Abstract
Methylation of the adenine base at the nitrogen 6 position (m6A) is the most common post-transcriptional epigenetic modification of RNA, and it plays a very important role in regulating gene expression. To investigate the role of m6A methylation in the expression of non-coding RNA and miRNA, we used a system of adenine base editors (ABEs). Here, we mutated regions up- and downstream of miRNA 675 m6A modification sites in the H19 locus using HEK293T, L02, MHCC97L, MHCC97H, A549, and SGC-7901 cells. Our results showed that a T–A base transversion had occurred in all cell lines. Moreover, mutation of the regions upstream of the miRNA 675 m6A modification site led to reduced expression of H19 and the induction of cell apoptosis in HEK293T cells. To further confirm our results, L02 and MHCC97L cells were detected using ABEs system. The results indicated increased cell apoptosis and reduced expression of miR675 as well as H19. To confirm the relationship between H19 and miR675 expression, overexpression and knockdown studies were performed. The results showed that reduced HI9 expression induced cell apoptosis through miR675. Taken together, these results indicate that m6A modification can regulate the expression of H19 and miR675 which induce cell apoptosis.
Keywords: cell apoptosis, gene expression, H19, m6A modification, miR675
Introduction
The long noncoding RNA (lncRNA) H19 plays a crucial role in the development of cancer [1]. miR675, derived from exon 1 of H19, has been shown to have an oncogenic role in liver cancers [2,3]. Our previous data suggest that reduced expression of H19 could induce cell apoptosis in A549, a lung cancer cell line [4]. Moreover, previous studies have shown that the H19/miR675 axis can regulate cell apoptosis [5]. These results demonstrate that altered expression of H19 or miR675 can influence tumor cell behavior.
Recent reports have suggested that m6A (methylation of the adenine base at the nitrogen 6 position) methylation plays an important role in the post-transcriptional modification of RNA [6], and it is known that this modification is regulated by adenosine methyltransferases and demethylases [7,8]. As ‘writers,’ the m6A methyltransferases METTL3, METTL14, and WTAP methylate the N6 position of adenosine [9,10]. As ‘erasers,’ the m6A demethylases FTO and ALKBH5 reverse the RNA methylation process [11,12]. Finally, YTHDF2, as an m6A ‘reader,’ recognizes m6A sites on target mRNAs and regulates the mRNAs’ fate [13–15]. Indeed, there is evidence that m6A modification in microRNA and lncNA affects cell development and fate [16]. These data indicate that m6A modification might have a role in noncoding RNA as well as miRNA.
Currently, the CRISPR/Cas9 system is the most widely used gene-editing tool. It uses an RNA-guide Cas9 protein combined with a short RNA (sgRNA) to induce double-strand breaks in target genomic DNA [17]. The adenine base editors (ABEs) system, which is based on the CRISPR/Cas9 platform, efficiently converts targeted A•T base pairs into G•C [18]. In the present study, the ABE7.10 system was used to analyze m6A modification of miRNA 675 in the H19 locus. Moreover, cell apoptosis and m6A expression levels were evaluated in HEK293T, L02, and MHCC97L cells. The role of m6A modification in the expression patterns of miRNA and lncRNA was analyzed using the ABEs system.
Materials and methods
Cell culture
HEK293T, L02, MHCC97L, MHCC97H, SGC-7901, and A549 cells were cultured in Dulbecco’s modified Eagle’s medium, high glucose (Gibco, U.S.A.), supplemented with 10% fetal bovine serum (Gibco, U.S.A.). The cells were maintained at 37°C in 5% CO2.
Construction and transfection of the plasmids
ABE7.10 plasmids were obtained from Addgene (102919). The m6A modification of miR675 in the H19 locus (upstream of position: chr11:2018320 and downstream of position: chr11:2017630) was analyzed using the online software tool m6AVar (http://m6avar.renlab.org).
Protocols for sgRNA design and the procedures required for in vitro transcription have been described previously [17]. The sgRNA-oligo sequences used in the present study are listed in Supplementary Table S1.
For cell transfection, HEK293T, L02, MHCC97L, MHCC97H, SGC-7901, and A549 cells were seeded into 48-well poly-d-lysine-coated plates (Corning) in the absence of any antibiotic. Twelve to fifteen hours after plating, cells were transfected with 750 ng of base-editor plasmid and 250 ng of guide RNA plasmid in the presence of 1 µl of Lipofectamine 2000 (Thermo Fisher Scientific).
Knockdown and overexpression of H19 and miR675
Synthetic RNA oligonucleotides targeting H19 were obtained from RiboBio (Guangzhou, China). The siRNA target sequence was GCGGGTCTGTTTCTTTACT. pcDNA3.1-H19 was purchased from GenePharma (Shanghai, China). miR675-3p-mimics and miR675-3p-inhibitor were obtained from RiboBio (Guangzhou, China). HEK293T cells were transfected with si-H19, pcDNA3.1-H19, miR675-3p-mimics, and miR675-3p-inhibitor for 48 h, respectively. Control cells were transfected with nonspecific, scrambled siRNA.
Gene expression analysis
Total RNA was extracted from cells using the AllPrep DNA/RNA Micro Kit (QIAGEN, Germany) according to the manufacturer’s instructions. cDNA was synthesized using the First-Strand cDNA Synthesis kit (Promega, U.S.A.). Quantitative real-time PCR (qRT-PCR) was performed to determine H19, miR675, and m6A-related gene expression using the BioEasy SYBR Green I Real-Time PCR Kit on Bio-Rad iQ5 Multicolor Real-Time PCR Detection System (Bioer Technology, China). The miR675 3p and 5p sequences are listed in Supplementary Table S2, and the miRNA primer sequences are listed in Supplementary Table S3. The primer sequences of m6A-related genes and H19 are listed in Supplementary Table S4. For PCR, the initial denaturation was conducted at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 15 s, and extension at 72°C for 30 s. The 2−ΔΔCT method was used to determine relative gene expression. The experiments were performed at least in triplicates.
Cell apoptosis analysis
The procedure for cell apoptosis detection has been previously described [19]. Briefly, HEK293T, L02, and MHCC97L cells were used for Annexin V-FITC/PI staining after treatment with ABE7.10 plasmids, si-H19, pcDNA3.1-H19 and miR675-3p-mimics and inhibitor for 48 h. Following incubation, the cells were washed twice with PBS and pooled at a concentration of 1 × 106 cells/ml. For each treated cell sample, Annexin V-FITC and PI were added according to the manufacturer’s instructions. These cells were incubated for 30 min and then analyzed with an Accuri™ C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, U.S.A.).
Immunofluorescence staining
Briefly, the cells were washed three times in PBS and then fixed with 4% paraformaldehyde for 30 min at room temperature. After fixation, the cells were washed again with PBS containing 0.2% Triton X-100 for 30 min. The cells were then incubated in PBS containing 1% bovine serum albumin (BSA) for 1 h. Next, the cells were probed with m6A (1:500, Abcam) antibodies and incubated at 4°C overnight. Following this, the cells were washed three times with PBS for 10 min each followed by incubation with Alexa Fluor 488–conjugated secondary (anti-rabbit) antibodies for 1 h at room temperature. DNA was stained with 10 ng/ml Hoechst 33342 (Thermo Scientific) for 15–20 min. The cells were then washed thrice with PBS for 10 min each, air-dried, and mounted on a coverslip and a glass slide using an antifade mounting medium (BOSTER, China). A confocal laser scanning microscope was used for imaging.
Statistical analysis
All data were analyzed using GraphPad Prism 5.0 (GraphPad Software, Inc., San Diego, CA). A t test (Unpaired t test) was used to analyze the data. A P-value <0.05 was considered statistically significant.
Results
Targeted point mutations of m6A modification sites induce cell apoptosis
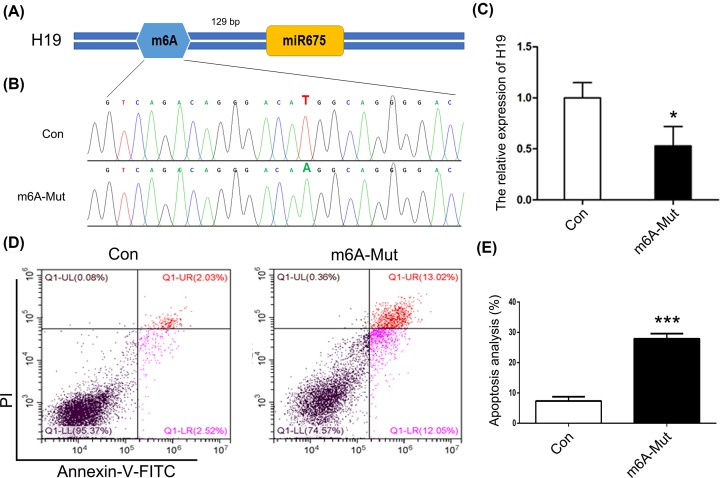
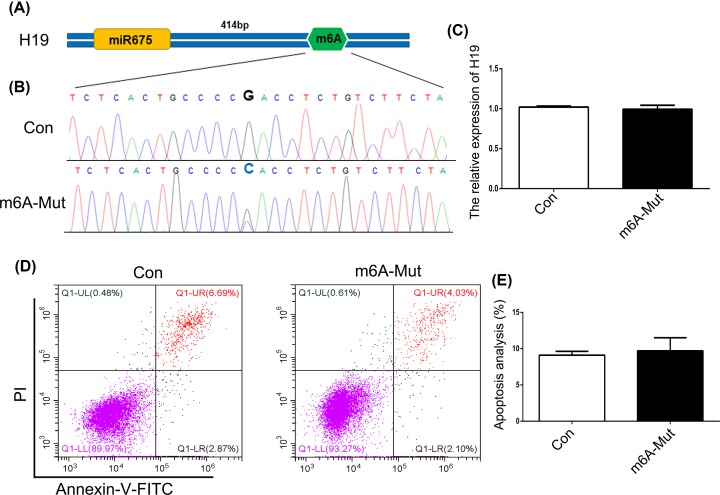
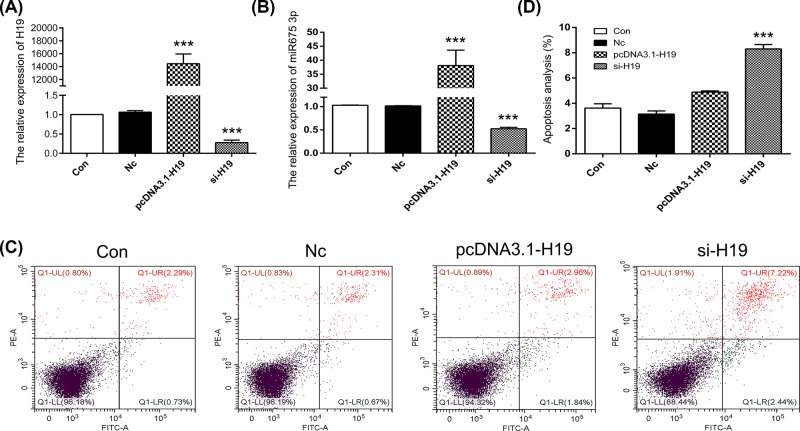
To investigate the role of m6A modification in the expression of H19, the ABE7.10 system was used. The m6A modification site 129 bp upstream of miR675 in the H19 locus was mutated in HEK293T cells (Figure 1A). Results of Sanger sequencing suggested T–A base transversion (Figure 1B). To confirm these results, similar tests were carried out with MHCC97H, SGC-7901, and A549 cells. The results confirmed T–A base transversion (Supplementary Figure S1). qPCR results showed decreased expression of H19 in the m6A-Mut group compared with that in the Con group (Figure 1C). To decipher the biological impact of m6A modification, we examined cell apoptosis. Our results indicated an increased apoptosis rate in the m6A-Mut group (Figure 1D,E). To further confirm the importance of m6A modification to H19 as well as to miR675, we mutated the m6A modification site 414 bp downstream of miR675 (Figure 2A). Results of Sanger sequencing suggested G–C base transversion (Figure 2B). qPCR results showed no difference in H19 expression between the Con and m6A-mut groups (Figure 2C). Moreover, the cell apoptosis rate did not increase after point mutations of the m6A modification sites (Figure 2D,E). To further analyze H19 expression patterns in apoptotic cells, knockdown or overexpression of H19 was performed in HEK293T cells. The results showed that H19 expression was reduced after transfection with si-H19 and was increased following transfection with pcDNA3.1-H19 (Figure 3A). miR675-3p expression was analyzed by qPCR. The results showed that the miR675-3p expression was similar to the expression pattern of H19 (Figure 3B). The cell apoptosis results showed that reduced expression of H19 induced cell death (Figure 3C,D). These results indicate that m6A modification regulates H19 expression which may induce cell apoptosis.
Figure 1. The role of m6A modification upstream of miR675.
(A) The schematic of m6A modification. (B) Sequencing analysis of the m6A modification site. (C) H19 expression analyzed by qPCR. (D) Cell apoptosis was analyzed after mutation of the m6A modification site. (E) Statistical analysis of apoptotic cell percentage. The data are presented as the mean ± SD. * (P<0.05) and *** (P<0.005) indicate statistically significant differences.
Figure 2. The role of m6A modification downstream of miR675.
(A) The schematic of m6A modification. (B) Sequencing analysis of the m6A modification site. (C) H19 expression was analyzed by qPCR. (D) Cell apoptosis was analyzed after mutation of the m6A modification site. (E) Statistical analysis of apoptotic cell percentage. The data are presented as the mean ± SD.
Figure 3. The expression pattern of H19 in apoptosis.
Expression of H19 (A) and miR675-3p (B) were analyzed by qPCR. (C) Cell apoptosis was analyzed after mutation of the m6A modification site. (D) Statistical analysis of apoptotic cell percentage. The data are presented as the mean ± SD. *** (P<0.005) indicates a statistically significant difference.
Targeted point mutations of m6A modification sites in liver cancer cells
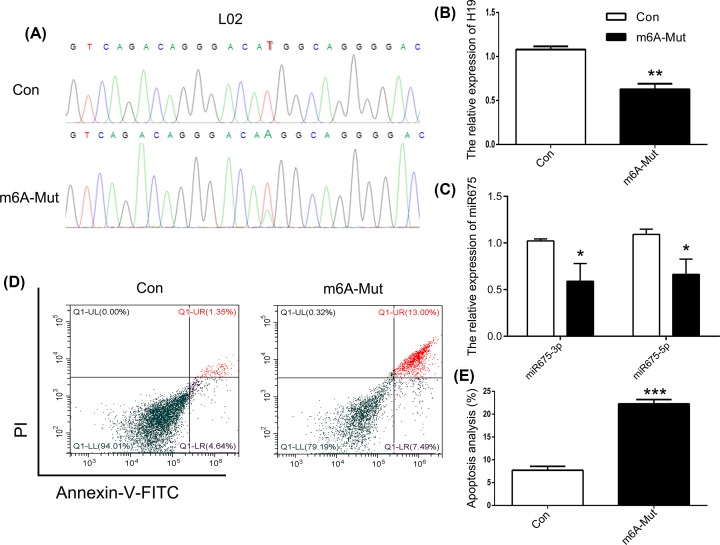
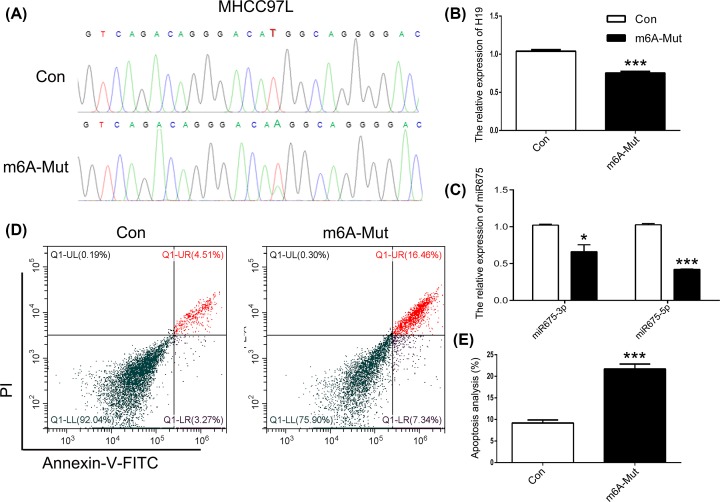
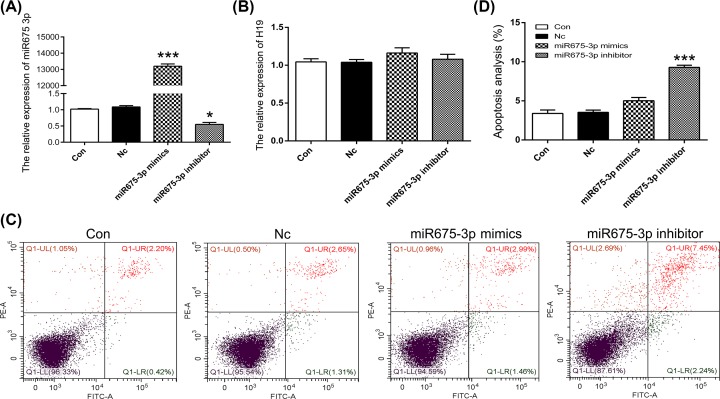
To further confirm that point mutations of m6A modification sites 129 bp upstream of miR675 induce cell apoptosis, we used L02 and MHCC97L cells. Results of Sanger sequencing showed identical point mutation patterns in both HEK293T and L02 cells (Figure 4A). qPCR results showed declined H19 expression in the m6A-Mut group (Figure 4B). To investigate the effects of m6A modification on the expression patterns of miRNA, the expression of miR675 was analyzed. The results showed decreased expression of both miR675-3p and miR675-5p (Figure 4C). In addition, an increased cell apoptosis rate was observed in the m6A-Mut group (Figure 4D,E). To confirm that point mutations do induce cell apoptosis, the MHCC97L cell line (liver cancer cells) was used. As observed with HEK293T and L02 cells, a T–A base transversion was observed in MHCC97L cells (Figure 5A). Moreover, decreased expression of H19, miR675-3p, and miR675-5p was noted in MHCC97L cells in the m6A-Mut group (Figure 5B,C). An increased cell apoptosis rate was observed in MHCC97L cells after the introduction of point mutations as observed in HEK293T and L02 cells (Figure 5D,E). To further analyze miR675 expression patterns in apoptotic cells, HEK293T cells were treated with a mimic or inhibitor of miR675-3p. The results showed that reduced miR675-3p expression induced cell apoptosis (Figure 6). These results suggest that targeted point mutations of m6A modification sites 129 bp upstream of miR675 induced cell apoptosis through reduced expression of H19.
Figure 4. Mutated m6A modification of miR675 expression in L02 cells.
(A) Sequencing analysis of m6A modification site. The expression of H19 (B) and miR675 (C) were analyzed by qPCR. (D) Cell apoptosis was analyzed after mutation of the m6A modification site. (E) Statistical analysis of apoptotic cell percentage. The data are presented as the mean ± SD. * (P<0.05), ** (P<0.01) and *** (P<0.005) indicate statistically significant differences.
Figure 5. Mutated m6A modification of miR675 in MHCC97L cells.
(A) Sequencing analysis of the m6A modification site. The expression of H19 (B) and miR675 (C) were analyzed by qPCR. (D) Cell apoptosis was analyzed after mutation of the m6A modification site. (E) Statistical analysis of apoptotic cell percentage. The data are presented as the mean ± SD. * (P<0.05) and *** (P<0.005) indicate statistically significant differences.
Figure 6. The expression pattern of miR675-3p in apoptosis.
The expression of miR675-3p (A) and H19 (B) were analyzed by qPCR. (C) Apoptosis was analyzed after mutation of the m6A modification site. (D) Statistical analysis of apoptotic cell percentage. The data are presented as the mean ± SD. * (P<0.05) and *** (P<0.005) indicate statistically significant differences.
m6A-related genes expression analysis by targeted point mutation
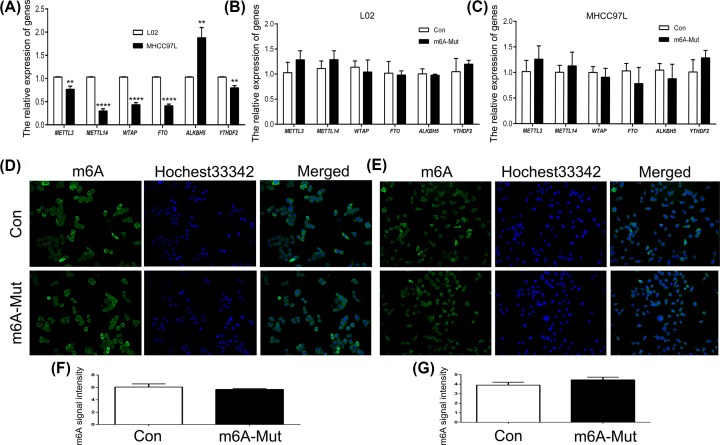
We further explored the expression patterns of m6A-related genes after introducing point mutations in L02 and MHCC97L cells. qPCR results showed increased expression of ALKBH5 and decreased expression of METTL3, METTL14, WTAP, FTO, and YTHDF2 in MHCC97L cells compared with L02 cells (Figure 7A). This result suggested that compared with L02, there is abnormal expression of m6A genes in MHCC97L cells. However, the expression of m6A-related genes was not changed by point mutations in L02 and MHCC97L cells (Figure 7B,C). m6A expression level was analyzed using immunoflorescence (IF). The results showed that m6A expression was not altered in either L02 and MHCC97L cells (Figure 7D,E). Also, the statistical analysis confirmed the IF data (Figure 7F,G). These results indicated that targeted point mutations of miR675 did not change the global m6A expression levels in L02 and MHCC97L cells.
Figure 7. Expression pattern of m6A-related genes.
The expression of m6A-related genes was analyzed in L02 and MHCC97L cells (A). The expression of m6A-related genes analyzed by point mutations in L02 (B) and MHCC97L (C) cells. IF localization of m6A in L02 (D) and MHCC97L (E) cells. The fluorescence intensities of m6A were measured in L02 (F) and MHCC97L (G) cells. ** (P<0.01) and **** (P<0.001) indicate statistically significant differences.
Discussion
Previous reports have indicated that targeted point mutations result in C-to-T (BE3) or A-to-G (ABE7.10) conversions [18,20]. In this study, our results showed that a T–A base transversion occurred upstream of miR675 in HEK293T, L02, and MHCC97L cells. While the expected result was an A•T to G•C conversion, our data showed that an A•T to A•A conversion had occurred. These results indicate the partial effectiveness of the ABE7.10 system, which might have induced cell apoptosis. To confirm these data, we transfected the ABE7.10 system into A549 (lung cancer cells), SGC7901 (gastric cancer cells), and MHCC97H (liver cancer cells) cells. The result was in accordance with our previous data. In addition, a G-to-A conversion was observed which might have indicated incomplete mutation downstream of miR675. These results suggested the presence of the T–A base conversion pattern, which might have a role in cell apoptosis.
To further investigate the role of m6A modification in apoptosis, the expression patterns of H19 and miR675 were analyzed. A previous study suggested that m6A modification was important for the expression of lncRNA and miRNA [21]. In our study, regions upstream and downstream of the m6A modification site of miR675 in the H19 locus were evaluated. The results demonstrated that mutations in the regions upstream of the m6A modification site could suppress the expression of H19 and miR675, whereas mutations in the regions downstream of the m6A modification site have no effect on the expression of H19 and miR675. To confirm the expression patterns of H19 and miR675 in apoptotic cells, overexpression and knockdown of H19 and miR675 were examined. Previous reports showed that the expression of H19 and miR675 was associated with cell apoptosis in cancer cells [22,23]. Our data suggest that reduced H19 expression induced cell apoptosis through miR675, which was confirmed in human colorectal cancer cells [24]. These results indicate that regions upstream of the m6A modification site play a role in regulating the expression of H19 and miR675 which can induce cell apoptosis.
There is evidence that reduced H19 and miR675 expression led to increased p53 protein expression, which regulates cell apoptosis [25]. Our data showed that mutation of the regions upstream of the m6A modification site inhibited miR675 and H19 expression, inducing cell apoptosis, possibly through the p53 protein. Moreover, an abnormal expression pattern of m6A-related genes was observed in liver cancer cells, which was in accordance with previous data [26]. A previous report suggested that ALKBH5 overexpression promotes invasion and metastasis in gastric cancer cells [27]. Indeed, metastasis is the major factor for HCC. This indicates that ALKBH5 expression may regulate the demethylated process and play a role in HCC metastasis. In addition, the global m6A expression was maintained after mutations of the regions upstream of the m6A modification site in L02 and MHCC97L cells. These results suggested that miR675 was regulated by m6A modification and has a role in H19 expression, which in turn influences the fate of the cell.
Conclusion
In summary, the ABE7.10 system resulted in efficient T–A base conversion of the m6A site upstream of miR675 in the H19 locus. The expression of H19 and miR675 was reduced by targeted point mutations. These mutations also induced cell apoptosis. Overall, our data suggest that m6A modification plays a role in gene expression and cell apoptosis.
Supplementary Material
Abbreviations
- ABE
adenine base editor
- IF
immunofluorescence
- lncRNA
long noncoding RNA
- m6A
methylation of the adenine base at the nitrogen 6 position
- PI
Propidium Iodide
- qPCR
Quantitative Real-time PCR
Contributor Information
Ziping Jiang, Email: waterjzp@jlu.edu.cn.
Dongxu Wang, Email: wang_dong_xu@jlu.edu.cn.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 31601003]; the China Postdoctoral Science Foundation [grant numbers 018T110250, 2016M601384]; the Fundamental Research Funds for the Central Universities [grant number 2019JCKT-70]; the Natural Science Foundation [grant number 2018SCZWSZX-045]; the Jilin Education Department Program [grant number JJKH20200950KJ]; and the Jilin Scientific and Technological Development Program [grant numbers 20190103071JH, 20180101254JC, 20170623093-TC].
Author Contribution
Dongxu Wang designed the experiments and wrote the manuscript. Jindong Hao, Chengshun Li, Yang Hao and Yidan Xia performed cell experiment and gene expression analysis. Xianfeng Yu and Ziping Jiang contributed reagents and materials. Fei Gao and Chao Lin analyzed the data and prepared figures. All authors reviewed the manuscript.
References
- 1.Schwarzenbach H. (2016) Biological and clinical relevance of H19 in colorectal cancer patients. EBioMedicine 13, 9–10 10.1016/j.ebiom.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hao Y., Crenshaw T., Moulton T., Newcomb E. and Tycko B. (1993) Tumour-suppressor activity of H19 RNA. Nature 365, 764–767 10.1038/365764a0 [DOI] [PubMed] [Google Scholar]
- 3.Li H., Li J., Jia S., Wu M., An J., Zheng Q. et al. (2015) miR675 upregulates long noncoding RNA H19 through activating EGR1 in human liver cancer. Oncotarget 6, 31958–31984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao Y., Wang G., Lin C., Li D., Ji Z., Gao F. et al. (2017) Valproic acid induces decreased expression of H19 promoting cell apoptosis in A549 cells. DNA Cell Biol. 36, 428–435 10.1089/dna.2016.3542 [DOI] [PubMed] [Google Scholar]
- 5.Li X., Wang H., Yao B., Xu W., Chen J. and Zhou X. (2016) lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci. Rep. 6, 36340 10.1038/srep36340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao B.S., Wang X., Beadell A.V., Lu Z., Shi H., Kuuspalu A. et al. (2017) m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478 10.1038/nature21355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M. et al. (2015) Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- 8.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S. et al. (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 9.Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J. et al. (2014) Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H.B., Tong J., Zhu S., Batista P.J., Duffy E.E., Zhao J. et al. (2017) m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548, 338–342 10.1038/nature23450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng C., Liu Y., Wang G., Deng Z., Zhang Q., Wu W. et al. (2014) Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J. Biol. Chem. 289, 11571–11583 10.1074/jbc.M113.546168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Q., Hou J., Zhou Y., Li Z. and Cao X. (2017) The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 18, 1094–1103 10.1038/ni.3830 [DOI] [PubMed] [Google Scholar]
- 13.Nguyen L.H., Robinton D.A., Seligson M.T., Wu L.W., Li L., Rakheja D. et al. (2014) Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 26, 248–261 10.1016/j.ccr.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H. et al. (2018) Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X. and He C. (2014) Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 11, 669–672 10.4161/rna.28829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fazi F. and Fatica A. (2019) Interplay between N-6-Methyladenosine (m(6)A) and non-coding RNAs in cell development and cancer. Front. Cell Dev. Biol. 7, 10.3389/fcell.2019.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I. et al. (2017) Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.William-Faltaos S., Rouillard D., Lechat P. and Bastian G. (2006) Cell cycle arrest and apoptosis induced by oxaliplatin (L-OHP) on four human cancer cell lines. Anticancer Res. 26, 2093–2099 [PubMed] [Google Scholar]
- 20.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A. and Liu D.R. (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coker H., Wei G. and Brockdorff N. (2019) m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 310–318 10.1016/j.bbagrm.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 22.Zhu M., Chen Q., Liu X., Sun Q., Zhao X., Deng R. et al. (2014) lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 281, 3766–3775 10.1111/febs.12902 [DOI] [PubMed] [Google Scholar]
- 23.Zhuang M., Gao W., Xu J., Wang P. and Shu Y. (2014) The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem. Biophys. Res. Commun. 448, 315–322 10.1016/j.bbrc.2013.12.126 [DOI] [PubMed] [Google Scholar]
- 24.Tsang W.P., Ng E.K., Ng S.S., Jin H., Yu J., Sung J.J. et al. (2010) Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 31, 350–358 10.1093/carcin/bgp181 [DOI] [PubMed] [Google Scholar]
- 25.Zheng Z.H., Wu D.M., Fan S.H., Zhang Z.F., Chen G.Q. and Lu J. (2019) Upregulation of miR-675-5p induced by lncRNA H19 was associated with tumor progression and development by targeting tumor suppressor p53 in non-small cell lung cancer. J. Cell. Biochem. 120, 18724–18735 10.1002/jcb.29182 [DOI] [PubMed] [Google Scholar]
- 26.Ma J.Z., Yang F., Zhou C.C., Liu F., Yuan J.H., Wang F. et al. (2017) METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary microRNA processing. Hepatology 65, 529–543 10.1002/hep.28885 [DOI] [PubMed] [Google Scholar]
- 27.Zhang J., Guo S., Piao H.Y., Wang Y., Wu Y., Meng X.Y. et al. (2019) ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J. Physiol. Biochem. 75, 379–389 10.1007/s13105-019-00690-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.