Abstract
The cuticle, a hydrophobic layer of cutin and waxes synthesized by plant epidermal cells, is the major barrier to water loss when stomata are closed at night and under water-limited conditions. Elucidating the genetic architecture of natural variation for leaf cuticular conductance (gc) is important for identifying genes relevant to improving crop productivity in drought-prone environments. To this end, we conducted a genome-wide association study of gc of adult leaves in a maize inbred association panel that was evaluated in four environments (Maricopa, AZ, and San Diego, CA, in 2016 and 2017). Five genomic regions significantly associated with gc were resolved to seven plausible candidate genes (ISTL1, two SEC14 homologs, cyclase-associated protein, a CER7 homolog, GDSL lipase, and β-D-XYLOSIDASE 4). These candidates are potentially involved in cuticle biosynthesis, trafficking and deposition of cuticle lipids, cutin polymerization, and cell wall modification. Laser microdissection RNA sequencing revealed that all these candidate genes, with the exception of the CER7 homolog, were expressed in the zone of the expanding adult maize leaf where cuticle maturation occurs. With direct application to genetic improvement, moderately high average predictive abilities were observed for whole-genome prediction of gc in locations (0.46 and 0.45) and across all environments (0.52). The findings of this study provide novel insights into the genetic control of gc and have the potential to help breeders more effectively develop drought-tolerant maize for target environments.
Keywords: Cuticle, Cuticular conductance, Genome-wide association study, RNA sequencing, Whole-genome prediction
The cuticle is a hydrophobic layer covering the surface of the shoot, which limits transpiration, in addition to protecting shoot tissues from UV radiation, heat, and pathogen attack (Shepherd and Wynne Griffiths 2006; Xue et al. 2017). Cuticles have two major lipid components: cutin and waxes. Cutin is a polymer composed of fatty acid derivatives and glycerol, and is highly cross-linked to form an insoluble matrix (Pollard et al. 2008; Fich et al. 2016). Soluble waxes are deposited into and on top of this matrix and consist mainly of aliphatic compounds derived from very-long-chain fatty acids including alkanes, aldehydes, alcohols, ketones, and wax esters (Yeats and Rose 2013). The major pathways for both wax and cutin monomer biosynthesis have been elucidated via genetic and biochemical studies conducted mainly on model plant systems (Yeats and Rose 2013; Lee and Suh 2015; Fich et al. 2016). Many transcriptional regulators of cuticle biosynthesis have also been identified (Borisjuk et al. 2014). Pathways for delivery of cuticle lipids from the intracellular membranes where they are synthesized to the extracellular environment, and to their final destinations external to the cell wall, have been partially elucidated. Golgi-mediated vesicle trafficking (McFarlane et al. 2014), ATP BINDING CASSETTE TRANSPORTER G (ABCG) family proteins (Pighin et al. 2004; Bird et al. 2007; Panikashvili et al. 2010; Bessire et al. 2011; Chen et al. 2011), and extracellular lipid transfer proteins (DeBono et al. 2009; Kim et al. 2012) are required for accumulation of multiple classes of cuticle lipids at the shoot surface, but how these transport processes work together is not well understood.
Cuticle composition and structure varies widely among species and tissue types (Jetter et al. 2008), and its permeability to water can vary as much as three orders of magnitude (Kerstiens 2006). Relationships between cuticle composition, structure, and water barrier function are complex. The simple idea that cuticle impermeability to water increases with increased wax load or cuticle thickness has been repeatedly refuted; instead, cuticle composition and the organization of components appear to determine water barrier function (Riederer and Schreiber 2001). Cuticle composition is also modulated by environmental factors. Light, temperature, and osmotic stress influence both the quantity and composition of cuticular waxes (Shepherd and Wynne Griffiths 2006; Kosma and Jenks 2007). While high relative humidity usually suppresses wax production (Sutter and Langhans 1982; Koch et al. 2006), drought stress has been found to change cuticular wax composition (Panikashvili et al. 2007; Kosma et al. 2009) and increase wax deposition in several crop species (Shepherd and Wynne Griffiths 2006; Cameron et al. 2006; Kosma and Jenks 2007).
In addition to modulation of its composition by drought stress, a variety of other observations support a role for the cuticle in drought tolerance. Most mutations and other genetic modifications affecting cuticle composition or wax load increase its permeability to water, and this has often been associated with decreased drought tolerance (Zhou et al. 2013; Zhu and Xiong 2013; Li et al. 2019). In a few cases, overexpression of cuticle lipid biosynthetic enzymes, or their transcriptional regulators, increased cuticular lipid abundance and drought tolerance (Aharoni et al. 2004; Zhang et al. 2005; Bourdenx et al. 2011; Wang et al. 2012). Glaucousness (a visible trait resulting from abundant accumulation of epicuticular wax crystals on shoot tissue surfaces) was selected for during the domestication of wheat and involves genes regulating wax biosynthesis (Hen-Avivi et al. 2016; Bi et al. 2016); analyses of heritable variation in glaucousness in wheat and barley has revealed positive correlations between this trait and drought tolerance (Febrero et al. 1998; Guo et al. 2016). These findings point to the potential relevance of cuticle modification for increasing drought tolerance in cereal crops.
There is a longstanding interest in breeding for cuticle-related traits such as drought tolerance, but the complexity of the relationship between cuticle composition and resistance to dehydration, and the lack of methods amenable to high-throughput phenotyping for cuticle-associated traits, has hampered progress in this area (Petit et al. 2017). Genome-wide association studies (GWAS) (Yu et al. 2006; Zhang et al. 2010; Lipka et al. 2015), taking advantage of natural variation in cuticle permeability and historical recombination events within a single species, offer an attractive approach to identifying genes and alleles that can decrease cuticle permeability. Additionally, findings from GWAS of cuticle-related phenotypes could be used to better inform the application of genomic selection strategies (Meuwissen et al. 2001; Lorenz et al. 2011) for increasing drought tolerance in crop species.
In this study, we utilized GWAS combined with laser microdissection RNA sequencing (LM-RNAseq) of epidermal tissue samples, from zones of the expanding maize leaf where the cuticle develops, to implicate candidate genes controlling the water barrier function of the maize leaf cuticle. Methods directly measuring permeability of the cuticle to water (Valeska Zeisler-Diehl et al. 2017) are not amenable to the high-throughput phenotyping needed for GWAS involving analysis of thousands of samples. Thus, we utilized a phenotyping strategy that indirectly measures cuticle water barrier function by calculating the drying rates of detached leaves placed in the dark to close stomata (Ristic and Jenks 2002). While some water may be lost via stomata that are not completely sealed, the sealing of stomata is thought to depend on cuticular flaps lining the stomatal pore that overlap when stomata are closed (Zhao and Sack 1999; Kosma and Jenks 2007). Thus, we used this phenotyping strategy with the expectation that it primarily measures cuticle-dependent water loss, which we refer to as adult leaf cuticular conductance (gc). Maize is most sensitive to drought stress at flowering (Grant et al. 1989), when juvenile leaves have already senesced and only adult leaves remain. Thus, we utilized adult leaves of field-grown plants for this analysis. Moreover, since cuticle composition is known to vary depending on the growth environment (Shepherd and Wynne Griffiths 2006), we utilized plants grown in two contrasting environments: the cooler and more humid environment of San Diego, CA, and the hotter and more arid environment of Maricopa, AZ. We aimed to (i) detect genomic regions associated with natural variation in gc; (ii) enhance selection of candidate genes impacting this trait through an LM-RNAseq analysis of the expanding maize adult leaf; and (iii) evaluate whole-genome prediction models to facilitate genomic selection on this trait, providing tools for potential improvement of drought-tolerance in maize for target environments.
Materials and Methods
Plant materials and experimental design
A set of 468 maize inbred lines from the Wisconsin Diversity panel (Hansey et al. 2011) was evaluated for adult leaf cuticular conductance (gc). The inbred lines were planted at the Maricopa Agricultural Center, Maricopa, AZ, and University of California San Diego, San Diego, CA, in 2016 and 2017. The layout of the experiment in each of the four environments (location × year combination) was arranged as an augmented incomplete block design. Within each of the 26 incomplete blocks, the incomplete block of 18 experimental lines was augmented by the addition of two checks (B73 and Mo17, 2016; N28Ht and Mo17, 2017) in random positions, for a total of 20 entries. The entire experiment of 468 unique inbred lines and 52 repeated checks was grown as a single replicate in each environment. Edge effects were reduced by planting a locally adapted maize inbred line around the perimeter of the experiment. Experimental units were one-row plots of 3.05 m (Maricopa) and 4.88 m (San Diego) in length with 1.016 m inter-row spacing. There was a 0.91 m alley at the end of each plot. In each plot, 12 kernels were planted, followed by thinning the stand as needed to limit plant-to-plant competition. We obtained and summarized meteorological data (Table S1) from automated weather stations located at a distance of ∼200 m or less from the experimental field sites in Maricopa, AZ (Arizona Meteorological Network; http://ag.arizona.edu/azmet/index.html) and San Diego, CA (Scripps Hydroclimate Weather Station).
gc evaluation and phenotypic data analysis
Initially, a porometer (model SG-1, Decagon Devices, Pullman, WA) was used to measure diffusion conductance for adult leaves collected from the field in the morning (at the same time of day that leaves were harvested for gc analysis, within a week of their anthesis dates), and transferred to a dark room where they were kept with cut leaf bases immersed in water throughout the analysis. Diffusion conductance was measured at a series of timepoints for a selection of lines that had previously been demonstrated to have high gc, and B73 as a reference standard harvested each sampling day. As shown in Figure S1, most lines reached minimum conductance values, indicating stomatal closure, within 30 min, and all lines reached minimum values within 90 min. Based on these findings, we concluded that 2 h in the dark was sufficient to achieve stomatal closure, even for lines with high gc.
The findings from the porometer study were used to inform our phenotyping of the Wisconsin Diversity panel. To inform sampling times and help control for maturity differences, flowering time (days to anthesis, DTA) was recorded as the number of days from planting to initiation of pollen shed for 50% of the plants in a plot. For each plot, gc was evaluated on an adult leaf from five plants when 50% of the plants in the plot were at anthesis. From each of five plants, the leaf subtending the uppermost ear, or the leaf immediately above or below it, was excised at 2.5 cm below the ligule. If there were fewer than five plants to measure in a plot, two, three, or four plants were evaluated to represent the gc of that plot. Cut ends were immersed in water and leaves were incubated in a dark, well-ventilated room for 2 h at 20-22° and 55–65% RH to close stomata while ensuring that the leaves were fully hydrated. Subsequently, each leaf was hung on a line from a boot clip in the same dark, temperature- and humidity-controlled room, after carefully wiping the leaves to remove excess water. In 2017, the cut leaf end was wrapped in parafilm when hung to further reduce the incidence of non-cuticular transpiration. The wet weight of each leaf was then recorded every 45 min over a time period of 3 h, for a total of five measurements per leaf. Leaves were subsequently dried at 60° for 4 d in a forced-air oven, and afterward weighed.
To investigate the degree to which dry weight approximates surface area, we constructed an imaging table that consisted of a digital camera (Canon PowerShot S110) mounted at a fixed distance and folding table top to help flatten the adult maize leaves. In 2017, each experimental leaf that had completed the gc phenotyping process was flattened, imaged, and then oven-dried as earlier described and weighed. Each image contained 4 to 7 leaves, depending on their sizes. Next, images were individually analyzed with a custom ImageJ (Schneider et al. 2012) macro to estimate the surface area (mm2) of each leaf. Even though every attempt was made to flatten the leaves, there were frequent occurrences of where wavy leaves could not be completely flattened. Very strong Pearson’s correlations were observed between the plot-level averages of leaf dry weight and surface area in MA (r = 0.93) and SD (r = 0.92) in 2017 (Figure S2; Table S2). Given these strong correlations and challenges in measuring surface area, leaf dry weight was considered to be a reasonable approximation of leaf surface area, and was used in the calculation of cuticular conductance as described below.
Adult leaf cuticular conductance (gc) from unit surface area was calculated as follows:
where b (g·h-1) is the coefficient of the linear regression of leaf wet weight (g) on time (h), and dry weight (g) is an approximation of leaf surface area.
To screen the phenotypic data (flowering time or gc) for significant outliers, univariate mixed linear models that enabled the estimation of genetic effects independent of field design effects were fitted as follows: (1) each single environment; (2) the two environments in each location; and (3) all four environments. The model terms included grand mean and check as fixed effects and environment, genotype, genotype-by-environment (G×E) interaction (only for Models 2 and 3), incomplete block within environment, and column within environment as random effects. The Studentized deleted residuals (Neter et al. 1996) generated from these mixed linear models were assessed and significant (α = 0.05) outliers removed. For each outlier screened phenotype, an iterative mixed linear model fitting procedure was conducted for each of the three (1 to 3) full models in ASReml-R version 3.0 (Gilmour et al. 2009). All random terms that were not significant at α = 0.05 in a likelihood ratio test (Littell et al. 2006) were removed from the model, allowing a final best-fit model to be obtained for each phenotype. These final models (Table S3) were used to generate a best linear unbiased predictor (BLUP) for gc and DTA for each line (Table S4).
Variance component estimates from the fitted full models (Table S5) were used to estimate heritability on a line-mean basis (Holland et al. 2003; Hung et al. 2012) for each phenotype in a location (Model 2) and across all four environments (Model 3). Standard errors of the heritability estimates were calculated with the delta method (Lynch et al. 1998; Holland et al. 2003). Pearson’s correlation coefficients were used to evaluate the strength of correlation between BLUP values for gc and DTA in all pairwise combinations. The significance of correlations (α = 0.05) was tested using the function “cor.test” in R version 3.5.1 (R core team 2018).
DNA extraction and genotyping
For each inbred line, total genomic DNA was isolated from a leaf tissue sample consisting of bulked young leaves from a single plant. The leaf tissue samples were lyophilized and ground using a GenoGrinder (Spex SamplePrep, Metuchen, NJ, USA), followed by isolation of total genomic DNA using the DNeasy 96 Plant Kit (Qiagen Incorporated, Valencia, CA, USA). The genotyping-by-sequencing (GBS) procedure was conducted on the DNA samples following Elshire et al. (2011) with ApeKI as the restriction enzyme at the Cornell Biotechnology Resource Center (Cornell University, Ithaca, NY, USA). The constructed 192-plex GBS libraries were sequenced on a NextSeq 500 (Illumina Incorporated, San Diego, CA, USA).
We called genotypes at 955,690 high-confidence single-nucleotide polymorphism (SNP) loci in B73 RefGen_v2 coordinates following the method of Baseggio et al. (2019). Briefly, the raw SNP genotype calls were initially filtered to exclude singleton and doubleton SNPs (a minor allele observed in only a single line) and retain only biallelic SNPs having a SNP call rate greater than 10% and minimum inbreeding coefficient of 0.8 per Romay et al. (2013). Additionally, only lines with a call rate greater than 40% were retained. Missing SNP genotypes were partially imputed using FILLIN (Swarts et al. 2014) with a set of maize haplotype donors files having a 4 kb window size (AllZeaGBSv2.7impV5_AnonDonors4k.tar.gz, available at panzea.org). Given that missing genotype data still remained following partial imputation, the imputed genotype data set was further filtered to retain SNPs with a minimum call rate of 0.6, a minimum inbreeding coefficient of 0.8, and a minimum minor allele frequency of 5%. The final complete set contained 235,004 high-quality SNP markers scored on 451 lines that had a BLUP value for gc in one or more environments. The genome coordinates of the SNP loci were uplifted by aligning 101 bp context sequences containing target SNPs (± 50 bp) to the B73 RefGen_AGPv4 reference genome with Vmatch (Kurtz 2003).
Population structure analysis
We estimated population structure from the 451 line × 235,004 SNP genotype matrix in fastSTRUCTURE version 1.0 (Raj et al. 2014), with the number of ancestral populations (K) varying from 1 to 10 using the simple prior. To complement population structure inference with fastSTRUCTURE, a principal component analysis (PCA) was performed on the identical genotype matrix with the prcomp function in R version 3.5.1 (R core team 2018). Next, results from fastSTRUCTURE and PCA were jointly interpreted and complemented with pedigree information (Hansey et al. 2011) to choose K = 6 as the number of subpopulations. Additionally, lines were classified following group assignments of Hansey et al. (2011) (Figure S3A). In fastSTRUCTURE, the simple prior approach was used with K = 6 to calculate the subpopulation composition of each line (Figure S3B). Lines with an assignment value of Q ≥ 0.50 were assigned to subpopulations (1, 2, 3, 4, 5, or 6), while those with assignment values of Q < 0.50 for all six subpopulations were classified as admixed (Figure S3C; Table S6). This information was used to inform a stratified sampling approach used for whole-genome prediction (WGP).
Genome-wide association study
To identify associations between each of the 235,004 SNP markers and the BLUP values of gc from the 451 inbreds, a GWAS was conducted with a univariate mixed linear model that employed population parameters previously determined (P3D) approximation (Zhang et al. 2010) in the R package Genome Association and Prediction Integrated Tool (GAPIT) version 3.0 (Lipka et al. 2012). To control for phenotypic variation due to maturity, population structure, and unequal relatedness, the fitted mixed linear models included BLUPs of flowering time (DTA) of corresponding environments, principal components (PCs), and a genomic relationship (kinship) matrix. The kinship matrix based on the centered IBS method (Endelman and Jannink 2012) implemented in TASSEL 5.0 (Bradbury et al. 2007) was calculated from a subset of 41,259 SNPs remaining after linkage disequilibrium (LD) pruning (r2 ≤ 0.2) of the complete marker data set in PLINK version 1.09_beta5 (Purcell et al. 2007). The optimal number of covariates (BLUPs of DTA and PCs based on the genotypic matrix) to include in the mixed linear model was determined with the Bayesian information criterion (BIC) (Schwarz 1978). Prior to conducting the GWAS, the remaining missing genotypes for all SNP loci were conservatively imputed as a heterozygote in GAPIT. A likelihood-ratio-based R2 statistic (R2LR) (Sun et al. 2010) was used to estimate the amount of phenotypic variation explained by the model with or without a tested SNP. The false discovery rate (FDR) was controlled at 10% with the Benjamini-Hochberg procedure (Benjamini and Hochberg 1995).
Linkage disequilibrium
The squared allele-frequency correlation (r2) method (Hill and Weir 1988) was used to estimate LD between all pairs of SNPs on a chromosome in TASSEL 5.0 (Bradbury et al. 2007). The complete set of 235,004 high-quality SNPs was used for LD estimation; however, the missing SNP genotypes remaining after FILLIN imputation were not imputed with a heterozygote value prior to LD analysis. The estimates of LD were used to approximate the physical distance at which LD decayed to genome-wide background levels (r2 < 0.10) and assess the pattern of LD surrounding GWAS-detected SNPs.
Candidate gene identification
We searched for candidate genes within ±200 kb of the most significant SNP (peak SNP) for each detected genomic region associated with gc. Gene functional annotations (B73_RefGen_v4) of identified candidate genes were obtained from Gramene (http://www.gramene.org/). Additionally, the Gramene Mart tool was used to identify homologs of candidate genes in Arabidopsis thaliana (L.) Heynh. (Columbia-0 ecotype) and rice (Oryza sativa L. ssp. Japonica cv. ‘Nipponbare’). Gene functional annotations for the Arabidopsis and rice homologs were obtained from TAIR (https://www.arabidopsis.org/) and RAP-DB (https://rapdb.dna.affrc.go.jp/), respectively.
Transcript abundance analysis for candidate genes
To investigate the patterns of transcript abundance for candidate genes potentially regulating gc of maize leaves, we analyzed the LM-RNAseq dataset of Qiao et al. (2019) preprint generated from epidermal and internal tissues that were LM from seven 2-cm-long intervals (six intervals from 2-14 cm, and one interval from 20-22 cm) from the expanding leaf 8 of maize inbred B73 with three biological replicates and three plants per replicate. The generated RNAseq reads were aligned to B73 genome RefGen_v4 with HiSAT2 (Kim et al. 2015) and counted with HTSeq (Anders et al. 2015). Transcript abundance levels for canonical protein coding genes were characterized as counts per million (cpm) after normalizing against library sizes with the R package edgeR 3.3.2 (Robinson et al. 2010). To identify potential outlier samples, genes with >1 cpm in two or more samples were retained. Next, a PCA was conducted on the cpm values of all the retained genes on all samples sequenced (Figure S4), and the epidermal and internal samples from interval 4-6 cm in the 3rd replicate were removed as outliers (circled samples in Figure S4). With the remaining epidermis samples, a gene was declared expressed in the epidermis if its cpm was greater than one for at least three out of the 20 samples.
Whole-genome prediction
WGP of gc was conducted using a maximum likelihood approach to ridge regression in the ‘rrBLUP’ R package (Endelman 2011). The final complete set of 235,004 SNPs with post-FILLIN missing genotype data imputed as a heterozygote was used for WGP. We trained WGP models with gc BLUP values from either MA (two environments), SD (two environments), or AllEnv (four environments), then each of the three models were separately used to predict gc for MA, SD, and AllEnv. Collectively, this resulted in a total of nine prediction scenarios. Fivefold cross-validation as described in Owens et al. (2014) was performed to evaluate the predictive ability for gc in each of the nine scenarios by calculating the Pearson’s correlation between observed BLUP values and genomic estimated breeding values. We used a stratified sampling approach to account for the presence of population structure, enabling each fold to have the proportion of subpopulations (1 to 6 and admixed) observed for the whole panel (Table S6). The procedure was conducted 50 times for each scenario, and the predictive ability was calculated as a mean of the correlations.
Data availability
The raw GBS sequence data were deposited in the National Center of Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number SRP160407 and in BioProject under accession PRJNA489924. The BLUP values of the phenotypes are provided in Table S4. The FILLIN partially imputed SNP genotype data for the 235,004 loci scored on the 451 inbred lines, weather data from Maricopa and San Diego in 2016 and 2017, and leaf image data from Maricopa and San Diego in 2017 are available at CyVerse (http://datacommons.cyverse.org/browse/iplant/home/shared/GoreLab/dataFromPubs/Lin_LeafCuticle_2019). The ImageJ macro for image analysis is available at Github (https://github.com/GoreLab/Maize_leaf_cuticle/blob/master/GWAS_CE/cliveMacro_2.4.txt). RNAseq reads of epidermal cells along the developmental gradient of the expanding leaf 8 of maize inbred B73 were deposited in the NCBI SRA under accession number SRP116320 and in BioProject under accession PRJNA400334, as described in Qiao et al. (2019) preprint. Supplemental material available at figshare: https://doi.org/10.25387/g3.10262951.
Results
Phenotypic variability
To establish the feasibility of GWAS for gc, the extent of phenotypic variation for adult leaf gc was assessed in the Wisconsin Diversity panel of more than 450 maize inbred lines that was grown in two field locations (SD, San Diego, CA; and MA, Maricopa, AZ) in 2016 and 2017. The climatically contrasting environments of the SD and MA locations (Table S1) had the lowest and highest average gc estimates, respectively (Table 1). There was a moderately strong correlation for gc between years in a location (r = 0.52 and 0.54), whereas correlations were slightly weaker (r = 0.36 to 0.48) between locations in and across years (Figure S5). With respect to flowering time, gc tended to have a moderately weak negative correlation with days to anthesis (DTA) in each environment (r = -0.14 to -0.50), with the exception of a negative correlation of moderate strength (-0.50) in the MA17 environment (Figure S5). Comparatively, the first two principal components (PCs) but not the third PC calculated from the SNP genotype matrix (PC1, 3.68%; PC2, 1.83%; and PC3, 1.62%), which infers population structure from genetic data, showed comparable correlations with gc across all four environments (|r| = 0.14 to 0.31). Heritability on a line-mean basis across (AllEnv) locations was 0.71, with a range of 0.63 to 0.67 in locations (Table 1). These findings suggest that most phenotypic variation for gc is due to the genetic variation among individuals in this diversity panel.
Table 1. Averages and ranges for best linear unbiased predictions (BLUPs) of adult maize leaf cuticular conductance (gc) in a location (MA and SD) and across all four environments (AllEnv), and estimated heritabilities on a line-mean basis.
| Environment | No. lines | BLUPs in g·h-1·g-1 | Heritability | |||||
|---|---|---|---|---|---|---|---|---|
| Average | Min | Max | S.D.a | C.V.b | Estimate | S.E.c | ||
| MA | 410 | 0.20 | 0.15 | 0.29 | 0.03 | 0.15 | 0.63 | 0.04 |
| SD | 445 | 0.13 | 0.08 | 0.21 | 0.02 | 0.15 | 0.67 | 0.03 |
| AllEnv | 451 | 0.18 | 0.13 | 0.25 | 0.02 | 0.11 | 0.71 | 0.02 |
Standard deviation of the BLUPs.
Coefficient of variation of the BLUPs.
Standard error of the heritability.
Genome-wide association study
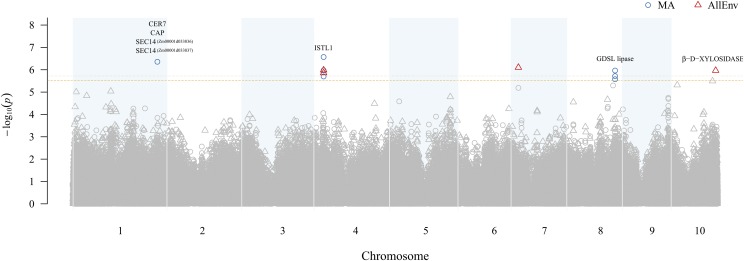
The genetic basis of natural variation for gc was examined in the Wisconsin Diversity panel that had been genotyped with 235,004 high-quality SNP markers. We conducted GWAS in (MA or SD) and across locations (AllEnv) using a mixed linear model that controls for population structure (PCs) and unequal relatedness (kinship). In addition, flowering time, which was found to have high heritability (0.88 to 0.93) for MA, SD, and AllEnv (Table S7) and mostly moderate weak correlations with gc (Figure S5), was retained as a covariate for GWAS of gc from MA and AllEnv based on the BIC values, thus attenuating the confounding effect of maturity when estimating allelic effects. A total of nine unique SNPs localized to five different genomic regions were significantly associated with gc from MA and/or AllEnv at a genome-wide FDR of 10% (Figures 1 and S6). Of these nine SNPs, two were associated with gc in both MA and AllEnv, for a total of 11 significant marker-trait associations (Table 2). No significant associations were detected for SNP markers with gc from SD at the 10% FDR level (Figure S6).
Figure 1.
Manhattan plot of results from a genome-wide association study (GWAS) of adult maize leaf cuticular conductance (gc) conducted in Maricopa (MA) and across all four environments (AllEnv). The -log10 P-value of each SNP tested in a mixed linear model analysis of gc is plotted as a point against its physical position (B73 RefGen_v4) for the 10 chromosomes of maize. The least significant single-nucleotide polymorphism (SNP) at a genome-wide false discovery rate of 10% in MA and AllEnv is indicated by a dashed horizontal orange line and a dotted horizontal orange line, respectively. SNPs significantly associated with gc in MA and AllEnv are represented by blue circles and red triangles, respectively. The most plausible candidate genes within ± 200 kb of the significantly associated SNPs are listed above their corresponding GWAS signal.
Table 2. Significant SNPs detected at false discovery rate (FDR) of 10% through a genome-wide association study of adult maize leaf cuticular conductance (gc) in a location (Maricopa, MA; San Diego, SD) and across all four environments (AllEnv).
| SNP IDa | Chr | SNP position (bp)b | P-value | FDR-adjusted P-valuec | Minor allele frequency | Allelic effect estimated | R2LRe | R2LR-SNPf | Environment |
|---|---|---|---|---|---|---|---|---|---|
| S1_270551534 | 1 | 275,318,146 | 4.37E-07 | 0.05 | 0.13 | 0.009 | 0.28 | 0.33 | MA |
| S4_30201406 | 4 | 31,733,780 | 1.39E-06 | 0.08 | 0.07 | 0.009 | 0.30 | 0.34 | AllEnv |
| 4 | 31,733,780 | 1.06E-06 | 0.06 | 0.07 | 0.011 | 0.28 | 0.33 | MA | |
| S4_30201436 | 4 | 31,733,810 | 2.68E-07 | 0.05 | 0.12 | 0.010 | 0.28 | 0.33 | MA |
| S4_30201599 | 4 | 31,733,973 | 1.04E-06 | 0.08 | 0.13 | 0.007 | 0.30 | 0.34 | AllEnv |
| 4 | 31,733,973 | 1.99E-06 | 0.08 | 0.13 | 0.008 | 0.28 | 0.33 | MA | |
| S2_231152916 | 7 | 23,984,279 | 7.88E-07 | 0.08 | 0.06 | 0.011 | 0.30 | 0.34 | AllEnv |
| S8_152967929 | 8 | 157,645,450 | 1.92E-06 | 0.08 | 0.20 | 0.008 | 0.28 | 0.33 | MA |
| S8_152967952 | 8 | 157,645,473 | 1.08E-06 | 0.06 | 0.19 | 0.008 | 0.28 | 0.33 | MA |
| S8_152967966 | 8 | 157,645,487 | 2.64E-06 | 0.09 | 0.20 | 0.007 | 0.28 | 0.32 | MA |
| S10_144686998 | 10 | 145,399,603 | 1.08E-06 | 0.08 | 0.23 | 0.011 | 0.30 | 0.34 | AllEnv |
SNP name is provided as “S” following by chromosome number and physical position in base pair from B73 Refgen_v2. The genomic position of S2_231152916 changed from chromosome 2 to 7 after uplifting from B73 RefGen_v2 to RefGen_v4.
Genomic position (bp) of the SNP from B73 Refgen_v4.
False discovery rate adjusted P-value.
Additive effect (g·h-1·g-1) of the minor allele for leaf cuticular conductance.
R2LR likelihood ratio value of model without SNP.
R2LR-SNP likelihood ratio value of model with SNP.
In the Wisconsin Diversity panel, genome-wide LD decayed to background levels (r2 < 0.1) by 213 kb at the 90% percentile of the r2 distribution (Figure S7). This percentile cutoff of the distribution was used to sample the large variance in LD structure presumably caused by rare variants (Wallace et al. 2014a) and allow for the inclusion of potential distant regulatory elements as has been observed for cloned quantitative trait loci (QTL) in maize (Salvi et al. 2007; Studer et al. 2011). Therefore, the genomic search space to identify candidate genes was restricted to ± 200 kb window of the five peak SNPs that each tagged a unique genomic interval, resulting in the inclusion of 76 annotated genes (Table S8). Of these 76 genes, 57 are canonical protein coding genes, while the other 19 encode non-coding RNAs.
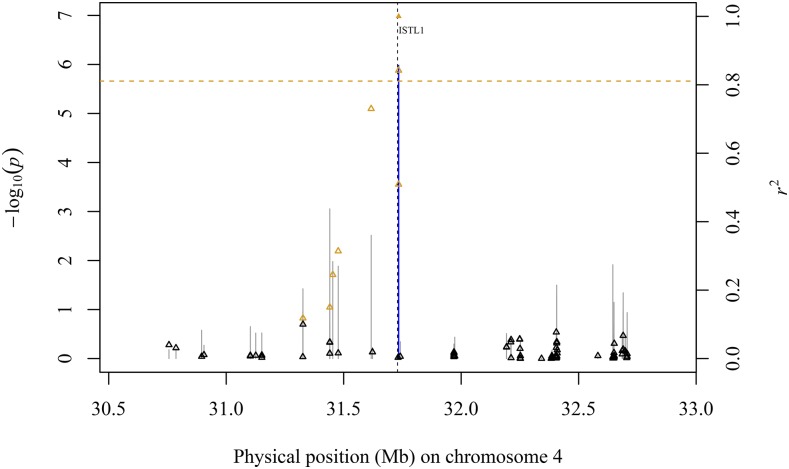
The strongest association signal was identified on chromosome 4 (Figure 1), with SNPs S4_30201436 and S4_30201599 comprising the peak associations with gc in MA (P-value 2.68 × 10−7) and AllEnv (P-value 1.04 × 10−6), respectively. The peak SNP S4_30201599 resides within a microRNA locus (zma-MIR2275a) and is located 1,146 bp from a gene (Zm00001d049479) encoding an INCREASED SALT TOLERANCE1-LIKE1 (ISTL1) protein (Figure 2; Table S8). ISTL1 is the most probable candidate gene underlying this association signal because it is predicted to function together with the endosomal sorting complex required for transport machinery (ESCRT) and ATPase VACUOLAR PROTEIN SORTING4 (VPS4) in the formation of multivesicular bodies (MVBs) (Hill and Babst 2012; Buono et al. 2016).
Figure 2.
Association of SNP markers with adult maize leaf cuticular conductance (gc) across a genomic region on chromosome 4. Scatter plot of association results from a mixed linear model analysis of gc conducted across all four environments (AllEnv) and linkage disequilibrium (LD) estimates (r2) for a genomic region that contains a gene encoding an INCREASED SALT TOLERANCE1-LIKE1 (ISTL1) protein (Zm00001d049479). The -log10 P-values of tested single-nucleotide polymorphisms (SNPs) are represented by vertical lines. Blue vertical lines are SNPs that are statistically significant at a false discovery rate (FDR) of 10%. The r2 values of each SNP relative to the peak SNP (indicated by a solid orange triangle) at 31,733,973 bp (B73 RefGen_v4) are indicated by triangles. Open orange triangles represent SNPs with r2 > 0.1 relative to the peak SNP. The least significant SNP at a genome-wide FDR of 10% is indicated by a dashed horizontal orange line. The black dashed vertical line indicates the genomic position of the ISTL1 protein.
The genomic region associated with gc on chromosome 1 was detected in MA, with the signal peak defined by SNP S1_27055151534 (P-value 4.37 × 10−7) (Table 2). The peak SNP is located 8,190 bp from the nearest gene (Zm00001d033835), but this gene encodes an uncharacterized protein. Two genes adjacent to this one encode closely related SEC14 homologs, separated by about 50 kb (Zm00001d033836, ∼50 kb from peak SNP; and Zm00001d033837, ∼100 kb from peak SNP) (Figure S8; Table S8). SEC14 proteins in plants and other eukaryotes function in the transfer of phosphoinositides between different cellular membranes (Huang et al. 2016) and are noteworthy candidates for regulators of gc because of their function in stimulating vesicle formation from the trans Golgi network in yeast (Kf de Campos and Schaaf 2017), given the role of Golgi-dependent vesicle trafficking in cuticle biosynthesis in plants (McFarlane et al. 2014). Additional plausible candidates on chromosome 1 encode a homolog of ECERIFERUM7 (CER7) (Zm00001d033842) and a CYCLASE-ASSOCIATED PROTEIN (CAP) (Zm00001d033830) located (+) 179,198 bp and (-) 152,746 bp from the peak SNP, respectively (Figure S8). CER7, a 3′-to-5′ exoribonuclease family protein, is a core subunit of the RNA degrading exosome that positively regulates the expression of CER3/WAX2/YRE, a key wax biosynthesis gene in Arabidopsis (Kurata et al. 2003; Chen et al. 2003). CAP, a regulator of actin cytoskeleton dynamics, is highly conserved in plants, yeast, flies, and mammals (Balcer et al. 2003; Ono 2013).
An additional association with gc was detected in MA on chromosome 8, defined by S8_152967952 as the peak SNP (P-value 1.92 × 10−6) along with two neighboring SNPs (S8_152967929 and S8_152967966). The closest gene (Zm00001d011655) is located 4,067 bp from the peak SNP, and it encodes a MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE18. A gene encoding a GLY-ASP-SER-LEU ESTERASE/LIPASE (GDSL lipase) (Zm00001d011661) was identified 170,643 bp from the peak SNP (Figure S9; Table S8), and it was identified as the most plausible candidate in this region based on the known functions of GDSL lipases in lipid biosynthesis (Girard et al. 2012; Yeats et al. 2012b).
Finally, in AllEnv, gc was also significantly associated with genomic regions on chromosomes 7 and 10 with peak SNPs S2_231152916 (P-value 7.88 × 10−7) and S10_144686998 (P-value 1.08 × 10−6), respectively. The peak SNP on chromosome 7 is located within a gene encoding CYTOKININ-N-GLUCOSYLTRANSFERASE1 (Zm00001d019250) (Figure S10; Table S8). The peak SNP on chromosome 10 is located within a gene that encodes β-D-XYLOSIDASE 4 (Zm00001d026415), a xylan-degrading enzyme (Figure S11; Table S8). The latter is a plausible candidate given that the outer epidermal cell wall and cuticle are closely connected and polysaccharides, for example, pectins and hemicelluloses like xylan or xyloglucans, can be found embedded in some parts of the cuticle (Yeats and Rose 2013; Hama et al. 2019). Therefore, sugar- or cell wall-modifying enzymes might directly or indirectly influence cuticle composition and permeability.
Transcript abundance analysis of identified candidate genes
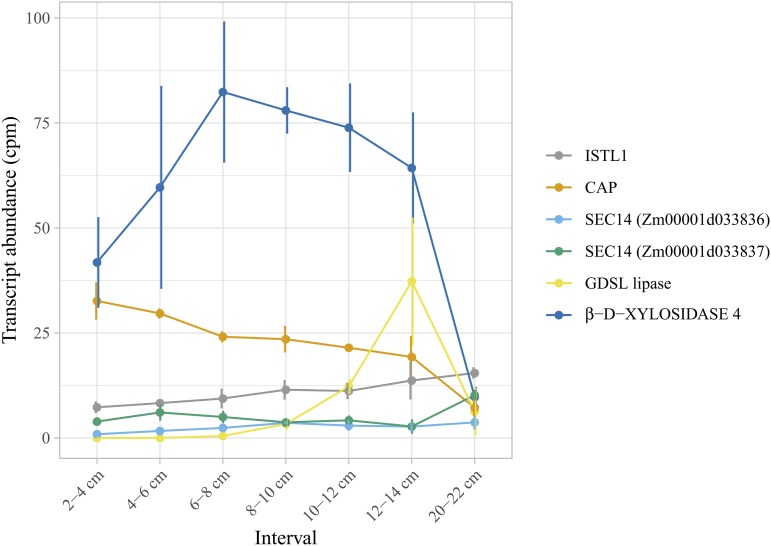
We analyzed the transcript abundance of the 57 candidate genes that encode a protein to help further prioritize which of them may be involved in the genetic control of gc. Given that the leaf cuticle is produced by epidermal cells, we analyzed LM-RNAseq data of epidermal cells from seven 2-cm intervals excised from the expanding leaf 8 of maize inbred B73 (Qiao et al. 2019 preprint), representing sequential stages in cuticle maturation (Bourgault et al. 2020). Of the 57 candidate genes, 24 were found to be expressed in at least one of the seven sampled intervals (Figure S12).
Of the seven promising candidate genes contributing to the genetic control of gc through potential influence on cuticle development, those encoding CAP, ISTL1 protein, GDSL lipase, β-D-XYLOSIDASE 4, and the two SEC14-like proteins were found to be epidermally expressed (Figure 3), while the homolog of CER7 was not declared to be expressed in the leaf epidermis based on its transcript abundance (Table S9). In general, transcript abundance increased for the ISTL1 protein and GDSL lipase along the leaf developmental gradient from the base (youngest, 2-4 cm) toward the tip (oldest, 20-22 cm) of the leaf, with the exception of a reduced transcript abundance for GDSL lipase in the 20-22 cm interval, at which time cuticle maturation is already completed (Bourgault et al. 2020). In contrast, CAP showed the opposite trend, with decreases in transcript abundance across the gradient from 2-4 cm to 20-22 cm intervals. The two genes encoding the SEC14-like proteins, Zm00001d033836 and Zm00001d033837, had relatively constant transcript abundances from the base to the tip, with a slightly increased abundance at the 20-22 cm interval for Zm00001d033837. The gene encoding β-D-XYLOSIDASE 4 had a peak abundance at the 6-8 cm interval, followed by a progressive decrease in abundance toward the leaf tip. These observed transcript abundance patterns are compatible with roles in cuticle maturation.
Figure 3.
Transcript abundance of six candidate genes in maize leaf epidermal cells. Transcript abundance (counts per million, cpm) of each gene is plotted against the developmental gradient (six intervals from 2–14 cm, and one interval from 20–22 cm) of expanding adult leaf 8 from maize inbred B73. Error bars represent the standard deviation.
Whole-genome prediction of gc
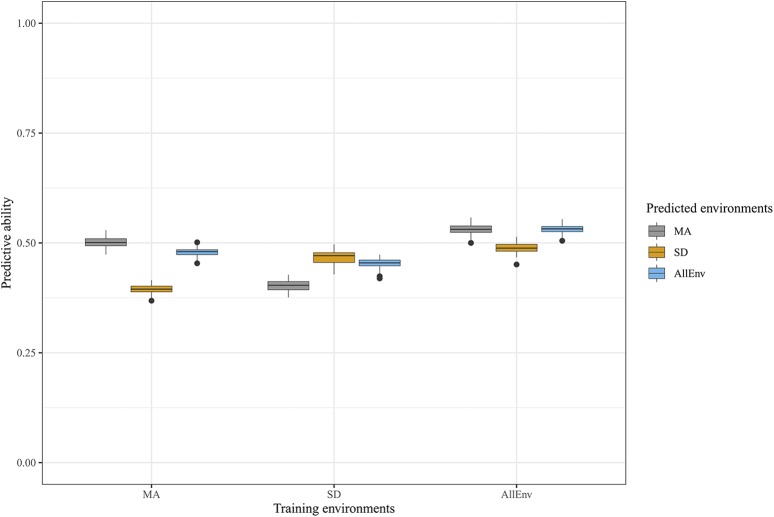
Whole-genome prediction (WGP) was performed with the complete marker data set of 235,004 SNPs to assess the feasibility of implementing genomic selection for the genetic improvement of gc in maize breeding populations. Predictive ability was evaluated on gc from MA, SD, and AllEnv in a scheme that used each of the three as a training set. The overall predictive ability was 0.48 across all of the nine training-prediction combinations, with individual predictive abilities ranging from 0.40 to 0.54 (Figure 4; Table S10). Not unexpectedly, the highest average predictive ability (0.52) was achieved when AllEnv was the training set, followed by MA (0.46) and SD (0.45). Indicative of contrasting environmental conditions between field locations, MA and SD showed lower predictive abilities for each other compared to when MA or SD was used to predict itself. However, AllEnv as the training set resulted in more similar prediction abilities for gc in MA (0.53) and SD (0.49).
Figure 4.
Predictive abilities of whole-genome prediction for adult maize leaf cuticular conductance (gc) in locations (Maricopa, MA; and San Diego, SD) and across all four environments (AllEnv) in a scheme that used each of the three as a training set. Gray, orange and blue represent the predictive abilities for gc in MA, SD, and AllEnv, respectively.
Discussion
The cuticle, a crucial barrier to plant water loss (Shepherd and Wynne Griffiths 2006; Xue et al. 2017), has been widely studied using single gene loss- and gain-of-function mutants, and interspecies crosses (Kosma and Jenks 2007; Yeats et al. 2012a; Beisson et al. 2012; Yeats and Rose 2013). However, no prior study has explored natural variation in a single species to elucidate the genetic control of cuticle function as a water barrier on a genome-wide scale. In this study, we focused on leaf cuticular conductance (gc), a trait of potential agronomic value related to drought tolerance. To that end, we evaluated the phenotypic variation of gc using detached adult leaves in a maize diversity panel in two climatically contrasting field locations (Table S1) and conducted a GWAS and WGP to explore the genetic architecture and the potential genetic gains that could be expected under selection in a breeding program for gc.
Analysis was performed to evaluate the repeatability of gc in and across locations, as well as how gc related to flowering time (DTA). Moderately strong correlations (r) of gc measurements were observed between years in a location, while relatively weaker correlations existed between locations (Figure S5). Given the lower relative humidity and higher solar radiation and air temperature of MA compared to SD during maize growing seasons (Table S1), the weaker correlative patterns between locations imply that gc is moderately responsive to environmental conditions. Noticeably, weak to moderately strong negative correlations (MA16, -0.33; MA17, -0.50; SD16, -0.14; SD17, -0.20) existed between gc and flowering time collected from the same year in each location. These findings suggest low to moderate levels of confounding between gc and maturity depending on the environment, with the prolonged exposure of the developing cuticle of plants to more extreme weather variables possibly explaining the stronger correlations in the MA field location. Even though these results implicated the need to assess for the confounding effect of flowering time when conducting GWAS, the ∼twofold range in gc variation, calculated as the ratio between the maximum and minimum BLUP values, in (MA and SD) and across (AllEnv) locations, and the moderately high heritabilities of gc (Table 1) indicate that this phenotype should be very amenable to genetic dissection and prediction in maize.
We conducted a GWAS that resulted in the identification of five genomic regions associated with gc in the Wisconsin Diversity panel. Of these five genomic regions, two were detected only in MA (chromosomes 1 and 8), two in AllEnv (chromosomes 7 and 10), and one (chromosome 4) was detected in both MA and AllEnv (Figure 1; Table 2). Notably, no significant associations at the 10% FDR level were detected only in SD, implying that G×E exists for gc as shown by the between-location correlation analysis (Figure S5). This is also further supported by comparing the ratio of genetic variance to G×E variance, which was 3.95, 2.57, and 1.81 for SD, MA, and AllEnv, respectively (Table S5). All together, these observations are consistent with those of previous studies showing that both the content and composition of cuticular waxes can be influenced by environmental factors such as ultraviolet (UV) radiation, temperature, humidity, and drought stress (Shepherd and Wynne Griffiths 2006; Xue et al. 2017). It has been reported that drought stress and low humidity increased wax loads in many plant species (Sutter and Langhans 1982; Shepherd and Wynne Griffiths 2006; Koch et al. 2006; Kosma and Jenks 2007), but a different combination of UV intensity and temperature resulted in more complex patterns of wax products (Baker 1974; Riederer and Schneider 1990; Shepherd et al. 1997). Although it is not straightforward to explain how each environmental factor affected gc through wax biosynthesis and deposition, the differences in GWAS results in MA and SD suggests that environment-specific factors in MA such as high temperature, low humidity, and elevated UV radiation (Table S1) might be inducing compositional features that affect water loss relative to plants in SD.
As the major barrier of water vapor diffusion from the intercellular air space to the atmosphere when stomata are closed, the composition and structure of the cuticle can affect water evaporation resistance, thus causing variation in gc. The candidate genes we propose for gc via GWAS encode proteins with functions that can be related to cuticle formation.
Three of the candidate genes implicated by our GWAS in control of gc encode proteins with likely functions in membrane trafficking, a process needed for deposition of cuticle lipids at the cell surface as well as for delivery of lipid transport proteins to the plasma membrane (McFarlane et al. 2014). Two adjacent genes, likely resulting from a recent tandem duplication, encode closely related SEC14 homologs. These belong to a subset of SEC14 homologs in maize lacking the GOLD (Golgi dynamics) and nodulin domains present in SEC14s that have been functionally characterized in plants (Huang et al. 2016; Kf de Campos and Schaaf 2017). Nevertheless, SEC14 domains are well known to function in movement of phosphoinositides (signaling lipids) between cellular membranes; in yeast this function stimulates vesicle formation from the trans-Golgi network (Kf de Campos and Schaaf 2017). Thus, the maize SEC14 proteins we identified may function in vesicle trafficking as well.
Another membrane-trafficking-related candidate encodes an IST1 family protein closely related to Arabidopsis ISTL1 (Buono et al. 2016). Yeast IST1 functions with the ATPase VPS4 to regulate the assembly and function of ESCRTIII complexes, which facilitate the formation of vesicles inside MVBs (Hill and Babst 2012). MVBs are late endosomal compartments that function in degradation of proteins retrieved via endocytosis from the plasma membrane. A function for ISTL1 proteins in degradation of plasma membrane proteins and formation of MVBs in Arabidopsis has been demonstrated (Buono et al. 2016). Thus, maize ISTL1 could function in an endosomal pathway critical for maintenance of plasma membrane-associated proteins that deliver cuticle lipids to the extracellular environment such as ABCG transporters and lipid transfer proteins. Moreover, MVB-like organelles have been implicated as the source of extracellular vesicles that deliver certain proteins and small RNAs to the extracellular environment in plants (Rutter and Innes 2018). This suggests the additional possibility that maize ISTL1 could impact gc because of a role in the formation of MVB vesicles containing cuticle lipids or GDSL lipases required for cutin polymerization, which may be released into the extracellular environment via fusion of MVBs with the plasma membrane. Confirmation of a role for MVBs in cuticle formation would reveal new aspects of membrane trafficking supporting cuticle formation.
CYCLASE-ASSOCIATED PROTEIN1, encoded by the candidate gene on chromosome 1 near peak SNP S1_270551534, has not been associated with cuticle development in previous studies. CAPs are actin monomer-binding proteins that regulate actin dynamics (Qualmann et al. 2000; Hubberstey and Mottillo 2002). Functions for CAPs in actin regulation and actin-dependent processes in Arabidopsis have been demonstrated through genetic and biochemical studies (Barrero et al. 2002; Chaudhry et al. 2007; Deeks et al. 2007). Actin dynamics have been implicated in multiple aspects of membrane trafficking in eukaryotic cells including vesicle formation and vesicle movement (Lanzetti 2007). Thus, CAP could impact gc in maize via regulation of membrane trafficking events supporting cuticle formation such as Golgi-mediated secretion (McFarlane et al. 2014; Luo et al. 2019) and/or endocytosis.
Two of the candidate genes we identified have predicted functions in cuticle formation. One of these is CER7, which functions in Arabidopsis to regulate the biosynthesis of alkanes, one of the major classes of cuticle waxes in this species (Jenks et al. 2002) and also in maize adult leaves(Bourgault et al. 2020). CER7 encodes an exosomal 3′-to-5′ exoribonuclease that regulates the expression level of CER3 (Hooker et al. 2007). The CER3 protein acts together with CER1 to catalyze the formation of alkanes (Bernard et al. 2012). The other candidate with a predicted function in cuticle formation encodes a GDSL lipase (Zm00001d011661). GDSL lipases catalyze the hydrolysis of mono-, di- and triglycerols and release free fatty acids and alcohols (Angkawidjaja and Kanaya 2006). Although GDSL lipases are involved in a large number of biological processes in plants, their function in the biosynthesis of cutin is well-recognized (Takahashi et al. 2010; Girard et al. 2012; Yeats et al. 2012b). Moreover, a homolog of Zm00001d011661 in Arabidopsis, AT3G16370, was down-regulated along with other cuticle associated genes in a transgenic DESPERADO/ABCG11 silenced line with changes in levels of cutin monomers (Panikashvili et al. 2010), whereas its homolog in Citrus clementina was reported as the top differentially expressed gene in the fruit epidermis (Matas et al. 2010).
The final candidate gene identified encodes the cell wall-modifying enzyme β-D-XYLOSIDASE 4 (Zm00001d026415). Sometimes the cuticle is described as a specialized lipidic modification of the cell wall, and polysaccharides are known to be deposited in some regions of the cuticle (Yeats and Rose 2013). While compounds like pectins are found to be confined to the cuticular layer close to the cell wall-cuticle border, transmission electron microscopy and non-destructive polarization modulation-infrared reflection-absorption spectroscopy localized hemicelluloses such as xylan and xyloglucan toward the surface of the cuticle, within the cuticle proper of several species (Guzmán et al. 2014; Hama et al. 2019). Therefore, we speculate that cell wall- or sugar-modifying enzymes such as a β-D-xylosidase could have an influence on the polysaccharide content of the cuticle, or might impact cuticle composition or organization by affecting the transit of cuticle lipids across the wall to the cuticle.
Genes encoding CAP, ISTL1, GDSL lipase, β-D-XYLOSIDASE 4, and both SEC14 homologs were declared expressed in epidermal cells of developing maize leaves (Figure 3), where the cuticle matures, indicating that these genes are involved in metabolic and other cellular processes in maize leaf epidermal cells and possibly associated with cuticle development. The abundance of the CER7 homolog transcript, a core subunit of the exosome that regulates wax biosynthesis (Hooker et al. 2007), was too low to be declared expressed, with cpm values ranging from 0.033 to 0.137 in 6 out of the 20 samples. Similarly, very low abundances of this transcript were observed by RNAseq in a number of different tissues from maize inbred B73 (Stelpflug et al. 2016). The increasing/decreasing trends in transcript abundance along the developmental gradient (Figure 3) are consistent with regulatory roles in cuticle maturation; however, these trends are equally consistent with roles in other developmental changes taking place at the same time. Additional work will be needed to definitively link changes in expression levels of these genes with variation in gc.
In our GWAS results, even after controlling for the confounding impact of flowering time, each locus significantly associated with gc explained 4–6% (R2LR-SNP - R2LR) of the variation in the panel (Table 2), thus only accounting for a minor fraction of the estimated heritability. If the genetic architecture of gc is predominated by a large number of loci with small allelic effect sizes, then it is possible that our association population was not of a sample size sufficient to offer the statistical power needed to detect functional variants with small effects (Long and Langley 1999). With only a total of 235,004 SNPs with common minor allele frequencies (5% or greater) scored on the diversity panel, the ’missing heritability’ could be also partially attributed to not having a density of SNP markers required to achieve strong LD (r2 > 0.80) with unscored causal variants that are potentially rare in frequency (Myles et al. 2009; Buckler et al. 2009). Therefore, increasing SNP marker density and the mapping population size are likely important for explaining the missing heritability of gc, which is possibly controlled by a large number of small-effect alleles in maize considering the complex cuticle biosynthesis network and transport mechanisms (Yeats and Rose 2013), as well as factors that affect leaf water trafficking. Such a hypothesis would cohere with findings from the powerful maize nested association mapping panel where genetic models consisting of many loci with small additive effects explain a majority of the genetic variance for a number of complex traits (Wallace et al. 2014a). However, we still cannot exclude the possible importance of rare variants of large effects, genotype-by-environment interaction, epigenetics, epistasis, incomplete penetrance, and other genetic mechanisms (Gibson 2012; Wallace et al. 2014b) in the genetic control of gc in the Wisconsin Diversity panel.
Genomics-assisted breeding approaches for improving plant water-use efficiency and drought tolerance include the use of genetic markers to select for increased water uptake through roots and diminished water loss through stomata and cuticles (Ruggiero et al. 2017). Since a lowered gc provides additional protection to the plant after stomatal closure under water-limited conditions, this potentially genetically complex trait is a candidate for genomic selection, enabled by WGP, in maize breeding programs with a focus on rainfed production environments that are prone to low rainfall. In the WGP analyses that we performed, the predictive abilities for gc were moderately high for the tested nine training-prediction combinations (Figure 4; Table S10), indicating that genetic gain for gc could be accelerated with WGP in maize breeding populations. Presumably due to influence of contrasting environmental conditions on the phenotypic data collected at the two field locations (Figure S5; Tables S1 and S4), the prediction abilities were lowest when the MA and SD training sets predicted gc each other (Figure 4; Table S10). However, AllEnv showed similar predictive abilities for MA and SD as for themselves, because AllEnv contained phenotypic information from both locations. Given the range in phenotypic variation observed for gc across locations, it is highly recommended that phenotyping and selection occur in the specific target breeding environments, especially if the focus is on hot, arid environments such as MA. Furthermore, if gc does ultimately have a polygenic genetic basis, genomic selection would be the optimal breeding strategy for gc in maize breeding populations rather than a marker-assisted selection approach designed for only a few loci with large-effects (Meuwissen et al. 2001; Lorenz et al. 2011).
Acknowledgments
We especially thank Aaron Waybright, Christine Caron, Angel Mendoza, Albert Nguyen, Indira Oueralta Castillo, Anastasia Zagordo, and the BICD 101 students at UCSD in the summer of 2016 and 2017, for collecting phenotypic data. We thank Mark Millard and Candice Gardner of the USDA-ARS North Central Regional Plant Introduction Station (NCRPIS) in Ames, Iowa, and Candice Hirsch at the University of Minnesota for providing seed of the Wisconsin Diversity panel. We also thank Bill Luckett, Andrew French, Kelly Thorp, Alison Thompson, John Dyer and others at the U.S. Arid-Land Agricultural Research Center in Maricopa, AZ, for their assistance with planting and providing a facility for phenotyping. Additionally, we thank Clint Jones, Greg Main, Russell Noon, Rick Ward and others at the University of Arizona, Maricopa Agricultural Center in Maricopa, AZ, for the management of the Arizona field trials. This research was supported by the National Science Foundation IOS1444507.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.10262951.
Communicating editor: R. Wisser
Literature Cited
- Aharoni A., Dixit S., Jetter R., Thoenes E., van Arkel G. et al. , 2004. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480. 10.1105/tpc.104.022897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., and Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angkawidjaja C., and Kanaya S., 2006. Family I.3 lipase: bacterial lipases secreted by the type I secretion system. Cell. Mol. Life Sci. 63: 2804–2817. 10.1007/s00018-006-6172-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E. A., 1974. The influence of environment on leaf wax development in Brassica oleracea var. gemmifera. New Phytol. 73: 955–966. 10.1111/j.1469-8137.1974.tb01324.x [DOI] [Google Scholar]
- Balcer H. I., Goodman A. L., Rodal A. A., Smith E., Kugler J. et al. , 2003. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13: 2159–2169. 10.1016/j.cub.2003.11.051 [DOI] [PubMed] [Google Scholar]
- Barrero R. A., Umeda M., Yamamura S., and Uchimiya H., 2002. Arabidopsis CAP regulates the actin cytoskeleton necessary for plant cell elongation and division. Plant Cell 14: 149–163. 10.1105/tpc.010301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseggio M., Murray M., Magallanes-Lundback M., Kaczmar N., Chamness J. et al. , 2019. Genome-wide association and genomic prediction models of tocochromanols in fresh sweet corn kernels. Plant Genome 12: 1–17. 10.3835/plantgenome2018.06.0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F., Li-Beisson Y., and Pollard M., 2012. Solving the puzzles of cutin and suberin polymer biosynthesis. Curr. Opin. Plant Biol. 15: 329–337. 10.1016/j.pbi.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., and Hochberg Y., 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300. [Google Scholar]
- Bernard A., Domergue F., Pascal S., Jetter R., Renne C. et al. , 2012. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24: 3106–3118. 10.1105/tpc.112.099796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessire M., Borel S., Fabre G., Carraça L., Efremova N. et al. , 2011. A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 23: 1958–1970. 10.1105/tpc.111.083121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi H., Luang S., Li Y., Bazanova N., Morran S. et al. , 2016. Identification and characterization of wheat drought-responsive MYB transcription factors involved in the regulation of cuticle biosynthesis. J. Exp. Bot. 67: 5363–5380. 10.1093/jxb/erw298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird D., Beisson F., Brigham A., Shin J., Greer S. et al. , 2007. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 52: 485–498. 10.1111/j.1365-313X.2007.03252.x [DOI] [PubMed] [Google Scholar]
- Borisjuk N., Hrmova M., and Lopato S., 2014. Transcriptional regulation of cuticle biosynthesis. Biotechnol. Adv. 32: 526–540. 10.1016/j.biotechadv.2014.01.005 [DOI] [PubMed] [Google Scholar]
- Bourdenx B., Bernard A., Domergue F., Pascal S., Léger A. et al. , 2011. Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156: 29–45. 10.1104/pp.111.172320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgault R., Matschi S., Vasquez M., Qiao P., Sonntag A. et al. , 2020. Constructing functional cuticles: Analysis of relationships between cuticle lipid composition, ultrastructure and water barrier function in developing adult maize leaves. Ann. Bot. (Lond.) 125: 79–91. 10.1093/aob/mcz143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y. et al. , 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J. et al. , 2009. The genetic architecture of maize flowering time. Science 325: 714–718. 10.1126/science.1174276 [DOI] [PubMed] [Google Scholar]
- Buono R. A., Paez-Valencia J., Miller N. D., Goodman K., Spitzer C. et al. , 2016. Role of SKD1 regulators LIP5 and IST1-LIKE1 in endosomal sorting and plant development. Plant Physiol. 171: 251–264. 10.1104/pp.16.00240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron K. D., Teece M. A., and Smart L. B., 2006. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 140: 176–183. 10.1104/pp.105.069724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry F., Guérin C., von Witsch M., Blanchoin L., and Staiger C. J., 2007. Identification of Arabidopsis cyclase-associated protein 1 as the first nucleotide exchange factor for plant actin. Mol. Biol. Cell 18: 3002–3014. 10.1091/mbc.e06-11-1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Goodwin S. M., Boroff V. L., Liu X., and Jenks M. A., 2003. Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185. 10.1105/tpc.010926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Komatsuda T., Ma J. F., Nawrath C., Pourkheirandish M. et al. , 2011. An ATP-binding cassette subfamily G full transporter is essential for the retention of leaf water in both wild barley and rice. Proc. Natl. Acad. Sci. USA 108: 12354–12359. 10.1073/pnas.1108444108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kf de Campos M. K., and Schaaf G., 2017. The regulation of cell polarity by lipid transfer proteins of the SEC14 family. Curr. Opin. Plant Biol. 40: 158–168. 10.1016/j.pbi.2017.09.007 [DOI] [PubMed] [Google Scholar]
- DeBono A., Yeats T. H., Rose J. K. C., Bird D., Jetter R. et al. , 2009. Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21: 1230–1238. 10.1105/tpc.108.064451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks M. J., Rodrigues C., Dimmock S., Ketelaar T., Maciver S. K. et al. , 2007. Arabidopsis CAP1 - a key regulator of actin organisation and development. J. Cell Sci. 120: 2609–2618. 10.1242/jcs.007302 [DOI] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K. et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman J. B., 2011. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4: 250–255. 10.3835/plantgenome2011.08.0024 [DOI] [Google Scholar]
- Endelman J. B., and Jannink J.-L., 2012. Shrinkage estimation of the realized relationship matrix. G3 (Bethesda) 2: 1405–1413. 10.1534/g3.112.004259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febrero A., Fernandez S., Molina-Cano J. L., and Araus J. L., 1998. Yield, carbon isotope discrimination, canopy reflectance and cuticular conductance of barley isolines of differing glaucousness. J. Exp. Bot. 49: 1575–1581. 10.1093/jxb/49.326.1575 [DOI] [Google Scholar]
- Fich E. A., Segerson N. A., and Rose J. K. C., 2016. The plant polyester cutin: biosynthesis, structure, and biological roles. Annu. Rev. Plant Biol. 67: 207–233. 10.1146/annurev-arplant-043015-111929 [DOI] [PubMed] [Google Scholar]
- Gibson G., 2012. Rare and common variants: twenty arguments. Nat. Rev. Genet. 13: 135–145. 10.1038/nrg3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour A. R., Gogel B. J., Cullis B. R., Thompson R., Butler D. et al. , 2009. ASReml user guide release 3.0, VSN International Ltd, Hemel Hempstead, UK. [Google Scholar]
- Girard A.-L., Mounet F., Lemaire-Chamley M., Gaillard C., Elmorjani K. et al. , 2012. Tomato GDSL1 is required for cutin deposition in the fruit cuticle. Plant Cell 24: 3119–3134. 10.1105/tpc.112.101055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R. F., Jackson B. S., Kiniry J. R., and Arkin G. F., 1989. Water deficit timing effects on yield components in maize. Agron. J. 81: 61–65. 10.2134/agronj1989.00021962008100010011x [DOI] [Google Scholar]
- Guo J., Xu W., Yu X., Shen H., Li H. et al. , 2016. Cuticular wax accumulation is associated with drought tolerance in wheat near-isogenic lines. Front. Plant Sci. 7: 1809 10.3389/fpls.2016.01809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P., Fernández V., García M. L., Khayet M., Fernández A. et al. , 2014. Localization of polysaccharides in isolated and intact cuticles of eucalypt, poplar and pear leaves by enzyme-gold labelling. Plant Physiol. Biochem. 76: 1–6. 10.1016/j.plaphy.2013.12.023 [DOI] [PubMed] [Google Scholar]
- Hama T., Seki K., Ishibashi A., Miyazaki A., Kouchi A. et al. , 2019. Probing the molecular structure and orientation of the leaf surface of Brassica oleracea L. by polarization modulation-infrared reflection-absorption spectroscopy. Plant Cell Physiol. 60: 1567–1580. 10.1093/pcp/pcz063 [DOI] [PubMed] [Google Scholar]
- Hansey C. N., Johnson J. M., Sekhon R. S., Kaeppler S. M., and de Leon N., 2011. Genetic diversity of a maize association population with restricted phenology. Crop Sci. 51: 704–715. 10.2135/cropsci2010.03.0178 [DOI] [Google Scholar]
- Hen-Avivi S., Savin O., Racovita R. C., Lee W.-S., Adamski N. M. et al. , 2016. A metabolic gene cluster in the wheat W1 and the barley Cer-cqu loci determines β-diketone biosynthesis and glaucousness. Plant Cell 28: 1440–1460. 10.1105/tpc.16.00197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. P., and Babst M., 2012. Structure and function of the membrane deformation AAA ATPase Vps4. Biochim. Biophys. Acta 1823: 172–181. 10.1016/j.bbamcr.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., and Weir B. S., 1988. Variances and covariances of squared linkage disequilibria in finite populations. Theor. Popul. Biol. 33: 54–78. 10.1016/0040-5809(88)90004-4 [DOI] [PubMed] [Google Scholar]
- Holland J. B., Nyquist W. E., and Cervantes-Martínez C. T., 2003. Estimating and interpreting heritability for plant breeding: an update. Plant Breed. Rev. 22: 9–111. 10.1002/9780470650202.ch2 [DOI] [Google Scholar]
- Hooker T. S., Lam P., Zheng H., and Kunst L., 2007. A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 19: 904–913. 10.1105/tpc.106.049304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ghosh R., and Bankaitis V. A., 2016. Sec14-like phosphatidylinositol transfer proteins and the biological landscape of phosphoinositide signaling in plants. Biochim. Biophys. Acta 1861: 1352–1364. 10.1016/j.bbalip.2016.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubberstey A. V., and Mottillo E. P., 2002. Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. FASEB J. 16: 487–499. 10.1096/fj.01-0659rev [DOI] [PubMed] [Google Scholar]
- Hung H.-Y., Shannon L. M., Tian F., Bradbury P. J., Chen C. et al. , 2012. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 109: E1913–E1921. 10.1073/pnas.1203189109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks M. A., Eigenbrode S. D., and Lemieux B., 2002. Cuticular waxes of Arabidopsis. Arabidopsis Book 1: e0016 10.1199/tab.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetter R., Kunst L., and Samuels A. L., 2008. Composition of plant cuticular waxes. Bio. Plant Cuticle 23: 145–181. [Google Scholar]
- Kerstiens G., 2006. Water transport in plant cuticles: an update. J. Exp. Bot. 57: 2493–2499. 10.1093/jxb/erl017 [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S. L., 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Lee S. B., Kim H. J., Min M. K., Hwang I. et al. , 2012. Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 53: 1391–1403. 10.1093/pcp/pcs083 [DOI] [PubMed] [Google Scholar]
- Koch K., Hartmann K. D., Schreiber L., Barthlott W., and Neinhuis C., 2006. Influences of air humidity during the cultivation of plants on wax chemical composition, morphology and leaf surface wettability. Environ. Exp. Bot. 56: 1–9. 10.1016/j.envexpbot.2004.09.013 [DOI] [Google Scholar]
- Kosma D. K., Bourdenx B., Bernard A., Parsons E. P., Lü S. et al. , 2009. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 151: 1918–1929. 10.1104/pp.109.141911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma D. K., and Jenks M. A., 2007. Eco-physiological and molecular-genetic determinants of plant cuticle function in drought and salt stress tolerance, pp. 91–120 in Advances in Molecular Breeding Toward Drought and Salt Tolerant Crops, edited by Jenks M. A., Hasegawa P. M., and Jain S. M.. Springer, Dordrecht: 10.1007/978-1-4020-5578-2_5 [DOI] [Google Scholar]
- Kurata T., Kawabata-Awai C., Sakuradani E., Shimizu S., Okada K. et al. , 2003. The YORE-YORE gene regulates multiple aspects of epidermal cell differentiation in Arabidopsis. Plant J. 36: 55–66. 10.1046/j.1365-313X.2003.01854.x [DOI] [PubMed] [Google Scholar]
- Kurtz S., 2003. The Vmatch large scale sequence analysis software. Ref Type: Computer Program 412: 297. [Google Scholar]
- Lanzetti L., 2007. Actin in membrane trafficking. Curr. Opin. Cell Biol. 19: 453–458. 10.1016/j.ceb.2007.04.017 [DOI] [PubMed] [Google Scholar]
- Lee S. B., and Suh M. C., 2015. Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 34: 557–572. 10.1007/s00299-015-1772-2 [DOI] [PubMed] [Google Scholar]
- Li L., Du Y., He C., Dietrich C. R., Li J. et al. , 2019. Maize glossy6 is involved in cuticular wax deposition and drought tolerance. J. Exp. Bot. 70: 3089–3099. 10.1093/jxb/erz131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka A. E., Kandianis C. B., Hudson M. E., Yu J., Drnevich J. et al. , 2015. From association to prediction: statistical methods for the dissection and selection of complex traits in plants. Curr. Opin. Plant Biol. 24: 110–118. 10.1016/j.pbi.2015.02.010 [DOI] [PubMed] [Google Scholar]
- Lipka A. E., Tian F., Wang Q., Peiffer J., Li M. et al. , 2012. GAPIT: genome association and prediction integrated tool. Bioinformatics 28: 2397–2399. 10.1093/bioinformatics/bts444 [DOI] [PubMed] [Google Scholar]
- Littell R. C., Milliken G. A., Stroup W. W., Wolfinger R. D., and Schabenberger O., 2006. Appendix 1: Linear mixed model theory, pp. 733–756 in SAS for Mixed Models. SAS Institute Inc., Cary, NC. [Google Scholar]
- Long A. D., and Langley C. H., 1999. The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res. 9: 720–731. [PMC free article] [PubMed] [Google Scholar]
- Lorenz A. J., Chao S., Asoro F. G., Heffner E. L., Hayashi T. et al. , 2011. Genomic selection in plant breeding: knowledge and prospects, pp. 77–123 in Advances in agronomy, edited by Sparks D. L. Elsevier Inc., Amsterdam. [Google Scholar]
- Luo Z., Tomasi P., Fahlgren N., and Abdel-Haleem H., 2019. Genome-wide association study (GWAS) of leaf cuticular wax components in Camelina sativa identifies genetic loci related to intracellular wax transport. BMC Plant Biol. 19: 187 10.1186/s12870-019-1776-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., B. Walsh, and Others, 1998 Genetics and analysis of quantitative traits. Sinauer Sunderland, MA.
- Matas A. J., Agustí J., Tadeo F. R., Talón M., and Rose J. K. C., 2010. Tissue-specific transcriptome profiling of the citrus fruit epidermis and subepidermis using laser capture microdissection. J. Exp. Bot. 61: 3321–3330. 10.1093/jxb/erq153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane H. E., Watanabe Y., Yang W., Huang Y., Ohlrogge J. et al. , 2014. Golgi- and trans-golgi network-mediated vesicle trafficking is required for wax secretion from epidermal cells. Plant Physiol. 164: 1250–1260. 10.1104/pp.113.234583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H., Hayes B. J., and Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S., Peiffer J., Brown P. J., Ersoz E. S., Zhang Z. et al. , 2009. Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21: 2194–2202. 10.1105/tpc.109.068437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neter J., Kutner M. H., Nachtsheim C. J., and Wasserman W., 1996. Applied linear statistical models, McGraw-Hill, Boston, MA. [Google Scholar]
- Ono S., 2013. The role of cyclase-associated protein in regulating actin filament dynamics–more than a monomer-sequestration factor. J. Cell Sci. 126: 3249–3258. 10.1242/jcs.128231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens B. F., Lipka A. E., Magallanes-Lundback M., Tiede T., Diepenbrock C. H. et al. , 2014. A foundation for provitamin A biofortification of maize: genome-wide association and genomic prediction models of carotenoid levels. Genetics 198: 1699–1716. 10.1534/genetics.114.169979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D., Savaldi-Goldstein S., Mandel T., Yifhar T., Franke R. B. et al. , 2007. The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol. 145: 1345–1360. 10.1104/pp.107.105676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D., Shi J. X., Bocobza S., Franke R. B., Schreiber L. et al. , 2010. The Arabidopsis DSO/ABCG11 transporter affects cutin metabolism in reproductive organs and suberin in roots. Mol. Plant 3: 563–575. 10.1093/mp/ssp103 [DOI] [PubMed] [Google Scholar]
- Petit J., Bres C., Mauxion J. P., and Bakan B., 2017. Breeding for cuticle-associated traits in crop species: traits, targets, and strategies. J. Exp. Bot. 68: 5369–5387. [DOI] [PubMed] [Google Scholar]
- Pighin J. A., Zheng H., Balakshin L. J., Goodman I. P., Western T. L. et al. , 2004. Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704. 10.1126/science.1102331 [DOI] [PubMed] [Google Scholar]
- Pollard M., Beisson F., Li Y., and Ohlrogge J. B., 2008. Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci. 13: 236–246. 10.1016/j.tplants.2008.03.003 [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R. et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, P., R. Bourgault, M. Mohammadi, L. G. Smith, M. A. Gore et al., 2019 Network analyses implicate a role for PHYTOCHROME-mediated light signaling in the regulation of cuticle development in plant leaves. bioRxiv. 10.1101/812107 (Preprint posted October 21, 2019). [DOI]
- Qualmann B., Kessels M. M., and Kelly R. B., 2000. Molecular links between endocytosis and the actin cytoskeleton. J. Cell Biol. 150: F111–F116. 10.1083/jcb.150.5.F111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2018 R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
- Ruggiero A., Punzo P., Landi S., Costa A., Van Oosten M. J. et al. , 2017. Improving plant water use efficiency through molecular genetics. Horticulturae 3: 31 10.3390/horticulturae3020031 [DOI] [Google Scholar]
- Raj A., Stephens M., and Pritchard J. K., 2014. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197: 573–589. 10.1534/genetics.114.164350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer M., and Schneider G., 1990. The effect of the environment on the permeability and composition of Citrus leaf cuticles. Planta 180: 154–165. 10.1007/BF00193990 [DOI] [PubMed] [Google Scholar]
- Riederer M., and Schreiber L., 2001. Protecting against water loss: analysis of the barrier properties of plant cuticles. J. Exp. Bot. 52: 2023–2032. 10.1093/jexbot/52.363.2023 [DOI] [PubMed] [Google Scholar]
- Ristic Z., and Jenks M. A., 2002. Leaf cuticle and water loss in maize lines differing in dehydration avoidance. J. Plant Physiol. 159: 645–651. 10.1078/0176-1617-0743 [DOI] [Google Scholar]
- Robinson M. D., McCarthy D. J., and Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romay M. C., Millard M. J., Glaubitz J. C., Peiffer J. A., Swarts K. L. et al. , 2013. Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol. 14: R55 10.1186/gb-2013-14-6-r55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter B. D., and Innes R. W., 2018. Extracellular vesicles as key mediators of plant–microbe interactions. Curr. Opin. Plant Biol. 44: 16–22. 10.1016/j.pbi.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Salvi S., Sponza G., Morgante M., Tomes D., Niu X. et al. , 2007. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 104: 11376–11381. 10.1073/pnas.0704145104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., and Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G., 1978. Estimating the dimension of a model. Ann. Stat. 6: 461–464. 10.1214/aos/1176344136 [DOI] [Google Scholar]
- Shepherd T., Robertson G. W., Griffiths D. W., and Bircht A. N. E., 1997. Effects of environment on the composition of epicuticular wax esters from kale and swede. Phytochemistry 46: 83–96. 10.1016/S0031-9422(97)00272-0 [DOI] [Google Scholar]
- Shepherd T., and Wynne Griffiths D., 2006. The effects of stress on plant cuticular waxes. New Phytol. 171: 469–499. 10.1111/j.1469-8137.2006.01826.x [DOI] [PubMed] [Google Scholar]
- Stelpflug S. C., Sekhon R. S., Vaillancourt B., Hirsch C. N., Buell C. R. et al. , 2016. An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9 10.3835/plantgenome2015.04.0025 [DOI] [PubMed] [Google Scholar]
- Studer A., Zhao Q., Ross-Ibarra J., and Doebley J., 2011. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43: 1160–1163. 10.1038/ng.942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Zhu C., Kramer M. H., Yang S.-S., Song W. et al. , 2010. Variation explained in mixed-model association mapping. Heredity 105: 333–340. 10.1038/hdy.2010.11 [DOI] [PubMed] [Google Scholar]
- Sutter E., and Langhans R. W., 1982. Formation of epicuticular wax and its effect on water loss in cabbage plants regenerated from shoot-tip culture. Can. J. Bot. 60: 2896–2902. 10.1139/b82-350 [DOI] [Google Scholar]
- Swarts K., Li H., Romero Navarro J. A., An D., Romay M. C. et al. , 2014. Novel methods to optimize genotypic imputation for low-coverage, next-generation sequence data in crop plants. Plant Genome 7: 3 10.3835/plantgenome2014.05.0023 [DOI] [Google Scholar]
- Takahashi K., Shimada T., Kondo M., Tamai A., Mori M. et al. , 2010. Ectopic expression of an esterase, which is a candidate for the unidentified plant cutinase, causes cuticular defects in Arabidopsis thaliana. Plant Cell Physiol. 51: 123–131. 10.1093/pcp/pcp173 [DOI] [PubMed] [Google Scholar]
- Valeska Zeisler-Diehl V., Migdal B., and Schreiber L., 2017. Quantitative characterization of cuticular barrier properties: methods, requirements, and problems. J. Exp. Bot. 68: 5281–5291. 10.1093/jxb/erx282 [DOI] [PubMed] [Google Scholar]
- Wallace J. G., Bradbury P. J., Zhang N., Gibon Y., Stitt M. et al. , 2014a Association mapping across numerous traits reveals patterns of functional variation in maize. PLoS Genet. 10: e1004845 10.1371/journal.pgen.1004845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J. G., Larsson S. L., and Buckler E. S., 2014b Entering the second century of maize quantitative genetics. Heredity 112: 30–38. 10.1038/hdy.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wan L., Zhang L., Zhang Z., Zhang H. et al. , 2012. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 78: 275–288. 10.1007/s11103-011-9861-2 [DOI] [PubMed] [Google Scholar]
- Xue D., Zhang X., Lu X., Chen G., and Chen Z.-H., 2017. Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front. Plant Sci. 8: 621 10.3389/fpls.2017.00621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats T. H., Buda G. J., Wang Z., Chehanovsky N., Moyle L. C. et al. , 2012a The fruit cuticles of wild tomato species exhibit architectural and chemical diversity, providing a new model for studying the evolution of cuticle function. Plant J. 69: 655–666. 10.1111/j.1365-313X.2011.04820.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats T. H., Martin L. B. B., Viart H. M.-F., Isaacson T., He Y. et al. , 2012b The identification of cutin synthase: formation of the plant polyester cutin. Nat. Chem. Biol. 8: 609–611. 10.1038/nchembio.960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats T. H., and Rose J. K. C., 2013. The formation and function of plant cuticles. Plant Physiol. 163: 5–20. 10.1104/pp.113.222737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W. H., Vroh Bi I., Yamasaki M. et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. 10.1038/ng1702 [DOI] [PubMed] [Google Scholar]
- Zhang J.-Y., Broeckling C. D., Blancaflor E. B., Sledge M. K., Sumner L. W. et al. , 2005. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 42: 689–707. 10.1111/j.1365-313X.2005.02405.x [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ersoz E., Lai C.-Q., Todhunter R. J., Tiwari H. K. et al. , 2010. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42: 355–360. 10.1038/ng.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., and Sack F. D., 1999. Ultrastructure of stomatal development in Arabidopsis (Brassicaceae) leaves. Am. J. Bot. 86: 929–939. 10.2307/2656609 [DOI] [PubMed] [Google Scholar]
- Zhou L., Ni E., Yang J., Zhou H., Liang H. et al. , 2013. Rice OsGL1–6 is involved in leaf cuticular wax accumulation and drought resistance. PLoS One 8: e65139 10.1371/journal.pone.0065139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., and Xiong L., 2013. Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice. Proc. Natl. Acad. Sci. USA 110: 17790–17795. 10.1073/pnas.1316412110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw GBS sequence data were deposited in the National Center of Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number SRP160407 and in BioProject under accession PRJNA489924. The BLUP values of the phenotypes are provided in Table S4. The FILLIN partially imputed SNP genotype data for the 235,004 loci scored on the 451 inbred lines, weather data from Maricopa and San Diego in 2016 and 2017, and leaf image data from Maricopa and San Diego in 2017 are available at CyVerse (http://datacommons.cyverse.org/browse/iplant/home/shared/GoreLab/dataFromPubs/Lin_LeafCuticle_2019). The ImageJ macro for image analysis is available at Github (https://github.com/GoreLab/Maize_leaf_cuticle/blob/master/GWAS_CE/cliveMacro_2.4.txt). RNAseq reads of epidermal cells along the developmental gradient of the expanding leaf 8 of maize inbred B73 were deposited in the NCBI SRA under accession number SRP116320 and in BioProject under accession PRJNA400334, as described in Qiao et al. (2019) preprint. Supplemental material available at figshare: https://doi.org/10.25387/g3.10262951.