The role of somatic SMAD3 mutations in melorheostosis pathogenesis
Abstract
In the current issue of JEM, Kang et al. (https://doi.org/10.1084/jem.20191499) describe somatic mutations in the SMAD3 gene causing endosteal melorheostosis. Using osteoblast models, the identified mutations are demonstrated to exert a gain-of-function mechanism, augmenting transforming growth factor (TGF) β signaling. These findings provide further insights into the genetic etiology of melorheostosis and consolidate the importance of the TGFβ pathway in skeletal disorders.
Melorheostosis is a rare, sporadic disorder that asymmetrically affects the bones of the limbs and adjacent soft tissues. The disease is characterized by excessive bone formation on the bone surface in the classic “dripping candle wax” pattern, leading to limb deformities, pain, and functional impairment. For a long time, the disease etiology remained largely elusive. A first breakthrough, about 15 yr ago, linked germline loss-of-function (LOF) mutations in LEMD3, encoding for an inner nuclear membrane protein MAN1, to the osteopoikilosis-associated melorheostosis phenotype as well as Buschke-Ollendorf syndrome (Hellemans et al., 2004). Over time, it was increasingly recognized that a somatic “second hit” mutation is necessary to evoke melorheostosis in LEMD3 mutation carriers. It took until 2017 to identify such a second hit. A somatic mutation in the KRAS gene, coding for a RAS/MAPK pathway component, was described in affected tissues from a syndromic LEMD3 mutation carrier presenting with melorheostosis (Whyte et al., 2017). Non-syndromic sporadic melorheostosis patients still remained genetically unexplained, though. Most recently, Kang et al. (2018) showed that MAP2K1 postzygotic mutations explain about half of the sporadic melorheostosis cases. In the latter study, it was observed that a subgroup of MAP2K1-negative patients had a distinct endosteal pattern of melorheostosis, with hyperostosis being restricted to the cortical boundaries of the bone.

Insights from Aline Verstraeten, Joe Davis Velchev, and Bart Loeys.
The current work of Kang et al. in this issue of JEM is an exciting continuation of this previous study. Based on a somatic hit hypothesis, whole-genome sequencing of affected and unaffected bone segments of endosteal MAP2K1-negative melorheostosis patients was performed to find novel genes. After variant filtering and gene prioritization, Kang et al. (2020a) discovered that one patient carried an interesting novel SMAD3 p.Ser264Tyr (c.791C>A) variant at 24% variant allele frequency. Using droplet digital PCR and an amplicon-based targeted sequencing assay, three additional somatic mutation carriers (variant allele frequency range: 1.7–8.2%) were identified, all affecting the identical p.Ser264 amino acid residue and leading either to SMAD3 p.Ser264Tyr (c.791C>A) or p.Ser264Phe (c.791C>T). Both variations were absent from healthy population databases, predicted to be pathogenic by prediction software tools and located within the functionally important MH2 domain. The recurrence and presence of variants at the identical amino acid residue suggested a gain-of-function (GOF) mechanism.
To elucidate the downstream consequences of the identified SMAD3 missense variants, the authors used a multitude of functional studies. First, increased mineralization was shown in affected tissue of SMAD3 mutation carriers compared to paired unaffected bone and MAP2K1-positive affected bone. Second, in TGFβ-stimulated primary osteoblasts derived from affected tissue and a murine osteoblast cell line (MC3T3-E1) overexpressing Smad3 p.Ser264Tyr, markedly higher levels of phosphorylated SMAD3 (pSMAD3) as well as a corresponding increase in the expression of the TGFβ/SMAD target genes, was demonstrated. Remarkably, the increase in nuclear localization was proven to be independent of TGFβ stimulation. These findings confirm the suggested GOF mechanism underlying these SMAD3 mutations (p.Ser264Tyr and p.Ser264Phe). Third, to further unravel the cellular pathways and processes involved in SMAD3-related melorheostosis, Kang et al. (2020a) performed RNA sequencing on SMAD3 mutant patient cells undergoing osteogenic differentiation. Besides the TGFβ pathway, markers for osteogenesis (e.g., Osterix) as well as β-catenin (i.e., a key mediator of the Wnt canonical signaling pathway) were found to be significantly up-regulated in affected cells. Genes associated with the interferon signaling pathway were also enriched in affected cells, which is in accordance with previous findings demonstrating that SMAD3 regulates the expression of certain interferon genes (Qing et al., 2004). Finally, upon BMP2 stimulation of patient cells, osteogenic dampening was observed, suggesting that constitutive activation of the TGFβ signaling cascade may disproportionally inhibit BMP-responsive gene expression and, hence, osteogenesis. Future studies are needed to fully decipher the interactions between TGFβ and BMP signaling, which can potentially lead to the development of medical treatments for SMAD3-related melorheostosis.
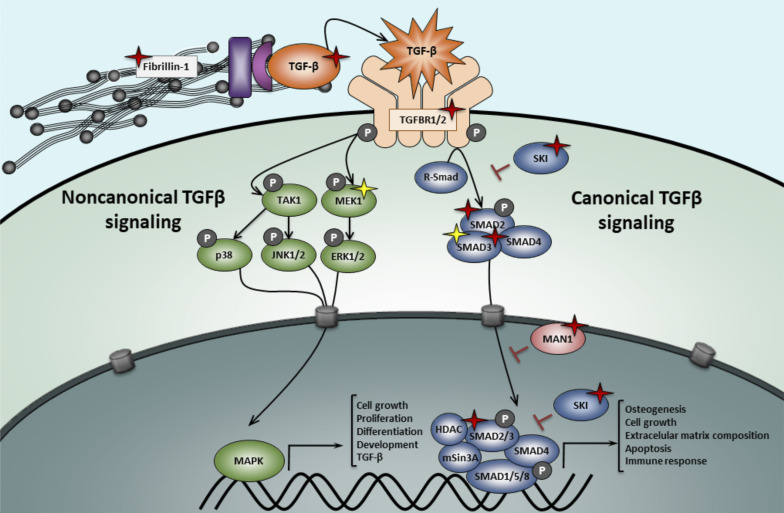
With the identification of somatic SMAD3 GOF mutations, a firm role for dysregulated TGFβ signaling in the etiology of melorheostosis is now established. All currently identified melorheostosis genes can be connected to the TGFβ pathway in some way. MAN1 (encoded by LEMD3) inhibits TGFβ signaling by binding to both SMAD2 and SMAD3. LOF LEMD3 mutations result in excessive TGFβ signaling, demonstrated by significantly elevated nuclear pSMAD2/3 levels (Cohen et al., 2007). KRAS is also known to crosstalk with TGFβ signaling. More precisely, Ras acts upstream of TGFβ-related activation of ERKs, autoinducing the SMAD pathway (Mulder, 2000). Finally, MAP2K1 has been shown to be an important regulator of SMAD3 expression (Ross et al., 2007) and activates the noncanonical TGFβ signaling cascade through ERK activation.
Components of the canonical and noncanonical TGFβ signaling pathway. Germline LOF mutations are represented with red star symbols. Somatic activating mutations are indicated with a yellow star symbol. In MFS patients, mutations in FBN1 (fibrillin-1) lead to an up-regulation of both canonical and noncanonical TGFβ pathways due to an uncontrolled release of TGFβ ligands. LOF mutations in the TGFβ ligands, receptors, or R-Smad effectors lead to a paradoxical increase in TGFβ signaling in LDS patients. Haploinsufficiency in LEMD3 (MAN1), a TGFβ-signaling antagonist, cause osteopoikilosis and Buschke-Ollendorf syndrome. Activating mutations in MAP2K1 (MEK1) and SMAD3 cause distinct types of melorheostosis.
Germline LOF SMAD3 mutations (i.e., truncating or missense mutations affecting the functionally important MH1/MH2 domains) are a well-established cause of Loeys-Dietz syndrome (LDS, type 3; van de Laar et al., 2011), an autosomal dominant connective tissue disorder characterized by hypertelorism, bifid uvula and/or cleft palate, and arterial tortuosity with widespread aortic/arterial aneurysm and/or dissection, as well as skeletal findings including joint laxity and pectus deformity. As one could predict, the GOF SMAD3 mutations identified by Kang et al. (2020a) have not been described in LDS patients, suggesting that LDS and melorheostosis are caused by opposite types of mutations (i.e., somatic GOF versus germline LOF). Besides SMAD3, mutations in five more genes involved in the canonical TGFβ signaling pathway (TGFΒR1, TGFΒR2, TGFΒ2, TGFΒ3, SMAD2) cause LDS (Verstraeten et al., 2017). Intriguingly, LDS-causing LOF mutations in the genes coding for these TGFβ ligands, receptors, and downstream effectors lead to a paradoxical increase in signaling at the tissue level, represented by an increase in pSMAD2/3 levels in aortic wall tissue. Different pathomechanisms have been suggested to explain this paradox, including a compensatory overdrive of the noncanonical MAPK/ERK pathway, a shift in TGFβ ligands, and non-cell autonomous signaling driven by intrinsic differences in sensitivity to TGFβ signaling between aortic cell types that are adjacent in the vessel wall but derived from different embryonic progenitors (Lindsay and Dietz, 2011). Losartan, an antihypertensive drug with TGFβ-antagonizing capacities, has been shown to perform at least equally well as β-blockers with respect to attenuation of arterial aneurysm progression in LDS and related syndromic presentations (i.e., Marfan syndrome [MFS]; Kang et al., 2020b). Given the fact that increased TGFβ signaling has been linked to melorheostosis, exploration of the use of TGFβ-blocking agents such as losartan in the treatment of melorheostosis patients might represent an interesting therapeutic avenue.
SMAD3 is not the only gene in which germline and somatic LOF and/or GOF mutations have been linked to phenotypically very distinct disorders. SMAD4 and GNAS are two other informative examples. Germline LOF SMAD4 mutations lead to juvenile polyposis, predisposing affected individuals to gastrointestinal cancer, and somatic LOF SMAD4 mutations are found in about half of the pancreatic cancers and 15% of colorectal cancers (Maurice et al., 2001). On the other hand, specific GOF SMAD4 mutations (at amino acids p.Arg496 and p.Ile500) are associated with Myhre syndrome, a skeletal dysplasia characterized by short stature, cognitive deficits with behavioral problems, and deafness (Caputo et al., 2014). Inactivating germline mutations in GNAS lead to Albright’s hereditary osteodystrophy, characterized by obesity, short stature, subcutaneous ossification, and mental retardation. Activating somatic GNAS mutations, on the other hand, cause McCune-Albright syndrome, which comes with fibrous dysplasia, café-au-lait pigmentation, and precocious puberty (Turan and Bastepe, 2015). Similar to SMAD3 GOF mutations, GNAS-activating mutations have not been reported to be vertically transmitted and are believed to be incompatible with life due to embryonic lethality. Two important essons can be derived from these observations. First, upon filtering of next-generation sequencing data for genetic diseases, it is very important to keep an open mind and even consider genes that have previously been linked to a very distinct disorder as candidate genes for the phenotype under investigation. Second, somatic mutations are now being implicated in a growing number of diseases. Their identification through next-generation sequencing approaches can be confounded by the extremely low variant frequency. Higher-depth sequencing, as well as more dedicated bioinformatics algorithms for somatic mosaicism, will be key for improved mutation detection sensitivity and specificity.
In syndromes caused by germline mutations in the TGFβ-related pathway genes, such as MFS and LDS, the most common cause of morbidity and mortality is the presence of aortic aneurysms leading to sudden dissections and rupture. These events are associated with a loss of extracellular matrix homeostasis and disturbed mechano-sensing due to altered matrix-vascular smooth muscle cell interaction. Mutations in these genes also explain non-syndromic presentations of thoracic aortic aneurysm and dissection. However, there is still a significant proportion of individuals (up to 70%) presenting with non-syndromic aneurysms that are not genetically explained (Verstraeten et al., 2017). By analogy to the bone phenotypes, this might indicate the possibility that these patients bear a somatic mutation in the aortic tissue affecting either the TGFβ or MAPK/ERK pathways. Although it may be less obvious to obtain unaffected aortic tissue, whole genome sequencing–based comparison between blood (unaffected) and aortic tissue (affected) may be an interesting avenue for further research in genetically unresolved aortic aneurysm and dissection patients.
References
- Caputo, V., et al. 2014. Am. J. Med. Genet. A. 10.1002/ajmg.a.36544 [DOI] [Google Scholar]
- Cohen, T.V., et al. 2007. Development. 10.1242/dev.02816 [DOI] [Google Scholar]
- Hellemans, J., et al. 2004. Nat. Genet. 10.1038/ng1453 [DOI] [PubMed] [Google Scholar]
- Kang, H., et al. 2018. Nat. Commun. 10.1038/s41467-018-03720-z [DOI] [Google Scholar]
- Lindsay, M.E., and Dietz H.C.. 2011. Nature. 10.1038/nature10145 [DOI] [Google Scholar]
- Maurice, D., et al. 2001. J. Biol. Chem. 10.1074/jbc.M105895200 [DOI] [Google Scholar]
- Mulder, K.M. 2000. Cytokine Growth Factor Rev. 10.1016/s1359-6101(99)00026-x [DOI] [PubMed] [Google Scholar]
- Qing, J., et al. 2004. Mol. Cell. Biol. 10.1128/MCB.24.3.1411-1425.2004 [DOI] [Google Scholar]
- Ross, K.R., et al. 2007. Cell. Signal. 10.1016/j.cellsig.2006.11.008 [DOI] [PubMed] [Google Scholar]
- Turan, S., and Bastepe M.. 2015. Curr. Osteoporos. Rep. 10.1007/s11914-015-0268-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H., et al. 2020a. J. Exp. Med. 10.1084/jem.20191499 [DOI] [Google Scholar]
- Kang, Y.-N., et al. 2020b. J. Formos. Med. Assoc. 10.1016/j.jfma.2019.03.018 [DOI] [Google Scholar]
- van de Laar, I.M., et al. 2011. Nat. Genet. 10.1038/ng.744 [DOI] [Google Scholar]
- Verstraeten, A., et al. 2017. Nat. Rev. Cardiol. 10.1038/nrcardio.2016.211 [DOI] [PubMed] [Google Scholar]
- Whyte, M.P., et al. 2017. Bone. 10.1016/j.bone.2017.04.010 [DOI] [PubMed] [Google Scholar]