Abstract
Over the past few years, long non-coding RNAs (lncRNAs) are recognized as key regulators of gene expression at chromatin, transcriptional and posttranscriptional level with pivotal roles in various biological and pathological processes, including cancer. Hypoxia, a common feature of the tumor microenvironment, profoundly affects gene expression and is tightly associated with cancer progression. Upon tumor hypoxia, the central regulator HIF (hypoxia-inducible factor) is upregulated and orchestrates transcription reprogramming, contributing to aggressive phenotypes in numerous cancers. Not surprisingly, lncRNAs are also transcriptional targets of HIF and serve as effectors of hypoxia response. Indeed, the number of hypoxia-associated lncRNAs (HALs) identified has risen sharply, illustrating the expanding roles of lncRNAs in hypoxia signaling cascade and responses. Moreover, through extra-cellular vesicles, lncRNAs could transmit hypoxia responses between cancer cells and the associated microenvironment. Notably, the aberrantly expressed cellular or exosomal HALs can serve as potential prognostic markers and therapeutic targets. In this review, we provide an update of the current knowledge about the expression, involvement and potential clinical impact of lncRNAs in tumor hypoxia, with special focus on their unique molecular regulation of HIF cascade and hypoxia-induced malignant progression.
Keywords: Tumor hypoxia, Long non-coding RNA, lncRNA, HIF-1α, Hypoxia-associated lncRNAs, HAL, Extracellular vesicles
Background
Hypoxia-associated lncRNAs (HALs) emerging as newly driving factors in tumorigenesis
In rapidly growing solid tumors, hypoxia is a common, microenvironmental characteristics, caused by insufficient vascularization, and the high tumor metabolic demands [1]. Accumulating evidence has demonstrated that tumor hypoxia is involved in the initial oncogenic transformation, but is also tightly linked to aggressive cancer phenotypes, such as metastases, recurrences and resistance to therapy [2–4]. Upon hypoxia, to survive, cancer cells co-opt the fundamental adaptive responses to this stress through modulating the central mediator of hypoxic response, the hypoxia-inducible factor-1 (HIF-1) complex.
The HIF-1 complex is a heterodimeric assembly of bHLH-PAS (basic helix-loop-helix DNA binding proteins of the PER-ARNT-SIM family) transcriptional factors, comprised of a constitutively expressed, stable HIF-1β subunit and an oxygen-sensitive HIF-1α subunit that determines HIF-1 activity [5, 6]. In mammals, two HIF-1α homologs, HIF-2α and HIF-3α (also known as IPAS-1; inhibitory PAS (Per/Arnt/Sim) domain protein), have been identified. Similar to HIF-1α, HIF-2α is also sensitive to oxygen concentration and can interact with HIF-1β to form the HIF-2 heterodimeric complex. Due to the structural similarity in DNA binding and dimerization domains as well as the difference in their transactivation domains, HIF-1α and HIF-2α regulate both common as well as distinct sets of target genes. Meanwhile, HIF-3α, an isoform lacking the transactivation domain, has a dominant negative effect on HIF-dependent gene transcription [7, 8].
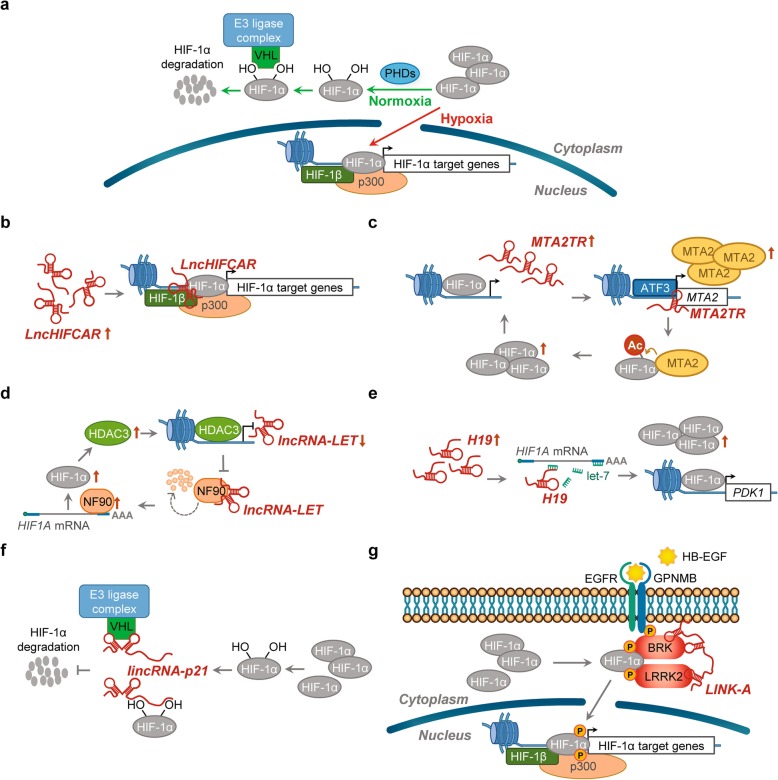
In the presence of sufficient oxygen, HIF-1α subunits are post-translationally modified by a family of dioxygenases (prolyl hydroxylase domain-containing dioxygenases PHD1, 2 and 3, also known as EGLN1-3, Egl-9 family hypoxia inducible factor 1-3,). Upon hydroxylation, HIF-1α subunits are recognized by the E3 ubiquitin ligase, VHL (von Hippel-Lindau tumor suppressor protein), leading to the poly-ubiquitination and subsequent rapid degradation through the ubiquitin-proteasome pathway (Fig. 1a). Under hypoxic conditions, the PHD dioxygenase activity is inhibited, and the accumulated HIF-1α subunit translocates into the nucleus, dimerizing with HIF-1β and binding to the HREs (hypoxia response elements; the consensus 5′-(A/G)CGTG-3′ nucleotide sequence) within the promoter regions of HIF target genes to stimulate downstream transcriptional activation of multiple hypoxia responsive genes (Fig. 1a), eliciting a wide spectrum of cellular adaptations, such as decreased apoptosis, enhanced angiogenesis, proliferation, migration and invasion [1, 9–11]. In addition to protein coding genes, it has been widely acknowledged that the non-coding transcriptome is also responsive to hypoxia and play critical roles in the hypoxic response and HIF-1 associated cancer progression [12–16].
Fig. 1.
Regulations of HIF-1 activity by HALs. a Regulation of HIF-1. Under normoxia (green arrows), HIF-1α subunit is hydroxylated by PHDs (prolyl hydroxylase domain proteins). Hydroxylation residues within HIF-1α facilitates interaction of HIF-1α with the E3 ubiquitin ligase VHL protein, targeting HIF-1α for polyubiquitination and subsequent proteasome-dependent degradation. Upon hypoxia (red arrows), the PHDs and other prolyl hydroxylases are inhibited, leading to HIF-1α stabilization and translocation into nucleus. After dimerization with its transcriptional partner HIF-1β and recruitment of co-activators (e.g. CBP/p300), the HIF-1 heterodimer binds the HRE (hypoxia response element) of target genes to regulate transcription. b Transcriptional co-activator. Hypoxia-induced LncHIFCAR could directly interact with HIF-1α and facilitate the recruitment of HIF-1α and p300 cofactor to the target loci, thereby upregulating HIF-1 target genes. c Recruitment of transcription factor. HIF-1α-induced LncRNA-MTA2TR could recruit ATF3 to the promoter area of MTA2, thereby transcriptionally upregulating the expression of oncogenic MTA2. MTA2 can subsequently enhance HIF-1α protein accumulation via deacetylation, forming a feedback loop to amplify HIF-1 signaling. d mRNA stability control. The expression of lncRNA-LET is repressed through hypoxia-induced HDAC3, which reduces the histone H3 and H4 acetylation at the LncRNA-LET promoter. Decreased lncRNA-LET expression reduces the lncRNA-LET–mediated degradation of HIF-1α negative regulator, NF90, leading to HIF-1α accumulation. e ceRNA/miRNA sponge. Hypoxia-induced H19 could upregulate HIF-1α expression by absorbing miRNA let-7 and nullifying let-7-mediated HIF1A mRNA suppression. f Molecular decoy. lincRNA-p21 is able to disrupt the interaction between HIF-1α and its negative regulator VHL via separate binding to both HIF-1α and VHL, thereby blocking VHL-dependent HIF-1α degradation. g Complex scaffold. LINK-A-mediated recruitment and enzymatic activation of BRK and LRRK2 kinases could facilitate phosphorylation of HIF-1α at specific residues. These phosphorylation modifications prevent subsequent HIF-1α degradation and enhance the association between HIF-1α and cofactor p300, thereby upregulating HIF-1 target genes. See text for a more detailed discussion
With recent advances in high-throughput sequencing, it is recognized that only a small fraction (< 2%) of the transcriptional output encodes proteins whereas the vast majority encode a variety of non-coding RNAs. Among these non-coding RNA species, long (> 200 bp) non-coding RNAs (lncRNAs) are a large class of regulatory transcripts [17], including lincRNAs (long intergenic RNAs), long intronic ncRNAs, pseudogenes, TCRs (transcribed ultra-conserved regions), asRNAs (antisense RNAs) and eRNAs (enhancer RNAs) [18]. According to the latest human genome annotation (GRch38, GENCODE release 33, January 2020; www.gencodegenes.org), 48,438 transcripts originating from 17,952 loci were identified as lncRNAs. Although less than 1% has been functionally annotated, growing evidence suggested the vital roles of these lncRNAs in regulation of gene expression at various stages, such as imprinting, transcription, RNA interference, RNA splicing, and translation control [19–23]. It is now believed that the distinctive RNA biochemical properties, such as base-pairing ability, dynamic expression and flexible structure, endow these lncRNAs with multi-functionality [24–28]. Collectively, it is now well appreciated that, through acting as signals, decoys, guides or scaffolds, lncRNA could act as a crucial player of biological regulation [23–25, 27, 29–33].
Over the last few years, a large number of dysregulated lncRNAs have been associated with numerous diseases, including cancer [34–37]. While a few cancer-associated lncRNAs have been well characterized [27, 38], the functions of most remain largely unknown. Dysregulation of many cancer-associated lncRNAs is linked to both clinicopathological features and survival outcomes of patients, suggesting that functional annotation of these lncRNAs will eventually identify new venues for early diagnosis and therapy of cancer [39]. Several studies have shown that the modulation of lncRNAs in response to hypoxia could play a regulatory role in HIF signaling cascade [14–16, 40, 41]. Here, we refer to these unique transcripts as “hypoxia-associated lncRNAs” (HALs). These RNA molecules are involved in multiple hypoxia-driven cancer progression pathways. In this review, we provide an updated summary of the tumor HALs, with a specific emphasis on the crosstalk between these lncRNA species and cellular hypoxia response (Table 1 and Additional file 1: Table S1). We address current models describing the functional involvement of these new players in cancer progression, highlighting their relevant clinical potential as cancer biomarkers or therapeutic targets. Our discussion is centered on tumor hypoxia. For the functional roles of lncRNAs in hypoxia-induced kidney/hepatic/myocardial injury and neuromuscular or cardiovascular diseases, interested readers are referred to a number of comprehensive reviews published in recent years [127–132].
Table 1.
| HAL-mediated HIF signaling control and cancer progression
| lncRNA | Status upon hypoxia | HIF involvement | Cancer Types | Clinical association | Functional Impact | Interactor | Target/Effect | Mechanistic Classification | Refs |
|---|---|---|---|---|---|---|---|---|---|
|
aHIF (HIF1A-AS2) |
Not further induced in nonpapillary disease, but can be induced in lymphocytes |
N.D. (2 Putative HREs) |
Renal carcinoma | • Up-regulated in non-papillary clear-cell renal carcinoma | N.D. | HIF1A mRNA | HIF1A mRNA stability |
mRNA stability control (Binding of HIF1A-AS2 to the HIF1A mRNA 3′-UTR could possibly expose AU-rich elements and thus increase the degradation of HIF1A mRNA) |
[42, 43] |
| Up-regulated | N.D. | Human umbilical vein endothelial cells (HUVECs) | • Up-regulated in HUVECs in hypoxia |
HUVECs viability ↑ Migration ability ↑ Tube formation ↑ |
miR-153-3p | The expression of HIF-1α |
Sequestration of miRNAs (Down-regulation of miR-153-3p-mediated repression of HIF-1α expression) |
[44] | |
| Up-regulated | N.D. | Bladder cancer | • Upregulated in bladder cancer after cisplatin treatment | Cisplatin resistance ↑ | N.D. | Promoting HMGA1 expression |
Transcriptional regulation (HIF1A-AS2 promoting the expression of HMGA1, which physically interacts with p53, p63, and p73, and therefore inhibits their transcriptional activity on Bax) |
[45] | |
| Up-regulated |
HIF-1α and/or HIF-2α dependent (2 HREs identified) |
Mesenchymal Glioblastoma Stem-like Cells (M-GSCs) |
• Upregulated in M-GSCs |
Growth of M-GSCs ↑ Neurosphere-forming capacity of M-GSCs ↑ Glioblastoma tumor growth ↑ |
IGF2BP2 and DHX9 | Maintenance of expression of HMGA1 |
Complex scaffold (The direct interaction among HIF1A-AS2, IGF2BP2 and DHX9 is needed for HMGA1 expression) |
[46, 47] | |
| Up-regulated | N.D. | Epithelial ovarian cancer (EOC) | • Up-regulated in EOC |
Cell apoptosis ↓ Cell proliferation ↑ Tumorigenesis ↑ Tumor growth ↑ |
N.D. | N.D. |
Unclear mechanism (May partially through the aHIF-mediated regulation of certain key mitochondrial apoptosis pathway-related genes, including Bcl-2, Bax, Caspase-7, and Caspase-9) |
[48] | |
| AGAP2-AS1 | Up-regulated | N.D. | Hepatocellular carcinoma (HCC) |
• Up-regulated in HCC • Correlated with adverse clinical features and poor prognosis of HCC |
Cell proliferation ↑ Migration and invasion ↑ EMT progression ↑ Apoptosis ↓ |
miR-16-5p | The expression of ANXA11 |
Sequestration of miRNAs (Down-regulation of miR-16-5p-mediated repression of ANXA11) |
[49] |
| ANRIL (CDKN2B-AS1) | Up-regulated |
HIF-1α dependent (1 HRE identified) |
Osteosarcoma | • Up-regulated in osteosarcoma |
Hypoxic viability ↑ Hypoxia-induced Invasion ↑ Hypoxia-induced apoptosis ↓ |
N.D. | N.D. |
Unclear mechanism (Possibly through epigenetic modification) |
[50] |
| BC005927 | Up-regulated |
HIF-1α dependent (2 HREs identified) |
Gastric cancer (GC) |
• Up-regulated in GC • Correlated with higher tumor-node-metastasis stages and poorer prognoses |
Metastasis ↑ | N.D. | N.D. |
Transcriptional regulation (The neighboring gene, EPHB4, a metastasis-related gene, is regulated by BC005927) |
[51] |
| BX111887 (ZEBTR) | Up-regulated |
HIF-1α dependent (1 HRE identified) |
Pancreatic cancer (PC) |
• Upregulated in PC • Correlated with late TNM stage, lymphatic invasion and distant metastasis |
Proliferation ↑ Migration ↑ Invasion ↑ |
YB1 | ZEB1 promoter |
Transcriptional regulation (BX111 promotes ZEB1 transcription by recruiting YB1 to ZEB1 promoter) |
[52] |
| CASC9 | N.D. | N.D. | Nasopharyngeal carcinoma (NPC) | Up-regulated in NPC tissues |
Glycolysis and tumorigenesis ↑ Cell growth ↑ |
HIF-1α | The stability of HIF-1α |
Protein Stability (CASC9 interacts with HIF-1α and enhances the stabilization of HIF-1α) |
[53] |
|
CF129 (lncRNA-CF129145.1) |
Down-regulated | Downregulated by binding of HIF-1α/HDAC1 complex to CF129 promoter | Pancreatic cancer (PC) |
• Down-regulated in PC • Low CF129 expression predicted short overall survival |
Invasion and metastasis ↓ | p53 and E3 ligase MKRN1 | FOXC2 transcription |
Post-Translational modification (CF129 directly binds to p53 and E3 ligase MKRN1, inducing p53 protein ubiquitination and degradation, and thereby suppressing FOXC2 transcription) |
[54] |
| CPS1-IT1 |
Down-regulated (treatment of hypoxia mimetic, CoCl2) |
N.D. | Colorectal cancer | Down-regulated in colorectal cancer | EMT and autophagy ↓ | N.D. | N.D. |
Unclear mechanism (May partially through suppressing expression levels of HIF-1α, LC3-I, LC3-II, Beclin-1 and EMT associated proteins under hypoxia) |
[55] |
|
CRPAT4 (RP11-225B17) |
Down-regulated | HIF-1α dependent, HIF-2α independent |
Clear cell renal cell carcinoma (ccRCC) |
• Up-regulated in ccRCC • Associated with poor overall survival and progression-free survival |
Cell migration ↑ Proliferation ↑ |
N.D. | N.D. |
Unclear mechanism (May partially through the CRPAT4-mediated regulation of migration-associated gene AVL9 expression) |
[56] |
| DANCR | N.D. | N.D. | Nasopharyngeal carcinoma (NPC) |
• Up-regulated in NPC • Associated with poor prognosis |
Metastasis ↑ Invasion ↑ |
NF90/NF45 complex |
HIF-1α mRNA stability |
mRNA stability control (DANCR could increase HIF-1α mRNA stability through interacting with the NF90/NF45 complex) |
[57] |
| DARS-AS1 | Up-regulated | HIF-1α dependent, But HIF-2α independent (2 HREs identified) | Myeloma |
• Up-regulated in myeloma • Correlated with poor prognosis |
Survival ↑ Tumorigenesis ↑ |
RBM39 | RBM39 stability |
Post-Translational modification (The interaction between DARS-AS1 and RNA-binding protein 39 (RBM39) impedes the interaction between RBM39 and its E3 ubiquitin ligase RNF147, preventing RBM39 from degradation) |
[58] |
| EIF3J-AS1 (EIF3J-DT) | Up-regulated | N.D. | Hepatocellular carcinoma (HCC) |
• Up-regulated in HCC tissues • Correlated with tumor size, vascular invasion, tumor stage and poor prognosis |
Cell proliferation ↑ Migration ↑ Invasion ↑ |
miR-122-5p | The expression of CTNND2 |
Sequestration of miRNAs (Down-regulation of miR-122-5p-mediated repression of CTNND2) |
[59] |
| ENST00000480739 (RPL13AP23) | N.D. | N.D. |
Pancreatic ductal adenocarcinoma (PDAC) |
• Down-regulated in PDAC • Associated with tumor node metastasis (TNM) stage and lymph node metastasis • Independent risk factor for PDAC survival following surgery |
Invasion ↓ OS-9 mRNA & protein ↑ |
N.D. | Transcription of OS-9 (Negative regulation of HIF-1α) |
Epigenetic and transcriptional regulation (ENST00000480739 induces OS-9 expression at the transcriptional level, possibly through modifying the H3K27 acetylation level of OS9 gene promoter) |
[60] |
| FALEC | Up-regulated | HIF-1α inducible | Prostate cancer (PCa) |
• Up-regulated in PCa • Independent prognostic factor |
Cell proliferation ↑ Migration and invasion ↑ |
N.D. | N.D. |
Unclear mechanism (May partially through the FALEC-mediated regulation of p21 and its downstream components expression) |
[61] |
| FAM201A | N.D. | N.D. |
Non-small cell lung cancer (NSCLC) |
• Up-regulated in tissues obtained from NSCLC patients resistant to radiotherapy |
Cell proliferation ↑ Apoptosis (under X-ray irradiation) ↓ |
miR-370 | The expression of EGFR |
Sequestration of miRNAs (Down-regulation of miR-370-mediated repression of EGFR) |
[62] |
| FEZF1-AS1 | N.D. | N.D. | Pancreatic cancer | • Upregulated in pancreatic cancer |
Cell proliferation ↑ Invasion ↑ |
miR-142 and miR-133a | The expression of HIF-1α and EGFR |
Sequestration of miRNAs (Down-regulation of miR-142- and miR-133a-mediated repression of HIF-1α and EGFR expression) |
[63] |
| GAPLINC | Up-regulated |
HIF-1α (2 HREs identified) (2 HREs) |
Gastric cancer |
• Upregulated in GC • High expression of GAPLINC correlates with poorer survival • GAPLINC correlates with CD44 activation |
Proliferation ↑ Apoptosis ↓ Invasion ↑ Migration ↑ |
miR-211-3p | The expression of CD44 |
Sequestration of miRNAs (Down-regulation of miR-211-3p-mediated repression of CD44) |
[64, 65] |
| H19 | Up-regulated | N.D. | Breast cancer stem cells (BCSCs) | • H19 expression strongly correlates with PDK1 in primary breast carcinomas |
Glycolysis ↑ BCSC maintenance ↑ |
let-7 | The expression of HIF-1α |
Sequestration of miRNAs (Down-regulation of let-7-mediated repression of HIF-1α expression) |
[66] |
| Up-regulated | N.D. | Multiple Myeloma (MM) | N.D. |
The expression of the hypoxia induced genes ↑ Adhesion on stromal cells ↑ |
N.D. | N.D. |
HIF-1α nuclear translocation (H19 is required for HIF-1α nuclear translocation and the expression of the hypoxia-induced genes, such as CXCR4 and Snail) |
[67] | |
| Up-regulated |
HIF-1α dependent (3 HREs identified) |
Glioblastoma (GBM) |
• Up-regulated in GBM • Correlated with poor prognosis • The HIF-1α levels were positively correlated with H19 levels in GBM specimens |
Migration and invasion ↑ Tumor growth ↑ EMT ↑ |
miR-181d | The expression of β-catenin |
Sequestration of miRNAs (Down-regulation of miR-181d-mediated repression of β-catenin expression) |
[68–71] | |
| Up-regulated | N.D. | Prostate Cancer |
• Upregulated by estrogen or hypoxia • Reduced upon combined treatment |
Cell motility ↓ Invasion ↓ |
N.D. | Repression of beta3 and beta4 Integrins |
Unclear mechanism (Combined Estrogen and Hypoxia treatment could cause H19 down-regulation, followed by up-regulation of both β3 and β4 Integrins and E-cadherin) |
[72] | |
| Up-regulated | N.D. | Breast cancer, Non-small cell lung carcinoma (NSCLC) |
• Up-regulated in NSCLC with chronic obstructive pulmonary disease (COPD) • Up-regulated in all common metastatic sites tested |
Migration and invasion ↑ Tumor growth ↑ EMT ↑ |
N.D. | Up-regulation of miR-675-5p |
Unclear mechanism (H19 could induce upregulation of miR-675-5p, whereas P53 is a target gene of miR-675-5p and P53 downstream target genes involved in EMT, survival and tumorigenesis are thereby repressed) |
[73, 74] | |
| HAS2-AS1 | Up-regulated | HIF-1α dependent (1 HRE identified) | Oral squamous cell carcinoma (OSCC) | • Up-regulated in OSCC | EMT ↑ | N.D. | N.D. |
Unclear mechanism (HAS2-AS1-mediated hypoxia-induced EMT is dependent on cell-adhesion molecule CD44 and RHAMM) |
[75] |
| HIF2PUT | N.D. | N.D. | Osteosarcoma | • Expression of HIF2PUT is correlated with HIF2A mRNA |
Cell proliferation and migration ↓ Expression of CSC marker CD133 ↓ Sphere-forming ability ↓ |
N.D. | Transcription of HIF2A |
Transcriptional regulation (HIF-2α was positively regulated by lncRNA HIF2PUT) |
[76] |
| N.D, | N.D. | Osteosarcoma cancer stem cell |
• Down-regulated in osteosarcoma cell lines • A strong positive correlation between relative HIF2PUT and HIF-2α level in osteosarcoma cancer tissues |
Proliferation ↓ Migration and invasion ↓ Sphere-formation ↓ |
N.D. | N.D. |
Unclear mechanism (May partly through HIF2PUT-mediated regulation of HIF-2 expression) |
[77] | |
| HINCUT-1 (uc.475) | Up-regulated |
HIF-1α dependent (3 HREs identified) |
Colon and breast cancer cell lines | N.D. | Hypoxic cell proliferation ↑ | N.D. | N.D. |
Transcriptional regulation (HINCUT-1 is required for the expression of OGT mRNA expression and global O-GlcNAcylation of proteins) |
[78] |
| HOTAIR | N.D. | N.D. | Renal cell carcinoma | • Upregulated and correlated with tumor progression |
RCC proliferation ↑ Migration and EMT ↑ Apoptosis ↓ |
miR-217 | The expression of HIF-1α/AXL |
Sequestration of miRNAs (Down-regulation of miR-217-mediated repression of HIF-1α/AXL expression) |
[79] |
| Up-regulated |
HIF-1α dependent (1 HRE identified) |
Non-small cell lung carcinoma (NSCLC) | • High level of HOTAIR is associated with poor clinical outcome in multiple cancers |
Cell proliferation under hypoxia ↑ Invasion & migration under hypoxia ↑ Apoptosis under hypoxia ↓ |
N.D. | N.D. |
Unclear mechanism (Possibly through HOTAOR-mediated epigenetic modification) |
[80, 81] | |
| HOTTIP | Up-regulated | HIF-1α dependent | Glioma |
• Up-regulated in glioma • Associated with metastasis and poor patient survival |
EMT ↑ Invasion ↑ Migration ↑ |
miR-101 | The expression of ZEB1 |
Sequestration of miRNAs (Down-regulation of miR-101-mediated repression of ZEB1) |
[82] |
| IDH1-AS1 | N.D. |
N.D. (c-Myc-mediated repression) |
Multiple cell lines (HeLa, HCT116, H1299, P493 and 293 T) | N.D. | Glycolysis ↓ | IDH1 | IDH1 dimerization |
Protein Dimerization (IDH1-AS1 interacts with IDH1 and promotes Its Homo-dimerization) |
[83] |
| LINC01436 | Up-regulated | N.D. |
Non-small cell lung cancer (NSCLC) |
• Up-regulated in NSCLC • Associated with poor overall survival |
Cell growth ↑ Migration and invasion ↑ |
miR-30a-3p | The expression of EPAS1 |
Sequestration of miRNAs (Down-regulation of miR-30a-3p-mediated repression of EPAS1) |
[84] |
| lincRNA-p21 (TP53COR1) | Up-regulated |
HIF-1α dependent & preference (2 HREs identified) |
Cervical, lung and breast cancer cell lines | N.D. |
Hypoxic glycolysis ↑ Tumor growth ↑ |
HIF-1α and VHL | The disruption of the VHL-HIF-1α interaction |
Protein-Protein Interaction Decoy (Stabilization of HIF-1α by disrupting the VHL-HIF-1α Interaction) |
[85] |
| Up-regulated | N.D. | Hepatoma, glioma | N.D. |
Apoptosis ↓ Cell proliferation and motility ↑ Autophagy ↑ |
N.D. | N.D. |
Unclear mechanism (LincRNA-p21 could promote autophagy of hypoxic tumor cells by up-regulating HIF-1α protein levels and suppressing Akt/mTOR/P70S6K signaling pathways) |
[86] | |
| linc-ROR | Up-regulated | N.D. | Hepatocellular cancer | Up-regulated in malignant liver cancer cells |
Cell viability during hypoxia ↑ Tumor growth ↑ |
miR-145 | The expression of p70S6K1 (RPS6KB1) |
Sequestration of miRNAs (Down-regulation of miR145-mediated repression of p70S6K1 expression) |
[87] |
|
LINK-A (LINC01139) |
N.D. | N.D. | Triple-negative breast cancer |
• Upregulated in TNBC • High levels of LINK-A correlated with unfavorable recurrence-free survival for breast cancer patients |
Glycolysis ↑ Tumor growth ↑ |
BRK and LRRK2 kinase | HIF-1α phosphorylation |
Complex scaffold (LINK-A facilitates the recruitment of BRK and LRRK2 kinase activation, thereby causing HIF-1α stabilization, HIF-1α/p300 interaction, and activation of HIF-1α transcriptional programs under normoxic conditions) |
[88] |
| LncHIFCAR (MIR31HG) | Up-regulated | HIF-1α dependent | Oral cancer |
• Up-regulated in oral cancer • High levels of LncHIFCAR predicted worse overall survival and recurrence-free survival |
Hypoxic glycolysis ↑ Tumor metastasis ↑ Invasion and migration ↑ Hypoxic cell proliferation ↑ Sphere-forming ability ↑ |
HIF-1α | Activation of HIF-1 signaling |
Transcriptional regulation (LncHIFCAR acts as HIF-1α coactivator) |
[89] |
| lncRNA-AK058003 | Up-regulated | N.D. | Gastric cancer | Up-regulated in GC |
Invasion & migration ↑ Metastasis ↑ |
N.D. | N.D. |
Epigenetic regulation (AK058003 expression is positively correlated with SNCG expression and SNCG promoter demethylation) |
[90] |
| lncRNA-EFNA3 | Up-regulated |
HIF-1α dependent (1 HRE identified) |
Breast cancer | A strong correlation between high EFNA3 expression and shorter metastasis-free survival in breast cancer patients |
Cell extravasation ↑ Metastatic dissemination ↑ |
miR-210 | The expression of EFNA3 |
Sequestration of miRNAs (Down-regulation of miR-210-mediated repression of EFNA3) |
[91] |
|
lncRNA-HAL (lnc-METTL16-2) |
Up-regulated |
HIF-1α dependent (3 putative HREs found) |
Breast cancer | Up-regulated in triple negative breast cancer |
Migration ↑ Cancer stem cell phenotype ↑ Mammospheres ↑ Clonogenic growth ↑ |
Histones and hnRNPs. | N.D. |
Unclear mechanism (The binding of lncRNA-HAL to histones and hnRNPs may suggest a participation at the chromatin level and transcriptional regulation) |
[92] |
| lncRNA-LET (NPTN-IT1) | Down-regulated |
HIF-1α dependent (Indirect: Histone deacetylation) |
Lung squamous-cell cancer (LSCC), hepatocellular carcinoma (HCC) and colorectal cancer (CRC) |
• Down-regulated in in LSCC, HCC and CRC • Correlated with hypoxia, histone acetylation disorder and metastasis in HCC |
Metastasis ↓ Invasion ↓ |
NF90 (RNA-binding protein) | HIF1A mRNA stability |
mRNA stability control (The association between lncRNA-LET and NF90 protein enhanced the degradation of NF90, thereby decreasing HIF1A mRNA) |
[93] |
|
lncRNA-SARCC (lnc-P2RY1-1) |
VHL-dependent |
HIF-2α dependent (1 HRE identified) |
Renal cell carcinoma | Differentially regulated by hypoxia in a von Hippel-Lindau (VHL)-dependent manner in RCC clinical specimens |
Hypoxic cell cycle progression (VHL-restored RCC cells) ↑ Hypoxic cell cycle progression (VHL-mutant RCC cells) ↓ |
AR (androgen receptor) | AR ubiquitination and degradation |
Post-Translational modification (lncRNA-SARCC could promote AR degradation via ubiquitin-mediated proteolysis to suppress AR/HIF-2α/C-MYC signals) |
[94] |
| lncTCF7 (WSPAR) | Up-regulated | N.D. | Glioma |
• Up-regulated in glioma • Associated with WHO grade and tumor size |
Cell migration ↑ Proliferation ↑ Tumorigenicity ↑ |
N.D. | N.D. |
Unclear mechanism (LncTCF7 could promote the migration and proliferation of glioma cell partially through activating the Wnt signalling pathway) |
[95] |
| MALAT1 | Up-regulated |
HIF-2α dependent & preference (1HRE) |
Hepatocellular carcinoma | N.D. |
Cell growth ↑ Glycolysis ↑ Migration & invasion ↑ Vasculature formation ↑ Metastasis ↑ |
N.D. | N.D. |
Post-Translational modification (MALAT1 decreases hydroxylation of HIF-1α/HIF-2α, possibly through disassociation of the VHL protein from HIF-1α/HIF-2α) |
[96, 97] |
| Up-regulated | N.D. | Lung adenocarcinoma | N.D. |
Proliferation ↑ Migration ↑ Invasion ↑ |
PTB-associated splicing factor (PSF) | GAGE6 promoter |
Transcriptional regulation (The physical interaction of MALAT1 and PSF released the binding of PSF to GAGE6 promoter) |
[98, 99] | |
| Up-regulated | N.D. | Hepatocellular carcinoma | N.D. |
Proliferation ↑ Migration and invasion ↑ Apoptosis ↓ |
miR-200a | N.D. |
Sequestration of miRNAs (Down-regulation of miR-200a) |
[100] | |
| MEG3 | Up-regulated | N.D. | Pheochromocytoma | N.D. | Hypoxia-induced PC12 cell injury ↑ | Methylation proteins (DNMT3a, DNMT3b, and MBD1) | TIMP2 promoter methylation |
Epigenetic regulation (MEG3 recruited methylation proteins DNMT3a, DNMT3b, and MBD1 and accelerated TIMP2 promoter methylation, which in turn inhibited its expression) |
[101] |
| MTA2TR | Up-regulated |
HIF-1α dependent (1 HRE identified) |
Pancreatic cancer (PC) | Upregulated in PC tissues |
Cell proliferation ↑ Invasion ↑ |
Activating transcription factor 3 (ATF3) |
The expression of MTA2 (MTA2 stabilizes the HIF-1α via deacetylation) |
Transcriptional regulation (MTA2TR transcriptionally upregulates MTA2 expression by recruiting ATF3 to the promoter area of MTA2) |
[102] |
| NEAT1 | Up-regulated | HIF-2α dependent | Non-small cell lung cancer (NSCLC) |
• Up-regulated in NSCLC • Associated with TNM stage and metastasis |
Cell proliferation ↑ Migration and invasion ↑ |
miR-101-3p | SOX9/Wnt/β-catenin signaling pathway |
Sequestration of miRNAs (Down-regulation of miR-101-3p-mediated repression of SOX9/Wnt/β-catenin signaling pathway) |
[103] |
| Up-regulated | HIF-2α dependent & preference | Breast cancer | High expression of NEAT1 is associated with poor survival of breast cancer patients |
Proliferation ↑ Apoptosis ↓ Clonogenic survival ↑ Paraspeckle formation ↑ |
N.D. | N.D. |
Complex scaffold (Induces paraspeckle formation, thereby enhancing cancer cell survival in hypoxia) |
[12, 104–106] | |
|
NDRG-OT1 (lnc-NDRG1-1) |
Up-regulated | N.D. | Breast cancer | • N.D. | N.D. | NDRG1 | NDRG1 degradation |
Post-Translational modification (NDRG-OT1 could promote NDRG1 degradation via ubiquitin-mediated proteolysis) |
[107] |
| NORAD | Up-regulated | N.D. | Pancreatic cancer (PC) |
• Upregulated in PC • Correlated with shorter overall survival |
Migration ↑ Invasion ↑ EMT ↑ Metastasis ↑ |
miR-125a-3p | The expression of RhoA |
Sequestration of miRNAs (Down-regulation of miR-125a-3p-mediated repression of RhoA) |
[108] |
|
NUTF2P3-001 (NUTF2P3) |
Up-regulated |
HIF-1α dependent (1 HRE identified) |
Pancreatic cancer |
• Upregulated in pancreatic cancer • A positive correlation between NUTF2P3 and KRAS • Associated with tumor stage and prognosis |
Cell viability, proliferation ↑ Invasion ↑ KRAS expression ↑ Metastasis ↑ |
miR-3923 | The expression of KRAS |
Sequestration of miRNAs (Down-regulation of miR-3923-mediated repression of KRAS) |
[109] |
| PCGEM1 | Up-regulated | N.D. | Gastric cancer (GC) | Up-regulated in GC | Invasion and metastasis ↑ | N.D. | N.D. |
Unclear mechanism (Partially through regulating SNAI1, a key transcription factor of EMT) |
[110] |
| PVT1 | N.D. | N.D. | Nasopharyngeal carcinoma (NPC) |
• Up-regulated in NPC • Up-regulation is associated with a poor prognosis in NPC patients |
NPC cell proliferation ↑ Colony formation ↑ In vivo tumorigenesis ↑ |
KAT2A (chromatin modification factor) | Transcription of NF90 (RNA-binding protein) |
Epigenetic regulation (PVT1 serves as a scaffold for KAT2A, which mediates H3K9 acetylation, recruiting the nuclear receptor binding protein TIF1β to activate NF90 transcription, thereby increasing HIF-1α mRNA stability) |
[111] |
| N.D. | N.D. | Hepatocellular carcinoma (HCC) | Up-regulated in HCC tissues and cell lines |
Cell proliferation ↑ Migration ↑ Invasion and iron uptake ↑ Apoptosis ↓ |
miR-150 | The expression of HIG2 (Hypoxia-inducible protein 2) |
Sequestration of miRNAs (Down-regulation of miR-150-mediated repression of HIG2) |
[112] | |
| N.D. | N.D. | Gastric cancer |
• Upregulated in GC tissues and cell lines • High expression levels correlated with advanced tumor stage and lymph node metastasis |
GC cell proliferation ↑ GC cell invasion ↑ |
miR-186 | The expression of HIF-1α |
Sequestration of miRNAs (Down-regulation of miR-186-mediated repression of HIF-1α expression) |
[113] | |
| Up-regulated | N.D. | Non-small cell lung cancer (NSCLC) |
• Up-regulated in HIF-1α high group compared with HIF-1α low group • Negatively correlated with miR-199a-5p expression in NSCLC tissues |
Cell proliferation ↑ | miR-199a-5p | The expression of HIF-1α |
Sequestration of miRNAs (Down-regulation of miR-199a-5p-mediated repression of HIF-1α expression) |
[114] | |
|
Up-regulated treatment of hypoxia mimetic CoCl2) |
N.D. | Cervical Cancer |
• Up-regulated in Cervical cancer • Correlates with poorer overall survival |
Cell proliferation ↑ Migration and invasion ↑ Apoptosis ↓ Cisplatin resistance ↑ |
N.D. | N.D. |
Unclear mechanism (Possible involvement of the interaction with nucleolin) |
[115] | |
|
RERT-lncRNA (RAB4B-EGLN2) |
N.D. | N.D. |
Hepatocellular carcinoma (HCC) |
The expression levels of RERT-lncRNA and EGLN2 were significantly correlated in HCC | EGLN2 expression ↑ | N.D. | N.D. |
Transcriptional regulation (RERT-lncRNA induces EGLN2/PHD1 expression at the transcriptional level) |
[116] |
| UBE2CP3 | N.D. | N.D. | Hepatocellular carcinoma (HCC) |
• Up-regulated in HCC, especially in high EV (endothelial vessel) density tissues • UBE2CP3 expression combined with EV density is associated with HCC patient prognosis |
Proliferation ↑ Migration ↑ Tube formation ↑ |
N.D. | N.D. |
Unclear mechanism (May partially through UBE2CP3-induced increase in the secretion of VEGFA into the supernatant via activation of the ERK/HIF-1α signaling pathway) |
[117] |
| UCA1 | Up-regulated | HIF-1α-dependent | Estrogen receptor (ER)-positive breast cancer | N.D. | Tamoxifen resistance ↑ | miR-18a | The expression of HIF-1α |
Sequestration of miRNAs (Down-regulation of miR-18a-mediated repression of HIF-1α expression) |
[118] |
| Up-regulated | N.D. | Hypoxia-resistant gastric cancer (HRGC) | Upregulated in HRGC cells | Migration ↑ | miR-7-5p | The expression of EGFR |
Sequestration of miRNAs (Down-regulation of miR-7-5p-mediated repression of EGFR) |
[119] | |
| Up-regulated | N.D. | Acute myeloid leukemia (AML) | Upregulated following ADR (adriamycin)-based chemotherapy |
Cytotoxic effect of ADR ↓ HIF-1α-dependent glycolysis ↑ |
miR-125a | The expression of HK2 |
Sequestration of miRNAs (Down-regulation of miR-125a-mediated repression of HK2) |
[120] | |
| Up-regulated |
HIF-1α dependent (2 HREs) |
Bladder cancer |
• Upregulated in bladder cancer • UCA1 expression associated with the clinical stage and histologic grade of bladder cancer |
Cell proliferation under hypoxia ↑ Invasion & migration under hypoxia ↑ Apoptosis under hypoxia ↓ |
N.D. | N.D. |
Unclear mechanism (UCA1 could modulate the expression of several genes involved in tumorigenic potential, drug resistance and embryonic development) |
[121, 122] | |
| Up-regulated |
HIF-1α dependent (1 HRE identified) |
Osteosarcoma | N.D. | Cell growth ↑ | N.D. | N.D. |
Unclear mechanism (May partially through inactivating the PTEN/AKT signaling pathway) |
[123] | |
| WT1-AS | Up-regulated |
HIF-1 dependent (DNA demethylation of the CpG island) |
Myeloid Leukemia |
• Upregulated in Wilms’ tumors • Aberrant WT1-AS splicing often found in acute myeloid leukemia |
N.D. | N.D. | N.D. |
Epigenetic regulation (WT1-AS mediates hypoxia-induced WT-1 mRNA upregulation through modulating histone methylation) |
[124, 125] |
| ZEB2-AS1 | Up-regulated | HIF-1α dependent |
Gastric cancer (GC) |
• Upregulated in GC • Correlated with poor differentiation, lymph node metastasis and distant metastasis |
Cell proliferation and growth ↑ Invasion ↑ In vivo tumor growth ↑ |
miR-143-5p | The expression of HIF-1α |
Sequestration of miRNAs (Down-regulation of miR-143-5p-mediated repression of HIF-1α expression) |
[126] |
Abbreviation: CRC colorectal cancer, CSC cancer stem cell, EMT epithelial–mesenchymal transition, GC Gastric cancer, HCC hepatocellular cancer, HRE hypoxia response element, HUVECs human umbilical vein endothelial cells, ICC Immunocytochemistry, LC lung cancer, M-GSCs Mesenchymal glioblastoma multiforme stem-like cells, N.D. Not determined, NSCLC non-small cell lung carcinoma, OSCC Oral squamous cell carcinoma, PDAC pancreatic ductal adenocarcinoma, RCC Renal Cell Carcinoma, RNP ribonucleic protein, TNM tumor, node, metastasis, VHL von Hippel-Lindau protein, WHO World Health Organization
Review
LncRNAs as emerging driving forces in cancer progression upon tumor hypoxia
Given the pivotal roles of lncRNA in hypoxia-associated tumorigenesis pathways, multiple approaches have been applied in the identification of hypoxia-regulated lncRNAs [87, 90]. A comprehensive analysis coupling RNA-seq with ChIP-seq [12] revealed the extensive involvement of HIF-1α and HIF-2α in the transcriptional regulation of lncRNAs upon hypoxia. In recent years, the rapid expansion of research on lncRNAs has provided additional insights into those associated with cellular hypoxia response. Table 1 presents an updated list of these hypoxia-associated lncRNAs (HALs). Upon hypoxia, most HALs are up-regulated. HIF could directly promote the expression of these hypoxia-inducible lncRNAs through binding to the HREs (hypoxia response elements) located in their promoter (Table 1) [41]. lncRNA-LET [93], CF129 [54] and CRPAT4 [56] are among the few which are down-regulated in hypoxic conditions. Notably, lncRNA-SARCC is able to respond to hypoxic stress differentially in a VHL-dependent manner [94].
Most of the HALs identified have impacts on cancer progression, although the mechanistic details are not all clear. Table 1 shows an overview of the tumor HALs. We summarize in the table, their potential molecular target related to hypoxic responses as well as their reported functions and signaling pathways. These HALs may also have hypoxia-independent functions. For the sake of conciseness, those targets are not included in the table. In addition, some of these lncRNAs can be captured by exosomes and transmitted to tumor microenvironment to exert their functions and further propagate the hypoxic responses (Table 2). Notably, several HALs, such as UCA1, PVT1, H19 and MALAT1, might adapt more than one action mode in different cancer types. In the discussion below, we highlight the selected few HALs to illustrate their mechanisms of actions.
Table 2.
| HALs identified extracellularly
| LncRNA | Extracellular space identified | Cell to Cell Transfer | Functional Impact | Mechanism | Ref |
|---|---|---|---|---|---|
|
aHIF (HIF1A-AS2) |
Serum (aHIF level in serum correlates with its expression in matched ectopic endometria) |
Endometriotic cyst stromal cells (ECSCs)-derived exosomes to human umbilical vein endothelial cells (HUVECs) | Elicits proangiogenic behavior in HUVECs, thus facilitating endometriosis angiogenesis. | Activates VEGF-A, VEGF-D, and b-FGF in HUVECs | [133] |
| CCAT2 | Exosomes secreted from cultured glioma cells | U87-MG glioma cells to HUVECs | Promotes HUVEC angiogenesis and inhibits apoptosis induced by hypoxia | Promotes VEGF-A, TGF-β and Bcl2 expression. Inhibits BAX and caspase 3 expression | [134] |
| HISLA (LINC01146) | Extracellular vesicles secreted by tumor associated fibroblasts (TAMs) | TAMs to breast cancer cells | Enhances aerobic glycolysis and apoptotic resistance of cancer cells | Stabilizes HIF-1α | [135] |
| PVT1 | Exosomes secreted from cultured colon cancer cells. Cancer cells with more aggressive phenotypes have more extracellular PVT1 | Not determined | Promotes cell proliferation and inhibits apoptosis. | [136] | |
| linc-ROR | Exosomes secreted from cultured hepatocellular carcinoma cells | HCC cancer cells to cancer cells | Promotes cell survival of recipient cells | Through a miR-145–HIF-1α signaling module to increase HIF-1α expression | [87] |
| UCA1 | Exosomes secreted from cultured bladder cancer cells & serum | Bladder cancer 5637 cells with high expression of UCA1 to bladder cancer UMUC2 cells with low expression of UCA1 |
Promotes cell proliferation, migration and invasion of recipient cells Promotes xenograft growth |
Through regulating the expression of genes involved in EMT (E-cad, MMP9, vimentin) | [137] |
HAL-mediated epigenetic and transcriptional regulation
A large number of lncRNAs are localized in the nucleus, participating in various biological processes, including chromatin organization, nuclear structure, transcriptional and post-transcriptional regulation of gene expression. With regard to chromatin organization, the pangenomic investigations of RNA–protein interactions have shown that two hypoxia-inducible, oncogenic antisense RNAs ANRIL (also known as CDKN2B antisense RNA 1) and HOTAIR (HOX transcript antisense RNA) [50, 80] could interact with different histone-modifying complexes, and have thus been proposed to impact the chromatin modification and transcriptional state [138]. However, whether these two antisense RNAs are involved in modulating gene expression in response to hypoxia via epigenetic modification or chromatin re-organization remains to be characterized. In addition, WT1-AS could mediate hypoxia-induced upregulation of oncogenic transcription factor WT-1 in cis through modulating histone H3K4 and H3K9 methylation around the transcription start site of WT1 mRNA, contributing to acute myeloid leukemia (AML) progression [124]. Similarly, in gastric cancer, lncRNA-AK058003, which could be profoundly induced by hypoxia, resides upstream of SNCG (synuclein gamma, a synuclein family member, promotes migration, invasion and metastasis) and enhances SNCG expression in cis through demethylation of SNCG promoter CpG islands, thereby driving hypoxia-induced metastasis [90]. In the context of nasopharyngeal carcinoma (NPC), up-regulated PVT1 could serve as a scaffold for a transcriptional activator, the histone acetyltransferase KAT2A, to activate transcription of NF90. NF90, a RNA-binding protein, has been reported to stabilize many target mRNAs, including HIF1A mRNA. Indeed, the upregulated NF90 increased HIF1A mRNA stability and promoted malignant transformation of NPC cells [111]. In addition, in hypoxia-injured pheochromocytoma cells, up-regulated MEG3 (maternally expressed gene 3) could recruit methylation proteins DNMT3a, DNMT3b and MBD1 to facilitate TIMP2 promoter methylation, which in turn inhibited the expression of this cell cycle arrest inducer TIMP2. Moreover, a HIF-1α negative regulator, OS-9, is reported to facilitate HIF-1α hydroxylation and subsequent proteasomal degradation through tethering the interaction between HIF-1α and prolyl hydroxylases (PHDs) [139]. Interestingly, in pancreatic ductal adenocarcinoma (PDAC), another lncRNA ENST00000480739 could inhibit HIF-1α by up-regulating OS9 (osteosarcoma amplified-9) expression through enhancing the acetylation of H3K27 within OS9 gene promoter [60]. Of note, in PDAC, the level of ENST00000480739 is markedly downregulated, and negatively correlated with lymph node metastasis, in agreement with its negative regulatory role in HIF-1 signaling [60]. As ENST00000480739 resides upstream of the OS9 promoter region, this lncRNA also act in cis to induce OS9 transcription.
Apart from chromatin structure remodeling, a series of HALs could modulate transcription and thereby fine-tune the HIF network. For instance, lncRNA HIF2PUT (HIF-2α promoter upstream transcript), RERT-lncRNA and hypoxia-inducible BC005927 are all found to act in cis to up-regulate neighboring protein-coding genes HIF2A (encodes HIF-2α), EGLN2 (encodes prolyl hydroxylase PHD1) and EPHB4 (encodes Ephrin type-B receptor 4, a metastasis-related gene), at the transcriptional level, respectively [51, 76, 116].
Moreover, HALs could directly act on specific transcription factors through physical interactions to modulate their transactivation activities. We recently identified a hypoxia-inducible lncRNA LncHIFCAR (long noncoding HIF-1α co-activating RNA, also known as MIR31HG) acting as a HIF-1α co-activator via direct interaction with HIF-1α, thereby enhancing the binding of HIF-1α and cofactor p300 to the target loci (Fig. 1b). As the abundance of the HIF complex increases, the hypoxia-induced HIF-1 signaling cascade is augmented to further promote subsequent cancer progression [89]. Meanwhile, in pancreatic cancer, HIF-1α-induced lncRNA-MTA2TR (MTA2 transcriptional regulator RNA) transcriptionally up-regulates the expression of oncogenic MTA2 (metastasis associated protein 2) by recruiting ATF3 (activating transcription factor 3) to the promoter area of MTA2 [102]. Subsequently, MTA2 can enhance the accumulation of HIF-1α protein via MTA2-mediated HIF-1α deacetylation and stabilization, which further activates HIF-1α transcriptional activity, forming feedback loops to augment HIF-1 signaling [102] (Fig. 1c). In addition, through binding to PSF (PTB-associated splicing factor), hypoxia-induced lncRNA MALAT1 released PSF from its downstream proto-oncogene GAGE6 (proto-oncogene G antigen 6) and activated its transcription, thereby promoting proliferation, migration and invasion of lung adenocarcinoma cells [98, 99]. Given the extraordinary variety of transcriptional regulatory machinery discovered in the cell, it is anticipated that more lncRNAs-mediated regulation on hypoxia-induced transcriptional program will be unraveled in the imminent future.
HAL-mediated post-transcriptional control
HALs also participate in post-transcriptional regulation including mRNA stability and miRNA-mediated gene silencing.
mRNA stability control
Three HALs, lncRNA-LET (Long noncoding RNA Low Expression in Tumor), DANCR (Differentiation Antagonizing Non-Protein Coding RNA) and HIF1A-AS2 (HIF1A Antisense RNA 2; also known as aHIF), have all been reported to affect HIF1A mRNA stability. lncRNA-LET expression is generally suppressed in various types of tumors, whereas hypoxia-induced HDAC3 (histone deacetylase 3) could repress its expression by reducing the histone acetylation of the lncRNA-LET promoter region [93, 140]. Mechanistically, lncRNA-LET is bound to NF90 (nuclear factor 90), which increases NF90 degradation by the proteasome. As RNA binding protein NF90 could stabilize HIF1A mRNA [93, 141], the downregulation of lncRNA-LET upon hypoxia plays a key role in the stabilization of NF90 protein, thereby increasing HIF-1A mRNA stability upon hypoxia and accordingly hypoxia-induced cancer cell invasion [93] (Fig. 1d). Likewise, in nasopharyngeal carcinoma, another oncogenic lncRNA DANCR was up-regulated and associated with lymph lode metastasis and poor survival [57]. Through interaction with the NF90/NF45 complex, DANCR could increase HIF1A mRNA stability, leading to metastasis and disease progression.
In addition, another hypoxia-inducible antisense lncRNA HIF1A-AS2, was shown to be up-regulated in various tumors [42, 43, 46, 142, 143] and could differentially regulate HIF-1α and HIF-2α expression during long-term hypoxic conditions [43, 47]. Upon acute hypoxia, HIF-1α and HIF-2α were similarly induced. Interestingly, during prolonged hypoxia, these two proteins were differentially regulated as HIF-1α protein level gradually decreased due to a reduction in its mRNA stability, whereas HIF-2α protein remained upregulated. Meanwhile, long-term hypoxia also induced an increase in HIF1A-AS2, whose gene promoter harbors functional HREs. During prolonged hypoxia, HIF1A-AS2 could bind to its sense counterpart, the HIF-1A mRNA 3′-UTR, and possibly expose the AU-rich elements in this region, thereby destabilizing HIF-1A mRNA to convey target gene specificity [43, 47]. Paradoxically, HIF1A-AS2 was also shown to sequester miR153-3p (see next section) to enhance HIF-1A expression [44]. Thus, the mode of action of HIF1A-AS2 is complex and likely context-dependent.
miRNA sponges
A wealth of lncRNAs adapt a well-characterized, common mechanism, “ceRNA (competing endogenous RNA)” or “RNA sponges”, to repress miRNA-mediated gene silencing. The ceRNAs compete for shared miRNAs, sequester these miRNAs and diminish their silencing effect on target mRNAs.
Functional manipulations have demonstrated that several HALs, such as lincRNA-ROR [87], PVT1 [113, 114], HIF1A-AS2 [44], UCA1 [118], HOTAIR [79], FEZF1-AS1 [63], ZEB2-AS1 [126] and H19 [66], could act as a ‘ceRNA’ to reduce individual specific miRNA-mediated HIF1A mRNA destabilization and thereby restoring HIF-1α levels and consequently promote cancer progression (Table 1). Specifically, in breast cancer stem cells, by absorbing endogenous miRNA let-7 and aborting let-7-mediated HIF1A mRNA suppression, hypoxia-induced H19 could stimulate HIF-1α expression [66] (Fig. 1e). In addition, in glioblastoma, hypoxia-induced H19 up-regulation has been shown to confer an aggressive behavior by sequestering miR-181d and nullifying its suppression on an oncogenic EMT-associated factor, β-catenin [68].
In a similar way, certain HALs could act as a ceRNA to modulate other hypoxia-responsive regulators than HIF-1α. In gastric cancer, GAPLINC (Gastric Adenocarcinoma Associated, Positive CD44 Regulator, Long Intergenic Non-Coding RNA) is a HIF-1α direct, transcriptional downstream target, and could promote invasive tumor progression [64]. Mechanistically, GAPLINC could serve as a decoy for miR-211-3p to restore the levels of cancer stem cell marker CD44, enhancing tumor progression [65]. Aside from GAPLINC, NORAD [108], UCA1 [119, 120], HOTTIP [82], EIF3J-AS1 [59], MALAT1 [100], FAM201A [62], AGAP2-AS1 [49], LINC01436 [84], NEAT1 [103], NUTF2P3 [109] lncRNAs were shown to function in this way (Table 1). Collectively, in response to hypoxia, the crosstalk among the lncRNA and miRNA transcriptomes build a reciprocal repression feedback network, eliciting concordant shift to transcriptional reprogram. Further exploration of this pertinent co-working group of lncRNAs and miRNAs under hypoxic conditions would help appreciate this emerging additional layer of post-transcriptional regulation governed by HALs.
HAL-mediated control of protein activity, stability and/or higher-order complex formation
In addition to acting as ceRNAs to modulate gene expression through interaction with miRNAs, HALs have multiple molecular modes to act at the protein level to further modulate gene expression. One of the hypoxia-induced lncRNAs, PVT1 (plasmacytoma variant translocation 1), was implicated in cervical cancer progression, likely through its interaction with a multifunctional shuttling protein, nucleolin [115]. In multiple cancer cell lines, HIF-1-induced lincRNA-p21 provides another example as to how HALs modulate hypoxia response by protein sequestration. Through separate binding to HIF-1α and VHL, lincRNA-p21 could increase HIF-1α accumulation by disruption of the VHL/HIF-1α interaction and subsequent attenuation of VHL-mediated HIF-1α ubiquitination and degradation [85] (Fig. 1f). Another HIF-1α binding lncRNA CASC9 (cancer susceptibility candidate 9) is highly expressed in nasopharyngeal carcinoma (NPC) tissues. CASC9 could interact with and stabilize HIF-1α, promoting the glycolysis and tumorigenesis of NPC cells [53].
Nevertheless, in addition to fine-tuning the activity of one single protein, HALs can also dynamically modulate higher-order protein organizations by serving as scaffolds or molecular decoys. In mesenchymal glioblastoma stem-like cells, through direct binding to two RNA binding proteins, DHX9 (ATP-dependent RNA helicase A) and IGF2BP2 (insulin-like growth factor 2 mRNA-binding protein 2), lncRNA HIF1A-AS2 could facilitate the interaction between this protein complex and their mRNA target HMGA1 (high mobility group AT-hook 1), thereby enhancing HMGA1 expression as well as the downstream molecular response to hypoxic stress [46, 47].
In triple-negative breast cancer (TNBC), LINK-A (long intergenic non-coding RNA for kinase activation) has a critical role in the growth factor-induced HIF-1α signaling under normoxic conditions [88]. LINK-A is required for the recruitment of BRK (breast tumor kinase) and subsequent enzymatic activation, which is stimulated by HB-EGF (Heparin-binding EGF-like growth factor) signal. HB-EGF mediates the heterodimerization of EGFR (epidermal growth factor receptor) and GPNMB (transmembrane glycoprotein NMB) to form ‘EGFR:GPNMB’ complex. Due to its direct interaction with BRK and LRRK2 (leucine-rich repeat kinase 2), LINK-A could recruit these two kinases to EGFR:GPNMB heterodimer, thereby inducing their kinase activities, resulting in HIF-1α phosphorylation: BRK-mediated HIF-1α phosphorylation at Tyr565, a phosphorylation preventing the adjacent Pro564 hydroxylation of HIF-1α and subsequent HIF-1α degradation under normoxic conditions; and LRRK2-mediated HIF-1α phosphorylation at Ser797, which facilitates the interaction of HIF-1α with the transcriptional cofactor p300 [88] (Fig. 1g). In TNBC samples, both LINK-A abundance and HIF-1 signaling activation are correlated with cancer progression and shorter survival, revealing potential therapeutic targets for TNBC [88].
An additional novel function of lncRNAs is their structural role in the assembly of nuclear domains. For instance, MALAT1 (metastasis-associated lung adenocarcinoma transcript 1, also known as NEAT2) and NEAT1 (nuclear enriched abundant transcript 1) are located in two well-characterized nuclear bodies, nuclear speckles and paraspeckles, respectively. Also known as SC35 splicing domains, nuclear speckles are membrane-less compartments and their formation involves “phase-separation” mediated by aggregated lncRNAs and proteins. Being an abundant component of the nuclear speckles, MALAT1 associates with numerous splicing factors and other SR (serine/arginine-rich) proteins, and is required for their correct localization to the nuclear speckles, although the overall nuclear speckle assembly is not dependent on the abundance of MALAT1 [144, 145]. So far, the functional involvement of MALAT1 in RNA splicing in response to hypoxia remains to be determined. In contrast, lncRNA NEAT1 is shown to be an essential architectural component of nuclear paraspeckles [144, 145]. The precise function of paraspeckles remains largely elusive, but proposed to regulate gene expression via the retention of hyper-edited RNA and other multifunctional factors in the nucleus [104]. Given the functional involvement of both MALAT1 and NEAT1 in nuclear structure, further investigation of the extent to which these nuclear structures and their associated transcription reprogramming respond to hypoxia will deepen our understanding of the cellular dynamic response to hypoxia.
HAL-mediated control of hypoxia response via unclear mechanism
As listed in Table 1, most of the HALs identified with profound impact on tumorigenesis have not yet been examined in mechanistic detail. However, other reports regarding the same lncRNA with functional characterization might reveal clues about their biological roles in response to hypoxia. For instance, lncRNA PCGEM1 was found to be overexpressed in gastric cancer, and could be induced by hypoxia [110]. In gastric cancer cells, PCGEM1 could promote the invasion and metastasis through activating the expression of SNAI1, a key transcription factor of EMT, though the underlying mechanism remains elusive [110]. Notably, in prostate cancer, our group previously reported that the oncogenic PCGEM1 could promote chromatin recruitment of c-Myc and enhances its transactivation activity through direct physical interaction [146]. As SNAI1 is a well-characterized downstream gene of c-Myc, the possible functional role of the PCGEM1/c-Myc/SNAI1 signaling axis in hypoxia-associated cancer progression warrants further investigation.
In summary, as noted in the above sections, given the relatively large size and the structural flexibility of lncRNAs, it is to be expected that they interact with multiple RNA or protein components and have multi-functions, perhaps in a context-dependent manner. As such, their roles in hypoxia responses and in tumor progression may differ appreciably in different cancer types.
LncRNAs as predictive biomarkers and therapeutic targets for hypoxic tumor
Extracellular vesicles-containing HALs and their biologic effects on tumorigenesis
Extracellular vesicles are effective devices for transporting biomolecules among various cells types [147, 148]. Based on the difference in size and biogenesis, cell-derived extracellular vesicles can be broadly divided into two main categories: exosomes (30–100 nm in diameter) and microvesicles. Together with proteins and other non-coding RNAs, emerging evidence has shown that lncRNAs are packaged into exosomes [149, 150], and the abundance of lncRNAs in exosomes correlates with their expression level in the cell of origin [151]. Through exosomal transfer, several lncRNAs are shown to potentiate cell responses to hypoxia between cancer cells [87], as well as between cancer cell and the associated microenvironment [150]. Table 2 summarizes hypoxia-associated lncRNAs identified extracellularly. For example, linc-ROR was found abundant in tumor cells as well as in exosomes derived from tumor cells [87]. It is increased both in cells or exosomes during hypoxia, and it up-regulates HIF-1α expression by absorbing miR-145. By co-culture systems, linc-ROR-containing exosomes increase HIF-1a transcription in recipient cells [87]. Hypoxia can shape and fine tune specific macrophage phenotypes in the tumor milieu that are known to promote tumor progression [152]. Chen et al found lncRNA HISLA (also known as LINC01146), secreted by tumor-associated macrophages, stabilized HIF-1α and enhanced aerobic glycolysis in cancer cells, leading to contagious metabolic reprogramming within tumor regions [150]. PVT1, a lncRNA that often co-amplifies with c-myc and functions as miRNA sponge to upregulate HIF-1α expression [153, 154], is another example of exosomal transfer between TAMs (tumor associated macrophages) and cancer cells. PVT1 is detected in exosomes derived from colon cancer cells, particularly in more aggressive cells [136]. In granulocytic myeloid-derived suppressor cells (G-MDSCs), PVT1 was up-regulated by HIF-1α under hypoxia and contributed to immunosuppression, given its depletion reduced the suppression of these cells on T-cells and delayed tumor progression [155]. Other exosomal-transferred lncRNAs that are implicated in cancer cells during hypoxia include UCA1 in bladder cancer for promoting tumor growth and EMT [137], and CCAT2 for glioma’s resistance to apoptosis and angiogenesis [134].
The functions of lncRNAs in exosomes for tumor progression await to be explored given a significant level of non-coding RNAs are revealed in exosomes (and elevated upon hypoxia) whereas only a small fraction has been studied [149, 150, 156]. Accordingly, it is conceivable that multiple tumor phenotypes and signaling pathways are affected upon exosomal loading. Indeed, by microarray analyses, Mao et al showed hundreds of lncRNAs, together with other transcripts, are changed in endothelial cell recipients of exosomes derived from squamous cancer cells [157]. Importantly, they found exosomes obtained from hypoxic condition facilitate angiogenesis and metastasis better than those obtained from normoxic condition in a xenograft model. Similar effects between normoxic exosomes and hypoxic exosomes on angiogenesis were found in a mouse xenograft model of glioblastoma, with additional effect on accelerating tumor expansion at later stage [158]. The elevation in transcripts by exosomes could result from direct gene transfer, or sequential effects mediated by the transferred genes. By which mechanism lncRNAs are selected to be packaged in the exosomes upon stimuli is not known; nevertheless, these studies revealed exosomes as a means by which hypoxia in the tumor microenvironment facilitates tumor cells to spread and progress.
Diagnostic potential of HALs
Several HALs with known oncogenic functions have been detected in patient-derived exosomes, including H19 in serum from patients with bladder cancer [159], HOTAIR in urinary exosomes from patients with urothelial bladder cancer [156], UCA1 in serum from bladder cancer patients [137], and HIF1A-AS2 in patients with endometriosis [133]. Future studies aimed at identifying hypoxia-responsive transcripts in extracellular vesicles would surely reveal more players in this aspect. Bearing differential expression patterns between normal and malignant stages and/or tumor size, oncogenic lncRNAs that can be detected extracellularly would potentially serve as non-invasive biomarkers for early detection, prognosis prediction, and disease surveillance. PCA3, up-regulated in > 90% of men with prostate cancer, is an example of this [160]. A urine-based assay has been approved by the United States Food and Drug Administration (FDA) since 2012 as an alternative diagnostic test for patients undergoing repeat prostate biopsy or with previous negative prostate biopsy.
As described above, there is considerable evidence indicating hypoxia as a progression factor for tumor development [161]. Hypoxia promotes angiogenesis, tumor metastasis, immune evasion and therapy-resistance. The oxygenation status of tumor was reported to influence local tumor response to radiation treatment, as well as overall survival in a variety of tumors [162–164]. Chemotherapeutic drugs, such as Docetaxel and Sorafenib, also tend to be more effective in normoxic conditions [165, 166]. The hypoxic regions in tumors are infiltrated with cells which promote tumor tolerance (regulatory T-cells, myeloid-derived suppressor cells, and macrophages), while antitumor T-cells are devoid and inhibited by HIF-1α-mediated accumulation of extracellular adenosine [167–169]. PD-L1 (Programmed death-ligand 1), a ligand expressed by tumor cells or myeloid-derived suppressor cells to suppress T-cell’s anti-tumor immunity, is up-regulated by and a direct target of HIF-1α during hypoxia [170]. It has become increasingly apparent that hypoxia in tumors fosters immune suppression and prevents effective immunotherapy. Considering the ill-effects of hypoxia, it is important to detect and to overcome tumor hypoxia even before therapy starts, for the best of patient care.
By far, while there has been a great deal of interest in methodologies to measure hypoxia in patients, an efficient, non-invasive, while sensitive method to detect small regions of hypoxia that frequently occur in the tumors is still lacking [163]. A few metabolic markers (HIF-1α, HIF-2, CA9 and GLUT1) have been used to assess low oxygen tensions by immunohistochemistry [171, 172]; however, the application of them in clinic is limited given that their expressions can be triggered by factors other than hypoxia and that biopsies only represent a small sampling of the tumor. As exosome composition mirrors the hypoxia status of tumors [158], a hypoxia signature may be formulated based on the exosomal hypoxia-responsive transcripts including HALs to evaluate oxygenation in the body for clinical exploitation, once our knowledge is advanced.
Therapeutic potential of HALs (targeting hypoxia in cancer therapy, a lncRNA perspective)
Several approaches have been proposed to target hypoxia in tumor [161, 163]. These include drugs that induce cell death selectively in hypoxic cells, e.g. hypoxia-activated prodrugs, or drugs sensitizing hypoxic cells to radiation. Since the adaptive response to hypoxia mainly orients from the transactivation of HIF signaling, some approaches seek to block hypoxia-induced responses by targeting HIFs and the related signaling, or to target pathways that also play pivotal roles in hypoxia adaptation, such as signaling involving mTOR, DNA damage response, and the unfolded protein response. In that regard, HALs that are elevated upon hypoxia and contribute to tumor progression in pre-clinical studies could potentially serve as molecular targets, e.g. PVT, LncHIFCAR, etc. (see Table 1) [41]. By contrast, HALs that are repressed in order to magnify hypoxia response, such as lncRNA-LET, could be induced for therapeutic intervention.
Various strategies have been developed to modulate RNAs. Silencing lncRNAs by small interfering RNAs, antisense oligonucleotides (ASOs), or ribozymes and deoxynucleotides are well demonstrated in pre-clinical studies. Until now, three ASOs and one aptamer therapies have been approved by the FDA for diseases and a handful of others are in clinical trials. The development of short oligonucleotides that fold into three-dimensional structures, aptamers, offers a greater specificity as they target specific structure regions to either mediate RNA degradation or disrupt functional interactions between binding partners [173]. Small molecules that bind to lncRNA and hinder its interaction surface have similar advantages. Additionally, peptide nucleic acids (PNA)-based approach against lncRNAs have been described. HOTAIR-targeting PNAs conjugated with pH-low insertion peptide (pHLIP) successfully delivered the anti-lncRNA to the acidic tumor. It blocked the interaction between HOTAIR and EZH2, subsequently inhibited HOTAIR-EZH2 activity and re-sensitized resistant ovarian tumors to platinum [174].
In any case, an issue all hypoxia-based therapeutics need to consider is the poorly perfused tissue in tumors. In response to their rapid growth, tumor cells secret pro-angiogenic factors such as VEGF to induce vascular formation, yet the constant stimulation leaves tumor vasculature ill-formed and leaky [175]. Simultaneous blockade of HIFs and pro-angiogenic factors has been proposed for targeting tumor hypoxia, in that targeting the angiogenic factors may allow vasculature to mature, resulting more effective blood supply and drug delivery. Another strategy is to relieve oxygen demand by drugs that alleviates oxygen tension in tumors. Papaverine, an FDA-approved drug as a smooth muscle relaxant, was found to inhibit mitochondria complex I and enhance the response to radiotherapy, while well-oxygenated normal tissues were not sensitized [176]. Accordingly, lncRNAs that regulate mitochondria respiration may be considered for targeting tumor hypoxia as an adjuvant treatment.
Conclusion and future perspectives
Decades of intensive scientific research on hypoxia and HIF biology has greatly contributed to our understanding of oxygen homeostasis. Over the past few years, a substantial increase in our knowledge of the noncoding transcriptomes, while putting on an additional layer of complexity in hypoxia regulation and responses, has advanced our comprehension of hypoxic biology. This review has presented an update of our current insights regarding lncRNAs involved in hypoxia-associated processes, highlighting the diverse mechanisms and functions of hypoxia-associated lncRNAs (HALs). These novel action modes unveil the unanticipated predominance of HALs in the regulation of gene expression under hypoxic conditions and outline the elaborate network among the different types of RNA transcripts, chromatin DNA and protein factors. However, advancement in analytical methodologies and in structural and genomic technologies of RNA are required to open up new important directions for in-depth investigation. For the state-of-the-art methodologies to unveil the functions of lncRNAs, readers are directed to two excellent recent reviews [177, 178], as well as those in this special issue.
The role of HIF in hypoxia responses has been the central topic of most investigations. Indeed, HIF has been shown to be a central regulator of the coding and non-coding transcriptome and tightly associated with cancer risk [12, 179–181]. Most HALs, in particular, are highly responsive to hypoxia and HIF and, in turn, participate in the regulation of the protein-coding genome either in cis or in trans to offer multiple routes to HIF-mediated gene regulation, implementing both positive and negative feedback loops that either strengthen or repress the hypoxia response. Most notably, the extracellular vesicles-containing HALs could evoke peculiar response to specific cell population, affecting nearby cells and those at a great distance, diversifying the hypoxia response far beyond the previously recognized. The cellular adaptation to hypoxia requires the precisely coordinated regulatory network to cope with the acute, transient and dynamic oxygen deprivation stress in local regions, whereas lncRNAs, with their flexible structure for interaction and quick biogenesis nature, could be uniquely suited to provide rapid, precise and reversible responses to this insult. It is clear that HALs and their downstream targets are shown to confer a series of biological effective responses to hypoxia. Feasibly, this extensive molecular crosstalk between lncRNA and hypoxic signaling cascades may undergo co-evolution to maintain such an exquisite, orchestrated program. Thus, for a comprehensive understanding of hypoxia-associated tumor biology, it is of relevance to characterize the long non-coding transcriptome involved in hypoxia adaptation.
Given the prominent pathological roles of HALs in hypoxia-associated cancer progression, these RNAs could be exploited as useful indicators to define the cancer intrinsic subtypes to aid in precision medicine. Importantly, HALs are often tissue specific and respond to hypoxia in a cell context dependent manner. As such, they are excellent markers for tissue and tumor hypoxia responses. Compared with other bio-molecules, lncRNAs are ideal biomarkers that provide specificity, stability, sensitivity and easy accessibility [38]. Most notably, cell-free lncRNAs or those packed in extracellular vesicles can be detectable in various body fluids [182]. Hence, the genome-wide annotation of tissue-specific HAL signatures could guide development of promising, non-invasive biomarkers for early diagnosis, prognosis and prediction. Although most lncRNA-targeted treatments are still in their infancy stages, the recent success in RNA-based therapeutics holds promises for future technical innovations. With in-depth characterization of the interplay among hypoxia microenvironment and lncRNA function, more HALs could surely accelerate the design of therapeutics for tumor patients, enabling the targeting of the previously undruggable transcriptome in the near future.
Supplementary information
Additional file 1: Table S1. Hypoxia-associated lncRNAs.
Acknowledgements
None.
Abbreviations
- ASO
Antisense oligonucleotide
- bHLH-PAS
Basic helix-loop-helix DNA binding proteins of the PER-ARNT-SIM family
- BRK
Breast tumor kinase
- ceRNA
competitive endogenous RNA
- CRC
Colorectal cancer
- CSC
Cancer stem cell
- DHX9
ATP-dependent RNA helicase A
- EGFR
epidermal growth factor receptor
- EMT
Epithelial-mesenchymal transition
- FDA
Food and Drug Administration
- GC
Gastric cancer
- G-MDSCs
Granulocytic myeloid-derived suppressor cells
- GPNMB
Transmembrane glycoprotein NMB
- GSC
Glioblastoma stem-like cells
- HB-EGF
Heparin-binding EGF
- HCC
Hepatocellular cancer
- HMGA1
High mobility group AT-hook 1
- HAL
Hypoxia-associated lncRNAs
- HUVECs
Human umbilical vein endothelial cells
- IGF2BP2
Insulin-like growth factor 2 mRNA-binding protein 2
- LC
Lung cancer
- lncRNA
long non-coding RNA
- LRRK2
Leucine-rich repeat kinase 2
- M-GSCs
Mesenchymal glioblastoma multiforme stem-like cells
- ncRNA
non-coding RNA
- N.D.
Not determined
- NF90
Nuclear factor 90
- NSCLC
Non-small cell lung carcinoma
- OGT
O-linked N-acetylglucosamine transferase
- OS-9
Osteosarcoma amplified-9
- OSCC
Oral squamous cell carcinoma
- PDAC
Pancreatic ductal adenocarcinoma
- PD-L1
Programmed death-ligand 1
- pHLIP
pH-low insertion peptide
- PNA
Peptide nucleic acids
- RCC
Renal Cell Carcinoma
- SNCG
Synuclein gamma
- TNBC
Triple-negative breast cancer
- UTR
Untranslated region
- VEGF
Vascular endothelial growth factor
- VHL
Von Hippel-Lindau protein
Authors’ contributions
TCK, JWS and HJK wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by the “TMU Research Center of Cancer Translational Medicine” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (to JWS and HJK). It is also supported by grants MOST108-2321-B-010-013-MY2, MOST108-2320-B-038-011 and MOST107-2320-B-038-055-MY3 from Ministry of Science and Technology of Taiwan (to HJK); TMU106-AE1-B52 from Taipei Medical University and MOST108-2320-B-038-009-MY2 from Ministry of Science and Technology of Taiwan (to JWS). TCK was supported by an independent research scholar grant from Ministry of Science and Technology of Taiwan (MOST 105-2321-B-400-011-MY3), and is indebted to Ting-Feng Tsai (National Yang-Ming University) for her support after the end of the grant.
Availability of data and materials
Not Applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12929-020-00654-x.
References
- 1.Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 2.Liu Q, Liu L, Zhao Y, Zhang J, Wang D, Chen J, He Y, Wu J, Zhang Z, Liu Z. Hypoxia induces genomic DNA demethylation through the activation of HIF-1alpha and transcriptional upregulation of MAT2A in hepatoma cells. Mol Cancer Ther. 2011;10:1113–1123. doi: 10.1158/1535-7163.MCT-10-1010. [DOI] [PubMed] [Google Scholar]
- 3.Jackson AL, Zhou B, Kim WY. HIF, hypoxia and the role of angiogenesis in non-small cell lung cancer. Expert Opin Ther Targets. 2010;14:1047–1057. doi: 10.1517/14728222.2010.511617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison LR, Micha D, Brandenburg M, Simpson KL, Morrow CJ, Denneny O, Hodgkinson C, Yunus Z, Dempsey C, Roberts D, Blackhall F, Makin G, Dive C. Hypoxic human cancer cells are sensitized to BH-3 mimetic-induced apoptosis via downregulation of the Bcl-2 protein Mcl-1. J Clin Invest. 2011;121:1075–1087. doi: 10.1172/JCI43505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makino Y, Kanopka A, Wilson WJ, Tanaka H, Poellinger L. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3alpha locus. J Biol Chem. 2002;277:32405–32408. doi: 10.1074/jbc.C200328200. [DOI] [PubMed] [Google Scholar]
- 8.Maynard MA, Evans AJ, Hosomi T, Hara S, Jewett MA, Ohh M. Human HIF-3alpha4 is a dominant-negative regulator of HIF-1 and is down-regulated in renal cell carcinoma. FASEB J. 2005;19:1396–1406. doi: 10.1096/fj.05-3788com. [DOI] [PubMed] [Google Scholar]
- 9.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhry H, Schodel J, Oikonomopoulos S, Camps C, Grampp S, Harris AL, Ratcliffe PJ, Ragoussis J, Mole DR. Extensive regulation of the non-coding transcriptome by hypoxia: role of HIF in releasing paused RNApol2. EMBO Rep. 2014;15:70–76. doi: 10.1002/embr.201337642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gee HE, Ivan C, Calin GA, Ivan M. HypoxamiRs and cancer: from biology to targeted therapy. Antioxid Redox Signal. 2014;21:1220–1238. doi: 10.1089/ars.2013.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang YN, Zhang K, Hu ZM, Qi HX, Shi ZM, Han XH, Han YW, Hong W. Hypoxia-regulated lncRNAs in cancer. Gene. 2016;575:1–8. doi: 10.1016/j.gene.2015.08.049. [DOI] [PubMed] [Google Scholar]
- 15.Choudhry H, Harris AL, McIntyre A. The tumour hypoxia induced non-coding transcriptome. Mol Aspects Med. 2016;47-48:35–53. doi: 10.1016/j.mam.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Dong J, Xu J, Wang X, Jin B. Influence of the interaction between long noncoding RNAs and hypoxia on tumorigenesis. Tumour Biol. 2016;37:1379–1385. doi: 10.1007/s13277-015-4457-0. [DOI] [PubMed] [Google Scholar]
- 17.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker M. Long noncoding RNAs: the search for function. Nat Methods. 2011;8:379–383. doi: 10.1038/nmeth0511-379. [DOI] [Google Scholar]
- 20.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 23.Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bohmdorfer G, Wierzbicki AT. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 2015;25:623–632. doi: 10.1016/j.tcb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt AM, Chang HY. Long noncoding RNAs in Cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 33.Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7:582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 39.Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34:613–620. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 40.Choudhry H, Harris AL. Advances in hypoxia-inducible factor biology. Cell Metab. 2018;27:281–298. doi: 10.1016/j.cmet.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Shih JW, Kung HJ. Long non-coding RNA and tumor hypoxia: new players ushered toward an old arena. J Biomed Sci. 2017;24:53. doi: 10.1186/s12929-017-0358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J Natl Cancer Inst. 1999;91:143–151. doi: 10.1093/jnci/91.2.143. [DOI] [PubMed] [Google Scholar]
- 43.Rossignol F, Vache C, Clottes E. Natural antisense transcripts of hypoxia-inducible factor 1alpha are detected in different normal and tumour human tissues. Gene. 2002;299:135–140. doi: 10.1016/S0378-1119(02)01049-1. [DOI] [PubMed] [Google Scholar]
- 44.Li L, Wang M, Mei Z, Cao W, Yang Y, Wang Y, Wen A. lncRNAs HIF1A-AS2 facilitates the up-regulation of HIF-1alpha by sponging to miR-153-3p, whereby promoting angiogenesis in HUVECs in hypoxia. Biomed Pharmacother. 2017;96:165–172. doi: 10.1016/j.biopha.2017.09.113. [DOI] [PubMed] [Google Scholar]
- 45.Chen X, Liu M, Meng F, Sun B, Jin X, Jia C. The long noncoding RNA HIF1A-AS2 facilitates cisplatin resistance in bladder cancer. J Cell Biochem. 2019;120:243–252. doi: 10.1002/jcb.27327. [DOI] [PubMed] [Google Scholar]
- 46.Mineo M, Ricklefs F, Rooj AK, Lyons SM, Ivanov P, Ansari KI, Nakano I, Chiocca EA, Godlewski J, Bronisz A. The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep. 2016;15:2500–2509. doi: 10.1016/j.celrep.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, Clerici C. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J Biol Chem. 2004;279:14871–14878. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 48.Qiu JJ, Lin XJ, Zheng TT, Tang XY, Hua KQ. Natural antisense transcript of hypoxia-inducible factor 1 regulates hypoxic cell apoptosis in epithelial ovarian cancer. Onco Targets Ther. 2018;11:9101–9110. doi: 10.2147/OTT.S173816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z, Wang Y, Wang L, Yao B, Sun L, Liu R, Chen T, Niu Y, Tu K, Liu Q. Long non-coding RNA AGAP2-AS1, functioning AS a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16-5p and promotes proliferation and metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:194. doi: 10.1186/s13046-019-1188-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Wei X, Wang C, Ma C, Sun W, Li H, Cai Z. Long noncoding RNA ANRIL is activated by hypoxia-inducible factor-1alpha and promotes osteosarcoma cell invasion and suppresses cell apoptosis upon hypoxia. Cancer Cell Int. 2016;16:73. doi: 10.1186/s12935-016-0349-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Liu X, Wang Y, Sun L, Min J, Liu J, Chen D, Zhang H, Zhang H, Zhang H, Zhou Y, Liu L. Long noncoding RNA BC005927 upregulates EPHB4 and promotes gastric cancer metastasis under hypoxia. Cancer Sci. 2018;109:988–1000. doi: 10.1111/cas.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng SJ, Chen HY, Ye Z, Deng SC, Zhu S, Zeng Z, He C, Liu ML, Huang K, Zhong JX, Xu FY, Li Q, Liu Y, Wang CY, Zhao G. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018;37:5811–5828. doi: 10.1038/s41388-018-0382-1. [DOI] [PubMed] [Google Scholar]
- 53.Su X, Li G, Liu W. The long noncoding RNA Cancer susceptibility candidate 9 promotes nasopharyngeal carcinogenesis via stabilizing HIF1alpha. DNA Cell Biol. 2017;36:394–400. doi: 10.1089/dna.2016.3615. [DOI] [PubMed] [Google Scholar]
- 54.Liu M, Zhong J, Zeng Z, Huang K, Ye Z, Deng S, Chen H, Xu F, Li Q, Zhao G. Hypoxia-induced feedback of HIF-1alpha and lncRNA-CF129 contributes to pancreatic cancer progression through stabilization of p53 protein. Theranostics. 2019;9:4795–4810. doi: 10.7150/thno.30988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang W, Yuan W, Song J, Wang S, Gu X. LncRNA CPS1-IT1 suppresses EMT and metastasis of colorectal cancer by inhibiting hypoxia-induced autophagy through inactivation of HIF-1alpha. Biochimie. 2018;144:21–27. doi: 10.1016/j.biochi.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 56.Zhang W, Wang J, Chai R, Zhong G, Zhang C, Cao W, Yan L, Zhang X, Xu Z. Hypoxia-regulated lncRNA CRPAT4 promotes cell migration via regulating AVL9 in clear cell renal cell carcinomas. Onco Targets Ther. 2018;11:4537–4545. doi: 10.2147/OTT.S169155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wen X, Liu X, Mao YP, Yang XJ, Wang YQ, Zhang PP, Lei Y, Hong XH, He QM, Ma J, Liu N, Li YQ. Long non-coding RNA DANCR stabilizes HIF-1alpha and promotes metastasis by interacting with NF90/NF45 complex in nasopharyngeal carcinoma. Theranostics. 2018;8:5676–5689. doi: 10.7150/thno.28538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tong J, Xu X, Zhang Z, Ma C, Xiang R, Liu J, Xu W, Wu C, Li J, Zhan F, Wu Y, Yan H. Hypoxia-induced long non-coding RNA DARS-AS1 regulates RBM39 stability to promote myeloma malignancy. Haematologica. 2019;104:218289. [DOI] [PMC free article] [PubMed]
- 59.Yang X, Yao B, Niu Y, Chen T, Mo H, Wang L, Guo C, Yao D. Hypoxia-induced lncRNA EIF3J-AS1 accelerates hepatocellular carcinoma progression via targeting miR-122-5p/CTNND2 axis. Biochem Biophys Res Commun. 2019;518:239–245. doi: 10.1016/j.bbrc.2019.08.039. [DOI] [PubMed] [Google Scholar]
- 60.Sun YW, Chen YF, Li J, Huo YM, Liu DJ, Hua R, Zhang JF, Liu W, Yang JY, Fu XL, Yan T, Hong J, Cao H. A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1alpha in pancreatic ductal adenocarcinoma. Br J Cancer. 2014;111:2131–2141. doi: 10.1038/bjc.2014.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao R, Sun F, Bei X, Wang X, Zhu Y, Jiang C, Zhao F, Han B, Xia S. Upregulation of the long non-coding RNA FALEC promotes proliferation and migration of prostate cancer cell lines and predicts prognosis of PCa patients. Prostate. 2017;77:1107–1117. doi: 10.1002/pros.23367. [DOI] [PubMed] [Google Scholar]
- 62.Liu AM, Zhu Y, Huang ZW, Lei L, Fu SZ, Chen Y. Long noncoding RNA FAM201A involves in radioresistance of non-small-cell lung cancer by enhancing EGFR expression via miR-370. Eur Rev Med Pharmacol Sci. 2019;23:5802–5814. doi: 10.26355/eurrev_201907_18319. [DOI] [PubMed] [Google Scholar]
- 63.Ou ZL, Zhang M, Ji LD, Luo Z, Han T, Lu YB, Li YX. Long noncoding RNA FEZF1-AS1 predicts poor prognosis and modulates pancreatic cancer cell proliferation and invasion through miR-142/HIF-1alpha and miR-133a/EGFR upon hypoxia/normoxia. J Cell Physiol. 2019;234:15407–19. [DOI] [PubMed]
- 64.Liu L, Zhao X, Zou H, Bai R, Yang K, Tian Z. Hypoxia promotes gastric Cancer malignancy partly through the HIF-1alpha dependent transcriptional activation of the long non-coding RNA GAPLINC. Front Physiol. 2016;7:420. doi: 10.3389/fphys.2016.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, Chen H, Hong J, Zou W, Chen Y, Xu J, Fang JY. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890–6902. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- 66.Peng F, Wang JH, Fan WJ, Meng YT, Li MM, Li TT, Cui B, Wang HF, Zhao Y, An F, Guo T, Liu XF, Zhang L, Lv L, Lv DK, Xu LZ, Xie JJ, Lin WX, Lam EW, Xu J, Liu Q. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene. 2018;37:1062–1074. doi: 10.1038/onc.2017.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corrado C, Costa V, Giavaresi G, Calabrese A, Conigliaro A, Alessandro R. Long non coding RNA H19: a new player in hypoxia-induced multiple myeloma cell dissemination. Int J Mol Sci. 2019;20:801. [DOI] [PMC free article] [PubMed]
- 68.Wu W, Hu Q, Nie E, Yu T, Wu Y, Zhi T, Jiang K, Shen F, Wang Y, Zhang J, You Y. Hypoxia induces H19 expression through direct and indirect Hif-1alpha activity, promoting oncogenic effects in glioblastoma. Sci Rep. 2017;7:45029. doi: 10.1038/srep45029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, Fellig Y, Degroot N, Galun E, Hochberg A. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta. 2010;1803:443–451. doi: 10.1016/j.bbamcr.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 71.Voellenkle C, Garcia-Manteiga JM, Pedrotti S, Perfetti A, De Toma I, Da Silva D, Maimone B, Greco S, Fasanaro P, Creo P, Zaccagnini G, Gaetano C, Martelli F. Implication of long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA-sequencing. Sci Rep. 2016;6:24141. doi: 10.1038/srep24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bacci L, Aiello A, Ripoli C, Loria R, Pugliese D, Pierconti F, Rotili D, Strigari L, Pinto F, Bassi PF, Mai A, Grassi C, Pontecorvi A, Falcioni R, Farsetti A, Nanni S. H19-dependent transcriptional regulation of beta3 and beta4 Integrins upon estrogen and hypoxia favors metastatic potential in prostate Cancer. Int J Mol Sci. 2019;20:4012. [DOI] [PMC free article] [PubMed]
- 73.Matouk IJ, Raveh E, Abu-lail R, Mezan S, Gilon M, Gershtain E, Birman T, Gallula J, Schneider T, Barkali M, Richler C, Fellig Y, Sorin V, Hubert A, Hochberg A, Czerniak A. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta. 2014;1843:1414–1426. doi: 10.1016/j.bbamcr.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 74.Zheng ZH, Wu DM, Fan SH, Zhang ZF, Chen GQ, Lu J. Upregulation of miR-675-5p induced by lncRNA H19 was associated with tumor progression and development by targeting tumor suppressor p53 in non-small cell lung cancer. J Cell Biochem. 2019;120:18724–18735. doi: 10.1002/jcb.29182. [DOI] [PubMed] [Google Scholar]
- 75.Zhu G, Wang S, Chen J, Wang Z, Liang X, Wang X, Jiang J, Lang J, Li L. Long noncoding RNA HAS2-AS1 mediates hypoxia-induced invasiveness of oral squamous cell carcinoma. Mol Carcinog. 2017;56:2210–2222. doi: 10.1002/mc.22674. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y, Yao J, Meng H, Yu Z, Wang Z, Yuan X, Chen H, Wang A. A novel long non-coding RNA, hypoxia-inducible factor-2alpha promoter upstream transcript, functions as an inhibitor of osteosarcoma stem cells in vitro. Mol Med Rep. 2015;11:2534–2540. doi: 10.3892/mmr.2014.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao D, Wang S, Chu X, Han D. LncRNA HIF2PUT inhibited osteosarcoma stem cells proliferation, migration and invasion by regulating HIF2 expression. Artif Cells Nanomed Biotechnol. 2019;47:1342–1348. doi: 10.1080/21691401.2019.1596934. [DOI] [PubMed] [Google Scholar]
- 78.Ferdin J, Nishida N, Wu X, Nicoloso MS, Shah MY, Devlin C, Ling H, Shimizu M, Kumar K, Cortez MA, Ferracin M, Bi Y, Yang D, Czerniak B, Zhang W, Schmittgen TD, Voorhoeve MP, Reginato MJ, Negrini M, Davuluri RV, Kunej T, Ivan M, Calin GA. HINCUTs in cancer: hypoxia-induced noncoding ultraconserved transcripts. Cell Death Differ. 2013;20:1675–1687. doi: 10.1038/cdd.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hong Q, Li O, Zheng W, Xiao WZ, Zhang L, Wu D, Cai GY, He JC, Chen XM. LncRNA HOTAIR regulates HIF-1alpha/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis. 2017;8:e2772. doi: 10.1038/cddis.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Zhou C, Ye L, Jiang C, Bai J, Chi Y, Zhang H. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1alpha activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumour Biol. 2015;36:9179–9188. doi: 10.1007/s13277-015-3453-8. [DOI] [PubMed] [Google Scholar]
- 81.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang S, Wang W, Liu G, Xie S, Li Q, Li Y, Lin Z. Long non-coding RNA HOTTIP promotes hypoxia-induced epithelial-mesenchymal transition of malignant glioma by regulating the miR-101/ZEB1 axis. Biomed Pharmacother. 2017;95:711–720. doi: 10.1016/j.biopha.2017.08.133. [DOI] [PubMed] [Google Scholar]
- 83.Xiang S, Gu H, Jin L, Thorne RF, Zhang XD, Wu M. LncRNA IDH1-AS1 links the functions of c-Myc and HIF1alpha via IDH1 to regulate the Warburg effect. Proc Natl Acad Sci U S A. 2018;115:E1465–E1E74. doi: 10.1073/pnas.1711257115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yuan S, Xiang Y, Wang G, Zhou M, Meng G, Liu Q, Hu Z, Li C, Xie W, Wu N, Wu L, Cai T, Ma X, Zhang Y, Yu Z, Bai L, Li Y. Hypoxia-sensitive LINC01436 is regulated by E2F6 and acts as an oncogene by targeting miR-30a-3p in non-small cell lung cancer. Mol Oncol. 2019;13:840–856. doi: 10.1002/1878-0261.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 86.Shen Y, Liu Y, Sun T, Yang W. LincRNA-p21 knockdown enhances radiosensitivity of hypoxic tumor cells by reducing autophagy through HIF-1/Akt/mTOR/P70S6K pathway. Exp Cell Res. 2017;358:188–198. doi: 10.1016/j.yexcr.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 87.Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci. 2014;127:1585–1594. doi: 10.1242/jcs.141069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y, Wei Y, Ma G, Park PK, Zhou J, Zhou Y, Hu Z, Zhou Y, Marks JR, Liang H, Hung MC, Lin C, Yang L. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shih JW, Chiang WF, Wu ATH, Wu MH, Wang LY, Yu YL, Hung YW, Wang WC, Chu CY, Hung CL, Changou CA, Yen Y, Kung HJ. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1alpha co-activator driving oral cancer progression. Nat Commun. 2017;8:15874. doi: 10.1038/ncomms15874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Y, Liu X, Zhang H, Sun L, Zhou Y, Jin H, Zhang H, Zhang H, Liu J, Guo H, Nie Y, Wu K, Fan D, Zhang H, Liu L. Hypoxia-inducible lncRNA-AK058003 promotes gastric cancer metastasis by targeting gamma-synuclein. Neoplasia. 2014;16:1094–1106. doi: 10.1016/j.neo.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gomez-Maldonado L, Tiana M, Roche O, Prado-Cabrero A, Jensen L, Fernandez-Barral A, Guijarro-Munoz I, Favaro E, Moreno-Bueno G, Sanz L, Aragones J, Harris A, Volpert O, Jimenez B, del Peso L. EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination. Oncogene. 2015;34:2609–2620. doi: 10.1038/onc.2014.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Garcia-Venzor A, Mandujano-Tinoco EA, Lizarraga F, Zampedri C, Krotzsch E, Salgado RM, Davila-Borja VM, Encarnacion-Guevara S, Melendez-Zajgla J, Maldonado V. Microenvironment-regulated lncRNA-HAL is able to promote stemness in breast cancer cells. Biochim Biophys Acta Mol Cell Res. 2019;1866:118523. doi: 10.1016/j.bbamcr.2019.118523. [DOI] [PubMed] [Google Scholar]
- 93.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 94.Zhai W, Sun Y, Jiang M, Wang M, Gasiewicz TA, Zheng J, Chang C. Differential regulation of LncRNA-SARCC suppresses VHL-mutant RCC cell proliferation yet promotes VHL-normal RCC cell proliferation via modulating androgen receptor/HIF-2alpha/C-MYC axis under hypoxia. Oncogene. 2016;35:4866–4880. doi: 10.1038/onc.2016.19. [DOI] [PubMed] [Google Scholar]
- 95.Gao X, Guo X, Xue H, Qiu W, Guo X, Zhang J, Qian M, Li T, Liu Q, Shen J, Deng L, Li G. lncTCF7 is a negative prognostic factor, and knockdown of lncTCF7 inhibits migration, proliferation and tumorigenicity in glioma. Sci Rep. 2017;7:17456. doi: 10.1038/s41598-017-17340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Luo F, Sun B, Li H, Xu Y, Liu Y, Liu X, Lu L, Li J, Wang Q, Wei S, Shi L, Lu X, Liu Q, Zhang A. A MALAT1/HIF-2alpha feedback loop contributes to arsenite carcinogenesis. Oncotarget. 2016;7:5769–5787. doi: 10.18632/oncotarget.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luo F, Liu X, Ling M, Lu L, Shi L, Lu X, Li J, Zhang A, Liu Q. The lncRNA MALAT1, acting through HIF-1alpha stabilization, enhances arsenite-induced glycolysis in human hepatic L-02 cells. Biochim Biophys Acta. 1862;2016:1685–1695. doi: 10.1016/j.bbadis.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 98.Hu L, Tang J, Huang X, Zhang T, Feng X. Hypoxia exposure upregulates MALAT-1 and regulates the transcriptional activity of PTB-associated splicing factor in A549 lung adenocarcinoma cells. Oncol Lett. 2018;16:294–300. doi: 10.3892/ol.2018.8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li L, Feng T, Lian Y, Zhang G, Garen A, Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci U S A. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao ZB, Chen F, Bai XF. Long noncoding RNA MALAT1 regulates hepatocellular carcinoma growth under hypoxia via sponging MicroRNA-200a. Yonsei Med J. 2019;60:727–734. doi: 10.3349/ymj.2019.60.8.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng XY, Lei BX, Lin YY, Sui MH, Zhang ML, Zhuang ZQ, Dong JT, Jin DM, Yan TB. Long noncoding RNA MEG3 silencing protects against hypoxia-induced pheochromocytoma-12 cell injury through inhibition of TIMP2 promoter methylation. J Cell Physiol. 2020;235:1649–1662. doi: 10.1002/jcp.29085. [DOI] [PubMed] [Google Scholar]
- 102.Zeng Z, Xu FY, Zheng H, Cheng P, Chen QY, Ye Z, Zhong JX, Deng SJ, Liu ML, Huang K, Li Q, Li W, Hu YH, Wang F, Wang CY, Zhao G. LncRNA-MTA2TR functions as a promoter in pancreatic cancer via driving deacetylation-dependent accumulation of HIF-1alpha. Theranostics. 2019;9:5298–5314. doi: 10.7150/thno.34559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kong X, Zhao Y, Li X, Tao Z, Hou M, Ma H. Overexpression of HIF-2alpha-dependent NEAT1 promotes the progression of non-small cell lung Cancer through miR-101-3p/SOX9/Wnt/beta-catenin signal pathway. Cell Physiol Biochem. 2019;52:368–381. doi: 10.33594/000000026. [DOI] [PubMed] [Google Scholar]
- 104.Choudhry H, Mole DR. Hypoxic regulation of the noncoding genome and NEAT1. Brief Funct Genomics. 2016;15:174–185. doi: 10.1093/bfgp/elv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choudhry H, Albukhari A, Morotti M, Haider S, Moralli D, Smythies J, Schodel J, Green CM, Camps C, Buffa F, Ratcliffe P, Ragoussis J, Harris AL, Mole DR. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4482–4490. doi: 10.1038/onc.2014.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lelli A, Nolan KA, Santambrogio S, Goncalves AF, Schonenberger MJ, Guinot A, Frew IJ, Marti HH, Hoogewijs D, Wenger RH. Induction of long noncoding RNA MALAT1 in hypoxic mice. Hypoxia (Auckl) 2015;3:45–52. doi: 10.2147/HP.S90555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin HC, Yeh CC, Chao LY, Tsai MH, Chen HH, Chuang EY, Lai LC. The hypoxia-responsive lncRNA NDRG-OT1 promotes NDRG1 degradation via ubiquitin-mediated proteolysis in breast cancer cells. Oncotarget. 2018;9:10470–10482. doi: 10.18632/oncotarget.23732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li H, Wang X, Wen C, Huo Z, Wang W, Zhan Q, Cheng D, Chen H, Deng X, Peng C, Shen B. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer. 2017;16:169. doi: 10.1186/s12943-017-0738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li X, Deng SJ, Zhu S, Jin Y, Cui SP, Chen JY, Xiang C, Li QY, He C, Zhao SF, Chen HY, Niu Y, Liu Y, Deng SC, Wang CY, Zhao G. Hypoxia-induced lncRNA-NUTF2P3-001 contributes to tumorigenesis of pancreatic cancer by derepressing the miR-3923/KRAS pathway. Oncotarget. 2016;7:6000–6014. doi: 10.18632/oncotarget.6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang J, Jin HY, Wu Y, Zheng ZC, Guo S, Wang Y, Yang D, Meng XY, Xu X, Zhao Y. Hypoxia-induced LncRNA PCGEM1 promotes invasion and metastasis of gastric cancer through regulating SNAI1. Clin Transl Oncol. 2019;21:1142–1151. doi: 10.1007/s12094-019-02035-9. [DOI] [PubMed] [Google Scholar]
- 111.Wang Y, Chen W, Lian J, Zhang H, Yu B, Zhang M, Wei F, Wu J, Jiang J, Jia Y, Mo F, Zhang S, Liang X, Mou X, Tang J. The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1alpha. Cell Death Differ. 2020;27:695–710. doi: 10.1038/s41418-019-0381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu Y, Luo X, He W, Chen G, Li Y, Li W, Wang X, Lai Y, Ye Y. Long non-coding RNA PVT1/miR-150/ HIG2 Axis regulates the proliferation, invasion and the balance of Iron metabolism of hepatocellular carcinoma. Cell Physiol Biochem. 2018;49:1403–1419. doi: 10.1159/000493445. [DOI] [PubMed] [Google Scholar]
- 113.Huang T, Liu HW, Chen JQ, Wang SH, Hao LQ, Liu M, Wang B. The long noncoding RNA PVT1 functions as a competing endogenous RNA by sponging miR-186 in gastric cancer. Biomed Pharmacother. 2017;88:302–308. doi: 10.1016/j.biopha.2017.01.049. [DOI] [PubMed] [Google Scholar]
- 114.Wang C, Han C, Zhang Y, Liu F. LncRNA PVT1 regulate expression of HIF1alpha via functioning as ceRNA for miR199a5p in nonsmall cell lung cancer under hypoxia. Mol Med Rep. 2018;17:1105–1110. doi: 10.3892/mmr.2017.7962. [DOI] [PubMed] [Google Scholar]
- 115.Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, Rader JS. The lncRNA PVT1 contributes to the cervical Cancer phenotype and associates with poor patient prognosis. PLoS One. 2016;11:e0156274. doi: 10.1371/journal.pone.0156274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhu Z, Gao X, He Y, Zhao H, Yu Q, Jiang D, Zhang P, Ma X, Huang H, Dong D, Wan J, Gu Z, Jiang X, Yu L, Gao Y. An insertion/deletion polymorphism within RERT-lncRNA modulates hepatocellular carcinoma risk. Cancer Res. 2012;72:6163–6172. doi: 10.1158/0008-5472.CAN-12-0010. [DOI] [PubMed] [Google Scholar]
- 117.Lin J, Cao S, Wang Y, Hu Y, Liu H, Li J, Chen J, Li P, Liu J, Wang Q, Zheng L. Long non-coding RNA UBE2CP3 enhances HCC cell secretion of VEGFA and promotes angiogenesis by activating ERK1/2/HIF-1alpha/VEGFA signalling in hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:113. doi: 10.1186/s13046-018-0727-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li X, Wu Y, Liu A, Tang X. Long non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer cells through a miR-18a-HIF1alpha feedback regulatory loop. Tumour Biol. 2016;37:14733–14743. doi: 10.1007/s13277-016-5348-8. [DOI] [PubMed] [Google Scholar]
- 119.Yang Z, Shi X, Li C, Wang X, Hou K, Li Z, Zhang X, Fan Y, Qu X, Che X, Liu Y. Long non-coding RNA UCA1 upregulation promotes the migration of hypoxia-resistant gastric cancer cells through the miR-7-5p/EGFR axis. Exp Cell Res. 2018;368:194–201. doi: 10.1016/j.yexcr.2018.04.030. [DOI] [PubMed] [Google Scholar]
- 120.Zhang Y, Liu Y, Xu X. Knockdown of LncRNA-UCA1 suppresses chemoresistance of pediatric AML by inhibiting glycolysis through the microRNA-125a/hexokinase 2 pathway. J Cell Biochem. 2018;119:6296–6308. doi: 10.1002/jcb.26899. [DOI] [PubMed] [Google Scholar]
- 121.Xue M, Li X, Li Z, Chen W. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1alpha-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumour Biol. 2014;35:6901–6912. doi: 10.1007/s13277-014-1925-x. [DOI] [PubMed] [Google Scholar]
- 122.Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582:1919–1927. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 123.Li T, Xiao Y, Huang T. HIF1alphainduced upregulation of lncRNA UCA1 promotes cell growth in osteosarcoma by inactivating the PTEN/AKT signaling pathway. Oncol Rep. 2018;39:1072–1080. doi: 10.3892/or.2018.6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McCarty G, Loeb DM. Hypoxia-sensitive epigenetic regulation of an antisense-oriented lncRNA controls WT1 expression in myeloid leukemia cells. PLoS One. 2015;10:e0119837. doi: 10.1371/journal.pone.0119837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dallosso AR, Hancock AL, Malik S, Salpekar A, King-Underwood L, Pritchard-Jones K, Peters J, Moorwood K, Ward A, Malik KT, Brown KW. Alternately spliced WT1 antisense transcripts interact with WT1 sense RNA and show epigenetic and splicing defects in cancer. RNA. 2007;13:2287–2299. doi: 10.1261/rna.562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu F, Gao H, Liu K, Gao B, Ren H, Li Z, Liu F. The lncRNA ZEB2-AS1 is upregulated in gastric cancer and affects cell proliferation and invasion via miR-143-5p/HIF-1alpha axis. Onco Targets Ther. 2019;12:657–667. doi: 10.2147/OTT.S175521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol. 2016;12:360–373. doi: 10.1038/nrneph.2016.51. [DOI] [PubMed] [Google Scholar]
- 128.El Azzouzi H, Doevendans PA, Sluijter JP. Long non-coding RNAs in heart failure: an obvious lnc. Ann Transl Med. 2016;4:182. doi: 10.21037/atm.2016.05.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kaur P, Liu F, Tan JR, Lim KY, Sepramaniam S, Karolina DS, Armugam A, Jeyaseelan K. Non-coding RNAs as potential Neuroprotectants against ischemic brain injury. Brain Sci. 2013;3:360–395. doi: 10.3390/brainsci3010360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jin Q, Zhao Z, Zhao Q, Yu X, Yan L, Zhang Y, Luo Q, Liu Z. Long noncoding RNAs: emerging roles in pulmonary hypertension. Heart Fail Rev. 2019. 10.1007/s10741-019-09866-2. [DOI] [PubMed]
- 131.Zhou H, Wang B, Yang YX, Jia QJ, Zhang A, Qi ZW, Zhang JP. Long noncoding RNAs in pathological cardiac remodeling: a review of the update literature. Biomed Res Int. 2019;2019:7159592. doi: 10.1155/2019/7159592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Haddad G, Kolling M, Lorenzen JM. The hypoxic kidney: pathogenesis and noncoding RNA-based therapeutic strategies. Swiss Med Wkly. 2019;149:w14703. doi: 10.4414/smw.2019.14703. [DOI] [PubMed] [Google Scholar]
- 133.Qiu JJ, Lin XJ, Zheng TT, Tang XY, Zhang Y, Hua KQ. The Exosomal long noncoding RNA aHIF is upregulated in serum from patients with endometriosis and promotes angiogenesis in endometriosis. Reprod Sci. 2019;26:1590–1602. doi: 10.1177/1933719119831775. [DOI] [PubMed] [Google Scholar]
- 134.Lang HL, Hu GW, Zhang B, Kuang W, Chen Y, Wu L, Xu GH. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol Rep. 2017;38:785–798. doi: 10.3892/or.2017.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, Tu Q, Yin D, Lin D, Wong P-P, Huang D, Xing Y, Zhao J, Li M, Liu Q, Su F, Su S, Song E. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21:498–510. doi: 10.1038/s41556-019-0299-0. [DOI] [PubMed] [Google Scholar]
- 136.Guo K, Yao J, Yu Q, Li Z, Huang H, Cheng J, Wang Z, Zhu Y. The expression pattern of long non-coding RNA PVT1 in tumor tissues and in extracellular vesicles of colorectal cancer correlates with cancer progression. Tumour Biol. 2017;39:1010428317699122. doi: 10.1177/1010428317699122. [DOI] [PubMed] [Google Scholar]
- 137.Xue M, Chen W, Xiang A, Wang R, Chen H, Pan J, Pang H, An H, Wang X, Hou H, Li X. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16:143. doi: 10.1186/s12943-017-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 139.Baek JH, Mahon PC, Oh J, Kelly B, Krishnamachary B, Pearson M, Chan DA, Giaccia AJ, Semenza GL. OS-9 interacts with hypoxia-inducible factor 1alpha and prolyl hydroxylases to promote oxygen-dependent degradation of HIF-1alpha. Mol Cell. 2005;17:503–512. doi: 10.1016/j.molcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 140.Ma MZ, Kong X, Weng MZ, Zhang MD, Qin YY, Gong W, Zhang WJ, Quan ZW. Long non-coding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol Carcinog. 2015;54:1397–1406. doi: 10.1002/mc.22215. [DOI] [PubMed] [Google Scholar]
- 141.Kuwano Y, Pullmann R, Jr, Marasa BS, Abdelmohsen K, Lee EK, Yang X, Martindale JL, Zhan M, Gorospe M. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 2010;38:225–238. doi: 10.1093/nar/gkp861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bertozzi D, Iurlaro R, Sordet O, Marinello J, Zaffaroni N, Capranico G. Characterization of novel antisense HIF-1alpha transcripts in human cancers. Cell Cycle. 2011;10:3189–3197. doi: 10.4161/cc.10.18.17183. [DOI] [PubMed] [Google Scholar]
- 143.Chen WM, Huang MD, Kong R, Xu TP, Zhang EB, Xia R, Sun M, De W, Shu YQ. Antisense long noncoding RNA HIF1A-AS2 is upregulated in gastric Cancer and associated with poor prognosis. Dig Dis Sci. 2015;60:1655–1662. doi: 10.1007/s10620-015-3524-0. [DOI] [PubMed] [Google Scholar]
- 144.Ip JY, Nakagawa S. Long non-coding RNAs in nuclear bodies. Develop Growth Differ. 2012;54:44–54. doi: 10.1111/j.1440-169X.2011.01303.x. [DOI] [PubMed] [Google Scholar]
- 145.Gutschner T, Hammerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 146.Hung CL, Wang LY, Yu YL, Chen HW, Srivastava S, Petrovics G, Kung HJ. A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci U S A. 2014;111:18697–18702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 148.Yousefi H, Maheronnaghsh M, Molaei F, Mashouri L, Reza Aref A, Momeny M, Alahari SK. Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene. 2020;39:953–974. doi: 10.1038/s41388-019-1040-y. [DOI] [PubMed] [Google Scholar]
- 149.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, Tu Q, Yin D, Lin D, Wong PP, Huang D, Xing Y, Zhao J, Li M, Liu Q, Su F, Su S, Song E. Extracellular vesicle-packaged HIF-1alpha-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21:498–510. doi: 10.1038/s41556-019-0299-0. [DOI] [PubMed] [Google Scholar]
- 151.Naderi-Meshkin H, Lai X, Amirkhah R, Vera J, Rasko JEJ, Schmitz U. Exosomal lncRNAs and cancer: connecting the missing links. Bioinformatics. 2019;35:352–360. doi: 10.1093/bioinformatics/bty527. [DOI] [PubMed] [Google Scholar]
- 152.Henze AT, Mazzone M. The impact of hypoxia on tumor-associated macrophages. J Clin Invest. 2016;126:3672–3679. doi: 10.1172/JCI84427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Derderian C, Orunmuyi AT, Olapade-Olaopa EO, Ogunwobi OO. PVT1 signaling is a mediator of Cancer progression. Frontiers Oncol. 2019;9:502. doi: 10.3389/fonc.2019.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang C, Han C, Zhang Y, Liu F. LncRNA PVT1 regulate expression of HIF1α via functioning as ceRNA for miR-199a-5p in non-small cell lung cancer under hypoxia. Mol Med Rep. 2017;17:1105–1110. doi: 10.3892/mmr.2017.7962. [DOI] [PubMed] [Google Scholar]
- 155.Zheng Y, Tian X, Wang T, Xia X, Cao F, Tian J, Xu P, Ma J, Xu H, Wang S. Long noncoding RNA Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Mol Cancer. 2019;18:61. doi: 10.1186/s12943-019-0978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Berrondo C, Flax J, Kucherov V, Siebert A, Osinski T, Rosenberg A, Fucile C, Richheimer S, Beckham CJ. Expression of the long non-coding RNA HOTAIR correlates with disease progression in bladder Cancer and is contained in bladder Cancer patient urinary exosomes. PLoS One. 2016;11:e0147236. doi: 10.1371/journal.pone.0147236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mao Y, Wang YM, Dong LX, Zhang YJ, Zhang YQ, Wang C, Zhang Q, Yang S, Cao LY, Zhang XY, Li X, Fu ZZ. Hypoxic exosomes facilitate angiogenesis and metastasis in esophageal squamous cell carcinoma through altering the phenotype and transcriptome of endothelial cells. J Exp Clin Canc Res. 2019;38:389. doi: 10.1186/s13046-019-1384-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringner M, Morgelin M, Bourseau-Guilmain E, Bengzon J, Belting M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci U S A. 2013;110:7312–7317. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang J, Yang K, Yuan W, Gao Z. Determination of serum Exosomal H19 as a noninvasive biomarker for bladder Cancer diagnosis and prognosis. Med Sci Monitor. 2018;24:9307–9316. doi: 10.12659/MSM.912018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wei JT, Feng Z, Partin AW, Brown E, Thompson I, Sokoll L, Chan DW, Lotan Y, Kibel AS, Busby JE, Bidair M, Lin DW, Taneja SS, Viterbo R, Joon AY, Dahlgren J, Kagan J, Srivastava S, Sanda MG. Can urinary PCA3 supplement PSA in the early detection of prostate Cancer? J Clin Oncol. 2014;32:4066–4072. doi: 10.1200/JCO.2013.52.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Qiu GZ, Jin MZ, Dai JX, Sun W, Feng JH, Jin WL. Reprogramming of the tumor in the hypoxic niche: the emerging concept and associated therapeutic strategies. Trends Pharmacol Sci. 2017;38:669–686. doi: 10.1016/j.tips.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 162.Dewhirst MW. A potential solution for eliminating hypoxia as a cause for radioresistance. Proc Natl Acad Sci U S A. 2018;115:10548–10550. doi: 10.1073/pnas.1814212115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hughes VS, Wiggins JM, Siemann DW. Tumor oxygenation and cancer therapy-then and now. Br J Radiol. 2019;92:20170955. doi: 10.1259/bjr.20170955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Milosevic M, Warde P, Menard C, Chung P, Toi A, Ishkanian A, McLean M, Pintilie M, Sykes J, Gospodarowicz M, Catton C, Hill RP, Bristow R. Tumor hypoxia predicts biochemical failure following radiotherapy for clinically localized prostate cancer. Clin Cancer Res. 2012;18:2108–2114. doi: 10.1158/1078-0432.CCR-11-2711. [DOI] [PubMed] [Google Scholar]
- 165.Strese S, Fryknas M, Larsson R, Gullbo J. Effects of hypoxia on human cancer cell line chemosensitivity. BMC Cancer. 2013;13:331. doi: 10.1186/1471-2407-13-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Teicher BA, Lazo JS, Sartorelli AC. Classification of antineoplastic agents by their selective toxicities toward oxygenated and hypoxic tumor cells. Cancer Res. 1981;41:73–81. [PubMed] [Google Scholar]
- 167.Hatfield SM, Sitkovsky M. A2A adenosine receptor antagonists to weaken the hypoxia-HIF-1alpha driven immunosuppression and improve immunotherapies of cancer. Curr Opin Pharmacol. 2016;29:90–96. doi: 10.1016/j.coph.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hatfield SM, Kjaergaard J, Lukashev D, Schreiber TH, Belikoff B, Abbott R, Sethumadhavan S, Philbrook P, Ko K, Cannici R, Thayer M, Rodig S, Kutok JL, Jackson EK, Karger B, Podack ER, Ohta A, Sitkovsky MV. Immunological mechanisms of the antitumor effects of supplemental oxygenation. Sci Transl Med. 2015;7:277ra30. doi: 10.1126/scitranslmed.aaa1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Facciabene A, Peng X, Hagemann IS, Balint K, Barchetti A, Wang LP, Gimotty PA, Gilks CB, Lal P, Zhang L, Coukos G. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and T(reg) cells. Nature. 2011;475:226–230. doi: 10.1038/nature10169. [DOI] [PubMed] [Google Scholar]
- 170.Noman MZ, Desantis G, Janji B, Hasmim M, Karray S, Dessen P, Bronte V, Chouaib S. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Medicine. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gillies RM, Robinson SP, McPhail LD, Carter ND, Murray JF. Immunohistochemical assessment of intrinsic and extrinsic markers of hypoxia in reproductive tissue: differential expression of HIF1alpha and HIF2alpha in rat oviduct and endometrium. J Mol Histol. 2011;42:341–354. doi: 10.1007/s10735-011-9338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kim BW, Cho H, Chung JY, Conway C, Ylaya K, Kim JH, Hewitt SM. Prognostic assessment of hypoxia and metabolic markers in cervical cancer using automated digital image analysis of immunohistochemistry. J Transl Med. 2013;11:185. doi: 10.1186/1479-5876-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Darfeuille F, Reigadas S, Hansen JB, Orum H, Di Primo C, Toulme JJ. Aptamers targeted to an RNA hairpin show improved specificity compared to that of complementary oligonucleotides. Biochemistry-us. 2006;45:12076–12082. doi: 10.1021/bi0606344. [DOI] [PubMed] [Google Scholar]
- 174.Özeş AR, Wang Y, Zong X, Fang F, Pilrose J, Nephew KP. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci Rep-uk. 2017;7:894. doi: 10.1038/s41598-017-00966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Alfranca A. VEGF therapy: a timely retreat. Cardiovasc Res. 2009;83:611–612. doi: 10.1093/cvr/cvp228. [DOI] [PubMed] [Google Scholar]
- 176.Benej M, Hong X, Vibhute S, Scott S, Wu J, Graves E, Le QT, Koong AC, Giaccia AJ, Yu B, Chen CS, Papandreou I, Denko NC. Papaverine and its derivatives radiosensitize solid tumors by inhibiting mitochondrial metabolism. Proc Natl Acad Sci U S A. 2018;115:10756–10761. doi: 10.1073/pnas.1808945115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cao M, Zhao J, Hu G. Genome-wide methods for investigating long noncoding RNAs. Biomed Pharmacother. 2019;111:395–401. doi: 10.1016/j.biopha.2018.12.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ortiz-Barahona A, Villar D, Pescador N, Amigo J, del Peso L. Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res. 2010;38:2332–2345. doi: 10.1093/nar/gkp1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Grampp S, Platt JL, Lauer V, Salama R, Kranz F, Neumann VK, Wach S, Stohr C, Hartmann A, Eckardt KU, Ratcliffe PJ, Mole DR, Schodel J. Genetic variation at the 8q24.21 renal cancer susceptibility locus affects HIF binding to a MYC enhancer. Nat Commun. 2016;7:13183. doi: 10.1038/ncomms13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Grampp S, Schmid V, Salama R, Lauer V, Kranz F, Platt JL, Smythies J, Choudhry H, Goppelt-Struebe M, Ratcliffe PJ, Mole DR, Schodel J. Multiple renal cancer susceptibility polymorphisms modulate the HIF pathway. PLoS Genet. 2017;13:e1006872. doi: 10.1371/journal.pgen.1006872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Van Roosbroeck K, Pollet J, Calin GA. miRNAs and long noncoding RNAs as biomarkers in human diseases. Expert Rev Mol Diagn. 2013;13:183–204. doi: 10.1586/erm.12.134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Hypoxia-associated lncRNAs.
Data Availability Statement
Not Applicable.