Abstract
The microtubule (MT)-depolymerizing activity of MCAK/Kif2C can be quantified by expressing the motor in cultured cells and measuring tubulin fluorescence levels after enough hours have passed to allow tubulin autoregulation to proceed. This method allows us to score the impact of point mutations within the motor domain. We found that, despite their distinctly different activities, many mutations that impact transport kinesins also impair MCAK/Kif2C’s depolymerizing activity. We improved our workflow using CellProfiler to significantly speed up the imaging and analysis of transfected cells. This allowed us to rapidly interrogate a number of MCAK/Kif2C motor domain mutations documented in the cancer database cBioPortal. We found that a large proportion of these mutations adversely impact the motor. Using green fluorescent protein–FKBP-MCAK CRISPR cells we found that one deleterious hot-spot mutation increased chromosome instability in a wild-type (WT) background, suggesting that such mutants have the potential to promote tumor karyotype evolution. We also found that increasing WT MCAK/Kif2C protein levels over that of endogenous MCAK/Kif2C similarly increased chromosome instability. Thus, endogenous MCAK/Kif2C activity in normal cells is tuned to a mean level to achieve maximal suppression of chromosome instability.
INTRODUCTION
MCAK/Kif2C is a member of the kinesin-13 family of microtubule (MT)-depolymerizing kinesins. This kinesin shares high amino acid identity and conservation within the ATP-hydrolyzing motor domain with minus end– and plus end–directed transport kinesins (Supplemental Figure S1, A and B). MCAK/Kif2C uses its depolymerizing activity to suppresses erroneous MT attachments to chromosomes during cell division.
Mechanistically, this is accomplished by controlling MT assembly rates and lengths in all cellular MTs (Domnitz et al., 2012; Ertych et al., 2014; Wordeman et al., 2016) and also directly by end modulation and turnover of MTs at kinetochores (Wordeman et al., 2007; Bakhoum et al., 2009). MCAK/Kif2C’s depolymerizing activity has been extensively studied in vitro (Helenius et al., 2006; Cooper et al., 2010; Friel and Howard, 2011; Gardner et al., 2011), but it can also be quantified in cells by measuring tubulin levels using anti-tubulin antibodies (Ovechkina et al., 2002; Ogawa et al., 2004; Manning et al., 2007; Montenegro Gouveia et al., 2010; Domnitz et al., 2012; Parker et al., 2018). This is because the release of tubulin dimers from MT polymer in cells will suppress tubulin expression, leading to a decrease in overall tubulin fluorescence in cells over the course of several hours. This effect can be mimicked by exposing cells to short versus long time-course exposure to a MT-depolymerizing drug such as nocodazole. A short (15 min) treatment that depolymerizes all the MTs in the cell will not lead to a significant overall decrease in tubulin fluorescence as detected by anti-tubulin antibodies compared with untreated cells with an intact MT array. However, a longer (12 h) exposure to nocodazole will result in lower detectable tubulin fluorescence (Supplemental Figure S1C). Although this quantification of MCAK/Kif2C’s depolymerizing activity occurs through the tubulin autoregulation pathway (Pachter et al., 1987), it can be surprisingly consistent, as there are only a modest number of key tubulin genes that respond to MT stabilization versus destabilization (Gasic et al., 2019). This property has enabled us to measure the depolymerizing activity of a number of MCAK/Kif2C mutants and kinesin-13 family members in a consistent manner. In addition, we have previously determined that this in vivo quantification matches closely with in vitro measures of activity (Ovechkina et al., 2002). The principal disadvantage of our previously published method is that it is manually performed and not well suited to large screens. We sought to improve this analysis using the free software CellProfiler (McQuin et al., 2018) so that we could further investigate the functional consequences of point mutations within the MCAK/Kif2C motor domain and also interrogate the naturally occurring genomic alterations of MCAK/Kif2C reported in tumors.
Initially, we used our manual method of analysis to evaluate mutations in the MCAK/Kif2C MT-depolymerizing kinesin that had conservation with and had been well studied in motile kinesins (Woehlke et al., 1997; Song and Endow, 1998; Rice et al., 1999). We were able to infer a great deal of useful information from the crystal structure of Kif2C as well (Ogawa et al., 2004, 2017). These background data informed our analysis and allowed us to confirm and extend some of the proposed structural mechanisms, at least with respect to their impact on depolymerization activity. Given how well the activity mirrored expectations for many of the mutants, we endeavored to use a similar approach to evaluate missense mutations in MCAK/Kif2C that appear in some tumors.
Web-based oncogenomic portals such as COSMIC (Tate et al., 2019) and cBioPortal (Cerami et al., 2012) enable researchers to investigate mutations and copy-number alterations for specific genes in multiple types of cancers using genome-wide analyses of hundreds of cancer samples (Klonowska et al., 2016). Less well developed are assays to investigate the cellular consequences of genetic alterations that arise in cancers. This is because each assay must, by necessity, be gene product specific, and many proteins exhibit complex activities that are difficult to quantify in cells. With regard to MCAK/Kif2C, many studies have implicated changes in expression with a poor prognosis for cancer recovery (Duan et al., 2016; Chen et al., 2017; van Dam et al., 2018; Bai et al., 2019; Cai et al., 2019; Gan et al., 2019; Pan et al., 2019). While we have a reliable in vivo assay for MCAK/Kif2C activity, our manual method would likely be too slow to use to evaluate the missense mutations in the cancer databases, many of which might be less interesting “passenger” mutations (Hess et al., 2019). However, it has been proposed that mutations that appear with low or modest frequency in tumors (such as those recorded for MCAK/Kif2C) can act as drivers in some cancers in which they appear (Wood et al., 2007). For this reason, we hypothesized that it would be useful to interrogate these mutations. Using high-throughput CellProfiler pipelines, we were able to rapidly assay some of the mutational hot spots identified in the MCAK/Kif2C motor domain. We found that many of the mutations inhibited the kinesin’s activity and promoted chromosome instability. Surprisingly, we found that excess MCAK/Kif2C with normal activity had the same effect, suggesting that both amplifications and mutations have the potential to facilitate rapid karyotype evolution in tumors.
RESULTS
Using a manual method of analysis, we have previously investigated the impact of the positively charged “neck” of MCAK/Kif2C N-terminal to the motor domain on the protein’s MT-depolymerizing activity. We found that cellular MT-depolymerizing activity decreased with loss of positive charges, and this correlated well with in vitro activity measured with purified mutant protein (Ovechkina et al., 2002). We used the same manual method to investigate point mutations within the motor domain corresponding to residues of identity and conservation with the kinesin superfamily and the kinesin-13 family within the motor domain (Figure 1A). Briefly, fixed cells labeled for tubulin and expressing measured levels of green fluorescent protein (GFP) fluorescence were selected and imaged. Selection criteria were normalized to include only cells expressing GFP fluorescence levels that would result in close to all MTs depolymerized in GFP-WT-MCAK/Kif2C (GFP-WT-MCAK/Kif2C)–expressing cells. Additionally, a similar-sized untransfected cell in the same field was used to measure reference tubulin fluorescence. This controlled for lighting anomalies such as lamp flicker on a cell by cell basis. This method, while reliable, resulted in very slow workflow for both the imaging, which required two similar-sized cells per field (one transfected and one reference cell), and for the measurements, which required hand tracing of each cell to measure fluorescence. For technical reasons, the nuclear fluorescence was included in both measurements, which has the potential to introduce error, as some mutations can influence the nuclear transport of MCAK/Kif2C (Wordeman et al., 1999).
FIGURE 1:
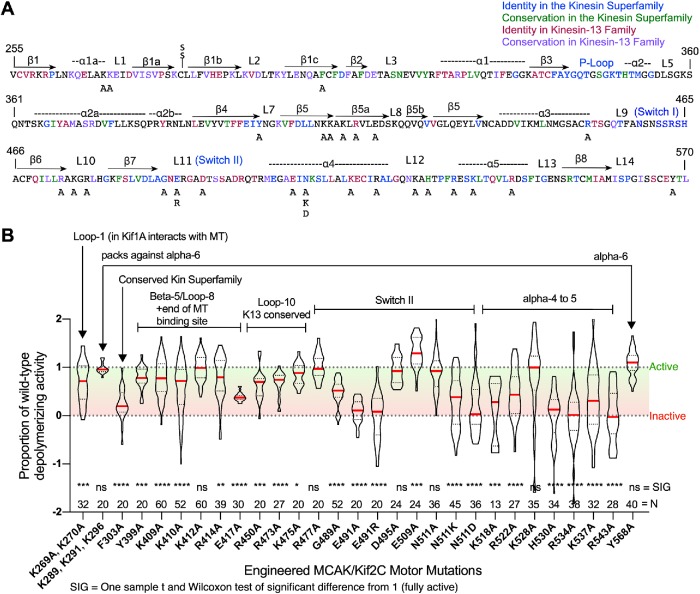
Point mutations engineered into the MCAK/Kif2C motor domain and analyzed manually exhibit altered depolymerizing activity. (A) The sequence of the MCAK/Kif2C annotated for structural elements and colored to indicate residues exhibiting high identity in the kinesin superfamily (blue), high conservation in the kinesin superfamily (green), high identity in the kinesin-13 family (red), and high conservation in the kinesin-13 family (purple). Mutations to alanine (“A”) or other residues are also indicated. (B) Proportion of MT-depolymerizing activity normalized to GFP-WT-MCAK/Kif2C. N = number of cells individually measured. Violin plots show the median activity (red line).
Over the course of several years, we were able to use our manual analysis method to investigate a large number of point mutations within the motor domain of MCAK/Kif2C that were associated with conserved structural features (such as ATP hydrolysis or MT binding) and also residues unique to kinesin-13 family members (Figure 1A). We transfected each of these mutants into CHO cells and measured the tubulin fluorescence using the method by hand. The results of this survey are shown in Figure 1B with annotations indicating the structural features of the motor domain that they are expected to impact. For reference, many of these structural features are modeled in Supplemental Figure S2. Most of the mutations were deleterious to MCAK/Kif2C activity by design, as they targeted residues previously identified to be important for motile kinesin function or highly conserved within the kinesin-13 family. However, one mutant (E509A) appeared to exhibit significantly enhanced activity (p = 0.0001). We cannot presently explain this enhanced activity, so it would be worthy of further study.
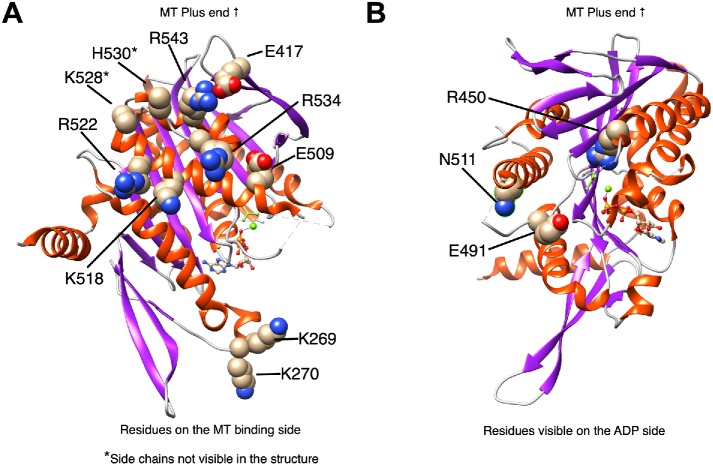
Figure 2 illustrates the position of select residues that significantly impact MACK/Kif2C activity superimposed on the ADP-BeFx structure (middle prehydrolysis state) of Kif2C (Ogawa et al., 2017). Residues likely to impact MT binding are shown in Figure 2A, which looks down on the MT binding surface. Residue E491, which is directly involved in ATP hydrolysis (Rice et al., 1999), and residue N511, which blocks coordination between MT binding and ATP hydrolysis (Song and Endow, 1998; Yun et al., 2001, 2003), are shown in Figure 2B. Another residue (G489) that is directly involved in ATP hydrolysis (Rice et al., 1999) is not shown, because its side chain is not visible in the crystal structure. Mutation of each of these residues also profoundly impairs MCAK/Kif2C MT-depolymerizing activity.
FIGURE 2:
Structural map of MCAK/Kif2C motor domain mutants. (A) Residues involved in the MT binding site with the caveat that the side chains of the starred (*) residues are not visible in the structure, so they are only predicted to interact with MTs. Structure used is the ADP-BeFx map of Kif2C (5XJB). (B) Residues visible on the ADP side of the structure.
While informative, our manual analysis of MCAK/Kif2C activity proved to be extremely slow. Each mutant took approximately 1 wk to image and analyze, and this analysis had to be performed individually for each mutant, because reference positive and negative controls were required for each mutant analyzed to control for microscope imaging variation. We hypothesized that CellProfiler would allow us to speed up both the imaging and analysis of MCAK/Kif2C mutants, because we could image and analyze fields of cells without having to select reference cells or control for expression levels. To speed up the workflow, we imaged successive fields of transfected cells at lower power (20×) without controlling precisely for a particular level of GFP. CellProfiler then measured mean tubulin fluorescence in successive fields of cells consisting of summed z-stacks (see Materials and Methods). Previously, the manual analysis of MCAK/Kif2C activity represented a bottleneck in MCAK/Kif2C mutant analysis. We hypothesized that, using CellProfiler, we could quantify the cellular effects of MCAK/Kif2C missense mutations as rapidly as they could be engineered. If this proved to be true, then it would be possible for researchers to quantify the activity of the 164 MCAK/Kif2C missense mutations recorded in the curated set of nonredundant cancer-sequencing studies (cBioPortal.org).
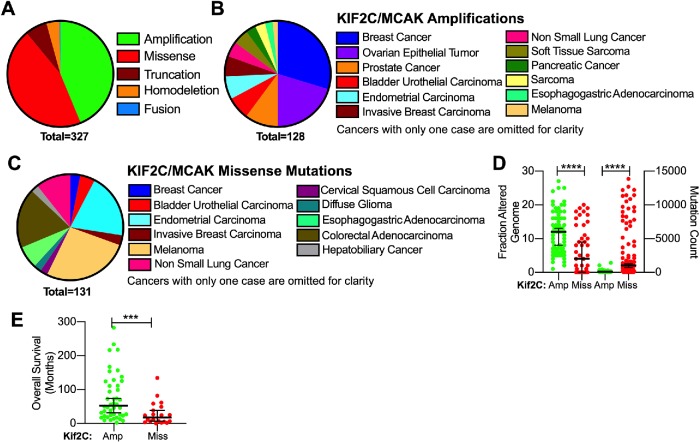
We found that cBioPortal predicted that MCAK/Kif2C alterations manifest primarily either as missense mutations or amplifications in cancer patients and that these two classifications of MCAK/Kif2C alterations did not overlap (Figure 3A). Amplifications were prevalent in breast, ovarian, and prostate cancer consistent with published reports (Figure 3B). Missense mutations in MCAK/Kif2C were found primarily in melanoma, colorectal adenocarcinoma, and endometrial carcinoma samples (Figure 3C). Not surprisingly, amplifications were associated with samples displaying highly altered genomes (Taylor et al., 2018) in contrast to samples displaying a high mutation count (Figure 3D). This suggests that cancers exhibiting MCAK/Kif2C amplifications may differ fundamentally from those exhibiting MCAK/Kif2C missense mutations in ways we do not fully understand. Overall survival in months was higher in cancer patients with MCAK/Kif2C amplifications rather than mutations (Figure 3E). MCAK/Kif2C has the potential to suppress chromosome instability when present and promote chromosome instability when lost (Bakhoum et al., 2009; Stolz et al., 2010). For this reason, we wanted to know the effect of these alterations on MCAK activity in cells. Previously, we have identified single point mutations that can either decrease or (more rarely) increase MT-depolymerizing activity (Figure 1B). Our improved high-throughput analysis now allows us to rapidly investigate the cellular activity of some of these MCAK/Kif2C cancer mutations.
FIGURE 3:
Summary of 327 cancer patients with MCAK/Kif2C mutations. (A) Patients with missense mutations in MCAK/Kif2C do not overlap with patients exhibiting amplification of the MCAK/Kif2C alleles. (B) Cancers represented in patients with MCAK/Kif2C amplifications. (C) Cancers exhibiting MCAK/Kif2C point mutations. (D) MCAK/Kif2C amplifications are seen in patients exhibiting a higher fraction of genome alterations (left axis), whereas missense mutations were found in patients with a higher overall mutation count in many genes (right axis). (E) Overall survival was higher for patients exhibiting amplifications of MCAK/Kif2C vs. missense mutations. ***p < 0.001; ****p < 0.0001.
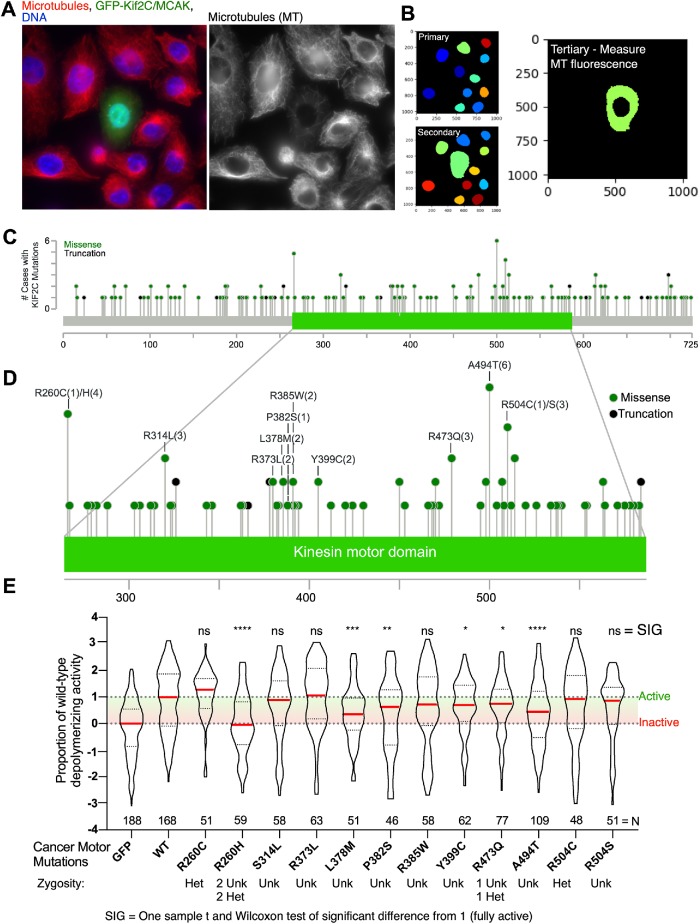
A typical field image is shown in Figure 4A. Transfection of cultured cells with GFP-MCAK/Kif2C results in an overall reduction in tubulin fluorescence in cells commensurate with a decrease of MT polymer (Figure 4A, right). To measure the tubulin fluorescence in these transfected cells, we constructed a pipeline using CellProfiler that will identify nuclei as primary objects and GFP-transfected cells as secondary objects. Subtraction of the primary from the secondary object results in a measurement of cytoplasmic tubulin fluorescence (Figure 4B). Although we did not report this here, this method can also be used to correlate cytoplasmic GFP fluorescence levels with tubulin fluorescence levels. Also, because MCAK/Kif2C accumulates in both the nucleus and cytoplasm, it is preferable to isolate cytoplasmic fluorescence from total cell fluorescence. In this way, the pipeline controls for mutants that may alter nuclear transport (Wordeman et al., 1999) leaving us with isolated cytoplasmic fluorescence measurements specific to the MT compartment of the cell. In this way, we can rapidly image and measure tubulin levels in cells transfected with GFP-MCAK mutants and compare them with GFP-WT-MCAK/Kif2C (full activity) and plain GFP (no activity) transfected cells.
FIGURE 4:
CellProfiler improves the workflow for the analysis of MCAK/Kif2C cancer mutations. (A) A field of cells transfected with GFP-WT-MCAK/Kif2C. GFP, green; MTs, red; DNA, blue. (B) CellProfiler pipeline to analyze cytoplasmic tubulin levels in transfected cells. (C) Missense mutations and truncations mapped to full-length MCAK/Kif2C (cBioPortal). (D) Missense mutations that we engineered were biased toward hot spots (defined as more than two patients). (E) Proportion of MT-depolymerizing activity measured in engineered cancer mutations. N = successive 20× fields of cells containing between one and three transfected cells. Violin plots show the median activity (red line). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Supplemental Figure S3A compares the positive and negative control measurements between the manual and automated method. In the manual method, a transfected cell (either GFP or WT MCAK/Kif2C) is imaged for GFP and tubulin fluorescence. The tubulin fluorescence is also measured in an untransfected cell in the same field and used to calculate the “MT ratio,” which is the ratio of arbitrary tubulin fluorescence of the transfected cell divided by the reference cell. Because the reference cell must be of the same size and shape as the transfected cell, the workflow to collect the images is fairly slow. The advantage, however, is that a single image plane can be used for the fluorescence measurement. The level of GFP fluorescence is normalized at the time the images are acquired in the manual method. Importantly, the cells chosen to represent WT MCAK/Kif2C tubulin levels were manually restricted to only those cells in which all MT polymer is gone. This slows the workflow and requires selection by eye. In contrast, the automated method consists of imaging fields of cells for GFP, DNA, and tubulin. The tubulin fluorescence for both the negative (GFP) and positive (WT MCAK/Kif2C) controls are normalized to the negative control mean tubulin fluorescence without regard for the expression level of the transfected protein. The distribution of fluorescence measurements for the manual and automated analysis is remarkably similar for the GFP control measurements (Supplemental Figure S3A, black and blue). Importantly, and as expected, the distribution of the tubulin fluorescence in the automated measurements for cells transfected with WT MCAK/Kif2C exhibits a higher median, because cells expressing all levels of GFP-MCAK/Kif2C were included in the analysis. The disadvantage of this method is it afforded us a smaller range of fluorescence to measure partially active constructs (Supplemental Figure S3A, arrows) and a greater spread in the distribution of tubulin fluorescence for the positive control cells (Supplemental Figure S3A, light blue points). A key advantage is that the method requires no visual selection, is accordingly faster, and can include more cells without a significant increase in effort. We estimate that, using our manual method, analysis of one mutant construct requires a week of full-time effort (assuming all steps go well). In contrast, with Cell Profiler, the imaging and analysis of several constructs can be accomplished in a little less than 5 h: 4 h for the imaging (depending on the automation potential of the microscope) and about 15 min for CellProfiler analysis of the images (300 fields). Of course, this method also requires 2 or 3 h to create the original CellProfiler pipeline that will be use in multiple experiments. We used this new method to analyze MCAK/Kif2C mutations found in tumor samples isolated from cancer patients using the cBioPortal.
Figure 4C maps the point mutations found in 131 patient samples. We noted that there were a few hot spots for mutations in the motor domain and, we prepared constructs that would express these mutant versions of MCAK/Kif2C (Figure 4D). CellProfiler analysis of fields of cells transfected with these mutants revealed that slightly more than half of these mutants exhibited impaired MT-depolymerizing activity, while the rest were not significantly different from WT (Figure 4E). The CellProfiler analysis allows us to subtract the nucleus from the quantification and measure only the cytoplasm. Then, the entire data set is normalized to zero (the median of the tubulin values from GFP-transfected cells) and one (the median of the tubulin values from GFP-WT-MCAK/Kif2C–transfected cells). This method considerably increases the speed of the imaging and analysis, but it also increases the spread of the tubulin fluorescence measurements around the median (Figure 4E) relative to the more labor-intensive manual measurements (Figure 1B).
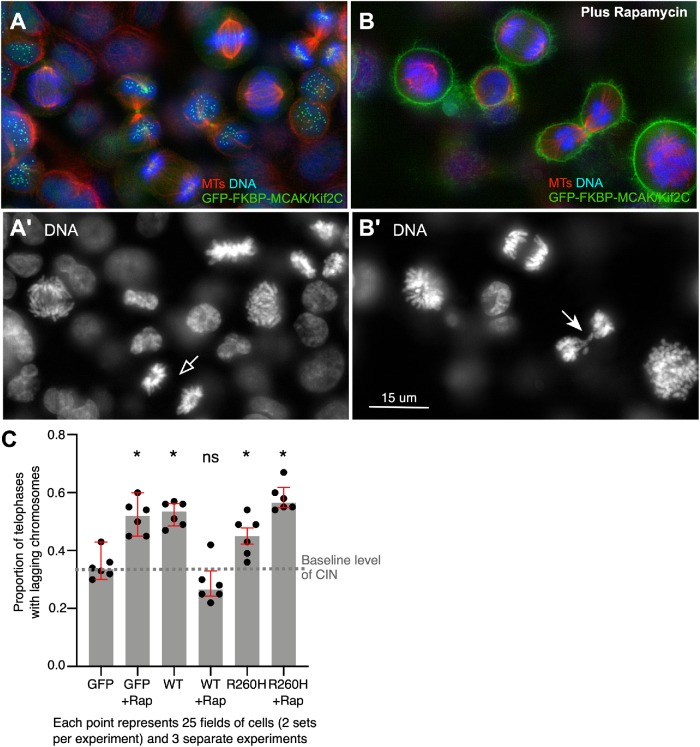
Because impaired activity would be expected to increase chromosome instability, we tested one of the more highly represented and severely impaired mutations (R260H) for its effect on the appearance of lagging chromosomes in telophase cells. We hypothesize that the highly inactive R260H mutant would exhibit impaired ATP binding, because histidine would be sterically incompatible with ATP binding (Supplemental Figure S3B). To test the effect of this mutant in dividing cells, we employed a GFP-FKBP-MCAK/Kif2C CRISPR-engineered transgenic cell line that enables us to rapidly remove MCAK/Kif2C from spindle structures and localize it to the cell membrane (Figure 5, A and B). Removal of MCAK/Kif2C from spindle structures in rapamycin results in an increase in lagging chromosomes in cells transfected with GFP alone (Figure 5C, GFP and GFP+Rap). This effect could be rescued by transfection of GFP-WT-MCAK/Kif2C, which will not relocalize in rapamycin (Figure 5C, WT+Rap). Surprisingly, in the absence of rapamycin, addition of excess GFP-MCAK/Kif2C led to an increase in lagging chromosomes. Transfection of inactive R260H into cells resulted in increased lagging chromosomes that was further exacerbated with the removal of endogenous GFP-FKBP-MCAK/Kif2C possessing normal MT-depolymerizing activity (Figure 5C, R260H and R260H+Rap). These data lead us to hypothesize that both amplifications and mutations in MCAK/Kif2C have the potential to increase chromosome instability in tumors, although we do not, at this time, know the background within which this activity operates in patient cells.
FIGURE 5:
MCAK/Kif2C levels must be tuned to suppress chromosome instability. (A) A field of CRISPR cells expressing endogenous levels of GFP-FKPB-MCAK/Kif2C (green) and labeled for tubulin (red) and DNA (blue). (A′) DNA only showing late anaphase cell (open arrow). (B) A field of CRISPR cells expressing endogenous levels of GFP-FKPB-MCAK/Kif2C (green) and labeled for tubulin (red) and DNA (blue) in 200 nM rapamycin. (B′) DNA only showing a telophase cell with lagging chromosomes (arrow). (C) Relocalization of endogenous GFP-FKBP-MCAK increases the proportion of lagging chromosomes in GFP-transfected control cells. This can be rescued by cotransfection of GFP-MCAK/Kif2C, but this operation increases lagging chromosomes when performed in a background of properly localized endogenous protein. The inactive R260H mutant increases chromosome instability regardless of the status of endogenous protein. Each point represents measurements for 25 fields of cells. Medians and interquartile range (red bars) are plotted. Significance is calculated from the median of the points using the one-sample t and Wilcoxon’s tests of significant difference from the baseline level of chromosome instability (CIN); *p < 0.05.
DISCUSSION
We previously developed a method to manually quantify the MT depolymerization activity of MCAK/Kif2C in live cells (Ovechkina et al., 2002). Using this manual assay, we interrogated a collection of point mutations within the motor domain of Kif2C/MCAK. Several of our point mutations (R534A, K537A, H530A) were shown by Woehlke et al. (1997) to strongly reduce MT affinity when equivalent residues were mutated in motile kinesin-1, Kif5B. Mutant R543 is conserved in the kinesin-13 family and also likely to contribute to MT affinity. Loop-8 has been postulated to serve as a sensor for the different nucleotide states of kinesin-13s to form a warped surface along the core MT binding surface (Ogawa et al., 2004, 2017). This might explain the reduced activity of most of the beta-5/loop-8 mutants. Mutations within the Switch II domain (G489A, E491A, E491R) correspond, respectively, to the pre- and post-powerstroke mutations in kinesin-1 (Rice et al., 1999). In kinesin-13s, they are quite interesting, as the E491 mutants are able to detach tubulin from MT but unable to free themselves from the detached tubulin dimer to continue their depolymerization cycle (Wagenbach et al., 2008). The slightly impaired R450A mutant may have slowed ATP hydrolysis as predicted by the analogous mutation in Kar3 (Yun et al., 2001). Finally, the N511K mutation has been shown to uncouple ATP hydrolysis from MT-dependent motility in Ncd and Kar3 (Song and Endow, 1998; Yun et al., 2001, 2003). The thinking is that this residue may be crucial to transmit the state of MT binding to the ATPase. These studies, which were performed using motile kinesins, also strongly impact the MT-depolymerizing kinesin MCAK/Kif2C.
The manual assay used to quantify the different MCAK/Kif2C mutants for activity is very reproducible, but it is labor intensive. We improved the speed and workflow of this assay by employing CellProfiler pipelines to measure cytoplasmic tubulin fluorescence in cells transfected with GFP-MCAK/Kif2C displaying mutations catalogued from cancer patients. When we tested a number of point mutations associated with cancer patients, we found that most were either deleterious or not significantly altered in activity. Interestingly, two hot spots (meaning that the same mutation was found in more than two patients) were significantly inhibited. We tested one of these and found it increased the proportion of lagging chromosomes whether endogenous WT protein was present or absent. Zygosity data were not available for all the mutants that we tested, but where they were, the mutant was heterozygous. We have previously found that exogenously expressed MCAK/Kif2C, which is a homodimer, will heterodimerize with endogenous protein (Maney et al., 2001) and that monomeric MCAK/Kif2C is impaired in tubulin dimer removal from MTs (Cooper et al., 2010); thus it is likely that heterozygotic cells in which one allele of MCAK/Kif2C is impaired will exhibit some level of impaired MT depolymerization activity. Interestingly, we also saw increased chromosome instability in cells expressing GFP-WT-MCAK/Kif2C over endogenous protein, although the excess protein could rescue chromosome instability when endogenous protein is removed. This suggests that MCAK/Kif2C protein level and activity must be at the correct intermediate level for proper chromosome segregation.
However, one thing that we cannot test at this time is how the sum total effect of background genetic alterations in patient tumors affect MT dynamics and how alterations of MCAK would play into this. For example, exogenously expressed MCAK/Kif2C can rescue chromosome instability induced by other genetic lesions that impact MT dynamics such as BRCA1 knockdown (Stolz et al., 2010). For tumors with amplified MCAK/Kif2C genes, we do not know whether the amplification contributes compensatory activity to lower chromosome instability in the face of other genetic alterations or, alternatively, whether it contributes to the overall level of genetic instability in those tumors. Regardless, high-throughput assays such as this will prove useful in the future for evaluating MT regulators, drivers that impact MT pathways, and drugs directed at MT regulators.
MATERIALS AND METHODS
Constructs and cell lines
GFP-WT-MCAK in pEGFP-C1 was expressed as described (Ovechkina et al., 2002). Point mutations were introduced either by the method of Kunkel (1985) or by overlapping PCR and confirmed by DNA sequencing. Genomic EGFP-FKBP fusion to the MCAK/Kif2C gene was constructed using CRISPR Cas9 cleavage and homology-directed repair (HDR). Chromosomal sequences were analyzed with the program CRISPRdirect (http://crispr.dbcls.jp) to identify target sites. Custom oligonucleotides encoding CRISPR target sites (IDT, Skokie, IL) were ligated to pX459 version 2 (AddGene, Boston, MA) to create the CRISPR targeting vector. Synthetic HDR template DNA (BioBasic, Montreal, QC, Canada) was made containing the first exon of the MCAK/Kif2C gene fused to fluorescent protein and FKBP sequences, flanked by ∼600 base pairs of genomic sequence from each side of the exon. HCT116 cells were cotransfected with linearized HDR template and a CRISPR targeting plasmid and then treated from 24 to 72 h posttransfection with 0.4 μg/ml puromycin to transiently select cells containing the targeting vector. Surviving cells containing fluorescent fusion proteins were selected, grown to homogeneous populations, and analyzed by PCR of genomic DNA and Western blots to identify populations in which both alleles of the locus were recombinant. Cells were subsequently transfected with FRB-BFP (Wordeman et al., 2016), and stably integrated cell lines were selected with G418.
Cell transfection and immunofluorescence
CHO AA8 Tet-OffTM cells (Clontech Laboratories) were grown in 90% alpha-MEM, 10% FBS, and 100 μg/ml G418 at 37°C, 5% CO2. Cells were removed from plates using 5% trypsin-EDTA and transfected with constructs using a Nuclofector II (Lonza). Cells were then plated overnight and fixed the following day in either 1% paraformaldehyde (PFA) in precooled methanol for 10 min (motor mutants) or 3% PFA in 37°C phosphate-buffered saline (PBS) for 10 min. The cells were then incubated with mouse anti–tubulin DM1alpha (Sigma-Aldrich) at either 1:50 dilution for 1 h or 1:100 overnight at 4°C and either Texas red anti–mouse antibodies (Jackson ImmunoResearch Laboratories) at 1:100 dilution in PBS or Alexa Fluor 560 anti–mouse antibodies (Jackson ImmunoResearch Laboratories) plus 0.1% Triton X-100 and 1% bovine serum albumin for 1 h.
Quantitation of MT depolymerization activity in vivo
Imaging and manual quantification of activity has been described (Ovechkina et al., 2002). For the automated analysis, successive fields of cells were imaged for nuclei (4′,6-diamidino-2-phenylindole), MTs, and GFP. A consistent series of twelve 0.5-micron z-planes was imaged using a DeltaVision system (GE Healthcare, Issaquah, WA) equipped with either a CoolSNAP HQCCD camera (Photometrics, Tucson, AZ) or an sCMOS camera (Photometrics,Tucson, AZ), SoftWorx software (GE Healthcare, Issaquah, WA), and a 25×/0.75 NA objective (Olympus Tokyo, Japan). Summed stacks were analyzed using CellProfiler in which primary objects (nuclei, identified as 100–500 pixels in diameter discarded from the borders) were subtracted from secondary objects (GFP identified using propagation, global threshold by minimum cross entropy with a threshold correction factor of 1.0, no smoothing, lower and upper bounds between 0.0 and 1.0 on threshold, a regularization factor of 0.05, and holes filled in identified objects). The remaining tertiary objects (cytoplasmic MTs) were scored for mean fluorescence intensity. Successive fields of cells, in which each field possessed between one and three transfected cells (N for automated CellProfiler analysis; Figure 4E) or individually measured single cells (N for manual analysis; Figure 1B) were plotted using Prism 8 (GraphPad) as violin plots showing median (red) and interquartile range (dotted). Mutants were tested for significant differences from the median of cells transfected with WT MCAK/Kif2C using the one-sample t and Wilcoxon’s tests. All those scored as significant were between a 95 and a 97% confidence interval. Positions of the residues that significantly affect MCAK/Kif2C activity were modeled using Chimera v. 1.13.1 (Pettersen et al., 2004).
Rescue of rapamycin-dependent MCAK/Kif2C delocalization
GFP-FKBP-MCAK/Kif2C CRISPR cells were transfected with GFP, GFP-MCAK/Kif2C, or GFP-R260H and plated onto two 18-mm2 coverslips for each treatment. One set was placed in 200 nM rapamycin and one in dimethyl sulfoxide (DMSO). After 16 h, the coverslips were fixed and labeled for DNA and MTs as described earlier. For each repetition of the experiment, 50 fields of cells were imaged at 20× and twelve 0.5-micron z-planes. Transfected telophases were identified and scored for the presence or absence of lagging chromosomes. Three separate experiments were performed in this way and analyzed in two sets of imaging on different days resulting in six data points that were mean proportions of transfected telophases exhibiting lagging chromosomes for 50 successive fields of cells. Significant differences of the median of the experiments of the proportion of telophases with lagging chromosomes from the GFP-transfected control cells not exposed to rapamycin were tested using the one-sample t and Wilcoxon’s tests. All those scored as significant were at the 97% confidence interval.
Supplementary Material
Acknowledgments
We thank Heather Dungan for assistance with mutant data analysis, Donelson Smith for assistance in the preparation of the CRISPR cell line, and Justin Decarreau for many helpful discussions This work was supported by National Institute of General Medical Sciences grant GM069429.
Abbreviations used:
- CIN
chromosome instability
- DMSO
dimethyl sulfoxide
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- HDR
homology-directed repair
- MT
microtubule
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-09-0503) on November 20, 2019.
REFERENCES
- Bai Y, Xiong L, Zhu M, Yang Z, Zhao J, Tang H. (2019). Co-expression network analysis identified KIF2C in association with progression and prognosis in lung adenocarcinoma. Cancer Biomark , 371–382. [DOI] [PubMed] [Google Scholar]
- Bakhoum SF, Genovese G, Compton DA. (2009). Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol , 1937–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Mei J, Xiao Z, Xu B, Jiang X, Zhang Y, Zhu Y. (2019). Identification of five hub genes as monitoring biomarkers for breast cancer metastasis in silico. Hereditas , 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov , 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li S, Zhou S, Cao S, Lou Y, Shen H, Yin J, Li G. (2017). Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma. J Cancer Res Ther , 651–659. [DOI] [PubMed] [Google Scholar]
- Cooper JR, Wagenbach M, Asbury CL, Wordeman L. (2010). Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat Struct Mol Biol , 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domnitz SB, Wagenbach M, Decarreau J, Wordeman L. (2012). MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochore attachment. J Cell Biol , 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Zhang X, Wang FX, Cai MY, Ma GW, Yang H, Fu JH, Tan ZH, Fu XY, Ma QL, et al (2016). KIF-2C expression is correlated with poor prognosis of operable esophageal squamous cell carcinoma male patients. Oncotarget , 80493–80507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertych N, Stolz A, Stenzinger A, Weichert W, Kaulfuss S, Burfeind P, Aigner A, Wordeman L, Bastians H. (2014). Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat Cell Biol , 779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friel CT, Howard J. (2011). The kinesin-13 MCAK has an unconventional ATPase cycle adapted for microtubule depolymerization. EMBO J , 3928–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H, Lin L, Hu N, Yang Y, Gao Y, Pei Y, Chen K, Sun B. (2019). KIF2C exerts an oncogenic role in nonsmall cell lung cancer and is negatively regulated by miR-325–3p. Cell Biochem Funct , 424–431. [DOI] [PubMed] [Google Scholar]
- Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. (2011). Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell , 1092–1103. [DOI] [PubMed] [Google Scholar]
- Gasic I, Boswell SA, Mitchison TJ. (2019). Tubulin mRNA stability is sensitive to change in microtubule dynamics caused by multiple physiological and toxic cues. PLoS Biol , e3000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. (2006). The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature , 115–119. [DOI] [PubMed] [Google Scholar]
- Hess JM, Bernards A, Kim J, Miller M, Taylor-Weiner A, Haradhvala NJ, Lawrence MS, Getz G. (2019). Passenger hotspot mutations in cancer. Cancer Cell , 288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonowska K, Czubak K, Wojciechowska M, Handschuh L, Zmienko A, Figlerowicz M, Dams-Kozlowska H, Kozlowski P. (2016). Oncogenomic portals for the visualization and analysis of genome-wide cancer data. Oncotarget , 176–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. (1985). Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA , 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T, Wagenbach M, Wordeman L. (2001). Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J Biol Chem , 34753–34758. [DOI] [PubMed] [Google Scholar]
- Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. (2007). The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell , 2970–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, et al (2018). CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol , e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro Gouveia S, Leslie K, Kapitein LC, Buey RM, Grigoriev I, Wagenbach M, Smal I, Meijering E, Hoogenraad CC, Wordeman L, et al (2010). In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr Biol , 1717–1722. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Nitta R, Okada Y, Hirokawa N. (2004). A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell , 591–602. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Saijo S, Shimizu N, Jiang X, Hirokawa N. (2017). Mechanism of catalytic microtubule depolymerization via KIF2-tubulin transitional conformation. Cell Rep , 2626–2638. [DOI] [PubMed] [Google Scholar]
- Ovechkina Y, Wagenbach M, Wordeman L. (2002). K-loop insertion restores microtubule depolymerizing activity of a neckless MCAK mutant. J Cell Biol , 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachter JS, Yen TJ, Cleveland DW. (1987). Autoregulation of tubulin expression is achieved through specific degradation of polysomal tubulin mRNAs. Cell , 283–292. [DOI] [PubMed] [Google Scholar]
- Pan S, Zhan Y, Chen X, Wu B, Liu B. (2019). Identification of biomarkers for controlling cancer stem cell characteristics in bladder cancer by network analysis of transcriptome data stemness indices. Front Oncol , 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker AL, Teo WS, Pandzic E, Vicente JJ, McCarroll JA, Wordeman L, Kavallaris M. (2018). β-Tubulin carboxy-terminal tails exhibit isotype-specific effects on microtubule dynamics in human gene-edited cells. Life Sci Alliance , e201800059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem , 1605–1612. [DOI] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, et al (1999). A structural change in the kinesin motor protein that drives motility. Nature , 778–784. [DOI] [PubMed] [Google Scholar]
- Song H, Endow SA. (1998). Decoupling of nucleotide- and microtubule-binding sites in a kinesin mutant. Nature , 587–590. [DOI] [PubMed] [Google Scholar]
- Stolz A, Ertych N, Bastians H. (2010). Loss of the tumour-suppressor genes CHK2 and BRCA1 results in chromosomal instability. Biochem Soc Trans , 1704–1708. [DOI] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al (2019). COSMIC: the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res , D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, et al (2018). Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell , 676–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam PA, Rolfo C, Ruiz R, Pauwels P, Van Berckelaer C, Trinh XB, Ferri Gandia J, Bogers JP, Van Laere S. (2018). Potential new biomarkers for squamous carcinoma of the uterine cervix. ESMO Open , e000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenbach M, Domnitz S, Wordeman L, Cooper J. (2008). A kinesin-13 mutant catalytically depolymerizes microtubules in ADP. J Cell Biol , 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD. (1997). Microtubule interaction site of the kinesin motor. Cell , 207–216. [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al (2007). The genomic landscapes of human breast and colorectal cancers. Science , 1108–1113. [DOI] [PubMed] [Google Scholar]
- Wordeman L, Decarreau J, Vicente JJ, Wagenbach M. (2016). Divergent microtubule assembly rates after short- versus long-term loss of end-modulating kinesins. Mol Biol Cell , 1300–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L, Wagenbach M, Maney T. (1999). Mutations in the ATP-binding domain affect the subcellular distribution of mitotic centromere-associated kinesin (MCAK). Cell Biol Int , 275–286. [DOI] [PubMed] [Google Scholar]
- Wordeman L, Wagenbach M, von Dassow G. (2007). MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J Cell Biol , 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M, Bronner CE, Park CG, Cha SS, Park HW, Endow SA. (2003). Rotation of the stalk/neck and one head in a new crystal structure of the kinesin motor protein, Ncd. EMBO J , 5382–5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M, Zhang X, Park CG, Park HW, Endow SA. (2001). A structural pathway for activation of the kinesin motor ATPase. EMBO J , 2611–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.