Zhan et al. reveal that glioma stem-like cells (GSCs) evade type I IFN suppression through MBD3/NuRD complex–mediated STAT1 downregulation, thereby allowing GSCs to initiate a tumor in the inhospitable microenvironment in glioblastoma (GBM).
Abstract
Type I interferons (IFNs) are known to mediate antineoplastic effects during tumor progression. Type I IFNs can be produced by multiple cell types in the tumor microenvironment; however, the molecular mechanisms by which tumor cells evade the inhibition of immune microenvironment remain unknown. Here we demonstrate that glioma stem-like cells (GSCs) evade type I IFN suppression through downregulation of STAT1 to initiate tumor growth under inhospitable conditions. The downregulation of STAT1 is mediated by MBD3, an epigenetic regulator. MBD3 is preferentially expressed in GSCs and recruits NuRD complex to STAT1 promoter to suppress STAT1 expression by histone deacetylation. Importantly, STAT1 overexpression or MBD3 depletion induces p21 transcription, resensitizes GSCs to IFN suppression, attenuates GSC tumor growth, and prolongs animal survival. Our findings demonstrate that inactivation of STAT1 signaling by MBD3/NuRD provides GSCs with a survival advantage to escape type I IFN suppression, suggesting that targeting MBD3 may represent a promising therapeutic opportunity to compromise GSC tumorigenic potential.
Graphical Abstract
Introduction
Cancer stem cells (CSCs) are an aggressive population of tumor cells that have been identified in many malignant tumors, with high capacity for self-renewal, therapeutic resistance, and driving tumor progression (Batlle and Clevers, 2017; Bleau et al., 2009; Eramo et al., 2006; Kreso and Dick, 2014; Saygin et al., 2019). In addition to their intrinsic programs, CSCs are tightly regulated by the tumor microenvironment, which plays crucial roles in CSC maintenance through secreted factors from different types of stroma cells (Lathia et al., 2011). Notably, the tumor microenvironment generates not only the supportive signals but also the unfavorable factors, including certain inflammatory signals that suppress the malignant progression of tumor cells (Junttila and de Sauvage, 2013; Zitvogel et al., 2015). It remains unclear how CSCs persist and promote tumor growth and malignant progression under these inhospitable conditions.
Type I IFNs are a series of pleiotropic cytokines believed to protect against tumor propagation by intrinsic impact on tumor cells through inducing differentiation or inhibiting proliferation or survival, or by extrinsic effects on tumor development through regulation of immune response. In the tumor microenvironment, type I IFNs are produced by immune cells, stromal cells, and even tumor cells and act in an autocrine or paracrine manner (Dunn et al., 2006; Parker et al., 2016; Zitvogel et al., 2015). Downregulation of the type I IFN receptor, IFNAR1, has been shown in colorectal cancer cells, stimulating colorectal tumorigenesis (Katlinski et al., 2017). The defect of IFN pathway genes in melanoma tumor cells also contributes to tumor progression and therapeutic resistance (Gao et al., 2016). Furthermore, the attenuated IFN-α response mediated by the LCOR pathway promotes the maintenance of breast CSCs (Celià-Terrassa et al., 2017). In glioblastoma (GBM), the most deadly primary brain tumor (Stupp et al., 2009; Wen and Kesari, 2008), several studies have shown that the expression of TLR4 and activation of IFN regulatory factor 3 are higher in differentiated glioma cells relative to glioma stem-like cells (GSCs; Alvarado et al., 2017; Pencheva et al., 2017), indicating that the IFN signaling in GSCs might be reduced. However, how GSCs respond to IFNs and survive this environmental pressure remains to be elucidated.
The binding of IFNs to its membrane receptors leads to the phosphorylation of JAKs and STATs, which activate the IFN signaling pathway (Ivashkiv and Donlin, 2014; Parker et al., 2016). STAT1 is essential for biological effects of IFN signaling, but the roles of STAT1 in tumor progression are controversial. Originally, STAT1 was classified as a tumor suppressor, since STAT1 deletion in mice promoted tumor development (Badgwell et al., 2004; Lesinski et al., 2003). A recent study showed that STAT1 promoted leukemia development by maintaining high MHC class I expression (Kovacic et al., 2006), indicating varied roles of STAT1 in different types of tumors. Nonetheless, the function of STAT1 in CSCs of GBM remains elusive.
The methyl-CpG–binding domain 3 (MBD3) is an essential scaffold protein of the nucleosome remodeling and deacetylase (NuRD) complex, which plays well-documented roles in transcription, chromatin assembly, and genomic stability (Hu and Wade, 2012; Lai and Wade, 2011; Le Guezennec et al., 2006). Although MBD3/NuRD has been shown to regulate stem cell pluripotency, its effects are still controversial and might be tissue dependent (dos Santos et al., 2014; Kaji et al., 2006; Rais et al., 2013). In this study, we found that GSCs evaded the suppression of type I IFNs through downregulation of STAT1 mediated by the MBD3/NuRD complex. We demonstrated that MBD3 was preferentially expressed in GSCs and recruited the NuRD complex to the STAT1 promoter to suppress STAT1 expression by histone deacetylation. STAT1 overexpression or MBD3 silencing significantly inhibited GSC proliferation through inducing p21 expression, resensitized GSCs to type I IFN suppression, and attenuated GSC tumor progression. These results demonstrate that inactivation of STAT1 signaling is a crucial mechanism by which GSCs escape from the suppression of the immune microenvironment.
Results
GSCs display less sensitivity to type I IFN suppression
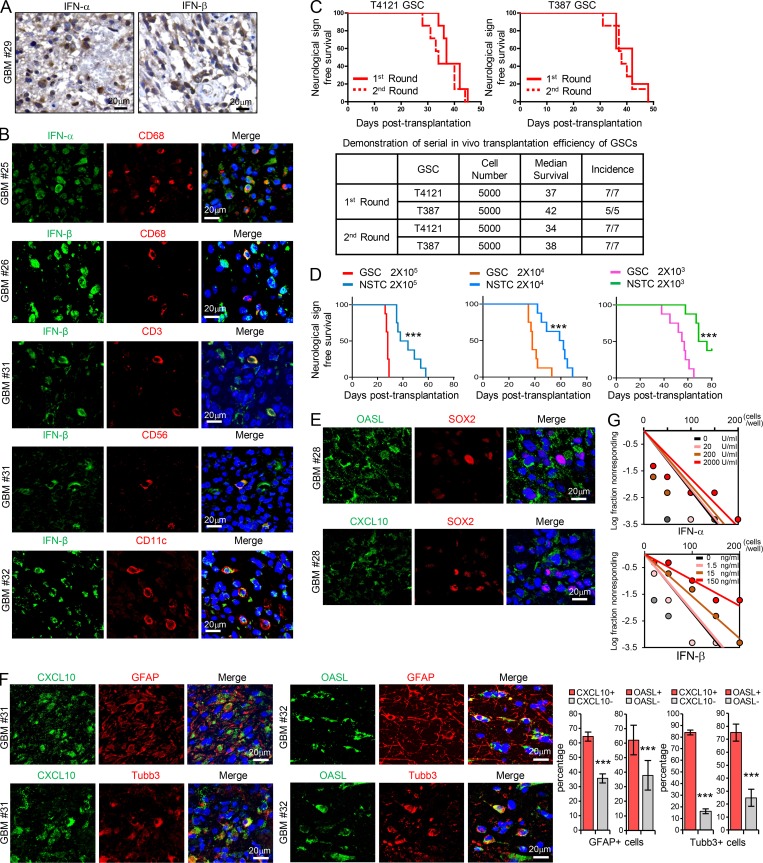
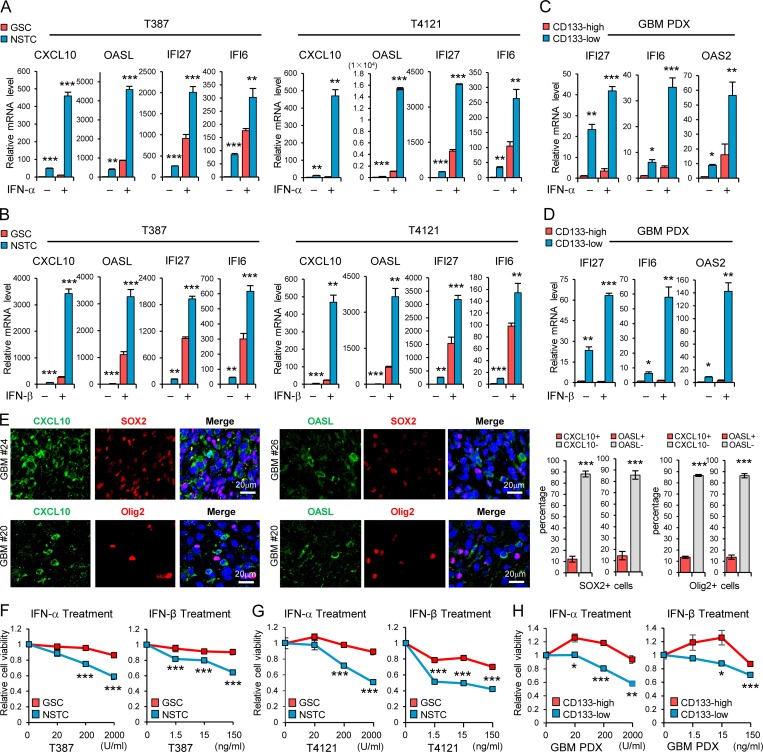
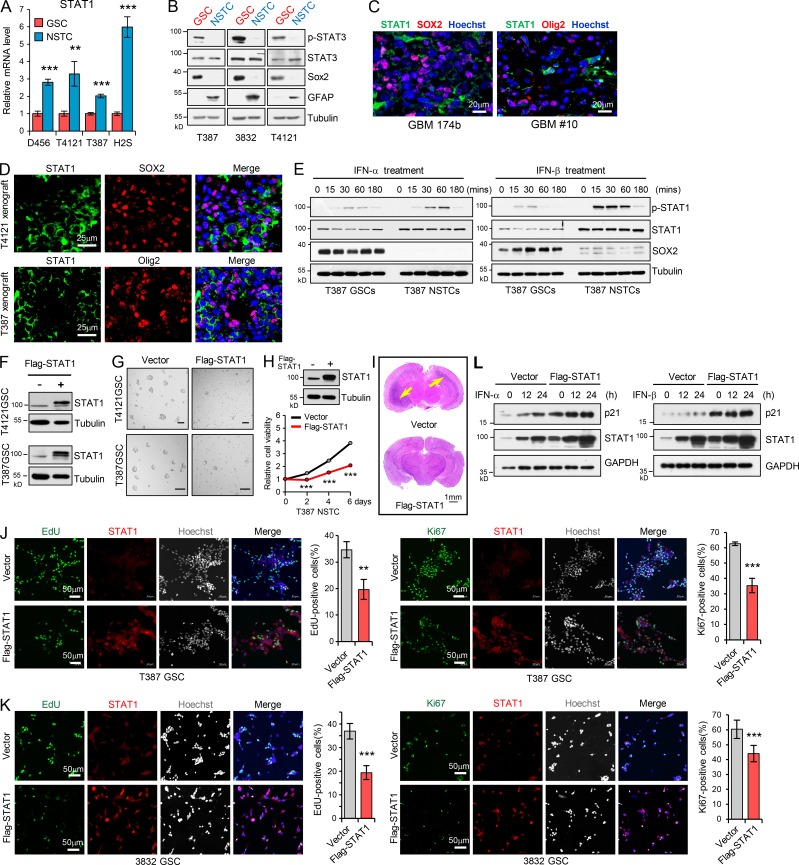
Because type I IFNs generated in the tumor microenvironment have intrinsic inhibitory effects on tumor cells and malignant progression (Parker et al., 2016; Zitvogel et al., 2015), we were interested in understanding whether CSCs in GBM could evade the immune inhibitory signals to propagate tumor. To this end, we initially examined the production of type I IFNs in human GBM specimens by immunohistochemistry (IHC) analysis and immunofluorescence (IF) staining. We found that IFN-α and IFN-β were expressed by many cells including macrophages (CD68+), tumor-infiltrating T cells (CD3+), natural killer cells (CD56+), and dendritic cells (CD11c+) in GBM tumor tissues (Fig. S1, A and B), as reported in other solid tumors (Celià-Terrassa et al., 2017; Parker et al., 2016). To directly assess the functional importance of type I IFN signaling in GSC regulation, we treated GSCs and non-stem tumor cells (NSTCs), with IFN-α or IFN-β and then analyzed the expression of IFN-regulated genes (IRGs). GSCs and the matched NSTCs used in the study were derived from GBM patients or patient-derived xenografts (PDXs), as previously described (Eyler et al., 2011; Man et al., 2018; Singh et al., 2004; Zhou et al., 2017), and were functionally validated by using in vivo serial transplantation assays and in vivo limiting dilution assays in this study (Fig. S1, C and D). By using these cells, we found that the mRNA levels of IRGs including CXCL10, OASL, IFI27, and IFI6 were significantly upregulated in NSTCs compared with GSCs (Fig. 1, A and B). IFN-α or IFN-β treatment induced the expression of these IRGs in GSCs, but to a much higher level in NSTCs (Fig. 1, A and B). As CD133 can be used for the enrichment of GSCs from GBM (Son et al., 2009), we collected tumor cells expressing high and low levels of CD133 by FACS from PDXs originating from human GBM. Consistently, IRGs including IFI27, IFI6, and OAS2 were significantly increased and preferentially induced by type I IFNs in CD133low tumor cells relative to CD133high tumor cells (Fig. 1, C and D). CXCL10 and OASL were not detected in CD133high PDX tumor cells even upon treatment with IFN-α or IFN-β.
Figure S1.
GSCs display less sensitivity to type I IFNs suppression. (A) IHC staining of IFN-α and IFN-β in human GBM specimens. Section was counterstained with hematoxylin. (B) Co-IF staining of IFN-α or IFN-β (green) with macrophage marker CD68, T cell marker CD3, natural killer cell marker CD56, and dendritic cell marker CD11c (red) in human GBM specimens. Nuclei were counterstained with Hoechst (blue). (C) GSCs tumor initiation in the in vivo serial transplantation. 5 × 103 GSCs (T4121 or T387) were intracranially injected into the brains of nude mice (nu/nu) for the primary xenograft. All recipients developed tumor and died within 50 d, and these tumor cells (5 × 103) could be serially transplanted at the second round, suggesting the long-term self-renewal and tumorigenicity of GSCs. Kaplan–Meier survival curves of mice implanted with GSCs are shown (top). Summary of the serial transplantation experiment is shown (bottom). (D) In vivo limiting dilution assays were performed with T387 GSCs and matched NSTCs. Kaplan–Meier survival plots are shown. n = 8 for each group, log-rank test. (E and F) Co-IF staining of OASL or CXCL10 (green) with SOX2 (red; E) or with GFAP or b3-tubulin (red; F) in human GBM specimens. Nuclei were counterstained with Hoechst (blue). Quantification are shown (percentage of CXCL10+/− or OASL+/− cells in GFAP+ cells, n = 10; percentage of CXCL10+/− cells in Tubb3+ cells, n = 10; percentage of OASL+/− cells in Tubb3+ cells, n = 15; unpaired Student’s t test). Data are represented as mean ± SD. (G) Limiting dilution analysis of the effect of IFN-α or IFN-β on T387 GSCs. Cells were plated in a limiting dilution manner (1–200 cells per well, 10 wells per condition). 10 d later, each well was evaluated for the presence or absence of tumorsphere. ***, P < 0.001.
Figure 1.
GSCs display less sensitivity to type I IFN suppression. (A and B) GSCs and matched NSTCs (T387 and T4121) were treated with IFN-α (200 U/ml; A, n = 3) or IFN-β (15 ng/ml; B, n = 3) for 12 h. The mRNA levels of IRGs CXCL10, OASL, IFI27, and IFI6 were analyzed by real-time qPCR. (C and D) CD133high and CD133low glioma cells isolated from GBM PDX were treated with IFN-α (200 U/ml; C, n = 3–4) or IFN-β (15 ng/ml; D, n = 3–4) for 12 h. The mRNA levels of IRGs IFI27, IFI6 and OAS2 were analyzed by real-time qPCR. Data were normalized to untreated GSC group that was set to 1. The y axis represents fold changes (A–D). (E) Co-IF staining of CXCL10 or OASL (green) and putative stem cell markers (SOX2 or Olig2, red) in human GBM specimens. Nuclei were counterstained with Hoechst (blue). Representative images are shown (left). Quantifications are shown (right, n = 3). (F–H) T387 GSCs (F, n = 3) or T4121 GSCs (G, n = 3) and the matched NSTCs, or the CD133high/CD133low glioma cells isolated from GBM PDX (H, n = 4), were treated with indicated dose of IFN-α or IFN-β for 3 d. Cell viability was assessed at day 3 and normalized to the untreated control. Data are represented as mean ± SD (A, B, and E–G) or mean ± SEM (C, D, and H). *, P < 0.05; **, P < 0.01; ***, P < 0.001, as assayed by unpaired Student’s t test or Welch’s t test.
We further performed coimmunofluorescence (co-IF) staining of CXCL10/OASL with SOX2/Olig2 (putative stem cell markers; Ludwig and Kornblum, 2017), GFAP (an astrocyte marker), or β3-tubulin (a neuronal marker) in human GBM tissues. The results showed that fractions of IRG-expressing cells were increased in NSTCs relative to those in GSCs in tumors (Fig. 1 E and Fig. S1, E and F), suggesting that type I IFN signal is higher in NSTCs than in GSCs in vivo. Importantly, treatment with type I IFNs significantly inhibited NSTC viability in a dose-dependent manner but showed limited impact on GSC growth (Fig. 1, F and G). These results were further validated in CD133low and CD133high tumor cells from GBM PDXs (Fig. 1 H). In addition, during long-term treatment, the self-renewal of GSCs was slightly inhibited by high doses of type I IFNs (Fig. S1 G). Taken together, our results suggest that GSCs show low signal of type I IFNs and less sensitivity to type I IFN suppression relative to NSTCs.
STAT1 is preferentially downregulated in GSCs and is associated with GSC tumorigenicity
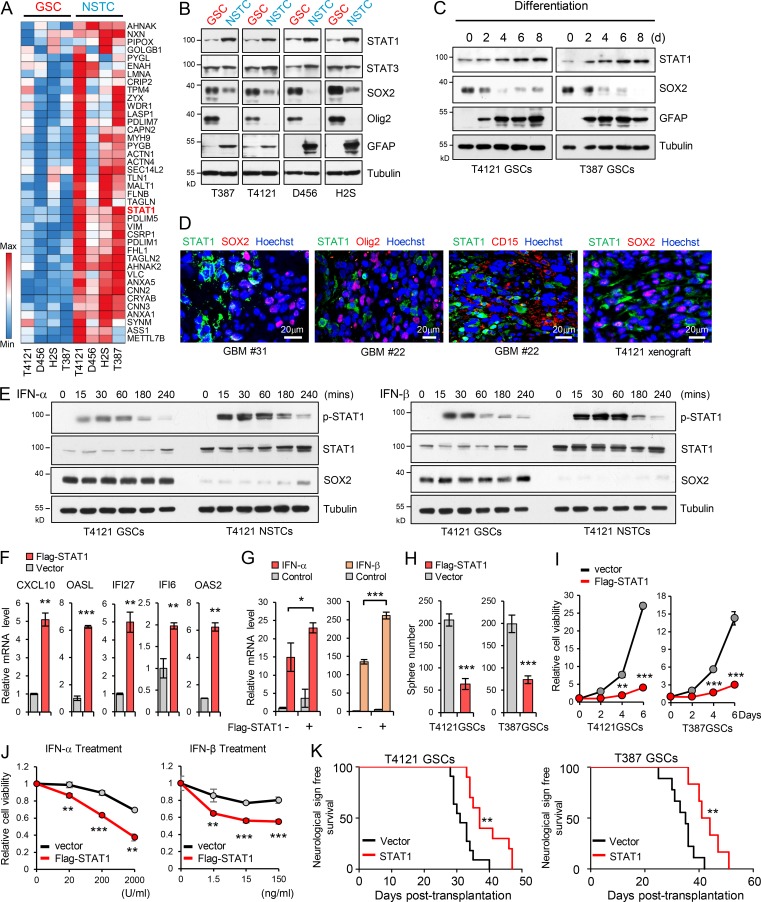
To determine the potential regulator of the observed phenotype, we performed a mass spectrometry (MS) screening to identify proteins differentially expressed between GSCs and NSTCs. STAT1 was identified as one of the 40 upregulated proteins, with the average change more than twofold in four pairs of NSTCs relative to the matched GSCs (Fig. 2 A). Since STAT1 has been demonstrated as the key signal transducer of the IFN signaling pathway to mediate IFN-induced gene activation (Ivashkiv and Donlin, 2014; Levy and Darnell, 2002; Silvennoinen et al., 1993), we focused on the potential roles of STAT1 in GSCs. We found that both mRNA and protein levels of STAT1 were significantly decreased in GSCs relative to NSTCs (Fig. 2 B and Fig. S2 A). We also examined STAT3, another STAT family member protein with critical roles in maintaining GSC tumorigenic potential (Kim et al., 2013; Sherry et al., 2009). Although expression of STAT3 was equal in two paired GSCs and NSTCs (T387 and H2S) and slightly upregulated in T4121 and D456 NSTCs, phosphorylation of STAT3 was significantly increased in GSCs relative to matched NSTCs (Fig. 2 B and Fig. S2 B). A gradual increase in the expression of STAT1 was observed during GSC differentiation (Fig. 2 C). Importantly, STAT1 and the putative GSC markers were expressed exclusively in the distinct populations of tumor cells in human primary GBM specimens and GBM xenografts (Fig. 2 D and Fig. S2, C and D), suggesting a specific suppression of STAT1 expression in GSCs. Together, these data suggest that STAT1 is preferentially downregulated in GSCs.
Figure 2.
Low STAT1 expression is associated with GSC proliferation and tumorigenicity. (A) Heatmap representation of the 40 upregulated proteins (average change more than twofold) screened by MS in four NSTCs compared with matched GSCs derived from human GBM tumors. Raw data were log2 transformed. A relative color scheme uses the minimum and maximum values in each row to convert values to colors. Red is high expression and blue is low. (B) Immunoblot (IB) analysis of STAT1, STAT3, SOX2, Olig2, and GFAP (astrocyte marker) in four GSCs and matched NSTCs. (C) IB analysis of STAT1, SOX2, and GFAP during GSC differentiation induced by serum (10% FBS). (D) Co-IF staining of STAT1 (green) and SOX2, Olig2, and CD15 (red) in human GBM specimens and mouse GBM orthotopic xenografts. Nuclei were counterstained with Hoechst (blue). (E) GSCs and matched NSTCs (T4121) were treated with IFN-α (200 U/ml) or IFN-β (15 ng/ml) for indicated times. Phosphorylated-STAT1, STAT1, SOX2, and tubulin were examined by IB analysis. (F and G) Flag-vector or Flag-STAT1–overexpressing GSCs (T4121) were untreated (F, n = 3) or treated with IFN-α (200 U/ml) or IFN-β (15 ng/ml; G, n = 3) for 12 h. mRNA levels of indicated genes were analyzed by real-time qPCR. Data were normalized to untreated GSC group that was set to 1. The y axis represents fold changes. Unpaired Student’s t test or Welch’s t test. (H and I) Overexpression of Flag-STAT1 impaired sphere formation of GSCs (H) and inhibited cell viability of GSCs (I). Quantification of sphere number (per 2,000 cells) formed by GSCs are shown (H, n = 3). Cell viability of GSCs was assessed at indicated time and normalized to day 0 in each group (I, n = 3). Unpaired Student’s t test. (J) T4121 GSCs expressing vector or Flag-STAT1 were treated with indicated dose of IFN-α (n = 3) or IFN-β (n = 5) for 3 d. Cell viability was assessed and normalized to the untreated control. Unpaired Student’s t test. (K) GSCs expressing vector or Flag-STAT1 were transplanted into brains of nude mice (2 × 104 cells/mouse, nu/nu nude mice). Kaplan–Meier survival curves of mice implanted with T4121 GSCs (vector, n = 11; Flag-STAT1, n = 10; left) or T387 GSCs (vector, n = 9; Flag-STAT1, n = 6; right) are shown. Log-rank test. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure S2.
Low STAT1 expression is associated with GSC proliferation and tumorigenicity. (A) mRNA levels of STAT1 in 4 GSCs and matched NSTCs were analyzed by real-time qPCR (n = 3). (B) IB analysis of p-STAT3 (Tyr705), STAT3, SOX2, Olig2, and GFAP in GSCs and matched NSTCs. (C and D) Co-IF staining of STAT1 (green) and SOX2 or Olig2 (red) in human GBM specimens (C) and mouse xenografts (D). Nuclei were counterstained with Hoechst (blue). (E) GSCs and matched NSTCs (T387) were treated with IFN-α (200 U/ml) or IFN-β (15 ng/ml) for indicated times. Phosphorylated-STAT1, STAT1, SOX2, and tubulin were examined by IB analysis. (F) IB analysis of Flag-STAT1 overexpression in GSCs. (G) Tumorsphere formation assay of GSCs expressing vector or Flag-STAT1. Tumorsphere images were assessed by bright-field microscopy. Scale bar represents 100 μm. (H) Cell viability of NSTCs (T387) expressing vector or Flag-STAT1 (n = 3). (I) Representative images of cross sections (H&E stain) of mouse brains (nu/nu) 31 d after transplantation with T4121 GSC expressing vector or Flag-STAT1. (J and K) Overexpression of Flag-STAT1 inhibited GSC proliferation. EdU incorporation assay (J and K, left) and Ki67 staining (J and K, right) in T387 GSCs (J, n = 3) and 3832 GSCs (K, n = 6) expressing vector or Flag-STAT1. Representative images are shown. The percentage of EdU+ or Ki67+ cells was quantified. (L) Flag-vector- or Flag-STAT1–overexpressing GSCs (T4121) were treated with IFN-α (200 U/ml) or IFN-β (15 ng/ml) for the indicated times. The indicated protein levels were analyzed by IB. Data are represented as mean ± SD. **, P < 0.01; ***, P < 0.001, as assayed by unpaired Student’s t test.
To further determine the activation of STAT1 in GSCs and NSTCs, we treated GSCs and matched NSTCs with IFN-α/IFN-β and examined the phosphorylation of STAT1. Phosphorylated STAT1 was significantly induced 15 min after IFN-α or IFN-β treatment, which was maintained at high levels until 1 h, and then gradually decreased in both GSCs and NSTCs. Notably, phosphorylation of STAT1 induced by type I IFNs was much higher in NSTCs compared with GSCs (Fig. 2 E and Fig. S2 E), suggesting the lower activation of STAT1 in GSCs. In addition, ectopic expression of Flag-STAT1 in GSCs strongly induced the expression of IRGs (Fig. 2 F and Fig. S2 F), which was further increased by type I IFN treatment (Fig. 2 G). Forced STAT1 expression significantly inhibited GSCs tumorsphere formation (Fig. 2 H and Fig. S2 G) and decreased cell viability of GSCs (Fig. 2 I) but showed a relatively lower efficiency in NSTCs (Fig. S2 H). Importantly, STAT1 overexpression resensitized GSCs to type I IFN–induced suppression (Fig. 2 J). These results suggest that the low expression of STAT1 is crucial for GSC maintenance and less sensitivity to type I IFN suppression.
To examine the effect of ectopic expression of STAT1 on GSC tumorigenicity, we established GBM orthotopic xenografts in nude mice (nu/nu) by intracranial injection of GSCs expressing vector or Flag-STAT1. A subset of mice in each group was sacrificed for histological analysis when the first few control mice developed neurological signs. STAT1 overexpression significantly decreased GSC-driven tumor growth (Fig. S2 I) and extended median survival of mice (Fig. 2 K). The antitumor effects of STAT1 overexpression may not be mediated through the immune microenvironment, since an immunocompromised mouse model was used here. Together, these findings suggest that the lower sensitivity of GSCs to the inhibition of type I IFNs is mediated by downregulation of STAT1, and low levels of STAT1 promote GSC tumor initiation.
Low STAT1 expression is essential for GSC proliferation through p21 regulation
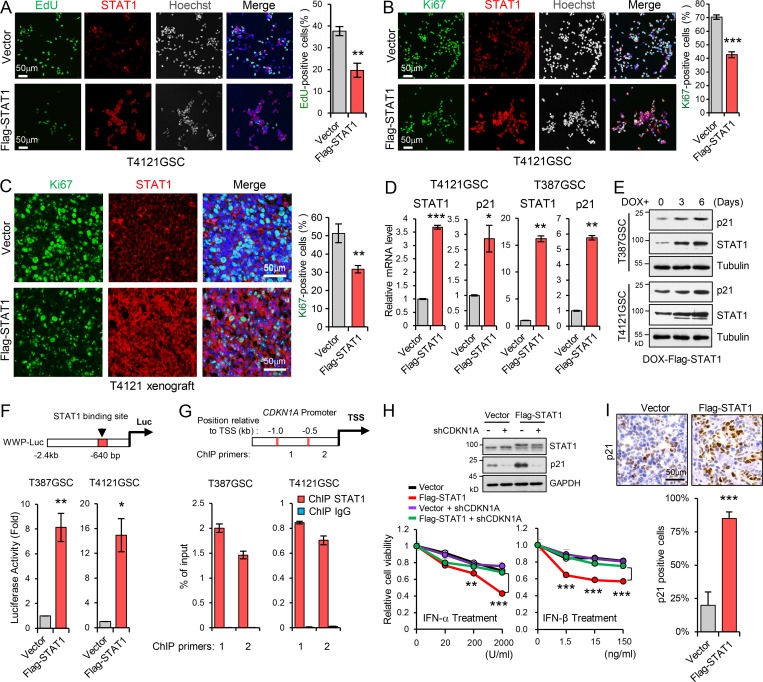
To gain mechanistic insight into the suppression of STAT1 on the GSC phenotype, we examined the expression of multiple markers for stem cells and regulators of apoptosis and proliferation in GSCs expressing Flag-STAT1. Overexpression of STAT1 had little effect on the expression of pluripotency genes (SOX2, Olig2, Oct4, and Nanog) and genes involved in cell apoptosis (data not shown) but dramatically decreased proliferation of GSCs, which was assessed by EdU (5-ethynyl-2′-deoxyuridine) incorporation and Ki67 staining in vitro (Fig. 3, A and B; and Fig. S2, J and K) and in vivo (Fig. 3 C).
Figure 3.
Low STAT1 expression is essential for GSC proliferation through p21 regulation. (A and B) Overexpression of Flag-STAT1 inhibited GSC proliferation. EdU incorporation assay (A, n = 3) and Ki67 staining (B, n = 3) in T4121 GSCs expressing vector or Flag-STAT1. Representative images are shown (left). The percentage of EdU+ or Ki67+ cells was quantified (right). (C) Co-IF staining of Ki67 (green) and STAT1 (red) in GBM xenografts derived from T4121 GSCs expressing vector or Flag-STAT1 (left). The percentage of Ki67+ cells was quantified (n = 3, right). (D) Real-time qPCR analysis of STAT1 and p21 expression in T4121 and T387 GSC expressing vector or Flag-STAT1 (n = 3). (E) IB analysis of STAT1 and p21 expression in T4121 and T387 GSCs expressing Dox-inducible-Flag-STAT1. GSCs were treated with 100 ng/ml Dox for 0, 3, and 6 d. (F) The p21 promoter-reporter construct (WWP-Luc) is shown on top. Transcriptional activation of p21 was measured using a p21 promoter luciferase reporter assay (bottom). Luciferase activity was measured 60 h after transfection, and activity was normalized to the level of control vector expression (n = 3). (G) ChIP analyses on p21 (CDKN1A) promoter. Assays were performed with STAT1 antibody, and immunoprecipitates were subjected to qPCR analyses. Schematic showing the ChIP primer location with respect to the TSS of the p21 promoter (n = 3). (H) Relative cell viability of GSCs transduced with shCDKN1A in the setting of Flag-STAT1 overexpression. Cells were treated with indicated dose of IFN-α or IFN-β for 3 d. IB analyses of STAT1 and p21 expression in GSCs are shown (top). Data were normalized to untreated GSC group (n = 3). (I) IHC staining of p21 in GBM xenografts derived from T4121 GSCs expressing vector or Flag-STAT1 (top). The percentage of p21+ cells was quantified (bottom, n = 3). Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as assayed by unpaired Student’s t test or Welch’s t test.
Previously, STAT1 has been shown to upregulate endogenous inhibitors of cyclin-dependent kinase, such as p21 (Abbas and Dutta, 2009; Xiong et al., 1993), and to inhibit cell proliferation in response to multiple stimulation (Chin et al., 1996; Huang et al., 2000; Johnson et al., 1998). Thus we investigated the impact of STAT1 on p21 expression in GSCs. Overexpression of STAT1 strongly increased p21 mRNA and protein expression in GSCs (Fig. 3, D and E), and the expression of p21 was further induced by type I IFN treatment in GSCs (Fig. S2 L). To further determine whether the increased level of p21 by STAT1 was the result of p21 transcriptional upregulation, we tested for the increased activation of p21 promoter by using a WWP-luciferase (WWP-luc) reporter, as the promoter contains a potential binding site for STAT1 (640 bp 5′ of the TATA box, Fig. 3 F, top; Chin et al., 1996). Our results showed that STAT1 overexpression significantly increased the WWP-luc activity in GSCs (Fig. 3 F, bottom). Through a chromatin immunoprecipitation (ChIP) assay with STAT1 antibody followed by quantitative PCR (qPCR) analysis, we found that STAT1 bound to p21 (CDKN1A) promoter mainly between −1.0 kb and −0.5 kb relative to the p21 transcriptional start site (TSS) in GSCs (Fig. 3 G). Importantly, knockdown of p21 significantly compromised the inhibition of IFN-α/IFN-β on STAT1-overexpressing GSCs (Fig. 3 H). Moreover, ectopic expression of Flag-STAT1 strongly induced p21 expression in GBM orthotopic xenografts (Fig. 3 I). Together, these data suggest that low STAT1 expression is essential for GSC proliferation through regulation of p21 transcription.
The MBD3/NuRD complex promotes H3K27 deacetylation on STAT1 promoter to inhibit STAT1 expression in GSCs
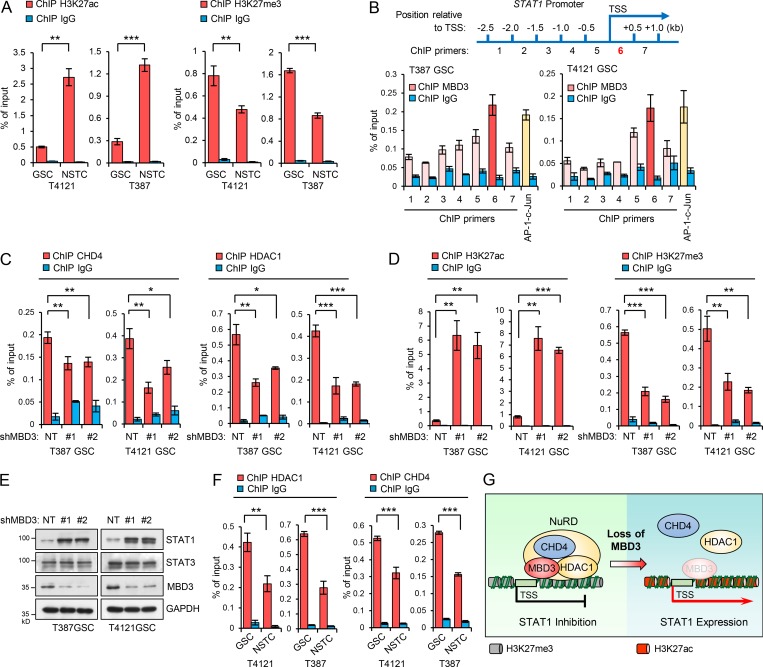
To understand the molecular mechanisms underlying the preferential downregulation of STAT1 in GSCs, we analyzed the modification of STAT1 promoter by using UCSC Genome Browser Database (http://genome.ucsc.edu). Interestingly, we found that there was a very weak occupation of acetylated histone 3 at lysine 27 (H3K27ac) on STAT1 promoter in human embryonic stem cells compared with other mature cell lines (Fig. S3 A). It is well established that histone acetylation and methylation play critical roles in regulating gene expression during stem cell differentiation (Easwaran et al., 2014; Mack et al., 2016; Suvà et al., 2013). To determine whether the modification of H3K27 contributes to STAT1 downregulation in GSCs, we analyzed the acetylation and methylation of H3K27 on STAT1 promoter. Through a ChIP assay with H3K27ac or H3K27me3 antibody followed by qPCR analysis, we found significantly decreased H3K27ac but increased H3K27me3 occupation on the promoter of STAT1 in GSCs relative to NSTCs (Fig. 4 A). As a control, enrichment of H3 on STAT1 promoter was not changed between GSCs and NSTCs (Fig. S3 B).
Figure S3.
The MBD3/NuRD complex promotes H3K27 deacetylation on STAT1 promoter to inhibit STAT1 expression in GSCs. (A) The UCSC Genome Browser shows the acetylation of H3K27 on the promoter of STAT1 in GM12878 (B-lymphocyte), hESC (human embryonic stem cells), HSMM (skeletal muscle myoblasts), HUVEC (human umbilical vein endothelial cell), K562 (leukemia), NHEK (epidermal keratinocytes), and NHLF (lung fibroblasts) cells. (B and C) ChIP analyses on STAT1 promoter in GSCs/NSTCs or GSCs expressing shNT/shMBD3s. Assays were performed with the H3 antibody, and immunoprecipitates were subjected to qPCR analyses (n = 3). (D) IB analysis of the indicated genes in 4 GSCs and matched NSTCs derived from 4 human GBM tumors. (E) Liquid chromatography-tandem MS (LC MS/MS) analysis of the purified MBD3/NuRD complex in GSC. Flag IP was performed in T387 GSC expressing Flag-MBD3. The components of NuRD complex were identified with MS. (F) IP of MBD3 was performed in T4121GSCs (left) and T387GSCs (right). The IB for CDH4, HDAC1, MBD3, and MBD2 are shown. IgG was used as an antibody control for IPs. Asterisks indicate nonspecific bands. (G) Real-time qPCR analysis of mRNA levels of MBD3, STAT1, and STAT3 in T4121GSCs or T387GSCs expressing shNT or shMBD3s (n = 3). (H) Knockdown of MBD3 increased the expression of STAT1 in both mRNA and protein in D456 GSCs (n = 3). (I) IB analysis of STAT1, p21, and H3K27ac in GSCs treated with SAHA for the indicated times. (J and K) T387 GSCs (J, n = 3) and T4121 GSCs (K, n = 3) were treated with the indicated dose of IFN-α/IFN-β in the absence or presence of SAHA (2 mM) for 3 d. Cell viability was assessed and normalized to the untreated control. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as assayed by unpaired Student’s t test or Welch’s t test. ns, not significant.
Figure 4.
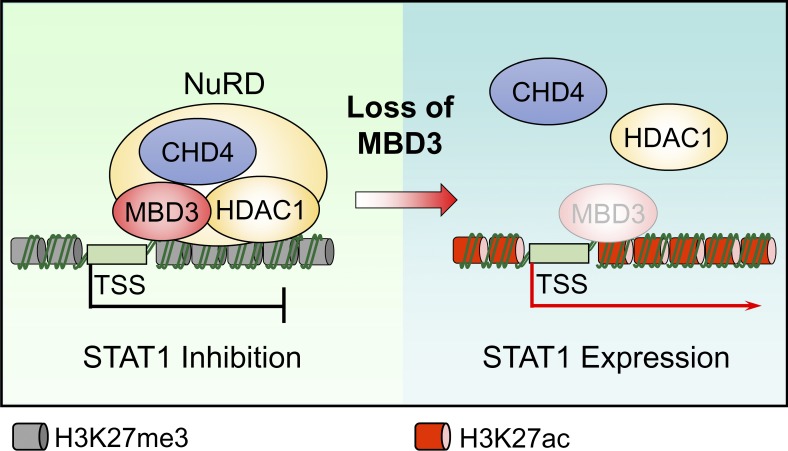
The MBD3/NuRD complex promotes H3K27 deacetylation on STAT1 promoter to inhibit STAT1 expression in GSCs. (A) ChIP analyses on STAT1 promoter. Assays were performed with the H3K27ac (left, n = 3) and H3K27me3 (right, n = 3) antibodies, and immunoprecipitates were subjected to qPCR analyses. (B) ChIP analysis with MBD3 antibody showing the enrichment of MBD3 at the promoter of STAT1 (around primer 6) in T4121GSCs and T387GSCs. Schematic showing the ChIP primer location with respect to the TSS of the STAT1 promoter (top). (C and D) ChIP analysis on the promoter of STAT1 in T4121GSCs (n = 3) and T387GSCs (n = 3) expressing shNT or two independent shMBD3s. Assays were performed with the indicated antibodies, and immunoprecipitates were subjected to qPCR analyses (primer 6). (E) IB analysis of STAT1, STAT3, and MBD3 in T387GSCs and T4121GSCs expressing shNT or two independent shMBD3s. (F) ChIP analyses on STAT1 promoter in GSCs and matched NSTCs. Assays were performed with the HDAC1 (left, n = 3) and CHD4 (right, n = 3) antibodies, and immunoprecipitates were subjected to qPCR analyses (primer 6). (G) Proposed model for MBD3/NuRD-mediated regulation of STAT1 transcription. In GSCs, MBD3 is highly expressed and binds to STAT1 promoter, recruits the NuRD complex (including CHD4 and HDAC1) to suppress STAT1 expression by H3K27 deacetylation. Loss of MBD3 disassembles the NuRD complex, increases H3K27 acetylation, and promotes STAT1 transcription. Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as assayed by unpaired Student’s t test or Welch’s t test.
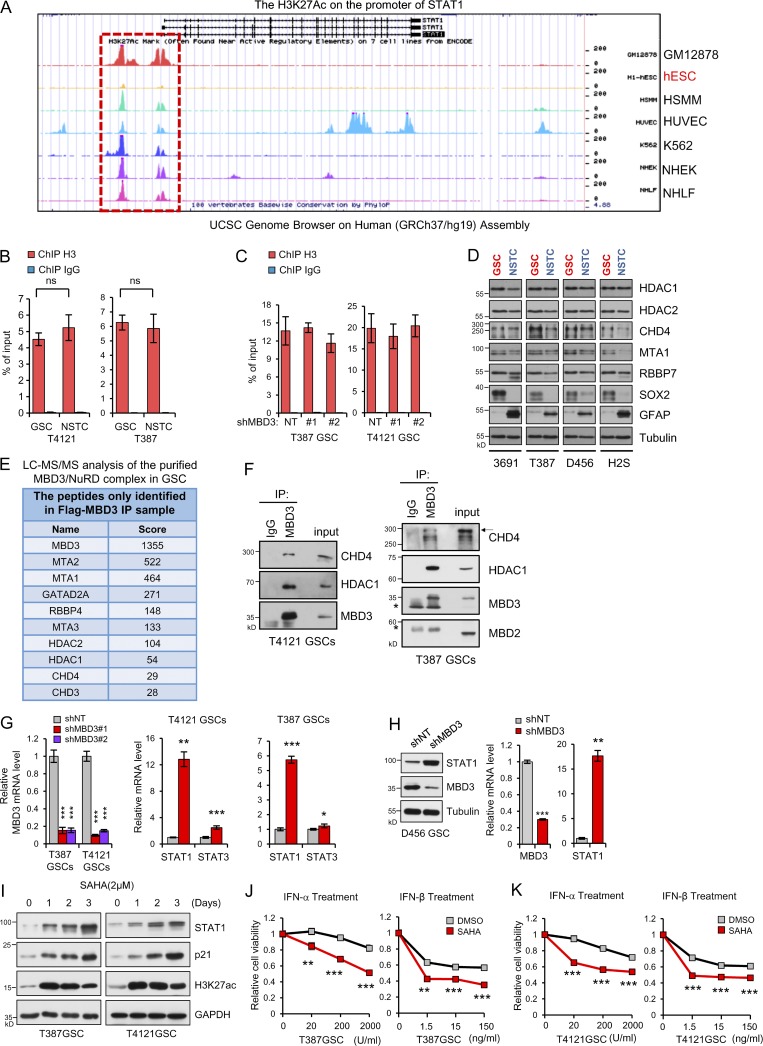
Histone deacetylases (HDACs) are major regulators of histone deacetylation and transcriptional repression (Laugesen and Helin, 2014). However, we did not find obvious differences in the expression of HDACs, such as HDAC1 or HDAC2, between GSCs and NSTCs (Fig. S3 D). As HDAC1 and HDAC2 are also major components of the NuRD complex, a type of ATP-dependent chromatin remodeling complex that controls the balance between acetylation and methylation of H3K27 (Hu and Wade, 2012; Reynolds et al., 2012b), we examined whether NuRD complex regulated the occupation of HDACs on STAT1 promoter in GSCs. Because MBD3 is the key scaffold protein that recruits and assembles the NuRD complex on DNA (dos Santos et al., 2014; Lai and Wade, 2011; Menafra and Stunnenberg, 2014; Rais et al., 2013), we then investigated the binding of MBD3 on STAT1 promoter in GSCs. The ChIP assay with MBD3 antibody in the vicinity of the promoter region of STAT1 was performed (Fig. 4 B, top). c-Jun, a known target of MBD3 (Aguilera et al., 2011), was used as the positive control. Our results showed that MBD3 was significantly enriched at the downstream of STAT1 TSS in GSCs (Primer 6, Fig. 4 B, bottom).
To further assess if MBD3 recruits the NuRD complex to STAT1 promoter, we first validated the association of MBD3 with other major components of the NuRD complex in GSCs. We stably expressed Flag-MBD3 in GSCs and performed flag immunoprecipitation (IP) and MS. All components of the NuRD complex including the core chromatin remodeling ATPase CHD3/4, HDAC1/2, and other members (Lai and Wade, 2011; Zhang et al., 1999) were detected in the co-IP (Fig. S3 E). The interactions were further confirmed by IP of endogenous MBD3 in GSCs (Fig. S3 F). Importantly, the major components of NuRD, such as CHD4 and HDAC1, were enriched on STAT1 promoter, and the enrichments were significantly reduced after MBD3 knockdown in GSCs (Fig. 4 C and Fig. S3 G, left), suggesting the recruitment of NuRD complex by MBD3 to STAT1 promoter. Consequently, knockdown of MBD3 markedly increased the enrichment of H3K27ac but reduced H3K27me3 on STAT1 promoter in GSCs (Fig. 4 D). However, binding of H3 on STAT1 promoter was not affected by MBD3 depletion (Fig. S3 C). These data suggest that MBD3 binds to STAT1 promoter and recruits NuRD complex to promote H3K27 deacetylation at STAT1 promoter in GSCs.
To further evaluate if MBD3/NuRD complex controls the expression of STAT1 in GSCs, we knocked down MBD3 and found that both mRNA and protein levels of STAT1 were significantly increased in GSCs (Fig. 4 E and Fig. S3, G and H). Notably, knockdown of MBD3 had no obvious impact on the expression of STAT3 (Fig. 4 E and Fig. S3 G). Consistently, our data showed that the NuRD components CHD4 and HDAC1 were strongly enriched on the STAT1 promoter in GSCs, and the enrichment was significantly decreased in NSTCs (Fig. 4 F). Some epigenetic inhibitors can block H3K27 deacetylation. To assess whether these inhibitors increased STAT1 levels in GSCs and resensitized GSCs to type I IFNs suppression, we examined the effect of a pan-HDAC inhibitor, suberoylanilide hydroxamic acid (SAHA), on the expression of STAT1 and p21 in GSCs. SAHA treatment significantly increased acetylation of H3K27 and upregulated STAT1 and p21 expression in a time-dependent manner in GSCs (Fig. S3 I). Importantly, administration of SAHA significantly resensitized GSCs to IFN-α and IFN-β treatment (Fig. S3, J and K). These results suggested that targeting NuRD complex by epigenetic inhibitors may improve the antitumor effects of type I IFNs on GSCs. Collectively, our data suggest that MBD3 recruits NuRD complex to STAT1 promoter to repress STAT1 expression by H3K27 deacetylation in GSCs (Fig. 4 G).
MBD3 is preferentially expressed in GSCs
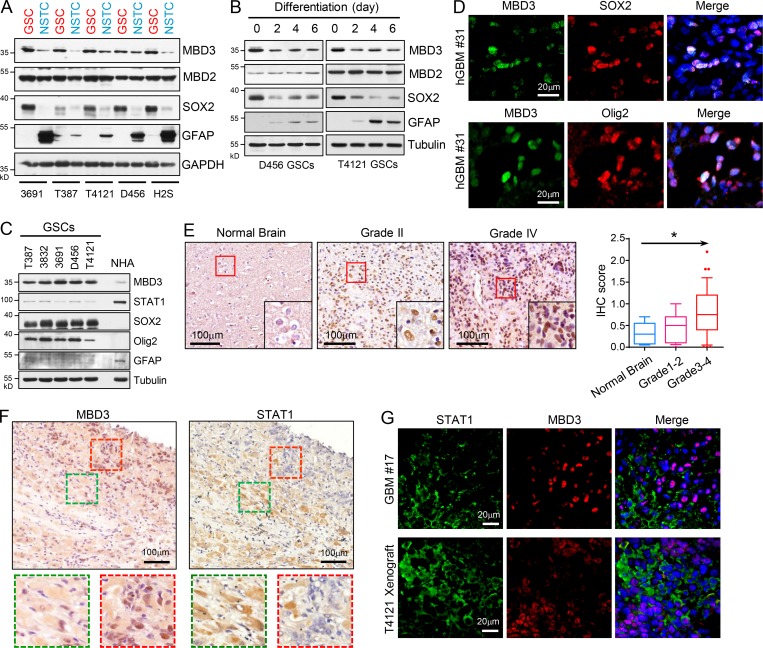
We then investigated why NuRD complex differentially suppressed STAT1 transcription in GSCs. First, we assessed the expression of MBD3 in GSCs and matched NSTCs and found that MBD3 was significantly upregulated in GSCs compared with NSTCs (Fig. 5 A). However, other major components of NuRD complex, including CHD4, HDAC1/2, MTA1, and RBBP7 (Lai and Wade, 2011; Zhang et al., 1999), were comparably expressed between GSCs and NSTCs (Fig. S3 D). We found that GSCs rapidly lost MBD3 expression during differentiation (Fig. 5 B). Interestingly, MBD2, another MBD family protein that forms the MBD2/NuRD complex (Le Guezennec et al., 2006), was equally expressed between GSCs and NSTCs (Fig. 5, A and B), indicating a specific role of the MBD3/NuRD complex in GSCs. Interestingly, MBD3 was rarely expressed in normal human astrocytes (NHAs), but STAT1 was significantly upregulated in NHAs relative to GSCs (Fig. 5 C). Importantly, the glioma cells with a high level of MBD3 were found to be enriched for SOX2, Olig2, or NESTIN in human GBM specimens and GBM xenografts (Fig. 5 D and Fig, S4, A–C). These data suggest that MBD3 is preferentially expressed in GSCs and mediates the assembly of NuRD complex on STAT1 promoter in GSCs.
Figure 5.
MBD3 is preferentially expressed in GSCs. (A) IB analysis of MBD3, MBD2, SOX2, and GFAP in GSCs and matched NSTCs derived from five human GBM tumors. (B) IB analysis of MBD3, MBD2, SOX2, and GFAP during GSC differentiation. (C) IB analysis of MBD3, STAT1, SOX2, and Olig2 in GSCs and NHAs. (D) Co-IF staining of MBD3 (green) and SOX2/Olig2 (red) in human GBM specimens. Nuclei were counterstained with Hoechst (blue). (E) IHC staining of MBD3 in brain tumor tissue microarray. Section was counterstained with hematoxylin (left). Box plot of histoscore of MBD3 (right). Normal brain tissue (n = 5), low-grade gliomas (I–II, n = 15), and high-grade gliomas (III–IV, n = 39). One-way ANOVA; *, P < 0.05. (F) IHC staining of MBD3 (left) and STAT1 (right) in serial sections of human GBM specimens. Sections were counterstained with hematoxylin. (G) Co-IF staining of STAT1 (green) and MBD3 (red) in human GBM specimen and mouse GBM orthotopic xenograft. Nuclei were counterstained with Hoechst (blue).
Figure S4.
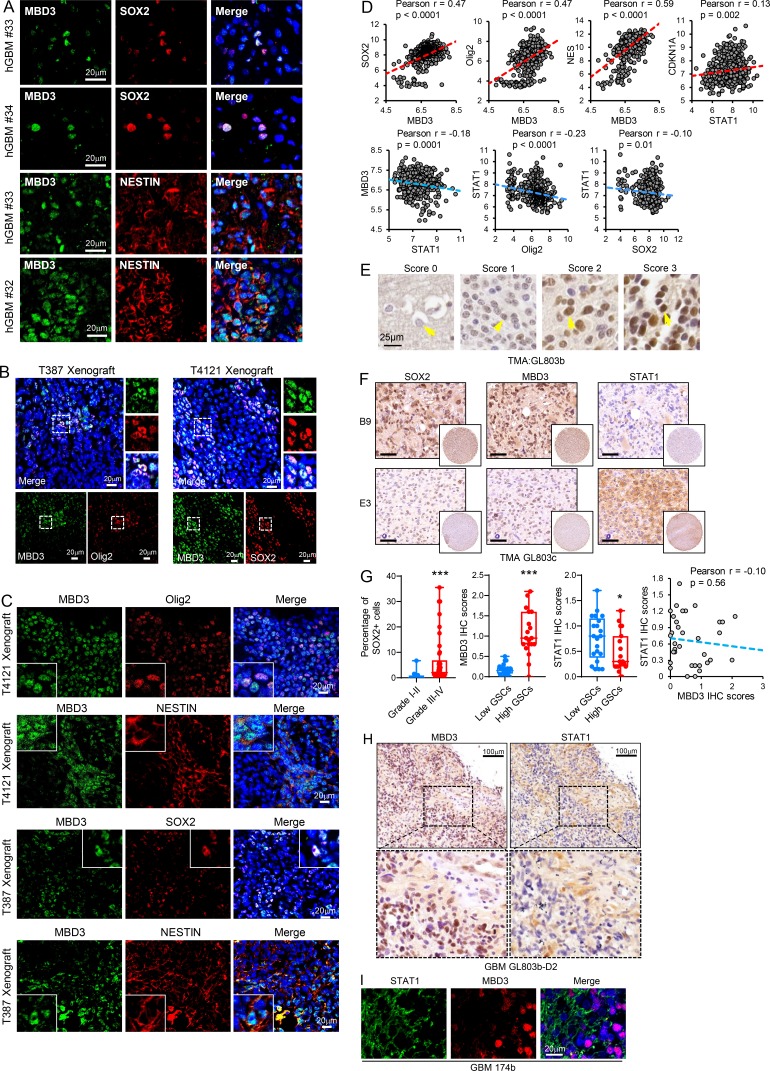
MBD3 is preferentially expressed in GSCs. (A) Co-IF staining of MBD3 (green) and SOX2, NESTIN (red) in human GBM specimens. Nuclei were counterstained with Hoechst (blue). (B and C) Co-IF staining of MBD3 (green) and SOX2, Olig2, NESTIN (red) in mouse GBM xenografts. Nuclei were counterstained with Hoechst (blue). (D) Pairwise correlation analysis of the indicated genes in TCGA GBM database. Pearson correlation coefficient (r) value and P value are shown (n = 538). (E–H) IHC staining of SOX2, MBD3, and STAT1 in the serial sections of human glioma tissue microarrays. Sections were counterstained with hematoxylin (E, F, and H). IHC score of MBD3 in brain tumor tissue microarray (E). Boxplot (G, left) and correlation analysis (G, right; n = 35) of histoscores of the tissue microarray stained for indicated proteins are shown. Low-grade gliomas (I–II, n = 13) and high-grade gliomas (III–IV, n = 42). SOX2+ cells were quantified to imply the fraction of GSCs in tumor (G; low GSCs, n = 21; high GSCs, n = 19). The scale bar represents 50 μm (F). *, P < 0.05; ***, P < 0.001, as assayed by unpaired Student’s t test. (I) Co-IF staining of STAT1 (green) and MBD3 (red) in human GBM specimens. Nuclei were counterstained with Hoechst (blue).
To gain insight into the relevance of MBD3 and STAT1 expression in human GBM, we first queried the TCGA GBM database and found that expression of MBD3 was significantly and positively correlated with levels of SOX2, Olig2, and NESTIN in human GBM (Fig. S4 D). Moreover, expression of STAT1 positively correlated with p21 levels but inversely correlated with MBD3 or stem cell markers in human GBM (Fig. S4 D). We further performed IHC staining with STAT1, MBD3, or SOX2 antibodies in serial sections of glioma tissue microarray. SOX2+ cells were quantified to indicate the fraction of GSCs in tumor. We found that the fraction of SOX2+ cells increased in high-grade gliomas compared with low-grade gliomas (Fig. S4 G, left), which was consistent with previous reports (Chen et al., 2015; Singh et al., 2004). The results showed that MBD3 expression was upregulated in gliomas compared with normal brain tissue and was largely increased in high-grade gliomas (Fig. 5 E and Fig. S4 E). In addition, MBD3 expression was positively correlated with fractions of GSCs in high-grade gliomas (Fig. S4, F and G, left). However, levels of STAT1 were relatively lower and inversely correlated with MBD3 in GBM tissues with high fractions of GSCs (Fig. S4, F and G). The P value of the correlation was not significant (Fig. S4 G, right), which might be due to the limited number of samples. Interestingly, we found that tumor cells highly expressing STAT1 or MBD3 were localized in distinct areas (Fig. 5 F and Fig. S4 H) in the same tumor specimen. Co-IF staining showed that the expression of STAT1 or MBD3 was exclusive in distinct tumor cells (Fig. 5 G and Fig. S4 I). Collectively, these data suggest that MBD3 inhibits STAT1 expression in vivo and that the levels of MBD3 may indicate the fraction of GSCs in human GBM.
Depletion of MBD3 attenuates GSC growth and tumorigenic potential
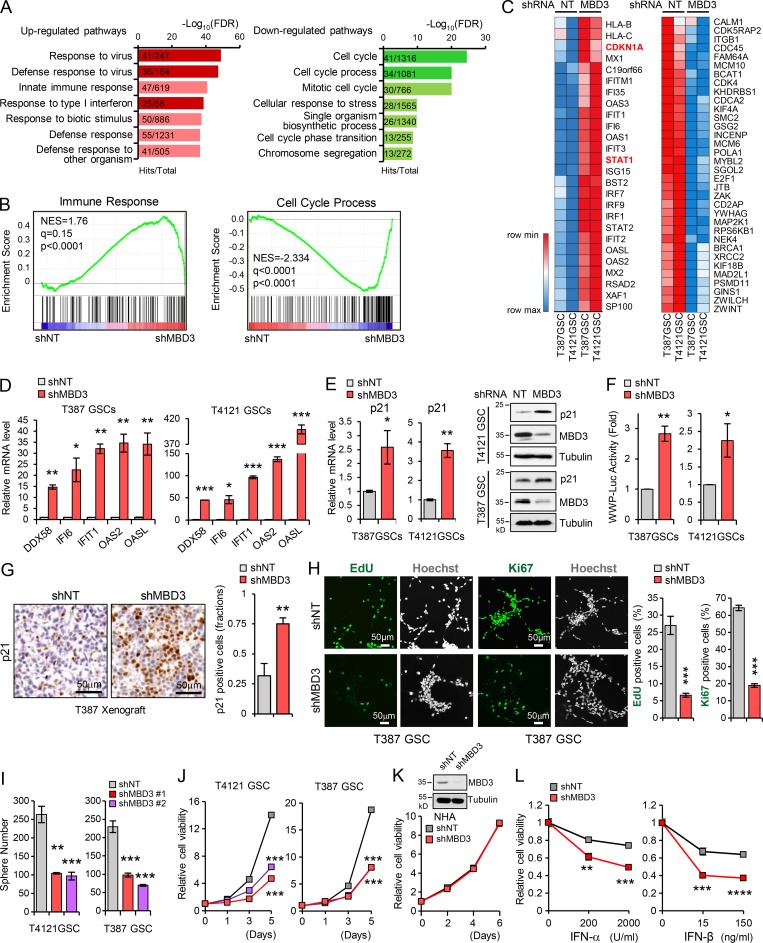
To assess whether MBD3 contributed to the reduced sensitivity of GSCs to type I IFN suppression and promoted GSC proliferation through STAT1 signaling, we profiled gene expression in GSCs expressing MBD3 shRNA or nontargeting (NT) shRNA using RNA sequencing. Gene ontology (Fig. 6 A) and gene set enrichment analysis (Fig. 6 B) revealed that MBD3 silencing resulted in enrichment of gene sets related to upregulated type I IFN response and immune response to virus (Fig. 6, A and B, left) and downregulated cell cycle process (Fig. 6, A and B, right). Moreover, the upregulation of STAT1, p21 (CDKN1A), and IRG genes (Fig. 6 C, left) and the downregulation of cell proliferation–related genes (Fig. 6 C, right) were shown in MBD3 knockdown GSCs. The upregulation of IRGs (DDX58, IFI6, IFIT1, OAS2, and OASL) and downregulation of cell proliferation–related genes (MCM10, POLA1, CDK4, and CDC45) were further confirmed by qPCR in MBD3 knockdown GSCs (Fig. 6 D and Fig. S5 A).
Figure 6.
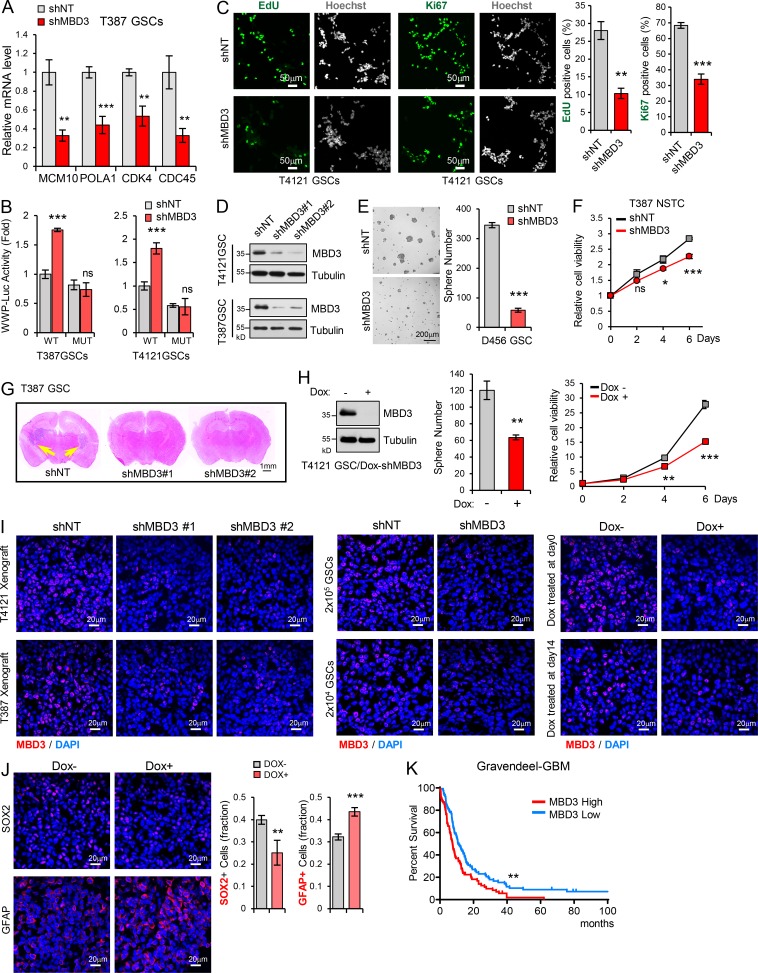
Depletion of MBD3 leads to upregulation of IFN signaling and growth inhibition in GSCs. (A) Overrepresented gene ontology terms among upregulated gene sets (left) and downregulated gene sets (right) in shMBD3-GSCs compared with the shNT-GSCs. (B) Gene set enrichment analysis shows the enrichment of gene sets positive related to immune response (left) and negative related to cell cycle process (right) in shMBD3-GSCs compared with the shNT-GSCs. (C) Heatmap representation of upregulated genes involved in IFN response (left) and downregulated genes involved in cell cycle process (right) in shMBD3-GSCs compared with the shNT-GSCs. (D) Real-time qPCR analysis of mRNA level of IRGs in T387 or T4121 GSCs expressing shNT or shMBD3 (n = 3). (E) Real-time qPCR (left) and IB (right) analysis of p21 expression in T387 or T4121 GSCs expressing shNT or shMBD3 (n = 3). (F) p21 promoter (WWP-Luc) luciferase reporter assay showed that knockdown of MBD3 induced the transcription activation of p21 in GSCs (n = 3). (G) IHC staining of p21 in GBM xenografts derived from T387 GSCs expressing shNT or shMBD3 (left). The percentage of p21+ cells was quantified (right; n = 3). (H) Knockdown of MBD3 impaired GSC proliferation assessed by EdU incorporation assay and Ki67 staining in T387 GSCs. Representative images are shown (left). The percentage of EdU+ or Ki67+ cells was quantified (right; n = 3). (I and J) Knockdown of MBD3 with two shRNA sequences inhibited GSC sphere formation (I) and cell viability (J; n = 3). (K) Knockdown of MBD3 had no obvious effect on cell viability of NHA (n = 3). (L) T4121 GSCs expressing shNT or shMBD3 were treated with indicated dose of IFN-α or IFN-β for 3 d, and cell viability was assessed and normalized to the untreated control (n = 3). Data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as assayed by unpaired Student’s t test or Welch’s t test.
Figure S5.
Depletion of MBD3 upregulates IFN signaling and inhibits GSC growth. (A) Real-time qPCR analysis of mRNA levels of MCM10, POLA1, CDK4, and CDC45 in T387GSCs expressing shNT or shMBD3 (n = 3). (B) CDKN1A promoter (WWP-Luc) luciferase reporter assay showed that MBD3 depletion had no effect on the reporter with STAT1 binding site mutation. Binding sites of STAT1 on CDKN1A promoter was mutated from 5′-TTCCCGGAA-3′ to 5′-AAGCTTGAA-3′ (n = 3). (C) Knockdown of MBD3 inhibited GSC proliferation assessed by EdU incorporation assay and Ki67 staining in T4121 GSCs expressing shNT or shMBD3. Representative images are shown (left). The percentage of EdU+ or Ki67+ cells was quantified (right; n = 3). (D) IB analysis showed the knockdown of MBD3 with two different shRNAs in T4121GSCs and T387GSCs. (E) Knockdown of MBD3 inhibited D456 GSC sphere formation. (F) Cell viability of T387 NSTCs expressing shNT or shMBD3 (n = 3). (G) Representative images of cross sections (H&E stain) of mouse brains (nu/nu) 38 d after transplantation with T387 GSC expressing shNT, shMBD3#1, or shMBD3#2. (H) T4121 GSCs transduced with Tet-on-shMBD3 were treated with Dox (100 ng/ml) or vehicle control. IB analysis showed the knockdown of MBD3 in T4121 GSCs (left). Inducible knockdown of MBD3 inhibited T4121 GSCs tumorsphere formation (middle) and cell viability (right; n = 3). (I) IF staining of MBD3 (red) in xenograft tissues to assess the efficiency of MBD3 knockdown in vivo in Fig. 7 (A–D), respectively. (J) IF staining of SOX2 or GFAP (red) in xenografts of T4121 GSCs (Dox-shMBD3) implanting mice (nu/nu) treated with or without Dox. Quantification of SOX2 or GFAP percentage are shown (right, n = 5). (K) Kaplan–Meier survival analysis of patients with high (n = 76) and low (n = 79) expression of MBD3 in Gravendeel GBM dataset. Log-rank test. For A–J, data are represented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as assayed by unpaired Student’s t test or Welch’s t test.
We then validated the upregulation of p21 in MBD3 knockdown GSCs at both mRNA and protein levels (Fig. 6 E). Similar to ectopic expression of STAT1 in GSCs, silencing of MBD3 significantly induced the WWP-luc reporter activity in vitro (Fig. 6 F) and increased the p21 expression in vivo (Fig. 6 G). However, MBD3 depletion did not increase the activity of the reporter with mutated STAT1 binding site (Fig. S5 B), suggesting that the regulation of MBD3 on p21 was mediated by STAT1. Consistently, MBD3 knockdown strongly inhibited cell proliferation, tumorsphere formation, and cell viability of GSCs (Fig. 6, H–J; and Fig. S5, C–E). However, MBD3 silencing had no obvious effect on NHA growth (Fig. 6 K) and slightly inhibited cell viability of NSTCs (Fig. S5 F). Importantly, we found that disruption of MBD3 resensitized GSCs to IFN treatment (Fig. 6 L). Together, these results suggest that MBD3 promotes GSC proliferation through inhibiting the STAT1 signaling pathway.
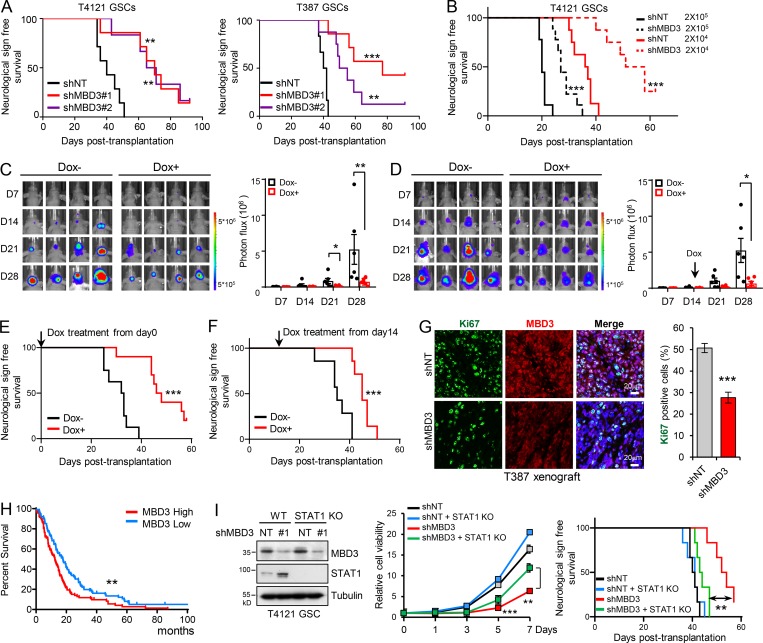
We next investigated the impact of MBD3 on GSC tumorigenicity in vivo. Silencing MBD3 with two independent shRNAs significantly decreased GSC-driven tumor growth and extended the median survival of mice (Fig. 7 A and Fig. S5, G and I, left). We further performed an in vivo limiting dilution assay and found that knockdown of MBD3 potently inhibited GSC tumorigenic activity in vivo (Fig. 7 B and Fig. S5 I, middle). To rule out the artificial effect of pretreating cells in vitro, we employed a doxycycline (Dox)-inducible shRNA system to silence endogenous MBD3 expression and further evaluated its roles in GSC tumorigenesis. A pool of GSCs stably expressing the luciferase and Dox-inducible shMBD3 were generated. Dox treatment of GSCs decreased MBD3 expression, inhibited GSC tumorsphere formation, and reduced cell viability (Fig. S5 H). Next, we intracranially transplanted the Dox-shMBD3/luciferase GSCs into mice and treated them with Dox from day 0. Inducible knockdown of MBD3 significantly inhibited GSC tumorigenesis, as shown by bioluminescent imaging (Fig. 7 C), and prolonged animal survival (Fig. 7 E and Fig. S5 I, right). Furthermore, knockdown of MBD3 significantly decreased the proliferation of tumor cells (Fig. 7 G) and the fraction of SOX2+ cells but increased the percentage of GFAP+ cells in vivo (Fig. S5 J). To assess the role of MBD3 in established tumors, we treated the mice bearing Dox-inducible shMBD3 GSCs with Dox when all tumors reached a similar size (day 14 after GSC implantation; Fig. 7 D). Depletion of MBD3 in the established tumors strongly suppressed tumor growth and extended survival (Fig. 7, D and F; and Fig. S5 I, right). Moreover, GBM database analysis showed that high expression of MBD3 correlated with poor survival in GBM patients (Fig. 7 H and Fig. S5 K).
Figure 7.
Highly expressed MBD3 promotes GSC malignant progression. (A) GSCs expressing shNT or shMBD3s were transplanted into brains of nude mice (5 × 104 cells/mouse). Kaplan–Meier survival curves of mice implanted with T4121 GSCs (shNT, n = 7; shMBD3#1, n = 6; shMBD3#2, n = 7) or T387 GSCs (shNT, n = 6; shMBD3#1, n = 8; shMBD3#2, n = 7) are shown. Log-rank test. (B) T4121 GSCs expressing shNT or shMBD3 were transplanted into brains of nude mice in a limiting dilution manner (2 × 105 or 2 × 104 cells/mouse, n = 9 or n = 8, respectively). Kaplan–Meier survival plots are shown. Log-rank test. (C–F) Luciferase-labeled T4121GSCs were transduced with the Tet-on-inducible shMBD3 and then transplanted into the brains of nude mice (2 × 104 cells/mouse). Mice were treated with vehicle control or Dox (2 mg/ml in drinking water) to induce expression of shMBD3 from day 0 (C and E) or day 14 (D and F). GBM xenografts were tracked by bioluminescence, and the representative images from animals at the indicated time are shown (C and D, left). Bioluminescent quantification indicated that induced knockdown of MBD3 inhibited GSC tumor initiation and growth (C and D, right). Kaplan–Meier survival plots of mice are shown (E, shNT, n = 8; shMBD3, n = 10; F, n = 7 for each group). Unpaired Student’s t test for C and D. Log-rank test for E and F. (G) Co-IF staining of Ki67 and MBD3 in GBM xenografts derived from T387 GSCs expressing shNT or shMBD3 (left). The percentage of Ki67+ cells was quantified (right, n = 3). Data are represented as mean ± SD (unpaired Student’s t test). (H) Kaplan–Meier survival analysis of patients with high (n = 93) and low expression (n = 88) of MBD3 in REMBRANDT GBM dataset. Log-rank test. (I) Knockout of STAT1 rescued the inhibition of MBD3 depletion on GSC viability and tumor initiation. IB of WT and STAT1 KO GSCs transduced with shNT or shMBD3 (left). Cell viability was assessed with GSCs as indicated (middle, n = 3, unpaired Student’s t test). The indicated GSCs were transplanted into brains of nude mice (2 × 104 cells/mouse). Kaplan–Meier survival curves of mice are shown (n = 6 for each group). Log-rank test. Data are represented as mean ± SD (G and I) or mean ± SEM (C and D). *, P < 0.05; **, P < 0.01; ***, P < 0.001. nu/nu nude mice were used in the animal experiments.
To assess whether the effect of MBD3 in GSCs was mediated by STAT1, we knocked down MBD3 in STAT1 knockout GSCs, which were generated using the CRISPR/Cas9 system (Fig. 7 I, left). Compared with GSCs containing WT STAT1, loss of STAT1 expression largely rescued the suppression of GSC viability (Fig. 7 I, middle) and the inhibition of GSC-derived tumor initiation by MBD3 depletion (Fig. 7 I, right). These data suggest that MBD3 depletion inhibits GSC growth, at least partially, through increasing STAT1 expression. Collectively, these results suggest that the expression of MBD3 correlates with the malignant behavior of gliomas, indicating that targeting MBD3 may represent a potential therapeutic opportunity to compromise GSC tumorigenic potential.
Discussion
Type I IFNs secreted by multiple cell types have intrinsic inhibitory effects on tumor cells in the tumor microenvironment (Parker et al., 2016; Zitvogel et al., 2015). In GBM, previous studies have shown that type I IFNs are produced in the tumor microenvironment (Larsson et al., 1978; Silginer et al., 2017; Tada and de Tribolet, 1993). Indeed, we found that several types of cells including macrophages, dendritic cells, and tumor-infiltrating lymphocytes express type I IFNs in the tumor microenvironment of GBM. Compared with NSTCs, GSCs show more resistance to type I IFN–induced suppression. In consonance with previous findings (Du et al., 2017), our results suggest that only long-term treatment with high-dose type I IFNs results in a light inhibition of GSC tumorsphere formation, supporting the reduced sensitivity of GSCs to type I IFN–induced antitumor effects. However, how GSCs evade the IFN suppression to propagate tumor is poorly understood. Our results demonstrate that GSCs could evade the inhibitory pressure of IFNs in the tumor microenvironment to maintain the CSC phenotype, through the downregulation of STAT1.
STAT1 is the core regulator of the IFN signaling pathway, and its function in cancer progression is controversial across different cancer types (Kovacic et al., 2006; Lesinski et al., 2003; Parker et al., 2016). Our study demonstrates that STAT1 is preferentially downregulated in GSCs, which is essential for the reduced sensitivity of GSCs to type I IFN inhibition through controlling the expression of p21, a crucial inhibitor of cyclin-dependent kinase. Thus, the downregulation of STAT1 is an important mechanism driving GSC growth in the tumor microenvironment to promote malignant progression, as overexpression of STAT1 presents strong inhibitory effects on GSC tumorigenicity. Interestingly, recent studies have shown that STAT1 is a mediator of stemness induced by CD95/Fas in breast cancer cells and was enriched in breast CSCs and CD95 highly expressing mesenchymal GSCs (Qadir et al., 2017). The GSCs used in our study were a classical subtype of GSCs (Flavahan et al., 2013; Venere et al., 2015). This suggests that the inconsistent roles of STAT1 in maintaining stemness or inhibiting growth of CSCs might be tissue or subtype dependent, which remains to be further explored. In addition, although the clinical trials show that administration of type I IFNs as an adjuvant treatment enhances the effect of temozolomide in human GBM therapy, the benefits are limited (Motomura et al., 2011; Shen et al., 2015). We hypothesize that GSCs with deficiency in IFN signaling may be responsible for the therapeutic resistance.
Our findings suggest that the lower level of STAT1 in GSCs is mediated by MBD3/NuRD, a chromatin remodeling complex. We found that the NuRD complex is recruited to STAT1 promoter by MBD3, where it facilitates the deacetylation of H3K27, thus inhibiting STAT1 transcription in GSCs (modeled in Fig. 4 G). These results are consistent with the model that NuRD complex mediates gene repression through deacetylation of histone by its component HDACs (Hu and Wade, 2012; McDonel et al., 2009; Reynolds et al., 2012b). MBD3 is an essential component of the NuRD complex, which has been implicated in the regulation of differentiation, development, and cancer progression (Lai and Wade, 2011). Recently, the opposite roles of MBD3/NuRD complex in stem cell reprogramming have been reported (dos Santos et al., 2014; Rais et al., 2013), suggesting a cell context–dependent manner of MBD3 function. However, the role of MBD3 in GSC tumorigenesis has not been elucidated. Our data demonstrate that MBD3, but not other components of the NuRD complex, is preferentially expressed in GSCs to promote GSC tumorigenicity. In contrast to our results, a recent study reported that MBD3 inhibits proliferation of the established GBM cell line SF767 (Cui et al., 2016). We speculate that different culture conditions contribute to the inconsistent roles of MBD3 in malignant glioma cells. Decades of culture in serum-containing medium may have altered the genetic and epigenetic landscape of the established tumor cell lines. Compared with these lines, GSCs recapitulate the genotype and in vivo biology of GBM and have been demonstrated as a reliable model of primary human tumors (Lee et al., 2006).
In disagreement with recent reports (Rais et al., 2013; Reynolds et al., 2012a), we did not find the regulation of multiple pluripotency genes by MBD3 in GSCs. Our results showed that depletion of MBD3 led to a significant activation of type I IFN response and immune response of GSCs and further inhibited GSC proliferation by upregulating p21. Notably, the expression of MBD3 is very low in normal brain tissue and human astrocytes and significantly correlates with the malignant progression of gliomas and the survival of GBM patients, suggesting that targeting MBD3 may be a promising therapeutic opportunity to compromise GSC tumorigenic potential.
Recent studies show that STAT3 may represent an inhibitory effect on STAT1 expression and type I IFN response (Tsai et al., 2019). STAT3 is preferentially activated in GSCs, suggesting that STAT3 activation may contribute to the lower sensitivity of GSCs to type I IFN–induced suppression. Whether blockade of STAT3 signaling improves the efficiency of IFN treatment on GSCs remains to be investigated. In addition, our data showed that MBD2, another MBD family protein that has also been identified as one of the core subunits of NuRD complex (Le Guezennec et al., 2006; Menafra and Stunnenberg, 2014), is equally expressed between GSCs and NSTCs and exists in a mutually exclusive NuRD complex in GSCs. It seems that more than one type of NuRD complex exists in GSCs, as we did not find any association between MBD3 and MBD2 in GSCs. Knockdown of MBD2 also resulted in the inhibition of tumorsphere formation and cell viability of GSCs but had no effect on the expression of STAT1 (not depicted), suggesting that different NuRD complexes may show multiple roles in GSCs.
Recently, immunotherapy has been shown to be promising in various types of tumors. However, the effects of immunotherapy on GBM are limited because of the severe immunosuppression and T cell exhaustion in GBMs (Lim et al., 2018). The lowered response of GSCs to IFNs might be one of the reasons, as the IFN signaling pathway is crucial for antitumor immunity (Gao et al., 2016; Zitvogel et al., 2015). Combination immunotherapy with IFNs might provide a potential approach to overcoming immunosuppression and increasing GBM patient responsiveness. Taken together, our study results demonstrate that the defective response to IFNs is crucial for GSCs to evade the suppressive pressure of the tumor microenvironment in GBMs. The MBD3-STAT1-p21–dependent mechanism may not only protect GSCs but also promote GSC-driven tumor growth.
Materials and methods
Cell culture and reagents
GSCs and NSTCs were isolated and functionally characterized from GBM surgical specimens or xenografts as previously described (Cheng et al., 2013; Man et al., 2018; Zhou et al., 2017). A Papain Dissociation System (Worthington Biochemical) was used to dissociate tumors according to the manufacturer’s instructions. Cells were recovered in stem cell medium (Neurobasal-A medium with B27 supplement, 10 ng/ml epidermal growth factor [EGF], and 10 ng/ml fibroblast growth factor [FGF]) overnight, then sorted by magnetic cell sorting using the surface marker CD133 (Miltenyi Biotec, CD133high cells), cultured in stem cell medium as described above, and validated by stem cell marker expression (SOX2, Olig2, and Nestin). GSCs were assessed for sphere-forming ability and tumor propagation by in vivo limiting dilution assay. Differentiation of GSCs was induced by exposure to DMEM with 10% FBS, and cells were maintained in serum culture conditions (Bao et al., 2006; Cheng et al., 2013; Man et al., 2018) and assayed for gain of differentiation markers (GFAP) and loss of stem cell markers (SOX2 and Olig2). To rule out the impact of serum on experiments, NSTCs were washed and cultured in serum-free conditions as GSCs for at least 24 h each time before IFN-α/β treatment.
GSCs were from the following GBM patients: 3691, 59-yr-old female; T4121, 53-yr-old male (recurrent GBM); T387, 69-yr-old, sex not available; D456, 60-yr-old, sex not available; 3832, 75-yr-old female; and H2S, age and sex not available. For EdU incorporation assay, GSCs were incubated with EdU (Click-iT EdU Imaging Kits, Invitrogen) for 100 min. Cells were fixed and permeabilized, and EdU was detected according to the manufacturer’s directions. The NHA cell line was purchased from BeNa Culture Collection (BNCC 341796) and cultured in DMEM with 10% FBS. The cells were maintained at 37°C in a humidified incubator with 5% CO2.
FBS was purchased from Macgene, protease inhibitors were purchased from Roche, and Dox was purchased from Sigma-Aldrich. EGF, βFGF, and IFN-α were purchased from R&D Systems, IFN-β was purchased from Peprotech, and all were prepared according to the manufacturers’ instructions.
Tissue acquisition and tissue microarray analysis
GBM surgical specimens were collected for this study in accordance with the First Hospital of Jilin University Review Board–approved protocol.Informed consent was obtained from all subjects. GBM tissues were obtained from newly diagnosed patients. All patients were sufficiently healthy to undergo surgery. The glioma tissue microarray was purchased from US Biomax (GL803b and GL803c). Histological diagnosis of the tissue microarray cores was reviewed by a pathologist. The tissue microarray GL803b included tumors from four patients with grade I astrocytoma, two females and two males, ages 15–52 yr; 11 patients with grade II astrocytoma, three females and eight males, ages 14–73 yr; seven patients with grade III astrocytoma, two females and five males, ages 10–71 yr; 32 patients with GBM, 15 females, 17 males, ages 6–68 yr; and five adjacent normal brain tissue from two females and three males, ages 30–52 yr. The tissue microarray GL803c included tumors from three patients with grade I astrocytoma, three males, ages 25–66 yr; 10 patients with grade II astrocytoma, three females and seven males, ages 14–53 yr; two patients with grade III astrocytoma, one female and one male, ages 19–51 yr; and 40 patients with GBM, 17 females, 23 males, ages 1–66 yr.
Orthotopic mouse xenografts
All animal experiments were performed in accordance with the National Institutes of Health guide for the care and use of laboratory animals and with the approval of the Institutional Animal Care and Use Committee of the National Center of Biomedical Analysis. Mice used in our studies were 4-wk-old female NU/NU nude mice purchased from Beijing Vital River Laboratory Animal Technology. Animal care was monitored daily by certified veterinary staff and laboratory personnel. Every effort was made to minimize discomfort, distress, pain, or injury to the mice. A maximum of five mice per cage was allowed. Intracranial transplantation of GSCs to establish GBM xenografts was performed as described (Man et al., 2018). Briefly, GSCs were transduced with different plasmids as described in this paper through lentiviral infection. 48 h later, cells were selected in culture containing puromycin (2 µg/ml) for 48 h. GSCs were implanted into the right frontal lobes of mice. For the survival experiments, animals were maintained until manifestation of neurological signs or 120 d after transplantation. Mice were euthanized by exsanguination using transcardiac perfusion with PBS under deep anesthesia, when exhibiting signs of declining neurological status or performance status. Mouse brains were fixed in 4% paraformaldehyde for 24 h at 4°C, followed by 48 h of cryoprotection with 30% sucrose before sectioning at a thickness of 7 µm.
Antibodies and Western blots
Antibodies used in the Western blots are p-STAT1(Tyr701) (1:1,000, 7649S, Cell Signaling, RRID:AB_10950970), SOX2 (1:1,000, MAB4423, Millipore, RRID:AB_11213224), Olig2 (1:1,000, sc-48817, Santa Cruz, RRID:AB_2157550), Nestin (1:1,000, 611659, BD, RRID:AB_399177), GFAP (1:1,000, Z0344, Dako, RRID:AB_10013482), MBD3 (1:1,000, 14258-1-AP, Protein Tech group, RRID:AB_2139745), STAT1 (1:1,000, 14994S, Cell Signaling, RRID:AB_2737027), MBD2 (1:1,000, ab38646, Abcam, RRID:AB_2139612), Tubulin (1:5,000, T5168, Sigma-Aldrich, RRID:AB_477579), Flag (1:5,000, F3165, Sigma-Aldrich, RRID:AB_259529), STAT3 (1:1,000, sc-7179, Santa Cruz, RRID:AB_661407), p-STAT3(Tyr705) (1:1,000, 9145S, Cell Signaling, RRID:AB_2491009), CHD4 (1:1,000, 11912, Cell Signaling, RRID:AB_2751014), HDAC1 (1:1,000, 5356, Cell Signaling, RRID:AB_10612242), HDAC2 (1:1,000, 5113, Cell Signaling, RRID:AB_10624871), MTA1 (1:1,000, 5647, Cell Signaling, RRID:AB_10705601), and RBBP7 (1:1,000, 6882, Cell Signaling, RRID:AB_10830730). Western blot analyses were performed as previously described (Man et al., 2018). Briefly, cells were lysed in M2 buffer (20 mM Tris HCl, pH 7.0, 0.5% NP-40, 250 mM NaCl, 3 mM EDTA and 3 mM EGTA, 1× protease inhibitor cocktail, 2 mM DTT, 0.5 mM PMSF, 20 mM β-glycerol phosphate, and 1 µM sodium vanadate). Protein samples were loaded on SDS-PAGE gels and transferred to polyvinyl difluoride membranes. Blots were incubated with primary antibodies overnight at 4°C followed by HRP-conjugated species-specific antibodies (Jackson Immuno Research, 1:5,000) at room temperature.
IF staining
IF staining of cells and tissue sections was performed as previously described (Man et al., 2018). Cultured cells or human surgical specimens were fixed in 4% paraformaldehyde for 10 min at room temperature. Samples were blocked with 1% BSA and 0.3 M glycine in PBS + 0.05% Tween for 1 h at room temperature, and then incubated with primary antibodies overnight at 4°C followed by incubation with appropriate secondary antibody for 1 h at room temperature. Nuclei were counterstained with Hoechst (Invitrogen). Images were acquired with a 20×/0.5 objective or 40×/0.75 objective on a Zeiss LSM880 system; the acquisition software was Zen 2.1 SP2. ImageJ (National Institutes of Health) was used for image processing after data acquisition. The following antibodies were used: SOX2 (1:50, sc-17320, Santa Cruz, RRID:AB_2286684), Olig2 (1:50, AF2418, R&D Systems, RRID:AB_2157554), Nestin (1:100, 611659, BD, RRID:AB_399177), CD15 (1:50, ab754, Abcam, RRID:AB_305962), MBD3 (1:100, ab188401, Abcam), STAT1 (1:100, 14994, CST, RRID:AB_2737027; 1:50, ab155933, Abcam), Ki67 (1:100, 9449S, Cell Signaling, RRID:AB_2715512), OASL (1:100, ab38325, Abcam, RRID:AB_776890), CXCL10 (1:50, ab9807, Abcam, RRID:AB_308792), IFN-α (1:100, ab193055, Abcam), IFN-β (1:100, ab84258, Abcam, RRID: AB_1859645), CD68 (1:100, ZM-0060, Zhong-shan Golden Bridge), GFAP (1:200, Z0344, Dako, RRID:AB_10013482; 1:50, A21282, Thermo Fisher Scientific, RRID: AB_2535827), TUBB3 (1:50, 4466S, Cell Signaling, RRID:AB_1904176), CD3 (1:50, ab699, Abcam, RRID:AB_305686), CD56 (1:50, 304601, BioLegend, RRID:AB_314443), and CD11c (1:50, 60258-1-lg, ProteinTech group).
IHC
IHC staining of tissue sections was performed as previously described (Man et al., 2018). Tissue microarrays including normal brain and low-grade and high-grade gliomas were purchased from US Biomax. The staining intensity of MBD3 was scored using a modified histochemical scoring (H-score) system to assess both the intensity of the staining and the percentage of positively stained cells. Briefly, for the intensity, a score of 0 to 3 (corresponding to negative, weak, moderate, or strong staining) was recorded, and the percentage of positively stained cells at each intensity was estimated. The H-score was calculated as [1 × (weak %)] + [2 × (moderate %)] + [3 × (strongly stained %)]. Images were acquired with a 20×/0.75 objective on a Hamamatsu 2.0 HT digital slide scanner; the acquisition software was Nanozoomer 2.0 HT. The following antibodies were used: MBD3 (1:50, 14258-1-AP, Protein Tech group, RRID:AB_2139745), STAT1 (1:500, 14994, Cell Signaling, RRID:AB_2737027), IFN-α (1:500, ab193055, Abcam), IFN-β (1:1,000, ab84258, Abcam, RRID:AB_1859645), p21 (1:100, 2947, Cell Signaling, RRID:AB_823586), and SOX2 (1:100, sc-365823, Santa Cruz, RRID:AB_10842165).
ChIP assay
ChIP was performed according to the manufacturer’s instructions (Cell Signaling, 9002S). Briefly, cells were cross-linked with 1% formaldehyde and stopped by adding glycine. Chromatin was digested by micrococcal nuclease, and the lysate was sonicated with several pulses to break the nuclear membrane. The digested chromatin was incubated with antibodies against MBD3 (Abcam, ab91458, RRID:AB_2049907), CHD4 (Cell Signaling, 12011S, RRID: AB_2734702), HDAC1 (Cell Signaling, 34589S, RRID:AB_2756821), H3K27me3 (Cell Signaling, 9733S, RRID:AB_2616029), and H3K27ac (Millipore, 17–683, RRID:AB_1977529) or rabbit IgG (Cell Signaling, 2729) as a control. Complexes were isolated with protein G agarose beads. Following elution, cross-links were reversed by Proteinase K for 2 h at 65°C. The released DNA was purified with a column and analyzed by qPCR using SYBR Green (Applied Biosystems) on the Real-Time PCR Detection System (Applied Biosystems) with the specific ChIP primers. Quantification of binding to the promoter was defined as the percentage of the whole-cell lysate input DNA. Primers used for ChIP-PCR assay at STAT1 and p21 (CDKN1A) promoters are included in Table S1.
Inducible knockdown
The inducible shMBD3 construct was constructed by insertion of MBD3 shRNA (5′-CAAGATGCTGATGAGCAAGAT-3′) into Tet-pLKO-puro (Addgene). GSCs stably expressing firefly luciferase were infected with lentivirus carrying inducible pLKO-Tet-On-shMBD3. The transduced cells were cultured in 0.1 µg/ml Dox for 7 d before cell lysates were collected to confirm inducible knockdown of MBD3. After validation of knockdown efficiency in vitro, GSCs were transplanted intracranially into athymic nude mice and supplied with water containing 2 mg/ml Dox or control water. Growth of orthotopic GBM tumors was monitored by bioluminescence imaging using the Xenogen IVIS Spectrum in vivo imaging system.
Inducible expression
To generate the inducible-expressed lentiviral construct, the CMV promoter and the copGFP on pCDH-CMV-MCS-EF1α-GreenPuro vector (System Biosciences, CD513B-1) was replaced by the TetO-mCMV inducible promoter (Addgene, 14901) and rtTA3 transactivator (Addgene 26429), separately. Then, the inducible-expressed STAT1 plasmid was constructed by insertion of STAT1 cDNA sequence into the modified tetracycline-inducible expression vector.
DNA constructs and lentiviral transfection
Human Flag-STAT1 and Flag-MBD3 were generated by PCR and cloned into the pCDH-MCS-T2A-Puro-MSCV lentiviral vector (System Biosciences). Lentiviral clones expressing MBD3 shRNA (#1, 5′-CAAGATGCTGATGAGCAAGAT-3′; #2, 5′-CCTGTGCAAAGCCTTCATGGT-3′) were prepared according to the modified protocols from Addgene (https://www.addgene.org/tools//protocols/plko/). Viral particles were produced in HEK293T cells by cotransfection of lentiviral vectors, pSPAX2, and pVSVG. Two of five shRNAs that displayed high knockdown efficiency (>80% reduction) were used for all related experiments. For knockdown of p21 (CDKN1A), we generated a vector expressing shRNA sequence (5′-CGCTCTACATCTTCTGCCTTA-3′) targeting p21 in the lentiviral pLKO.1 vector.
For gene knockout, the fragment for STAT1 gRNA (5′-GGTGGCAAATGAAACATCAT-3′) was inserted into the lentiGuide-Puro vector (Sanjana et al., 2014; Shalem et al., 2014). Lentivirus was produced in HEK293T cells by cotransfection of lentiviral vectors (lentiGuide-Puro), pSPAX2 and pVSVG. GSCs-cas9 (cas9 was integrated into GSCs with lentiCas9-Blast, a lentiviral construct that delivers hSpCas9 and blasticidin resistance) were transduced with lentivirus encoding STAT1 sgRNA and selected with 2 µg/ml puromycin.
The WWP-luc reporter with mutated STAT1 binding site was made by PCR amplification of the WWP-luc plasmid using primers 5′-TCTCCAATTCCCTCCAAGCTTCAAGCATGTGA-3′ and 5′-GAAGCTTGGAGGGAATTGGAGAGACTACCAAA-3′ followed by digestion of the template with DpnI (Thermo Fisher Scientific).
Cell viability assays
Cell viability assays were conducted by plating cells at a density of 2 × 103 cells/well in 96-well plates in triplicate. Cell titers were measured after the indicated number of days using the Cell Titer-Glo Luminescent Cell Viability Assay kit (Promega). All data were normalized to the initial reading at day 0 and presented as mean ± SD.
Luciferase assay
A total of 3 × 105 cells per well were placed in 24-well plates for cell transfection. The WWP-luc vector (p21-luciferase reporter, kindly provided by Dr. Qinong Ye, Beijing Institute of Biotechnology, Collaborative Innovation Center for Cancer Medicine, China) and pRL-TK internal control vector (Promega) were cotransfected into GSCs with STAT1 overexpression or MBD3 knocked down by using Lipofectamine 3000 (Invitrogen). After incubation for 60 h, luciferase reporter assay was performed according to the manufacturer’s instructions (Promega). The luciferase activity was normalized to pRL-TK activity.
Spectrometry analysis
For the differentially expressed protein screening between GSCs and NSTCs, cells were lysed in lysis buffer (50 mM Hepes, 6 M urea, 2 M thiourea, and 1× protease inhibitor cocktail) followed by sonication on ice. The samples were centrifugated at 14,000 g/4°C for 30 min, and the supernatant was collected. The concentration of protein was measured with the Bradford protein assay (Bio-Rad). Proteins from each sample were directly digested with trypsin using filter-aided sample preparation without being separated on gel. After digestion, the peptides were dried in a vacuum for MS analysis. Proteins were quantified by SWATH-MS (sequential window acquisition of all theoretical mass spectra), a label-free quantification approach to compare protein expression (Gillet et al., 2012). For the MBD3 binding proteins identification, GSCs were transduced with Flag-MBD3 or vector control through lentiviral infection. Cells were subjected to lysis in IP buffer (50 mM Hepes, 500 mM NaCl, 1% NP-40, and 5 mM EDTA) supplemented with protease inhibitors. The MBD3 complexes were pulled down by anti-Flag-M2 beads and separated on SDS-PAGE followed by silver staining. Gel fragments were excised, detained in 50% ethanol and 5% acetic acid, dehydrated in acetonitrile, dried in a Speed vacuum, and digested with trypsin. The peptides were extracted from the polyacrylamide and subjected to LC-MS analysis.
IP
GSCs were collected and lysed in IP buffer (50 mM Hepes, 500 mM NaCl, 1% NP-40, and 5 mM EDTA) supplemented with protease inhibitors, incubated on ice for 15 min, and followed by centrifugation at 14,000 g for 10 min at 4°C. The supernatant was subjected to IP with MBD3 antibody (Abcam, ab91458, RRID: AB_2049907) or isotype control antibody (Cell Signaling, 2779) overnight at 4°C. The precipitates were extensively washed four times with IP buffer and once with Buffer D (20 mM Hepes, 15% glycerol, 250 mM NaCl, 0.2 mM EDTA, and 0.1% NP-40), boiled with SDS loading buffer, and subjected to SDS-PAGE.
RNA isolation and real-time PCR
Cells were collected, and the total RNA was extracted with TRIzol (Sigma-Aldrich), then reversely transcribed to cDNA with PrimeScript RT Master Mix (Takara Bio). Real-time PCR was performed with SYBR Green Master Mix (Applied Biosystems) on a cycler (Applied Biosystems). The primer pairs used to detect the mRNA levels are listed in Table S2. GAPDH was used for normalization.
RNA sequencing
Total RNA was isolated from cells using Trizol (Sigma-Aldrich). Strand-specific libraries were generated using the Illumina TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold. Paired-end 125-bp reads were generated on an Illumina HiSeq 2500 instrument at the Oebiotech.corp. Reads were aligned to the GRCh38.p7 genome using TopHat v2.1.1 with the library type option set to first strand. Fragments per kilobase of transcript per million mapped reads of known genes were calculated using eXpress v1.5.1.
Statistics
All grouped data are presented as mean ± SD from studies performed at least in triplicate unless otherwise specified. For bar graphs, the unpaired Student’s t test (two-tailed) was used to compare two groups with equal variances and Welch’s t test for the comparison of two groups with unequal variances. One-way ANOVA was applied for multigroup data comparison. A probability value <0.05 was considered significant. For the survival analysis, Kaplan–Meier survival curves were analyzed by using log-rank statistics comparing the different patient or mouse groups. For the analysis of genes in glioma patients, the freely available data were downloaded from TCGA GBM, REMBRANDT gliomas, and Gravendeel gliomas database (http://gliovis.bioinfo.cnio.es). For the figures presented in box-and-whisker format, the center line represents the median, and the lower and upper limits of the box represent the 25th and 75th percentiles. The 5th to 95th percentiles are connected to the center box through the vertical lines (whiskers). Prism Software (GraphPad Software) was used for all statistical analysis.
Online supplemental material
Fig. S1 shows the expression of IFN-α/IFN-β in primary GBM tissues, the characterization of GSCs, and the expression of IRGs in GSCs or NSTCs in primary GBM tissues. Fig. S2 shows the expression and activation of STAT1 or STAT3 in GSCs or GSCs treated with type I IFNs, the impact of Flag-STAT1 on cell proliferation of GSCs, and the effects of Flag-STAT1 on p21 expression in GSCs. Fig. S3 shows the acetylation status on STAT1 promoter, the association of MBD3 and NuRD complex components, and the effects of a HDAC1 inhibitor on STAT1 expression and sensitivity of GSCs to type I IFN suppression. Fig. S4 shows the coexpression of MBD3 and stem cell markers in primary GBM tissues and xenografts, the clinical relevance of MBD3 in GBM database, and the correlation of STAT1 and MBD3 in primary GBM tissues. Fig. S5 shows the effects of MBD3 depletion on GSCs proliferation and NSTCs growth, the efficiency of MBD3 knockdown in GBM xenografts, the effects of MBD3 depletion on GSC fraction in vivo, and the correlation of MBD3 levels and survival of GBM patients. Table S1 lists the PCR primer sequences used for ChIP analysis. Table S2 lists the PCR primer sequences used for qPCR analysis.
Supplementary Material
lists primer sequences used for ChIP analysis.
lists primer sequences used for real-time qPCR analysis.
Acknowledgments
We appreciate Dr. Xiuwu Bian (Third Military Medical University, China) for the GBM PDX and Dr. Qinong Ye (Beijing Institute of Biotechnology, Collaborative Innovation Center for Cancer Medicine, China) for the WWP-luc reporter.
This research was supported by grants from the National Key Research and Development Program of China (2017YFA0505602, 2017YFC1601100, 2017YFC1601101, 2017YFC1601102, and 2017YFC1601104) and National Natural Science Foundation of China (81521064, 81872408, 81572889, 31570837, 81790252, 31430021, and 81672275).
Author contributions: J. Man, X. Zhan, and S. Guo designed the experiments, analyzed the data, and prepared the manuscript with contributions from all authors. J. Man, A. Li, and X. Zhang supervised the project and acquired funding for the study. X. Zhan, S. Guo, Y. Li, H. Ran, H. Huang, L. Mi, J. Wu, X. Wang, D. Xiao, L. Chen, D. Li, S. Zhang, and Q. Han performed the experiments. J. Man, X. Zhan, and Tingting Li performed database analyses. J. Cui, X. Yan, and Y. Yu prepared GBM specimens. J.N. Rich, S. Bao, Tao Li, T. Zhou, K. He, and X. Zhang provided scientific advice for the manuscript. S. Bao helped edit the manuscript.
References
- Abbas T., and Dutta A.. 2009. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 9:400–414. 10.1038/nrc2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera C., Nakagawa K., Sancho R., Chakraborty A., Hendrich B., and Behrens A.. 2011. c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature. 469:231–235. 10.1038/nature09607 [DOI] [PubMed] [Google Scholar]
- Alvarado A.G., Thiagarajan P.S., Mulkearns-Hubert E.E., Silver D.J., Hale J.S., Alban T.J., Turaga S.M., Jarrar A., Reizes O., Longworth M.S., et al. . 2017. Glioblastoma Cancer Stem Cells Evade Innate Immune Suppression of Self-Renewal through Reduced TLR4 Expression. Cell Stem Cell. 20:450–461.e4. 10.1016/j.stem.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badgwell B., Lesinski G.B., Magro C., Abood G., Skaf A., and Carson W. III. 2004. The antitumor effects of interferon-alpha are maintained in mice challenged with a STAT1-deficient murine melanoma cell line. J. Surg. Res. 116:129–136. 10.1016/j.jss.2003.09.005 [DOI] [PubMed] [Google Scholar]
- Bao S., Wu Q., Sathornsumetee S., Hao Y., Li Z., Hjelmeland A.B., Shi Q., McLendon R.E., Bigner D.D., and Rich J.N.. 2006. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 66:7843–7848. 10.1158/0008-5472.CAN-06-1010 [DOI] [PubMed] [Google Scholar]
- Batlle E., and Clevers H.. 2017. Cancer stem cells revisited. Nat. Med. 23:1124–1134. 10.1038/nm.4409 [DOI] [PubMed] [Google Scholar]
- Bleau A.M., Hambardzumyan D., Ozawa T., Fomchenko E.I., Huse J.T., Brennan C.W., and Holland E.C.. 2009. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell. 4:226–235. 10.1016/j.stem.2009.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celià-Terrassa T., Liu D.D., Choudhury A., Hang X., Wei Y., Zamalloa J., Alfaro-Aco R., Chakrabarti R., Jiang Y.Z., Koh B.I., et al. . 2017. Normal and cancerous mammary stem cells evade interferon-induced constraint through the miR-199a-LCOR axis. Nat. Cell Biol. 19:711–723. 10.1038/ncb3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.H., McGowan L.D., Cimino P.J., Dahiya S., Leonard J.R., Lee D.Y., and Gutmann D.H.. 2015. Mouse low-grade gliomas contain cancer stem cells with unique molecular and functional properties. Cell Rep. 10:1899–1912. 10.1016/j.celrep.2015.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Huang Z., Zhou W., Wu Q., Donnola S., Liu J.K., Fang X., Sloan A.E., Mao Y., Lathia J.D., et al. . 2013. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 153:139–152. 10.1016/j.cell.2013.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin Y.E., Kitagawa M., Su W.C., You Z.H., Iwamoto Y., and Fu X.Y.. 1996. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 272:719–722. 10.1126/science.272.5262.719 [DOI] [PubMed] [Google Scholar]
- Cui Y., Li J., Weng L., Wirbisky S.E., Freeman J.L., Liu J., Liu Q., Yuan X., and Irudayaraj J.. 2016. Regulatory landscape and clinical implication of MBD3 in human malignant glioma. Oncotarget. 7:81698–81714. 10.18632/oncotarget.13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos R.L., Tosti L., Radzisheuskaya A., Caballero I.M., Kaji K., Hendrich B., and Silva J.C.. 2014. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell. 15:102–110. 10.1016/j.stem.2014.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Cai C., Sims M., Boop F.A., Davidoff A.M., and Pfeffer L.M.. 2017. The effects of type I interferon on glioblastoma cancer stem cells. Biochem. Biophys. Res. Commun. 491:343–348. 10.1016/j.bbrc.2017.07.098 [DOI] [PubMed] [Google Scholar]
- Dunn G.P., Koebel C.M., and Schreiber R.D.. 2006. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 6:836–848. 10.1038/nri1961 [DOI] [PubMed] [Google Scholar]
- Easwaran H., Tsai H.C., and Baylin S.B.. 2014. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell. 54:716–727. 10.1016/j.molcel.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo A., Ricci-Vitiani L., Zeuner A., Pallini R., Lotti F., Sette G., Pilozzi E., Larocca L.M., Peschle C., and De Maria R.. 2006. Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ. 13:1238–1241. 10.1038/sj.cdd.4401872 [DOI] [PubMed] [Google Scholar]
- Eyler C.E., Wu Q., Yan K., MacSwords J.M., Chandler-Militello D., Misuraca K.L., Lathia J.D., Forrester M.T., Lee J., Stamler J.S., et al. . 2011. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 146:53–66. 10.1016/j.cell.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan W.A., Wu Q., Hitomi M., Rahim N., Kim Y., Sloan A.E., Weil R.J., Nakano I., Sarkaria J.N., Stringer B.W., et al. . 2013. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat. Neurosci. 16:1373–1382. 10.1038/nn.3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Shi L.Z., Zhao H., Chen J., Xiong L., He Q., Chen T., Roszik J., Bernatchez C., Woodman S.E., et al. . 2016. Loss of IFN-γ Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell. 167:397–404.e9. 10.1016/j.cell.2016.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet L.C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., and Aebersold R.. 2012. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 11:016717 10.1074/mcp.O111.016717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., and Wade P.A.. 2012. NuRD and pluripotency: a complex balancing act. Cell Stem Cell. 10:497–503. 10.1016/j.stem.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.Q., Li J.J., and Karpatkin S.. 2000. Thrombin inhibits tumor cell growth in association with up-regulation of p21(waf/cip1) and caspases via a p53-independent, STAT-1-dependent pathway. J. Biol. Chem. 275:6462–6468. 10.1074/jbc.275.9.6462 [DOI] [PubMed] [Google Scholar]
- Ivashkiv L.B., and Donlin L.T.. 2014. Regulation of type I interferon responses. Nat. Rev. Immunol. 14:36–49. 10.1038/nri3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.R., Valentine C., Basilico C., and Mansukhani A.. 1998. FGF signaling activates STAT1 and p21 and inhibits the estrogen response and proliferation of MCF-7 cells. Oncogene. 16:2647–2656. 10.1038/sj.onc.1201789 [DOI] [PubMed] [Google Scholar]
- Junttila M.R., and de Sauvage F.J.. 2013. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 501:346–354. 10.1038/nature12626 [DOI] [PubMed] [Google Scholar]
- Kaji K., Caballero I.M., MacLeod R., Nichols J., Wilson V.A., and Hendrich B.. 2006. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 8:285–292. 10.1038/ncb1372 [DOI] [PubMed] [Google Scholar]
- Katlinski K.V., Gui J., Katlinskaya Y.V., Ortiz A., Chakraborty R., Bhattacharya S., Carbone C.J., Beiting D.P., Girondo M.A., Peck A.R., et al. . 2017. Inactivation of Interferon Receptor Promotes the Establishment of Immune Privileged Tumor Microenvironment. Cancer Cell. 31:194–207. 10.1016/j.ccell.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Kim M., Woo D.H., Shin Y., Shin J., Chang N., Oh Y.T., Kim H., Rheey J., Nakano I., et al. . 2013. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 23:839–852. 10.1016/j.ccr.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacic B., Stoiber D., Moriggl R., Weisz E., Ott R.G., Kreibich R., Levy D.E., Beug H., Freissmuth M., and Sexl V.. 2006. STAT1 acts as a tumor promoter for leukemia development. Cancer Cell. 10:77–87. 10.1016/j.ccr.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Kreso A., and Dick J.E.. 2014. Evolution of the cancer stem cell model. Cell Stem Cell. 14:275–291. 10.1016/j.stem.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Lai A.Y., and Wade P.A.. 2011. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer. 11:588–596. 10.1038/nrc3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson I., Landström L.E., Larner E., Lundgren E., Miörner H., and Strannegård L.. 1978. Interferon production in glia and glioma cell lines. Infect. Immun. 22:786–789. 10.1128/IAI.22.3.786-789.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia J.D., Heddleston J.M., Venere M., and Rich J.N.. 2011. Deadly teamwork: neural cancer stem cells and the tumor microenvironment. Cell Stem Cell. 8:482–485. 10.1016/j.stem.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A., and Helin K.. 2014. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell. 14:735–751. 10.1016/j.stem.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Le Guezennec X., Vermeulen M., Brinkman A.B., Hoeijmakers W.A., Cohen A., Lasonder E., and Stunnenberg H.G.. 2006. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 26:843–851. 10.1128/MCB.26.3.843-851.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M., Pastorino S., Purow B.W., Christopher N., Zhang W., et al. . 2006. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403. 10.1016/j.ccr.2006.03.030 [DOI] [PubMed] [Google Scholar]
- Lesinski G.B., Anghelina M., Zimmerer J., Bakalakos T., Badgwell B., Parihar R., Hu Y., Becknell B., Abood G., Chaudhury A.R., et al. . 2003. The antitumor effects of IFN-alpha are abrogated in a STAT1-deficient mouse. J. Clin. Invest. 112:170–180. 10.1172/JCI16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.E., and Darnell J.E. Jr. 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3:651–662. 10.1038/nrm909 [DOI] [PubMed] [Google Scholar]
- Lim M., Xia Y., Bettegowda C., and Weller M.. 2018. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 15:422–442. 10.1038/s41571-018-0003-5 [DOI] [PubMed] [Google Scholar]
- Ludwig K., and Kornblum H.I.. 2017. Molecular markers in glioma. J. Neurooncol. 134:505–512. 10.1007/s11060-017-2379-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack S.C., Hubert C.G., Miller T.E., Taylor M.D., and Rich J.N.. 2016. An epigenetic gateway to brain tumor cell identity. Nat. Neurosci. 19:10–19. 10.1038/nn.4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man J., Yu X., Huang H., Zhou W., Xiang C., Huang H., Miele L., Liu Z., Bebek G., Bao S., and Yu J.S.. 2018. Hypoxic Induction of Vasorin Regulates Notch1 Turnover to Maintain Glioma Stem-like Cells. Cell Stem Cell. 22:104–118.e6. 10.1016/j.stem.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P., Costello I., and Hendrich B.. 2009. Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int. J. Biochem. Cell Biol. 41:108–116. 10.1016/j.biocel.2008.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menafra R., and Stunnenberg H.G.. 2014. MBD2 and MBD3: elusive functions and mechanisms. Front. Genet. 5:428 10.3389/fgene.2014.00428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura K., Natsume A., Kishida Y., Higashi H., Kondo Y., Nakasu Y., Abe T., Namba H., Wakai K., and Wakabayashi T.. 2011. Benefits of interferon-β and temozolomide combination therapy for newly diagnosed primary glioblastoma with the unmethylated MGMT promoter: A multicenter study. Cancer. 117:1721–1730. 10.1002/cncr.25637 [DOI] [PubMed] [Google Scholar]
- Parker B.S., Rautela J., and Hertzog P.J.. 2016. Antitumour actions of interferons: implications for cancer therapy. Nat. Rev. Cancer. 16:131–144. 10.1038/nrc.2016.14 [DOI] [PubMed] [Google Scholar]
- Pencheva N., de Gooijer M.C., Vis D.J., Wessels L.F.A., Würdinger T., van Tellingen O., and Bernards R.. 2017. Identification of a Druggable Pathway Controlling Glioblastoma Invasiveness. Cell Rep. 20:48–60. 10.1016/j.celrep.2017.06.036 [DOI] [PubMed] [Google Scholar]
- Qadir A.S., Ceppi P., Brockway S., Law C., Mu L., Khodarev N.N., Kim J., Zhao J.C., Putzbach W., Murmann A.E., et al. . 2017. CD95/Fas Increases Stemness in Cancer Cells by Inducing a STAT1-Dependent Type I Interferon Response. Cell Rep. 18:2373–2386. 10.1016/j.celrep.2017.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y., Zviran A., Geula S., Gafni O., Chomsky E., Viukov S., Mansour A.A., Caspi I., Krupalnik V., Zerbib M., et al. . 2013. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 502:65–70. 10.1038/nature12587 [DOI] [PubMed] [Google Scholar]
- Reynolds N., Latos P., Hynes-Allen A., Loos R., Leaford D., O’Shaughnessy A., Mosaku O., Signolet J., Brennecke P., Kalkan T., et al. . 2012a NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 10:583–594. 10.1016/j.stem.2012.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N., Salmon-Divon M., Dvinge H., Hynes-Allen A., Balasooriya G., Leaford D., Behrens A., Bertone P., and Hendrich B.. 2012b NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 31:593–605. 10.1038/emboj.2011.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saygin C., Matei D., Majeti R., Reizes O., and Lathia J.D.. 2019. Targeting Cancer Stemness in the Clinic: From Hype to Hope. Cell Stem Cell. 24:25–40. 10.1016/j.stem.2018.11.017 [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., and Zhang F.. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343:84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D., Guo C.C., Wang J., Qiu Z.K., Sai K., Yang Q.Y., Chen Y.S., Chen F.R., Wang J., Panasci L., and Chen Z.P.. 2015. Interferon-α/β enhances temozolomide activity against MGMT-positive glioma stem-like cells. Oncol. Rep. 34:2715–2721. 10.3892/or.2015.4232 [DOI] [PubMed] [Google Scholar]
- Sherry M.M., Reeves A., Wu J.K., and Cochran B.H.. 2009. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 27:2383–2392. 10.1002/stem.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silginer M., Nagy S., Happold C., Schneider H., Weller M., and Roth P.. 2017. Autocrine activation of the IFN signaling pathway may promote immune escape in glioblastoma. Neuro-oncol. 19:1338–1349. 10.1093/neuonc/nox051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvennoinen O., Ihle J.N., Schlessinger J., and Levy D.E.. 1993. Interferon-induced nuclear signalling by Jak protein tyrosine kinases. Nature. 366:583–585. 10.1038/366583a0 [DOI] [PubMed] [Google Scholar]
- Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., and Dirks P.B.. 2004. Identification of human brain tumour initiating cells. Nature. 432:396–401. 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- Son M.J., Woolard K., Nam D.H., Lee J., and Fine H.A.. 2009. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 4:440–452. 10.1016/j.stem.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., et al. National Cancer Institute of Canada Clinical Trials Group . 2009. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10:459–466. 10.1016/S1470-2045(09)70025-7 [DOI] [PubMed] [Google Scholar]
- Suvà M.L., Riggi N., and Bernstein B.E.. 2013. Epigenetic reprogramming in cancer. Science. 339:1567–1570. 10.1126/science.1230184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M., and de Tribolet N.. 1993. Recent advances in immunobiology of brain tumors. J. Neurooncol. 17:261–271. 10.1007/BF01049981 [DOI] [PubMed] [Google Scholar]
- Tsai M.H., Pai L.M., and Lee C.K.. 2019. Fine-Tuning of Type I Interferon Response by STAT3. Front. Immunol. 10:1448 10.3389/fimmu.2019.01448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venere M., Horbinski C., Crish J.F., Jin X., Vasanji A., Major J., Burrows A.C., Chang C., Prokop J., Wu Q., et al. . 2015. The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Sci. Transl. Med. 7:304ra143 10.1126/scitranslmed.aac6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen P.Y., and Kesari S.. 2008. Malignant gliomas in adults. N. Engl. J. Med. 359:492–507. 10.1056/NEJMra0708126 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Hannon G.J., Zhang H., Casso D., Kobayashi R., and Beach D.. 1993. p21 is a universal inhibitor of cyclin kinases. Nature. 366:701–704. 10.1038/366701a0 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ng H.H., Erdjument-Bromage H., Tempst P., Bird A., and Reinberg D.. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924–1935. 10.1101/gad.13.15.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Chen C., Shi Y., Wu Q., Gimple R.C., Fang X., Huang Z., Zhai K., Ke S.Q., Ping Y.F., et al. . 2017. Targeting Glioma Stem Cell-Derived Pericytes Disrupts the Blood-Tumor Barrier and Improves Chemotherapeutic Efficacy. Cell Stem Cell. 21:591–603.e4. 10.1016/j.stem.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L., Galluzzi L., Kepp O., Smyth M.J., and Kroemer G.. 2015. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 15:405–414. 10.1038/nri3845 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists primer sequences used for ChIP analysis.
lists primer sequences used for real-time qPCR analysis.