Abstract
Cotton is an important crop that has made significant gains in production over the last century. Emerging pests such as the reniform nematode have threatened cotton production. The rare African diploid species Gossypium longicalyx is a wild species that has been used as an important source of reniform nematode immunity. While mapping and breeding efforts have made some strides in transferring this immunity to the cultivated polyploid species, the complexities of interploidal transfer combined with substantial linkage drag have inhibited progress in this area. Moreover, this species shares its most recent common ancestor with the cultivated A-genome diploid cottons, thereby providing insight into the evolution of long, spinnable fiber. Here we report a newly generated de novo genome assembly of G. longicalyx. This high-quality genome leveraged a combination of PacBio long-read technology, Hi-C chromatin conformation capture, and BioNano optical mapping to achieve a chromosome level assembly. The utility of the G. longicalyx genome for understanding reniform immunity and fiber evolution is discussed.
Keywords: Gossypium longicalyx, nematode resistance, cotton fiber, genome sequence, PacBio
Cotton (genus Gossypium) is an important crop that provides the largest natural source of fiber. Colloquially, the term cotton refers to one of four domesticated species, primarily the tetraploid G. hirsutum (n = 26), which is responsible for over 98% of cotton production worldwide (Kranthi 2018). Gossypium contains over 50 additional wild species related to the domesticated cottons (partitioned into groups of closely related species designated “A-G” and “K” for diploids; “AD” for tetraploids), which serve as potential sources of disease and pest resistance. Among these, Gossypium longicalyx J.B. Hutch. & B.J.S. Lee (n = 13) is the only representative of the diploid F-genome (Wendel and Grover 2015) and the only species with immunity to reniform nematode infection (Yik and Birchfield 1984). Discovered only 60 years ago (Hutchinson and B. J. S. Lee 1958), it is both cytogenetically differentiated from members of the other genome groups (Phillips 1966) and morphologically isolated (Fryxell 1971, 1992). Importantly, G. longicalyx is sister to the A-genome cottons (Wendel and Albert 1992; Wendel and Grover 2015; Chen et al. 2016), i.e., G. arboreum and G. herbaceum (both n = 13), the only diploids with long, spinnable fiber.
Interest in the genome of G. longicalyx is twofold. First, broad-scale screening of the cotton germplasm collection indicates that domesticated cotton lacks appreciable natural resistance to reniform nematode (Birchfield and Brister 1963; Yik and Birchfield 1984), and while several other species exhibit degrees of resistance, only G. longicalyx exhibits immunity to infection (Yik and Birchfield 1984). This is significant as reniform nematode has emerged as a major source of cotton crop damage, reducing cotton production by over 205 million bales per year (Lawrence et al. 2015) and accounting for ∼11% of the loss attributable to pests (Khanal et al. 2018). Current reniform resistant lines are derived from complex breeding schemes which are required to introgress reniform immunity from the diploid G. longicalyx into polyploid G. hirsutum (Bell and Robinson 2004; Dighe et al. 2009; Khanal et al. 2018); however, undesirable traits have accompanied this introgression (Nichols et al. 2010) extreme stunting of seedlings and plants exposed to dense nematode populations, prohibiting commercial deployment (Zheng et al. 2016).
The genome of G. longicalyx is also valuable because it is phylogenetically sister to the only diploid clade with spinnable fiber (Wendel and Albert 1992; Wendel and Grover 2015; Chen et al., 2016), the A-genome species, which contributed the maternal ancestor to polyploid cotton. Consequently, there has been interest in this species as the ancestor to spinnable fiber (Hovav et al. 2008; Paterson et al. 2012), although progress has been limited due to lack of genomic resources in G. longicalyx. Comparisons between the G. longicalyx genome and other cotton genomes, including the domesticated diploids (Du et al. 2018), may provide clues into the evolutionary origin of “long” fiber.
Here we describe a high-quality, de novo genome sequence for G. longicalyx, a valuable resource for understanding nematode immunity in cotton and possibly other species. This genome also provides a foundation to understand the evolutionary origin of spinnable fiber in Gossypium.
Methods & Materials
Plant material and sequencing methods
Leaf tissue of mature G. longicalyx (F1-1) was collected from a Brigham Young University (BYU) greenhouse. DNA was extracted using CTAB techniques (Kidwell and Osborn 1992), and the amount recovered was measured via Qubit Fluorometer (ThermoFisher, Inc.). The sequencing library was constructed by the BYU DNA Sequencing Center (DNASC) using only fragments >18 kb, which were size selected on the BluePippen (Sage Science, LLC) and verified in size using a Fragment Analyzer (Advanced Analytical Technologies, Inc). Twenty-six PacBio cells were sequenced from a single library on the Pacific Biosciences Sequel system. Resulting reads were assembled using Canu V1.6 using default parameters (Koren et al. 2017) to create a sequence assembly called Longicalyx_V1.0, composed of 229 large contigs (Supplemental Figure 1). This draft assembly was combined with Chicago HiRise (Koch et al. 2016; Putnam et al. 2016) + Dovetail Hi-C libraries by Dovetail Genomics to produce an intermediate assembly, Longicalyx_V3.0.
High-molecular weight DNA was extracted from young G. longicalyx leaves and subsequently purified, nicked, labeled, and repaired according to Bionano Plant protocol and standard operating procedures for the Irys platform. BssSI was used in conjunction with the IrysSolve pipeline to assemble an optical map on the BYU Fulton SuperComputing cluster. The resulting optical map was aligned to the assembly named Longicalyx_V3.0 using an in silico labeled reference sequence. Bionano maps linked large contigs present in this assembly, producing 17 large scaffolds (Longicalyx_V4.0).
Minion sequencing libraries were created and sequenced following the standard protocol from Oxford Nanopore. Scaffolds from Longicalyx_V4.0 were polished (Supplemental File 1) with existing Illumina (SRR1174179 and SRR1174182 from the NCBI Short Read Archive) and the newly generated Minion data for G. longicalyx using both PBjelly (English et al. 2012) and GapFiller (Boetzer and Pirovano 2012) to produce the final assembly, Longicalyx_V5.0.
Repeat and gene annotation
Repeats were identified using two methods. The first is a homology-based approach, i.e., a combination of RepeatMasker (Smit et al. 2015) and “One code to find them all” (Bailly-Bechet et al. 2014), whereas the second method (i.e., RepeatExplorer; (Novák et al. 2010) clusters reads based on sequence similarity and automatically annotates the most abundant cluster using RepeatMasker. Each RepeatMasker run used a custom library, which combines Repbase 23.04 repeats (Bao et al. 2015) with cotton-specific repeats. Default parameters were run, except the run was “sensitive” and was set to mask only TEs (no low-complexity). Parameters are available https://github.com/Wendellab/longicalyx. “One code to find them all” was used to aggregate multiple hits from the first method (RepeatMasker) into TE models using default parameters. The resulting output was aggregated and summarized in R/3.6.0 (R Core Team 2017) using dplyr /0.8.1(Wickham et al. 2015). Cluster results were obtained from (Grover et al. 2019) and https://github.com/IGBB/D_Cottons_USDA, and these were parsed in R/3.6.0 (R Core Team 2017). All code is available at https://github.com/Wendellab/longicalyx.
RNA-Seq libraries were generated from G. longicalyx leaf (CL), floral (FF), and stem tissues (FS) to improve genome annotation. RNA-seq libraries were independently constructed by BGI Americas (Davis, CA) using Illumina TruSeq reagents and subsequently sequenced (single-end, 50 bp). The newly sequenced G. longicalyx RNA-seq was combined with existing RNA-seq from G. longicalyx (SRR1174179) as well as two closely related species, i.e., G. herbaceum (developing fibers and seed; PRJNA595350 and SRR959585, respectively) and G. arboreum (5 seed libraries and 1 seedling; SRR617075, SRR617073, SRR617068, SRR617067, SRR959590, and SRR959508). RNA-seq libraries were mapped to the hard-masked G. longicalyx genome using hisat2 [v2.1.0] (Kim et al. 2015). BRAKER2 [v2.1.2] (Hoff et al. 2019) was used in conjunction with GeneMark [v4.36] (Borodovsky and Lomsadze 2011) generated annotations to train Augustus [v3.3.2] (Stanke et al. 2006). Mikado [v1.2.4] (Venturini et al. 2018) was used to produce high quality RNA-seq based gene predictions by combining the RNA-seq assemblies produced by StringTie [v1.3.6] (Pertea et al. 2015) and Cufflinks [v2.2.1] (Ghosh and Chan 2016) with a reference-guided assembly from Trinity [v2.8.5] (Grabherr et al. 2011) and a splice junction analysis from Portcullis [v1.2.2] (Mapleson et al. 2018). The Trinity assembly was formatted using GMAP [v2019-05-12] (Wu and Watanabe 2005). MAKER2 [v2.31.10] (Holt and Yandell 2011; Campbell et al. 2014) was used to integrate gene predictions from (1) BRAKER2 trained Augustus, (2) GeneMark, and (3) Mikado, also using evidence from all Gossypium ESTs available from NCBI (nucleotide database filtered on “txid3633” and “is_est”) and a database composed of all curated proteins in Uniprot Swissprot [v2019_07] (UniProt Consortium 2008) combined with the annotated proteins from the G. hirsutum (https://www.cottongen.org/species/Gossypium_hirsutum/jgi-AD1_genome_v1.1) and G. raimondii (n = 13; Paterson et al. 2012) genomes. Maker scored each gene model using the annotation edit distance (AED - (Eilbeck et al. 2009; Holt and Yandell 2011; Yandell and Ence 2012) metric based on EST and protein evidence provided. Gene models with an AED greater than 0.47 were removed from further analyses, and the remaining gene models were functionally annotated using InterProScan [v5.35-74.0] (Jones et al. 2014) and BlastP [v2.9.0+] (Camacho et al. 2009) searches against the Uniprot SwissProt database. Orthologs between the G. longicalyx annotations and the existing annotations for G. arboreum (Du et al. 2018), G. raimondii (Paterson et al. 2012), G. hirsutum (Hu et al. 2019), and G. barbadense (n = 26; Hu et al. 2019) were predicted by OrthoFinder using default settings (Emms and Kelly 2015, 2019). All genomes are hosted through CottonGen (https://www.cottongen.org; Yu et al. 2014) and running parameters are available from https://github.com/Wendellab/longicalyx.
ATAC-seq and data analysis
ATAC-seq was performed as described previously (Lu et al. 2017). For each replicate, approximately 200 mg freshly collected leaves or flash frozen leaves were immediately chopped with a razor blade in ∼1 ml of pre-chilled lysis buffer (15 mM Tris-HCl pH 7.5, 20 mM NaCl, 80 mM KCl, 0.5 mM spermine, 5 mM 2-mercaptoethanol, 0.2% Triton X-100). The chopped slurry was filtered twice through miracloth and once through a 40 μm filter. The crude nuclei were stained with DAPI and loaded into a flow cytometer (Beckman Coulter MoFlo XDP). Nuclei were purified by flow sorting and washed in accordance with Lu et al. (Lu et al. 2017). The sorted nuclei were incubated with 2 μl Tn5 transposomes in 40 μl of tagmentation buffer (10 mM TAPS-NaOH ph 8.0, 5 mM MgCl2) at 37° for 30 min without rotation. The integration products were purified using a Qiagen MinElute PCR Purification Kit or NEB Monarch DNA Cleanup Kit and then amplified using Phusion DNA polymerase for 10-13 cycles. PCR cycles were determined as described previously (Buenrostro et al. 2013). Amplified libraries were purified with AMPure beads to remove primers. ATAC-seq libraries were sequenced in paired-end 35 bp at the University of Georgia Genomics & Bioinformatics Core using an Illumina NextSeq 500 instrument.
Reads were adapter and quality trimmed, and then filtered using “Trim Galore” [v0.4.5] (Krueger 2015). Clean reads were subsequently aligned to the Longicalyx_V5.0 assembly using Bowtie2 [v2.3.4] (Langmead and Salzberg 2012) with the parameters “–no-mixed–no-discordant–no-unal–dovetail”. Duplicate reads were removed using Picard [v2.17.0] with default parameters (http://broadinstitute.github.io/picard/). Only uniquely mapped read pairs with a quality score of at least 20 were kept for peak calling. Phantompeakqualtools [v1.14] (Landt et al. 2012) was used to calculate the strand cross-correlation, and deepTools [v2.5.2] (Ramírez et al. 2016) was used to calculate correlation between replicates. The peak calling tool from HOMER [v4.10] (Heinz et al. 2010), i.e., findpeaks, was run in “region” mode and with the minimal distance between peaks set to 150 bp. MACS2 [v2.1.1] (Zhang et al. 2008) callpeak, a second peak-calling algorithm, was run with the parameter “-f BAMPE” to analyze only properly paired alignments, and putative peaks were filtered using default settings and false discovery rate (FDR) < 0.05. Due to the high level of mapping reproducibility (Pearson’s correlation r = 0.99 and Spearman correlation r = 0.77 by deepTools), peaks were combined and merged between replicates for each tool using BEDTools [v2.27.1] (Quinlan 2014). BEDTools was also used to intersect HOMER peaks and MACS2 peaks to only retain peak regions identified by both tools as accessible chromatin regions (ACRs) for subsequent analyses.
ACRs were annotated in relation to the nearest annotated genes in the R environment [v3.5.0] as genic (gACRs; overlapping a gene), proximal (pACRs; within 2 Kb of a gene) or distal (dACRs; >2 Kb from a gene). Using R package ChIPseeker [v1.18.0] (Yu et al. 2015), the distribution of ACRs was calculated around transcription start sites (TSS) and transcription termination sites (TTS), and peak distribution was visualized with aggregated profiles and heatmaps. To compare GC contents between ACRs and non-accessible genomic region, the BEDTools shuffle command was used to generate the distal (by excluding genic and 2 Kb flanking regions) and genic/proximal control regions (by including genic and 2 Kb flanking regions), and the nuc command was used to calculate GC content for each ACR and permuted control regions.
Identification of the RenLon region in G. longicalyx
Previous research (Dighe et al. 2009; Zheng et al. 2016) identified a marker (BNL1231) that consistently cosegrates with resistance and that is flanked by the SNP markers Gl_168758 and Gl_072641, which are all located in the region of G. longicalyx chromosome 11 referred to as “RenLon”. These three markers were used as queries of gmap (Wu and Watanabe 2005) against the assembled genome to identify the genomic regions associated with each. The coordinates identified by gmap were placed in a bed file; this file was used in conjunction with the G. longicalyx annotation and bedtools intersect (Quinlan 2014) to identify predicted G. longicalyx genes contained within RenLon. Samtools faidx (Li et al. 2009) was used to extract the 52 identified genes from the annotation file, which were functionally annotated using blast2go (blast2go basics; biobam) and including blastx (Altschul et al. 1990), gene ontology (The Gene Ontology Consortium 2019), and InterPro (Jones et al. 2014). Orthogroups containing each of the 52 RenLon genes were identified from the Orthofinder results (see above).
Comparison between G. arboreum and G. longicalyx for fiber evolution
Whole-genome alignments were generated between G. longicalyx and either G. arboreum, G. raimondii, G. turneri (Udall et al. 2019), G. hirsutum (A-chromosomes), and G. barbadense (A-chromosomes) using Mummer (Marçais et al. 2018) and visualized using dotPlotly (https://github.com/tpoorten/dotPlotly) in R (version 3.6.0) (R Development Core Team and Others 2011). Divergence between G. longicalyx and G. arboreum or G. raimondii was calculated using orthogroups that contain a single G. longicalyx gene with a single G. arboreum and/or single G. raimondii gene. Pairwise alignments between G. longicalyx and G. arboreum or G. raimondii were generated using the linsi from MAFFT (Katoh and Standley 2013). Pairwise distances between G. longicalyx and G. arboreum and/or G. raimondii were calculated in R (version 3.6.0) using phangorn (Schliep 2011) and visualized using ggplot2 (Wickham 2016). To identify genes unique to species with spinnable fiber (i.e., G. arboreum and the polyploid species), we extracted any G. arboreum gene contained within orthogroups composed solely of G. arboreum or polyploid A-genome gene annotations, and subjected these to blast2go (as above). Syntenic conservation of genes contained within the RenLon region, as compared to G. arboreum, was evaluated using GEvo as implemented in SynMap via COGE (Lyons and Freeling 2008; Haug-Baltzell et al. 2017).
Data availability
The assembled genome sequence of G. longicalyx is available at NCBI under PRJNA420071 and CottonGen (https://www.cottongen.org/). The raw data for G. longicalyx are also available at NCBI PRJNA420071 for PacBio and Minion, and PRJNA420070 for RNA-Seq. Supplemental material available at figshare: https://doi.org/10.25387/g3.11865921.
Results and Discussion
Genome assembly and annotation
We report a de novo genome sequence for G. longicalyx. This genome was first assembled from ∼144x coverage (raw) of PacBio reads, which alone produced an assembly consisting of 229 contigs with an N50 of 28.8MB (Table 1). The contigs were scaffolded using a combination of Chicago Highrise, Hi-C, and BioNano to produce a chromosome level assembly consisting of 17 contigs with an average length of 70.4 Mb (containing only 8.4kb of gap sequence). Thirteen of the chromosomes were assembled into single contigs. Exact placement of the three unscaffolded contigs (∼100 kb) was not determined, but these remaining sequences were included in NCBI with the assembled chromosomes. The final genome assembly size was 1190.7 MB, representing over 90% of the estimated genome size (Hendrix and Stewart 2005).
Table 1. Statistics for assembly versions.
| G. longicalyx assemblies* | ||||
|---|---|---|---|---|
| Longicalyx V1.0 | Longicalyx V3.0 | Longicalyx V4.0 | Longicalyx V5.0 | |
| Method | PacBio/Canu | +Chicago HighRise+HiC | +BioNano | +IllumiNa+Minion |
| Coverage | 79.45 | |||
| Total Contig Number | 229 | 135 | 17 | 17 |
| Assembly Length** | 1196.17 Mb | 1196.19 Mb | 1190.66 Mb | 1190.67 Mb |
| Average Contig Length | 5.22 Mb | 8.86 Mb | 70.04 Mb | 70.04 Mb |
| Total Length of Ns | 0 | 18200 | 18000 | 8488 |
| N50 value is | 28.88 Mb | 95.88 Mb | 95.88 Mb | 95.88 Mb |
| N90 value is | 7.58 Mb | 76.48 Mb | 76.48 Mb | 76.29 Mb |
Statistics for Longicalyx_V2.0 not calculated.
Genome size for G. longicalyx is 1311 Mb (Hendrix and Stewart 2005).
To assess genome assembly, we performed a BUSCO analysis of the completed genome (Waterhouse et al. 2017), which recovered 95.8% complete BUSCOs (from the total of 2121 BUSCO groups searched; Table 2). Most BUSCOs (86.5%) were both complete and single copy, with only 9.3% BUSCOs complete and duplicated. Less than 5% of BUSCOs were either fragmented (1.4%) or missing (2.8%), indicating a general completeness of the genome. Genome contiguity was independently verified using the LTR Assembly Index (LAI) (Ou et al. 2018), which is a reference-free method to assess genome contiguity by evaluating the completeness of LTR-retrotransposon assembly within the genome. This method, applied to over 100 genomes in Phytozome, suggests that an LAI between 10 and 20 should be considered “reference-quality”; the G. longicalyx genome reported here received an LAI score of 10.74. Comparison of the G. longicalyx genome to published cotton genomes (Table 2) suggests that the quality of this assembly is similar or superior to other currently available cotton genomes.
Table 2. BUSCO and LAI scores for the G. longicalyx genome compared to existing cotton genomes.
| Complete BUSCO | Incomplete BUSCO | LAI score | Reference | ||||
|---|---|---|---|---|---|---|---|
| Total | Single | Duplicated | Fragmented | Missing | |||
| G. longicalyx | 95.80% | 86.50% | 9.30% | 1.40% | 2.80% | 10.74 | |
| G. turneri | 95.80% | 86.00% | 9.80% | 1.00% | 3.20% | 8.51 | (Udall et al. 2019) |
| G. raimondii (BYU) | 92.80% | 85.10% | 7.70% | 2.70% | 4.50% | 10.57 | (Udall et al. 2019) |
| G. raimondii (JGI) | 98.00% | 87.30% | 10.70% | 0.70% | 1.30% | 8.51 | (Paterson et al. 2012) |
| G. arboreum (CRI) | 94.70% | 85.20% | 9.50% | 1.00% | 4.30% | 12.59 | (Du et al. 2018) |
| G. barbadense 3-79 (HAU v2) | 96.30% | 12.20% | 84.10% | 0.80% | 2.90% | 10.38 | (Wang et al. 2019) |
| G. hirsutum TM1 (HAU v1) | 97.70% | 14.50% | 83.20% | 0.50% | 1.80% | 10.61 | (Wang et al. 2019) |
Genome annotation produced 40,181 transcripts representing 38,378 unique genes. Comparatively, the reference sequences for the related diploids G. raimondii (Paterson et al. 2012) and G. arboreum (Du et al. 2018) recovered 37,505 and 40,960 genes, respectively. Ortholog analysis between G. longicalyx and both diploids suggests a simple 1:1 relationship between a single G. longicalyx gene and a single G. raimondii or G. arboreum gene for 67–68% of the G. longicalyx genes (25,637 and 26,249 genes, respectively, out of 38,378 genes; Table 3). Approximately 7–8% of the G. longicalyx genome (i.e., 2,615 and 3,158 genes) are in “one/many” (Table 3) relationships whereby one or more G. longicalyx gene model(s) matches one or more G. raimondii or G. arboreum gene model(s), respectively. The remaining 5,009 genes were not placed in orthogroups with any other cotton genome, slightly higher than the 2,016 - 2,556 unplaced genes in the other diploid species used here. While this could be partly due to genome annotation differences in annotation pipelines, it is also likely due to differences in the amount of RNA-seq available for each genome.
Table 3. Orthogroups between G. longicalyx and two related diploid species. Numbers of genes are listed and percentages within species are in parentheses. Relationships listed in the last four lines of the table represent one/many G. longicalyx genes relative to one or many genes from G. arboreum or G. raimondii.
| G. longicalyx | G. arboreum | G. raimondii* | |
|---|---|---|---|
| Number of genes | 38,378 | 40,960 | 37,223 |
| Genes in orthogroups | 33,369 (86.9%) | 38,404 (93.8%) | 35,207 (94.6%) |
| Unassigned genes | 5,009 (13.1%) | 2,556 (6.2%) | 2,016 (5.4%) |
| Orthogroups containing species** | 26,591 (78.5%) | 29,763 (87.8%) | 29,153 (86.0%) |
| Genes in species-specific orthogroups** | 74 (0.2%) | 0 | 8 (0.0%) |
| 1-to-1 relationship | 26,249 (70.5%) | 25,637 (68.9%) | |
| 1-to-many relationship | 1,207 (3.2%) | 1,153 (3.1%) | |
| many-to-1 relationship | 1,438 (3.9%) | 1,172 (3.1%) | |
| many-to-many relationship | 513 (1.4%) | 290 (0.8%) | |
| * only designated primary transcripts were included | 3158 | 8.23% | |
| ** orthogroups may contain one or more genes per species | 2615 | 6.81% | |
Repeats
Transposable element (TE) content was predicted for the genome, both by de novo TE prediction (Bailly-Bechet et al. 2014; Smit et al. 2015) and repeat clustering (Novák et al. 2010). Between 44–50% of the G. longicalyx genome is inferred to be repetitive by RepeatMasker and RepeatExplorer, respectively. While estimates for TE categories (e.g., DNA, Ty3/gypsy, Ty1/copia, etc.) were reasonably consistent between the two methods (Supplemental Table 1), RepeatExplorer recovered nearly 100 additional megabases of putative repetitive sequences, mostly in the categories of Ty3/gypsy, unspecified LTR elements, and unknown repetitive elements. Interestingly, RepeatMasker recovered a greater amount of sequence attributable to Ty1/copia and DNA elements (Supplemental Table 1); however, this only accounted for a total of 22 Mb (less than 20% of the total differences over all categories). The difference between methods with respect to each category and the total TE annotation is relatively small and may be attributable to a combination of methods (homology-based TE identification method vs. similarity clustering), the under-exploration of the cotton TE population, and sensitivity differences in each method with respect to TE age/abundance.
Because the RepeatExplorer pipeline allows simultaneous analysis of multiple samples (i.e., co-clustering), we used that repeat profile for both description and comparison to the closely related sister species, G. herbaceum and G. arboreum (from subgenus Gossypium). Relative to other cotton species, G. longicalyx has an intermediate amount of TEs, as expected from its intermediate genome size (1311 Mb; genome size range for Gossypium diploids = 841 - 2778 Mb). Approximately half of the genome (660 Mb) is composed of repetitive sequences, somewhat less than the closely related sister (A-genome) clade, whose species are slightly bigger in total size and have slightly more repetitive sequence (∼60% repetitive; Table 4). Over 80% of the G. longicalyx repetitive fraction is composed of Ty3/gypsy elements, a similar proportion to the proportion of Ty3/gypsy in subgenus Gossypium genomes. Most other element categories were roughly similar in total amount and proportion between G. longicalyx and the two subgenus Gossypium species (Supplemental Figure 2).
Table 4. Transposable element content in G. longicalyx vs. the sister clade (subsection Gossypium).
| Subsection Longiloba F-genome | Subsection Gossypium A-genome | ||
|---|---|---|---|
| G. longicalyx | G. herbaceum | G. arboreum | |
| Genome Size | 1311 | 1667 | 1711 |
| LTR/Gypsy (Ty3) | 557 | 876 | 943 |
| LTR/Copia (Ty1) | 39 | 43 | 41 |
| LTR, unspecified | 44 | 62 | 57 |
| DNA (all element types) | 2.3 | 2.7 | 2.4 |
| unknown | 18 | 27 | 25 |
| Total repetitive clustered | 660 | 1011 | 1067 |
| % genome is repet | 50% | 61% | 62% |
| % genome is gypsy | 42% | 53% | 55% |
| % repet is gypsy | 84% | 87% | 88% |
Chromatin accessibility in G. longicalyx
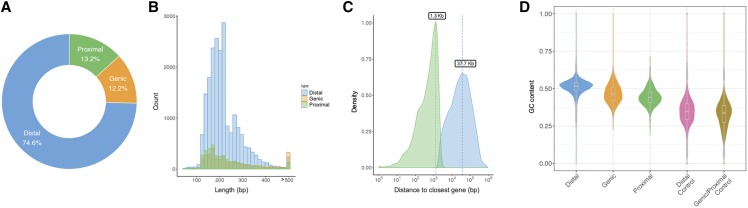
We performed ATAC-seq to map accessible chromatin regions (ACRs) in leaves. Two replicated ATAC-seq libraries were sequenced to ∼25.7 and ∼45.0 million reads per sample. The strand cross-correlation statistics supported the high quality of the ATAC-seq data, and the correlation of mapping read coverages (Pearson r = 0.99 and Spearman r = 0.77) suggested a high level of reproducibility between replicates (Supplemental Table 2). A total of 28,030 ACRs (6.4 Mb) were identified ranging mostly from 130 bp to 400 bp in length, which corresponds to ∼0.5% of the assembled genome size (Supplemental Table 3). The enrichment of ACRs around gene transcription start sites (Supplemental Figure 3) suggested that these regions were functionally important and likely enriched with cis-regulatory elements. Based on proximity to their nearest annotated genes, these ACRs were categorized as genic (gACRs; overlapping a gene), proximal (pACRs; within 2 Kb of a gene) or distal (dACRs; >2 Kb from a gene). The gACRs and pACRs represented 12.2% and 13.2% of the total number of ACRs (952 Kb and 854 Kb in size, respectively), while approximately 75% (4.6 Mb) were categorized as dACRs, a majority of which were located over 30 Kb from the nearest gene (Figure 1). This high percentage of dACRs is greater than expected (∼40% of 1 GB genome) given previous ATAC-seq studies in plants (Lu et al. 2019; Ricci et al. 2019) and may reflect challenges in annotating rare transcripts. While more thorough, species-specific RNA-seq will improve later annotation versions and refine our understanding of ACR proximity to genes, we do note that our observation of abundant dACRs and potentially long-range cis-regulatory elements is consistent with previous results (Lu et al. 2019; Ricci et al. 2019) The dACRs discovered here were the most GC-rich, followed by gACRs and pACRs (52%, 46%, and 44%, respectively), all of whom had GC contents significantly higher than randomly selected control regions with the same length distribution (Figure 1d). Because high GC content is associated with several distinct features that can affect the cis-regulatory potential of a sequence (Landolin et al. 2010; Wang et al. 2012), these results support the putative regulatory functions of ACRs.
Figure 1.
Accessible chromatin regions (ACRs) in the G. longicalyx genome. a. Categorization of ACRs in relation to nearest gene annotations - distal dACRs, proximal pACRs, and genic gACRs. b. Length distribution of ACRs that were identified by both HOMER and MACS2 contained within various genomic regions. c. Distance of gACRs and pACRs to nearest annotated genes. d. Boxplot of GC content in ACRs and control regions.
Genomics of G. longicalyx reniform nematode resistance
Reniform nematode is an important cotton parasite that results in stunted growth, delayed flowering and/or fruiting, and a reduction in both yield quantity and quality (Robinson 2007; Khanal et al. 2018). While domesticated cotton varieties are largely vulnerable to reniform nematode (Robinson et al. 1997), nematode resistance is found in some wild relatives of domesticated cotton, including G. longicalyx, which is nearly immune (Yik and Birchfield 1984). Recent efforts to elucidate the genetic underpinnings of this resistance in G. longicalyx (i.e., RENlon) identified a marker (BNL1231) that consistently cosegrates with resistance and is flanked by the SNP markers Gl_168758 and Gl_072641 (Dighe et al. 2009; Zheng et al. 2016). Located in chromosome 11, this region contains one or more closely-linked nearly dominant gene(s) (Dighe et al. 2009) that confer hypersensitivity to reniform infection (Khanal et al. 2018), resulting in the “stunting” phenotype; however, the possible effects of co-inherited R-genes has not been eliminated. Because the introgressed segment recombines at reduced rates in interspecific crosses, it has been difficult to fine-map the gene(s) of interest. Additionally, progress from marker-assisted selection has been lacking, as no recombinants have possessed the desired combination of reniform resistance and “non-stunting” (Zheng et al. 2016). Therefore, more refined knowledge of the position, identity of the resistance gene(s), mode(s) of immunity, and possible causes of “stunting” will likely catalyze progress on nematode resistance.
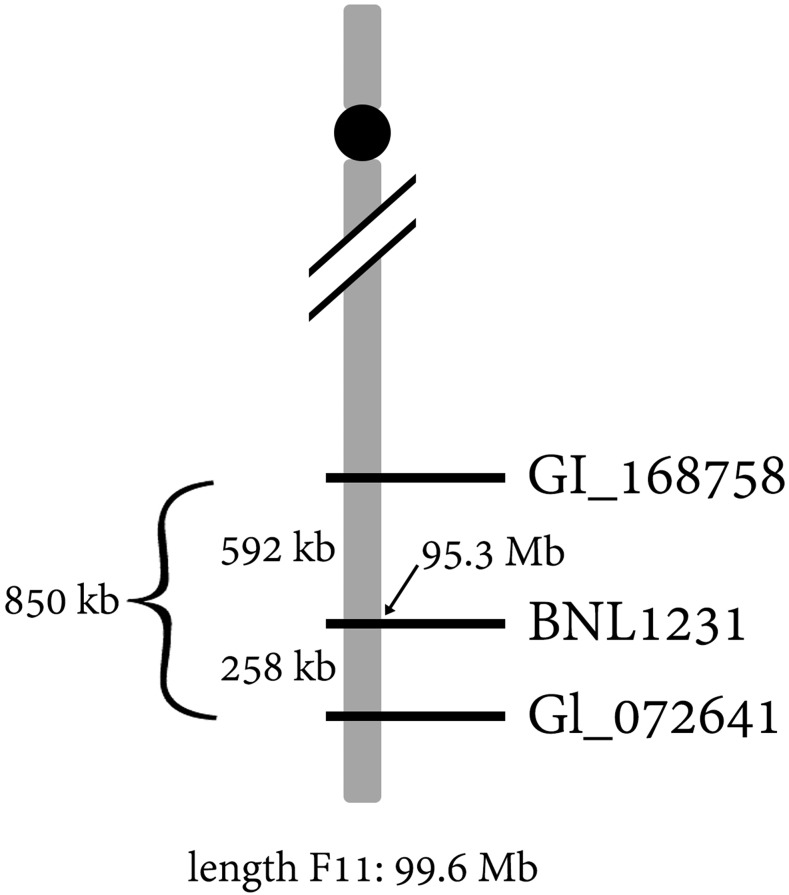
BLAST analysis of the three RENlon-associated markers (above) to the assembled G. longicalyx genome identifies an 850 kb region on chromosome F11 (positions 94747040..95596585; Figure 2) containing 52 predicted genes (Supplemental Table 4). Functional annotation reveals that over half of the genes (29, or 56%) are annotated as “TMV resistance protein N-like” or similar. In tobacco, TMV resistance protein N confers a hypersensitive response to the presence of the tobacco mosaic virus (TMV; (Erickson et al. 1999). Homologs of this gene in different species can confer resistance to myriad other parasites and pathogens, including aphid and nematode resistance in tomato (Rossi et al. 1998); fungal resistance in potato (Hehl et al. 1999) and flax (Ellis et al. 2007); and viral resistance in pepper (Guo et al. 2017). Also included in this region are 6 genes annotated as strictosidine synthase-like (SSL), which may also function in immunity and defense (Sohani et al. 2009). While the six SSL-like genes are tandemly arrayed without disruption, several other genes are intercalated within the array of TMV resistance-like genes, including the 6 SSL-like genes (Supplemental Table 4).
Figure 2.
Diagram of the RENlon region in G. longicalyx. Marker BNL1231, which co-segregates with nematode resistance, is located at approximately 95.3 Mb on chromosome F11.
Because there is agronomic interest in transferring nematode resistance from G. longicalyx to other species, we generated orthogroups between G. longicalyx, the two domesticated polyploid species (i.e., G. hirsutum and G. barbadense), and their model diploid progenitors (G. raimondii and G. arboreum; Supplemental Table 5; Supplemental File 2). Interestingly, many of the defense-relevant G. longicalyx genes in the RENlon region did not cluster into orthogroups with any other species (15 out of 38; Table 5), including 11 of the 29 TMV resistance-related genes in the RENlon region, and fewer were found in syntenic positions in G. arboreum. Most of the TMV resistance-related genes that cluster between G. longicalyx and other Gossypium species are present in a single, large orthogroup (OG0000022; Table 5), whereas the remaining TMV-resistance like genes from G. longicalyx are commonly in single gene orthogroups. Since disease resistance (R) proteins operate by detecting specific molecules elicited by the pathogen during infection (Martin et al. 2003), the increased copy number and variability among the G. longicalyx TMV-resistance-like genes may suggest specialization among copies.
Table 5. Orthogroup identity (by Orthofinder) for defense-related genes in the RenLon region and the copy number per species. In G. longicalyx, this number includes genes found outside of the RenLon region. G. hirsutum and G. barbadense copy numbers are split as genes found on the A or D chromosomes, or on scaffolds/contigs not placed on a chromosome.
| Description | Orthogroup | G. longicalyx gene in RenLon region | G. longicalyx | G. arboreum | G. raimondii | G. hirsutum | G. barbadense |
|---|---|---|---|---|---|---|---|
| adenylyl-sulfate kinase 3-like | OG0053444 | Golon.011G359300* | 1 | — | — | — | — |
| L-type lectin-domain containing receptor kinase IV.2-like | OG0053450 | Golon.011G361200 | 1 | — | — | — | — |
| T-complex protein 1 subunit theta-like | OG0053447 | Golon.011G360400 | 1 | — | — | — | — |
| protein STRICTOSIDINE SYNTHASE-LIKE 10-like | OG0000242 | Golon.011G363400 | 6 | 4 | 2 | 6 A | 9 A, 5 scaffold |
| Golon.011G363500 | |||||||
| Golon.011G363600* | |||||||
| Golon.011G363700 | |||||||
| Golon.011G363800 | |||||||
| OG0053454 | Golon.011G363300 | 1 | — | — | — | — | |
| TMV resistance protein N-like | OG0000022 | Golon.011G360100 | 25 | 22 | 5 | 10 A, 22 D | 12 A, 21 D, 1 scaffold |
| Golon.011G360300 | |||||||
| Golon.011G360500 | |||||||
| Golon.011G360700 | |||||||
| Golon.011G360800 | |||||||
| Golon.011G361000 | |||||||
| Golon.011G361100 | |||||||
| Golon.011G361400 | |||||||
| Golon.011G361900 | |||||||
| Golon.011G362000 | |||||||
| Golon.011G362400 | |||||||
| Golon.011G362700 | |||||||
| Golon.011G362800 | |||||||
| Golon.011G362900* | |||||||
| Golon.011G364000 | |||||||
| OG0028874** | Golon.011G359900 | 4 | — | — | — | — | |
| Golon.011G362600 | |||||||
| OG0028544 | Golon.011G363200 | 3 | — | — | 1 A | — | |
| OG0030067 | Golon.011G360200 | 1 | — | — | 2 A | — | |
| OG0030069 | Golon.011G362500 | 1 | — | — | 1 A | 1 A | |
| OG0053445 | Golon.011G359800 | 1 | — | — | — | — | |
| OG0053446 | Golon.011G360000 | 1 | — | — | — | — | |
| OG0053448 | Golon.011G360600 | 1 | — | — | — | — | |
| OG0053451 | Golon.011G361700 | 1 | — | — | — | — | |
| OG0053452 | Golon.011G361800 | 1 | — | — | — | — | |
| OG0053453 | Golon.011G362100 | 1 | — | — | — | — | |
| TMV resistance protein N-like isoform X1 | OG0053449 | Golon.011G360900 | 1 | — | — | — | — |
| TMV resistance protein N-like isoform X2 | OG0028874** | Golon.011G362300 | 4 | — | — | — | — |
| OG0033549 | Golon.011G363900 | 1 | — | — | — | 1 A |
This gene is syntenically conserved with G. arboreum in the COGE-GEVO analysis.
This orthogroup is split between two related, but separately named, annotations.
Comparative genomics and the evolution of spinnable fiber
Cotton fiber morphology changed dramatically between G. longicalyx and its sister clade, composed of the A-genome cottons G. arboreum and G. herbaceum. Whereas G. longicalyx fibers are short and tightly adherent to the seed, A-genome fibers are longer and suitable for spinning. Accordingly, there has been interest in the changes in the A-genome lineage that have led to spinnable fiber (Hovav et al. 2008; Paterson et al. 2012). Progress here has been limited by the available resources for G. longicalyx, relying on introgressive breeding (Nacoulima et al. 2012), microarray expression characterization (Hovav et al. 2008), and SNP-based surveys (Paterson et al. 2012) of G. longicalyx genes relative to G. herbaceum. As genomic resources and surveys for selection are becoming broadly available for the A-genome cottons, our understanding of the evolution of spinnable fiber becomes more tangible by the inclusion of G. longicalyx.
Whole-genome alignment between G. longicalyx and the closely related G. arboreum (domesticated for long fiber) shows high levels of synteny and overall sequence identity (Figure 3). In general, these two genomes are largely collinear, save for scattered rearrangements and several involving chromosomes 1 and 2; these latter may represent a combination of chromosomal evolution and/or misassembly in one or both genomes. Notably, comparison of G. longicalyx to other recently published genomes (Supplemental Figures 4-7) suggests that an inversion in the middle of G. longicalyx Chr01 exists relative to representatives of the rest of the genus; however, the other structural rearrangements are restricted to G. arboreum and its derived A subgenome in G. hirsutum and G. barbadense, suggesting that these differences are limited to comparisons between G. longicalyx and A-(sub)genomes.
Figure 3.
Synteny between G. longicalyx and domesticated G. arboreum. Mean percent identity is illustrated by the color (93–94% identity from blue to red), including intergenic regions. Lower right inset: Distribution of pairwise p-distances between coding regions of predicted orthologs (i.e., exons only, start to stop) between G. longicalyx and either G. arboreum (blue) or G. raimondii (green). Only orthologs with <5% divergence are shown, which comprises most orthologs in each comparison.
Genic comparisons between G. longicalyx and G. arboreum suggests a high level of conservation. Orthogroup analysis finds a one-to-one relationship between these two species for over 70% of genes. Most of these putative orthologs exhibit <5% divergence (p-distance) in the coding regions, with over 50% of all putative orthologs exhibiting less than 1.5% divergence. Comparatively, the median divergence for putative orthologs between G. longicalyx and the more distantly related G. raimondii is approximately 2%, with ortholog divergence generally being higher in the G. raimondii comparison (Figure 3, inset).
Because G. longicalyx represents the ancestor to spinnable fiber, orthogroups containing only G. arboreum or polyploid A-genome gene annotations may represent genes important in fiber evolution. Accordingly, we extracted 705 G. arboreum genes from orthogroups composed solely of G. arboreum or polyploid (i.e., G. hirsutum or G. barbadense) A-genome gene annotations for BLAST and functional annotation. Of these 705 genes, only 20 represent genes known to influence fiber, i.e., ethylene responsive genes (10), auxin responsive genes (5), and peroxidase-related genes (5 genes; Supplemental Table 5). While other genes on this list may also influence the evolution of spinnable fiber, identifying other candidates will require further study involving comparative coexpression network analysis or explicit functional studies.
Conclusion
While several high-quality genome sequences are available for both wild and domesticated cotton species, each new species provides additional resources to improve both our understanding of evolution and our ability to manipulate traits within various species. In this report, we present the first de novo genome sequence for G. longicalyx, a relative of cultivated cotton. This genome not only represents the ancestor to spinnable fiber, but also contains the agronomically desirable trait of reniform nematode immunity. This resource forms a new foundation for understanding the source and mode of action that provides G. longicalyx with this valuable trait, and will facilitate efforts in understanding and exploiting it in modern crop species.
Acknowledgments
We thank Emma Miller and Evan Long for technical assistance. We thank the National Science Foundation Plant Genome Research Program (Grant #1339412) and Cotton Inc. for their financial support. This research was funded, in part, through USDA ARS Agreements 58-6066-6-046 and 58-6066-6-059. Support for R.J.S and Z.L. was provided by NSF IOS-1856627 and the Pew Charitable Trusts. We thank BYU Fulton SuperComputer lab for their resources and generous support. We also thank ResearchIT for computational support at Iowa State University. We thank Rise Services for office accommodations in Orem, UT.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11865921.
Communicating editor: E. Akhunov
Literature Cited
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bailly-Bechet M., Haudry A., and Lerat E., 2014. “One code to find them all”: a perl tool to conveniently parse RepeatMasker output files. Mob. DNA 5: 13 10.1186/1759-8753-5-13 [DOI] [Google Scholar]
- Bao W., Kojima K. K., and Kohany O., 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6: 11 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, A., and A. F. Robinson, 2004 Development and characteristics of triple species hybrids used to transfer reniform nematode resistance from Gossypium longicalyx to Gossypium hirsutum, pp. 422–426 in Proceedings of the Beltwide Cotton Conferences, naldc.nal.usda.gov.
- Birchfield W., and Brister L. R., 1963. Susceptibility of cotton and relatives to reniform nematode in Louisiana. Plant Dis. Rep. 47: 990–992. [Google Scholar]
- Boetzer M., and Pirovano W., 2012. Toward almost closed genomes with GapFiller. Genome Biol. 13: R56 10.1186/gb-2012-13-6-r56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky M., and Lomsadze A., 2011. Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Curr. Protoc. Bioinformatics Chapter 4: Unit 4.6.1–Unit 4.6.10. 10.1002/0471250953.bi0406s35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., and Greenleaf W. J., 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J. et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M. S., Holt C., Moore B., and Yandell M., 2014. Genome Annotation and Curation Using MAKER and MAKER-P. Curr. Protoc. Bioinformatics 48 10.1002/0471250953.bi0411s48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Feng K., Grover C. E., Li P., Liu F. et al. , 2016. Chloroplast DNA Structural Variation, Phylogeny, and Age of Divergence among Diploid Cotton Species. PLoS One 11: e0157183 10.1371/journal.pone.0157183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dighe N. D., Robinson A. F., Bell A. A., Menz M. A., Cantrell R. G. et al. , 2009. Linkage Mapping of Resistance to Reniform Nematode in Cotton following Introgression from Gossypium longicalyx (Hutch. & Lee). Crop Sci. 49: 1151–1164. 10.2135/cropsci2008.03.0129 [DOI] [Google Scholar]
- Du X., Huang G., He S., Yang Z., Sun G. et al. , 2018. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 50: 796–802. 10.1038/s41588-018-0116-x [DOI] [PubMed] [Google Scholar]
- Eilbeck K., Moore B., Holt C., and Yandell M., 2009. Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics 10: 67 10.1186/1471-2105-10-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J. G., Dodds P. N., and Lawrence G. J., 2007. Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu. Rev. Phytopathol. 45: 289–306. 10.1146/annurev.phyto.45.062806.094331 [DOI] [PubMed] [Google Scholar]
- Emms D. M., and Kelly S., 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20: 238 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D. M., and Kelly S., 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16: 157 10.1186/s13059-015-0721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English A. C., Richards S., Han Y., Wang M., Vee V. et al. , 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 7: e47768 10.1371/journal.pone.0047768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson F. L., Holzberg S., Calderon-Urrea A., Handley V., Axtell M. et al. , 1999. The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 18: 67–75. 10.1046/j.1365-313X.1999.00426.x [DOI] [PubMed] [Google Scholar]
- Fryxell P. A., 1992. A revised taxonomic interpretation of Gossypium L (Malvaceae). Rheeda 2: 108–165. [Google Scholar]
- Fryxell P. A., 1971. Phenetic analysis and the phylogeny of the diploid species of Gossypium L. (Malvaceae). Evolution 25: 554–562. 10.1111/j.1558-5646.1971.tb01916.x [DOI] [PubMed] [Google Scholar]
- Ghosh S., and Chan C.-K. K., 2016. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol. Biol. 1374: 339–361. 10.1007/978-1-4939-3167-5_18 [DOI] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A. et al. , 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover C. E., Arick M. A. 2nd, Thrash A., Conover J. L., Sanders W. S. et al. , 2019. Insights into the evolution of the new world diploid cottons (Gossypium, Subgenus Houzingenia) based on genome sequencing. Genome Biol. Evol. 11: 53–71. 10.1093/gbe/evy256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Wang S., Liu J., Pan B., Diao W. et al. , 2017. Rapid identification of QTLs underlying resistance to Cucumber mosaic virus in pepper (Capsicum frutescens). Theor. Appl. Genet. 130: 41–52. 10.1007/s00122-016-2790-3 [DOI] [PubMed] [Google Scholar]
- Haug-Baltzell A., Stephens S. A., Davey S., Scheidegger C. E., and Lyons E., 2017. SynMap2 and SynMap3D: web-based whole-genome synteny browsers. Bioinformatics 33: 2197–2198. 10.1093/bioinformatics/btx144 [DOI] [PubMed] [Google Scholar]
- Hehl R., Faurie E., Hesselbach J., Salamini F., Whitham S. et al. , 1999. TMV resistance gene N homologues are linked to Synchytrium endobioticum resistance in potato. Theor. Appl. Genet. 98: 379–386. 10.1007/s001220051083 [DOI] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C. et al. , 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix B., and Stewart J. M., 2005. Estimation of the nuclear DNA content of Gossypium species. Ann. Bot. 95: 789–797. 10.1093/aob/mci078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff K. J., Lomsadze A., Borodovsky M., and Stanke M., 2019. Whole-genome annotation with BRAKER. Methods Mol. Biol. 1962: 65–95. 10.1007/978-1-4939-9173-0_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C., and Yandell M., 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491 10.1186/1471-2105-12-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav R., Udall J. A., Chaudhary B., Hovav E., Flagel L. et al. , 2008. The evolution of spinnable cotton fiber entailed prolonged development and a novel metabolism. PLoS Genet. 4: e25 10.1371/journal.pgen.0040025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Chen J., Fang L., Zhang Z., Ma W. et al. , 2019. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51: 739–748. 10.1038/s41588-019-0371-5 [DOI] [PubMed] [Google Scholar]
- Hutchinson J. B., and Lee B. J. S., 1958. Notes from the East African Herbarium: IX: A New Species of Gossypium from Central Tanganyika. Kew Bull. 13: 221–223. 10.2307/4109517 [DOI] [Google Scholar]
- Jones P., Binns D., Chang H.-Y., Fraser M., Li W. et al. , 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., and Standley D. M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanal C., McGawley E. C., Overstreet C., and Stetina S. R., 2018. The Elusive Search for Reniform Nematode Resistance in Cotton. Phytopathology 108: 532–541. 10.1094/PHYTO-09-17-0320-RVW [DOI] [PubMed] [Google Scholar]
- Kidwell K. K., and Osborn T. C., 1992. Simple plant DNA isolation procedures, pp. 1–13 in Plant Genomes: Methods for Genetic and Physical Mapping, edited by Beckmann J. S., and Osborn T. C.. Springer Netherlands, Dordrecht: 10.1007/978-94-011-2442-3_1 [DOI] [Google Scholar]
- Kim D., Langmead B., and Salzberg S. L., 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch L., 2016. Chicago HighRise for genome scaffolding. Nat. Rev. Genet. 17: 194.26900024 [Google Scholar]
- Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H. et al. , 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranthi K. R., 2018. Cotton production practices: snippets from global data 2017. The ICAC Recorder XXXVI: 4–14. [Google Scholar]
- Krueger, F., 2015 Trim galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files.
- Landolin J. M., Johnson D. S., Trinklein N. D., Aldred S. F., Medina C. et al. , 2010. Sequence features that drive human promoter function and tissue specificity. Genome Res. 20: 890–898. 10.1101/gr.100370.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt S. G., Marinov G. K., Kundaje A., Kheradpour P., Pauli F. et al. , 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22: 1813–1831. 10.1101/gr.136184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence K., Olsen M., Faske T., Hutmacher R., Muller J. et al. , 2015. Cotton disease loss estimate committee report, 2014, pp. 188–190 in Proceedings of the 2015 Beltwide Cotton Conferences, San Antonio, TX. Cordova: National Cotton Council [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Hofmeister B. T., Vollmers C., DuBois R. M., and Schmitz R. J., 2017. Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res. 45: e41 10.1093/nar/gkw1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Marand A. P., Ricci W. A., Ethridge C. L., Zhang X. et al. , 2019. The prevalence, evolution and chromatin signatures of plant regulatory elements. Nat. Plants 5: 1250–1259. 10.1038/s41477-019-0548-z [DOI] [PubMed] [Google Scholar]
- Lyons E., and Freeling M., 2008. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 53: 661–673. 10.1111/j.1365-313X.2007.03326.x [DOI] [PubMed] [Google Scholar]
- Mapleson D., Venturini L., Kaithakottil G., and Swarbreck D., 2018. Efficient and accurate detection of splice junctions from RNA-seq with Portcullis. Gigascience 7: 1–11. 10.1093/gigascience/giy131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Delcher A. L., Phillippy A. M., Coston R., Salzberg S. L. et al. , 2018. MUMmer4: A fast and versatile genome alignment system. PLOS Comput. Biol. 14: e1005944 10.1371/journal.pcbi.1005944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. B., Bogdanove A. J., and Sessa G., 2003. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54: 23–61. 10.1146/annurev.arplant.54.031902.135035 [DOI] [PubMed] [Google Scholar]
- Nacoulima N., Baudoin J. P., and Mergeai G., 2012. Introgression of improved fiber fineness trait in G. hirsutum L. from G. longicalyx Hutch. &. Lee. Commun. Agric. Appl. Biol. Sci. 77: 207–211. [PubMed] [Google Scholar]
- Nichols R. L., Bell A., Stelly D., Dighe N., Robinson F. et al. , 2010. Phenotypic and genetic evaluation of LONREN germplasm, pp. 798–799 in Proc. Beltwide Cotton Conf. New Orleans, LA [Google Scholar]
- Novák P., Neumann P., and Macas J., 2010. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11: 378 10.1186/1471-2105-11-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S., Chen J., and Jiang N., 2018. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res. 46: e126 10.1093/nar/gky730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Wendel J. F., Gundlach H., Guo H., Jenkins J. et al. , 2012. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492: 423–427. 10.1038/nature11798 [DOI] [PubMed] [Google Scholar]
- Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T. et al. , 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips L. L., 1966. The cytology and phylogenetics of the diploid species of Gossypium. Am. J. Bot. 53: 328–335. 10.1002/j.1537-2197.1966.tb07343.x [DOI] [Google Scholar]
- Putnam N. H., O’Connell B. L., Stites J. C., Rice B. J., Blanchette M. et al. , 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26: 342–350. 10.1101/gr.193474.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., 2014. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinformatics 47: 11.12.1–, 11.12.34.. 10.1002/0471250953.bi1112s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F. et al. , 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2017. R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- R Development Core Team, R., and Others, 2011 R: A language and environment for statistical computing.
- Ricci W. A., Lu Z., Ji L., Marand A. P., Ethridge C. L. et al. , 2019. Widespread long-range cis-regulatory elements in the maize genome. Nat. Plants 5: 1237–1249. 10.1038/s41477-019-0547-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. F., 2007. Reniform in U.S. cotton: when, where, why, and some remedies. Annu. Rev. Phytopathol. 45: 263–288. 10.1146/annurev.phyto.45.011107.143949 [DOI] [PubMed] [Google Scholar]
- Robinson A. F., Inserra R. N., Caswell-Chen E. P., Vovlas N., and Troccoli A., 1997. Rotylenchulus Species: Identification, Distribution, Host Ranges, and Crop Plant Resistance | Nematropica. Nematropica 27: 127–180. [Google Scholar]
- Rossi M., Goggin F. L., Milligan S. B., Kaloshian I., Ullman D. E. et al. , 1998. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA 95: 9750–9754. 10.1073/pnas.95.17.9750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep K. P., 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27: 592–593. 10.1093/bioinformatics/btq706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. F. A., R. Hubley, and P. Green, 2015 RepeatMasker Open-4.0. 2013–2015.
- Sohani M. M., Schenk P. M., Schultz C. J., and Schmidt O., 2009. Phylogenetic and transcriptional analysis of a strictosidine synthase-like gene family in Arabidopsis thaliana reveals involvement in plant defence responses. Plant Biol. 11: 105–117. 10.1111/j.1438-8677.2008.00139.x [DOI] [PubMed] [Google Scholar]
- Stanke M., Keller O., Gunduz I., Hayes A., Waack S. et al. , 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34: W435–W439. 10.1093/nar/gkl200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium , 2019. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47: D330–D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall J. A., Long E., Hanson C., Yuan D., Ramaraj T. et al. , 2019. De novo genome sequence assemblies of Gossypium raimondii and Gossypium turneri. G3 (Bethesda) 9: 3079–3085. 10.1534/g3.119.400392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium , 2008. The universal protein resource (UniProt). Nucleic Acids Res. 36: D190–D195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini L., Caim S., Kaithakottil G. G., Mapleson D. L., and Swarbreck D., 2018. Leveraging multiple transcriptome assembly methods for improved gene structure annotation. Gigascience 7: 1–12. 10.1093/gigascience/giy093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhuang J., Iyer S., Lin X., Whitfield T. W. et al. , 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 22: 1798–1812. 10.1101/gr.139105.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R. M., Seppey M., Simão F. A., Manni M., Ioannidis P. et al. , 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35: 543–548. 10.1093/molbev/msx319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J. F., and Albert V. A., 1992. Phylogenetics of the Cotton Genus (Gossypium): Character-State Weighted Parsimony Analysis of Chloroplast-DNA Restriction Site Data and Its Systematic and Biogeographic Implications. Syst. Bot. 17: 115–143. 10.2307/2419069 [DOI] [Google Scholar]
- Wendel J. F., and Grover C. E., 2015. Taxonomy and Evolution of the Cotton Genus, Gossypium, pp. 25–44 in Cotton. Agronomy Monograph, American Society of Agronomy, Inc., Crop Science Society of America, Inc., and Soil Science Society of America, Inc., Madison, WI. [Google Scholar]
- Wickham, H., 2016 ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York. [Google Scholar]
- Wickham, H., R. Francois, L. Henry, K. Müller, and Others, 2015 dplyr: A grammar of data manipulation. R package version 0. 4 3.:
- Wu T. D., and Watanabe C. K., 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875. 10.1093/bioinformatics/bti310 [DOI] [PubMed] [Google Scholar]
- Yandell M., and Ence D., 2012. A beginner’s guide to eukaryotic genome annotation. Nat. Rev. Genet. 13: 329–342. 10.1038/nrg3174 [DOI] [PubMed] [Google Scholar]
- Yik C. P., and Birchfield W., 1984. Resistant Germplasm in Gossypium Species and Related Plants to Rotylenchulus reniformis. J. Nematol. 16: 146–153. [PMC free article] [PubMed] [Google Scholar]
- Yu J., Jung S., Cheng C.-H., Ficklin S. P., Lee T. et al. , 2014. CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42: D1229–D1236. 10.1093/nar/gkt1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., and He Q.-Y., 2015. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31: 2382–2383. 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S. et al. , 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Hoegenauer K. A., Quintana J., Bell A. A., Hulse-Kemp A. M. et al. , 2016. SNP-Based MAS in cotton under depressed-recombination for Renlon–flanking recombinants: results and inferences on wide-cross breeding strategies. Crop Sci. 56: 1526–1539. 10.2135/cropsci2015.07.0436 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The assembled genome sequence of G. longicalyx is available at NCBI under PRJNA420071 and CottonGen (https://www.cottongen.org/). The raw data for G. longicalyx are also available at NCBI PRJNA420071 for PacBio and Minion, and PRJNA420070 for RNA-Seq. Supplemental material available at figshare: https://doi.org/10.25387/g3.11865921.