Yang et al. show that IL-21 is the major extrinsic factor that inhibits the generation of IgE+ B cells under multiple conditions. IL-21, IL-4, and CD40 combinatorially regulate class switch recombination to IgE in both mouse and human B cells.
Abstract
IgE antibodies may elicit potent allergic reactions, and their production is tightly controlled. The tendency to generate IgE has been thought to reflect the balance between type 1 and type 2 cytokines, with the latter promoting IgE. Here, we reevaluated this paradigm by a direct cellular analysis, demonstrating that IgE production was not limited to type 2 immune responses yet was generally constrained in vivo. IL-21 was a critical negative regulator of IgE responses, whereas IFN-γ, IL-6, and IL-10 were dispensable. Follicular helper T cells were the primary source of IL-21 that inhibited IgE responses by directly engaging the IL-21 receptor on B cells and triggering STAT3-dependent signaling. We reconciled previous discordant results between mouse and human B cells and revealed that the inhibition of IgE class switch recombination by IL-21 was attenuated by CD40 signaling, whereas IgG1 class switch recombination was potentiated by IL-21 in the context of limited IL-4. These findings establish key features of the extrinsic regulation of IgE production by cytokines.
Introduction
IgE is the major antibody isotype associated with allergic diseases, including atopic dermatitis, allergic rhinitis, asthma, and food allergy (Gould and Wu, 2018). IgE can trigger potent immediate-type hypersensitivity reactions mediated by mast cells and basophils. When these reactions occur systemically, a dangerous life-threatening condition known as systemic anaphylaxis may ensue. However, despite the increasing prevalence of allergic diseases, the majority of individuals do not experience systemic anaphylaxis (Tanno et al., 2018). Given the multitude of exposures of the immune system to diverse stimuli such as allergens, pathogens, and vaccination, IgE responses must normally be limited in magnitude. Indeed, IgE is typically the rarest antibody isotype in serum (Gould and Wu, 2018). While numerous factors limit the amount of serum IgE, including a short-half life and clearance mechanisms, there is significant evidence that the number of cells producing IgE is also limited (Gould and Wu, 2018; Yang et al., 2014).
Due to technical limitations in the detection of the cells producing IgE, most early studies relied on indirect measurements of secreted IgE. Recently, improved methodology, including IgE reporter mice and flow cytometry staining methods, have enabled the direct detection of IgE-expressing (IgE+) B cells and plasma cells (PCs; He et al., 2013; Talay et al., 2012; Yang et al., 2012). These studies revealed that IgE+ B cells only appeared transiently and in limited numbers in germinal centers (GCs). IgE responses showed evidence of constrained affinity maturation, and memory IgE responses appear to be initiated by IgG+ rather than IgE+ memory B cells (He et al., 2017; Turqueti-Neves et al., 2015; Yang et al., 2014). The majority of IgE+ B cells were found to differentiate into PCs that were primarily short-lived (Yang et al., 2014). The various distinct features of IgE responses have been attributed to unique properties of the IgE B cell receptor (BCR; Achatz-Straussberger et al., 2008; Haniuda et al., 2016; Laffleur et al., 2015; Tong et al., 2017; Vanshylla et al., 2018; Yang et al., 2016).
While the IgE BCR is an important cell intrinsic regulator of the fate of IgE+ B cells, extrinsic factors also are known to regulate IgE responses. The production of IgE is associated with type 2 immune responses and is thought to be dysregulated in the context of allergic diseases (Gould and Wu, 2018). A prevalent model has been that the tendency to develop allergic sensitization represents an inappropriate balance between type 1 and type 2 immune responses as a result of increased hygiene in modernized societies, a so-called hygiene hypothesis (Renz and Herz, 2002; Romagnani, 2004). The initial impetus for this paradigm can be traced back to cell culture studies that revealed the dichotomous polarization of T cells into T helper type 1 (Th1) versus Th2 cells (Coffman, 2006). The prototypical cytokine made by Th1 cells, IFN-γ, was found to inhibit the production of IgE, whereas the prototypical cytokine made by Th2 cells, IL-4, was found to promote the production of IgE. This led to the belief that reduced exposure to viral and bacterial pathogens in early life could shift the balance from Th1 to Th2 cells and thus promote IgE production in response to allergen exposure (Renz and Herz, 2002; Romagnani, 2004). However, despite the simplicity of this model, further consideration has indicated that IgE responses must also be regulated by additional mechanisms (Lambrecht and Hammad, 2017).
Other cytokines have also been implicated in the regulation of IgE responses. IL-10 is produced by regulatory T cells and B cells and has been associated with a shift from IgE to IgG4 production (Akdis and Akdis, 2014; Jeannin et al., 1998). IL-21 has been associated with both the suppression and induction of IgE in divergent studies. Early studies of IL-21 receptor (IL-21R)–deficient mice revealed exaggerated IgE responses (Kasaian et al., 2002; Ozaki et al., 2002), and IL-21 was reported to inhibit IgE production in mouse B cell cultures (Harada et al., 2006; Suto et al., 2002). However, mixed results were reported regarding the impact of IL-21 on human IgE production in cell culture, with findings that IL-21 could either inhibit or promote IgE production (Avery et al., 2008; Caven et al., 2005; Harada et al., 2006; Pène et al., 2006; Wood et al., 2004). The inhibition of IgE production by IL-21 was generally observed in cultures of total peripheral blood mononuclear cells, whereas IL-21 promoted IgE production in purified B cell cultures (Pène et al., 2006; Wood et al., 2004). This was thought to indicate that IL-21 promoted IgE production by B cells but that IL-21 induced other factors, such as IFN-γ in T cells, that could suppress IgE production in mixed cultures (Pène et al., 2006). These findings have led some to presume that IL-21 may have a different impact on the IgE responses of mouse versus human B cells.
Here, we characterized the extrinsic regulation of IgE responses in mice as well as cultured mouse and human B cells. By a direct cellular analysis in mice, we established that IgE+ B cells and PCs were generated under a wide range of immunization conditions that were not restricted to type 2 immune responses yet were generally very limited in number. We demonstrated that IL-21 was a major broad negative regulator of IgE responses, whereas IFN-γ, IL-6, and IL-10 were not required for normal IgE regulation. T follicular helper (Tfh) cells were the main cellular source of IL-21 that inhibited IgE class switch recombination (CSR) by binding the IL-21R and triggering STAT3 activation in B cells. We identified CD40 and IL-4 signals as key modulators of the suppression of IgE CSR and the promotion of IgG1 CSR by IL-21 in both mouse and human B cells. Our findings indicate that IgE CSR is predominantly controlled by IL-21 rather than the type 1/type 2 axis.
Results
IgE+ B cells are generated in a variety of immune responses
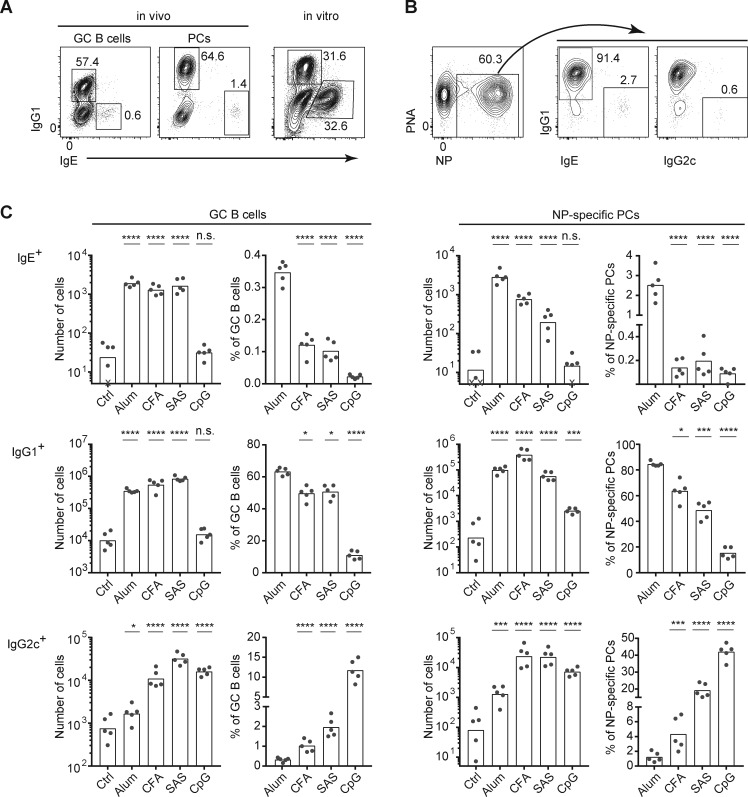
IgE production is generally considered a key feature of type 2 immune responses, yet our direct flow cytometric analyses have shown a low frequency and number of IgE+ cells in mouse models of type 2 immunity, including immunizations with alum adjuvant or infection with helminths (Yang et al., 2012). For example, representative FACS profiles of GC B cells and PCs in the draining LNs of mice immunized subcutaneously with a T cell–dependent antigen, 4-hydroxy-3-nitrophenylacetyl conjugated to chicken gamma globulin (NP-CGG), in alum adjuvant, at the peak of the response are shown in Fig. 1 A. Both IgE+ GC B cells and PCs are typically on the order of 50-fold less abundant than their IgG1+ counterparts. In contrast to the small IgE response in vivo, comparable frequencies of IgE+ and IgG1+ B cells were generated by culturing primary mouse B cells with anti-CD40 antibody and recombinant IL-4 to mimic B cell activation by T cells (Fig. 1 A). This striking contrast in the generation of IgE+ cells in vivo versus in cell culture suggests that IgE responses are largely suppressed in vivo, even in the context of type 2 immune responses.
Figure 1.
The IgE response is broadly suppressed in vivo under multiple adjuvant conditions. (A) Representative flow cytometry of GC B cells and PCs from the draining LNs from a mouse immunized with NP-CGG in alum adjuvant (left) versus splenocytes cultured for 4 d after in vitro activation with anti-CD40 and IL-4 (right). (B) Representative flow cytometry to show the gating strategy used to identify NP-specific PCs and enumerate the relative frequencies of PCs expressing each isotype from the draining LNs of a mouse immunized with NP-CGG in alum adjuvant. (C) Cell number and frequency of IgE+, IgG1+, or IgG2c+ GC B cells and NP-specific PCs from the draining LNs of mice immunized with NP-CGG with the indicated adjuvants or no adjuvant (Ctrl). WT mice (B6/J from our colony [A] or purchased NCI B6 CD45.1 congenics [B and C]) were analyzed 7 d after subcutaneous immunization. Cells cultured in A were from a B6/J mouse bred in our colony. GC B cells were gated as B220hi CD138− PNAhi CD38lo IgD− cells, and PCs were gated as B220lo CD138+ CD38lo IgD− cells, as shown previously (Yang et al., 2012). NP-specific PCs were identified by total (intracellular and surface) staining with NP-APC. Isotype-specific cells were identified by total (IgG1 and IgG2c) or intracellular (IgE) staining (see Yang et al., 2012 and Materials and methods). IgM+ cells were excluded from the analysis in B and C by total IgM staining. Dots represent individual mice. Xs represent cell counts that were below the limit of detection. Bars represent the mean (frequency plots) or geometric mean (cell number plots). n.s., not significant; *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA with Dunnett’s post-test comparing each group to the leftmost control group). Data in A and B are representative of >10 and 5 experiments, respectively. Similar IgE and IgG1 responses as in C were observed in a separate experiment when mice were immunized with NP-KLH in alum, CFA, and SAS (data not shown). PNA, peanut agglutinin.
Our findings above prompted us to revisit the paradigm that IgE is induced only during type 2 immune responses. We assessed the frequency and number of GC B cells and PCs of various BCR isotypes by flow cytometry with intracellular staining for IgE, as we reported previously (Yang et al., 2012, 2016), in mice immunized with different adjuvants. In detail, we subcutaneously immunized mice with NP-CGG together with the type 2 adjuvant alum, compared with various adjuvants that elicit a predominantly type 1 response, specifically CFA, Sigma Adjuvant System (SAS), and deoxy-cytidylate-phosphate-deoxy-guanylate (CpG)–DNA, or without adjuvant as a control (Fig. 1 B). Surprisingly, similar numbers of IgE GC B cells were generated following immunization with alum, CFA, and SAS (Fig. 1 C). However, as CFA and SAS induced a stronger total GC B cell response than alum (Fig. S1), the relative frequency of IgE+ cells within the GC was lower with CFA and SAS than with alum. IgE PCs were also clearly induced with CFA and SAS compared with no adjuvant, although the magnitude of these responses was lower than with alum (Fig. 1 C). CFA and SAS also induced GC B cells and PCs of the IgG2c isotype, whereas minimal IgG2c responses were observed with alum, as expected (note that IgG2c in the C57BL/6 strain is sometimes denoted IgG2ab and is induced in type 1 immune responses in a similar manner to IgG2a on other mouse strains). CpG, a highly type 1–polarizing adjuvant, induced a strong IgG2c response but did not induce an IgE response (Fig. 1 C). Thus, while highly polarized type 2 and type 1 responses may be induced by alum and CpG, respectively, CFA and SAS induced a mixed response including IgE, IgG1, and IgG2c isotypes. These data indicate that the generation of IgE GC B cells and PCs is not limited to type 2–polarized responses. Remarkably, under all of these conditions, the frequency of IgE+ cells was very small compared with IgG1+ cells, suggesting general mechanisms restrain the generation of IgE+ cells in vivo.
Figure S1.
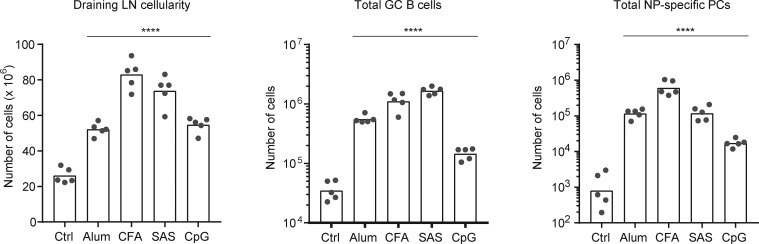
Total cell counts in draining LNs from mice immunized with various adjuvants. Related to Fig. 1. Cell counts are shown for the experiment shown in Fig. 1 C (see Fig. 1 legend for details). Mice were analyzed 7 d after subcutaneous immunization with NP-CGG and the indicated adjuvants or no adjuvant (Ctrl). Dots represent individual mice. Bars represent the mean (draining LN cellularity plot) or geometric mean (total GC B and NP-specific PCs plots). ****, P < 0.0001 (one-way ANOVA with Dunnett’s post-test comparing each group to the leftmost control group).
IL-21, but not IFN-γ, broadly inhibits IgE responses in mice
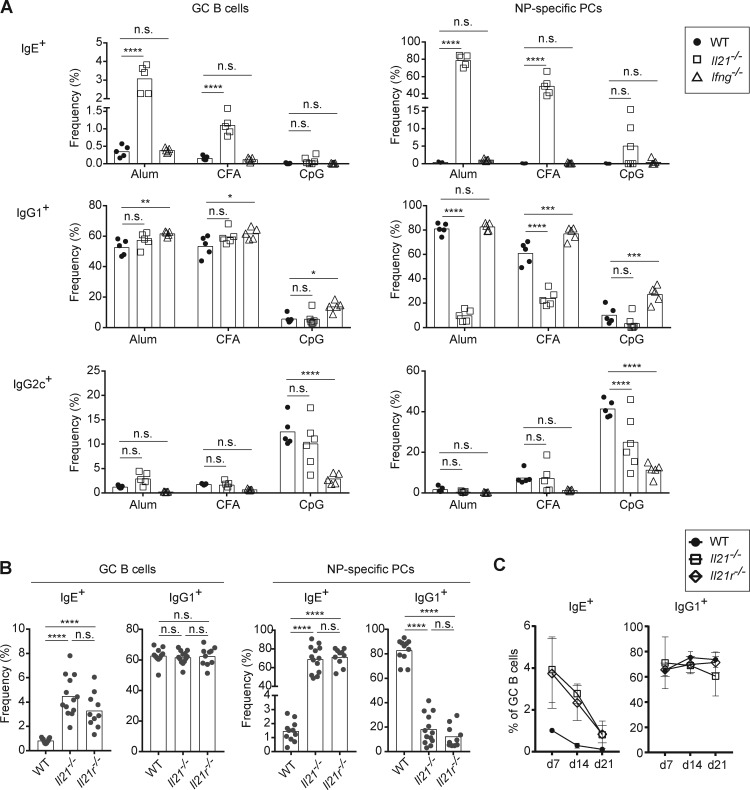
The low frequency and number of IgE+ B cells observed in a wide range of in vivo immune responses suggests that general extrinsic factors tightly regulate in vivo IgE responses. As discussed above (see Introduction), IL-21 and IFN-γ have been reported in some previous studies to negatively regulate IgE production. A challenge, however, in the interpretation of such studies is that these cytokines are known to affect various general properties of B cells such as proliferation, apoptosis, and PC differentiation that could have an indirect impact on measurements of secreted IgE (Moens and Tangye, 2014). We therefore reexamined the direct impact of these cytokines on IgE responses by flow cytometry to assess the generation of IgE GC B cells and PCs in gene-targeted mice immunized with NP-CGG in the context of three different adjuvants (alum, CFA, and CpG; Figs. 2 A and S2) in light of our earlier findings above. Strikingly, IL-21–deficient mice showed increased frequencies of IgE GC B cells and PCs after immunization with either alum or CFA (Fig. 2 A). Even in the context of CpG adjuvant, a small IgE response was noted in some IL-21–deficient mice. In contrast, no significant alterations in the IgE response were observed in IFN-γ–deficient mice (Fig. 2 A). The marked reduction in the IgG2c response in the IFN-γ knockout mice confirmed that this cytokine is necessary for IgG2c CSR (Finkelman et al., 1990). Taken together, these results indicate that IL-21, but not IFN-γ, is the major factor that suppresses IgE response in vivo.
Figure 2.
IL-21, rather than IFN-γ, is a major suppressor of IgE responses in vivo. (A–C) Mice were immunized subcutaneously with NP-CGG with the indicated adjuvants (A) or alum adjuvant (B and C). Draining LNs were collected and analyzed 7 d (A and B) or 7–21 d (C) after immunization, and then cells were analyzed by flow cytometry to enumerate isotype-specific GC B cells and NP-specific PCs as in Fig. 1. (A) Frequency of IgE+, IgG1+, and IgG2c+ cells in the GC B cell and NP-specific PC compartments in WT, Il21−/−, and Ifng−/− mice. (B) Frequency of IgE+ and IgG1+ cells in the GC B cell and NP-specific PC compartments in WT, Il21−/−, and Il21r−/− mice. (C) Kinetic analysis of the frequency of IgE+ and IgG1+ B cells among GC B cells in WT, Il21−/−, and Il21r−/− mice. WT mice were B6/J (purchased from The Jackson Laboratory; A) or a mixture of purchased B6/J mice and WT mice on a B6 background bred in our colony (B and C). Dots represent individual mice. Bars represent the mean. n.s., not significant; *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 (two-way ANOVA with Dunnett’s post-test comparing each knockout to the WT group [A] or one-way ANOVA with Dunnett’s post-tests for multiple comparisons [B]). Similar IgE and IgG1 responses as in A were observed in a separate experiment when mice were immunized with NP-CGG in alum (data not shown). Data in B are compiled from three experiments. Data in C at day 7 were from one experiment represented in B.
Figure S2.
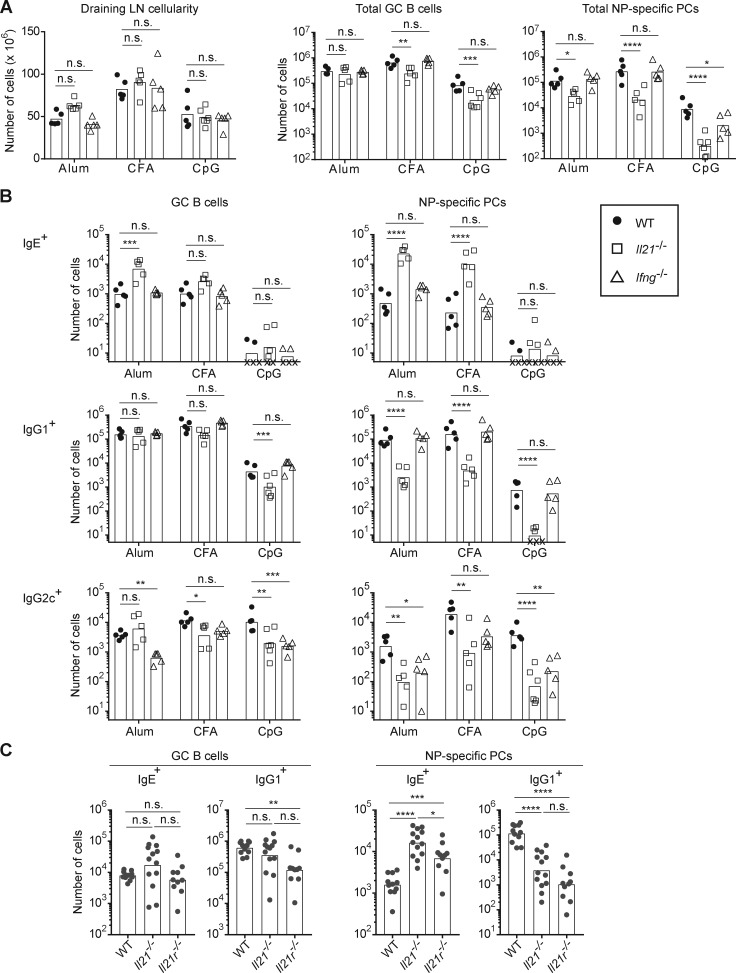
IL-21 selectively suppresses IgE responses in vivo. Related to Fig. 2. Cell counts are shown for the experiments in Fig. 2 (see Fig. 2 legend for details). (A–C) Mice were immunized subcutaneously with NP-CGG with the indicated adjuvants (A and B) or alum adjuvant (C). Draining LNs were collected and analyzed 7 d after immunization. (A and B) Total cell counts in draining LNs and enumeration of the number of total GC B cells and NP-specific PCs (A) and the number of IgE+, IgG1+, and IgG2c+ cells in the GC B cell and NP-specific PC compartments (B) in immunized WT, Il21−/−, and Ifng−/− mice. (C) Quantification of the number of IgE+ and IgG1+ cells in the GC B cell and NP-specific PC compartments in WT, Il21−/−, and Il21r−/− mice. Dots represent individual mice. Xs represent cell counts that were below the limit of detection. Bars represent the mean (draining LN cellularity plot) or geometric mean (cell number plots). n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (two-way ANOVA with Dunnett’s post-test comparing each knockout to the WT group [A and B] and one-way ANOVA with Dunnett’s post-test for multiple comparisons [C]).
To confirm that the enhanced IgE response observed in Il21−/− mice was in fact due to a loss of IL-21 signaling, we also analyzed IL-21R–deficient mice in the context of immunization with alum adjuvant. We observed a similar elevation in the frequency of IgE GC B cells and PCs in Il21r−/− mice as in Il21−/− mice (Fig. 2 B). These findings confirm that IL-21 is a major negative regulator of IgE responses and exclude a role for potential indirect effects of the targeted Il21 or Il21r loci on neighboring genes (McGuire et al., 2015).
Differential impact of IL-21 on IgE GC versus PC responses
It was notable that in both Il21−/− and Il21r−/− mice, the increased IgE PC response was substantially more pronounced than the increase in IgE GC B cells. Indeed, while IgE PCs normally comprise <5% of the total NP-binding PCs after immunization, in Il21−/− and Il21r−/− mice nearly 80% of NP-binding PCs were IgE+ (Fig. 2 B). The absolute number of IgE PCs was also substantially elevated in Il21−/− and Il21r−/− mice (Fig. S2, B and C). This result sharply contrasted with the reduced numbers of IgG1 and IgG2c PCs in the Il21−/− and Il21r−/− mice, consistent with previous reports showing defective IgG responses in these mice (Moens and Tangye, 2014). While the relative proportion of GC B cells that were IgE+ was increased in Il21−/− and Il21r−/− mice (Fig. 2 B), the majority of GC B cells still expressed the IgG1 isotype, and the absolute number of IgE GC B cells was not consistently elevated (Fig. S2, B and C). In addition, when we analyzed the kinetics of the IgE GC B cell response, we found that the frequency of IgE+ B cells within GCs rapidly declined over time in Il21−/− and Il21r−/− mice, whereas the frequency of IgG1+ B cells within GCs was stable, similar to the trends observed in WT mice (Fig. 2 C). These results suggest that IgE GC B cell responses are largely constrained in an IL-21–independent manner. Reciprocally, the propensity of IgE+ B cells to differentiate into PCs also seems to be IL-21 independent.
IL-21 inhibits IgE responses through the IL-21R in B cells
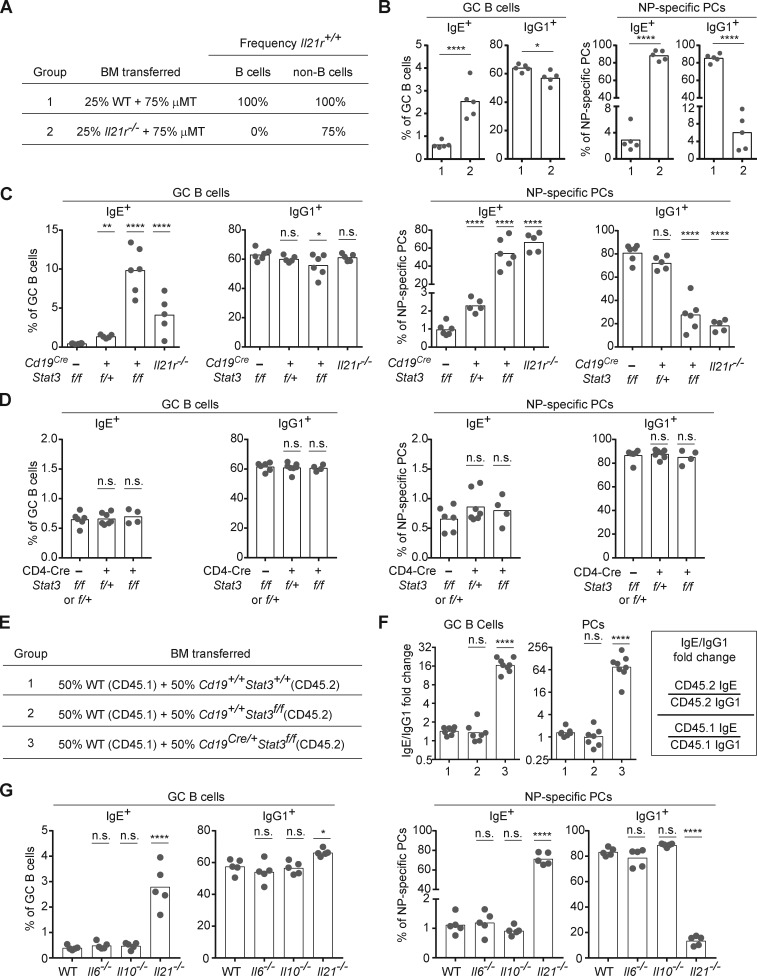
IL-21 has been shown to exert biological functions in numerous cell types, and the IL-21R is broadly expressed (Spolski and Leonard, 2014). To test whether the inhibition of IgE response by IL-21 is a consequence of its binding to the IL-21R on B cells, we generated mixed bone marrow (BM) chimeras in which all B cells are selectively Il21r−/−, whereas the majority of other cell types are Il21r+/+ (Fig. 3 A). After reconstitution and immunization with NP-CGG in alum, we observed that selective deficiency of Il21r in B cells recapitulated the results observed in the full Il21r−/− mice, including a relative increase in the frequency of IgE+ B cells in GCs as well as the vast majority of PCs expressing the IgE isotype (Fig. 3 B). These observations indicate that the inhibition of IgE responses by IL-21 is primarily mediated by the direct action of IL-21 on B cells.
Figure 3.
IL-21 suppresses IgE responses through the IL-21R–STAT3 signaling axis in B cells. Draining LNs were collected 7 d after subcutaneous immunization with NP-CGG in alum adjuvant, and then cells were analyzed by flow cytometry to enumerate isotype-specific GC B cells and NP-specific PCs as in Fig. 1. (A) Groups of BM chimeras generated to test the impact of selective deficiency of the IL-21R in B cells. The indicated combinations of BM cells were transplanted into lethally irradiated μMT recipient mice. WT BM was obtained from a B6/J mouse bred in our colony. The expected frequencies of cells capable of IL-21R expression (frequency Il21r+/+) are indicated. (B) Frequency of IgE+ and IgG1+ cells in the GC B cell and NP-specific PC compartments in BM chimeras generated as in A. (C and D) Frequency of IgE+ and IgG1+ cells in the GC B cell and NP-specific PC compartments in mice with conditional Stat3 deletion in B cells (C) or T cells (D). Control mice were littermates. Il21r−/− mice were included for comparison in C. (E) Groups of BM chimeras generated to test the cell intrinsic role of STAT3 in B cells. The WT CD45.1 BM donor was a purchased NCI B6 CD45.1 mouse. The Cd19+/+ Stat3+/+ BM was from a B6 mouse bred in our colony. The Cd19+/+ Stat3f/f BM and Cd19Cre/+ Stat3f/f BM were from littermates. The indicated mixtures of BM cells were transferred into lethally irradiated CD45.1 WT recipients (NCI B6 CD45.1). (F) Fold change in the ratio of IgE+ cells to IgG+ cells (IgE/IgG1) among CD45.2 cells compared with CD45.1 cells in the GC B cell and PC compartments in the mixed BM chimeras generated as in E. The calculation of the ratio is indicated in the box on the right. Note that total PCs (rather than NP-specific PCs) were enumerated to allow for quantification of CD45.1+ vs. CD45.2+ cells. (G) Frequency of IgE+ and IgG1+ cells in GC B cell and NP-specific PC compartments in WT, Il6−/−, Il10−/−, and Il21−/− mice. WT mice were B6/J bred in our colony. Dots represent individual mice. Bars represent the mean (frequency plots) or geometric mean (relative IgE/IgG1 plots). n.s., not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (t tests with the Holm-Sidak correction for multiple comparisons [B] or one-way ANOVA with Dunnett’s post-test comparing each group to the leftmost control group [C, D, F, and G]). The B cell–intrinsic role shown in A, B, E, and F was confirmed in a separate set of chimeras with a mixture of Il21r+/+ and Il21r−/− B cells (data not shown). Data in C and D are representative of three experiments. The comparison of WT, Il10−/−, and Il21−/− mice in G was confirmed in a separate experiment.
STAT3 signaling in B cells mediates the inhibitory function of IL-21 on the IgE response
To further test whether IL-21R signaling in B cells negatively regulates IgE responses, we considered the cellular requirement for its major downstream signaling adapter, STAT3 (Spolski and Leonard, 2014). Specifically, we used our direct cellular assays to enumerate IgE+ B cells in mice with conditional deletion of Stat3 in B cells by Cd19Cre versus in T cells with a Cd4-Cre transgene. Conditional deletion of Stat3 in B cells (Fig. 3 C), but not in T cells (Fig. 3 D), led to elevated IgE GC B cell and PC responses, as we had observed in Il21r−/− mice. To further test whether STAT3 signaling negatively regulates IgE in a B cell–intrinsic manner, we generated mixed BM chimeras in which half of B cells were Stat3 deficient (CD45.2+) and half had normal Stat3 (CD45.1+; Fig. 3 E). In this competitive setting, Stat3-deficient B cells showed a relatively poorer contribution to the total GC and PC compartments compared with Stat3+/+ B cells as expected (data not shown). To normalize for this difference in precursor frequency and to specifically consider the impact on the IgE response, we determined whether there was a change in the IgE/IgG1 ratio among Stat3-deficient (CD45.2) cells relative to the IgE/IgG1 ratio among control (CD45.1) cells. This analysis revealed increases in the IgE/IgG1 ratios in Stat3-deficient B cells of ∼16-fold among GC B cells and 64-fold among PCs, whereas the relative IgE/IgG1 ratios observed in control mixed chimeras were close to 1 (Fig. 3 F). We conclude that the negative regulation of the IgE response by STAT3 occurs in a B cell–intrinsic manner.
IL-6 and IL-10 are not essential for IgE regulation in mice
Our analysis of GC B cells in mice with conditional deletion of Stat3 in B cells showed a somewhat more pronounced relative increase in IgE+ GC B cells compared with Il21r−/− mice (Fig. 3 C), possibly implicating additional cytokines that signal via STAT3 in the suppression of IgE responses. In addition to IL-21, the cytokines IL-6 and IL-10 also signal via STAT3 (Moens and Tangye, 2014; Vinuesa et al., 2016) and have been implicated in the regulation of IgE responses (Akdis and Akdis, 2014; Fukushima et al., 2007; Jabara et al., 1990; Mäkelä et al., 2000; Neveu et al., 2009; Noble and Zhao, 2016; Tournoy et al., 2000). However, we observed no change in the relative frequency of IgE GC B cells and PCs in Il6−/− and Il10−/− mice immunized with NP-CGG and alum, whereas Il21−/− mice showed clearly enhanced IgE responses as in our above experiments (Fig. 3 G). The more pronounced IgE GC response in the Cd19Cre/+ Stat3flox/flox mice than in the Il21r−/− mice could instead be due to the synergistic impact of Cd19 heterozygosity, which was previously found to selectively enhance the frequency of IgE GC B cells (Haniuda et al., 2016; Yang et al., 2016). While we cannot exclude minor or redundant contributions from IL-6 and/or IL-10, our data suggest that IL-21 is the major cytokine that negatively regulates IgE responses via IL-21R–mediated STAT3 signals in B cells.
Tfh cells are a major source of IL-21 that regulates IgE responses
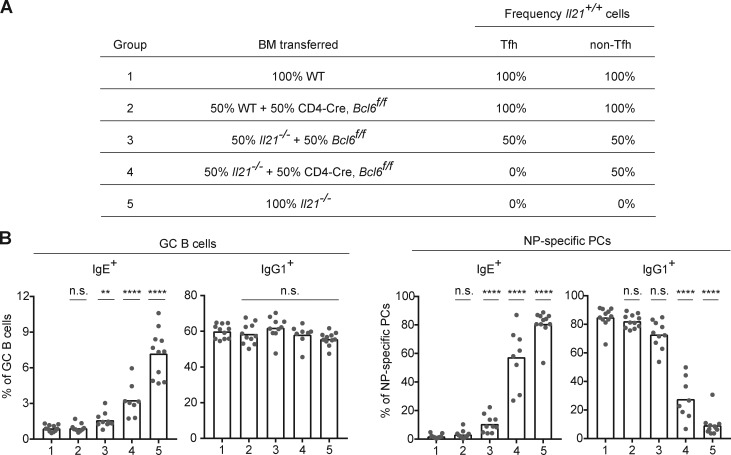
We next sought to identify the cellular source of IL-21 that suppresses IgE responses in vivo. Previous studies have implicated Tfh cells, Th17 cells, and natural killer T cells as the major producers of IL-21 (Spolski and Leonard, 2014). A subset of Th1 cells that expresses IL-21 was also reported to promote IgG2 antibody production (Miyauchi et al., 2016). Among these subsets, given the broad role of Tfh cells in B cell responses (Crotty, 2015; Vinuesa et al., 2016), we reasoned that Tfh cells were the most likely candidates to suppress IgE. To test the role of Tfh cells, we generated mixed BM chimeras in which Tfh cells were selectively Il21−/− (Fig. 4 A). As a source of donor BM deficient in Tfh cell differentiation, but capable of differentiating into other T cell subsets, we conditionally deleted the transcription factor Bcl6 in T cells in Cd4-Cre Bcl6flox/flox mice as reported (Hollister et al., 2013; Kaji et al., 2012; Kobayashi et al., 2017; Meli et al., 2017). BM from Cd4-Cre Bcl6flox/flox BM was mixed with Il21−/− BM such that Tfh cells would only be derived from the Il21−/− BM, whereas non-Tfh cells derived from the Cd4-Cre Bcl6flox/flox BM would be capable of IL-21 expression (group 4; Fig. 4 A). After immunization, we observed that selective deficiency of IL-21 in Tfh cells (group 4) resulted in increased frequencies of IgE+ GC B cells and PCs (Fig. 4 B). Though the magnitude of the IgE response with selective Tfh cell deficiency in IL-21 (group 4) was somewhat less pronounced than with the 100% Il21−/− (group 5), overall, the group 4 response was clearly significantly elevated compared with the control groups (Fig. 4 B), particularly when considering the frequency of IgE+ PCs. Taken together, these data suggest that Tfh cells are the major cellular source of IL-21 that negatively regulates the IgE response, with perhaps minor contributions from other T cell subsets.
Figure 4.
IgE responses in vivo are inhibited by IL-21 derived primarily from Tfh cells. Draining LNs were collected 7 d after subcutaneous immunization with NP-CGG in alum adjuvant, and then cells were analyzed by flow cytometry to enumerate isotype-specific GC B cells and NP-specific PCs as in Fig. 1. (A) Groups of BM chimeras generated to test the impact of selective deficiency of IL-21 in Tfh cells. The indicated combinations of BM cells were transplanted into lethally irradiated Il21−/− mice. The expected frequencies of cells capable of IL-21 expression (frequency Il21+/+) in the Tfh and non-Tfh cell compartments in the resultant chimeras are indicated. Donor frequencies were confirmed in the chimeras by congenic markers as follows: WT BM was from a Boy/J (CD45.1) mouse bred in our colony. The Il21−/− BM was CD45.1, derived from backcrossing Il21−/− B6 to Boy/J congenic mice. The CD4-Cre, Bcl6f/f BM, and control Bcl6f/f BM were from littermates (with the CD45.2 congenic marker). The recipient mice were Il21−/− CD45.1/CD45.2. (B) Frequency of IgE+ and IgG1+ cells in the GC B cell and NP-specific PC compartments of the mixed BM chimeras generated as in A. Dots represent individual mice. Bars represent the mean. n.s., not significant; **, P < 0.01; ****, P < 0.0001 (one-way ANOVA with Dunnett’s post-test comparing each group to group 1). Data in B were pooled from two experiments.
The inhibition of IgE responses by IL-21 is attenuated by CD40 signaling
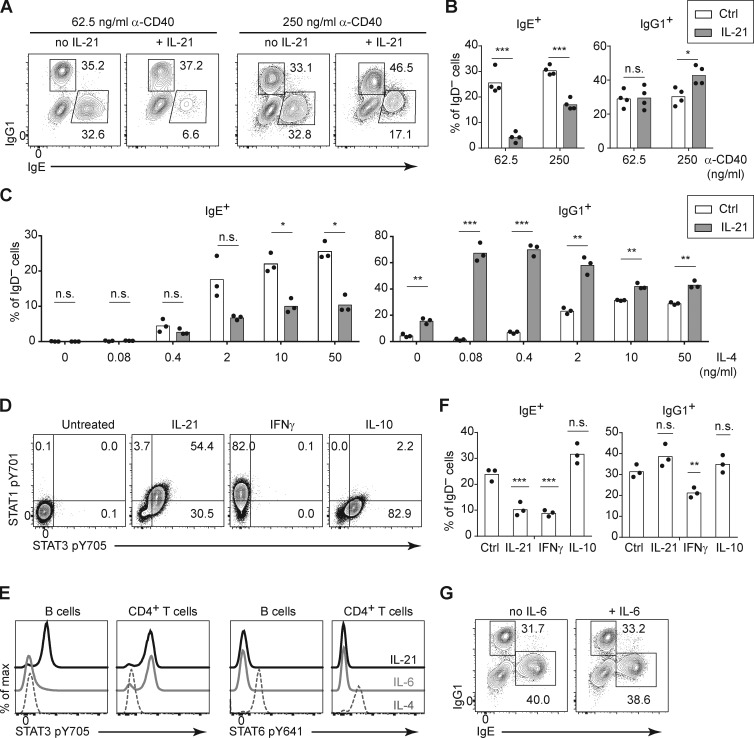
Having defined the major cell types involved in the suppression of IgE responses by IL-21 in vivo, we sought to further characterize this suppression in the context of a simplified cell culture model. IL-21 was reported to inhibit IgE production when mouse B cells were cultured with LPS and IL-4 (Suto et al., 2002); however, in pilot experiments, we found that the addition of IL-21 in these cultures led to the loss of nearly all viable B cells (data not shown), precluding a meaningful cellular analysis. It was previously reported that, in addition to promoting apoptosis, IL-21 strongly induced the growth arrest of mouse B cells stimulated with LPS, whereas IL-21 enhanced the proliferation of mouse B cells stimulated with anti-CD40 (Caven et al., 2005; Jin et al., 2004). As we have extensively studied mouse IgE+ B cell differentiation when B cells were cultured with anti-CD40 and IL-4 in previous studies (Yang et al., 2012, 2016), we tested the impact of IL-21 in this context. In our initial experiments, we observed a relatively modest impact of IL-21 on the frequency of IgE+ cells when mouse B cells were cultured with anti-CD40 and IL-4 (data not shown), which was surprising given our evidence above in mice that IL-21 suppressed IgE through the IL-21R and STAT3 in B cells. We considered the possibility that the stimulation conditions might affect the suppression of IgE by IL-21. We had previously observed that IgE PC differentiation in vitro was modulated by the concentration anti-CD40 antibody (Yang et al., 2016). We therefore tested whether the concentration of anti-CD40 antibody affected the B cell response to IL-21, with a fixed high concentration of IL-4. We observed that when mouse B cells were cultured with a low concentration of anti-CD40, then the addition of IL-21 promoted a >5-fold reduction in the frequency of IgE+ cells (Fig. 5, A and B). In contrast, in the context of a higher concentration of anti-CD40, the addition of IL-21 caused a <2-fold reduction in the frequency of IgE+ cells (Fig. 5, A and B). Therefore, strong CD40 stimulation counteracts the inhibition of IgE by IL-21.
Figure 5.
IL-21 inhibits IgE and promotes IgG1 responses of cultured mouse B cells depending on the strength of CD40 and IL-4 signals. (A and B) Representative flow cytometry of IgD− activated B cells (A) and quantification of the frequency of IgE+ and IgG1+ cells (B). Purified B cells were cultured for 4 d with a fixed concentration of IL-4 (12.5 ng/ml) and the indicated concentrations of anti-CD40 (α-CD40) in the presence or absence of IL-21 (50 ng/ml). (C) Quantification of the frequency of IgE+ and IgG1+ cells among IgD– activated B cells. Splenocytes were cultured for 4 d with a fixed concentration of anti-CD40 (125 ng/ml) and the indicated concentrations of IL-4 in the presence or absence of IL-21 (25 ng/ml). (D) Representative flow cytometry of B cells (gated as B220+) after splenocytes were incubated with the indicated cytokines (IL-21, IFN-γ, or IL-10; 100 ng/ml each) for 20 min at 37°C. (E) Representative flow cytometry of B cells (gated as B220+) and CD4+ T cells (gated as CD4+) after splenocytes were incubated with the indicated cytokines (IL-21, IL-6, or IL-4; 50 ng/ml each) for 15 min at 37°C. (F) Quantification of the frequency of IgE+ and IgG1+ cells among IgD− activated B cells. Purified B cells were cultured for 4 d with anti-CD40 (125 ng/ml) and IL-4 (12.5 ng/ml) alone (Ctrl) or with the addition of the indicated cytokines (IL-21 50 ng/ml, IFN-γ 25 ng/ml, or IL-10 25 ng/ml). (G) Representative flow cytometry of IgE+ and IgG1+ B cells within the population of class-switched B cells (B220+ IgD− IgM−) after splenocytes were cultured for 4 d with anti-CD40 (62.5 ng/ml) and IL-4 (25 ng/ml) in the presence or absence of IL-6 (81 ng/ml). Cells were from WT mice on a B6 background or Boy/J CD45.1 congenic mice bred in our colony. Dots represent data points of cells from individual mice, and bars represent the mean (B, C, and F). n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 (t tests with the Holm-Sidak correction for multiple comparisons [B and C] or one-way ANOVA followed by Dunnett’s post-test comparing each cytokine treatment to the control [F]). Data were pooled from four experiments (B) or three experiments (F) or are representative of two experiments (C–E) or three experiments (G).
IL-21 selectively promotes IgG1 responses in the context of limited IL-4
In addition to the impact of CD40 stimulation, we also considered the possibility that the strength of IL-4 stimulation could also have an impact on the response of B cells to IL-21. Notably, the type 1 IL-4R and the IL-21R signal via the shared cytokine receptor common γ chain (Spolski and Leonard, 2014); thus, signals via the IL-4R and IL-21R could be competitive. We cultured mouse B cells with an intermediate concentration of anti-CD40, together with a broad range of IL-4 concentrations, in the presence or absence of IL-21. Surprisingly, the IL-21–mediated reduction in the frequency of IgE+ B cells was relatively similar across a range of IL-4 concentrations, until we reached IL-4 concentrations too low (≤0.4 ng/ml) to stimulate a good IgE response (Fig. 5 C). IgG1 responses were also minimal in the presence of anti-CD40 with low IL-4 alone (Fig. 5 C). Strikingly, however, at these low concentrations of IL-4, the addition of IL-21 selectively enhanced the frequency of IgG1+ cells (Fig. 5 C). This result was particularly notable when we cultured mouse B cells with 0.08 ng/ml of IL-4, which in the absence of IL-21 led to virtually no detectable IgE+ or IgG1+ cells, but in the presence of IL-21, >60% of cells expressed IgG1, whereas IgE+ cells were undetectable (Fig. 5 C). Thus, culturing B cells with a small amount of IL-4 together with IL-21 led to the majority of cells expressing IgG1 and only a small fraction expressing IgE. Therefore, in the context of weak IL-4 stimulation, IL-21 selectively promoted an IgG1 response. Taken together, these findings indicate that signals via CD40 and the IL-4R are important modulators of the B cell response to IL-21. When mouse B cells were stimulated with limiting doses of anti-CD40 antibody or IL-4, we found that IL-21 suppressed IgE and/or promoted IgG1 responses, respectively, recapitulating our findings in vivo.
Characterization of the effects of IFN-γ, IL-10, and IL-6 on the IgE responses of cultured mouse B cells
Given that we observed IL-21 was a major inhibitor of IgE responses in vivo yet mice deficient in other cytokines such as IFN-γ, IL-10, and IL-6 did not have a substantial impact on the IgE response, we examined the effects of these cytokines on mouse B cells cultured in vitro compared with IL-21. We first confirmed the activity of the cytokines by measuring phosphorylation of distinct STAT proteins. Consistent with prior reports (Moens and Tangye, 2014), IL-21 induced phosphorylation of both STAT1 and STAT3 in B cells (Fig. 5 D). IFN-γ induced phosphorylation of STAT1, but not STAT3; conversely, IL-10 induced phosphorylation of STAT3, but not STAT1 (Fig. 5 D). IL-6 did not induce STAT3 phosphorylation in B cells but did induce STAT3 phosphorylation in CD4 T cells (Fig. 5 E), likely due to low expression of the IL-6R on B cells (Dienz et al., 2009). In contrast, IL-4 induced STAT6 phosphorylation in both B cells and CD4 T cells as expected (Fig. 5 E; Geha et al., 2003). Consistent with classical studies (Coffman, 2006; Geha et al., 2003; Moens and Tangye, 2014), in cultures of mouse B cells with anti-CD40 and IL-4, the addition of IFN-γ led to a reduction in the frequency of IgE+ cells (Fig. 5 F). However, the addition of IL-10 (Fig. 5 F) and IL-6 (Fig. 5 G) had a minimal impact on the frequency of IgE+ cells when B cells were cultured with anti-CD40 and IL-4. We found that IFN-γ, but not IL-10 or IL-6, also reduced the frequency of IgG1+ B cells in these cultures (Fig. 5, F and G). Taken together, these data indicate that IL-21 selectively inhibits IgE and, under some circumstances (as described above), can enhance IgG1 responses, whereas IFN-γ inhibits both IgE and IgG1 responses of cultured mouse B cells. However, in immunized mice, IFN-γ had a minimal impact on the IgE response (Fig. 2 A), suggesting that the effects of IL-21 were dominant. We elaborate below on the implications of these findings in vitro and in vivo (see Discussion).
IL-21 selectively suppresses IgE responses by inhibiting IgE CSR rather than inducing apoptosis
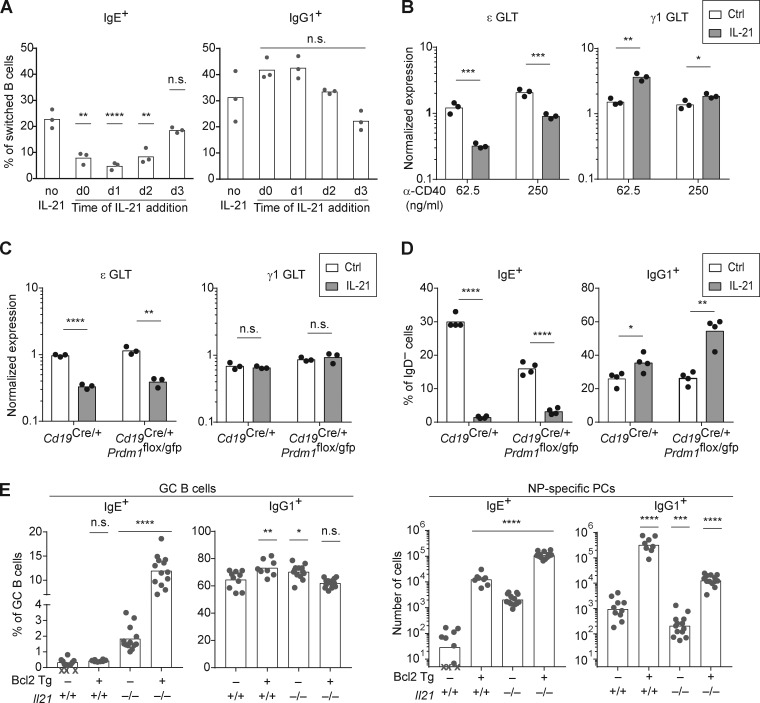
We next considered potential mechanisms for the IL-21–mediated reduction in IgE+ cells. In one study, IL-21 was reported to suppress IgE germline transcript (GLT) expression, which is necessary for IgE CSR (Suto et al., 2002). However, this result was obtained in the context of LPS stimulation, which we had found led to the loss of nearly all viable B cells in combination with IL-21 (data not shown), as had been previously observed (Jin and Malek, 2006). Another study reported that IL-21 promoted the selective apoptosis of IgE+ B cells (Harada et al., 2006). We tested the impact of adding IL-21 to B cells cultured with anti-CD40 and IL-4 at different time points, ranging from before CSR to IgE occurs until after IgE+ B cells have been generated. In this culture system, IgE+ B cells first begin to appear around day 3 and reach peak frequencies by day 4 (data not shown), indicating CSR occurs around days 2–3. Interestingly, if IL-21 was added on day 0, 1, or 2, substantial reductions in the frequency of IgE+ B cells were noted, which was most pronounced at day 1 (Fig. 6 A). However, if IL-21 was added to the culture at day 3, then no significant reduction in IgE+ cells was observed (Fig. 6 A). Thus, IL-21 only had an impact when added on days 0–2, before CSR to IgE, but had no effect when added on day 3, when IgE+ B cells had already been generated in culture. These data strongly suggest that IL-21 suppresses IgE responses by preventing IgE CSR rather than by inducing apoptosis of IgE+ B cells.
Figure 6.
IL-21 acts independently of apoptosis and inhibits IgE germline transcription. (A) Quantification of the frequency of IgE+ and IgG1+ B cells among class-switched B cells (B220+ IgD− IgM−) 4 d after mouse splenocytes were cultured with anti-CD40 (62.5 ng/ml) and IL-4 (12.5–25 ng/ml), with IL-21 (25 ng/ml) added at different time points (compared with no IL-21 control). (B) Relative quantification of IgE (ε) and IgG1 (γ1) GLTs from purified mouse B cells cultured for 3 d with IL-4 (12.5 ng/ml) and the indicated concentrations of anti-CD40 in the presence or absence of IL-21 (50 ng/ml). GLT expression was normalized by endogenous Hprt transcripts. (C) Relative quantification of ε and γ1 GLT from purified B cells of the indicated genotypes that were cultured with anti-CD40 (125 ng/ml) and IL-4 (12.5 ng/ml) for 1 d before the addition of IL-21 (50 ng/ml) to the indicated samples. RNA was isolated 8 h after the addition of IL-21, and GLT expression was normalized by endogenous Hprt transcripts. (D) Quantification of the frequency of IgE+ and IgG1+ B cells among activated B cells (B220+ IgD−) 4 d after purified B cells of the indicated genotypes were cultured with anti-CD40 (62.5 ng/ml) and IL-4 (12.5 ng/ml). IL-21 (50 ng/ml) was added on day 1 to the indicated samples. (E) Quantification of the frequency of IgE+ and IgG1+ cells in the GC B cell and NP-specific PC compartments from draining LNs of mice with the indicated genotypes 14 d after subcutaneous immunization with NP-CGG in alum adjuvant. Isotype-specific GC B cells and NP-specific PCs were enumerated by flow cytometry as in Fig. 1. Cells in A and B were from WT mice on a B6 background or Boy/J CD45.1 congenic mice bred in our colony, and cells in C and D were from BM chimeras that were generated by injecting BM from donors with the indicated genotypes (CD45.2), bred in our colony, into irradiated NCI B6 CD45.1 congenic recipients. In E, for each given Il21 genotype (Il21+/+ or Il21−/−), the Bcl2 Tg− and Tg+ mice were littermates. Dots represent data points derived from individual mice. Xs represent cell counts that were below the limit of detection. Bars represent the mean (A–D and E, left panel) or the geometric mean (E, right panel). n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA followed by Dunnett’s post-test [A and E] comparing each condition to the leftmost column or t tests with the Holm-Sidak correction for multiple comparisons [B–D] comparing the presence or absence of IL-21 within each group). Cells were from three experiments (A and B) or two experiments (D), or data were pooled from two experiments (E). Similar results as in C were observed in WT mice in a separate experiment.
To further assess the impact of IL-21 on IgE CSR, we quantified IgE and IgG1 GLTs in B cells cultured for 3 d with anti-CD40 and IL-4 in the presence or absence of IL-21. Interestingly, we observed that IL-21 inhibited IgE GLT expression, which was much more pronounced with the low dose of anti-CD40 (Fig. 6 B). In contrast, IL-21 promoted IgG1 GLT expression, which again was more pronounced with the low dose of anti-CD40 (Fig. 6 B). These data suggest that IL-21 suppresses IgE responses by inhibiting IgE germline transcription, thereby preventing IgE CSR.
We next considered the possibility that the reduction in IgE germline transcription after IL-21 treatment might be secondary to other effects of IL-21, such as the induction of PC differentiation. We thus quantified IgE and IgG1 GLTs at an early time point, only 8 h after the addition of IL-21. To prevent PC differentiation, as in our previous study (Yang et al., 2012), we cultured B cells from BM chimeras with conditional deficiency in Prdm1, the gene encoding BLIMP-1 (Shapiro-Shelef et al., 2003). We observed that IL-21 treatment led to a marked reduction in IgE GLTs after 8 h in both control and Prdm1-deficient B cells (Fig. 6 C). Consistent with this result, we found that after 4 d of cell culture, IL-21 treatment led to a reduction in the frequency of IgE+ B cells even when PC differentiation was prevented by Prdm1-deficiency (Fig. 6 D), similar to our observations in WT B cells.
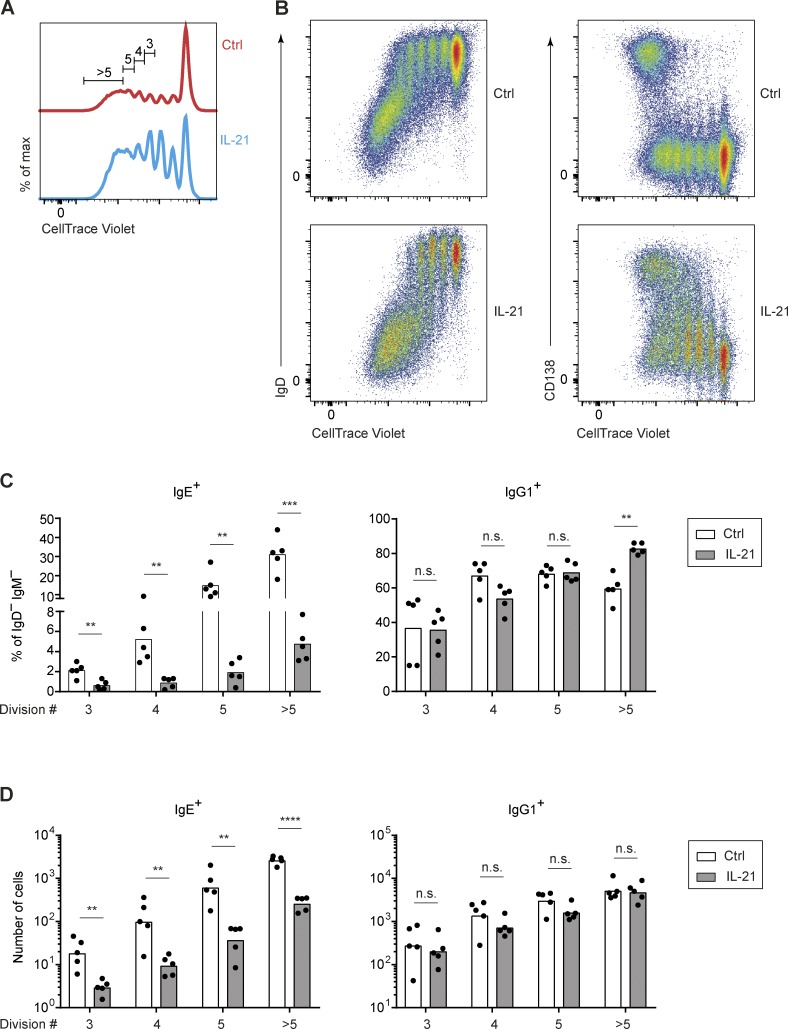
Since IL-21 is also known to induce the proliferation of B cells (Moens and Tangye, 2014), and both CSR and PC differentiation events require multiple cell divisions (Hasbold et al., 1998, 2004), we further evaluated the relationship between proliferation and CSR to IgE and IgG1. After labeling B cells with CellTrace Violet dye to measure cell divisions by dye dilution, we observed, as expected, that IL-21 treatment led to an increased proportion of divided cells (Fig. S3 A), although apoptosis also increased (data not shown) such that the net number of divided cells was fairly similar. Class-switched IgD−, IgE+, and IgG1+ cells began to appear after three cell divisions and increased with subsequent divisions (Figs. S3, B–D). The reduction of IgE+ B cells after IL-21 treatment was already apparent after three cell divisions and maintained through subsequent cell divisions, and this was reflected in both the frequency and absolute number of IgE+ B cells (Fig. S3, C and D). In contrast, the frequency and absolute number of IgG1+ B cells were relatively unaffected by IL-21 treatment in this setting, with the exception of an increase in the frequency of IgG1+ B cells among cells that had undergone more than five divisions (Fig. S3, C and D). These data suggest that IL-21 has a direct impact on the initial appearance of IgE+ B cells, consistent with the model that IL-21 inhibits IgE CSR. Indeed, PC differentiation, detected by staining for the marker CD138, did not reach a peak until cells had undergone more than five divisions (Fig. S3 B), indicating this event is temporally dissociated from the effects of IL-21 on the abundance of IgE+ B cells. Taken together, the above cell culture data provide evidence that IL-21 inhibits IgE CSR independent of the effects of IL-21 on proliferation, differentiation, and apoptosis.
Figure S3.
The inhibition of the generation of IgE-producing B cells by IL-21 in vitro is independent of cell division. Related to Fig. 6. Purified mouse B cells, labeled with CellTrace Violet, were cultured for 4 d with anti-CD40 (62.5 ng/ml) and IL-4 (12.5 ng/ml), with the addition of IL-21 (50 ng/ml) to the indicated samples on day 1. (A) Representative flow cytometry of cell divisions shown by dilution of CellTrace Violet dye. Cells that had undergone three to six divisions were gated for further analysis. (B) Representative flow cytometry showing B cell activation and CSR (IgD−, left) and PC differentiation (CD138+, right) in relation to CellTrace Violet dilution. (C and D) Quantification of the frequency (C) and number (D) of IgE+ and IgG1+ cells among IgD−IgM− cells in different cell divisions. Cells were from WT mice on a B6 background bred in our colony. Dots represent data points from individual mice (C and D). Bars represent the mean (C) or the geometric mean (D). n.s. not significant; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (t tests with the Holm-Sidak correction for multiple comparisons [C and D]). Data in C and D awere pooled from four experiments.
As apoptosis in vivo may differ from in vitro cell culture models, we further tested whether apoptosis was involved in the inhibition of IgE responses by IL-21 in vivo. Eμ-Bcl2-22 transgenic (Bcl2 Tg) mice overexpress the antiapoptotic protein Bcl-2 in the B cell lineage, rendering B cells relatively resistant to apoptosis (Strasser et al., 1991). We previously reported a large expansion in IgE PCs but only a small increase in the frequency of IgE GC B cells in Bcl2 Tg mice 14 d after immunization, suggesting most IgE+ B cells differentiate into short-lived PCs in vivo, whose survival can be rescued by Bcl-2 overexpression (Yang et al., 2012). We also observed a modest increase in the frequency of IgE GC B cells and a large expansion of IgE PCs in Il21−/− mice. Previous studies demonstrated that B cell apoptosis induced by IL-21 could be counteracted by Bcl2 overexpression (Jin et al., 2004; Mehta et al., 2003). Consequently, if the inhibition of the IgE response by IL-21 is mediated by apoptosis, then we would expect that the combination of Bcl2 overexpression and IL-21 deficiency would have no additive effect on the abundance of IgE GC B cells and PCs. However, if IL-21 acts by a mechanism distinct from inducing apoptosis, such as preventing IgE CSR, then the combination of Bcl2 overexpression and IL-21 deficiency would result in a further enhanced IgE response. After immunization, we observed that the combined overexpression of Bcl2 together with IL-21 deficiency led to a remarkable increase in the frequency of IgE+ GC B cells and a further expansion of IgE PCs compared with the Bcl-2 Tg or IL-21 deficiency alone (Fig. 6 E). Specifically, the frequency of IgE+ GC B cells in Bcl-2 Tg, Il21−/− mice was on average 12% and reached as high as 18%, and the number of IgE PCs exceeded 105 cells per mouse. These data indicate that the suppression of IgE responses by IL-21 in vivo occurs by an independent mechanism distinct from IgE+ cell apoptosis, consistent with the cell culture results above demonstrating that IL-21 inhibits IgE CSR.
IL-21 inhibits IgE CSR and promotes IgG1 CSR in human B cells
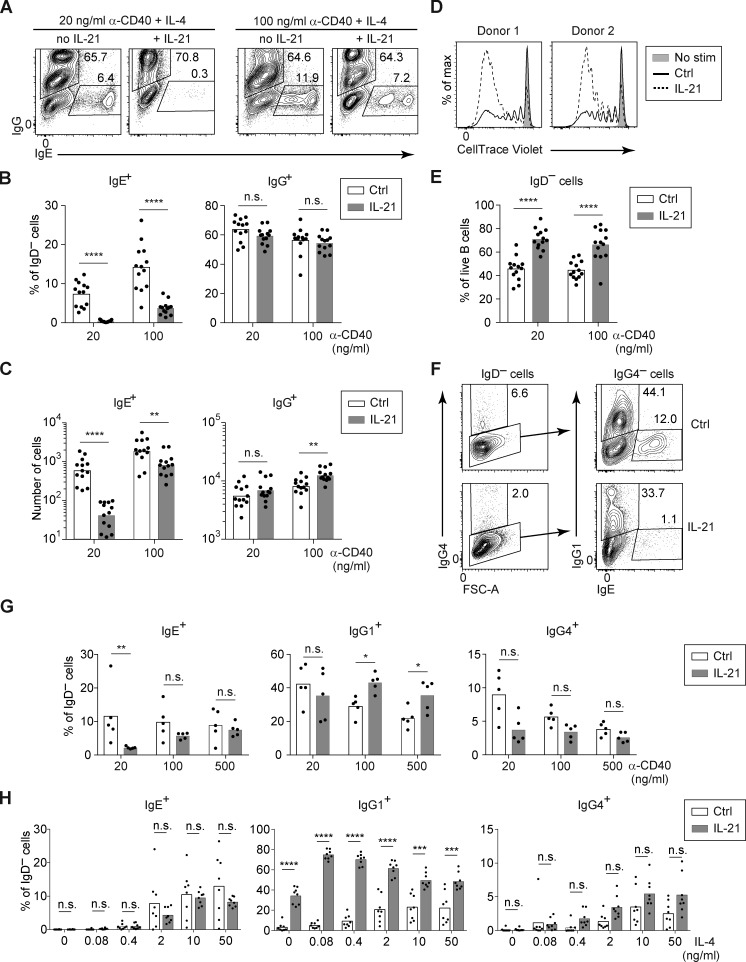
Whereas our above findings in mice demonstrated a clear inhibition of IgE responses by IL-21, some prior reports on human B cells concluded that IL-21 stimulated IgE antibody production (Avery et al., 2008; Caven et al., 2005; Pène et al., 2006; Wood et al., 2004). However, these human B cell studies measured as their primary readout the IgE secreted into the cell culture supernatants, which could be affected by a multitude of factors including cell proliferation and PC differentiation. We instead opted for a cellular analytic approach, in which we specifically identified human IgE+ B cells and PCs by intracellular staining for IgE, followed by flow cytometry, similar to the methodology we previously described with mouse B cells (Yang et al., 2012). We induced CSR to IgE by culturing purified primary B cells from human tonsils in the presence of anti-CD40 antibody and IL-4 for 7 d, with or without IL-21. In light of our findings above with mouse B cells that the inhibitory effect of IL-21 on IgE CSR was attenuated with stronger CD40 stimulation, we varied the concentration of anti-CD40 antibody. Indeed, treatment of human tonsil B cells with IL-21 led to a reduction in the frequency and number of IgE+ cells, which was substantially more pronounced in the context of low anti-CD40 stimulation (Fig. 7, A–C).
Figure 7.
IL-21 inhibits IgE and promotes IgG1 responses of cultured human B cells depending on the strength of CD40 and IL-4 signals. Total B cells (A–E) or naive B cells (F–H) were purified from human tonsils and cultured for 7 d with anti-CD40 (as indicated) and IL-4 (20 ng/ml, except in H), in the presence or absence of IL-21 (20 ng/ml). (A–C) Representative flow cytometry (A) and quantification of the frequency (B) and total number (C) of IgG+ and IgE+ cells among IgD− cells from cultures of total B cells with the indicated concentrations of anti-CD40 and a fixed concentration of IL-4 (20 ng/ml) in the presence or absence of IL-21. (D) Representative flow cytometry showing proliferation of total B cells cultured from human tonsils with anti-CD40 (100 ng/ml) and IL-4, in the presence or absence of IL-21, as measured by dilution of CellTrace Violet. (E) Frequency of activated (IgD−) cells among total tonsil B cells after culture with anti-CD40 (as indicated) and IL-4, in the presence or absence of IL-21, quantified as a percentage of live cells. (F–H) Representative flow cytometry (F) and quantification (G and H) of the frequency of IgG4+, IgG1+, and IgE+ B cells, quantified as a percentage of IgD− cells, after culturing naive B cells. (F) Naive B cells were cultured with anti-CD40 (20 ng/ml) and IL-4 in the presence or absence of IL-21. Cells were pregated as IgD– (left panels) to identify IgG4+ cells; gates for IgG1+ and IgE+ cells were then drawn within the IgG4− population (right panels). IgG1+ and IgG4+ populations were gated sequentially to account for cross-reactivity of the anti-IgG1 antibody with IgG4. (G) Naive B cells were cultured with a variable concentration of anti-CD40 as indicated with a fixed concentration of IL-4 (20 ng/ml), in the presence or absence of IL-21. (H) Naive B cells were cultured with a fixed concentration of anti-CD40 (100 ng/ml) and a variable concentration of IL-4, as indicated, in the presence or absence of IL-21. Dots represent data points from individual donors. Bars represent arithmetic (B, E, G, and H) or geometric (C) means. n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (t tests with the Holm-Sidak correction for multiple comparisons). Data were pooled from 10 experiments (B, C, and E), five experiments (G), or two experiments (H). In D, each donor is from a different experiment, and the results for each donor were replicated in another experiment.
The discrepancy between our results that IL-21 led to a reduction in the frequency of IgE+ cells versus prior reports of increased IgE antibody production could be a consequence of IL-21 generally promoting B cell proliferation and activation (Moens and Tangye, 2014). In the context of anti-CD40 and IL-4, the addition of IL-21 led to substantially increased B cell division, as measured by dilution of CellTrace Violet dye (Fig. 7 D), as well as increased activation (frequency IgD−; Fig. 7 E), which were much more pronounced than our observations in mouse B cells (Fig. S3). Increased B cell division and activation were likely the dominant effects of IL-21 in most published cell cultures studies of human B cells, whereas the inhibition of IgE CSR by IL-21 was likely attenuated by strong stimulation of CD40 with high concentrations of anti-CD40 antibody or with CD40L.
Due to their constant exposure to foreign antigens, tonsil B cells represent a population with mixed activation and differentiation states, including GC B cells and memory B cells, many of which express IgG isotypes. To control for this heterogeneity, we isolated naive B cells from human tonsils and then cultured them with anti-CD40 and IL-4 for 7 d in the presence or absence of IL-21. To better characterize the effects of IL-21 on different isotypes, we additionally stained for two subclasses of IgG, IgG1 and IgG4. Human IgG1 is the most abundant subclass of IgG, yet IgG4 has been posited to be more similar to mouse IgG1 since it is induced by IL-4 (Aalberse et al., 2016). Culture of human naive B cells with anti-CD40 and IL-4 resulted in CSR to IgG1, IgG4, and IgE (Fig. 7 F). The addition of IL-21 to human naive B cells led to a reduction in the frequency of IgE+ cells in the context of weak anti-CD40 stimulation (Fig. 7 G), similar to our findings with total tonsil B cells above (Fig. 7 B). We observed a trend toward decreased frequencies of IgG4+ cells when IL-21 was added, though this did not reach statistical significance. We also considered the impact of IL-21 in the context of limiting amounts of IL-4 with high amounts of anti-CD40 as in our studies of mouse cells above. We found that the addition of IL-21 together with a very small amount of IL-4 led to dramatically increased frequencies of IgG1+ cells, but not IgE+ cells, analogous to our above findings with IgG1 and IgE in mice (Fig. 7 H). Taken together, these findings indicate a similar impact of IL-21 on CSR in human B cells as we had observed with mouse B cells. Specifically, IL-21 inhibits IgE CSR in the context of weak CD40 stimulation, whereas IL-21 promotes IgG1 CSR in the context of limiting amounts of IL-4.
Discussion
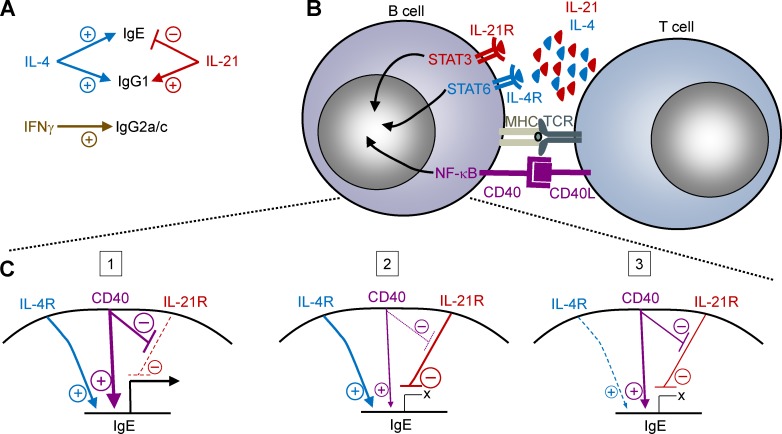
Our observations that IgE responses were highly constrained in vivo compared with in vitro cell culture suggested that one or more major negative regulators of IgE responses are present in vivo. Here, we demonstrated that IL-21 is the major cytokine that negatively regulates IgE responses in vivo. IL-21 limited IgE responses across a range of immunization conditions in mice, whereas IFN-γ, IL-10, and IL-6 were dispensable. Tfh cells were the major source of IL-21, which acted on B cells via the IL-21R and STAT3 to suppress IgE. We found that quantitative changes in the strength of CD40 and IL-4 signals determined whether IL-21 inhibited IgE CSR or promoted IgG1 CSR. Our comparable results with mouse and human B cells indicate that this fine-tuned regulation of IgE and IgG1 CSR is highly conserved. We have provided model diagrams depicting these findings in Fig. 8.
Figure 8.
Models of the regulation of IgE CSR. (A) Our data indicate that while IL-4 promotes both IgE and IgG1 CSR, IL-21 inhibits IgE CSR but promotes IgG1 CSR. IL-21 appears to be the key negative regulator of IgE under a broad range of conditions in both mouse and human B cells. We observed that IFN-γ was required for IgG2a/c CSR but in vivo had no physiological impact on IgE and IgG1 CSR under the conditions tested. (B) Model of the interaction of a T cell (such as a Tfh cell) with a B cell. Our data indicate that IL-21 produced by T cells, signaling via the IL-21R and STAT3 in B cells, inhibits IgE germline transcription, thereby preventing IgE CSR. Conversely, IL-4, signaling via the IL-4R and STAT6; and CD40L, signaling via CD40 and NF-κB; are known to directly promote IgE germline transcription in B cells (Geha et al., 2003). The model does not exclude the possibility that other signaling pathways downstream of these receptors may also contribute to IgE regulation. The hatched lines show an expanded view of the regulation of IgE germline transcription by these receptors in C. (C) Model of the regulation of IgE germline transcription by the relative strength of IL-4R, CD40, and IL-21R signaling in B cells. Quantitative differences in signals from these receptors may depend on the extent and/or duration of receptor ligation (such as differences in the relative amounts of IL-4, IL-21, and CD40L expressed by T cells, versus the duration of T cell–B cell contacts). Three cases are provided for consideration: (1) IgE germline transcription is promoted in the context of strong IL-4R and CD40 signals. The CD40 signals attenuate the inhibitory signals downstream of the IL-21R. (2) When CD40 signals are weaker, strong IL-21R signals can inhibit IgE germline transcription, even in the presence of strong IL-4R signals. (3) When IL-4R signals are too weak, only minimal IgE germline transcription would occur regardless of the relative strength of CD40 and IL-21R signals. Note that in all three cases, IgG1 germline transcription would be promoted by these signals, and thus cases 2 or 3 would lead to a bias toward IgG1 CSR rather than IgE CSR.
Early studies had shown that IgE production in cell culture was suppressed by IFN-γ but promoted by IL-4, the prototypical Th1 and Th2 cytokines, respectively (Coffman, 2006). We confirmed that IFN-γ was able to inhibit IgE and IgG1 CSR when added to cultures of mouse B cells together with IL-4 and anti-CD40. The exogenous administration of IFN-γ was also reported to reduce IgE in mouse models (Finkelman et al., 1990; Hofstra et al., 1998; Lack et al., 1994) and some human patients (Finkelman et al., 1990). Surprisingly, however, there have been relatively few studies on the impact of genetic deficiencies in IFN-γ on IgE production in mice. Mice deficient in IFN-γ or its receptor were reported to have elevated serum IgE after immunization with ovalbumin in alum adjuvant or Leishmania major infection (Coyle et al., 1996; Wang et al., 1994), although variable results were observed in other studies with ovalbumin immunization (Bruselle et al., 1997; Hofstra et al., 1998). Another study also concluded that IL-21, but not IFN-γ, regulated IgE in mice, though the flow cytometry analysis of IgE surface staining in this study likely reflected IgE captured on basophils and naive B cells rather than IgE-producing cells (Shang et al., 2006). By a robust cellular analysis of IgE+ GC B cells and PCs, our data in IFN-γ–deficient mice indicated that this cytokine is largely dispensable for IgE regulation under multiple immunization conditions. We speculate that the limited impact of IFN-γ on IgE CSR in vivo might be due to either the frequency or kinetics of B cell interactions with Tfh cells or Th1 cells that express IFN-γ. Tfh cells have been reported to express IL-21 at early stages of the immune response (Gonzalez et al., 2018; Weinstein et al., 2016), and IL-21 is broadly expressed among Tfh cells (Vinuesa et al., 2016); thus, this cytokine may play a more dominant role in IgE regulation. The B cells that receive IFN-γ signals may be the subset that undergo CSR to IgG2a/IgG2c, which was further supported by our data here showing that IFN-γ–deficient C57BL/6 mice had a marked reduction in IgG2c+ GC B cells and PCs in the context of mixed or type 1 immunizations. It was reported that substantially less exogenous IFN-γ was needed to stimulate IgG2a than to inhibit IgE responses in mice (Finkelman et al., 1990), which together with our results here suggests that the quantitative exposure of B cells to IFN-γ in vivo may often be limited.
IgE responses are also thought to be suppressed by regulatory T cells. IL-10 is a cytokine produced by regulatory T cells and B cells, as well as in the context of type 2 immune responses, that has been implicated in the inhibition of IgE CSR (Akdis and Akdis, 2014). IL-10 deficiency was reported to have an impact on serum IgE in immunized mice in some studies, but not in others (Fukushima et al., 2007; Mäkelä et al., 2000; Tournoy et al., 2000). We found that IL-10 was dispensable for the regulation of IgE GC B cell and PC responses in mice immunized with NP-CGG and alum adjuvant and that IL-10 did not inhibit IgE CSR in cell culture. Since IL-10 and IL-21 both promoted STAT3 phosphorylation in B cells, our data suggest that STAT3-mediated signaling is necessary, but not sufficient, for the inhibition of IgE CSR. Alternatively, the extent and/or kinetics of STAT3 phosphorylation induced by IL-10 versus IL-21 could differ. We observed that IL-6 induced STAT3 phosphorylation in T cells, but not in B cells. Although IL-6 has also been reported to promote IL-21 production by mouse T cells (Dienz et al., 2009) and Tfh cell differentiation (Vinuesa et al., 2016), we found no impact of IL-6 deficiency on the IgE response in mice immunized with NP-CGG and alum adjuvant, in contrast to the clear increase in IgE in IL-21–deficient mice.
Our mixed chimera data indicated that the majority of the IL-21 production relevant for inhibiting IgE responses was derived from Bcl6-dependent T cells. While these data do not exclude contributions from other T cell subsets, our results implicate Tfh cells as the major subset contributing to the negative regulation of IgE CSR by IL-21. Conversely, recent evidence from studies in helminth infection and asthma models showed a contribution of Bcl6-dependent T cells to IgE production (Kobayashi et al., 2017; Meli et al., 2017). Studies of IL-4 enhancer mutants (Harada et al., 2012; Vijayanand et al., 2012) and an IL-6R mutant (Noble and Zhao, 2016) have also implicated Tfh cells in IgE production. A recent study described a subpopulation of Tfh cells that produces IL-13 in addition to IL-4 yet produces relatively low amounts of IL-21, potentially implicating these “Tfh13” cells in IgE production (Gowthaman et al., 2019). These Tfh13 cells have been detected in mouse models of repetitive allergen inhalation (Clement et al., 2019; Gowthaman et al., 2019), but not in the context of infection with the helminth Nippostrongylus brasiliensis that strongly induces IgE production, suggesting the potential role of Tfh13 cells in IgE CSR may be limited to particular types of immune responses. Indeed, the production of IgE in N. brasiliensis infection was found to be normal in mice in which IL-13–producing T cells were depleted, suggesting that IL-4–producing Tfh cells drive IgE CSR in helminth infection (Liang et al., 2011). Overall, Tfh cells appear to be the primary negative and positive regulators of IgE CSR through the production of the cytokines IL-21 and IL-4, respectively.
The relative amount of IL-4 versus IL-21 produced by a Tfh cell, as it interacts with a B cell, has been proposed to determine the isotype to which the B cell may switch (Crotty, 2015). In support of this model, our cell culture data with both mouse and human B cells showed that very low concentrations of IL-4 together with high concentrations of IL-21 promoted IgG1 CSR, with minimal IgE CSR. These data support previous findings that IL-21 can promote IgG1 CSR (Moens and Tangye, 2014) and interestingly suggest some similarity of the induction of mouse IgG1 and human IgG1. The degree to which IL-4 is limiting during in vivo Tfh cell–B cell interactions, however, is unclear. Haploinsufficiency of IL-4 in mice led to markedly reduced IgE responses, suggesting that the concentration of IL-4 at the immunological synapse may be close to the threshold needed for IgE CSR (Robinson et al., 2017). Yet, in numerous studies in different immunization models, the majority of Tfh cells seem to express IL-4 (Bao and Reinhardt, 2015). One study reported that there is a progression from IL-21 production alone to dual IL-4 and IL-21 production followed by IL-4 production alone (Weinstein et al., 2016). One might thus expect IgE CSR to be favored by IL-4 production alone at later stages of the immune response. In contrast to this timeline, however, previous studies indicated that the IgE response is dominated by the relatively early appearance of IgE PCs and GC B cells, both of which decline rapidly thereafter, suggesting ongoing CSR to IgE is minimal at later stages of the B cell response (Yang et al., 2014). IL-21 was reported to inhibit the sequential switching of IgG1+ B cells to IgE (Erazo et al., 2007), which may be an ongoing mechanism for GC B cells and memory B cells to maintain IgG1 expression.
Our work also demonstrated that the suppression of IgE CSR by IL-21 was inversely correlated with the strength of CD40 stimulation. This result suggests that IgE CSR may require extended or frequent contacts with T cells expressing CD40L in order to overcome the inhibition by IL-21. Some indications that CD40 signaling could modify responses to IL-21 were noted in earlier studies (Mehta et al., 2003; Parrish-Novak et al., 2000; Suto et al., 2002; Wood et al., 2004). However, somewhat paradoxically in relation to our findings here, CD40 signaling was reported to up-regulate IL-21R expression (de Totero et al., 2006; Good et al., 2006). Future studies may reveal how the signals downstream of the IL-21R and CD40 interact to regulate IgE CSR.
Our findings showing that CD40 signaling can override the inhibition of IgE CSR by IL-21, together with our direct assessment of IgE+ B cells by flow cytometry, allowed us to reveal that IL-21 can suppress IgE CSR in cultured human B cells. Previous observations that IL-21 promoted IgE production were likely due to strong CD40 stimulation coupled with the general induction of B cell proliferation and PC differentiation by IL-21, leading to an increase in the total amount of secreted IgE antibody. It was thought that the inhibition of IgE by IL-21 in human cell cultures involved the production of factors from other cells such as IFN-γ by T cells (Pène et al., 2006), but here, we showed that IL-21 could directly inhibit IgE CSR of purified human B cells. Consistent with our findings reported here, polymorphisms in the IL21 and IL21R genes have been associated with serum IgE levels (Hecker et al., 2003; Li et al., 2013; Pène et al., 2006), and several patients with IL21 or IL21R loss-of-function mutations were reported to have elevated serum IgE (Erman et al., 2015; Kotlarz et al., 2014). Dominant negative mutations in the downstream signaling adapter STAT3 are prevalent in patients with hyper-IgE syndrome (Moens and Tangye, 2014), although there has been confusion as to the cellular basis for the increased IgE. An early study concluded that STAT3 was dispensable in mouse B cells for the regulation of IgE (Fornek et al., 2006), yet this has been contradicted by two more recent studies (Dascani et al., 2018; Kane et al., 2016). Through the use of mixed chimeras as well as conditional deletion of STAT3, our experiments confirmed that both the IL-21R and STAT3 were required in B cells in a cell-intrinsic manner for the suppression of IgE CSR by IL-21. Conversely, conditional deletion of STAT3 in T cells had no impact on the generation of IgE+ B cells and PCs. As conditional deletion of STAT3 in T cells inhibits Th17 cell generation (Korn et al., 2009), our findings also imply that Th17 cells are dispensable for the regulation of IgE CSR. Interestingly, similar to our studies in mice, a human patient was identified with a somatic mutation of STAT3 in B cells but not in T cells, which led to hyper-IgE but normal IL-17A production (Alcántara-Montiel et al., 2016). Taken together, in light of these studies as well as our findings here, we propose that the excessive IgE production in hyper-IgE syndrome occurs as a result of the STAT3 mutations causing defective IL-21R signaling in B cells.
IL-21R signaling is also known to promote B cell proliferation, apoptosis, and PC differentiation (Moens and Tangye, 2014). Our data presented here indicate that these effects of IL-21 can be dissociated from the impact of IL-21 on IgE CSR. Interestingly, it appears that unlike B cells expressing other isotypes, IgE-switched B cells can readily undergo PC differentiation in the absence of IL-21. This was apparent in our studies of immunized Il21−/− and Il21r−/− mice, in which most PCs were of the IgE isotype, yet IgG1 PCs were greatly reduced in number. Conversely, though there was an increase in the frequency of IgE+ GC B cells in Il21−/− and Il21r−/− mice, most GC B cells were still IgG1+. Previous studies have indicated that the IgE BCR has constitutive signaling activity that promotes differentiation into short-lived PCs and limits IgE GC B cell responses (Haniuda et al., 2016; Yang et al., 2016). The IgE BCR has also been reported to directly promote apoptosis in some studies (Haniuda et al., 2016; Laffleur et al., 2015). The cell-intrinsic regulation by the IgE BCR is likely independent from the extrinsic regulation of IgE CSR by IL-21. We propose that IL-21 limits the initial generation of IgE+ B cells, whereas the IgE BCR constrains the fate of those IgE+ B cells that are generated. Consistent with this model, we found that preventing apoptosis with a Bcl2 transgene, in the context of IL-21 deficiency, led to a remarkable combined increase in the number of IgE GC B cells and PCs.
In summary, we conclude that IL-21 is a major negative regulator of the generation of IgE+ B cells by suppressing IgE CSR. Conversely, IL-4 is known to be the major positive regulator of IgE CSR (Finkelman et al., 1990). Tfh cells, which are abundant producers of both of these cytokines as well as CD40L (Crotty, 2015; Vinuesa et al., 2016), are thus heavily implicated in the tight control of IgE CSR. The likelihood of a B cell undergoing CSR to IgE may depend on several features of B cell–T cell interactions, such as their frequency and duration, which would affect the extent of CD40 ligation as well as the amounts of IL-4 and IL-21 secreted. These interactions are likely to be heterogeneous, with particular characteristics (see Fig. 8) leading a small number of B cells to undergo CSR to IgE. It will be interesting in future studies to assess the features of B cell–T cell interactions that lead to IgE production specific for allergens in the context of allergic sensitization.
Materials and methods
Mice and chimeras
All mice in this study were on a C57BL/6 (B6) background (backcrossed ≥10 generations). Mice for experiments were sex and age matched as much as possible. The origins of the control mice are indicated in the figure legends. B6/J mice (000664; C57BL/6J), Boy/J CD45.1 mice (002014; B6.SJL-Ptprca Pepcb/BoyJ), Bcl6flox mice (023727; B6.129S(FVB)-Bcl6tm1.1Dent/J), Cd4-Cre mice (022071; B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ), Prdm1flox mice (008100; B6.129-Prdm1tm1Clme/J), Stat3flox mice (016923; B6.129S1-Stat3tm1Xyfu/J), and μMT mice (002288; B6.129S2-Ighmtm1Cgn/J) were originally purchased from The Jackson Laboratory and then bred in our colony. Some B6/J mice, Ifng−/− mice (002287; B6.129S7-Ifngtm1Ts/J), Il6−/− mice (002650; B6.129S2-Il6tm1Kopf/J), and Il10−/− mice (002251; B6.129P2-Il10tm1Cgn/J) were purchased directly from The Jackson Laboratory for experiments. Il21−/− mice (032800-UCD; B6.129S-Il21tm1Lex/Mmucd) were obtained from the Mutant Mouse Resource and Research Centers, where they were originally deposited by Lexicon Pharmaceuticals, and then bred in our colony. Cd19Cre mice (Rickert et al., 1995), Eμ-Bcl2-22 Tg mice (Strasser et al., 1991), Il21r−/− mice (Ozaki et al., 2002), and Prdm1gfp mice (Kallies et al., 2004) were maintained in our colony on a B6 background. Otherwise, B6 CD45.1 congenic mice, which were used as WT mice in some experiments as indicated, were purchased from the National Cancer Institute (NCI)/Charles River (01B96; B6-Ly5.2/Cr, later renamed to B6-Ly5.1/Cr). Mice were housed in specific pathogen–free facilities, and protocols were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco (UCSF). The preparation of BM from donor, irradiation, and reconstitution of recipients were as described previously (Yang et al., 2012). We allowed at least 6 wk for reconstitution of the chimeras before immunization or cell culture studies.
Immunizations
To prepare antigen for immunization for each mouse, 50 µg NP-CGG in 60 µl PBS was mixed with equal volume of PBS or one of the following adjuvants: alum (Alhydrogel; Accurate Chemical and Scientific), CFA (Sigma-Aldrich), SAS (Sigma-Aldrich), or CpG-DNA for mouse (50 µg; Hycult Biotech) according to the manufacturer’s instructions. 30 µl antigen mix per site was injected into the upper flanks and above the shoulders to generate responses in the draining axillary and brachial LNs.
Human tonsil B cell purification
Human tonsil specimens were acquired from patients undergoing routine tonsillectomies under a protocol approved by the UCSF Human Research Protection Program and Institutional Review Board. Tonsil specimens were deidentified and subsequent studies were classified as not human subjects research according to the guidelines from the UCSF Institutional Review Board.
Tonsillar tissue was dissociated, a single cell suspension was prepared and then cells were cryopreserved as described previously (Wu et al., 2018). Total B cells were purified with the Dynabeads Untouched Human B cells kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Naive B cells were purified by flow cytometric sorting (for Fig. 7 G; see details below) or magnetic bead depletion (for Fig. 7 H) using the Mojosort Human Naive B Cell Isolation kit (BioLegend) with the following modifications to deplete GC B cells: ∼20 million tonsil lymphocytes were resuspended in 0.2 ml PBS with 1% FBS, and then 20 µl antibody mix from the kit, a 1:100 dilution of anti-human CD10-biotin (clone: SN5c; eBioscience), and a 1:20 dilution of anti-human IgG1-biotin (clone IS11-12.E4.23.20; Miltenyi Biotec) were added to the sample. The mixture was incubated on ice for 20 min, then 20 µl Mojosort beads were added and incubated on ice for 10 min. The mixture was then transferred to a 5-ml tube with 3 ml ice-cold PBS with 1% FBS, and the tube was then put in a MojoSort magnet (BioLegend) on ice for 5 min. The supernatant was then decanted into a new 5-ml tube and centrifuged at 450 ×g for 5 min. The cell pellet was resuspended in 100 µl complete IMDM medium for cell culture.
Cell culture
Primary mouse B cells were purified as described previously (Sullivan et al., 2011). Purified splenic B cells or crude splenocytes were cultured as described previously (Yang et al., 2012, 2016). Briefly, 1 × 105 cells were cultured in complete RPMI media with rat anti-mouse CD40 antibody (FGK-45; Miltenyi Biotec) and the indicated cytokine(s) in 96-well Microtest U-bottom plates (BD Falcon) with a volume of 200 µl per well. Recombinant mouse IL-4 (Peprotech), IL-6 (R&D Systems), IL-10 (Peprotech), IL-21 (R&D Biosystems), or IFN-γ (Peprotech) were added to cultures as indicated. Cells were cultured for 4 d in a humidified incubator at 37°C in 5% CO2.
Total or naive B cells from human tonsils were isolated and purified as described above. Cells were then cultured as described previously (Wu et al., 2018) for 7 d with anti-human CD40 antibody (clone G28.5; Bio-X-Cell), recombinant human IL-4 (Peprotech), and/or recombinant human IL-21 (Peprotech or R&D Biosystems).
Cell proliferation assay
To track cell divisions in vitro, B cells were labeled with CellTrace Violet (Thermo Fisher Scientific) before culture. Cells were resuspended in PBS containing 2% FBS to a concentration between 2 and 20 million cells/ml. CellTrace Violet was diluted in PBS to a working concentration of 2 μM and then added at a 1:1 ratio to the cell suspension, achieving a final concentration of 1 μM. The mixture was incubated at 37°C for 20 min, agitating every 5 to 10 min to prevent cells from settling. Cells were then washed twice with PBS containing 2% FBS. Prior to centrifugation, FBS was carefully layered at the bottom of the tube at 1/10 of the suspension volume. Finally, cells were resuspended in complete media and cultured as described above.
Flow cytometry
Cell suspensions were prepared from LNs or cell culture and stained with antibodies (Table S1andTable S2) essentially as described previously (Yang et al., 2012, 2016). Cell counts were obtained from total LN cells on a Coulter Counter Z2 instrument equipped with a 100-µm ampoule aperture tube, using a size threshold of 22.45 fl (μm3) to 113.10 fl after treatment of samples diluted in ISOTON II with ZAP-OGLOBIN II lytic reagent. Cell counts for cultured cells were determined by flow cytometry by adding a fixed amount of AccuCount Ultra Rainbow Fluorescent Particles (5.0–5.9 μm; Spherotech). Flow cytometry data were collected on an LSR Fortessa (BD Biosciences) and analyzed with FlowJo v10. Nonviable cells were excluded by labeling with fixable viability dye eFluor780 (eBioscience). All samples were gated on FSC-A versus SSC-A over a broad range of FSC-A to include blasting lymphocytes, followed by FSC-W versus FSC-H and then SSC-W versus SSC-H gates to exclude doublets.
Human intracellular IgE staining was analogous to our method for mouse intracellular IgE staining (Yang et al., 2012). Briefly, cell surface IgE was blocked with an excess of unconjugated anti-human IgE antibody clone MHE-18 (BioLegend) during surface staining. Mouse gamma globulin (Jackson ImmunoResearch) was used as a blocking agent during surface staining of human B cells (rather than “Fc block,” which we found interfered with subsequent staining with anti-human IgG antibodies). Cells were then fixed and permeabilized using a Cytofix/Cytoperm Fixation/Permeabilization solution kit (BD Biosciences). Finally, intracellular IgE was stained with fluorescently conjugated MHE-18 together with antibodies to detect total (surface and intracellular) IgG or specific IgG isotypes (IgG1 and IgG4) and total IgM (Table S2).
For PhosFlow analysis of phosphorylation of STAT proteins, cells were incubated with eFluor780 viability dye at room temperature for 10 min, washed once with PBS, and then resuspended in 100 µl culture medium. Cytokines were added, and cells were incubated at 37°C for 20 min; then, 100 µl of 4% PFA was added, and the sample was incubated at room temperature for 10 min to stop cytokine signaling and fix the cells. The cells were washed once with FACS buffer and stained with surface markers at room temperature for 20 min. The cells were washed once with FACS buffer and resuspended in 120 µl FACS buffer, and then 80 µl 4% PFA was added, followed by incubation at room temperature for 5 min. The cells were then centrifuged at 1,000 ×g at room temperature for 2 min, resuspended in the residual liquid, and cooled on ice for 2 min. Then 3 × 30 µl ice-cold 90% methanol were added to the cell samples, while mixing the samples by tapping after each addition. The samples were incubated on ice for 30 min before 150 µl FACS buffer was added, and then the samples were further incubated on ice for 5 min. The cells were then pelleted down by centrifugation at 1,000 ×g for 3 min and washed again with FACS buffer. Cell pellets were resuspended in FACS staining buffer with pSTAT-specific antibodies (Table S1) and then incubated at room temperature for 30 min. Cells were washed twice with FACS buffer before analysis.
For sorting of naive B cells from human tonsils, cryopreserved tonsil lymphocytes were thawed and resuspended in sorting media consisting of Hanks’ balanced salt solution containing 1% FBS, 0.5% (wt/vol) bovine serum albumin fraction V (Roche), 10 mM Hepes (Gibco), and 2 mM EDTA (Thermo Fisher Scientific) that had been filter sterilized using a 0.2-µm syringe filter (Corning). Cells were first blocked by incubating with 0.1 mg/ml mouse gamma-globulin (Jackson ImmunoResearch) for 15 min; stained with anti-CD3, anti-CD27, and anti-CD38 at a 1/50 dilution (see Table S2) for 30 min on ice; and then washed with an excess of sorting media. Stained cells were resuspended in sorting media to a concentration of 30 million cells per milliliter and sorted on a FACS Aria IIIu (BD Biosciences) cell sorter. Sorted cells were collected in a 1:1 (vol/vol) mixture of FBS and IMDM.
Transcriptional analysis
Purified B cells were cultured under the conditions indicated in Fig. 6, B and C, as indicated, and then five wells of cells of each treatment were pooled and harvested by centrifugation at 750 ×g for 2 min. Cells were washed once with PBS, and the cell pellet was immediately lysed in 0.25 ml lysis buffer LBP of the Nucleospin RNA Plus kit (Macherey-Nagel) and processed for total RNA preparation using the kit according to manufacturer’s instructions. Total RNA from each sample was eluted in 30 µl RNase-free water, and 6 µl RNA from each sample was used as template for cDNA synthesis using the ProtoScript First Strand cDNA Synthesis Kit (NEB) following the manufacturer’s manual. The resultant cDNA samples were used as templates for quantitative PCR (qPCR) assays using SYBR Premix Ex Taq Mastermix (Takara) in a 10-µl volume in a 384-well plate on an ABI7900HT machine. The following specific primers were used for the qPCR assays: ε GLT (forward: 5′-TCGAATAAGAACAGTCTGGCC-3′; reverse: 5′-TCACAGGACCAGGGAAGTAG-3′); γ1 GLT (forward: 5′-CAGGTTGAGAGAACCAAGGAAG-3′; reverse: 5′-AGGGTCACCATGGAGTTAGT-3′); and internal control Hprt (forward: 5′-TGACACTGGCAAAACAATGCA-3′; reverse: 5′-GGTCCTTTTCACCAGCAAGCT-3′). The final concentration of each primer was 250 nM. The qPCR thermal cycler conditions were 95°C 30 s; 95°C 8 s, 60°C 60 s for 40 cycles. A relative standard curve for cDNA derived from each transcript was constructed by using serially diluted cDNA from B cells cultured with anti-CD40 plus IL-4 as qPCR standards, and the relative quantity of cDNA of each gene was calculated based on the corresponding standard curve. The relative quantity of ε and γ1 GLT was finally normalized by the quantity of Hprt cDNA in the same sample.
Statistical analysis
GraphPad Prism v7 was used for statistical analyses, with appropriate tests chosen based on experimental design after consulting the GraphPad Statistics Guide. Data approximated a log-normal distribution and thus were log transformed for statistical tests. In cases where samples had zero events detected, data were either not log transformed for frequency calculations or the cell count was set to a value of five, which was approximately twofold below our limit of detection (denoted as “X” in corresponding plots). All tests were two tailed. To achieve sufficient power to discern meaningful differences, experiments were performed with multiple biological replicates and/or multiple times, with details provided in each individual figure legend. The number of samples chosen for each comparison was determined based on past similar experiments or by performing pilot experiments to assess the expected magnitude of differences.
Online supplemental material
Fig. S1 provides the total cell counts for the analyses shown in Fig. 1 C. Fig. S2 provides the cell counts for the experiments in Fig. 2. Fig. S3 shows an analysis of the differentiation of IgE+ versus IgG1+ B cells, as well as PCs, with respect to the cell divisions of mouse B cells cultured with or without IL-21. Table S1 is a list of reagents used for flow cytometric analysis of mouse samples. Table S2 is a list of reagents used for flow cytometric analysis of human samples.
Supplementary Material
lists the reagents used for flow cytometric analysis of mouse samples.
lists the reagents used for flow cytometric analysis of human samples.
Acknowledgments
We thank C. Cho for technical assistance; J. Baron (UCSF, San Francisco, CA) for originally providing Il21r−/− mice; Y. Kamimura and L. Lanier (UCSF, San Francisco, CA) for providing some Ifng−/− mice; K. Rosbe (UCSF, San Francisco, CA) and the UCSF Human Biospecimen Repository for providing tonsil samples; and K.M. Ansel, S. Crotty, W. Leonard, R. Locksley, M. Robinson, and G. Smith for helpful advice.
This work was supported by the Pew Charitable Trusts (C.D.C. Allen is a Pew Scholar in the Biomedical Sciences) as well as the Sandler Asthma Basic Research Center and the Cardiovascular Research Institute at UCSF. C-A.M. Wu received support from the Howard Hughes Medical Institute Medical Research Fellows and UCSF Pathways to Discovery programs. S. Targ was funded by the AOA Carolyn L. Kuckein Student Research Fellowship program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Author contributions:Z. Yang performed all mouse studies and part of the human B cell studies. C-A.M. Wu performed most human B cell studies. S. Targ provided insights and performed some preliminary experiments. C.D.C. Allen supervised the research. Z. Yang, C-A.M. Wu, and C.D.C. Allen analyzed data and wrote the manuscript.
References
- Aalberse R.C., Platts-Mills T.A., and Rispens T.. 2016. The Developmental History of IgE and IgG4 Antibodies in Relation to Atopy, Eosinophilic Esophagitis, and the Modified TH2 Response. Curr. Allergy Asthma Rep. 16:45 10.1007/s11882-016-0621-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achatz-Straussberger G., Zaborsky N., Königsberger S., Luger E.O., Lamers M., Crameri R., and Achatz G.. 2008. Migration of antibody secreting cells towards CXCL12 depends on the isotype that forms the BCR. Eur. J. Immunol. 38:3167–3177. 10.1002/eji.200838456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akdis M., and Akdis C.A.. 2014. Mechanisms of allergen-specific immunotherapy: multiple suppressor factors at work in immune tolerance to allergens. J. Allergy Clin. Immunol. 133:621–631. 10.1016/j.jaci.2013.12.1088 [DOI] [PubMed] [Google Scholar]
- Alcántara-Montiel J.C., Staines-Boone T., López-Herrera G., Berrón-Ruiz L., Borrego-Montoya C.R., and Santos-Argumedo L.. 2016. Somatic mosaicism in B cells of a patient with autosomal dominant hyper IgE syndrome. Eur. J. Immunol. 46:2438–2443. 10.1002/eji.201546275 [DOI] [PubMed] [Google Scholar]
- Avery D.T., Ma C.S., Bryant V.L., Santner-Nanan B., Nanan R., Wong M., Fulcher D.A., Cook M.C., and Tangye S.G.. 2008. STAT3 is required for IL-21-induced secretion of IgE from human naive B cells. Blood. 112:1784–1793. 10.1182/blood-2008-02-142745 [DOI] [PubMed] [Google Scholar]
- Bao K., and Reinhardt R.L.. 2015. The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. Cytokine. 75:25–37. 10.1016/j.cyto.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruselle G.G., Kips J.C., Peleman R.A., Joos G.F., Devos R.R., Tavernier J.H., and Pauwels R.A.. 1997. Role of IFN-γ in the inhibition of the allergic airway inflammation caused by IL-12. Am. J. Respir. Cell Mol. Biol. 17:767–771. 10.1165/ajrcmb.17.6.2820 [DOI] [PubMed] [Google Scholar]
- Caven T.H., Shelburne A., Sato J., Chan-Li Y., Becker S., and Conrad D.H.. 2005. IL-21 dependent IgE production in human and mouse in vitro culture systems is cell density and cell division dependent and is augmented by IL-10. Cell. Immunol. 238:123–134. 10.1016/j.cellimm.2006.03.001 [DOI] [PubMed] [Google Scholar]
- Clement R.L., Daccache J., Mohammed M.T., Diallo A., Blazar B.R., Kuchroo V.K., Lovitch S.B., Sharpe A.H., and Sage P.T.. 2019. Follicular regulatory T cells control humoral and allergic immunity by restraining early B cell responses. Nat. Immunol. 20:1360–1371. 10.1038/s41590-019-0472-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman R.L. 2006. Origins of the T(H)1-T(H)2 model: a personal perspective. Nat. Immunol. 7:539–541. 10.1038/ni0606-539 [DOI] [PubMed] [Google Scholar]
- Coyle A.J., Tsuyuki S., Bertrand C., Huang S., Aguet M., Alkan S.S., and Anderson G.P.. 1996. Mice lacking the IFN-gamma receptor have impaired ability to resolve a lung eosinophilic inflammatory response associated with a prolonged capacity of T cells to exhibit a Th2 cytokine profile. J. Immunol. 156:2680–2685. [PubMed] [Google Scholar]
- Crotty S. 2015. A brief history of T cell help to B cells. Nat. Rev. Immunol. 15:185–189. 10.1038/nri3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascani P., Ding C., Kong X., Tieri D., Hu X., Zhang H.G., Kitamura D., Bolli R., Rouchka E.C., and Yan J.. 2018. Transcription Factor STAT3 Serves as a Negative Regulator Controlling IgE Class Switching in Mice. Immunohorizons. 2:349–362. 10.4049/immunohorizons.1800069 [DOI] [PubMed] [Google Scholar]
- de Totero D., Meazza R., Zupo S., Cutrona G., Matis S., Colombo M., Balleari E., Pierri I., Fabbi M., Capaia M., et al. 2006. Interleukin-21 receptor (IL-21R) is up-regulated by CD40 triggering and mediates proapoptotic signals in chronic lymphocytic leukemia B cells. Blood. 107:3708–3715. 10.1182/blood-2005-09-3535 [DOI] [PubMed] [Google Scholar]
- Dienz O., Eaton S.M., Bond J.P., Neveu W., Moquin D., Noubade R., Briso E.M., Charland C., Leonard W.J., Ciliberto G., et al. 2009. The induction of antibody production by IL-6 is indirectly mediated by IL-21 produced by CD4+ T cells. J. Exp. Med. 206:69–78. 10.1084/jem.20081571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erazo A., Kutchukhidze N., Leung M., Christ A.P., Urban J.F. Jr., Curotto de Lafaille M.A., and Lafaille J.J.. 2007. Unique maturation program of the IgE response in vivo. Immunity. 26:191–203. 10.1016/j.immuni.2006.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erman B., Bilic I., Hirschmugl T., Salzer E., Çagdas D., Esenboga S., Akcoren Z., Sanal O., Tezcan I., and Boztug K.. 2015. Combined immunodeficiency with CD4 lymphopenia and sclerosing cholangitis caused by a novel loss-of-function mutation affecting IL21R. Haematologica. 100:e216–e219. 10.3324/haematol.2014.120980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelman F.D., Holmes J., Katona I.M., Urban J.F. Jr., Beckmann M.P., Park L.S., Schooley K.A., Coffman R.L., Mosmann T.R., and Paul W.E.. 1990. Lymphokine control of in vivo immunoglobulin isotype selection. Annu. Rev. Immunol. 8:303–333. 10.1146/annurev.iy.08.040190.001511 [DOI] [PubMed] [Google Scholar]
- Fornek J.L., Tygrett L.T., Waldschmidt T.J., Poli V., Rickert R.C., and Kansas G.S.. 2006. Critical role for Stat3 in T-dependent terminal differentiation of IgG B cells. Blood. 107:1085–1091. 10.1182/blood-2005-07-2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A., Sumi T., Fukuda K., Kumagai N., Nishida T., and Ueno H.. 2007. Endogenous interleukin-10 produced by antigen-irrelevant cells promotes the development of experimental murine allergic conjunctivitis. Int. Arch. Allergy Immunol. 144:79–84. 10.1159/000102618 [DOI] [PubMed] [Google Scholar]
- Geha R.S., Jabara H.H., and Brodeur S.R.. 2003. The regulation of immunoglobulin E class-switch recombination. Nat. Rev. Immunol. 3:721–732. 10.1038/nri1181 [DOI] [PubMed] [Google Scholar]
- Gonzalez D.G., Cote C.M., Patel J.R., Smith C.B., Zhang Y., Nickerson K.M., Zhang T., Kerfoot S.M., and Haberman A.M.. 2018. Nonredundant Roles of IL-21 and IL-4 in the Phased Initiation of Germinal Center B Cells and Subsequent Self-Renewal Transitions. J. Immunol. 201:3569–3579. 10.4049/jimmunol.1500497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good K.L., Bryant V.L., and Tangye S.G.. 2006. Kinetics of human B cell behavior and amplification of proliferative responses following stimulation with IL-21. J. Immunol. 177:5236–5247. 10.4049/jimmunol.177.8.5236 [DOI] [PubMed] [Google Scholar]
- Gould H.J., and Wu Y.B.. 2018. IgE repertoire and immunological memory: compartmental regulation and antibody function. Int. Immunol. 30:403–412. 10.1093/intimm/dxy048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowthaman U., Chen J.S., Zhang B., Flynn W.F., Lu Y., Song W., Joseph J., Gertie J.A., Xu L., Collet M.A., et al. 2019. Identification of a T follicular helper cell subset that drives anaphylactic IgE. Science. 365:eaaw6433 10.1126/science.aaw6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniuda K., Fukao S., Kodama T., Hasegawa H., and Kitamura D.. 2016. Autonomous membrane IgE signaling prevents IgE-memory formation. Nat. Immunol. 17:1109–1117. 10.1038/ni.3508 [DOI] [PubMed] [Google Scholar]
- Harada M., Magara-Koyanagi K., Watarai H., Nagata Y., Ishii Y., Kojo S., Horiguchi S., Okamoto Y., Nakayama T., Suzuki N., et al. 2006. IL-21-induced Bepsilon cell apoptosis mediated by natural killer T cells suppresses IgE responses. J. Exp. Med. 203:2929–2937. 10.1084/jem.20062206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y., Tanaka S., Motomura Y., Harada Y., Ohno S., Ohno S., Yanagi Y., Inoue H., and Kubo M.. 2012. The 3′ enhancer CNS2 is a critical regulator of interleukin-4-mediated humoral immunity in follicular helper T cells. Immunity. 36:188–200. 10.1016/j.immuni.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Hasbold J., Lyons A.B., Kehry M.R., and Hodgkin P.D.. 1998. Cell division number regulates IgG1 and IgE switching of B cells following stimulation by CD40 ligand and IL-4. Eur. J. Immunol. 28:1040–1051. [DOI] [PubMed] [Google Scholar]
- Hasbold J., Corcoran L.M., Tarlinton D.M., Tangye S.G., and Hodgkin P.D.. 2004. Evidence from the generation of immunoglobulin G-secreting cells that stochastic mechanisms regulate lymphocyte differentiation. Nat. Immunol. 5:55–63. 10.1038/ni1016 [DOI] [PubMed] [Google Scholar]
- He J.S., Meyer-Hermann M., Xiangying D., Zuan L.Y., Jones L.A., Ramakrishna L., de Vries V.C., Dolpady J., Aina H., Joseph S., et al. 2013. The distinctive germinal center phase of IgE+ B lymphocytes limits their contribution to the classical memory response. J. Exp. Med. 210:2755–2771. 10.1084/jem.20131539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.S., Subramaniam S., Narang V., Srinivasan K., Saunders S.P., Carbajo D., Wen-Shan T., Hidayah Hamadee N., Lum J., Lee A., et al. 2017. IgG1 memory B cells keep the memory of IgE responses. Nat. Commun. 8:641 10.1038/s41467-017-00723-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M., Bohnert A., König I.R., Bein G., and Hackstein H.. 2003. Novel genetic variation of human interleukin-21 receptor is associated with elevated IgE levels in females. Genes Immun. 4:228–233. 10.1038/sj.gene.6363954 [DOI] [PubMed] [Google Scholar]
- Hofstra C.L., Van Ark I., Hofman G., Nijkamp F.P., Jardieu P.M., and Van Oosterhout A.J.. 1998. Differential effects of endogenous and exogenous interferon-gamma on immunoglobulin E, cellular infiltration, and airway responsiveness in a murine model of allergic asthma. Am. J. Respir. Cell Mol. Biol. 19:826–835. 10.1165/ajrcmb.19.5.3027 [DOI] [PubMed] [Google Scholar]
- Hollister K., Kusam S., Wu H., Clegg N., Mondal A., Sawant D.V., and Dent A.L.. 2013. Insights into the role of Bcl6 in follicular Th cells using a new conditional mutant mouse model. J. Immunol. 191:3705–3711. 10.4049/jimmunol.1300378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabara H.H., Fu S.M., Geha R.S., and Vercelli D.. 1990. CD40 and IgE: synergism between anti-CD40 monoclonal antibody and interleukin 4 in the induction of IgE synthesis by highly purified human B cells. J. Exp. Med. 172:1861–1864. 10.1084/jem.172.6.1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannin P., Lecoanet S., Delneste Y., Gauchat J.F., and Bonnefoy J.Y.. 1998. IgE versus IgG4 production can be differentially regulated by IL-10. J. Immunol. 160:3555–3561. [PubMed] [Google Scholar]
- Jin H., and Malek T.R.. 2006. Redundant and unique regulation of activated mouse B lymphocytes by IL-4 and IL-21. J. Leukoc. Biol. 80:1416–1423. 10.1189/jlb.0206096 [DOI] [PubMed] [Google Scholar]
- Jin H., Carrio R., Yu A., and Malek T.R.. 2004. Distinct activation signals determine whether IL-21 induces B cell costimulation, growth arrest, or Bim-dependent apoptosis. J. Immunol. 173:657–665. 10.4049/jimmunol.173.1.657 [DOI] [PubMed] [Google Scholar]
- Kaji T., Ishige A., Hikida M., Taka J., Hijikata A., Kubo M., Nagashima T., Takahashi Y., Kurosaki T., Okada M., et al. 2012. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J. Exp. Med. 209:2079–2097. 10.1084/jem.20120127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Hasbold J., Tarlinton D.M., Dietrich W., Corcoran L.M., Hodgkin P.D., and Nutt S.L.. 2004. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J. Exp. Med. 200:967–977. 10.1084/jem.20040973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane A., Lau A., Brink R., Tangye S.G., and Deenick E.K.. 2016. B-cell-specific STAT3 deficiency: Insight into the molecular basis of autosomal-dominant hyper-IgE syndrome. J. Allergy Clin. Immunol. 138:1455–1458.e3. 10.1016/j.jaci.2016.05.018 [DOI] [PubMed] [Google Scholar]
- Kasaian M.T., Whitters M.J., Carter L.L., Lowe L.D., Jussif J.M., Deng B., Johnson K.A., Witek J.S., Senices M., Konz R.F., et al. 2002. IL-21 limits NK cell responses and promotes antigen-specific T cell activation: a mediator of the transition from innate to adaptive immunity. Immunity. 16:559–569. 10.1016/S1074-7613(02)00295-9 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Iijima K., Dent A.L., and Kita H.. 2017. Follicular helper T cells mediate IgE antibody response to airborne allergens. J. Allergy Clin. Immunol. 139:300–313.e7. 10.1016/j.jaci.2016.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Oukka M., and Kuchroo V.K.. 2009. IL-17 and Th17 Cells. Annu. Rev. Immunol. 27:485–517. 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- Kotlarz D., Ziętara N., Milner J.D., and Klein C.. 2014. Human IL-21 and IL-21R deficiencies: two novel entities of primary immunodeficiency. Curr. Opin. Pediatr. 26:704–712. 10.1097/MOP.0000000000000160 [DOI] [PubMed] [Google Scholar]
- Lack G., Renz H., Saloga J., Bradley K.L., Loader J., Leung D.Y., Larsen G., and Gelfand E.W.. 1994. Nebulized but not parenteral IFN-gamma decreases IgE production and normalizes airways function in a murine model of allergen sensitization. J. Immunol. 152:2546–2554. [PubMed] [Google Scholar]
- Laffleur B., Duchez S., Tarte K., Denis-Lagache N., Péron S., Carrion C., Denizot Y., and Cogné M.. 2015. Self-Restrained B Cells Arise following Membrane IgE Expression. Cell Reports. 10:900–909. 10.1016/j.celrep.2015.01.023 [DOI] [PubMed] [Google Scholar]
- Lambrecht B.N., and Hammad H.. 2017. The immunology of the allergy epidemic and the hygiene hypothesis. Nat. Immunol. 18:1076–1083. 10.1038/ni.3829 [DOI] [PubMed] [Google Scholar]
- Li N., Zhu Q., Li Z., Han Q., Chen J., Lv Y., Wang Y., Zeng X., Chen Y., Yang C., and Liu Z.. 2013. IL21 and IL21R polymorphisms and their interactive effects on serum IL-21 and IgE levels in patients with chronic hepatitis B virus infection. Hum. Immunol. 74:567–573. 10.1016/j.humimm.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Liang H.E., Reinhardt R.L., Bando J.K., Sullivan B.M., Ho I.C., and Locksley R.M.. 2011. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat. Immunol. 13:58–66. 10.1038/ni.2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä M.J., Kanehiro A., Borish L., Dakhama A., Loader J., Joetham A., Xing Z., Jordana M., Larsen G.L., and Gelfand E.W.. 2000. IL-10 is necessary for the expression of airway hyperresponsiveness but not pulmonary inflammation after allergic sensitization. Proc. Natl. Acad. Sci. USA. 97:6007–6012. 10.1073/pnas.100118997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire H.M., Vogelzang A., Warren J., Loetsch C., Natividad K.D., Chan T.D., Brink R., Batten M., and King C.. 2015. IL-21 and IL-4 Collaborate To Shape T-Dependent Antibody Responses. J. Immunol. 195:5123–5135. 10.4049/jimmunol.1501463 [DOI] [PubMed] [Google Scholar]
- Mehta D.S., Wurster A.L., Whitters M.J., Young D.A., Collins M., and Grusby M.J.. 2003. IL-21 induces the apoptosis of resting and activated primary B cells. J. Immunol. 170:4111–4118. 10.4049/jimmunol.170.8.4111 [DOI] [PubMed] [Google Scholar]
- Meli A.P., Fontés G., Leung Soo C., and King I.L.. 2017. T Follicular Helper Cell-Derived IL-4 Is Required for IgE Production during Intestinal Helminth Infection. J. Immunol. 199:244–252. 10.4049/jimmunol.1700141 [DOI] [PubMed] [Google Scholar]
- Miyauchi K., Sugimoto-Ishige A., Harada Y., Adachi Y., Usami Y., Kaji T., Inoue K., Hasegawa H., Watanabe T., Hijikata A., et al. 2016. Protective neutralizing influenza antibody response in the absence of T follicular helper cells. Nat. Immunol. 17:1447–1458. 10.1038/ni.3563 [DOI] [PubMed] [Google Scholar]
- Moens L., and Tangye S.G.. 2014. Cytokine-Mediated Regulation of Plasma Cell Generation: IL-21 Takes Center Stage. Front. Immunol. 5:65 10.3389/fimmu.2014.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu W.A., Allard J.B., Dienz O., Wargo M.J., Ciliberto G., Whittaker L.A., and Rincon M.. 2009. IL-6 is required for airway mucus production induced by inhaled fungal allergens. J. Immunol. 183:1732–1738. 10.4049/jimmunol.0802923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble A., and Zhao J.. 2016. Follicular helper T cells are responsible for IgE responses to Der p 1 following house dust mite sensitization in mice. Clin. Exp. Allergy. 46:1075–1082. 10.1111/cea.12750 [DOI] [PubMed] [Google Scholar]
- Ozaki K., Spolski R., Feng C.G., Qi C.F., Cheng J., Sher A., Morse H.C. III, Liu C., Schwartzberg P.L., and Leonard W.J.. 2002. A critical role for IL-21 in regulating immunoglobulin production. Science. 298:1630–1634. 10.1126/science.1077002 [DOI] [PubMed] [Google Scholar]
- Parrish-Novak J., Dillon S.R., Nelson A., Hammond A., Sprecher C., Gross J.A., Johnston J., Madden K., Xu W., West J., et al. 2000. Interleukin 21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature. 408:57–63. 10.1038/35040504 [DOI] [PubMed] [Google Scholar]
- Pène J., Guglielmi L., Gauchat J.F., Harrer N., Woisetschläger M., Boulay V., Fabre J.M., Demoly P., and Yssel H.. 2006. IFN-gamma-mediated inhibition of human IgE synthesis by IL-21 is associated with a polymorphism in the IL-21R gene. J. Immunol. 177:5006–5013. 10.4049/jimmunol.177.8.5006 [DOI] [PubMed] [Google Scholar]
- Renz H., and Herz U.. 2002. The bidirectional capacity of bacterial antigens to modulate allergy and asthma. Eur. Respir. J. 19:158–171. 10.1183/09031936.02.00254202 [DOI] [PubMed] [Google Scholar]
- Rickert R.C., Rajewsky K., and Roes J.. 1995. Impairment of T-cell-dependent B-cell responses and B-1 cell development in CD19-deficient mice. Nature. 376:352–355. 10.1038/376352a0 [DOI] [PubMed] [Google Scholar]
- Robinson M.J., Prout M., Mearns H., Kyle R., Camberis M., Forbes-Blom E.E., Paul W.E., Allen C.D., and Le Gros G.. 2017. IL-4 Haploinsufficiency Specifically Impairs IgE Responses against Allergens in Mice. J. Immunol. 198:1815–1822. 10.4049/jimmunol.1601434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnani S. 2004. Immunologic influences on allergy and the TH1/TH2 balance. J. Allergy Clin. Immunol. 113:395–400. 10.1016/j.jaci.2003.11.025 [DOI] [PubMed] [Google Scholar]
- Shang X.Z., Ma K.Y., Radewonuk J., Li J., Song X.Y., Griswold D.E., Emmell E., and Li L.. 2006. IgE isotype switch and IgE production are enhanced in IL-21-deficient but not IFN-gamma-deficient mice in a Th2-biased response. Cell. Immunol. 241:66–74. 10.1016/j.cellimm.2006.07.011 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M., Lin K.-I., McHeyzer-Williams L.J., Liao J., McHeyzer-Williams M.G., and Calame K.. 2003. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 19:607–620. 10.1016/S1074-7613(03)00267-X [DOI] [PubMed] [Google Scholar]
- Spolski R., and Leonard W.J.. 2014. Interleukin-21: a double-edged sword with therapeutic potential. Nat. Rev. Drug Discov. 13:379–395. 10.1038/nrd4296 [DOI] [PubMed] [Google Scholar]
- Strasser A., Whittingham S., Vaux D.L., Bath M.L., Adams J.M., Cory S., and Harris A.W.. 1991. Enforced BCL2 expression in B-lymphoid cells prolongs antibody responses and elicits autoimmune disease. Proc. Natl. Acad. Sci. USA. 88:8661–8665. 10.1073/pnas.88.19.8661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.M., Liang H.E., Bando J.K., Wu D., Cheng L.E., McKerrow J.K., Allen C.D., and Locksley R.M.. 2011. Genetic analysis of basophil function in vivo. Nat. Immunol. 12:527–535. 10.1038/ni.2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto A., Nakajima H., Hirose K., Suzuki K., Kagami S., Seto Y., Hoshimoto A., Saito Y., Foster D.C., and Iwamoto I.. 2002. Interleukin 21 prevents antigen-induced IgE production by inhibiting germ line C(epsilon) transcription of IL-4-stimulated B cells. Blood. 100:4565–4573. 10.1182/blood-2002-04-1115 [DOI] [PubMed] [Google Scholar]
- Talay O., Yan D., Brightbill H.D., Straney E.E., Zhou M., Ladi E., Lee W.P., Egen J.G., Austin C.D., Xu M., and Wu L.C.. 2012. IgE+ memory B cells and plasma cells generated through a germinal-center pathway. Nat. Immunol. 13:396–404. 10.1038/ni.2256 [DOI] [PubMed] [Google Scholar]
- Tanno L.K., Bierrenbach A.L., Simons F.E.R., Cardona V., Thong B.Y., Molinari N., Calderon M.A., Worm M., Chang Y.S., Papadopoulos N.G., et al. on behalf the Joint Allergy Academies . 2018. Critical view of anaphylaxis epidemiology: open questions and new perspectives. Allergy Asthma Clin. Immunol. 14:12 10.1186/s13223-018-0234-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong P., Granato A., Zuo T., Chaudhary N., Zuiani A., Han S.S., Donthula R., Shrestha A., Sen D., Magee J.M., et al. 2017. IgH isotype-specific B cell receptor expression influences B cell fate. Proc. Natl. Acad. Sci. USA. 114:E8411–E8420. 10.1073/pnas.1704962114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournoy K.G., Kips J.C., and Pauwels R.A.. 2000. Endogenous interleukin-10 suppresses allergen-induced airway inflammation and nonspecific airway responsiveness. Clin. Exp. Allergy. 30:775–783. 10.1046/j.1365-2222.2000.00838.x [DOI] [PubMed] [Google Scholar]
- Turqueti-Neves A., Otte M., Schwartz C., Schmitt M.E., Lindner C., Pabst O., Yu P., and Voehringer D.. 2015. The Extracellular Domains of IgG1 and T Cell-Derived IL-4/IL-13 Are Critical for the Polyclonal Memory IgE Response In Vivo. PLoS Biol. 13:e1002290 10.1371/journal.pbio.1002290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanshylla K., Opazo F., Gronke K., Wienands J., and Engels N.. 2018. The extracellular membrane-proximal domain of membrane-bound IgE restricts B cell activation by limiting B cell antigen receptor surface expression. Eur. J. Immunol. 48:441–453. 10.1002/eji.201747196 [DOI] [PubMed] [Google Scholar]
- Vijayanand P., Seumois G., Simpson L.J., Abdul-Wajid S., Baumjohann D., Panduro M., Huang X., Interlandi J., Djuretic I.M., Brown D.R., et al. 2012. Interleukin-4 production by follicular helper T cells requires the conserved Il4 enhancer hypersensitivity site V. Immunity. 36:175–187. 10.1016/j.immuni.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa C.G., Linterman M.A., Yu D., and MacLennan I.C.. 2016. Follicular Helper T Cells. Annu. Rev. Immunol. 34:335–368. 10.1146/annurev-immunol-041015-055605 [DOI] [PubMed] [Google Scholar]
- Wang Z.E., Reiner S.L., Zheng S., Dalton D.K., and Locksley R.M.. 1994. CD4+ effector cells default to the Th2 pathway in interferon gamma-deficient mice infected with Leishmania major. J. Exp. Med. 179:1367–1371. 10.1084/jem.179.4.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J.S., Herman E.I., Lainez B., Licona-Limón P., Esplugues E., Flavell R., and Craft J.. 2016. TFH cells progressively differentiate to regulate the germinal center response. Nat. Immunol. 17:1197–1205. 10.1038/ni.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood N., Bourque K., Donaldson D.D., Collins M., Vercelli D., Goldman S.J., and Kasaian M.T.. 2004. IL-21 effects on human IgE production in response to IL-4 or IL-13. Cell. Immunol. 231:133–145. 10.1016/j.cellimm.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Wu C.M., Roth T.L., Baglaenko Y., Ferri D.M., Brauer P., Zuniga-Pflucker J.C., Rosbe K.W., Wither J.E., Marson A., and Allen C.D.C.. 2018. Genetic engineering in primary human B cells with CRISPR-Cas9 ribonucleoproteins. J. Immunol. Methods. 457:33–40. 10.1016/j.jim.2018.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Sullivan B.M., and Allen C.D.. 2012. Fluorescent in vivo detection reveals that IgE(+) B cells are restrained by an intrinsic cell fate predisposition. Immunity. 36:857–872. 10.1016/j.immuni.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Yang Z., Robinson M.J., and Allen C.D.. 2014. Regulatory constraints in the generation and differentiation of IgE-expressing B cells. Curr. Opin. Immunol. 28:64–70. 10.1016/j.coi.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Robinson M.J., Chen X., Smith G.A., Taunton J., Liu W., and Allen C.D.. 2016. Regulation of B cell fate by chronic activity of the IgE B cell receptor. eLife. 5:e21238 10.7554/eLife.21238 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the reagents used for flow cytometric analysis of mouse samples.
lists the reagents used for flow cytometric analysis of human samples.