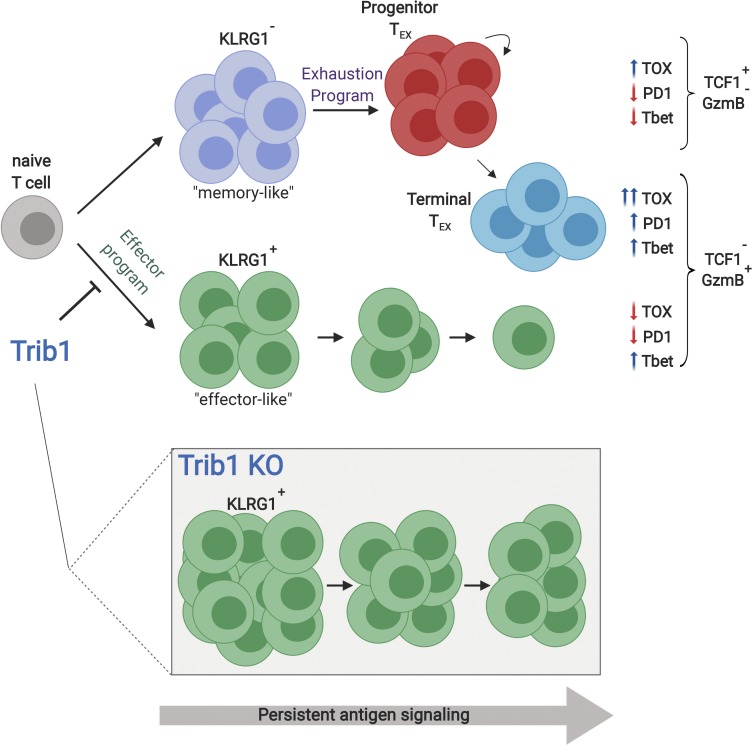
Rome et al. show that Trib1 controls effector versus exhausted T cell differentiation and function by restraining effector programming during persistent infection, revealing a new regulatory axis that could be targeted to recover waning T cell responses during chronic disease.
Abstract
In chronic infections, the immune response fails to control virus, leading to persistent antigen stimulation and the progressive development of T cell exhaustion. T cell effector differentiation is poorly understood in the context of exhaustion, but targeting effector programs may provide new strategies for reinvigorating T cell function. We identified Tribbles pseudokinase 1 (Trib1) as a central regulator of antiviral T cell immunity, where loss of Trib1 led to a sustained enrichment of effector-like KLRG1+ T cells, enhanced function, and improved viral control. Single-cell profiling revealed that Trib1 restrains a population of KLRG1+ effector CD8 T cells that is transcriptionally distinct from exhausted cells. Mechanistically, we identified an interaction between Trib1 and the T cell receptor (TCR) signaling activator, MALT1, which disrupted MALT1 signaling complexes. These data identify Trib1 as a negative regulator of TCR signaling and downstream function, and reveal a link between Trib1 and effector versus exhausted T cell differentiation that can be targeted to improve antiviral immunity.
Graphical Abstract
Introduction
The adaptive immune response is characterized by the rapid expansion and differentiation of cytolytic and/or cytokine-producing effector T cells that are required to clear pathogens. These processes are initiated in response to signaling via multiple receptors including the TCR, costimulatory receptors and cytokine receptors. Following activation, T cells transiently express inhibitory receptors (IRs) that initiate signaling pathways to restrain activation and promote resolution of the immune response. However, in the context of chronic antigen exposure, such as in persistent infections or cancer, antigen is never cleared, and T cells gradually lose effector function, up-regulate and sustain expression of multiple IRs, and become “exhausted” (Wherry and Kurachi, 2015). Immune checkpoint blockade (ICB), such as PD-1 inhibition, has emerged as a strategy for bolstering the immune response during chronic disease by suppressing signaling through IRs. Despite early success, treatment responses vary among patients and are often transient (Pauken and Wherry, 2015), demonstrating a critical need to identify more robust and durable therapies. While ICB aims to reverse T cell exhaustion by targeting IRs, identifying other mechanisms controlling T cell function, such as effector cell differentiation, offers alternative strategies for augmenting T cell immunity to chronic disease. Indeed, augmenting T cell responses by targeting effector response regulators, such as IL-2 or glucocorticoid-induced tumor necrosis factor receptor family member, promotes T cell immunity during chronic infection (Blattman et al., 2003; Clouthier et al., 2015).
Recent work has begun to elucidate the heterogeneity of the exhausted CD8 T cell (TEX) population and has identified a progenitor subset that can repopulate more terminally differentiated TEX cells that are short-lived but often possess residual effector function; combined, these populations maintain steady-state control of chronic infection long-term (He et al., 2016; Im et al., 2016; Leong et al., 2016; Paley et al., 2012; Utzschneider et al., 2016; Wu et al., 2016). These exhausted subsets develop and differentiate from the pool of KLRG1−CD127+ cells generated early in infection (Angelosanto et al., 2012), which in the context of an acute infection would define memory precursor effector cells (MPEC), which go on to form the memory pool, as compared with the short-lived effector cell population (SLEC) marked by KLRG1+CD127− expression. During chronic disease, the potential to form memory is progressively lost, and the KLRG1+ pool contracts as exhaustion progresses. Multiple recent studies identified the DNA-binding factor TOX as a central driver of the exhaustion program (Alfei et al., 2019; Khan et al., 2019; Scott et al., 2019; Seo et al., 2019; Yao et al., 2019). In the absence of TOX, there is a dramatic expansion of KLRG1+ cells, but also a concomitant inability to form TEX subsets or sustain an antiviral response, and TOX KO cells are rapidly lost following infection. The contraction of this effector-like lineage is directed by the transcription factor TCF1, which represses the KLRG1+ population and promotes KLRG1− TEX progenitor cells (Chen et al., 2019). Despite these new insights, the role of cells marked by effector molecules such as KLRG1 and how these cells fit into the landscape of cells responding during chronic disease is not well understood. Identifying pathways that can enhance and, more importantly, sustain effector activity within an exhausted setting may offer novel strategies to improve the control of chronic diseases.
CD4 effector cells also expand following infection and provide CD4 help to the CD8 response, promoting CD8 effector T cell expansion, survival, and function (Snell et al., 2016; Swain et al., 2012). Mouse models of CD4 T cell deficiency showed that CD4 T cells are critical for controlling viral titers (Aubert et al., 2011; Matloubian et al., 1994; Zajac et al., 1998), and defective CD4 T cell responses correlate with persistent infection in human disease (Schulze Zur Wiesch et al., 2012). However, the cellular and molecular mechanisms underlying these phenomena are not clear. Therefore, identifying regulatory factors that control both CD4 and CD8 T cell responses will reveal important insights into the dynamic interplay between these responses during chronic disease.
Trib1 is a member of the Tribbles (Trib) pseudokinase family, an evolutionarily conserved protein family that is implicated in diverse cellular processes including proliferation, survival, metabolism, differentiation, and oncogenic transformation (Hegedus et al., 2007; Lohan and Keeshan, 2013). The three mammalian homologues of Trib, Trib1, Trib2, and Trib3, are characterized by a conserved pseudokinase domain (Hegedus et al., 2007) and a C-terminal E3 ligase-binding domain (Qi et al., 2006). In multiple contexts, Trib proteins function as scaffolding molecules that facilitate protein degradation via a proteasome-dependent mechanism. Important targets of degradation include C/EBPα, C/EBPβ, and acetyl CoA carboxylase. Proteasomal degradation of these proteins relies on the direct association of the E3 ligase COP1 with the Trib C terminus (Dedhia et al., 2010; Keeshan et al., 2006; Murphy et al., 2015; Naiki et al., 2007; Qi et al., 2006; Uljon et al., 2016). Trib proteins are also implicated in COP1-independent signaling pathways, including Akt (Du et al., 2003; Hill et al., 2017; Naiki et al., 2007) and MAPK (Kiss-Toth et al., 2004; Yokoyama et al., 2010), that are inhibited by their interaction with Trib. The physiological significance of these COP1-independent interactions is poorly understood. KO mouse studies suggest different functions for Trib1–3 in the immune system. Trib1 KO mice have normal lymphoid development but defective myelopoiesis (Mack et al., 2019; Satoh et al., 2013). Trib2 (Stein et al., 2016) and Trib3 (Okamoto et al., 2007) KO mice have no overt defects in immune cell development, though subtle/transient defects in thymic T cell development were described in one strain of Trib2-deficient mice (Liang et al., 2016). The basis for the different functions of Trib1–3 is unknown, and the role of Trib proteins in T cell responses is uncharacterized.
Here, we identify Trib1 as a central regulator of T cell–mediated antiviral immunity. We found that Trib1 was up-regulated in T cells ex vivo following stimulation, as well as during the progression of chronic infection. To understand the significance of this finding, we generated T cell–specific Trib1 cKO mice. These mice generated significantly more effective CD4 and CD8 T cell responses during chronic lymphocytic choriomeningitis virus (LCMV) infection, resulting in accelerated control of viral replication. We found that Trib1 functions to restrain the accumulation, persistence, and function of effector-like cells during chronic infection, demonstrating a new regulatory event in the progression of a chronic T cell response. Using single-cell RNA-sequencing (scRNA-seq), we further defined the effector-like population of CD8 T cells enriched in the absence of Trib1 as transcriptionally distinct from both progenitor and terminal TEX subsets. We also identified a molecular mechanism of T cell signaling whereby Trib1 restricts the function of MALT1, a crucial mediator of TCR signaling and T cell function (Meininger and Krappmann, 2016). Together, these data demonstrate that Trib1 expression limits effector T cell responses during chronic disease, and reveal a new regulatory mechanism restraining pro-effector T cell signaling complexes via an inhibitory Trib1:MALT1 interaction.
Results
Trib1 controls antiviral T cell immunity
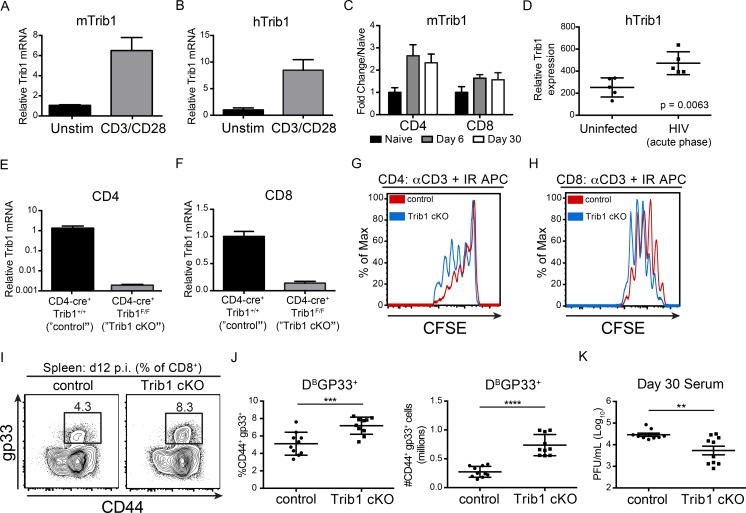
To determine if Trib1 expression is regulated downstream of T cell activation, we stimulated naive mouse CD4 T cells (Fig. 1 A) and human Jurkat T cells (Fig. 1 B) with anti-CD3/CD28 and measured Trib1 expression. Trib1/TRIB1 mRNA was induced by 4 h after stimulation, suggesting a role for Trib1 in T cell function after activation. In the context of chronic disease, we find that Trib1 is also up-regulated during persistent viral infection. Trib1 expression was induced and sustained in both CD4 and CD8 T cells from mice infected with clone 13 (Fig. 1 C), a chronic strain of LCMV characterized by sustained viral replication and progressive T cell exhaustion (Wherry et al., 2003). Similarly, TRIB1 expression was elevated in human CD4 T cells from chronically infected (HIV) patients (Fig. 1 D; Hyrcza et al., 2007). These data suggested that Trib1 might regulate T cell responses during chronic disease.
Figure 1.
Trib1 negatively regulates T cell proliferation, antiviral T cell expansion, and viral control. (A and B) Relative Trib1/TRIB1 expression measured by qPCR from (A) sorted naive murine CD4 T cells and (B) human Jurkat T cells following 4 h ± anti-CD3/CD28 stimulation. CT (cycle threshold) values normalized to 18s ribosomal RNA. (C) Quantification of microarray data from clone 13 infected mice. Expression at day 6 and day 30 p.i. in tetramer+ CD4 (gp66) or CD8 (gp33) T cells is displayed as fold change relative to naive (GSE41870). (D) Relative TRIB1 expression in CD4 T cells from uninfected or HIV-infected human patient samples (GSE6740). “Acute phase” denotes the clinical stage of HIV infection and not an acute infection. (E and F) Relative Trib1 expression measured by qPCR from MACs-purified CD4 T cells (E) or purified CD8 T cells from control and Trib1 cKO mice (F). CT values normalized to 18s ribosomal RNA. (G and H) CFSE dilution measured by flow cytometry from FACS-sorted naive CD4+CD25− T cells (G) or naive CD8 T cells cultured with anti-CD3 and congenic irradiated APCs (CD45.1 spleen) for 3 d (H; gated on CD45.2+). (I and J) Representative flow plots (I) and summary of frequency (left) and absolute number (right) of splenic LCMV-specific CD8 (gp33+) T cells at day 12 p.i. (J; clone 13; gated on CD8+). (K) Viral titers 30 d p.i. in sera from mice infected with clone 13 LCMV. Data in E and F are representative of more than five mice per genotype. Data in G and H are representative of more than three independent experiments. Data in I and J are representative of two independent experiments each with 7–10 mice per genotype. Data in K are representative of three independent experiments each with 7–10 mice per genotype. Error bars are ± SD (A–J) or ± SEM (K). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired Student’s t test (A–J) or by unpaired Student’s t test with Welch’s correction (K). Control: CD4-cre+ Trib1+/+; Trib1 cKO: CD4-cre+ Trib1F/F. Unstim, unstimulated; max, maximum.
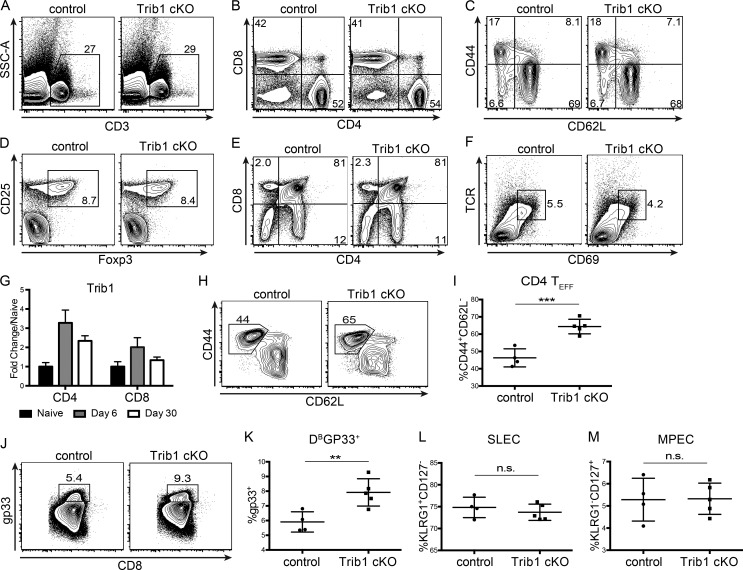
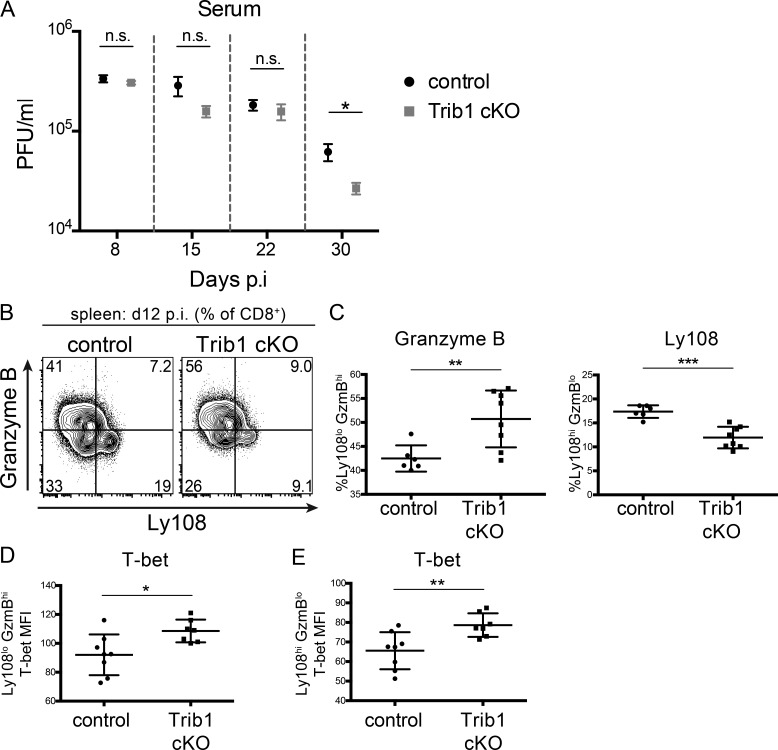
To determine the role of Trib1 in the T cell immune response during infection, we bred CD4-cre mice (Lee et al., 2001) to Trib1F/F mice (Bauer et al., 2015) to generate CD4-cre Trib1F/F mice that conditionally ablate Trib1 expression in the T cell compartment (Fig. 1, E and F). These conditional Trib1-deficient mice (referred to as “Trib1 cKO”) have no overt phenotype in the naive state and have expected T cell frequencies in the spleen and thymus (Fig. S1, A–F). Ex vivo, we found that Trib1 restrained T cell proliferation following activation of either naive CD4 or CD8 T cells (Fig. 1, G and H), suggesting that Trib1 functions intrinsically to suppress function in both cell types. To measure in vivo T cell responses, we infected Trib1 cKO mice with clone 13 and tracked a subset of the LCMV-specific CD8 response using the MHC class I gp33 (DbGP33-41) tetramer. Consistent with the increased proliferation observed ex vivo, expansion of the gp33+ population in Trib1 cKO mice was increased compared with Trib1+/+ CD4-cre+ controls (referred to as “control”; Fig. 1, I and J). In accordance with an enhanced CD8 response, viral titers at day 30 post-infection (p.i.) were significantly reduced in Trib1 cKO mice compared with controls (Fig. 1 K), with no significant difference in viral titers at days 8–22 p.i. (Fig. S2 A). Of note, Trib1 expression is also induced during acute infection in T cells from mice infected with LCMV Armstrong (Fig. S1 G). Trib1 cKO mice infected with Armstrong had increased expansion of the CD4 effector (CD44+CD62L−; Fig. S1, H and I) and CD8 gp33+ populations (Fig. S1, J and K), suggesting Trib1 broadly restrains T cell responses during both acute and chronic infection. However, loss of Trib1 did not alter SLEC (KLRG1+CD127−) or MPEC (KLRG1−CD127+) differentiation during acute infection (Fig. S1, L and M).
Figure S1.
Trib1 is dispensable for T cell development and restrains CD4 and CD8 T cell responses to acute infection without altering effector/memory differentiation. (A–D) Representative flow plots of the frequency of CD3+ T cells (A), CD4 and CD8 subsets (B), CD4 effector cells (C; CD44+CD62L−, gated on CD4+), and T reg cells (CD25+Foxp3+, gated on CD4+) from the spleens of naive 6–10-wk-old mice (D). (E and F) Representative flow plots of the frequency of CD4 and CD8 single-positive (SP) and double-positive (DP) thymocytes (E) and the frequency of TCRhiCD69+ DP thymocytes from naive 6–10-wk-old mice (F). (G) Quantification of microarray data from Armstrong-infected mice. Expression at day 6 and day 30 p.i. in tetramer+ CD4 (gp66) or CD8 (gp33) T cells is displayed as fold change relative to naive (GSE41870). (H and I) Representative flow plot (H) and summary of the frequency of CD4 effector T cells (CD44+CD62L−, gated on CD4+) at day 8 after Armstrong infection (I). (J and K) Representative flow plot (J) and summary of the frequency of LCMV-specific CD8 (gp33+) T cells at day 8 after Armstrong infection (K). (L and M) Summary of the frequency of SLECs (L; KLRG1+CD127−) and MPECs (KLRG1−CD127+; gated on CD8+) at day 8 post Armstrong infection (M). Data in A–F are representative of more than five mice per genotype. Data in H–K represent an experiment with four or five mice per genotype. Data in L and M are representative of two independent experiments each with four or five mice per genotype. Error bars are mean ± SD. **, P < 0.01; ***, P < 0.001 by unpaired Student’s t test. Control: CD4-cre+ Trib1+/+; Trib1 cKO: CD4-cre+ Trib1F/F. SSC-A, side scatter area.
Figure S2.
Trib1 cKO TEX subset phenotype is established early in chronic infection and before differences in viral burden between control and Trib1 cKO mice. (A) Viral titers 8, 15, 22, and 30 d p.i. in sera from mice infected with clone 13 LCMV. Data represent one independent experiment with 9 or 10 mice per genotype. (B and C) Representative flow plots (B) and summary of the frequency of Ly108/GranzymeBhi/lo CD8 T cells (gated on CD8+) from the spleens of mice at day 12 p.i. (C). (D and E) Summary of T-bet expression (measured by MFI) in splenic Ly108loGranzymeBhi (D) or Ly108hiGranzymeBlo CD8 T cells at day 12 p.i. (E). Day 30 titers are representative of three independent experiments with a second experimental cohort (of the three) represented in Fig. 1 K. Data in B and C are representative of two independent experiments each with 6-10 mice per genotype. Data in D and E represent an experiment with seven or eight mice per genotype. Error bars are ± SEM in A and ± SD in B–E. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by unpaired Student’s t test. Control: CD4-cre+Trib1+/+; Trib1 cKO: CD4-cre+Trib1F/F.
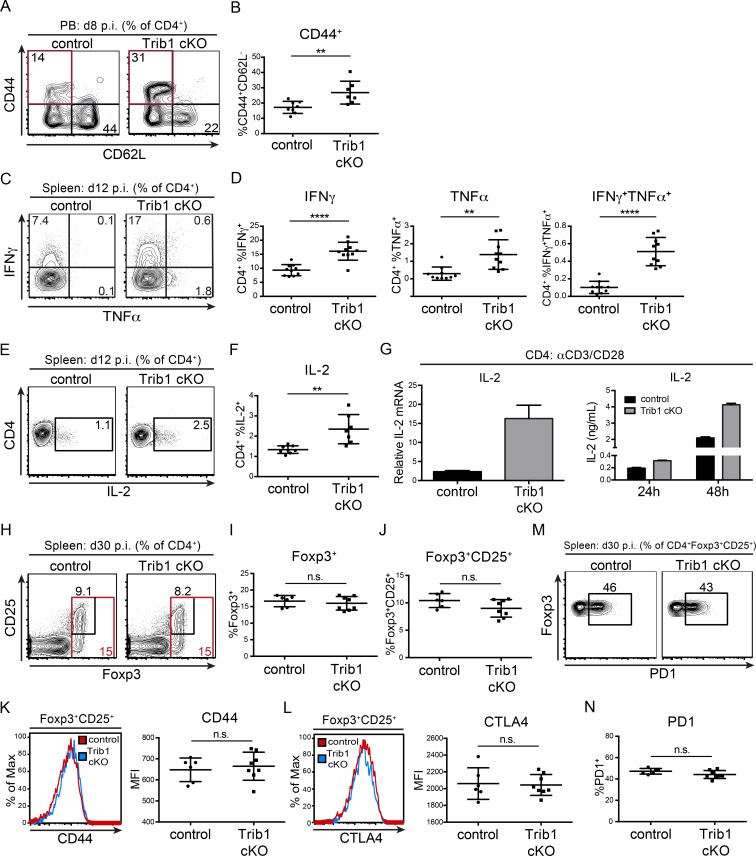
CD4 T cells play an important role in chronic infection and are necessary to maintain viral control. Similar to acute infection and consistent with the elevated CD4 proliferation observed in culture, the CD4 effector cell population (marked by CD44+CD62L−) was expanded in Trib1 cKO mice at day 8 p.i. with clone 13 (Fig. 2, A and B). Trib1 cKO CD4 T cells also produced more IFN-γ and TNF-α following ex vivo PMA/ionomycin stimulation (Fig. 2, C and D), demonstrating an increased functional capacity. During an immune response, IL-2 produced by CD4 T cells promotes the differentiation and proliferation of CD8 T cells. At day 12 p.i. with clone 13, CD4 T cells from Trib1 cKO mice produced significantly more IL-2 than controls (Fig. 2, E and F). These data were consistent with the ability of Trib1-deficient CD4 T cells to produce more IL-2 following TCR/CD28 stimulation ex vivo (Fig. 2 G), further validating that Trib1 inhibits IL-2 production and CD4 function. CD4 regulatory T (T reg) cells also play a less characterized but important role in regulating antiviral immunity (Gangaplara et al., 2018; Park et al., 2015; Penaloza-MacMaster et al., 2014; Veiga-Parga et al., 2013); however, we did not observe alterations in the relative abundance of T reg cells or markers of T reg cell activation in Trib1 cKO mice during clone 13 infection (Fig. 2, H–N).
Figure 2.
Trib1 deficiency promotes elevated CD4 effector cell expansion and function in response to chronic infection. (A and B) Representative flow plots (A) and summary of the frequency of CD4 effector T cells (B; CD44+CD62L−, gated on CD4+) from peripheral blood (PB) at day 8 after clone 13 infection. (C and D) Representative flow plots (C) and summary of cytokine production in splenic CD4 T cells harvested from mice at day 12 p.i. with clone 13 and stimulated ex vivo with PMA and ionomycin for 5 h (D; gated on CD4+). (E and F) Representative flow plots (E) and summary of IL-2 cytokine production in splenic CD4 T cells harvested from mice at day 12 p.i. and stimulated as in C and D (F). (G) Relative Il-2 expression measured by qPCR from naive CD4 T cells cultured for 24 h (left) or IL-2 secretion measured by ELISA (right) from CD4 T cells cultured for the indicated time points ± anti-CD3/CD28 stimulation. (H–J) Representative flow plots (H) and summary of the frequency of Foxp3+ (I) and Foxp3+CD25+ T reg cells in the spleen at day 30 after clone 13 infection (J). (K) Representative histogram (left) and summary (right) of the MFI of CD44 expression on splenic T reg cells at day 30 after clone 13 infection. (L) Representative histogram (left) and summary (right) of the MFI of CTLA4 expression on splenic T reg cells at day 30 after clone 13 infection. (M and N) Representative flow plots (M) and summary of the frequency of PD1+ T reg cells in the spleen of mice at day 30 p.i. (N). Data in A and B are representative of four independent experiments each with 6–10 mice per genotype. Data in C–F are representative of two independent experiments each with 7–10 mice per genotype. Data in G are representative of two (left) and more than five (right) independent experiments. Data in I are representative of two independent experiments each with 6–10 mice per genotype. Data in J–N represent an experiment with 6–8 mice per genotype. CT values normalized to 18s ribosomal RNA. Error bars are mean ± SD. **, P < 0.01; ****, P < 0.0001 by unpaired Student’s t test. Control: CD4-cre+ Trib1+/+; Trib1 cKO: CD4-cre+ Trib1F/F. n.s., not significant.
Trib1 deficiency enforces and sustains an effector-like T cell phenotype in response to chronic infection
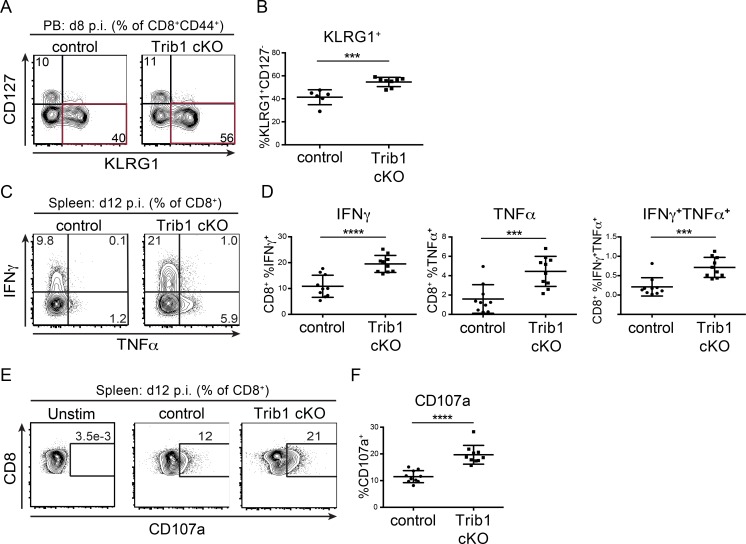
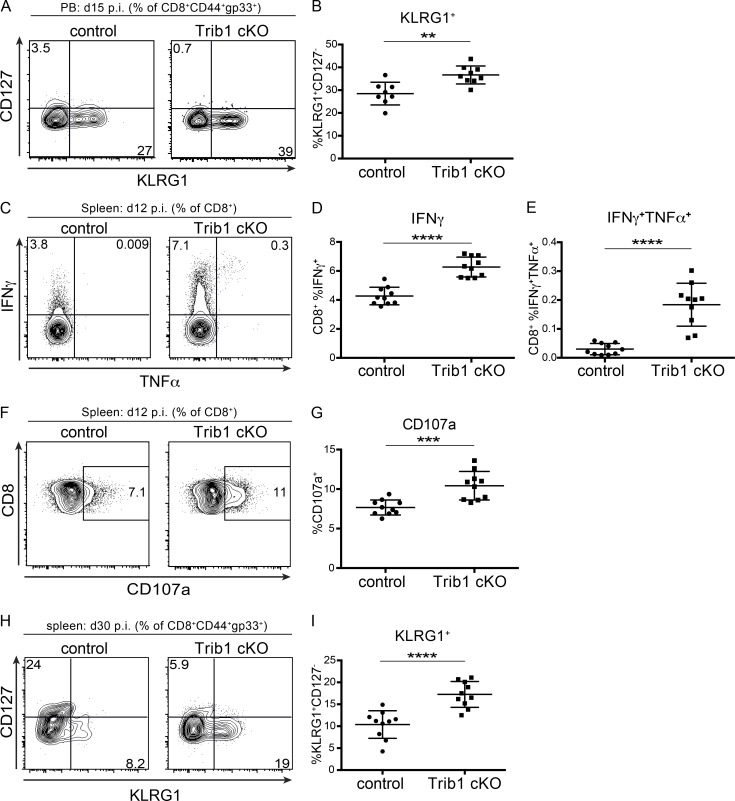
To identify features of the CD8 T cell response in Trib1 cKO mice that may contribute to improved viral control, we tracked markers of CD8 differentiation and function over the course of clone 13 infection. Given that gp33+ CD8 T cells account for only a portion of the LCMV-specific CD8 response, we analyzed both “bulk” activated CD8 T cells (CD44+) as well as gp33+ CD8 T cells in order to characterize the broad T cell response to chronic infection. Early in the response (day 8–15 p.i.), we found that Trib1 deficiency significantly increased the frequency of KLRG1+ effector-like cells in bulk CD8+ as well as gp33+ CD8 T cell populations (Fig. 3, A and B; and Fig. S3, A and B), suggesting that Trib1 restricts CD8 effector-like cell differentiation during chronic infection.
Figure 3.
Trib1 deficiency promotes CD8 KLRG1+ effector-like cell expansion and CD8 function in response to chronic infection. (A and B) Representative flow plots (A) and summary of the frequency of CD8 effector-like T cells (KLRG1+CD127−, gated on CD8+CD44+) from peripheral blood (PB) at day 8 after clone 13 infection (B). (C and D) Representative flow plots (C) and summary of cytokine production in splenic CD8 T cells harvested from mice at day 12 p.i. with clone 13 and stimulated ex vivo with PMA and ionomycin for 5 h (D; gated on CD8+). (E and F) Representative flow plots (E) and summary of CD107a staining on splenic CD8 T cells harvested from mice at day 12 p.i. and stimulated ex vivo as in C and D (F). Data in A and B are representative of four independent experiments each with 6–10 mice per genotype. Data in C–F are representative of two independent experiments each with 7–10 mice per genotype. Error bars are mean ± SD. ***, P < 0.001; ****, P < 0.0001 by unpaired Student’s t test. Control: CD4-cre+ Trib1+/+; Trib1 cKO: CD4-cre+ Trib1F/F.
Figure S3.
Trib1 deficiency promotes DBGP33+ T cell effector-like responses in chronic infection. (A and B) Representative flow plots (A) and summary of the frequency of peripheral blood (PB) LCMV-specific CD8 effector-like T cells (KLRG1+CD127−, gated on CD8+CD44+gp33+) at day 15 after clone 13 infection (B). (C–E) Representative flow plots (C) and summary of cytokine production in splenic CD8 T cells harvested from mice at day 12 p.i. and stimulated ex vivo with gp33 peptide for 5 h (D and E). (F and G) Representative flow plots (F) and summary of CD107a staining in splenic CD8 T cells harvested from mice at day 12 p.i. and stimulated ex vivo with gp33 peptide for 5 h (G). (H and I) Representative flow plots (H) and summary of the frequency of splenic LCMV-specific CD8 effector-like T cells (KLRG1+CD127−, gated on CD8+CD44+gp33+) at day 30 after clone 13 infection (I). Data in A and B are representative of three independent experiments with 7–10 mice per genotype. Data in C–G are representative of two independent experiments each with 7–10 mice per genotype. Data in H and I are representative of three independent experiments each with 8–10 mice per genotype. Error bars are mean ± SD. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired Student’s t test. Control: CD4-cre+ Trib1+/+; Trib1 cKO: CD4-cre+ Trib1F/F.
Our data show that Trib1 suppresses the accumulation of gp33+ cells and KLRG1+ short-lived effector-like T cells, and that loss of Trib1 improves viral control. Given these findings, we assessed whether Trib1 deficiency also results in elevated CD8 T cell function. At 12 d p.i., CD8 T cells from Trib1 cKO mice displayed an enhanced capacity to produce cytokines when stimulated ex vivo with PMA/ionomycin (Fig. 3, C and D) or gp33 peptide (Fig. S3, C–E) and possessed increased CD8 T cell cytolytic potential as measured by CD107a expression (Fig. 3, E and F; and Fig. S3, F and G). Combined, these data show that Trib1 limits effector-like cell frequency and function in CD8 T cells during chronic infection.
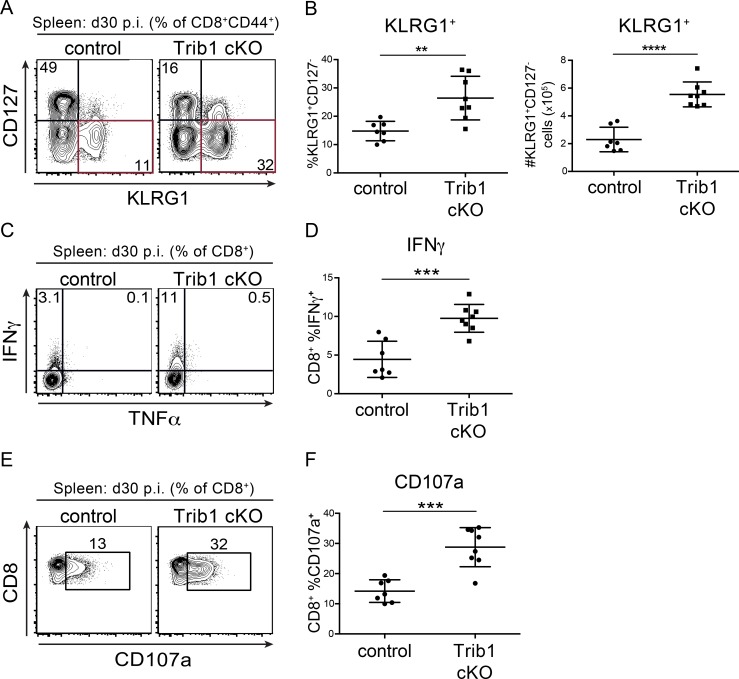
As a chronic infection progresses, short-lived CD8 T cell populations contract. To determine if Trib1 regulates the persistence of the short-lived effector-like KLRG1+ population that was elevated at day 8–15 p.i., we measured the abundance of these cells at day 30 following clone 13 infection. We found that Trib1 cKO mice sustain expanded KLRG1+ populations in bulk CD8+ as well as gp33+ CD8 T cells until at least 30 d p.i. (Fig. 4, A and B; and Fig. S3, H and I). In addition to the progressive erosion of effector-like cell numbers during clone 13 infection, there is a concomitant decline (exhaustion) in T cell function (Wherry and Kurachi, 2015). While we did not observe a significant difference in function following ex vivo gp33 peptide stimulation at day 30 p.i. (data not shown), we found that CD8 T cells from Trib1 cKO mice still produced more IFN-γ and had elevated cytolytic capacity following ex vivo PMA/ionomycin stimulation (Fig. 4, C–F), suggesting an enhanced functional capacity across the bulk CD8+ response even later into chronic infection.
Figure 4.
Trib1 cKO mice sustain elevated CD8 effector-like T cells and function during chronic infection. (A and B) Representative flow plots (A) and summary of the frequency (left) and absolute number (right) of splenic CD8 effector-like T cells (KLRG1+CD127−, gated on CD8+ CD44+) at day 30 after clone 13 infection (B). (C and D) Representative flow plots (C) and summary of cytokine production measured by frequency of cytokine-producing cells in splenic CD8 T cells harvested from mice at day 30 p.i. and stimulated ex vivo as in Fig. 3, C–F (D). (E and F) Representative flow plots (E) and summary of CD107a staining on splenic CD8 T cells harvested from mice at day 30 p.i. and stimulated ex vivo as in C and D (F). Data in A and B are representative of four independent experiments each with 8–10 mice per genotype. Data in C–F are representative of three or four independent experiments each with 6–10 mice per genotype. Error bars are mean ± SD. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired Student’s t test. Control: CD4-cre+ Trib1+/+; Trib1 cKO: CD4-cre+ Trib1F/F.
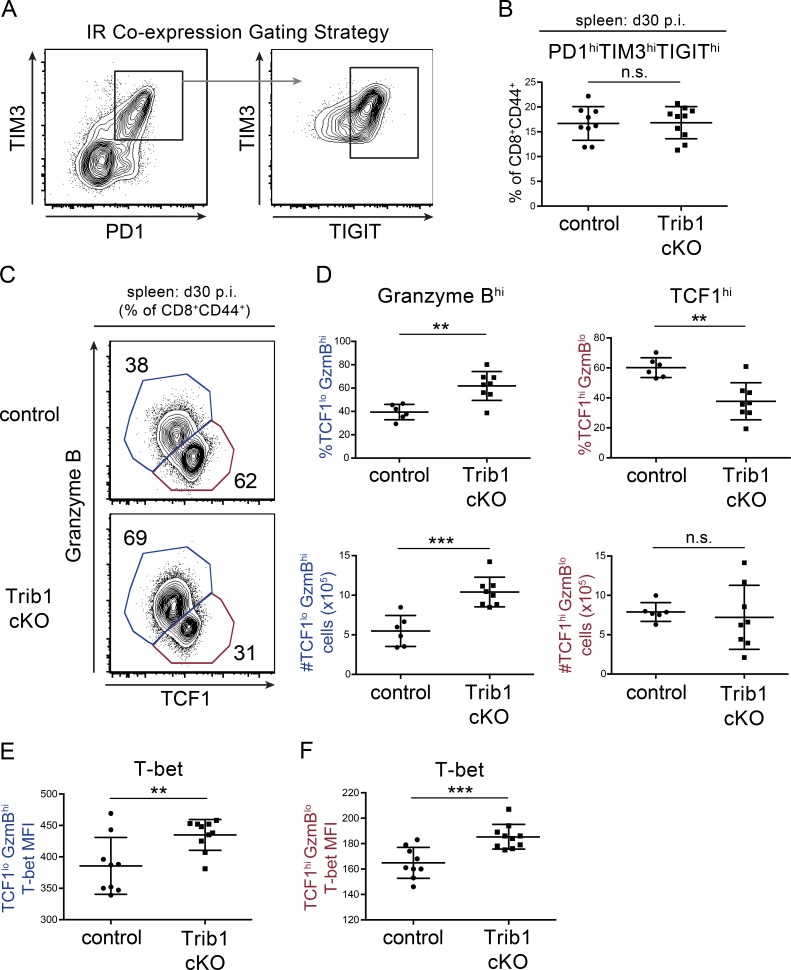
Loss of Trib1 promotes the expansion of TCF1loGzmBhi CD8 T cells while retaining TCF1hi progenitor TEX cells
Given that Trib1 loss enhanced CD8 T cell function and viral control, we asked whether Trib1 increased CD8 T cell exhaustion by promoting IR expression during clone 13 infection. However, we did not find a significant reduction in IR expression when we measured the coexpression of PD-1, TIM-3, and T cell immunoglobulin and ITIM domain (TIGIT) on Trib1 cKO CD8 T cells at day 30 p.i. (Fig. 5, A and B). We next asked whether Trib1 influenced TEX cell subset differentiation. Recent work elucidated a critical role for TCF1 in this process. TCF1hi progenitors can be distinguished from their terminally differentiated progeny marked by TCF1lo expression and increased granzyme B (GzmB) expression (He et al., 2016; Im et al., 2016; Leong et al., 2016; Utzschneider et al., 2016; Wu et al., 2016). Trib1 cKO mice at day 30 p.i. had significantly more terminally differentiated TCF1loGzmBhi CD8 T cells by both frequency and absolute number compared with controls (Fig. 5, C and D), suggesting an important role for Trib1 in regulating TEX subsets. Further, while the relative frequency of the TCF1hi population was reduced in Trib1 cKO mice, the absolute number of these cells was similar to controls. Together, these data indicate that Trib1 loss promotes terminal cell differentiation and persistence while also retaining the TCF1hi progenitor TEX subset.
Figure 5.
Trib1 regulates TEX cell subsets in chronic infection. (A and B) Gating strategy (A) and summary of the frequency of coexpression of the IRs PD-1, TIM-3, and TIGIT on splenic CD8 T cells (gated on CD8+CD44+) at day 30 p.i. with clone 13 (B). (C and D) Representative flow plots (C) and summary of the frequency (top) or absolute number (bottom) of TCF1/GzmBhi/lo splenic CD8 T cells (gated on CD8+CD44+) at day 30 p.i. (D). (E and F) Summary of T-bet expression measured by MFI in TCF1loGzmBhi (“terminal”; E) or TCF1hiGzmBlo (“progenitor”) splenic CD8 T cells at day 30 p.i. (F). Data in B–D are representative of two independent experiments each with 6–10 mice per genotype. Data in E and F represent an experiment with 9 or 10 mice per genotype. Error bars are mean ± SD. **, P < 0.01; ***, P < 0.001 by unpaired Student’s t test. Control: CD4-cre+ Trib1+/+; Trib1 cKO: CD4-cre+ Trib1F/F.
During chronic infection, there is also a loss of CD8+ T-bet expression, which contributes to CD8 T cell dysfunction (Kao et al., 2011; Paley et al., 2012). We examined T-bet expression in both terminal and progenitor TEX subsets and found that Trib1 deficiency enhanced T-bet expression in both populations at day 30 p.i. (Fig. 5, E and F). These data demonstrate that Trib1 restrains T-bet expression broadly across CD8 TEX cell subsets.
To determine if the differences in TEX subsets between Trib1 cKO and controls are established earlier in chronic infection, we looked at the frequency of these populations at day 12 p.i. Consistent with day 30, Trib1 cKO mice also had an elevated frequency of terminally differentiated CD8 T cells at day 12 p.i. as marked by an increase in the percentage of GzmBhiLy108lo expressing CD8 T cells (Ly108 used here as a surrogate for TCF1 expression; Fig. S2, B and C; Utzschneider et al., 2016). Similarly, T-bet expression was significantly increased on both terminal and progenitor CD8 TEX populations from Trib1 cKO mice at day 12 p.i. (Fig. S2, D and E). These data show that Trib1 loss promoted a terminal effector-like phenotype in CD8 TEX subsets early p.i. when viral loads were not significantly different between control and Trib1 cKO mice, indicating a direct role for Trib1 in regulating these CD8 T cell subsets.
scRNA-seq analysis reveals a KLRG1+ effector population that is transcriptionally distinct from TEX subsets and is enriched in the absence of Trib1
To understand more deeply the nature of the effector-like CD8 population that is expanded in Trib1 cKO mice and to gain a broader perspective on how Trib1 regulates the landscape of T cells responding to a chronic infection, we performed an unbiased single-cell transcriptome analysis of control and Trib1 cKO T cells following clone 13 infection. We sorted bulk activated (CD44+) T cells (TCRβ+) from the spleens of control or Trib1 cKO mice at day 15 post-clone 13 infection and measured the transcriptome of individual cells by scRNA-seq (10x Genomics).
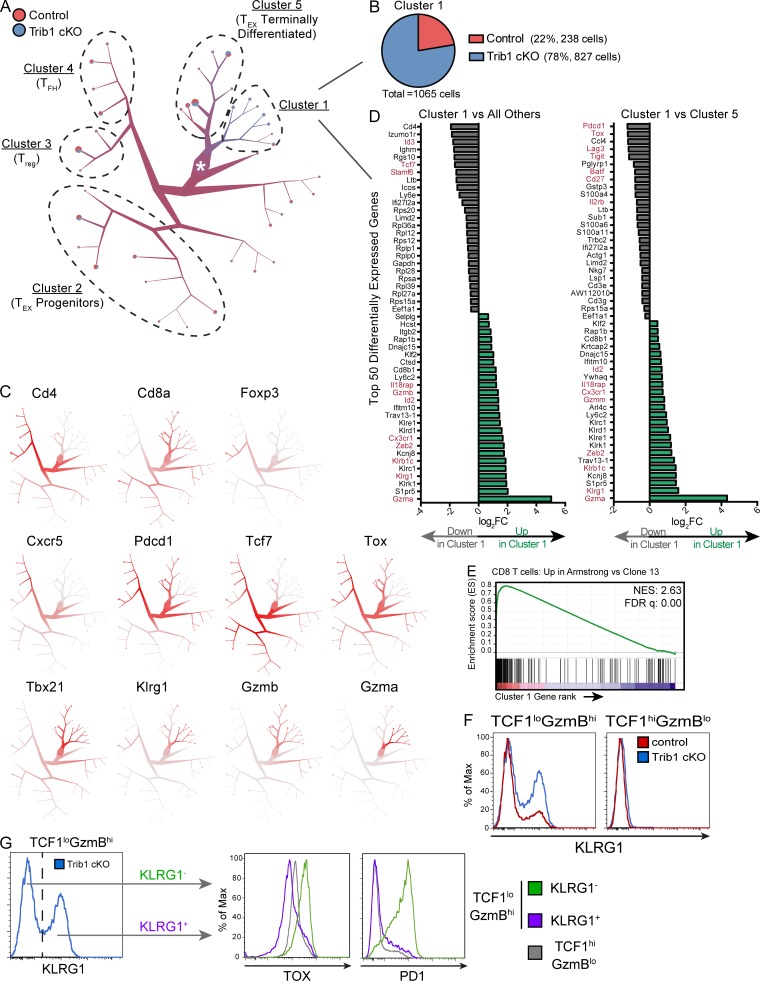
We used TooManyCells, a scRNA-seq hierarchal spectral clustering and visualization tool (Schwartz et al., 2020), to stratify transcriptionally distinct subpopulations of control and Trib1 cKO cells. We observed a distinct cluster (“Cluster 1,” Fig. 6 A) with a predominance of Trib1 cKO cells (Fig. 6 B). To further identify the identity of cells separated in each cluster, we queried the expression of a number of key T cell transcriptional markers (Fig. 6 C). Cd4 and either Foxp3 or Cxcr5 expression patterns suggested that CD4 T reg cells and CD4 T follicular helper cells mostly segregated to “Cluster 3” and “Cluster 4,” respectively. Based on the expression of genes including Cd8, Cxcr5, Pdcd1, Tcf7, Tox, Tbx21, and Gzmb, TEX progenitors appeared to group to “Cluster 2,” whereas terminally differentiated TEX cells predominantly segregated to “Cluster 5.” Cluster 1 distinctly consisted of a preponderance of cells expressing high levels of Klrg1, in line with the elevated abundance of KLRG1+ cells observed in Trib1 cKO mice by flow cytometry (Fig. 3 and Fig. 4). Projection of cells onto uniform manifold approximation and projection (UMAP) scatter plot confirmed these results (Fig. S4 A; Becht et al., 2018).
Figure 6.
scRNA-seq analysis reveals a KLRG1+ effector population that is transcriptionally distinct from TEX subsets and is enriched in the absence of Trib1. Activated T cells (TCRβ+CD44+) were isolated by FACS from spleens of either control or Trib1 cKO mice at day 15 after clone 13 infection and prepared for scRNA-seq analyses. (A) Visualization of single-cell clustering using TooManyCells from 7,611 TCRβ+CD44+ cells. All cells begin at the central node (white star) and are recursively bipartitioned based on transcriptional similarity. To this end, TooManyCells produces a hierarchy of nested cell clusters where each inner node is a cluster at a given resolution and a leaf node is a finer-grain cluster where any additional bipartitioning would be as good as randomly bipartitioning the cells. Branches connect similar groups of cells and their width and color correspond to the number and mixed annotation of cells within the connected clusters, respectively. Terminal branches contain pie charts delineating the fraction of cells from either control or Trib1 cKO in each terminal branch. Clusters of interest and clusters with predominance of known T cell populations based on expression of known markers are denoted. (B) Number and composition of cells in Cluster 1. (C) Normalized gene expression per cell (red = higher expression, gray = lower expression). (D) Top 50 differentially expressed genes in Cluster 1 vs. all other cells (left) or Cluster 1 vs. Cluster 5 (right). Red genes denote genes associated with effector, terminal TEX, or progenitor TEX cells. FC, fold-change. (E) GSEA of the transcriptional signature associated with CD8 T cells from Armstrong vs. cl13 infection (GSE30962) compared with the differentially expressed genes in Cluster 1 vs. all other cells. NES, normalized enrichment score; FDR, false discovery rate. (F) Representative histograms of KLRG1 expression in TCF1loGzmBhi and TCF1hiGzmBlo CD8 T cells (gated on CD8+CD44+ and applicable TCF1/GzmB gating as defined in Fig. 5) from the spleens of control or Trib1 cKO mice at day 30 after clone 13 infection measured by flow cytometry. (G) Gating strategy (left) of KLRG1+ and KLRG1− cells from TCF1loGzmBhi CD8 T cells in Trib1 cKO mice at day 30 p.i. (same cells as presented in panel F, left, blue) for analysis of TOX and PD1 expression within each of these subsets (right). Expression of TOX and PD1 on TCF1hiGzmBlo cells from the same Trib1 cKO mouse are shown in gray for comparison. scRNA-seq data are representative of two biologically independent pooled samples per genotype. Data in F and G are representative of an experiment with six to eight mice per genotype. Control: CD4-cre+Trib1+/+; Trib1 cKO: CD4-cre+Trib1F/F.
Figure S4.
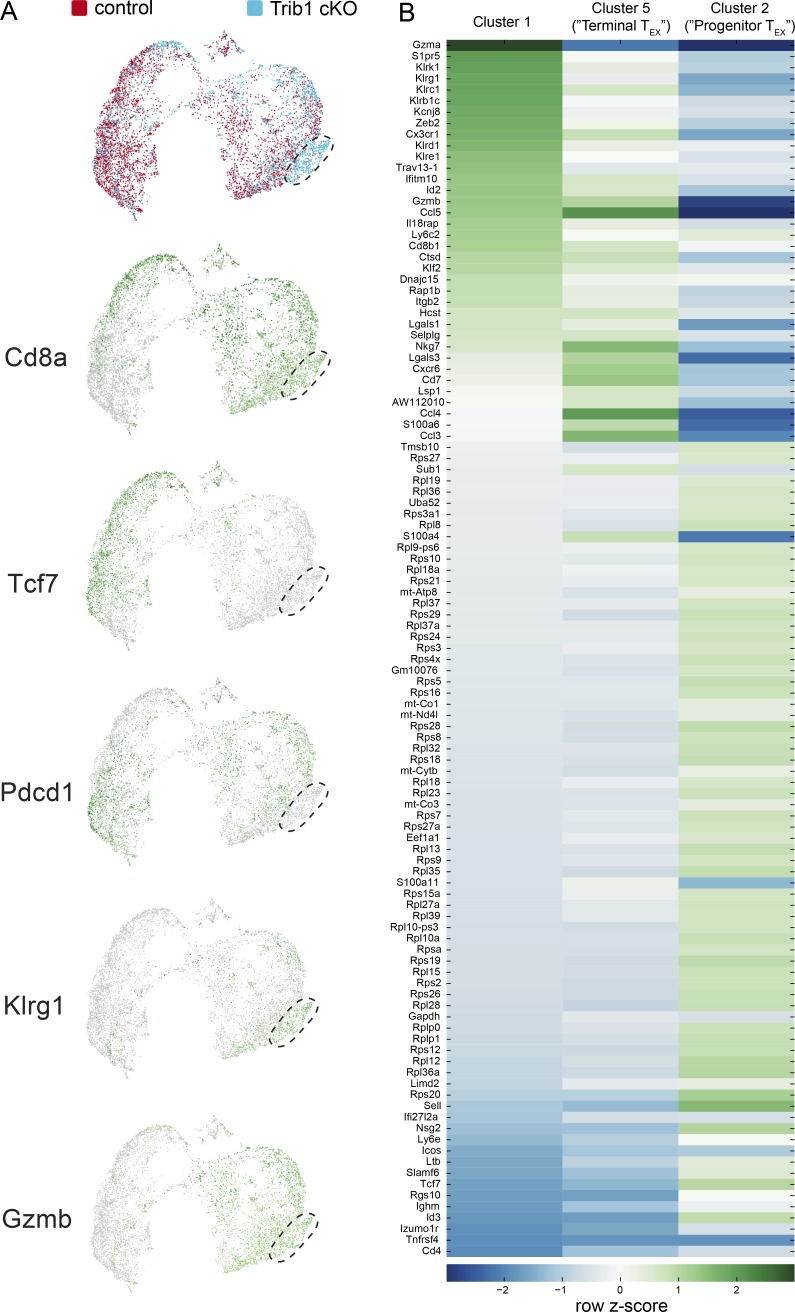
Extended characterization of scRNA-seq data from activated T cells in control and Trib1 cKO mice infected with clone 13. (A) UMAP of control and Trib1 cKO scRNA-seq data from experiment in Fig. 6. Cluster of cells enriched for Trib1 cKO cells denoted. Each dot represents one cell. Top projection displays control (red) vs. Trib1 cKO (blue) cells. Lower projections display gene expression across cells (green = higher expression, gray = lower expression). (B) Heat map of the top 50 most differentially expressed genes (by P value) in Clusters 1, 2, and 5 from Fig. 6. Heat map was generated using row-normalized z-scores derived from log2 differential expression values for each gene in each indicated cluster compared with all other clusters.
Visualization of inter-cluster relationships showed transcriptional proximity between Klrg1-expressing (Cluster 1) and terminally differentiated TEX cells (Cluster 5), suggesting some overlap in their transcriptional programs, albeit with some critical differences (Fig. 6 D and Fig. S4 B). Similar to terminally differentiated TEX cells, Cluster 1 cells expressed relatively low levels of Tcf7, Cxcr5, and Slamf6, as well as relatively high levels of Gzmb and Tbx21. However, cells in Cluster 1 had lower expression of Tox. Also, notably different from Cluster 5 and consistent with reduced expression of Tox, Cluster 1 had significantly reduced expression of IRs including Pdcd1, Lag3, and Tigit, as well as reduced expression of Cd27. In addition to reduced expression of exhaustion-associated genes, cells in Cluster 1 had elevated expression of genes associated with effector T cell and cytotoxic programming including Gzma, Klrb1c, IL18rap, and Cx3cr1. Further, Gene Set Enrichment Analysis (GSEA) showed that the gene signature of cells in Cluster 1 (compared with all other clusters) was significantly enriched for the signature of CD8 T cells generated during an Armstrong LCMV infection compared with clone 13 (Fig. 6 E). Combined, these data show that Trib1 restrains effector cell programming during chronic infection, and reveal that loss of Trib1 restores a population of KLRG1+TOXlo CD8 T cells characterized by an effector T cell program distinct from that of TEX cell subsets.
Using scRNA-seq, we demonstrated that Trib1 loss enriches a population of KLRG1+ cells that are transcriptionally distinct from TEX cells, but more similar to the terminal TEX subset as compared with TEX progenitors. We validated some of these findings by flow cytometry and found an expanded KLRG1+ population exclusively in the TCF1loGzmBhi subset of CD8 T cells from clone 13–infected Trib1 cKO mice (Fig. 6 F). In line with the scRNA-seq data and the TOX literature, the KLRG1+ subset expressed considerably less TOX than both KLRG1− terminal TEX cells and progenitor TEX cells, as well as significantly less PD-1 than KLRG1− terminal TEX cells (comparable expression to TEX progenitors; Fig. 6 G). These data provide further evidence that the KRLG1+ subset expanded and sustained during chronic infection in Trib1 cKO mice is distinct from TEX cells.
Trib1 interacts with MALT1 to disrupt TCR signaling complexes and limit T cell activation
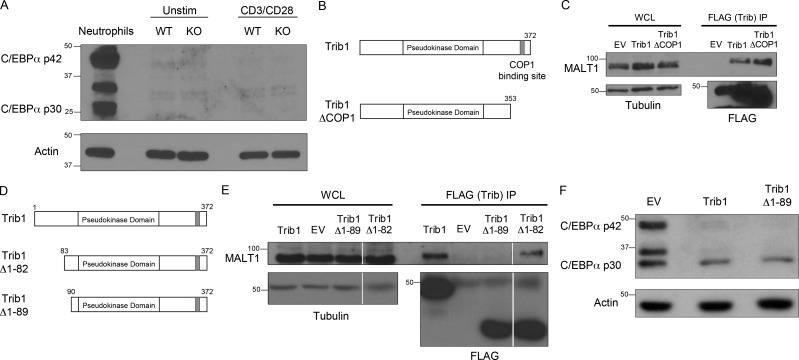
Given the role of Trib1 in regulating T cell responses and T cell activation, we sought to identify the molecular mechanisms by which Trib1 regulates T cell function. In multiple biological systems, including myeloid development and leukemia, Trib1 promotes degradation of the transcription factor C/EBPα via interaction with the E3 ubiquitin ligase COP1 (Dedhia et al., 2010; Satoh et al., 2013). While C/EBPα is predominantly associated with myeloid development (Ma et al., 2014; Zhang et al., 1997, 2004), it has also been linked to IFN-γ expression in T helper cells (Tanaka et al., 2014) as well as age-dependent CD4 T cell senescence (Shimatani et al., 2009). However, C/EBPα was not enriched in Trib1-deficient T cells with or without TCR stimulation ex vivo (Fig. S5 A).
Figure S5.
Trib1 interaction with MALT1 requires the Trib1 N terminus between amino acids 82 and 89. (A) Western blot of lysates from sorted WT or Trib1 cKO CD4 T cells ± stimulation with 1 µg/ml anti-CD3/CD28 for 24 h. Neutrophil whole-cell lysates (WCL) were used as a positive control for C/EBPα expression. (B) Schematic of full-length Trib1 (top) and Trib1-ΔCOP1 (bottom). (C) HEK293T cells were transfected with FLAG-tagged empty-vector control, full-length Trib1, or Trib1-ΔCOP1. FLAG-tagged proteins were immunoprecipitated using anti-FLAG agarose beads, and MALT1 binding was assessed by Western blot. (D) Schematic of full-length Trib1 (top), Trib1-Δ1-82 (middle), and Trib1-Δ1-89 (bottom). (E) HEK293T cells were transfected with FLAG-tagged empty-vector control, full-length Trib1, Trib1-Δ1-82, or Trib1-Δ1-89. FLAG-tagged proteins were immunoprecipitated using anti-FLAG agarose beads, and MALT1 binding was assessed by Western blot. Each row is from the same blot and exposure (white lines: cut unrelated lanes). (F) 32D cells (myeloid cell line expressing C/EBPα) were retrovirally transduced with MigR1 empty vector control, full-length Trib1, or Trib1-Δ1-89. Transduced cells were sorted by GFP, and C/EBPα degradation was assessed by Western blot. WT: CD4-cre+Trib1+/+; KO: CD4-cre+Trib1F/F.
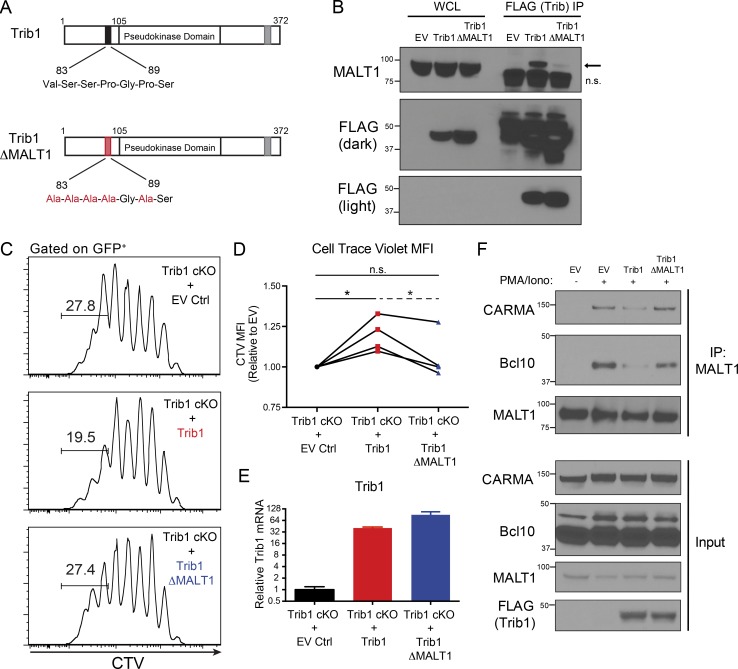
To identify Trib1 interacting proteins that might provide insight into Trib1-regulated signaling pathways, we performed a proteomic screen and identified the TCR signaling molecule MALT1 as a potentially relevant interaction partner (Uljon et al., 2016). We also detected this interaction by pull-down of FLAG-tagged Trib1 followed by immunoblot analysis for recovery of MALT1 in HEK293 cells and used a combination of deletion mapping and mutational analysis to show that the interaction required the N terminus of Trib1. This analysis informed the construction of a MALT1-defective binding mutant (Trib1-ΔMALT1) in which five residues between amino acid residues 83 and 89 were mutated to alanines (Fig. 7, A and B; and Fig. S5, B–E). While disrupting the N terminus abrogated binding to MALT1, deletion of N-terminal residues 1–89 from Trib1 did not disrupt the ability of Trib1 to promote C/EBPα degradation in 32D cells (Fig. S5 F), supporting the conclusion that the structural integrity of the kinase and C-terminal domains remained intact in our Trib1 N-terminal mutants (Murphy et al., 2015; Uljon et al., 2016).
Figure 7.
Trib1 interacts with MALT1 to disrupt MALT1 signaling complexes and suppress T cell signaling and function. (A) Schematic of full-length Trib1 (top) and Trib1-ΔMALT1 (bottom) with mutated MALT1-binding site highlighted in red. (B) HEK293T cells were transfected with FLAG-tagged empty-vector control MigR1, full-length Trib1, or Trib1-ΔMALT1. FLAG-tagged proteins were immunoprecipitated using anti-FLAG agarose beads, and MALT1 binding was assessed by Western blot (arrow: MALT1, n.s., nonspecific band). (C) Representative plots of CTV dilution measured by flow cytometry from purified Trib1 cKO naive CD8+ T cells stained with CTV, stimulated with anti-CD3 and congenic irradiated APCs (splenocytes), transduced with empty-vector control MigR1, full-length Trib1, or Trib1-ΔMALT1, and cultured for 72 h after transduction (gated on GFP+-transduced CD8 T cells). Gate indicates frequency of cells that have undergone more than five divisions. (D) Summary of CTV MFI (of GFP+ CD8 T cells) from four independent experiments (normalized to empty vector, EV) of which a representative experiment is shown in C. Solid brackets represent one-sample t tests to test the null hypothesis that the normalized CTV MFI of the group mean (Trib1 or Trib1-ΔMALT1) is 1 (i.e., that the CTV MFI of the group and of the EV control are not significantly different). Dashed bracket indicates a paired t test using differences to control for the day-effect to test whether the difference between the Trib1 and the Trib1-ΔMALT1 group means are significantly different from 0. (E) Representative plot of Trib1 expression at 72 h after transduction measured by qPCR from cultures in C. (F) Jurkat-E T cells were retrovirally transduced with FLAG-tagged empty vector control MigR1, full-length Trib1, or Trib1-ΔMALT1 and sorted by GFP. Sorted GFP+ cells were expanded in culture and stimulated for 30 min ± PMA and ionomycin. MALT1 was immunoprecipitated, and Bcl10 and CARMA1 coimmunoprecipitation was assessed by Western blot. Western blots in B and F are representative of more than three independent experiments. Data in C and E are representative of four independent experiments, summarized in D. CT values normalized to 18s ribosomal RNA. Error bars are mean ± SD. *, P < 0.05. Control: CD4-cre+Trib1+/+; Trib1 cKO: CD4-cre+Trib1F/F.
To test the importance of the Trib1:MALT1 interaction in T cell activation, we measured the proliferative capacity of Trib1-deficient T cells with enforced expression of either WT Trib1 or Trib1-ΔMALT1. Purified naive CD8 T cells from Trib1 cKO mice were stained with CellTrace Violet (CTV) dye, stimulated with anti-CD3 and irradiated splenocytes (source of APCs), and transduced with Trib1, Trib1-ΔMALT1, or vector control. The proliferation of transduced T cells (GFP+) was measured by flow cytometry analysis of CTV dilution. Cells expressing Trib1 showed reduced proliferation (less CTV dilution/higher CTV mean fluorescent intensity [MFI]) relative to cells expressing the vector control, whereas cells expressing Trib1-ΔMALT1 proliferated better than those expressing WT Trib1 and were similar to those expressing vector control (Fig. 7, C and D). Exogenous Trib1 expression was comparable between WT and Trib1-ΔMALT1 constructs (Fig. 7 E). These results suggest that Trib1 suppresses T cell activation at least in part through its interaction with MALT1.
Following TCR activation, MALT1 is recruited to CARMA1 via Bcl10 to form the CARMA1/Bcl10/MALT1 (CBM) complex. Formation of this complex facilitates downstream signaling events, including NF-κB and JNK activation, required for optimal T cell activation and function (Meininger and Krappmann, 2016; Paul and Schaefer, 2013; Thome et al., 2010). To determine the effect of Trib1 on MALT1 signaling, we measured CBM complex formation by immunoblot following immunoprecipitation (IP) of MALT1 in stimulated Jurkat-E T cells transduced with Trib1, Trib1-ΔMALT1, or vector control. Trib1 expression inhibited CBM complex formation (measured by Bcl10 and CARMA1 coimmunoprecipitation), whereas expression of Trib1-ΔMALT1 did not (Fig. 7 F). These data identify Trib1 as a negative regulator of TCR-induced CBM complex formation and provide insight into how Trib1 regulates T cell function.
Discussion
There are two predominant factors that contribute to the magnitude and duration of a T cell response to chronic antigen: the robustness of the effector T cell response and the progression toward exhaustion as marked by IR signaling and waning T cell function. Significant work in the field has characterized the cellular, genetic, and epigenetic processes associated with T cell exhaustion (Crawford et al., 2014; McLane et al., 2019; Pauken et al., 2016; Wherry et al., 2007; Wherry and Kurachi, 2015) and identified multiple promising therapeutic targets; however, very little is understood about effector response regulation as exhaustion develops. In this study, we delineate a novel regulator of effector differentiation during the response to chronic infection, providing insight into alternative differentiation programs that can be targeted to bolster T cell immunity.
We identify Trib1 as a central regulator of the antiviral T cell response whereby Trib1 restrains an effector-like T cell phenotype within the exhausted setting of persistent infection. Specifically, we observed an increase in the expansion and persistence of the short-lived effector-like KLRG1+ T cell population in Trib1 cKO mice infected with clone 13. In an acute infection, KLRG1 expression marks a population of SLECs that expand to clear infection. However, the role of KLRG1+ cells in a chronic infection is less clear. A population of KLRG1+ CD8 T cells can be found early in chronic infection and rapidly decreases as the infection progresses, while KLRG1lo CD8 T cells persist and give rise to the TEX pool (Angelosanto et al., 2012). The effect of maintaining a pool of KLRG1+ cells later into chronic infection and throughout the progression of exhaustion, as we observe in the Trib1 cKO mice, has not been well-characterized, but our data suggest that sustaining this population could improve viral control. The characteristically short half-life of these cells in WT mice could suggest that Trib1 deficiency increases effector cell half-life. Alternatively, Trib1 loss might sustain a precursor population that can generate and sustain KLRG1+ effector cells, as has been found in other models of infection (Chu et al., 2016).
Despite elevated CD8 T cell function, we did not observe an overall net decrease in IR expression on CD8 T cells in Trib1 cKO mice. Recent studies have defined a terminally differentiated exhausted CD8 T cell population in chronic infection marked by loss of TCF1 expression. We observed an expansion of TCF1loGzmBhi CD8 T cells early in chronic infection in Trib1 cKO mice, and this phenotype persisted until at least 30 d p.i. By measuring the transcriptome of individual cells, we found that the KLRG1+ subset expanded in Trib1 cKO mice was transcriptionally most similar to terminally differentiated TEX cells. However, this effector population was also notably distinct from both terminal and progenitor TEX cells, having significantly lower IR expression and importantly lower TOX expression, an essential driver of T cell exhaustion. Given the dramatic reduction in IR and particularly TOX expression, these data suggest an effector differentiation program regulated by Trib1 that is mutually exclusive to T cell exhaustion. This is supported by the enrichment of acute over chronic CD8 response genes in the Klrg1-expressing population, as well as by a recent report that characterized KLRG1+ cells as an independent fate of exhausted T cells following chronic infection (Chen et al., 2019).
We also observe a significant increase in T-bet expression in both the terminal (TCF1lo) and progenitor (TCF1hi) TEX subsets from Trib1 cKO mice. T-bet is an important regulator of effector cell programming, and loss of T-bet in chronic infection is associated with increased T cell dysfunction. As we observed a broad increase in T-bet expression across both CD8 TEX cell subsets, this likely represents a general shift toward increased “pro-effector” programming within the Trib1-deficient response. Broadly increased T-bet expression is also consistent with the accumulation of KLRG1+ cells observed in the Trib1 cKO mice, as T-bet directs the differentiation of this population in acute infection (Joshi et al., 2007). Interestingly, Trib1 did not restrict SLEC differentiation during acute infection, suggesting additional differences in the regulation of KLRG1+ populations between acute and chronic immune responses. Similar to TCF1, T-bet expression marks a progenitor pool of TEX cells with self-renewal capacity (Paley et al., 2012); however, T-bet is also expressed in both TCF1hi and TCFlo TEX subsets, with higher expression in the latter. The lack of alignment between the TCFhi progenitor pool and the T-bethi progenitor pool has been suggested as evidence of a heterogeneous TEX progenitor population (McLane et al., 2019). Our data suggest a similar heterogeneity in TEX cell subsets whereby Trib1 deficiency enforces an effector-like phenotype while simultaneously maintaining TCF1hi progenitor cell pools with broadly enriched T-bet expression.
Here, we use the Trib1 cKO mouse model as a lens by which to understand how effector cells fit into the exhausted T cell landscape. This work provides important insights into the regulation of effector versus exhausted differentiation, and the relationship between these two programs in chronic infection. Trib1 deficiency expands and sustains a KLRG1+ effector population, while also maintaining progenitor and terminal TEX cell subsets. This is in contrast to TOX-deficient T cells that also significantly expand a KLRG1+ effector-like population, but have a dramatic defect in generating TEX cells and thus cannot sustain an antiviral response to chronic antigen. Whether Trib1 regulates TOX expression (either directly or indirectly) is unknown, but it is possible that loss of Trib1 might partially limit or suppress TOX expression within specific T cell subsets. A partial loss of TOX function might account for the expanded effector-like population, sustained T cell response, and improved viral control seen in Trib1 cKO mice, and is supported by observations that T cells partially deficient for TOX have improved tumor control (Khan et al., 2019). However, the differences between the Trib1 and TOX phenotypes during chronic infection may also suggest that Trib1 functions independently of TOX regulation. Our data also show that Trib1 functions to disrupt CBM signaling complexes downstream of TCR activation; however, the downstream targets of CBM or the effects of engaging this pathway long-term have not been characterized in the setting of T cell exhaustion. Trib1 may also regulate T cell differentiation in vivo via additional, including CBM-independent, mechanisms. While additional molecular mechanisms underlying our findings may still be identified, our data show that Trib1 can be targeted to direct effector programming and improve effector T cell function, while retaining TEX cell populations and sustaining the immune response to chronic antigen.
In addition to CD8 T cells, the effector functions of CD4 T cells are also critical for controlling viral burden during chronic infection. Here we demonstrate that deleting Trib1 promotes both the CD4 and CD8 arms of the T cell response. IL-2 produced by CD4 T cells promotes CD8 effector differentiation and function, and increased IL-2 signaling enhances CD8 effector responses (Kalia et al., 2010; Pipkin et al., 2010) and synergizes with PD-1 blockade to control viral titers in chronically infected mice (West et al., 2013). Our data indicate that manipulating Trib1 signaling can boost CD4 responses during infection and also suggest that inhibiting Trib1 in conjunction with ICB may result in more durable T cell responses and improve clinical outcomes. Our data are also consistent with a recent report demonstrating that CD4 T cell function is essential for the formation of a cytolytic effector CD8 T cell population, characterized by expression of CX3CR1, KLRG1, T-bet, and Zeb-2, that is protective during chronic disease (Zander et al., 2019).
In summary, our work suggests a model whereby Trib1 restrains effector programming and differentiation in CD8 T cells within an exhausted setting, and this regulatory axis can be targeted to enhance and sustain effector-like cell frequency and T cell function. Importantly, these data uncover new insights into effector versus exhausted differentiation during chronic infection. We also show that Trib1 expression is elevated in human T cells from chronically infected patients, further suggesting that Trib1 or Trib1-regulated pathways may be viable targets in T cell–based immune therapies in human chronic disease and an alternate strategy to targeting IR signaling and TEX subsets. Collectively, we demonstrate the value in targeting T cell effector differentiation during persistent infection and identify new regulatory mechanisms underlying antiviral immunity.
Materials and methods
Mice
Conditional Trib1 mice (C57BL/6-Trib1tm1.1 mrl, Taconic no. 10265; Bauer et al., 2015) were crossed to Cd4-Cre transgenic mice (JAX) to generate Trib1 cKO mice. Control mice in all experiments were age-matched Cd4-Cre+ with the WT Trib1 allele. Homozygous control and Trib1 cKO mice were routinely regenerated from heterozygotes to prevent genetic drift between genotypes. All mice were on the C57BL/6 background and were analyzed between 5 and 12 wk of age. Control and Trib1 cKO mice were sex-matched, and both female and male mice were used in experiments. Congenic C57BL/6 CD45.1/SJL mice (for APCs in ex vivo cultures) were purchased from Charles River Laboratories. Animals were housed in a specific pathogen–free facility at the University of Pennsylvania. Experiments were performed according to the guidelines from the National Institutes of Health with approved protocols from the University of Pennsylvania Animal Care and Use Committee.
Cell lines
Jurkat, HEK293T, and 32D cell lines were obtained from the American Tissue Culture Collection. Jurkat-E cells (Wolkowicz et al., 2005) were provided by Garry Nolan (Stanford University, Stanford, CA). Cell lines were regularly tested for mycoplasma.
Constructs
Trib1 cDNA (N terminus FLAG tag) was cloned into the MigR1 plasmid (GFP+) to generate the MigR1-Trib1 construct. Trib1-ΔMALT1, Trib1-Δ1-82, Trib1-Δ1-89, and Trib1-ΔCOP1 constructs were generated by mutating MigR1-Trib1 with PCR site-directed mutagenesis using the QuikChange II kit (Agilent) per the manufacturer’s instructions.
Cell culture
Primary mouse T cells were cultured in T cell media (TCM) that consisted of IMDM supplemented with 10% FBS (HyClone), 50 µM 2-ME (Sigma-Aldrich), 1% L-glutamine (Gibco), and 1% penicillin/streptomycin (Gibco). Jurkat and Jurkat-E cells were cultured in RPMI 1640 supplemented with 10% FBS (HyClone), 50 µM 2-ME, 1% L-glutamine (Gibco), and 1% penicillin/streptomycin (Gibco). HEK293T were cultured in DMEM supplemented with 10% FBS, 1% L-glutamine (Gibco), and 1% penicillin/streptomycin (Gibco). 32D cells were cultured in IMDM supplemented with 10% FBS (HyClone), 10% WEHI-conditioned media (Lee et al., 1982), 1% L-glutamine (Gibco), and 1% penicillin/streptomycin (Gibco). All cells were cultured at 37°C and 5% CO2.
Quantitative PCR (qPCR) from T cell cultures
Trib1 expression was measured by qPCR as we could not identify a Trib1 antibody that is both specific and capable of detecting endogenous mouse Trib1 protein. Murine Trib1 (mTrib1) expression was measured in purified CD4 T cells (purified by magnetic-activated cell sorting [MACs]) or in naive CD4 T cells FACS sorted by CD4+CD44−CD62L+CD25− and stimulated for 4 h with platebound anti-CD3 and anti-CD28 (2 µg/ml each). Murine Il-2 (mIL-2) expression was measured from naive CD4 T cells (sorted or purified using the EasySep Mouse naive CD4 T cell Isolation Kit, STEMCELL) cultured for 24 h ± platebound anti-CD3 and anti-CD28 (0.5 µg/ml each). Mouse T cells were isolated from spleen and lymph nodes. Human TRIB1 (hTRIB1) expression was measured in Jurkat T cells ± 4 h stimulation with human T-Activator CD3/CD28 Dynabeads (per the manufacturer’s instructions). For both mouse and human T cells, RNA was extracted using TRIzol (Ambion), followed by cDNA synthesis (SuperScript III kit; Invitrogen). qPCR was performed using TaqMan PCR master mix (Applied Biosystems) on a ViiA 7 system (ABI), and mRNA quantities were normalized to 18s. Primer/probe sets were 18s (4318839), mTrib1 (Mm00454875_m1), hTRIB1 (Hs00179769_m1), and mIL-2 (Mm00434256_m1) that were purchased from Life Technologies.
Flow cytometry and cell sorting
Surface staining was performed in PBS containing 2% FBS using antibodies against TCRβ (H57-597, eBioscience), CD3 (17A2, BD), CD8 (53–6.7, eBioscience), CD4 (RM4-5, BioLegend), CD44 (IM7, BioLegend and BD), CD62L (MEL-14, BD), CD25 (PC61.5, eBioscience), CD69 (H1.2F3, BD), KLRG1 (2F1, eBioscience), CD127 (A7R34, BioLegend and eBioscience), PD-1 (RMP1-30, eBioscience and BioLegend), TIM-3 (RMT3-23, BioLegend), TIGIT (1G9, BD), Ly108 (13G3, BD), and CD45.2 (104, BioLegend). Biotinylated monomers specific for H2-Db–restricted gp33–41 of LCMV were obtained from the National Institutes of Health Tetramer Core Facility and tetramerized using their published protocol. Zombie Violet (BioLegend), Live/Dead Aqua (Invitrogen), Propidium Iodide (eBioscience), or DAPI (Sigma-Aldrich) was used for live/dead discrimination. For all analysis and sorting, doublets were excluded. Intracellular cytokine staining was performed following 5 h ex vivo stimulation with either PMA (20 ng/ml) and ionomycin (200 ng/ml) or 0.4 µg/ml gp3333-41 peptide in the presence of brefeldin A (Sigma-Aldrich), monensin (BD), and anti-CD107a (1D4B, BD) using antibodies against IL-2 (JES6-5H4, BD), IFN-γ (XMG1.2, eBioscience), and TNF-α (MP6-XT22, BD) using the BD Cytofix/Cytoperm kit per the manufacturer’s instructions. Staining for Foxp3 (FJK-16s, eBioscience), CTLA4 (UC10-4F10-11, BD), T-bet (4B10, eBioscience), TCF1 (C63D9, Cell Signaling), TOX (REA473, Miltenyi), and GzmB (GB11, Invitrogen) was performed using the Foxp3/Transcription Factor Staining kit (eBioscience) per the manufacturer’s instructions. Cells were analyzed on an LSR II or LSR Fortessa flow cytometer (BD), and data were analyzed with FlowJo software v.9.7 (TreeStar). Cell sorting was performed on a FACSAria II (BD) using a 70-µm nozzle at 70 psi. Numbers on flow plots denote frequency of cells within the gate (as a percentage).
LCMV infection and calculation of viral loads
For acute infection, 6–8-wk-old mice were infected i.p. with 2 × 105 PFUs LCMV Armstrong virus. For all clone 13 studies, 5–6-wk-old mice were infected i.v. with 4 × 106 PFUs LCMV clone 13 virus. For calculation of viral titers in serum, serum was collected from peripheral blood, and 10-fold dilutions were incubated on adherent Vero cells for 1 h. Cells were overlaid with a 1:1 mixture of medium and 1% agarose and cultured for 4 d. PFUs were counted after overlaying with a 1:1:20 mixture of medium:1% agarose:neutral red for 16 h.
ELISA
Mouse CD4 T cells were CD4 MACs-purified or purified using the EasySep Mouse naive CD4 T cell Isolation Kit (STEMCELL Technologies) from spleen and lymph nodes and cultured in TCM ± platebound anti-CD3 and anti-CD28 (0.5 µg/ml each) in a 96-well U-bottom plate. Cell-free supernatants were assessed for IL-2 cytokine production by standard sandwich ELISA using purified rat anti-mouse IL-2 capture antibody (JES6-1A12, BD) and biotin rat anti-mouse IL-2 detection antibody (JES6-5H4, BD; 2 µg/ml of each).
T cell proliferation assays
Naive CD4 T cells (FACS sorted by CD4+CD44−CD62L+CD25−) or naive CD8 T cells (purified using the EasySep Mouse CD8+ T cell Isolation Kit, STEMCELL Technologies) were isolated from spleens and lymph nodes of mice, stained with 5 µM CellTrace CFSE (Invitrogen) or CTV (Invitrogen) per the manufacturer’s instructions, and cultured for 72 h in TCM with 1 µg/ml soluble anti-CD3 and irradiated (2,500 rad) congenic splenocytes (2 × 104 T cells and 2 × 105 irradiated splenocytes per well) in a 96-well U-bottom plate.
Retroviral (RV) transduction experiments
High titer RV supernatants were produced in HEK293T cells as described (Stein et al., 2016). For RV transduction of 32D cells, cells were transduced with RV at a concentration of 2 million cells/ml in a 6-well dish in the presence of 4 µg/ml polybrene during spin infection at 2,000 g for 90 min at 30°C. For RV transduction of Jurkat-E T cells, cells were transduced with RV at a concentration of 2 million cells/ml in a 6-well dish in the presence of 4 µg/ml polybrene during spin infection at 2,000 g for 90 min at 30°C. 48 h after spinfection, the top 30% of GFP-expressing cells were FACS sorted, expanded in culture, and used for experiments within 2 wk of spinfection. For RV transduction of primary murine CD8 T cells (protocol adapted from Kurachi et al. [2017]), CD8 T cells were purified from spleens of mice using the EasySep Mouse CD8+ T cell Isolation Kit (STEMCELL Technologies), stained with CTV and stimulated in vitro for 24 h with 1 µg/ml soluble anti-CD3 and irradiated (2,500 rad) congenic splenocytes (5 × 105 T cells and 5 × 106 irradiated splenocytes per well) in a 6-well dish. Activated CD8 T cell cultures were counted and transduced with RV at a concentration of 1–3 million cells/ml in a 6-well dish in the presence of 4 µg/ml polybrene during spin infection at 2,000 g for 90 min at 30°C. Following spinfection, cells were cultured at 37°C for 4 h and then resuspended in a 96-well U-bottom plate at 1 million cells/ml in the enriched TCM from the pretransduction 24 h activation cultures.
Transfection
HEK293T cells were transfected at 60% confluence using the FuGENE HD Transfection reagent (Promega) per the manufacturer’s instructions.
Western blot and IP
Whole-cell lystates were prepared with radioimmunoprecipitation assay buffer (50 mM Tris, pH 8.0, 1 mM EDTA, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, and 0.1% SDS) supplemented with 10 mM NaF, 1 mM Na3VO4, and EDTA-free proteinase inhibitor cocktail cOmplete (Roche, no. 11836170001). Protein concentration was determined by Bradford Assay using the Bio-Rad protein assay dye reagent. For detection of C/EBPα in primary mouse cells, cells were directly lysed in 2× SDS sample buffer (10% SDS; BioRad) and boiled for 10 min. Proteins were separated using SDS-PAGE and wet-transferred to polyvinylidene difluoride membranes. Blots were visualized with SuperSignal west pico or femto chemiluminescence substrate (Thermo Fisher Scientific). We used the following antibodies: C/EBPα (8178, Cell Signaling), β-actin (A5316, Sigma-Aldrich), FLAG (M2, Sigma-Aldrich), MALT1 (B-12, Santa Cruz), β-tubulin (DM1A, Abcam), CARMA/CARD11 (1D12, Cell Signaling), and Bcl10 (C78F1, Cell Signaling). IP was performed with 0.5–1 mg of fresh protein lysate using either 25 µl of anti-FLAG agarose beads (M2, Sigma-Aldrich) or 2 µg anti-MALT1 (B-12, Santa Cruz) incubated overnight at 4°C with rotation. Following overnight rotation, MALT1 IP samples were then incubated with G Agarose beads (Invitrogen) for 4 h at 4°C with rotation. Beads were washed four times in IP Wash Buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, and 0.5% NP-40). Protein was eluted from beads by boiling at 95°C for 15 min in 2× SDS sample buffer.
scRNA-seq
Activated (CD44+) T cells (TCRβ+) were FACS-sorted from the spleens of control or Trib1 cKO mice at day 15 post-clone 13 infection into 1.5ml Lo-Bind Eppendorf tubes containing complete RPMI (10% FBS). Two biological replicates were pooled for each genotype to create one sample per genotype. 750,000 cells per genotype were sorted, collected, washed twice with RPMI, diluted in RPMI to 1,000 cells/μl, and loaded onto the Chromium single cell sorting system (10x Genomics) with a standard loading targeting 5,000 cells for recovery. Library construction was performed using the Chromium Single Cell 5′ Library & Gel Bead Kit according to the manufacturer’s protocol. The final pooled library (containing control and Trib1 cKO samples) was sequenced on a NextSeq 550 using paired-end sequencing on one high output FlowCell (Illumina).
scRNA-seq analysis
Raw sequencing files were aligned, filtered, and barcoded using Cell Ranger software (10x Genomics). Mitochondrial genes were removed, and cells with ≥250 UMI counts were included in the analysis. Data were analyzed using TooManyCells with default normalization of term frequency-inverse document frequency on raw gene expression counts (Schwartz et al., 2020). The tree projection was pruned by one median absolute deviation away from the median of all node sizes of the original tree for visualization purposes using the TooManyCells smart-cutoff argument. Gene overlays were generated using the TooManyCells draw-leaf argument and represent the relative gene expression across the tree normalized by upper-quartile scaling. Differential gene expression between cell clusters was obtained using the TooManyCells differential entry point and plotted in GraphPad Prism v.7. (Fig. 6 D) or in python using the pandas software library to arrange the data for the heatmap in Fig. S4 B, which was plotted using the seaborn package. UMAP projections in Fig. S4 A were generated by umap-learn python library.
GSEA
GSEA was performed on a preranked list of all genes included in the single-cell analysis (ranked by differential expression, log2 fold change, in Cluster 1 vs. all other clusters) using the Broad GSEA software and MSigDB database (Mootha et al., 2003; Subramanian et al., 2005).
Statistical analysis
Analysis in Fig. 1 D was performed using the GEO2R web tool on the Gene Expression Omnibus. Analyses in Fig. 7 D were performed using SAS. All other statistical analyses were performed using GraphPad Prism v.7. Statistical tests are described in the figure legends. Unless otherwise indicated in figure legend, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired Student’s t test.
Data availability
All sequencing data that support the findings of this study have been deposited in GEO under accession no. GSE143802.
Online supplemental material
Fig. S1 assesses the role of Trib1 in T cell development and acute infection. Fig. S2 presents the viral load in control and Trib1 cKO mice throughout infection and shows that characteristics of the Trib1 phenotype are present at earlier time points in chronic infection (before differences in viral load). Fig. S3 characterizes the gp33-specific response in control and Trib1 cKO mice during chronic infection. Fig. S4 provides further characterization of the scRNA-sequencing analysis from Fig. 6. Fig. S5 characterizes C/EBPα expression in control and Trib1 cKO T cells and describes the generation and validation of the Trib1-ΔMALT mutant.
Acknowledgments
We thank the University of Pennsylvania Flow Cytometry and Cell Sorting Core facility for assistance with cell sorting (National Institutes of Health P30CA016520), the University of Pennsylvania mouse husbandry core (University Laboratory Animal Resources), Daniel Rader (University of Pennsylvania, Philadelphia, PA) for the gift of the Trib1F/F mice, Garry Nolan for providing Jurkat-E cells, Shannon Carty, Kristen Pauken, and Ryan Staupe for advice on LCMV infection experiments, Zeyu Chen for advice on single-cell analyses of LCMV-infected T cells, Maximilian Wengyn, Jorge Gutierrez, Yeqiao Zhou, and Roshni Kailar for technical assistance, Will Bailis for technical advice and for reviewing the manuscript, Ivan Maillard and Josephine Giles for reviewing the manuscript, Scott Rome for assistance with scRNA-seq data analysis and presentation, and the entire Pear laboratory for thoughtful discussions. Our graphical abstract was created using the BioRender tool.
This study was supported by grants from National Institutes of Health grants R01AI047833 (M.S. Jordan and W.S. Pear), R35CA220340 (S.C. Blacklow), F31CA189661 (K.S. Rome), T32CA009140 (K.S. Rome, S.J. Stein, and G.W. Schwartz), T32HL743937 and F30HL136127 (E.A. Mack), K08CA166227 (S. Uljon), P30CA016520 (P.A. Gimotty), T32CA009615 (S.E. McClory), the National Science Foundation Graduate Research Fellowship DGE-1321851 and the Patel Family Scholars Award (K.S. Rome), the American Cancer Society PF-15-065-01-TBG (S.J. Stein), and grants from the Samuel Waxman Cancer Research Foundation (W.S. Pear and S.C. Blacklow) and Alex’s Lemonade Stand Foundation for Childhood Cancer (W.S. Pear).
Author contributions: K.S. Rome conceived, designed, and performed experiments, and wrote the manuscript; S.J. Stein conceived, designed, and performed experiments; M. Kurachi designed and performed experiments; J. Petrovic performed experiments; G.W. Schwartz performed data processing and analyses; E.A. Mack performed experiments; S. Uljon performed experiments; W.W. Wu performed experiments; A.G. DeHart performed experiments; S.E. McClory performed experiments; L. Xu performed experiments; P.A. Gimotty performed statistical analyses; S.C. Blacklow conceived and designed experiments, and wrote the manuscript; R.B. Faryabi conceived and designed experiments, and wrote the manuscript; E.J. Wherry conceived and designed experiments, and wrote the manuscript; M.S. Jordan conceived and designed experiments, and wrote the manuscript; and W.S. Pear conceived and designed experiments, and wrote the manuscript.
References
- Alfei F., Kanev K., Hofmann M., Wu M., Ghoneim H.E., Roelli P., Utzschneider D.T., von Hoesslin M., Cullen J.G., Fan Y., et al. 2019. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 571:265–269. 10.1038/s41586-019-1326-9 [DOI] [PubMed] [Google Scholar]
- Angelosanto J.M., Blackburn S.D., Crawford A., and Wherry E.J.. 2012. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J. Virol. 86:8161–8170. 10.1128/JVI.00889-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert R.D., Kamphorst A.O., Sarkar S., Vezys V., Ha S.J., Barber D.L., Ye L., Sharpe A.H., Freeman G.J., and Ahmed R.. 2011. Antigen-specific CD4 T-cell help rescues exhausted CD8 T cells during chronic viral infection. Proc. Natl. Acad. Sci. USA. 108:21182–21187. 10.1073/pnas.1118450109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer R.C., Sasaki M., Cohen D.M., Cui J., Smith M.A., Yenilmez B.O., Steger D.J., and Rader D.J.. 2015. Tribbles-1 regulates hepatic lipogenesis through posttranscriptional regulation of C/EBPα. J. Clin. Invest. 125:3809–3818. 10.1172/JCI77095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G., Ginhoux F., and Newell E.W.. 2018. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Blattman J.N., Grayson J.M., Wherry E.J., Kaech S.M., Smith K.A., and Ahmed R.. 2003. Therapeutic use of IL-2 to enhance antiviral T-cell responses in vivo. Nat. Med. 9:540–547. 10.1038/nm866 [DOI] [PubMed] [Google Scholar]
- Chen Z., Ji Z., Ngiow S.F., Manne S., Cai Z., Huang A.C., Johnson J., Staupe R.P., Bengsch B., Xu C., et al. 2019. TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity. 51:840–855.e5. 10.1016/j.immuni.2019.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.H., Chan S.W., Gosling J.P., Blanchard N., Tsitsiklis A., Lythe G., Shastri N., Molina-París C., and Robey E.A.. 2016. Continuous Effector CD8(+) T Cell Production in a Controlled Persistent Infection Is Sustained by a Proliferative Intermediate Population. Immunity. 45:159–171. 10.1016/j.immuni.2016.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier D.L., Zhou A.C., Wortzman M.E., Luft O., Levy G.A., and Watts T.H.. 2015. GITR intrinsically sustains early type 1 and late follicular helper CD4 T cell accumulation to control a chronic viral infection. PLoS Pathog. 11:e1004517 10.1371/journal.ppat.1004517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford A., Angelosanto J.M., Kao C., Doering T.A., Odorizzi P.M., Barnett B.E., and Wherry E.J.. 2014. Molecular and transcriptional basis of CD4+ T cell dysfunction during chronic infection. Immunity. 40:289–302. 10.1016/j.immuni.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedhia P.H., Keeshan K., Uljon S., Xu L., Vega M.E., Shestova O., Zaks-Zilberman M., Romany C., Blacklow S.C., and Pear W.S.. 2010. Differential ability of Tribbles family members to promote degradation of C/EBPalpha and induce acute myelogenous leukemia. Blood. 116:1321–1328. 10.1182/blood-2009-07-229450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K., Herzig S., Kulkarni R.N., and Montminy M.. 2003. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 300:1574–1577. 10.1126/science.1079817 [DOI] [PubMed] [Google Scholar]
- Gangaplara A., Martens C., Dahlstrom E., Metidji A., Gokhale A.S., Glass D.D., Lopez-Ocasio M., Baur R., Kanakabandi K., Porcella S.F., and Shevach E.M.. 2018. Type I interferon signaling attenuates regulatory T cell function in viral infection and in the tumor microenvironment. PLoS Pathog. 14:e1006985 10.1371/journal.ppat.1006985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Hou S., Liu C., Zhang A., Bai Q., Han M., Yang Y., Wei G., Shen T., Yang X., et al. 2016. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature. 537:412–428. 10.1038/nature19317 [DOI] [PubMed] [Google Scholar]
- Hegedus Z., Czibula A., and Kiss-Toth E.. 2007. Tribbles: a family of kinase-like proteins with potent signalling regulatory function. Cell. Signal. 19:238–250. 10.1016/j.cellsig.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Hill R., Madureira P.A., Ferreira B., Baptista I., Machado S., Colaço L., Dos Santos M., Liu N., Dopazo A., Ugurel S., et al. 2017. TRIB2 confers resistance to anti-cancer therapy by activating the serine/threonine protein kinase AKT. Nat. Commun. 8:14687 10.1038/ncomms14687 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hyrcza M.D., Kovacs C., Loutfy M., Halpenny R., Heisler L., Yang S., Wilkins O., Ostrowski M., and Der S.D.. 2007. Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells. J. Virol. 81:3477–3486. 10.1128/JVI.01552-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im S.J., Hashimoto M., Gerner M.Y., Lee J., Kissick H.T., Burger M.C., Shan Q., Hale J.S., Lee J., Nasti T.H., et al. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 537:417–421. 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., and Kaech S.M.. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia V., Sarkar S., Subramaniam S., Haining W.N., Smith K.A., and Ahmed R.. 2010. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 32:91–103. 10.1016/j.immuni.2009.11.010 [DOI] [PubMed] [Google Scholar]
- Kao C., Oestreich K.J., Paley M.A., Crawford A., Angelosanto J.M., Ali M.A., Intlekofer A.M., Boss J.M., Reiner S.L., Weinmann A.S., and Wherry E.J.. 2011. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat. Immunol. 12:663–671. 10.1038/ni.2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeshan K., He Y., Wouters B.J., Shestova O., Xu L., Sai H., Rodriguez C.G., Maillard I., Tobias J.W., Valk P., et al. 2006. Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell. 10:401–411. 10.1016/j.ccr.2006.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan O., Giles J.R., McDonald S., Manne S., Ngiow S.F., Patel K.P., Werner M.T., Huang A.C., Alexander K.A., Wu J.E., et al. 2019. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature. 571:211–218. 10.1038/s41586-019-1325-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Toth E., Bagstaff S.M., Sung H.Y., Jozsa V., Dempsey C., Caunt J.C., Oxley K.M., Wyllie D.H., Polgar T., Harte M., et al. 2004. Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J. Biol. Chem. 279:42703–42708. 10.1074/jbc.M407732200 [DOI] [PubMed] [Google Scholar]
- Kurachi M., Kurachi J., Chen Z., Johnson J., Khan O., Bengsch B., Stelekati E., Attanasio J., McLane L.M., Tomura M., et al. 2017. Optimized retroviral transduction of mouse T cells for in vivo assessment of gene function. Nat. Protoc. 12:1980–1998. 10.1038/nprot.2017.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.C., Hapel A.J., and Ihle J.N.. 1982. Constitutive production of a unique lymphokine (IL 3) by the WEHI-3 cell line. J. Immunol. 128(6):2393–2398. [PubMed] [Google Scholar]
- Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774. 10.1016/S1074-7613(01)00227-8 [DOI] [PubMed] [Google Scholar]
- Leong Y.A., Chen Y., Ong H.S., Wu D., Man K., Deleage C., Minnich M., Meckiff B.J., Wei Y., Hou Z., et al. 2016. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 17:1187–1196. 10.1038/ni.3543 [DOI] [PubMed] [Google Scholar]
- Liang K.L., O’Connor C., Veiga J.P., McCarthy T.V., and Keeshan K.. 2016. TRIB2 regulates normal and stress-induced thymocyte proliferation. Cell Discov. 2:15050 10.1038/celldisc.2015.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohan F., and Keeshan K.. 2013. The functionally diverse roles of tribbles. Biochem. Soc. Trans. 41:1096–1100. 10.1042/BST20130105 [DOI] [PubMed] [Google Scholar]
- Ma O., Hong S., Guo H., Ghiaur G., and Friedman A.D.. 2014. Granulopoiesis requires increased C/EBPα compared to monopoiesis, correlated with elevated Cebpa in immature G-CSF receptor versus M-CSF receptor expressing cells. PLoS One. 9:e95784 10.1371/journal.pone.0095784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack E.A., Stein S.J., Rome K.S., Xu L., Wertheim G.B., Melo R.C.N., and Pear W.S.. 2019. Trib1 regulates eosinophil lineage commitment and identity by restraining the neutrophil program. Blood. 133:2413–2426. 10.1182/blood.2018872218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matloubian M., Concepcion R.J., and Ahmed R.. 1994. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J. Virol. 68:8056–8063. 10.1128/JVI.68.12.8056-8063.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLane L.M., Abdel-Hakeem M.S., and Wherry E.J.. 2019. CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu. Rev. Immunol. 37:457–495. 10.1146/annurev-immunol-041015-055318 [DOI] [PubMed] [Google Scholar]
- Meininger I., and Krappmann D.. 2016. Lymphocyte signaling and activation by the CARMA1-BCL10-MALT1 signalosome. Biol. Chem. 397:1315–1333. 10.1515/hsz-2016-0216 [DOI] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Murphy J.M., Nakatani Y., Jamieson S.A., Dai W., Lucet I.S., and Mace P.D.. 2015. Molecular Mechanism of CCAAT-Enhancer Binding Protein Recruitment by the TRIB1 Pseudokinase. Structure. 23:2111–2121. 10.1016/j.str.2015.08.017 [DOI] [PubMed] [Google Scholar]
- Naiki T., Saijou E., Miyaoka Y., Sekine K., and Miyajima A.. 2007. TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPbeta. J. Biol. Chem. 282:24075–24082. 10.1074/jbc.M701409200 [DOI] [PubMed] [Google Scholar]
- Okamoto H., Latres E., Liu R., Thabet K., Murphy A., Valenzeula D., Yancopoulos G.D., Stitt T.N., Glass D.J., and Sleeman M.W.. 2007. Genetic deletion of Trb3, the mammalian Drosophila tribbles homolog, displays normal hepatic insulin signaling and glucose homeostasis. Diabetes. 56:1350–1356. 10.2337/db06-1448 [DOI] [PubMed] [Google Scholar]
- Paley M.A., Kroy D.C., Odorizzi P.M., Johnnidis J.B., Dolfi D.V., Barnett B.E., Bikoff E.K., Robertson E.J., Lauer G.M., Reiner S.L., and Wherry E.J.. 2012. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 338:1220–1225. 10.1126/science.1229620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.J., Park J.S., Jeong Y.H., Son J., Ban Y.H., Lee B.H., Chen L., Chang J., Chung D.H., Choi I., and Ha S.J.. 2015. PD-1 upregulated on regulatory T cells during chronic virus infection enhances the suppression of CD8+ T cell immune response via the interaction with PD-L1 expressed on CD8+ T cells. J. Immunol. 194:5801–5811. 10.4049/jimmunol.1401936 [DOI] [PubMed] [Google Scholar]
- Pauken K.E., and Wherry E.J.. 2015. Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 36:265–276. 10.1016/j.it.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauken K.E., Sammons M.A., Odorizzi P.M., Manne S., Godec J., Khan O., Drake A.M., Chen Z., Sen D.R., Kurachi M., et al. 2016. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 354:1160–1165. 10.1126/science.aaf2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., and Schaefer B.C.. 2013. A new look at T cell receptor signaling to nuclear factor-κB. Trends Immunol. 34:269–281. 10.1016/j.it.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster P., Kamphorst A.O., Wieland A., Araki K., Iyer S.S., West E.E., O’Mara L., Yang S., Konieczny B.T., Sharpe A.H., et al. 2014. Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. J. Exp. Med. 211:1905–1918. 10.1084/jem.20132577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin M.E., Sacks J.A., Cruz-Guilloty F., Lichtenheld M.G., Bevan M.J., and Rao A.. 2010. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 32:79–90. 10.1016/j.immuni.2009.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., Heredia J.E., Altarejos J.Y., Screaton R., Goebel N., Niessen S., Macleod I.X., Liew C.W., Kulkarni R.N., Bain J., et al. 2006. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science. 312:1763–1766. 10.1126/science.1123374 [DOI] [PubMed] [Google Scholar]
- Satoh T., Kidoya H., Naito H., Yamamoto M., Takemura N., Nakagawa K., Yoshioka Y., Morii E., Takakura N., Takeuchi O., and Akira S.. 2013. Critical role of Trib1 in differentiation of tissue-resident M2-like macrophages. Nature. 495:524–528. 10.1038/nature11930 [DOI] [PubMed] [Google Scholar]
- Schulze Zur Wiesch J., Ciuffreda D., Lewis-Ximenez L., Kasprowicz V., Nolan B.E., Streeck H., Aneja J., Reyor L.L., Allen T.M., Lohse A.W., et al. 2012. Broadly directed virus-specific CD4+ T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J. Exp. Med. 209:61–75. 10.1084/jem.20100388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz G.W., Zhou Y., Petrovic J., Fasolino M., Xu L., Shaffer S.M., Pear W.S., Vahedi G., and Faryabi R.B.. 2020. TooManyCells identifies and visualizes relationships of single-cell clades. Nat. Methods. 10.1038/s41592-020-0748-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A.C., Dündar F., Zumbo P., Chandran S.S., Klebanoff C.A., Shakiba M., Trivedi P., Menocal L., Appleby H., Camara S., et al. 2019. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 571:270–274. 10.1038/s41586-019-1324-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H., Chen J., González-Avalos E., Samaniego-Castruita D., Das A., Wang Y.H., López-Moyado I.F., Georges R.O., Zhang W., Onodera A., et al. 2019. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. USA. 116:12410–12415. 10.1073/pnas.1905675116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimatani K., Nakashima Y., Hattori M., Hamazaki Y., and Minato N.. 2009. PD-1+ memory phenotype CD4+ T cells expressing C/EBPalpha underlie T cell immunodepression in senescence and leukemia. Proc. Natl. Acad. Sci. USA. 106:15807–15812. 10.1073/pnas.0908805106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell L.M., Osokine I., Yamada D.H., De la Fuente J.R., Elsaesser H.J., and Brooks D.G.. 2016. Overcoming CD4 Th1 Cell Fate Restrictions to Sustain Antiviral CD8 T Cells and Control Persistent Virus Infection. Cell Rep. 16:3286–3296. 10.1016/j.celrep.2016.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S.J., Mack E.A., Rome K.S., Pajcini K.V., Ohtani T., Xu L., Li Y., Meijerink J.P., Faryabi R.B., and Pear W.S.. 2016. Trib2 Suppresses Tumor Initiation in Notch-Driven T-ALL. PLoS One. 11:e0155408 10.1371/journal.pone.0155408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain S.L., McKinstry K.K., and Strutt T.M.. 2012. Expanding roles for CD4+ T cells in immunity to viruses. Nat. Rev. Immunol. 12:136–148. 10.1038/nri3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Tanaka K., Magnusson F., Chung Y., Martinez G.J., Wang Y.H., Nurieva R.I., Kurosaki T., and Dong C.. 2014. CCAAT/enhancer-binding protein α negatively regulates IFN-γ expression in T cells. J. Immunol. 193:6152–6160. 10.4049/jimmunol.1303422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome M., Charton J.E., Pelzer C., and Hailfinger S.. 2010. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb. Perspect. Biol. 2:a003004 10.1101/cshperspect.a003004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uljon S., Xu X., Durzynska I., Stein S., Adelmant G., Marto J.A., Pear W.S., and Blacklow S.C.. 2016. Structural Basis for Substrate Selectivity of the E3 Ligase COP1. Structure. 24:687–696. 10.1016/j.str.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzschneider D.T., Charmoy M., Chennupati V., Pousse L., Ferreira D.P., Calderon-Copete S., Danilo M., Alfei F., Hofmann M., Wieland D., et al. 2016. T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity. 45:415–427. 10.1016/j.immuni.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Veiga-Parga T., Sehrawat S., and Rouse B.T.. 2013. Role of regulatory T cells during virus infection. Immunol. Rev. 255:182–196. 10.1111/imr.12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West E.E., Jin H.T., Rasheed A.U., Penaloza-Macmaster P., Ha S.J., Tan W.G., Youngblood B., Freeman G.J., Smith K.A., and Ahmed R.. 2013. PD-L1 blockade synergizes with IL-2 therapy in reinvigorating exhausted T cells. J. Clin. Invest. 123:2604–2615. 10.1172/JCI67008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J., and Kurachi M.. 2015. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15:486–499. 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J., Blattman J.N., Murali-Krishna K., van der Most R., and Ahmed R.. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77:4911–4927. 10.1128/JVI.77.8.4911-4927.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J., Ha S.J., Kaech S.M., Haining W.N., Sarkar S., Kalia V., Subramaniam S., Blattman J.N., Barber D.L., and Ahmed R.. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 27:670–684. 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Wolkowicz R., Jager G.C., and Nolan G.P.. 2005. A random peptide library fused to CCR5 for selection of mimetopes expressed on the mammalian cell surface via retroviral vectors. J. Biol. Chem. 280:15195–15201. 10.1074/jbc.M500254200 [DOI] [PubMed] [Google Scholar]
- Wu T., Ji Y., Moseman E.A., Xu H.C., Manglani M., Kirby M., Anderson S.M., Handon R., Kenyon E., Elkahloun A., et al. 2016. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1:eaai8593 10.1126/sciimmunol.aai8593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Sun H.W., Lacey N.E., Ji Y., Moseman E.A., Shih H.Y., Heuston E.F., Kirby M., Anderson S., Cheng J., et al. 2019. Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat. Immunol. 20:890–901. 10.1038/s41590-019-0403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama T., Kanno Y., Yamazaki Y., Takahara T., Miyata S., and Nakamura T.. 2010. Trib1 links the MEK1/ERK pathway in myeloid leukemogenesis. Blood. 116:2768–2775. 10.1182/blood-2009-10-246264 [DOI] [PubMed] [Google Scholar]
- Zajac A.J., Blattman J.N., Murali-Krishna K., Sourdive D.J., Suresh M., Altman J.D., and Ahmed R.. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188:2205–2213. 10.1084/jem.188.12.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander R., Schauder D., Xin G., Nguyen C., Wu X., Zajac A., and Cui W.. 2019. CD4+ T Cell Help Is Required for the Formation of a Cytolytic CD8+ T Cell Subset that Protects against Chronic Infection and Cancer. Immunity. 51(6):1028–1042.e4. 10.1016/j.immuni.2019.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.E., Zhang P., Wang N.D., Hetherington C.J., Darlington G.J., and Tenen D.G.. 1997. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. USA. 94:569–574. 10.1073/pnas.94.2.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Iwasaki-Arai J., Iwasaki H., Fenyus M.L., Dayaram T., Owens B.M., Shigematsu H., Levantini E., Huettner C.S., Lekstrom-Himes J.A., et al. 2004. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 21:853–863. 10.1016/j.immuni.2004.11.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequencing data that support the findings of this study have been deposited in GEO under accession no. GSE143802.