Immunoassays make use of highly specific antigen-antibody binding and provide sensitive ways to detect a wide range of biomacromolecules, bacteria, viruses, and small molecules. There are a range of types of immunoassay systems including single analyte sensors, 96-well plate formats, arrays, microfluidic sensors, microfluidic arrays, etc. A big target is encompassed by medical diagnostic biomarkers, which are “molecules that can be measured objectively as indicators of normal or disease processes and responses to therapeutic intervention”.1 Accurate, low-cost measurements of multiple proteins are major applications of immunoarrays that are critical for future early detection and monitoring of cancer and other diseases. Multiplexing is very important, since panels of biomarker proteins, as opposed to single biomarkers, are required to provide sufficient information content for reliable disease diagnostics.
Development of new immunosensors and immunoarrays faces stiff competition from the many available commercial immunoassays. Enzyme-linked immunosorbent assay (ELISA) is the gold standard with limits of detection (LODs) of 3–10 pg/mL for many proteins1–3 but has well-known limitations in sensitivity, analysis time, and multiplexing. However, the newer commercial multiprotein immunoassays can be expensive and technically demanding and usually do not achieve detection below pg/mL levels.4 Many commercial methods feature microbead technologies with optical or electrochemiluminescence (ECL) readout and 1–10 pg/mL LODs,1,5 including mesoscale ECL6 and Luminex7 fluorescent bead systems, Quansys Q-Plex multiplexed ELISA.8 An exception is the newer Simoa HD-19 protein counting system from Quanterix that has achieved protein LODs of 4–200 fg/mL.10 Simoa-HD-1 detected Prostate Specific Antigen (PSA) down to 14 fg/mL (0.4 fM) in serum of prostate cancer patients after prostate removal, illustrating an important application of low-abundance protein detection, since PSA increases after surgery when cancer returns. Despite major advances, fast, cost-effective immunoassay measurements of multiple proteins below ~4 fg/mL (~10 aM) with commercial kits and hardware remain problematic.11–13
Larger analytes such as proteins, viruses, antibodies, and large peptides for which two antibodies that bind to different epitopes on the analyte molecule exist can be detected by sandwich immunoassays (Figure 1). This review covers research literature in new multiplexed immunoassays published between May 1, 2017 and October 1, 2019. Assays detecting new panels of biomolecules with established commercial technologies have not been included, although a few novel applications or modifications of commercial platforms are included. Our focus here is mainly on new validated approaches to multiplexed immunoassays that have paid proper attention to analytical figures of merit and accuracy benchmarking. Important focus issues include multiplexing, speed, low cost, sensitivity, and for diagnostics, ease of use and technical simplicity for point-of-care applications.
Figure 1.
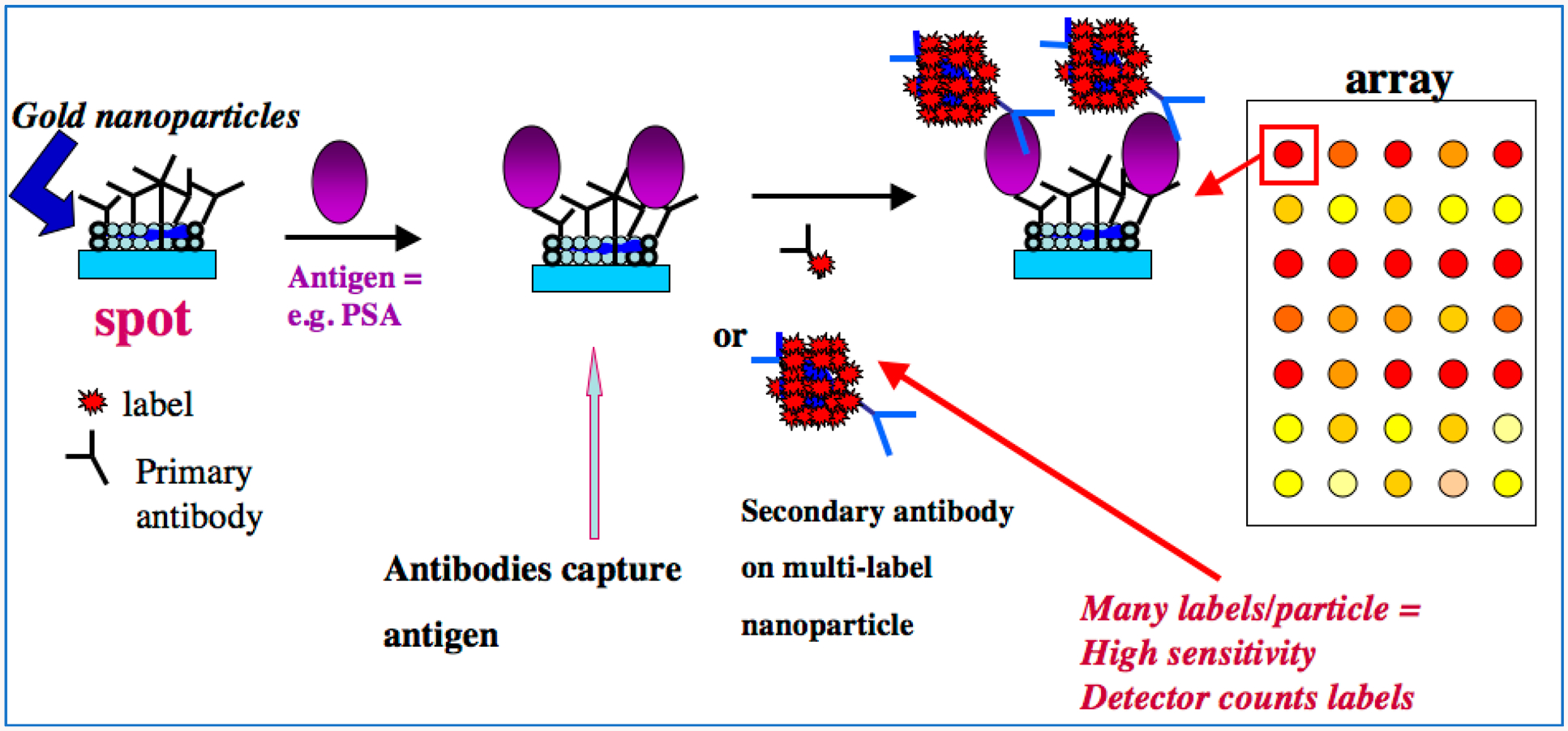
Example of a modern approach to sandwich immunoarrays. On left, a gold nanoparticle-decorated spot (to achieve high surface area) on the array is represented with attached primary antibodies (Ab1). Sample is delivered to the array, which can have a number of different Ab1 spots to capture a range of different antigens in a multiplexed assay. For our example SPOT, the antigen is prostate specific antigen, a biomarker protein for prostate cancer.2 The antigens are captured by Ab1’s on their specific spots, usually during an incubation period. After washing, secondary or detection antibodies (Ab2) are introduced, shown here by two examples. The conventional approach employs a single labeled antibody, while a more sensitive assay can be designed with multiple labels to amplify the signals.1 This step is followed by another incubation period, washing, and detection. These kinds of arrays can be integrated with microfluidics for sample and reagent delivery and automation.
ELECTROCHEMICAL SENSING
Electrochemical methods have long been used to facilitate sensitive measurements with low-cost instrumentation but usually need separate electrical connections to an array of sensor for multiplexing. New forms of carbon such as carbon nanotubes, graphene, and graphene oxide have become popular nanomaterials for immunosensor development. Wei et al. developed an electrochemical immunosensor by fabrication on a glassy carbon electrode by loading graphene oxides attached to redox probes and covalently attaching of secondary antibodies.14 Multiplexed detection of proteins IL-6, IL-1b, and TNF-α in the low pg/mL range was achieved by labeling their secondary antibodies with methylene blue, Nile blue, and ferrocene, one label for each different antibody. Amperometric detection of three cytokines was attained with pg/mL LODs, good specificity and accurate spike recovery. Tuteja and co-workers reported a dual sensing electrochemical platform for fatty acids and beta hydroxy butyrate (bHBA) as critical biomarkers for early diagnosis of negative energy balance in dairy cows. They used electroreduced graphene oxide (E-rGO) deposited on screen printed carbon electrodes.15 Antibodies were conjugated into the E-rGO surface for label free detection, with LODs from 0.1 mM to 10 mM for both analytes within response time less than a minute. Wang and Li et al., developed a gold-reduced graphene oxide nanocomposite (Au/r-GO) with gold nanocages serving as carriers for secondary antibodies and redox probes to detect autophagic biomarkers.16 The assay enabled detecting 2 autophagic biomarkers at clinically useful levels, Beclin-1 and LC3B-II, with LODs ~0.03 ng/mL.
Graphene oxide has also been used for electrochemical protein detection in microfluidic immunoarrays. In our group, Sharafeldin et al. made a composite of Fe3O4 nanoparticles loaded onto graphene oxide nanosheets (Fe3O4@GO) to detect protein biomarkers for prostate cancer.17 Antibodies were attached onto the paramagnetic Fe3O4@GO, captured their specific binding partner proteins, and delivered them to a screen-printed carbon array. This system simultaneously enabled ultrasensitive mediator-free electrochemical detection of PSA with LOD 15 fg/mL and PSMA at LOD 5 fg/mL, 1000-fold better than commercial assays at reagent cost of $0.85/assay. Methods were presented to tune the assays to meet dynamic range requirements of samples to be analyzed. Alizadeh et al. reported an immunoassay that traps Fe3O4 nanoparticles modified with capture antibodies into an indium tin oxide (ITO) electrode attached to a magnet.18 They incubated the immunosensor with carcinoembryonic antigen (CEA) and α-fetoprotein (AFP) proteins followed by detection of signal labels on Ab2 attached to Au-nanoparticles, i.e., AbCEA-Au-thionine and AbAFP-Au-ferrocene. LODs were 12 pg mL−1 for CEA and 18 pg mL−1 for AFP.
Screen printed carbon electrodes (SPCE) and arrays continue to be used extensively for immunoarray development. Zhao et al. used an SPCE array with individualized electroactive labels in a hydrogel on array electrodes.19 Multiplexing of four cancer biomarkers demonstrated proof of concept with LODs in the low pg mL−1 range. Pingarrón’s group used immobilized capture antibodies on carbon electrodes via a p-aminobenzoic acid linkage using a graphene quantum dot-multiwalled carbon nanotube (MWCNTs) composite with detection antibodies and horseradish peroxidase (HRP) to detect IL-13sRα2 and CDH-17 with sub- to low-ng/mL LODs.20 In collaboration with Bartosik, this group developed a novel SPCE immunoarray for microRNAs (miRNAs) miR-21, let-7a, and miR-31 associated with cervical cancer (Figure 2). Magnetic bead-antibodies conjugates were used to capture the miRNAs hybridized with a DNA probe which is then bound to two biotin-labeled auxiliary DNA probes via a hybridization chain reaction (HCR). Biotin units bind streptavidin-HRP labels for amperometric detection upon adding H2O2 and hydroquinone mediator.21 The method discriminates well against single mismatches and was used to determine miRNAs for cancer diagnostics. Marques et al. used a AuNP-coated SPCE for sandwich immunoassays with LODs in the ng mL−1 range to detect breast cancer biomarker proteins HER2 and CA-15–3 using detection antibodies labeled with alkaline phosphatase and Ag+ for detection.22 Mercer et al. developed an automated microfluidic system for amperometric immunoassay of up to eight proteins on an SPCE array.23
Figure 2.
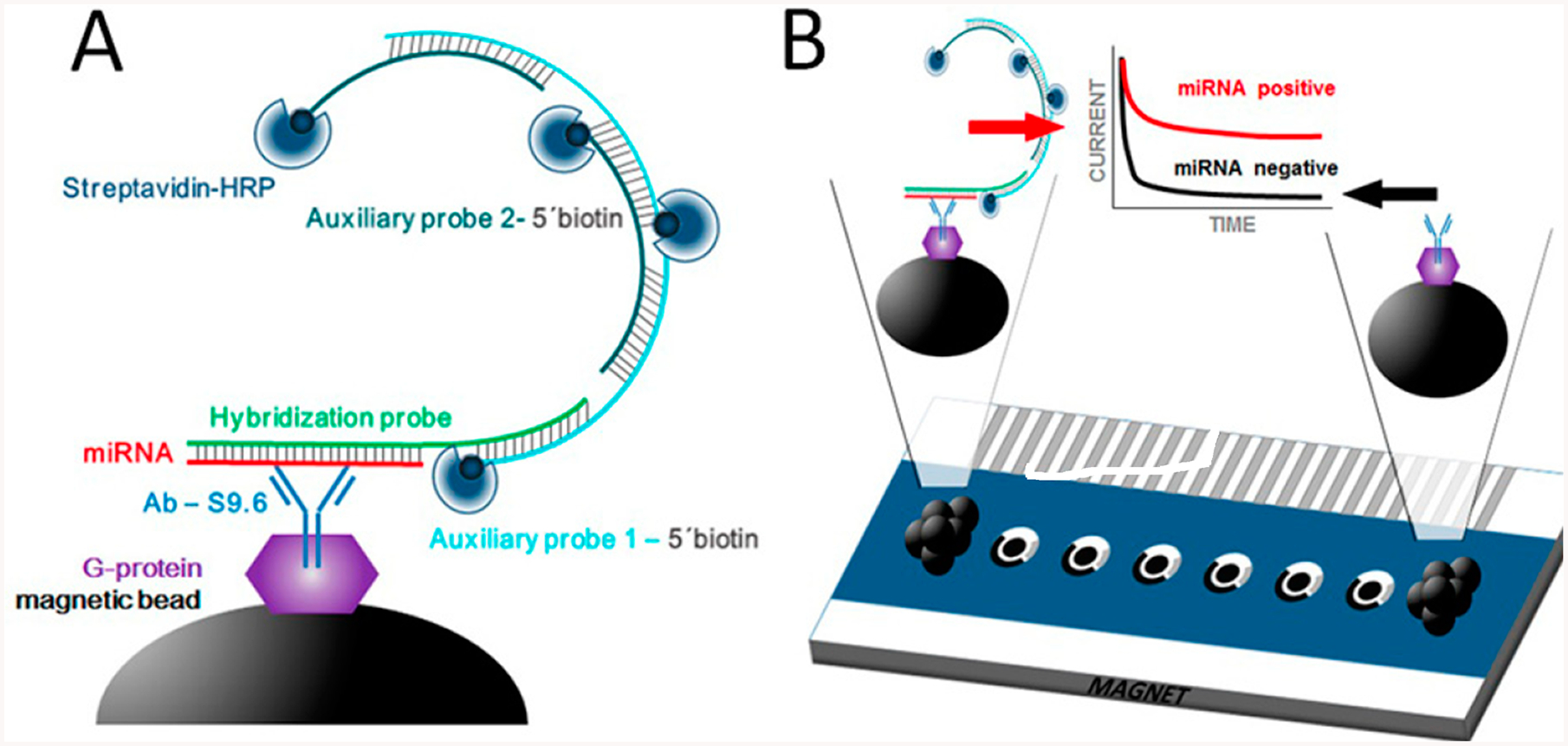
Schematic display of HCR-based miRNA immunosensing: (A) preparation of MBs including modification with AbS9.6, selective capture of the target miRNA/HP heteroduplex, HCR at the MBs by hybridization of the HP with AP1 and AP2 probes, and labeling of the biotin moieties with Strep-HRP. (B) Magnetic attraction of the modified MBs to the surface of the working electrodes (via magnet placed below the array) and chronoamperometric measurement of the enzymatic reduction of H2O mediated by hydroquinone. The cathodic current increased with the concentration of the target miRNA. Reproduced from Multiplexed Immunosensing Platform Coupled to Hybridization Chain Reaction for Electrochemical Determination of Micro RNAs in Clinical Samples, Jirakova, L.; Hrstka, R.; Campuzano, S.; Pingarrón, J.M.; Bartosik, M. Electroanalysis, Vol. 31, Issue 2 (ref 21). Copyright 2019 Wiley.
Eissa et al. used an SPCE for a competitive immunoassay to detect metabolites of drugs of abuse with LODs 1.2–8.0 pg mL−1.24 This group also developed a competitive immunoassay for corona virus from Middle East respiratory syndrome (MERS-CoV) with AuNP-coated carbon electrodes using square wave voltammetry (SWV) and a ferrocyanide probe with LODs 0.4 pg mL−1 for human corona virus (HCoV) and 1.0 pg mL−1 for MERS-CoV.25 They also used similar AuNP-carbon electrodes to detect DOCK8, PGM3, and STAT3 protein biomarkers with LODs ~3 pg mL−1 aimed at diagnosis of Hyper-Immunoglobulin E syndrome.26 They used disposable carbon fiber immunosensors to detect survival motor neuron 1, cystic fibrosis transmembrane conductance regulator, and Duchenne muscular dystrophy proteins.27
Reports have appeared combining electrochemical immunosensing with a second approach for different kinds of analytes, as well as using novel approaches to electrode materials or detection. Joe Wang’s team at UC San Diego developed the first dual-marker electrochemical chip with integrated enzyme-and immunodetection.28 A common Ag/AgCl reference/counter electrode and two Au sensor electrodes were used to detect glucose and insulin in a dual diabetes biomarker assay (Figure 3). Glucose is detected by reaction with glucose oxidase, while insulin detection relies on an HRP-labeled sandwich immunoassay. Clinically relevant LODs of 1 pg mL−1 for insulin and millimolar levels for glucose were achieved, and direct measurements in blood and saliva were demonstrated. Kim et al. employed membrane-based microwave-mediated electrochemical immunoassay based on fast conjugation of antibodies with target antigens in a three-dimensional nylon membrane to detect osteoporosis biomarkers including type I collagen peptides, Osteocalcin (OC), and parathyroid hormone (PTH).29 Microwave radiation was used here to greatly shorten incubation time for efficient antibody-antigen binding. Detection employed an alkaline phosphatase label that produced p-aminophenol for differential pulse voltammetry measurement by a separate sensor electrode. A related capture antibody-membrane approach without microwaves was used by Arya and Estrala,30 with separate antibody-membrane and offline electrochemical sensor, for bladder cancer protein markers. Poly(methyl methacrylate) sheets and polycarbonate membranes were equipped with appropriate antibodies for analyte capture. Alkaline phosphatase-detection antibodies and 4-aminophenyl phosphate were added, and the electroactive product was delivered to a gold sensor electrode for amperometry. LODs in the ng/mL range for nuclear mitotic apparatus protein 1 and complement factor H-related 1 protein.
Figure 3.
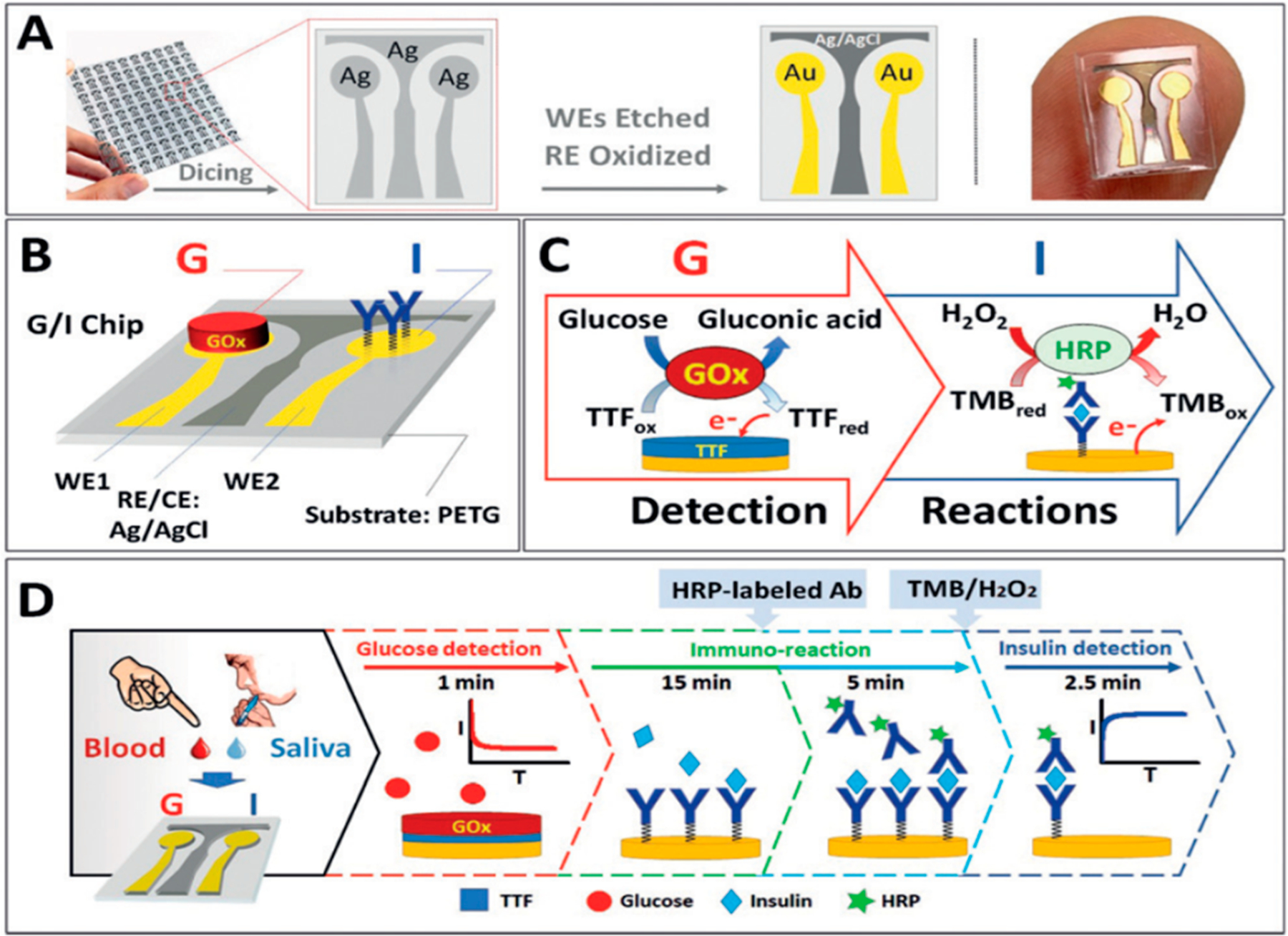
Dual G/I biosensor chip. (A) The G/I sensor chip array showing the two Au WEs for glucose and insulin biosensors, with Ag/AgCl electrode reference. (B) Localized detection of G and I on a sensor chip, showing the immobilized GOx and insulin antibody bioreceptors. (C) Recognition and redox processes in G/I sensing. Glucose detection is amperometric +0.2 V on AuWE1 with TTF-mediated biocatalytic (GOx) oxidation of G. Insulin is detected on Au WE2 at @0.1 V by sandwich immunoreaction assay by measuring H2O2 reduction current catalyzed HRP and mediated by 3,3′,5,5′-tetramethylbenzidine (TMB). (D) Glucose is measured followed by 15 min sample incubation and subsequent 5 min incubation with HRP-labeled antibody. Finally, insulin is monitored after washing the chip and adding mediator and H2O2. Reproduced from Enzymatic/Immunoassay Dual-Biomarker Sensing Chip: Toward Decentralized Insulin/Glucose Detection, Vargas, E.; Teymourian, H.; Tehrani, F.; Eksin, E.; Sánchez-Tirado, E.; Warren, P.; Erdem, A.; Dassau, E.; Wang, J. Angew. Chem. Int. Ed., Vol. 58, Issue 19 (ref 28). Copyright 2019 Wiley.
Nanomaterials continue to be used in sensor electrode materials and as components of detection labels. Shanmugam et al. used nanostructured ZnO sensors made by low-temperature methods to achieve high surface areas that increased the sensitivity of electrochemical impedance spectroscopy (EIS) assays for cardiac biomarkers troponin-T, troponin I, and natriuretic peptide with LOD ~1 pg/mL and 5 decades dynamic range.31 Kalyoncu et al. used individual nanoparticle labels (nanotags) PbAu@γFe2O3, CuAu@γFe2O3, and ZnAu@ γFe2O3 with different redox potentials in a brief study of simultaneous detection of cancer biomarker proteins CEA, VEGF, and AFP.32 Detection relied on the blocking effect of these nanotags monitored by differential pulse voltammetry and EIS on SPCEs using Fe(CN)62+ as an oxidizable probe. Putnin et al. used polyethylenimine-coated gold nanoparticles (PEIAuNPs) for surface modification of SPCEs and bonding to antibodies.33 Moreover, it has properties advantageous in chelating heavy metal ions. Specific detection antibodies were attached to PEI-AuNPs, then metal ions were bound to make PEI-AuNPs-anti-AFP-Pb2+, PEI-AuNPs-anti-CEA-Cu2+, PEIAuNPs-anti-PSA-Cd2+, and PEI-AuNPs-anti-IL-8-Ag+. These labels were used to detect AFP, CEA, PSA, and IL-8 in serum with LODs of 1–2 pg mL−1. Tang, Ren, and Lu detected human IgG (HIgG) and rabbit IgG (RIgG) using antibody-labeled Cu and Cd metal nanocrystals.34 Electrodes were modified with AuNPs linked with Fab fragments of capture antibodies. Detection at pH 3.5 by square-wave anodic stripping voltammetry gave LODs of 3.4 pg mL−1.
ELECTROCHEMILUMINESCENT DETECTION
Electrochemiluminescence (ECL) generates light via an electro-chemically initiated redox pathway.35,36 Some commercial sandwich immunoassays use the dye Ru(bpy)32+ (RuBPY) as a label on antibodies (Ab2). Once the RuBPY-Ab2 binds to proteins captured on an array, ECL is initiated by oxidizing coreactant tripropylamine (TPrA), typically at about 1 V vs SCE. TPrA oxidation products react with Ru(bpy)33+ to yield electronically excited [Ru(bpy)32+]* that emits 610 nm light. ECL is well suited to immunoarrays due to inherent high sensitivity from light measured against a very low background and the lack of a need for individually connected sensor electrodes or a light source. Larger microwell patterned electrodes can be used.37 Quantum dot nanoparticles can also be used to emit ECL.38 Recent research has also focused on high quantum yield organic ECL emitters such as bifunctional spirofluorene derivatives that can be tuned to emit blue to green light.39 Although these molecules are insoluble in water, they were recently attached to surfaces to extend-capabilities of multicolor ECL.40
Several recent reports have addressed ECL multiplexing by using potential- or wavelength-resolution. Guo et al. synthesized new iridium(III) and ruthenium(II) ECL complexes to obtain a multicolor ECL immunoassay system.41 These new ECL dyes were used for detection of biomarkers CEA, α-fetoprotein (AFP), and β-human chorionic gonadotropin (β-HCG). ECL complexes were loaded onto carboxylate polystyrene beads separately and with specific detection antibodies for CEA, AFP, and β-HCG via biotin-streptavidin complexation to form dual-coded beads, Ab-M@PSB (M = Ru or Ir). The immunoassay is performed in three steps: homogeneous sandwich immuno-reaction, magnetic separation, and swelling of beads to release ECL dyes for detection. LODs were not reported, but the assay used cutoff concentrations of 5 ng/mL for CEA, 25 ng/mL for AFP, and 5 mIU/mL for β-HCG. CdTe776 and CdSe550 were used to detect CEA (LOD 1 pg/mL) and AFP (LOD 10 fg/mL).42 J. Zhou et al. used semiconductor nanocrystals (CdSe550, CdTe650, and CdTe776) for a tricolor ECL immunoassay to detect CEA at 1 pg/mL, PSA at 10 pg/mL, and AFP at 0.01 pg/mL LODs.43 AFP and CA125 were detected at pg/mL levels using nanocrystal labels and a dichromic mirror to separate the different wavelengths.44 CdTe@CdS and RuBPY were used as dual labels to detect CA125 and CA15–3 at low μU/mL levels.45 A related approach using CdS quantum dots, luminol, and carbon quantum dots was used46 to detect three diagnostic biomarkers for latent tuberculosis infection with ~2 pg/mL LODs.
Several papers appeared that extend the number of proteins that can be determined in a multiplexed microfluidic immunoassay. Our laboratory developed a fully automated 3D-printed microfluidic immunoarray demonstrated to detect 8 proteins in duplicate.47 Samples, regents, wash buffers, and RuBPY-silica-Ab2 nanoparticles are delivered by micropumps controlled by a low-cost microprocessor to a detection chamber for CCD camera measurement of ECL in a dark box (Figure 4). The detection chamber features computer printed 10 nm deep microwells containing upright single wall carbon nanotubes with Ab1 attached. An eight-protein prostate cancer biomarker panel (IGF-1, PSA, PF-4, CD-14, VEGF-D, GOLM-1, PSMA, and IGFBP-3) was simultaneously detected at levels as low as 80–110 fg mL−1 in diluted serum. A similar 3D-printed microfluidic immunoarray was interfaced with a multichannel programmable syringe pump for automation to run 3 assays at once for a smaller number of proteins analytes.48
Figure 4.
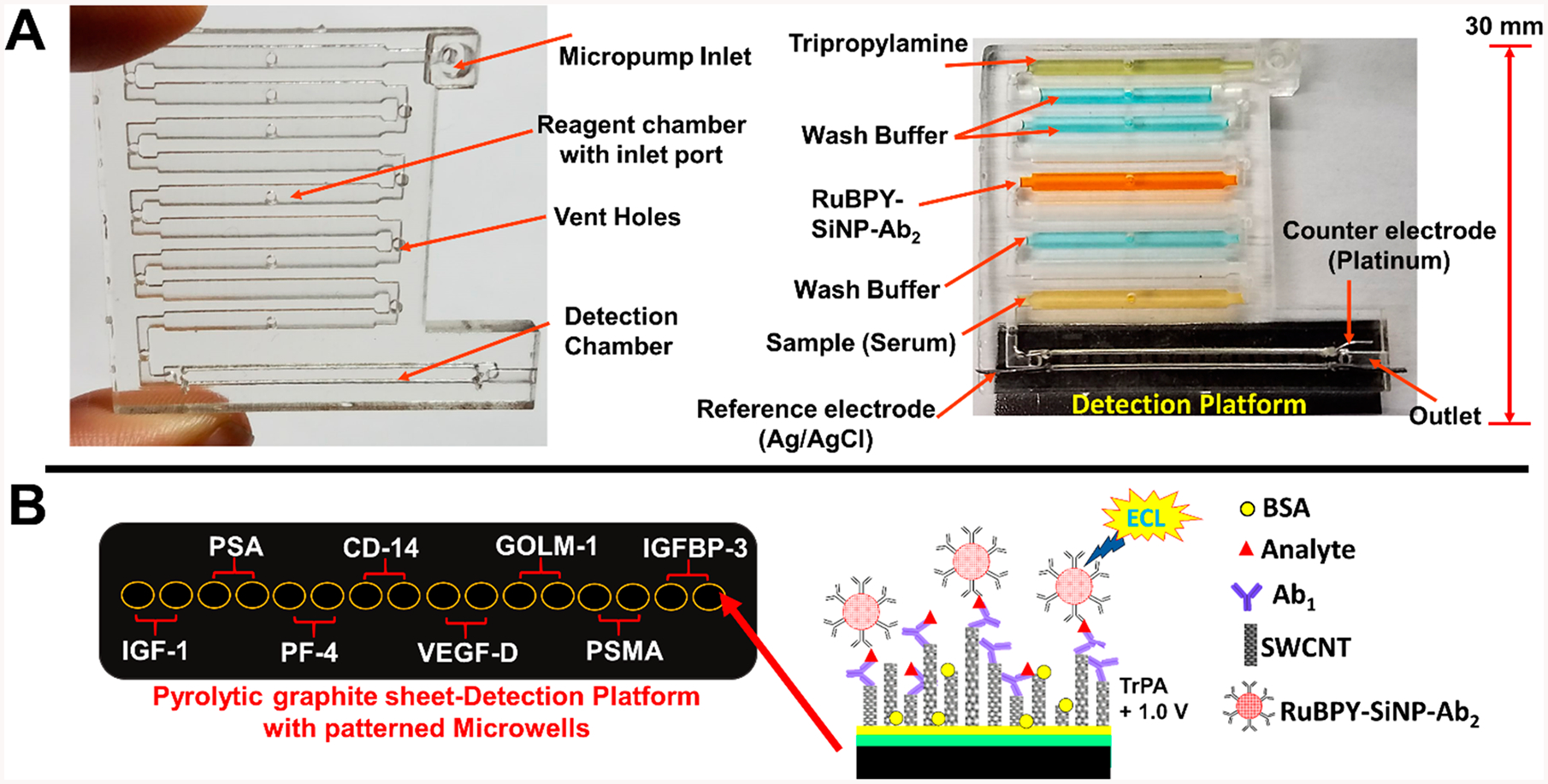
Representations of 3D-printed immunoarray: (A) 3D printed microfluidic array with chambers to hold sample, wash buffers, detection nanoparticles, and coreactant for ECL generation. Array is shown on the left without detection chip, and on the right bonded to a pyrolytic graphite sheet (PGS) microwell detection chip with reagent and sample chambers filled with dye solutions for visualization. (B) Representative disposable PGS chip with heat transferred microwells printed using hydrophobic toner ink. Inset illustrates a sandwich immunoassay on a single wall carbon nanotube forest (SWCNT) in one microwell. Reproduced from Kadimisetty, K.; Malla, S.; Bhalerao, K. S.; Mosa, I. M.; Bhakta, S.; Lee, N. H.; Rusling, J. F. Anal. Chem. 2018, 90, 7569–7577 (ref 47). Copyright 2018 American Chemical Society.
Croner et al. used the MesoScale Discovery system to analyze 27 plasm proteins in 4435 patient samples with LODs in the 10–500 pg/mL range to develop a new colorectal cancer blood test for at-risk patients.49 Chakraborty et al. used the MesoScale platform to create an assay for six enterotoxigenic Escherichia coli antigens to help develop enterotoxigenic E. coli vaccines.50 Dillon and co-workers used the MesoScale platform to validate a panel of 16 protein biomarkers for advanced adenoma and colorectal cancer with LODs 0.12–2000 pg/mL.51
CHEMILUMINESCENCE DETECTION
Typical arrangements for chemiluminescence (CL) detection in sandwich immunoassays employ secondary antibodies (Ab2) labeled with luminol or horseradish peroxidase (HRP). After the analyte binds to primary antibody (Ab1), labeled Ab2 is introduced. If luminol is the label, HRP followed by H2O2 are added to emit CL. If HRP is the label, a luminol-H2O2 mixture can be added to develop CL.
Click chemistry labeling of Ab2 using trans-cyclooctene (TCO) and 1,2,4,5-tetrazine (Tz) for controlled attachment of HRP labels at different levels to obtain tunable signal amplification and dynamic ranges for different proteins (Figure 5).52 Multiplexed detection of inflammation biomarker proteins interleukin 6 (IL-6), procalcitonin (PCT), and C-reactive protein (CRP) was achieved using H2O2 and luminol in a labon-a chip for detecting low- and high-abundance proteins in a single assay. LODs adjusted for abundance of the proteins in serum were 0.47 pg mL−1 for IL-6, 2.6 pg mL−1 for PCT, and 40 ng mL−1 for CRP.
Figure 5.
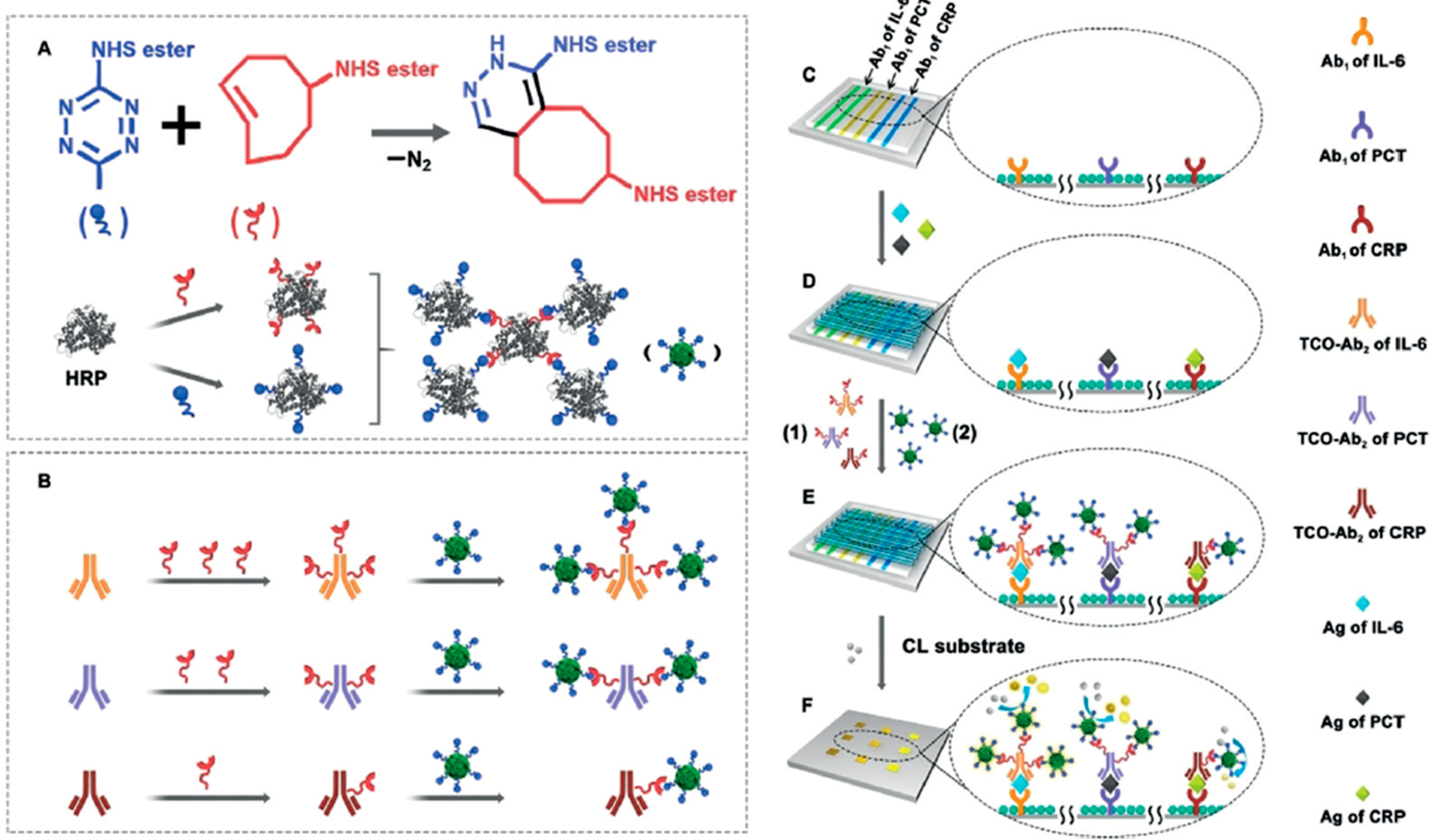
Click-chemistry-mediated assembly of multiple HRPs and detection antibodies for simultaneous multiplexed bioassays with a tunable detection range. Cyclooctene can be quantitatively modified on the detection antibodies to react with Tz-functionalized enzyme assemblies for simultaneous immunoassays of PCT, IL-6, and CRP, each of which requires a different sensitivity. Reproduced from Controllable Assembly of Enzymes for Multiplexed Lab-on-a-Chip Bioassays with a Tunable Detection Range, Xianyu, Y.; Wu, J.; Chen, Y.; Zheng, W.; Xie, M.; Jiang, X. Angew. Chem. Int. Ed., Vol. 57, Issue 25 (ref 52). Copyright 2018 Wiley.
Mou et al. developed an automated microchip assay to detect CRP, PCT and IL-6 with different dynamic ranges simultaneously in undiluted human serum samples.53 The hierarchically structured microchip has a top polydimethylsiloxane (PDMS) layer and a bottom poly(L-lactide-co-ε-caprolactone) layer of electrically spun fibers with patterned capture antibodies used for detection with chemiluminescent probes. LODs tailored for sample characteristics were 1.9 μg/mL for CRP, 0.17 ng/mL for PCT, and 50 pg/mL for IL-6.
From our group, Sharafeldin et al. described multiplexed ELISA-in-a-pipet tip that is more sensitive, faster, and requires less sample than traditional ELISA.54 Data acquisition by an iPhone enables electronic delivery of the results to care providers. Optically clear multipipet tip arrays were designed and 3D printed by stereolithography to fit commercial 8-tippipets. Capture antibodies (Ab1) were immobilized on inner pipet tip walls, and samples and reagents are introduced and expelled by pipetting. CL or colorimetric detection employed iPhone camera imaging, CCD camera imaging, or a spectroscopic plate reader. Four prostate cancer biomarkers proteins were measured in diluted serum with LODs better than microplate ELISA at 25% the cost. CCD camera detection of CL using femto-luminol gave LODs down to 0.5 pg/mL. Patient sample analyses for three proteins gave good correlations with a microfluidic electrochemical immunoassay and with ELISA.
CCD camera imaging of CL was also used in a DNA microarray to detect breast cancer biomarkers CEA, AFP, and CA-125 developed by Xiao et al.55 Aldehyde functionalized glass was spotted with different functional hairpin DNAs. A sandwich immunocomplex forms when complementary DNA-labeled antibodies hybridize with the hairpin DNAs on the glass chip. Opening the hairpin structure triggers the HCR and using streptavidin-biotin conjugation, and numerous HRP enzymes were coupled on the HCR assemblies to amplify the CL signal from luminol and H2O2. LODs were 50 pg mL−1 for CEA, 60 pg mL−1 for AFP, and 0.02 U mL−1 for CA-125. Similarly, CCD imaging was used for a CL immunosensor array developed for measuring chicken cytokines.56 Capture antibodies were assembled on a disposable glass chip when HRP and antibody-conjugated AuNPs were used for multienzyme amplification. Simultaneous measurement of chicken IL-4 and interferon-γ (IFN- γ) gave LODs of 2–3 pg mL−1. This group also reported a CL imaging immunoassay using CuS nanoparticles as peroxidase mimetics for signal development, but LODs for IFN-γ and IL-4 were still 2–3 pg mL−1.57
An immunoarray was developed by Li et al. for food contaminating mycotoxins. Polymeric array spots were fabricated on a planar substrate using UV copolymerization of a cross-linker to make antibody-functional monomer bioconju-gates.58 HRP-labeled mycotoxins were used in displacement assays with CL measured with a luminescence detector. LODs were 13–36 ng mL−1 for mycotoxins ZEN, DON, T-2, HT-2, AFS, OTA, and FB1. A seven-plex prototype assay using proprietary Q-plex technology was utilized to determine micronutrient deficiencies in pregnant women from Africa.59 The system uses quantitative CL and simultaneously measures up to seven different analytes in a mixture. It uses a 96-well plate format and requires a minimum 5 μL sample, which is diluted to 50 μL with the necessary reagents. The micronutrients and biomarkers and LODs included thyroglobulin (0.0244 μg/mL), ferritin (0.206 μg/mL), soluble transferrin receptor (0.192 mg/mL), retinol binding protein 4 (0.0054 μmol/L), α−1-acid glycoprotein (0.0019 g/L), C-reactive protein (90.1 mg/mL), and histidine-rich protein II (0.0017 μg/L). Assay performance was assessed with international reference standards and verified with patient plasma samples. The correlation between each biomarker was good except for thyroglobulin and resulted in sensitivities from 74 to 93% and specificities from 81 to 98%. The low concentrated proteins were detected by the sandwich immunoassay and the lowest ones by a competitive assay. These assays were recommended for population screening of nutrient deficiencies and malaria infection, applicable under limited resource conditions.60
MAGNETIC BEAD- AND MICROSPHERE-ASSISTED ASSAYS
Many commercial assays summarized earlier in this review use bead-based technologies; however, new approaches continue to be reported. One promising area starting to receive more attention is giant magneto resistance (GMS) sensing. Gao et al. developed a giant magneto resistance (GMR) sensor chip integrated with a microfluidic device and magnetic nanobead labels for multiprotein detection.61 Cancer biomarker proteins were detected by sandwich immunoassay using biotinylated-Ab2 and streptavidin-linked magnetic beads (Figure 6). This GMS was used to quantify 12 cancer biomarker proteins in serum, AFP, CEA, CYFRA21–1, NSF, SCC, PGI, PGII, CA 19–9, total PSA, free PSA, free-β-hCG, and Tg, in 15 min with LODs 0.02–1 ng mL−1. The assay offers fast, low-cost detection, and LODs might be improved in the future by decreasing nonspecific binding (NSB). A similar detection approach was used with a microprocessor-controlled microfluidic GMR sensor chip that features an array of sensors that detect magnetic nanoparticles.62 Antibodies are immobilized on the GMR using a robotic spotter, magnetic bead labeled secondary antibodies were used, and the change in resistance upon binding was measured by a smart phone-assisted detector. This device detects human immunodeficiency virus (HIV) in saliva with an accuracy of 80% and leukocytosis in plasma with an accuracy of 90% and is aimed for diagnostic use in personalized medicine.
Figure 6.
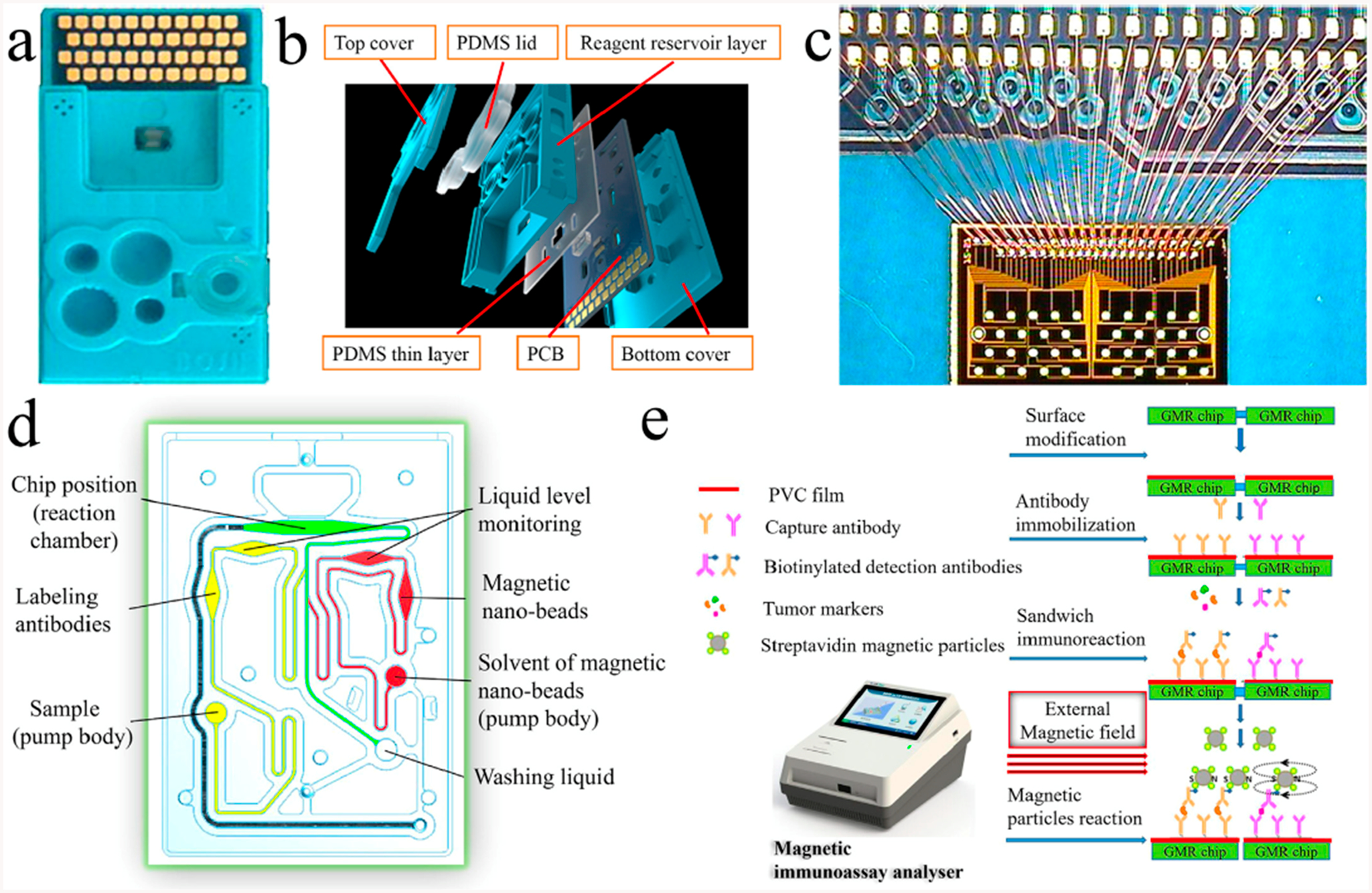
Giant magneto resistance sensor: (a) test card, (b) multilayer structure of the test card, (c) GMR chip and the connection between the GMR chip and PCB, (d) structure of the microchannel system, and (e) reaction process of the GMR multibiomarker immunoassay. Reprinted from Biosensors and Bioelectronics, Vol. 123, Gao, Y.; Huo, W.; Zhang, L.; Lian, J.; Tao, W.; Song, C.; Tang, J.; Shi, S.; Gao, Y. Multiplexed measurement of 12 tumor markers using a GMR multibiomarker immunoassay biosensor, pp. 204–210 (ref 61). Copyright 2019, with permission from Elsevier.
Bauer et al. used metal affinity magnetic beads attached to histidine-rich peptides linked to antibodies to make “Capture and Release” (CaR) antibodies that can be used to enrich protein biomarkers in solution63 and improve specificity of immunocapture for multibiomarker detection. CaR capture proteins, then release them at the appropriate stage of sample preparation using an imidazole salt elution buffer for preconcentration and downstream detection. The CaRs improved rapid diagnostics in tests for malaria targets histidine-rich protein II (HRPII) and plasmodium lactate dehydrogenase (pLDH). A 17.5-fold improvement in LOD to 6.7 pM for pLDH and 1.8 pM HRPII was found using CaRs with ELISA kits.
Ricks et al. used virus like particles (VLPs) and a MAGPIX platform for multiplexed detection of alpha and chikungunya viruses.64 The VLPs were functionalized with the antigen of the virus in question and then conjugated to the magnetic beads to form VLP-conjugated microspheres (VCMs) for detection on a MAGPIX platform. These VCMs detected IgM and IgG antibodies in primate and human serum giving a 2-fold log increase in sensitivity.
Hong et al. reported an inductively coupled plasma-MS approach for proteins and applied it to gynecological cancer biomarkers.65 Antibodies were selectively labeled with Eu3+, Lu3+, or TM3+ to distinguish carbohydrate antigen (CA15–3), HER-2, and human epididymis protein 4 (HE4). Limits of detection were 1.62 U mL−1 for CA15–3, 4 ng mL−1 for HER-2, and 0.06 pg mL−1 for HE4, and results were well correlated with a standard luminescence immunoassay. Some bead-based immunoassays have been developed for IgG antibodies and bacteria. A multiplexed magnetic microsphere immunoassay was developed to measure mouse serum antibodies against B. Pertussis antigens, pertussis toxin, filamentous hemagglutinin, pertactin, tetanus, and diphtheria for testing of combination animal vaccines.66 Antigens were linked to luminescent-labeled magnetic microspheres, and fluorescent output was monitored. Appropriate limits of quantitation (LOQs) for the practical measurement of antibodies in vaccines were obtained. Janus microbeads were designed to be encoded and decoded by a magnetic field by coating with a plasmonic layer of gold nanoislands.67 Encoding and decoding in this way gave 180-fold increase in fluorescence intensity. Proof of concept measurements on rabbit and human IgGs gave LODs of ~15 pg/mL. A multiplexed magnetic microsphere luminescence immunoassay was developed to detect IgG antibodies for six human corona viruses.68 Paired human sera was used to screen IgG reactivity against the six viruses to obtain 86% sensitivity and 84% specificity for diagnosis. A microfluidic plasmonic microarray was developed with gold nanohole sensors for sensitive analysis of sexually transmitted bacterial infections, Chlamydia trachomatis and Neisseria gonorrheae. The gold nanohole sensor enabled extraordinary optical transmission to enhance sensitivity.69 Each nanohole is functionalized with a specific antibody and detection is by spectroscopic imaging (Figure 7). Bacteria could be detected simultaneously or separately in urine in real time without extraction or amplification of DNA. C. trachomatis had LOD 300 CFU/mL and N. gonorrheae 1500 CFU/mL.
Figure 7.
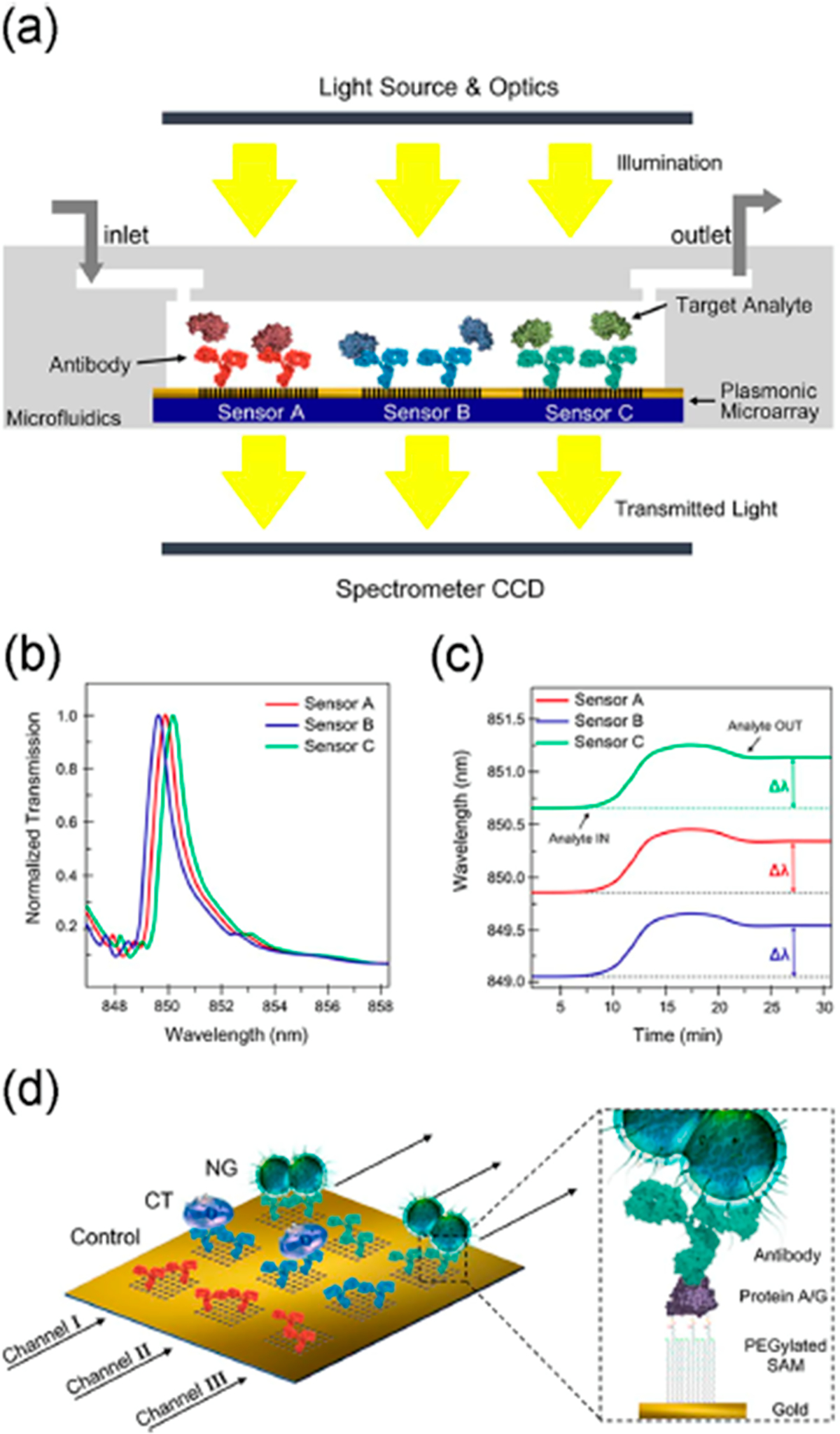
Nanohole biosensor: (a) cross-sectional view, (b) graph illustrating the three EOT spectral peaks acquired simultaneously from the three sensor arrays, (c) graph illustrating the multiplexed real-time monitoring of the EOT wavelength shift, and (d) schematics of sensor surface biofunctionalization. Different nanohole arrays are decorated with different antibodies: anti-Neisseria gonorrheae (NG) (green), anti-Chlamydia trachomatis (CT) (blue), and a control antibody (red). Each microfluidic channel (black arrows) cover three inline sensor arrays. The zoomed-in section illustrates the surface chemistry for the sensor functionalization. Reprinted from Biosensors and Bioelectronics, Vol. 94, Sole, M.; Belushkin, A.; Cavallini, A.; Kebbi-Beghdadi, C.; Greub, G.; Altug, H. Multiplexed nanoplasmonic biosensor for one-step simultaneous detection of Chlamydia trachomatis and Neisseria gonorrheae in urine, pp. 560–567 (ref 69). Copyright 2017, with permission from Elsevier.
Fluorescent bead assays have remained very popular due to sensitivity, ease of use and commercial availability and have been coupled with unique technologies for new immunoassays. Berger et al. developed an assay to detect five Sarovar antibodies of A. pleuropneumoniae using antigens attached to magnetic fluorescent beads and using a BioPlex-200 plate reader with flow cytometry capabilities.70 The multiplexed assay had better throughput and sensitivity than single antibody ELISA and complementary fixation tests. Bo Zhang reported incorporating sandwich immunoassay reactions into a dual-mode magnetic fluorescent immunochromatography lateral flow test strip (mFICTS).71 Antibody-decorated polymer-iron oxide super-paramagnetic beads capable of efficient fluorescent quenching first selectively captures the analytes in a test tube. Then, on a strip, a fluorescent-tagged secondary antibody captures analyte-bead conjugates, leading to a bioconjugate that is preconcentrated by the quenching iron oxide-polymer beads. Aggregation of the black quenching beads on the test line enables rapid screening by eye. Dual fluorescence quenching and colorimetry was used for quantitation. The mFICTS detected breast cancer biomarkers with LODs of 60 pg mL−1 for CEA and 0.09 U mL−1 for CA153. An assay using antigen-coupled fluorescent microspheres was used to detect antibodies against HIV and validated according to Good Clinical Laboratory Practice (GCLP) guidelines.72 LODs ranged from 1 to 126 ng/mL for three antibodies against HIV-1/SIV. Bilan et al. reported a suspension immunoassay employing fluorescent microspheres of different sizes that were encoded with Q-dots.73 This approach was used to measure lung cancer biomarker proteins α−1-microglobulin/bikunin precursor (AMBP), peroxiredoxin 2 (PRDX2), and Parkinson disease protein 7 (PARK7) in bronchoalveolar lavage fluid (BALF). The quantum dot encoded-microsphere assay was well correlated with an alternative commercial Luminex xMAP. Statistical analysis of preclinical validation predicted high diagnostic utility. A novel smart hydrogel approach to detect secreted proteins from single cells was been reported by M. N. Hsu et al.74 Single cell protein secretions are captured in a hydrogel particle featuring specific antibodies for proteins of interest in a microfluidic device. These particles were then collected and mixed with analyte-specific fluorescent-labeled detection antibodies. Fluorescence was enhanced by measuring with a fluorescence microscope above the hydrogel’s lower critical solution temperature of 38.5 °C. LODs in pM were 35 for IL-6 (0.78 mg/mL), 25.0 (0.30 ng/mL) for IL-8, and 30.8 for MCP-1 (0.38 ng/mL) at 50 °C. Another suspension array was reported by Luan et al., who used plasmonic-encoded reverse opal microbeads integrated into a special microfluidic chip.75 These so-called solvent-responsive breathing microbeads were used in a specially designed capillary microfluidic chip to give better performance than silica colloidal crystal bead arrays. The authors claimed an order of magnitude improvement in sensitivity for photonic encoded microbead arrays, but the method was used only to detect a single analyte, the protein AFP with 60 pg/mL LOD. In general, LODs by most of these fluorescent bead methods were often not as good as ELISA.
FLUORESCENCE DETECTION
Bead-based immunoassays with fluorescent detection were discussed in the previous section. We continue here reporting on fluorescence detection using other approaches. Quantum dots (Q-dots) have been used as labels by several groups, some described above in the bead section. Belolazove et al. developed water-dispersible core/shell InP/ZnS quantum dots coated with silica to encapsulate fluorescent labels.76 These Cd-free nontoxic labels were used to simultaneously detect two mycotoxins, zearalenone (ZEN) and deoxynivalenol (DON), in maize and wheat by lateral flow immunoassay. Cutoff levels were 50 pg/g of ZEN and 500 pg/g for DON. S. Chen and co-workers optimized spectral properties and minimized nonspecific binding using fluorescent quantum dot labels measured by flow cytometry.77 Tyrosine kinase receptors (RTKs) including VEGFR1 and VEGFR2, their coreceptor neuropilin1 (NRP1), and platelet-derived growth factor receptors (PDGFRα and PDGFRβ) were measured in cell cultures. Multiplexed quantification of these receptors was achieved at the single-cell level with a dynamic range of 800–20 000 receptors per cell. A flow cytometry immunoassay (FCI) was also developed by using template-free DNA extension on the surface of the magnetic beads.78 PSA was detected individually, and CEA and AFP were measured simultaneously with LODs of 0.5 pg/mL.
A quantum dot encoded bead array was described79 in which optical tweezers trap and decode biomolecule-conjugated immunomicrobeads that are then decoded and identified using spectrophotometry, resulting in LODs for various anti-IgGs in the low pM range. Food contaminants clenbuterol (Clen) and ractopamine (RAC) in pig urine were detected using a lateral flow immunoassay employing newly synthesized, highly luminescent, green-emitting gold nanoclusters.80 The assay is suitable for farmyard and food preparation sites and takes 18 min per assay with LODs for Clen of 3.0 pg/mL and RAC of 2.3 pg/mL.
Lam et al. developed a microfluidic device to measure immune cell secretion of cytokines in real time.81 The chip integrates isolation, culture, and stimulation to serve as an experimental workstation. Real-time monitoring of cytokine secretions was done by using cytometric fluorescent microbeads. Immune cell secretory cytokines IL-6, IL-8, and tumor necrosis factors of human peripheral blood mononuclear cells were simultaneously quantified with LODs ~20 pg/mL. Y. Ji and co-workers developed patterned antibody barcodes for secreted extracellular vesicles (EV) and cytokines.82 Over 1 000 cells can be analyzed concurrently. This platform enables high-throughput, multiplexed profiling of single-cell EV secretions and combines a high-density microchamber array, with a spatially resolved barcode glass slide read by fluorescence. Single-cell EV analysis was done on human oral squamous cell carcinoma cell lines for validation. Ma used anti-dsDNA antibody-Fe3O4 magnetic nanoparticles and fluorescently labeled DNA. Fluorescent signal increases when the labeled DNAs are bound by specific transcription factor; Oct4, SoX2, and Nanog were detected at 35–800 pg/mL levels. The assay detects target analytes in one-step and reveals interactions between transcription factors and DNA.
Liang’s team developed several proximity immunoassays, which feature pairs of polynucleotides with complementary sequences labeled with antibodies and detection by a DNA amplification strategy. They developed a four-color, one-step fluorescent proximity immunoassay utilizing this approach for detection of PSA, AFP, VEGF, and CEA.83 The quenching ability of gold nanoparticles and amplification by ribonuclease H release of the dye labels resulted in LODs in pg/mL of 2.9 for CEA, 1.25 for PSA, 0.65 for AFP, and 2.3 for VEGF. They also introduced isothermal quadratic amplification using a 3-color polydopamine nanoparticle proximity immunoassay.84 Analytes PSA, CEA, and AFP are introduced and bind to specific pairs of DNA tethered-antibodies as above and form proximate complexes. The isothermal amplification step recycles many hairpin probes in this rather complex scheme, substantially increasing the fluorescent signal resulting in attomolar biomarker detection. LODs for target proteins were substantially better than their previously described proximity assay at 20 fg/mL for AFP, 14 fg/mL for CEA, and 27 fg/mL for PSA.
A number of new fluorescent immunoassays were developed to detect toxins and toxic bacteria. Soares et al. used an array of Si:H thin-film photodiodes to capture fluorescence signals in a multiplexed bead-based microfluidic immunoarray for highly toxic mycotoxins.85 LODs in food assays for aflatoxin B1, ochratoxin A, and deoxynivalenol were below 1 ng/mL. Another fluorescent protein microarray was developed for the simultaneous immunodetection of four common mycotoxins, T-2 toxin (T-2), aflatoxin B1 (AFB1), ochratoxin A (OTA), and zearalenone (ZEN), in food.86 Detection can be accomplished in 30 min resulting in LODs of 0.12 ng/mL for AFB1, 0.03 ng/mL for T-2, 1.24 ng/mL for OTA, and 0.58 ng/mL for ZEN in spiked water and corn. Wu et al. developed a digital immunoassay for multiplex virus detection.87 Here, fluorescent magnetic multifunctional nanospheres were optimized as capture antibody carriers and signal labels for target separation, which enables subsequent single particle counting in an array. The assay detected three avian influenza viruses, H9N2, H1N1, and H7N9, with 0.02 pg/mL LODs. A novel translucent ZnO nanorod array was used to detect avian influenza virus (AIV) at levels 22 times below conventional ELISA using fluorescent labels that are enhanced in the nanostructure ZnO sensor.88 A biochip with a dithiobis (succinimidylundecanoate)-activated gold surface was used for serological screening of three B. burgdorferi antigens and three borrelial VlsE protein IR6 peptides with screening potential for Lyme disease.89 LODs were 0.39 μg/mL for VlsE and OspC and 0.78 μg/mL for flagellin. An immunosensing system was developed for multiplex detection of pathogens S. aureus and E. coli.90 The sensor integrates fluorescence with renewable microcolumns and bead injection analysis for concentration measurements. Analysis takes 20 min with LODs of <50 CFU/mL for E. coli and 100 CFU/mL S. aureus in milk, comparable to standard microbiological tests. Silicon discs with binary coding bits on their perimeter were equipped with antibodies for fluorescent detection of Influenza A nucleoprotein.91 Antibodies Ab11, Ab12, and Ab66 were assessed for binding affinity to Influenza A nucleoprotein. Ab66 had the lowest dissociation constant and gave an LOD of 0.04 ng/mL.
Unique applications of polymers are also being pursued in fluorescent immunoassays. Semiconducting polymer dots were used to build a multiplexed immunochromatographic test strip (ICTS) to measure PSA, AFP, and CEA. Detection took 10 min with LODs 2.1 pg/mL for PSA, 3.3 pg/mL for AFP, and 4.9 pg/mL for CEA.92 An ELISA type immunoassay was developed using encoded hydrogel microparticles.93 Antibodies are attached to surface groups on the microparticles to achieve high surface loading densities. LODs were 18 pg/mL for PSA and PIGF 4.2 pg/mL, and 4 pg/mL for CG beta. Three biomarkers related to acute myocardial infarction and heart failure were detected using porous hydrogel-encapsulated photonic crystal barcode suspension array.94 A sandwich assay fluorescence platform with labeled antibodies gave LODs 9 pg/mL for cardiac troponin I, 0.084 pg/mL for B-type natriuretic peptide (BNP), and 0.68 ng/mL for myoglobin. A multiplex immunoassay using aptamers was designed using morphologically different magnetic beads to detect Alzheimer’s disease biomarkers, Aβ42 and tau441.95 A fluorophore label was measured by a total internal reflection fluorescence microscopy electron-multiplying charge-coupled device imaging system. LODs were 4 fM for Aβ42 and 15 fM for tau441. A porous layer open tubular capillary was used for multiplexed detection of AFP and CEA.96 The 3D porous structures of the capillary increase surface-area-to-volume ratio which increases the detection sensitivity. LOQs were of 0.05 ng mL−1 for AFP and 0.1 ng mL−1 for CEA.
OPTICAL ABSORBANCE AND COLORIMETRY
Multiparametric ELISA was analyzed for its ability to diagnose autoimmune blistering dermatoses in Slavic populations.97 Two multiparametric ELISAs were evaluated, one with bullous pemphigoid 180 (BP180) and bullous pemphigoid 230 (BP230) and another with desmoglein 1 (DSG1) and desmoglein 3 (DSG3) biomarkers. Multiparametric ELISA for BP180, BP230, DSG1, and DSG3 showed diagnostic statistics of 87.5%, 80%, 50%, and 80% sensitivity, 87.5%, 91%, 100%, and 80% specificity, 87.5%, 87.5%, 82%, and 92% reliability, respectively. ELISA for BP180 and BP230 was a better multiparametric system than ELISA with DSG1 and DSG3. This analysis was recommended for diagnosis of autoimmune blistering dermatoses with patients suffering from autoimmune dermatoses.
A colorimetric immunoassay was developed in a 96-well plate for 20 different phosphorylated peptide biomarkers.98 It was successfully applied over a widely tunable range from pg/mL to μg/mL of the peptide biomarkers. The approach exploited peptide-mediated controlled aggregation of negatively charged AuNPs. The tunability of the range comes from the removal of the phosphate group from the phosphorylated peptide by alkaline phosphatase, yielding a positively charged peptide, which triggered AuNP aggregation. The color of the peptide/AuNP upon aggregation change from blue to red, enabling visual detection of the biomarker or in a spectrometer. Three biomarkers, PCT, IL-6, and CRP, were tested for validation of the assay, with LODs 3.15 μg mL−1 for CRP, 0.24 ng mL−1 for PCT, and 12.51 pg mL−1 for IL-6. These biomarkers were also measured in clinical serum samples.
A cotton-swab based colorimetric sandwich assay was developed to detect pathogenic bacteria, Salmonella typhimurium, Salmonella enteritidis, Staphylococcus aureus, and Campylobacter jejuni.99 Antibodies that are specific to each of these bacteria were immobilized on cotton swabs, and cells were collected from contaminated surfaces of a facility or meat samples. The bacteria are picked up by the swab upon binding to the corresponding antibodies, and detection involves immersing the cotton swab in colored beads carrying detection antibodies. The swabs bound with different colored nanobeads form a sandwich assay and the colors correspond to different bacteria bound. Qualitative assay of the color intensity of the nanobeads gave an estimate of bacterial levels (LOD = 10 cfu mL−1). This has the potential to be fully automated by using a smartphone as detector and wireless reporter.
An assay similar to the one above used lactoferrin immobilized on the cotton swab, instead of the antibodies.100 The cotton swab was used to collect different bacteria on the swab, preconcentrate them, then lactoferrin-bound bacteria were detected in a sandwich assay. The detection was via the binding of different colored microbeads with particular antibodies on them, each generating a particular color on the swab. The color intensity increased with bacterial concentration, and the detection limit was as low as 10 CFU/mL for Salmonella typhimurium and Campylobacter jejuni, 100 CFU/mL for Salmonella enteritidis, and 100 CFU/mL for Staphylococcus aureus on chicken meat surfaces. Quantitative measurements were done by determining the intensity of each color by using ImageJ software on smartphones. The above two assays are inexpensive, applicable to a wide variety of pathogens, and suitable for implementing on a large number of samples for food analysis.
A multiplexed sandwich assay was developed using six different signal detection schemes, with the entire system mounted on a chip which can be scanned with an office scanner, for point of care applications.101 The detection antibody was labeled with different reagents: (1) AuNPs, (2) carbon NPs, (3) oxidized carbon NPs and biotinylated detection antibody, (4) neutravidin-carbon, (5) streptavidin-gold, and (6) streptavidin-HRP. Using IL-8 as a model analyte, oxidized carbon NPs worked best. Three biomarkers, IL8, decorin (DCN), and VEGF in synthetic urine had the best LODs < 15 ng/mL. This report compares performance of different detection labels using the same type of assay and the same analyte under the same conditions through a lab-on a chip sandwich immunoassay that can be implemented in a doctor’s office.
A competitive immunoassay, usually used for small molecules not amenable to sandwich assays, was reported using a microfluidic lab-on-a-chip.102 The design consisted of three main sections: autonomous fluid delivery, reaction chamber, and capillary pumps to control liquid flow rate. The assay was performed in less than 10 min, and had detection limits of <40 ng/mL for ochratoxin A, 0.1–0.2 ng/mL for aflatoxin B1, and <10 ng/mL for deoxynivalenol. The immunoassay was done using competitive binding and displacement with 3,3′,5,5′-tetramethylbenzidine (TMB) to generate color. The measurements were taken on a standard smartphone camera and gray scale quantification procedure, making this a simple and quick method for the analysis of food contamination.
Products of proteolysis of gluten in beers, breads, vinegars, and sauces were followed using nine different antibodies of gluten, each specific for a particular epitope.103 A competitive multiplex ELISA, for example, was developed to measure gliadins, glutenins, and deamidated-gliadin specific epitopes in wheat and barley beers, soy, teriyaki and Worcestershire sauces, vinegar, and sourdough bread by using multiple epitope antibodies. Cluster analysis of the gluten concentrations in the samples indicated high potential to divverentiate protein/peptide profile characteristics from different processing methods. LODs ranged from 0.85 to 2.9 μg/mL.
A novel optical biosensor array based on silicon light sources and broadband Mach-Zenhder interferometers (MZI) was used to simultaneously detect different toxins in beer, in less than 15 min.104 The device could be reused for up to 20 times without loss of sensitivity. The detection employed immobilized mycotoxin-specific antibodies, followed by secondary immuno-reactions between the immunoadsorbed antibodies and goat anti-mouse IgG antibody. LODs were 0.8 ng/mL for aflatoxin B1, 5.6 ng/mL for fumonisin B1, and 24 ng/mL for deoxynivalenol. This is a simple, inexpensive food protection device that can detect multiple toxins for point of service.
A novel suspension particle array assay featured the high multiplexing capacity and scalability of simple colorimetric readout suitable for POC tests.105 Nearly millimeter size “bricks” (900 μm) are lithographically coded with four unique characters, allowing more than two million codes covalently coupled to binder molecules. Detection of up to 1900 particles was done using a flatbed scanner by depositing the particles on a transparent plastic sheet. Antigens were covalently coupled to the surface, with one antigen per code. With this array, a sandwich immunoassay was constructed using antibiotin gold nanoparticles which change color based on analyte concentrations. As proof of concept, autoantibodies toward anoctamin 2 were detected. Analytical sensitivity of 4 ng/mL was obtained with this milliarray.
An optical biosensor chip was made for rapid, simultaneous detection of food allergens in bovine milk protein, peanut protein, soy protein, and gliadin.106 The sensor chip consisted of monolithic silicon coupled with 10 broad-band Mach-Zehnder interferometers and broad band LED light sources. Interferometer surfaces were biofunctionalized to form a sandwich assay with the target antigen antibodies, antigens, and their corresponding antispecies-specific antibodies. Change on the chip surface due to the immunoreaction of the antigens is detected by a miniaturized spectrometer. LODs were 0.04 μg/mL for bovine milk protein, 1.0 μg/mL for peanut protein, 0.80 μg/mL for soy protein, and 0.10 μg/mL for gliadin. The results agreed with the corresponding ELISAs, but with the fast detection time and small footprint, this is an attractive tool for on-site food analysis.
A biochemical breadboard was built which consisted of a number of paper fluidic constructs that could be assembled, arranged, and configured for a particular analysis.107 Such breadboards could lower the time and effort spent in making paper fluidic devices. A predesigned block library (LEGO) is used to construct a functional lateral flow immunoassay (LFI) unit (Figure 8). A library of blocks was utilized for tailoring different setups for various immunoassays without laser cutters or 3D printers and accommodates LFI for positive and negative controls. The method was validated by measuring glucose, nitrile detection, and multiplexed cancer biomarkers (DNP, fluorescein amidite (FAM), or AF488 peptide analytes). This qualitative assay had no reported LOD values.
Figure 8.
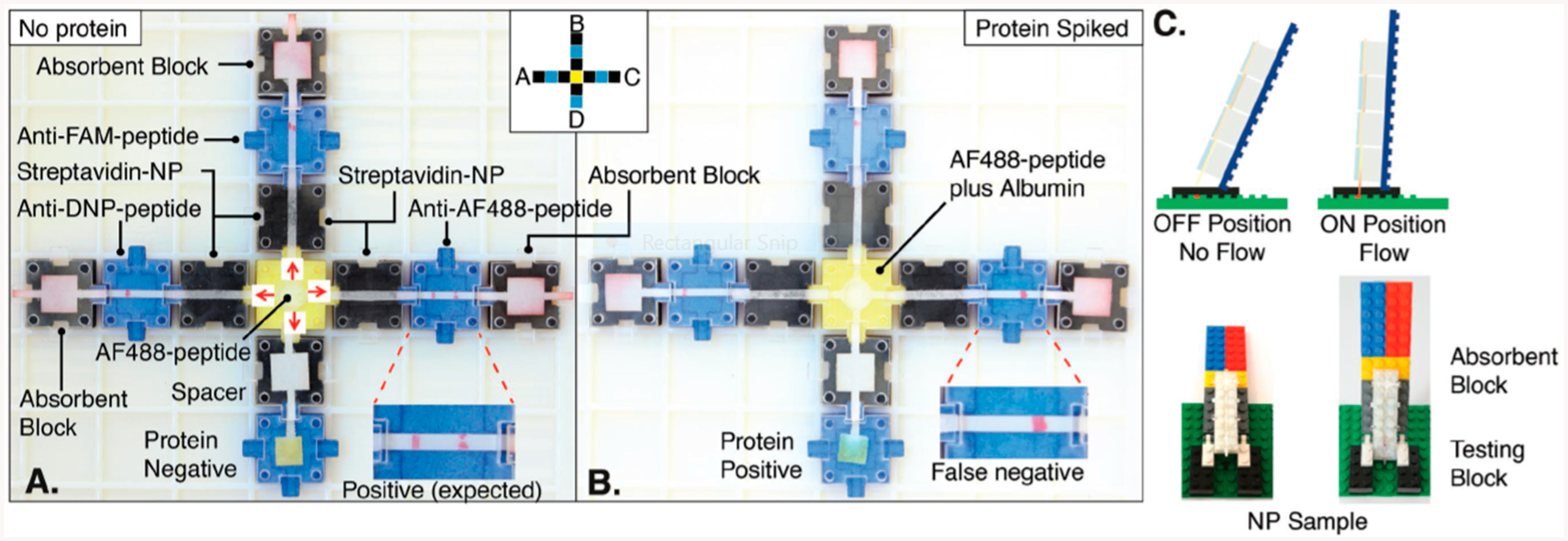
Biochemical breadboard made of a library of linear flow paper fluidic devices that can be put together by the user to construct modular detection systems. Reproduced from Ampli: A Construction Set for Paperfluidic Systems, Phillips, E.A., Young, A.K., Albarran, N., Butler, J., Lujan, K., Hamad-Schifferli, K. and Gomez-Marquez, J. Advanced Healthcare Materials, Vol. 7, Issue 14 (ref 107). Copyright 2018 Wiley.
A silica based porous layer open tubular capillary was modified by biphasic sol-gel reaction and an aldehyde active cetyltrimethylammonium bromide (CTAB) surface has been created. Capture antibodies were immobilized on this surface for multiplexed detection.108 Two such PLOT tubes containing anti-AFP and anti-CEA were connected via a PEEK MicroTight Connector. Fluorescence detection at 520 nm with excitation by a 488 nm laser led to simultaneous identification of AFP and CEA. The LODs for these in clinical serum samples were 0.05 and 0.1 ng/mL, respectively.
A novel multiplexed immune assay was developed to simultaneously detect multiple plasmodium antigens and CRP from asymptomatic patients confirmed with malaria.109 This was carried out with a 4-plex ELISA kit from Quansys Biosciences. The detection limits were 0.2 pg/mL for Plasmodium falciparum antigen histidine-rich protein 2 (HRP2), 1.5 pg/mL for malaria-conserved antigen lactate dehydrogenase (pLDH) for P. vivax, 9.3 pg/mL for all-malaria LDH (pan LDH), and 26.6 ng/mL for CRP. The concentrations of pLDH correlated closely with parasite density, while HRP2 levels did not. Colorimetry was also a detection option in 3D printed ELISA-in-a pipet tip described earlier under chemiluminescent detection.54 Color development was analyzed by a cell phone and reported LODs were 25 pg/mL for PSA, 25 pg/mL for CD14, 2.5 pg/mL for VEGF with the lowest LOD of 0.5 pg/mL for IGF-1. The assay is faster and LODs are lower than traditional ELISA and feature a 25% reduction in cost.
SURFACE-ENHANCED RAMAN SCATTERING
Interference-free mixing SERS emission (m-SERS) was used to simultaneously detect three liver cancer proteins AFP, CEA, and ferritin (FER).110 Sandwich antibody architecture was used to form microscale core-satellite assemblies between magnetic beads and triple coded SERS tags, enabling single SERS emission for each target. This immunoassay offers fast enrichment and separation abilities, with LODs of 0.15 pg/mL for AFP, 20 pg/mL for CEA, and 4 pg/mL for FER.
A SERS based immunoassay was developed for evaluating the risk of pancreatic cancer (PC) using proteins CA19–9, MMP7, and MUC4 as biomarkers.111 The system was first optimized in terms of the size of the gold particles used, the gap between particles, and the substrate material. This was followed by the design of a micropatterning approach for multiplex detection of the analytes. LOD determined using spiked human sera was as low as 2 ng mL−1.
SERS immunosensors also give amplified signals arising from plasmonic resonance effects on molecules positioned close to gold or silver surfaces. Using plasmonic metal nanoparticles decorated with antibodies confers biorecognition specificity for SERS immunosensor signals. Nonspecific binding was minimized, and antibodies of wild type p53 and p53R175H were assayed,112 enabling LODs of both targets in the attomolar range in buffer and femtomolar range in human serum.
A vertical flow assay based on SERS was developed for simultaneous detection of cancer biomarkers, PSA, CEA, and AFP.113 Strong Raman-active dyes encoded with core-shell SERS nanotags were used and a high surface to volume ratio of the porous sensing membrane. This configuration enhanced signal capabilities of the nanostructured sensor surface. The biosensor has wide dynamic range and LODs of 0.37 pg/mL for PSA, 0.43 pg/mL for CEA, and 0.26 pg/mL for AFP.
A SERS immunoassay for prostate cancer diagnostics was developed employing magnetic beads and SERS nanotags.114 This approach enabled simultaneous detection of free (f-PSA) and complexed (c-PSA) PSA. Utilizing magnetic sandwich immunocomplexes, the assay analyzed 30 clinical samples and the data was correlated with ECL assays of the same samples. The SERS assay correlated well with ECL with LODs of 0.012 ng/mL for f-PSA and 0.15 ng/mL for c-PSA. Also, an automated droplet-based microfluidic platform with SERS detection was designed for simultaneous analysis of free PSA and total PSA.115 The magnetic immunocomplexes of the analytes and the supernatant were introduced by way of a Y-junction of two daughter droplets from a parent microdroplet, where magnetic separation is achieved. This two-channel format allowed for SERS signal analysis of sequential droplets resulting in LODs below 0.1 ng mL−1 for both cancer biomarkers.
A panel of soluble cancer protein biomarkers was detected by SERS without the need for monoclonal antibodies.116 This was achieved by replacing the antibodies with nanoyeast single-chain variable fragments (Figure 9). The protein biomarker panel included soluble programmed death 1 (sPD-1), soluble programmed death-ligand 1 (sPD-L1), and soluble epithermal growth factor receptor (sEGFR). High sensitivity was achieved by the use of gold-silver alloy nanoboxes with characteristic Raman spectral enhancements. Within 1 h, the panel of biomarkers were detected with LODs of 6.17 pg/mL for sPD-1, 0.68 pg/mL for PD-L1, and 69.86 pg/mL for sEGFR.
Figure 9.
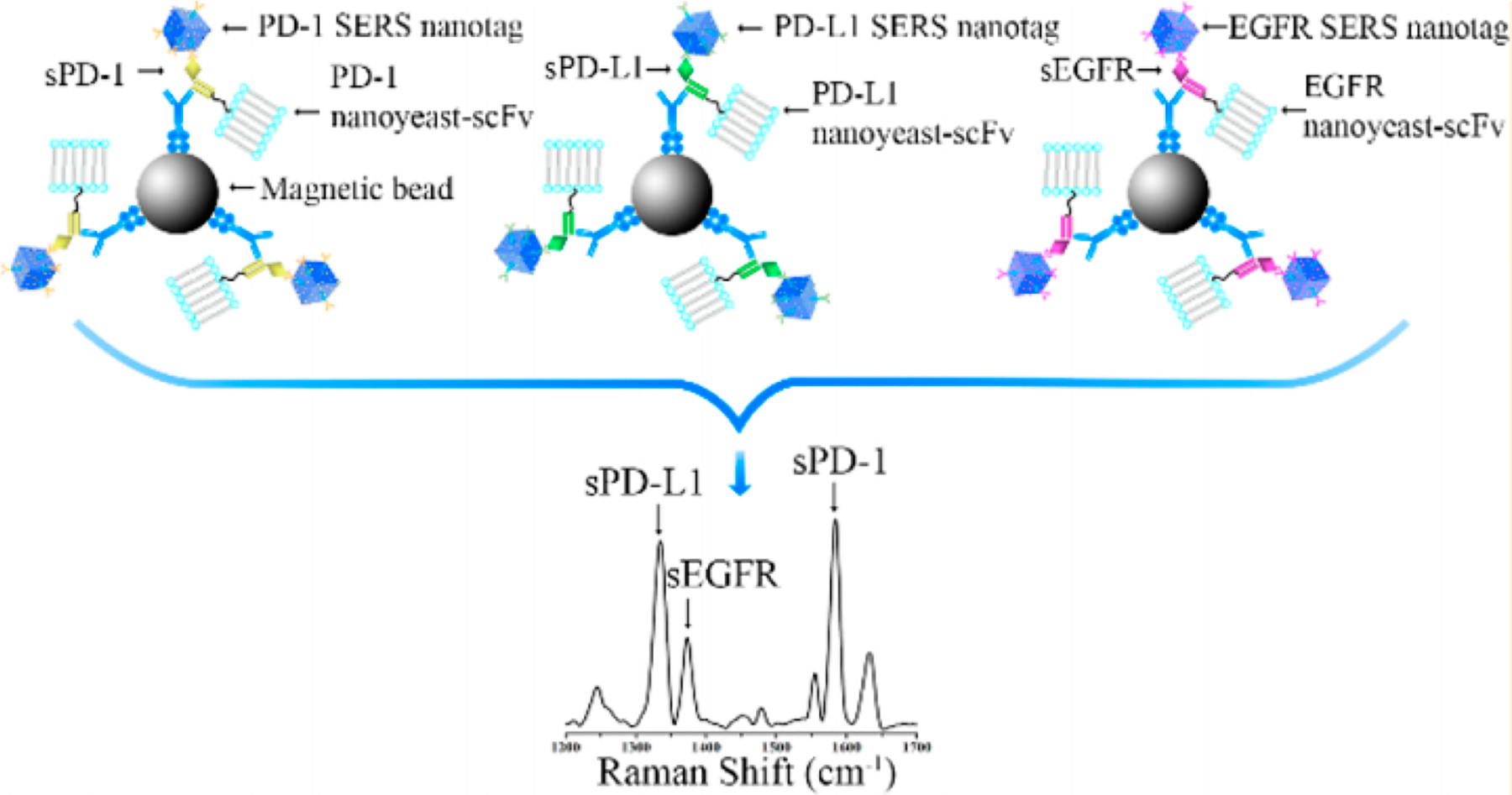
Replacement of antibodies by nanoyeast single chain variable fragments for high sensitivity detection of multiple biomarkers. Reproduced from Li, J.; Wang, J.; Grewal, Y.S.; Howard, C.B.; Raftery, L.J.; Nahler, S.; Wang, Y.; Trau, M. Anal. Chem. 2018, 90, 10377–10384 (ref 116). Copyright 2018 American Chemical Society.
A single porous hydrogel bead with buried gold-silver core-shell SERS nanotags encoded with Raman dyes was used for multiplexed immunoassays.117 In combination with SERS, increased binding of biomarkers on the porous hydrogel further increased the detectable concentration range of analytes. LODs of tumor biomarkers were 3.6 fg mL−1 for AFP and 12.6 fg mL−1 for CEA. These LODs are improvements over previous methods.
A novel concept for early diagnosis of hepatocellular carcinoma by SERS immunochips was based on Raman frequency shift and intensity change.118 The frequency shift of 4-mercaptobenzoic acid (MBA) due to interactions of protein AFP with its antibody attached to the MBA-modified silver SERS immunochip was first monitored. This was followed by the intensity changes of the SERS signal when AFP-L3 binds to its antibody attached to SERS chip containing 5,5′-dithiobis-(succinimidyl-2-nitrobenzenoate)-modified immunogold in a sandwich assay. The combination of orthogonal measurements simplified the estimation % AFP-L3 and total AFP, with lowest detectable signal of 0.5 ng/mL for both AFP-L3 and AFP.
A SERS-based immunoassay utilizing magnetic capture of silica encapsulated SERS nanotags was developed to simultaneously detect pathogenic viruses.119 West Nile virus, Rift Valley fever virus, and Yersinia pestis were assayed with improved assay time and reaction kinetics by using colloidal particles instead of the traditional solid surface system. A 100-fold decrease in LOD (10 pg/mL) was achieved for all biomarkers when compared to a single antigen capture assay. In a related approach, simultaneous detection of Zika and Dengue fever viruses was achieved by SERS encoded-gold nanostars conjugated to specific antibodies with increased sensitivity, when SERS was coupled with lateral flow immunoassay.120 The limits of detection were 0.72 ng/mL for Zika NS1 and 7.67 ng/mL for Dengue NS1 were reported. A SERS immunoassay was also developed for simultaneous and single sample detection of Ebola, Lassa, and malaria.121 The SERS nanotags were improved upon to be stable at extreme environments, negating the need for cold-chain transport and storage, while simplifying the processing and analysis time by eliminating sample preparation and wash steps. The system provided results in less than 30 min and an LOD of 105–106 PFU/mL with sensitivity and specificity between 90% and 100%.
A multiplexed platform was developed integrating SERS with alternating current electrohydrodynamic fluid flow and graphene oxide functionalization for enhanced surface capture. The approach simultaneously determines circulating cancer cells and cancer biomarkers in the same analysis, providing a more robust measurement.122 Expression profiling of cell surface biomarkers and simultaneous detection of cells and proteins within the same system are two major innovations. Single-cell resolution was achieved with protein LODs of 100 fg mL−1 for both human epidermal growth factor receptor 2 (EGFR-2) and Mucin 16.
Simultaneous SERS and fluorescence detection was developed using a programmable immunosensor featuring Raman active fluorogens and an enzyme sensitive linker.123 The sensor nanoprobes included a cathepsin-sensitive peptide linker (Phe-Lys-Cys) for the release of the fluorogen that quenches the SERS signal. Three lung cancer biomarkers napsin-A (Nap), cytokeratin-19 (CK), and EGFR were detected.
A three-dimensional macroporous Au-plasmonic SERS platform was developed using SERS reporter tags on Ag-Au nanostars.124 Characterization of the plasmonic array revealed the generation of “hot spots” causing enhancement in the SERS signals. These, in turn, enabled ultrasensitive detection of biomarkers with LODs of 0.76 fg/mL for cardiac troponin I, 0.53 fg/mL for N-terminal prohormone of brain natriuretic peptide, and 0.41 fg/mL neutrophil gelatinase-associated lipocalin. A nanopillar plasmonic substrate was used in a SERS mapping technique for multiplexed detection of three mycotoxins.125 The leaning of the nanopillars decreases gaps among them, which creates hot spots for SERS and improves sensitivity. Furthermore, the densely packed nonpillars offer high uniformity with minimized spot-to-spot fluctuation in Raman signal providing highly reproducible assay detection. LODs were 5.09 pg mL−1 for ochratoxin, 5.1 pg mL−1 for fumonisin B, and 6.07 pg mL−1 for aflatoxin B1.
A SERS immunoassay was developed for ultrasensitive multiplexed detection of tumor markers PSA and AFP.126 The assay was designed using SiO2@Ag immuno probes functionalized with 4-MBA and 4-NTP for differential detection of the targets. A gold-film hemisphere array was used as the immune substrate. LODs of 3.4 fg mL−1 for PSA and 4.1 fg mL−1 for AFP were reported. A SERS multiplexed immunoassay was developed for the simultaneous detection of two illegal growth-promoting drugs, clenbuterol and ractopamine.127 Two different SERS nanoprobes were developed, 4′-dipyridyl (DP) immobilized with clenbuterol antibody and 2,2′-dipyridyl immobilized with ractopamine antibody. The method has a wide dynamic range with a LOD of 1 pg mL−1, providing a simple platform for drug analysis.
A lateral flow assay was developed for multiplexed quantitative detection with core-shell SERS nanotags for early diagnosis of acute myocardial infarction.128 Three cardiac biomarkers Myo, cTnI, and CK-MB were detected using Nile Blue encapsulated silver-gold core-shell bimetallic particles as SERS nanotags. The assay enabled a wide dynamic range and LODs of 3.2 pg/mL for Myo, 0.44 pg/mL for cTnI, and 0.55 pg/mL for CK-MB. A microfluidic chip was used for the development of a SERS immunoassay for early detection of breast cancer biomarkers.129 The immuno-Ag aggregates were labeled with Raman reporters specific to biomarkers and the LODs were 0.01 U/mL for both CA153 and CA125 and 1 pg/mL for CEA. SERS detection of multiple miRNA biomarkers specific to hepatocellular carcinoma was developed following two different approaches.130 In the first, three types of Raman-active dye-coded Au nanoparticles with narrow intrananogap structures were functionalized with probe DNA strands to recognize target miRNAs. In the second, micrometer-sized hollow silver microspheres (Ag-HMSs) with Lactococcus lactis subsp. cremoris as templates were synthesized and used in the sandwich assays by immobilizing DNA strands on the surface. The LODs for the target miRNAs reported were 2.72 pM for miR-21, 0.24 pM for miR-122, and 2.68 pM for miR-223.
Using SERS analysis for PSA in patient samples, a unique classification system was developed for prostate cancer diagnosis.131 Detection involved a combination of SERS immunoassay using silver nanoparticles as immune probes and SiC@Ag@Ag-NPs SERS as immune substrates. The patients are classified as being healthy, having benign prostate hyperplasia or having prostate cancer using new algorithms. This complex immunoassay had an LOD of 0.46 fg mL−1 for PSA, 1.1 fg mL−1 for prostate-specific membrane antigen, and 0.7 fg mL−1 for human kallikrein 2. Blood plasma levels of Type 1 cytokine were monitored by SERS immunoassay.132 The method incorporated SERS sensors into a microfluidic device and had LODs of 3.8 pg mL−1 for IL-6, 7.5 pg mL−1for IL-8, and 5.2 pg mL−1 for IL-18.
SURFACE PLASMON RESONANCE
Early diagnostic tests for Shiga toxin-producing E. coli (STEC) is important for diagnosis and positive therapy outcomes and to minimize the spread of the disease.133 SERS immunosensors were capable of rapid multiplexed detection of a variety of relevant ligands. Image-based SPR array sensing (SPRi) allowed a high degree of multiplexing. SPRi quantified the O-antigen serogroups of up to 11 O-antigens in 188 isolates of SETC, with an overall sensitivity of 98.9% LODs 1.1 to 17.6 × 106 CFU mL−1. Scanning-angle SPRi is a variation that utilizes different incident angles for imaging.134 A first application of multiplexed, label free detection of pancreatic human hormones by scanning angle SPRi reported LODs of 1 nM for insulin, 4 nM for glucagon, and 246 nM for somatostatin. The chip had an antifouling sensing surface comprised by a mixed self-assembled monolayer of CH3O-PEG-SH and 16-mercaptohexadecanoic acid, facilitating operation in complex matrixes.
Castiello and Tabrizian compared three different configurations of gold nanoparticles: (1) AuNPs immobilized on the sensor surface, (2) AuNPs conjugated with primary antibodies (AuNP-Ab1), and (3) AuNPs conjugated with a secondary antibody (AuNP-Ab2) for detection of hormones insulin, glucagon, and somatostatin.135 AuNP-Ab2 gave the best LOD for the three proteins with LODs of 0.15 nM for insulin, 0.39 nM for glucagon, and 1.22 nM for somatostatin. A first attempt in detecting mycotoxins using AuNP enhanced SPRi also used AuNPs bound to Ab2.136 In a competitive inhibition format, AuNP-Ab2 increased the bound mass, increasing the SPR signal substantially to enable detecting small amounts of mycotoxin directly. The chip can be used 46 times without significant signal loss (<12%, 46 reuses). LODs were 15 μg/kg for deoxynivalenol, 24 μg/kg for zearalenone, and 12 μg/kg for T-2 toxin.
A feasibility study for the detection of nine different antibodies in body fluids by SPRi using principal component analysis was reported.137 Antibodies were printed on the SPRi chips in a hydrogel using active ester groups and a continuous flow microspotter. The potential of SPRi to serve as a novel multiplex body fluid identifier for forensic applications was demonstrated.
Localized surface plasmon resonance (LSPR) using dispersed nanoparticles in solution was used. A recent report describing on-chip microfluidic coupled for simultaneous detection of four representative cancer biomarkers.138 Gold nanorods were deposited on the surface of a glass chip and a sandwich immunoassay constructed by the immobilization of mercaptoundecanoic acid. LSPR sensing enabled the sensing volume to be brought down to the molecular scale and used direct coupling with propagating light, unlike in conventional SPR where an index-matching fluid is used. The reported LODs were 0.21 U/mL for CA15–3, 0.139 U/mL for CA 125, 1.021 U/mL for carcinoembryonic antigen (CEA), and 3.9 ng/mL for HER2.
Our group reported microfluidic-assisted surface plasmon resonance imaging (SPRi) immunoassay for differentiating IgE antibodies by epitope-resolved detection.139 Capture of IgE antibodies by magnetic beads modified with IgE ε-chain-specific antibodies was the first step of the protocol. Then, epitopes of peanut allergen glycoprotein Arachis hypogaea h2 were added to the array. Differential epitope responses were mapped in 45 min assays and verified with human patient serum samples. LODs of 0.5–5.0 pg/mL were obtained, a 50-fold improvement over the corresponding ImmunoCAP assays.
A variation on SPR was developed as a new technology based on photonic crystal (PC) recording of the total internal reflection angle and the excitation angle of the PC surface wave.140 A one-dimensional PC consisting of seven-layered SiO2/Ta2O5 is the sensor chip, and light is reflected off the chip due to total internal reflection (Figure 10). Chips are coated with amine functional groups and stored for later use for subsequent biomolecule conjugation. Ovarian CA-125 was detected with an LOD of 0.55 U/mL, HEGFR-2 had an LOD of 620 fg/mL, while CA-15 had a LOD of 1.84 U/mL.
Figure 10.
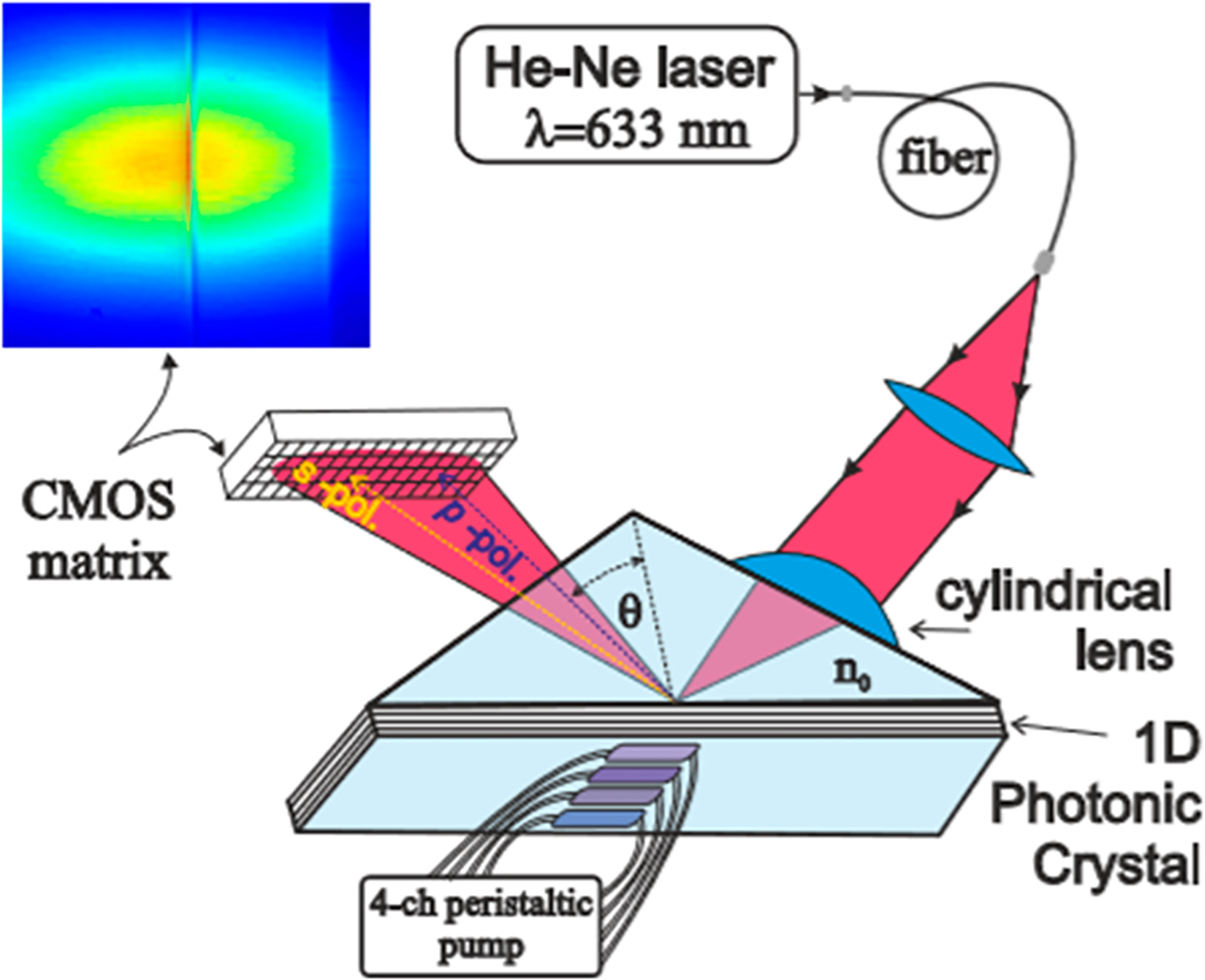
Schematic of the photonic crystal sensor, analogous to that of an SPR chip, and detects signals as molecules on the crystal surface interact with the analytes. Changes in the environment adjacent to the crystal surface changes the intensity of the light with total internal reflection. Reprinted with permission from Petrova, I.; Konopsky, V.; Nabiev, I.; Sukhanova, A. Label-Free Flow Multiplex Biosensing via Photonic Crystal Surface Mode Detection. Sci. Rep. 2019, Vol. 9, 8745 (ref 140). Copyright 2019 Springer Nature.
MISCELLANEOUS APPROACHES
In this section, we discuss new approaches to multiplexed immunoassays that do not fit well in the other categories. An interferometric sensor chip featuring of 10 Mach-Zehnder interferometric sensor along with individual accompanying LED sources in a microfluidic device was developed for competitive protein immunoassays.141 Transmission spectra were continuously recorded while flowing antibody-sample mixtures across standard protein spots. Data processing by Fourier transform analysis provided polarization phase shifts for each sensor. This system was used for simultaneous detection of four common food allergens with LODs of 40 ng/mL for bovine k-casein, 1.0 μg/mL for peanut protein, 0.80 μg/mL for soy protein, and 0.10 μg/mL for gliadin. Analysis of food processing samples with this 7 min assay were well correlated with the much slower single-protein ELISA.
Several new approaches were designed for microarray capture of analytes. A biosensor array was developed by immobilizing haptens onto a dextran network planar waveguide microarray platform coated with hydrophobic lipases.142 These haptens bind to the lipases and in turn were used to bind antibodies to effect a competitive fluorescent immunoassay for water pollutant toxins atrazine (ATR), enrofloxacin (ENRO), and microcystin LR (MCLR) detected simultaneously at LODs in the 0.03–13 ng/mL range. Microporous track-etched polycarbonate membranes with immobilized antibodies were used to develop a radiometric microarray immunoassay for thyroid cancer prognosis.143 Using 125I-labeled detection antibodies, thyroid cancer biomarkers were measured at LODs 0.07 μIU/mL for thyrotropin and 0.13 ng/mL for thyroid stimulating hormone. A graphically encoded silica array of suspended planar particles was developed for a microplate multiplex immunoassay.144 The flat silica microparticles fabricated by photolithography were 25 by 14 μm silica with high-reflection “barcode” markings on their surfaces in a 128-plex assay. Antibodies are attached to the barcode particles that can be identified after analyte and fluorescent-labeled secondary antibody capture in microplate wells with multiple optical mode readout. The system and associated interpretive software was used to determine inflammation biomarkers related to Type I diabetes and gave LODs in pg/mL of 0.08 for TNF-α, 0.21 for IFNγ, 0.08 for IL-1β, and for 0.17 FGF-19.
CONCLUSIONS
In this review, we summarized research on new methodologies for immunoassays published between May 1, 2017 and October 1, 2019. Our focus is on multiplexing, novel approaches, novel applications, and high sensitivity. Applications include measurement of disease biomarkers, viruses, toxins, and environmental pollutants, and many novel, innovative approaches have been reported. By far, the main application in these 2 years was new approaches to cancer diagnostics by measuring panels of biomarkers. However, new methodology for ultrahigh sensitivity detection were few, and many of the papers we discuss do not achieve lower LODs or better sensitivity than commercial single-protein ELISA kits. Protein detection in biological samples at concentrations less than about 1 fg/mL were reported in only 2 cases, but general multiplexed protein determination at such levels remain problematic. Effective reproducible protein detection at lower LOD ranges needs to be addressed in the future so that more of the human proteome can be quantified and perhaps used as more effective biomarkers.
At the same time, the ability and possibility to measure multiple analytes as biomarkers for disease states by immunoassay to impact early detection of diseases such as cancer is apparent. Unfortunately, multiplexed immunoassay approaches have not yet found wide diagnostic usage in hospitals and clinics, despite their potential for saving lives. It is our hope that we realize more widespread progress in immunoarray use for clinical disease diagnosis in the near future.
ACKNOWLEDGMENTS
Preparation of this review was supported by Grant No.EB016707 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB), NIH to J.F.R., by an Academic Plan Grant to J.F.R., and by a Research Excellence grant to C.V.K from University of Connecticut.
PROTEIN AND VIRUS ACRONYM GLOSSARY
- IL
interleukin
- HRP
horseradish peroxidase
- HRP2
histidine-rich protein 2
- TNF-α
tumor necrosis factor-α
- LDH
lactate dehydrogenase
- MYO
myoglobin
- CTn
cardiac troponin
- CK-MB
creatine kinase
- FER
ferritin
- MUC
mucin
- MMP
matrix metalloproteinase
- LC3B
microtubule-associated protein 1 light chain 3B
- PSA
prostate specific antigen
- PSMA
prostate specific membrane antigen
- CEA
carcinoembryonic antigen
- CRP
C-reactive protein
- PCT
procalcitonin
- AFP
α-fetoprotein
- CDH17
cadherin human 17
- HER2
human epidermal growth factor receptor-2
- CA
cancer antigen
- MERS-CoV
corona virus from Middle East respiratory syndrome
- STAT3
signal transducer and activator of transcription 3
- DOCK8
dedicator of cytokinesis 8
- PGM3
phosphoglucomutase 3
- OC
osteocalcin
- PTH
parathyroid hormone
- VEGF
vascular endothelial growth factor
- VEGF-D
vascular endothelial
- VEGFR
vascular endothelial growth factor receptor
- β-HCG
β-human chorionic gonadotropin growth factor-D
- ERG
ETS related gene protein
- IGF-1
insulin-like growth factor-1
- GOLM-1
Golgi membrane protein 1
- IGFPB-3
insulin-like growth factor binding protein 3
- CD-14
serum monocyte differentiation antigen CD-14
- IFN- γ
, interferon-γ
- CEA
carcinoembryonic antigen
- CYFRA
1cytokeratin fragment
- NSE
neuron specific enolase
- free-β-hCG
free β-subunit of human chorionic gonadotropin
- SCC
squamous cell carcinoma protein
- PG
pepsinogen
- Tg
thyroglobulin
- pLDH
plasmodium lactate dehydrogenase
- CA15–3
carbohydrate antigen 15–3
- HE4
human epididymis protein 4
- AMBP
α-1-microglobulin/bikunin precursor
- PRDX2
peroxiredoxin 2
- PARK7
Parkinson disease protein 7
- NRP1
neuropilin1
- PDGFR
platelet-derived growth factor receptor
- PIGF
phosphatidylinositol-glycan biosynthesis class F
- CG-β
chorionic gonadotropin beta
- BNP
B-type natriuretic peptide
- Aβ
amyloid beta
- DSG
desmoglein
- BP
bullous pemphigoid
- PTC
phenylthiocarbamide
- DCN
decorin
- sPD-1
soluble programmed death 1
- sPD-L1
soluble programmed death-ligand 1
- sEGFR
soluble epithermal growth factor receptor
- Nap
napsin-A
- CK-19
cytokeratin-19
Biographies
Abby Jones graduated with a B.Sc. degree in Chemistry from University of New England in 2012. She joined the group of Prof. James F. Rusling at the University of Connecticut and is currently completing her Ph.D. degree in Analytical Chemistry. She has been actively working in development of new biosensors on various platforms with her research focused on multiplexed cancer biomarker detection for biomedical diagnostics.
Lasangi Dhanapala graduated with a B.Sc. degree in Chemistry (First class Honors) from University of Peradeniya, Sri Lanka in 2017 and joined the Ph.D. program at the University of Connecticut the same year under the patronage of Prof. James F. Rusling. She is currently working on development of immunoarrays with amperometric detection for ultrasensitive detection of proteins for prostate cancer biomarkers. Her main focus lies in development of a sensitive immunoarray aimed at testing for recurrence of cancer in patients that have had surgical prostatectomy.
Rumasha N. T. Kankanamage graduated with a B.Sc. degree in Biochemistry and Molecular Biology from University of Colombo, Sri Lanka, in 2015 and then entered the Ph.D. program in Prof. James F. Rusling’s group at University of Connecticut in 2016. Currently, she is working on the development of a biosensor to evaluate genotoxicity of carcinogens using electrochemiluminescence and the elucidation of mechanistic pathways for cyt P450 mediated metabolism of carcinogenic chemicals using LC-MS.
Challa V. Kumar obtained his Ph.D. from the Indian Institute of Technology Kanpur, India, in 1982 and worked as a postdoctoral associate at the University of Notre Dame for 2 years and then as a research associate at Columbia University from 1984 to 1988. He joined the University of Connecticut in 1988 and is currently a Professor of Biological and Physical Chemistry. He has coauthored over 182 peer-reviewed publications and 7 books. His research interests include Sustainable Biological Materials, DNA/RNA binders, Bio-catalytic Nanomaterials, and Protein Photoscissors.
James F. Rusling obtained his B.Sc. in Chemistry from Drexel University and Ph.D. from Clarkson University. He is a Professor of Chemistry at the University of Connecticut and Professor of Surgery and Member of the Neag Cancer Center at UConn Health. Current research includes development of nanoscience based and 3D-printed immunoarrays for cancer biomarkers, exploration of new protein and miRNA biomarkers for disease diagnostics, and electro-optical and LC-MS/MS arrays for toxicity screening, and fundamental bioelectrochemistry. He has over 400 research papers, several books, and is an accomplished musician interested in Irish and American folk styles.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Rusling JF; Kumar CV; Gutkind JS; Patel V Analyst 2010, 135, 2496–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Barry MJN Engl. J. Med 2001, 344, 1373–1377. [DOI] [PubMed] [Google Scholar]
- (3).Ward AM; Catto J; Hamdy F Ann. Clin. Biochem 2001, 38, 633–651. [DOI] [PubMed] [Google Scholar]
- (4).Srinivas PR; Kramer BS; Srivastava S Lancet Oncol. 2001, 2, 698–704. [DOI] [PubMed] [Google Scholar]
- (5).http://www.quansysbio.com/ Last accessed September 1, 2019.
- (6).http://www.bioagilytix.com/landing-pages/meso-scale-discovery-electrochemiluminescence/ Last accessed September 1, 19.
- (7).https://www.thermofisher.com/us/en/home/life-science/protein-biology/protein-assays-analysis/luminex-assays.html Last accessed 09/01/19.
- (8).http://quansysbio.com/multiplex/multiplex-assays/ Last accessed October 4, 19.
- (9).Rissin DM; Kan CW; Campbell TG; et al. Nat. Biotechnol 2010, 28, 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).(a) Rissin DM; Kan CW; Song L; et al. Lab Chip 2013, 13, 2902–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) https://www.quanterix.com/products-technology/instruments/hd-x Last accessed September 1, 19.; (c) Walt D Personal communication. [Google Scholar]
- (11).Bensmail H; Haoudi A J. Biomed. Biotechnol 2003, 2003 (4), 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Sydor JR; Nock S Proteome Sci. 2003, 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Rusling JF Bioanalysis 2010, 2, 847–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wei H; Ni S; Cao C; Yang G; Liu G ACS Sensors 2018, 3, 1553–1561. [DOI] [PubMed] [Google Scholar]
- (15).Tuteja SK; Duffield T; Neethirajan SJ Mater. Chem. B 2017, 5, 6930–6940. [DOI] [PubMed] [Google Scholar]
- (16).Wang G; Li Y; Liu J; Yuan Y; Shen Z; Mei X Sci. Rep 2017, 7, 2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Sharafeldin M; Bishop GW; Bhakta S; El-SAwy A; Suib SL; Rusling JF Biosens. Bioelectron 2017, 91, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Alizadeh N; Salimi A; Hallaj R J. Electroanal. Chem 2018, 811, 8–15. [Google Scholar]
- (19).Zhao L; Han H; Ma Z Biosens. Bioelectron 2018, 101, 304–310. [DOI] [PubMed] [Google Scholar]
- (20).Serafín V; Valverde A; Garranzo-Asensio M; Barderas R; Campuzano S; Yáñez-Sedeño P; Pingarron JM Microchim. Acta 2019, 186, 411. [DOI] [PubMed] [Google Scholar]
- (21).Jirakova L; Hrstka R; Campuzano S; Pingarrón JM; Bartosik M Electroanalysis 2019, 31, 293–302. [Google Scholar]
- (22).Marques RC; Costa-Rama E; Viswanathan S; Nouws HP; Costa-Garcia A; Delerue-Matos C; González-García MB Sens. Actuators, B 2018, 255 (255), 918–925. [Google Scholar]
- (23).Mercer C; Jones A; Rusling JF; Leech D Electroanalysis 2019, 31, 208–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Eissa S; Almthen RA; Zourob M Microchim. Acta 2019, 186, 523. [DOI] [PubMed] [Google Scholar]
- (25).Layqah LA; Eissa S Microchim. Acta 2019, 186, 224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Eissa S; Abdulkarim H; Dasouki M; Al Mousa H; Arnout R; Al Saud B; Rahman AA; Zourob M Biosens. Bioelectron 2018, 117, 613–619. [DOI] [PubMed] [Google Scholar]
- (27).Eissa S; Alshehri N; Abduljabbar M; Rahman AMA; Dasouki M; Nizami IY; Al-Muhaizea MA; Zourob M Biosens. Bioelectron 2018, 117, 84–90. [DOI] [PubMed] [Google Scholar]
- (28).Vargas E; Teymourian H; Tehrani F; Eksin E; Sánchez-Tirado E; Warren P; Erdem A; Dassau E; Wang J Angew. Chem., Int. Ed 2019, 58, 6376–6379. [DOI] [PubMed] [Google Scholar]
- (29).Kim H; Sato S; Takenaka S; Lee MH Sensors 2018, 18, 2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Arya SK; Estrela P Biosens. Bioelectron 2018, 117, 620–627. [DOI] [PubMed] [Google Scholar]
- (31).Shanmugam NR; Muthukumar S; Tanak AS; Prasad S Future Cardiol. 2018, 14, 131–141. [DOI] [PubMed] [Google Scholar]
- (32).Kalyoncu D; Buyuksunetci YT; Anık Ü Mater. Sci. Eng., C 2019, 101, 88–91. [DOI] [PubMed] [Google Scholar]
- (33).Putnin T; Ngamaroonchote A; Wiriyakun N; Ounnunkad K; Laocharoensuk R Microchim. Acta 2019, 186, 305–317. [DOI] [PubMed] [Google Scholar]
- (34).Tang D; Ren J; Lu M Analyst 2017, 142, 4794–4800. [DOI] [PubMed] [Google Scholar]
- (35).Bard AJ, Ed. Electrogenerated Chemiluminescence; Marcel Dekker: New York, 2004. [Google Scholar]
- (36).Forster RJ; Bertoncello P; Keyes TE Annu. Rev. Anal. Chem 2009, 2, 359–385. [DOI] [PubMed] [Google Scholar]
- (37).Kadimisetty K; Malla S; Sardesai N; Joshi AA; Faria RC; Lee N; Rusling JF Anal. Chem 2015, 87, 4472–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Chen X; Liu Y; Ma QJ Mater. Chem. C 2018, 6, 942–959. [Google Scholar]
- (39).Rizzo F; Polo F; Bottaro G; Fantacci S; Antonello S; Armelao L; Quici S; Maran F J. Am. Chem. Soc 2017, 139, 2060–2069. [DOI] [PubMed] [Google Scholar]
- (40).Kudruk S; Villani E; Polo F; Lamping S; Körsgen M; Arlinghaus HF; Paolucci F; Ravoo BJ; Valenti G; Rizzo F Chem. Commun 2018, 54, 4999–5002. [DOI] [PubMed] [Google Scholar]
- (41).Guo W; Ding H; Gu C; Liu Y; Jiang X; Su B; Shao YJ Am. Chem. Soc 2018, 140, 15904–15915. [DOI] [PubMed] [Google Scholar]
- (42).Zou G; Tan X; Long X; He Y; Miao W Anal. Chem 2017, 89, 13024–13029. [DOI] [PubMed] [Google Scholar]
- (43).Zhou J; Nie L; Zhang B; Zou G Anal. Chem 2018, 90, 12361–12365. [DOI] [PubMed] [Google Scholar]
- (44).Zhang F; He Y; Fu K; Fu L; Zhang B; Wang H; Zou G Biosens. Bioelectron 2018, 115, 77–82. [DOI] [PubMed] [Google Scholar]
- (45).Babamiri B; Hallaj R; Salimi A Biosens. Bioelectron 2018, 99, 353–360. [DOI] [PubMed] [Google Scholar]
- (46).Zhou B; Zhu M; Hao Y; Yang P ACS Appl. Mater. Interfaces 2017, 9, 30536–30542. [DOI] [PubMed] [Google Scholar]
- (47).Kadimisetty K; Malla S; Bhalerao KS; Mosa IM; Bhakta S; Lee NH; Rusling JF Anal. Chem 2018, 90, 7569–7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Kadimisetty K; Spak AP; Bhalerao KS; Sharafeldin M; Mosa IM; Lee NH; Rusling JF Anal. Methods 2018, 10, 4000–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Croner LJ; Dillon R; Kao A; Kairs SN; Benz R; Christensen IJ; Nielsen HJ; Blume JE; Wilcox B Clin. Proteomics 2017, 14, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Chakraborty S; Brubaker J; Harro C; Weirzba T; Sack DJ Immunol. Methods 2019, 470, 6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Dillon R; Croner LJ; Bucci J; Kairs SN; You J; Beasley S; Blimline M; Carino RB; Chan VC; Cuevas D; Diggs J J. Pharm. Biomed. Anal 2018, 154, 85–94. [DOI] [PubMed] [Google Scholar]
- (52).Xianyu Y; Wu J; Chen Y; Zheng W; Xie M; Jiang X Angew. Chem., Int. Ed 2018, 57, 7503–7507. [DOI] [PubMed] [Google Scholar]
- (53).Mou L; Dong R; Hu B; Li Z; Zhang J; Jiang X Lab Chip 2019, 19, 2750–2757. [DOI] [PubMed] [Google Scholar]
- (54).Sharafeldin M; Kadimisetty K; Bhalerao KR; Bist I; Jones A; Chen T; Lee NH; Rusling JF Anal. Chem 2019, 91, 7394–7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Xiao Q; Wu J; Dang P; Ju H Anal. Chim. Acta 2018, 1032, 130–137. [DOI] [PubMed] [Google Scholar]
- (56).Zhong Y; Wu X; Li J; Lan Q; Jing Q; Min L; Ren C; Hu X; Lambert A; Cheng Q; Yang Z Anal. Chim. Acta 2019, 1049, 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Zhong Y; Tang X; Li J; Lan Q; Min L; Ren C; Hu X; Torrente-Rodríguez RM; Gao W; Yang Z Chem. Commun 2018, 54, 13813–13816. [DOI] [PubMed] [Google Scholar]
- (58).Li L; Xia L; Zhao Y; Wang M; Jiang XJ Sci. Food Agric 2017, 97, 2426–2435. [DOI] [PubMed] [Google Scholar]
- (59).Brindle E; Lillis L; Barney R; Hess SY; Wessells KR; Ouédraogo CT; Stinca S; Kalnoky M; Peck R; Tyler A; Lyman C; Boyle DS PLoS One 2017, 12, e0185868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Brindle E; Lillis L; Barney R; Bansil P; Lyman C; Boyle DS PLoS One 2019, 14, e0210212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Gao Y; Huo W; Zhang L; Lian J; Tao W; Song C; Tang J; Shi S; Gao Y Biosens. Bioelectron 2019, 123, 204–210. [DOI] [PubMed] [Google Scholar]
- (62).Ng E; Yao C; Shultz TO; Ross-Howe S; Wang SX Nanomedicine 2019, 16, 10–19. [DOI] [PubMed] [Google Scholar]
- (63).Bauer WS; Gulka CP; Silva-Baucage L; Adams NM; Haselton FR; Wright DW Anal. Chem 2017, 89, 10216–10223. [DOI] [PubMed] [Google Scholar]
- (64).Ricks KM; Shoemaker CJ; Dupuy LC; Flusin O; Voorhees MA; Fulmer AN; Badger CV; Schmaljohn CS; Schoepp RJ J. Virol. Methods 2019, 270, 12–17. [DOI] [PubMed] [Google Scholar]
- (65).Hong W; Sun G; Zhang Y; Xing Z; Huang B; Zhang S; Zhang X Anal. Methods 2017, 9, 2546–2552. [Google Scholar]
- (66).Kadam L; Patel K; Gautam M; Thorat S; Kale P; Ghule AK; Gairola A; Rao H; Shinde Y; Shaligram U; Gairola S Methods 2019, 158, 33–43. [DOI] [PubMed] [Google Scholar]
- (67).Sheng T; Xie Z; Liu P; Chen J; Chen S; Ding H; Deng J; Yuan Y; Deng D Adv. Mater. Interfaces 2018, 5, 1800343. [Google Scholar]
- (68).Trivedi SU; Miao C; Sanchez JE; Caidi H; Tamin A; Haynes L; Thornburg NJ Sci. Rep 2019, 9, 1390–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Soler M; Belushkin A; Cavallini A; Kebbi-Beghdadi C; Greub G; Altug H Biosens. Bioelectron 2017, 94, 560–567. [DOI] [PubMed] [Google Scholar]
- (70).Berger SS; Lauritsen KT; Boas U; Lind P; Andresen LO J. Vet. Diagn. Invest 2017, 29, 797–804. [DOI] [PubMed] [Google Scholar]
- (71).Zhang B; Ma W; Li F; Gao W; Zhao Q; Peng W; Piao J; Wu X; Wang H; Gong X; Chang J Nanoscale 2017, 9, 18711. [DOI] [PubMed] [Google Scholar]
- (72).Brown EP; Weiner JA; Lin S; Natarajan H; Normandin E; Barouch DH; Alter G; Sarzotti-Kelsoe M; Ackerman ME J. Immunol. Methods 2018, 455, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Bilan R; Ametzazurra A; Brazhnik K; Escorza S; Fernandez D; Uribarri M; Nabiev I; Sukhanova A Sci. Rep 2017, 7, 44668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Hsu MN; Wei S; Guo S; Phan D; Zhang Y; Chen C Small 2018, 14, 1802918. [DOI] [PubMed] [Google Scholar]
- (75).Luan C; Xu Y; Chen B; Yang Z Sens. Actuators, B 2017, 242, 1259–1264. [Google Scholar]
- (76).Beloglazova NV; Sobolev AM; Tessier MD; Hens Z; Goryacheva IY; De Saeger S Methods 2017, 116, 141–148. [DOI] [PubMed] [Google Scholar]
- (77).Chen S; Imoukhuede PI Anal. Chem 2019, 91, 7603–7612. [DOI] [PubMed] [Google Scholar]
- (78).Zhu L; Chen D; Lu X; Qi Y; He P; Liu C; Li Z Chem. Sci 2018, 9, 6605–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).He Q; Chen X; He Y; Guan T; Feng G; Lu B; Wang B; Zhou X; Hu L; Cao D Biosens. Bioelectron 2019, 129, 107–117. [DOI] [PubMed] [Google Scholar]
- (80).Peng T; Wang J; Zhao S; Zeng Y; Zheng P; Liang D; Mari GM; Jiang H Anal. Chim. Acta 2018, 1040, 143–149. [DOI] [PubMed] [Google Scholar]
- (81).Cui X; Liu Y; Hu D; Qian W; Tin C; Sun D; Chen W; Lam R Lab Chip 2018, 18, 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Ji Y; Qi D; Li L; Su H; Li X; Luo Y; Sun B; Zhang F; Lin B; Liu T; Lu Y Proc. Natl. Acad. Sci. U. S. A 2019, 116, 5979–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Xu J; Shi M; Chen W; Huang Y; Fang L; Yao L; Zhao S; Chen Z; Liang H Chem. Commun 2018, 54, 2719–2722. [DOI] [PubMed] [Google Scholar]
- (84).Yao L; Xu J; Shi M; Huang Y; Fang L; Zhao S; Chen Z; Liang H Sens. Actuators, B 2019, 282, 626–635. [Google Scholar]
- (85).Soares RRG; Santos DR; Pinto IF; Azevedo AM; Aires-Barros MR; Chu V; Conde JP Lab Chip 2018, 18, 1569–1580. [DOI] [PubMed] [Google Scholar]
- (86).Chen Y; Meng X; Zhu Y; Shen M; Lu Y; Cheng J; Xu Y Talanta 2018, 186, 299–305. [DOI] [PubMed] [Google Scholar]
- (87).Wu Z; Zeng T; Guo W; Bai Y; Pang D; Zhang Z ACS Appl. Mater. Interfaces 2019, 11, 5762–5770. [DOI] [PubMed] [Google Scholar]
- (88).Yu X; Xia Y; Tang Y; Zhang W; Yeh Y; Lu H; Zheng S Small 2017, 13, 1700425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Huang N; Ye L; Lv H; Du Y; Schneider M; Fan L; Du W Clin. Chim. Acta 2017, 472, 13–19. [DOI] [PubMed] [Google Scholar]
- (90).Juronen D; Kuusk A; Kivirand K; Rinken A; Rinken T Talanta 2018, 178, 949–954. [DOI] [PubMed] [Google Scholar]
- (91).Leirs K; Leblebici P; Lammertyn J; Spasic D Anal. Chim. Acta 2017, 982, 193–199. [DOI] [PubMed] [Google Scholar]
- (92).Fang C; Chou C; Yang Y; Wei-Kai T; Wang Y; Chan Y Anal. Chem 2018, 90, 2134–2140. [DOI] [PubMed] [Google Scholar]
- (93).Lee HJ; Roh YH; Kim HU; Kim SM; Bong KW Lab Chip 2019, 19, 111–119. [DOI] [PubMed] [Google Scholar]
- (94).Ji J; Lu W; Zhu Y; Jin H; Yao Y; Zhang H; Zhao Y 2019 2019, 4, 1384–1390. [DOI] [PubMed] [Google Scholar]
- (95).Chan HN; Xu D; Ho SL; He D; Wong MS; Li HW Theranostics 2019, 9 (10), 2939–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Song X; Nie R; Liu X; Chen Y; Yang L Anal. Chim. Acta 2018, 1043, 1–10. [DOI] [PubMed] [Google Scholar]
- (97).Gornowicz-Porowska J; Seraszek-Jaros A; Bowszyc-Dmochowska M; Bartkiewicz P. ł; Kaczmarek E; Dmochowski M J. Clin Lab Anal 2018, 32, No. e22336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Ran B; Zheng W; Dong M; Xianyu Y; Chen Y; Wu J; Qian Z; Jiang X Anal. Chem 2018, 90, 8234–8240. [DOI] [PubMed] [Google Scholar]
- (99).Alamer S; Eissa S; Chinnappan R; Zourob M Microchim. Acta 2018, 185, 164. [DOI] [PubMed] [Google Scholar]
- (100).Alamer S; Eissa S; Chinnappan R; Herron P; Zourob M Talanta 2018, 185, 275–280. [DOI] [PubMed] [Google Scholar]
- (101).Gogalic S; Sauer U; Doppler S; Preininger C Biosensors 2018, 8, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Machado JM; Soares RR; Chu V; Conde JP Biosens. Bioelectron 2018, 99, 40–46. [DOI] [PubMed] [Google Scholar]
- (103).Panda R; Boyer M; Garber EAE Anal. Bioanal. Chem 2017, 409, 6959–6973. [DOI] [PubMed] [Google Scholar]
- (104).Pagkali V; Petrou PS; Makarona E; Peters J; Haasnoot W; Jobst G; Moser I; Gajos K; Budkowski A; Economou A; Misiakos K; Raptis I; Kakabakos SE J. Hazard. Mater 2018, 359, 445–453. [DOI] [PubMed] [Google Scholar]
- (105).Svedberg G; Jeong Y; Na H; Jang J; Nilsson P; Kwon S; Gantelius J; Svahn HA Lab Chip 2017, 17, 549–556. [DOI] [PubMed] [Google Scholar]
- (106).Angelopoulou M; Petrou PS; Makarona E; Haasnoot W; Moser I; Jobst G; Goustouridis D; Lees M; Kalatzi K; Raptis I; Misiakos K; Kakabakos SE Anal. Chem 2018, 90, 9559–9567. [DOI] [PubMed] [Google Scholar]
- (107).Phillips EA; Young AK; Albarran N; Butler J; Lujan K; Hamad-Schifferli K; Gomez-Marquez J Adv. Healthcare Mater 2018, 7, 1800104. [DOI] [PubMed] [Google Scholar]
- (108).Song X; Nie R; Liu X; Chen Y; Yang L Anal. Chim. Acta 2018, 1043, 1–10. [DOI] [PubMed] [Google Scholar]
- (109).Jang IK; Tyler A; Lyman C; Kahn M; Kalnoky M; Rek JC; Arinaitwe E; Adrama H; Murphy M; Imwong M; Ling CL J. Clin. Microbiol 2019, 57, No. e00948–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Bai X; Wang L; Ren J; Bai X; Zeng L; Shen A; Hu J Anal. Chem 2019, 91, 2955–2963. [DOI] [PubMed] [Google Scholar]
- (111).Banaei N; Foley A; Houghton JM; Sun Y; Kim B Nanotechnology 2017, 28, 455101. [DOI] [PubMed] [Google Scholar]
- (112).Bizzarri AR; Moscetti I; Cannistraro S Anal. Chim. Acta 2018, 1029, 86–96. [DOI] [PubMed] [Google Scholar]
- (113).Chen F; Liu B; Ni H; Chang N; Luan C; Ge Q; Dong J; Zhao X Analyst 2019, 144, 4051. [DOI] [PubMed] [Google Scholar]
- (114).Cheng Z; Choi N; Wang R; Lee S; Moon KC; Yoon S; Chen L; Choo J ACS Nano 2017, 11, 4926–4933. [DOI] [PubMed] [Google Scholar]
- (115).Gao R; Cheng Z; Wang X; Yu L; Guo Z; Zhao G; Choo J Biosens. Bioelectron 2018, 119, 126–133. [DOI] [PubMed] [Google Scholar]
- (116).Li J; Wang J; Grewal YS; Howard CB; Raftery LJ; Mahler S; Wang Y; Trau M Anal. Chem 2018, 90, 10377–10384. [DOI] [PubMed] [Google Scholar]
- (117).Liu B; Zhang D; Ni H; Wang D; Jiang L; Fu D; Han X; Zhang C; Chen H; Gu Z; Zhao X ACS Appl. Mater. Interfaces 2018, 10, 21–26. [DOI] [PubMed] [Google Scholar]
- (118).Ma H; Sun X; Chen L; Cheng W; Han XX; Zhao B; He C Anal. Chem 2017, 89, 8877–8883. [DOI] [PubMed] [Google Scholar]
- (119).Neng J; Li Y; Driscoll AJ; Wilson WC; Johnson PA J. Agric. Food Chem 2018, 66, 5707–5712. [DOI] [PubMed] [Google Scholar]
- (120).Sánchez-Purrà M; Carré-Camps M; de Puig H; Bosch I; Gehrke L; Hamad-Schifferli K ACS Infect. Dis 2017, 3, 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Sebba D; Lastovich AG; Kuroda M; Fallows E; Johnson J; Ahouidi A; Honko AN; Fu H; Nielson R; Carruthers E; Diedhiou C; Ahmadou D; Soropogui B; Ruedas J; Peters K; Bartkowiak M; Magassouba N’F; Mboup S; Ben Amor Y; Connor JH; Weidemaier K Sci. Transl. Med 2018, 10, No. eaat0944. [DOI] [PubMed] [Google Scholar]
- (122).Reza KK; Dey S; Wuethrich A; Sina AAI; Korbie D; Wang Y; Trau M Nanoscale 2018, 10, 18482. [DOI] [PubMed] [Google Scholar]
- (123).Saranya G; Joseph MM; Karunakaran V; Nair JB; Saritha VN; Veena VS; Sujathan K; Ajayaghosh A; Maiti KK ACS Appl. Mater. Interfaces 2018, 10, 38807–38818. [DOI] [PubMed] [Google Scholar]
- (124).Su Y; Xu S; Zhang J; Chen X; Jiang L; Zheng T; Zhu J Anal. Chem 2019, 91, 864–872. [DOI] [PubMed] [Google Scholar]
- (125).Wang X; Park S; Ko J; Xiao X; Giannini V; Maier SA; Kim D; Choo J Small 2018, 14, 1801623. [DOI] [PubMed] [Google Scholar]
- (126).Wang Z; Yang H; Wang M; Petti L; Jiang T; Jia Z; Xie S; Zhou J Colloids Surf., A 2018, 546, 48–58. [Google Scholar]
- (127).Yu M; Hu Y; Liu J New J. Chem 2017, 41, 10407. [Google Scholar]
- (128).Zhang D; Huang L; Liu B; Ni H; Sun L; Su E; Chen H; Gu Z; Zhao X Biosens. Bioelectron 2018, 106, 204–211. [DOI] [PubMed] [Google Scholar]
- (129).Zheng Z; Wu L; Li L; Zong S; Wang Z; Cui Y Talanta 2018, 188, 507–515. [DOI] [PubMed] [Google Scholar]
- (130).Zhou W; Tian Y; Yin B; Ye B Anal. Chem 2017, 89, 6120–6128. [DOI] [PubMed] [Google Scholar]
- (131).Zhou L; Liu Y; Wang F; Jia Z; Zhou J; Jiang T; Petti L; Chen Y; Xiong Q; Wang X Talanta 2018, 188, 238–244. [DOI] [PubMed] [Google Scholar]
- (132).Kamińska A; Winkler K; Kowalska A; Witkowska E; Szymborski T; Janeczek A; Waluk J Sci. Rep 2017, 7, 10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Nakano S; Nagao M; Yamasaki T; Morimura H; Hama N; Iijima Y; Shinomiya H; Tanaka M; Yamamoto M; Matsumura Y; Miyake S; Ichiyama SJ Infect. Chemother 2018, 24, 443–448. [DOI] [PubMed] [Google Scholar]
- (134).Castiello FR; Tabrizian M Anal. Chem 2018, 90, 3132–3139. [DOI] [PubMed] [Google Scholar]
- (135).Castiello FR; Tabrizian M Analyst 2019, 144, 2541–2549. [DOI] [PubMed] [Google Scholar]
- (136).Hossain MZ; Maragos CM Biosens. Bioelectron 2018, 101, 245–252. [DOI] [PubMed] [Google Scholar]
- (137).Stravers CS; Gool EL; van Leeuwen TG; Aalders MC; van Dam A Sens. Actuators, B 2019, 283, 355–362. [Google Scholar]
- (138).Yavas O; Aćimović SS; Garcia-Guirado J; Berthelot J; Dobosz P; Sanz V; Quidant R ACS sensors 2018, 3, 1376–1384. [DOI] [PubMed] [Google Scholar]
- (139).Shen M; Joshi AA; Vannam R; Dixit CK; Hamilton RG; Kumar CV; Rusling JF; Peczuh MW ChemBioChem 2018, 19, 199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Petrova I; Konopsky V; Nabiev I; Sukhanova A Sci. Rep 2019, 9, 8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Angelopoulou M; Petrou PS; Makarona E; Haasnoot W; Moser I; Jobst G; Goustouridis D; Lees M; Kalatzi K; Raptis I; Misiakos K; Kakabakos SE Anal. Chem 2018, 90, 9559–9567. [DOI] [PubMed] [Google Scholar]
- (142).Herranz S; Marciello M; Marco MP; Garcia-Fierro JL; Guisan JM; Moreno-Bondi MC Sens. Actuators, B 2018, 257, 256–262. [Google Scholar]
- (143).Jain B; Kumarasamy J; Gholve C; Kulkarni S; Rajan MGR Indian J. Clin. Biochem 2017, 32, 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Zheng K; Chen C; Chen X; Xu M; Chen L; Hu Y; Bai Y; Liu B; Yan C; Wang H; Li J Biosens. Bioelectron 2019, 132, 47–54. [DOI] [PubMed] [Google Scholar]


