Abstract
Several techniques have been developed to study specific gene function in loss-of-function situations. In Drosophila melanogaster, RNAi and the generation of mutant clones are widely used. However, both techniques have the limitation that there is a significant time lag before gene function is abolished. Given the relatively rapid development of Drosophila, such perdurance is a serious impediment to study gene function. Here we describe the adaptation of the anchor-away technique for use in Drosophila. Anchor-away was originally developed in yeast to quickly and efficiently abrogate the function of nuclear proteins by sequestering - anchoring - them away in a different cellular compartment. The required components are present in the cells, and the system is triggered by the addition of rapamycin, resulting in a rapid generation of a loss-of-function situation. We provide here proof of principle for the system by producing loss-of-function situations for two nuclear proteins – Pygopus and Brinker. The system allows to study the requirement of any protein during any time window, and at the same time circumvents difficulties, such as off-target effects or variable phenotypes, which are inherent in other techniques, for example RNAi.
Keywords: Drosophila melanogaster, mTOR, rapamycin, pygopus, brinker
Loss-of-function (LOF) experiments have been performed for decades to study gene function. In Drosophila, several methods have been developed and extensively used (Cooley et al. 1998, Adams & Sekelsky 2002). Mutagenesis screens have led to the discovery of the function of hundreds of proteins and were integral in the quest for identifying pathway components in the embryo (Nüsslein-Volhard & Wieschaus 1980; St Johnston 2002; Jenny & Basler 2014). To facilitate the study of the function of an essential gene at later stages, techniques were developed that interfered with gene function only regionally (induction of genetic mosaics) or only transiently (conditional alleles). The preeminent approach that allowed the generation of mosaic LOF situations in tissues was the generation of mitotic recombinant clones (Xu & Rubin 1993). RNAi (Fire et al. 1998) was the most widely adopted and extensively used method to transiently down-regulate gene expression in Drosophila; one reason for this was the generation of libraries where almost any gene in the genome could be targeted, allowing scientists to performing reverse genetics. Although powerful, these methods have a major drawback: they do not directly target the protein but act on the gene or the mRNA level and are thus sensitive to issues such as protein half-life, causing a delay before the LOF takes effect in the tissue (Boutros & Ahringer 2008).
To overcome this problem, several approaches have been developed to achieve a more rapid and efficient LOF by targeting the protein directly. These methods rely on targeted protein degradation, cleavage or sequestering (Harmansa et al. 2017; Haruki et al. 2008; Caussinus et al. 2011). One of these methods, developed in yeast, is the anchor-away technique. LOF is achieved by sequestering the target protein in another compartment of the cell where it is unable to perform its physiological function. This sequestering is triggered by the addition of rapamycin, allowing investigators to trigger the LOF at any time point. The effect of the anchor-away method is essentially instantaneous as all the necessary components are already present in the cell, and the loss of function is triggered by the addition of rapamycin (Haruki et al. 2008).
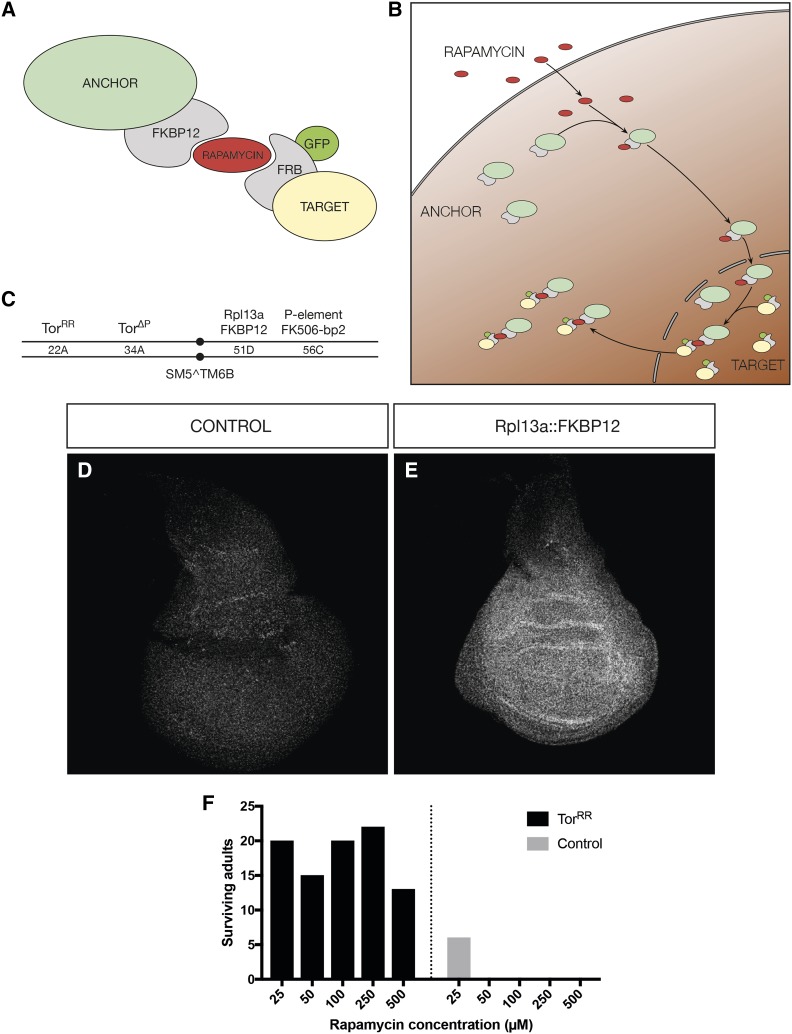
The technique is based on a binary system whose components have to be integrated beforehand: an anchor protein (by which the protein of interest will be sequestered) and an engineered target protein. The anchoring process is based on the interaction between the human FK506 binding protein (FKBP12), and the 11 kD FKBP12-rapamycin-binding (FRB) domain of the human mTor (Chen et al. 1995; Belshaw et al. 1996). Rapamycin binds to FKBP12, and this creates an interaction surface for FRB, which binds and forms a tight ternary complex (Chen et al. 1995). By tagging the anchor with FKBP12 and the target with FRB, the two proteins will bind strongly to each other after the addition of rapamycin. As a consequence, the target will be sequestered to the subcellular compartment where the anchor is located (Figure 1A).
Figure 1.
Adapting the anchor-away to Drosophila. A) Schematic of the anchor-away components. B) Schematic of the mechanism of action of the anchor-away upon rapamycin addition. The anchor first binds rapamycin, and this complex drives the capture of the target protein to the cytoplasm. C) Chromosomal localization of the anchor-away components in the second chromosome. The components are either homozygous or balanced over SM5^TM6b. D-E) Staining against human FKBP12 in control discs without Rpl13a::FKBP12 (D), or discs from larvae carrying Rpl13a::FKBP12 (E). F) Survival of animals upon different rapamycin treatments with and without TorRR.
There are various possibilities when anchoring proteins away, depending on the subcellular location of the protein of interest. For nuclear proteins, a cytoplasmic anchor is an obvious choice, and ribosomal proteins have been shown to be suitable for this, as once ribosomes are assembled, they will remain cytoplasmic (Haruki et al. 2008). In addition, ribosomal proteins translocate to the nucleus after biosynthesis, where they combine with the different rRNA molecules to assemble ribosomes. Afterward, the large and small ribosomal complexes are translocated to the cytoplasm (Zemp & Kutay 2007; Köhler & Hurt 2007). In this process, the target protein will also bind to the ribosomal protein anchor, and is subsequently translocated to the cytoplasm, where it is prevented from going back to the nucleus (Figure 1B). If the target protein is cytoplasmic, a membrane-bound anchor has been shown to be efficient (Tsuchiya et al. 2013). The development of a suitable anchor is key to this method and will depend mainly on the cellular localization of the target to be anchored away.
In the present work, we adapted the anchor-away technique to Drosophila: we devised a ribosomal protein anchor to be able to study LOF of Drosophila nuclear proteins. As a proof of principle, we have tested the technique with two nuclear factors of independent pathways – Pygopus (Wingless signaling) and Brinker (Decapentaplegic signaling). The LOF phenotypes confirmed the specificity and efficiency of this system.
Material and methods
Drosophila strains
The following fly stocks were used for the experiments: TorΔP and 22A-TorS1956T (Zhang et al. 2000), FK506-bp2 Kyoto stock 205244 (P{GSV6}GS10737) (Toba et al. 1999), brkM68 (Jaźwińska et al. 1999), 56C-Rpl13a::FKBP12, 86Fb-FRBGFP::Brk, FRBGFP::Pygo, C765-Gal4, Vienna RNAi stocks v19692, v19693, v100724, v2919 and v100824.
Cloning procedures
rpl13a was tagged with 2xFKBP12 under the control of its own promoter, endogenous 5′ and 3′ UTRs and supposedly all its endogenous regulatory regions. The resulting transgene was integrated via the C31 integrase system into the landing site at 51D (Bischof et al. 2007).
FRB-GFP::Brk was generated by fusing the FRB-GFP cassette into a BAC containing the whole brk genomic region by BAC recombineering (Warming et al. 2005). The resulting vector was integrated via the C31 integrase into the landing site at 86Fb (Bischof et al. 2007).
For FRBGFP::Pygo, CRISPR gRNA were cloned in pU6-BbsI-gRNA (Gratz et al. 2013). A donor plasmid (pFRBGFP) was generated by using the pDsRed-attP plasmid as a backbone. We replaced the fragment between the multiple cloning sites for the homology regions with an in frame FRBGFP DNA fragment, by digesting the plasmid with AarI and SapI and cloning a PCR fragment containing the FRB-GFP fragment in such a way that it will be in frame once the homology arms are cloned in the plasmid. gRNA and donor plasmid were co-injected into embryos expressing nos-Cas9 (Port et al. 2014). The F1 was screened by PCR to confirm the insertion of the FRBGFP fragment in the correct region.
Immunostaining
Third instar imaginal discs were dissected in PBS and fixed during 30 min with 4% Formaldehyde in PBS. Prior to antibody staining, discs were blocked with 2% heat inactivated goat serum (HINGS) and stained overnight with primary antibodies. The following antibodies and concentrations were used: guinea pig α-Sens (Nolo et al. 2000), 1:1000; guinea pig α-Brk (Doumpas et al. 2013), 1:500; Cell Signaling mouse α-FKBP12, 1:500. Secondary antibody staining was performed for 2 hr, using Thermo Fisher Alexa antibodies. Discs were mounted in Vectashield and images were taken with a Zeiss LSM710 confocal microscope.
Rapamycin culture ex vivo
Imaginal wing discs were dissected in Wing Medium 1 (WM1) (Restrepo et al. 2016) and transferred to reaction tubes. The solution was replaced by WM1 containing rapamycin 50 µM and incubated for 1 to 4 hr. After incubation, Rapamycin was removed and discs were fixed and stained as described in the prior section.
Data availability
All plasmids, fly strains and reagents used for the study are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25387/g3.11959209.
Results
Adapting the anchor-away system to Drosophila
To test and apply the anchor-away method in Drosophila melanogaster it was necessary to generate an anchor appropriate for a functional target. In addition, the modification of various genes was required (Figure 1C). First, we wanted to generate a genetic background in which Drosophila is insensitive to rapamycin. The primary target of rapamycin is Tor. We thus introduced a tor transgene with the mutation S1956T. This mutation renders Tor rapamycin resistant (Zhang et al. 2000). It was crossed into a tor null mutant background (tor∆P).
Second, to avoid potential competitive binding to rapamycin, we also abolished the expression of an endogenous FK506 binding protein, the Drosophila homolog of the yeast FPR1, FK506-bp2. For this we used a null allele, which carried a P-element insertion in the second exon of FK506-bp2 (Toba et al. 1999).
Next, we generated a protein anchor, which we wanted to be expressed ubiquitously and at high levels to ensure efficient sequestering of the target. We selected the ribosomal protein Rpl13a, the homolog of the protein used in the yeast system (Haruki et al. 2008). This protein has an exposed C-terminus (Haruki et al. 2008, Anger et al. 2013), allowing it to be fused to two copies of the human FKBP12 rapamycin-binding domain. These modifications were made in the context of a genomic rescue construct such that the gene was controlled by its endogenous regulatory elements. The construct was integrated in the second chromosome via attB/attP integration (Bischof et al. 2007), and proper localization of the protein was assessed by immunostaining (Figure 1D-E). In parallel, we generated and introduced a transgene, UAS-FKBP12::Rpl13a, that could potentially be used to restrict the anchoring to a subset of cells by using compartment- or tissue-specific Gal4 lines.
Once all transgenes were integrated, we assessed if the animals were viable when exposed to rapamycin. We transferred eggs into food containing rapamycin, from 20 to 500 µM to determine a concentration that will affect WT but not the rapamycin-resistant anchor-away larvae. Based on the viability, we concluded that a concentration of 50 µM rapamycin was optimal (Figure 1F), similar to what has been used in prior reports in which Tor signaling was studied in Drosophila (Oldham et al. 2000). Lower amounts of rapamycin allowed some WT larvae to develop, and very high concentrations affected even the transgenic anchor-away strain.
Generation of anchorable Brinker and Pygopus variants
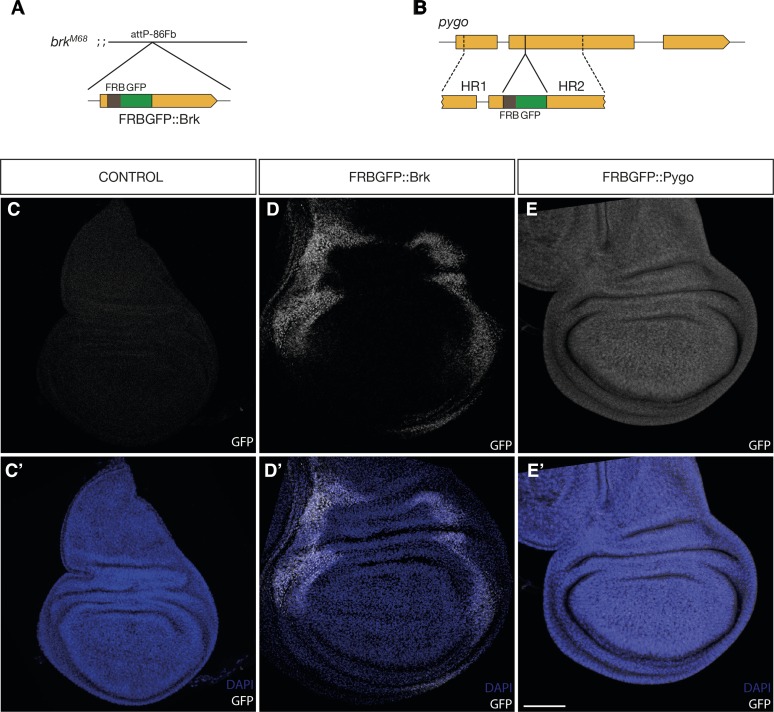
To test the feasibility of anchoring nuclear proteins in Drosophila, we generated FRB-tagged variants of two well-studied nuclear factors — Brinker (Brk) and Pygopus (Pygo). Brk is the main effector of the Decapentaplegic (Dpp) pathway in Drosophila. Its expression domain in the wing imaginal disc is restricted to the lateral regions, and a brk LOF produces a clear overgrowth phenotype and derepression of Dpp target genes (Jaźwińska et al. 1999). Pygo is one the binding partners of Armadillo, the fly homolog of β-catenin. Its recruitment is critical in the signal transduction of Wingless (Wg) target genes (Kramps et al. 2002). Mutants for pygo present severe undergrowth phenotypes. In addition to the rapamycin binding FRB tag, we also added the eGFP sequence (FRB-GFP), to be able to directly localize the anchored targets. The targets were engineered by using two different approaches.
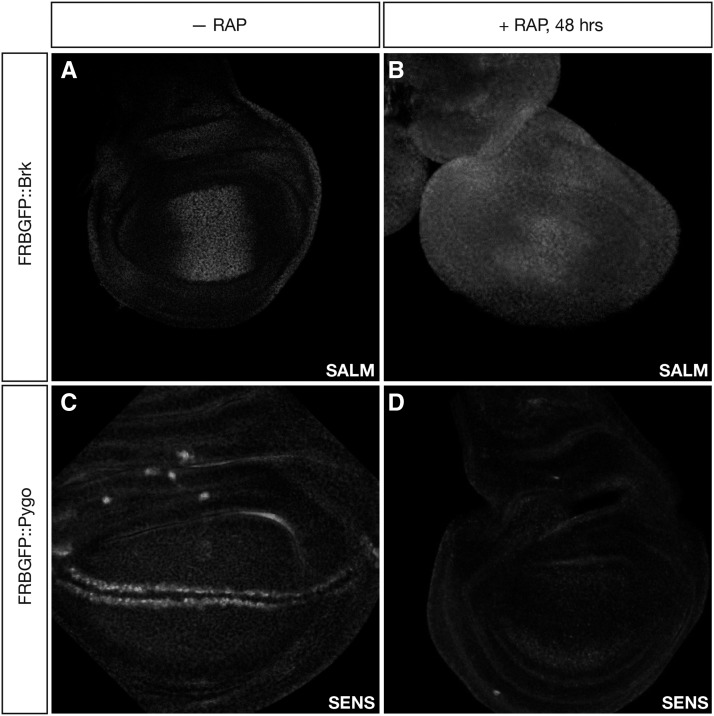
To generate FRB-GFP::brk, we introduced the FRB-GFP tag in the N-terminal end of brk, and cloned it in a BAC construct to integrate the whole genomic fragment via attB/attP in the third chromosome, in the attP-86Fb locus (Bischof et al. 2007), depicted in Figure 2A. Both the expression pattern and the subcellular localization of the fusion protein, assessed by GFP expression, were the same as those of the endogenous Brk protein (Figure 2D-D’). To assess the efficiency of the anchoring, the transgenic construct was crossed into a brkM68 null background (Jaźwińska et al. 1999), which also contained the other anchor-away transgenes. Anchoring of FRB-GFP::Brk by rapamycin exposure was lethal at the third larval stage. We assessed the effect of Brk sequestration by observing the phenotypes of third instar larval wing discs. Discs from rapamycin-treated animals exhibited overgrowth, resembling the phenotype of brk LOF. The effect in Dpp signaling was confirmed by immunostaining against the downstream target spalt major (salm). Discs where Brk was anchored away showed widespread derepression of Salm, as it is expected for a brk LOF situation (Figure 3A-B).
Figure 2.
Establishing anchorable Brk and Pygo. A) Schematic of the brk BAC rescue. This BAC was integrated on the third chromosome and coupled with the null allele brkM68. B) Schematic of the modification of the pygo locus by CRISPR/Cas9. FRBGFP was integrated in frame right after the 5′UTR in the second exon of pygo. C-E’) Detection of the GFP-tagged targets. C-C’) Control discs without anchor-away target. D-D’) Discs carrying FRBGFP::Brk over a brk-null background. The shape of the disc is normal and Brk is localized in the normal expressing region and is present in the nucleus. E-E’) Discs carrying homozygous FRBGFP::Pygo. Discs retain a WT-like shape and Pygo is produced ubiquitously and localizes in the nucleus as expected. Scalebar = 50 μM.
Figure 3.
Pygo and Brk are successfully anchored in vivo. A) FRBGFP::Brk disc without rapamycin treatment. B) FRBGFP::Brk disc dissected 48 hr after feeding rapamycin to larvae. C) Control disc carrying FRBGFP::Pygo, without rapamycin treatment. D) Disc carrying FRBGFP::Pygo dissected 48 hr after feeding rapamycin to larvae.
To generate a FRB-GFP-tagged Pygo, we used the CRISPR/Cas9 method (Port et al. 2014). We recombined the FRB-GFP fragment in frame at the beginning of the open reading frame of the endogenous pygo gene (Figure 2B). As with Brk, Pygo expression was unaffected by the modification and showed the expected localization (Figure 2E-E’). Homozygous FRB-GFP::pygo animals carrying the anchor-away transgenes were exposed to rapamycin. This caused a developmental arrest, and larvae were not able to pupariate. To determine the functional consequence of anchoring-away Pygo, we assayed the expression of the Wg target gene Senseless (Sens) by immunostaining. In larvae fed with rapamycin for 48 hr, Sens expression was completely abolished, confirming the inactivation of the Wg pathway (Figure 3C-D).
These results demonstrate that the Anchor-away method works in Drosophila to induce an acute LOF situation.
Anchoring away as a fast and efficient knock-down system
We next wanted to assess how rapidly the anchor-away method creates a LOF situation. Due to the ubiquitous localization of Pygo in the wing disc, we used FRB-GFP::Pygo to measure how fast the cytoplasmic anchor traps a nuclear target in Drosophila.
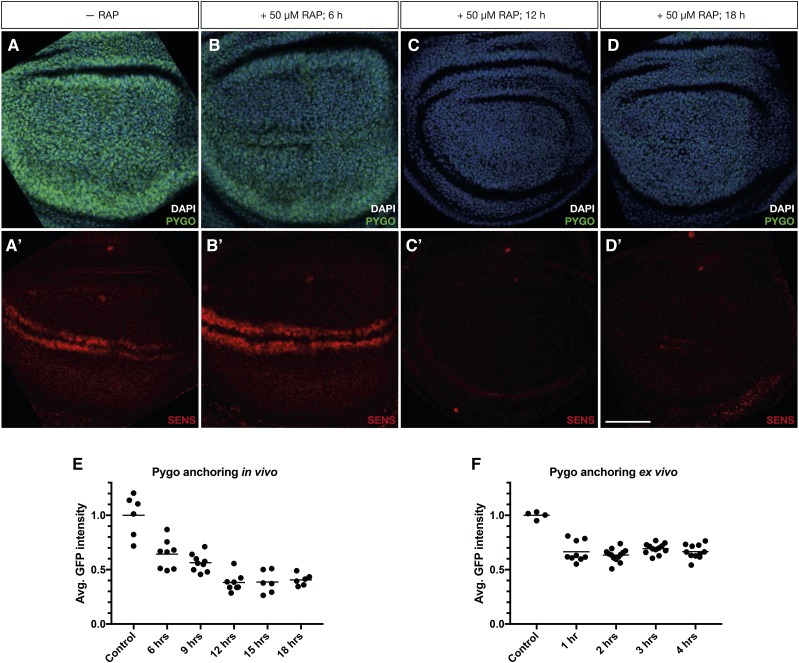
We first assessed how quickly after feeding larvae with rapamycin relocalization of FRB-GFP::Pygo occurs. We collected eggs in regular food and let the ensuing larvae develop until third instar. We then transferred them to rapamycin-containing food and dissected them at defined time points. To determine when the effect of Pygo anchoring away affected target genes, we again used Sens protein levels as a readout (Figure 4A’-D’). We found that 12 hr after treatment Sens protein was not detectable anymore (Figure 4C’). Pygo localization was also affected (Figure 4A-D, E). As early as 6 hr after rapamycin feeding, there is a clear decay in the nuclear FRB-GFP::Pygo signal (Figure 4B, E). At 12 hr after treatment, the Pygo signal was much lower than the control without treatment (Figure 4C,E). We hypothesize that the decay in signal is due to the sequestering of Pygo to the cytoplasm. As the protein is now more diffuse, the fluorescent signal is also more delocalized. We measured the amount of Pygo by Western blot to find out if cytoplasmically anchored Pygo is degraded at a higher rate by the proteasome, causing the decrease of the signal. However, Pygo levels did not change over the course of 18 hr (Suppl. Figure 1), in contrast with the decay in fluorescence signal (≥ 50%).
Figure 4.
Pygo anchoring in vivo achieves a LOF phenotype in 12 hr. A-D) Pygo (green) fluorescence decays over time after rapamycin treatment. A’-D’) Sens is undetectable 12 hr after rapamycin treatment. A, A’) Control discs without rapamycin treatment, B-D’) Representative discs from larvae fed with rapamycin for 6 hr (B, B’), 12 hr (C, C’) and 18 hr (D, D’). E) Decay in GFP::Pygo fluorescence after feeding larvae with rapamycin. The minimal intensity is achieved already at 6 hr after rapamycin treatment. F) Decay in GFP::Pygo fluorescence after treating dissected discs with 50 mM rapamycin. The maximal decay happens in the timeframe of 1 hr and is maintained thereafter. Scalebar = 50 µM.
In conclusion, the effect of the anchor-away in vivo is detectable as early as 6 hr after rapamycin feeding, and the pathway is inhibited 12 hr after treatment.
To assess how rapidly Pygo is anchored ex vivo, we dissected third instar discs and applied rapamycin-containing Wing Medium 1 (Restrepo et al. 2016). We then measured the anchoring efficiency ex vivo by the fluorescence decay due to protein diffusion to the cytoplasm, as it was the fastest readout of the rapamycin-induced anchoring in vivo (Figure 4E). When cultured in rapamycin, most nuclear Pygo was anchored already 1 hr after exposure to rapamycin, and Pygo signal was low for at least 4 hr of culture (Figure 3H). Therefore, anchoring proteins away is also feasible and highly efficient in Drosophila cultured discs.
Depletion via anchor-away is more effective than RNAi downregulation
RNAi-mediated downregulation (Fire et al. 1998) is widely used in Drosophila, in part due to the existence of collections targeting all the genes in the genome. Other advantage is the possibility of spatio-temporal control of the knock-down (Dietzl et al. 2007; Ni et al. 2009). Despite these benefits, RNAi downregulation has drawbacks, such as off-target effects, that can confound analyses and high variability or delayed repression of targets (Boutros & Ahringer 2008).
The anchor-away technique is a viable alternative to RNAi, especially for studies of a small number of genes. One of the major impediments of RNAi is that its efficiency can vary greatly (Boutros & Ahringer 2008). We tested two different RNAi lines which target pygo and two RNAi lines targeting brk and compared the efficiency of their knock-down functionally examining the effect on Sens and Salm levels, respectively (Suppl. Figure 2). We triggered expression of the RNAi transgene for 48 hr by using the disc driver C765-Gal4. Although both RNAi targeting pygo were able to decrease its levels up to the point where Sens was barely detectable (Suppl. Figure 2A-C), in some discs Sens levels were still high, or not affected at all (Suppl. Figure 2B’-C’). In addition, the RNAi constructs against brk were not able to decrease Brk function significantly when driven by C765-Gal4, as salm expression was unaffected (Suppl. Figure 2D-F). This illustrates the variability inherent in the RNAi method.
The effects of the anchor-away system were more reliable, as Sens staining was never detected after rapamycin treatment in several independent tests of the system (Figure 5B-B’). Only for the situations in which RNAi worked, it was as effective as the anchor-away method (Figure 5D-D’, Suppl. Figure 2B). However, the variability exhibited by the different RNAi lines poses a real problem to perform LOF experiments.
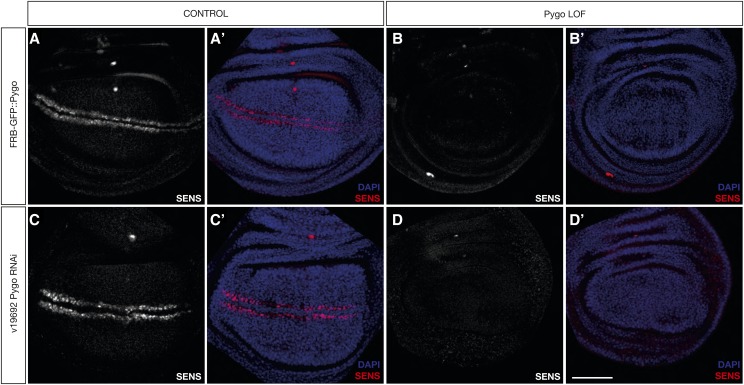
Figure 5.
The Anchor-away yields LOF phenotypes as efficiently as RNAi. A-A’) FRBGFP::Pygo discs without rapamycin treatment. B-B’) FRBGFP::Pygo discs dissected 48 hr after rapamycin treatment. C-C’) Discs without induction of pygo RNAi. D-D’) Discs expressing pygo RNAi during 48 hr. Scalebar = 50 μM
All in all, our results show that the anchor-away is an efficient alternative method in Drosophila to perform LOF analyses.
Discussion
The anchor-away technique has been developed in yeast (Haruki et al. 2008; Ding et al. 2014; Tsuchiya et al. 2013). In the current work, we successfully adapted the system to the model organism Drosophila melanogaster. The required transgenes were properly expressed, localized in their respective subcellular compartments, and preserved their molecular functions. We confirmed the efficiency of the technique to trap nuclear proteins. The strength of the LOF phenotypes indicates that complete inhibition was reached. The method also delivered a very fast effect, which was detectable within only a few hours (1 hr ex vivo, 6 hr in vivo).
RNAi knock-down will continue to be widely used for its simplicity and possibility to screen many genes. As a screening technique, it is thus unmatched thanks to the existing libraries (Dietzl et al. 2007; Ni et al. 2009) and simplicity of use, allowing for fast data acquisition and selection of candidate gene for further studies. However, when analyzing the function of a specific gene, the variability of the RNAi method has been a constant problem for researchers (DasGupta et al. 2007, Green et al. 2014). Off-target effects, or even dominant phenotypic effects, can affect the results of screens and LOF studies. The anchor-away method therefore represents a useful tool to rapidly induce a LOF in Drosophila.
Since its discovery, CRISPR/Cas9 has changed the time required to generate new alleles. It allows efficient generation of new transgenic strains or perform screens in a fraction of the time needed before (Hsu et al. 2014). Combined with CRISPR/Cas9, the anchor-away system can be adapted to any protein target in a time scale of weeks. CRISPR/Cas9 has also been used in Drosophila to perform genome-wide mutagenesis or overexpression screens (Ewen-Campen et al. 2017; Bassett et al. 2015). Following a similar approach, one could devise a genome-wide application of the anchor-away method, where potentially large libraries of FRB-tagged genes could be generated for screens in Drosophila.
By utilizing different anchors, other protein families could be sequestered from their subcellular compartments of residence. For example, cytoplasmic proteins could be sequestered to the plasma membrane (Tsuchiya et al. 2013). The anchor-away method could also be used to relocate proteins to different compartments and thereby force them to perform a secondary function, providing versatility to the technique. In summary, our adaption of the anchor-away system represents a useful addition to the toolbox of Drosophila researchers.
Acknowledgments
We would like to thank George Hausmann for critical reading of the manuscript, Eliane Escher for technical support with BAC recombineering and Marc Debrunner for technical assistance with embryonic microinjection. We are grateful to Richard S. Mann and Aurelio Telemann for the Sens and Brk antibodies, respectively. We would like to thank Thomas P. Neufeld, the Bloomington Drosophila Stock Center, Kyoto Stock Center and Vienna Drosophila Resource Center for providing fly stocks. This work was supported by the Swiss National Science Foundation and the Canton of Zurich.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11959209.
Communicating editor: B. Reed
Literature Cited
- Adams M. D., and Sekelsky J. J., 2002. From sequence to phenotype: reverse genetics in drosophila melanogaster. Nat. Rev. Genet. 3: 189–198. 10.1038/nrg752 [DOI] [PubMed] [Google Scholar]
- Anger A. M., Armache J.-P., Berninghausen O., Habeck M., Subklewe M. et al. , 2013. Structures of the human and Drosophila 80S ribosome. Nature 497: 80–85. 10.1038/nature12104 [DOI] [PubMed] [Google Scholar]
- Bassett A. R., Kong L., and Liu J.-L., 2015. A Genome-Wide CRISPR Library for High-Throughput Genetic Screening in Drosophila Cells. J. Genet. Genomics 42: 301–309. 10.1016/j.jgg.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw P. J., Ho S. N., Crabtree G. R., and Schreiber S. L., 1996. Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc. Natl. Acad. Sci. USA 93: 4604–4607. 10.1073/pnas.93.10.4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., and Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific C31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M., and Ahringer J., 2008. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 9: 554–566. 10.1038/nrg2364 [DOI] [PubMed] [Google Scholar]
- Caussinus E., Kanca O., and Affolter M., 2011. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 19: 117–121. 10.1038/nsmb.2180 [DOI] [PubMed] [Google Scholar]
- Chen J., Zheng X. F., Brown E. J., and Schreiber S. L., 1995. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA 92: 4947–4951. 10.1073/pnas.92.11.4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L., Kelley R., and Spradling A., 1988. Insertional mutagenesis of the Drosophila genome with single P elements. Science 239: 1121–1128. 10.1126/science.2830671 [DOI] [PubMed] [Google Scholar]
- DasGupta R., Nybakken K., Booker M., Mathey-Prevot B., Gonsalves F. et al. , 2007. A case study of the reproducibility of transcriptional reporter cell-based RNAi screens in Drosophila. Genome Biol. 8: R203 10.1186/gb-2007-8-9-r203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y. et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Ding L., Laor D., Weisman R., and Forsburg S. L., 2014. Rapid regulation of nuclear proteins by rapamycin-induced translocation in fission yeast. Yeast 31: 253–264. 10.1002/yea.3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumpas N., Ruiz Romero M., Blanco E., Edgar B., Corominas M. et al. , 2013. Brk regulates wing disc growth in part via repression of Myc expression. EMBO Rep. 14: 261–268. 10.1038/embor.2013.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen-Campen B., Yang-Zhou D., Fernandes V. R., González D. P., Liu L.-P. et al. , 2017. Optimized strategy for in vivo Cas9-activation in Drosophila. Proc. Natl. Acad. Sci. USA 114: 9409–9414. 10.1073/pnas.1707635114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E. et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K. et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. 10.1534/genetics.113.152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E. W., Fedele G., Giorgini F., and Kyriacou C. P., 2014. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat. Methods 11: 222–223. 10.1038/nmeth.2856 [DOI] [PubMed] [Google Scholar]
- Harmansa S., Alborelli I., Bieli D., Caussinus E., and Affolter M., 2017. A nanobody-based toolset to investigate the role of protein localization and dispersal in Drosophila. eLife 6: e22549 10.7554/eLife.22549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H., Nishikawa J., and Laemmli U. K., 2008. The Anchor-Away Technique: Rapid, Conditional Establishment of Yeast Mutant Phenotypes. Mol. Cell 31: 925–932. 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S., and Zhang F., 2014. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157: 1262–1278. 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaźwińska A., Kirov N., Wieschaus E., Roth S., and Rushlow C., 1999. The Drosophila Gene brinker Reveals a Novel Mechanism of Dpp Target Gene Regulation. Cell 96: 563–573. 10.1016/S0092-8674(00)80660-1 [DOI] [PubMed] [Google Scholar]
- Jenny F. H., and Basler K., 2014. Powerful Drosophila screens that paved the wingless pathway. Fly (Austin) 8: 218–225. 10.4161/19336934.2014.985988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A., and Hurt E., 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8: 761–773. 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- Kramps T., Peter O., Brunner E., Nellen D., Froesch B. et al. , 2002. Wnt/Wingless Signaling Requires BCL9/Legless-Mediated Recruitment of Pygopus to the Nuclear β-Catenin-TCF Complex. Cell 109: 47–60. 10.1016/S0092-8674(02)00679-7 [DOI] [PubMed] [Google Scholar]
- Ni J.-Q., Liu L.-P., Binari R., Hardy R., Shim H.-S. et al. , 2009. A Drosophila Resource of Transgenic RNAi Lines for Neurogenetics. Genetics 182: 1089–1100. 10.1534/genetics.109.103630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R., Abbott L. A., and Bellen H. J., 2000. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102: 349–362. 10.1016/S0092-8674(00)00040-4 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., and Wieschaus E., 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- Oldham S., Montagne J., Radimerski T., Thomas G., and Hafen E., 2000. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 14: 2689–2694. 10.1101/gad.845700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H.-M., Lee T., and Bullock S. L., 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111: E2967–E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo S., Zartman J. J., and Basler K., 2016. Cultivation and Live Imaging of Drosophila Imaginal Discs. Drosophila 1478: 203–213. [DOI] [PubMed] [Google Scholar]
- St Johnston D., 2002. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 3: 176–188. 10.1038/nrg751 [DOI] [PubMed] [Google Scholar]
- Toba G., Ohsako T., Miyata N., Ohtsuka T., Seong K. H. et al. , 1999. The gene search system. A method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics 151: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya H., Arai N., Tanaka K., and Saeki Y., 2013. Cytoplasmic proteasomes are not indispensable for cell growth in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 436: 372–376. 10.1016/j.bbrc.2013.05.105 [DOI] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D. L., Jenkins N. A., and Copeland N. G., 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33: e36 10.1093/nar/gni035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., and Rubin G. M., 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117: 1223–1237. [DOI] [PubMed] [Google Scholar]
- Zemp I., and Kutay U., 2007. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 581: 2783–2793. 10.1016/j.febslet.2007.05.013 [DOI] [PubMed] [Google Scholar]
- Zhang H., Stallock J. P., Ng J. C., Reinhard C., and Neufeld T. P., 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14: 2712–2724. 10.1101/gad.835000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All plasmids, fly strains and reagents used for the study are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25387/g3.11959209.