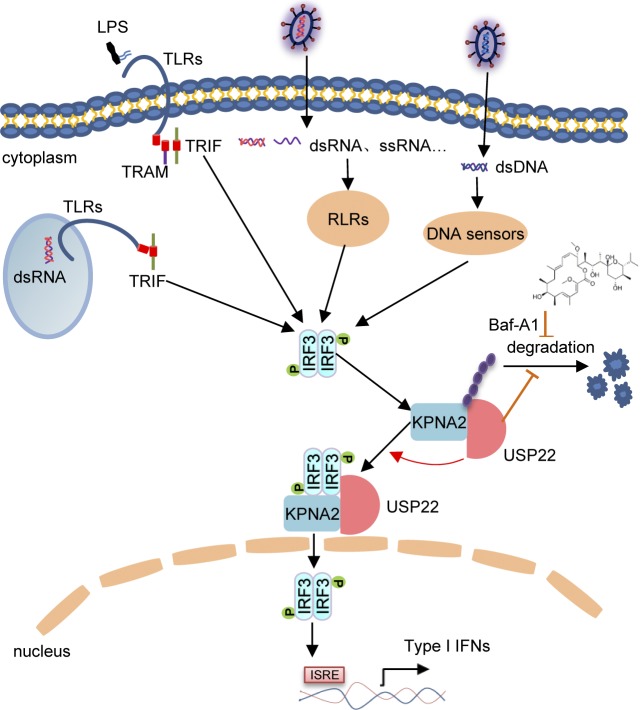
USP22 is a cytoplasmic and nuclear deubiquitinating enzyme. Cai et al. show that the cytoplasmic USP22 deubiquitinates and stabilizes the importin protein KPNA2 after viral infection, which promotes nuclear translocation of IRF3 and cellular antiviral responses.
Abstract
USP22 is a cytoplasmic and nuclear deubiquitinating enzyme, and the functions of cytoplasmic USP22 are unclear. Here, we discovered that cytoplasmic USP22 promoted nuclear translocation of IRF3 by deubiquitianting and stabilizing KPNA2 after viral infection. Viral infection induced USP22-IRF3 association in the cytoplasm in a KPNA2-depedent manner, and knockdown or knockout of USP22 or KPNA2 impaired IRF3 nuclear translocation and expression of downstream genes after viral infection. Consistently, Cre-ER Usp22fl/fl or Lyz2-Cre Usp22fl/fl mice produced decreased levels of type I IFNs after viral infection and exhibited increased susceptibility to lethal viral infection compared with the respective control littermates. Mechanistically, USP22 deubiquitinated and stabilized KPNA2 after viral infection to facilitate efficient nuclear translocation of IRF3. Reconstitution of KPNA2 into USP22 knockout cells restored virus-triggered nuclear translocation of IRF3 and cellular antiviral responses. These findings define a previously unknown function of cytoplasmic USP22 and establish a mechanistic link between USP22 and IRF3 nuclear translocation that expands potential therapeutic strategies for infectious diseases.
Graphical Abstract
Introduction
Detection of pathogen-associated molecular patterns by host pattern-recognition receptors represents the first step to initiate innate immune signaling (Akira et al., 2006). Viral nucleic acids, including RNAs and DNAs, are classical pathogen-associated molecular patterns that are recognized by TLRs, retinoic acid-inducible gene I (RIG-I)–like receptors, and cytosolic DNA sensors (Tan et al., 2018; Wu and Chen, 2014). These receptors either recruit adaptor proteins or catalyze synthesis of second messengers to directly or indirectly induce the oligomerization of adaptor proteins. For example, upon binding to double-stranded RNA, RIG-I directly interacts with the mitochondrial adaptor protein VISA (also known as MAVS, IPS-1, and CARDIF) and induces VISA to form prion-like oligomers (Hou et al., 2011; Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005), whereas cyclic AMP-GMP synthase binds to dsDNA and catalyzes synthesis of 2′-3′ cyclic AMP-GMP, which binds to the ER and mitochondrial adaptor protein MITA (also known as STING, ERIS, and MYPS) and induces oligomerization of MITA (Ishikawa and Barber, 2008; Sun et al., 2013; Sun et al., 2009; Wu et al., 2013; Zhong et al., 2008). The oligomerized adaptor proteins further recruit kinases to phosphorylate and activate transcription factors that enter into nucleus to initiate transcription of a large array of downstream genes. It has been shown that MITA recruits and activates TANK-binding kinase 1 (TBK1) to phosphorylate the transcription factor IRF3 at the C-terminal cluster of serine residues (Sharma et al., 2003; Zhang et al., 2019). Phosphorylated IRF3 undergoes dimerization and enters the nucleus to initiate transcription of downstream genes including type I IFNs and other cytokines.
Nuclear translocation of IRF3 are strictly controlled by the nuclear export sequence (NES) and the NLS (Kumar et al., 2000). Mutation of the NES of IRF3 fails to bind to the shuttling receptor CRM1 and results in a predominant localization in the nucleus before or after viral infection. Viral infection induces the binding of IRF3 to CBP/p300, which masks the NES of IRF3 and promotes the nuclear accumulation of IRF3 (Kumar et al., 2000). Two bipartite NLSs have been identified in IRF3, and mutation either of them results in failure to translocate into nucleus after viral infection (Zhu et al., 2015). More importantly, simultaneous mutation both NLS and NES of IRF3 leads to the retention in the cytoplasm with or without viral infection, indicating that the presence of a functional NLS activity plays a predominant role in the active transport and accumulation of IRF3 in the nucleus.
Macromolecules containing NLS shuttle from cytoplasm to nucleoplasm in a manner dependent on a group of importin proteins (also known as karyopherin, KPN) that form heterodimers with an α and a β subunit (known as KPNA or KPNB; Görlich and Mattaj, 1996; Nigg, 1997). The α subunit recognizes the NLS of the cargo proteins through the NLS-binding site, and binds to the β subunit through its N-terminal importin-β binding domain. The β subunit docks to the nuclear pore complex and subsequently facilitates translocation of the α subunit-cargo protein complex into nucleus (Miyamoto et al., 2016). In vitro binding assays suggest that KPNA3 and KPNA4 bind to IRF3 dependently on the NLS of IRF3 and might be responsible for the constitutive nuclear accumulation of NES-mutated IRF3 (Kumar et al., 2000). It is unclear whether these or other KPNA proteins are involved in virus-triggered nuclear translocation of IRF3 and how such a process is regulated.
Deubiquitinating enzymes (DUBs) remove poly- or mono-ubiquitin conjugates from the target proteins and thereby regulate their stability or activity (Mevissen and Komander, 2017). Ubiquitin-specific protease (USP) 22 belongs to the USP subfamily of DUBs and is implicated in placenta development of mice and cancers in humans (Kim et al., 2017; Koutelou et al., 2019; Melo-Cardenas et al., 2016). USP22 contains an NLS and shuttles between cytoplasm and nucleoplasm (Xiong et al., 2014). It has been reported that USP22 interacts with ENY2 and ATXN7L3 and forms the transcriptional coactivator Spt-Ada-Gcn5-acetyltransferase (SAGA) complex to regulate global levels of H2B monoubiquitination, thereby promoting antibody class switch recombination by facilitating nonhomologous end joining (Atanassov et al., 2016; Li et al., 2018; Ramachandran et al., 2016). Recently, USP22 has been implicated in the control of cell cycle by deubiquitination of the G1 cyclin D1 independently of the SAGA complex (Gennaro et al., 2018). The cytoplasmic functions of USP22 are unknown, and whether and how USP22 is involved in viral infection-triggered signaling remains to be investigated.
In this study, we found an essential cytoplasmic role of USP22 in virus-triggered nuclear translocation of IRF3 and cellular antiviral responses. USP22 interacted with IRF3 after viral infection in a KPNA2-dependent manner. USP22 deubiquitinated and stabilized KPNA2, which facilitated virus-triggered nuclear translocation of IRF3 and subsequent expression of downstream genes. Knockout of USP22 impaired nuclear accumulation of IRF3 after viral infection and potentiated viral replication in vitro and in vivo, which was restored by complementation of KPNA2. Collectively, these findings define a previously unknown function of cytoplasmic USP22 and establish a mechanistic link between USP22 and IRF3 nuclear translocation that expands potential therapeutic strategies for infectious diseases.
Results
USP22 is associated with IRF3 after viral infection
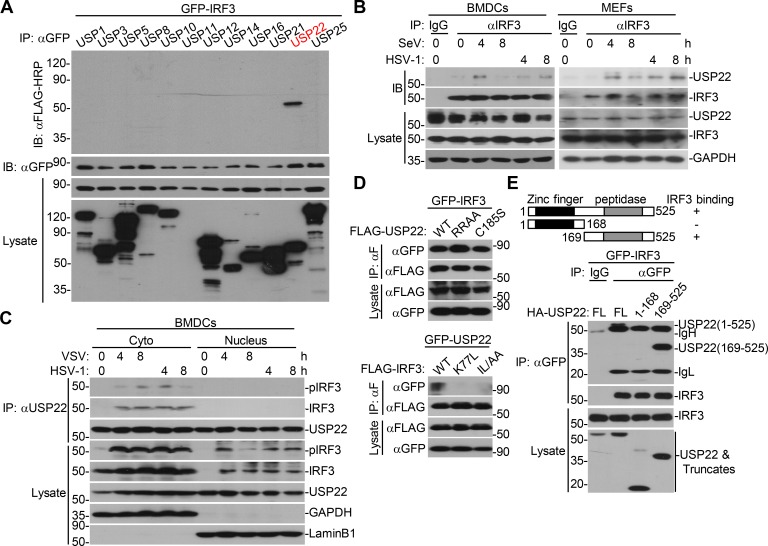
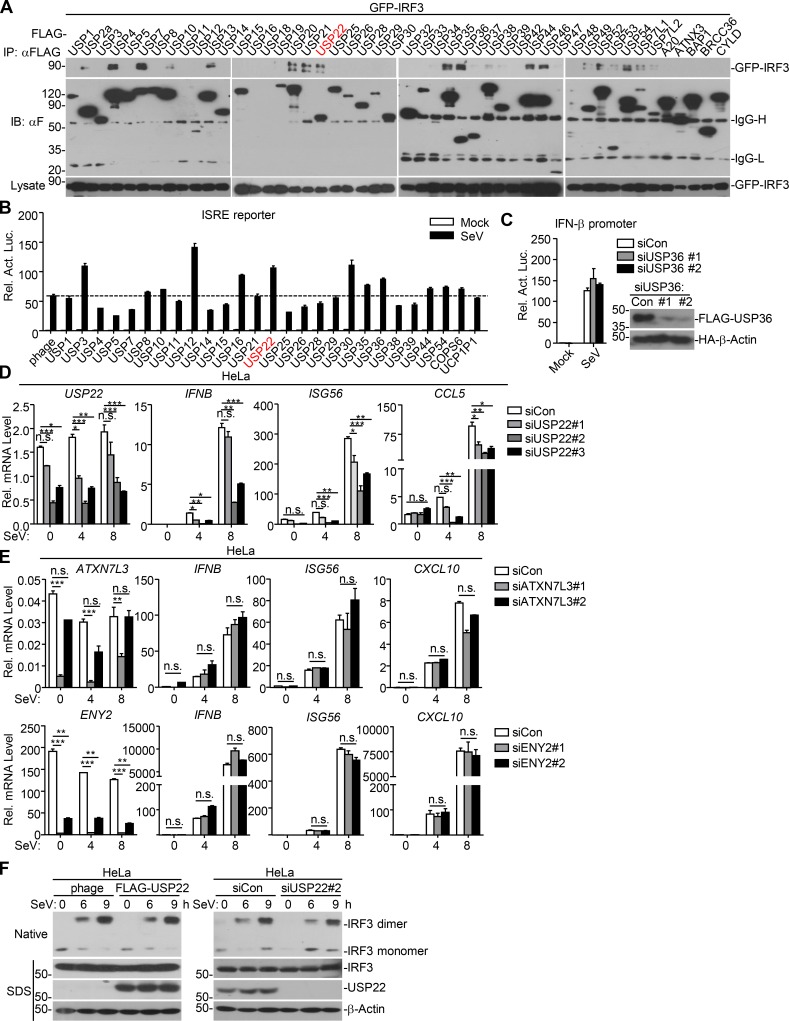
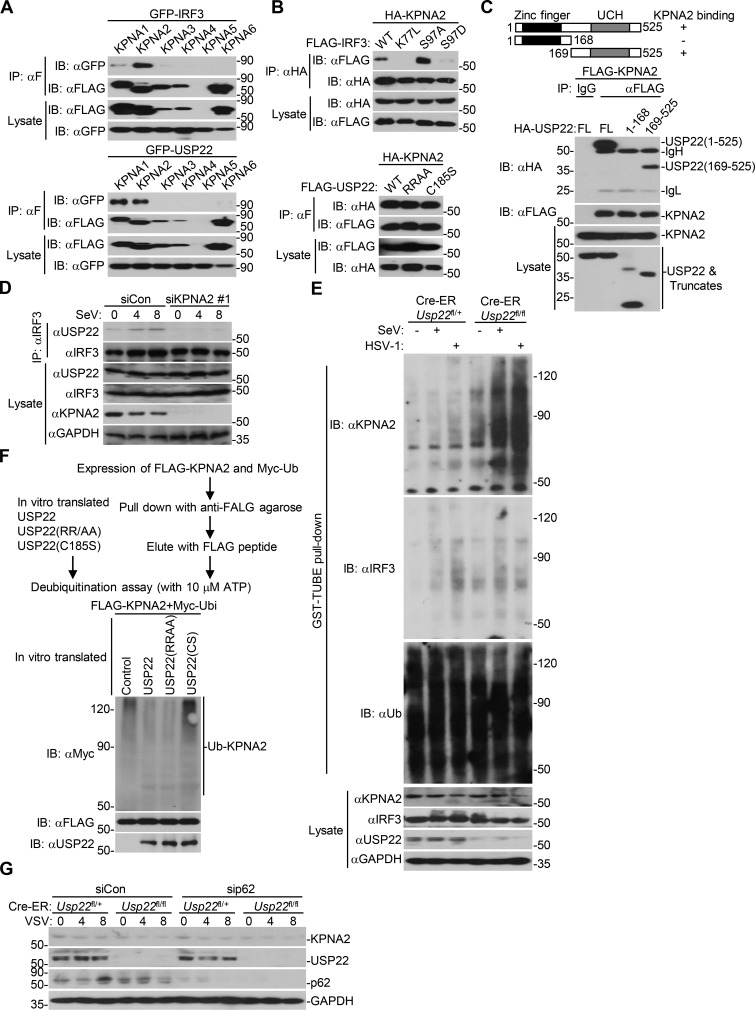
IRF3 is a transcriptional factor that mediates expression of a large array of genes after viral infection. We hypothesized that DUBs might regulate the activity or stability of IRF3 and thereby facilitate or inhibit cellular antiviral responses. To test this hypothesis, we screened the DUBs that interacted with IRF3 by cotransfection of individual FLAG-tagged DUBs and GFP-IRF3 into HEK293 cells followed by coimmunoprecipitation and immunoblot assays. This led to the identification of USP22 as an IRF3-interacting DUB in multiple screening assays (Fig. 1 A and Fig. S1 A). In a parallel reporter screening assay, USP22 activated Sendai virus (SeV)–induced activation of interferon stimulated response element (ISRE) promoter (Fig. S1 B). We have found that USP35 interacted with MITA/STING (stimulator of interferon genes) and VISA/MAVS in similar screening (Liuyu et al., 2019; Zhang et al., 2016), indicating that USP35 lacks specificity and might not target IRF3. We also found that USP36 interacted with IRF3 and potentiated SeV-induced activation of ISRE. However, loss-of-function assay with siUSP36 showed that knockdown of USP36 did not affect SeV-induced IFN-β promoter luciferase activity (Fig. S1 C). In contrast, USP22 is localized both in the cytoplasm and nucleus, and knockdown of USP22 impaired virus-triggered expression of downstream genes (described below). There is a higher possibility for USP22 than other DUBs regarding the regulation of IRF3. We thus chose USP22 for further study.
Figure 1.
USP22 interacts with IRF3 after viral infection. (A) Immunoprecipitation (IP, with anti-GFP) and immunoblot (IB, with anti-FLAG-HRP and anti-GFP and goat anti-mouse IgG[H+L] antibody) analysis of HEK293 cells that were transfected with plasmids encoding GFP-IRF3 and FLAG-tagged DUBs for 24 h. Cell lysates were analyzed by immunoblot with anti-FLAG-HRP or anti-GFP. (B) Immunoprecipitation (with control IgG or mouse anti-IRF3) and immunoblot (with rabbit anti-USP22, anti-IRF3, and goat anti-rabbit IgG[H+L] antibody) of BMDCs (upper panels) and MEFs (lower panels) that were left uninfected or infected with SeV or HSV-1 for 4–8 h. Cell lysates were analyzed by immunoblot with antibodies against the indicated proteins. (C) Immunoprecipitation (with mouse anti-USP22) and immunoblot (with rabbit anti-USP22, anti-IRF3, anti-pIRF3 and goat anti-rabbit IgG[H+L] antibody) of cytoplasmic (Cyto) and nuclear extracts from BMDCs that were left uninfected or infected with VSV or HSV-1 for 4–8 h. The cytoplasmic and nuclear extracts were analyzed by immunoblot with antibodies against the indicated proteins. (D) Immunoprecipitation analysis (with anti-FLAG) and immunoblot analysis (with anti-FLAG-HRP or anti-GFP and goat anti-mouse IgG[H+L] antibody) of HEK293 cells transfected with the indicated plasmids for 24 h. (E) Immunoprecipitation analysis (with anti-GFP) and immunoblot analysis (with anti-HA or anti-GFP and goat anti-mouse IgG[H+L] antibody) of HEK293 cells transfected with plasmids encoding GFP-IRF3 and HA-USP22 or truncations for 24 h. RRAA, USP22(RR164/165AA); ILAA, IRF3(IL139/140AA). Data are representative of three (A and B) or two (C–E) independent experiments.
Figure S1.
USP22 interacts with IRF3. (A) Immunoprecipitation (IP, with anti-GFP) and immunoblot (IB, with anti-FLAG and anti-GFP and goat anti-mouse IgG[H+L] antibody) analysis of HEK293 cells that were transfected with plasmids encoding GFP-IRF3 and FLAG-tagged DUBs for 24 h. Cell lysates were analyzed by immunoblot with anti-GFP. (B) Luciferase reporter assays analysis of ISRE activity of HEK293 cells that were transfected with plasmids encoding FLAG-tagged USPs for 24 h followed by infection with SeV for 8 h. Rel. Act. Luc., relative actvity of luciferase. (C) Luciferase reporter assays of IFN-β promoter activity of HEK293 cells that were transfected with control siRNA or siUSP36 for 36 h followed by infection with SeV for 8 h. Immunoblot analysis of FLAG-USP36 and HA-β-actin in HEK293 cells that were transfected with plasmids encoding FLAG-USP36 and HA-β-actin and control siRNA or siUSP36 for 36 h. (D) qRT-PCR analysis of IFNB, ISG56, CCL5, and USP22 mRNA in HeLa cells transfected with control siRNA (siCon) or siRNA targeting USP22 (siUSP22#1, siUSP22#2, or siUSP22#3) for 36 h followed by infection with SeV for 0–8 h. (E) qRT-PCR analysis of IFNB, ISG56, CXCL10, and ATXN7L3 or ENY2 mRNA in HeLa cells transfected with control siRNA (siCon) or siRNA targeting ATXN7L3 (siATXN7L3#1 or siATXN7L3#2) or siRNA targeting ENY2 (siENY2#1 or siENY2#2) for 36 h followed by SeV infection with for 0–8 h. (F) Immunoblot analysis of IRF3 dimer in HEK293 cells transfected with FLAG-USP22 for 24 h (left) or siUSP22#2 for 36 h (right) followed by infection with SeV for 0–9 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant (two-way ANOVA followed by Bonferroni post-test). Data are representative of two (A–C and E) or three (D and F) independent experiments (graphs show mean ± SD, n = 3).
We next examined the endogenous association between IRF3 and USP22, and immunoblot analysis revealed that USP22 interacted with IRF3 in mouse bone marrow–derived dendritic cells (BMDCs) and MEFs after SeV or HSV-1 infection (Fig. 1 B). USP22 has been reported to be localized in cytoplasm and nucleoplasm (Xiong et al., 2014). We observed that USP22 interacted with IRF3 and phosphorylated IRF3 (pIRF3) in the cytoplasm but not in the nucleus after vesicular stomatitis virus (VSV) or HSV-1 infection in BMDCs (Fig. 1 C). The N-terminal four basic amino acid residues (163-KRRK-166) are an NLS essential for the nuclear localization of USP22 (Xiong et al., 2014). Interestingly, we found that the USP22(RR164/165AA) was still associated with IRF3 (Fig. 1 D). In contrast, two IRF3 mutants, IRF3(K77L) and IRF3(IL139/140AA), which lost their NLS and NES signals and are localized in cytoplasm and nucleus, respectively, failed to associated with USP22 (Fig. 1 D). Results from domain mapping analysis suggested that the C-terminal ubiquitin peptidase domain (aa169–525) of USP22 was responsible for their association (Fig. 1 E). These data suggest that USP22 interacts with IRF3 in the cytoplasm after viral infection.
Knockdown of USP22 inhibits IRF3 nuclear accumulation in human cell lines
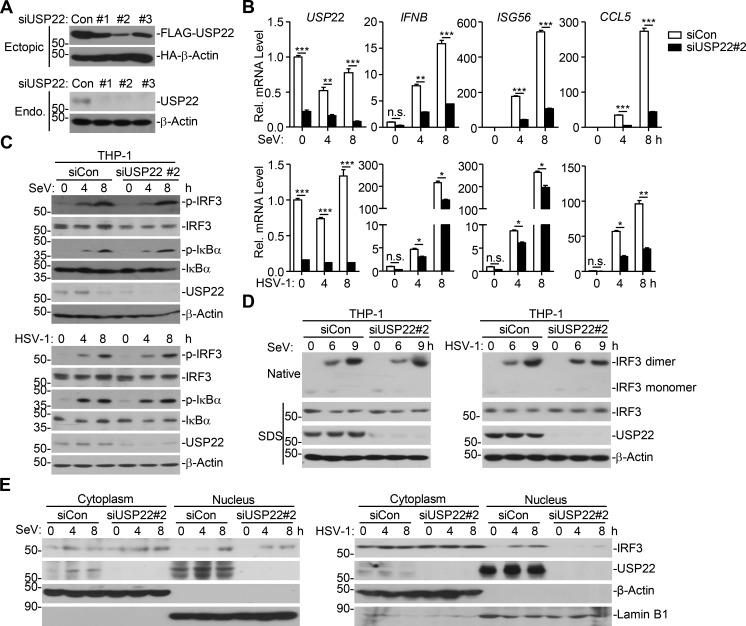
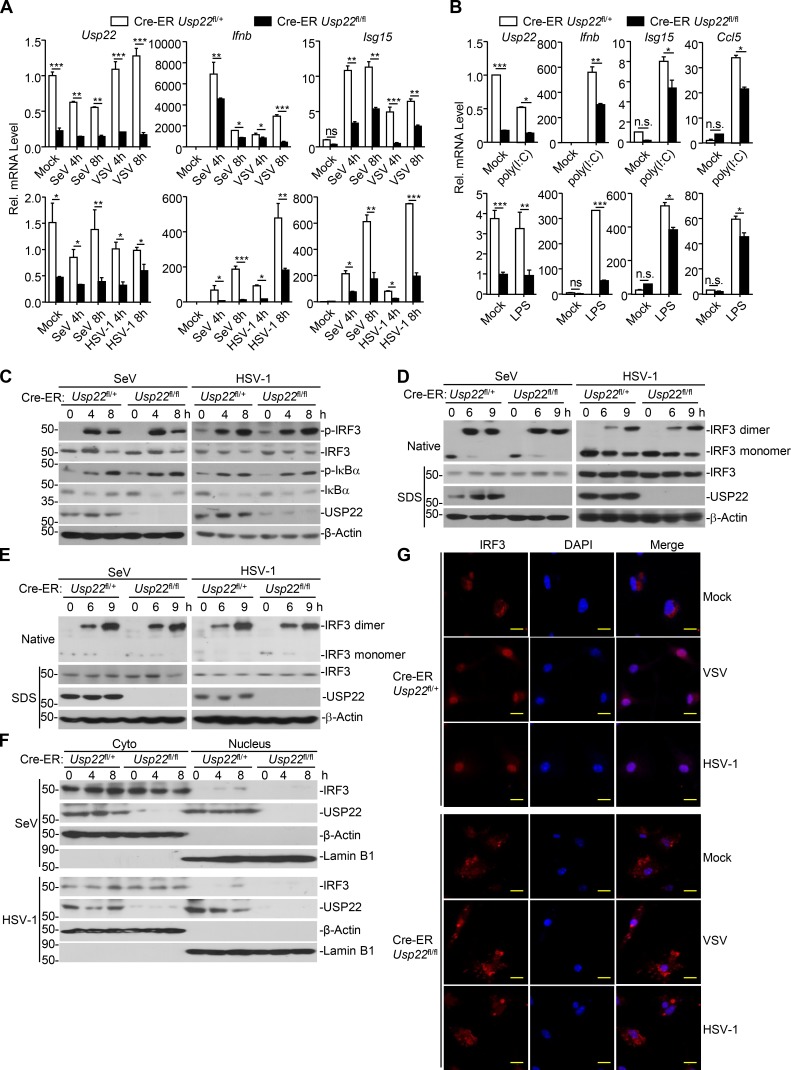
To determine whether USP22 is a physiological regulator for IRF3, we designed three siRNAs targeting USP22, two of which potently down-regulated the protein levels of ectopic and endogenous USP22 (Fig. 2 A). The #2 siRNA was used for the experiments described below, and similar results were obtained with #3 siRNA. Results from quantitative reverse transcription PCR (qRT-PCR) analysis suggested that knockdown of USP22 inhibited SeV- or HSV-1–induced expression of IFNB, ISG56, and CCL5 in THP-1 cells and SeV-induced expression of IFNB, ISG56, and CCL5 in HeLa cells (Fig. 2 B and Fig. S1 D). Previous study has shown that USP22 interacts with ENY2 and ATXN7L3 and forms the transcriptional coactivator SAGA complex to regulate global levels of H2B monoubiquitination (Atanassov et al., 2016). However, knockdown of ENY2 or ATXN7L3 had no obvious effect on SeV-induced expression of IFNB, ISG56, and CXCL10 in HeLa cells (Fig. S1 E), indicating that USP22 regulates viruses-triggered expression of downstream genes in human cell lines in a manner independent of SAGA complex formation.
Figure 2.
Knockdown of USP22 inhibits virus-triggered IRF3 nuclear accumulation. (A) Immunoblot analysis (with anti-FLAG or anti-HA) of HEK293T cells transfected for 36 h with plasmids encoding FLAG-USP22 and HA-β-actin and either USP22-targeting siRNA (#1, #2, or #3) or control siRNA (siCon; upper panels). Immunoblot analysis (with anti-USP22 or anti-β-actin) of THP-1 cells transfected with siCon siUSP22 (#1, #2, or #3; lower panels) for 36 h. (B) qRT-PCR analysis of IFNB, ISG56, CCL5, and USP22 mRNA in THP-1 cells transfected with siCon or siUSP22#2 for 36 h followed by infection with SeV or HSV-1 for 4–8 h. Rel., relative. (C and D) Immunoblot analysis of total and pIκBα and IRF3, total USP22, and β-actin (C) or IRF3 dimer (D) in THP-1 cells transfected with siCon or siUSP22#2 for 36 h followed by infection with SeV or HSV-1 for 4–8 or 6–9 h. (E) Immunoblot analysis of cytoplasmic and nuclear IRF3 in THP-1 cells in C. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and n.s., not significant (two-way ANOVA followed by Bonferroni post-test). Data are representative of four (A) or three (B–E) independent experiments (graphs show mean ± SD, n = 3).
Phosphorylation and dimerization of IRF3 are two hallmarks for the activation of IRF3 after viral infection. However, SeV- or HSV-1–induced phosphorylation of IRF3 was not impaired by knockdown of USP22 in THP-1 cells (Fig. 2 C). In addition, neither overexpression nor knockdown of USP22 affected virus-induced dimerization of IRF3 in THP-1 or HeLa cells (Fig. 2 D and Fig. S1 F). Surprisingly, instead, we found that knockdown of USP22 substantially impaired nuclear accumulation of IRF3 in THP-1 cells after SeV or HSV-1 infection (Fig. 2 E). These data together suggest that USP22 is involved in nuclear accumulation of IRF3 after viral infection.
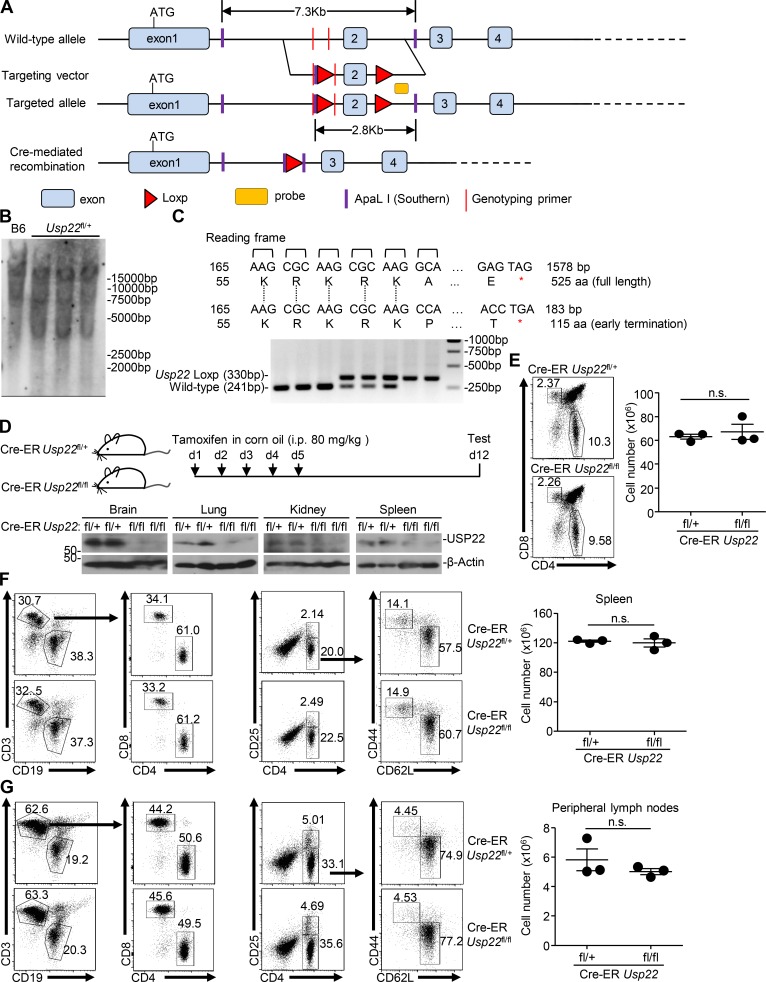
Knockout of USP22 in mouse primary cells impairs virus-triggered signaling
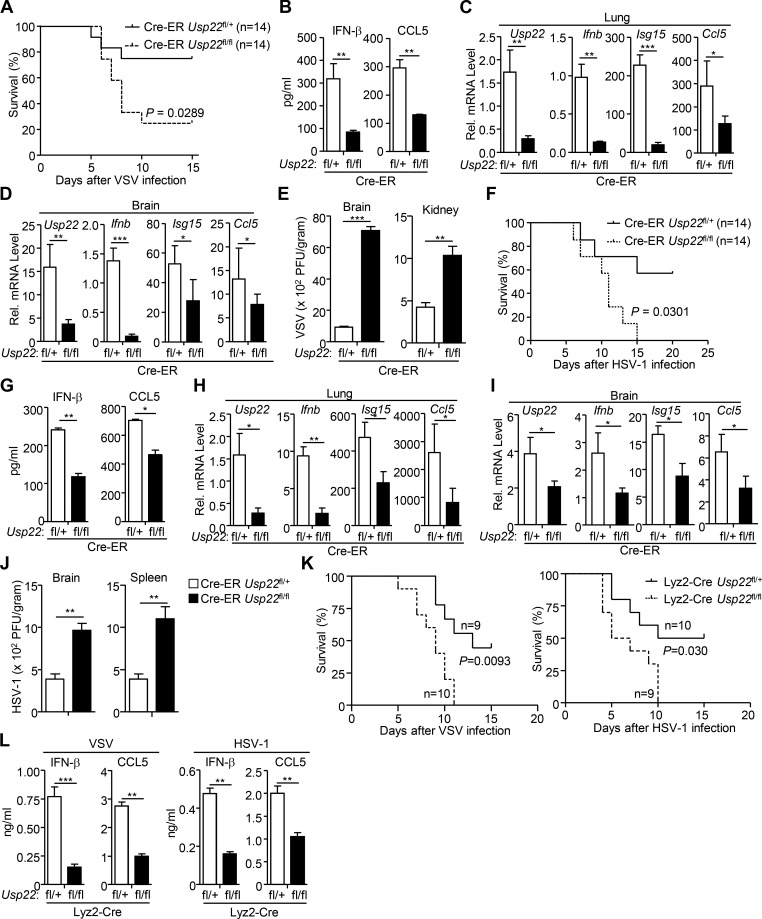
USP22 has been implicated in early embryonic development of mice and neurodegenerative diseases, and cancers in human and germline deletion of USP22 in mice leads to early embryonic defects (Lin et al., 2012). To further investigate the role of USP22 in antiviral signaling in vivo, we generated Usp22fl/+ mice by CRISPR/Cas9-mediated genome editing (Fig. 3 A). Southern blot analysis suggested that the targeting vector was successfully recombined with the wild-type allele (Fig. 3 B). Cre recombinase–mediated deletion of exon 2 with flanking loxp sites would result in an early translational termination of USP22(aa1-115) or Usp22 mRNA instability (Fig. 3 C). The Usp22fl/+ mice were crossed with ROSA26-CreERT2 mice (here referred to Cre-ER) to obtain Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl mice. Intraperitoneal injection of tamoxifen led to efficient deletion of USP22 in various organs including brain, lung, kidney, and spleen (Fig. 3 D), suggesting that the strategy to knock out USP22 in mice is reliable and successful. Results from cytometry analysis showed that the immune cell numbers or composition in the thymus, spleen, or peripheral lymph nodes was comparable between Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl mice (Fig. 3, E–G), indicating that knockout of USP22 does not affect immune cell homeostasis.
Figure 3.
Generation of Usp22fl/fl mice. (A) A scheme for CRIPSR/Cas9-mediated genome editing of the Usp22 gene locus. (B) Southern blot analysis of the F1 Usp22fl/+ mice. (C) Genotyping of Usp22+/+, Usp22fl/+, and Usp22fl/fl mice. (D) Immunoblot analysis of USP22 and β-actin in brains, lungs, kidneys, and spleens from 8-wk-old Cre-ER Usp22fl/+ (n = 2) and Cre-ER Usp22fl/fl (n = 2) mice that were intraperitoneally injected with tamoxifen for 5 successive days and rested for 7 d. (E–G) Flow cytometry analysis of thymus (E), spleen (F), and peripheral lymph nodes (G) of 8-wk-old Cre-ER Usp22fl/+ (n = 3) and Cre-ER Usp22fl/fl (n = 3) mice treated as in D. Data are representative of two independent experiments (D–G; graphs show mean ± SD). n.s., not significant.
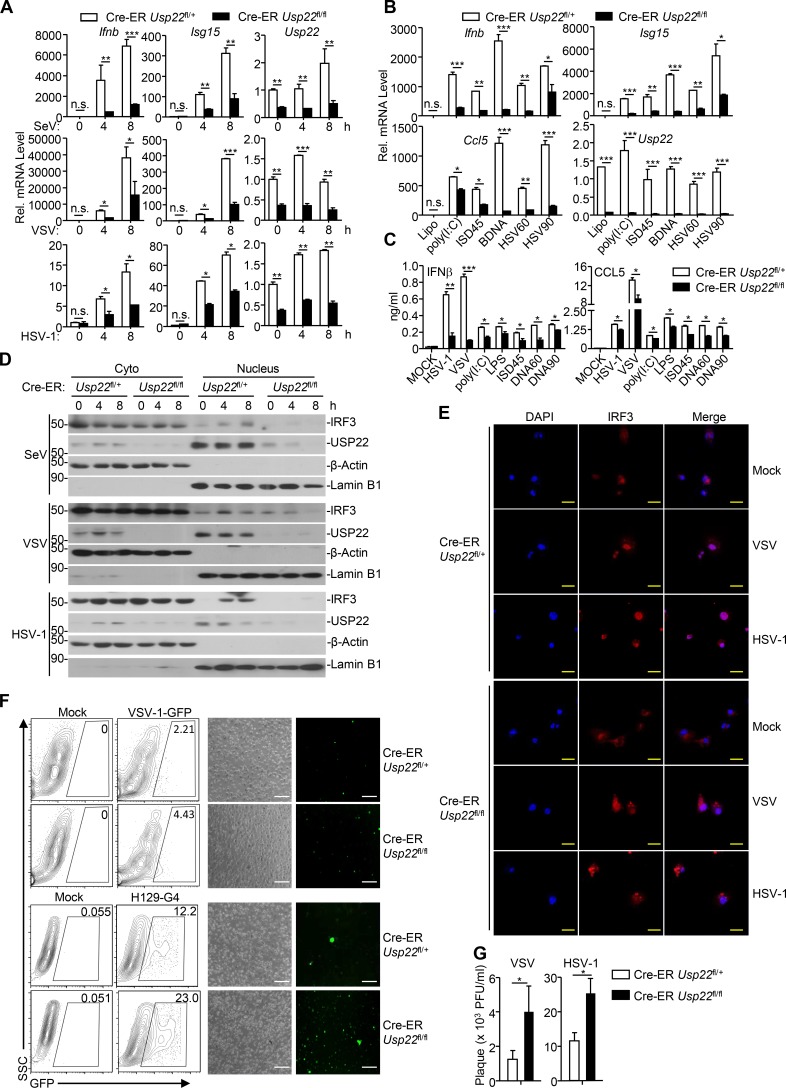
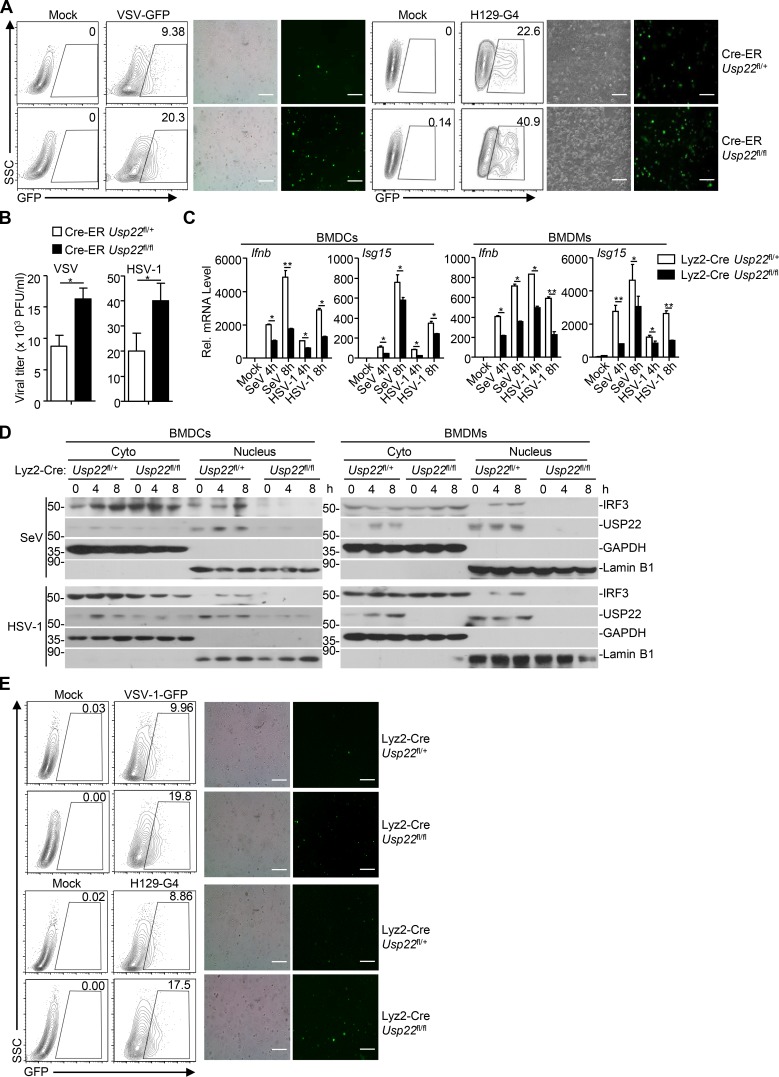
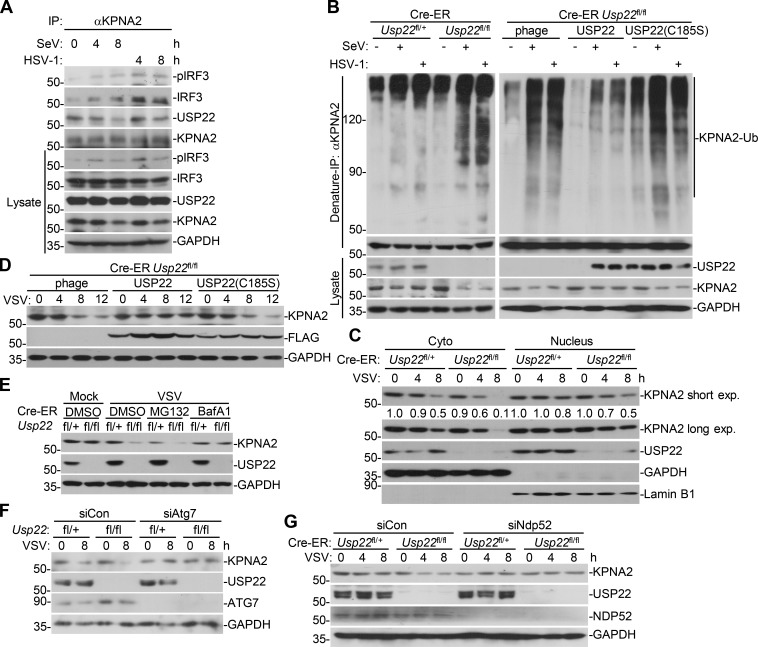
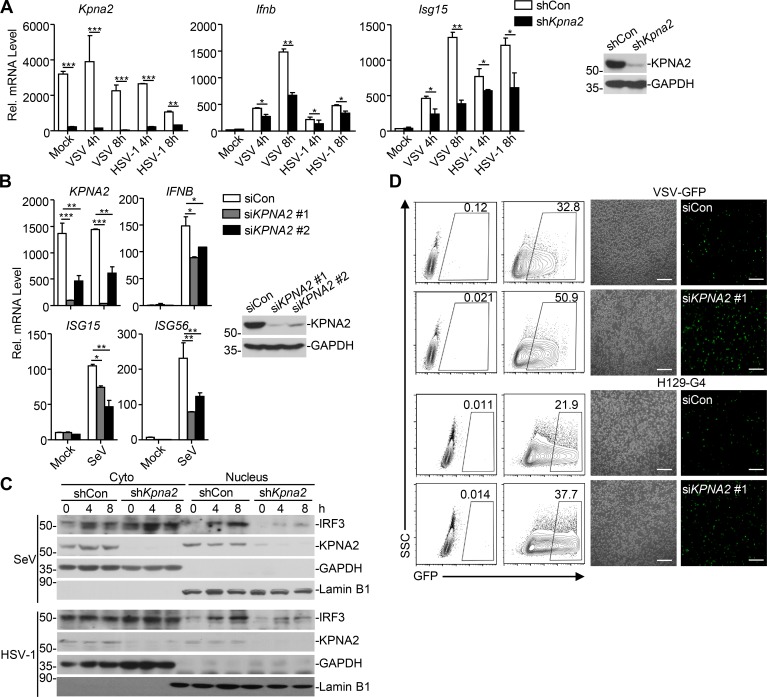
We next examined virus-triggered induction of downstream genes in various 4-hydroxytamoxifen (4-OHT)–treated Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl cells. As expected, the expression of Usp22 was almost diminished in 4-OHT–treated Cre-ER Usp22fl/fl BMDCs and bone marrow–derived macrophages (BMDMs) compared with the Cre-ER Usp22fl/+ counterparts (Fig. 4 A and Fig. S2 A). Results from qRT-PCR analysis suggested that knockout of USP22 significantly impaired SeV-, VSV-, HSV-1–, or transfected poly(I:C)-, cytoplasmic DNA-, and LPS-induced expression of Ifnb, Isg15, and Ccl5 in BMDCs and BMDMs (Fig. 4, A and B; and Fig. S2, A and B). In addition, knockout of USP22 in BMDCs substantially impaired VSV-, HSV-1–, or ligand-induced production of IFN-β and CCL5 in the supernatants of cell cultures (Fig. 4 C). Consistent with the results observed in human cell lines, SeV- or HSV-1–induced phosphorylation or dimerization of IRF3 was not affected by knockout of USP22 in BMDCs or MEFs (Fig. S2, C–E). Instead, knockout of USP22 in BMDCs or MEFs substantially inhibited the accumulation of IRF3 in the nucleus (Fig. 4, D and E; and Fig. S2, F and G), suggesting that USP22 promotes virus-triggered nuclear translocation of IRF3. Consistent with this notion, knockout of USP22 in BMDCs or MEFs significantly promoted wild-type or GFP-tagged VSV or HSV-1 replication as determined by the plaque assays and flow cytometry analyses (Fig. 4, F and G; and Fig. 5, A and B).
Figure 4.
Knockout of USP22 impairs virus-triggered IRF3 nuclear translocation. (A and B) Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl BMDCs treated with 4-OHT (1 µM) for 3 d. The cells were infected with SeV, VSV, or HSV-1 for 4–8 h (A) or transfected with poly(I:C), ISD45, BDNA, HSV60, or HSV90 for 4 h (B) followed by qRT-PCR analysis of Ifnb, Isg15, Ccl5, and Usp22 mRNA. (C) ELISA analysis of IFN-β and CCL5 in the supernatants of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl BMDCs treated as in A that were infected with VSV, HSV-1, or LPS for 12 h, or transfected with ligands for 6 h. (D) Immunoblot analysis of cytoplasmic and nuclear IRF3 in Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl BMDCs treated as in A that were infected with SeV, VSV, or HSV-1 for 0–8 h. (E) Immunofluorescence staining (with anti-IRF3) and microscopy imaging of BMDCs cells Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl BMDCs treated as in A that were infected with VSV or HSV-1 infection for 0–8 h. (F) Flow cytometry analysis (left graph) and microscopy imaging (right graph) of the replication of VSV-GFP or H129-G4 in Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl BMDCs treated as in A that were infected with VSV-GFP (multiplicity of infection [MOI] = 0.5) or H129-G4 (MOI = 1) for 1 h followed by PBS wash twice and cultured in full medium for 24 h. Numbers adjacent to the outlined areas indicate percentages of GFP+ BMDCs. (G) Plaque assay of the supernatants of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl BMDCs treated as in A that were infected with VSV (MOI = 0.5) or HSV-1 (MOI = 1) for 1 h followed by PBS wash twice and cultured in full medium for 24 h. *, P < 0.05; **, P < 0.01; and ***, P < 0.001; n.s., not significant (two-way ANOVA followed by Bonferroni post-test or two-tailed t test). Scale bars represent 10 µm (yellow bars, E) or 200 µm (white bars, F). Data are representative of three (A–C) or two (D–G) independent experiments (graphs show mean ± SD, n = 3). SSC, side scatter.
Figure S2.
Knockout of USP22 inhibits virus-triggered IRF3 nuclear translocation. (A and B) Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl BMDMs treated with 4-OHT (1 µM) for 3 d. The cells were infected with SeV, VSV, or HSV-1 for 4–8 h (A) or stimulated with poly(I:C) or LPS for 6 h (B) followed by qRT-PCR analysis of Ifnb, Isg15, Ccl5, and Usp22 mRNA. (C) Immunoblot analysis of total and pIκBα, IRF3, total USP22 and β-actin in Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl BMDMs treated as in A that were infected with SeV or HSV-1 for 4–8 h. (D and E) Immunoblot analysis of IRF3 dimer in Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl BMDMs (D) or MEFs (E) treated as in A that were infected with SeV or HSV-1 for 6–9 h. (F) Immunoblot analysis of cytoplasmic and nuclear IRF3 in Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl BMDMs treated as in A that were infected with SeV or HSV-1 for 4–8 h. (G) Immunofluorescence staining (with anti-IRF3) and microscopy imaging analysis of BMDMs treated as in A that were infected with VSV or HSV-1 infection for 0–8 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant (two-way ANOVA followed by Bonferroni post-test). Scale bars, 10 µm. Data are representative of three (A and B) or two (C–G) independent experiments (graphs show mean ± SD, n = 3).
Figure 5.
USP22 is required for optimal induction of type I INFs after viral infection. (A) Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated with 4-OHT (1 µM) for 3 d. These cells were infected with VSV-GFP (MOI = 1) or H129-G4 (MOI = 1) for 1 h followed by PBS wash twice and cultured in full medium for 24 h. The replication of VSV-GFP and H129-G4 was measured by flow cytometry analysis (left) and microscopy imaging analysis (right). (B) Plaque assay of the supernatants of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated as in A that were infected with VSV (MOI = 1) or HSV-1 (MOI = 1) for 1 h followed by PBS wash twice and cultured in full medium for 24 h. (C) Lyz2-Cre Usp22fl/+ and Lyz2-Cre Usp22fl/fl BMDCs (left two graphs) or BMDMs (right two graphs) were infected with SeV or HSV-1 for 4–8 h followed by qRT-PCR analysis of Ifnb and Isg15 mRNA. (D) Immunoblot analysis of cytoplasmic and nuclear IRF3 in Lyz2-Cre Usp22fl/+ and Lyz2-Cre Usp22fl/fl BMDCs (left panels) or BMDMs (right panels) infected with SeV or HSV-1 for 4–8 h. (E) Lyz2-Cre Usp22fl/+ and Lyz2-Cre Usp22fl/fl BMDCs were infected with VSV-GFP (MOI = 1) or H129-G4 (MOI = 1) for 1 h followed by PBS wash twice and cultured in full medium for 24 h. The replication of VSV-GFP and H129-G4 was measured by flow cytometry analysis (left) and microscopy imaging analysis (right). *, P < 0.05; **, P < 0.01 (Student’s t test or two-way ANOVA followed by Bonferroni post-test). Scale bars represent 200 µm (A and E). Data are representative of two independent experiments (graphs show mean ± SD, n = 3). SSC, side scatter.
We also obtained Lyz2-Cre Usp22fl/+ and Lyz2-Cre Usp22fl/fl mice by crossing Usp22fl/+mice and Lyz2-Cre mice and generated Lyz2-Cre Usp22fl/+ and Lyz2-Cre Usp22fl/fl BMDCs or BMDMs followed by viral infection and qRT-PCR analysis. Similar to the findings in Cre-ER Usp22fl/fl cells, Lyz2-Cre–mediated deletion of USP22 significantly impaired SeV- or HSV-1–induced expression of Ifnb and Isg15 in BMDCs (Fig. 5 C). In addition, SeV- or HSV-1–induced nuclear translocation of IRF3 was inhibited in Lyz2-Cre Usp22fl/fl BMDCs or BMDMs compared with the Lyz2-Cre Usp22fl/+ counterparts (Fig. 5 D). Consistent with these observations, the replication of VSV-GFP or H129-G4 was substantially potentiated in Lyz2-Cre Usp22fl/fl BMDCs compared with Lyz2-Cre Usp22fl/+ BMDCs as determined by the flow cytometry and fluorescent microscopy imaging (Fig. 5 E). Together, these data suggest that USP22 is essential for virus-triggered nuclear translocation of IRF3 and subsequent cellular antiviral responses.
USP22-deficient mice exhibit increased susceptibility to viral infection
To characterize the role of USP22 in RNA virus infection in vivo, we intraperitoneally injected tamoxifen (80 mg/kg) into Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl mice for 5 successive days, and 7 d later, the mice were infected with VSV or HSV-1 via tail vein or intraperitoneal injection. As shown in Fig. 6 A, the Cre-ER Usp22fl/fl mice were more susceptible to lethal VSV infection than the control littermates. Moreover, the concentrations of IFN-β and CCL5 were significantly decreased in the sera of Cre-ER Usp22fl/fl mice compared with their respective control littermates at 12 h after VSV infection (Fig. 6 B). The expression of Ifnb, Isg15, and Ccl5 was severely impaired in lungs or brains from Cre-ER Usp22fl/fl mice compared with Cre-ER Usp22fl/+ mice at 24 h or 4 d after VSV infection, respectively (Fig. 6, C and D). Consistently, the VSV titers were significantly higher in the brains or kidneys of Cre-ER Usp22fl/fl mice than in those of Cre-ER Usp22fl/+ mice at 4 d after intraperitoneal injection of VSV (Fig. 6 E). Similarly, we observed that knockout of USP22 led to increased susceptibility to lethal HSV-1 infection and decreased production of IFN-β and CCL5 in the sera after HSV-1 infection (Fig. 6, F and G). In addition, the expression of Ifnb, Isg15, and Ccl5 was severely impaired in lungs or brains from Cre-ER Usp22fl/fl mice compared with Cre-ER Usp22fl/+ mice at 24 h or 4 d after HSV-1 infection, respectively (Fig. 6, H and I). The HSV-1 titers were significantly increased in the brains or spleens by knockout of USP22 (Fig. 6 J). Similarly, deletion of USP22 by Lyz2-Cre led to increased susceptibility to lethal VSV or HSV-1 infection (Fig. 6 K) and decreased production of IFN-β and CCL5 in the sera after VSV or HSV-1 infection (Fig. 6 L). These results collectively suggest that USP22 positively regulates virus-induced expression of downstream genes and is essential for host defense against viruses in vivo.
Figure 6.
USP22-deficient mice exhibit increased susceptibility to viral infection. (A and F) Cre-ER Usp22fl/+ (n = 14) and Cre-ER Usp22fl/fl (n = 14) mice were intraperitoneally injected with tamoxifen (80 µg/g dissolved in corn oil) for 5 consecutive days and rested for 7 d followed by various experiments. Survival (Kaplan–Meier curve) analysis of these mice intravenously injected with VSV (2 × 107 PFU per mouse; A) or HSV-1 (3 × 106 PFU per mouse; F). (B and G) ELISA analysis of IFN-β and CCL5 in the sera of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl mice treated as in A and intravenously injected with VSV (2 × 107 PFU per mouse, n = 6; B) or HSV-1 (3 × 106 PFU per mouse, n = 6; G) for 12 h. (C, D, H, and I) qRT-PCR analysis of Ifnb, Isg15, Ccl5, or Usp22 mRNA in the lungs (C and H, n = 6 per group) and brains (D and I, n = 6 per group) from Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl mice treated as in A followed by intravenous injection with VSV (2 × 107 PFU per mouse) or HSV-1 (5 × 106 PFU per mouse) for 24 h (C and H) or 4 d (D and I). (E and J) Plaque assays analyzing VSV or HSV-1 titers in the brains and kidneys (E) or brains and spleens (J) from Cre-ER Usp22fl/+and Cre-ER Usp22fl/fl mice treated as in A followed by intraperitoneal injection with VSV (2 × 107 PFU per mouse, n = 6) or HSV-1 (5 × 106 PFU per mouse, n = 6) for 4 d. (K) Survival (Kaplan–Meier curve) analysis of Lyz2-Cre Usp22fl/+ (n = 9, left; n = 10, right) and Lyz2-Cre Usp22fl/fl (n = 10, left; n = 9, right) mice intravenously injected with VSV (2 × 107 PFU per mouse; left) or HSV-1 (3 × 106 PFU per mouse; right). (L) ELISA analysis of IFN-β and CCL5 in the sera of Lyz2-Cre Usp22fl/+ and Lyz2-Cre Usp22fl/fl mice intravenously injected with VSV (2 × 107 PFU per mouse, n = 5) or HSV-1 (3 × 106 PFU per mouse, n = 5) for 12 h. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (Student’s t test). Data are a combination (A, F, and K) or representative (B–E, G–J, L, and M) of two independent experiments (graphs show mean ± SD).
The enzyme activity of USP22 is required for antiviral signaling
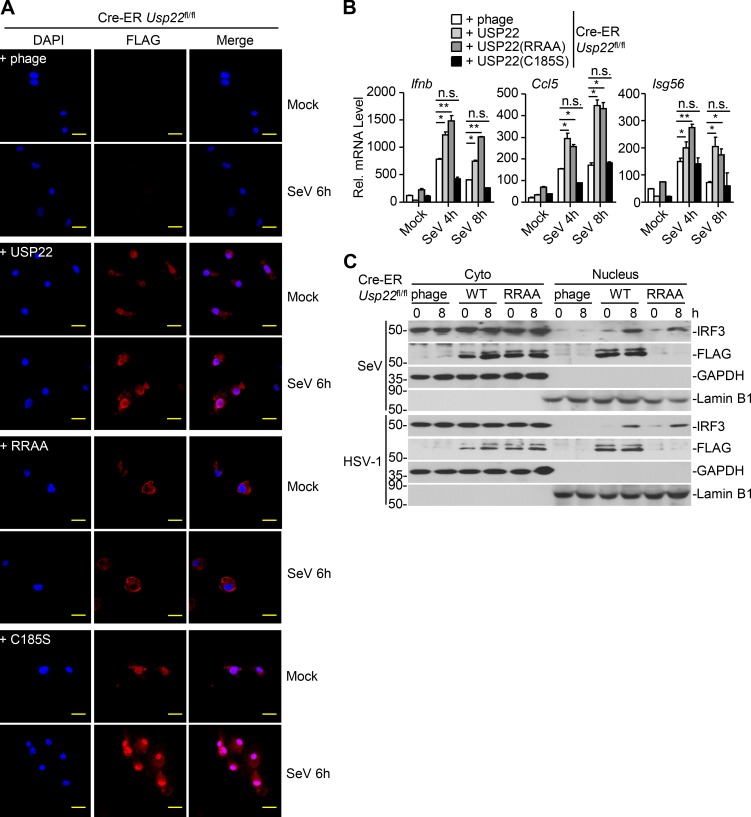
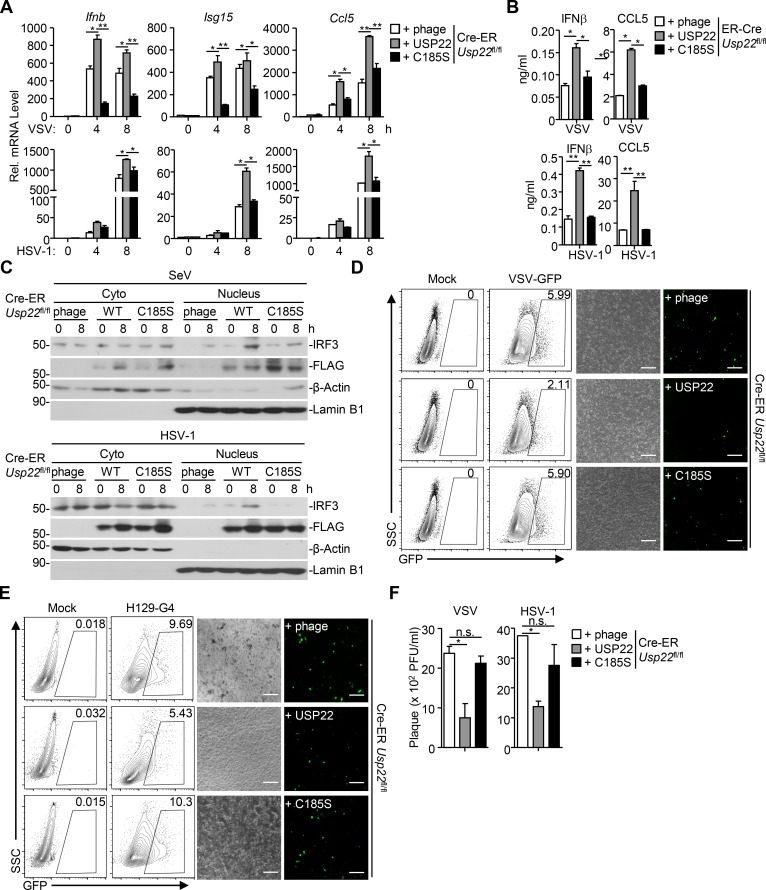
We next examined whether the nuclear localization or the deubiquitinating activity was required for USP22-mediated activation of antiviral signaling. The empty vector, USP22, the NLS mutant USP22(RR164/165AA), or the enzymatic inactive mutant USP22(C185S) was reconstituted into Cre-ER Usp22fl/fl cells followed by 4-OHT treatment and SeV, VSV, or HSV-1 infection (Fig. S3 A; Xiong et al., 2014). Results from qRT-PCR and ELISA analysis suggested that virus-induced expression of Ifnb, Isg15, Ccl5, or Isg56 and production of IFN-β and CCL5 were substantially rescued in Cre-ER Usp22fl/fl MEFs reconstituted with USP22 or USP22(RR/AA) but not in those reconstituted with USP22(C185S) (Fig. 7, A and B; and Fig. S3 B), indicating that the enzyme activity but not the nuclear localization of USP22 is required for optimal virus-triggered signaling. In addition, SeV- or HSV-1–induced nuclear translocation of IRF3 was increased by the reconstitution of USP22 or USP22(RR/AA) but not USP22(C185S) into Cre-ER Usp22fl/fl MEFs (Fig. 7 C and Fig. S2 C). Consistently, replication of VSV and HSV-1 was decreased in Cre-ER Usp22fl/fl MEFs reconstituted with USP22 but not in those reconstituted with USP22(C185S) as monitored by GFP signals or determined by plaque assays (Fig. 7, D–F). Taken together, these data suggest that USP22-mediated virus-triggered signaling requires its deubiquitinating enzymatic activity.
Figure S3.
The enzyme activity of USP22 is required for virus-triggered IRF3 nuclear translocation. (A) Immunofluorescence staining (with anti-FLAG) and microscopy imaging of Cre-ER Usp22fl/fl BMDMs treated with 4-OHT (1 µM) for 3 d that were reconstituted with the empty vector (phage), FLAG-tagged USP22, USP22 (RR164/165AA), or USP22 (C185S) followed by infection with SeV for 6 h. (B) qRT-PCR analysis of Ifnb, Ccl5, and Isg56 mRNA in Cre-ER Usp22fl/fl BMDMs treated as in A followed by infection with SeV for 0–8 h. (C) Immunoblot analysis of cytoplasmic and nuclear IRF3 in Cre-ER Usp22fl/fl MEFs treated with 4-OHT (1 µM) for 3 d that were reconstituted with the empty vector (phage), FLAG-tagged USP22, or USP22 (RR164/165AA) followed by infection with SeV or HSV-1 for 0–8 h. *, P < 0.05; **, P < 0.01 (two-way ANOVA followed by Bonferroni post-test). Scale bars, 10 µm. Data are representative of two independent experiments (graphs show mean ± SD, n = 3).
Figure 7.
USP22 functions dependently on its enzyme activity. (A) qRT-PCR analysis of Ifnb, Isg15, or Ccl5 mRNA in Cre-ER Usp22fl/fl MEFs treated with 4-OHT (1 µM) for 3 d and reconstituted with the empty vector (phage), USP22, USP22 (C185S) followed by infection with VSV, or HSV-1 for 0–8 h. (B) ELISA analysis of IFN-β and CCL5 in the supernatants of Cre-ER Usp22fl/fl MEFs treated as in A followed by infection with VSV or HSV-1 for 24 h. (C) Immunoblot analysis of cytoplasmic and nuclear IRF3 in cells obtained in A followed by infection with SeV or HSV-1 for 0–8 h. (D and E) Flow cytometry analysis (left graph) and microscopy imaging (right graph) of the replication of GFP-VSV (D) or H129-G4 (E) in cells obtained in A infected with VSV-GFP (MOI = 0.5) or HSV-1-GFP (MOI = 0.5). SSC, side scatter. (F) Plaque assays analyzing VSV or HSV-1 titers in the supernatants of cells obtained in A that were infected with VSV (MOI = 0.5) or HSV-1 (MOI = 0.5) for 24 h. *, P < 0.05; **, P < 0.01; n.s., no significant (two-way ANOVA followed by Bonferroni post-test). Scale bars represent 200 µm (D and E). Data are representative of two independent experiments (graphs show mean ± SD, n = 3).
USP22 functions at IRF3 level but does not target IRF3 for deubiquitination
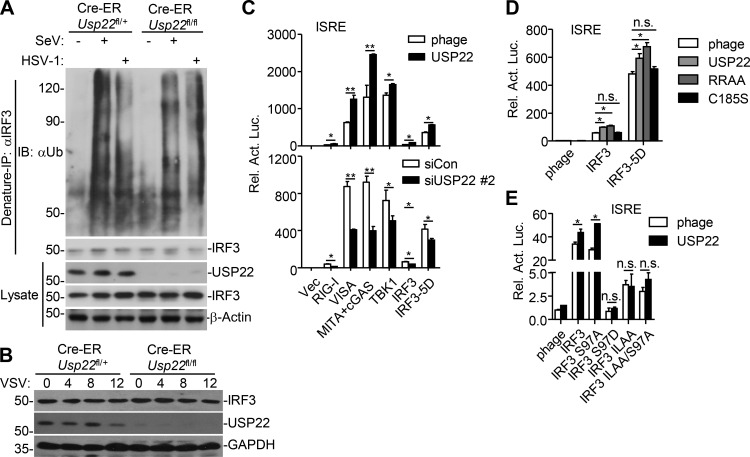
Because USP22-mediated virus-triggered signaling requires its enzyme activity and USP22 interacts with IRF3, we hypothesized that USP22 might catalyze deubiquitination of IRF3 and regulate the nuclear translocation of IRF3. However, we found that SeV- or HSV-1-induced ubiquitination of IRF3 was comparable between 4-OHT-treated Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs (Fig. S4 A). In addition, knockout of USP22 had minimal effect on the stability of IRF3 after VSV infection (Fig. S4 B). Overexpression or knockdown of USP22 potentiated or inhibited IRF3(5D)-mediated activation of the ISRE reporter, respectively, and overexpression of USP22(RR/AA) but not USP22(C185S) promoted IRF3- or IRF3(5D)-mediated activation of ISRE (Fig. S4, C and D; Lin et al., 1998), indicating a predominant role of USP22 enzyme activity in the cytoplasm for promoting the virus-triggered signaling. Previous study has shown that phosphatase and tensin homolog (PTEN)–mediated dephosphorylation of IRF3 at Ser97 facilitates its nuclear translocation (Li et al., 2016). We found that overexpression of USP22 potentiated IRF3(S97A)- but not IRF3(S97D)-, IRF3(IL139/140AA)- or IRF3(IL139/140AA, S97A)-mediated activation of ISRE (Fig. S4 E). These data together suggest that USP22 facilitates virus-induced nuclear translocation of IRF3 in the cytoplasm in a manner dependent on its enzyme activity.
Figure S4.
USP22 does not target IRF3 for deubiquitination. (A) Denature-IP (with rabbit anti-IRF3) and immunoblot analysis (with mouse anti-ubiquitin, anti-IRF3, anti-β-actin, or rabbit anti-USP22, goat anti-rabbit and anti-mouse IgG[H+L] antibody) of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated with 4-OHT for 3 d followed by infection with SeV or HSV-1 for 8 h. (B) Immunoblot analysis (with anti-IRF3, anti-USP22, or anti-GAPDH) of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated as in A followed by infection with VSV for 0–12 h. (C) Luciferase reporter assays analyzing ISRE activity of HEK293 cells transfected with plasmids encoding HA-tagged RIG-I, VISA, MITA, cyclic AMP-GMP synthase, TBK1, IRF3, IRF3-5D, and FLAG-tagged USP22 (upper) for 24 h or siUSP22#2 (lower) for 36 h. (D) Luciferase reporter assays analyzing ISRE activity of HEK293 cells transfected with plasmids encoding IRF3 or IRF3-5D and the empty vector (phage), USP22, USP22 (RR164/165AA), or USP22 (C185S) for 24 h. (E) Luciferase reporter assays analyzing ISRE promoter activity of HEK293 cells transfected with plasmids encoding the empty vector (phage) or USP22 and IRF3 or the indicated IRF3 mutants for 24 h. *, P < 0.05; **, P < 0.01; ns, not significant (two-way ANOVA followed by Bonferroni post-test). Data are representative of two (A and B) or three (C–E) independent experiments (graphs show mean ± SD, n = 3).
USP22 deubiquitinates and stabilizes KPNA2
Because USP22 does not directly target IRF3, we hypothesized that USP22 might directly target an intermediate protein for deubiquitination to regulate nuclear translocation of IRF3. Such an intermediate protein should associate with both IRF3 and USP22 and promote virus-triggered signaling by facilitating the nuclear translocation of IRF3. KPNA proteins are shuttling receptors for NLS and play essential roles in nuclear translocation of the NLS-containing proteins (Görlich and Mattaj, 1996; Miyamoto et al., 2016; Nigg, 1997). We first examined the associations between KPNAs and USP22 or IRF3 and found that KPNA2 was precipitated with IRF3 and USP22 (Fig. S5 A). Mutation of the NLS of IRF3 (IRF3-K77L) failed to be precipitated with KPNA2 (Fig. S5 B), indicating that the IRF3-KPNA2 association depends on the NLS of IRF3. Interestingly, IRF3(S97A) and IRF3(S97D) exhibited increased and decreased binding to KPNA2, respectively (Fig. S5 B), indicating that PTEN-mediated dephosphorylation of IRF3 at Ser97 modulates its association with KPNA2. In contrast, mutation of USP22 NLS (USP22-RR/AA) did not affect its association with KPNA2 (Fig. S5 B), suggesting that USP22 interacts with KPNA2 independently of its NLS. Consistent with this notion, results from domain mapping analysis suggested that the C-terminal peptidase domain but not the N-terminal NLS domain of USP22 was associated with KPNA2 (Fig. S5 C).
Figure S5.
USP22 interacts with and deubiquitinates KPNA2. (A) Immunoprecipitation (IP, with anti-FLAG) and immunoblot (IB, with anti-FLAG, anti-GFP, and goat anti-mouse IgG[H+L] antibody) analysis of HEK293 cells that were transfected with plasmids encoding FLAG-tagged KPNAs and GFP-IRF3 (upper panels) or GFP-USP22 (lower panels) for 24 h. Cell lysates were analyzed by immunoblot with anti-FLAG or anti-GFP. (B) Immunoprecipitation (with anti-FLAG) and immunoblot (with anti-FLAG-HRP and anti-GFP and goat anti-mouse IgG[H+L] antibody) analysis of HEK293 cells that were transfected with plasmids encoding HA-KPNA2 and FLAG-tagged IRF3 or IRF3 mutants (upper panels) or FLAG-tagged USP22 or USP22 mutants (lower panels) for 24 h. Cell lysates were analyzed by immunoblot with anti-FLAG or anti-HA. (C) Immunoprecipitation (with anti-FLAG) and immunoblot (with anti-FLAG, anti-HA, and goat anti-mouse IgG[H+L] antibody) analysis of HEK293 cells that were transfected with plasmids encoding FLAG-tagged KPNA2 and HA-tagged USP22 or USP22 truncations for 24 h. Cell lysates were analyzed by immunoblot with anti-FLAG-HRP or anti-HA. UCH, ubiquitin carboxyl-terminal hydrolase. (D) Immunoprecipitation (with mouse anti-IRF3) and immunoblot (with rabbit anti-IRF3, anti-USP22, and goat anti-rabbit and anti-mouse IgG[H+L] antibody) analysis of MEFs that were transfected with control shCon or shKpna2 followed by infection with SeV for 0–8 h. Cell lysates were analyzed by immunoblot with anti-IRF3, anti-USP22, and anti-KPNA2. (E) GST-TUBE pull-down assay (with GST beats and TUBE) and immunoblot analysis (with anti-ubiquitin, anti-KPNA2, anti-USP22, anti-IRF3, or anti-GAPDH) of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated as in D followed by infection with SeV or HSV-1 for 8 h. (F) In vitro deubiquitination analysis of ubiquitin-modified KPNA2 eluted from anti-FLAG precipitates by FLAG peptide of HEK293 cells transfected with FLAG-KPNA2 and Myc-ubiquitin incubated with in vitro–generated USP22, USP22 (RR164/165AA), or USP22 (C185S) obtained from an in vitro transcription and translation kit. (G) Immunoblot analysis of KPNA2, USP22, p62, and GAPDH of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated with 4-OHT for 3 d and transfected with sip62 or siNDP52 for 36 h followed by infection with VSV for 0–8 h. Data are representative of two independent experiments.
Results from endogenous immunoprecipitation assays suggest that KPNA2 interacted with USP22 constitutively but interacted with IRF3 or pIRF3 in THP-1 cells in a manner dependent on viral infection (Fig. 8 A). Knockdown of KPNA2 impaired virus-induced USP22-IRF3 association in MEFs (Fig. S5 D), indicating that USP22 interacts with IRF3 through KPNA2 after viral infection. We next examined ubiquitination of KPNA2 in USP22-sufficient or -deficient MEFs in the presence or absence of viral infection. The results showed that virus-induced ubiquitination of KPNA2 was substantially potentiated by knockout of USP22, and reconstitution of USP22 but not USP22(C185S) into USP22-deficient MEFs diminished the ubiquitination of KPNA2 (Fig. 8 B and Fig. S5 E). In addition, USP22 or USP22(RR164/165AA) but not USP22(C185S) deubiquitinated KPNA2 in vitro (Fig. S5 F), indicating that USP22 directly catalyzes deubiquitination of KPNA2. As expected, the half-life of KPNA2 was shorter in USP22-deficient MEFs than in USP22-sufficient MEFs after VSV infection, which was reversed by reconstitution of USP22 but not USP22(C185S) (Fig. 8, C and D). The accelerated degradation of KPNA2 in USP22 knockout MEFs was fully rescued by Bafilomycin A1 (BafA1) or knockdown of ATG7 but not by MG132 (Fig. 8, E and F; Ohsumi, 2014), indicating that USP22 deubiquitinates and protects KPNA2 from autophagy-dependent but proteasome-independent degradation after viral infection. NDP52 and p62 are important selective autophagy receptors that recognize and transport polyubiquitin-modified targets to autophagosome (Thurston et al., 2009; Zheng et al., 2009). Interestingly, knockdown of NDP52 but not p62 completely inhibited virus-triggered degradation of KPNA2 in USP22-deficient MEFs (Fig. 8 G and Fig. S5 G). These data together support the notion that USP22-mediated deubiquitination of KPNA2 inhibits the degradation of KPNA2 through the NDP52-mediated selective autophagy pathway.
Figure 8.
USP22 deubiquitinates and stabilizes KPNA2. (A) Immunoprecipitation (IP, with rabbit anti-KPNA2) and immunoblot (with mouse anti-USP22, anti-IRF3, and rabbit anti-KPNA2 or anti-pIRF3, goat anti-rabbit, and anti-mouse IgG[H+L] antibody) of THP-1 cells that were left uninfected or infected with SeV or HSV-1 for 4–8 h. Cell lysates were analyzed by immunoblot with antibodies against the indicated proteins. (B) Denature-IP (with anti-KPNA2) and immunoblot analysis (with anti-Ub, anti-KPNA2, anti-USP22 or anti-GAPDH) of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated with 4-OHT (1 µM) for 3 d and reconstituted with empty vector (phage), USP22, or USP22(C185S) followed by infection with SeV or HSV-1 for 0–8 h. (C) Immunoblot analysis of KPNA2, USP22, GAPDH, and Lamin B1 in the cytoplasmic (30 µg) and nuclear (45 µg) extracts of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated as in B followed by infection with VSV for 0–8 h. (D) Immunoblot analysis of KPNA2, FLAG-USP22, and GAPDH in Cre-ER Usp22fl/fl MEFs treated as in B followed by infection with VSV for 0–12 h. (E) Immunoblot analysis of KPNA2, USP22, and GAPDH in Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated as in B followed by infection with VSV for 12 h in the presence of MG132 or BafA1 for 11 h. (F and G) Immunoblot analysis of KPNA2, USP22, ATG7, and GAPDH of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated with 4-OHT for 3 d and transfected with siATG7 (F) or siNDP52 (G) for 36 h followed by infection with VSV for 8 h. Data are representative of two independent experiments.
KPNA2 mediates virus-triggered nuclear translocation of IRF3
We next examined the effect of KPNA2 on virus-triggered signaling and nuclear translocation of IRF3 and found that knockdown of KPNA2 significantly inhibited VSV- or HSV-1–induced expression of Ifnb and Isg15 in primary MEFs or in THP-1 cells (Fig. 9, A and B). As expected, knockdown of KPNA2 substantially impaired nuclear translocation of IRF3 in MEFs after SeV or HSV-1 infection and promoted the replication of VSV or HSV-1 in THP-1 cells (Fig. 9, C and D). These data suggest that KPNA2 is essential for virus-induced nuclear translocation of IRF3 and cellular antiviral responses.
Figure 9.
Knockdown of KPNA2 inhibits virus-triggered IRF3 nuclear translocation. (A) qRT-PCR of Ifnb, Isg15, or Kpna2 mRNA (left graphs) and immunoblot analysis (with anti-KPNA2 or anti-GAPDH; right panels) of MEFs stably transfected with an empty shRNA vector (shCon), or shRNA targeting KPNA2 (shKpna2). (B) qRT-PCR of IFNB, ISG15, ISG56, or KPNA2 mRNA (left graphs) and immunoblot analysis (with anti-KPNA2 or anti-GAPDH; right panels) of THP-1 cells transfected with an empty siRNA vector (siCon), or siRNA targeting KPNA2 (siKPNA2 #1 or #2). (C) Immunoblot analysis of cytoplasmic and nuclear IRF3 in THP-1 cells obtained in A that were infected with SeV or HSV-1 for 0–8 h. (D) Flow cytometry analysis (left) and microscopy imaging (right) of THP-1 cells obtained in B that were infected with VSV-GFP or H129-G4 for 24 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-way ANOVA followed by Bonferroni post-test). Scale bars represent 200 µm (D). Data are representative of three (A and B) or two (C and D) independent experiments (graphs show mean ± SD, n = 3). SSC, side scatter.
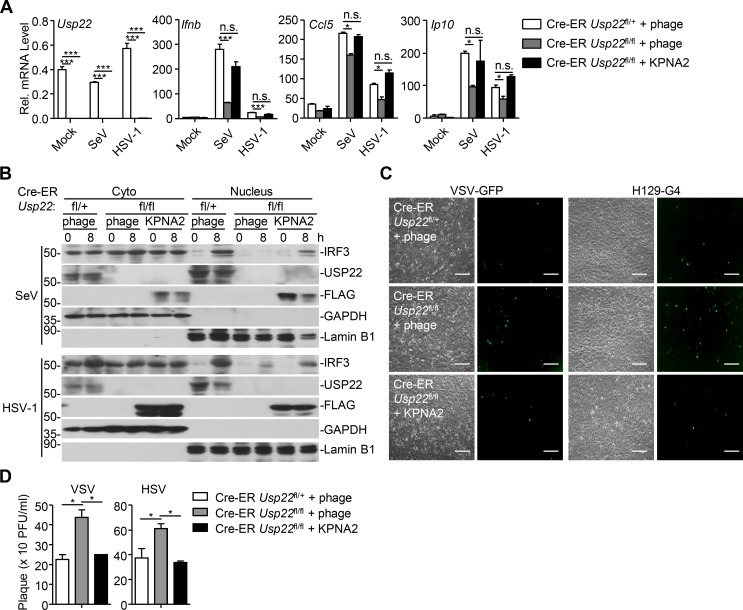
Because USP22 regulates the stability of KPNA2, which facilitates nuclear translocation of IRF3, we speculated that complementation of KPNA2 into USP22-deficient cells would restore cellular antiviral responses. As expected, reconstitution of KPNA2 into USP22 knockout MEFs restored the expression of Ifnb and Ccl5 and the nuclear translocation of IRF3 after SeV or HSV-1 infection (Fig. 10, A and B). Consistently, the replication of VSV and HSV-1 was significantly inhibited in USP22 knockout cells reconstituted with KPNA2 but not in those reconstituted with empty vector (Fig. 10, C and D). Together, these data suggest that USP22 promotes virus-induced nuclear translocation of IRF3 by deubiquitinating and stabilizing KPNA2.
Figure 10.
Reconstitution of KPNA2 into USP22 knockout cells restores virus-triggered IRF3 nuclear translocation. (A) qRT-PCR analysis of Ifnb, Ccl5, or Usp22 mRNA in Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated with 4-OHT (1 µM) for 3 d and reconstituted with empty vector (phage) or KPNA2 followed by infection with SeV or HSV-1 for 0–8 h. (B) Immunoblot analysis of cytoplasmic and nuclear IRF3 in Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated as in A followed by infection with SeV or HSV-1 for 0–8 h. (C) Microscopy imaging of the replication of GFP-VSV or H129-G4 in cells obtained in A infected with VSV-GFP (MOI = 0.5) or HSV-1-GFP (MOI = 0.5). (D) Plaque assay of the supernatants of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl MEFs treated as in A followed by infection with VSV (MOI = 0.5) or HSV-1 (MOI = 0.5) for 1 h followed by twice PBS wash and cultured in full medium for 24 h. *, P < 0.05; ***, P < 0.001; n.s., not significant (two-way ANOVA followed by Bonferroni post-test). Scale bars represent 200 µm (C). Data are representative of three (A) or two (B–D) independent experiments (graphs show mean ± SD, n = 3).
Discussion
USP22 is located both in the cytoplasm and the nucleus. Previous studies have reported essential roles of the nucleic USP22 in deconjugation of ubiquitin from monoubiquitin-modified H2B and polyubiquitin-modified CCND1 to regulate DNA damage repair and cell cycle progression, respectively (Atanassov et al., 2016; Gennaro et al., 2018; Ramachandran et al., 2016). In addition, it has been reported that USP22 interacts strongly with and functions at the level of IRF3 (Liu et al., 2018b). It is thus enigmatic that USP22 catalyzes the removal of K27-linked ubiquitin chains from MITA/STING in a transient transfection and overexpression artificial system, though USP22 interacts weakly with MITA/STING in the same system (Liu et al., 2018b). In this study, by using multiple USP22 knockout mouse models or cells, we identified a previously uncharacterized role of the cytoplasmic USP22 in facilitating virus-induced nuclear translocation of IRF3. Knockout of USP22 impaired virus-induced nuclear translocation of IRF3 without affecting its phosphorylation (at Sre396) or dimerization. Reconstitution of USP22(RR164/165AA) but not USP22(C185S) restored the expression of type I IFNs after viral infection, suggesting that the cytoplasmic USP22 was sufficient to support cellular antiviral responses. In this context, we observed that IRF3(IL139/140AA), which is predominantly located in the nucleus, did not interact with USP22, and USP22(RR164/165AA) interacted with IRF3, as did wild-type USP22. Together, these findings suggest an essential role of cytoplasmic USP22 in promoting IRF3 nuclear translocation after viral infection.
USP22 is a DUB, and its enzyme activity is critical for its function in deubiquitinating H2B and CCND1 (Atanassov et al., 2016; Gennaro et al., 2018). We also found that the enzyme activity of USP22 was required for its promotion of IRF3 nuclear translocation after viral infection. Although IRF3 is extensively modified by ubiquitination (Lin and Zhong, 2015; Li and Zhong, 2018), USP22 deficiency did not affect the ubiquitination or stability of IRF3 in the presence or absence of viral infection, indicating that USP22 might not directly target IRF3 for deubiquitination to facilitate its nuclear translocation. Instead, we found that the protein levels of KPNA2 were substantially decreased and the ubiquitination of KPNA2 was potentiated in USP22 knockout cells compared with USP22-sufficient cells after viral infection. Reconstitution of wild-type USP22 but not USP22(C185S) into USP22 knockout cells diminished the ubiquitination of and stabilized KPNA2. The down-regulation of KPNA2 in USP22 knockout cells was rescued by the autophagy inhibitor BafA1 but not the proteasome inhibitor MG132, indicating that USP22 protects KPNA2 from degradation in a manner independent of the proteasome pathway. The type(s) of polyubiquitin chains on KPNA2 targeted by USP22 and how such a regulation is linked to autophagy-mediated degradation require further investigations.
It is acknowledged that viral infection–induced activation of IRF3 involves at least three sequential steps, i.e., phosphorylation, dimerization, and translocation from cytoplasm into nucleus. While the phosphorylation and dimerization of IRF3 have been extensively studied, the mechanism and regulation of nuclear translocation of IRF3 are less clear. Although KPNA3 and PKNA4 have been reported to bind to IRF3 in vitro (Kumar et al., 2000), we observed that KPNA2 but not KPNA3 or KPNA4 was associated with IRF3 in HEK293 cells in our transient transfection and coimmunoprecipitation assays. Intriguingly, it has been observed that p65 differentially binds to KPNA2 or KPNA3/4 in vivo and in vitro in a fashion similar to IRF3 (discussed below; Liang et al., 2013). In addition, IRF3(K77L), which lost its NLS, failed to interact with KPNA2, indicating that IRF3 interacts with KPNA2 via its NLS. A recent study has shown that PTEN-mediated dephosphorylation of IRF3 at Ser97 is critical for its nuclear translocation (Li et al., 2016). Interestingly, we found that compared with wild-type IRF3, IRF3(S97A) and IRF3(S97D) exhibited increased and decreased binding to KPNA2, respectively, suggesting that dephosphorylation of IRF3 at Ser97 facilitates its binding to KPNA2, which promotes its nuclear translocation. In this context, we found that overexpression of USP22 promoted IRF3(S97A)- but not IRF3(IL139/140AA)- or IRF3(S97D)-mediated activation of ISRE. Together with the observations that knockdown of KPNA2 impaired IRF3-USP22 associations after viral infection and that reconstitution of KPNA2 into USP22 knockout cells restored virus-induced nuclear translocation of IRF3 and expression of downstream genes, we concluded that USP22 promotes nuclear translocation of IRF3 primarily through deubiquitinating and stabilizing KPNA2.
Previously it has been shown that bacterially expressed or baculovirus-expressed KPNA3 and KPNA4 bind to p65 in TNFα-stimulated A549 cells (Fagerlund et al., 2005, 2008). However, another study shows that KPNA2 and KPNB1 bind to and participate in the nuclear translocation of p65 (Wong et al., 2009). Results from a more recent study suggest that knockdown of KPNA2 but not KPNA3 or KPNA4 inhibits TNF-α–induced KPNB1-p65 binding and nuclear translocation of p65 in A549 cells (Liang et al., 2013), indicating that KPNA2 is a primary transporter for p65 nuclear translocation in cells. The mechanisms behind the discrepancies are unclear. The simplest explanation for this is that the cellular KPNA2 might undergo posttranslational modifications that render different behaviors from the bacterially expressed or baculovirus-expressed KPNA2, which awaits more detailed investigations. In our study, we found that knockout of USP22 impaired virus-triggered nuclear translocation of p65 and induction of p65-target genes such as Tnf and Il6, which was restored by reconstitution of KPNA2, suggesting a role of the USP22–KPNA2 axis in NF-κB activation after viral infection (data not shown). In this context, it has been demonstrated that knockdown of KPNA2 or overexpression of miR-302 cluster (KPNA2-targeting microRNAs) significantly inhibits the nuclear translocation of p65 and the expression of p65 target genes after EV71 infection (Peng et al., 2018).
Given the importance of the USP22–KPNA2–IRF3 axis in the innate antiviral signaling, many viral proteins may target KPNA2 to block the function of KPNA2 as a strategy to counteract host immune responses. For example, Japanese encephalitis virus NS5 protein binds to KPNA2 and blocks its association with IRF3 and p65 to inhibit type I IFN induction, and hepatitis C virus NS3/4A protein cleaves and inactivates KPNA2 (Wang et al., 2014; Ye et al., 2017). So far, we do not know how KPNA2 is ubiquitinated after viral infection. It is possible that some viral proteins work alone or cooperate with host E3s to catalyze the ubiquitination of KPNA2. In this context, it has been reported that vaccina virus A55 protein recruits cullin-3 to mediate ubiquitination and degradation of KPNA2 (Pallett et al., 2019). However, we also found that knockout of USP22 inhibited poly(I:C)-, LPS-, or transfected DNA-induced expression of Ifnb, which might be due to accelerated degradation of KPNA2 and impaired nuclear translocation of IRF3. It is thus more likely that host E3s alone are responsible for the ubiquitination and degradation of KPNA2 during viral infection or ligand stimulation. We reason that these E3s interact with KPNA2 in a stimulation (viral infection or ligand treatment)–dependent manner that might tightly regulate the expression and/or activity of the E3s. Such a strategy is helpful for avoiding excessive harmful immune responses during infection while simultaneously benefiting viral infection. Further studies are required to fully address this point. Nonetheless, it would be beneficial for viral infection–related diseases by developing strategies that release KPNA2 and restore KPNA2–IRF3 or KPNA2–p65 associations.
Materials and methods
Mice
Usp22fl/+ mice were generated by GemPharmatech Co. Ltd. In brief, guide RNAs (5′-GCAACTATGAATATAGTGGTAGG-3′ and 5′-GTCTGTTACATGCAGTGGTGGGG-3′) were obtained through in vitro transcription and purification. The guide RNAs were incubated with purified Cas9 protein and injected into the fertilized eggs (at the one-cell stage) together with the targeting vector with two loxp sites flanking the exon 2 of the Usp22 gene. The injected fertilized eggs were cultured to the two-cell stage followed by transplantation into pseudopregnant mice. The targeted genomes of F0 mice were amplified by PCR and sequenced, and the chimeras were crossed with wild-type C57BL/6 mice to obtain F1 Usp22fl/+ mice. Southern blot analysis was conducted with the tail DNA from F1 mice to confirm correct recombination and exclude random insertions of the targeting vector. Lyz2-Cre mice were purchased from the Nanjing Biomedical Research Institute of Nanjing University. Cre-ER mice (B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj) were from the Jackson Laboratory and were kindly provided by Dr. C. Dong (Tsinghua University, Beijing, China). Usp22fl/+ mice were crossed with Cre-ER mice or Lyz2-Cre mice to obtain Cre-ER Usp22fl/+ and Lyz2-Cre Usp22fl/+ mice, respectively. Age- and sex-matched Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl littermates or Lyz2-Cre Usp22fl/+ and Lyz2-Cre Usp22fl/fl littermates were used for all the described experiments. All mice were housed in the specific pathogen–free animal facility at Wuhan University, and all animal experiments were in accordance with protocols approved by the Institutional Animal Care and Use Committee of Wuhan University. Mice genotypes were determined by PCR analysis of tail DNA, and the genotyping primers are as follows (wild-type allele is of 241 bp; floxed allele is of 330 bp; positive Cre-ER is of 825 bp; positive Lyz2-Cre is of 750 bp): Usp22 forward: 5′-GCTACCAAGAGTCCACAAAGCAG-3′, Usp22 reverse: 5′-CGCCGAAGTTTGGCTGAGTGTT-3′; Cre-ER forward: 5′-AAAGTCGCTCTGAGTTGTTAT-3′, Cre-ER reverse: 5′-CCTGATCCTGGCAATTTCG-3′; Lyz2-Cre forward: 5′-CTTGGGCTGCCAGAATTTCTC-3′, and Lyz2-Cre reverse: 5′-CCCAGAAATGCCAGATTACG-3′.
Tamoxifen-mediated knockout of USP22
To achieve conditional knockout of USP22, 9–10-wk-old Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl mice were injected intraperitoneally with tamoxifen (80 mg/kg body weight, dissolved in corn oil; Sigma-Aldrich, T5648) for 5 successive days. 7 d later, mice were either subjected to euthanasia to test the knockout efficiency or infected with VSV or HSV-1. To knock out USP22 in cultured cells, Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl cells were treated with 4-OHT (1 µM; Sigma-Aldrich, H6278) for 2 or 3 d. Cells were then reseeded into culture dishes or plated in 4-OHT–free medium and rested for 24 h followed by infection with SeV, VSV, or HSV-1, or transfection with poly(I:C) or double-stranded DNA or treatment with LPS.
Preparation of cytoplasmic and nuclear proteins
Cells were harvested with ice-cold PBS and lysed by douncing 20 times in 500 µl membrane lysis buffer (10 mM, pH 7.9, Hepes, 10 mM KCl, 0.1 mM EDTA, 0.4% Nonidet P-40) containing protease inhibitors. The homogenate was centrifuged at 500 g for 10 min. The supernatant was saved as cytosol, and the pellet was saved as crude nuclei. The crude nuclei were washed twice with 500 µl membrane lysis buffer and resuspended in 20–50 µl of extract buffer (20 mM, pH 7.9, Hepes, 0.4 M NaCl, 1 mM EDTA) and shaken vigorously every 30 s for 15 min, followed by centrifugation at 15,000 g for 10 min. The supernatants containing nuclear proteins were saved for subsequent analysis.
Plaque assay
The supernatants of cell cultures or tissue homogenates (or the serial dilutions) from infected Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl mice (tamoxifen treated) were used to infect monolayers of Vero cells. 1 h later, the supernatants or the homogenates or dilutions were removed, and the infected Vero cells were washed with prewarmed PBS twice followed by incubation with DMEM containing 2% methylcellulose for 48 h. The cells were fixed with 4% paraformaldehyde for 15 min and stained with 1% crystal violet for 30 min before counting the plaques.
Reagents, antibodies, and constructs
Poly(I:C) and LPS were described previously (Liuyu et al., 2019; Lu et al., 2017; Zhang et al., 2016). Mouse control IgG (Santa Cruz Biotechnology, sc-2025) and rabbit control IgG (Millipore, 12–370), HRP-conjugated goat-anti mouse or rabbit IgG (Thermo Fisher Scientific, PA1-86717 and SA1-9510), mouse anti-FLAG (Sungene, KM8002), anti-FLAG-HRP (Sigma-Aldrich, A8592-1MG), anti-GAPDH (Sungene, KM9002), anti–β-actin (KM9001), anti-tubulin (F0601), anti-HA (COVANCE, MMS-101R), anti-pIκBα (9246L), anti-ubiquitin (sc-8017), anti-IRF3 (sc-33641), anti-USP22 (sc-390585), rabbit anti-lamin B1 (Proteintech, 12987–1-AP), anti-p62 (Cell Signaling Technologies, 39749S), anti-ATG7 (Cell Signaling Technologies, 8558S), anti-IRF3 (sc-9082), anti–p-IRF3 (Cell Signaling Technologies, 4947S), anti-IκBα (sc-371), anti-IκBα (Cell Signaling Technologies, 9246L), and anti-USP22 (Abcam, ab-195289) were purchased from the indicated manufacturers. Rabbit anti-sera to NDP52 were raised against recombinant murine NDP52(1–160). The ISRE promoter luciferase reporter constructs, mammalian expression plasmids for RIG-I, VISA, MITA, TBK1, IRF3, IRF3 truncations, and ubiquitin were previously described (Liuyu et al., 2019; Sun et al., 2017; Zhang et al., 2016). Mammalian expression plasmids for USP22 and USP22 mutants and truncations were constructed by standard molecular biology techniques. Plasmids encoding KPNA proteins were kindly provided by Dr. B. Ge (Tongji University, Shanghai, China; Liu et al., 2018a). Plasmids encoding IRF3 mutants were gifts from Dr. Y. Chen (Wuhan University, Wuhan, China; Li et al., 2016; Zhu et al., 2015). Staining antibodies against CD3, CD4, CD8, CD19, CD25, CD44, and CD62L were purchased from Biolegend. Flow cytometry analysis was performed with a BD FACSCelesta system.
Quantitative real-time PCR and ELISA
Total RNA was extracted from cells using TRIzol (Invitrogen), and the first-strand cDNA was reverse-transcribed with All-in-One cDNA Synthesis SuperMix (Biotool). Gene expression was examined with a Bio-Rad CFX Connect system by a fast two-step amplification program with 2× SYBR Green Fast qPCR Master Mix (Biotool). The value obtained for each gene was normalized to that of the gene encoding β-actin. Gene-specific primers have been described previously (Liuyu et al., 2019). The ELISA kits for IFN-β and CCL5 (Biolegend) were used to detect the indicated cytokines in the sera.
Co-immunoprecipitation and immunoblot analysis
The experiments were performed as previously described (Liuyu et al., 2019; Lu et al., 2017; Zhang et al., 2016). In brief, cells were lysed in Nonidet P-40 lysis buffer containing 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, and 1% protease and phosphatase inhibitor cocktail (Biotool). Cell lysates were subjected to Native-PAGE or SDS-PAGE, and immunoblot analysis was performed with the appropriate antibodies. For immunoprecipitation assays, the lysates were immunoprecipitated with IgG or the appropriate antibodies, and the precipitants were washed three times with lysis buffer containing 500 mM NaCl, followed by immunoblot analysis. The antibodies were diluted in 3–5% (wt/vol) fat-free milk (BD Biosciences) or 1% BSA (Sigma-Aldrich) in TBS (1:500–1:2,000).
In vivo and in vitro deubiquitination assays
These experiments were performed as previously described (Liuyu et al., 2019; Sun et al., 2017; Zhang et al., 2016). For deubiquitination assays in cells, cells were lysed with the lysis buffer (100 µl), and the supernatants were denatured at 95°C for 5 min in the presence of 1% SDS by lysates. The denatured lysates were diluted with lysis buffer until the concentration of SDS was reduced below 0.1% followed by immunoprecipitation (denature-IP) with the indicated antibodies. The immunoprecipitants were subject to immunoblot analysis with anti-ubiquitin. For in vitro deubiquitination, FLAG-tagged KPNA2 and Myc-tagged ubiquitin were cotransfected into HEK 293 cells. Denature-IP was performed, and the precipitants were eluted by 3× FLAG peptide (Sigma-Aldrich) to obtain ubiquitin-modified IRF3. USP22, USP22(RR164/165AA), and USP22(C185S) were obtained by an in vitro transcription and translation kit (Promega). The ubiquitinated KPNA2 was incubated with in vitro synthesized proteins at 37°C for 2 h followed by overnight incubation at 16°C in the presence of 1 µM ATP. The mixture was analyzed by immunoblot with the indicated antibodies.
The plasmid encoding GST-Ub-Tandem Ubiquitin Binding Entity (TUBE) was previously described and kindly provided by Dr. M. Gyrd-Hansen (University of Oxford, Oxford, UK; Fiil et al., 2013). GST-TUBE was expressed in Escherichia coli (DE3) and purified with a GST column as previously described (Ye et al., 2019; Zhao et al., 2018). To analyze ubiquitinated KPNA2 or IRF3, GST-TUBE was incubated with cell lysates and pulled down by GST beads. The GST beads were washed with lysis buffer containing 500 mM NaCl three times and subject to PAGE electrophoresis and immunoblot analysis.
Transfection and luciferase reporter assay
HEK293 cells were cotransfected with ISRE promoter firefly luciferase reporter plasmid and a TK-Renilla luciferase reporter, together with vector or various DUBs, or control siRNA or siRNA targeting USP22. 24 h later, cells were infected with SeV for 8 h. Luciferase activity was measured with a Dual-Luciferase Reporter Assay System (Promega).
Cell culture
MEFs were prepared from E14.5 embryos. Bone marrow cells were isolated from mouse femur. The cells were cultured in DMEM containing 20% fetal bovine serum, 1% streptomycin and penicillin, and 10 µM β-mercaptoethanol, with M-CSF (10 ng/ml, Peprotech) for BMDM differentiation or GM-CSF (20 ng/ml, Peprotech) for BMDC differentiation. Primary cells of Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl were treated with 4-OHT (1 µM dissolved in ethyl alcohol; Sigma-Aldrich, H6278).
siRNA and shRNA
The siRNAs were synthesized and transfected with Lipofectamine 2000 according to the manufacturer’s manual. 24 h after transfection, cells were harvested or stimulated followed by immunoblot, quantitative PCR, or deubiquitination assays. Oligo DNAs targeting mouse Kpna2 were synthesized, annealed, and inserted into the pLenti-GFP vector. The siRNA and shRNA sequences are listed as follows: Control, 5′-UUCUCCGAACGUGUCACGUTT-3′; siUSP22#1: 5′-CCUGCCUCUACUGUGUCUUTT-3′; siUSP22#2, 5′-GGAGAAAGAUCACCUCGAATT-3′; siUSP22#3, 5′-GCUACCAGGAGUCCACAAATT-3′; siUSP36#1, 5′-GCAAGAGCUUCUCCUACCATT-3′; siUSP36#2, 5′-GCACACGUAUGAGAGCUGUTT -3′; siKPNA2#1: 5′-GCAUCAUGAUGAUCCAGAATT-3′; siKPNA2#2, 5′-GGAGCUUCUGAAUUGCCAATT-3′; siATXN7L3#1, 5′-GGACAUCUUUGGACAGGUUTT-3′; siATXN7L3#2, 5′-GCCUAGGUUCCAACAAGAATT-3′; siENY2#1, 5′-GGUUAGCAAGAUGAACAAATT-3′; siENY2#2, 5′-GAGCUCCUACAAAGAAUAATT-3′; siATG7#1, 5′-CAGACAAGAAGCUCCUUCUTT-3′; siATG7#2, 5′-CAGCCUGGCAUUUGAUAAATT-3′; sip62#1, 5′-GGAACUCGCUAUAAGUGCATT-3′; sip62#2, 5′-GAUGACUGGACACAUUUGUTT-3′; siNDP52#1, 5′-GGAGAUAUCAUGUGCUAUUTT-3′; siNDP52#2, 5′-GCGCUACCAGUUCUGCUAUTT-3′; and shKpna2: 5′-GCAGATTCTTCCTACGTTA-3′.
Viral infection
For viral replication assays, cells (2–5 × 105) were infected with VSV, HSV-1, VSV-GFP, or H129-G4. 1 h later, the supernatants were removed, and cells were washed with prewarmed PBS (1 ml) twice followed by culture in full medium for 24 h. Viral replication was analyzed by flow cytometry analysis or standard plaque assays. For mice infection, age- and sex-matched Cre-ER Usp22fl/+ and Cre-ER Usp22fl/fl littermates were injected with VSV (2 × 107 PFU per mouse) or with HSV-1 (3 × 106 PFU per mouse), and the survival of animals was monitored every day. The lungs or brains were collected for qRT-PCR analysis and plaque assays at 24 h or 4 d after infection, respectively.
Immunofluorescence labeling and confocal microscopy
Cells were fixed for 10 min with 4% paraformaldehyde in PBS and then were permeabilized for 15 min with 0.2% Triton X-100 in PBS. After blockade of nonspecific binding by incubation of cells for 30 min with 1% BSA in PBS, coverslips were incubated with the appropriate primary antibodies (identified above) and then sequentially with FITC–goat anti–rabbit immunoglobulin G (sc-2012, Santa Cruz Biotechnology) and then were visualized with a Leica confocal microscope under a 40× objective.
Lentivirus-mediated gene transfer
HEK293 cells were transfected with phage-6tag-IRF3, phage-6tag-USP22, phage-6tag-USP22(RR164/165AA), phage-6tag-USP22(C185S), phage-6tag-KPNA2, pLenti-shKpna2, or the empty vector along with the packaging vectors pSPAX2 and pMD2G. The medium was changed with fresh full medium (10% FBS, 1% streptomycin-penicillin, and 10 µM β-mercaptoethanol) after 8 h. 40 h later, the supernatants were harvested to infect MEFs followed by various analyses.
Statistical analysis
Differences between experimental and control groups were tested using Student’s t test or two-way ANOVA with Bonferroni post-test. P values <0.05 were considered statistically significant. For animal survival analysis, the Kaplan–Meier method was adopted to generate graphs, and the survival curves were analyzed with log-rank analysis.
Online supplemental material
Fig. S1 characterizes USP22 as an IRF3-interacting protein, and shows knockdown of USP22 inhibits SeV-induced expression of downstream genes without affecting the dimerization of IRF3. Fig. S2 shows knockout of USP22 inhibits virus-triggered expression of downstream genes and nuclear translocation of IRF3 but does not affects the phosphorylation or dimerization of IRF3 in BMDMs or MEFs. Fig. S3 provides evidence that the enzyme activity but not the nuclear localization of USP22 is essential for virus-triggered expression of downstream genes and nuclear translocation of IRF3. Fig. S4 demonstrates USP22 deficiency does not affect ubiquitination or stability of IRF3. Fig. S5 describes KPNA2 as a substrate and interacting protein of USP22.
Acknowledgments
We thank Drs. Chen Dong (Tsinghua University), Yu Chen (Wuhan University), Baoxue Ge (Tongji University), and Mads Gyrd-Hansen (University of Oxford) for reagents, and members of the Zhong laboratory and the core facilities of Medical Research Institute for technical help.
This study was supported by grants from National Key Research and Development Program of China (2018YFE0204500 and 2018YFC1004601), the National Natural Science Foundation of China (31671454 and 31930040), the Natural Science Foundation of Hubei Province (2018CFA016), and the Medical Science Advancement Program (Basic Medical Sciences) of Wuhan University (TFJC2018004 and 2042019kf0323).
Author contributions: B. Zhong designed and supervised the study; Z. Cai performed the major experiments; M.-X. Zhang helped with the animal studies; Z. Tang and Q. Zhang purified GST-TUBE and performed GDT pull-down analysis; J. Ye, Z.-D. Zhang and T.-C. Xiong helped with cell culture and viral infections; Z. Cai and B. Zhong wrote the paper; and all the authors analyzed the data.
References
- Akira S., Uematsu S., and Takeuchi O.. 2006. Pathogen recognition and innate immunity. Cell. 124:783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Atanassov B.S., Mohan R.D., Lan X., Kuang X., Lu Y., Lin K., McIvor E., Li W., Zhang Y., Florens L., et al. 2016. ATXN7L3 and ENY2 Coordinate Activity of Multiple H2B Deubiquitinases Important for Cellular Proliferation and Tumor Growth. Mol. Cell. 62:558–571. 10.1016/j.molcel.2016.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlund R., Kinnunen L., Köhler M., Julkunen I., and Melén K.. 2005. NF-kappaB is transported into the nucleus by importin alpha3 and importin alpha4. J. Biol. Chem. 280:15942–15951. 10.1074/jbc.M500814200 [DOI] [PubMed] [Google Scholar]
- Fagerlund R., Melén K., Cao X., and Julkunen I.. 2008. NF-kappaB p52, RelB and c-Rel are transported into the nucleus via a subset of importin alpha molecules. Cell. Signal. 20:1442–1451. 10.1016/j.cellsig.2008.03.012 [DOI] [PubMed] [Google Scholar]
- Fiil B.K., Damgaard R.B., Wagner S.A., Keusekotten K., Fritsch M., Bekker-Jensen S., Mailand N., Choudhary C., Komander D., and Gyrd-Hansen M.. 2013. OTULIN restricts Met1-linked ubiquitination to control innate immune signaling. Mol. Cell. 50:818–830. 10.1016/j.molcel.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro V.J., Stanek T.J., Peck A.R., Sun Y., Wang F., Qie S., Knudsen K.E., Rui H., Butt T., Diehl J.A., and McMahon S.B.. 2018. Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells. Proc. Natl. Acad. Sci. USA. 115:E9298–E9307. 10.1073/pnas.1807704115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., and Mattaj I.W.. 1996. Nucleocytoplasmic transport. Science. 271:1513–1518. 10.1126/science.271.5255.1513 [DOI] [PubMed] [Google Scholar]
- Hou F., Sun L., Zheng H., Skaug B., Jiang Q.X., and Chen Z.J.. 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 146:448–461. 10.1016/j.cell.2011.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., and Barber G.N.. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 455:674–678. 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., and Akira S.. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988. 10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- Kim D., Hong A., Park H.I., Shin W.H., Yoo L., Jeon S.J., and Chung K.C.. 2017. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J. Cell. Physiol. 232:3664–3676. 10.1002/jcp.25841 [DOI] [PubMed] [Google Scholar]
- Koutelou E., Wang L., Schibler A.C., Chao H.P., Kuang X., Lin K., Lu Y., Shen J., Jeter C.R., Salinger A., et al. 2019. USP22 controls multiple signaling pathways that are essential for vasculature formation in the mouse placenta. Development. 146:dev174037 10.1242/dev.174037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K.P., McBride K.M., Weaver B.K., Dingwall C., and Reich N.C.. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 20:4159–4168. 10.1128/MCB.20.11.4159-4168.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., and Zhong B.. 2018. Regulation of Cellular Antiviral Signaling by Modifications of Ubiquitin and Ubiquitin-like Molecules. Immune Netw. 18:e4 10.4110/in.2018.18.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhu M., Pan R., Fang T., Cao Y.Y., Chen S., Zhao X., Lei C.Q., Guo L., Chen Y., et al. 2016. The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat. Immunol. 17:241–249. 10.1038/ni.3311 [DOI] [PubMed] [Google Scholar]
- Li C., Irrazabal T., So C.C., Berru M., Du L., Lam E., Ling A.K., Gommerman J.L., Pan-Hammarström Q., and Martin A.. 2018. The H2B deubiquitinase Usp22 promotes antibody class switch recombination by facilitating non-homologous end joining. Nat. Commun. 9:1006 10.1038/s41467-018-03455-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Zhang H., Wang G., Li S., Cong S., Luo Y., and Zhang B.. 2013. KPNB1, XPO7 and IPO8 mediate the translocation ofNF-κB/p65 into the nucleus. Traffic. 14:1132–1143. [DOI] [PubMed] [Google Scholar]
- Lin D., and Zhong B.. 2015. Regulation of cellular innate antiviral signaling by ubiquitin modification. Acta Biochim. Biophys. Sin. (Shanghai). 47:149–155. 10.1093/abbs/gmu133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Heylbroeck C., Pitha P.M., and Hiscott J.. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986–2996. 10.1128/MCB.18.5.2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Yang H., Kong Q., Li J., Lee S.M., Gao B., Dong H., Wei J., Song J., Zhang D.D., and Fang D.. 2012. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell. 46:484–494. 10.1016/j.molcel.2012.03.024 [DOI] [PubMed] [Google Scholar]
- Liu H., Zhang H., Wu X., Ma D., Wu J., Wang L., Jiang Y., Fei Y., Zhu C., Tan R., et al. 2018a Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 563:131–136. 10.1038/s41586-018-0629-6 [DOI] [PubMed] [Google Scholar]
- Liu Q., Wu Y., Qin Y., Hu J., Xie W., Qin F.X., and Cui J.. 2018b Broad and diverse mechanisms used by deubiquitinase family members in regulating the type I interferon signaling pathway during antiviral responses. Sci. Adv. 4:r2824 10.1126/sciadv.aar2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuyu T., Yu K., Ye L., Zhang Z., Zhang M., Ren Y., Cai Z., Zhu Q., Lin D., and Zhong B.. 2019. Induction of OTUD4 by viral infection promotes antiviral responses through deubiquitinating and stabilizing MAVS. Cell Res. 29:67–79. 10.1038/s41422-018-0107-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Ren Y., Sun X., Han C., Wang H., Chen Y., Peng Q., Cheng Y., Cheng X., Zhu Q., et al. 2017. Induction of INKIT by Viral Infection Negatively Regulates Antiviral Responses through Inhibiting Phosphorylation of p65 and IRF3. Cell Host Microbe. 22:86–98.e4. 10.1016/j.chom.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Melo-Cardenas J., Zhang Y., Zhang D.D., and Fang D.. 2016. Ubiquitin-specific peptidase 22 functions and its involvement in disease. Oncotarget. 7:44848–44856. 10.18632/oncotarget.8602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen T.E.T., and Komander D.. 2017. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 86:159–192. 10.1146/annurev-biochem-061516-044916 [DOI] [PubMed] [Google Scholar]
- Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., and Tschopp J.. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 437:1167–1172. 10.1038/nature04193 [DOI] [PubMed] [Google Scholar]
- Miyamoto Y., Yamada K., and Yoneda Y.. 2016. Importin α: a key molecule in nuclear transport and non-transport functions. J. Biochem. 160:69–75. 10.1093/jb/mvw036 [DOI] [PubMed] [Google Scholar]
- Nigg E.A. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 386:779–787. 10.1038/386779a0 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. 2014. Historical landmarks of autophagy research. Cell Res. 24:9–23. 10.1038/cr.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallett M.A., Ren H., Zhang R.Y., Scutts S.R., Gonzalez L., Zhu Z., Maluquer de Motes C., and Smith G.L.. 2019. Vaccinia Virus BBK E3 Ligase Adaptor A55 Targets Importin-Dependent NF-κB Activation and Inhibits CD8+ T-Cell Memory. J. Virol. 93:e00051-19 10.1128/JVI.00051-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng N., Yang X., Zhu C., Zhou L., Yu H., Li M., Lin Y., Wang X., Li Q., She Y., et al. 2018. MicroRNA-302 Cluster Downregulates Enterovirus 71-Induced Innate Immune Response by Targeting KPNA2. J. Immunol. 201:145–156. 10.4049/jimmunol.1701692 [DOI] [PubMed] [Google Scholar]
- Ramachandran S., Haddad D., Li C., Le M.X., Ling A.K., So C.C., Nepal R.M., Gommerman J.L., Yu K., Ketela T., et al. 2016. The SAGA Deubiquitination Module Promotes DNA Repair and Class Switch Recombination through ATM and DNAPK-Mediated γH2AX Formation. Cell Reports. 15:1554–1565. 10.1016/j.celrep.2016.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., and Chen Z.J.. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 122:669–682. 10.1016/j.cell.2005.08.012 [DOI] [PubMed] [Google Scholar]
- Sharma S., tenOever B.R., Grandvaux N., Zhou G.P., Lin R., and Hiscott J.. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science. 300:1148–1151. 10.1126/science.1081315 [DOI] [PubMed] [Google Scholar]
- Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., and Jiang Z.. 2009. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA. 106:8653–8658. 10.1073/pnas.0900850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., and Chen Z.J.. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 339:786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Zhang Q., Jing Y.Y., Zhang M., Wang H.Y., Cai Z., Liuyu T., Zhang Z.D., Xiong T.C., Wu Y., et al. 2017. USP13 negatively regulates antiviral responses by deubiquitinating STING. Nat. Commun. 8:15534 10.1038/ncomms15534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Sun L., Chen J., and Chen Z.J.. 2018. Detection of Microbial Infections Through Innate Immune Sensing of Nucleic Acids. Annu. Rev. Microbiol. 72:447–478. 10.1146/annurev-micro-102215-095605 [DOI] [PubMed] [Google Scholar]
- Thurston T.L., Ryzhakov G., Bloor S., von Muhlinen N., and Randow F.. 2009. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10:1215–1221. 10.1038/ni.1800 [DOI] [PubMed] [Google Scholar]
- Wang L., Huang J., Jiang M., Chen Q., Jiang Z., and Feng H.. 2014. CAMK1 phosphoinositide signal-mediated protein sorting and transport network in human hepatocellular carcinoma (HCC) by biocomputation. Cell Biochem. Biophys. 70:1011–1016. 10.1007/s12013-014-0011-8 [DOI] [PubMed] [Google Scholar]
- Wong C.H., Chan H., Ho C.Y., Lai S.K., Chan K.S., Koh C.G., and Li H.Y.. 2009. Apoptotic histone modification inhibits nuclear transport by regulating RCC1. Nat. Cell Biol. 11:36–45. 10.1038/ncb1810 [DOI] [PubMed] [Google Scholar]
- Wu J., and Chen Z.J.. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32:461–488. 10.1146/annurev-immunol-032713-120156 [DOI] [PubMed] [Google Scholar]
- Wu J., Sun L., Chen X., Du F., Shi H., Chen C., and Chen Z.J.. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 339:826–830. 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Wang Y., Gong Z., Liu J., and Li W.. 2014. Identification of a functional nuclear localization signal within the human USP22 protein. Biochem. Biophys. Res. Commun. 449:14–18. 10.1016/j.bbrc.2014.04.133 [DOI] [PubMed] [Google Scholar]
- Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., and Shu H.B.. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 19:727–740. 10.1016/j.molcel.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Ye J., Chen Z., Li Y., Zhao Z., He W., Zohaib A., Song Y., Deng C., Zhang B., Chen H., and Cao S.. 2017. Japanese Encephalitis Virus NS5 Inhibits Type I Interferon (IFN) Production by Blocking the Nuclear Translocation of IFN Regulatory Factor 3 and NF-κB. J. Virol. 91:e00039-17 10.1128/JVI.00039-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L., Zhang Q., Liuyu T., Xu Z., Zhang M.X., Luo M.H., Zeng W.B., Zhu Q., Lin D., and Zhong B.. 2019. USP49 negatively regulates cellular antiviral responses via deconjugating K63-linked ubiquitination of MITA. PLoS Pathog. 15:e1007680 10.1371/journal.ppat.1007680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zhang M.X., Zhang Q., Zhu G.F., Yuan L., Zhang D.E., Zhu Q., Yao J., Shu H.B., and Zhong B.. 2016. USP18 recruits USP20 to promote innate antiviral response through deubiquitinating STING/MITA. Cell Res. 26:1302–1319. 10.1038/cr.2016.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Shang G., Gui X., Zhang X., Bai X.C., and Chen Z.J.. 2019. Structural basis of STING binding with and phosphorylation by TBK1. Nature. 567:394–398. 10.1038/s41586-019-1000-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang X., Wang Q., Deng Y., Li K., Zhang M., Zhang Q., Zhou J., Wang H.Y., Bai P., et al. 2018. USP2a Supports Metastasis by Tuning TGF-β Signaling. Cell Reports. 22:2442–2454. 10.1016/j.celrep.2018.02.007 [DOI] [PubMed] [Google Scholar]
- Zheng Y.T., Shahnazari S., Brech A., Lamark T., Johansen T., and Brumell J.H.. 2009. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 183:5909–5916. 10.4049/jimmunol.0900441 [DOI] [PubMed] [Google Scholar]
- Zhong B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., and Shu H.B.. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 29:538–550. 10.1016/j.immuni.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Zhu M., Fang T., Li S., Meng K., and Guo D.. 2015. Bipartite Nuclear Localization Signal Controls Nuclear Import and DNA-Binding Activity of IFN Regulatory Factor 3. J. Immunol. 195:289–297. 10.4049/jimmunol.1500232 [DOI] [PubMed] [Google Scholar]