9 |
| Sex | ||||
| Female | 5 | 7 | 3 | 5 |
| Male | 7 | 3 | 8 | 4 |
| Clinical form at diagnosis, WHO | ||||
| MB | 11 | 2 | 10 | 1 |
| PB | 1 | 8 | 1 | 8 |
| Age range at diagnosis, years | 10 to 43 | 8 to 79 | 10 to 49 | 10 to 69 |
| Mean age at first diagnosis, years (SD) | 24.5 (11.7) | 32.2 (22.7) | 26.3 (12.3) | 34 (17.6) |
Abbreviations: MB, multibacillary; PB, paucibacillary; SD, standard deviation; WHO, World Health Organization.
Mycobacterium leprae Resistance and Molecular Epidemiology Analyses
Skin lesion biopsies were obtained and processed for diagnosis and estimation of the number of acid-fast bacilli. Mycobacterium leprae suspensions were inoculated in BALB/c mouse footpads for in vivo DDS and RIF resistance testing. Mycobacterium leprae DNA was obtained for molecular testing. Specific DRDR of genes folP1, rpoB, and gyrA were genotyped by Sanger sequencing; 17 short tandem repeats were selected and genotyped by fluorescence-based capillary electrophoresis. The individual discriminatory power of each short tandem repeat marker was calculated using the Hunter-Gaston discriminatory index (HGDI). An unweighted pair group method with arithmetic mean–based similarity matrix was created for the generation of a dendrogram and minimum spanning trees [22]. Detailed descriptions of these experimental procedures are available in the Supplementary Methods.
RESULTS
Clinical and Epidemiological Data
In the first stage, 117 suspected cases of leprosy relapse and 85 contacts were evaluated; 12 cases of relapse were confirmed and 10 household contacts with active leprosy were identified. A total of 611 individuals were evaluated during the second stage of recruitment (55 of these were evaluated both in 2009 and 2013), and 20 cases of active leprosy were detected: 11 relapses and 9 new cases. Of the relapse cases, 5 had already been diagnosed with a leprosy relapse in 2009. Table 1 summarizes the epidemiological data of all diagnosed patients and Table 2 details the data of all diagnosed patients; information on previous treatments was available for all patients (Supplementary Table 1).
Table 2.
Clinical Information and Resistance Status of the Isolates
| Family | ID | Recruitmenta | Disease Status | Clinical Form at Diagnosis, WHO | Resistanceb | Resistance Genotype | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dapsone | Rifampin | Ofloxacin | folP1 | rpoB | gyrA | |||||
| A | PA-043 | 2009 and 2013 | Relapse | MB | Resistantc | Resistantc | Susceptible | CCC55CGC (Pro-Arg) | TCG456ATG (Ser-Met)/ GAT441TAT (Asp-Tyr)d | WT |
| A | PA-073 | 2009 | New case | PB | Resistant | Resistant | Susceptible | CCC55CGC (Pro-Arg) | TCG456ATG (Ser-Met) | WT |
| A | PA-074 | 2009 | New case | PB | Resistant | Resistant | Susceptible | CCC55CGC (Pro-Arg) | TCG456ATG (Ser-Met) | WT |
| A | PA-220 | 2013 | New case | PB | Resistant | Resistant | Unknown | CCC55CGC (Pro-Arg) | TCG456ATG (Ser-Met) | PCR neg |
| A | PA-221 | 2013 | Relapse | MB | Susceptible | Susceptible | Susceptible | WT | WT | WT |
| B | PA-084 | 2009 and 2013 | Relapse | MB | Resistantc | Resistantc | Susceptible | CCC55CGC (Pro-Arg) | TCG456ATG (Ser-Met) | WT |
| B | PA-085 | 2009 and 2013 | Relapse | PB | Resistant | Resistant | Unknown | CCC55CGC (Pro-Arg) | TCG456ATG (Ser-Met) | PCR neg |
| C | PA-058 | 2009 | New case | PB | Susceptible | Susceptible | Susceptible | WT | WT | WT |
| C | PA-103 | 2009 | New case | PB | Resistant | Unknown | Unknown | CCC55CGC (Pro-Arg) | PCR neg | PCR neg |
| D | PA-118 | 2009 | New case | PB | Susceptible | Susceptible | Susceptible | WT | WT | WT |
| D | PA-119 | 2009 | New case | PB | Unknown | Resistant | Susceptible | PCR neg | TCG456ATG (Ser-Met) | WT |
| E | PA-192 | 2009 and 2013 | Relapse | MB | Resistant | Resistant | Susceptible | CCC55CGC (Pro-Arg) | TCG456ATG (Ser-Met) | WT |
| E | PA-199 | 2009 | New case | MB | Unknown | Unknown | Unknown | PCR neg | PCR neg | PCR neg |
| … | PA-006 | 2009 | Relapse | MB | Susceptible | Susceptible | Susceptible | WT | WT | WT |
| … | PA-012 | 2009 and 2013 | Relapse | MB | Susceptiblec | Susceptiblec | Susceptible | PCR neg | PCR neg | WT |
| … | PA-016 | 2009 | Relapse | MB | Resistant | Resistant | Susceptible | ACC53GCC (Thr-Ala) | TCG456TTG (Ser-Leu) | WT |
| … | PA-034 | 2013 | Relapse | MB | Susceptible | Susceptiblec | Susceptible | WT | CTA470CAA (Leu-Gln) | WT |
| … | PA-040 | 2009 | Relapse | MB | Resistant | Resistant | Susceptible | CCC55CGC (Pro-Arg) | TCG456ATG (Ser-Met) | WT |
| … | PA-067 | 2009 | Relapse | MB | Resistant | Resistant | Susceptible | CCC55CGC (Pro-Arg) | TCG456ATG (Ser-Met) | WT |
| … | PA-070 | 2009 | New case | PB | Unknown | Susceptible | Susceptible | PCR neg | WT | WT |
| … | PA-131 | 2009 | Relapse | MB | Susceptible | Susceptible | Susceptible | WT | WT | WT |
| … | PA-146 | 2009 | Relapse | MB | Resistant | Susceptible | Susceptible | CCC55CGC (Pro-Arg) | WT | WT |
| … | PA-155 | 2009 | Relapse | MB | Resistantc | Susceptible | Susceptible | CCC55CTC (Pro-Leu) | WT | WT |
| … | PA-164 | 2009 | New case | PB | Unknown | Unknown | Unknown | PCR neg | PCR neg | PCR neg |
| … | PA-167 | 2009 | New case | MB | Unknown | Unknown | Susceptible | PCR neg | PCR neg | WT |
| … | PA-208 | 2013 | Relapse | MB | Resistant | Resistant | Susceptible | CCC55CGC (Pro-Arg) | TCG456ATG (Ser-Met) | WT |
| … | PA-213 | 2013 | New case | PB | Unknown | Unknown | Unknown | PCR neg | PCR neg | PCR neg |
| … | PA-214 | 2013 | Relapse | MB | Susceptible | Susceptible | Unknown | WT | WT | TTA97TTT (Leu-Phe) |
| … | PA-215 | 2013 | New case | PB | Unknown | Unknown | Unknown | PCR neg | PCR neg | PCR neg |
| … | PA-232 | 2013 | New case | PB | Unknown | Unknown | Unknown | PCR neg | PCR neg | PCR neg |
| … | PA-235 | 2013 | New case | PB | Resistant | Resistant | Susceptible | CCC55CGC (Pro-Arg) | TCG456ATG (Ser-Met) | WT |
| … | PA-240 | 2013 | Relapse | MB | Unknown | Unknown | Unknown | PCR neg | PCR neg | PCR neg |
| … | PA-243 | 2013 | New case | PB | Susceptible | Unknown | Unknown | WT | PCR neg | TTA97TTT (Leu-Phe) |
| … | PA-245 | 2013 | New case | PB | Susceptible | Unknown | Susceptible | WT | PCR neg | WT |
| … | PA-246 | 2013 | New case | MB | Susceptible | Unknown | Susceptible | WT | PCR neg | WT |
| … | PA-248 | 2013 | Relapse | MB | Unknown | Susceptible | Susceptible | CGG100TGG (Arg- Trp) | WT | WT |
| … | PA-254 | 2013 | New case | PB | Susceptible | Susceptible | Susceptible | WT | WT | WT |
Abbreviations: ID, identification number; MB, multibacillary; PB, paucibacillary; PCR neg, not amplified polymerase chain reaction; WHO,World Health Organization classification; WT, wild type.
aFirst step in 2009 and second step in 2013.
bResistance inferred from mutations in drug resistance–determining regions that have been proved to cause resistance.
cResistance confirmed on mice test (in vivo).
dVariant GAT441TAT (Asp-Tyr) has not been detected in any other members of the family.
In 2017, the research team located the clinical records of 27 of the 37 leprosy patients diagnosed during the study. According to these records, 23 had received regular MDT; of these, 11 had been characterized as drug sensitive or inconclusive; 9 were resistant to DDS and RIF; 2 were resistant to DDS; and 1 was resistant to RIF. Also, of these 23 patients, 17 had completed the therapeutic course; 2 had died; 1 was still under treatment; and 3 did not have treatment information available. Our team of clinicians examined 5 out of these 23 patients after treatment: 1 sensitive and 4 resistant. All 5 presented full remission of the lesions. Out of the 27, 4 follow-up patients had received RIF, ofloxacin, and minocycline therapy: 1 was sensitive to all antibiotics tested and dropped out of treatment, and 3 were MDR, had completed the therapeutic course, and were clinically inactive after completion of treatment.
In Vivo Resistance
In 2009, skin biopsies were obtained from the 12 relapse cases and used for mouse footpad inoculation. There were 6 samples that yielded sufficient bacilli: 2 (PA-043 and PA-084) were resistant to both DDS and RIF; 1 (PA-155) was resistant to DDS; and 2 (PA-006 and PA-012) were susceptible to both DDS and RIF. The sixth sample, PA-192, did not show any growth (Supplementary Table 2). Skin biopsies for mouse footpad inoculation were also obtained from 8 relapses and 6 new cases detected in 2013. Of these, 3 presented sufficient bacilli for inoculation: 2 (PA-012 and PA-208) did not grow in any mouse group and 1 (PA-034) showed susceptibility to both DDS and RIF (Supplementary Table 2).
Genotyping of Mycobacterium leprae Drug Resistance–Associated Genes
A detailed description of the variants detected is available in Table 2. In summary, MDR M. leprae was detected in 40.9% (9/22) of the leprosy cases detected in 2009: 7 relapses and 2 new cases. In addition, a single rpoB mutation was detected in 1 new case, and a single folP1 mutation was detected in 2 relapses and 1 new case. No gyrA gene variants were detected in any of the patients (Table 2). Out of the additional 15 leprosy cases identified only in 2013, 1 relapse case and 2 new cases presented MDR. Mutations outside of DRDR regions were detected: CGG100TGG in folP1 (PA-248), CTA470CAA in rpoB (PA-034), and TTA97TTT in gyrA (PA-214 and PA-243; Table 2). Of note, 100% concordance on sensitivity/resistance statuses was observed across the molecular and in vivo analyses.
Familial Occurrences for Mycobacterium leprae Resistance Genes
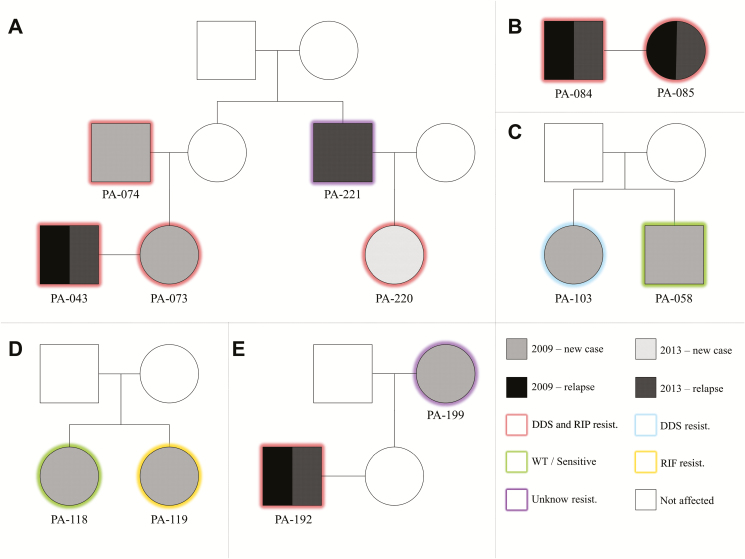
Our strategy to include household contacts as primary targets of the surveys allowed the detection of 5 multiplex pedigrees displaying M. leprae–resistant isolates (Figure 1):
Figure 1.
Male and female individuals are represented by squares and circles, respectively. Unfilled shapes are unexamined individuals. Labels under the squares/circles describe patients’ IDs. Different fills indicate the year of diagnosis and whether it is a case or a relapse. The border color indicates the molecular resistance profile. Abbreviations: DDS, dapsone; ID, identification number; RIF, rifampicin; WT, wild-type.
In the most remarkable case of familial clustering of M. leprae resistance, patient PA-043 was diagnosed with leprosy relapse in 2009. A contact examination revealed that his wife (PA-073) and his father-in-law (PA-074) were also affected. In 2013, the brother-in-law of PA-074 (PA-221) was diagnosed with leprosy relapse; an examination of his contacts led to the diagnosis of his daughter (PA-220) as a new leprosy case. Cases PA-043, PA-073, PA-074, and PA-220 were MDR, with PA-043 also showing MDR in vivo. Patient PA-221 did not yield a polymerase chain reaction product for DRDR analysis (Figure 1A).
Patient PA-084 and his wife, PA-085, were both diagnosed with relapses in 2009 and 2013, both with MDR strains (Figure 1B).
In 2009, 2 brothers, PA-103 (CCC55CGC in folP1) and PA-058 (wild-type), were diagnosed as new leprosy cases (Figure 1C).
In 2009, 2 sisters, PA-118 (wild-type) and PA-119 (TCG456ATG mutation in rpoB), were diagnosed as new leprosy cases (Figure 1D).
An MDR patient, PA-192, was identified as a relapse in both 2009 and 2013. Upon a contact examination in 2009, his mother-in-law (PA-199) was diagnosed as a new leprosy case; unfortunately, none of the DRDR genes of PA-199 yielded polymerase chain reaction product for DR/MDR investigation (Figure 1E).
Multiple-Locus Variable Number of Tandem Repeat Analysis Typing
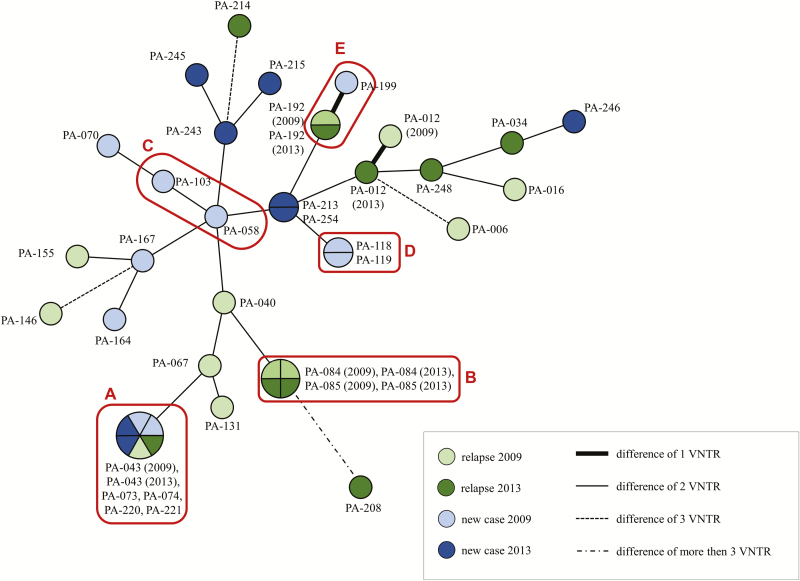
A total of 42 samples, derived from 37 different patients (5 patients presented leprosy both in 2009 and 2013), were genotyped for the variable number tandem repeat markers. All 17 markers were successfully genotyped in 29 samples; the remaining samples had 9 to 16 genotyped markers (Suppl. Table 3). Of the evaluated markers, 5 were nonpolymorphic (HGDI = 0): AC8b, GGT5, 6-3A, 21–3, and 23- 3. The largest allelic variability was detected for markers TA18, AT15, and AT17, with 14, 10, and 9 alleles, respectively, and HGDI scores between 0.77–0.89. The remaining markers presented HGDI scores ranging between 0.10–0.70 and have been ordered by decreasing variability as follows: GTA9 (6 alleles), AC8a and GGA21 (both 5 alleles), TA10 (4 alleles), AC9 and 6–7 (both with 3 alleles), 27-5 and 12–5 (both with 2 alleles), and 18-8 (2 alleles; Supplementary Table 3).
An analysis revealed the existence of 5 clusters (Figure 2): 1 included all 6 samples of 5 patients belonging to pedigree A; 1 corresponded to 4 samples from the 2 patients of pedigree B; 1 included the 2 samples of pedigree D; 1 was composed of both samples obtained from patient PA-192 (2009 and 2013); and 1 included patients PA-213 and PA-254. Of note, for all 5 cases that had samples collected in 2009 and 2013, only 1 (PA-012) did not have the 2 samples included in the same cluster, due to a difference in the AT15 marker.
Figure 2.
The lengths of the branches of the tree represent the distances between the genetic standards. Red boxes define individuals from the same pedigree, as labeled by the letters A (pedigree A of Figure 1) to E (pedigree E of Figure 1). Abbreviation: VNTR, variable number tandem repeats.
DISCUSSION
Today, antimicrobial resistance is arguably the main challenge in fighting human infection. Yet, little is known about antimicrobial resistance in leprosy and most of the information on leprosy resistance to treatment comes from sporadic reports of resistant cases, reinforced by a few investigations of larger samples [16, 18, 23] and, more recently, a large study of global reach [19, 20]. These studies show a proportion of resistant M. leprae significantly higher than those reported before by the WHO [19, 24], indicating that the real extent of the problem of M. leprae resistance to antibiotics is still unknown. This becomes even more critical in the scenario of a high hidden prevalence of leprosy, as recently suggested [25].
Here, we present the results of the first population-based, systematic report of M. leprae resistance in an isolated, hyperendemic population with decades of leprosy history. Our combined strategy of relapse monitoring and active searches for new cases has resulted in the identification of 37 leprosy cases, 16 (43.2%) of them showing some degree of resistance and 12 (32.4%) of which were resistant to both DDS and RIF. These high proportions of resistant M. leprae are likely due to the peculiar nature of the studied population, which has been heavily exposed to leprosy for almost 100 years: a combination of increased natural susceptibility and environmental factors likely favors the emergence, persistence, and propagation of resistant M. leprae through undiagnosed cases and asymptomatic carriers. Also, some relapse patients were first treated with DDS monotherapy in the 1960s and some have records of up to 4 rediagnoses, due to relapses or reinfections. Thus, the high level of resistance observed in the Prata Village is likely exceptional and not necessarily extensive to other populations. Still, the Prata Village, if seen as a natural experiment and interpreted accordingly, alerts us to the existence of a possible, elusive scenario of emergence of M. leprae–resistant strains that, due to disease characteristics such as an extremely long incubation period, may manifest as a public health problem only years ahead.
The combined molecular analysis of resistance and phylogenetic markers between isolates obtained from members of the same Prata Village families strongly suggests transmission along the pedigrees, in particular, of resistant strains characterizing primary resistance. This is particularly evident for pedigree A, which displayed at least 3 cases of primary, double-resistant leprosy (Figure 1) and phylogenetic data positioning all M. leprae isolates into a single cluster (Figure 2). Complete concordance of both resistance and phylogenetic profiles was also observed for pedigree B. For pedigree D, phylogenetic data placed both isolates into the same cluster; however, the resistance profiles are distinct, likely due to the emergence of RIF resistance in 1 of the sisters. For pedigree C, distinct phylogenetic and resistance profiles suggest different isolates caused leprosy independently across siblings. For pedigree E, phylogenetic data differed only by a single AT17 variant; unfortunately, it was not possible to obtain resistance information for the primary case, which would have been particularly valuable since the index case is a multiple-relapse, double-resistant patient. Since marker AT17 is highly unstable and polymorphic, it is possible that the discordance is due to natural molecular changes observed within the same isolate. A similar explanation may apply to the single divergence (marker AT15, patient PA-012) among the 5 patients for whom samples were obtained both in 2009 and 2013. This great similarity between strains of the same patients, collected at different stages, indicates that the disease in these cases was caused by the same strain at both moments. Of note, a complementary phylogenetic analysis indicated the presence of M. leprae Single Nucleotide Polymorphism types 3 and 4 in the Prata Village—with a 76.2% predominance of the later—that is compatible with the mixed ethnic background of the Prata Village [21] (data not shown).
3 of the alleles in the DRDR region of the genes detected in the study haven’t been previously associated with M. leprae resistance: (1) CTA470CAA (Leu-Gln) in rpoB, detected in 1 relapse patient; (2) CGG100TGG (Arg-Trp) in folP1, also detected in 1 relapse patient; and (3) TTA97TTT (Leu-Phe) in gyrA, detected in 1 relapse and 1 new case. All isolates harboring these mutations were inoculated in the mouse footpad; however, only rpoB CTA470CAA (Leu-Gln) yielded positive growth, showing sensitivity to DDS and RIF in vivo. Thus, both folP1 and gyrA mutations remain as natural candidates for further investigation of a potential role in leprosy resistance.
The long-term follow-up indicates that a minor proportion of the patients resistant do DDS, RIF, or both had received regular MDT, perhaps following outdated guidelines, combined with complex administrative and bureaucratic procedures, in order to guarantee treatment. The impact of such deleterious effects upon the emergence and maintenance of M. leprae resistance is unknown.
We are aware of the limitations of our study. First, despite important technological advances, working with M. leprae still poses challenges, such as obtaining enough quantities from biological samples to successfully achieve bacterial growth in mice—particularly difficult for paucibacillary patients—in order to allow in vivo testing of resistance and access to enough DNA to perform molecular analyses. To address this limitation, molecular data have been re-checked by a reference laboratory with 100% concordance. Second, a recent genomic analysis suggests the existence of novel targets involved in antimicrobial resistance in leprosy that haven’t yet been tested in the Prata population [26, 27]; including these novel markers through a follow-up study involving whole-genome sequencing would produce a more comprehensive description of the resistance molecular profile and a higher resolution pattern of transmission, possibly increasing resistance rates even further.
In summary, the results of our systematic study revealed the underdetection of primary and secondary resistance and dissemination of M. leprae–resistant strains in the context of a very unique population. It is unlikely that the scenario observed in the Prata Village also applies to less atypical contexts; however, we expect that our data can raise awareness for a potential, alarming scenario that demands immediate action from leprosy control authorities in this particular region of Brazil and calls for better surveillance in other regions of the globe.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the patients for agreeing to participate in the study, as well as Masanori Matsuoka and Masanori Kai from the National Institute of Health in Japan for quality control of sequenced samples.
Financial support. This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (grant numbers MCT/CNPq/MS-SCTIE-DECIT 35/2005, MCT/CNPq/CT-Saúde/MS/SCTIE/DECIT Nº 034/2008, and MCTI/CNPq/MS-SCTIE – Decit N° 40/2012).
Potential conflict of interests. M. T. M. was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (grant number CNPq-PQ 2). H. R. S. D. was supported by Fundação Araucária (grant number FA 247/2015). P. N. S. was supported by the Damien Foundation. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Smith CS, Aerts A, Saunderson P, Kawuma J, Kita E, Virmond M. Multidrug therapy for leprosy: a game changer on the path to elimination. Lancet Infect Dis 2017; 17:e293–7. [DOI] [PubMed] [Google Scholar]
- 2. Pettit JH, Rees RJ. Sulphone resistance in leprosy. an experimental and clinical study. Lancet 1964; 2:673–4. [DOI] [PubMed] [Google Scholar]
- 3. Pearson JM, Haile GS, Rees RJ. Primary dapsone-resistant leprosy. Lepr Rev 1977; 48:129–32. [DOI] [PubMed] [Google Scholar]
- 4. Jacobson RR, Hastings RC. Rifampin-resistant leprosy. Lancet 1976; 2:1304–5. [DOI] [PubMed] [Google Scholar]
- 5. World Health Organization. Chemotherapy of leprosy for control programmes. World Health Organ Tech Rep Ser 1982; 675:1–33. [PubMed] [Google Scholar]
- 6. Shetty VP, Uplekar MW, Antia NH. Primary resistance to single and multiple drugs in leprosy–a mouse footpad study. Lepr Rev 1996; 67:280–6. [DOI] [PubMed] [Google Scholar]
- 7. Cambau E, Perani E, Guillemin I, Jamet P, Ji B. Multidrug-resistance to dapsone, rifampicin, and ofloxacin in Mycobacterium leprae. Lancet 1997; 349:103–4. [DOI] [PubMed] [Google Scholar]
- 8. Machado D, Lecorche E, Mougari F, Cambau E, Viveiros M. Insights on Mycobacterium leprae efflux pumps and their implications in drug resistance and virulence. Front Microbiol 2018; 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yew WW, Liang D, Chan DP, Shi W, Zhang Y. Molecular mechanisms of clofazimine resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 2017; 72:2943–4. [DOI] [PubMed] [Google Scholar]
- 10. Shepard CC. The experimental disease that follows the injection of human leprosy bacilli into foot-pads of mice. J Exp Med 1960; 112:445–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Contreras Mejía Mdel C, Porto Dos Santos M, Villarouco da Silva GA, et al. Identification of primary drug resistance to rifampin in Mycobacterium leprae strains from leprosy patients in Amazonas State, Brazil. J Clin Microbiol 2014; 52:4359–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsuoka M. Drug resistance in leprosy. Jpn J Infect Dis 2010; 63:1–7. [PubMed] [Google Scholar]
- 13. Williams DL, Gillis TP. Drug-resistant leprosy: monitoring and current status. Lepr Rev 2012; 83:269–81. [PubMed] [Google Scholar]
- 14. da Silva Rocha A, Cunha Md, Diniz LM, et al. Drug and multidrug resistance among Mycobacterium leprae isolates from Brazilian relapsed leprosy patients. J Clin Microbiol 2012; 50:1912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. World Health Organization. Guidelines for global surveillance of drug resistance in Leprosy. New Delhi, India: WHO Regional Office for South-East Asia, 2009. [Google Scholar]
- 16. Beltrán-Alzate C, López Díaz F, Romero-Montoya M, et al. Leprosy drug resistance surveillance in Colombia: the experience of a sentinel country. PLOS Negl Trop Dis 2016; 10:e0005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Avanzi C, Busso P, Benjak A, et al. Transmission of drug-resistant leprosy in Guinea-Conakry detected using molecular epidemiological approaches. Clin Infect Dis 2016; 63:1482–4. [DOI] [PubMed] [Google Scholar]
- 18. Lavania M, Singh I, Turankar RP, et al. Molecular detection of multidrug-resistant Mycobacterium leprae from Indian leprosy patients. J Glob Antimicrob Resist 2018; 12:214–9. [DOI] [PubMed] [Google Scholar]
- 19. Cambau E, Saunderson P, Matsuoka M, et al. Antimicrobial resistance in leprosy: results of the first prospective open survey conducted by a WHO surveillance network for the period 2009–2015. Clin Microbiol Infect 2018; 24:1305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cambau E, Saunderson P, Matsuoka M, et al. ; World Health Organization Surveillance Network of Antimicrobial Resistance in Leprosy. Antimicrobial resistance in leprosy: results of the first prospective open survey conducted by a WHO surveillance network for the period 2009-15. Clin Microbiol Infect 2018; 24:1305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lázaro FP, Werneck RI, Mackert CC, et al. A major gene controls leprosy susceptibility in a hyperendemic isolated population from north of Brazil. J Infect Dis 2010; 201:1598–605. [DOI] [PubMed] [Google Scholar]
- 22. Fontes ANB, Lima LNGC, Mota RMS, et al. Genotyping of Mycobacterium leprae for better understanding of leprosy transmission in Fortaleza, Northeastern Brazil. PLOS Negl Trop Dis 2017; 11:e0006117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Avanzi C, Del-Pozo J, Benjak A, et al. Red squirrels in the British Isles are infected with leprosy bacilli. Science 2016; 354:744–7. [DOI] [PubMed] [Google Scholar]
- 24. World Health Organization. A guide for surveillance of antimicrobial resistance in leprosy: 2017 update. New Delhi, India: WHO Regional Office for South-East Asia, 2017. [Google Scholar]
- 25. Salgado CG, Barreto JG, da Silva MB, et al. Are leprosy case numbers reliable? Lancet Infect Dis 2018; 18:135–7. [DOI] [PubMed] [Google Scholar]
- 26. Singh P, Benjak A, Carat S, et al. Genome-wide re-sequencing of multidrug-resistant Mycobacterium leprae Airaku-3. Clin Microbiol Infect 2014; 20:O619–22. [DOI] [PubMed] [Google Scholar]
- 27. Benjak A, Avanzi C, Singh P, et al. Phylogenomics and antimicrobial resistance of the leprosy bacillus Mycobacterium leprae. Nat Commun 2018; 9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.