Somatic mosaicism for MAP2K1-activating mutations causes radiographical “dripping candle wax” melorheostosis. Kang et al. report somatic mosaicism for SMAD3 mutations in bone lesions of endosteal pattern melorheostosis. The SMAD3 mutations increase TGF-β signaling and stimulate osteoblast differentiation and matrix mineralization.
Abstract
Melorheostosis is a rare sclerosing dysostosis characterized by asymmetric exuberant bone formation. Recently, we reported that somatic mosaicism for MAP2K1-activating mutations causes radiographical “dripping candle wax” melorheostosis. We now report somatic SMAD3 mutations in bone lesions of four unrelated patients with endosteal pattern melorheostosis. In vitro, the SMAD3 mutations stimulated the TGF-β pathway in osteoblasts, enhanced nuclear translocation and target gene expression, and inhibited proliferation. Osteoblast differentiation and mineralization were stimulated by the SMAD3 mutation, consistent with higher mineralization in affected than in unaffected bone, but differing from MAP2K1 mutation–positive melorheostosis. Conversely, osteoblast differentiation and mineralization were inhibited when osteogenesis of affected osteoblasts was driven in the presence of BMP2. Transcriptome profiling displayed that TGF-β pathway activation and ossification-related processes were significantly influenced by the SMAD3 mutation. Co-expression clustering illuminated melorheostosis pathophysiology, including alterations in ECM organization, cell growth, and interferon signaling. These data reveal antagonism of TGF-β/SMAD3 activation by BMP signaling in SMAD3 mutation–positive endosteal melorheostosis, which may guide future therapies.
Graphical Abstract
Introduction
Melorheostosis is a rare, sporadic, sclerotic bone dysostosis with asymmetric distribution (Online Mendelian Inheritance in Man #155950). The bone lesions in this progressive hyperostotic disorder are associated with pain, functional impairment, joint contracture, and deformity. First described by Leri and Joanny (1922), the etiology of isolated sporadic melorheostosis was long elusive until recent findings of somatic mosaicism for the genes in the RAS-MEK-ERK pathway. Postzygotic mosaicism for a mutated KRAS proto-oncogene GTPase (p.Q61H) was identified in dermatoses from a boy with polyostotic melorheostosis, who also has familial osteopoikilosis (OPK; Whyte et al., 2017). The same KRAS mutation was found in the cervical lymphatic malformation and hyperpigmented skin from a girl with polyostotic melorheostosis and vascular anomalies (Seidel, V., E. Guillén, V.M. Martínez-Glez, Á.M. Lancharro Zapata, F. Ballesteros Tejerizo, V.A. Parra Blanco, A. García Martín, A. Salcedo Posadas, A., and M. Campos Domínguez. 2018. European Society of Human Genetics Conference. Abstr. P11.051C). The definitive connection of somatic mutations to bone lesions of patients with the classic form of melorheostosis, with a radiographical “dripping candle wax” appearance, involved mutations in MAP2K1. Half of 15 melorheostosis patients biopsied in our study had somatic mosaicism for activating mutations in MAP2K1 in affected, but not unaffected, bone and skin (Fratzl-Zelman et al., 2019; Jha et al., 2019; Kang et al., 2018).
Melorheostosis can also coexist with OPK (“spotted bone”) or Buschke–Ollendorff syndrome (BOS; dermatoosteopoikilosis), a benign autosomal dominant sclerosing bone dysplasia with symmetric distribution. Germline heterozygous loss-of-function mutations in LEMD3 (Hellemans et al., 2006; Hellemans et al., 2004; Mumm et al., 2007) cause OPK and BOS. LEMD3 encodes MAN1, an integral protein of the inner nuclear membrane, which interacts directly with phosphatase PPM1A and receptor-regulated SMADs (R-SMADs; SMAD2/3 or SMAD1/5) to antagonize TGF-β and bone morphogenetic protein (BMP) signaling (Bourgeois et al., 2013; Lin et al., 2006). Although somatic mutations in LEMD3 have not been reported in isolated sporadic melorheostosis (Mumm et al., 2007), as melorheostosis often coexists with OPK in the same individual or within a family with LEMD3 mutations, the TGF-β/SMAD pathway may contribute to melorheostosis pathogenesis.
TGF-β subfamily ligands transduce cellular signals through cell surface serine/threonine kinase receptors to intracellular SMADs. The TGFβRI kinase phosphorylates serine residues within the conserved SSxS motif at the carboxy-terminus of R-SMADs, SMAD2 and SMAD3. Phosphorylation of the carboxy-terminus of R-SMADs mediates oligomerization between R-SMADs and the coSMAD SMAD4. The complex translocates into the nucleus, where it functions to activate or repress expression of specific sets of TGF-β/SMAD target genes, depending on spatial and temporal tissue contexts (David and Massagué, 2018).
The TGF-β/SMAD pathway is crucial for skeletal embryonic development and postnatal homeostasis (Wu et al., 2016). Dysregulation of the TGF-β signaling pathway is associated with a spectrum of osseous defects as seen in several dominant genetic disorders. In Marfan syndrome, excessive SMAD-independent TGF-β signaling via ERK/MAPK activation induces aortic aneurysms (Holm et al., 2011), as well as tall stature and scoliosis. In Loeys-Dietz syndrome (LDS), skeletal involvement is caused by pathogenic variants of TGF-β signaling pathway components, including SMAD3 (Loeys and Dietz, 1993; Schepers et al., 2018). Activating germline mutations in the TGFB1 gene and uncontrolled activation of the TGF-β pathway cause Camurati-Engelmann disease, a progressive diaphyseal hyperostosis (Janssens et al., 2000; Kinoshita et al., 2000).
We noted distinctive features in the clinical, radiographical, and bone phenotypes between MAP2K1 mutation–positive versus –negative patients in the National Institutes of Health (NIH) cohort of 15 biopsied melorheostosis individuals (Jha et al., 2019). We hypothesized that different melorheostosis radiographical patterns may be associated with distinct molecular and cellular mechanisms caused by postzygotic mutations in different genes. In our cohort, some MAP2K1 mutation–negative patients showed an endosteal hyperostosis with asymmetric distribution. Here, we report four melorheostosis patients with an endosteal radiographical pattern whose affected, but not unaffected, bone samples contain novel somatic missense mutations causing substitution of serine 264 in the MH2 domain of SMAD3. The mutations increase SMAD3 activity and drive increased osteogenesis. Functional studies and RNA sequencing (RNA-Seq)–based transcriptome analysis implicate the TGF-β/SMAD pathway in the pathogenesis of endosteal melorheostosis and connect this form of melorheostosis to the TGF-β pathway activation that occurs with LEMD3 loss-of-function mutations causing OPK and BOS.
Results
Clinical findings
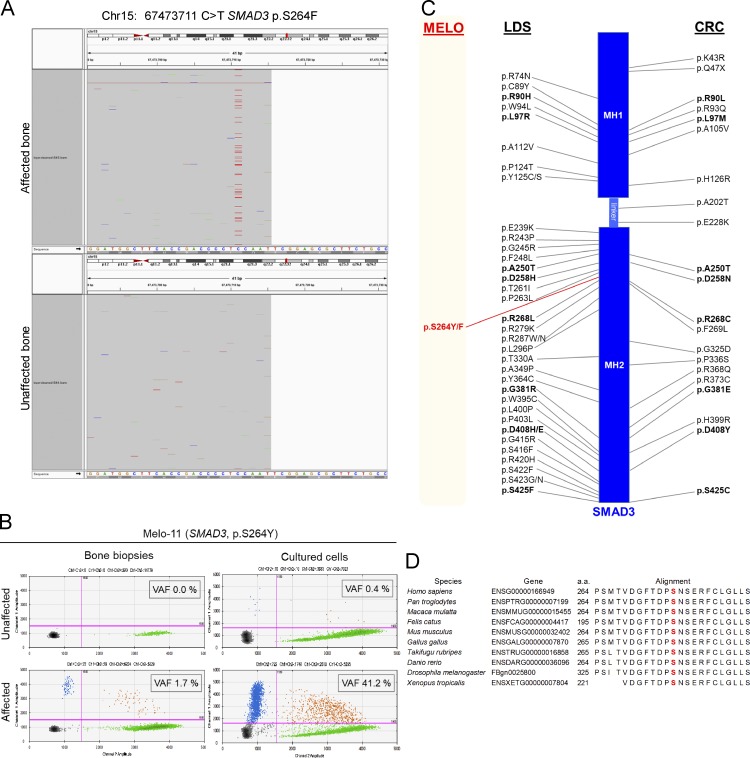
Seven patients in a cohort of 15 melorheostosis patients who underwent paired bone biopsies had an endosteal pattern of melorheostosis (Fig. 1, A–C; and Fig. S1). We are using the new term “endosteal melorheostosis” to describe the radiographical pattern seen in our patients, although a previous report used the term “osteoma-like” to categorize a similar radiographical pattern seen in 7 of 23 cases (Freyschmidt, 2001). Distinct from osteomas—which are small, have a radiolucent nidus, and tend to expand beyond the cortices—the abnormal bone tends to fill the intramedullary and intracortical space, so endosteal melorheostosis is a more descriptive term. This pattern is defined by hyperostosis that does not extend outside the bone cortical margin and has eccentric location at or in the bone. Lesions are oriented in the long axis of the involved bone and have a diameter ≥5 cm. One or more bones are involved, or if only one bone is involved, there is a circumscribed skin or subcutaneous fibrosis above the involved skeleton (Table S1; Freyschmidt, 2001). The lesions noted on radiographs corresponded to increased uptake on positron emission tomography/computed tomography using sodium fluoride tracer (Fig. 1 D), suggestive of increased bone turnover and vascularity of the lesions. This endosteal pattern of melorheostosis contrasts with the classic “dripping candle wax” radiographical appearance of melorheostosis seen in patients with MAP2K1 mutation–positive melorheostosis (Kang et al., 2018).
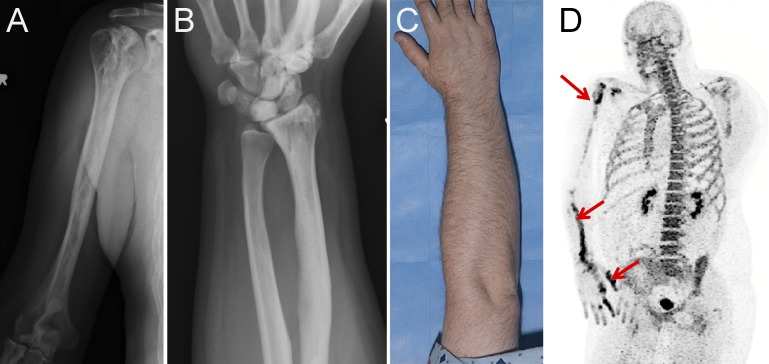
Figure 1.
Clinical findings in SMAD3 mutation–positive melorheostosis. (A and B) Radiographs of the (A) humerus and (B) forearm of patient Melo-17 with melorheostosis of the right upper extremity. Note increased density and intramedullary sclerosis extending across joints into lunate and capitate bones following an endosteal pattern without dripping candle wax or heterotopic bone. (C) Clinical appearance of patient Melo-17 with melorheostosis. Note that there is no visible thickening of the affected extremity and normal-appearing skin. The affected bone was found to harbor a SMAD3 mutation, not found in paired unaffected bone from the contralateral extremity. (D) Maximum intensity projection 18F-NaF positron emission tomography image of the skull and torso showing abnormally increased metabolic activity in the right scapula, humeral head, radius, and hand. Red arrows show areas of increased tracer uptake on NaF bone scan.
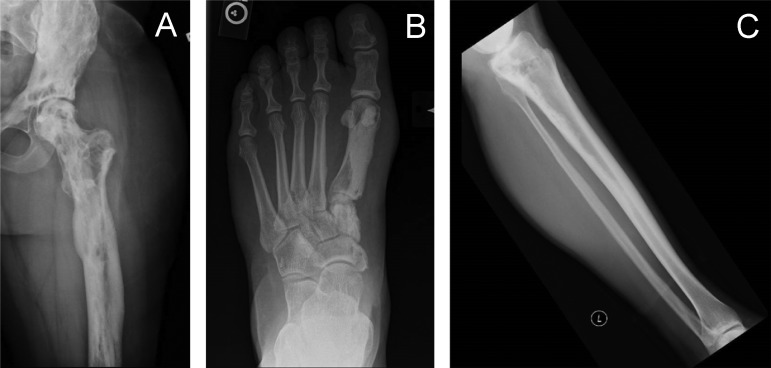
Figure S1.
Radiographs of three patients with SMAD3 mutation–positive melorheostosis demonstrating the endosteal appearance. (A) Left femur of patient Melo-12. (B) First metatarsal, left foot of patient Melo-8. (C) Left tibia of patient Melo-11.
In four of the seven patients (Table 1), somatic mutations were detected in DNA from affected bone at the codon for the same residue in SMAD3 (Fig. 2). The remaining three patients with an endosteal pattern of melorheostosis were negative for mutations in either MAP2K1 or SMAD3. Relevant clinical findings from patients with SMAD3 variants are summarized (Table S1). Disease duration ranged from 6 to 43 yr; the two patients with higher variant allele frequency (VAF) in bone tissue (Melo-12 and Melo-17) presented in childhood. No cutaneous changes were noted in any of the four patients. Melo-12 also had a history of renal artery stenosis ipsilateral to her melorheostosis skeletal lesion. Due to uncontrolled hypertension, she required revascularization and bypass at 8 yr of age. In addition, Melo-12 incurred multiple stress fractures in her foot, the majority of which occurred on the side affected with melorheostosis. There was no history of prior fractures or vascular problems in the remaining three patients.
Table 1. VAFs of SMAD3 mutations.
| Patient | Mutation | Whole exome | Amplicon | ddPCR | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Aff. bone | Unaff. bone | Aff. bone | Unaff. bone | Aff. bone | Unaff. bone | Skin overlying aff. bone | Skin overlying unaff. bone | Blood | ||
| Melo-8 | S264Y | ND | 0% | - | - | 2.30% | 0% | 0.29% | 0% | 0% |
| Melo-11 | S264Y | ND | 0% | 1.60% | 0% | 1.70% | 0% | 0.15% | 0% | 0% |
| Melo-12a | S264Y | 24% | 0% | - | - | 13.70% | 0% | 1.30% | 0% | 0% |
| Melo-17 | S264F | ND | 0% | 7.00% | 0% | 8.20% | 0% | 1.50% | 0% | 0% |
Amplicon sequencing identified two cases (Melo-11 and Melo-17). All mutations were confirmed by ddPCR and were seen in affected bone and overlying skin. Aff., affected; ND, not detectable; Unaff., unaffected.
Patient Melo-12 was identified by whole exome sequencing.
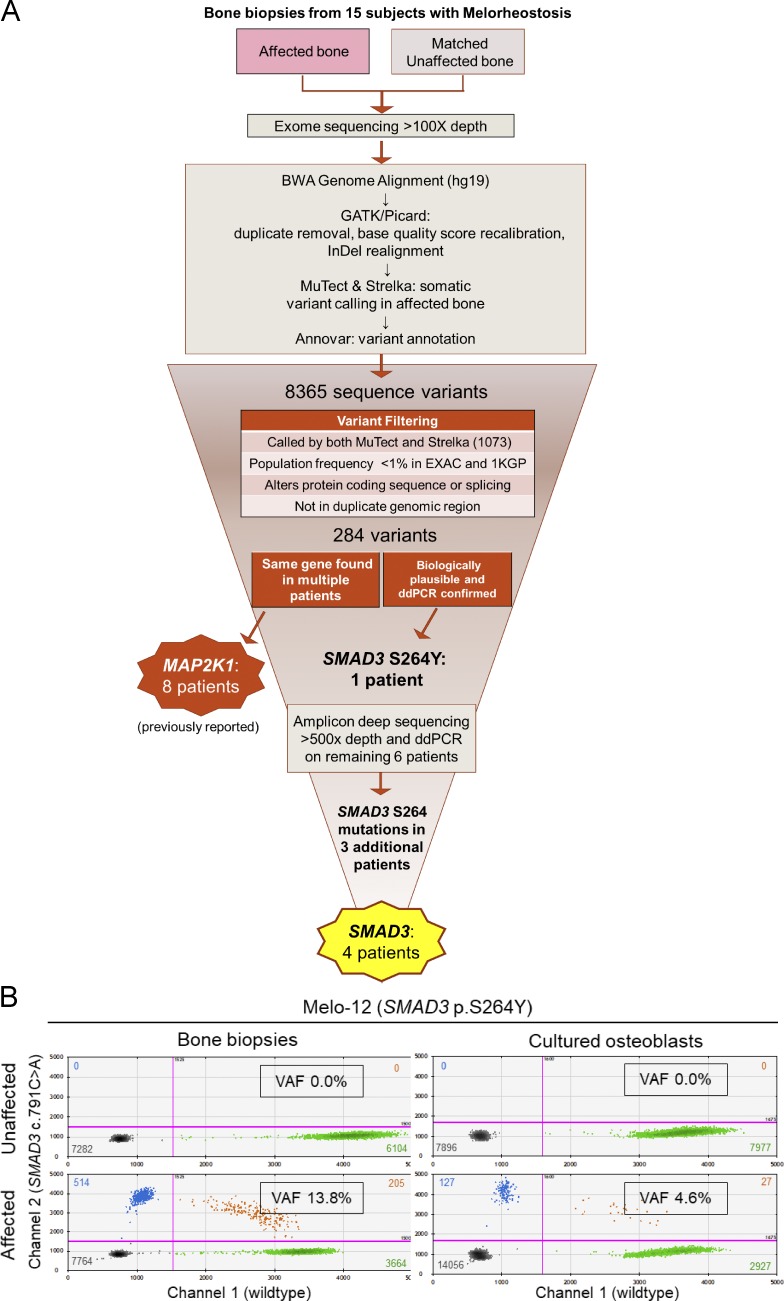
Figure 2.
Identification of SMAD3 somatic mutations in four individuals with melorheostosis. (A) Flow chart illustrating genetic analysis strategy leading to identification of SMAD3 somatic mutations in the affected bone of melorheostosis patients. (B) Quantification of mutant allele abundance by ddPCR in unaffected and affected bone biopsies (left), as well as in cultured osteoblasts (passage 1) from the respective biopsies (right), from patient Melo-12 identified by WES to have a SMAD3 p.S264Y mutation in affected bone. Each dot in the 2D fluorescence amplitude plots represents a droplet, with blue being mutant positive, green being wild-type positive, orange being positive for both, and black being negative for both. Color-matched numbers correspond to count of droplets per quadrant. The boxed number (VAF) is the calculated fractional abundance of mutant allele in each sample. Magenta lines are thresholds of negative vs. positive populations.
As germline mutations in SMAD3 have been described in LDS, characterized by vascular (tortuosity, aneurysms, and/or dissections), skeletal (pectus excavatum, scoliosis, or joint laxity), craniofacial (widely spaced eyes, strabismus, and cleft palate), and cutaneous findings (velvety and translucent skin, easy bruising, and dystrophic scars; Loeys and Dietz, 1993), the four patients were evaluated for features of LDS. No significant LDS findings were noted on physical exam or on computed tomography of the chest and abdomen with contrast (Table S1). Echocardiograms revealed normal aortic root and ascending aorta dimensions.
Biochemical markers of bone turnover were normal, except one patient (Melo-11) had decreased urinary N-telopeptide, a marker of bone resorption, and osteopenia. Melo-12 had osteopenia with uncertain etiology.
Identification of SMAD3 somatic mutations in melorheostosis
After applying the filtering strategy shown in Fig. 2 A, 284 potential somatic mutations were present in affected, but not unaffected, bone in 15 patients with melorheostosis. Among them was a novel SMAD3 p.S264Y (c.791C>A) variant at 24% VAF in patient Melo-12. While this variant was present in only one patient, the SMAD3 gene was prioritized based on the relatively high VAF and biological relevance of the TGF-β pathway to bone disease.
After validation of the initial SMAD3 p.S264Y variant by droplet digital PCR (ddPCR), we screened several MAP2K1 mutation–negative patients for this mutation by ddPCR and identified a second individual (Melo-8) with a SMAD3 p.S264Y mutation at 2.3% VAF. Prompted by these findings, we developed a custom amplicon-based targeted sequencing assay, which included all exons of SMAD3 as well as other genes potentially related to melorheostosis (KRAS, MAP2K1, MAP2K2, and MAP3K3). We sequenced the remaining MAP2K1 mutation–negative patients and identified two more patients (Melo-11 and Melo-17) with SMAD3 variants at the same residue: one, p.S264Y (c.791C>A), at 1.7% and one, p.S264F (c.791C>T), at 8.2% VAF (Fig. 2 B and Fig. S2, A and B). Both variants were confirmed by ddPCR (Table 1). Although skin overlying affected bone appeared normal in our patients, mosaicism for the SMAD3 mutation was also found in skin overlying affected bone in all four patients, at much lower allele frequencies than in bone (skin VAF ≈10% of bone VAF), but not in skin from the contralateral extremity. DNA from peripheral blood leukocytes tested negative for these mutations in all four patients (Table 1).
Figure S2.
Identification of SMAD3 somatic mutations in melorheostosis. (A) Targeted sequencing of Melo-17 (SMAD3 p.S264F). (B) Quantification of mutant allele abundance by ddPCR. Genomic DNAs isolated from bone biopsies and cultured cells from Melo-11 (SMAD3 p.S264Y) were subjected to ddPCR, and VAF was determined. Each dot represents a droplet, with blue being mutant positive, green being wild-type positive, orange being positive for both, and black being negative for both. Magenta lines are thresholds of negative vs. positive populations. (C) Schematic representation of human SMAD3 protein structure. Boxes represent MH1 domain at the N-terminus and MH2 domain at the C-terminus, connected by linker region of SMAD3 protein. On each side of the schematic are the previously reported SMAD3 missense mutations in LDS and CRC. Comprehensive lists of the SMAD3 mutations can be found in Schepers et al. (2018) and Fleming et al. (2013). The SMAD3 mutations identified in this melorheostosis study (MELO) are marked in red. Positions of the mutations are not to scale. (D) SMAD3 protein sequence alignment among the indicated species (http://www.mutationtaster.org) around the sites of the missense mutations (p.S264Y or p.S264F; highlighted in red).
The SMAD3 serine264 residue is highly conserved across species (van de Laar et al., 2011). Both SMAD3 mutations (p.S264Y and p.S264F) are absent from population databases such as gnomAD and 1000G. Pathogenicity prediction software tools (SIFT, PolyPhen2, MutationTaster, MutationAssessor, and CADD [Combined Annotation Dependent Depletion]) assigned both mutations as damaging or disease-causing. The CADD scores for the two mutations are 32 for S264Y and 29.7 for S264F.
Increased matrix mineralization and distinct histomorphometry in melorheostotic bone
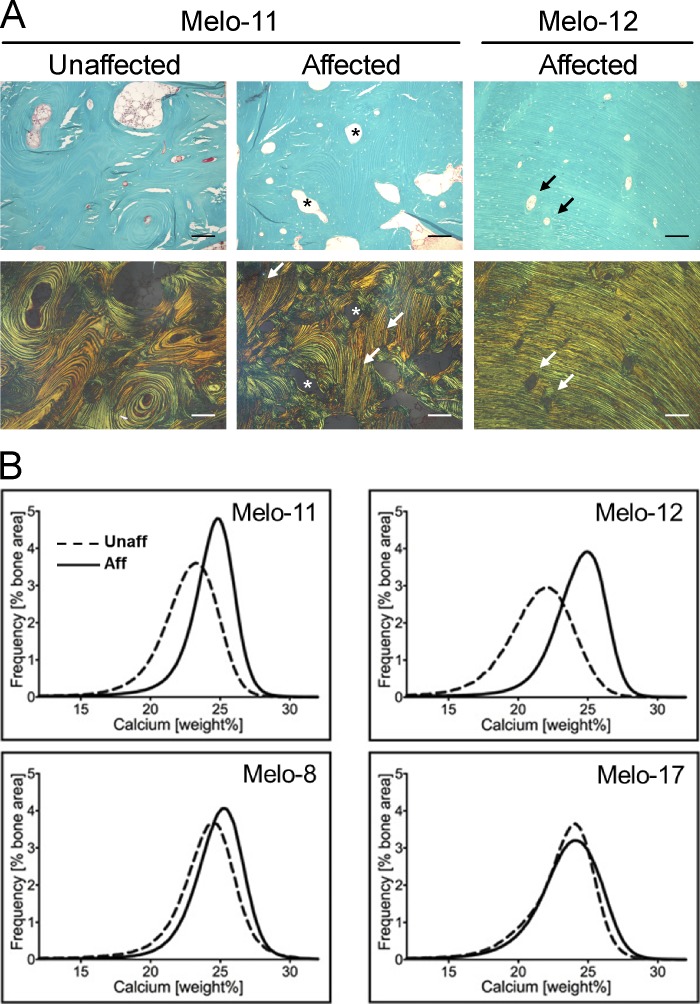
Histological evaluation revealed that the bone biopsy samples from melorheostotic lesions consisted of a mixture of osteonal-like remodeled bone, with concentric lamellae around vascular channels, and areas of aligned parallel lamellar bone (Fig. 3 A). Affected bone from Melo-12 showed mostly multiple layers of long parallel lamellae, indicating intense periosteal bone apposition associated with the endosteal radiographical pattern of melorheostosis. As in unaffected bone, osteonal bone surfaces in affected bone were characterized by low osteoid formation, sparse eroded surfaces, and absence of osteoblasts (Fig. 3 A and Table S2). Unaffected bone samples contained mostly trabecular bone (Table S2). Histomorphometric indices of bone formation and bone resorption were not significantly altered between affected and unaffected bone (Table S2). These results contrast markedly with our previous findings in melorheostotic lesions caused by somatic-activating mutations in MAP2K1 that showed intense bone remodeling and increased osteoid (Jha et al., 2019; Kang et al., 2018). In comparison, affected bone from patients with the SMAD3 mutations has significantly lower osteoid thickness (−62%, P = 0.003), osteoid surface per bone surface (−92%, P < 0.0001), and eroded surface per bone surface (−84%, P = 0.0016) than affected bone from patients with MAP2K1 mutations. Active bone-forming osteoblasts were not viewed in the selected areas; osteoclast indices are not statistically different between these groups.
Figure 3.
Histology of melorheostotic bones. (A) Light microscopy images of cortical bone from patients Melo-11 and Melo-12 (upper row). Goldner’s trichrome staining is viewed under polarized light (lower row). Green is mineralized bone. Note the vascular channels with concentric arranged bone lamellae (asterisks) in Melo-11 unaffected and affected bone. The arrows point toward parallel lamellar bone, which is only viewed in affected bone. In Melo-11, there is a mixture of concentric lamellar structure and parallel lamellar structure. In contrast, in Melo-12, the bone lamellae are perfectly parallel-oriented. Note that the vascular channels are rather small. Scale bars = 150 µm. (B) BMDD curves obtained by qBEI. The BMDD curves from affected bone from Melo-11 and Melo-12 shifted toward a higher degree of mineralization compared with unaffected bone from the same patient. In Melo-8 and Melo-17, the degree of mineralization in affected and unaffected bone is similar. Aff, affected; Unaff, unaffected.
Bone mineralization density distribution (BMDD) of biopsy samples from SMAD3 mutation–positive melorheostosis patients are shown in Fig. 3 B. Compared with unaffected bone, the BMDD curves from affected bone shifted toward higher mineral content in Melo-11 and Melo-12, while that of Melo-8 and Melo-17 were within normal limits. Statistical analysis supported an overall increase in mineralization of affected bone with the SMAD3 mutations, compared with both paired unaffected biopsies and MAP2K1 mutation–positive affected bone (Table S2). Of the BMDD variables, only CaHigh was significantly altered in the set of biopsies with SMAD3 mutations. CaHigh was nearly doubled in SMAD3 mutation–positive melorheostotic lesions compared with unaffected bone (P = 0.0240). There was also a strong trend to higher average mineral content in affected bone (CaMean: +6.7%, P = 0.0596).
Compared with MAP2K1 mutation–positive affected bone, lesions with SMAD3 mutations were more highly mineralized, with CaMean +7.8%, P = 0.002 and CaPeak +4.0%, P = 0.0048. CaHigh values were very variable in both groups; on average, however, the value was doubled in the SMAD3 mutation–positive group (P = 0.0005). In contrast, CaLow was markedly reduced (−65%, P = 0.0012) compared with MAP2K1 mutation–positive affected bone (+89%, P = 0.009; Fratzl-Zelman et al., 2019).
Increased TGF-β signaling by the novel gain-of-function mutation in SMAD3
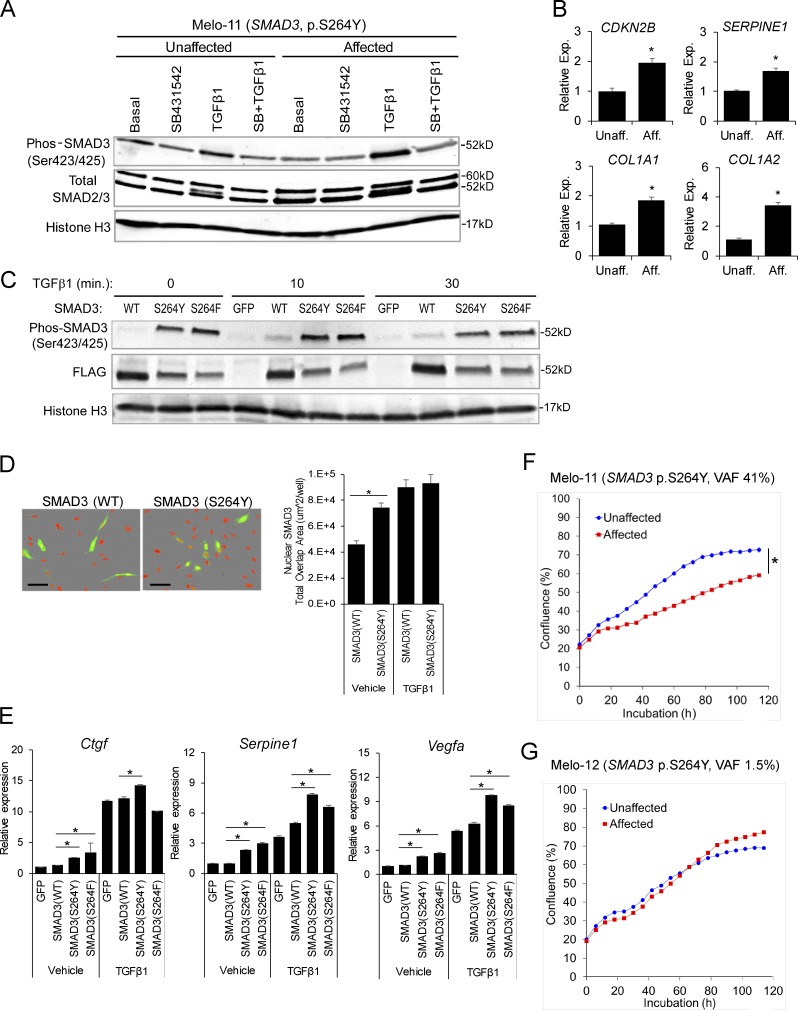
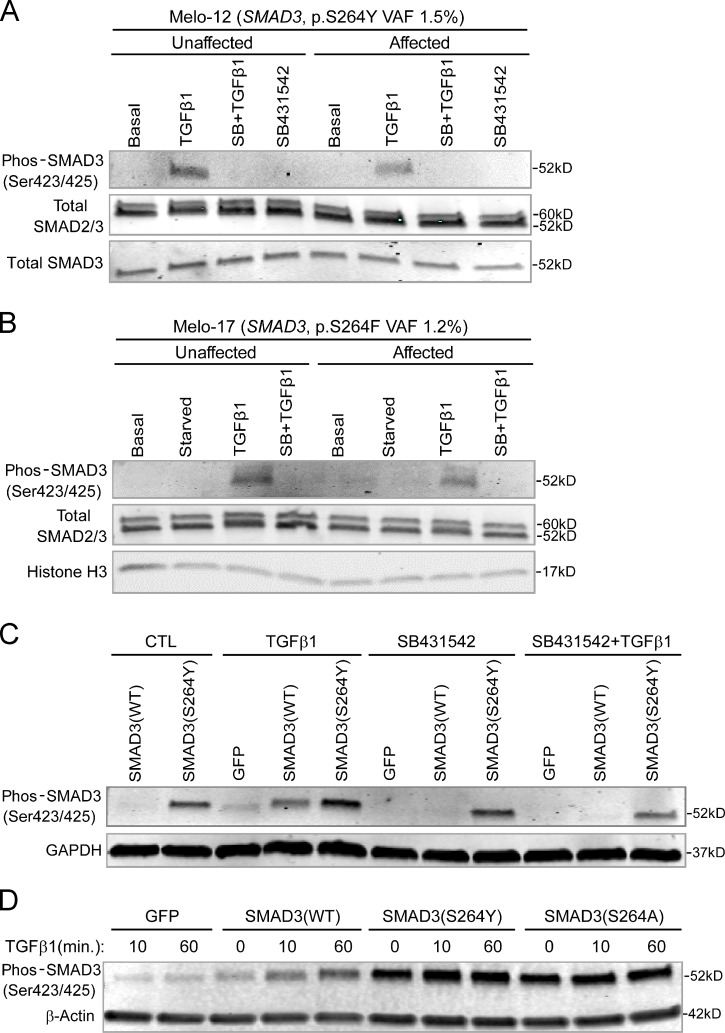
The identified SMAD3 missense mutations occurred in exon 6 of the gene, substituting serine at residue 264 with either tyrosine (p.S264Y) in three patients or phenylalanine (p.S264F) in one patient. To investigate the biological consequences of the novel SMAD3 mutations, primary osteoblasts isolated from affected and unaffected bone tissues of the same individual were subjected to Western blot analysis for SMAD3 activation upon TGF-β stimulation. Cells from affected bone showed higher SMAD3 carboxy-terminal SSxS motif phosphorylation relative to their unaffected counterparts (Fig. 4 A). This was detectable in affected cell cultures containing relatively high VAF (41%). In affected cells with relatively low VAF (∼1–2%), there were marginal differences between affected and unaffected cells (Fig. S3, A and B).
Figure 4.
Endosteal melorheostosis caused by gain-of-function SMAD3 mutations. (A) SMAD3 phosphorylation upon TGF-β stimulation with or without TGFβRI inhibitor (SB431542) pretreatment was assessed by Western blotting (Melo-11, SMAD3 p.S264Y). Total SMAD2/3 and histone H3 were internal controls. After quantification and normalization against histone H3, band intensities of phos-SMAD3 relative to basal control (lane 1) were presented in the table below the images. (B) TGF-β pathway target gene expression examined by real-time qPCR (Melo-17, SMAD3 p.S264F; n = 3; *, P < 0.05; Student's t test). (C) Mutant SMAD3 (p.S264Y and p.S264F) and wild-type SMAD3 (all Flag-tagged) were overexpressed in MC3T3-E1, and SMAD3 phosphorylation was examined by Western blotting. Histone H3 was used as internal control. After quantification and normalization against histone H3, band intensities of phos-SMAD3 and FLAG-SMAD3 relative to basal control (lane 1) were presented in the table below the images. (D) SMAD3 p.S264Y and wild-type SMAD3 were overexpressed in MC3T3-E1, and nuclear localization was detected by immunofluorescence and quantitated. Scale bars = 100 µm (n = 4; *, P < 0.05; two-way ANOVA). (E) SMAD3 mutants (p.S264Y and p.S264F) and wild-type SMAD3 were overexpressed in MC3T3-E1, and transcriptional response was analyzed by real-time qPCR (n = 3; *, P < 0.05; Student's t test). Error bars: SD of replicates. (F) Primary cells isolated from affected and unaffected bone of melorheostosis patient Melo-11 (SMAD3 p.S264Y, VAF 41%) were subjected to live-cell imaging to measure cell proliferation rate (n = 8; *, P < 0.05; two-way ANOVA). (G) Cell proliferation rate between affected and unaffected cells from a melorheostosis patient with relatively low VAF (Melo-12, SMAD3 p.S264Y, VAF 1.5%) was comparable (n = 8; n.s., not significant; two-way ANOVA). Presented are representative data from at least three independent experiments. Aff, affected; Exp, expression; Unaff, unaffected.
Figure S3.
Effects of SMAD3 mutations on TGF-β signaling. (A and B) Effect of the mutant SMAD3 with low VAF on TGF-β signaling activation. SMAD3 phosphorylation in affected cells of melorheostotic bone in response to TGF-β stimulation was comparable to that in unaffected cells. (A) Melo-12 (SMAD3 p.S264Y, VAF 1.5%). (B) Melo-17 (SMAD3 p.S264F, VAF 1.2%). Total SMAD2/3 and Histone H3 were probed as internal controls. (C) SMAD3 p.S264Y resistant to TGF-β receptor kinase inhibitor. SMAD3 p.S264Y and wild-type SMAD3 were overexpressed in MC3T3-E1 cells. Cells were stimulated with recombinant TGF-β1 with or without pretreatment of TGF-β I receptor inhibitor, SB431542. Western blotting showed the increased SMAD3 phosphorylation in SMAD3 (p.S264Y)–expressing cells compared with wild-type SMAD3–expressing cells. SB431542 significantly decreased phos-SMAD3 in wild-type SMAD3, but not in SMAD3 p.S264Y. GAPDH was used as internal control. (D) Increased SMAD3 phosphorylation preserved in SMAD3 p.S264A. The SMAD3 mutants (p.S264Y or p.S264A) and wild-type SMAD3 were overexpressed in MC3T3-E1 cells. Cells were stimulated with recombinant TGF-β1 for the indicated time. Western blotting showed increased SMAD3 phosphorylation in wild-type SMAD3 over time in response to TGF-β1 stimulation. However, SMAD3 p.S264Y– and SMAD3 p.S264A–expressing cells displayed constitutive activation of SMAD3, even in the absence of TGF-β1 stimulation. β-actin was used as internal control. Data are representative of at least two independent experiments.
To further examine the effect of the mosaic SMAD3 mutation on the canonical TGF-β/SMAD pathway, cells were stimulated with TGF-β and target gene expression was examined by real-time quantitative PCR (qPCR). In agreement with the increased level of SSxS motif phosphorylated (activated) SMAD3, expression of TGF-β/SMAD pathway target genes, such as CDKN2B, SERPINE1, COL1A1, and COL1A2, was higher in affected than in unaffected cells (Fig. 4 B). These results indicate that the SMAD3 mutation is activating for the TGF-β/SMAD pathway.
Expression of SMAD3 (p.S264Y) and SMAD3 (p.S264F) in osteoblastic cells MC3T3-E1 reveals constitutive activity
Functional activity of the SMAD3 mutations (p.S264Y and p.S264F) was further investigated by transfecting plasmid vectors encoding wild-type and mutant SMAD3 into MC3T3-E1 (Fig. 4, C–E). In the absence of TGF-β stimulation, the basal level of SMAD3 phosphorylation was significantly higher in SMAD3 S264 mutants than wild-type SMAD3. Both SMAD3 mutants showed markedly higher levels of carboxy-terminal SSxS motif phosphorylation than wild-type SMAD3 upon TGF-β stimulation (Fig. 4 C). Under these conditions, the increased phosphorylation of SMAD3 S264 mutants appears to be constitutive, as it did not respond to further stimulation by TGF-β and was resistant to inhibition of the kinase activity of TGF-β receptor (Fig. S3 C). The hydroxyl group of tyrosine residues may be subject to post-translational modification, such as phosphorylation or glycosylation. However, substitution of the Tyr264 residue by alanine (p.S264Y to p.S264A) did not abolish the increased SMAD3 phosphorylation (Fig. S3 D), implying that loss of serine at residue 264 is critical to SMAD3 activation.
We noted that when SMAD3 was overexpressed, SMAD3 mutant had lower total SMAD3 than wild-type SMAD3 on Western blots, indicative of a higher rate of SMAD3 turnover or lower stability of mutant than wild-type SMAD3 (Fig. 4 C). Since activated SMAD3 undergoes rapid turnover by proteasome-mediated degradation as a part of the mechanism that regulates duration of TGF-β/SMAD signaling (Massagué, 2000), this indicates that levels of phosphorylated SMAD3 are increased in overexpressed mutants even in the context of lower total SMAD3.
SMAD3 p.S264Y increases nuclear translocation and TGF-β pathway target gene expression
Nuclear SMAD complexes (e.g., SMAD2/3/4) partner with transcription coactivators or corepressors to execute their TGF-β signaling responses (David and Massagué, 2018). Due to the mosaic nature of the SMAD3 mutations in cells of melorheostotic bones, we relied on a plasmid vector–based transfection system to assess nuclear translocation of the SMAD3 mutants. SMAD3 p.S264Y was expressed in MC3T3-E1, and nuclear translocation of SMAD3 was assessed by immunofluorescence. Consistent with its increased phosphorylation, SMAD3 p.S264Y displayed increased nuclear localization compared with wild-type SMAD3, even without TGF-β stimulation (Fig. 4 D). Further increase in nuclear translocation upon TGF-β stimulation appears to represent activated endogenous SMAD3.
The increased phosphorylation and nuclear translocation of mutant SMAD3 resulted in the expected increase in transcription of TGF-β/SMAD pathway target genes Ctgf, Serpine1, and Vegfa (Fig. 4 E). Up-regulation of target gene expression in SMAD3 mutant cells was significant, even without TGF-β stimulation. TGF-β stimulation further increased the target gene expression, but these enhanced levels likely represent the response of the endogenous wild-type SMAD3 rather than of plasmid vector–expressed SMAD3, since the extent of up-regulation was similar between wild-type and mutant SMAD3. These results suggest that the SMAD3 mutations (p.S264Y and p.S264F) are gain-of-function mutations augmenting signaling of the TGF-β/SMAD pathway.
Mosaic gain-of-function mutation in SMAD3 inhibits cell growth
SMAD3 plays a critical role in TGF-β–mediated regulation of cell growth and osteoblast differentiation (Sowa et al., 2002). Temporal cell cycle arrest in the G1 phase is a prerequisite for cell differentiation (Kaji et al., 2006). Overexpression of SMAD3 has both a cytostatic effect and a stimulatory effect on bone matrix protein expression (Kaji et al., 2006). To examine the effect of the mosaic-activating SMAD3 mutations on cell growth, the cell proliferation rate of osteoblasts isolated from affected and unaffected bone of a melorheostosis patient was assessed by live-cell imaging. Osteoblast cultures from affected bone with relatively high VAF (37%) of SMAD3 p.S246Y displayed a significantly decreased cell growth rate compared with unaffected cells (Fig. 4 F). Proliferation of affected cells with very low VAF (1–2%) of SMAD3 p.S264Y was comparable to that of unaffected cells (Fig. 4 G).
Melorheostosis-causing SMAD3 mutation promotes osteogenic differentiation and extracellular matrix (ECM) mineralization
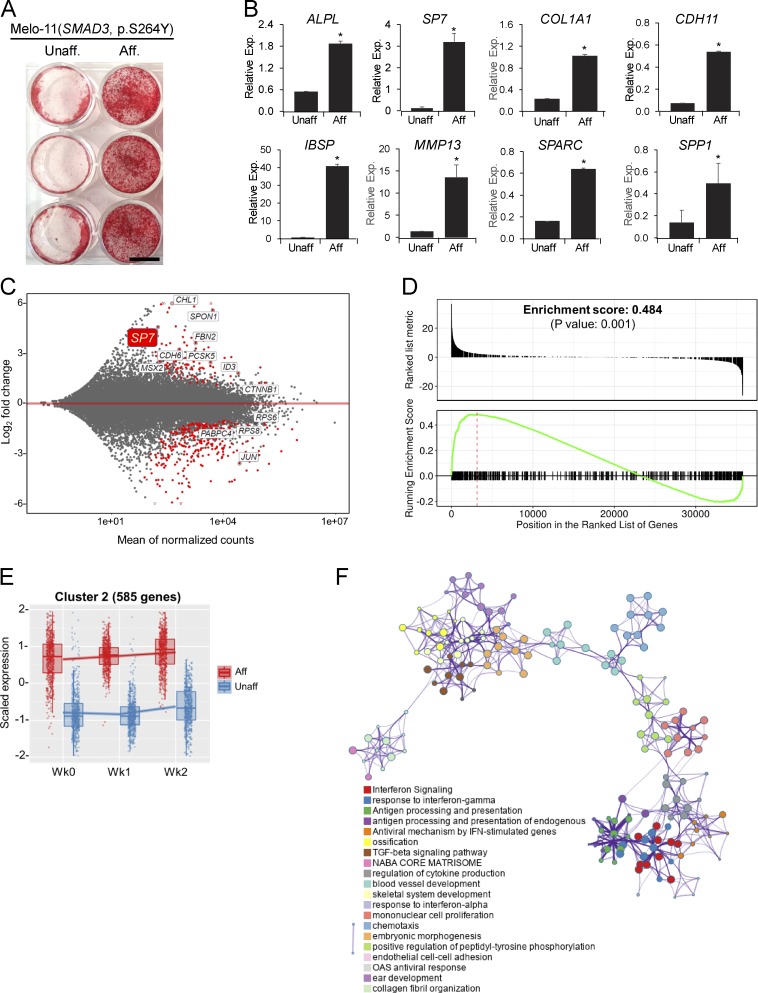
To examine the impact of the mosaic SMAD3 mutation on osteogenesis in vitro, cells from affected and unaffected bone tissue of a melorheostosis patient (Melo-11, SMAD3 p.S264Y, VAF 41% at passage 3) were cultured in conventional osteogenic media. Osteogenic differentiation and ECM mineralization were significantly increased in affected compared with unaffected cells (Fig. 5 A). A concomitant increase in osteogenesis was confirmed by significantly higher expression of osteogenic marker genes in cells from affected bone (Fig. 5 B).
Figure 5.
Osteogenesis stimulation by SMAD3 gain-of-function mutation in affected cells. (A) Alizarin Red staining of ECM mineralization in osteoblasts (Melo-11, SMAD3 p.S264Y). After 6 wk in osteogenic media containing L-ascorbate, β-glycerophosphate, and dexamethasone, significantly increased mineralization was observed in affected compared with unaffected cells. Scale bar = 10 mm. (B) Real-time qPCR of osteogenic marker expression in osteoblasts (n = 3; *, P < 0.05; Student's t test). Error bars indicate the SD of replicates. (C) MA plot of DEGs from RNA-Seq (Melo-11, SMAD3 p.S264Y). Red dots represent significantly up- or down-regulated DEGs (FDR < 1e-20). (D) GSEA plot for skeletal genes, with black bars indicating genes ranked by log2-fold change. (E) Cluster 2 from degPatterns analysis of RNA-Seq data. Genes in cluster 2 showed relatively higher expression in affected cells (red dots) than in unaffected cells (blue dots) throughout the time points (week 0, week 1, and week 2). (F) Functional and pathway enrichment analysis of cluster 2 (585 genes) with Metascape. Enriched pathway terms depicted as an interaction network and listed according to their P values (<0.01). Circle nodes represent enriched functions for an annotated ontology term, and the circle size indicates the number of genes that fall into that term. Presented are representative data from at least two independent experiments. Aff, affected; Exp, expression; Unaff, unaffected.
To further illuminate cellular pathways and biological processes influenced by the mosaic SMAD3 mutation, transcriptome profiles were acquired by RNA-Seq on cells harvested at 1-wk intervals during osteogenic differentiation in culture. Totals of 261, 172, and 250 genes were found to be differentially expressed with more than twofold changes (false discovery rate [FDR] < 1e-20) at 0-, 1-, and 2-wk time points, respectively (Fig. S4 A). Osteogenic markers and transcription factors, represented by SP7 (Osterix), and other skeletal-related genes (Ho et al., 2000) were among the up-regulated genes in affected cells (Fig. 5, C and D). Examination of the differentially expressed genes (DEGs) by Ingenuity Pathway Analysis identified β-catenin (CTNNB1), the signal transducer of the canonical Wnt pathway, as one of the top upstream regulators (KMT2A, POLG, KAT6A, CTNNB1, and HOXB3) of the DEGs, further accounting for the increased osteogenesis in affected cells (Table S3).
Figure S4.
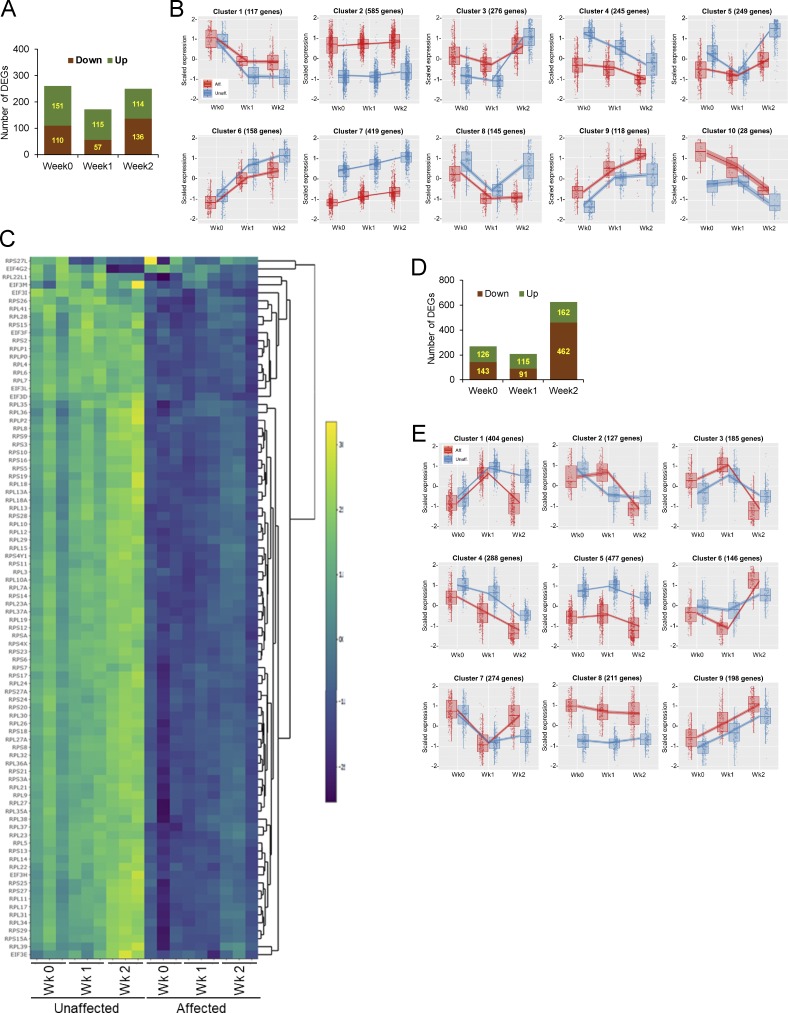
Transcriptome profiling by RNA-Seq. (A–C) BMP2(−) condition. (A) A bar graph shows total number of DEGs with more than twofold changes (FDR < 1e-20) at 0-, 1-, and 2-wk time points in conventional osteogenic media without recombinant BMP2. (B) Clusters from degPatterns analysis of RNA-Seq data. The number of member genes in each cluster is also shown. (C) Cell growth inhibition by SMAD3 gain-of-function mutation. Heatmap of gene sets in eIF2 pathway displays significant down-regulation of ribosomal protein components (RPS [ribosomal protein S] and RPL (ribosomal protein L]) in affected cells. (D and E) BMP2(+) condition. (D) A bar graph shows total number of DEGs with more than twofold changes (FDR < 1e-20) at 0-, 1-, and 2-wk time points in conventional osteogenic media supplemented with recombinant BMP2. (E) 9 clusters from degPatterns analysis of RNA-Seq data. The number of member genes in each cluster is also shown. Aff, affected; Unaff, unaffected.
Since genes sharing similar expression patterns during osteogenic differentiation may also be involved in similar cellular processes, coexpression clustering of the DEGs identified 10 clusters, each of which displayed distinct expression trends over the time points (Fig. S4 B and Data S1). To gain insight into putative cellular pathways that were changed by the mosaic SMAD3 mutation, gene ontology and pathway enrichment analyses were performed on the DEGs residing in each coexpression cluster (Data S1 and Data S3). Further exhibiting functionality of the activating SMAD3 mutation, the TGF-β pathway was enriched in cluster 2, with gene transcript levels increased at baseline and subsequently (Fig. 5 E). Up-regulation of the TGF-β pathway is not attributable to LEMD3 (MAN1) action, as our RNA-Seq data indicated that transcript levels of LEMD3 and PPM1A, whose protein products form a complex to dephosphorylate R-SMADs and terminate TGF-β signaling, remained steady during osteoblast differentiation and were not altered by the SMAD3 mutations. Intriguingly, the interferon signaling pathway and cytokine signaling in the immune system were also enriched in cluster 2, with significantly higher expression in affected than in unaffected cells. The set of 21 genes whose expression was previously validated in systemic lupus erythematosus for interferon-inducible gene signature was found to have high- to medium-fold increase in affected cells of Melo-11 (Table S4; Yao et al., 2009). The genes in cluster 3, with increased transcripts in affected cells at baseline and week 1, were associated with adipogenesis and inflammatory response, in addition to ossification (Fig. S4 B and Data S3). Enrichment of genes for ECM organization was found in clusters 4, 5, 6, and 8, in which expression is down-regulated in affected cells. Genes related to translation and ribosome biogenesis were over-represented in cluster 7. Given that ribosomal proteins have a significant role in ribosome biogenesis, protein synthesis, and cell growth (Zimmermann, 2003), down-regulation of ribosomal protein subunit genes contributes to the reduced cell growth in affected cells (Fig. S4 C and Data S3).
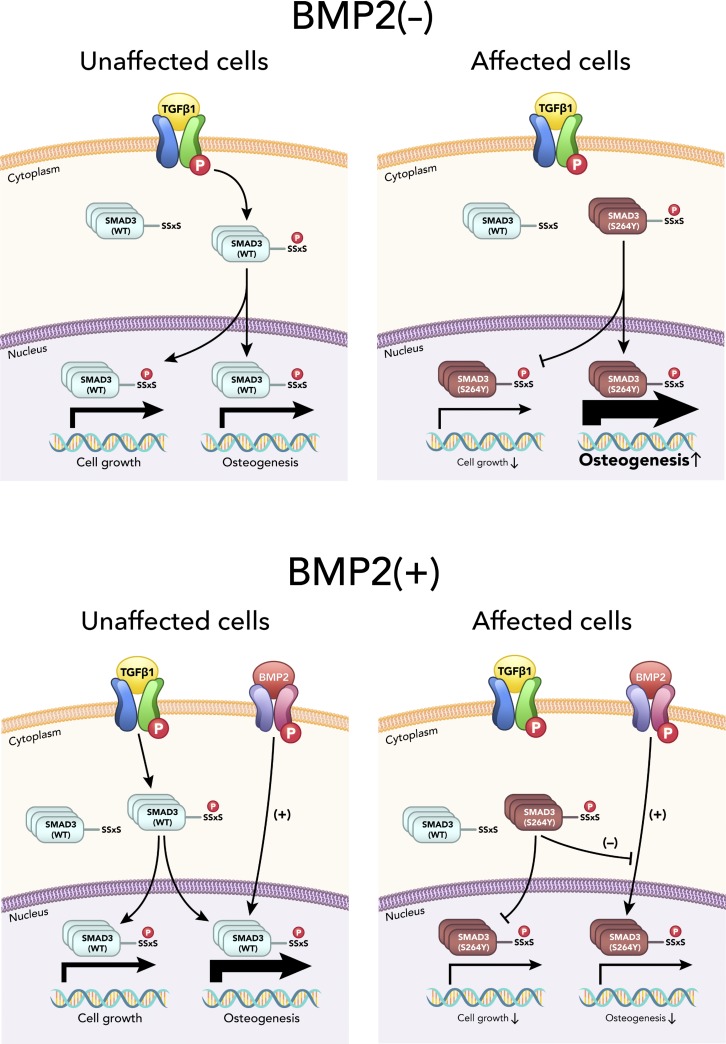
Constitutively active SMAD3 mutant dampens pro-osteogenic property of BMP2
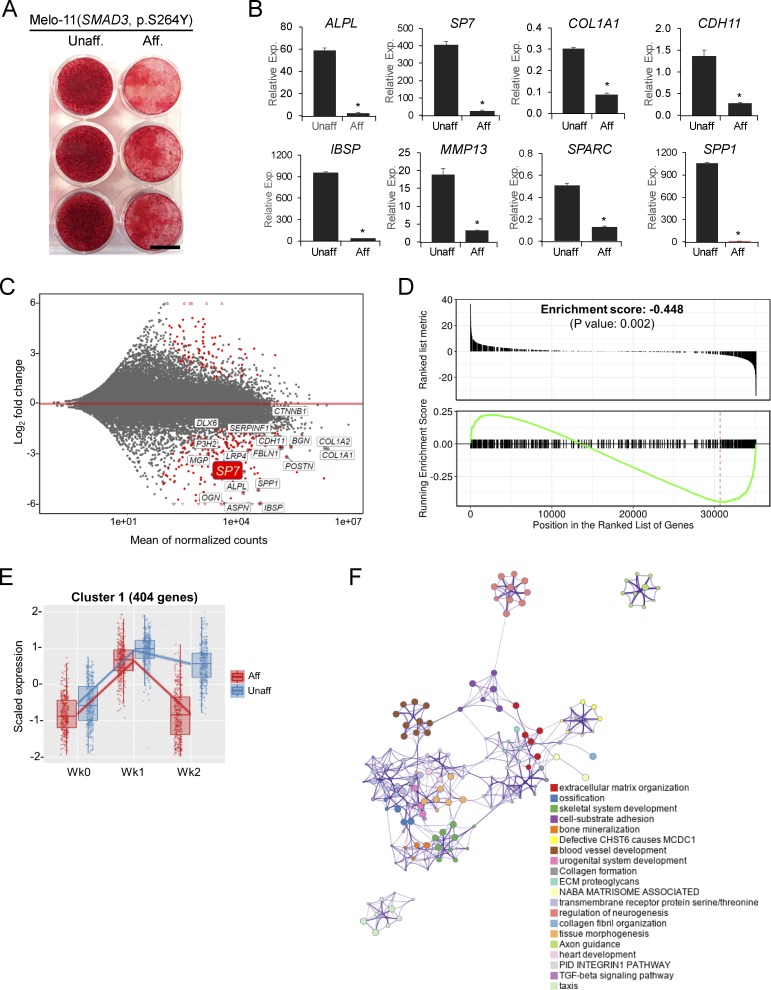
Surprisingly, BMP2 supplementation in osteogenic culture media strikingly dampened the mutant SMAD3’s stimulatory effect on osteogenesis (Fig. 6 A). Concordantly, osteogenic marker genes elevated in untreated affected cells (Fig. 5 B) were significantly down-regulated with BMP2-treated compared with unaffected cells (Fig. 6 B). These results suggest that activated SMAD3 inhibited cellular signaling events activated by BMP2—including those associated with osteogenic differentiation and ECM mineralization.
Figure 6.
BMP2-stimulated osteogenesis dampened by SMAD3-activating mutation. (A) Alizarin Red staining of mineralization in osteoblasts (Melo-11, SMAD3 p.S264Y). After 6 wk in osteogenic media (L-ascorbate, β-glycerophosphate, and dexamethasone) supplemented with BMP2, significantly decreased mineralization was observed in affected compared with unaffected cells. Scale bar = 10 mm. (B) Real-time qPCR of osteogenic marker expression in osteoblasts undergoing differentiation in the presence of BMP2 (n = 3; *, P < 0.05, Student's t test). Error bars indicate the SD of replicates. (C) MA plot of DEGs from RNA-Seq (Melo-11, SMAD3 p.S264Y). Genes in red dots are significantly up- or down-regulated (FDR < 1e-20). (D) GSEA plot for skeletal genes, with black bars indicating genes ranked by log2-fold change. (E) Cluster 1 from degPatterns analysis of RNA-Seq data (affected shown in red dots and unaffected in blue dots; time points: week 0, week 1, and week 2). (F) Functional and pathway enrichment analysis of cluster 1 (404 genes). Enriched pathway terms depicted as an interaction network and listed according to their P values (<0.01). Circle nodes represent enriched functions for an annotated ontology term, and the circle size indicates the number of genes that fall into that term. Presented are representative data from at least two independent experiments. Aff, affected; Exp, expression; Unaff, unaffected.
Transcriptome profiling by RNA-Seq on Melo-11’s affected and unaffected cells, cultured in the presence of BMP2 and compared pairwise at each time point, identified 269, 206, and 624 DEGs with more than twofold changes at 0, 1, and 2 wk, respectively (FDR < 1e-20; Fig. S4 D). Supporting the antagonistic effect of BMP2, osteogenic marker genes, as exemplified by SP7 in MA plot, were significantly down-regulated in affected cells in the presence of BMP2 (Fig. 6, C and D).
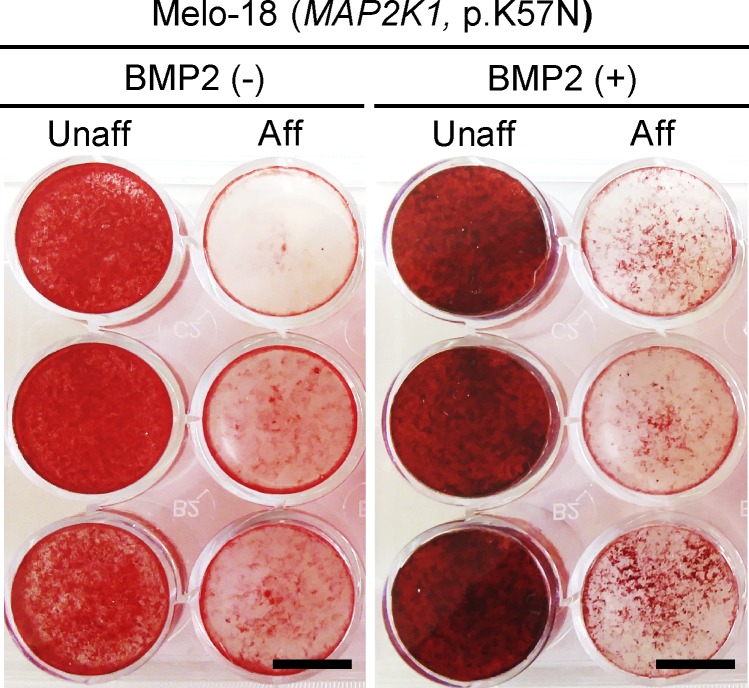
Analysis of coexpressed genes identified nine clusters in the BMP2(+) condition (Data S2 and Data S4). Gene ontology analysis and pathway enrichment of each cluster revealed that genes in cluster 1 were associated with the TGF-β pathway, ECM organization, and ossification (Data S2 and Data S4). In agreement with the inhibitory effect of the SMAD3 mutation on osteogenic differentiation in the presence of BMP2, the transcriptional response of genes in cluster 1 was noticeably down-regulated at week 2 (Fig. 6 E). Genes in cluster 2 and cluster 8, with up-regulation at week 1 or at all time points, respectively, were implicated in interferon signaling and immune response (Fig. S4 E and Data S4). Genes for ribosome biogenesis and protein translation were over-represented in cluster 5, containing genes down-regulated in affected cells at baseline and subsequently. Interestingly, the transcriptional response of genes for interferon response (cluster 2, both BMP2[−] and BMP2[+]) and ribosomal proteins (cluster 7 in BMP2[−]; cluster 5 in BMP2[+]) appeared to be highly up-regulated and down-regulated, respectively, in affected cells regardless of BMP2 (Fig. S4, B and E; and Data S3 and Data S4). These results showed that BMP2 pathway activation in cells with mosaic-activating SMAD3 mutation decreased osteogenic differentiation and ECM mineralization (graphical summaries in Fig. 7). In contrast, the activating MAP2K1 mutations, which we identified in our previous report (Kang et al., 2018), significantly inhibited osteoblast differentiation and ECM mineralization regardless of the presence of BMP2 (Fig. S5). Along with the histology of melorheostotic bone with SMAD3 mutations, these results indicate that melorheostosis caused by SMAD3 mutations is clinically, pathologically, and mechanistically distinct from MAP2K1 mutation–positive melorheostosis.
Figure 7.
Graphical summaries. (A) Mosaic-activating SMAD3 mutation accelerates osteogenesis in affected cells cultured in conventional osteogenic media without BMP2. (B) Osteogenesis-stimulating effect of BMP2 is dampened by the mosaic-activating SMAD3 mutation in affected cells that were cultured in osteogenic media supplemented with BMP2. Cell growth–associated gene expression is significantly down-regulated by the gain-of-function SMAD3 mutation, regardless of BMP2 presence. P in red circle denotes phosphorylation on TGF-β receptors, BMP2 receptors, and C-terminal SxxS motif of SMAD3 protein. Thickness of arrows indicates levels of gene expression in response to signaling via TGF-β/SMAD and BMP2/SMAD pathways. Thicker arrows indicate higher gene expression.
Figure S5.
BMP2-independent inhibition of ECM mineralization by mosaic-activating MAP2K1 mutation. Affected and unaffected cells from Melo-18 (MAP2K1 p.K57N) were cultured in osteogenic media with or without recombinant BMP2. Alizarin Red S staining displays significant inhibition of ECM mineralization by mosaic-activating MAP2K1 mutation, regardless of the presence of BMP2 in osteogenic media. Data are representative of at least two independent experiments. Scale bar = 10 mm. Aff, affected; Unaff, unaffected.
Discussion
Melorheostosis is an asymmetrically distributed sclerosing bone dysostosis caused by postzygotic mosaicism in the osteoblast lineage. In our previous report, we identified activating mutations in MAP2K1 as causative for classic melorheostosis with a radiographical “dripping candle wax” appearance (Kang et al., 2018). While this form is best known, its prevalence is lower than that of other radiographical patterns of melorheostosis (Freyschmidt, 2001). In this study, we report somatic mosaicism in SMAD3 for two novel mutations recurring at the same residue (p.S264Y or p.S264F) in four individuals whose melorheostosis has an endosteal radiographical pattern. The serine 264 residue is located in the region within the MH2 domain that is evolutionarily conserved (van de Laar et al., 2011). These patients had tested negative for mutations in MAP2K1, KRAS, or LEMD3.
The mosaic SMAD3 p.S264 substitutions were demonstrated to cause gain of function, with a constitutive increase in TGF-β signaling. Nuclear translocation of mutant SMAD3 was increased, and expression of TGF-β target genes was enhanced. Further, mosaicism for the SMAD3 mutation inhibited the proliferation of affected cells, consistent with the known action of TGF-β/SMAD signaling to modulate cell proliferation and growth in a cell context–dependent manner (Muñoz et al., 2008). TGF-β stimulates cell growth in fibroblasts and epithelial cells by positively regulating the mTOR pathway (Rahimi et al., 2009; Wu and Derynck, 2009). The growth inhibitory function of TGF-β was observed in muscle atrophy and hematopoietic stem cells, in which TGF-β/SMAD signaling counterbalanced the Akt/mTOR pathway (Chabanon et al., 2008; Sartori et al., 2009). Likewise, our RNA-Seq data revealed that osteoblasts suppressed expression of genes related to growth pathways regardless of BMP2 stimulation. The proximal cause of decreased cell growth in melorheostotic cells may be down-regulation of multiple ribosomal protein genes, with their significant role in protein synthesis (Maguire and Zimmermann, 2001; Naora, 1999).
In melorheostotic cells, the SMAD3 mutation promoted osteogenic differentiation and ECM mineralization. This was consistent with the findings of generally increased mineralization in affected tissue determined by quantitative back-scattered electron imaging. Specifically, CaHigh reflects the percentage of bone mineralized above 25.30 weight percent calcium, and thus older tissue age; in SMAD3 mutation–affected versus –unaffected bone, the threefold increase in CaHigh is thus indicative of slow remodeling rates, leading to increased mineral content over time.
TGF-β has bi-functional effects on osteogenesis depending on the stage of cell differentiation. TGF-β inhibits differentiation of mature osteoblasts and ECM mineralization (Kaji et al., 2006) but stimulates osteoprogenitor proliferation and early-stage osteoblast differentiation by enhancing ECM deposition and anti-apoptotic activity. The stimulated osteogenesis of SMAD3 mutation–positive melorheostosis reflected increased expression of osteogenic transcription factors (Chen et al., 2012), such as SP7 and MSX2 in affected cells. SP7 (Osterix) is the master regulator of osteoblast differentiation acting downstream of RUNX2 and regulates cell differentiation from preosteoblast to mature osteoblasts and osteocytogenesis (Nakashima et al., 2002). TGF-β signaling regulates SP7 expression through a SMAD-dependent pathway (Choi et al., 2016). MSX2, a homeobox transcription factor, is a direct transcriptional target of TGF-β and inhibits RUNX2 DNA-binding activity (Shirakabe et al., 2001); mutations in MSX2 are associated with craniosynostosis and enlarged parietal foramina (Ma et al., 1996; Wilkie et al., 2000). Our results suggest that the constitutively active form of SMAD3 in melorheostotic bone augmented the output of the TGF-β/SMAD pathway and overrode stage-specific osteogenesis regulation.
The SMAD3 serine 264 residue, which is substituted in melorheostosis, is located in the MH2 domain of SMAD3 on the L1 loop at the periphery of the protein-protein interface and forms a second network of hydrogen bonds with residues of the adjacent monomer (Fig. S2; Wu et al., 2001). There is an overrepresentation of hits in the SMAD3 MH2 domain among somatic missense mutations causing colorectal cancers (CRCs; Fleming et al., 2013). Most of these mutations reduce protein stability or hinder SMAD complex formation, undermining the role of SMAD2/3/4 as tumor suppressors in these cells (Fleming et al., 2013). Substitution in CRC at p.D258, adjacent to SMAD3 p.S264, interferes with oligomerization and decreases transcription in luciferase reporter assays, while p.S266 is critical for binding of coregulators to SMAD3. In contrast, our functional study of the melorheostosis-causing mosaic SMAD3 mutations in osteoblastic cells revealed that SMAD3 p.S264Y and p.S264F are gain-of-function substitutions that elicit stimulatory effects on osteogenesis and an inhibitory effect on cell growth.
SMAD3 germline mutations have also been demonstrated in connective tissue disorders associated with increased TGF-β signaling and skeletal, articular, and aortic findings. LDS is caused by defects in SMAD3 as well as five other genes (SMAD2, TGFB2, TGFB3, TGFBR1, or TGFBR2) in the TGF-β signaling pathway. LDS manifestations of vascular tortuosity and craniofacial and skeletal abnormalities are often associated with excess activation of TGF-β signaling, but LDS-causing genetic variants themselves are mostly loss-of-function mutations (Pezzini et al., 2012). Of the 67 SMAD3 mutations reported in LDS (Schepers et al., 2018), the majority were in the MH2 domain, but no hotspot was found. Substitutions in SMAD3 in LDS spanned exon 6, including the p.D258 and p.S266 residues associated with CRC and residues flanking the p.S264 residue substituted in melorheostosis (Fleming et al., 2013). Families with aneurysms and early onset of a distinctive osteoarthritis (originally AOS, now reclassified as LDS3) have SMAD3 p.T262I and p.R287W substitutions in the MH2 domain (van de Laar et al., 2011). As in our melorheostosis-causing SMAD3 mutations, these LDS3-causing SMAD3 mutations are predicted to interfere with heterotrimer formation. The LDS3-causing mutations were associated with increased immunohistochemical labeling of phosphorylated SMAD3, as well as upstream ligand TGF-β, in aortic tissue (van de Laar et al., 2011).
Sclerotic bone phenotypes in OPK and BOS are attributable to augmented SMAD-dependent TGF-β/BMP signaling caused by heterozygous loss-of-function mutations in LEMD3 (MAN1; Hellemans et al., 2004). Smad3 KO mice are osteopenic secondary to reduced bone formation, caused by increased apoptosis of osteoblasts and osteocytes (Borton et al., 2001). They have reduced TGF-β signaling, leading to abnormal chondrocyte development and osteoarthritis, and the majority die from ruptured aneurysms (van der Pluijm et al., 2016). Since patients with melorheostosis caused by SMAD3 p.S264 substitutions do not have findings of LDS, melorheostosis expands the type of bone abnormality associated with abnormal TGF-β signaling. Dynamic expression and stage-specific function of SMAD3 in osteoblast differentiation (Lin et al., 2019) may explain the phenotypic discrepancy between SMAD3 overexpression and melorheostosis-causing mosaic SMAD3 mutations.
Surprisingly, addition of BMP2, also a pro-osteogenic factor, to the conventional osteogenic media caused affected cells to display inhibition of osteogenic differentiation and ECM mineralization in vitro. Several factors are relevant to the mechanism of this adverse influence on osteogenesis. First, although the pro-osteogenic transcription factor RUNX2 is activated by BMP signaling, SMAD3 inhibits RUNX2 activity by interfering with its DNA binding (Soltanoff et al., 2009). Second, TGF-β stimulation results in a decreased level of SMAD1/5–SMAD4 complexes formed in response to BMP and an increase in mixed R-SMAD complexes (SMAD1/5–SMAD2/3), which bind the BMP-responsive element and exert inhibitory effects (Grönroos et al., 2012). This TGF-β action depends on SMAD3 rather than on SMAD2. Thus, the SMAD3 mutation’s constitutive activation of TGF-β may augment the inhibition of BMP-responsive gene expression and contribute to decreased osteogenesis in the presence of BMP2. It will be important to obtain further insight into the crosstalk between TGF-β/SMAD3 and BMP pathways in affected cells, since this may impact future therapeutics for SMAD3 mutation–positive melorheostosis. For example, the distinct signaling characteristics of BMP type I receptors could be exploited. BMP2 signals through the hetero-oligomerization of two type I receptor subtypes, BMPRIA (ALK3) or BMPRIB (ALK6), and the type II receptor, BMPRII. BMPRIA and BMPRIB transmit different signals to bone-derived mesenchymal progenitors, dictating whether adipogenesis or osteogenesis is induced (Chen et al., 1998). In addition, the BMPRIA expression level was ∼10 times higher than that of BMPRIB in our RNA-Seq data from melorheostotic osteoblasts. Although further studies are needed, we speculate that BMP2 muteins may be designed to signal preferentially through BMPRIA and exert an antagonistic function on the pro-osteogenic process in affected cells with the activating SMAD3 mutation.
Interestingly, up-regulation of genes regulated by the interferon signaling pathway was apparent in affected cells, regardless of BMP2 presence in culture. Osteoblasts from affected melorheostotic bone showed strong up-regulation of interferon-inducible signature genes (Yao et al., 2009) in early differentiation (Table S4). Although TGF-β generally suppresses type I interferon production, SMAD3 has been shown to enhance the effects of IRF7 and promotes interferon pathway target gene expression (Qing et al., 2004). The important physiological anabolic effects of IFN-γ signaling in bone formation were demonstrated by reduced bone volume in IFNγR1 KO mice with subsequent rescue of the ovariectomy-induced osteoporosis by IFN-γ administration (Duque et al., 2011). Given that the up-regulation of interferon signal-regulated genes was not altered by BMP2, the interferon signaling pathway does not seem to have an antagonistic crosstalk with the BMP pathway in affected cells. Whether or how the augmentation of the interferon signaling pathway and up-regulation of downstream target genes in affected cells may contribute to pathological phenotypes by inducing immune responses within melorheostotic bone lesion remains to be investigated.
Our study has some limitations and generates intriguing questions about the distinctions and similarities of melorheostosis caused by recurrent mutations in specific residues in MAP2K1 and SMAD3. Our previous report showed that activating MAP2K1 mutations increased bone remodeling within melorheostotic bone lesion. This was attributed to highly proliferative osteoblasts stimulating RANKL expression and osteoclastogenesis (Kang et al., 2018). In melorheostosis caused by SMAD3 mutations, up-regulation of SP7 may increase the increased RANKL expression in cultured affected cells. Although histology of melorheostosis bone tissues showed a minor increase in osteoclasts, the sample size (n = 4) was too small to perform statistical analysis on osteoclastogenesis-associated phenotype. Likewise, only one of four melorheostosis patients with SMAD3 mutations had a high VAF, limiting our RNA-Seq to technical replicates and a compensatory stringent threshold of FDR < 1e-20.
Finally, the clinical, bone tissue, and cellular phenotypes of SMAD3 mutation–positive melorheostosis may be compared with MAP2K1 mutation–positive melorheostosis. Patients with SMAD3 mutation–positive endosteal pattern of melorheostosis on radiographs do not present with a visible bone overgrowth. While patients with MAP2K1 mutation–positive melorheostosis were likely to have cutaneous vascular changes in skin overlying affected bone, no cutaneous changes were noted in patients with SMAD3 mutation–positive melorheostosis. The growth-promoting effects of activating MAP2K1 mutations, which we observed in osteoblasts (Kang et al., 2018), may account for the bone overgrowth outside the cortical margins in these patients, and the vascular skin phenotype may be related to the known ability of MAP2K1 mutations to promote vascular malformations (Couto et al., 2017).
Both mutations in SMAD3 led to bone growth by periosteal apposition of multilayered compact parallel lamellar bone that was subsequently remodeled, although apparently at a slower rate than in the MAP2K1 cases. This provides evidence that the so-called “periosteal reaction” is a common response in both forms of melorheostosis, as well as in other benign or malignant lesions (Bisseret et al., 2015). Although the SMAD3 mutation decreases cell proliferation, there is bone overgrowth. We speculate that this is due to focal apposition of new bone formed by cells of the periosteum, which may not carry a SMAD3 mutation. Also, several cellular distinctions are noted between melorheostosis caused by MAP2K1 mutations and by SMAD3 mutations: (a) mineralization was inhibited in MAP2K1 mutation–positive melorheostosis, with osteoid accumulation and decreased mineral content of the bone matrix, while osteogenesis was increased in SMAD3 mutation–positive melorheostosis, with a shift of the BMDD peak toward higher mineral content; (b) cell growth was stimulated by MAP2K1 mutations but inhibited by SMAD3 mutations; and (c) effects of the activating SMAD3 mutation on osteogenic differentiation were in opposite directions depending on whether BMP2 stimulation was applied or not, but the inhibitory effect of the activating MAP2K1 mutations on osteogenic differentiation was independent of BMP2.
The study presented here demonstrated the critical role of dysregulated TGF-β/SMAD3 pathway signaling in the pathogenesis of melorheostosis with an endosteal radiographical pattern (graphical summaries in Fig. 7). The potential connections between the genes and pathways currently identified in melorheostosis are intriguing. The nature of the crosstalk between the TGF-β and BMP pathways in melorheostosis progression remains to be further elucidated. There was no evidence of alteration in LEMD3 expression by the SMAD3 or MAP2K1 mutations, and LEMD3 transcript level remained similar during osteoblast differentiation. The SMAD3 mutations (p.S264Y or p.S264F) would not be expected to impact physical interaction of SMAD3 and MAN1 (LEMD3 gene product), given that MAN1 recognizes the L3 loop of the MH2 domains of R-SMAD proteins within the helix-bundle region (Miyazono et al., 2018). However, it still remains to be determined whether the SMAD3 mutations have an influence on activities of MAN1 and PPM1A at the post-transcriptional level.
Together with our previous report (Kang et al., 2018), this study demonstrated that melorheostosis is a genetically heterogeneous disorder, with somatic mutations in different genes responsible for distinct radiographical patterns of melorheostosis through different molecular and cellular mechanisms. Our identification of MAP2K1 and SMAD3 as melorheostosis-causing genes accounts for 12 of 15 biopsied patients in our cohort. These data further underscore dysregulation of signals in the RAS/MEK/MAPK pathway and the TGF-β/SMAD pathway as the culprit in this group of skeletal disorders.
Materials and methods
Patients and bone biopsy
All studies were approved by NIH Intramural Institutional Review Board (protocol no. NCT02504879). Unrelated adults with a diagnosis of melorheostosis were recruited. Study subjects were not compensated for their participation. Informed consent was obtained from all patients/parents. The patients underwent open surgical biopsy of the melorheostotic lesion and of a contralateral, generally smaller, sample of unaffected bone, preferentially from the iliac crest (Jha et al., 2019; Kang et al., 2018). Two patients requested a different skeletal site for this elective procedure, and biopsy samples were obtained from the tibia and ulna (Melo-8 and Melo-17, respectively). Affected and unaffected bone samples were divided and immediately immersed either in culture media for genomic DNA extraction and cell culture or fixed in 70% ethanol before being embedded in polymethylmethacrylate for bone histology using standard procedures.
Whole exome sequencing
High-coverage (100×) whole exome sequencing was performed by a sequencing service provider (Otogenetics). Agilent SureSelect Human V5 (51 Mbp) kit was used for exome capture. After paired-end sequencing on Illumina HiSeq sequencers, FASTQ files were aligned to Human Genome build 37 (hg19) using Burrows-Wheeler Aligner. The standard PICARD-GATK pipeline was used to remove duplicate reads, refine alignment around indels, and recalibrate base quality scores. The resulting BAM files (one per sample) served as inputs to somatic mutation callers. Two somatic mutation callers (muTect v1.1.7 and Strelka v1.0.14) were used, with default parameters for whole exome sequencing data. Somatic variants called by either method were annotated with functional impact and population frequency using ANNOVAR. We filtered somatic variants for those that were not present in the unaffected bone, caused protein sequence changes or splicing changes, had <1% frequency in ExAC and 1000 Genomes Project databases, were not in a duplicated genomic region, and were called by both muTect and Strelka. The whole exome sequencing data have been deposited to the database of Genotypes and Phenotypes (dbGaP) under the accession code phs001976.v1.p1. To access these data, users may apply for access to the dbGaP data repository (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?login=&page=login).
Amplicon sequencing
In collaboration with Swift Biosciences and IDT, we designed a custom amplicon-based sequencing panel to screen genes of interest. The sensitivity of this platform can detect somatic variants with frequency as low as 0.5% VAF. Sequencing was performed to an average depth of ∼1,000× on the Illumina MiSeq instrument, with 2 × 150 bp paired-end reads. Alignment to the hg19 reference genome was done with Burrows-Wheeler Aligner mem aligner, reads were processed according to GATK best practices, and variants were called using LoFreq somatic variant caller. Subsequent filtering of variants excluded those with poor quality and common population variants.
ddPCR
ddPCR assays were designed for the SMAD3 p.S264Y and p.S264F mutations for the purposes of validation and variant allele quantification. The FAM- and HEX-labeled probes were specific to wild-type and mutant versions of the DNA, respectively. Droplet generation and signal quantification were performed by the Bio-Rad Q×200 ddPCR System. Calculation of VAF was performed by QuantaSoft software (Bio-Rad).
Bone histology and quantitative backscattered electron imaging (qBEI)
For histological evaluations, 4-µm sections were cut from the tissue block and stained with modified Goldner’s Trichrome using standard procedures (Glorieux et al., 2000). A light microscope equipped with a Zeiss AxioCam video camera was used to obtain digital images of the sections that were analyzed using NIH ImageJ software (Ver. 1.63). We evaluated osteoid thickness, osteoid surface per bone surface, osteoblast surface per bone surface, osteoclast surface per bone surface, and eroded surface per bone surface on four randomly chosen areas throughout each bone section. Bone lamellar organization was observed under polarized light. Subsequently, the residual blocks were prepared for qBEI by grinding and polishing to obtain plane parallel surfaces, then carbon coated (Roschger et al., 1998). The entire cross-sectioned bone sample area was imaged with a spatial resolution of 1.8 µm per pixel using a field emission scanning electron microscope (Zeiss Supra 40; Oberkochen) equipped with a four-quadrant semiconductor backscatter electron detector. The field emission scanning electron microscope was operated with an electron energy of 20 keV. The gray levels reflecting the mineral/calcium content were calibrated by the material contrast of pure carbon and aluminum. Gray-level histograms were deduced and transformed into calcium weight percent, resulting in a corresponding BMDD curve described by five variables: CaMean, the average calcium concentration (weighted mean); CaPeak, the most frequently occurring calcium concentration (the peak position of the BMDD) in the sample; CaWidth, the width of the BMDD distribution (full width at half maximum) reflecting the heterogeneity in matrix mineralization; CaLow, the percentage of low mineralized bone area, which is mineralized below 17.68 weight percent calcium, reflecting bone areas undergoing primary mineralization; and CaHigh, the percentage of highly mineralized bone matrix, having calcium content above 25.30 weight percent calcium (Roschger et al., 2008). Statistical evaluation was performed with GraphPad Prism 6.0h. Comparison of histomorphometric and BMDD indices was based either on paired t test for comparison of affected and unaffected bone from the same patient or on unpaired t test comparing affected tissue from the present study with affected tissue from patients with somatic activating in MAP2K1 reported previously (Fratzl-Zelman et al., 2019; Kang et al., 2018). Statistical significance was considered as P < 0.05.
Cell proliferation
Primary osteoblasts were grown from freshly minced bone chips as described previously (Kang et al., 2018). Osteoblasts were plated (3,000 cells/well) in 96-well tissue culture plates and imaged at 2-h intervals for 120 h at 10× magnification using the IncuCyte ZOOM Kinetic Imaging System (Essen Bioscience). During the live-cell imaging, cells were incubated in 37°C and 8% CO2. Percentage of confluence was calculated from the percentage of the well area occupied by cells. Doubling time was calculated using GraphPad Prism 6.0 using nonlinear regression. To analyze comparable populations of cells between groups, only wells with confluency at the first time point of image acquisition within one-half SD of the mean were included in the analysis. Statistical comparison was done by two-way ANOVA.
Plasmid constructs and site-directed mutagenesis
Constructs encoding the human SMAD3 (p.S264Y) and SMAD3 (p.S264F) mutants were generated by site-directed mutagenesis using the In-Fusion kit (Clontech). The primer pairs to introduce each mutation were as follows. SMAD3 (p.S264Y) forward: 5′-GGCTTCACCGACCCCTACAATTCGGAGCGCTTCTGCCTAG-3′, SMAD3 (p.S264Y) reverse: 5′-GAAGCGCTCCGAATTGTAGGGGTCGGTGAAGCCATCCACAGTC-3′; and SMAD3 (p.S264F) forward: 5′-GGCTTCACCGACCCCTTCAATTCGGAGCGCTTCTGCCTAG-3′, SMAD3 (p.S264F) reverse: 5′-GAAGCGCTCCGAATTGAAGGGGTCGGTGAAGCCATCCACAGTC-3′. Plasmid CS2-Flag-SMAD3 (WT) was used as a template for PCR cloning to generate plasmid vectors expressing the SMAD3 mutants described in the study. The plasmid constructs were verified by Sanger sequencing. CS2-Flag-SMAD3 was a gift from Joan Massagué (Cancer Biology and Genetics Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, NY; Addgene plasmid #14052; Kretzschmar et al., 1997).
Transfection of plasmids
MC3T3-E1 cells (American Type Culture Collection) were maintained in α-MEM base media supplemented with 10% FBS (GemCell, Gemini), 100 µg/ml streptomycin, and 100 U/ml penicillin (Gibco). Cells were cultured at 37°C and 5% CO2 and were sub-cultured upon reaching 80% confluence. For transfection, 8 × 104 cells per well were plated overnight in 12-well plates and transfected the next day with plasmid DNA constructs (1–2 µg/well) using X-tremeGENE HP DNA transfection reagent (Sigma). In some experiments, cells were serum starved 3 h with or without TGF-β receptor inhibitor SB431542 (10 µM; Sigma) and then incubated with recombinant TGF-β1 (R&D Systems) for various times as indicated.
Nuclear localization
MC3T3-E1 cells were transfected with plasmid DNA constructs expressing SMAD3 (wild-type and p.S264Y) using X-tremeGENE HP DNA Transfection Reagent. 24 h after transfection, cells were treated with or without TGF-β1 (10 ng/ml) for 30 min and fixed with 2% paraformaldehyde for 10 min at 37°C and washed five times with 1× PBS. Cells were permeabilized with 1% BSA/0.1% Triton X-100/0.2% Saponin in 1× PBS for 40 min at room temperature and washed five times with 1× PBS before incubating at 4°C overnight with primary antibodies (anti-FLAG; clone M2, #F1804; Sigma) or anti-SMAD3 (C67H9, rabbit, #9523; Cell Signaling Technology) diluted in 2% BSA/1× PBS. Cells were washed five times with 1× PBS before blocking with 5% goat serum diluted in 1× PBS for 30 min at room temperature. Cells were then stained with secondary antibodies and nuclear dye at 4°C for 1 h protecting from light. The following secondary antibodies and fluorescent dyes were used for imaging using the IncuCyte system: Alexa Fluor 568 goat anti-mouse IgG (H+L; #A11004; Invitrogen), Alexa Fluor 488 donkey anti-rabbit IgG (H+L; #A11008; Invitrogen), and Syto61 (Invitrogen).
Western blot analysis
Cultured and treated cells were lysed in radioimmunoprecipitation assay buffer and subjected to Western blot analysis, as described in the previous report (Kang et al., 2018). The following primary and secondary antibodies were used for Western blotting: phospho-Smad3 (Ser423/425; C25A9) rabbit mAb (#9520; Cell Signaling Technology), phospho-Smad3 (Ser423/425) rabbit polyclonal Ab (#600–401-919; Rockland Inc.), Smad2/3 (D7G7) XP rabbit mAb (#8685; Cell Signaling Technology), SMAD3 (C67H9) rabbit mAb (#9523; Cell Signaling Technology), β-Actin (AC-15) mouse mAb (#A5441; Sigma), GAPDH (D16H11) XP rabbit mAb (#5174; Cell Signaling Technology), histone H3 (D1H2) XP rabbit mAb (#4499; Cell Signaling Technology), and FLAG (M2) mouse mAb (#F1804; Sigma). Secondary antibodies (LI-COR Biosciences) were IRDye 680RD anti-rabbit IgG (#926-68071) or IRDye 800CW anti-mouse IgG (#926-32210).
RNA isolation and real-time qPCR
Total RNA was extracted from osteoblasts using the RNeasy Mini kit (Qiagen) following the manufacturer’s instruction. Genomic DNA was removed from RNA preps by on-column DNase digestion. Synthesis of cDNA was performed using 500 ng of total RNA and iScript reverse transcription Supermix (Bio-Rad), following the manufacturer’s instructions. Comparative real-time qPCR was performed in TaqMan Universal PCR Master Mix in triplicate using Applied Biosystems Prism 7500 Fast Sequence Detection System, according to the manufacturer’s protocol. TaqMan probes and primers were purchased from Applied Biosystems: Human COL1A1 (Hs00164004_m1), COL1A2 (Hs01028956_m1), CDKN2B (Hs00793225_m1), SERPINE1 (Hs00167155_m1), ALPL (Hs01029144_m1), SP7 (Hs01866874_s1), CDH11 (Hs00901479_m1), MMP13 (Hs00942584_m1), and SPP1 (Hs00959010_m1). Mouse Ctgf (Mm01192933_g1), Serpine1 (Mm00435858_m1), and Vegfa (Mm00437306_m1). Human GAPDH (Hs02786624_g1), human TBP (Hs00427620_m1), mouse Gapdh (Mm99999915_g1), and mouse Tbp (Mm00446971_m1) were used as endogenous controls for normalization of real-time qPCR. Relative expression was calculated using the comparative ΔΔCt method.
Osteoblast differentiation and ECM mineralization
Primary patient osteoblasts were plated at passage 2 with 50,000 cells/well in 12-well tissue culture plates and cultured to confluency in α-MEM supplemented with 10% FBS and antibiotics at 37°C and 8% CO2. After confluence, cells were further cultured in osteogenic media (50 µg/ml L-ascorbic acid, 2.5 mM β-glycerophosphate, and 10 nM dexamethasone, all from Sigma), with or without recombinant BMP2 (100 ng/ml; R&D Systems), refreshing osteogenic media every 3 d. To visualize ECM mineralization, cells were washed once with 1× PBS and fixed with 4% paraformaldehyde for 10 min at room temperature. Fixed cells were washed three times with 1× PBS and stained with 2% Alizarin Red S solution (pH 4.2) for 10 min at room temperature. Excess Alizarin Red S stain was removed by washing five times with distilled water. Cells were air-dried in the dark before imaging.
RNA-Seq and data analysis
Total RNA samples (∼1–4 µg) were purified with Poly-A extraction, and then purified mRNAs were used to construct RNA-Seq libraries with specific bar codes using the Illumina TruSeq Stranded mRNA Library Prep Kit. All the RNA-Seq libraries were pooled together and sequenced using Illumina HiSeq 2500 to generate ∼40 million 2 × 100 paired-end reads for each sample. Demultiplexed 100-bp paired-end fastq reads were aligned to GRCh38 human reference using GENCODE release 27 gene definitions using RNA STAR v2.5.4a (Dobin et al., 2013). Following alignment, reads were quantitated using the subread package featureCounts v1.5.2 (Liao et al., 2014) against GENCODE release 27 gene definitions, counting multi-mapping and multi-overlapping reads at the level of gene names. All downstream analyses were performed in R v3.5.1, Ingenuity Pathway Analysis (Qiagen Bioinformatics), and Metascape. For each of these conditions, we loaded the counts data into DESeq2 v1.20 (Love et al., 2014) and performed differential expression analysis between affected and unaffected samples at each time point. Since the samples are technical replicates of cells from one melorheostosis patient, this resulted in lower reported P values than biological replicates. To compensate for this, we chose a more stringent threshold of FDR < 1e-20 to select DEGs. Other thresholds return largely the same results.
To identify patterns of expression throughout the data, we used the degPatterns function from the DEGreport v1.16 Bioconductor package (Steinbaugh et al., 2018). Briefly, this analysis takes normalized counts from genes of interest, generates a correlation matrix of genes across condition and time, clusters that matrix, and cuts the resulting tree into modules of coexpressed genes. We used counts normalized with the DESeq2 variance stabilizing transformation and provided counts for the union of genes differentially expressed between affected and unaffected at any time point. Resulting clusters were merged together if the pairwise correlation coefficient was >0.5, and clusters with <15 genes after merging were dropped. The plots show normalized counts for the genes in each cluster. MA plots show the log of mean normalized counts for each gene on the x axis and the log2 fold change on the y axis. These plots reflect the default behavior of DESeq2 to perform fold-change shrinkage, shifting the reported log fold change of genes with low information toward zero. Heatmaps show normalized counts after applying the variance-stabilizing transform, with genes subsetted from the full normalized counts depending on the gene list of interest. Plots were generated with the heatmaply v0.15.2 package, scaling by row, disabling column clustering, and otherwise using defaults. Gene set enrichment analysis (GSEA) analysis was performed using the clusterProfiler v3.8.1 package’s GSEA function (Yu et al., 2012) after providing a custom annotation containing only the set of genes of interest. The RNA-Seq data have been deposited to dbGaP under the accession code phs001976.v1.p1. To access these data, users may apply for access to the dbGaP data repository (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?login=&page=login).
Statistical analyses
For bone histology, statistical evaluation was performed with GraphPad Prism 6.0h. Comparison of histomorphometric and BMDD indices was based either on paired t test for comparison of affected and unaffected bone from the same patient or on unpaired t test comparing affected tissue from the present study with affected tissue from MAP2K1 mutation–positive patients. Statistical significance was considered as P value <0.05. For cell proliferation assay and SMAD3 nuclear localization, statistical comparison was done by two-way ANOVA. For real-time qPCR data analysis, differences between groups were evaluated with the Student’s t test using a two-tailed distribution. Results were considered significant when P value was <0.05. Data are expressed as mean ± SD.
Online supplemental material
Fig. S1 shows radiographs of three patients with SMAD3 mutation–positive melorheostosis demonstrating the endosteal appearance. Fig. S2 shows the results of a genetic study identifying SMAD3 somatic mutations in melorheostosis. Fig. S2 also illustrates human SMAD3 missense mutations found in melorheostosis, CRC, and LDS. Fig. S3 shows effects of SMAD3 mutations on TGF-β signaling. Fig. S4 shows the results of transcriptome profiling by RNA-Seq. Fig. S5 shows BMP2-independent inhibition of ECM mineralization by mosaic-activating MAP2K1 mutation. Table S1 summarizes the clinical findings on SMAD3 mutation–positive melorheostosis patients. Table S2 summarizes the bone histomorphometry and qBEI analyses in melorheostosis bone. Table S3 shows β-catenin (CTNNB1) up-regulation and changes of downstream target genes in affected cells of Melo-11. Table S4 shows up-regulation of interferon-inducible signature genes (21 genes) in affected cells of melorheostosis (Melo-11, SMAD3, p.S264Y). Data S1 lists the genes in each cluster generated by degPatterns analysis of RNA-Seq data of BMP2(−) condition. Data S2 lists the genes in each cluster generated by degPatterns analysis of RNA-Seq data of BMP2(+) condition. Data S3 lists enriched pathway terms depicted as an interaction network for each cluster generated through the Metascape analysis (BMP2[−] condition). Data S4 lists enriched pathway terms depicted as an interaction network for each cluster generated through the Metascape analysis (BMP2[+] condition).
Supplementary Material
summarizes clinical findings on SMAD3 mutation–positive melorheostosis patients.
summarizes histomorphometry and qBEI analyses in melorheostosis bone.
shows β-catenin (CTNNB1) upregulation and changes of downstream target genes in affected cells of Melo-11.
lists up-regulation of interferon-inducible signature genes (21 genes) in affected cells of melorheostosis (Melo-11, SMAD3, p.S264Y).
lists the genes in each cluster generated by degPatterns analysis of RNA-Seq data of BMP2(−) condition.
lists the genes in each cluster generated by degPatterns analysis of RNA-Seq data of BMP2(+) condition.
lists enriched pathway terms depicted as an interaction network for each cluster generated through the Metascape analysis (BMP2[−] condition).
lists enriched pathway terms depicted as an interaction network for each cluster generated through the Metascape analysis (BMP2[+] condition).
Acknowledgments
The authors thank the Center for Cancer Research Genomics Core facility (National Cancer Institute) for conducting ddPCR assays and Daniela Gabriel, Petra Keplinger, Sonja Lueger, and Phaedra Messmer (Ludwig Boltzmann Institute of Osteology at the Hanusch Hospital of Wiener Gebietskrankenkasse and Allgemeine Unfallversicherungsanstalt Trauma Center Meidling) for careful sample preparations and qBEI measurements. We thank Tianwei Li and James Iben at the Molecular Genomics Core (National Institute of Child Health and Human Development) for RNA-Seq and primary data processing, and Nicole Swan (National Institute of Child Health and Human Development) for assistance with graphics. We are especially grateful to Kathleen Harper, the Melorheostosis Association, and the patients for their support of this study.
This work was supported by National Institutes of Health intramural funds to J.C. Marini (National Institute of Child Health and Human Development, 1ZIAHD008973-02), R.M. Siegel (National Institute of Arthritis and Musculoskeletal and Skin Diseases, AR041173-11, AR041207-03, and AR041180-11), and T. Bhattacharyya (National Institute of Arthritis and Musculoskeletal and Skin Diseases, 1ZIDAR041180-09), and the Allgemeine Unfallversicherungsanstalt (research funds of the Austrian Workers’ Compensation Board) and Wiener Gebietskrankenkasse (Vienna Regional Health Insurance Fund) to N. Fratzl-Zelman. A. Ivovic was supported by the National Institutes of Health-Oxford-Cambridge PhD Scholars Program.
Author contributions: T. Bhattacharyya, S. Jha, E. Lange, E.W. Cowen, and J. Katz wrote the clinical protocol and provided patient care. H. Kang, A. Ivovic, S. Jha, Z. Deng, N. Fratzl-Zelman, A. Mitra, W.A. Cabral, E.P. Hanson, P. Roschger, K. Klaushofer, R.K. Dale, and T. Bhattacharyya designed and conducted experiments and analyzed data. The manuscript was drafted by H. Kang and edited by J.C. Marini with significant input from A. Ivovic, S. Jha, Z. Deng, N. Fratzl-Zelman, R.M. Siegel, and T. Bhattacharyya.
References
- Bisseret D., Kaci R., Lafage-Proust M.H., Alison M., Parlier-Cuau C., Laredo J.D., and Bousson V.. 2015. Periosteum: characteristic imaging findings with emphasis on radiologic-pathologic comparisons. Skeletal Radiol. 44:321–338. 10.1007/s00256-014-1976-5 [DOI] [PubMed] [Google Scholar]
- Borton A.J., Frederick J.P., Datto M.B., Wang X.F., and Weinstein R.S.. 2001. The loss of Smad3 results in a lower rate of bone formation and osteopenia through dysregulation of osteoblast differentiation and apoptosis. J. Bone Miner. Res. 16:1754–1764. 10.1359/jbmr.2001.16.10.1754 [DOI] [PubMed] [Google Scholar]
- Bourgeois B., Gilquin B., Tellier-Lebègue C., Östlund C., Wu W., Pérez J., El Hage P., Lallemand F., Worman H.J., and Zinn-Justin S.. 2013. Inhibition of TGF-β signaling at the nuclear envelope: characterization of interactions between MAN1, Smad2 and Smad3, and PPM1A. Sci. Signal. 6:ra49 10.1126/scisignal.2003411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabanon A., Desterke C., Rodenburger E., Clay D., Guerton B., Boutin L., Bennaceur-Griscelli A., Pierre-Louis O., Uzan G., Abecassis L., et al. 2008. A cross-talk between stromal cell-derived factor-1 and transforming growth factor-beta controls the quiescence/cycling switch of CD34(+) progenitors through FoxO3 and mammalian target of rapamycin. Stem Cells. 26:3150–3161. 10.1634/stemcells.2008-0219 [DOI] [PubMed] [Google Scholar]
- Chen D., Ji X., Harris M.A., Feng J.Q., Karsenty G., Celeste A.J., Rosen V., Mundy G.R., and Harris S.E.. 1998. Differential roles for bone morphogenetic protein (BMP) receptor type IB and IA in differentiation and specification of mesenchymal precursor cells to osteoblast and adipocyte lineages. J. Cell Biol. 142:295–305. 10.1083/jcb.142.1.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Deng C., and Li Y.P.. 2012. TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci. 8:272–288. 10.7150/ijbs.2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Ahn Y.H., Kim T.H., Bae C.H., Lee J.C., You H.K., and Cho E.S.. 2016. TGF-β Signaling Regulates Cementum Formation through Osterix Expression. Sci. Rep. 6:26046 10.1038/srep26046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto J.A., Huang A.Y., Konczyk D.J., Goss J.A., Fishman S.J., Mulliken J.B., Warman M.L., and Greene A.K.. 2017. Somatic MAP2K1 Mutations Are Associated with Extracranial Arteriovenous Malformation. Am. J. Hum. Genet. 100:546–554. 10.1016/j.ajhg.2017.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C.J., and Massagué J.. 2018. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 19:419–435. 10.1038/s41580-018-0007-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque G., Huang D.C., Dion N., Macoritto M., Rivas D., Li W., Yang X.F., Li J., Lian J., Marino F.T., et al. 2011. Interferon-γ plays a role in bone formation in vivo and rescues osteoporosis in ovariectomized mice. J. Bone Miner. Res. 26:1472–1483. 10.1002/jbmr.350 [DOI] [PubMed] [Google Scholar]
- Fleming N.I., Jorissen R.N., Mouradov D., Christie M., Sakthianandeswaren A., Palmieri M., Day F., Li S., Tsui C., Lipton L., et al. 2013. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 73:725–735. 10.1158/0008-5472.CAN-12-2706 [DOI] [PubMed] [Google Scholar]
- Fratzl-Zelman N., Roschger P., Kang H., Jha S., Roschger A., Blouin S., Deng Z., Cabral W.A., Ivovic A., Katz J., et al. 2019. Melorheostotic Bone Lesions Caused by Somatic Mutations in MAP2K1 Have Deteriorated Microarchitecture and Periosteal Reaction. J. Bone Miner. Res. 34:883–895. 10.1002/jbmr.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyschmidt J. 2001. Melorheostosis: a review of 23 cases. Eur. Radiol. 11:474–479. 10.1007/s003300000562 [DOI] [PubMed] [Google Scholar]
- Glorieux F.H., Travers R., Taylor A., Bowen J.R., Rauch F., Norman M., and Parfitt A.M.. 2000. Normative data for iliac bone histomorphometry in growing children. Bone. 26:103–109. 10.1016/S8756-3282(99)00257-4 [DOI] [PubMed] [Google Scholar]
- Grönroos E., Kingston I.J., Ramachandran A., Randall R.A., Vizán P., and Hill C.S.. 2012. Transforming growth factor β inhibits bone morphogenetic protein-induced transcription through novel phosphorylated Smad1/5-Smad3 complexes. Mol. Cell. Biol. 32:2904–2916. 10.1128/MCB.00231-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Preobrazhenska O., Willaert A., Debeer P., Verdonk P.C., Costa T., Janssens K., Menten B., Van Roy N., Vermeulen S.J., et al. 2004. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat. Genet. 36:1213–1218. 10.1038/ng1453 [DOI] [PubMed] [Google Scholar]
- Hellemans J., Debeer P., Wright M., Janecke A., Kjaer K.W., Verdonk P.C., Savarirayan R., Basel L., Moss C., Roth J., et al. 2006. Germline LEMD3 mutations are rare in sporadic patients with isolated melorheostosis. Hum. Mutat. 27:290 10.1002/humu.9403 [DOI] [PubMed] [Google Scholar]
- Ho N.C., Jia L., Driscoll C.C., Gutter E.M., and Francomano C.A.. 2000. A skeletal gene database. J. Bone Miner. Res. 15:2095–2122. 10.1359/jbmr.2000.15.11.2095 [DOI] [PubMed] [Google Scholar]
- Holm T.M., Habashi J.P., Doyle J.J., Bedja D., Chen Y., van Erp C., Lindsay M.E., Kim D., Schoenhoff F., Cohn R.D., et al. 2011. Noncanonical TGFβ signaling contributes to aortic aneurysm progression in Marfan syndrome mice. Science. 332:358–361. 10.1126/science.1192149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens K., Gershoni-Baruch R., Guañabens N., Migone N., Ralston S., Bonduelle M., Lissens W., Van Maldergem L., Vanhoenacker F., Verbruggen L., and Van Hul W.. 2000. Mutations in the gene encoding the latency-associated peptide of TGF-beta 1 cause Camurati-Engelmann disease. Nat. Genet. 26:273–275. 10.1038/81563 [DOI] [PubMed] [Google Scholar]
- Jha S., Fratzl-Zelman N., Roschger P., Papadakis G.Z., Cowen E.W., Kang H., Lehky T.J., Alter K., Deng Z., Ivovic A., et al. 2019. Distinct Clinical and Pathological Features of Melorheostosis Associated With Somatic MAP2K1 Mutations. J. Bone Miner. Res. 34:145–156. 10.1002/jbmr.3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji H., Naito J., Sowa H., Sugimoto T., and Chihara K.. 2006. Smad3 differently affects osteoblast differentiation depending upon its differentiation stage. Horm. Metab. Res. 38:740–745. 10.1055/s-2006-955085 [DOI] [PubMed] [Google Scholar]
- Kang H., Jha S., Deng Z., Fratzl-Zelman N., Cabral W.A., Ivovic A., Meylan F., Hanson E.P., Lange E., Katz J., et al. 2018. Somatic activating mutations in MAP2K1 cause melorheostosis. Nat. Commun. 9:1390 10.1038/s41467-018-03720-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A., Saito T., Tomita H., Makita Y., Yoshida K., Ghadami M., Yamada K., Kondo S., Ikegawa S., Nishimura G., et al. 2000. Domain-specific mutations in TGFB1 result in Camurati-Engelmann disease. Nat. Genet. 26:19–20. 10.1038/79128 [DOI] [PubMed] [Google Scholar]
- Kretzschmar M., Liu F., Hata A., Doody J., and Massagué J.. 1997. The TGF-beta family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 11:984–995. 10.1101/gad.11.8.984 [DOI] [PubMed] [Google Scholar]
- Leri A., and Joanny J.P.. 1922. Une affection non décrite des os: Hyperostose “en coulée” sur toute la longueur d’un membre ou “melorhéostose”. Bulletins et Mémoires de la Société des Hôpitaux de Paris. 46:1141–1145. [Google Scholar]
- Liao Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lin X., Duan X., Liang Y.Y., Su Y., Wrighton K.H., Long J., Hu M., Davis C.M., Wang J., Brunicardi F.C., et al. 2006. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell. 125:915–928. 10.1016/j.cell.2006.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.T., Chen S.K., Guo J.W., Su I.C., Huang C.J., Chien C.C., and Chang C.J.. 2019. Dynamic expression of SMAD3 is critical in osteoblast differentiation of PDMCs. Int. J. Mol. Med. 43:1085–1093. [DOI] [PubMed] [Google Scholar]
- Loeys B.L., and Dietz H.C.. 1993. Loeys-Dietz syndrome. In GeneReviews®. Pagon R.A., Adam M.P., Ardinger H.H., Wallace S.E., Amemiya A., Bean L.J.H., Bird T.D., Ledbetter N., Mefford H.C., Smith R.J.H., and Stephens K., editors. GeneReviews [Internet], Seattle, WA. [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Golden S., Wu L., and Maxson R.. 1996. The molecular basis of Boston-type craniosynostosis: the Pro148-->His mutation in the N-terminal arm of the MSX2 homeodomain stabilizes DNA binding without altering nucleotide sequence preferences. Hum. Mol. Genet. 5:1915–1920. 10.1093/hmg/5.12.1915 [DOI] [PubMed] [Google Scholar]
- Maguire B.A., and Zimmermann R.A.. 2001. The ribosome in focus. Cell. 104:813–816. 10.1016/S0092-8674(01)00278-1 [DOI] [PubMed] [Google Scholar]
- Massagué J. 2000. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 1:169–178. 10.1038/35043051 [DOI] [PubMed] [Google Scholar]
- Miyazono K.I., Ohno Y., Wada H., Ito T., Fukatsu Y., Kurisaki A., Asashima M., and Tanokura M.. 2018. Structural basis for receptor-regulated SMAD recognition by MAN1. Nucleic Acids Res. 46:12139–12153. 10.1093/nar/gky925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm S., Wenkert D., Zhang X., McAlister W.H., Mier R.J., and Whyte M.P.. 2007. Deactivating germline mutations in LEMD3 cause osteopoikilosis and Buschke-Ollendorff syndrome, but not sporadic melorheostosis. J. Bone Miner. Res. 22:243–250. 10.1359/jbmr.061102 [DOI] [PubMed] [Google Scholar]
- Muñoz N.M., Baek J.Y., and Grady W.M.. 2008. TGF-beta has paradoxical and context dependent effects on proliferation and anoikis in human colorectal cancer cell lines. Growth Factors. 26:254–262. 10.1080/08977190802291667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J.M., Behringer R.R., and de Crombrugghe B.. 2002. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 108:17–29. 10.1016/S0092-8674(01)00622-5 [DOI] [PubMed] [Google Scholar]
- Naora H. 1999. Involvement of ribosomal proteins in regulating cell growth and apoptosis: translational modulation or recruitment for extraribosomal activity? Immunol. Cell Biol. 77:197–205. 10.1046/j.1440-1711.1999.00816.x [DOI] [PubMed] [Google Scholar]
- Pezzini A., Del Zotto E., Giossi A., Volonghi I., Costa P., and Padovani A.. 2012. Transforming growth factor β signaling perturbation in the Loeys-Dietz syndrome. Curr. Med. Chem. 19:454–460. 10.2174/092986712803414286 [DOI] [PubMed] [Google Scholar]
- Qing J., Liu C., Choy L., Wu R.Y., Pagano J.S., and Derynck R.. 2004. Transforming growth factor beta/Smad3 signaling regulates IRF-7 function and transcriptional activation of the beta interferon promoter. Mol. Cell. Biol. 24:1411–1425. 10.1128/MCB.24.3.1411-1425.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi R.A., Andrianifahanana M., Wilkes M.C., Edens M., Kottom T.J., Blenis J., and Leof E.B.. 2009. Distinct roles for mammalian target of rapamycin complexes in the fibroblast response to transforming growth factor-beta. Cancer Res. 69:84–93. 10.1158/0008-5472.CAN-08-2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschger P., Fratzl P., Eschberger J., and Klaushofer K.. 1998. Validation of quantitative backscattered electron imaging for the measurement of mineral density distribution in human bone biopsies. Bone. 23:319–326. 10.1016/S8756-3282(98)00112-4 [DOI] [PubMed] [Google Scholar]
- Roschger P., Paschalis E.P., Fratzl P., and Klaushofer K.. 2008. Bone mineralization density distribution in health and disease. Bone. 42:456–466. 10.1016/j.bone.2007.10.021 [DOI] [PubMed] [Google Scholar]
- Sartori R., Milan G., Patron M., Mammucari C., Blaauw B., Abraham R., and Sandri M.. 2009. Smad2 and 3 transcription factors control muscle mass in adulthood. Am. J. Physiol. Cell Physiol. 296:C1248–C1257. 10.1152/ajpcell.00104.2009 [DOI] [PubMed] [Google Scholar]
- Schepers D., Tortora G., Morisaki H., MacCarrick G., Lindsay M., Liang D., Mehta S.G., Hague J., Verhagen J., van de Laar I., et al. 2018. A mutation update on the LDS-associated genes TGFB2/3 and SMAD2/3. Hum. Mutat. 39:621–634. 10.1002/humu.23407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakabe K., Terasawa K., Miyama K., Shibuya H., and Nishida E.. 2001. Regulation of the activity of the transcription factor Runx2 by two homeobox proteins, Msx2 and Dlx5. Genes Cells. 6:851–856. 10.1046/j.1365-2443.2001.00466.x [DOI] [PubMed] [Google Scholar]
- Soltanoff C.S., Yang S., Chen W., and Li Y.P.. 2009. Signaling networks that control the lineage commitment and differentiation of bone cells. Crit. Rev. Eukaryot. Gene Expr. 19:1–46. 10.1615/CritRevEukarGeneExpr.v19.i1.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa H., Kaji H., Yamaguchi T., Sugimoto T., and Chihara K.. 2002. Smad3 promotes alkaline phosphatase activity and mineralization of osteoblastic MC3T3-E1 cells. J. Bone Miner. Res. 17:1190–1199. 10.1359/jbmr.2002.17.7.1190 [DOI] [PubMed] [Google Scholar]
- Steinbaugh M.J., Pantano L., Kirchner R.D., Barrera V., Chapman B.A., Piper M.E., Mistry M., Khetani R.S., Rutherford K.D., Hofmann O., et al. 2018. bcbioRNASeq: R package for bcbio RNA-seq analysis. F1000 Res. 6:1976 10.12688/f1000research.12093.2 [DOI] [Google Scholar]
- van de Laar I.M., Oldenburg R.A., Pals G., Roos-Hesselink J.W., de Graaf B.M., Verhagen J.M., Hoedemaekers Y.M., Willemsen R., Severijnen L.A., Venselaar H., et al. 2011. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat. Genet. 43:121–126. 10.1038/ng.744 [DOI] [PubMed] [Google Scholar]
- van der Pluijm I., van Vliet N., von der Thusen J.H., Robertus J.L., Ridwan Y., van Heijningen P.M., van Thiel B.S., Vermeij M., Hoeks S.E., Buijs-Offerman R.M.G.B., et al. 2016. Defective Connective Tissue Remodeling in Smad3 Mice Leads to Accelerated Aneurysmal Growth Through Disturbed Downstream TGF-β Signaling. EBioMedicine. 12:280–294. 10.1016/j.ebiom.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte M.P., Griffith M., Trani L., Mumm S., Gottesman G.S., McAlister W.H., Krysiak K., Lesurf R., Skidmore Z.L., Campbell K.M., et al. 2017. Melorheostosis: Exome sequencing of an associated dermatosis implicates postzygotic mosaicism of mutated KRAS. Bone. 101:145–155. 10.1016/j.bone.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie A.O., Tang Z., Elanko N., Walsh S., Twigg S.R., Hurst J.A., Wall S.A., Chrzanowska K.H., and Maxson R.E. Jr. 2000. Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nat. Genet. 24:387–390. 10.1038/74224 [DOI] [PubMed] [Google Scholar]
- Wu L., and Derynck R.. 2009. Essential role of TGF-beta signaling in glucose-induced cell hypertrophy. Dev. Cell. 17:35–48. 10.1016/j.devcel.2009.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.W., Hu M., Chai J., Seoane J., Huse M., Li C., Rigotti D.J., Kyin S., Muir T.W., Fairman R., et al. 2001. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol. Cell. 8:1277–1289. 10.1016/S1097-2765(01)00421-X [DOI] [PubMed] [Google Scholar]
- Wu M., Chen G., and Li Y.P.. 2016. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 4:16009 10.1038/boneres.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Higgs B.W., Morehouse C., de Los Reyes M., Trigona W., Brohawn P., White W., Zhang J., White B., Coyle A.J., et al. 2009. Development of Potential Pharmacodynamic and Diagnostic Markers for Anti-IFN-α Monoclonal Antibody Trials in Systemic Lupus Erythematosus. Hum. Genomics Proteomics. 2009:374312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., and He Q.Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R.A. 2003. The double life of ribosomal proteins. Cell. 115:130–132. 10.1016/S0092-8674(03)00804-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
summarizes clinical findings on SMAD3 mutation–positive melorheostosis patients.
summarizes histomorphometry and qBEI analyses in melorheostosis bone.
shows β-catenin (CTNNB1) upregulation and changes of downstream target genes in affected cells of Melo-11.
lists up-regulation of interferon-inducible signature genes (21 genes) in affected cells of melorheostosis (Melo-11, SMAD3, p.S264Y).
lists the genes in each cluster generated by degPatterns analysis of RNA-Seq data of BMP2(−) condition.
lists the genes in each cluster generated by degPatterns analysis of RNA-Seq data of BMP2(+) condition.
lists enriched pathway terms depicted as an interaction network for each cluster generated through the Metascape analysis (BMP2[−] condition).
lists enriched pathway terms depicted as an interaction network for each cluster generated through the Metascape analysis (BMP2[+] condition).