This study identifies USP22 as an essential regulator specifically for iNKT but not conventional T cell development. USP22 suppresses histone H2A monoubiquitination through its interaction with MED1 to partially control expression of genes required for iNKT cell development and differentiation.
Abstract
The ubiquitin pathway has been shown to regulate iNKT cell immunity, but the deubiquitinase involved in this process has not been identified. Herein we found that ubiquitin-specific peptidase 22 (USP22) is highly expressed in iNKT cells during their early developmental stage 1. USP22 deficiency blocked the transition from stage 1 to 2 during iNKT cell development in a cell-intrinsic manner. USP22 suppression also diminishes iNKT17 and iNKT1 differentiation but favors iNKT2 polarization without altering conventional T cell activation and differentiation. USP22 interacts with the Mediator complex subunit 1 (MED1), a transcription coactivator involved in iNKT cell development. Interestingly, while interacting with MED1, USP22 does not function as a deubiquitinase to suppress MED1 ubiquitination for its stabilization. Instead, USP22 enhances MED1 functions for IL-2Rβ and T-bet gene expression through deubiquitinating histone H2A but not H2B monoubiquitination. Therefore, our study revealed USP22-mediated histone H2A deubiquitination fine-tunes MED1 transcriptional activation as a previously unappreciated molecular mechanism to control iNKT development and functions.
Introduction
Invariant natural killer T (iNKT) cells play an important role in linking innate and adaptive immune responses and have been implicated in infectious disease, allergy, asthma, autoimmunity, and tumor surveillance. iNKT cells express a highly restricted TCR that specifically responds to CD1d-restricted lipid ligands. In contrast to the conventional T cells, which are selected by peptide antigens in complex with MHC class I or II molecules present on the surface of thymic epithelial cells, NKT cells develop following selection by self-glycolipid antigens in complex with the MHC class I–like molecule CD1d presented by CD4+CD8+ double-positive (DP) thymocytes (Bendelac et al., 2007). Upon activation, mature iNKT cells rapidly differentiate into NKT1, NKT2, and NKT17, and secrete a broad range of T cell lineage–specific cytokines, such as IFN-γ, IL-4, and IL-17, respectively (Engel et al., 2012; Kadowaki et al., 2001; Lee et al., 2013). The ubiquitin pathway has been shown to play important roles in regulating iNKT cell development and functions. The ubiquitin-modifying enzyme A20, an upstream regulator of TCR signaling in T cells, is an essential cell-intrinsic regulator of iNKT development (Drennan et al., 2016). A20 is differentially expressed during NKT cell development, regulates NKT cell development maturation, and specifically controls the differentiation and survival of NKT1 and NKT2 but not NKT17 subsets, possibly through modulating the transcriptional activation of NF-κB. The RING-finger containing E3 ubiquitin ligase Cbl-b has been identified to promote monoubiquitination of CARMA1, a critical signaling molecule in NF-κB activation, to suppress iNKT cell activation, and induces iNKT cell tolerance to tumor antigen (Kojo et al., 2009). In addition, the targeted deletion of Roquin E3 ligase impairs iNKT development and iNKT2 differentiation (Drees et al., 2017). However, the ubiquitin-specific peptidase that reverses the ubiquitin conjugation involved in iNKT cell development and activation remains to be identified.
Ubiquitin-specific peptidase 22 (USP22) was initially identified as a death from signature genes involved in cancer development, metastasis, and chemotherapy resistance (Glinsky et al., 2005). USP22 is ubiquitously expressed in adult mammalian tissues and is predominantly enriched within the nucleus (Lee et al., 2006), and its expression level is up-regulated in a variety types of tumors (Melo-Cardenas et al., 2018). USP22 is an evolutionarily conserved ubiquitin hydrolase, both in sequence and function, which deubiquitinates and stabilizes the histones and transcription factors to achieve its biological functions. USP22 can also be assembled into the Spt-Ada-USP22 acetyltransferase (SAGA) complex as a transcription coactivator for transcription of genes involved in cell proliferation and survival. The predominant function of USP22 and its orthologues, Nonstop (Drosophila melanogaster) and Ubp8 (Saccharomyces cerevisiae), is the removal of H2Bub1 to promote gene transcription (Atanassov et al., 2009; Zhang et al., 2008; Zhao et al., 2008). More recent studies show that USP22 also removes histone 2A monoubiquitination (H2Aub; Atanassov and Dent, 2011; Lang et al., 2011) and modulates the level of polyubiquitination of several nonhistone substrates such as Sirt1, cyclin B1, PU.1, and TRF6 (Atanassov and Dent, 2011; Lin et al., 2012, 2015; Melo-Cardenas et al., 2018). The tissue-specific physiological functions of USP22 remain largely unknown.
In the current study, we generated a strain of mice with a T cell–specific USP22 gene deletion and discovered that USP22 is essential for iNKT development. Loss of USP22 function diminished the transition of iNKT cells from stage 1 to stage 2 transition during iNKT development. We further discovered that USP22 regulates iNKT cell development through its interaction with and activation of the mediator of RNA polymerase II transcription subunit 1 (MED1), a transcription coactivator that is found to play an important role in the early stage of iNKT cell development through promoting the transcription of T-box transcription factor(T-bet) and IL-2 receptor β chain(IL-2Rβ; Yue et al., 2011). Our studies define a previously unappreciated epigenetic mechanism that drives iNKT cell development.
Results
USP22 is required for iNKT cell development
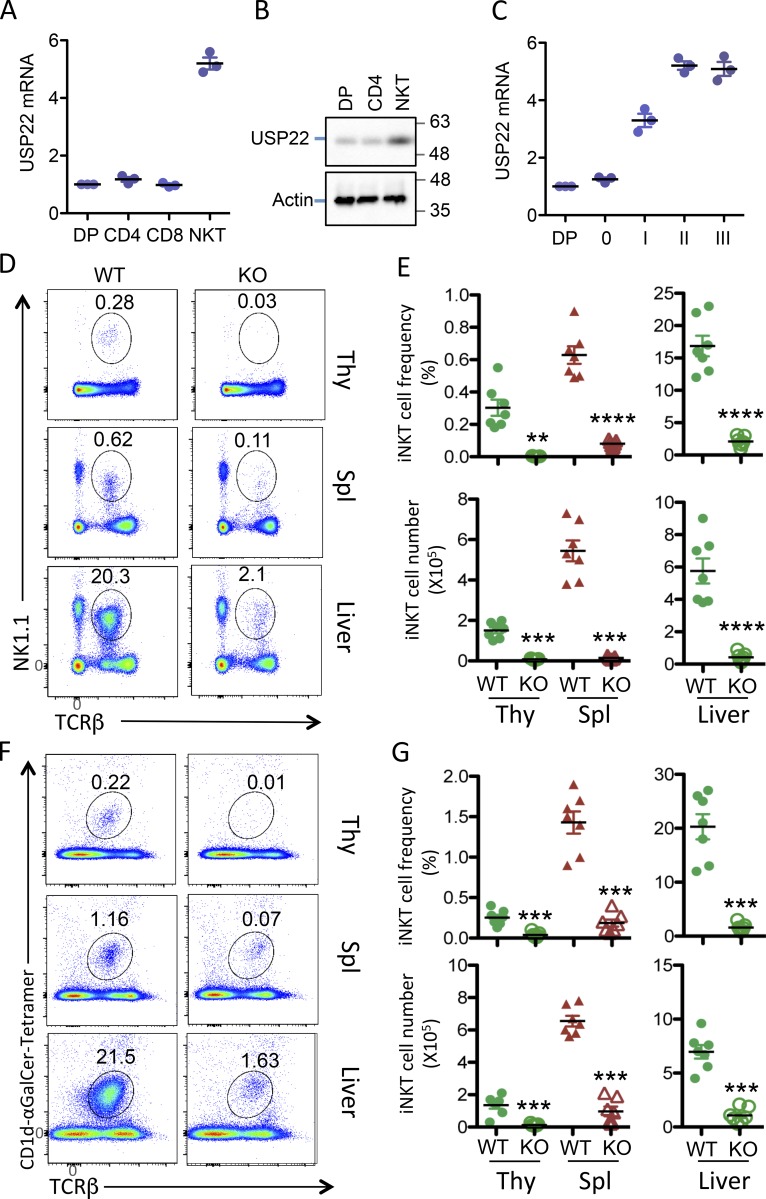
We first analyzed the expression of USP22 in T cells and iNKT cells and detected its highest expression in iNKT cells (Fig. 1, A and B). Further analysis demonstrated that USP22 expression is up-regulated during iNKT developmental stage 1, which is further increased at stage 2 (Fig. 1 C), indicating that USP22 may play important roles in iNKT immunity.
Figure 1.
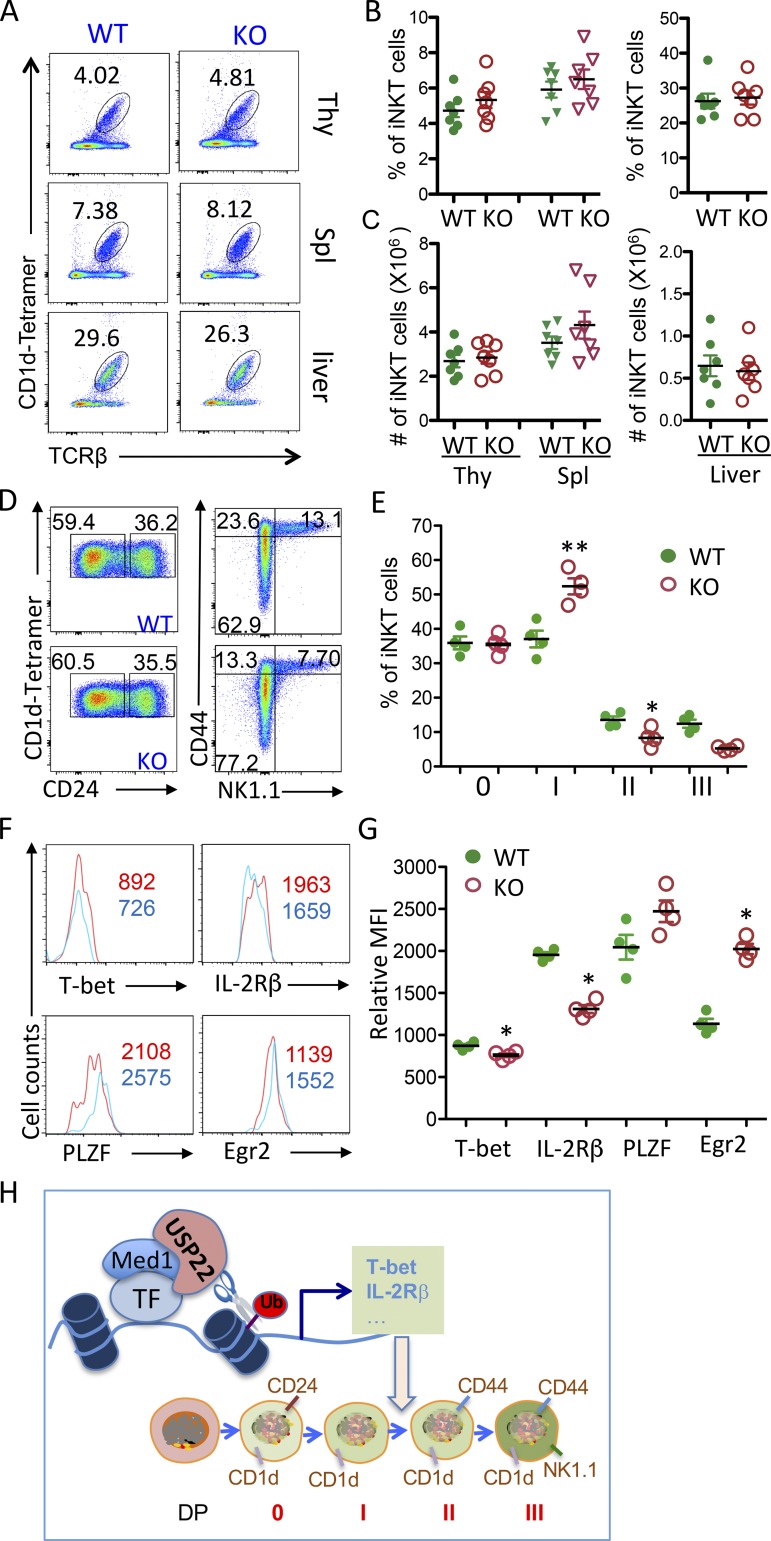
Impaired NKT cell development in USP22 cKO mice. Single-cell suspensions of thymus and spleen, as well as purified lymphocytes from liver tissue, were collected from WT and USP22 cKO mice. (A and B) The expression levels of USP22 in the sorted cells from thymus were determined by real-time RT-PCR (A) and Western blotting (B). (C) Cells at each indicated stage during iNKT development were sorted, the USP22 mRNA levels were analyzed. (D–G) Cells were labeled with antibodies specific to TCRβ together with anti-NK1.1, or (F and G) with CD1d-αGalCer tetramer, and then analyzed by flow cytometry. The representative images (D and F), the percentages (E and G, top panels), and absolute numbers (E and G, bottom panels) of iNKT cells from seven pairs of mice are shown. Each symbol (A, C, E, and G) represents an individual mouse. Thy, thymus; Spl, spleen. Error bars represent mean ± SD. Student’s t test was used for statistical analysis. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. In A–C, results are representative of three independent experiments; in D–G, data are pooled from three replicate experiments with seven mice in total.
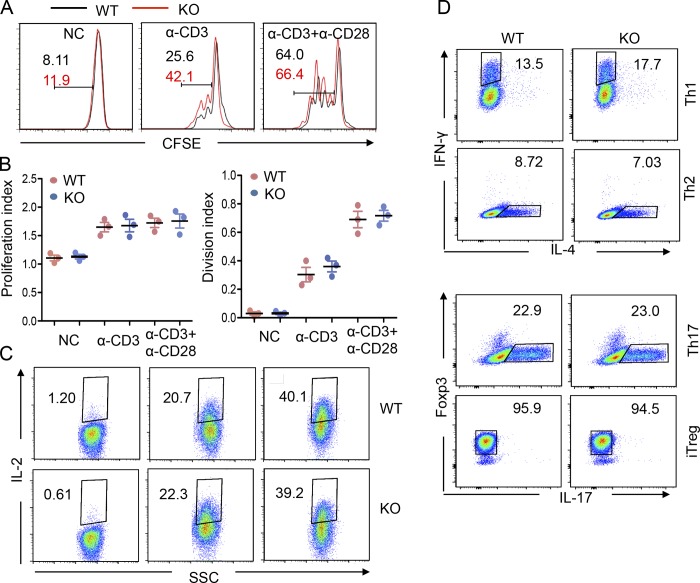
Our laboratory has recently generated a strain of USP22 conditional mutant (USP22 floxed) mice (Melo-Cardenas et al., 2018). To further investigate the role of USP22 in immune regulation, we generated the T cell-specific USP22 conditional KO (USP22 cKO) mice by breeding the USP22 floxed mice with Lck-Cre transgenic mice, whose Cre recombinase expression driven by the Lck promoter mediates USP22 deletion from the CD4/CD8 double-negative stage (Hennet et al., 1995). The obligation of USP22 protein expression in thymic T cells was validated by Western blotting (Fig. S1 A). Flow cytometry analysis did not detect any changes in the percentages and absolute numbers of cells at CD4/CD8 double-negative, DP, and single-positive T cells in the thymus of USP22 cKO mice (Fig. S1, B–D). Further intracellular staining analysis showed that the percentages of CD4+CD25+FoxP3+ regulatory T (T reg) cells in the thymus from WT and USP22 cKO mice were also comparable (Fig. S1 B). In addition, a comparable percentages and numbers of the CD4+ and CD8+ T cells in the spleen between USP22-null and WT littermate control mice were further confirmed (Fig. S1, E–G). The frequency of CD4+CD25+FoxP3+ T reg cells and γδ T cells were unaltered by USP22 deletion (Fig. S1, H and J). Further analysis of T cells by CD44 and CD62L cell surface expression did not detect any changes in chronic T cell activation (Fig. S1 I). As expected, both the frequency and numbers of B220+ B cells in the spleen of USP22 cKO mice were unaltered (Fig. S1, K and L). These results imply that USP22 is dispensable for conventional T cell development and maturation in mice.
Figure S1.
Analysis of lymphocyte development in USP22 cKO mice. (A) USP22 protein expression in the total thymocytes from WT and USP22 cKO mice was determined by Western blotting. (B–D) CD4 and CD8 expression of the thymocytes was analyzed by flow cytometry. The FoxP3+CD25+ T reg cells were analyzed on the gated CD4 single-positive T cells (B). Representative flow images are shown (B). The percentages (C) and absolute numbers (D) of each sub-population from seven pairs of mice are indicated. (E–G) Analysis of T cells in spleens of WT and USP22 cKO mice. The percentages and absolute numbers of CD4 and CD8 T cells are shown. (H and I) The FoxP3+CD25+ T reg cells (H) and memory T cells (I) were analyzed on the gated CD4 T cells. (J) The ratios of αβ and γδ T cells on the gated CD3ε+ T cells were analyzed. (K and L) The B220+ B cells in spleens were analyzed. Error bars represent data from six or seven pairs of mice. Student’s t test (unpaired) was used for statistical analysis. In A–L, data are pooled from three independent experiments. SSC, SSSC, side-scattered light.
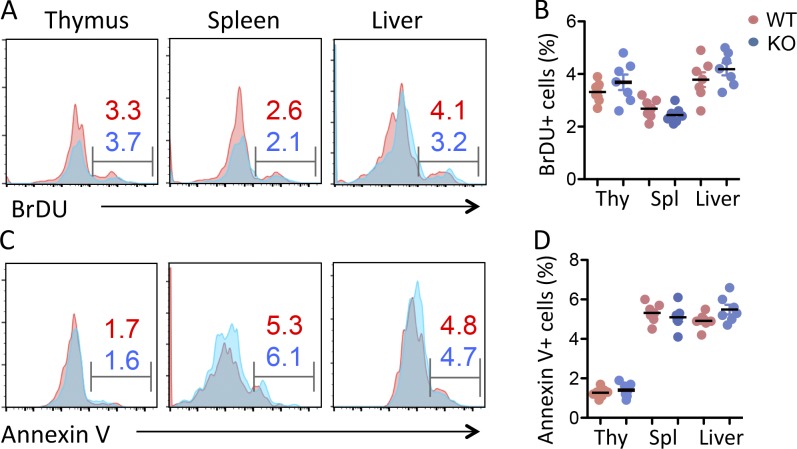
Interestingly, flow cytometry analysis detected a >90% reduction in the TCRβ- and NK1.1-positive iNKT cell population in the thymus of USP22 cKO mice compared with their WT littermate controls (Fig. 1, D and E). Further analysis using a CD1d-αGalCer tetramer confirmed the diminished iNKT cell population in thymus (Fig. 1, F and G), indicating that USP22 is essential for generation of iNKT cells in mice. This defect in iNKT development could not be compensated in the periphery, as indicated by a profound decrease in iNKT cell frequencies and numbers in the spleen and liver of USP22 cKO mice (Fig. 1). It has been shown that USP22 suppression suppresses cancer cell proliferation and induces their apoptosis (Lin et al., 2012). However, the impaired iNKT cell development was unlikely due to the reduction in cell proliferation and the elevated cell death, as neither the BrdU incorporation nor annexin V+ populations of iNKT cells in the thymus, spleen, and liver were altered by USP22 gene deletion (Fig. S2). Therefore, these results indicated that USP22 is required for the development of iNKT cells independent of cell growth and death in mice.
Figure S2.
USP22 suppression has no effects on iNKT proliferation and survival. Seven pairs of WT and USP22 cKO mice were injected with BrdU. 24 h after treatment, mice were euthanized, and single-cell suspensions from their thymus (Thy) were stained with anti-TCRβ, CD1d-αGalCer tetramer, anti-BrdU, and annexin V. The BrdU+ and annexin V+ populations were analyzed on the gated iNKT cells. (A–D) The BrdU incorporation and (C and D) annexin V+ cells were analyzed. Representative images (A and C) and data from seven pairs of mice (B and D) are shown. Student’s t test was used for statistical analysis. In A–D, data are pooled from three independent experiments. Spl, spleen.
USP22 regulates iNKT cell development in a cell-intrinsic manner
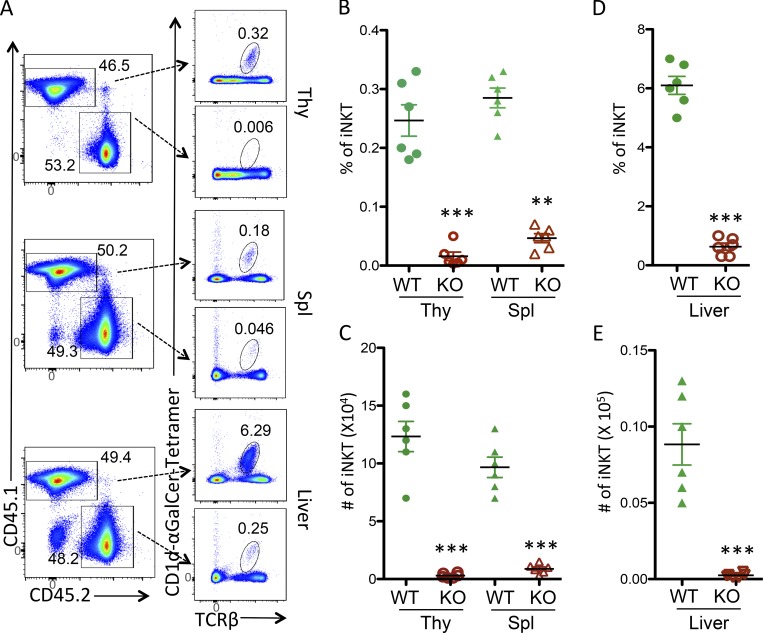
To determine whether USP22 regulates iNKT cell development in a cell autonomous manner, we generated bone marrow chimeras by reconstituting lethally irradiated CD45.1+ congenic WT recipients with a 1:1 mixture of CD45.1+ congenic WT and CD45.2+ USP22 cKO bone marrow cells. We analyzed chimeras 10 wk after the adoptive transfer to determine whether USP22 cKO CD4+CD8+ DP thymocytes developed into iNKT cells in the presence of normal DP thymocytes. As shown in Fig. 2, the donor bone marrow cells from USP22 cKO poorly reconstituted the iNKT cell compartment in thymus, spleen, and liver even in the presence of WT cells (Fig. 2 A). As a consequence, both the frequency and absolute numbers of CD45.2 iNKT cells are largely diminished in the thymus, spleen, and liver when compared with the CD45.1 iNKT cells in the same recipient (Fig. 2, B–E). These studies indicate that USP22 regulates iNKT development in a cell-intrinsic manner.
Figure 2.
USP22 regulation of iNKT cell development is cell intrinsic. Bone marrow cells from USP22 cKO mice and CD45.1-congenic B6/SJL mice were mixed in a 1:1 ratio, and adoptively transferred into the lethally irradiated B6/SJL mice. 10 wk after transfer, recipients were euthanized. iNKT cells in the gated CD45.1 (WT) and CD45.2 (USP22 cKO) populations from thymus (Thy), spleen (Spl), and liver were analyzed by CD1d-αGalCer tetramer and TCRβ. (A–E) The representative images (A), the percentages (B and C), and absolute numbers (D and E) of iNKT cells from six recipient mice are shown. Each symbol (B–E) represents an individual mouse. Error bars represent mean ± SD. Student’s t test was used for statistical analysis. **, P < 0.01; ***, P < 0.001. Data are pooled from three independent experiments (A–E).
USP22 promotes iNKT development during the maturation stage
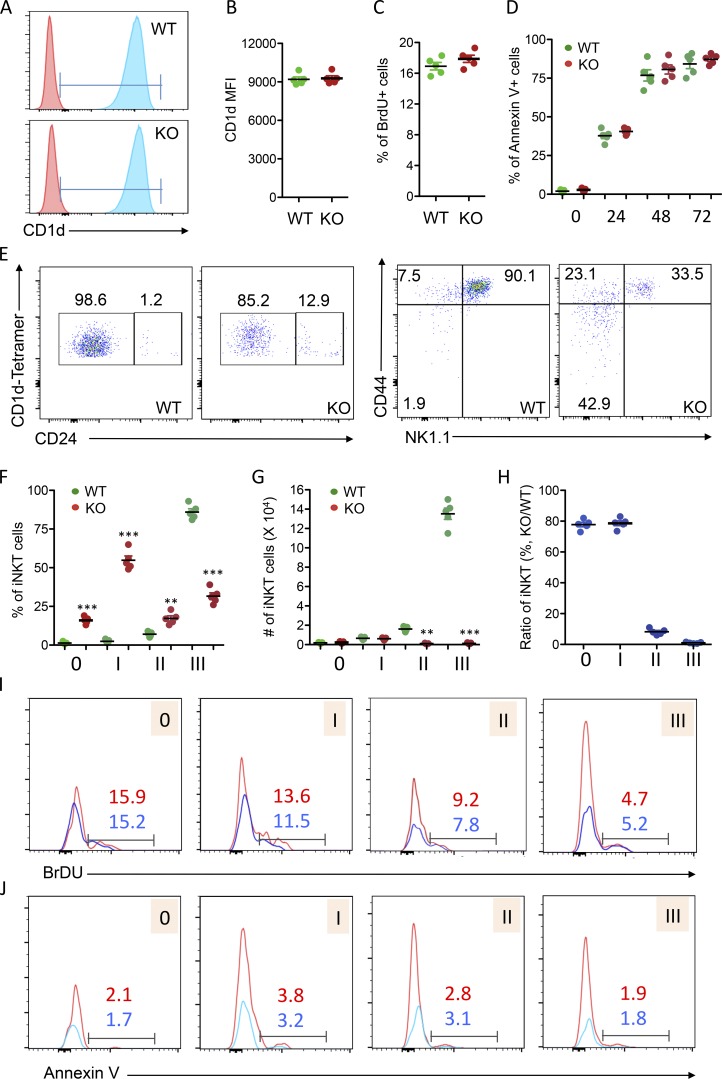
iNKT cells originate from CD4+CD8+ DP thymocytes and are positively selected by CD1d expression (Chiu et al., 1999; Exley et al., 1997). We then speculated that USP22 deficiency impairs iNKT cell development due to the reduced CD1d expression; however, the cell surface CD1d expression levels on the CD4+CD8+ thymocytes were comparable between WT and USP22 cKO mice (Fig. 3, A and B). In addition, since USP22 functions are critical for the proliferation and survival for cancer cells (Melo-Cardenas et al., 2016), we then questioned whether USP22 gene deletion resulted in the increased cell death and reduced growth of CD4+CD8+ iNKT processors. However, the percentage of annexin V+ CD4+CD8+ cells was not altered by targeted USP22 gene deletion even after 24–72 h in culture. In addition, the BrdU incorporation into CD4+CD8+ DP cells was unaltered by USP22 deletion in mice (Fig. 3, C and D). These results largely excluded the possibility that USP22 promotes iNKT cell development by promoting the growth or survival of iNKT precursors.
Figure 3.
Analysis of iNKT cell development and maturation in WT and USP22 cKO mice. (A–D) Five pairs of WT and USP22 cKO mice were injected with BrdU. 24 h after treatment, mice were euthanized, and single-cell suspensions from their thymus were isolated, stained, and analyzed by flow. CD4+CD8+ cells in the thymus of WT and USP22 cKO mice were gated, and expression of CD1d was analyzed by flow cytometry. The representative images (A) and average mean fluorescence intensity (MFI; B) are shown. The BrdU+ proportions of CD4+CD8+ cells were analyzed (C). The annexin V+ cells in the gated CD4+CD8+ population were analyzed after in vitro culture for indicated hours. The average percentages of annexin V+ cells from five independent experiments are shown (D). (E–H) iNKT development in the thymus of WT and USP22 cKO mice was analyzed by their expression of CD24 (E, left panels). CD24−CD1d tetramer+ populations were further analyzed for their expression of NK1.1 and CD44 (E, right panels). Representative images from seven pairs of mice are shown, and the average percentages are indicated. The percentages (F) and absolute numbers (G) of each stage as indicated in E were calculated. The percentages in USP22 cKO iNKT relative to WT iNKT cells from seven pairs of mice are indicated (H). (I and J) The BrdU incorporation (I) and apoptotic cells (J) were analyzed in four pairs of mice and representative data are shown. Each symbol (B–D and F–H) represents an individual mouse. Error bars represent mean ± SD. Student’s t test was used for statistical analysis. **, P < 0.01; ***, P < 0.001. Data are from four experiments with samples pooled from multiple mice (A–H) or are representative of at least four experiments (I and J).
iNKT cells undergo several well-defined developmental stages in the thymus through analyzing their cell surface expression profile of CD24, CD44, and NK1.1. The earliest detectable CD1d tetramer-positive cells are CD24hiCD44−NK1.1− (stage 0). Subsequently, the cells progress through three developmental stages: (1) CD24loCD44loNK1.1−, (2) CD24loCD44hiNK1.1−, and (3) CD24loCD44hiNK1.1+ (Benlagha et al., 2005; Bezbradica et al., 2005; Egawa et al., 2005). The presence of a few residual iNKT cells in the thymus of USP22 cKO mice allowed us to further characterize the transition from earlier to later stages during iNKT development. As shown in Fig. 3 E, in contrast to the WT thymus, in which ∼1% of CD1d tetramer-positive iNKT cells were at stage 0, an average of 15% of CD1d tetramer-positive iNKT cells were at stage 0 in the thymus of USP22 cKO mice (Fig. 3, E and F). However, despite this dramatic accumulation of iNKT cells at the stage 0 in thymus of USP22 knockout mice, their absolute number was not altered due to a >90% reduction in total iNKT cells (Fig. 1 and Fig. 3 G). Similarly, while the percentage of CD24loCD44loNK1.1− stage 1 iNKT cells was increased from 2.5 to 20.5%, their absolute numbers were comparable between WT and USP22 cKO mice. Importantly, even with a slight but statistically significant increase in the percentage of CD24loCD44hiNK1.1− stage 2 iNKT cells, their absolute numbers were decreased >80%. As a consequence, both the percentage and the absolute numbers of CD24loCD44hiNK1.1+ stage 3 iNKT cells are largely reduced (Fig. 3, E–H). Further analysis demonstrated that neither the BrdU incorporation nor the annexin V+ apoptotic cells at each developmental stage during iNKT cell development were altered by USP22 deletion in mice (Fig. 3, I and J). These findings indicated that USP22 function is essential for the transition from stage 1 to 2 during iNKT development.
USP22 functions are involved in regulating iNKT but not the conventional CD4 T cell differentiation
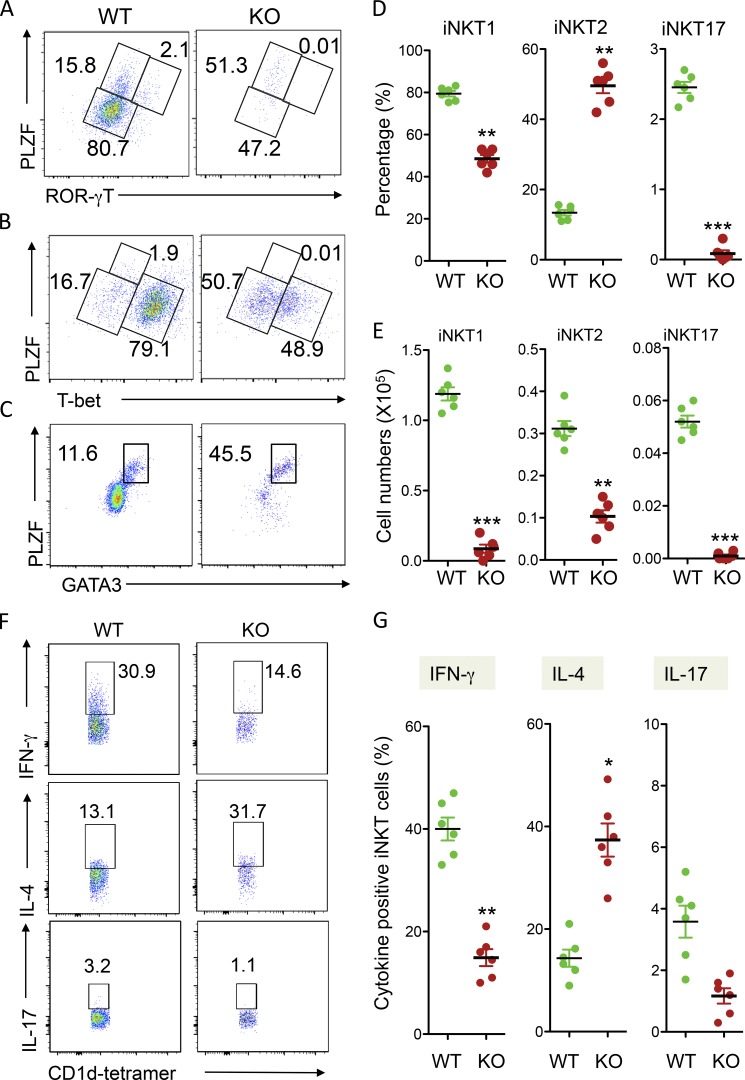
Next, we analyzed the effects of USP22 deficiency on iNKT differentiation. As reported, analysis of the expression levels of promyelocytic leukemia zinc finger (PLZF) and T helper (Th) 17 cell lineage-specific transcription factor ROR-γT in the gated TCRβ and CD1d tetramer-positive thymic iNKT cells by intracellular staining allowed us to define PLZFlowROR-γT− NKT1, PLZFhiROR-γT− NKT2, and PLZFhiROR-γT+ NKT17 cells (Wang et al., 2017). Interestingly, USP22 deficiency resulted in significant reductions in the percentages of NTK1 (from 80.7% to 47.2%) and NKT17 (from 2.1% to 0.01%). In contrast, the percentage of NKT2 cells was increased from 15.8 to 51.3%, suggesting that USP22 is required for NKT1 and NKT17 differentiation, but loss of USP22 functions facilitates iNKT cell differentiation toward NKT2 (Fig. 4, A and D). To support this notion, analysis the Th1 cell lineage-specific transcription factor T-bet by intracellular staining confirmed the significant reduction (from 79.1% to 48.9%) in PLZFlowTbet+ NKT1 cells in USP22 cKO mice (Fig. 4, B and D). In addition, intracellular staining of GATA3, the Th2 cell lineage-specific transcription factor, detected a significant increase of PLZFhiGATA3+ NKT2 cells in USP22 cKO mice (Fig. 4, C and D). Consistent to the reduced percentages, there is a significant reduction in the absolute numbers of NKT1 and NKT17 in USP22 cKO thymus. Despite a significant increase in NKT2 percentages, their absolute numbers were also decreased due to a >90% reduction in total NKT cell populations in USP22 cKO mice (Fig. 4, D and E). Further analysis of the lineage-specific cytokine production validated a significant reduction in IFN-γ production iNKT1 cells and increase in IL-4–producing iNKT2 cells in USP22-null mice. Since the percentage of iNKT17 is extremely low, while our IL-17 intracellular staining analysis detected a similar trend of decreased IL-17–producing iNKT17 cells by USP22 deficiency, statistical analysis did not reach significant changes from five pairs of mice (Fig. 4, F and G). However, USP22 gene deletion affected neither CD4 T cell proliferation (Fig. S3, A and B) nor their in vitro differentiation toward Th1, Th2, Th17, and T reg cells (Fig. S3, C and D). These results suggest an important role of USP22 in specifically reprogramming iNKT cell differentiation.
Figure 4.
Analysis of iNKT differentiation in USP22 cKO mice. (A–C) TCRβ and CD1d tetramer-positive iNKT cells in WT and USP22 cKO thymocytes were gated, and their expression of PLZF and ROR-γT (A), or T-bet (B), or GATA3 (C) was determined by intracellular staining and flow cytometry. (D and E) The average percentages (D) and absolute numbers (E) of iNKT1, iNKT2, and iNKT17 from six pairs of WT and USP22 cKO mice are shown. (F and G) The lineage-specific cytokine production in total splenic iNKT cells was determined by intracellular staining. Representative images (F) and data from six pairs of mice (G) are shown. Each symbol (D, E, and G) represents an individual mouse. Error bars represent mean ± SD. Student’s t test was used for statistical analysis. **, P < 0.01; ***, P < 0.001. In A–G, data are pooled from six independent experiments.
Figure S3.
USP22 suppression on CD4 T cell activation and differentiation. CD4+ T cells were sorted from the spleen of WT and USP22 cKO mice. (A and B) the sorted CD4+ T cells were stained with CFSE and cultivated with anti-CD3 or anti-CD3 plus anti-CD28. Cell proliferation was determined by CFSE dilution (A). The proliferation and division index were analyzed from four independent experiments (B). (C) The IL-2 production was determined by intracellular staining. (D) CD4+ T cells were cultured under each polarization condition and analyzed by flow cytometry. All experiments were repeated at least three times, and representative data are shown. NC, negative control; SSC, side-scattered light; iTreg, induced T regulatory cells.
USP22 suppresses H2A monoubiquitination for the expression of genes driving iNKT development
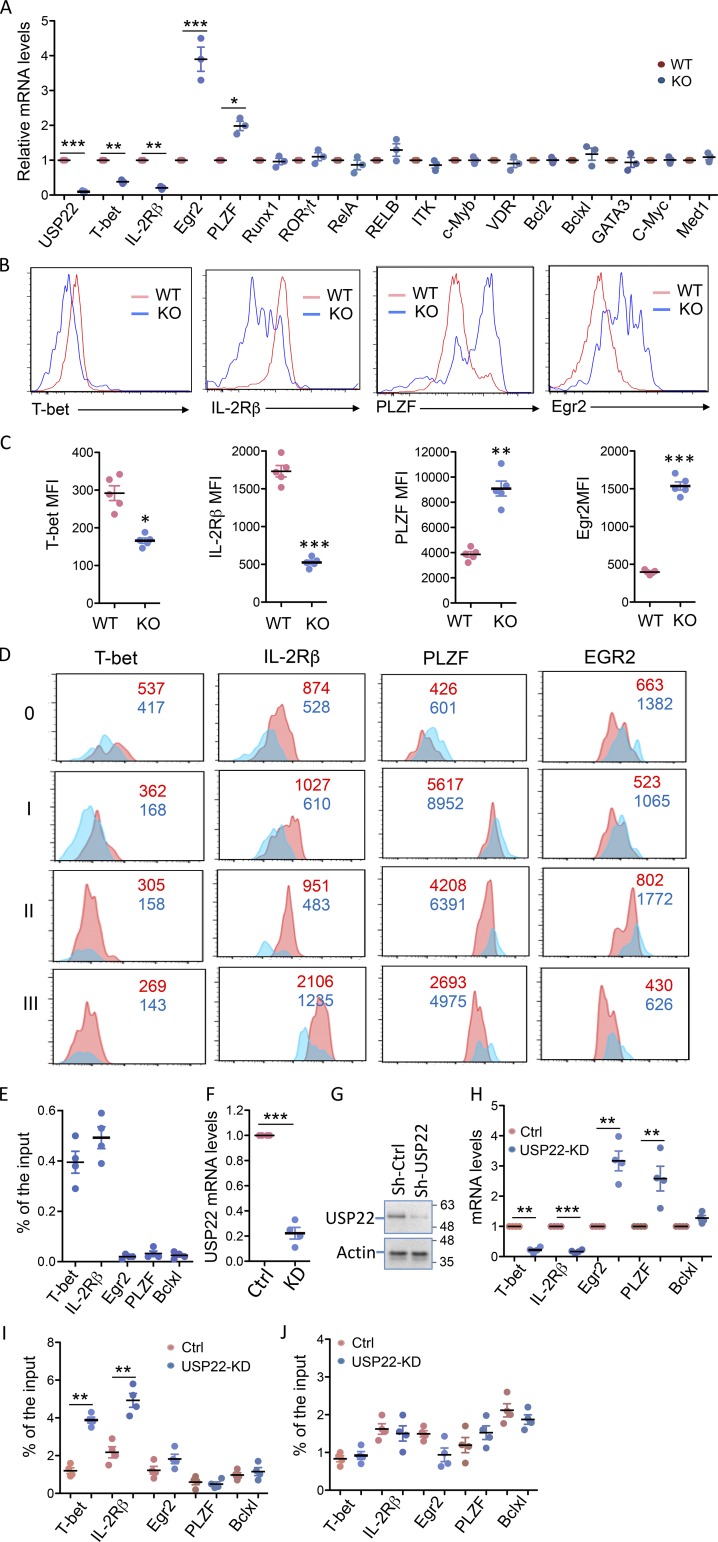
To define the molecular mechanisms underlying how USP22 regulates iNKT cell development and functions, we analyzed the expression levels of genes that have been shown to be critical for iNKT cell development, maturation, and survival (Godfrey et al., 2010). As shown in Fig. 5 A, real-time RT-PCR analysis detected significantly reduction in the expression of T-bet and IL-2Rβ in USP22 cKO thymic iNKT cells compared with WT iNKT cells. We also detected a significant increase in EGR2 and PLZF expression, both of which are involved in promoting early stages of iNKT cell development (Hu et al., 2010). The expression of several other transcription factors including RUNX1, ROR-γT, NF-κB family transcription factor RelA and p50, GATA3, VDR, and c-Myc, as well the Tec kinase ITK, the transcription coactivator MED1, and the anti-apoptotic factors Bcl2 and Bclxl, were not altered by loss of USP22 functions, indicating that USP22 specifically regulates a limited number of factors critical for iNKT development. As a control, the mRNA expression of USP22 was completely diminished in USP22 cKO iNKT cells, further confirming efficient USP22 gene deletion by Cre recombinase driven by the Lck promoter (Fig. 5 A). We then confirmed the differences in protein expression levels of T-bet, IL-2Rβ, PLZF, and EGR2 in WT and USP22 cKO iNKT cells by flow cytometry. Consistent with our real-time quantitative PCR (qPCR) analysis, the protein expression levels of T-bet and IL-2Rβ were significantly lower in USP22 cKO iNKT cells compared with WT cells (Fig. 5, B and C). We also confirmed that the protein expression levels of both PLZF and EGR2 in thymic iNKT cells were dramatically increased by USP22 deletion (Fig. 5, B and C). Both PLZF and EGR2 are enriched during the early stages of iNKT development, raising a possibility that their elevated expression is due to the developmental blocked to early stages by USP22 deletion (Benlagha et al., 2005; Seiler et al., 2012). However, analysis of the expression in T-bet, IL-2Rβ, PLZF, and EGR2 at each developmental stage further confirmed a similar reduction in T-bet and IL-2Rβ, as well as a similar increase in PLZF and EGR2 expression at each stage, largely excluded the possibility that the altered expression is a consequence of the blockade in the transition from stage 1 to 2 during iNKT development by USP22 deficiency (Fig. 5 D). Collectively, our data indicate that USP22 may promote iNKT cell development possibly through, at least partially, regulating the expression of T-bet, IL-2Rβ, PLZF, and EGR2.
Figure 5.
USP22 regulates iNKT cell gene expression through repressing histone H2A monoubiquitination. (A) TCRβ and CD1d tetramer-positive iNKT cells were sorted from the thymus of WT and USP22 cKO mice. mRNA expression levels of each indicated gene were analyzed by real-time RT-PCR. Error bars represent the standard error of the mean of three independent experiments. (B–D) The expression of T-bet, IL-2Rβ, PLZF, and EGR2 in thymic iNKT cells were analyzed. Representative images (B) and the average MFI (C) from five pairs of mice are shown. Their expression at each developmental stage is analyzed and representative data are shown (D). (E) Primary mouse iNKT cells were sorted, and the binding of USP22 to the promoter of each indicated gene was determined by ChIP analysis. (F and G) DN32.D3 cells were infected with virus that carries an USP22-specific shRNA. 3 d after infection, GFP+ cells were sorted. The expression levels of USP22 mRNA (F) and protein (G) were determined. (H) The expression levels in USP22 KD and control DN32.D3 iNKT cells were determined by real-time RT-PCR. (I and J) The levels of histone H2A (I) or H2B (J) at the promoter region of each indicated gene was analyzed by ChIP using specific antibodies. Error bars represent mean ± SD. Student’s t test was used for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data are representative of three experiments in A, three experiments with five pairs of mice for C, and four independent experiments for E, F, and H–J. KD, knockdown; Sh-Ctrl, small hairpin control.
As a transcription coactivator component, USP22 is often recruited to the promoter region of target genes (Lang et al., 2011; Zhang et al., 2008; Zhao et al., 2008). We then tested whether USP22 binds to the promoter of genes whose expression is altered in USP22-null iNKT cells. Indeed, the binding of USP22 to the promoters of both T-bet and IL-2Rβ gene was detected by chromatin immunoprecipitation (ChIP) assay (Fig. 5 E). In contrast, while USP22 deletion increased EGR2 and PLZF transcription, ChIP analysis failed to detect USP22 binding to their promoters, implying that USP22 suppresses EGR2 and PLZF transcription through an indirect mechanism. USP22 has been shown to regulate gene transcription through suppressing the monoubiquitination of histone H2A or H2B or both (Zhang et al., 2008; Zhao et al., 2008). While it would be ideal to analyze the levels of H2AUb or H2BUb at T-bet and IL-2Rβ promoters in USP22-null iNKT cells, it is technically difficult as the numbers of iNKT cells from USP22 cKO mice are extremely limited. To overcome this, we used an shRNA-mediated knockdown approach to knock down USP22 expression in DN32.D3 cells, a mouse iNKT hybridoma cell line. Real-time RT-PCR and Western blotting analysis confirmed that the shRNA efficiently inhibited >85% of USP22 mRNA and protein expression in DN32.D3 cells (Fig. 5, F and G). Further analysis confirmed that USP22 suppression resulted in a significant reduction in T-bet and IL-2Rβ but elevated EGR2 and PLZF expression, as we observed in primary USP22-null iNKT cells (Fig. 5 H). Importantly, USP22 suppression resulted in a dramatic increase in the levels of histone H2A but not H2B monoubiquitination at the promoter region of T-bet and IL-2Rβ (Fig. 5, I and J), indicating that USP22 promotes the expression of genes required for iNKT development through suppressing histone H2A monoubiquitination. Similar to our finding that USP22 is not recruited to both PLZF and EGR2 promoters, neither H2A nor H2B monoubiquitination levels on their promoters were altered by USP22 knockdown in DN32.D3 cells (Fig. 5, I and J).
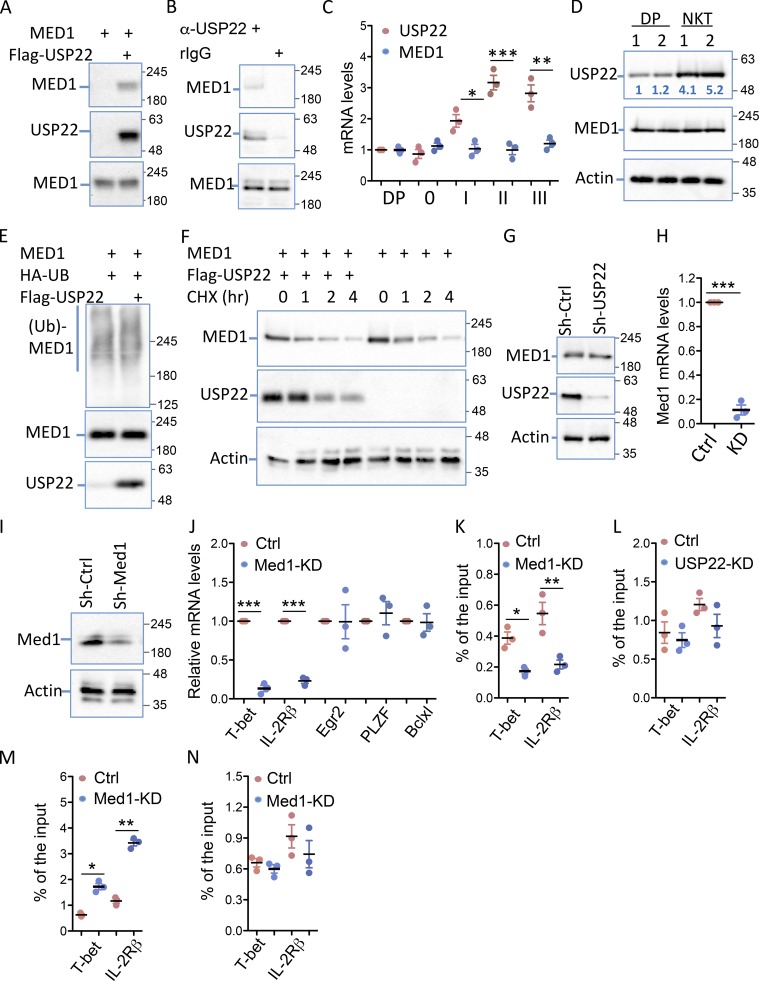
USP22 interacts with MED1 in iNKT cells
As a transcription coactivator, USP22 is often recruited to the specific gene promoter through interacting with transcription factors or coactivators. Indeed, we discovered that USP22 interacts with MED1, a transcription coactivator that has been shown to be critical for iNKT development (Yue et al., 2011), in transiently transfected HEK293 cells (Fig. 6 A). The interaction between the endogenous USP22 and MED1 was further validated in mouse primary iNKT cells because MED1 was detected in anti-USP22 immunoprecipitates but not the normal rabbit IgG controls (Fig. 6 B), indicating a possibility that USP22 regulates iNKT cell development through, at least partially, MED1 interaction. However, we are aware that, unlike USP22, whose deletion blocks iNKT cell development from stage 1 to 2 transition, MED1 promotes iNKT development from CD4+CD8+ processor to stage 0 transition (Yue et al., 2011). Interestingly, real-time RT-PCR analysis indicated that MED1 is constitutively expressed in all stages during iNKT development. In contrast, USP22 expression remained at a lower level during the early stages of iNKT development and is significantly up-regulated from stage 1, which is further increased at stages 2 and 3 (Fig. 6 C). Since it is technically challenging to obtain sufficient iNKT cells at each developmental stage (in particular in stages 0 and 1) due to their low frequency to analyze USP22 protein expression levels, we compared the USP22 protein expression in pan-iNKT cells with that in CD4+CD8+ cells. Consistent with our real-time RT-PCR analysis, an average of fourfold to fivefold increase in USP22 protein expression was confirmed in iNKT cells, but the expression levels of MED1 were comparable between iNKT and CD4+CD8+ cells (Fig. 6 D). These results partially explain the reason why USP22-deficency impairs iNKT development at a later stage comparing to that of MED1 loss of function. As a ubiquitin-specific peptidase, USP22 often stabilizes its interaction partners through suppressing their ubiquitination (Lin et al., 2012, 2015). However, USP22 expression failed to suppress the levels of MED1 ubiquitination (Fig. 6 E). Consistent with this observation, USP22 expression did not increase the half-life of MED1 protein (Fig. 6 F). In addition, MED1 protein expression levels were not increased by USP22 knockdown in iNKT cells (Fig. 6 G). Therefore, it is unlikely that USP22 regulates iNKT cell development through MED1 stabilization.
Figure 6.
USP22 interacts with MED1 to regulate T-bet and IL-2Rβ transcription in iNKT cells. (A) MED1 interaction with USP22 in transiently transfected HEK293 cells was determined by coimmunoprecipitation with anti-Flag and Western blotting with anti-MED1 (top panel). The same membrane was reprobed with anti-USP22 (middle panels), and the expression of MED1 in the whole cell lysate was detected by Western blotting with anti-MED1 (bottom panel). (B) The interaction of endogenous USP22 with MED1 in DN32.D3 cells was determined using anti-USP22 and control rabbit IgG as described in A. (C and D) The expression levels of USP22 and MED1 in sorted CD4+CD8+ DP cells and iNKT cells at each stage were determined by real-time RT-PCR (C). The protein expression levels of USP22 and MED1 in sorted CD4+CD8+ cells and iNKT cells were determined by Western blotting, and the numbers indicate the relative levels of USP22 protein expression (D). (E) MED1 and HA-ubiquitin (Ub) expression plasmids were cotransfected with or without USP22, and the ubiquitin conjugation on MED1 was determined by coimmunoprecipitation with anti-MED1 and Western blotting with anti-HA (top panel). The same membrane was reblotted with anti-MED1 (middle panel), and the levels of USP22 protein in the whole cell lysates were determined (bottom panel). (F) The effect of USP22 expression on MED1 protein stability was determined in HEK293 cells treated with cychloheximide (CHX) for different times. (G) The expression levels of MED1 in USP22 KD cells were determined with β-actin as a loading control. (H and I) MED1-specific knockdown DN32.D3 cells were generated and validated by real-time RT-PCR (H) and Western blotting (I). (J) The expression levels of each indicated genes in the MED1 knockdown (KD) and WT control (Ctrl) cells were determined by real-time RT-PCR. (K and L) The promoter-binding of USP22 in MED1 KD cells (K) and MED1 in USP22 KD (L) cells were determined by ChIP. (M and N) The levels of H2A (M) and H2B (N) monoubiquitination at the promoters of T-bet and IL-2Rβ were analyzed by ChIP. Error bars represent mean ± SD from three experiments (A–N). Student’s t test was used for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
USP22 regulates the expression of genes driving iNKT development partially through MED1
Since USP22 interacts with MED1, both of which are transcription coactivators involved in promoting T-bet and IL-2R gene transcription in iNKT cells, we then asked whether MED1 binds to the promoters of T-bet and IL-2Rβ in a USP22-dependent manner, or vice versa. To achieve this, we used an shRNA approach and generated the MED1 knockdown DN32.D3 cells, in which MED1 mRNA and protein expression were largely diminished (Fig. 6, H and I). Similar to that in USP22 knockdown DN32.D3 cells, the expression levels of both T-bet and IL-2Rβ genes were dramatically reduced by MED1 suppression (Fig. 6 J). However, unlike USP22 knockdown, MED1 suppression did not alter the expression of EGR2 and PLZF (Fig. 6 J). These results suggest that USP22 promotes T-bet and IL-2Rβ gene transcription in a MED1-dependent manner, but it suppresses EGR2 and PLZF expression independent of MED1.
Notably, MED1 knockdown significantly reduced the levels of USP22 binding to both T-bet and IL-2Rβ promoters (Fig. 6 K). In contrast, the binding of MED1 to T-bet and IL-2Rβ promoters was unaltered by USP22 suppression (Fig. 6 L). These results clearly indicate that USP22 is recruited to T-bet and IL-2Rβ promoters through MED1 interaction. To support this notion, we further detected a dramatic increase in the levels of histone H2A but not H2B monoubiquitination at the T-bet and IL-2Rβ promoters in MED1 knockdown DN32.D3 iNKT cells (Fig. 6, M and N). Collectively, our data indicate that USP22 regulates the expression of genes driving iNKT development partially through MED1 interaction.
Vα14-Jα18 TCR transgenic expression bypasses iNKT cell defects in USP22 cKO mice
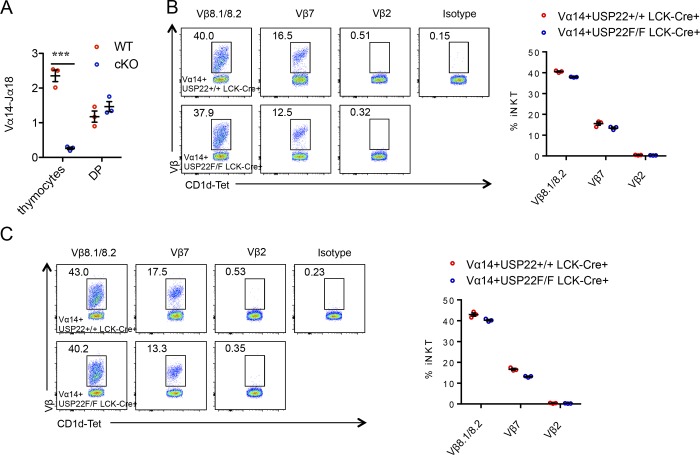
It has been shown that the developmental defect of iNKT cells in MED1-null mice can be rescued by the forced expression of an iNKT cell–specific Vα14-Jα18 TCR transgene (Yue et al., 2011). If USP22 promotes iNKT development through MED1, we would expect that the forced expression of an iNKT cell–specific Vα14-Jα18 TCR transgene could rescue iNKT development in USP22 cKO mice. Therefore, we crossed USP22 cKO mice with transgenic mice expressing Vα14-Jα18 TCR transgene under the control of CD4 promoter. Indeed, both the relative proportion and their absolute numbers of iNKT cells were largely restored in the thymus, spleen, and liver of USP22 cKO mice by the Vα14-Jα18 TCR transgene (Fig. 7, A–C). Vα14-Jα18 TCR transgenic expression resulted in accumulation of iNKT cells during the early developmental stages (Egawa et al., 2005), and further analysis detected a similar iNKT developmental pattern in the thymus of WT and USP22-null mice (Fig. 7, D and E). We also detected a slight but statistically significant decrease in T-bet and IL-2Rβ and increase in PLZF and EGR2 in USP22-null Vα14-Jα18 TCR transgenic iNKT cells (Fig. 7, F and G), but their changes by USP22 deficiency are rather modest compared to those in mice without the Vα14-Jα18 TCR transgene, implying that Vα14-Jα18 TCR transgenic expression largely rescued USP22 deficiency. To further delineate how Vα14-Jα18 TCR transgenic expression partially rescues iNKT cell development, we analyzed the TCRα rearrangement in USP22 KO mice in the absence of Vα14-Jα18 TCR transgene. However, the levels of Vα14-Jα18 TCR rearrangement were comparable in the iNKT precursor CD4 CD8 DP T cells between WT and USP22 KO mice (Fig. S4 A). In addition, analysis of the TCR Vβ repertoire did not detect any changes by USP22 deficiency in Vα14-Jα18 TCR transgenic mice (Fig. S4, B and C), suggesting that the rescue of USP22 deficiency by Vα14-Jα18 TCR transgenic expression is not due to the altered TCR Vβ repertoire.
Figure 7.
Vα14-Jα18 TCR transgenic expression restores iNKT cell development in USP22 cKO mice. (A–C) iNKT cells in the thymus (Thy), spleen (Spl), and liver from Vα14-Jα18/USP22+/+ and Vα14-Jα18/USP22−/− mice were determined by flow cytometry. Representative images (A), the percentages (B), and absolute numbers (C) of iNKT cells from seven pairs of mice are shown. (D and E) The developmental stages were analyzed as in Fig. 3. Representative images (D) and data from four pairs of mice (E) are shown. (F and G) The levels of T-bet, IL-2Rβ, PLZF, and EGR2 in thymic iNKT cells were analyzed. Representative images (F) and the average MFI (G) from four pairs of mice are shown. (H) A proposed model for USP22-mediated histone H2A deubiquitination through MED1 interaction to regulate genes critical for iNKT development. Each symbol (B, C, E, and G) represents an individual mouse. Error bars represent mean ± SD. Student’s t test was used for statistical analysis. *, P < 0.05; **, P < 0.01. Data are representative of five experiments (A–C) or three experiments (D–G). TF, transcriptional factor.
Figure S4.
Analysis of TCR rearrangement and repertoire of iNKT cells in Vα14 transgenic mice. (A) RNA was extracted from total thymocytes or sorted CD4+CD8+ cells from WT and USP22 KO mice. The levels of Vα14 Jα18 region were determined by real-time PCR. Data from three pairs of mice are shown. (B and C) The expression levels of Vb2, Vb7, and Vb8 on gated CD3+CD1d-tetramer+ iNKT cells from thymus (B) and spleens (C) of Vα14 and Vα14 USP22 KO mice were analyzed using isotype controls. Representative flow images and data from three pairs of mice are shown. Student’s t test was used for statistical analysis. ***, P < 0.001. (A–C) Data are representative of three independent experiments.
Based on the observation, we proposed a model for USP22 in regulating iNKT cell development and functions (Fig. 7 H). USP22 is recruited to the T-bet and IL-2Rβ promoters through its interaction with another transcription coactivator, MED1. Instead of deubiquitinating and stabilizing MED1, USP22 inhibits the histone H2A monoubiquitination to promote expression of genes that are required for iNKT development, including T-bet and IL-2Rβ. As a consequence, targeted deletion of USP22 gene resulted in iNKT developmental defect by largely blocking its transition from stage 1 to 2.
Discussion
Our current study has defined a previously unappreciated epigenetic mechanism essential for regulating iNKT development and differentiation by the deubiquitinase USP22-mediated suppression of histone H2A monoubiquitination. This conclusion is supported by the following innovative observations: first, genetic USP22 suppression specifically blocks iNKT cell development without affecting conventional T cell selection in a cell autonomous manner; second, the targeted deletion of USP22 resulted in the reduced NKT1 and NKT17 cells, in contrast the percentages of NKT2 cells are increased in the USP22 cKO mice; third, USP22 promotes the expression of genes required for iNKT development including T-bet and IL-2Rβ through suppressing target specific histone H2A but not H2B monoubiquitination; and last but not least, USP22 is recruited to specific promoters through its interaction with another transcription coactivator, MED1, the functions of which are also essential for iNKT development and functions.
USP22 appears to regulate iNKT cell development and functions through, at least partially, its interaction with another transcription coactivator, MED1. Both in USP22- and MED1-deficient iNKT cells, T-bet and IL-2Rβ expressions are markedly reduced, most probably accounting for the development block in the final maturation of thymic iNKT cells as well as iNKT1 differentiation. Indeed, T-bet deletion in mice blocks iNKT cell terminal maturation and hemostasis (Townsend et al., 2004; Yokoyama, 2004). At the molecular level, USP22 is recruited to T-bet as well as IL-2Rβ promoters through its interaction of MED1, as a consequence the levels of histone H2A but not H2B monoubiquitination is repressed to turn on T-bet and IL-2Rβ gene transcription. Despite this, USP22-null mice did not perfectly phenocopy MED1 knockout mice in iNKT cell development because USP22 suppression blocks iNKT development during stage 1 transition to 2, but loss of MED1 functions resulted in an earlier blockade at stage 0. This is partially because USP22 expression level is up-regulated at stage 1 during iNKT development; in contrast, MED1 is constitutively expressed at all stages. Loss of T-bet leads to a higher iNKT turnover as both proliferation (BrdU incorporation) and apoptosis (Townsend et al., 2004; Yokoyama, 2004). In contrast, neither MED1 nor USP22 deletion, while they lead to the reduced T-bet expression, affected iNKT cell turnover in mice.
USP22 deletion resulted in the elevated expression of EGR2 and PLZF, both of which are important for iNKT development and functions. However, MED1 suppression did not alter the expression of EGR2 and PLZF. These discrepancies suggest a MED1-independent mechanism underlying USP22 regulation of iNKT development. In fact, in addition to MED1 and T-bet, several transcription factors have been discovered critical for earlier stages of iNKT development, including EGR2 (Lazarevic et al., 2009), Runx1 (Egawa et al., 2005), PLZF (Savage et al., 2008), c-Myb (Hu et al., 2010), and some of the NF-κB proteins (Beraza et al., 2009; Elewaut et al., 2003; Schmidt-Supprian et al., 2004; Sivakumar et al., 2003; Vallabhapurapu et al., 2008; Yue et al., 2005). Further studies are needed to elucidate whether USP22 regulates the transcriptional activation of any of these transcription factors to control iNKT cell development and activation.
We have recently shown that the lysine acetyltransferase GCN5 plays a critical role in iNKT development and functions (Wang et al., 2017). Since both GCN5 and USP22 are components of the SAGA transcription coactivator complex, USP22 may regulate iNKT cell development and functions through SAGA-mediated transcriptional activation. Indeed, targeted deletion of either USP22 or GCN5 largely impaired both iNKT1 and iNKT17 differentiation but enhanced iNKT2 populations (Wang et al., 2017). However, GCN5 appears to promote iNKT development at an earlier stage. More importantly, GCN5 functions as an acetyltransferase of EGR2 to promote iNKT development. Loss of GCN5 functions resulted in the diminished EGR2 transcriptional activity without altering EGR2 expression. In contrast, USP22 suppression even significantly increased EGR2 expression. In addition, GCN5 is required for EGR2 target genes RUNX1 and PLZF expression, but USP22 deletion did not alter RUNX1 expression and even enhanced PLZF expression. Therefore, USP22 is likely to regulate iNKT development through a distinct molecular pathway from GCN5.
While both EGR2 and PLZF are up-regulated in USP22-null iNKT cells, USP22 suppresses EGR2 and PLZF though an indirect mechanism because USP22 binding to either EGR2 or PLZF promoter in iNKT cells was not detected. Since EGR2 regulates PLZF transcription upon GCN5-mediated acetylation (Wang et al., 2017), it is possible that the increased PLZF expression is a consequence of the elevated EGR2 expression in USP22-null iNKT cells. In addition, it has been shown that USP22 interacts with the c-Myc transcription factor in cancer cells (Zhang et al., 2008), and genetic c-Myc suppression impairs iNKT development but with increased PLZF expression (Dose et al., 2009; Mycko et al., 2009), raising the possibility that USP22 represses PLZF expression through its interaction with c-Myc. If in this the case, one would expect that USP22 would be recruited onto the promoter of the PLZF gene. However, ChIP analysis failed detect USP22 binding to PLZF promoter DNA, together with the fact that USP22 interaction with c-Myc enhances but not suppresses c-Myc transcriptional activity (Zhang et al., 2008), largely excluding the possibility that USP22 inhibits PLZF transcription through c-Myc. Further studies are need to dissect the molecular mechanisms underlying how USP22 represses EGR2 and PLZF expression as well as its functional consequences in iNKT cells.
Similar to that observed in MED1 KO mice (Lazarevic et al., 2009), the Vα14-Jα18 TCR transgene largely rescued USP22 deficiency in iNKT development. In addition, while the underlying molecular mechanisms remain largely unknown, Vα14-Jα18 TCR transgenic expression under the CD4 promoter could rescue iNKT developmental defects by targeted deletion of several genes. It has been shown that c-Myb suppression impairs TCR Vα14-Jα18 rearrangement; therefore, it can be rescued by transgenic expression of the prerearranged Vα14-Jα18 TCR (Hu et al., 2010). However, USP22 deficiency did not alter the TCR Vα14-Jα18 rearrangement in mice, suggesting the rescue of USP22 deficiency is likely through a different mechanism. In addition, USP22 appears not to regulate the iNKT cell TCRβ chain repertoire, largely excluding the possibility that TCR Vα14-Jα18 transgene rescues USP22 deficiency through altering the TCR repertoire. Further studies are needed to dissect the molecular mechanisms underlying how TCR Vα14-Jα18 transgene rescues USP22 deficiency in iNKT development.
In summary, our study identified a surprisingly lineage-specific function of genetic cofactor USP22 in iNKT development. USP22 appears to promote iNKT development through, at least partially, its interaction with another transcription coactivator, MED1. Specific inhibitors that suppress the deubiquitinase catalytic activity of USP22 may have therapeutic potential in the treatment of iNKT-mediated immune disorders, including autoimmune diseases.
Materials and methods
Mice
USP22 floxed mice were generated using C57/BL6/j ES cells and used as recently described (Melo-Cardenas et al., 2018). T cell–specific USP22-null mice (USP22 cKO) were then generated by breeding USP22 floxed mice with Lck-Cre transgenic mice. LCK-Cre transgenic (stock no. 003802) mice and CD45.1 congenic B6/SJL mice (stock no. 002014) were purchased from Jackson Laboratory. LCK-Cre+USP22+/f or LCK-Cre+USP22+/+ littermates were used as controls. Primers used for genotyping the genetically modified mice are listed in the Table S1 A. The Vα14Tg mice on a B6 background were a kind gift by A. Bendelac (University of Chicago, Chicago, IL) and used as reported (Egawa et al., 2005). Vα14Tg and T cell–specific USP22-null mice (Vα14+USP22f/f LCK-Cre+) were then generated by breeding USP22 cKO mice with Vα14Tg mice. Vα14+USP22f/f LCK-Cre+ and their littermate controls (Vα14+USP22+/+ LCK-Cre+) were used for the experiments. All mice used in this study were at the C57/Black6 genetic background and housed at the Northwestern University (Evanston, IL) mouse facility under pathogen-free conditions according to institutional guidelines. The animal study proposal has been approved by the Institutional Animal Care and Use Committee of Northwestern University.
Cells, antibodies, and plasmids
Human HEK293 cells were cultured in DMEM with 10% FBS. Mouse iNKT hybridoma DN32.D3 cells were cultivated in RPMI1640 with 10% FBS. Antibodies specifically against USP22, Flag, and HA were from Santa Cruz, and against MED1 and acetylated lysine were from Cell Signaling. Fluorescence-conjugated antibodies used for cell surface marker analysis and intracellular staining including CD3, CD4, CD8, NK1.1, CD24, CD45.1, CD45.2, CD44, IL-2Rβ/CD122, T-bet, PLZF, EGR2, and antibodies against each specific isotype of mouse immunoglobulin are listed in Table S1 C. USP22 expression plasmid was purchased from Addgene, and MED1 expression plasmids were used as reported previously (Chen et al., 2009).
Flow cytometry analysis
Single-cell suspensions of thymus or spleen were used for the analysis of the cell surface markers including CD3, TCRβ, NK1.1, CD24, and CD44 by labeling with fluorescence-labeled antibodies specific to each cell surface marker, followed by flow cytometry analysis. CD1d/PBS57 tetramers were obtained from the National Institutes of Health tetramer facility and used for identification of iNKT cells as reported previously (Zhao et al., 2014). For the analysis of transcription factors including T-bet, PLZF, GATA3, ROR-γT, and EGR2 in primary iNKT cells, intracellular staining was performed using a Biolegend intracellular staining kit. For the analysis of iNKT cells from liver, mice were euthanized, and the liver was infiltrated with collagenase. Liver tissues were homogenized and filtered, and the resident lymphocytes were isolated by Percoll purification as reported (Zhang et al., 2009). All the antibodies used for flow cytometry analysis are listed in the Table S1 B.
Bone marrow chimera and adoptive transfer
WT marrow cells were isolated from the CD45.1+ congenic Swiss Jim Lambert (SJL) mice and mixed with these isolated from USP22 cKO (CD45.2) mice at a 1:1 ratio. A total of 107 mixed cells were adoptively transferred into lethally irradiated SJL mice by intravenous injection. 10 wk after adoptive transfer, recipient mice were euthanized, and the iNKT cells in thymus, spleen, and liver were analyzed by flow cytometry using fluorescence-labeled CD45.1, CD45.2, TCRβ, and CD1d-tetramer.
Real-time qPCR analysis
Total RNA from the sorted mouse primary iNKT cells or DN32.D3 cells were extracted using Trizol (Invitrogen). mRNAs were reverse-transcribed using a cDNA synthesis kit purchased from Invitrogen. The expression levels of genes were analyzed by real-time qPCR. All primers used in the study are listed in Table S1 A.
Coimmunoprecipitation and Western blotting analysis
Experiments were performed as reported previously (Lin et al., 2012). Briefly, transient transfection of DN32.D3 cells was performed using Lipofectamine 2000 (Invitrogen). 2 d after transfection, cells were collected and lysed in NP-40 lysis buffer (1% NP-40, 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, and freshly added protease inhibitor cocktail). The cell lysates were precleaned and then incubated with antibodies (1 µg) for 2 h on ice, followed by the addition of 30 µl of fast-flow protein G–Sepharose beads (GE Healthcare Bioscience) overnight at 4°C. Immunoprecipitates were washed four times with NP-40 lysis buffer and boiled in 30 µl 2× Laemmli buffer. Samples were separated by 8% or 10% SDS-PAGE and electro-transferred onto nitrocellulose membranes (0.45 µm; Bio-Rad). Membranes were probed with the indicated primary antibodies, followed by horseradish peroxidase–conjugated secondary antibodies. Membranes were then washed and visualized with an enhanced chemiluminescence detection system (Bio-Rad). When necessary, membranes were stripped by incubating in stripping buffer (Thermo), washed, and then reprobed with other antibodies as indicated. All antibodies used for coimmunoprecipitation and Western blotting analysis are listed in Table S1 C.
ChIP
DN32.D3 cells were cross-linked with 10% formalin and subjected to ChIP using the Chromatin Immunoprecipitation Assay Kit (Millipore) as reported previously (Kong et al., 2015). In brief, 2 × 106 cells were lysed in SDS lysis buffer. Cell lysates were sonicated, and 3% of each sample was used to determine the total amount of target DNA. The remaining cell lysate was diluted in ChIP dilution buffer. Immunoprecipitation was performed with each of the indicated antibodies (3 μg) at 4°C overnight. Immune complexes were then mixed with a salmon sperm DNA/protein agarose 50% slurry at 4°C for 1 h. After immunoprecipitates were washed sequentially with low-salt buffer, high-salt buffer, LiCl wash buffer, and Tris EDTA, DNA–protein complexes were eluted with elution buffer, and cross-linking was reversed. Genomic DNA was extracted using phenol/chloroform, and ethanol-precipitated DNA was resuspended in Tris EDTA. qPCR was then performed with specific primers. All primers and the antibodies used for ChIP are listed in the Table S1.
Online supplemental material
Fig. S1 shows that USP22 deletion did not alter the congenital T lymphocyte development in mice. Fig. S2 shows that USP22 is not involved in iNKT cell proliferation and survival in vivo. Fig. S3 shows that USP22 is dispensable for in vitro activation and differentiation of congenital T cells. Fig. S4 shows that USP22 is not involved in regulating TCR rearrangement and repertoire of iNKT cells in Vα14 transgenic mice. Table S1 lists the primers and antibodies used.
Supplementary Material
lists primers used for the study, antibodies used for the flow cytometry analysis, and antibodies used for Western blotting, ChIP, and immunoprecipitation.
Acknowledgments
We thank Fang laboratory members for critical reading of the manuscript and constructive suggestions during our research.
This work was supported by National Institutes of Health R01 grants (AI079056, AI108634, AR006634, DK120330 and CA232347) to D. Fang as well the National Natural Science Foundation of China (81773061) to Z. Sun.
Author contributions: Y. Zhang, Y. Wang, B. Gao, Y. Sun, L. Cao, and S.M. Genardi performed the experiments and analyzed the data. C.R. Wang and Z. Sun contributed critical reagent and in experimental design. H. Li, Y. Yang, C.R. Wang, and D. Fang designed the study, analyzed the data, and wrote the manuscript.
References
- Atanassov B.S., and Dent S.Y.. 2011. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep. 12:924–930. 10.1038/embor.2011.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov B.S., Evrard Y.A., Multani A.S., Zhang Z., Tora L., Devys D., Chang S., and Dent S.Y.. 2009. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol. Cell. 35:352–364. 10.1016/j.molcel.2009.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A., Savage P.B., and Teyton L.. 2007. The biology of NKT cells. Annu. Rev. Immunol. 25:297–336. 10.1146/annurev.immunol.25.022106.141711 [DOI] [PubMed] [Google Scholar]
- Benlagha K., Wei D.G., Veiga J., Teyton L., and Bendelac A.. 2005. Characterization of the early stages of thymic NKT cell development. J. Exp. Med. 202:485–492. 10.1084/jem.20050456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraza N., Malato Y., Sander L.E., Al-Masaoudi M., Freimuth J., Riethmacher D., Gores G.J., Roskams T., Liedtke C., and Trautwein C.. 2009. Hepatocyte-specific NEMO deletion promotes NK/NKT cell- and TRAIL-dependent liver damage. J. Exp. Med. 206:1727–1737. 10.1084/jem.20082152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezbradica J.S., Hill T., Stanic A.K., Van Kaer L., and Joyce S.. 2005. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4+8+ stage of thymic ontogeny. Proc. Natl. Acad. Sci. USA. 102:5114–5119. 10.1073/pnas.0408449102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Gao B., Zhang J., McEwen T., Ye S.Q., Zhang D., and Fang D.. 2009. The HECT-type E3 ubiquitin ligase AIP2 inhibits activation-induced T-cell death by catalyzing EGR2 ubiquitination. Mol. Cell. Biol. 29:5348–5356. 10.1128/MCB.00407-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y.H., Jayawardena J., Weiss A., Lee D., Park S.H., Dautry-Varsat A., and Bendelac A.. 1999. Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J. Exp. Med. 189:103–110. 10.1084/jem.189.1.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dose M., Sleckman B.P., Han J., Bredemeyer A.L., Bendelac A., and Gounari F.. 2009. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc. Natl. Acad. Sci. USA. 106:8641–8646. 10.1073/pnas.0812255106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees C., Vahl J.C., Bortoluzzi S., Heger K.D., Fischer J.C., Wunderlich F.T., Peschel C., and Schmidt-Supprian M.. 2017. Roquin Paralogs Differentially Regulate Functional NKT Cell Subsets. J. Immunol. 198:2747–2759. 10.4049/jimmunol.1601732 [DOI] [PubMed] [Google Scholar]
- Drennan M.B., Govindarajan S., Verheugen E., Coquet J.M., Staal J., McGuire C., Taghon T., Leclercq G., Beyaert R., van Loo G., et al. 2016. NKT sublineage specification and survival requires the ubiquitin-modifying enzyme TNFAIP3/A20. J. Exp. Med. 213:1973–1981. 10.1084/jem.20151065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T., Eberl G., Taniuchi I., Benlagha K., Geissmann F., Hennighausen L., Bendelac A., and Littman D.R.. 2005. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 22:705–716. 10.1016/j.immuni.2005.03.011 [DOI] [PubMed] [Google Scholar]
- Elewaut D., Shaikh R.B., Hammond K.J., De Winter H., Leishman A.J., Sidobre S., Turovskaya O., Prigozy T.I., Ma L., Banks T.A., et al. 2003. NIK-dependent RelB activation defines a unique signaling pathway for the development of V alpha 14i NKT cells. J. Exp. Med. 197:1623–1633. 10.1084/jem.20030141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I., Zhao M., Kappes D., Taniuchi I., and Kronenberg M.. 2012. The transcription factor Th-POK negatively regulates Th17 differentiation in Vα14i NKT cells. Blood. 120:4524–4532. 10.1182/blood-2012-01-406280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exley M., Garcia J., Balk S.P., and Porcelli S.. 1997. Requirements for CD1d recognition by human invariant Valpha24+ CD4-CD8- T cells. J. Exp. Med. 186:109–120. 10.1084/jem.186.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky G.V., Berezovska O., and Glinskii A.B.. 2005. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 115:1503–1521. 10.1172/JCI23412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey D.I., Stankovic S., and Baxter A.G.. 2010. Raising the NKT cell family. Nat. Immunol. 11:197–206. 10.1038/ni.1841 [DOI] [PubMed] [Google Scholar]
- Hennet T., Hagen F.K., Tabak L.A., and Marth J.D.. 1995. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc. Natl. Acad. Sci. USA. 92:12070–12074. 10.1073/pnas.92.26.12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T., Simmons A., Yuan J., Bender T.P., and Alberola-Ila J.. 2010. The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat. Immunol. 11:435–441. 10.1038/ni.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki N., Antonenko S., Ho S., Rissoan M.C., Soumelis V., Porcelli S.A., Lanier L.L., and Liu Y.J.. 2001. Distinct cytokine profiles of neonatal natural killer T cells after expansion with subsets of dendritic cells. J. Exp. Med. 193:1221–1226. 10.1084/jem.193.10.1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojo S., Elly C., Harada Y., Langdon W.Y., Kronenberg M., and Liu Y.C.. 2009. Mechanisms of NKT cell anergy induction involve Cbl-b-promoted monoubiquitination of CARMA1. Proc. Natl. Acad. Sci. USA. 106:17847–17851. 10.1073/pnas.0904078106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong S., Dong H., Song J., Thiruppathi M., Prabhakar B.S., Qiu Q., Lin Z., Chini E., Zhang B., and Fang D.. 2015. Deleted in Breast Cancer 1 Suppresses B Cell Activation through RelB and Is Regulated by IKKα Phosphorylation. J. Immunol. 195:3685–3693. 10.4049/jimmunol.1500713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G., Bonnet J., Umlauf D., Karmodiya K., Koffler J., Stierle M., Devys D., and Tora L.. 2011. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol. Cell. Biol. 31:3734–3744. 10.1128/MCB.05231-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V., Zullo A.J., Schweitzer M.N., Staton T.L., Gallo E.M., Crabtree G.R., and Glimcher L.H.. 2009. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat. Immunol. 10:306–313. 10.1038/ni.1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.J., Kim M.S., Shin J.M., Park T.J., Chung H.M., and Baek K.H.. 2006. The expression patterns of deubiquitinating enzymes, USP22 and Usp22. Gene Expr. Patterns. 6:277–284. 10.1016/j.modgep.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Holzapfel K.L., Zhu J., Jameson S.C., and Hogquist K.A.. 2013. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14:1146–1154. 10.1038/ni.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Yang H., Kong Q., Li J., Lee S.M., Gao B., Dong H., Wei J., Song J., Zhang D.D., and Fang D.. 2012. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell. 46:484–494. 10.1016/j.molcel.2012.03.024 [DOI] [PubMed] [Google Scholar]
- Lin Z., Tan C., Qiu Q., Kong S., Yang H., Zhao F., Liu Z., Li J., Kong Q., Gao B., et al. 2015. Ubiquitin-specific protease 22 is a deubiquitinase of CCNB1. Cell Discov. 1:15028 10.1038/celldisc.2015.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Cardenas J., Zhang Y., Zhang D.D., and Fang D.. 2016. Ubiquitin-specific peptidase 22 functions and its involvement in disease. Oncotarget. 7:44848–44856. 10.18632/oncotarget.8602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Cardenas J., Xu Y., Wei J., Tan C., Kong S., Gao B., Montauti E., Kirsammer G., Licht J.D., Yu J., et al. 2018. USP22 deficiency leads to myeloid leukemia upon oncogenic Kras activation through a PU.1-dependent mechanism. Blood. 132:423–434. 10.1182/blood-2017-10-811760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mycko M.P., Ferrero I., Wilson A., Jiang W., Bianchi T., Trumpp A., and MacDonald H.R.. 2009. Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. J. Immunol. 182:4641–4648. 10.4049/jimmunol.0803394 [DOI] [PubMed] [Google Scholar]
- Savage A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., and Bendelac A.. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403. 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Supprian M., Tian J., Grant E.P., Pasparakis M., Maehr R., Ovaa H., Ploegh H.L., Coyle A.J., and Rajewsky K.. 2004. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-kappaB activation. Proc. Natl. Acad. Sci. USA. 101:4566–4571. 10.1073/pnas.0400885101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M.P., Mathew R., Liszewski M.K., Spooner C.J., Barr K., Meng F., Singh H., and Bendelac A.. 2012. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol. 13:264–271. 10.1038/ni.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar V., Hammond K.J., Howells N., Pfeffer K., and Weih F.. 2003. Differential requirement for Rel/nuclear factor kappa B family members in natural killer T cell development. J. Exp. Med. 197:1613–1621. 10.1084/jem.20022234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend M.J., Weinmann A.S., Matsuda J.L., Salomon R., Farnham P.J., Biron C.A., Gapin L., and Glimcher L.H.. 2004. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 20:477–494. 10.1016/S1074-7613(04)00076-7 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S., Powolny-Budnicka I., Riemann M., Schmid R.M., Paxian S., Pfeffer K., Körner H., and Weih F.. 2008. Rel/NF-kappaB family member RelA regulates NK1.1- to NK1.1+ transition as well as IL-15-induced expansion of NKT cells. Eur. J. Immunol. 38:3508–3519. 10.1002/eji.200737830 [DOI] [PubMed] [Google Scholar]
- Wang Y., Yun C., Gao B., Xu Y., Zhang Y., Wang Y., Kong Q., Zhao F., Wang C.R., Dent S.Y.R., et al. 2017. The Lysine Acetyltransferase GCN5 Is Required for iNKT Cell Development through EGR2 Acetylation. Cell Reports. 20:600–612. 10.1016/j.celrep.2017.06.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama W.M. 2004. Betting on NKT and NK cells. Immunity. 20:363–365. 10.1016/S1074-7613(04)00085-8 [DOI] [PubMed] [Google Scholar]
- Yue S.C., Shaulov A., Wang R., Balk S.P., and Exley M.A.. 2005. CD1d ligation on human monocytes directly signals rapid NF-kappaB activation and production of bioactive IL-12. Proc. Natl. Acad. Sci. USA. 102:11811–11816. 10.1073/pnas.0503366102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X., Izcue A., and Borggrefe T.. 2011. Essential role of Mediator subunit Med1 in invariant natural killer T-cell development. Proc. Natl. Acad. Sci. USA. 108:17105–17110. 10.1073/pnas.1109095108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.Y., Varthi M., Sykes S.M., Phillips C., Warzecha C., Zhu W., Wyce A., Thorne A.W., Berger S.L., and McMahon S.B.. 2008. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell. 29:102–111. 10.1016/j.molcel.2007.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Lee S.M., Shannon S., Gao B., Chen W., Chen A., Divekar R., McBurney M.W., Braley-Mullen H., Zaghouani H., and Fang D.. 2009. The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. J. Clin. Invest. 119:3048–3058. 10.1172/JCI38902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Lang G., Ito S., Bonnet J., Metzger E., Sawatsubashi S., Suzuki E., Le Guezennec X., Stunnenberg H.G., Krasnov A., et al. 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 29:92–101. 10.1016/j.molcel.2007.12.011 [DOI] [PubMed] [Google Scholar]
- Zhao J., Weng X., Bagchi S., and Wang C.R.. 2014. Polyclonal type II natural killer T cells require PLZF and SAP for their development and contribute to CpG-mediated antitumor response. Proc. Natl. Acad. Sci. USA. 111:2674–2679. 10.1073/pnas.1323845111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists primers used for the study, antibodies used for the flow cytometry analysis, and antibodies used for Western blotting, ChIP, and immunoprecipitation.