Abstract
The incidence of prostate cancer (PCa) is increasing, and it is currently the second most frequent cause of death by cancer in men. Despite advancements in cancer therapies, new therapeutic approaches are still needed for treatment-refractory advanced metastatic PCa. Cross-species analysis presents a robust strategy for the discovery of new potential therapeutic targets. This strategy involves the integration of genomic data from genetically engineered mouse models (GEMMs) and human PCa datasets. Considering the role of antioxidant pathways in tumor initiation and progression, we searched oxidative stress-related genes for a potential therapeutic target for PCa. First, we analyzed RNA-sequencing data from Pb-Cre4; Ptenf/f mice and discovered an increase in sulfiredoxin (Srxn1) mRNA expression in high-grade prostatic intraepithelial neoplasia (PIN), well-differentiated adenocarcinoma (medium-stage tumors), and poor-differentiated adenocarcinoma (advanced-stage prostate tumors). The increase of SRXN1 protein expression was confirmed by immunohistochemistry in mouse prostate tumor paraffin samples. Analyses of human databases and prostate tissue microarrays demonstrated that SRXN1 is overexpressed in a subset of high-grade prostate tumors and correlates with aggressive PCa with worse prognosis and decreased survival. Analyses in vitro showed that SRXN1 expression is also higher in most PCa cell lines compared to normal cell lines. Furthermore, siRNA-mediated downregulation of SRXN1 led to decreased viability of PCa cells LNCaP. In conclusion, we identified the antioxidant enzyme SRXN1 as a potential therapeutic target for PCa. Our results suggest that the use of specific SRXN1 inhibitors may be an effective strategy for the adjuvant treatment of castration-resistant PCa with SRXN1 overexpression.
1. Introduction
The incidence of prostate cancer (PCa) has progressively increased in the western world, representing the second most prevalent cancer with the second highest mortality rate in men [1–3]. Androgen receptor (AR) and circulating androgen are essential for normal prostate development [4], and AR is the main oncogenic driver of PCa initiation and progression. Therefore, therapeutic strategies against this type of tumor are usually aimed at inhibiting AR activity [5, 6]. If detected early, the chances of curing PCa are high, but more advanced PCa develops resistance to androgen deprivation therapies [7, 8]. These tumors are referred to as “castration-resistant PCa,” are highly heterogeneous in their molecular alterations [9–11], and are resistant to available therapies [12–14]. It is therefore crucial to identify new therapeutic targets and additional approaches to cure or at least increase the survival of patients with advanced PCa [15, 16].
Emerging technologies have allowed a deeper understanding of the cancer genome and the differential expression of genes involved in tumor development [17–19]. One recent example of the success of modern precision medicine for the treatment of cancer is the development of PARP1 inhibitors (PARPi). This specific adjuvant treatment benefits patients with defective DNA-damage repair such as the BRCA1 and BRCA2 mutations that frequently occur in breast and ovarian cancers [20, 21]. Studies involving PARPi for the treatment of PCa are already underway [22, 23]. In this context, one strategy for the discovery of new cancer biomarkers and/or potential therapeutic targets is to integrate genomic data from genetically engineered mouse models (GEMMs) and human cancer patients [24, 25]. Data generated by gene expression analysis of GEMM tumors help to search through genome-wide expression datasets generated from human prostate tumors. GEMMs are also indispensable in preclinical studies to test new drugs in immunocompetent animals [26]. Thus, cross-species analyses provide a powerful tool to pinpoint genes conserved across both species that are master regulators of tumor development [27–29].
Cancer is a complex disease involving many molecular variables. For PCa, oxidative stress is one of the main age-associated factors that influences the risk of developing this tumor [30, 31], such as alterations in GSTP1 expression by hypermethylation [32]. Several studies suggest that the prostate is exceptionally vulnerable to elevated reactive oxygen species (ROS) and oxidative stress [33, 34]. In normal cells, elevated ROS causes cumulative damage in lipids, proteins, and DNA, which may result in mutations and cancer initiation [35–37]. However, oxidative stress is also directly involved in cancer progression and metastasis [38–40]. Antioxidant pathways play an important cytoprotective role in tumors by preventing treatment-induced apoptosis and conferring chemoresistance [41–45]. Since these tumor cells are highly dependent on antioxidant mechanisms [38, 40], we aimed to identify genes involved in oxidative stress homeostasis that can be therapeutically targeted for the treatment of PCa. Using cross-species analyses of GEMMs and human data, we have investigated antioxidant genes with altered expression in different stages of PCa progression. In this study, we identified the antioxidant enzyme sulfiredoxin (SRXN1) as a potential target, and we validated its relevance in advanced PCa.
2. Materials and Methods
2.1. Genetically Engineered Mouse Model (GEMM)
We used RNAseq data and prostate samples from GEMM Ptenf/f, wild type (WT), and from the Pb-Cre4; Ptenf/f, which contains a deletion in both alleles of Pten, a tumor suppressor gene, exclusively in the prostate epithelium (conditional knockout). This model shows stages of tumor progression similar to human PCa, such as prostatic intraepithelial neoplasia (PIN), microinvasive and invasive well-differentiated adenocarcinoma (medium-stage tumors, MT), and fully invasive poorly differentiated adenocarcinoma (advanced-stage tumors, AT), with slow progression pace. Moreover, PTEN loss has been consistently associated with more aggressive disease features and worse prognosis, since PTEN loss range from less than 20% in clinically localized prostate tumors to more than 40% in metastatic castration-resistant PCa [46]. Although there are other interesting GEMMs for PCa, few studies have combined the stages of tumor progression with all prostatic lobes (anterior, AP; ventral, VP; lateral, LP; and dorsal prostate, DP) in a deep RNA-sequencing experiment. Additional details about this conditional knockout mice, histopathological analysis, and transcriptome data have been previously described [29].
RNA-sequencing data from samples of all prostatic lobes were accessed through the NCBI Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) platform, reference number GSE94574. Briefly, 72 samples were submitted to RNAseq analysis, including 20 WT prostatic lobes, 16 PIN, 20 MT, and 16 AT. At least four samples for each prostatic lobe and pathological condition were submitted to RNAseq analysis. First, we looked for the differentially expressed genes in each lobe and stages of tumor progression with adjusted p value < 0.05. We filtered all the deregulated genes using a known list of 84 genes involved in oxidative stress response [47, 48] and selected those which were altered in at least three prostatic lobes in PIN, MT, and/or AT. To select a potential therapeutic target, the consensus list of the upregulated genes involved in oxidative stress pathways was evaluated for clinical relevance in PCa using the cBioPortal for Cancer Genomics [49, 50], Cambridge Carcinoma of the Prostate App (CamcAPP) [51], and SurvExpress [52] databases.
The animal experiments used in this study were approved by the CRUK Institute Ethics Committee of the Cambridge University, UK, under design license 80/2435 and by the Ethics Committee on Animal Experimentation from the Institute of Biosciences of Botucatu, UNESP, Brazil, under protocol CEEA 613/2014.
2.2. Immunohistochemistry (IHC)
Paraffin blocks of all prostatic lobes containing WT and tumor samples (PIN, MT, and AT) from GEMMs were obtained by donation from David Neal's Uro-Oncology Group at the CRUK Cambridge Institute (University of Cambridge, UK). Histological sections (5 μm) of WT and tumor-bearing prostate lobes from the GEMMs (n = 5 per group) were deparaffinized, rehydrated, boiled for 30 min in 10 mM sodium citrate solution (pH 6.0) for antigen retrieval, and quenched in 3% H2O2 methanol solution. Prostate sections were blocked in 5% nonfat milk in phosphate-buffered saline (PBS) and incubated overnight at 4°C with a specific primary antibody against SRXN1 (Abcam, ab92298, 1 : 100), our chosen gene. Next, sections were incubated with a secondary peroxidase-conjugated antibody (Santa Cruz Biotechnology, 1 : 200), which was developed using diaminobenzidine (Sigma-Aldrich) as the chromogen. Slides were counterstained with Harris's hematoxylin. The negative control was obtained by excluding the primary antibody incubation step. The sections were visualized using a Leica DMLB 80 microscope.
2.3. Human Database Analyses
The antioxidant enzyme SRXN1 was chosen as a potential target by RNA-sequencing and IHC analysis of the prostate tumors from GEMMs. To validate the SRXN1 expression pattern in human PCa, we searched available databases from published studies. We used GEO (https://www.ncbi.nlm.nih.gov/geoprofiles/) to profile SRXN1 gene expression from prostate tumors of different grades, and from different normal and epithelial prostate tumor cell lines. We also investigated the SRXN1 gene expression pattern using the cBioPortal for Cancer Genomics database (http://www.cbioportal.org/), using studies from The Cancer Genome Atlas, TCGA (https://cancergenome.nih.gov/), the CamcAPP dataset (https://bioinformatics.cruk.cam.ac.uk/apps/camcAPP/), and the SurvExpress database (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp) to determine the association of SRXN1 gene alterations with patient clinical data, such as risk/prognosis and survival rates.
2.4. Human Prostate Tumor Tissue Microarrays (TMAs)
After validating the overexpression of SRXN1 in GEMM samples and human database analyses, we investigated the SRXN1 protein expression in human prostate samples. Human TMAs were constructed using the prostates of patients who underwent radical prostatectomy between 1980 and 2000. Of the patients, 104 samples originated from organ-confined tumors, and 16 samples originated from adjacent nonneoplastic tissue. One-tissue cores of 1 mm diameter were used for each sample. The TMA was donated to and analyzed by the consultant pathologist Flávio de Oliveira Lima at the Botucatu Medical School, UNESP, Brazil. This study was approved by the Medical Ethics Committee of the Botucatu Medical School, UNESP, Brazil (protocol number 3888/2011).
According to glandular histoarchitecture and histopathological staging, tumor samples from human TMAs were classified into five prognostic categories (1-5, from more differentiated to less differentiated), according to the Gleason score [53, 54] and the International Society of Urological Pathology (ISUP) grade [55, 56]. The prostate tumor classifications from TMAs are presented in Table 1.
Table 1.
Number of cases and tumor classification (Gleason score and prognosis category) from human prostate samples (tissue microarrays).
| Number of cases | Gleason score | Prognosis category |
|---|---|---|
| 16 | Adjacent nonneoplastic tissue | — |
| 31 | 6 (3 + 3) | 1 |
| 17 | 7 (3 + 4) | 2 |
| 20 | 7 (4 + 3) | 3 |
| 1 | 8 (3 + 5) | 4 |
| 19 | 8 (4 + 4) | |
| 1 | 8 (5 + 3) | |
| 6 | 9 (4 + 5) | 5 |
| 5 | 9 (5 + 4) | |
| 4 | 10 (5 + 5) |
SRXN1 protein expression in human TMAs was detected by immunohistochemistry following the previously described protocol. The results were quantified and evaluated as negative (no staining, score 0) or positive (staining present, score 1), independent of the staining intensities. This analysis was performed by two independent observers without access to clinical data. SRXN1 staining scores were associated with the clinical and pathological characteristics (Gleason score, prognosis category, and survival time). Details regarding the clinical data and the SRXN1 IHC score are available in Supplementary Table S1, and the association between Gleason score/prognosis and survival rates in Supplementary Figure S1.
2.5. Cell Lines and Culture Conditions
The following three prostate cell lines were used for the analyses: RWPE-1 (normal), LNCaP (tumor, androgen sensitive), and PC-3 (tumor, castration-resistant). The cells were obtained from the American Type Cell Culture (Manassas, Virginia, USA). LNCaP and PC-3 cell lines were cultured using RPMI 1640 medium supplemented with 2 mM L-glutamine, 10% fetal bovine serum, 50 μg/mL penicillin, 50 μg/mL streptomycin, and 0.5 μg/mL amphotericin B (GIBCO/Invitrogen). RWPE-1 cells were cultured with a Keratinocyte Serum Free Medium Kit supplemented with bovine pituitary extract and recombinant human epidermal growth factor (GIBCO/Invitrogen). The medium was changed twice per week, and the cells were monitored daily under an inverted microscope (Zeiss Axiovert). Cells were grown at 37°C and 5% CO2. For passaging, cells were detached with 0.25% trypsin (GIBCO/Invitrogen) for 5 min at 37°C, resuspended in a growth medium, and reseeded. Cell lines were authenticated by Short Tandem Repeat (STR) DNA profiling by the Biorepository Facility of the Institute of Biosciences, UNESP, Brazil. Mycoplasma testing was carried out at regular intervals throughout the experiments, and the results were negative.
2.6. RNA Extraction and RT-qPCR
Total RNA from prostate cell lines RWPE-1, LNCaP, and PC-3 was extracted using the RNeasy mini kit (QIAGEN) according to the manufacturer's instructions. RNA quantification was determined by a Nanovue Spectrophotometer (GE Healthcare). cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR reactions were performed using the QuantStudio 12K Flex Real-Time PCR system (Applied Biosystems). Relative gene expression was calculated using the 2-ΔΔCT method [57]. The gene target detected was SRXN1, and β-actin (ACTB) was used as a housekeeping gene. Details of primers used are given in Table 2.
Table 2.
Primers used in the RT-qPCR reactions.
| Genes | Primer forward | Primer reverse | Amplicon |
|---|---|---|---|
| SRXN1 | CAAGGTGCAGAGCCTCGT | CAGCCCCCAAAGGAGTAGAA | 105 |
| ACTB | GATTCCTATGTGGGCGACGA | TGTAGAAGGTGTGGTGCCAG | 124 |
2.7. Sulfiredoxin Knockdown In Vitro
To analyze the effects of SRNX1 suppression in vitro, we chose PCa cell line LNCaP as our model due to its elevated SRXN1 mRNA expression. LNCaP cells (1 × 105) were seeded in 6-well plates using a complete RPMI medium. The transfection was performed using the Lipofectamine RNAi MAX Transfection Reagent (Invitrogen) according to the manufacturer's instructions. Lipofectamine and siRNA targeting the mRNA of SRXN1 (MISSION esiRNA, Sigma-Aldrich) were individually diluted in an Opti-MEM medium (GIBCO/Invitrogen), mixed, and incubated for 5 min. Next, siRNA-lipid complex was added to cells and incubated for 48 h. The final siRNA concentration was 25 and 50 nM. In addition, siRNA targeting eGFP (MISSION esiRNA, Sigma-Aldrich) was used as negative control.
2.8. Cell Viability Assays
LNCaP cells (6 × 104) were seeded in 24-well plates. Once cells became 60% confluent, siRNA against the mRNA of SRXN1 was added as previously described. After 48, 72, and 96 h of transfection, cell viability was determined by the MTT (Thiazolyl Blue Tetrazolium Bromide, Sigma-Aldrich) reduction method according to the manufacturer's instructions [58, 59]. The reaction was transferred to a 96-well plate, and the absorbance (550 nm) was read by a spectrophotometer (ASYS HITECH GmbH, Eugendorf) to determine the percentage of cell viability relative to control cells.
2.9. Statistical Analysis
For parametric data, Student's t-test with Welch's Correction Factor or Factorial ANOVA was used. For nonparametric data, the Mann-Whitney or Kruskal-Wallis test was used. For association analyses, the Chi Square Contingency test and Kaplan-Meier/Log-Rank test were used. Differences were considered statistically significant when p < 0.05. Statistical analyses were performed using the GraphPad Prism program (version 5.0).
3. Results
3.1. Oxidative Stress Response Genes Are Deregulated during Prostate Tumor Progression in Pb-Cre4; Ptenf/f Mice
RNAseq data from GEMMs showed on average 2,000 genes differently expressed in each prostatic lobe (AP, VP, LP, and DP) at the tumor stages of PIN, MT, and AT. We filtered all these deregulated genes using a known list of 84 genes involved in oxidative stress response [47, 48]. We selected those genes which were changed in at least three prostatic lobes in PIN, MT, and/or AT, resulting in a final list with the top 19 deregulated oxidative stress-related genes, out of which 11 upregulated and 8 downregulated (Figure 1). We investigated the prognosis significance of the upregulated and druggable genes. Excluding those genes with no clinical relevance, and those which have already been associated with PCa, such as Ctsb, Gpx2, Idh1, and Nos2 [60–66], we selected the antioxidant enzyme Srxn1 that had a strong correlation with patient outcome and has no previous related functional studies in PCa.
Figure 1.
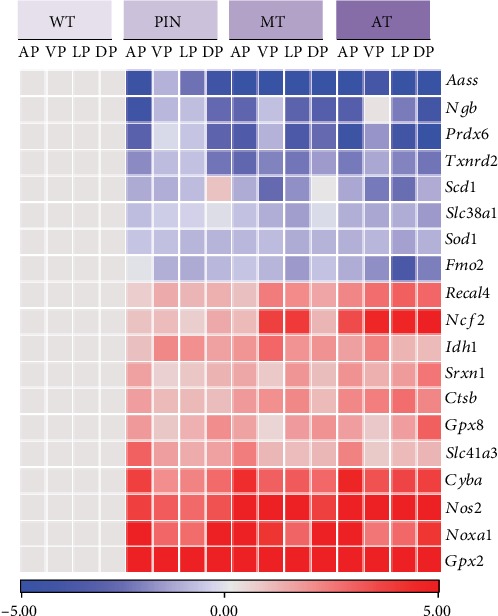
Consensus deregulated oxidative stress genes in prostate tumors from Pb-Cre4; Ptenf/f mice. Heat map of oxidative stress response genes in the four lobes (anterior, AP; ventral, VP; lateral, LP; and dorsal prostate, DP) of wild-type (WT) prostate and tumor samples (prostatic intraepithelial neoplasia, PIN; medium-stage tumors, MT; and advanced-stage tumors, AT) from GEMM Pb-Cre4; Ptenf/f. Relative gene expression level (median) is showed as log2 fold change related to WT. Values between -5 and 0 represent downregulated genes (blue gradient) and between 0 and 5 upregulated genes (red gradient).
3.2. SRXN1 Expression Is Increased in Prostate Tumors from Pb-Cre4; Ptenf/f Mice
Prostate transcriptome data from Pb-Cre4; Ptenf/f mice comparing WT to tumor samples (PIN, MT, and AT) showed that the relative mRNA expression of Srxn1 increases progressively and significantly during tumor progression in all prostatic lobes, with p < 0.001 (Figure 2(a)). To evaluate the pattern of SRXN1 protein expression in PCa, IHC analysis was performed, and we observed increased SRXN1 protein expression (higher intensity immunostaining) in prostate tumors compared to WT tissue (Figure 2(b)).
Figure 2.
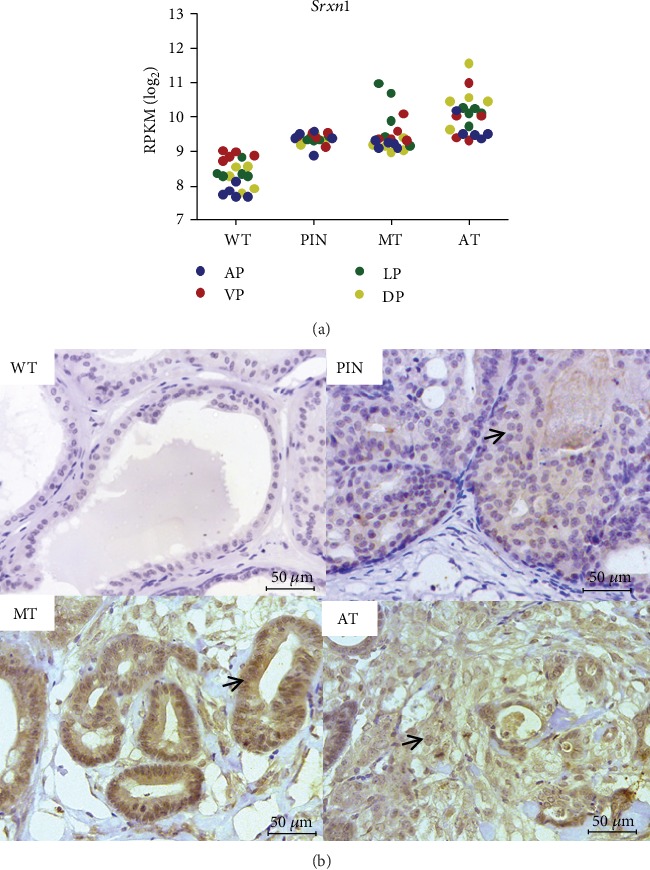
SRXN1 gene and protein expression is higher in prostate tumor tissue from Pb-Cre4; Ptenf/f mice compared to normal tissue. (a) Srxn1 expression levels in the four lobes (anterior, AP; ventral, VP; lateral, LP; and dorsal prostate, DP) of wild-type (WT) prostate and tumor samples (prostatic intraepithelial neoplasia, PIN; medium-stage tumors, MT; and advanced-stage prostate tumors, AT) from Pb-Cre4; Ptenf/f mice. Data are expressed as log2 of reads per kilobase per million (RPKM). The relative curve of Srxn1 mRNA expression increases significantly with p < 0.001. (b) Representative images of immunohistochemistry detecting SRXN1 protein in WT and prostate tumors in different stages of progression: PIN, MT, and AT from Pb-Cre4; Ptenf/f mice. Scale bars = 50 μM. Arrows indicate positively stained cells.
3.3. SRXN1 Is Overexpressed in High-Grade Human Prostate Tumors and Is Associated with Cancer Aggressiveness
Published and available data (database GEO profiles) on global gene expression in human PCa demonstrated increased expression of SRNX1 in patients with advanced tumors (Gleason scores 8 and 9) relative to control (Supplementary Figure S2a). Similarly, studies available in the SurvExpress database revealed that increased expression of SRXN1 correlates with a high-risk/worse prognosis of patients with PCa (Figure 3(a)). The same association was observed using several studies available in the CamcAPP dataset. This resource demonstrated an increase in SRXN1 expression in clusters of patients with poor prognosis (Supplementary Figure S2c, S2d, and S2e). Data from TCGA studies obtained from cBioPortal demonstrated that patients with alterations in SRXN1 (especially overexpression) have a lower disease-free/progression-free survival (Figure 3(b)). The same effect was observed using the CamcAPP dataset (Supplementary Figure S2f).
Figure 3.
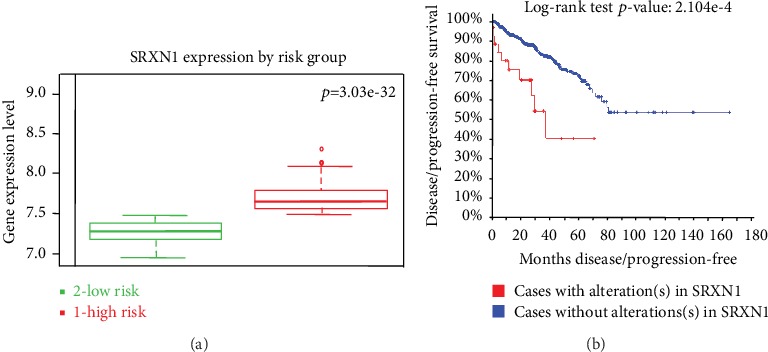
The increased expression of SRXN1 in patients with prostate cancer (PCa) is associated with high-risk/worse prognosis and lower disease-/progression-free survival. (a) The level of SRXN1 gene expression (median) in PCa patients with low-risk/better prognosis (green) and patients with high-risk/worse prognosis (red). Data and analyses were cataloged using the SurvExpress database [52] from a MSKCC study [18]. The difference between boxplots is statistically significant with p = 3.03−32. (b) Kaplan-Meier curve displaying disease-/progression-free survival of PCa patients with (red) or without (blue) SRXN1 alterations, cataloged using the cBioPortal human database [49, 50] from a provisional study of TCGA (https://cancergenome.nih.gov/). Curves are significantly different with p = 2.104−4.
3.4. SRXN1 Protein Expression Is Associated with Poor Prognosis and Lower Survival of PCa Patients
To further validate the association of SRXN1 with PCa aggressiveness, we performed IHC analyses on SRXN1 in TMAs of human prostate samples (benign and cancerous tissue). Protein expression was increased (positive immunostaining) in more than 70% of high-grade tumors (Figure 4(a)), representing patients with high Gleason score and worse prognosis.
Figure 4.
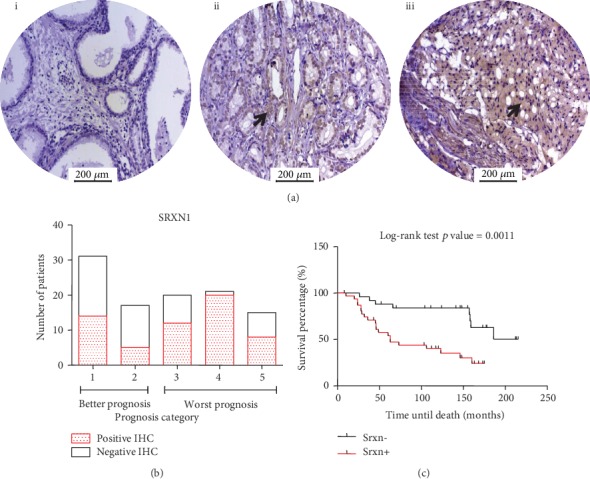
SRNX1 protein expression is increased in human advanced prostate cancer (PCa) and is associated with worse prognosis and decreased survival. (a) Representative image of SRXN1 immunohistochemistry (IHC) in (i) adjacent nonneoplastic tissue, (ii) medium-stage/low-grade tumor, and (iii) advanced-stage/high-grade tumor from tissue microarrays (TMAs) of human prostate samples. Arrows indicate positively stained cells. Scale bars = 200 μM. (b) Representative graph showing the association between SRXN1 protein expression (by IHC analysis in human prostate TMAs) and prognosis, which were divided into groups with good prognosis (categories 1 and 2) and worse prognosis (categories 3, 4, and 5), according to the International Society of Urological Pathology (ISUP) grade. White bars represent patients with negative SRXN1 immunostaining, and red bars represent patients with positive immunostaining. (c) Global survival curve of patients with PCa obtained from SRXN1 IHC analyses in human prostate samples (TMAs) associated with patient survival data. The Kaplan-Meier curve from patients with negative SRXN1 immunostaining is represented by black (Srxn-) and positive immunostaining by red (Srxn+). Curves are significantly different with p = 0.0011.
Additionally, the negative and positive immunostaining of SRNX1 in TMAs was associated with clinical data. It was observed that among the 48 patients with better outcome (prognosis categories 1 and 2), 39.5% expressed SRXN1. In comparison, among the 56 patients with worse prognosis (categories 3, 4, and 5), 71.4% expressed SRXN1, which differed significantly (p = 0.0015) (Figure 4(b)). Analysis of survival time (n = 69) revealed an association between SRXN1 protein expression and decreased survival (Figure 4(c)). Interestingly, the expression pattern of SRXN1 stratified patients as well as the prognosis score according to the ISUP classification, since patients with expression of SRXN1 had decreased survival similarly to patients with worse prognosis (Supplementary Figure S1).
3.5. PCa Cell Lines Express Higher Levels of SRXN1 Compared to Normal Cell Lines, and Inhibition of SRXN1 mRNA in the PCa Cell Line LNCaP Decreases Viability
Quantitative PCR showed that prostate tumor cell lines LNCaP and PC-3 express higher levels of SRXN1 than the normal prostate cell line RWPE-1 (Figure 5(a)). Among the prostate tumor cell lines, LNCaP has increased SRXN1 gene expression compared to PC-3 (Figure 5(a)). Database analysis of other studies from GEO profiles also showed increased expression of SRXN1 in LNCaP and PC-3, and in most prostate tumor cells (Supplementary Figure S2b).
Figure 5.
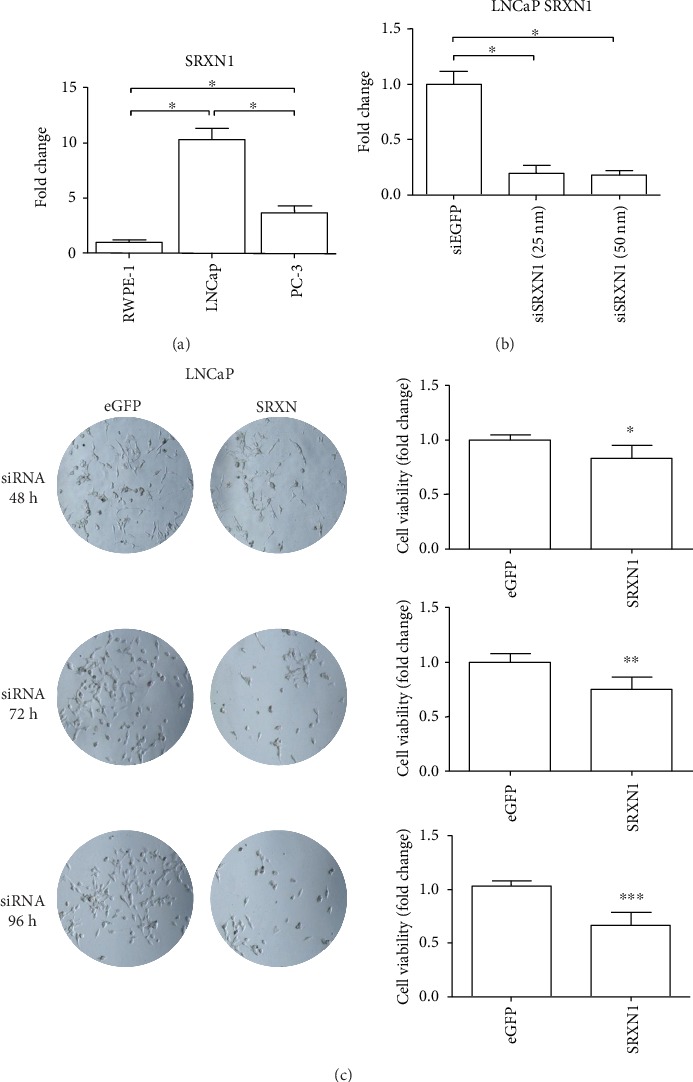
Prostate tumor cell lines overexpress SRXN1, and attenuation of SRXN1 mRNA levels in prostate cancer (PCa) cell line LNCaP decreases tumor cell viability. (a) SRXN1 gene expression level in prostate cell line RWPE-1 (normal), LNCaP (PCa, androgen sensitive), and PC-3 (PCa, castration-resistant). ∗Denotes statistical significance with p < 0.0001. Data are expressed as fold change normalized to ACTB expression. (b) qRT-PCR graph of SRXN1 gene expression in LNCaP cells after transfection (48 h) with siRNA targeting eGFP (control) or SRXN1 mRNA (25 and 50 nM). ∗Denotes statistical significance with p < 0.0001. (c) Cell viability of PCa cell line LNCaP without (control, eGFP) and with inhibition of SRXN1 mRNA by siRNA-mediated silencing (25 nM) after 48, 72, and 96 h. ∗Denotes statistical significance with p = 0.0043; ∗∗p = 0.0002; and ∗∗∗p < 0.0001.
To analyze the effects of decreased SRXN1 expression in vitro, we performed siRNA-mediated knockdown of SRXN1 mRNA followed by cell viability assays at three different time points after transfection (48, 72, and 96 h). PCa cell line LNCaP was chosen because of its high expression of SRXN1. After transfection, we observed a >80% reduction of the SRXN1 mRNA (Figure 5(b)). The silencing analyses demonstrated that the viability of LNCaP cells with decreased SRXN1 mRNA expression was lower compared to cells that expressed SRXN1 (eGFP siRNA, negative control). The viability of PCa cells significantly decreased in all analyzed time points after SRXN1 knockdown (Figure 5(c)) (p < 0.05 in all cases).
4. Discussion
Cross-species analyses from GEMMs and human data revealed that SRXN1 is overexpressed in PCa tissue and cell lines, with a remarked increase in advanced tumors. Cancer cells are more dependent on antioxidant mechanisms and more vulnerable to ROS-induced damage than normal cells [38, 40]. SRXN1 is an antioxidant enzyme induced by NRF2 and AP-1 and plays an important role in oxidative stress balance [67–69]. Specifically, SRXN1 acts in peroxiredoxin (I-IV) reactivation [70]. The peroxiredoxins are a group of peroxidase enzymes responsible for the reduction of ROS, such as hydrogen peroxide and organic peroxides [71], protecting the cell from high levels of ROS-induced oxidative stress. By reducing the hyperoxidized peroxiredoxins, SRXN1 protects them from degradation [72].
Several studies have demonstrated the protective role of SRXN1 against oxidative injury [73, 74]. The increased expression of SRXN1 has been observed especially in solid tumors, as we observed for PCa in this study. Wei et al. showed elevated SRXN1 protein expression in lung tumor samples by IHC analyses [75]. Another study from Wei et al. demonstrated high SRXN1 protein expression in human colon carcinoma and showed that Srxn1 knockout animals were resistant to carcinogenic induction [76]. In addition, SRXN1 expression is increased in different human skin malignancies [77, 78] and gastric cancer [79].
In addition to the overexpression of SRXN1 in PCa high-grade tumors, the current study also showed a close relationship between SRXN1 expression and both cancer aggressiveness and patient outcome. Researchers have already demonstrated that SRXN1 is essential for cancer cell proliferation [80], and its depletion decreases cell viability [74], suppresses cell migration, and inhibits tumor growth [75, 77], increasing the aggressiveness of cancers with SRXN1 upregulation. Polymorphisms in the SRXN1 gene have been shown to promote breast cancer development and influence patient survival [81]. SRXN1 expression is also associated with poor survival of patients with pancreatic adenocarcinoma [82]. Raatikainen et al. observed an inverse correlation between SRXN1 expression and worse PCa prognosis [83], in contrast to our results and the results of other studies.
Considering the pathogenic role of SRXN1 in human cancer, it has already been suggested as a new potential therapeutic target for different tumors, but not yet for PCa. In our study, we observed that decreasing SRXN1 mRNA in PCa cell line LNCaP decreased cell viability, reinforcing the importance of this antioxidant enzyme in PCa cells and suggesting that SRXN1 activity supports tumor cell survival and growth. In malignant human skin tumors, Wei et al. showed that SRXN1 inhibition (by the AP-1 pathway) may be a novel strategy for skin cancer prevention and treatment [77]. Kim et al. and Kim et al. demonstrated that SRXN1 inhibition by synthetic inhibitors (J14 and K27) selectively promotes the death of A549 pulmonary tumor cells [84, 85]. All these studies suggest that when SRXN1 activity is inhibited, the high ROS levels exceed cell antioxidant capacity, resulting in accumulated damage that selectively leads to tumor cell death. Thus, our work proposes that SRXN1 can be an interesting therapeutic target for further preclinical in vivo tests using an immunocompetent and clinically relevant GEMM. Patients with advanced PCa presenting SRXN1 overexpression may benefit from SRXN1 inhibition therapy, providing cancer patients with more personalized treatment.
5. Conclusions
SRXN1 is increased in a subset of PCa patients with high-grade tumors (advanced stage) and correlates with poor prognosis and worse survival. Thus, our cross-species analyses pinpoint SRXN1 as a potential therapeutic target for PCa, which plays an important role in protection of prostate tumor cells against oxidative stress. We hypothesize that the use of selective SRXN1 inhibitors can be an effective adjuvant treatment strategy for metastatic PCa with SRXN1 overexpression.
Acknowledgments
We thank David Neal's Uro-Oncology group (CRUK Cambridge Institute, University of Cambridge, UK) for providing formalin-fixed and paraffin-embedded prostate samples of the GEEMs and the TCGA Research Network for the results published here that are in part based upon data from their database: https://www.cancer.gov/tcga. We also thank the financial support received from the National Council for Scientific and Technological Development (CNPq), Brazil (grants #310805/2018-0 and #132849/2017-8) and São Paulo Research Foundation (FAPESP), Brazil (grants #2015/26097-6, #2016/09532-3, and #2016/25945-6). The present work was also carried out with the support of the “Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES)” - Finance Code 001. This article represents part of a master thesis developed by CNB at the Institute of Biology at Campinas State University (UNICAMP), Brazil.
Data Availability
The RNAseq data from the GEMM Pb-Cre4; Ptenf/f mouse used to support the findings of this study have been deposited in the NCBI Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/), reference number GSE94574. Previously reported human databases were used to support this study and are available at the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geoprofiles/), the cBioPortal for Cancer Genomics (http://www.cbioportal.org/), The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/), the Cambridge Carcinoma of the Prostate App (camcAPP dataset) (https://bioinformatics.cruk.cam.ac.uk/apps/camcAPP/), and the SurvExpress database (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp). These prior studies (and datasets) are cited at relevant places within the text as references [18, 19, 49–52] and Satake et al. (supplementary material [1]) and Zhao et al. (supplementary material [2]). The clinical data of the PCa patients from TMA samples used to support the findings of this study are included within the supplementary information files (Table S1).
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Supplementary Materials
Table S1: description of the patient clinical data used in the preparation of the TMAs, such as Gleason score, prognostic category, survival time, and patient outcome. Figure S1: patients with PCa grouped by prognosis category (1-5) show expected survival curves. (A) Global survival curve of patients with PCa associated to prognosis category (1-5, from more differentiated to less differentiated), according to the ISUP grade. Clinical data are from those patients which prostate samples were used to construct TMAs. Kaplan-Meier curves are statistically different with p < 0.0001. (B) Additional analysis of the same patients of Figure S1a, dividing them into groups with good prognosis (categories 1 and 2, black curve) and worse prognosis (categories 3, 4, and 5, red curve), according to the ISUP grade. Kaplan-Meier curves are significantly different with p < 0.0001. Figure S2: SRXN1 expression is increased in advanced PCa and in most prostate tumor cell lines, and its overexpression is associated with poor prognosis and lower disease-/progression-free survival. (A) Levels of SRXN1 expression in wild-type prostate (control) and advanced PCa samples (Gleason scores 8 and 9) from a study available on the GEO profile human database (reference series GSE5016) [1]. (B) SRXN1 gene expression in different prostate cell lines (androgen sensitive and castration-resistant) obtained from a study available on the GEO profile human database (reference series GSE4016) [2]. (C) Expression of SRXN1 (median) in five PCa iClusters generated by the Cambridge Carcinoma of the Prostate App (camcAPP dataset) [3] from an integrative study [4]. iClusters 1 (red), 3 (green), and 5 (orange) represent groups of patients with worse prognosis, while iClusters 2 (blue) and 4 (purple) represent groups with better prognosis. Boxplots are significantly different, with p = 5.7833−9. (D) Expression of SRXN1 (median) in five PCa iClusters generated by the Cambridge Carcinoma of the Prostate App (camcAPP dataset) [3] from an integrative study [4]. iClusters 1 (red), 3 (green), and 5 (orange) represent groups of patients with worse prognosis, while iClusters 2 (blue) and 4 (purple) represent groups with better prognosis. Boxplots are significantly different with p = 0.034473. (E) Expression of SRXN1 (median) in six PCa iClusters generated by the Cambridge Carcinoma of the Prostate App (camcAPP dataset) [3] from an integrative study [5]. iClusters 1 (salmon), 2 (dark yellow), 3 (green), and 4 (turquoise) are groups of patients with more favorable prognosis with minimal copy number alterations (CNA), while iClusters 5 (light blue) and 6 (lilac) include most of the metastatic tumors with substantial CNA. Boxplots are significantly different, with p = 3.42−6. (F) Kaplan-Meier curve displaying the probability of freedom from biochemical recurrence of PCa with (red) or without (blue) SRNX1 overexpression, cataloged by the Cambridge Carcinoma of the Prostate App (camcAPP dataset) [3] from an integrative study [5]. Curves are statistically different with p = 0.0079.
References
- 1.Center M. M., Jemal A., Lortet-Tieulent J., et al. International Variation in Prostate Cancer Incidence and Mortality Rates. European Urology. 2012;61(6):1079–1092. doi: 10.1016/j.eururo.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 2.Stewart B. W., Wild C. International Agency for Research on Cancer, and World Health Organization, World cancer report. 2014
- 3.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2018. CA: A Cancer Journal for Clinicians. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Huggins C., Hodges C. V. Studies on prostatic Cancer: I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA: A Cancer Journal for Clinicians. 1972;22(4):232–240. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- 5.Hsing A. W., Reichardt J. K., Stanczyk F. Z. Hormones and prostate cancer: current perspectives and future directions. Prostate. 2002;52(3):213–235. doi: 10.1002/pros.10108. [DOI] [PubMed] [Google Scholar]
- 6.Aragon-Ching J. B. The evolution of prostate cancer therapy: targeting the androgen receptor. Frontiers in Oncology. 2014;4:p. 295. doi: 10.3389/fonc.2014.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan T.-C., Lin F.-F., Veeramani S., Chen S.-J., Earp H. S., Lin M.-F. ErbB-2 via PYK2 upregulates the adhesive ability of androgen receptor- positive human prostate cancer cells. Oncogene. 2007;26(54):7552–7559. doi: 10.1038/sj.onc.1210570. [DOI] [PubMed] [Google Scholar]
- 8.Komiya A., Suzuki H., Imamoto T., et al. Neuroendocrine differentiation in the progression of prostate cancer. International Journal of Urology. 2009;16(1):37–44. doi: 10.1111/j.1442-2042.2008.02175.x. [DOI] [PubMed] [Google Scholar]
- 9.Boyd L. K., Mao X., Lu Y.-J. The complexity of prostate cancer: genomic alterations and heterogeneity. Nature Reviews Urology. 2012;9(11):652–664. doi: 10.1038/nrurol.2012.185. [DOI] [PubMed] [Google Scholar]
- 10.Grasso C. S., Wu Y. M., Robinson D. R., et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tolkach Y., Kristiansen G. The heterogeneity of prostate cancer: a practical approach. Pathobiology. 2018;85(1–2):108–116. doi: 10.1159/000477852. [DOI] [PubMed] [Google Scholar]
- 12.Makarov D. V., Loeb S., Getzenberg R. H., Partin A. W. Biomarkers for prostate cancer. Annual Review of Medicine. 2009;60(1):139–151. doi: 10.1146/annurev.med.60.042307.110714. [DOI] [PubMed] [Google Scholar]
- 13.Yuan X., Cai C., Chen S., Chen S., Yu Z., Balk S. P. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene. 2014;33(22):2815–2825. doi: 10.1038/onc.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakazawa M., Paller C., Kyprianou N. Mechanisms of therapeutic resistance in prostate cancer. Current Oncology Reports. 2017;19(2):p. 13. doi: 10.1007/s11912-017-0568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhury A. D., Eeles R., Freedland S. J., et al. The role of genetic markers in the management of prostate cancer. European Urology. 2012;62(4):577–587. doi: 10.1016/j.eururo.2012.05.054. [DOI] [PubMed] [Google Scholar]
- 16.Madan R. A., Gulley J. L. Prostate cancer immunotherapy: the path forward. Current Opinion in Supportive and Palliative Care. 2017;11(3):225–230. doi: 10.1097/SPC.0000000000000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glinsky G. V., Glinskii A. B., Stephenson A. J., Hoffman R. M., Gerald W. L. Gene expression profiling predicts clinical outcome of prostate cancer. The Journal of Clinical Investigation. 2004;113(6):913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor B. S., Schultz N., Hieronymus H., et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross-Adams H., Lamb A. D., Dunning M. J., et al. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: a discovery and validation cohort study. eBioMedicine. 2015;2(9):1133–1144. doi: 10.1016/j.ebiom.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livraghi L., Garber J. E. PARP inhibitors in the management of breast cancer: current data and future prospects. BMC Medicine. 2015;13(1):p. 188. doi: 10.1186/s12916-015-0425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.George A., Kaye S., Banerjee S. Delivering widespread BRCA testing and PARP inhibition to patients with ovarian cancer. Nature Reviews Clinical Oncology. 2017;14(5):284–296. doi: 10.1038/nrclinonc.2016.191. [DOI] [PubMed] [Google Scholar]
- 22.Stark T. W., Hensley P. J., Spear A., Pu H., Strup S. S., Kyprianou N. Predictive value of epithelial-mesenchymal-transition (EMT) signature and PARP-1 in prostate cancer radioresistance. Prostate. 2017;77(16):1583–1591. doi: 10.1002/pros.23435. [DOI] [PubMed] [Google Scholar]
- 23.Lai Y., Kong Z., Zeng T., et al. PARP1-siRNA suppresses human prostate cancer cell growth and progression. Oncology Reports. 2018;39(4):1901–1909. doi: 10.3892/or.2018.6238. [DOI] [PubMed] [Google Scholar]
- 24.Ellwood-Yen K., Graeber T. G., Wongvipat J., et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4(3):223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 25.Tompkins V. S., Han S. S., Olivier A., et al. Identification of candidate B-lymphoma genes by cross-species gene expression profiling. PLoS One. 2013;8(10, article e76889) doi: 10.1371/journal.pone.0076889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Day C. P., Merlino G., Van Dyke T. Preclinical mouse cancer models: a maze of opportunities and challenges. Cell. 2015;163(1):39–53. doi: 10.1016/j.cell.2015.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robles-Espinoza C. D., Adams D. J. Cross-species analysis of mouse and human cancer genomes. Cold Spring Harbor Protocols. 2014;2014(4, article pdb.top078824):pdb.top078358. doi: 10.1101/pdb.top078824. [DOI] [PubMed] [Google Scholar]
- 28.Aytes A., Mitrofanova A., Lefebvre C., et al. Cross-Species Regulatory Network Analysis Identifies a Synergistic Interaction between FOXM1 and CENPF that Drives Prostate Cancer Malignancy. Cancer Cell. 2014;25(5):638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurmeister S., Ramos-Montoya A., Sandi C., et al. Identification of potential therapeutic targets in prostate cancer through a cross-species approach. EMBO Molecular Medicine. 2018;10(3, article e8274) doi: 10.15252/emmm.201708274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeWeese T. L., Hruszkewycz A. M., Marnett L. J. Oxidative stress in chemoprevention trials. Urology. 2001;57(4):137–140. doi: 10.1016/s0090-4295(00)00959-6. [DOI] [PubMed] [Google Scholar]
- 31.Minelli A., Bellezza I., Conte C., Culig Z. Oxidative stress-related aging: a role for prostate cancer? Biochimica et Biophysica Acta. 2009;1795(2):83–91. doi: 10.1016/j.bbcan.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Meiers I., Shanks J. H., Bostwick D. G. Glutathione S-transferase pi (GSTP1) hypermethylation in prostate cancer: review 2007. Pathology. 2007;39(3):299–304. doi: 10.1080/00313020701329906. [DOI] [PubMed] [Google Scholar]
- 33.De Marzo A. M., Platz E. A., Sutcliffe S., et al. Inflammation in prostate carcinogenesis. Nature Reviews Cancer. 2007;7(4):256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khandrika L., Kumar B., Koul S., Maroni P., Koul H. K. Oxidative stress in prostate cancer. Cancer Letters. 2009;282(2):125–136. doi: 10.1016/j.canlet.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bostwick D. G., Alexander E. E., Singh R., et al. Antioxidant enzyme expression and reactive oxygen species damage in prostatic intraepithelial neoplasia and cancer. Cancer. 2000;89(1):123–134. [PubMed] [Google Scholar]
- 36.Itoh K., Mimura J., Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxidants & Redox Signaling. 2010;13(11):1665–1678. doi: 10.1089/ars.2010.3222. [DOI] [PubMed] [Google Scholar]
- 37.Hu R., Saw C. L.-L., Yu R., Kong A.-N. T. Regulation of NF-E2-related factor 2 signaling for cancer chemoprevention: antioxidant coupled with antiinflammatory. Antioxidants & Redox Signaling. 2010;13(11):1679–1698. doi: 10.1089/ars.2010.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang P., Feng L., Oldham E. A., Keating M. J., Plunkett W. Superoxide dismutase as a target for the selective killing of cancer cells. Nature. 2000;407(6802):390–395. doi: 10.1038/35030140. [DOI] [PubMed] [Google Scholar]
- 39.Cairns R. A., Harris I. S., Mak T. W. Regulation of cancer cell metabolism. Nature Reviews Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 40.Gorrini C., Harris I. S., Mak T. W. Modulation of oxidative stress as an anticancer strategy. Nature Reviews Drug Discovery. 2013;12(12):931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 41.Gu L., Cui T., Fan C., et al. Involvement of ERK1/2 signaling pathway in DJ-1-induced neuroprotection against oxidative stress. Biochemical and Biophysical Research Communications. 2009;383(4):469–474. doi: 10.1016/j.bbrc.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 42.Ma X., Zhang J., Liu S., Huang Y., Chen B., Wang D. Nrf2 knockdown by shRNA inhibits tumor growth and increases efficacy of chemotherapy in cervical cancer. Cancer Chemotherapy and Pharmacology. 2012;69(2):485–494. doi: 10.1007/s00280-011-1722-9. [DOI] [PubMed] [Google Scholar]
- 43.Grossi V., Peserico A., Tezil T., Simone C. p38α MAPK pathway: a key factor in colorectal cancer therapy and chemoresistance. World Journal of Gastroenterology. 2014;20(29, article 9744):9758. doi: 10.3748/wjg.v20.i29.9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellezza I., Scarpelli P., Pizzo S. V., Grottelli S., Costanzi E., Minelli A. ROS-independent Nrf2 activation in prostate cancer. Oncotarget. 2017;8(40):67506–67518. doi: 10.18632/oncotarget.18724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leone A., Roca M. S., Ciardiello C., Costantini S., Budillon A. Oxidative stress gene expression profile correlates with cancer patient poor prognosis: identification of crucial pathways might select novel therapeutic approaches. Oxidative Medicine and Cellular Longevity. 2017;2017:18. doi: 10.1155/2017/2597581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jamaspishvili T., Berman D. M., Ross A. E., et al. Clinical implications of PTEN loss in prostate cancer. Nature Reviews Urology. 2018;15(4):222–234. doi: 10.1038/nrurol.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Somaio Neto F., Ikejiri A. T., Bertoletto P. R., et al. Gene expression related to oxidative stress in the heart of mice after intestinal ischemia. Arquivos Brasileiros de Cardiologia. 2013;102(2):165–174. doi: 10.5935/abc.20130240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tahmasbpour E., Ghanei M., Qazvini A., Vahedi E., Panahi Y. Gene expression profile of oxidative stress and antioxidant defense in lung tissue of patients exposed to sulfur mustard. Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 2016;800-801:12–21. doi: 10.1016/j.mrgentox.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 49.Cerami E., Gao J., Dogrusoz U., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao J., Aksoy B. A., Dogrusoz U., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 2013;6(269):p. pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dunning M. J., Vowler S. L., Lalonde E., et al. Mining human prostate cancer datasets: the “camcAPP” shiny app. eBioMedicine. 2017;17:5–6. doi: 10.1016/j.ebiom.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aguirre-Gamboa R., Gomez-Rueda H., Martínez-Ledesma E., et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8(9, article e74250) doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Epstein J. I. An update of the Gleason grading system. The Journal of Urology. 2010;183(2):433–440. doi: 10.1016/j.juro.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 54.Mellinger G. T., Gleason D., Bailar J. The histology and prognosis of prostatic cancer. The Journal of Urology. 1967;97(2):331–337. doi: 10.1016/s0022-5347(17)63039-8. [DOI] [PubMed] [Google Scholar]
- 55.Egevad L., Delahunt B., Srigley J. R., Samaratunga H. International Society of Urological Pathology (ISUP) grading of prostate cancer - an ISUP consensus on contemporary grading. APMIS. 2016;124(6):433–435. doi: 10.1111/apm.12533. [DOI] [PubMed] [Google Scholar]
- 56.Epstein J. I., Amin M. B., Reuter V. E., Humphrey P. A. Contemporary Gleason grading of prostatic carcinoma: an update with discussion on practical issues to implement the 2014 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. The American Journal of Surgical Pathology. 2017;41(4):e1–e7. doi: 10.1097/PAS.0000000000000820. [DOI] [PubMed] [Google Scholar]
- 57.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C (T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 58.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. Journal of Immunological Methods. 1983;65(1–2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 59.Berridge M. V., Tan A. S. Characterization of the cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT): subcellular localization, substrate dependence, and involvement of mitochondrial electron transport in MTT reduction. Archives of Biochemistry and Biophysics. 1993;303(2):474–482. doi: 10.1006/abbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- 60.Ouyang X., DeWeese T. L., Nelson W. G., Abate-Shen C. Loss-of-function ofNkx3.1Promotes increased oxidative damage in prostate carcinogenesis. Cancer Research. 2005;65(15):6773–6779. doi: 10.1158/0008-5472.CAN-05-1948. [DOI] [PubMed] [Google Scholar]
- 61.Naiki T., Naiki-Ito A., Asamoto M., et al. GPX2 overexpression is involved in cell proliferation and prognosis of castration-resistant prostate cancer. Carcinogenesis. 2014;35(9):1962–1967. doi: 10.1093/carcin/bgu048. [DOI] [PubMed] [Google Scholar]
- 62.Burke A. J., Garrido P., Johnson C., Sullivan F. J., Glynn S. A. Inflammation and nitrosative stress effects in ovarian and prostate pathology and carcinogenesis. Antioxidants and Redox Signaling. 2017;26(18):1078–1090. doi: 10.1089/ars.2017.7004. [DOI] [PubMed] [Google Scholar]
- 63.Arora K., Barbieri C. E. Molecular subtypes of prostate cancer. Current Oncology Reports. 2018;20(8, article 707) doi: 10.1007/s11912-018-0707-9. [DOI] [PubMed] [Google Scholar]
- 64.Hinsch A., Brolund M., Hube-Magg C., et al. Immunohistochemically detected IDH1R132H mutation is rare and mostly heterogeneous in prostate cancer. World Journal of Urology. 2018;36(6):877–882. doi: 10.1007/s00345-018-2225-7. [DOI] [PubMed] [Google Scholar]
- 65.Tang K. D., Liu J., Jovanovic L., et al. Adipocytes promote prostate cancer stem cell self-renewal through amplification of the cholecystokinin autocrine loop. Oncotarget. 2016;7(4):4939–4948. doi: 10.18632/oncotarget.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar A., Dhar S., Campanelli G., et al. MTA1 drives malignant progression and bone metastasis in prostate cancer. Molecular Oncology. 2018;12(9):1596–1607. doi: 10.1002/1878-0261.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soriano F. X., Léveillé F., Papadia S., et al. Induction of sulfiredoxin expression and reduction of peroxiredoxin hyperoxidation by the neuroprotective Nrf2 activator 3H-1,2-dithiole-3-thione. Journal of Neurochemistry. 2008;107(2):533–543. doi: 10.1111/j.1471-4159.2008.05648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Soriano F. X., Baxter P., Murray L. M., Sporn M. B., Gillingwater T. H., Hardingham G. E. Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Molecules and Cells. 2009;27(3):279–282. doi: 10.1007/s10059-009-0050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mishra M., Jiang H., Wu L., Chawsheen H. A., Wei Q. The sulfiredoxin-peroxiredoxin (Srx-Prx) axis in cell signal transduction and cancer development. Cancer Letters. 2015;366(2):150–159. doi: 10.1016/j.canlet.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jonsson T. J., Lowther W. T. The peroxiredoxin repair proteins. Sub-Cellular Biochemistry. 2007;44:115–141. doi: 10.1007/978-1-4020-6051-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cox A. G., Winterbourn C. C., Hampton M. B. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. The Biochemical Journal. 2009;425(2):313–325. doi: 10.1042/BJ20091541. [DOI] [PubMed] [Google Scholar]
- 72.Woo H. A., Jeong W., Chang T. S., et al. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. The Journal of Biological Chemistry. 2005;280(5):3125–3128. doi: 10.1074/jbc.C400496200. [DOI] [PubMed] [Google Scholar]
- 73.Li Q., Yu S., Wu J., Zou Y., Zhao Y. Sulfiredoxin-1 protects PC12 cells against oxidative stress induced by hydrogen peroxide. Journal of Neuroscience Research. 2013;91(6):861–870. doi: 10.1002/jnr.23218. [DOI] [PubMed] [Google Scholar]
- 74.Zhou Y., Yu S., Wu J., Chen Y., Zhao Y. Sulfiredoxin-1 exerts anti-apoptotic and neuroprotective effects against oxidative stress-induced injury in rat cortical astrocytes following exposure to oxygen-glucose deprivation and hydrogen peroxide. International Journal of Molecular Medicine. 2015;36(1):43–52. doi: 10.3892/ijmm.2015.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei Q., Jiang H., Xiao Z., et al. Sulfiredoxin-peroxiredoxin IV axis promotes human lung cancer progression through modulation of specific phosphokinase signaling. Proceedings of the National Academy of Sciences. 2011;108(17):7004–7009. doi: 10.1073/pnas.1013012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wei Q., Jiang H., Baker A., et al. Loss of sulfiredoxin renders mice resistant to azoxymethane/dextran sulfate sodium-induced colon carcinogenesis. Carcinogenesis. 2013;34(6):1403–1410. doi: 10.1093/carcin/bgt059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei Q., Jiang H., Matthews C. P., Colburn N. H. Sulfiredoxin is an AP-1 target gene that is required for transformation and shows elevated expression in human skin malignancies. Proceedings of the National Academy of Sciences. 2008;105(50):19738–19743. doi: 10.1073/pnas.0810676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu L., Jiang H., Chawsheen H. A., et al. Tumor promoter-induced sulfiredoxin is required for mouse skin tumorigenesis. Carcinogenesis. 2014;35(5):1177–1184. doi: 10.1093/carcin/bgu035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang J., Si L., Wang G., Bai Z., Li W. Increased sulfiredoxin expression in gastric cancer cells may be a molecular target of the anticancer component diallyl trisulfide. BioMed Research International. 2019;2019:8. doi: 10.1155/2019/4636804.4636804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lei K., Townsend D. M., Tew K. D. Protein cysteine sulfinic acid reductase (sulfiredoxin) as a regulator of cell proliferation and drug response. Oncogene. 2008;27(36):4877–4887. doi: 10.1038/onc.2008.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hartikainen J. M., Tengström M., Kosma V. M., Kinnula V. L., Mannermaa A., Soini Y. Genetic polymorphisms and protein expression of NRF2 and sulfiredoxin predict survival outcomes in breast cancer. Cancer Research. 2012;72(21):5537–5546. doi: 10.1158/0008-5472.CAN-12-1474. [DOI] [PubMed] [Google Scholar]
- 82.Soini Y., Eskelinen M., Juvonen P., et al. Nuclear Nrf2 expression is related to a poor survival in pancreatic adenocarcinoma. Pathology, Research and Practice. 2014;210(1):35–39. doi: 10.1016/j.prp.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 83.Raatikainen S., Aaaltomaa S., Kärjä V., Soini Y. Increased peroxiredoxin 6 expression predicts biochemical recurrence in prostate cancer patients after radical prostatectomy. Anticancer research. 2015;35:6465–6470. [PubMed] [Google Scholar]
- 84.Kim H., Lee G. R., Kim J., et al. Sulfiredoxin inhibitor induces preferential death of cancer cells through reactive oxygen species-mediated mitochondrial damage. Free Radical Biology & Medicine. 2016;91:264–274. doi: 10.1016/j.freeradbiomed.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 85.Kim J., Lee G. R., Kim H., et al. Effective killing of cancer cells and regression of tumor growth by K27 targeting sulfiredoxin. Free Radical Biology & Medicine. 2016;101:384–392. doi: 10.1016/j.freeradbiomed.2016.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: description of the patient clinical data used in the preparation of the TMAs, such as Gleason score, prognostic category, survival time, and patient outcome. Figure S1: patients with PCa grouped by prognosis category (1-5) show expected survival curves. (A) Global survival curve of patients with PCa associated to prognosis category (1-5, from more differentiated to less differentiated), according to the ISUP grade. Clinical data are from those patients which prostate samples were used to construct TMAs. Kaplan-Meier curves are statistically different with p < 0.0001. (B) Additional analysis of the same patients of Figure S1a, dividing them into groups with good prognosis (categories 1 and 2, black curve) and worse prognosis (categories 3, 4, and 5, red curve), according to the ISUP grade. Kaplan-Meier curves are significantly different with p < 0.0001. Figure S2: SRXN1 expression is increased in advanced PCa and in most prostate tumor cell lines, and its overexpression is associated with poor prognosis and lower disease-/progression-free survival. (A) Levels of SRXN1 expression in wild-type prostate (control) and advanced PCa samples (Gleason scores 8 and 9) from a study available on the GEO profile human database (reference series GSE5016) [1]. (B) SRXN1 gene expression in different prostate cell lines (androgen sensitive and castration-resistant) obtained from a study available on the GEO profile human database (reference series GSE4016) [2]. (C) Expression of SRXN1 (median) in five PCa iClusters generated by the Cambridge Carcinoma of the Prostate App (camcAPP dataset) [3] from an integrative study [4]. iClusters 1 (red), 3 (green), and 5 (orange) represent groups of patients with worse prognosis, while iClusters 2 (blue) and 4 (purple) represent groups with better prognosis. Boxplots are significantly different, with p = 5.7833−9. (D) Expression of SRXN1 (median) in five PCa iClusters generated by the Cambridge Carcinoma of the Prostate App (camcAPP dataset) [3] from an integrative study [4]. iClusters 1 (red), 3 (green), and 5 (orange) represent groups of patients with worse prognosis, while iClusters 2 (blue) and 4 (purple) represent groups with better prognosis. Boxplots are significantly different with p = 0.034473. (E) Expression of SRXN1 (median) in six PCa iClusters generated by the Cambridge Carcinoma of the Prostate App (camcAPP dataset) [3] from an integrative study [5]. iClusters 1 (salmon), 2 (dark yellow), 3 (green), and 4 (turquoise) are groups of patients with more favorable prognosis with minimal copy number alterations (CNA), while iClusters 5 (light blue) and 6 (lilac) include most of the metastatic tumors with substantial CNA. Boxplots are significantly different, with p = 3.42−6. (F) Kaplan-Meier curve displaying the probability of freedom from biochemical recurrence of PCa with (red) or without (blue) SRNX1 overexpression, cataloged by the Cambridge Carcinoma of the Prostate App (camcAPP dataset) [3] from an integrative study [5]. Curves are statistically different with p = 0.0079.
Data Availability Statement
The RNAseq data from the GEMM Pb-Cre4; Ptenf/f mouse used to support the findings of this study have been deposited in the NCBI Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/), reference number GSE94574. Previously reported human databases were used to support this study and are available at the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geoprofiles/), the cBioPortal for Cancer Genomics (http://www.cbioportal.org/), The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/), the Cambridge Carcinoma of the Prostate App (camcAPP dataset) (https://bioinformatics.cruk.cam.ac.uk/apps/camcAPP/), and the SurvExpress database (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp). These prior studies (and datasets) are cited at relevant places within the text as references [18, 19, 49–52] and Satake et al. (supplementary material [1]) and Zhao et al. (supplementary material [2]). The clinical data of the PCa patients from TMA samples used to support the findings of this study are included within the supplementary information files (Table S1).


