Abstract
Cell division cycle protein (CDC20) has been observed to be expressed higher in various kinds of human cancers and was associated with poor prognosis. However, studies on role of CDC20 in breast cancer are seldom reported till now, most of which are not systematic and conclusive. The present study was performed to analyze the expression pattern, potential function, and distinct prognostic effect of CDC20 in breast cancer using several online databases including Oncomine, bc-GenExMiner, PrognoScan, and UCSC Xena. To verify the results from databases, we compared the mRNA CDC20 expression in breast cancer tissues and adjacent normal tissues of patients by real-time PCR. We found that CDC20 was expressed higher in different types of breast cancer, comparing with normal tissues. Moreover, the patients with a more advanced stage of breast cancer tended to express higher level CDC20. CDC20 was expressed higher in breast cancer tissues than normal tissues from patients in our hospital, consistent with the results from databases. Estrogen receptor (ER) and progesterone receptor (PR) status were negatively correlated with CDC20 level. Conversely, Scarff–Bloom–Richardson (SBR) grade, Nottingham prognostic index (NPI), epidermal growth factor receptor-2 (HER-2) status, basal-like status, and triple-negative status were positively related to CDC20 expression in breast cancer patients with respect to normal individuals. Higher CDC20 expression correlated with worse survival. Finally, a positive correlation between CDC20 and Targeting protein for Xenopus kinesin-like protein 2 (TPX2) expression was revealed. CDC20 could be considered as a potential predictive indicator for prognosis of breast cancer with co-expressed TPX2 gene.
Keywords: Bioinformatics analysis, Breast cancer, CDC20, Prognosis
Introduction
Breast cancer is the most common malignant tumor and remains a major cause of deaths in women [1]. With the development of treatment, including surgery, chemotherapy, radiotherapy, endocrine therapy, and target therapy, both disease-free survival and overall survival (OS) of breast cancer have been significantly improved. However, after systemic therapies, there are still some patients that died of breast cancer, especially for advanced breast cancer. Breast cancer is related with inactivation of a large number of tumor suppressor genes and oncogenes [2]. As we know, biomarkers are reported as surrogates of the clinical features for predicting outcomes. It is important to identify more effective, sensitive, and specific biomarkers for the prognosis of patients with breast cancer [3].
The cell-division cycle consists of a series of complex processes which are regulated by numbers of cell cycle regulatory proteins [4]. The cell division cycle protein (CDC) 20 (CDC20), acting as a regulatory protein, is a target molecule in the cell-cycle checkpoint [5]. It is also a key E3 ligase, which can activate adenomatous polyposis coli (APC) [5]. In addition to regulating cell cycle, recent evidence has demonstrated that CDC20 also plays an important role in carcinogenesis and cancer progression, having the potential to become a promising therapeutic target [6]. CDC20 has been observed as expressed higher in different kinds of human cancers and was associated with poor prognosis such as, oral squamous cell carcinoma [7], gastric cancer [8], urothelial bladder cancer [9], colorectal cancer [10], lung cancer [11], and pancreatic cancer [12].
Recently, CDC20 has been demonstrated to act as an oncogene in breast cancer progression [13], However, studies on role of CDC20 in breast cancer are seldom reported till now, most of which are not systematic and conclusive. Moreover, the CDC20 expression’s prognostic significance is also uncertain. Therefore, it is imperative to recognize that further study is necessary to determine the oncogenic role of CDC20 in breast tumorigenesis.
In the present study, we performed a deep bioinformatics analysis of the clinical parameters and survival data related to CDC20 in breast cancer patients using several online databases in order to evaluate the prognostic significance of CDC20 gene in breast cancer treatment. Moreover, we used 22 pairs of breast tissues from breast cancer patients in our hospital to compare the expression of CDC20 gene between the cancer tissues and normal tissues by real-time PCR.
Materials and methods
ONCOMINE data-mining analysis
ONCOMINE (www.oncomine.org), an online web-based cancer database for RNA and DNA sequences, was used to facilitate data-mining of the transcriptional expressions of genes in 20 types of cancer [14]. Data used in the present study from ONCOMINE were updated in April 2019. Transcriptional expressions of CDC20 in cancer samples were compared with those in normal individuals using Student’s t test. Statistically significant values and fold change were demarcated as P-value ≤ 1E-4 and 2, respectively. Genes co-expressed with CDC20 were analyzed by using online Oncomine analysis tools.
UALCAN
UALCAN (http://ualcan.path.uab.edu/) is a user-friendly, interactive web resource for analyzing transcriptome data of cancers from The Cancer Genome Atlas (TCGA) [15]. Data from UALCAN was updated in March 2019. The mRNA expression of CDC20 between breast cancer and normal tissues as well as different stages cancer was also detected using the UALCAN web portal (TCGA level 3 data).
bc-GenExMiner v4.2
Breast Cancer Gene-Expression Miner v4.2 (bc-GenExMiner v4.2) is a statistical mining tool that incorporates 264 independent datasets and three classical mining functions: expression, prognosis, and correlation [16,17]. Expression data were analyzed with clinical parameters such as age, nodal status, estrogen receptor (ER), progesterone receptor (PR), epidermal growth factor receptor-2 (HER-2), Scarff–Bloom–Richardson (SBR) grade, and Nottingham prognostic index (NPI). Correlation of CDC20 and TPX2 was assessed using the correlation module.
PrognoScan
The PrognoScan (http://www.prognoscan.org/) is an online database for assessing the biological relationship between gene expression and survival data including OS, distant metastasis-free survival, relapse-free survival (RFS), and disease-specific survival in breast cancer patients. The results are based on a collection of publicly available cancer microarray datasets [18]. Data used in the present study from PrognoScan were updated in April 2019. P-value, hazard ratio (HR), and 95% confidence intervals (CI) could be automatically calculated according to a certain gene expression. For statistical analysis and visualization, R packages (http://www.r-project.org) were used.
UCSC Xena
The UCSC Xena (http://xena.ucsc.edu/) is a popular genomics browser that provides visualization and integration for analyzing and viewing the public data hubs [19]. The heat map and correlation between CDC20 and TPX2 were generated by data mining in TCGA Breast Cancer using the UCSC Xena browser.
Breast tissue samples
Twenty-two pairs of breast tissue samples used in quantitative real-time PCR (RT-PCR) were obtained from the First Affiliated Hospital of Nanjing Medical University, China, between 2014 and 2016. The collection and use of the samples was reviewed and approved by the Institutional Ethics Committee of the First Affiliated Hospital of Nanjing Medical University.
RNA isolation and RT-PCR analysis
Total RNA was isolated using TRIzol reagent (TaKaRa, Japan) and 1000 ng RNA was reverse-transcribed into cDNA using Primescript RT Reagent (TaKaRa, Japan). The RT-PCR was performed using FastStart Universal SYBR Green Master (Roche, Switzerland) in an RT-PCR instrument (Applied Biosystems, U.S.A.), and β-actin was used as an endogenous control. The following PCR primers were used:
CDC20 forward, 5′-GCACAGTTCGCGTTCGAGA-3′
CDC20 reverse, 5′-CTGGATTTGCCAGGAGTTCGG-3′
β-actin forward, 5′-GCTGTGCT ATCCCTGTACGC-3′
β-actin reverse, 5′-TGCCTCAGGGCAGCGGAACC-3′.
Statistical analysis
RT-PCR were repeated in triplicate, unless otherwise specified. The data were analyzed using the SPSS 20.0 software (Chicago, U.S.A.). We analyzed the statistical significance of the differences between groups using Student’s t test, and a statistically significant difference was considered at the level of P<0.05.
Results
Increased expression of CDC20 gene in breast cancer patients’ tissues
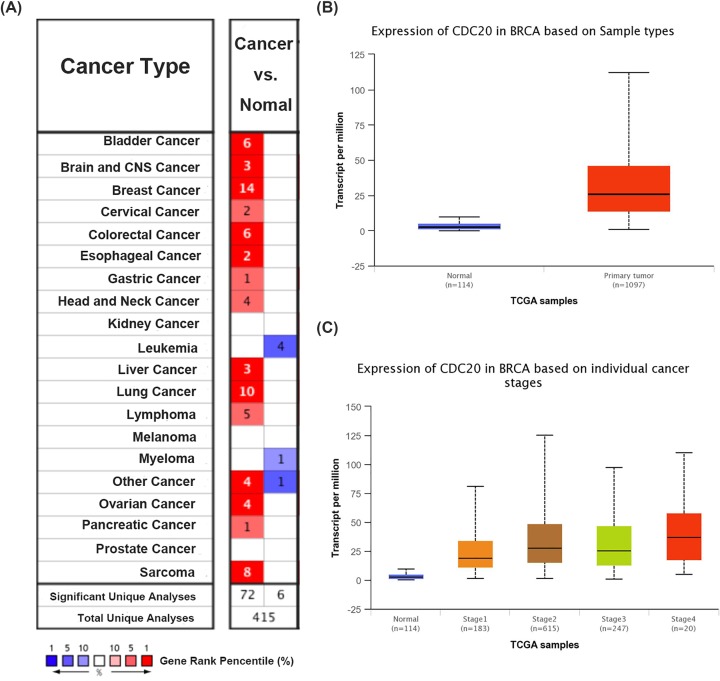
First, the expression of CDC20 gene in 20 types of cancer was measured and compared with normal tissues using the Oncomine online database (Figure 1A). We found that increased CDC20 (red) was observed in bladder cancer, brain and CNS cancer, cervical cancer, colorectal cancer, esophageal cancer gastric cancer, head and neck cancer, liver cancer, lung cancer, lymphoma, ovarian cancer, pancreatic cancer, sarcoma, and especially breast cancer, whereas, decreased level of CDC20 (blue) was observed in leukemia and myeloma. Consistently, using UALCAN website, we also found that higher mRNA CDC20 was expressed in breast cancer tissues than in normal tissues (Figure 1B, P<0.05). Next, we focused on whether mRNA expression of CDC20 was related to cancer stage in individual patients. As shown in Figure 1C, the results indicated that patients with a more advanced stage of breast cancer tended to express higher levels CDC20.
Figure 1. Increased expression of CDC20 gene in breast cancer tissues in public databases.
(A) Expression of CDC20 in 20 types of cancer vs. corresponding normal tissues using the Oncomine database with the threshold of fold change ≥ 2, P-value ≤1E-4, and gene rank ≥ top 10%. Red and blue, respectively, stand for the numbers of datasets with statistically significant (P<0.05) increased and decreased levels of CDC20 gene. (B) Higher mRNA CDC20 was expressed in breast cancer tissues than in normal tissues (P<0.05) using UALCAN website. (C) Patients with a more advanced stage of breast cancer tended to express higher levels of CDC20.
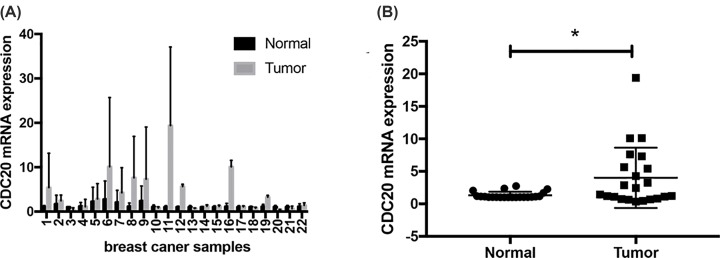
To verify the results above, we further compared the mRNA CDC20 expression in breast cancer tissues and adjacent normal tissues of patients in our hospital and found that CDC20 was expressed higher in breast cancer tissues, consistent with the results from databases (Figure 2, P<0.05).
Figure 2. Increased expression of CDC20 gene in breast cancer tissues from patients in our hospital.
(A) CDC20 mRNA expression in 22 pairs of breast cancer and adjacent tissues. (B) Average expression level of RBMS2 mRNA in 22 pairs of breast cancer tissues and adjacent normal breast tissues. Breast cancer tissues had higher expression of CDC20 than adjacent breast tissues (*, P<0.05).
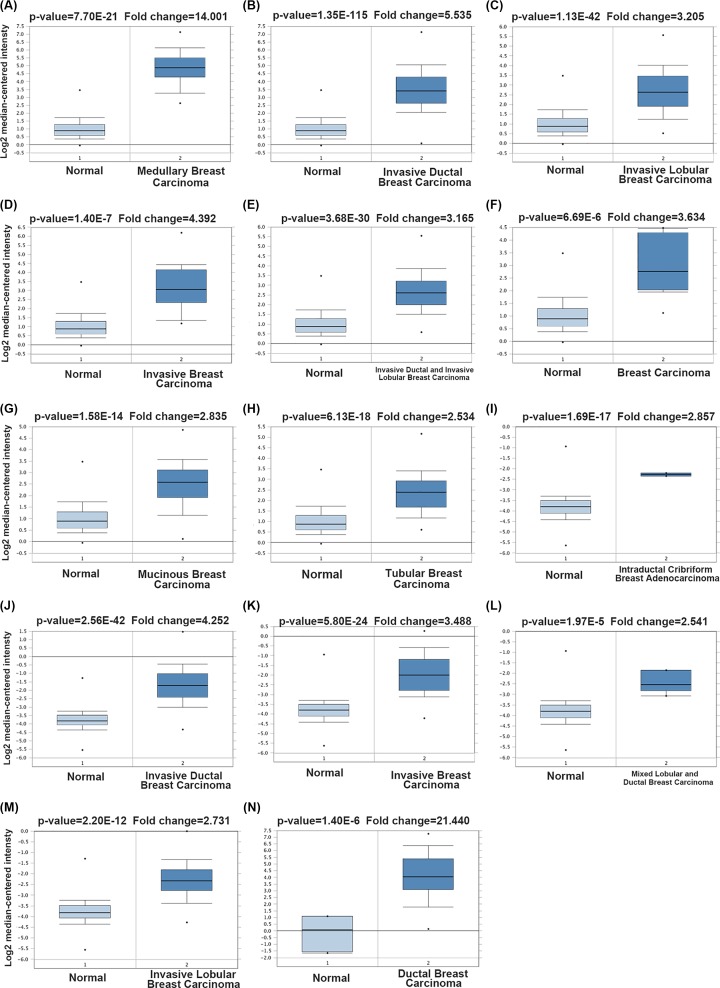
Oncomine analysis also revealed that CDC20 was significantly expressed higher in medullary breast carcinoma, invasive ductal breast carcinoma, invasive lobular breast carcinoma, invasive breast carcinoma, invasive ductal and invasive lobular breast carcinoma, breast carcinoma, mucinous breast carcinoma, tubular breast carcinoma, intraductal cribriform breast adenocarcinoma, invasive ductal breast carcinoma, invasive breast carcinoma, mixed lobular and ductal breast carcinoma, invasive lobular breast carcinoma, ductal breast carcinoma with respect to normal individuals (Figure 3 and Table 1).
Figure 3. Box plot comparing CDC20 expression in normal individuals and breast cancer patients obtained from the Oncomine database.
Analysis is shown for medullary breast carcinoma (A), invasive ductal breast carcinoma (B), invasive lobular breast carcinoma (C), invasive breast carcinoma (D), invasive ductal and invasive lobular breast carcinoma (E), breast carcinoma (F), mucinous breast carcinoma (G), tubular breast carcinoma (H), intraductal cribriform breast adenocarcinoma (I), invasive ductal breast carcinoma (J), invasive breast carcinoma (K), mixed lobular and ductal breast carcinoma (L), invasive lobular breast carcinoma (M), ductal breast carcinoma (N).
Table 1. The significant changes of CDC20 expression in transcription level between different types of breast cancer and normal tissues (ONCOMINE database).
| Subtype of breast cancer | P-value | Fold change | Rank (%) | Sample | Reference |
|---|---|---|---|---|---|
| Medullary breast carcinoma | 7.70E-21 | 14.001 | 1% | 176 | 22522925 |
| Invasive ductal breast carcinoma | 1.35E-115 | 5.535 | 1% | 1700 | 22522925 |
| Invasive lobular breast carcinoma | 1.13E-42 | 3.205 | 2% | 292 | 22522925 |
| Invasive breast carcinoma | 1.40E-7 | 4.392 | 2% | 165 | 22522925 |
| Invasive ductal and invasive lobular breast carcinoma | 3.68E-30 | 3.165 | 2% | 234 | 22522925 |
| Breast carcinoma | 6.69E-6 | 3.634 | 3% | 158 | 22522925 |
| Mucinous breast carcinoma | 1.58E-14 | 2.835 | 4% | 190 | 22522925 |
| Tubular breast carcinoma | 6.13E-18 | 2.534 | 7% | 211 | 22522925 |
| Intraductal cribriform breast adenocarcinoma | 1.69E-17 | 2.857 | 1% | 64 | TCGA |
| Invasive ductal breast carcinoma | 2.56E-42 | 4.252 | 1% | 450 | TCGA |
| Invasive breast carcinoma | 5.80E-24 | 3.488 | 2% | 137 | TCGA |
| Mixed lobular and ductal breast carcinoma | 1.97E-5 | 2.541 | 3% | 68 | TCGA |
| Invasive lobular breast carcinoma | 2.20E-12 | 2.731 | 3% | 97 | TCGA |
| Ductal breast carcinoma | 1.40E-6 | 21.440 | 4% | 47 | 16473279 |
CDC20 expression and clinical parameters of breast cancer patients
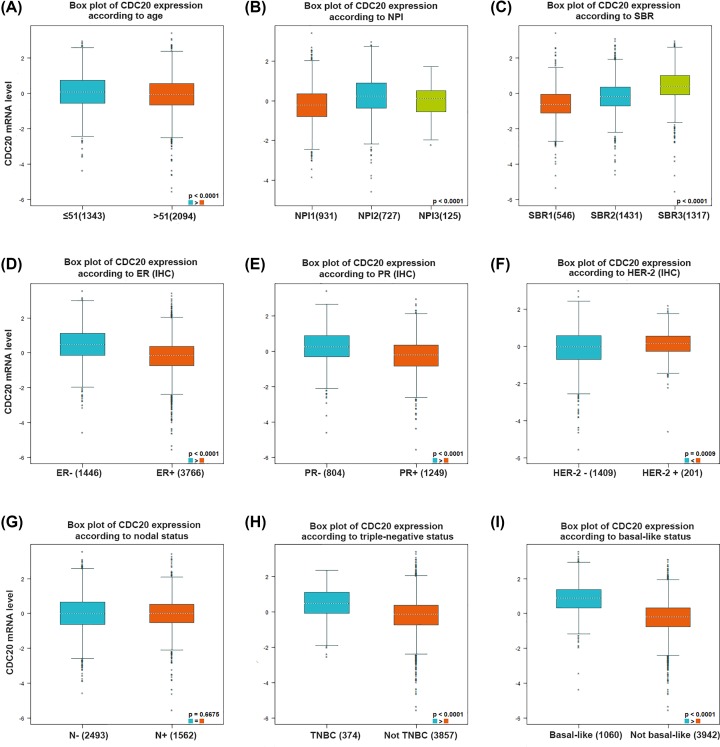
Using bc-GenExMiner v4.2 software, we implemented Welch’s test to compare the abnormal expression of CDC20 among different groups of patients according to clinical pathological features. For age criteria, CDC20 was significantly elevated in ≤51-year group with respect to >51-year group (Figure 4A and Table 2). As we know, the SBR is a histological grade that evaluates tubule formation, nuclear characteristics of pleomorphism, and mitotic index [20]. Based on tumor size, lymph node stage, and tumor grade, the NPI is used to stratify patients into additional prognostic groups. The SBR grade and NPI index are two wonderful prognostic models for breast cancer [21]. More advanced SBR grade and NPI index were associated with higher CDC20 level (Figure 4B,C and Table 2). ER-positive or PR-positive breast cancer patients tended to express lower CDC20 gene compared with ER-negative or PR-negative patients (Figure 4D,E and Table 2). Patients with HER-2-negative status showed reduced expression of CDC20 than HER-2-positive patients (Figure 4F and Table 2). Regarding nodal status, there was no significant difference between positive and negative group (Figure 4G and Table 2). Moreover, CDC20 was significantly reduced in non-triple-negative and non-basal-like breast cancer patients compared with triple-negative and basal-like breast cancer patients (Figure 4H,I and Table 2).
Figure 4. Box plot evaluating CDC20 expression among different groups of patients based on clinical parameters using the bc-GenExMiner 4.2.
Analysis is shown for age (A), NPI index (B), SBR grade (C), ER (D), PR (E), HER-2 (F), nodal status (G), triple-negative status (H) and basal-like status (I).
Table 2. The relationship between mRNA expression of CDC20 and clinicopathological parameters of breast carcinoma.
| Variables | Number of patients | CDC20 mRNA | P-value |
|---|---|---|---|
| Age | |||
| ≤51 | 1392 | ↑ | <0.0001 |
| >51 | 2210 | ||
| Nodal status | |||
| − | 2493 | 0.6675 | |
| + | 1562 | ||
| ER | |||
| − | 1446 | ↑ | <0.0001 |
| + | 3766 | ||
| PR | |||
| − | 804 | ↑ | <0.0001 |
| + | 1249 | ||
| HER2 | |||
| − | 1409 | 0.0009 | |
| + | 201 | ↑ | |
| SBR | |||
| 1 | 546 | <0.0001 | |
| 2 | 1431 | ↑ | |
| 3 | 1317 | ↑ | |
| NPI | |||
| 1 | 931 | <0.0001 | |
| 2 | 727 | ↑ | |
| 3 | 125 | ↑ | |
| Basal-like status | |||
| Not | 3492 | <0.0001 | |
| Basal-like | 1060 | ↑ | |
| Triple-negative status | |||
| Not | 3857 | ||
| TNBC | 374 | ↑ | <0.0001 |
CDC20 expression and survival data of breast cancer patients
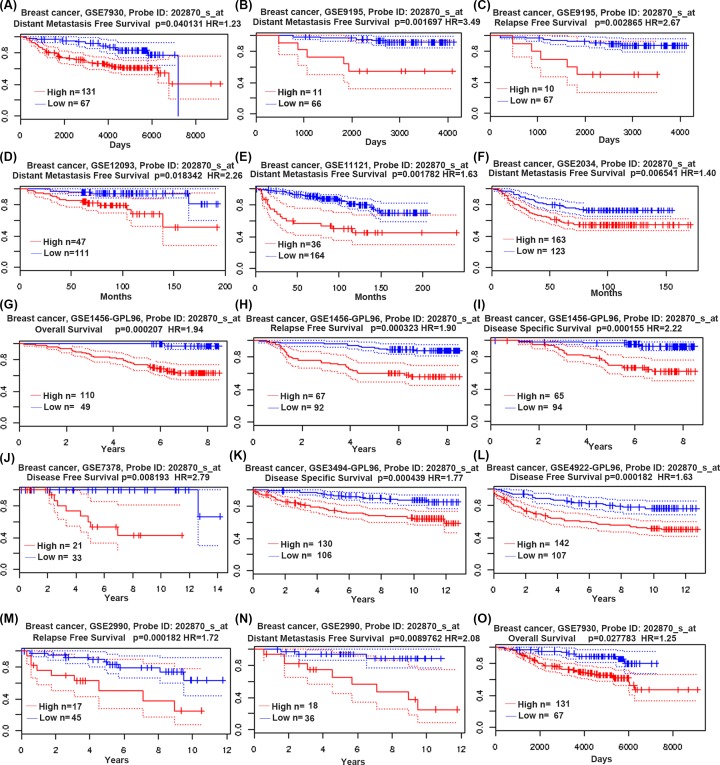
Then, we investigated the prognostic value of CDC20 gene using the PrognoScan database. Breast cancer patients with lower expression of CDC20 (blue) significantly showed preferable distant metastasis-free survival (Figure 5A,B,D–F,N and Table 3). Reduced CDC20 level (blue) was related to better RFS (Figure 5C,H,M and Table 3) and the cases with increased CDC20 gene presented worse disease-free survival (Figure 5J,L and Table 3). Moreover, down-regulated CDC20 gene (blue) was strongly associated with better disease-specific survival (Figure 5I,K and Table 3) and up-regulated CDC20 gene (red) was related to worse OS (Figure 5G,O and Table 3).
Figure 5. Survival curve evaluating the prognostic value of CDC20.
Survival curve using the PrognoScan Analysis is shown for distant metastasis-free survival (A,B,D–F,N), RFS for different probes (C,H,M), disease-free survival (J,L) disease-specific survival (I,K) and OS (G,O). Red and blue stand for high and low expression of CDC20 gene, respectively.
Table 3. The association of CDC20 expression and the survival in breast cancer patients.
| Dataset | End point | Probe ID | Location | n | Cox P-value | HR (95% CI) |
|---|---|---|---|---|---|---|
| GSE7390 | Distant metastasis-free survival | 202870_s_at | Uppsala, Oxford | 198 | 0.040131 | 1.23 (1.01–1.49) |
| GSE9195 | Distant metastasis-free survival | 202870_s_at | GUYT | 77 | 0.001697 | 3.49 (1.60–7.60) |
| GSE9195 | RFS | 202870_s_at | GUYT | 77 | 0.002865 | 2.67 (1.40–5.10) |
| GSE12093 | Distant metastasis-free survival | 202870_s_at | IO, NCI | 136 | 0.018342 | 2.26 (1.15–4.43) |
| GSE11121 | Distant metastasis-free survival | 202870_s_at | Mainz | 200 | 0.001782 | 1.63 (1.20–2.21) |
| GSE2034 | Distant metastasis-free survival | 202870_s_at | Rotterdam | 286 | 0.006541 | 1.40 (1.10–1.79) |
| GSE1456-GPL96 | OS | 202870_s_at | Stockholm | 159 | 0.000207 | 1.94 (1.37–2.74) |
| GSE1456-GPL96 | RFS | 202870_s_at | Stockholm | 159 | 0.000323 | 1.90 (1.34–2.70) |
| GSE1456-GPL96 | Disease-specific survival | 202870_s_at | Stockholm | 159 | 0.000155 | 2.22 (1.47–3.36) |
| GSE7378 | Disease-free survival | 202870_s_at | UCSF | 54 | 0.008193 | 2.79 (1.30–5.98) |
| GSE3494-GPL96 | Disease-specific survival | 202870_s_at | Uppsala | 236 | 0.000439 | 1.77 (1.29–2.44) |
| GSE4922-GPL96 | Disease-free survival | 202870_s_at | Uppsala | 249 | 0.000182 | 1.63 (1.26–2.11) |
| GSE2990 | RFS | 202870_s_at | Uppsala, Oxford | 62 | 0.014206 | 1.72 (1.11–2.65) |
| GSE2990 | Distant metastasis-free survival | 202870_s_at | Uppsala, Oxford | 54 | 0.008976 | 2.08 (1.20–3.59) |
| GSE7390 | OS | 202870_s_at | Uppsala, Oxford | 198 | 0.027783 | 1.25 (1.03–1.54) |
| GSE3143 | OS | 38414_at | Duke | 158 | 0.010445 | 1.67 (1.12–2.41) |
| E-TABM-158 | Disease-specific survival | 202870_s_at | UCSF | 117 | 0.038478 | 0.75 (0.57–0.98) |
Co-expression of CDC20 gene
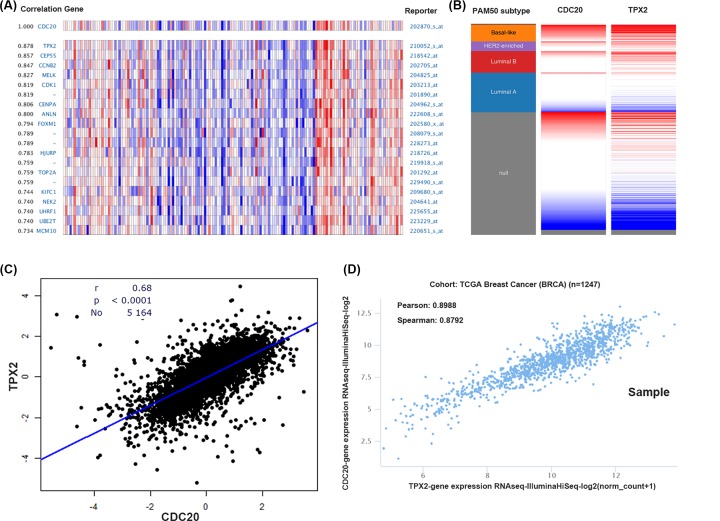
Finally, we investigated the co-expression of CDC20 gene using the Oncomine database. The co-expression profile of CDC20 was identified with a large cluster of 17779 genes across 159 breast cancer samples (Figure 6A). Targeting protein for Xenopus kinesin-like protein 2 (TPX2) is a top correlated gene, which is a microtubule-associated protein and encoded by a gene located on human chromosome band 20q11.1 [22]. A positive correlation between CDC20 and TPX2 expression was revealed using bc-GenExMiner 4.2 (Figure 6C). Moreover, after analyzing breast cancer patient data in the TCGA database using the UCSC Xena web-based tool, again we confirmed a positive correlation between CDC20 and TPX2 expression, as shown in the heat map (Figure 6B,D). These results suggested that CDC20 might be closely related to the TPX2 signaling pathway in breast cancer.
Figure 6. Co-expression of CDC20.
(A) CDC20 co-expression of genes analyzed using Oncomine. (B) Relationship between CDC20 and TPX2 in breast cancer analyzed using bc-GenExMiner v4.0. (C) Heat map of CDC20 expression and TPX2 mRNA expression across PAM50 breast cancer subtypes in the TCGA database, determined using UCSC Xena; D: correlation between CDC20 and TPX2 mRNA expression in the TCGA database, determined using UCSC Xena.
Discussion
Cell division cycle 20 (CDC20) is a vital molecule which acts as an important role in the cell cycle and an activator of the anaphase-promoting complex (APC/C) [23]. Higher expression of CDC20 has been observed in a variety of human cancers and is correlated with poor prognosis [7,24,25]. However, the significance of CDC20 expression in the development and prognosis of breast cancer remains largely unclear. To the best of our knowledge, this is one of the first study to identify CDC20 as a potential predictive biomarker for prognosis of breast cancer using comprehensive bioinformatics analysis.
In the present study, we performed a bioinformatics analysis of the clinical parameters and survival data related to CDC20 in breast cancer patients by pooling and analyzing several online tools. Comparing with normal tissues, Oncomine database revealed that CDC20 was expressed higher in different types of breast cancer including medullary breast carcinoma, invasive ductal breast carcinoma, invasive lobular breast carcinoma, and so on. CDC20 was also expressed higher in breast cancer tissues compared with adjacent normal tissues of patients in our hospital, confirming the results from databases online. Moreover, we also found patients with a more advanced stage of breast cancer tended to express higher levels CDC20. Consistently, Yuan et al. [26] reported that the mRNA and protein levels of CDC20 were significantly higher in breast cancer cells and high-grade primary breast cancer tissues.
For nodal status, there was no significant difference between positive and negative groups. ER and PR status were negatively correlated with CDC20 level. Conversely, SBR grade, NPI index, HER-2 status, basal-like status, and triple-negative status were positively related to CDC20 expression in breast cancer patients with respect to normal individuals. It is generally known that breast cancer patients with ER or PR positive, HER-2 negative, non-basal-like, or non-triple-negative status have a preferable outcome [27]. Therefore, these results indicated that lower expression of CDC20 may predict a better prognosis in breast cancer.
We further investigated the prognostic value of CDC20 in breast cancer using the PrognoScan database. These pooled results showed that higher CDC20 expression correlated with worse distant metastasis -ree survival, RFS, disease-free survival, disease-specific survival, and OS. These findings were in agreement with the notion of CDC20 as a tumor oncogene and a potential predictive biomarker for prognosis of breast cancer [13].
Finally, we checked the co-expression of CDC20 gene using the Oncomine, bc-GenExMiner, and UCSC Xena web-based tools and found that TPX2 was positively correlated with CDC20 expression. TPX2 is a microtubule-associated protein that is encoded by a gene located on human chromosome band 20q11.1 [22]. Overexpression of TPX2 has been observed in lung cancer, hepatic cancer, colon cancer [28–30]. Moreover, TPX2 is also a marker of poor tumor prognosis in several cancers [31]. Both CDC20 and TPX2 were related to the process of cell cycle [32]. Using bioinformatics analysis, Zhang et al. [33] found that elevated mRNA levels of CDC20 and TPX2 are associated with poor prognosis of lung adenocarcinoma. These observations, along with our findings of CDC20 in survival data, provided evidence that CDC20 gene might promote tumor progress associated with TPX2 expression.
In conclusion, the study was performed to comprehensively analyze the expression pattern, potential function, and distinct prognostic effect of CDC20 in breast cancer by pooling all currently available data online. CDC20 was highly expressed in different subtypes of breast cancer compared with normal tissues and was associated with several important clinical parameters. CDC20 could be considered as a potential predictive indicator for prognosis of breast cancer with co-expressed TPX2 gene. Over the past several decades, much research has focused on identifying new prognostic markers in order to make better clinical decisions and improve therapy and outcomes. More in-depth experiments are needed to validate the value of CDC20 for clinical decision-making in breast cancer.
Abbreviations
- bc-GenExMiner v4.2
Breast Cancer Gene-Expression Miner v4.2
- CDC20
cell division cycle protein
- ER
estrogen receptor
- HER-2
epidermal growth factor receptor-2
- NPI
Nottingham prognostic index
- OS
overall survival
- PR
progesterone receptor
- RFS
relapse-free survival
- RT-PCR
quantitative real-time PCR
- SBR
Scarff–Bloom–Richardson
- TCGA
The Cancer Genome Atlas
- TPX2
Targeting protein for Xenopus kinesin-like protein 2
Contributor Information
Ji-Fu Wei, Email: weijifu@hotmail.com.
Qiang Ding, Email: dingqiang@njmu.edu.cn.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Natural Science Foundation of China [grant numbers 81502294, 81702591]; the Project of Jiangsu Province Traditional Chinese Medicine Bureau [grant number YB201952]; the Natural Science Foundation of Jiangsu Province [grant number BK20170294]; and the Changzhou Sci&Tech Program [grant number CJ20159044].
Author Contribution
L.C., J.-F.W., and Q.D. conceived and designed the experiments. L.C., Y.-Z.H., and W.-X.C. performed the experiments. L.S., Z.L., X.Z., and X.-Y.D. analyzed the data. L.C. wrote the manuscript.
References
- 1.Fan L., Strasser-Weippl K., Li J.-J., St. Louis J., Finkelstein D.M., Yu K.-D. et al. (2014) Breast cancer in China. Lancet Oncol. 15, e279–e289 10.1016/S1470-2045(13)70567-9 [DOI] [PubMed] [Google Scholar]
- 2.Rennstam K., Baldetorp B., Kytola S., Tanner M. and Isola J. (2001) Chromosomal rearrangements and oncogene amplification precede aneuploidization in the genetic evolution of breast cancer. Cancer Res. 61, 1214–1219 [PubMed] [Google Scholar]
- 3.Duffy M.J., Walsh S., McDermott E.W. and Crown J. (2015) Biomarkers in breast cancer: where are we and where are we going? Adv. Clin. Chem. 71, 1–23 10.1016/bs.acc.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 4.Yu H. (2007) Cdc20: a WD40 activator for a cell cycle degradation machine. Mol. Cell 27, 3–16 10.1016/j.molcel.2007.06.009 [DOI] [PubMed] [Google Scholar]
- 5.Weinstein J., Jacobsen F.W., Hsu-Chen J., Wu T. and Baum L.G. (1994) A novel mammalian protein, p55CDC, present in dividing cells is associated with protein kinase activity and has homology to the Saccharomyces cerevisiae cell division cycle proteins Cdc20 and Cdc4. Mol. Cell. Biol. 14, 3350–3363 10.1128/MCB.14.5.3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smolders L. and Teodoro J.G. (2011) Targeting the anaphase promoting complex: common pathways for viral infection and cancer therapy. Expert Opin. Ther. Targets 15, 767–780 10.1517/14728222.2011.558008 [DOI] [PubMed] [Google Scholar]
- 7.Moura I.M., Delgado M.L., Silva P.M., Lopes C.A., do Amaral J.B., Monteiro L.S. et al. (2014) High CDC20 expression is associated with poor prognosis in oral squamous cell carcinoma. J. Oral Pathol. Med. 43, 225–231 [DOI] [PubMed] [Google Scholar]
- 8.Ding Z.Y., Wu H.R., Zhang J.M., Huang G.R. and Ji D.D. (2014) Expression characteristics of CDC20 in gastric cancer and its correlation with poor prognosis. Int. J. Clin. Exp. Pathol. 7, 722–727 [PMC free article] [PubMed] [Google Scholar]
- 9.Choi J.W., Kim Y., Lee J.H. and Kim Y.S. (2013) High expression of spindle assembly checkpoint proteins CDC20 and MAD2 is associated with poor prognosis in urothelial bladder cancer. Virchows Arch. 463, 681–687 10.1007/s00428-013-1473-6 [DOI] [PubMed] [Google Scholar]
- 10.Wu W.J., Hu K.S., Wang D.S., Zeng Z.L., Zhang D.S., Chen D.L. et al. (2013) CDC20 overexpression predicts a poor prognosis for patients with colorectal cancer. J. Transl. Med. 11, 142 10.1186/1479-5876-11-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato T., Daigo Y., Aragaki M., Ishikawa K., Sato M. and Kaji M. (2012) Overexpression of CDC20 predicts poor prognosis in primary non-small cell lung cancer patients. J. Surg. Oncol. 106, 423–430 10.1002/jso.23109 [DOI] [PubMed] [Google Scholar]
- 12.Chang D.Z., Ma Y., Ji B., Liu Y., Hwu P., Abbruzzese J.L. et al. (2012) Increased CDC20 expression is associated with pancreatic ductal adenocarcinoma differentiation and progression. J. Hematol. Oncol. 5, 15 10.1186/1756-8722-5-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karra H., Repo H., Ahonen I., Loyttyniemi E., Pitkanen R., Lintunen M. et al. (2014) Cdc20 and securin overexpression predict short-term breast cancer survival. Br. J. Cancer 110, 2905–2913 10.1038/bjc.2014.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D. et al. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B. et al. (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jezequel P., Campone M., Gouraud W., Guerin-Charbonnel C., Leux C., Ricolleau G. et al. (2012) bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res. Treat. 131, 765–775 10.1007/s10549-011-1457-7 [DOI] [PubMed] [Google Scholar]
- 17.Jezequel P., Frenel J.S., Campion L., Guerin-Charbonnel C., Gouraud W., Ricolleau G. et al. (2013) bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database 2013, bas060 10.1093/database/bas060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuno H., Kitada K., Nakai K. and Sarai A. (2009) PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genet. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casper J., Zweig A.S., Villarreal C., Tyner C., Speir M.L., Rosenbloom K.R. et al. (2018) The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 46, D762–D769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bansal C., Pujani M., Sharma K.L., Srivastava A.N. and Singh U.S. (2014) Grading systems in the cytological diagnosis of breast cancer: a review. J. Cancer Res. Ther. 10, 839–845 [DOI] [PubMed] [Google Scholar]
- 21.Lee A.H. and Ellis I.O. (2008) The Nottingham prognostic index for invasive carcinoma of the breast. Pathol. Oncol. Res. 14, 113–115 10.1007/s12253-008-9067-3 [DOI] [PubMed] [Google Scholar]
- 22.Neumayer G., Belzil C., Gruss O.J. and Nguyen M.D. (2014) TPX2: of spindle assembly, DNA damage response, and cancer. Cell. Mol. Life Sci. 71, 3027–3047 10.1007/s00018-014-1582-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang L., Zhang Z., Yang J., McLaughlin S.H. and Barford D. (2015) Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature 522, 450–454 10.1038/nature14471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y., Choi J.W., Lee J.H. and Kim Y.S. (2014) MAD2 and CDC20 are upregulated in high-grade squamous intraepithelial lesions and squamous cell carcinomas of the uterine cervix. Int. J. Gynecol. Pathol. 33, 517–523 10.1097/PGP.0000000000000082 [DOI] [PubMed] [Google Scholar]
- 25.Marucci G., Morandi L., Magrini E., Farnedi A., Franceschi E., Miglio R. et al. (2008) Gene expression profiling in glioblastoma and immunohistochemical evaluation of IGFBP-2 and CDC20. Virchows Arch. 453, 599–609 10.1007/s00428-008-0685-7 [DOI] [PubMed] [Google Scholar]
- 26.Yuan B., Xu Y., Woo J.H., Wang Y., Bae Y.K., Yoon D.S. et al. (2006) Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin. Cancer Res. 12, 405–410 10.1158/1078-0432.CCR-05-0903 [DOI] [PubMed] [Google Scholar]
- 27.Bagaria S.P., Ray P.S., Sim M.S., Ye X., Shamonki J.M., Cui X. et al. (2014) Personalizing breast cancer staging by the inclusion of ER, PR, and HER2. JAMA Surg. 149, 125–129 10.1001/jamasurg.2013.3181 [DOI] [PubMed] [Google Scholar]
- 28.Ma Y., Lin D., Sun W., Xiao T., Yuan J., Han N. et al. (2006) Expression of targeting protein for xklp2 associated with both malignant transformation of respiratory epithelium and progression of squamous cell lung cancer. Clin. Cancer Res. 12, 1121–1127 10.1158/1078-0432.CCR-05-1766 [DOI] [PubMed] [Google Scholar]
- 29.Liang B., Jia C., Huang Y., He H., Li J., Liao H. et al. (2015) TPX2 level correlates with hepatocellular carcinoma cell proliferation, apoptosis, and EMT. Dig. Dis. Sci. 60, 2360–2372 10.1007/s10620-015-3730-9 [DOI] [PubMed] [Google Scholar]
- 30.Wei P., Zhang N., Xu Y., Li X., Shi D., Wang Y. et al. (2013) TPX2 is a novel prognostic marker for the growth and metastasis of colon cancer. J. Transl. Med. 11, 313 10.1186/1479-5876-11-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomii C., Inokuchi M., Takagi Y., Ishikawa T., Otsuki S., Uetake H. et al. (2017) TPX2 expression is associated with poor survival in gastric cancer. World J. Surg. Oncol. 15, 14 10.1186/s12957-016-1095-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neumayer G. and Nguyen M.D. (2014) TPX2 impacts acetylation of histone H4 at lysine 16: implications for DNA damage response. PLoS ONE 9, e110994 10.1371/journal.pone.0110994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M.-Y., Liu X.-X., Li H., Li R., Liu X. and Qu Y.-Q. (2018) Elevated mRNA levels of AURKA, CDC20 and TPX2 are associated with poor prognosis of smoking related lung adenocarcinoma using bioinformatics analysis. Int. J. Med. Sci. 15, 1676–1685 10.7150/ijms.28728 [DOI] [PMC free article] [PubMed] [Google Scholar]