Abstract
Craniofacial morphogenesis is regulated in part by signaling from the Endothelin receptor type A (EDNRA). Pathogenic variants in EDNRA signaling pathway components EDNRA, GNAI3, PCLB4 and EDN1 cause Mandibulofacial Dysostosis with Alopecia (MFDA), Auriculocondylar syndrome (ARCND) 1, 2 and 3, respectively. However, cardiovascular development is normal in MFDA and ARCND individuals, unlike Ednra knockout mice. One explanation may be that partial EDNRA signaling remains in MFDA and ARCND, as mice with reduced, but not absent, EDNRA signaling also lack a cardiovascular phenotype. Here we report an individual with craniofacial and cardiovascular malformations mimicking the Ednra−/− mouse phenotype, including a distinctive micrognathia with microstomia and a hypoplastic aortic arch. Exome sequencing found a novel homozygous missense variant in EDNRA (c.1142A>C; p.Q381P). Bioluminescence resonance energy transfer assays revealed that this amino acid substitution in helix 8 of EDNRA prevents recruitment of G proteins to the receptor, abrogating subsequent receptor activation by its ligand, Endothelin-1. This homozygous variant is thus the first reported loss-of-function EDNRA allele, resulting in a syndrome we have named Oro-Oto-Cardiac Syndrome. Further, our results illustrate that EDNRA signaling is required for both normal human craniofacial and cardiovascular development, and that limited EDNRA signaling is likely retained in ARCND and MFDA individuals. This work illustrates a straightforward approach to identifying the functional consequence of novel genetic variants in signaling molecules associated with malformation syndromes.
Keywords: Neural crest cell, micrognathia, cardiovascular, Auriculocondylar syndrome, BRET
Introduction
Endothelin receptor type A (EDNRA, encoded by EDNRA [MIM 131243]) is a G protein-coupled receptor that is essential for development of the craniofacial complex and portions of the cardiovascular system (Clouthier et al., 1998; Nair, Li, Cornell, & Schilling, 2007). This process is mediated by EDNRA signaling-dependent patterning of the cranial and cardiac neural crest cells in the pharyngeal arches (Abe, Ruest, & Clouthier, 2007; Clouthier, Williams, Hammer, Richardson, & Yanagisawa, 2003; Clouthier et al., 2000; Miller, Yelon, Stainier, & Kimmel, 2003; Ozeki, Kurihara, Tonami, Watatani, & Kurihara, 2004; Ruest, Xiang, Lim, Levi, & Clouthier, 2004). In mice, genetic ablation of the receptor (Ednra−/−), ligand (Endothelin-1, Edn1−/−), ligand-processing enzyme (Endothelin converting enzyme 1, Ece1−/−), or primary signaling effectors (G protein α subunits Gq and G11, Gnaq−/−;Gna11−/−) result in severe defects in the mandible and ears and neonatal lethality from mechanical asphyxia (Clouthier et al., 1998; Dettlaff-Swiercz, Wettschureck, Moers, Huber, & Offermanns, 2005; Kurihara et al., 1994; Offermanns et al., 1998; Yanagisawa et al., 1999; Yanagisawa et al., 1998). A characteristic defect of endothelin signaling loss is the homeotic transformation of most mandible structures into maxillary-like structures (Ozeki et al., 2004; Ruest et al., 2004). These mice also exhibit cardiovascular defects that include interruption of the aorta and ventricular septal defects (VSD) (Clouthier et al., 1998, Yanagisawa et al., 1999; Yanagisawa et al., 1998). However, craniofacial and cardiovascular development appears differentially sensitive to loss of EDNRA signaling. In Ednra+/+ <->Ednra−/− chimeras, in which tagged Ednra−/− embryonic stem (ES) cells are injected into wild type blastocysts, the extent of craniofacial differences increased proportionally with the increasing contribution of Ednra−/− ES cell-derived cells to the developing embryo (Clouthier et al., 2003). However, cardiovascular differences were only seen in one embryo with the highest percentage of Ednra−/− ES cell-derived cells. These findings indicate that while EDNRA signaling is required for both craniofacial and cardiovascular development, cardiovascular development likely can withstand a greater disruption in EDNRA signaling (Clouthier et al., 2003).
In humans, pathogenic variants in signaling components of the EDNRA pathway are linked to craniofacial disorders characterized by mandible and ear malformations similar to Edn1/Ednra knockout mice, highlighting the evolutionary conservation of EDNRA signaling in craniofacial development (Table 1). Auriculocondylar syndrome 1 [ARCND1; MIM 602483] and ARCND2 [MIM 614669] are autosomal dominant disorders resulting from pathogenic variants in EDNRA downstream signaling components PLCB4 [MIM 600810] and GNAI3 [MIM 139370], respectively (Rieder et al., 2012). ARCND3 is an autosomal recessive disorder resulting from biallelic pathogenic variants in EDN1 (Gordon et al., 2013). Heterozygous pathogenic variants of EDN1 lead to the autosomal dominant disorder Question Mark Ears, Isolated (QME; MIM 612798). Mandibulofacial Dysostosis with Alopecia [MFDA; MIM 616367] results from gain-of-function variants in EDNRA and appears to be inherited in an autosomal dominant fashion (Gordon et al., 2015). The functional consequences of a given MFDA pathogenic variant is complex, as in the upper jaw, there is likely aberrant EDNRA signaling, while in the lower jaw, there appears to be disruption of EDNRA signaling (Gordon et al., 2015). However, unlike Edn1/Ednra knockout mice (Clouthier et al., 1998; Kurihara et al., 1994), individuals with ARCND and MFDA do not present with cardiovascular defects, suggesting that limited EDNRA signaling likely remains in affected individuals.
Table 1.
Phenotypes associated with mutations in Endothelin pathway genes
| Gene | MIM Name/Number | Inheritance Pattern | Clinical Features |
|---|---|---|---|
| GNAI3 | Auriculocondylar Syndrome 1, #602483 | Autosomal dominant | Micrognathia, mandibular ankylosis, mandibular condyle hypoplasia, microstomia, glossoptosis, malformed ears (question mark ears), prominent cheeks, palate abnormalities |
| PLCB4 | Auriculocondylar Syndrome 2, #614669 | Autosomal dominant | Micrognathia, mandibular ankylsosis, mandibular condyle hypoplasia, microstomia, glossoptosis, malformed ears (question mark ears), prominent cheeks, palate abnormalities |
| EDN1 | Auriculocondylar Syndrome 3, #615706 | Autosomal recessive | Micrognathia, mandibular hypoplasia, question mark ears, glossoptosis, bifid uvula, laryngeal cleft, lingual hamartomas |
| EDN1 | Question mark ears, isolated, #612798 | Autosomal dominant | Question mark ears |
| EDNRA | Mandibulofacial dysostosis with alopecia, #616367 | Autosomal dominant | Micrognathia, cleft palate, maxillary hypoplasia, dysplastic zygomatic arch, limited jaw mobility, thin or absent scalp hair, low set ears, dysplastic ears, sparse eyebrows and eyelashes, scant body hair, conductive hearing loss |
| EDNRA | N/A (this manuscript) | Autosomal recessive | Micrognathia, microstomia, microtia/anotia, absent middle ear structures, aglossia, oropharyngeal stenosis, absent soft palate and uvula, hypoplastic aortic arch, ventricular septal defect |
In this study, we have identified a homozygous variant in EDNRA inherited in an autosomal recessive manner that results in craniofacial and cardiovascular malformations leading to neonatal lethality. This is the first patient known to be affected with this condition, which more closely resembles the phenotype of Ednra−/− mice than MFDA and ARCND individuals. To determine the functional consequence of this variant, we used a variety of molecular and cell biological reporter assays including a series of bioluminescence resonance energy transfer (BRET) assays. We find that the variant disrupts EDNRA-G protein interactions but not protein folding or receptor localization. Our results provide a mechanistic basis for a novel pathogenic variant in EDNRA that likely results in significant reduction of EDNRA signaling in NCCs during craniofacial and cardiovascular development. Furthermore, our approach demonstrates the modularity and adaptability of BRET assays to probe functional consequences of novel GPCR variants in developmental disorders.
Materials and Methods
Editorial policies and ethical considerations.
The subject’s parents provided consent for publication of this case and photographs. No experimental procedures involving human and/or animal subjects were performed.
Clinical exome sequencing
Clinical exome sequencing was performed on the proband and both unaffected parents essentially as previously described (Gibson et al., 2018). Exome libraries were prepared using the SureSelect Clinical Research Exome kit (CRE v1), following the manufacturers protocol (Agilent Technologies, Santa Clara, CA). Sequencing was performed on a HiSeq2500 (Illumina, San Diego, CA). Following exome sequencing, reads were aligned to the reference human genome (GRCh37/hg19) and variants called with an in-house-developed bioinformatics pipeline that incorporated NovoAlign (Novacraft) for read alignment, Picard (broadinstitute.github.io/picard/) for duplicate marking and the Genome Analysis Toolkit (software.broadinstitute.org/gatk/) for variant calling. Variants exceeding a total minor allele frequency of 0.2% in the Exome Aggregation Consortium database were excluded from analysis, with the exception of a predefined list of known pathogenic variants in genes associated with autosomal recessive conditions that exceed these thresholds. All remaining variants were reviewed for overlap with the proband’s clinical phenotype. All de novo, hemizygous, homozygous and compound heterozygous variants received an additional level of clinical review, with special attention paid to variants residing in the ROH previously identified by array. The variant identified in EDNRA was confirmed by Sanger sequencing in the proband and both parents according to standard laboratory protocols.
Plasmids
The human EDNRA expression construct (pCMV6-XL5-EDNRA) was purchased from Origene. The p.Q381P variant was introduced into pCMV6-XL5-EDNRA using the QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) and primers 5’ GAAATTTAAAAATTGTTTCCCGTCATGCCTCTGCTGCTGC 3’ and 5’ GCAGCAGCAGAGGCATGACGGGAAACAATTTTTAAATTTC 3’ to change the A at nucleotide position 1142 to a C, thus changing the glutamine (Q) at aa 381 to a proline (P). These plasmids are referred to as pCMV-EDNRA (or wild type EDNRA) and pCMV-EDNRA p.Q381P. pEDNRA-RLuc8 was constructed by subcloning a myc-EDNRA fragment derived from pmyc-EDNRA-GFP (pmyc-ETA-GFP), a kind gift from Jeffery Walker, University of Wisconsin) using HindIII and AgeI. pEDNRA-RLuc8 p.Q381P was derived from EDNRA-RLuc8 using the QuickChange Lightning Site-Directed Mutagenesis Kit as described above. pcDNA3.1-VN-Gγ2, pcDNA3.1-VC-Gβ1, pNES-Venus-mG, pVenus-kras, pVenus-PTP1b, pVenus-giantin have been previously described (Hollins, Kuravi, Digby, & Lambert, 2009 Masuho, 2015 #2272; Wan et al., 2018). pcDNA3-Gαq-HA was a kind gift by P. Wedegaertner (Wedegaertner, Chu, Wilson, Levis, & Bourne, 1993). pcDNA3.1-masGRK3ct-Nluc was a kind gift from Kirill Martemyanov (Masuho, Ostrovskaya, et al., 2015). mCherry-MEM was purchased from Addgene (plasmid 55779, deposited by Catherine Berlot) (Yost, Mervine, Sabo, Hynes, & Berlot, 2007). All constructs were verified by Sanger sequencing.
Cell culture and transfection
MC3T3-E1 cells (ATCC) were cultured in MEM alpha (Invitrogen) supplemented with 20% fetal bovine serum (Sigma). HEK293 and HEK293T cells (ATCC) were cultured in DMEM (Corning) supplemented with 10% fetal bovine serum. All cell types were grown at 37°C with 5% CO2 in a humidified incubator. Transient transfections were performed with X-tremeGENE 9 (Roche) or linear polyethyleneimine; MW 25,000 (Polysciences Inc.). A 3:1 ratio of X-tremeGENE 9 (μl) to plasmid DNA (μg) was used, and 1μg of plasmid DNA was used for one well of a 6-well plate. Linear polyethyleneimine was used at an N/P ratio of 20, and up to 3 μg of plasmid DNA was transfected in one well of a 6-well plate. All transfections were monitored for relative transfection efficiency by qualitatively examining Venus fluorescence. To ensure similar transfection efficiencies between EDNRA-RLuc8 and EDNRA-RLuc8 p.Q381P, raw luciferase values were examined following each experiment, with no statistically significant difference observed between the two constructs (Supp. Figure 1A). In addition, by comparing the number of fluorescent cells vs total cells in representative transfections (n=3 for each condition), we found that the transfection efficiencies of mGsq-Venus, Gβγ-Venus, EDNRA-GFP and EDNRA-GFP p.Q381P was >67% for all vectors, with no statistically significant difference observed between the vectors (Supp. Figure 1B).
Quantitative real-time PCR assays
MC3T3-E1 cells were transiently transfected with pCMV6-EDNRA, pCMV6-EDNRA p.Q381P or pCS2 empty vector using X-tremeGENE9 for six hours before exchanging transfection media for serum free media. 24–36 hours after transfection, cells were treated with 5 nM EDN1 and 10 μM BQ-788 (EDNRB-specific antagonist) for 1 hour before collecting RNA with Direct-zol (Zymogen). cDNA was prepared from total RNA with the QuantiTect cDNA Synthesis Kit (Qiagen) and qRT-PCR performed with 5 ng of cDNA using the QuantiTect SYBR Green PCR kit (Qiagen) and QuantiTect Assay Primers for Actb, Dlx2 and Dlx5 (Qiagen) on a CFX Connect Real-time PCR Detection System (Bio-Rad). All assays were performed in triplicate at least three times. Statistical analysis was conducted using Prism, with significance calculated using an unpaired two-tailed t-test.
G protein activation assays:
HEK293T cells were transiently transfected with X-tremeGENE 9. pCMV6-EDNRA or pCMV6-EDNRA p.Q381P, along with pcDNA3-Gαq-HA, Venus155–239-Gβ1, Venus1–155-Gγ2 and masGRK3ct-Nanoluc were transfected at a 1:2:1:1:1 ratio (Masuho, Martemyanov, & Lambert, 2015). For control experiments, pcDNA3.1 empty vector was substituted for pCMV6-EDNRA. 24 hours later, cells were harvested and resuspended in reaction buffer (Tyrode’s salt solution with 0.1% glucose) as previously described (Masuho, Martemyanov, et al., 2015). 25,000 cells were added into individuals wells of an opaque 96-well plate (Perkin Elmer) and incubated with the Nanoluc luciferase substrate Furimazine (Promega) for 3 minutes prior to assay. BRET assays were performed in a Synergy 2 microplate reader (Biotek) equipped with emission filters for Venus (485/20 nm, reading 475–495 nm) and Nanoluc (528/20 nm, reading 518–538 nm). After a 1-minute measurement of basal BRET, EDN1 was added at a final concentration of 1 μM and BRET was measured for another 3 minutes. BRET measurements were made by automatic filter-switching every 2 seconds. Raw BRET signals were calculated as the emission intensity at 528/20 nm divided by the emission intensity at 485/20 nm. Delta BRET was calculated by subtracting the average raw BRET ratio signal before EDN1 from the individual raw BRET values. All assays were performed at least three times, with each assay measured in triplicate, with statistical analysis conducted using Prism.
Subcellular localization and miniG protein assays:
HEK293 cells were transfected with linear polyethyleneimine, and 12–48 hours later cells were washed with DPBS, harvested by trituration, and transferred to opaque black 96-well plates. Cells were exposed to either EDN1 (1 μM) or vehicle (1% bovine serum albumin). Coelenterazine h (5 μM; Nanolight, Pinetop, AZ) was added to all wells immediately prior to making measurements using a Mithras LB940 photon-counting plate reader (Berthold Technologies GmbH, Bad Wildbad, Germany). Raw BRET signals were calculated as the emission intensity at 520–545 nm divided by the emission intensity at 475–495 nm. Net BRET was calculated as this ratio minus the same ratio measured from cells expressing only the BRET donor. All assays were performed at least three times, with statistical analysis was conducted using Prism.
Live confocal microscopy
HEK293T cells were plated onto poly-D lysine coated glass bottom dishes (MatTek) and co-transfected with mCherry-MEM and pcDNA3.1 (mock), pCMV-EDNRA (wild type) or pCMV-EDNRA p.Q381P constructs using X-tremeGENE9. 6 hours later, growth medium was replaced with serum-free medium. 24 hours after initial transfection, live confocal microscopy was performed with a Leica TCS SP5 and 63X oil objective. Line scans were performed for 2 minutes prior and 10 minutes after addition of HiLyte Fluor 488-ET1 (HiLyte-Fluor 488-ET-1 (EDN1); 100 nM final concentration; Anaspec) to cells. HiLyte Fluor 488 is a proprietary fluorophore with similar excitation and emission spectra as fluorescein.
Fluorescent ligand binding assay
HEK293T cells were transfected in a 6-well dish with pcDNA3.1 empty vector, EDNRA or EDNRA p.Q381P constructs using X-tremeGENE9. 24 hours later, cells were dissociated and plated into individual wells of a poly-D lysine-coated black wall clear bottom 96-well dish (Corning) in serum-free growth medium at a concentration of 75,000 cells/well. 12–24 hours after plating, cells were incubated with or without 100 nM HiLyte-EDN1 in Tyrode’s Buffer (Sigma) supplemented with 0.1% glucose and 0.04% bovine serum albumin for 20 minutes at room temperature. Cells treated with the EDNRA-specific antagonist BQ-123 (Sigma) were incubated for 5 minutes prior to Hylite-488-EDN1 addition. Cells were washed once, replaced with Tyrode’s Buffer and assayed immediately in a Synergy 2 microplate reader fitted with a 485/20 nm excitation filter, 528/20 emission filter and 500 nm cut-off dichroic mirror. Specific fluorescence was calculated by subtracting background fluorescence (cells without HiLyte-EDN1) from raw fluorescence values. All assays were performed at least three times, with each experiment read in triplicate. Statistical analysis was performed using Prism.
Results
Clinical report
A 31-year-old Caucasian gravida 3 para 0 woman was initially seen at 25 weeks of gestation due to multiple fetal anomalies noted on ultrasound. The pregnancy was conceived through in vitro fertilization due to prior salpingectomy. There was a history of one first trimester pregnancy loss and one ectopic pregnancy. There were no unusual exposures during this pregnancy. Prenatal ultrasound at 19 weeks, 6 days gestation revealed micrognathia, polyhydramnios, choroid plexus cyst, and a suspected ventricular septal defect (VSD). Ultrasound at 25 weeks of gestation additionally found glossoptosis, elongated philtrum, small fetal stomach, microtia, suspected aortic stenosis, and mild pectus excavatum. Head size and extremities appeared normal. Fetal echocardiogram at 25 weeks was notable for a large VSD and hypoplastic aortic arch, though ventricular size and systolic function were normal. An amniocentesis was performed at 23 weeks of gestation and a single nucleotide polymorphism (SNP) microarray was performed from cultured amniocytes. While the microarray revealed no copy number abnormalities, a single region of homozygosity in 4q31.21-q31.23 arr[hg19](142,917,034–150,426,641) was present and covered 7.51 Mb. Family history was generally unremarkable; both parents were healthy and no consanguinity was known. Amnioreduction was required at 27 weeks, 2 days gestation. The pregnancy was also complicated by a maternal history of short cervix necessitating indomethacin treatment. Preterm labor subsequently commenced.
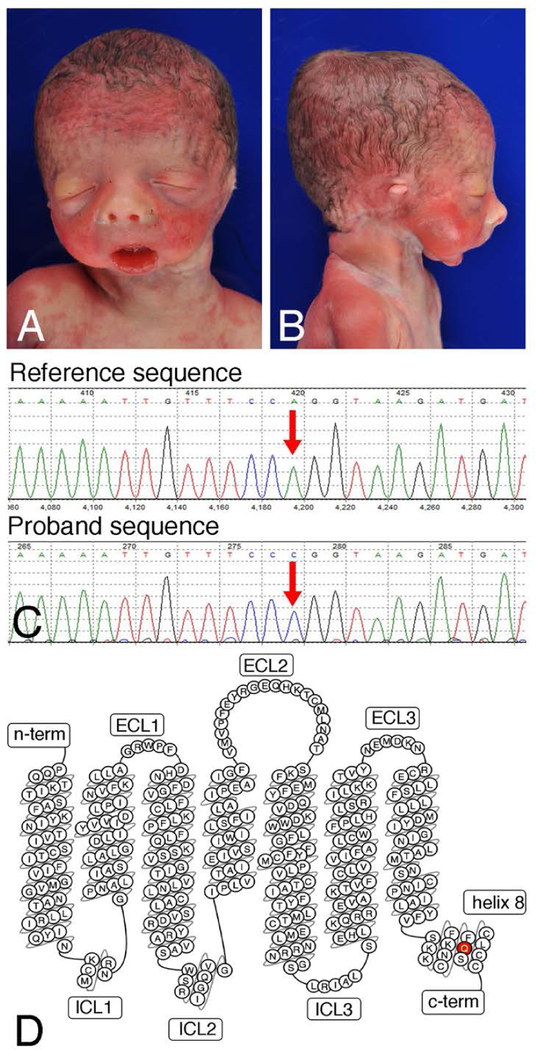
The male infant was born via vaginal delivery at 27 weeks, 3 days gestation weighing 879 grams (25th percentile for gestational age), with a length of 26 cm (30th percentile) and head circumference of 25 cm (25th percentile). The infant could not be orally intubated but was successfully nasally intubated. However, he suffered an acute decompensation event with bradycardia and hypoxemia that was unresponsive to resuscitation at 10 hours of life. Physical and autopsy examination found a wide variety of craniofacial and cardiovascular malformations (Fig. 1A, B and Table 2). These included distinctive micrognathia and microstomia with an inability to fully open the mouth, aglossia, and significant ear abnormalities. Computerized tomography (CT) scan of the mandible was not performed due to the short time frame in which the individual was in the neonatal intensive care unit. Cardiac outflow tract defects were also present (Table 2). Extremities were normal and no abnormalities of the liver, kidneys, genitalia, nor other intra-abdominal structures were noted. Brain autopsy was normal with exception of a germinal matrix hemorrhage, likely related to gestational age.
Figure 1. Phenotypic and genomic analysis.
(a) Anterior and (b) lateral views of the infant’s face demonstrating dysmorphic features including downslanting palpebral fissures, upturned nasal tip, thin upper lip, micrognathia, microstomia, and abnormal external ears. (c) Sanger sequencing confirmation of the c.1142A>C mutation in the proband, resulting in the p.Q381P variant. (d) Bubble diagram of the EDNRA (adaptedfromGPCRdb.org). The p.Q381P mutation in helix 8 is denoted in red. c-term, C-terminus; ECL, extracellular loop; ICL, intracellular loop; n-term, N-terminus.
Table 2.
Physical Examination and Autopsy Findings
| Organ System | Findings |
|---|---|
| Facial | Long, downslanting palpebral fissures |
| Increased outer canthal distance | |
| Fused eyelids | |
| Upturned nasal tip | |
| Upper lip hypoplasia | |
| Ears | Microtia/anotia |
| Absent external auditory canal | |
| Absent middle ear structures | |
| Mouth | Micrognathia |
| Microstomia Aglossia (no skeletal muscle present, only squamous mucosa) | |
| Maxillary-like appearance to lower jaw Severe posterior oropharyngeal stenosis | |
| Absent soft palate and uvula | |
| Grooved hard palate | |
| Cardiovascular | Hypoplastic aortic arch |
| VSD with posterior malalignment | |
| Bicuspid aortic valve | |
| Retroesophageal right subclavian artery | |
| Respiratory | Right lung with azygous lobe |
The initial differential diagnosis was limited, as the infant’s features did not fit previously described syndromes. The oral and jaw abnormalities were not consistent with Treacher Collins Syndrome (TCS1; MIM 154500). Agnathia-otocephaly complex (AGOTC; MIM 202650) was also considered, though the ears were not as displaced as may be expected. The dysmorphia seemed too significant to fall within the oculo-auriculo-vertebral spectrum (OAVS: MIM 164210), and the absence of question mark ear (QME) and presence of cardiovascular defects ruled out Auriculocondylar Syndrome 1–3 (ARCND1–3). In order to identify a possible underlying genetic etiology of this patient’s phenotype, trio clinical exome sequencing was performed.
Molecular studies
Clinical whole exome sequencing revealed rare homozygous variants in the EDNRA [MIM 131243] and FREM3 [MIM 608946] genes, both of which reside in the region of homozygosity on chromosome 4q31.21-q31.23. FREM3, encoding a basement membrane protein involved in the integrity of early embryonic skin (Pavlakis, Makrygiannis, Chiotaki, & Chalepakis, 2008), has not been associated with either Mendelian disease to date or in isolated craniofacial or cardiovascular development/disorders, though polymorphisms in FREM3 have been linked to major depressive disorders and differential susceptibility to severe malaria (Manjurano et al., 2015; Shi, Zhang, Wang, Shen, & Xu, 2012). In addition, the identified variant in the FREM3 transcript (NM_ 001168235.1, c.2192T>C; p.I1731T; dbSNP rs1450255129) is a conservative missense substitution at a weakly conserved residue, with threonine observed in multiple mammalian species (Lek et al., 2016) and with a minor allele frequency of 0.136% in the South Asian population (Karczewski et al., 2019). Thus, this variant was deemed not relevant to the clinical indication for the exome and was not included in the report.
The homozygous EDNRA variant (NM_00157.3, c.1142A>C; p.Q381P; dbSNP rs1219791712) results in a glutamine to proline change in helix 8 of EDNRA (Fig. 1C). Biparental inheritance was confirmed via exome sequencing, ruling out uniparental disomy as the cause of the region of homozygosity (Fig. 1D). While the variant resides at the −2 position from the exon 7 splice donor site, computational algorithms predict a weak to moderate effect on the splice donor site (data not shown). This variant is absent from the Exome Aggregation Consortium (ExAC) and Genome Aggregation Consortium (gnomAD) databases (data not shown) (Lek et al., 2016). Moreover, while not at the level of statistical significance, EDNRA may be intolerant of coding alterations and loss-of-function alterations (gnomAD z=2.93, pLI=0.99), making the identification of a novel missense variant highly significant. Based on the limited available data for this variant and absence of previous description of a craniofacial disorder caused by autosomal recessively inherited pathogenic variants in EDNRA, the p.Q381P variant was clinically interpreted by the diagnostic laboratory as a variant of uncertain significance. No other rare variants in genes that overlap with clinically verified disease-associated alleles were identified. Moreover, no other variants meeting a Mendelian mode of inheritance with plausible causality for the phenotypes described in this patient were identified, including homozygous, compound heterozygous, X-linked, de novo or variants in imprinted genes (data not shown). There were no variants noted in genes known to cause structural heart differences.
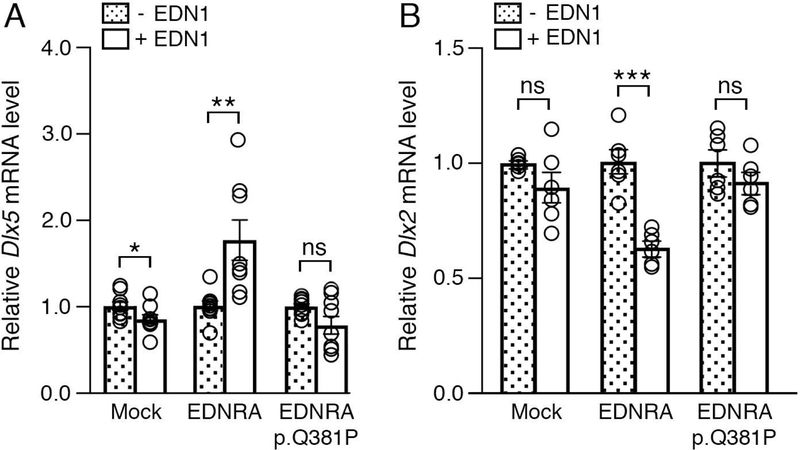
To determine whether this variant affects EDNRA function in a developmental context, we examined EDNRA-dependent gene expression. The earliest known gene expression regulatory event following EDNRA activation is induction of Dlx5 and Dlx6 expression in the ventral mandibular arches (Charité et al., 2001; Clouthier & Schilling, 2004; Clouthier et al., 2000; Tavares et al., 2012). Dlx genes encode homeobox transcription factors crucial for cranial neural crest cell (NCC) patterning (Depew, Simpson, Morasso, & Rubenstein, 2005), with Dlx5/Dlx6 expression required for establishing ventral NCC identity in the mandibular arches (Depew, Lufkin, & Rubenstein, 2002). This is in contrast to Dlx1/Dlx2, which are expressed in the dorsal mandibular arch but are repressed in the ventral arch by EDNRA signaling (Ruest et al., 2004). We transfected either wild type EDNRA or the p.Q381P variant into MC3T3 cells, a mouse pre-osteoblastic cell line with a genomic profile that closely resembles cranial NCC-derived mesenchymal cells (Tavares, Cox, Maxson, Ford, & Clouthier, 2017). Addition of EDN1 to the transfected cells expressing wild type EDRNA resulted in a 1.75-fold induction of Dlx5 (Fig. 2A), while Dlx5 expression after EDN1 addition was not induced in cells expressing the p.Q381P variant (Fig. 2A). In an opposite manner, addition of EDN1 to the transfected cells expressing wild type EDRNA resulted in a 1.58-fold reduction in Dlx2, while Dlx2 expression after EDN1 addition was not repressed in cells expressing the p.Q381P variant (Fig. 2B). Together, these results suggest that the p.Q381P variant represents a strong loss-of-function allele.
Figure 2. The EDNRA p.Q381P variant cannot induce EDNRA-dependent gene expression.
qRT-PCR analysis of Dlx5 (A) and Dlx2 (B) gene expression in MC3T3-E1 cells following transfection with empty vector (mock), EDNRA or EDNRA p.Q381P and treatment with EDN1. (a) While addition of EDN1 resulted in upregulation of Dlx5 expression in cells with wild type EDNRA, Dlx5 expression was not induced in cells with the EDNRA p.Q381P variant. (b) Addition of EDN1 resulted in downregulation of Dlx2 expression in cells with a wild type EDNRA but not in cells with the EDNRA p.Q381P variant (B). Assays were performed in duplicate or triplicate at least three times. Error bars represent SEM; two-tailed t-test; *p<0.05, **p< 0.005, ***p<0.001; ns., not significant.
Like other class A GPCRs, EDNRA is composed of an N-terminal extracellular domain, seven α-helical transmembrane (7-TM) domain and a C-terminal cytoplasmic domain that contains an eighth amphipathic helix (helix 8) that lies immediately after the seventh transmembrane domain. The role of helix 8 in EDNRA function has not been directly assessed, though helix 8 in other class A GPCRs is required for functions that include receptor localization(Feierler et al., 2011; Karpinsky-Semper et al., 2015), receptor desensitization and internalization (Faussner et al., 2005; Kirchberg et al., 2011) and G protein coupling (Delos Santos, Gardner, White, & Bahouth, 2006; Scheerer et al., 2008). Because a single proline substitution in an α helix can distort structure and disrupt normal function (Piela, Nemethy, & Scheraga, 1987; Yang et al., 1997), we hypothesized that the p.Q381P variant affects helix 8-dependent receptor properties.
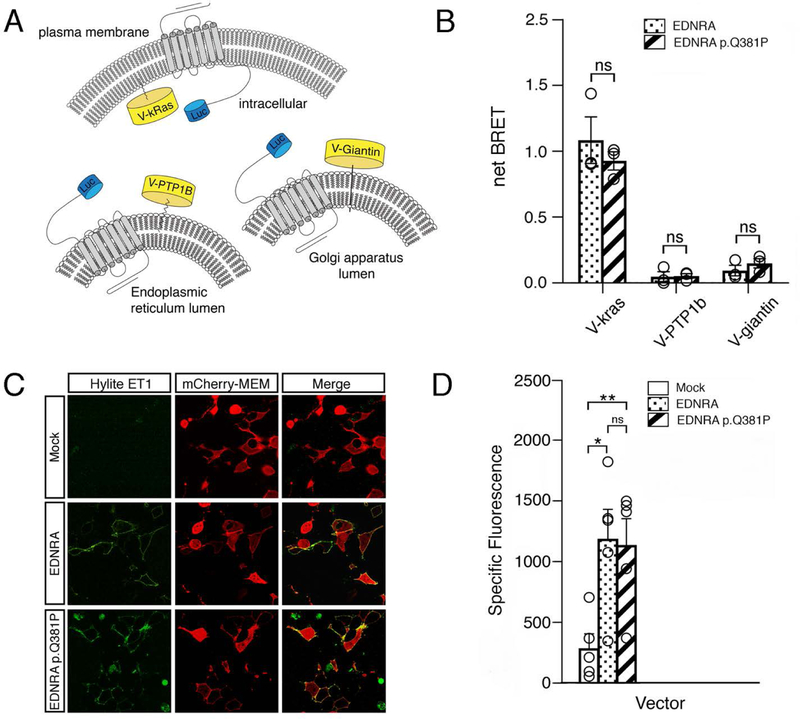
We first assessed whether the p.Q381P variant resulted in mislocalization of EDRNA by examining the relative distribution of EDNRA in the plasma membrane and endo-membrane compartments using a bioluminescence resonance energy transfer (BRET) assay (Lan, Liu, Li, Wu, & Lambert, 2012). BRET assays report the physical proximity of a fluorescent acceptor (in our assays, the yellow fluorescent protein Venus) and a bioluminescent protein donor (in our assays, either Renilla or Nanoluc luciferases (Promega))(Hamdan, Percherancier, Breton, & Bouvier, 2006). The emission wavelength of luciferase overlaps with the Venus excitation wavelength; therefore, in the presence of luciferase substrate, the close proximity of the donor-acceptor pair results in energy transfer and Venus fluorescence emission, which is referred to as BRET response (Fig. 3A and data not shown). By tagging Venus and luciferase to different proteins of interest and subsequently measuring BRET, many aspects of cellular processes can be examined. Cellular localization of EDNRA was examined using BRET donor-tagged membrane compartment markers Venus-kras (plasma membrane), Venus-PTP1b (endoplasmic reticulum) and Venus-giantin (Golgi apparatus) (Lan et al., 2012) (Fig. 3A). These markers were used in combination with Renilla luciferase variant 8-tagged EDNRA (EDNRA-RLuc8) as the BRET donor. BRET levels for wild type EDNRA and the p.Q381P variant were similar for all three cellular compartments, indicating that the p.Q381P variant did not impact overall cellular localization (Fig. 3B).
Figure 3. EDNRA subcellular localization and EDN1 binding is not disrupted by the p.Q381P variant.
(a) Schematic of the BRET assay for subcellular localization, in which EDNRA-Renilla luciferase 8 (RLuc8) is combined with Venus (V)-kras (plasma membrane), Venus-PTP1B (endoplasmic reticulum) or Venus-giantin (Golgi apparatus) to measure BRET in different cellular compartments. (b) Average net BRET of unstimulated HEK293 cells transfected with wildtype EDNRA-RLuc8 or EDNRA p.Q381P-RLuc8 and V-kras, V-PTPB1 or V-giantin. Assays were performed at least three times. Error bars represent SEM.; two-tailed t-test; n.s., not significant. (c) Confocal images of HEK293T cells transfected with mCherry-MEM and pcDNA3.1, wild type EDNRA or EDNRA p.Q381P and incubated with Hilyte fluor-488-ET-1. mCherry-MEM and Hilyte fluor-488-ET-1 co-localized within cells expressing wild type EDNRA or EDNRA p.Q381P but not in empty vector (mock)-transfected cells. Images are representative of ligand binding after a 5 minute incubation. Green and red channels represent Hilyte fluor-488-ET-1 and mCherry-MEM, respectively. (d) HEK293T cells transfected with empty vector (mock), wild type EDNRA or EDNRA p.Q381P were incubated with Hilyte fluor-488-ET-1 and quantitatively analyzed with a fluorescence microplate reader. Specific fluorescence was calculated by subtracting background fluorescence values (from empty wells treated with Hilyte fluor-488-ET-1) from the raw fluorescence values. Assays were performed in triplicate at least three times. Error bars represent SEM; two-tailed t-test *p < 0.05, **p< 0.01; n.s., not significant.
We next determined whether ligand binding was affected by the p.Q381P variant. EDN1 binds to the ligand-binding pocket in the 7-TM domain, which results in conformational changes to the transmembrane α-helices and cytoplasmic domains that facilitate G protein coupling and activation (Weis & Kobilka, 2018). Because of the allosteric mechanism that links the ligand-binding pocket and intracellular G protein coupling domains (DeVree et al., 2016; Gregorio et al., 2017; Huang et al., 2015; Yao et al., 2009), we examined whether the p.Q381P variant affects EDN1-binding. To assess this, we examined fluorophore-tagged EDN1 (HiLyte-ET1) binding with wild type EDNRA and the EDNRA p.Q381P variant. Using live confocal microscopy, we observed rapid membrane association of HiLyte-EDN1 following its addition in HEK293T cells expressing wild type EDNRA (Fig. 3C) (HEK293T cells do not express EDNRA; data not shown). Similarly, rapid membrane association was observed in cells expressing the p.Q381P variant (Fig. 3C). Further, when HEK293T cells were treated as above and assayed with a fluorescent plate reader, overall fluorescence was not statistically significant between cells expressing wild type or p.Q381P variant EDNRA constructs (Fig. 3D). These data indicate that both EDNRA and EDNRA p.Q381P are present at similar levels and that the p.Q381P variant does not affect EDN1 binding to EDNRA. Furthermore, these data suggest that general protein folding is not drastically altered by the p.Q381P variant.
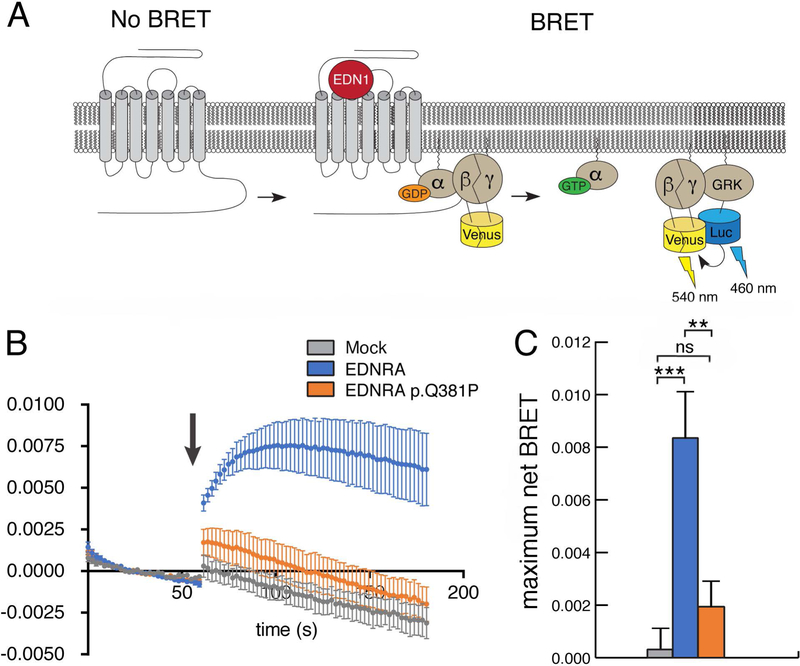
These results suggest that the p.Q381P variant affects EDNRA signaling downstream of ligand binding. GPCR signaling is mediated by a heterotrimeric G protein complex that is composed of a Gα subunit and an obligate Gβγ dimer (Gilman, 1987). Ligand binding to a GPCR initiates the rate-limiting step of G protein activation, in which GDP is exchanged for GTP in the Gα subunit. This exchange results in the dissociation of Gα and Gβγ and activation of their respective downstream signaling effectors (Gilman, 1987). Gq activates phospholipase C beta isoforms (PLCB) (Smrcka, Hepler, Brown, & Sternweis, 1991), while the Gβγ dimer binds to several different effectors as well as G protein-coupled receptor kinase (GRK)(Daaka et al., 1997). To determine whether the p.Q381P variant impairs G protein activation, we measured the Gαβγ dissociation event as an indirect reporter of EDNRA-mediated G protein activation. For our assay, we used Venus-Gβγ as the BRET acceptor, in which GβV1 and GγV2 encode complementary fragments that reconstitute Venus upon heterodimer formation of Gβγ (Hynes et al., 2004). The BRET donor was the Gβγ binding domain of G protein-coupled receptor kinase 3 (GRK3), fused to Nanoluc luciferase (masGRKct-Nanoluc, BRET donor) (Masuho, Ostrovskaya, et al., 2015) (Fig. 4A). Cells transfected with wild type EDRNA produced high BRET activity in response to addition of EDN1, indicating dissociation of Gαβγ and subsequent association of Venus-Gβγ with masGRK3ct-Nluc (Fig. 4B). However, cells expressing the p.Q381P variant produced significantly diminished BRET activity after EDN1 addition, indicating that Gq protein activation was impaired.
Figure 4. The EDNRA p.Q381P variant disrupts G protein activation.
(a) Schematic for the G protein activation BRET reporter assay. EDN1-induced EDNRA activation promotes dissociation of the G protein heterotrimer to Gαq and Gβγ-Venus. Subsequent interaction of Gβγ-Venus with mas-GRKct-nLuc produces BRET, which is an indirect reporter of G protein activation. (b) BRET assay in HEK293T cells transfected with the BRET components shown in (a) and empty vector (mock), wild type EDNRA or EDNRA p.Q381P. Baseline BRET was measured for 30 seconds, at which time cells were treated with EDN1 (indicated by arrow) and measured for another 2 minutes. A robust EDN1-induced BRET response was observed only in cells expressing wildtype EDNRA (blue trace). In the presence of EDNRA p.Q381P (orange trace), BRET was not statistically different from the presence of a mock vector (grey trace). Traces represent the average delta BRET of at least three independent transfections. Error bars at each timepoint represent SEM. (c) Maximum BRET response was quantified as the average of maximum delta BRET values elicited by EDN1. Assays were performed in triplicate at least three times. Error bars indicate SEM; two-tailed t-test; **p < 0.01, ***p< 0.005; n.s., not significant.
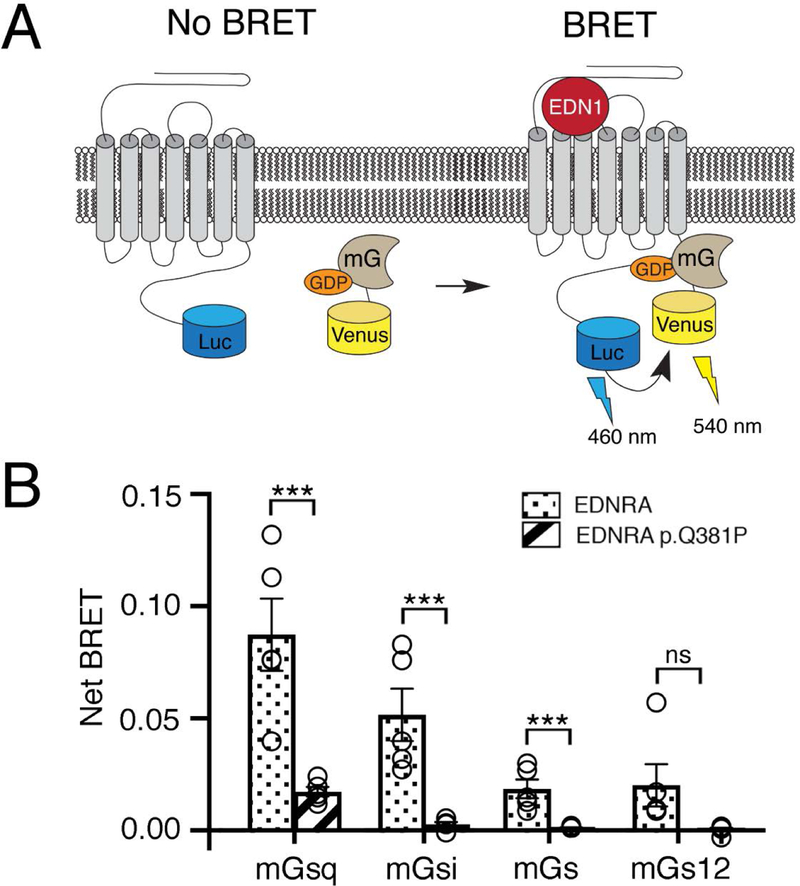
The EDNRA receptor couples promiscuously to several different Gα isoforms, including Gαq, Gαi, and Gα12 (IUPHAR database). We therefore tested whether coupling of several G protein isoforms to the EDRNA p.Q381P variant is disrupted. We took advantage of mini-G (mG) proteins, which lack the ability to bind Gβγ and contain a mutation that stabilizes the GPCR-mG association, making them ideal reporters for analyzing G protein-GPCR interactions (Wan et al., 2018). Gq interaction was examined using Venus-tagged mGsq (Venus-mGsq), a chimeric protein that has the stability of mGs but binding specificity of mGq (Wan et al., 2018), and wild type- or p.Q381P-EDNRA-RLuc8 (BRET donor) (Fig. 5A). Cells expressing Venus-mGsq and EDNRA-RLuc8 produced high BRET activity after EDN1 addition (Fig. 5B). However, cells expressing Venus-mGsq and EDNRA-RLuc8 p.Q381P produced significantly diminished BRET activity after EDN1 addition, suggesting that Gq-EDNRA interaction is disrupted by the p.Q381P variant (Fig. 5B). Additionally, using Venus-tagged mG proteins for Gi (mGi), Gs (mGs) and G12 (mG12), we found that cells expressing EDNRA-RLuc8 produced EDN1-induced BRET activity for all G protein classes, although they bound less well compared to mGsq (Fig. 5B). However, BRET activity was significantly diminished in cells expressing EDNRA-RLuc8 p.Q381P and mGi or mGs after EDN1 addition. Although EDN1-induced BRET activity was diminished for G12, no statistically significant difference was observed. Taken together, the results indicate that the p.Q381P variant disrupts overall G protein-EDNRA interactions.
Figure 5. The EDNRA p.Q381P variant disrupts G protein coupling.
(a) Schematic of the BRET assay for G protein coupling. A Venus-mini G protein can associate with a wild type EDNRA-RLuc8 after addition of EDN1, resulting in BRET. (b) The maximum BRET response was quantified for four classes of mini G proteins with wildtype EDNRA-RLuc8 or EDNRA-RLuc8 p.Q381P. Maximum BRET response for all mini G proteins was lower in cells expressing EDNRA-RLuc8 p.Q381P compared to wildtype EDNRA, though the change for mGs12 was not statistically significant (p=0.0678). Assays were performed in triplicate at least three times. Error bars indicate SEM; two-tailed t-test ***p<0.005; ns, not significant.
Discussion
Here we report the first known autosomal recessively inherited pathogenic variant in EDNRA that acts as a strong loss-of-function allele. This variant leads to malformation of craniofacial and cardiovascular structures consistent with early NCC patterning defects due to loss of EDNRA signaling. While we cannot rule out that one or more genetic variants not detected by exome sequencing contributed to the observed phenotype, based on the overall phenotypic similarities between this individual and Endothelin pathway mouse mutants, we believe that the observed defects in our individual result solely from the EDNRA variant. Furthermore, these phenotypic similarities highlight the evolutionary conservation of EDNRA signaling during patterning of the craniofacial complex (Clouthier, Garcia, & Schilling, 2010; Clouthier et al., 2013; Medeiros & Crump, 2012). It is possible that prior human pregnancies have been affected by loss-of-function variants of EDNRA but were never genetically investigated due to the lack of viability in the setting of airway stenosis. Notably this case constitutes a recognizable, unique phenotype that we have named Oro-Oto-Cardio syndrome.
EDNRA signaling is required for both cranial and cardiac NCC patterning (Clouthier et al., 1998; Kurihara et al., 1994; Yanagisawa et al., 1999). To date, only two other variants in EDNRA (p.Y129F, p.E303K) have been described that results in craniofacial anomalies, both in individuals with MFDA, though the absence of cardiovascular defects suggests that neither variant results in a complete loss of EDNRA signaling (Gordon et al., 2015). In actuality, the functional consequence of the p.Y129F variant in MFDA individuals is complex. The p.Y129 residue is responsible for limiting activation of EDNRA to EDN1, with a p.Y129F change leading to the ability of EDN3 to also bind to the receptor (Krystek et al., 1994; Lee et al., 1994). MDFA individuals have maxillary changes suggesting earlier inappropriate EDNRA signaling in the maxillary portion of the first arch, while retrognathia and middle ear abnormalities indicate loss of EDNRA signaling in the mandibular portion of arch one.(Gordon et al., 2015). Importantly, MDFA patients do not have cardiovascular defects exhibited by Edn1/Ednra/Ece1 knockout mice (Clouthier et al., 1998; Dettlaff-Swiercz et al., 2005; Kurihara et al., 1995; Offermanns et al., 1998; Yanagisawa et al., 1998). In stark contrast, the p.Q381P variant described in this study caused a severe reduction in Gq coupling (Fig. 5B) and activation (Fig. 4C), which likely resulted in a complete loss of EDNRA signaling in NCCs that give rise to the lower jaw structures, ears, and smooth muscle of the cardiac outflow tract.
Like MFDA, individuals with ARCND have facial phenotypes resembling those in Ednra−/− embryos but lack cardiovascular defects. The minimum level of EDNRA signaling required for cardiac development is not known. However, as described above, chimera analysis in mice, in which wild type and Ednra−/− ES cells are mixed, has provided some idea about the difference in sensitivity to loss of EDNRA signaling between cranial and cardiac NCCs (Clouthier et al., 2003). These experiments showed that EDNRA signaling occurs cell autonomously within both cranial and cardiac NCCs. However, all Ednra+/+<->Ednra−/− chimeric embryos had some degree of facial difference, with the severity higher in embryos with a higher contribution of Ednra−/− cells. In contrast, only one embryo that also had the highest number of Ednra−/− cells had a cardiovascular defect (Clouthier et al., 2003). This illustrates that cranial NCCs are far more sensitive to loss of EDNRA signaling than are cardiac NCCs, consistent with the findings in MFDA and ARCND individuals.
To date, variants within helix 8 of EDRRA in individuals with craniofacial malformations have not been identified. This may reflect sensitive or crucial functions of this helix that are not compatible with survival. In many class A GPCRs, including Endothelin receptor type B (EDNRB), ligand binding induces coordinated movements of transmembrane helix 7 and helix 8, along with other transmembrane helices, to expose the binding interface for the α5 helix of G protein α subunits (Weis & Kobilka, 2018). In the founding class A GPCR rhodopsin, the amino terminal end of helix 8 directly interacts with and activates Gα transducin (Scheerer et al., 2008). A potential consequence of the p.Q381P proline substitution is a distortion of the secondary structure or orientation of helix 8 that prevents EDN1-induced conformational transition of helix 7-helix 8. This model predicts that p.Q381P impairs the general G protein activation mechanism for EDNRA, accounting for the severe congenital anomalies.
In summary, we have found the first known homozygous loss-of-function variant in EDNRA. In vitro analysis of this allele illustrates that the variant disrupts G protein association with the receptor, which, in vivo, likely results in near-complete loss of EDNRA signaling and hence severe developmental defects in craniofacial and cardiovascular structures. From a clinical perspective, this case demonstrates a new, distinct phenotype that to our knowledge has not been previously described. The mandibular and oral findings have some overlap with syndromes caused by other genes in the endothelin pathway, including ARCND and MFDA, but the cardiac findings and severity of external ear differences may provide a clue to loss-of-function of EDNRA. This case highlights the utility of combining neonatal or prenatal whole exome or whole genome sequencing with functional analysis of relevant variants in cases where a genetic condition is suspected, as identification of an etiology can inform future reproductive decisions for a family and ameliorate some of the guilt of grieving parents. In this case, the family has proceeded with pre-implantation genetic diagnosis to ensure a fetus is not homozygous for this variant in EDNRA and has had two healthy children through in vitro fertilization. Prompt functional investigations into candidate genes as performed here are crucial given the limited accessibility of pre-implantation genetic diagnosis, especially in the case of a candidate gene. With the emergence of well-established, sensitive, and modular BRET sensors, it should be possible to rapidly assess genetic variants of G protein signaling components that arise in exome or genome sequencing to both identify potential candidates for further functional analysis and to understand how the variants function at the cellular level. Additionally, because BRET assays do not require sophisticated instruments outside of a microplate reader that detects fluorescence and bioluminescence, this approach should be widely accessible to researchers and clinicians. Currently, the time-consuming aspect of this approach is assembling and validating sensors required for specific aspects of G protein signaling. In addition, each new genetic variant has to be introduced into the appropriate cDNA. However, as these approaches become more widespread, the turnaround time from variant determination to confirmation of BRET results can be under one month. While getting Clinical Laboratory Improvement Amendment (CLIA) certification for these assays would take time, our results will hopefully encourage more wide-spread biochemical analysis of neonatal-lethal craniofacial malformations.
Supplementary Material
Acknowledgements
The authors would like to express remembrance and gratitude to Kasin Lyle Buchmyer and his family for allowing us to share his story and this work. The authors would also like to thank Holly N. Wood, Mark Dotseth and Tiffany Mundhenke for technical assistance and Philip Wedegaertner, Kirill Martemyanov and Jeffery Walker for plasmids. This work was supported in part by grant DE023050 from NIH/NIDCR (to D.E.C.) and grant GM130142 from NIH/NIGMS (to N.A.L.).
Footnotes
Declaration of Interests
The authors declare no conflicts of interests.
Accession Numbers
The EDNRA p.Q381P variant has been submitted to ClinVar (www.ncbi.nlm.nih.gov/clinvar/) and assigned the accession number SCV000924665.
References
- Abe M, Ruest L-B, & Clouthier DE (2007). Fate of cranial neural crest cells during craniofacial development in endothelin-A receptor deficient mice. International. Journal of Develeopmental. Biology, 51, 97–105. doi: 10.1387/ijdb.062237ma [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charité J, McFadden DG, Merlo GR, Levi G, Clouthier DE, Yanagisawa M, … Olson EN (2001). Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes and Development, 15, 3039–3049. doi: 10.1101/gad.931701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Garcia E, & Schilling TF (2010). Regulation of facial morphogenesis by endothelin signaling: insights from mouse and fish. American Journal of Medical Genetics, Part A., 152A, 2962–2973. doi: 10.1002/ajmg.a.33568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Hosoda K, Richardson JA, Williams SC, Yanagisawa H, Kuwaki T, … Yanagisawa M. (1998). Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development, 125, 813–824.9449664 [Google Scholar]
- Clouthier DE, Passos-Bueno MR, Tavares ALP, Lyonnet S, Amiel J, & Gordon CT (2013). Understanding the basis of Auriculocondylar syndrome: Insights from human, mouse and zebrafish studies. American Journal of Medical Genetics, Part C, 163, 306–317. doi: 10.1002/ajmg.c.31376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, & Schilling TF (2004). Understanding endothelin-1 function during craniofacial development in the mouse and zebrafish. Birth Defects Research (Part C), 72, 190–199. doi: 10.1002/bdrc.20007 [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Hammer RE, Richardson JA, & Yanagisawa M. (2003). Cell-autonomous and nonautonomous actions of endothelin-A receptor signaling in craniofacial and cardiovascular development. Developmental Biology, 261(2), 506–519. doi: 10.1016/s0012-1606(03)00128-3 [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, & Yanagisawa M. (2000). Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Developmental Biology, 217, 10–24. doi: 10.1006/dbio.1999.9527 [DOI] [PubMed] [Google Scholar]
- Daaka Y, Pitcher JA, Richardson M, Stoffel RH, Robishaw JD, & Lefkowitz RJ (1997). Receptor and Gβγ isoform-specific interactions with G protein-coupled receptor kinases. Proceedings of the National Acadamy of Sciences of the United States of America, 94(6), 2180–2185. doi: 10.1073/pnas.94.6.2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delos Santos NM, Gardner LA, White SW, & Bahouth SW (2006). Characterization of the residues in helix 8 of the human β1-adrenergic receptor that are involved in coupling the receptor to G proteins. Journal of Biological Chemistry, 281(18), 12896–12907. doi: 10.1074/jbc.M508500200 [DOI] [PubMed] [Google Scholar]
- Depew MJ, Lufkin T, & Rubenstein JL (2002). Specification of jaw subdivisions by Dlx genes. Science, 298, 381–385. doi: 10.1126/science.1075703 [DOI] [PubMed] [Google Scholar]
- Depew MJ, Simpson CA, Morasso M, & Rubenstein JLR (2005). Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. Journal of Anatomy, 207, 501–561. doi: 10.1111/j.1469-7580.2005.00487.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettlaff-Swiercz DA, Wettschureck N, Moers A, Huber K, & Offermanns S. (2005). Characteristic defects in neural crest cell-specific Gαq/Gα11- and Gα12/Gα13-deficient mice. Developmental Biology, 282(1), 174–182. doi: 10.1016/j.ydbio.2005.03.006 [DOI] [PubMed] [Google Scholar]
- DeVree BT, Mahoney JP, Velez-Ruiz GA, Rasmussen SG, Kuszak AJ, Edwald E, … Sunahara RK (2016). Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature, 535(7610), 182–186. doi: 10.1038/nature18324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faussner A, Bauer A, Kalatskaya I, Schussler S, Seidl C, Proud D, & Jochum M. (2005). The role of helix 8 and of the cytosolic C-termini in the internalization and signal transduction of B(1) and B(2) bradykinin receptors. Febs Journal, 272(1), 129–140. doi: 10.1111/j.1432-1033.2004.04390.x [DOI] [PubMed] [Google Scholar]
- Feierler J, Wirth M, Welte B, Schussler S, Jochum M, & Faussner A. (2011). Helix 8 plays a crucial role in bradykinin B(2) receptor trafficking and signaling. Journal of Biological Chemistry, 286(50), 43282–43293. doi: 10.1074/jbc.M111.256909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson KM, Nesbitt A, Cao K, Yu Z, Denenberg E, DeChene E, … Santani A. (2018). Novel findings with reassessment of exome data: implications for validation testing and interpretation of genomic data. Genetics in Medicine, 20(3), 329–336. doi: 10.1038/gim.2017.153 [DOI] [PubMed] [Google Scholar]
- Gilman AG (1987). G proteins: transducers of receptor-generated signals. Annual Review of Biochemistry, 56, 615–649. doi: 10.1146/annurev.bi.56.070187.003151 [DOI] [PubMed] [Google Scholar]
- Gordon CT, Petit F, Kroisel PM, Jakobsen L, Zechi-Ceide RM, Oufadem M, … Amiel J. (2013). Mutations in endothelin 1 cause recessive auriculocondylar syndrome and dominant isolated question-mark ears. American Journal of Human Genetics, 93(6), 1118–1125. doi: 10.1016/j.ajhg.2013.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CT, Weaver KN, Zechi-Ceide RM, Madsen EC, Tavares AL, Oufadem M, … Amiel J. (2015). Mutations in the endothelin receptor type A cause mandibulofacial dysostosis with alopecia. American Journal of Human Genetics, 96(4), 519–531. doi: 10.1016/j.ajhg.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio GG, Masureel M, Hilger D, Terry DS, Juette M, Zhao H, … Blanchard SC (2017). Single-molecule analysis of ligand efficacy in β2AR-G-protein activation. Nature, 547(7661), 68–73. doi: 10.1038/nature22354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Percherancier Y, Breton B, & Bouvier M. (2006). Monitoring protein-protein interactions in living cells by bioluminescence resonance energy transfer (BRET). Current Protocols in Neuroscience, Chapter 5, Unit 5 23. doi: 10.1002/0471142301.ns0523s34 [DOI] [PubMed] [Google Scholar]
- Hollins B, Kuravi S, Digby GJ, & Lambert NA (2009). The c-terminus of GRK3 indicates rapid dissociation of G protein heterotrimers. Cell Signaling, 21(6), 1015–1021. doi: 10.1016/j.cellsig.2009.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Manglik A, Venkatakrishnan AJ, Laeremans T, Feinberg EN, Sanborn AL, … Kobilka BK (2015). Structural insights into micro-opioid receptor activation. Nature, 524(7565), 315–321. doi: 10.1038/nature14886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes TR, Tang L, Mervine SM, Sabo JL, Yost EA, Devreotes PN, & Berlot CH (2004). Visualization of G protein βγ dimers using bimolecular fluorescence complementation demonstrates roles for both β and γ in subcellular targeting. Journal of Biological Chemistry, 279(29), 30279–30286. doi: 10.1074/jbc.M401432200 [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, … MacArthur DG (2019). Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv, 531210. doi: 10.1101/531210 [DOI] [Google Scholar]
- Karpinsky-Semper D, Tayou J, Levay K, Schuchardt BJ, Bhat V, Volmar CH, … Slepak VZ. (2015). Helix 8 and the i3 loop of the muscarinic M3 receptor are crucial sites for its regulation by the Gβ5-RGS7 complex. Biochemistry, 54(4), 1077–1088. doi: 10.1021/bi500980d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberg K, Kim TY, Moller M, Skegro D, Dasara Raju G, Granzin J, … Alexiev U. (2011). Conformational dynamics of helix 8 in the GPCR rhodopsin controls arrestin activation in the desensitization process. Proceedings of the National Acadamy of Sciences of the United States of America, 108(46), 18690–18695. doi: 10.1073/pnas.1015461108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystek SR Jr., Patel PS, Rose PM, Fisher SM, Kienzle BK, Lach DA, … Webb ML (1994). Mutation of peptide binding site in transmembrane region of a G protein-coupled receptor accounts for endothelin receptor subtype selectivity. Journal of Biological Chemistry, 269(17), 12383–12386. [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, Maemura K, Kuwaki T, Kumada M, & Yazaki Y. (1995). Impaired development of the thyroid and thymus in endothelin-1 knockout mice. J Cardiovascular Pharmacology, 26 Suppl 3, S13–16. [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, … Yazaki Y. (1994). Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature, 368, 703–710. [DOI] [PubMed] [Google Scholar]
- Lan TH, Liu Q, Li C, Wu G, & Lambert NA (2012). Sensitive and high resolution localization and tracking of membrane proteins in live cells with BRET. Traffic, 13(11), 1450–1456. doi: 10.1111/j.1600-0854.2012.01401.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Elliott JD, Sutiphong JA, Friesen WJ, Ohlstein EH, Stadel JM, … Peishoff CE (1994). Tyr-129 is important to the peptide ligand affinity and selectivity of human endothelin type A receptor. Proceedings of the National Acadamy of Sciences of the United States of America, 91(15), 7164–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, … MacArthur DG (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536(7616), 285–291. doi: 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjurano A, Sepulveda N, Nadjm B, Mtove G, Wangai H, Maxwell C, … Clark TG (2015). USP38, FREM3, SDC1, DDC, and LOC727982 Gene Polymorphisms and Differential Susceptibility to Severe Malaria in Tanzania. Journal of Infectious Diseases, 212(7), 1129–1139. doi: 10.1093/infdis/jiv192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuho I, Martemyanov KA, & Lambert NA (2015). Monitoring G Protein Activation in Cells with BRET. Methods in Molecular Biology, 1335, 107–113. doi: 10.1007/978-1-4939-2914-6_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuho I, Ostrovskaya O, Kramer GM, Jones CD, Xie K, & Martemyanov KA (2015). Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Science Signaling, 8(405), ra123. doi: 10.1126/scisignal.aab4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros DM, & Crump JG (2012). New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Developmental Biology, 371(2), 121–135. doi: 10.1016/j.ydbio.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stainier DY, & Kimmel CB (2003). Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development, 130(7), 1353–1365. doi: 10.1242/dev.00339 [DOI] [PubMed] [Google Scholar]
- Nair S, Li W, Cornell R, & Schilling TF (2007). Requirements for endothelin type-A receptors and endothelin-1 signaling in the facial ectoderm for the patterning of skeletogenic neural crest cells in zebrafish. Development, 134, 335–345. doi: 10.1242/dev.02704 [DOI] [PubMed] [Google Scholar]
- Offermanns S, Zhao LP, Gohla A, Sarosi I, Simon MI, & Wilkie TM (1998). Embryonic cardiomyocyte hypoplasia and craniofacial defects in G alpha q/G alpha 11-mutant mice. EMBO Journal, 17, 4304–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki H, Kurihara Y, Tonami K, Watatani S, & Kurihara H. (2004). Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mechanisms of Development, 121(4), 387–395. doi: 10.1016/j.mod.2004.02.002 [DOI] [PubMed] [Google Scholar]
- Pavlakis E, Makrygiannis AK, Chiotaki R, & Chalepakis G. (2008). Differential localization profile of Fras1/Frem proteins in epithelial basement membranes of newborn and adult mice. Histochemistry and Cell Biology, 130(4), 785–793. doi: 10.1007/s00418-008-0453-4 [DOI] [PubMed] [Google Scholar]
- Piela L, Nemethy G, & Scheraga HA (1987). Proline-induced constraints in alpha-helices. Biopolymers, 26(9), 1587–1600. doi: 10.1002/bip.360260910 [DOI] [PubMed] [Google Scholar]
- Rieder MJ, Green GE, Park SS, Stamper BD, Gordon CT, Johnson JM, … Cunningham ML (2012). A human homeotic transformation tesulting from mutations in PLCB4 and GNAI3 causes auriculocondylar syndrome. American Journal of Human Genetics, 90, 907–914. doi: 10.1016/j.ajhg.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Xiang X, Lim KC, Levi G, & Clouthier DE (2004). Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development, 131(18), 4413–4423. doi: 10.1242/dev.01291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, … Ernst OP (2008). Crystal structure of opsin in its G-protein-interacting conformation. Nature, 455(7212), 497–502. doi: 10.1038/nature07330 [DOI] [PubMed] [Google Scholar]
- Shi C, Zhang K, Wang X, Shen Y, & Xu Q. (2012). A study of the combined effects of the EHD3 and FREM3 genes in patients with major depressive disorder. American Journal of Medical Genetics, Part B, Neuropsychiatric Genetics, 159b(3), 336–342. doi: 10.1002/ajmg.b.32033 [DOI] [PubMed] [Google Scholar]
- Smrcka AV, Hepler JR, Brown KO, & Sternweis PC (1991). Regulation of polyphosphoinositide-specific phospholipase C activity by purified Gq. Science, 251(4995), 804–807. [DOI] [PubMed] [Google Scholar]
- Tavares ALP, Cox TC, Maxson RM, Ford HL, & Clouthier DE (2017). Negative regulation of Endothelin signaling by SIX1 is required for proper maxillary development. Development, 144(11), 2021–2031. doi: 10.1242/dev.145144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares ALP, Garcia EL, Kuhn K, Woods CM, Williams T, & Clouthier DE (2012). Ectodermal-derived Endothelin1 is required for patterning the distal and intermediate domains of the mouse mandibular arch. Developmental Biology, 371, 47–56. doi: 10.1016/j.ydbio.2015.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q, Okashah N, Inoue A, Nehme R, Carpenter B, Tate CG, & Lambert NA (2018). Mini G protein probes for active G protein-coupled receptors (GPCRs) in live cells. Journal of Biological Chemistry, 293(19), 7466–7473. doi: 10.1074/jbc.RA118.001975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedegaertner PB, Chu DH, Wilson PT, Levis MJ, & Bourne HR (1993). Palmitoylation is required for signaling functions and membrane attachment of Gq alpha and Gs alpha. Journal of Biological Chemistry, 268(33), 25001–25008. [PubMed] [Google Scholar]
- Weis WI, & Kobilka BK (2018). The Molecular Basis of G Protein-Coupled Receptor Activation. Annual Review of Biochemistry, 87, 897–919. doi: 10.1146/annurev-biochem-060614-033910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H, Hammer RE, Richardson JA, Williams SC, Clouthier DE, & Yanagisawa M. (1999). Role of Endothelin-1/Endothelin-A receptor-mediated signaling pathway in the aortic arch patterning in mice. Journal of Clinical Investigation, 102(1), 22–33. doi: 10.1172/JCI2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, Clouthier DE, … Hammer RE (1998). Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development, 125(5), 825–836.9449665 [Google Scholar]
- Yang JM, Yoneda K, Morita E, Imamura S, Nam K, Lee ES, & Steinert PM (1997). An alanine to proline mutation in the 1A rod domain of the keratin 10 chain in epidermolytic hyperkeratosis. Journal of Investigative Dermatology, 109(5), 692–694. doi: 10.1111/1523-1747.ep12338320 [DOI] [PubMed] [Google Scholar]
- Yao XJ, Velez Ruiz G, Whorton MR, Rasmussen SG, DeVree BT, Deupi X, … Kobilka B. (2009). The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proceedings of the National Acadamy of Sciences of the United States of America, 106(23), 9501–9506. doi: 10.1073/pnas.0811437106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost EA, Mervine SM, Sabo JL, Hynes TR, & Berlot CH (2007). Live cell analysis of G protein β5 complex formation, function, and targeting. Molecular Pharmacology, 72(4), 812–825. doi: 10.1124/mol.107.038075 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.