Abstract
In reverse transcription-quantitative polymerase chain reaction (RT-qPCR) studies, endogenous reference genes are routinely used to normalize the expression of target gene studies. In order to precisely evaluate the relative expression of genes in the cells of mice suffering from Kidney Yang Deficiency Syndrome (KYDS) in response to influenza A virus (IAV) H1N1 using RT-qPCR, it is crucial to identify reliable reference genes. In the present study, 15 candidate reference genes (Actb, β2m, Gapdh, Gusb, Tuba, Grcc10, Eif4h, Rnf187, Nedd8, Ywhae, 18S rRNA, Rpl13, Ubc, Rpl32, and Ppia) were investigated in lung cells from KYDS mice infected with IAV H1N1. NormFinder, BestKeeper, and GeNorm were used to assess the stability of reference genes. The results were authenticated over extended experimental settings by a group of 10 samples. In the present study, we explored a novel method using dual-gene combinations; the difference in gene expression between the model and normal control groups was statistically analyzed by an independent-samples t-test, and the difference in the mean value between the two groups was compared. A P value > 0.05 and the lowest absolute value of the difference indicated the optimal reference two-gene combination. Four additional host innate immune system-related genes (TLR3, TLR4, TLR7, and RIG-I) were analyzed together with the two treatment datasets to confirm the selected reference genes. Our results indicated that none of these 15 candidate reference genes can be used as reference gene individually for relative quantitative fluorescence PCR analysis; however, the combination of Grcc10 and Ppia, based on the process of calculating the higher P value and lower difference values between groups, was the best choice as a reference gene for the lung tissue samples in KYDS mice infected with IAV. This technique may be applied to promote the selection process of the optimal reference gene in other experiments.
1. Introduction
At present, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) is considered to be a sensitive, accurate, highly specific, and fast measurement of gene expression with absolute or relative quantification [1]. The absolute quantification for a particular transcript gene copy number is calculated by the standard curve, and the relative quantification for a target gene may be measured for the relative expression compared with the reference gene [2]. As most researchers focus on analyzing the discrepant changes in gene mRNA expression levels, relative quantification has become a widely applied method for amplifying and detecting gene expression [3].
Gene expression analyses rely on the use of reference genes, which are internal controls for varied samples and different experimental settings. The selection of an appropriate reference gene is crucial and stable. However, a number of studies have demonstrated that commonly employed reference genes are unstable, as their transcription is closely regulated in various experimental settings and varies among different tissues. For example, Gapdh is known as a key enzyme in glycolysis in mammalian cellular organisms, and it is frequently used as a stable reference gene. However, its expression is not always constant, particularly in cancer, since the expression of Gapdh changes markedly in rapidly dividing and proliferating tumor cells, and improper reference gene selection may lead to inaccurate results in the target gene expression levels [4, 5]. Thus, potential reference genes must be evaluated for their stability by several algorithms, such as GeNorm [6], which are currently available as part of qBase Plus [7], NormFinder [8], and Best Keeper [9, 10].
In traditional Chinese medicine (TCM), Kidney Yang Deficiency Syndrome (KYDS) is one of the classical syndrome patterns and is characterized by a set of complaints, mainly including tenderness and weakness in the knees and lumbar regions, mental fatigue, difficulty in urination, enuresis, female sterility, reduction of auditory function, and dental abnormalities [11]. It is the most common syndrome in old age [12]. Influenza is a contagious viral infection that affects millions of individuals annually worldwide, and pandemic outbreaks may cause significant deaths in the population aged >65 years, and up to 90% mortality associated with influenza occur in this age group [13–16]. These susceptible individuals belong to high-risk people for influenza infection, who are mostly similar to the KYDS according to the principles of TCM [12, 17]. An epidemiological investigation was conducted in 2137 healthy elderly above 60 years old; the results showed that the incidence rate of kidney deficiency was 78.80% [17]; kidney deficiency syndrome prevalence in participants showed an increasing trend with increasing age and deteriorating health status [18]. Another epidemiological study on 2067 adults aged >60 years revealed that 45.33% suffered from KYDS, showing that KYDS is the predominant TCM syndrome in high-risk populations [19], and these elderly groups are at increased risk for serious flu complications. KYDS or influenza A virus (IAV) infection (H1N1) causes universal variations in gene expression, which represent the molecular basis of the biological effects of KYDS after IAV infection [20–27].
To explain the mechanisms behind KYDS following IAV infection in order to develop an effective treatment for this disease, understanding the changes that occur during gene transcription is crucial. Our previous study demonstrated that certain common reference genes (for example, Gapdh or Actb) were not stable in lung tissue from mice with KYDS after IAV infection, compared with normal control mice. In fact, commonly used algorithms have been applied to evaluate the most reliable reference gene. However, through t-test analysis between groups, the single or multiple reference genes analyzed by these general algorithms exhibit significant variations in expression between the normal and model groups (P < 0.05). Therefore, a novel method for reference gene selection is needed.
In the present study, 15 candidate reference genes (Actb, β2m, Gapdh, Gusb, Tuba, Grcc10, Eif4h, Rnf187, Nedd8, Ywhae, 18S rRNA, Rpl13, Ubc, Rpl32, and Ppia) were selected, which may be used as internal control genes for normalizing gene expression. The stability of expression for the abovementioned reference genes in mice suffering from KYDS infected with IAV (H1N1) and their mock-inoculated controls were assayed by means of three statistical algorithms (BestKeeper, NormFinder, and GeNorm) and compared in mouse lung tissue using t-tests. Our analysis may facilitate the selection of the most accurate reference genes for calculating target gene expression to assess the KYDS-IAV correlation. This new technique may prove useful for selecting the optimal reference gene in other experiments.
2. Materials and Methods
2.1. Animals
This study was performed in accordance with the recommendations of the National Institutes of Health. The protocol was approved by the Ethics Committee in Shandong University of Traditional Chinese Medicine. The animals were regularly monitored by the veterinarian staff. No pathogens were found. To minimize animal suffering during surgery, isoflurane gas was used for anesthesia. SPF male BALB/c mice (weight, 18-20 g) were purchased from the Jinan Pengyue Experimental Animal Breeding Company, Ltd. After 3 days of acclimatization, all mice were randomly assigned to two groups (n = 10 per group) and treated for 2 weeks: estradiol benzoate (8 mg/kg) was administered intraperitoneally for 7 days to establish the KYDS mouse model as previously described [28], while normal control mice were intraperitoneally injected with normal saline (NS). The physical signs in mice were observed, and rectal temperature and body weight were measured and recorded. All animals were kept under a 12 h dark/light cycle at 21-23°C. Environmental humidity was controlled at 60-70%. On the 8th day, KYDS mice (model group) were anaesthetized with isoflurane and intranasally inoculated with 20 μl of viral suspension containing hemagglutination titer of 1 : 320 of influenza A/FM/1/47 (H1N1) virus, while the normal control group mice were inoculated with 20 μl NS. The mice were then allowed access to food and water ad libitum.
2.2. Tissue Preparation
On the 14th day, according to the approved protocol, the mice were sacrificed. The lungs were isolated and weighed for the organ index calculation. After harvesting, small tissue samples were immediately submerged in RNAstore Reagent (Tiangen) at a dilution ratio of 1 : 10 (w/v) and then stored at 4°C.
2.3. Extraction of Total RNA
Total RNA was extracted from 10 to 20 mg tissue samples using RNAprep Pure Tissue Kit (Tiangen) according to the manufacturer's instructions. The yield and purity of RNA were determined with the Quawell 5000 spectrophotometer (Quawell Technology). For further analysis, RNA samples with an absorbance OD 260/280 ratio of 1.8-2.0 were used. The assessment of RNA integrity was performed by agarose gel electrophoresis.
2.4. Synthesis of Reverse-Transcribed cDNA
Using a 20 μl reverse transcription system (FastQuant RT Kit with gDNase, Tiangen), first-strand cDNA was synthesized from 2 μg total RNA with random hexamer oligonucleotide primers and incubated at 42°C for 3 min to protect total RNA from genomic DNA interference. Using FastQuant RT Enzyme, reverse transcription was performed at 42°C for 15 min. cDNA was stored at -20°C until further use. For qPCR analysis, each sample of cDNA was diluted 10 times with nuclease-free water.
2.5. qPCR
The Bio-Rad CFX Connect Real-Time System was used to conduct qPCR. A total of 20 μl mixture containing 6 pmol each reverse and forward primer, 2 μl cDNA, and 10 μl 2Χ SuperReal PreMix Plus with SYBR Green I (Tiangen) was used for each reaction. The amplification conditions were as follows: 40 cycles at 95°C for 10 sec, 60°C for 32 sec, and 95°C for 15 min. Following amplification, the addition of a thermal denaturing cycle was performed to derive the dissociation curve of the PCR product to validate the specificity of the amplification. The measurement was performed three times for each sample.
For each primer, the efficiency of qPCR in exponential phase was measured employing standard curves (4-fold serial dilutions of pooled cDNA containing equal amounts of various sample sets). Mean quantification cycle (Cq) values of each serial dilution were plotted against the cDNA dilution factor logarithm, and the following equation was used to obtain its value: E = 10 (−1/slope) × 100. The determination of the linear dynamic range was performed by the correlation coefficient (R2), and the standard curve for each gene was created.
To obtain a standard for the SYBR assay, the IAV H1N1 pDZ-M plasmid was used as previously described [29] and quantified by spectrophotometric analysis. The copy numbers of the IAV H1N1 M gene were detected by the absolute quantitative method.
2.6. Primers
From “publically available” nucleotide sequences, 15 candidate reference genes were selected. The candidate genes were Gusb, Gapdh, Actb, β2m, Tuba, Grcc10, Eif4h, Rnf187, Nedd8, Ywhae, 18S rRNA, Rpl13, Ubc, Rpl32, and Ppia. Using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), the primers were identified from designed nucleotide sequences. Primers were obtained from Beijing Liuhe BGI Technology Company with their certificates of analysis. The characteristics of the selected reference genes are listed in Table 1. The primers for authentication of selected reference genes were as follows: toll-like receptor (TLR)3: forward CAGGATACTTGATCTCGGCCTT and reverse TGGCCGCTGAGTTTTTGTTC; TLR4: forward CATGGATCAGAAACTCAGCAAAGTC and reverse CATGCCATGCCTTGTCTTCA; TLR7: forward CTGGAGTTCAGAGGCAACCATT and reverse GTTATCACCGGCTCTCCATAGAA; and retinoic acid-inducible gene-I (RIG-I): forward GCAGGTTACTGTGGACTTTGTG and reverse TGCCATTCTCCCTTTAGTGTCT. The primers for the IVA H1N1 M gene were as follows: forward 5′-CTGAGAAGCAGATACTGGGC-3′ and reverse 5′-CTGCATTGTCTCCGAAGAAAT-3′.
Table 1.
Candidate reference genes and characteristics of RT-qPCR primers.
| Gene symbol | Gene name | Accession no. | Primer sequence | Amplicon size | Efficiency (%) | R 2 |
|---|---|---|---|---|---|---|
| Actb | Beta-actin | NM_007393.5 | F: CCTTCTTGGGTATGGAATCCTGT R: ACTGTGTTGGCATAGAGGTCTTTAC |
101 | 95.9 | 0.990 |
| β2m | Beta-2 microglobulin | NM_009735.3 | F: CATGGCTCGCTCGGTGAC R: CAGTTCAGTATGTTCGGCTTCC |
135 | 96.8 | 0.995 |
| Tuba | Alpha-tubulin | NM_011653.2 | F: TGTCCTGGACAGGATTCGC R: CTCCATCAGCAGGGAGGTG |
115 | 95.0 | 0.997 |
| Gusb | Beta-glucuronidase | NM_010368.1 | F: CCGACCTCTCGAACAACCG R: GCTTCCCGTTCATACCACACC |
169 | 96.8 | 0.993 |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | NM_008084.3 | F: TGCACCACCAACTGCTTAG R: GGATGCAGGGATGATGTTC |
177 | 94.2 | 0.996 |
| Grcc10 | Gene rich cluster, C10 gene | NM_013535.1 | F: GCGGAGGTGATTCAAGCG R: TGACCAGGCGGGCAAACT |
196 | 96 | 0.995 |
| Eif4h | Eukaryotic translation initiation factor 4H | NM_033561.2 | F: CCTTGGCTCGGGATTGTC R: TCCGCATTGGAGATGGATTA |
198 | 97.2 | 0.998 |
| Rnf187 | Ring finger protein 187 | NM_022423.2 | F: CTGGCACCACCCTTCATC R: ACAAGCCCGAGCACATTC |
155 | 95.1 | 0.991 |
| Ywhae | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein | NM_009536.4 | F: CCCATTCGTTTAGGTCTT R: TCCACAGCGTCAGGTTAT |
193 | 93.2 | 0.994 |
| Nedd8 | Neural precursor cell expressed, developmentally downregulated gene 8 | NM_008683.3 | F: TGGGAAGGAGATTGAGATAG R: TTGCTTGCCACTGTAGATG |
121 | 94 | 0.995 |
| 18S rRNA | 18S ribosomal RNA | NR_003278.2 | F: TTGACGGAAGGGCACCACCAG R: GCACCACCACCCACGGAATCG |
130 | 91.7 | 0.996 |
| Rpl13 | Ribosomal protein L-13 | NM_016738.5 | F: GTACGCTGTGAAGGCATCAA R: ATCCCATCCAACACCTTGAG |
135 | 91 | 0.997 |
| Ubc | Ubiquitin C | NM_019639 | F: GCCCAGTGTTACCACCAAGA R: CCCATCACACCCAAGAACA |
104 | 96.3 | 0.996 |
| Rpl32 | Ribosomal protein L32 | NM_172086.2 | F: GAACTGGCGGAAACCCA R: GGATCTGGCCCTTGAACCTT |
63 | 96.2 | 0.995 |
| Ppia | Peptide acyl preserved ammonia acyl isomerase A | NM_008907.1 | F: CGCTTGCTGCAGCCATGGTC R: CAGCTCGAAGGAGACGCGGC |
86 | 96.3 | 0.990 |
The correlation coefficients (R2) of the standard curve, efficiency of each pair of primers, length of the RT-qPCR transcripts, and primer sequences are indicated. RT-qPCR: reverse transcription-quantitative polymerase chain reaction.
2.7. Data Analysis
Threshold setting and baseline correction were performed by automatic calculation with the CFX Maestro software (Bio-Rad Laboratories, Inc.). Using this software in single threshold mode, the value of Cq was determined.
The stability of the reference genes was assessed by these data using three common statistical programs (NormFinder, GeNorm, and BestKeeper). The Cq values were changed into relative quantities by using 2-ΔCq (ΔCq = the corresponding Cq value − minimum Cq value). Calculations of NormFinder and GeNorm depend upon these converted values; BestKeeper was used to analyze the raw Cq. The relative expression level of each target gene was calculated according to the 2-ΔΔCq method for normalization of the gene to each potential reference gene [30]. An independent-samples t-test with SPSS statistical software was employed to assess the statistical significance of the differences in gene expression. P values < 0.05 were considered statistically significant. Microsoft Excel 2010 was used for variance analysis.
3. Results
3.1. Establishment of a KYDS-Virus Mouse Model
After the KYDS mice were inoculated through the nose with IAV H1N1 or NS, the body weight and rectal temperature in the KYDS-virus group were found to be significantly lower compared with those in the normal control group from the 6th to the 7th day (P < 0.01) (Figures 1(a) and 1(b)). As shown in Figure 1(c), in comparison with the normal group, the model group thymus and seminal vesicle indexes decreased significantly (P < 0.01), whereas the lung index increased significantly (P < 0.01), clearly indicating that the model group lung tissue sustained inflammatory injury. IAV H1N1 mRNA expression was also detected. Total RNA was isolated from lung tissue and converted into cDNA. The copy numbers of H1N1 M gene RNA were >6.1 × 106/ml, as determined using absolute quantification PCR by the standard curve (Figure 1(d)). These results demonstrated that the KYDS-virus model was successfully constructed.
Figure 1.
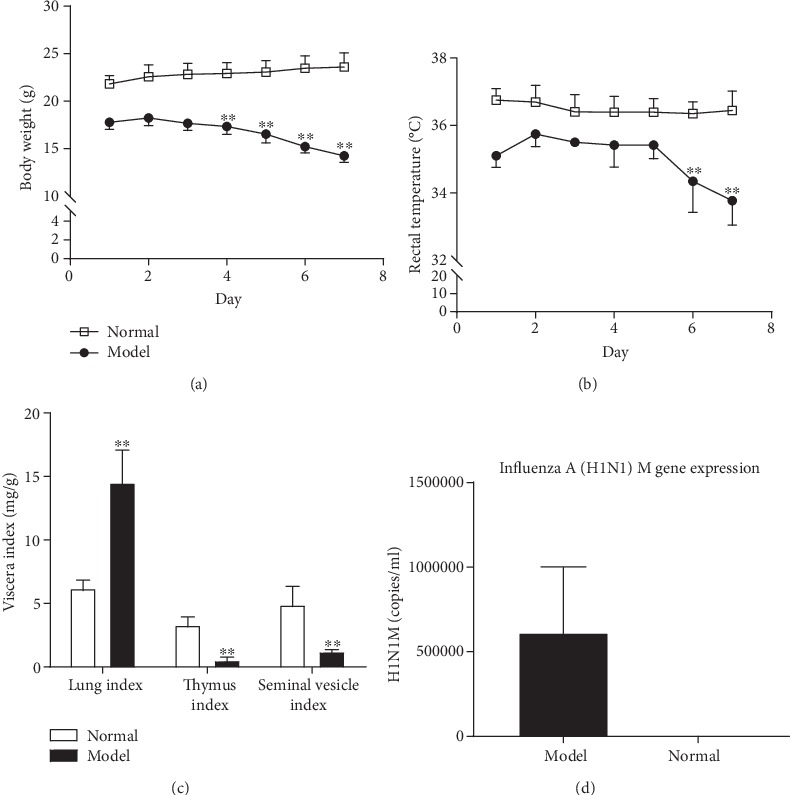
Assessment of the KYDS-virus model. (a) Whole body weight and (b) rectal temperature were measured after KYDS-virus model establishment for 7 days. (c) Visceral indices of different organs in mice. (d) Absolute quantity of influenza A virus (H1N1) M gene expression was calculated in the model group. ∗∗P < 0.01, compared with normal control; n = 10 in each group. KYDS: Kidney Yang Deficiency Syndrome.
3.2. Expression Profiles of Candidate Reference Genes
To accurately assess the patterns of the KYDS-virus expression studies, 15 candidate reference genes were finally selected on the basis of their common usage, as mentioned in the literature. Following qPCR analysis, each amplification primer set was tested for its performance. In all reactions, the amplification efficiency ranged from 91.0 to 97.2% and the standard curve correlation coefficients (R2) varied from 0.990 to 0.998 (Table 1). Strong correlation and very high efficiency were exhibited by all tested primer pairs. The amplification specificity of all transcripts was confirmed by melt curve analysis; as a single peak was produced, formation of primer dimers was not observed. Amplification was not observed in the absence of template (NTC control).
3.3. Analysis Using the BestKeeper, NormFinder, and GeNorm Algorithms
The GeNorm software was used for analysis of the expression stability of the 15 candidate reference genes. This software calculates gene expression stability (M) as the mean pairwise variation among all tested genes. The reference gene that yields the most similar results in all samples, regardless of the experimental circumstances, is considered the best reference gene. The reference gene with the smallest value of M was considered the most stable gene.
A sample with M value < 1.5 is usually considered a stably expressed gene (Figure 2(a)). For lung tissue, Ywhae and Tuba are the two most stable reference genes, with the lowest M values (M = 0.012), whereas the M values of Grcc10 and β2m (M = 0.044 and 0.049, respectively) were the highest, indicating that those genes were the most variably expressed.
Figure 2.
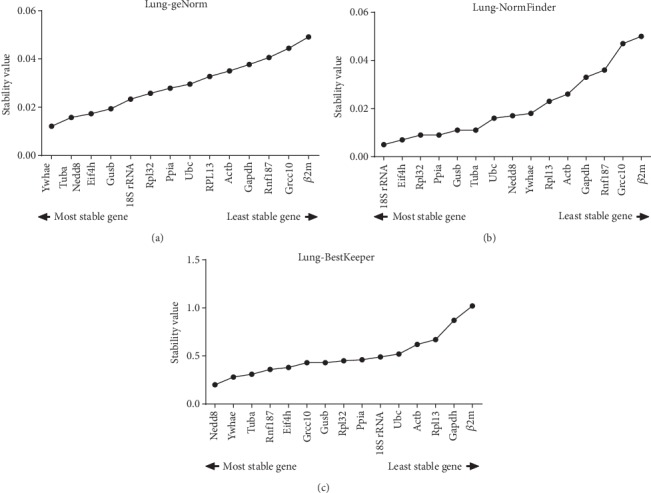
Mean expression stability (M) evaluated using GeNorm and NormFinder and mean expression stability standard deviation [SD (±CP)] evaluated using BestKeeper. M values and SD values rankings are indicated for mouse lung tissue under different conditions. (a) GeNorm, (b) NormFinder, and (c) BestKeeper.
NormFinder is used for the calculation of the gene stability index (M). The lowest M values have been observed in the most stably expressed genes (Figure 2(b)). The most stable gene was 18S rRNA, whereas the best gene combination was Eif4h and Rpl32 (Table 2).
Table 2.
NormFinder calculation results.
| Variables | Results |
|---|---|
| Best gene | 18S rRNA |
| Stability value | 0.005 |
| Best combination of two genes | Eif4h and Rpl32 |
| Stability value for the best combination of two genes | 0.004 |
BestKeeper software was employed as an expression standard of reference genes with the ranking of the standard deviation [SD (±CP)]. The most stable reference genes have the lowest SD values. The acceptable range of variation of SD values is <1 (Figure 2(c)). Nedd8 was ranked as the most stably expressed gene in lung tissue, with the lowest SD value by BestKeeper.
3.4. Independent-Samples t-Test with Combination of Pair Candidate Reference Genes
We analyzed the combination of pair candidate reference genes in the normal and model groups with the independent-samples t-test, and 15 combinations of P values > 0.05 were identified (Table 3). Subsequently, for these potential combinations of reference genes, the absolute difference in mean Cq values between the two groups was calculated, and the smallest absolute difference value indicated that the combination of the genes was the optimal stable candidate reference gene. Based on these results (Table 4), the best combination of genes was Grcc10 and Ppia.
Table 3.
P values of the independent-samples t-test for the 15 candidate reference gene combinations.
| Grcc10 | Actb | Gapdh | Eif4h | Nedd8 | Rnf187 | Ywhae | Gusb | Tuba | β2m | 18S rRNA | Rpl13 | Ubc | Rpl32 | Ppia | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grcc10 | 0.043 | 0.001 | 0.538 | 0.010 | 0.000 | 0.027 | 0.821 | 0.113 | 0.001 | 0.801 | 0.211 | 0.258 | 0.803 | 0.986 | |
| Actb | 0.043 | 0.000 | 0.000 | 0.000 | 0.009 | 0.001 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Gapdh | 0.001 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Eif4h | 0.538 | 0.000 | 0.000 | 0.000 | 0.293 | 0.003 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Nedd8 | 0.010 | 0.000 | 0.000 | 0.000 | 0.686 | 0.012 | 0.002 | 0.005 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | |
| Rnf187 | 0.000 | 0.009 | 0.001 | 0.293 | 0.686 | 0.810 | 0.266 | 0.712 | 0.001 | 0.059 | 0.023 | 0.010 | 0.045 | 0.145 | |
| Ywhae | 0.027 | 0.001 | 0.000 | 0.003 | 0.012 | 0.810 | 0.003 | 0.006 | 0.000 | 0.001 | 0.001 | 0.000 | 0.000 | 0.000 | |
| Gusb | 0.821 | 0.000 | 0.000 | 0.000 | 0.002 | 0.266 | 0.003 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Tuba | 0.113 | 0.001 | 0.000 | 0.001 | 0.005 | 0.712 | 0.006 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| β2m | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| 18S rRNA | 0.801 | 0.000 | 0.000 | 0.000 | 0.000 | 0.059 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Rpl13 | 0.211 | 0.000 | 0.000 | 0.000 | 0.000 | 0.023 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Ubc | 0.258 | 0.000 | 0.000 | 0.000 | 0.000 | 0.010 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Rpl32 | 0.803 | 0.000 | 0.000 | 0.000 | 0.000 | 0.045 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Ppia | 0.986 | 0.000 | 0.000 | 0.000 | 0.001 | 0.145 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Table 4.
Screening results of 15 reference gene combinations with P values, standard deviation, and absolute values of difference.
| Rank | Combination | P value | Standard deviation | Absolute value of difference |
|---|---|---|---|---|
| 1 | Grcc10+Ppia | 0.986 | 0.156 | 0.009 |
| 2 | Grcc10+Gusb | 0.821 | 0.221 | 0.0096 |
| 3 | Rnf187+Tuba | 0.712 | 0.228 | 0.018 |
| 4 | Grcc10+Rpl32 | 0.803 | 0.133 | 0.019 |
| 5 | Grcc10+18S rRNA | 0.801 | 0.180 | 0.0275 |
| 6 | Grcc10+Eif4h | 0.538 | 0.159 | 0.061 |
| 7 | Nedd8+Rnf187 | 0.686 | 0.187 | 0.074 |
| 8 | Rnf187+Ywhae | 0.810 | 0.226 | 0.075 |
| 9 | Grcc10+Ubc | 0.258 | 0.144 | 0.113 |
| 10 | Eif4h+Rnf187 | 0.293 | 0.228 | 0.1235 |
| 11 | Grcc10+Tuba | 0.113 | 0.154 | 0.1665 |
| 12 | Rnf187+Gusb | 0.266 | 0.325 | 0.175 |
| 13 | Rnf187+Ppia | 0.145 | 0.210 | 0.1935 |
| 14 | Rnf187+18S rRNA | 0.059 | 0.252 | 0.212 |
| 15 | Grcc10+Rpl13 | 0.211 | 0.232 | 0.237 |
3.5. Validation of the Selected Reference Genes
For validation of the reliability of selected reference genes, the changes in these gene expressions in the KYDS-virus samples were investigated. An analysis of the relative expression level of target genes was performed. The data confirmed the reliability of the recommended reference genes in mouse lung tissue. The expression of TLR3, TLR4, TLR7, and cytosolic RIG-I, which play important roles in the host's innate immune response, was examined. The interferon regulatory transcription factor is triggered by this interaction, and NF-κB is activated, which affects the expression of numerous chemokines and proinflammatory cytokines [31]. Thus, the expression of TLR3, TLR4, TLR7, and RIG-I was investigated in mouse lung tissue (Figure 3).
Figure 3.
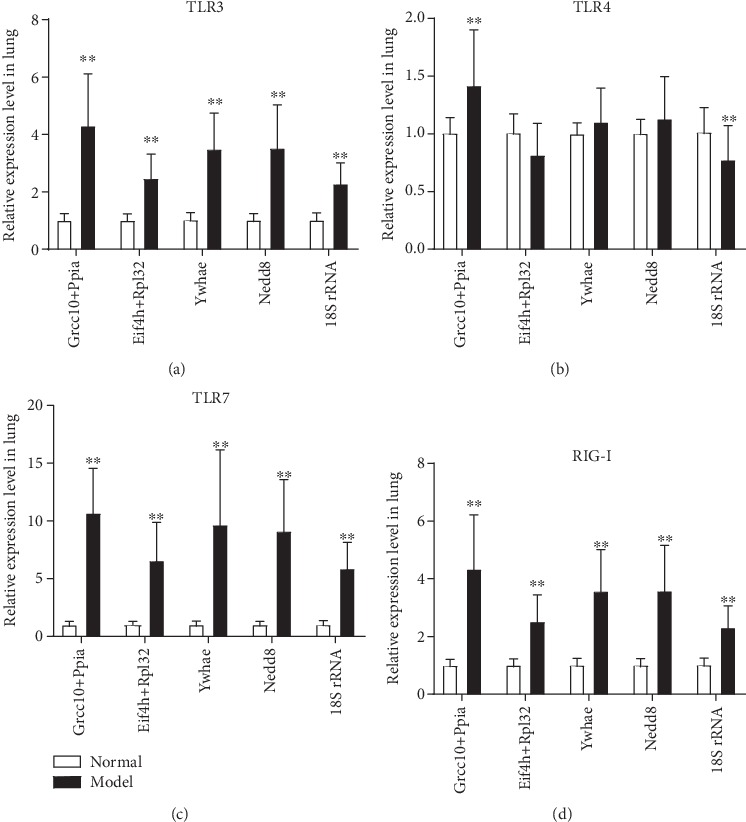
Relative expression levels of toll-like receptor (TLR)3, TLR4, TLR7, and retinoic acid-inducible gene-I (RIG-I) in lung tissues from normal and model mice were measured by reverse transcription-quantitative polymerase chain reaction and normalized to the indicated reference gene. The fold changes of model (KYDS-virus) mice are indicated next to the bars. The expression levels of normal mice were set at a relative expression of 1 (∗∗P < 0.01 vs. the normal group; n = 10 in each group, mean ± standard deviation). KYDS: Kidney Yang Deficiency Syndrome.
By using the recommended reference genes from the abovementioned analysis, the combination of Grcc10 and Ppia (independent-samples t-test), and the combination of Eif4h and Rpl32 (NormFinder), Ywhae (GeNorm), Nedd8 (BestKeeper), and 18S rRNA (NormFinder), we observed a significant increase of TLR3, TLR7, and RIG-I expression in the model group compared with the normal group (Figures 3(a), 3(c), and 3(d), P < 0.01). But the increased levels were different among these recommended reference genes, and the combination of Grcc10 and Ppia had the highest relative gene expression. For TLR4 study, the expressions of these candidate reference genes were very different, and only the combination of Grcc10 and Ppia and the single recommended gene 18S rRNA expression exhibited obvious changes in the model group compared with the normal group (Figure 3(b), P < 0.01). The combination of Grcc10 and Ppia exerted an upregulating effect; however, 18S rRNA exerted a downregulating effect on TLR4 gene expression. These results demonstrate the importance of selecting suitable reference genes for normalization to obtain reliable results in gene expression studies for the KYDS-virus treatment.
3.6. Diagram of Strategy for Screening Reference Gene
To summarize, the novel strategy for screening reference gene is mainly based on the independent-samples t-test and comparison of absolute value of difference for different groups. This novel strategy could determine the best single reference gene or the best combination of two genes (Figure 4).
Figure 4.
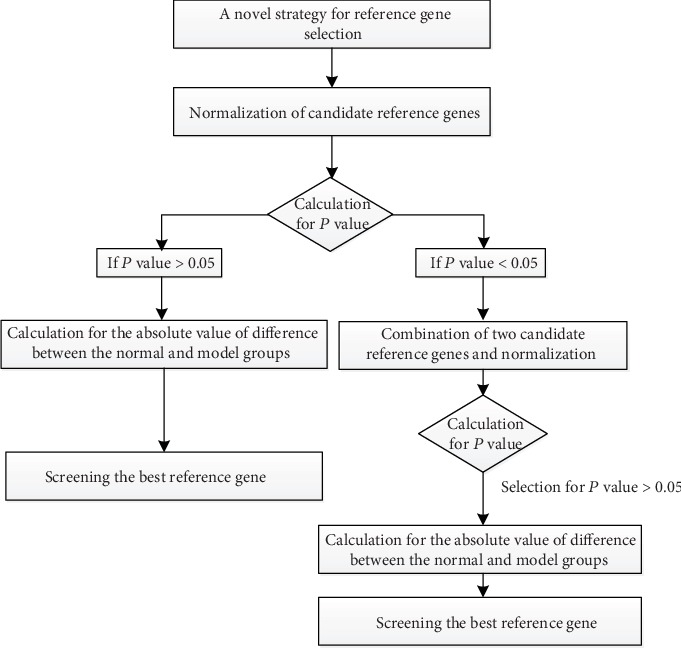
Diagram of strategy for screening reference gene.
4. Discussion
In molecular biological research, the most frequently used strategy to study genes is gene expression analysis. qPCR is generally employed to study relative gene expression due to its versatility and accuracy. The results obtained from qPCR are strongly affected by the stability of the reference gene selected to normalize the gene expression data; therefore, it is crucial to select stably expressed genes as internal references.
We therefore screened suitable genes for data normalization in KYDS-virus mouse lung tissue. The expression stability of 15 reference genes (Actb, β2m, Gapdh, Gusb, Tuba, Grcc10, Eif4h, Rnf187, Nedd8, Ywhae, 18S rRNA, Rpl13, Ubc, Rpl32, and Ppia) was analyzed using three very common software packages: GeNorm, NormFinder, and BestKeeper. These statistical algorithms have been employed to select and validate reference genes for RT-qPCR data normalization across numerous tissues, species, and different treatments. Grcc10, Eif4h, Rnf187, Nedd8, and Ywhae are all considered consistently and highly expressed genes that can be used as reference controls in expression profiling analysis by RNA-seq [32], particularly in mouse lung tissue. Additionally, two widely used reference genes (Gapdh and Actb) were selected. However, in numerous cases, the expression of these genes has been found to be variable in various tissues, cells, and experimental conditions, as discussed by Suzuki et al. [33]. In particular, cellular macromolecule synthesis (shut-off) is inhibited by influenza virus infection and causes global gene expression changes affecting the mRNA levels of several genes [34, 35].
Our previous work has found that traditionally used reference genes, for example, Gapdh or Actb, were not stable in lung tissue in KYDS after IAV infection compared with normal control mice (data not shown). It has also been reported that influenza virus infection strongly affects the expression levels of the Actb and Gapdh reference genes, and they are therefore not reliable [36].
Ywhae and Tuba were ranked as the most stable reference genes by GeNorm; 18S rRNA and the combination of Eif4h and Rpl32 were selected by NormFinder; and Nedd8 was recommended by BestKeeper. Although these software types are based on different algorithms, it was revealed that a single candidate reference gene was not suitable for relative gene expression (Figure 5(b)). We also proved that the combination of Eif4h and Rpl32 by NormFinder was not suitable as a reliable recommended reference gene. In the present study, a new strategy facilitating reference gene selection by RT-qPCR analysis in KYDS mice infected with IAV H1N1 was explored. An independent-samples t-test was used to evaluate the combination of candidate reference gene pairs between the normal and model groups, and 15 gene combinations were selected (P > 0.05). Subsequently, the absolute difference in mean Cq values for these potential combinations of reference genes was calculated in the two groups, and the smallest absolute difference value indicated that the combination of the genes was the best stable candidate reference gene. In view of these results (Table 4), the best combination of genes was Grcc10 and Ppia.
Figure 5.
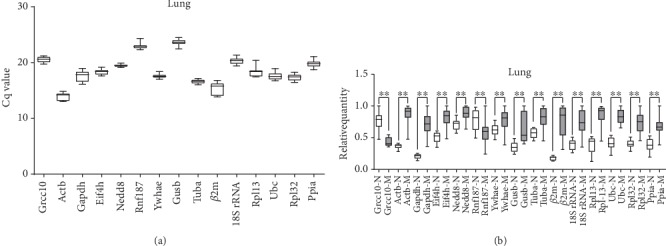
Distribution of quantification cycle (Cq) value and relative quantity for candidate reference genes. Box-whisker plot showing raw Cq values (a) and relative quantity (b) distribution of each reference gene in lung tissue. Boxes represent the quartiles with medians, whereas the maximum and minimum values are represented by whiskers.
Under various experimental conditions, unreliable conclusions have been reached due to the instability of the reference gene; it is therefore crucial to verify reference gene stability for each normalizing experiment that attempts to show little differences in mRNA abundance. By using the different recommended reference genes from different algorithms, we observed similar changes in TLR3, TLR7, and RIG-I after KYDS-virus model establishment. The outcomes normalized with different reference genes can be detrimental, as it can suggest that there is no change or even that the change is in the opposite direction with the TLR4 target gene. In the present study, we investigated this phenomenon when the data were normalized with a single or paired reference gene. In different treatment groups, the target gene expression could appear to be markedly different, yielding biased results. Our data demonstrated that it is crucial to apply validated reference genes and to study their expression stability when employed for normalization of RT-qPCR data.
In conclusion, to the best of our knowledge, this is the first study especially planned to assess the effects of KYDS-virus on mouse lung tissue using a set of reference genes for gene expression normalization with the help of RT-qPCR in a BALB/c mouse model. For statistical analysis, NormFinder, GeNorm, BestKeeper, and independent-samples t-test were used. The four algorithms yielded different results with respect to the ranking of these candidate reference genes, indicating the importance of using more than one software type to achieve the best result. It is also concluded that, among 15 candidate reference genes investigated, any individual reference gene is not suitable for KYDS-virus mice. The combination of reference genes is necessary, and the combination of Grcc10 and Ppia was found to be the most stable for the KYDS-virus mouse model. Validation of the selected reference genes with target genes (TLR3, TLR4, TLR7, and RIG-I) under certain experimental conditions was also performed. Selecting suitable reference genes is crucial for obtaining reliable results in gene expression research. The present study provides a basis for reference gene selection and practical strategies for future gene expression studies of KYDS combined with IAV infection. This method may be generally applied to promote the selection of the optimal reference gene in other experiments.
Acknowledgments
The present study was financially supported by the National Natural Science Foundation of China (grant nos. 81873220 and 81774167) and the Key Research and Development Program in Shandong Province (grant no. 2018CXGC1307). Ms. Li Liu is a technical support of Bio-Rad Company, she gave us very professional advice for qPCR data processing, and we are very appreciative of her help. We also need to thank Dr. Cheng-gang Wang of Shandong University of Traditional Chinese Medicine for his statistical analysis suggestion.
Contributor Information
Rong Rong, Email: rosierong@163.com.
Yong Yang, Email: yy7204@163.com.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
All the authors declare that they have no competing interests to disclose.
Supplementary Materials
Gapdh description: these are RT-qPCR raw data of Gapdh genes. The normal group and the KYDS model group both have good melt peak curves, but they have obvious differences in Ct values more than two times. The Ct values of Gapdh description: these are Ct values of Gapdh genes. The average of Ct values in the normal group is 21.90; the average of Ct values in the model group is 19.89. The difference value between the two groups is 2.02. The Ct values of 5 reference gene description: this excel file includes Ct values of the normal and model groups from 5 reference genes (Gusb, Gapdh, Actb, β2m, and Tuba). They were analyzed with statistical method by Student's t-test; all the paired groups have a significant difference (P < 0.01).
References
- 1.Derveaux S., Vandesompele J., Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50(4):227–230. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Leong D. T., Gupta A., Bai H. F., et al. Absolute quantification of gene expression in biomaterials research using real-time PCR. Biomaterials. 2007;28(2):203–210. doi: 10.1016/j.biomaterials.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 4.Guenin S., Mauriat M., Pelloux J., Van Wuytswinkel O., Bellini C., Gutierrez L. Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. Journal of Experimental Botany. 2009;60(2):487–493. doi: 10.1093/jxb/ern305. [DOI] [PubMed] [Google Scholar]
- 5.Artico S., Nardeli S. M., Brilhante O., Grossi-de-Sa M. F., Alves-Ferreira M. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biology. 2010;10:p. 49. doi: 10.1186/1471-2229-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandesompele J., De Preter K., Pattyn F., et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7, article RESEARCH0034) doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology. 2007;8(2, article R19) doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen C. L., Jensen J. L., Orntoft T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 9.Park S. C., Kim Y. H., Ji C. Y., et al. Stable internal reference genes for the normalization of real-time PCR in different sweetpotato cultivars subjected to abiotic stress conditions. PLoS One. 2012;7(12, article e51502) doi: 10.1371/journal.pone.0051502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnology Letters. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 11.Chen Q. Experimental Methodology of Pharmacological Research in Traditional Chinese Medicine. Beijing: People’s Health Publishing House; 1993. [Google Scholar]
- 12.Tang Z.-h., Chen H.-b., Li W.-h., et al. Comparative study of epidemiological investigation and expert-evaluation system of kidney-Yang insufficiency syndrome with old-aging. Liaoning Journal of Traditional Chinese Medicine. 2011;38:611–613. [Google Scholar]
- 13.Barker W. H., Borisute H., Cox C. A study of the impact of influenza on the functional status of frail older people. Archives of Internal Medicine. 1998;158(6):645–650. doi: 10.1001/archinte.158.6.645. [DOI] [PubMed] [Google Scholar]
- 14.Christenson B., Lundbergh P., Hedlund J., Ortqvist A. Effects of a large-scale intervention with influenza and 23-valent pneumococcal vaccines in adults aged 65 years or older: a prospective study. The Lancet. 2001;357(9261):1008–1011. doi: 10.1016/S0140-6736(00)04237-9. [DOI] [PubMed] [Google Scholar]
- 15.Thompson W. W., Shay D. K., Weintraub E., et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289(2):179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 16.Cox N. J., Subbarao K. Global epidemiology of influenza: past and present. Annual Review of Medicine. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- 17.Lu J.-b., Zhou R.-q., Liu R.-r., Xue R.-h. Investigation and analysis of kidney deficiency and patterns in the elderly. Journal of Shanghai University of Traditional Chinese Medicine. 2002;16(1):22–23. [Google Scholar]
- 18.Zhang Y., Liu Y., Li X., et al. Age-related changes in prevalence and symptom characteristics in kidney deficiency syndrome with varied health status: a cross-sectional observational study. Journal of Traditional Chinese Medical Sciences. 2014;1(1):20–27. doi: 10.1016/j.jtcms.2014.11.009. [DOI] [Google Scholar]
- 19.Huai-Chang F., Wei-hong L., Shi-hong Y., et al. Study on the diagnosis of kidney-"Yang" deficiency by 2067 cases of epidemiological survery. LIshizhen Medicine and Materia Medica Research. 2010;21:952–954. [Google Scholar]
- 20.Gong Y., Liu L., He X., et al. The th17/treg immune balance in ulcerative colitis patients with two different chinese syndromes: dampness-heat in large intestine and spleen and kidney yang deficiency syndrome. Evidence-based Complementary and Alternative Medicine. 2015;2015:10. doi: 10.1155/2015/264317.264317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding W. J., Yan S. L., Zeng Y. Z., et al. Insufficient activity of MAPK pathway is a key monitor of kidney-Yang deficiency syndrome. Journal of Alternative and Complementary Medicine. 2009;15(6):653–660. doi: 10.1089/acm.2008.0364. [DOI] [PubMed] [Google Scholar]
- 22.Yang J., Wang Y., Bao Y., Guo J. The total flavones from Semen cuscutae reverse the reduction of testosterone level and the expression of androgen receptor gene in kidney-yang deficient mice. Journal of Ethnopharmacology. 2008;119(1):166–171. doi: 10.1016/j.jep.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 23.Julkunen I., Sareneva T., Pirhonen J., Ronni T., Melen K., Matikainen S. Molecular pathogenesis of influenza A virus infection and virus-induced regulation of cytokine gene expression. Cytokine & Growth Factor Reviews. 2001;12(2-3):171–180. doi: 10.1016/s1359-6101(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 24.Zaas A. K., Chen M., Varkey J., et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host & Microbe. 2009;6(3):207–217. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wareing M. D., Lyon A. B., Lu B., Gerard C., Sarawar S. R. Chemokine expression during the development and resolution of a pulmonary leukocyte response to influenza A virus infection in mice. Journal of Leukocyte Biology. 2004;76(4):886–895. doi: 10.1189/jlb.1203644. [DOI] [PubMed] [Google Scholar]
- 26.Tang N., Liu L., Qiu H., Shi W., Mao D. Analysis of gene expression and functional changes of adrenal gland in a rat model of kidney yang deficiency syndrome treated with Sini decoction. Experimental and Therapeutic Medicine. 2018;16(4):3107–3115. doi: 10.3892/etm.2018.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z. X., Shao Y. Y., Ming L., Yi L., Ji R. Influence of bokhi on kidney-yang-deficiency syndrome in rats. Journal of Camel Practice and Research. 2017;24(3):281–290. doi: 10.5958/2277-8934.2017.00048.0. [DOI] [Google Scholar]
- 28.Rong R., Li R.-r., Hou Y.-b., et al. Mahuang-Xixin-Fuzi decoction reduces the infection of influenza a virus in kidney-Yang deficiency syndrome mice. Journal of Ethnopharmacology. 2016;192:217–224. doi: 10.1016/j.jep.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Li P., Cui Q., Wang L., et al. A simple and robust approach for evaluation of antivirals using a recombinant influenza virus expressing Gaussia luciferase. Viruses. 2018;10(6):p. 325. doi: 10.3390/v10060325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Z., Wang L., Yue D., et al. Evaluation of reference genes for normalization of RT-qPCR gene expression data for Trichoplusia ni cells during Antheraea pernyi (Lepidoptera: Saturniidae) multicapsid nucleopolyhedrovirus (AnpeNPV) infection. Journal of Insect Science. 2019;19(1):p. 4. doi: 10.1093/jisesa/iey133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawai T., Akira S. Signaling to NF-κB by toll-like receptors. Trends in Molecular Medicine. 2007;13(11):460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Li B., Qing T., Zhu J., et al. A comprehensive mouse transcriptomic BodyMap across 17 tissues by RNA-seq. Scientific Reports. 2017;7(1):p. 4200. doi: 10.1038/s41598-017-04520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki T., Higgins P. J., Crawford D. R. Control selection for RNA quantitation. BioTechniques. 2000;29(2):332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- 34.Reemers S. S., van Haarlem D. A., Groot Koerkamp M. J., Vervelde L. Differential gene-expression and host-response profiles against avian influenza virus within the chicken lung due to anatomy and airflow. The Journal of General Virology. 2009;90(9):2134–2146. doi: 10.1099/vir.0.012401-0. [DOI] [PubMed] [Google Scholar]
- 35.Kawada J., Kimura H., Kamachi Y., et al. Analysis of gene-expression profiles by oligonucleotide microarray in children with influenza. The Journal of General Virology. 2006;87, Part 6:1677–1683. doi: 10.1099/vir.0.81670-0. [DOI] [PubMed] [Google Scholar]
- 36.Kuchipudi S. V., Tellabati M., Nelli R. K., et al. 18S rRNA is a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virology Journal. 2012;9:p. 230. doi: 10.1186/1743-422x-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gapdh description: these are RT-qPCR raw data of Gapdh genes. The normal group and the KYDS model group both have good melt peak curves, but they have obvious differences in Ct values more than two times. The Ct values of Gapdh description: these are Ct values of Gapdh genes. The average of Ct values in the normal group is 21.90; the average of Ct values in the model group is 19.89. The difference value between the two groups is 2.02. The Ct values of 5 reference gene description: this excel file includes Ct values of the normal and model groups from 5 reference genes (Gusb, Gapdh, Actb, β2m, and Tuba). They were analyzed with statistical method by Student's t-test; all the paired groups have a significant difference (P < 0.01).
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


