Abstract
A myriad of transient, nanoscopic lipid- and protein-based interactions confer a steady-state organization of the plasma membrane in resting cells that is poised to orchestrate assembly of key signaling components upon reception of an extracellular stimulus. Although difficult to observe directly in live cells, these subtle interactions can be discerned by their impact on the diffusion of membrane constituents. Here, we quantified the diffusion properties of a panel of structurally distinct lipid, lipid-anchored, and transmembrane (TM) probes in RBL mast cells by imaging fluorescence correlation spectroscopy (ImFCS). We developed a statistical analysis of data combined from many pixels over multiple cells to characterize differences in diffusion coefficients as small as 10%, which reflect differences in underlying interactions. We found that the distinctive diffusion properties of lipid probes can be explained by their dynamic partitioning into Lo-like proteolipid nanodomains, which encompass a major fraction of the membrane and whose physical properties are influenced by actin polymerization. Effects on diffusion of functional protein modules in both lipid-anchored and TM probes reflect additional complexity in steady state membrane organization. The contrast we observe between different probes diffusing through the same membrane milieu represents the dynamic resting steady state, which serves as a baseline for monitoring plasma membrane remodeling that occurs upon stimulation.
INTRODUCTION
Cells typically exist in noisy environments, and their plasma membranes are predisposed to respond optimally to external chemical and physical stimuli, including specific chemical ligands (Simons and Sampaio, 2011), thermal shock (Tsvetkova et al., 2002), and electrical and mechanical forces (Akinlaja and Sachs, 1998; Verstraeten et al., 2010). For versatile and efficient responses, the membrane accommodates receptors and other structures that sense the external stimuli, as well as organizing surrounding lipids and proteins (Nicolson, 2014). Many of the underlying interactions are cooperative and weak, providing a dynamic steady state platform that has the capacity to respond to a specific stimulus over environmental noise and to regulate transmembrane signaling components (Brown, 2006; Trimble and Grinstein, 2015). Key to responsive membrane organization are structural configurations that can be modulated to selectively include/exclude other components. A prominent example is “lipid rafts,” an ill-defined term for dynamic nanodomains comprising proteins and lipids that resemble the liquid ordered (Lo) phase in model membranes (Goni, 2019). Although there is ample experimental support for participation of these Lo-like proteolipid nanodomains (the term we use here as our definition of “rafts”) in stimulated signaling (Brown, 2006; He and Marguet, 2008; Simons and Gerl, 2010; Kusumi et al., 2012b; Honigmann and Pralle, 2016; Sezgin et al., 2017), their physical nature has been difficult to define because of their diversity, their subresolution dimensions, and their transience (Eggeling et al., 2009; Sahl et al., 2010; Honigmann and Pralle, 2016; Sezgin et al., 2017; Winkler et al., 2017; Lyman et al., 2018; Nicovich et al., 2018; Raghunathan and Kenworthy, 2018; Kusumi et al., 2019). Over the years we have used a wide range of approaches to examine these nanodomains because of their clear participation in transmembrane signaling initiated by the high affinity receptor (FcεRI) for immunoglobulin E (IgE) on mast cells, as triggered by multivalent antigen in the allergic immune response (Holowka et al., 2005). Cells are sensitized to antigen when IgE antibodies bind to FcεRI, which diffuse as monomeric species in the plasma membrane (Menon et al., 1986; Shelby et al., 2013). Cell activation occurs only after addition of antigen, which cross-links the IgE-FcεRI to stabilize their association with Lo-like proteolipid nanodomains and consequently their functional coupling with Lyn tyrosine kinase. The stimulatory event depends on shifting the balance of phosphorylation and dephosphorylation of cross-linked IgE-FcεRI toward phosphorylation, leading to downstream signaling. This is facilitated by the capacity of the nanodomains to preferentially include the key kinase (Lyn), which is anchored to the inner leaflet by Lo-preferring fatty acid chains, and exclude a transmembrane phosphatase, which is accommodated more favorably in a liquid disordered (Ld)-like environment (Holowka and Baird, 2016). This example illustrates how the plasma membrane is predisposed or “poised” to respond to a specific stimulus by the steady state presence of Lo-like proteolipid nanodomains that are dynamic in nature but are stabilized and utilized when the stimulus arrives.
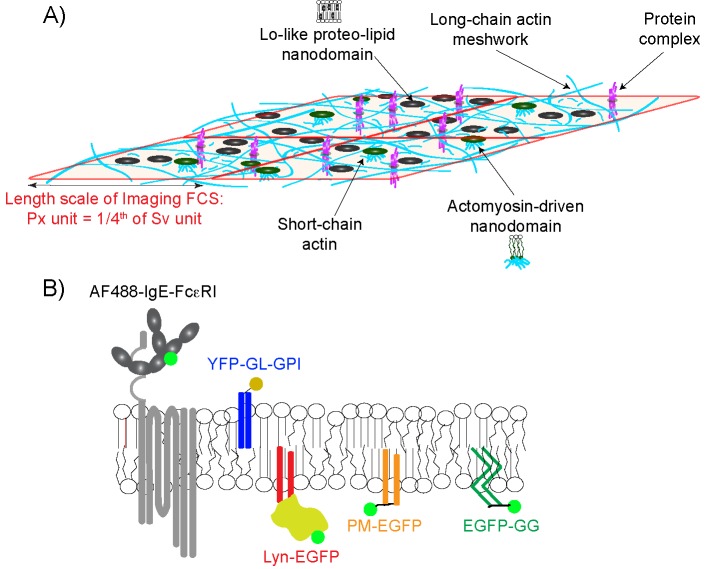
Coexistence of membrane structures that facilitate spatial compartmentalization underlies the “hierarchal model” proposed by Kusumi and colleagues, based primarily on their extensive ultra-high-speed single particle tracking (SPT) and scanning electron microscopy measurements, with additional features drawn from complementary studies in other laboratories (Figure 1A; Wieser et al., 2007; Kusumi et al., 2011, 2012a; Andrade et al., 2015; Sadegh et al., 2017; Chein et al., 2019). The hierarchal model builds on membrane compartments (corrals; 40–230 nm) defined by the long-chain actin meshwork (fence) with anchored transmembrane proteins (pickets). Importantly, such actin-based compartmentalization imposes fundamental membrane organization by preventing liquid ordered/liquid disordered (Lo/Ld) lipid phase separation (Machta et al., 2011; Gomez-Llobregat et al., 2013; Honigmann et al., 2014b; Vogel et al., 2017). Considerable evidence supports the view that nanoscale Lo-like channels align along the picket fences, either by the effects of critical behavior and pinned Lo-preferring components (Ehrig et al., 2011; Machta et al., 2011; Honigmann et al., 2014b) or by stabilization with Lo-preferring protein pickets (Dinic et al., 2013; Huang et al., 2015). In the hierarchical model, Lo-like nanodomains (2–20 nm) and protein complexes (3–10 nm) also exist within the corrals. As a further refinement, experiments and simulations underlying the “active-composite model” of Mayor, Rao, and colleagues showed that Lo-like nanodomains also arise from active myosin-driven asters of short actin chains that connect to inner leaflet lipids and cause alignment of tails of Lo-preferring lipids in the outer leaflet (Rao and Mayor, 2014; Raghupathy et al., 2015; Koster and Mayor, 2016).
FIGURE 1:
A composite of plasma membrane organization may be determined by monitoring the diffusion of structurally distinct probes. (A) The plasma membrane is organized at different length scales in a hierarchical scheme: a relatively static actin meshwork (cyan long chains); dynamic Lo-like proteolipid nanodomains (black circles) with variable physical properties within and across leaflets; transmembrane ordered lipids mediated by dynamic, myosin-driven assembly of short actin chains (green circles connected to short cyan chains); and stable or dynamic protein complexes. We note that Lo-like proteolipid nanodomains and protein complexes are much smaller than the dimensions of the actin meshwork and are not drawn to scale here. Interaction of a probe with these organizational features retards its diffusion, depending on that probe’s physicochemical properties. ImFCS measures diffusion coefficients (D) at the resolution of a Px unit (320 × 320 nm2), which has dimensions considerably larger than the actin meshwork; parameters τ0 and 1/Deff = Slope are measured in Sv units that comprise a square of 16 Px units. These measurements are illustrated in Figure 2. (B) Fluorescent lipid, lipid-anchored, and transmembrane (TM) probes are evaluated by ImFCS. AF488-IgE-FcεRI: transmembrane with seven TM regions; YFP-GL-GPI: outer-leaflet lipid probe with saturated acyl chain anchor and an extracellular consensus glycosylation site; Lyn-EGFP: inner leaflet probe with saturated acyl chain anchors and additional cytosolic protein modules; PM-EGFP: inner leaflet lipid probe with same saturated acyl chain anchors of Lyn-EGFP; EGFP-GG: inner leaflet lipid probe with unsaturated acyl chain anchors and membrane-proximal basic sequence. Previous studies showed that lipid probes YFP-GL-GPI, PM-EGFP dynamically partition into Lo-like proteolipid nanodomains, whereas EGFP-GG partitions less favorably into these nanodomains, preferring Ld-like regions.
Direct imaging of dynamic plasma membrane heterogeneity at the nanoscale is challenging, even with superresolution optical microcopy (Klotzsch and Schutz, 2013; van Zanten and Mayor, 2015). Fluorescence spectroscopy, which is often coupled with diffraction-limited microcopy, offers new possibilities for extracting dynamic properties of plasma membranes (Sarkar and Chattopadhyay, 2019). As described here, we employed imaging fluorescence correlation spectroscopy (ImFCS; Kannan et al., 2007), a camera-based modality of FCS (Magde et al., 1972), to examine the effects of membrane organization on diffusion of individual membrane components, based on the premise that local structures, mediated by proteins or lipids or both, curtail lateral diffusion differentially (Kenworthy et al., 2004; Kusumi et al., 2010; Klotzsch and Schutz, 2013). Within the refined hierarchical model described above, we take the basic view that the plasma membrane comprises Lo-like proteolipid nanodomains (along and within the boundaries of corrals) connected by Ld-like regions, and that this organization is modulated by long- and short-chain actin (Figure 1A). With ImFCS, we can quantify the diffusion of multiple, diverse probes that differentially interact with the constituents within the same plasma membrane and integrate these distinctive diffusion behaviors to create a composite picture of plasma membrane organization.
In contrast to conventional single-spot FCS measurements, the family of image-based fluctuation methods offers various kinds of spatial analysis in addition to evaluation of diffusion coefficients. The spatiotemporal information obtained from these methods depends on the resolution of the microscope, the scanning configuration, and the modes of fluctuation analysis (Digman and Gratton, 2011; Wiseman, 2012; Bag and Wohland, 2014). The ImFCS we employ was developed as an ensemble-averaged but single molecule–sensitive technique that provides a pixelated map of membrane diffusion properties (Kannan et al., 2007; Krieger et al., 2015). A continuous series of total internal reflection fluorescence microscopy (TIRFM) images of the ventral plane of fluorescently labeled live cells is captured by a fast, sensitive camera, which spatially divides the image into an array of submicrometer pixels (Bag and Wohland, 2014; Krieger et al., 2015). Each pixel of the camera corresponds to a diffraction-limited membrane spot. Autocorrelation function (ACF) analysis of temporal fluorescence fluctuations of each pixel yields a macroscopic Brownian diffusion coefficient (D) at that pixel. ImFCS data acquisition for a single cell typically contains hundreds of pixels, so that hundreds of parallel FCS experiments are carried out on that cell. When the pixel measurements for a single cell, or for multiple cells, can be combined into an ensemble, ImFCS can deliver much more robust estimates of diffusion coefficients than conventional FCS that is based on a single illumination volume. ImFCS addresses limitations of spatial resolution using spot variation FCS (svFCS), as developed by Lenne and colleagues for conventional FCS, which indirectly detects the existence of subresolution regions of confined diffusion (Wawrezinieck et al., 2005; He and Marguet, 2011). In an ImFCS experiment, the fluorescence fluctuations collected for each pixel can be used directly to perform svFCS analysis because pixel binning (i.e., summing over adjacent pixels) effectively generates spot areas of variable sizes (Bag et al., 2012). Combining inherent multiplexing capacity, straightforward implementation, and compatibility with a conventional live cell–imaging platform, ImFCS offers enhanced capabilities for directly evaluating macroscopic diffusion properties and indirectly assessing the possible influence of subresolution domains of confinement (Krieger et al., 2015).
Our goal in this study was to gain detailed knowledge of spatiotemporal organization in the “resting” steady state of the plasma membrane. We extended the capabilities of ImFCS by developing a straightforward statistical analysis to provide both spatial maps and highly precise values for diffusion coefficients and nanoscale confinement of membrane constituents. To achieve a composite picture, we comparatively evaluated the diffusion properties of a panel of well-established probes whose diffusion was modulated by distinctive physicochemical interactions with structural features in plasma membrane milieu of RBL cells (Figure 1B). Using fluorescent protein constructs and selected membrane anchors, we evaluated Lo-preferring lipids (palmitoyl–myristoyl [PM] and glycosylphosphatidylinositol (GPI]) and Ld-preferring lipid (geranyl–geranyl [GG]) in inner (PM, GG) and outer (GPI) leaflets of the plasma membrane. We also examined the diffusion properties of Lyn kinase, a 40-kDa protein that is anchored to the inner leaflet by PM chains as well as IgE–FcεRI, which we compared with other transmembrane proteins. Leveraging the unprecedented statistics offered by ImFCS, we detect distinctive populations of diffusants for each of the probes tested, and these characterize the micro- and nanoscopic membrane regions through which the probes travel. With a primary focus on the inner leaflet probes, our data also provide strong evidence for previous indications that Lo-like regions are the major component of RBL plasma membranes in the resting steady state. We further provide supporting evidence that filamentous actin regulates the membrane organization, as reflected by changes in the diffusion properties of membrane constituents.
RESULTS
Statistically robust analyses of ImFCS data at Px unit and Sv unit length scales quantify probe diffusion and thereby reveal subtle plasma membrane heterogeneity
In a typical ImFCS recording, the ventral plasma membrane is segmented into an array of Px units by the EMCCD camera chip (1 Px unit = 320 × 320 nm2; Figures 1A and 2A). In the context of the hierarchical model (Introduction; Figure 1A), each Px unit includes many protein complexes, Lo-like proteolipid nanodomains, and multiple corrals (Fujiwara et al., 2016). All of these heterogeneity features, even if nanoscopic and transient, may influence probe diffusion at the Px unit length scale. Among the membrane probes evaluated in our study (Figure 1B), lipid probes (PM-EGFP, EGFP-GG, YFP-GL-GPI) primarily undergo lipid-based interactions, but the extent of their partitioning into Lo-like proteolipid nanodomains differs (Baumgart et al., 2007; Sengupta et al., 2008). If a lipid-anchored probe also contains protein modules (e.g., Lyn-EGFP), these may contribute additionally to the level of confinement within nanodomains as well as to protein-based interactions outside. The case of transmembrane (TM) probes (e.g., AF488-IgE-FcεRI) is more complicated, as these may have various protein-based interactions in both leaflets (Trimble and Grinstein, 2015) and may be surrounded by a lipid “shell” that further affects diffusion and partitioning (Anderson and Jacobson, 2002; Jacobson et al., 2007; Corradi et al., 2018).
FIGURE 2:
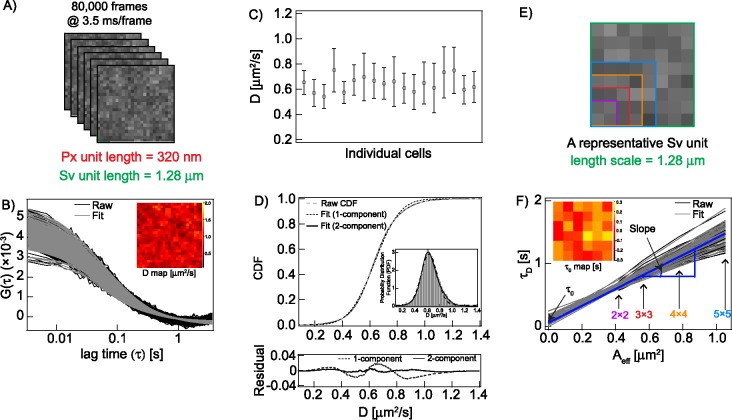
Very large data sets from ImFCS measurements are analyzed to examine spatially heterogeneous diffusion properties of plasma membrane probes, as exemplified by EGFP-GG. (A) A typical ImFCS measurement records image stack of 80,000 frames (3.5 ms/frame) from a region of interest (ROI) on the ventral plasma membrane. Representative first few images of 2 × 2 binned pixels (Px units) are shown. Representative Px unit (red box: 320 × 320 nm2) and Sv unit (green box: 1.28 × 1.28 μm2) in the image stack are shown. (B) Raw autocorrelation functions (ACFs; black) and respective fits (gray) for each Px unit using a single-component Brownian diffusion model (Eq. 1). Inset: spatial map of extracted diffusion coefficient (D) values at each Px unit of the image stack represented in A. (C) D values averaged over ROIs in 18 individual RBL cells expressing EGFP-GG; error bars are standard deviations about an average D for each cell. ROIs generally contain 400–625 Px units covering about 41–64 μm2 membrane area for each cell, yielding 10,527 total D values for 18 cells in this example. (D) Top: 10,527 D values obtained from ROIs in all cells are pooled and plotted as a normalized cumulative distribution function (CDF), which is fitted with one (Eq. 2) or two (Eq. 3) components, as indicated. The inset shows the same data for D plotted as a probability distribution function (PDF) with arbitrary binning of parameter values. Bottom: residual plots for one-component and two-component fits to CDF. (E) svFCS analysis is carried out on each Sv unit of the same raw data as depicted in A. A representative Sv unit is outlined in green, and four different sizes of observation area (Aeff) are created by pixel binning within the Sv unit (pink, red, orange, and blue boxes). (F) Linear svFCS plots (diffusion time [τD] vs. observation area [Aeff]; Eq. 5) are generated from all possible nonoverlapping Sv units (represented in A), yielding values for y-intercept (τ0) and Slope (1/Deff). Inset: spatial map of τ0 values determined from each Sv unit of an ROI represented in A.
We illustrate our ImFCS data analysis with EGFP-GG: enhanced green fluorescent protein (EGFP) tagged to a short amino acid sequence that includes a polybasic motif and an acylation site for unsaturated geranyl–geranyl (GG), causing its localization to the membrane inner leaflet (Pyenta et al., 2001). EGFP-GG is known to prefer Ld-like regions of the plasma membrane and partitions relatively weakly into Lo-like regions (Pyenta et al., 2001; Baumgart et al., 2007). In our experiments, time-dependent fluorescence fluctuations of diffusing EGFP-GG at each Px unit of an image series (80,000 frames and 3.5 ms/frame; Figure 2A) are autocorrelated to obtain individual raw ACFs. Although we expect features of membrane heterogeneity to cause anomalous diffusion (Bouchaud and Georges, 1990; Machta et al., 2011; Levental and Veatch, 2016), Eggeling et al. (2009) showed that this can be detected directly only at length scales well below 200 nm and that diffusion appears to be Brownian as the length scale approaches the diffraction limit of the microscope. Consistent with this observation, we found that our raw ACFs are well fit by a single-component Brownian diffusion model (Eq. 1). Figure 2B shows fitting of individual raw ACFs, from which a spatial map of the diffusion coefficient (D) for EGFP-GG is generated (Figure 2B, inset). The length scale of this D map is the Px unit (320 × 320 nm2).
The same raw data set can be used to evaluate temporal heterogeneity in the spatially resolved D maps in shorter time windows by dividing the entire raw image series into four equal 70-s segments and conducting ACF analysis on each segment. As shown in Supplemental Figure S1A (top), the D maps for EGFP-GG show moderate temporal fluctuations; that is, the values of each of these parameters for any given Px unit change somewhat across the time segments. However, there are no obvious regions of the maps that look distinctly different at this level of spatial resolution, and the D distributions across all pixels for each segment remain very similar (Supplemental Figure S1B, left). Therefore, we take the spatial heterogeneity of D values to represent the ensemble exhibited by the probe as it explores the plasma membrane of a cell.
The D values for 18 EGFP-GG–expressing RBL cells show little cell-to-cell variation (Figure 2C), which allowed us to combine data from multiple cells for further analysis (Figure 2D). The measured region of interest (ROI) for each cell includes 400-625 Px units, such that pooling all D values for these many cells gives adequate data statistics (>10,000 D values; Npx in Table 1) for EGFP-GG (or another probe) that is not biased from a subset of cells. The statistically robust arithmetic average of EGFP-GG (Table 1), Dav = 0.64 ± 0.002 μm2/s (mean ± SEM), is comparable to those obtained from other types of diffusion measurements (Pyenta et al., 2003; Edwald et al., 2014). The unusually small SEM reflects the high precision of these ImFCS measurements.
TABLE 1:
Parameters of diffusion properties for membrane probes shown in Figure 1B are determined by fitting very large ImFCS data sets with Eq. 3 (D values) or Eq. 5 (τ0 and Slope = 1/Deff).
| Probes | Membrane association | Treatment | Dfast (μm2/s)a | Dslow (μm2/s)a | Fslow | <DCDF> (μm2/s)b | Dav (μm2/s)c | τ0,av (s)c | Slopeav (s/μm2)c | NPx/Nsv (no. of cells)d |
| EGFP-GG | Lipid probe, Inner leaflet, Ld-preferring | No | 0.66 ± 0.19 | 0.61 ± 0.09 | 0.41 | 0.64 ± 0.12 | 0.64 ± 0.002 | 0.19 ± 0.005 | 1.18 ± 0.01 | 10,527/648 (18) |
| CytoD | 0.71 ± 0.24 | 0.66 ± 0.13 | 0.76 | 0.67 ± 0.15 | 0.67 ± 0.002 | 0.18 ± 0.005 | 1.14 ± 0.01 | 9366/539 (16) | ||
| PM-EGFP | Lipid probe, Inner leaflet, Lo-preferring | No | 0.67 ± 0.24 | 0.58 ± 0.13 | 0.63 | 0.61 ± 0.12 | 0.61± 0.002 | 0.27 ± 0.006 | 1.05 ± 0.02 | 9375/540 (15) |
| CytoD | 0.74 ± 0.19 | 0.56 ± 0.13 | 0.84 | 0.59 ± 0.11 | 0.59 ± 0.001 | 0.27 ± 0.002 | 1.15 ± 0.01 | 12,432/720 (20) | ||
| YFP-GL-GPI | Lipid probe, Outer leaflet, Lo-preferring | No | 0.40 ± 0.10 | 0.30 ± 0.06 | 0.72 | 0.33 ± 0.05 | 0.33 ± 0.001 | 0.72 ± 0.009 | 1.56 ± 0.02 | 12,892/719 (21) |
| CytoD | 0.36 ± 0.08 | 0.28 ± 0.05 | 0.76 | 0.30 ± 0.04 | 0.30 ± 0.001 | 0.73 ± 0.01 | 1.84 ± 0.02 | 11,074/661 (19) | ||
| Lyn-EGFP | Lipid-anchored probe, Inner leaflet, Lo-preferring |
No | 0.57 ± 0.15 | 0.45 ± 0.11 | 0.69 | 0.49 ± 0.09 | 0.49 ± 0.001 | 0.44 ± 0.009 | 1.13 ± 0.02 | 10,000/575 (16) |
| CytoD | 0.62 ± 0.17 | 0.44 ± 0.10 | 0.80 | 0.48 ± 0.09 | 0.48 ± 0.001 | 0.30 ± 0.006 | 1.55 ± 0.02 | 12,904/753 (21) | ||
| AF488-IgE-FcεRI | TM probe, 7 TMD |
No | 0.22 ± 0.07 | 0.14 ± 0.04 | 0.69 | 0.16 ± 0.03 | 0.17 ± 0.001 | 1.63 ± 0.01 | 2.56 ± 0.04 | 27,707/1644 (46) |
| CytoD | 0.23 ± 0.09 | 0.13 ± 0.03 | 0.63 | 0.17 ± 0.04 | 0.17 ± 0.001 | 1.93 ± 0.03 | 2.39 ± 0.07 | 10,624/558 (17) | ||
| YFP-GL-GT46 | TM probe, 1 TMD |
No | 0.32 ± 0.09 | 0.21 ± 0.04 | 0.76 | 0.24 ± 0.05 | 0.24 ± 0.001 | 1.09 ± 0.01 | 1.99 ± 0.04 | 6475/385 (12) |
| CytoD | — | |||||||||
| EGFR-EGFP | TM probe, 1 TMD |
No | 0.37 ± 0.11 | 0.24 ± 0.05 | 0.76 | 0.27 ± 0.04 | 0.27 ± 0.001 | 1.16 ± 0.02 | 1.23 ± 0.04 | 6440/384 (12) |
| CytoD | — | |||||||||
| AcGFP-Orai1 | TM probe, 4 TMD |
No | 0.18 ± 0.05 | 0.13 ± 0.03 | 0.70 | 0.15 ± 0.03 | 0.15 ± 0.001 | 2.56 ± 0.03 | 0.99 ± 0.04 | 6875/334 (11) |
| CytoD | — | |||||||||
TMD = transmembrane domain.
a± values are standard deviations of the fitted Gaussian distributions (Eq. 3).
b± values are weighted standard deviations calculated as the propagation of the errors of Dfast and Dslow.
c± values are standard error of the mean (SEM) for the arithmetic average.
dNPx = number of Px units at which D values are determined and Nsv = number of Sv units at which τ0 and slope values are determined.
We extract more detailed information about diffusion heterogeneity across Px units by compiling the pooled D values as a cumulative distribution function (CDF; Figure 2D, dashed line). The same data may be plotted as a probability distribution function (PDF; Figure 2D, inset), which allows easier visualization of underlying populations. CDFs, which are mathematically equivalent to PDFs but do not require arbitrary range binning of parameter values, are particularly useful for statistical analyses. We fitted the CDFs of D values with either one- or two-component Gaussian distribution models (Eqs. 2 and 3) and determined the best fit by comparing residuals and reduced chi-squared values (Figure 2D and Supplemental Figure S2). If the D CDF of a given probe has two components, then one corresponds to the population of Px units containing membrane features that interact more strongly with this probe, causing its slower diffusion. The second population is Px units exhibiting weaker interactions, such that this probe diffuses faster through these regions. In this case, the CDF fit parameters are reported as Dslow and Dfast, representing the average diffusion coefficients of probes moving through interaction-rich and interaction-poor Px units, respectively, in the plasma membrane. Fslow is the fraction of interaction-poor Px units; the fraction of interaction-rich Px units is Ffast = (1-Fslow). In the case of one-component CDF, the probe does not distinguish interaction-poor and interaction-rich Px units; that is, it undergoes a similar degree of interaction throughout the plasma membrane. In this case, Dfast = Dslow and Ffast = Fslow. We expand further upon the relationship between ImFCS diffusion parameters and constraining features within Px units in the Supplemental Appendix.
We find that the D CDF of EGFP-GG is fitted significantly better with a two-component than with a one-component Gaussian distribution model (Figure 2D). Because EGFP-GG is a lipid probe, its diffusion is most likely influenced by lipid-based interactions, primarily weak dynamic partitioning into Lo-like proteolipid nanodomains. Correspondingly, we interpret the two-component D CDFs as differences in nanodomain coverage among Px units that cause this probe’s diffusion properties to group into two populations. Although distinguishable, the values of the two D components do not differ by much for EGFP-GG (Table 1, Figure 3): Dslow = 0.61 ± 0.09 μm2/s (Fslow = 0.41) and Dfast = 0.66 ± 0.19 μm2/s (Ffast = 0.59), where the ± values are standard deviations (σ) of the fitted Gaussian distributions (Figure 2D; Eq. 3). As we demonstrate in the Supplemental Material (Supplemental Table S1 and Supplemental Figure S2), very large pooled data sets (NPx ∼ 10,000; Table 1) allow distinctions as small as 8% to be made for Dfast and Dslow components. The weighted average (Eq. 4), FfastDfast + FslowDslow = <DCDF> = 0.64 μm2/s, which is the same as Dav, the arithmetic average of the total pooled D values for EGFP-GG.
FIGURE 3:
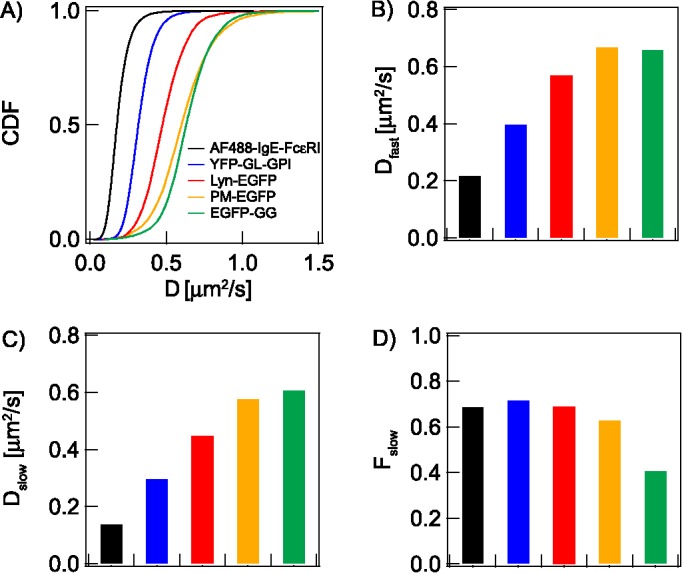
Diffusion parameters are determined from the statistical analyses of D CDF for lipid, lipid-anchored, and TM probes depicted in Figure 1B. (A) CDF of D values for indicated probes. Fitting of the respective CDFs yields: (B) Dfast: average diffusion coefficient of probes in Px unit population with less dynamic confinement (nanodomain-poor for lipid probes); (C) Dslow: average diffusion coefficient of probes in Px unit population with more dynamic confinement (nanodomain-rich for lipid probes); (D) Fslow: fraction of Px units exhibiting Dslow. Probes EGFP-GG, PM-EGFP, Lyn-EGFP, and YFP-GL-GPI are primarily subject to lipid-based interactions, whereas AF488-IgE-FcεRI (and other TM probes) depends on protein-based interactions. The color code in A identifies the probes in all panels. Numerical values of all parameters with defined errors are provided in Table 1. Because of very large data sets and robust statistics, all pairwise comparisons within each panel are significantly different (though the differences may be small): In B, PM-EGFP and EGFP-GG are different with p < 0.05, as determined by unpaired Student’s t test; all other comparisons within B and other panels A–D are different with p < 0.0001 (see Materials and Methods).
To explore the spatial distribution of slower and faster EGFP-GG diffusers and their connectivity, we made contour maps of D values on individual cells. Contour maps for one cell divided into 70-s time segments (as described above) confirm the dynamic heterogeneity of the plasma membrane as sensed by this probe (Supplemental Figure S3A). Contour maps of EGFP-GG D values determined from the standard (280-s) data acquisition period and compared for different single cells show diversity in distributions and connectivity, although the Dav values for each cell are very similar (Figure 2C and Supplemental Figure S3B).
Although the spatial scale of Px units precludes direct observation of Lo-like proteolipid nanodomains and other nanoscopic features that retard diffusion, their effects can be further quantified with svFCS carried out on the same data set for each probe (Introduction). For this spatial analysis, we define nonoverlapped groups of 8 × 8 pixels within the ROI (1 Sv unit = 1.28 × 1.28 μm2; Figure 2E). This is sufficient to create four observation areas of variable sizes (Aeff = 0.42–1.05 μm2, shown as colored boxes within the Sv unit in Figure 2E) by integrating the fluorescence signal from adjoining pixels in each size group and correlating the fluctuations (Bag et al., 2016). The resulting ACFs are fitted to obtain a single diffusion time (τD) for each Aeff, and the four points, τD versus Aeff, are fitted with a linear model, which assumes Brownian diffusion on this micrometer scale (Eq. 5). The constant Slope is the inverse effective diffusion coefficient (1/Deff) for each Sv unit, which is proportional to the apparent viscosity experienced by this probe on this length scale (Lenne et al., 2006; He and Marguet, 2011). Nanoscale information comes from extrapolation of τD versus Aeff to Aeff = 0, yielding a τ0 value for each Sv unit. The value of τ0 is expected to be zero for a probe that also diffuses freely on the nanoscale, and the degree to which τ0 is greater than zero provides a relative measure of confinement of that probe in domains on that length scale (much smaller than a Px unit). Repeating svFCS analysis over all possible nonoverlapping Sv units yields a τ0 map (Figures 2F, inset, and Supplemental Figure S1A) and a Slope = I/Deff map (Supplemental Figure S1A) for that ROI. Values of τ0 and Slope compiled over many cells can also be plotted as a CDF or PDF and fitted. However, the statistics are not as robust as the D values (∼36 Sv units/cell, ∼500 values for ∼15 cells), and we use the arithmetic averages τ0,av and Slopeav to compare probes. The averaged τ0 value for EGFP-GG (τ0,av = 0.19 ± 0.12 s) falls just at the point that can be distinguished from zero experimentally (0 < τ0 < 0.2 s; Ng et al., 2016), reflecting relatively little retardation by nanodomains. Together, the Dfast, Dslow, and τ0,av values for EGFP-GG reveal the presence of Lo-like nanodomains can be detected by our measurements. Ffast and Dfast represent the larger of two populations of Px units that are sensed by this probe as nanodomain-poor, further consistent with EGFP-GG partitioning relatively weakly into nanodomains. Diffusion parameters determined from Px units and Sv units for lipid probe EGFP-GG and all probes evaluated in this study are compiled in Table 1.
EGFP-GG, PM-EGFP, and Lyn-EGFP diffuse differentially in the membrane inner leaflet
Lyn kinase, a protein anchored to the inner leaflet of the plasma membrane by saturated palmitoyl (P) and myristoyl (M) chains, is involved in the earliest stage of transmembrane signaling triggered by antigen cross-linking of IgE-FcεRI (Introduction). PM-EGFP, which is constructed from the short amino acid sequence of Lyn that is acylated, has been established as an inner leaflet lipid probe that partitions preferentially into Lo-like environments of the plasma membrane, significantly more favorably than EGFP-GG (Pyenta et al., 2001; Sengupta et al., 2008). Lyn’s additional cytosolic protein modules include SH3, SH2, and kinase modules (Ingley, 2012). We compared PM-EGFP directly to EGFP-GG and to Lyn-EGFP to determine how lipid-based and protein-based interactions influence the diffusion properties of inner leaflet probes, including differential confinement in nanodomains (Figure 3).
As averaged over many Px units in multiple cells, Dav for PM-EGFP is 0.61 μm2/s, which is within the range of literature reports for similar probes (Ike et al., 2003; Douglass and Vale, 2005; Hammond et al., 2009; Golebiewska et al., 2011) and slower than that for EGFP-GG, which partitions relatively weakly into Lo-like environments (Table 1). The CDF of D values for PM-EGFP is satisfactorily fitted with two population components, with the larger fraction having the lower diffusion coefficient: Dslow = 0.58 μm2/s (Fslow = 0.63) and Dfast = 0.67 μm2/s (Ffast = 0.37) (Figure 3, B–D). We assume that PM-EGFP interacts with the plasma membrane by means of its saturated fatty acyl chains and that these chains are slowed in their lateral diffusion by the extent of their interactions with nanodomains. Consistent with their differential preference for Lo-like environments, their respective Fslow values (Table 1 and Figure 3D) indicate that diffusing PM-EGFP is more sensitive than EGFP-GG to the presence of nanodomains. Contour maps of D values for PM-EGFP and EGFP-GG for individual cells show consistent results that regions of slower diffusion are more pronounced for PM-EGFP than for EGFP-GG (Figure S3B). Moreover, the Fslow,cell values determined from contour maps of individual cells show significantly higher values for PM-EGFP than for EGFP-GG, consistent with differences determined from fitting the CDF of the data ensemble (Supplemental Figure S3C; Table 1)
The τ0,av of PM-EGFP (0.27 s) is larger than that of EGFP-GG (0.19 s; Table 1 and Figure 4E), similarly corresponding to greater confinement of PM-EGFP in nanodomains. The results for both PM-EGFP and EGFP-GG can be explained by a membrane model with nanodomain-rich and nanodomain-poor regions (Fslow, Ffast), and differences in F, D, and τ0 values reflect the degree of nanodomain confinement experienced by a particular probe (Supplemental Appendix, Scheme A3). The value of Slopeav for PM-EGFP (1.05 s/μm2) as compared with that for EGFP-GG (1.18 s/μm2) indicates that the apparent micrometer-scale viscosity is somewhat greater for EGFP-GG, suggesting that this probe is more slowed than PM-EGFP by interactions outside nanodomains.
FIGURE 4:
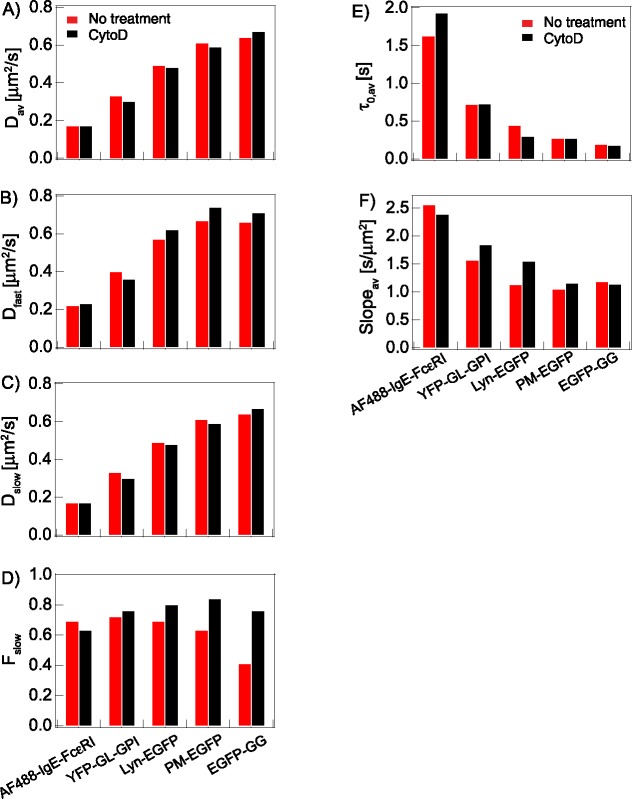
Inhibition of actin polymerization modulates plasma membrane organization and affects probe diffusion properties, as shown without (red) and with (black) CytoD (1 μM) treatment for probes depicted in Figure 1B: (A) Dav; (B) Dfast; (C) Dslow; (D) Fslow; (E) τ0,av; (F) Slopeav. D CDFs yielding values for B–D are shown in Supplemental Figure S4. Numerical values of all parameters with defined errors are provided in Table 1.
The Dav for Lyn-EGFP (0.49 μm2/s) is markedly lower than that for PM-EGFP (Table 1) and within the range reported previously for Lyn and other src family kinases (Ike et al., 2003; Douglass and Vale, 2005; Shvartsman et al., 2007). The CDF of D values for Lyn-EGFP resolves into two populations, with the larger fraction of Px units experienced by this probe as interaction-rich: Dslow = 0.45 μm2/s (Fslow = 0.69); Dfast = 0.57 μm2/s (Figure 3, Table 1). These values, in contrast to those of PM-EGFP, are consistent with Lyn-EGFP interacting more strongly with Lo-like proteolipid nanodomains so that this probe diffuses more slowly in both nanodomain-rich and nanodomain-poor Px units (Supplemental Appendix, Scheme A3). The substantially higher τ0,av value for Lyn-EGFP (0.46 s) than for PM-EGFP and EGFP-GG (Table 1 and Figure 4E) further indicates that Lyn-EGFP has more interactions to increase confinement on the nanoscale. The Slopeav for Lyn-EGFP (1.13 s/μm2) is between those for PM-EGFP and EGFP-GG (Table 1; Figure 4F). Given the clear differences in diffusional properties and assuming that the saturated PM acyl chains for both PM-EGFP and Lyn-EGFP are similarly restricted by Lo-like nanodomains, the cytosolic protein modules of Lyn-EGFP appear to be interacting additionally with proteins inside and possibly outside the nanodomains. As described in a subsequent section, we used inhibition of actin polymerization to further distinguish these contributions.
Lipid probes in the outer leaflet diffuse differently from those in the inner leaflet
We evaluated YFP-GL-GPI, a fluorescently labeled glycosylphosphatidylinositol (GPI), as an outer leaflet lipid probe that partitions favorably into Lo-like environments (Pralle et al., 2000). We find that Dav = 0.33 μm2/s (Table 1), which is within the range of values previously reported for this probe (Kenworthy et al., 2004; Meder et al., 2006; Golebiewska et al., 2011; Edwald et al., 2014; Albrecht et al., 2015). Notably, Dav is markedly lower and τ0,av (0.72 s) is markedly higher than for the inner leaflet lipid probe PM-EGFP (Table 1; Figure 4, A and E). These observations point to substantive differences in the physical properties that affect diffusion in the inner versus outer leaflet, particularly as related to confining Lo-like nanodomains. The D CDF for YFP-GL-GPI resolves into two populations of Px units, with the larger population showing the lower diffusion coefficient for this probe (Table 1; Figure 3): Dslow = 0.30 μm2/s (Fslow = 0.72) and Dfast = 0.40 μm2/s. The fractional amounts indicate that YFP-GL-GPI, similarly to PM-EGFP and Lyn-EGFP, exhibits slower diffusion in the bulk (60–70%) of the membrane sensed by the lipid probes as nanodomain-rich Px units. Consistent with the lower D and higher τ0,av, the Slopeav value for YFP-GL-GPI is larger than for the inner-leaflet Lo-preferring probes PM-EGFP and Lyn-EGFP (Table 1; Figure 3F), further reflecting differences in the interactions that retard diffusion for each probe.
Inhibition of actin polymerization affects diffusion of Lyn-EGFP and lipid probes differentially
The dynamic actin cytoskeleton has been shown to interact, directly or indirectly, with membrane-localized proteins, affecting their functions (Viola and Gupta, 2007; Shelby et al., 2016). Therefore, to gain insight into additional interactions of Lyn-EGFP compared with those of its lipid anchor alone (PM-EGFP), we evaluated effects of cytochalasin D (CytoD), which acutely inhibits actin polymerization (Figure 4 and Supplemental Figure S4; Table 1). We determined that RBL cells treated with 1 μM CytoD undergo no detectable morphological change, and found that this relatively mild treatment causes Dav values to decrease modestly for both Lyn-EGFP and PM-EGFP, with similar trends in Dslow (slight decrease) and Dfast (increase) (Figure 4, A–C, and Supplemental Figure S4A; Table 1). Fslow increases, indicating that more Px units are sensed as interaction-rich by both probes (Figure 4D). Changes in their respective τ0,av and Slopeav values after CytoD treatment clearly differentiate Lyn-EGFP from PM-EGFP. Whereas τ0,av decreases from 0.46 to 0.31 s and Slopeav increases from 1.13 to 1.55 s/μm2 for Lyn-EGFP, the value of these parameters stay about the same (τ0,av) or increase much less (Slopeav) for PM-EGFP (Figure 4, E and F, and Supplemental Figure S4, B and C; Table 1). Thus, it appears that values observed for Lyn-EGFP in untreated cells depend in part on protein-mediated interactions, which in turn depend on cytoskeletal organization that is perturbed by inhibition of actin polymerization. Notably, the τ0,av values for PM-EGFP (0.27 s) and Lyn-EGFP (0.31 s) in CytoD-treated cells are similar, suggesting that the two probes are confined similarly by Lo-like nanodomains under these conditions, as driven largely by their lipid components. The increased Slopeav for Lyn-EGFP indicates that CytoD treatment increases this probe’s protein-based interactions outside nanodomains, resulting in an increase in apparent membrane viscosity on the micrometer length scale.
The D, τ0,av, and Slopeav values for EGFP-GG, without and with CytoD treatment provide additional information about changes occurring in the membrane inner leaflet (Table 1). Whereas treatment with CytoD causes Dav values to decrease for Lo-preferring Lyn-EGFP and PM-EGFP, these value increase for Ld-preferring EGFP-GG (Figure 4A and Supplemental Figure S4A). For EGFP-GG, Dslow with treatment is the same as Dfast without treatment (0.66 μm2/s) and Dfast becomes even faster (0.71 μm2/s) with treatment, while Fslow increases from 0.41 to 0.76 (Figure 4, B–D). These changes suggest that EGFP-GG partitions even less into nanodomains after treatment, thereby diffusing faster in Px units sensed as nanodomain-poor, even as the fraction of Px units sensed as nanodomain-rich (Fslow) increases. Similarly to PM-EGFP, values for τ0,av remain about the same for EGFP-GG before and after CytoD treatment (Figure 4E and Supplemental S4B), but the value for Slopeav decreases by a small amount, rather than increasing as for PM-EGFP and Lyn-EGFP (Figure 4C and Supplemental Figure S4C). Collectively, our results indicate that CytoD treatment causes Lo-like nanodomains to become more ordered, and more Px units to become relatively nanodomain-rich in the membrane inner leaflet, and also that this treatment alters Lyn-EGFP’s protein-based interactions inside and outside of nanodomains.
YFP-GL-GPI in the outer leaflet diffuses more slowly than PM-EGFP in the inner leaflet, but CytoD treatment causes Dav to decrease and Fslow to increase for of all of the Lo-preferring probes (Figure 4, A–D, and Supplemental Figure S4A; Table 1), suggesting the ordered lipid character of nanodomains and their coverage area increases in both leaflets of the plasma membrane. The τ0,av value is similarly unchanged for both YFP-GL-GPI and PM-EGFP after treatment (Supplemental Figure S4B), indicating that the net level of confinement remains similar. However, Slopeav increases markedly for YFP-GL-GPI (Supplemental Figure S4B), pointing to additional interactions such that the apparent membrane viscosity on the micrometer scale increases after treatment as experienced by this probe.
AF488-IgE-FcεRI and other transmembrane proteins exhibit additionally restricted diffusion
TM proteins have additional potential for interacting with other proteins, and the types and strengths of these interactions (specific, nonspecific, steric) are likely to be complex. In most cases, protein-based interactions probably dominate over tendencies to partition into Lo-like nanodomains, although palmitoylation, for example, may modulate these interactions (Lorent et al., 2017). We monitor the TM receptor FcεRI as a complex with Alexafluor 488–labeled IgE (AF488-IgE-FcεRI; Figure 1B). As quantified with Dav (0.17 μm2/s) and τ0,av (1.65 s), FcεRI diffuses more slowly on the scale of Px units and is more confined nanoscopically than Lyn-EGFP and all the inner and outer leaflet lipid probes (Table 1). This Dav value agrees well with previous reports (Thomas et al., 1992; Larson et al., 2005; Lidke et al., 2007). The D CDF of AF488-IgE-FcεRI is fitted with two population components: Dslow = 0.14 μm2/s (Fslow = 0.69) and Dfast = 0.22 μm2/s (Figure 3). We observed small but significant changes in these values after treatment with CytoD (Table 1; Figure 4 and Supplemental Figure S4)
We also evaluated three other TM protein probes in resting RBL cells: epidermal growth factor receptor (EGFR-EGFP, Normanno et al., 2006), GT46 (YFP-GL-GT46, Pralle et al., 2000), and Ca2+ channel Orai1 (AcGFP-Orai1, Prakriya, 2013). In monomeric form, EGFR and GT46 (the TM segment of the LDL receptor and the cytoplasmic tail of CD46) have a single TM segment, whereas Orai1 has four and FcεRI has seven TM segments. The CDFs of D, τ0,av, and Slopeav values for all four TM probes are shown with key parameters summarized in Table 1 and Supplemental Figure S5. As expected, the Dav of these protein probes is consistently lower than that of the lipid and lipid-anchored probes, and those with a single TM segment diffuse somewhat faster: YFP-GT46 (0.26 μm2/s) and EGFR-EGFP (0.24 μm2/s), compared with AcGFP-Orai1 (0.15 μm2/s) and AF488-IgE-FcεRI (0.17 μm2/s). We also observed bimodal D CDFs with Fslow = 0.70–0.76 for these other TM probes. Thus, all TM protein probes tested distributed detectably into two diffusing populations, reflecting differences in membrane environments at the spatial scale of Px units.
All the protein probes show substantial nanoscale confinement as represented by relatively high τ0,av values: 1.14 s (YFP-GT46), 1.41 s (EGFR-EGFP), 1.63 s (AF488-IgE-FcεRI), and 2.56 s (AcGFP-Orai1) (Table 1; Supplemental Figure S5). There are multiple possible sources for confinement of each of these TM probes, including protein-based interactions with cellular constituents proximal to and within the plasma membrane. The Slopeav values show no obvious trends related to number of TM segments: among these four probes, AF488-IgE-FcεRI (2.56 s/μm2) experiences the highest apparent viscosity on the micrometer scale, AcGFP-Orai1 (0.99 s/μm2) the lowest, and those with a single TM segment are in between. Although the particular contributions of protein-based interactions cannot be discerned by comparing these probes, our results confirm expectations that proteins with TM segments are more restricted in diffusion than lipid-anchored membrane probes and provide quantitative details of their distinctive diffusion properties.
DISCUSSION
As exemplified by RBL mast cells, the plasma membrane is predisposed to respond to a specific stimulus (Introduction). Our study demonstrates the versatility and quantitative rigor of ImFCS measurements of diffusion properties and has two primary purposes: 1) to characterize the dynamic heterogeneity of the resting plasma membrane as sensed by a panel of structurally distinct probes including lipid, lipid-anchored, and TM probes; 2) to establish a foundation for elucidating subtle changes that occur when this poised plasma membrane responds to a stimulus and initiates transmembrane signaling. We are motivated by compelling evidence that IgE-FcεRI couples with lipid-anchored Lyn kinase in Lo-like proteolipid nanodomains that are stabilized by antigen engagement (Holowka and Baird, 2016).
We measured the diffusion coefficient (D) for each membrane probe at the largest spatial scale of ImFCS (Px unit = 320 × 320 nm2). The multiplexing capability of ImFCS and small cell-to-cell variability in D values for a given probe enabled us to combine data from multiple cells and thereby achieve exceptional data statistics (∼10,000 D values from that many Px units; Figure 2, A–C). This statistical robustness yields unusually high precision (e.g., very small SEM of Dav; Table 1). We further processed the pooled D values to construct CDFs and developed simple statistical distribution analysis criteria to distinguish two populations of Px units represented by Dfast and Dslow (<Dfast) (Eqs. 2 and 3; Figure 2D and Supplemental Figure S2), with respective values that characterize the probe. Remarkably, data-intensive ImFCS has the capacity to distinguish the Px unit populations when the average values of Dfast and Dslow differ by only 8% (Supplemental Figure S2). More or less subtle differences in Dfast and Dslow arise from variable degrees of weak nanoscopic interaction within Px units. We define populations of Px units corresponding to Dfast and Dslow as interaction-poor and interaction-rich, respectively. Fslow is the fraction of Px units sensed by the probe as interaction-poor. The diffusion-law equation (Eq. 5) applied to ImFCS measurements in Sv units (1.28 × 1.28 μm2) provides indirect information about a probe’s diffusion properties at both nanometer (τ0) and micrometer (Slope = 1/Deff) length scales.
Lipid probes PM-EGFP, EGFP-GG, and YFP-GL-GPI have acyl chains that partition differentially into Lo-like environments and no functional protein domains. We interpret our data for these probes simply in terms of a model that considers interactions with Lo-like proteolipid nanodomains that reside within regions of Ld-preferring lipids and proteins (Figure 1; Supplemental Appendix). This view fits within the refined “hierarchal model” for plasma membrane organization proposed previously (Introduction). The distinction between nanodomain-rich and nanodomain-poor Px units may be, for example, respectively higher and lower densities of cortical actin meshwork (Kusumi et al., 2011) or of actomyosin asters (Rao and Mayor, 2014). Lipid-anchored Lyn-EGFP has acyl chains (PM) that partition preferentially into Lo-like proteolipid nanodomains, and also protein modules that may interact with proteins inside and outside of these membrane structures. In general, the diffusion of TM protein probes may be more or less influenced by heterogeneities in lipid composition, depending on relative levels of lipid-based and protein-based interaction and how these are distributed in the membrane.
Inner leaflet probes sense the membrane differentially
We integrate the diffusion behavior of EGFP-GG, PM-EGFP, and Lyn-EGFP into a common inner-leaflet model (Figure 5, A and B). A higher Dav value and a lower τ0,av value for EGFP-GG compared with the other two probes (Figure 3, B, D, and E) is consistent with the view that the unsaturated geranyl–geranyl acyl chains are less dynamically confined in Lo-like nanodomains than are saturated palmitate–myristoylate chains. Correspondingly, a smaller fraction of Px units have sufficient nanodomains to yield the slower diffusing population (Fslow, Dslow) for EGFP-GG than for PM-EGFP (Figure 5A; Supplemental Appendix, Scheme A3). Precise numerical details in Table 1 provide additional insight into these subtle interactions. For example, values for Dfast and Dslow are not very different for these two probes and are more similar to each other for Dfast, as might be expected when both probes are diffusing in more Ld-like Px units (Figure 5A, top).
FIGURE 5:
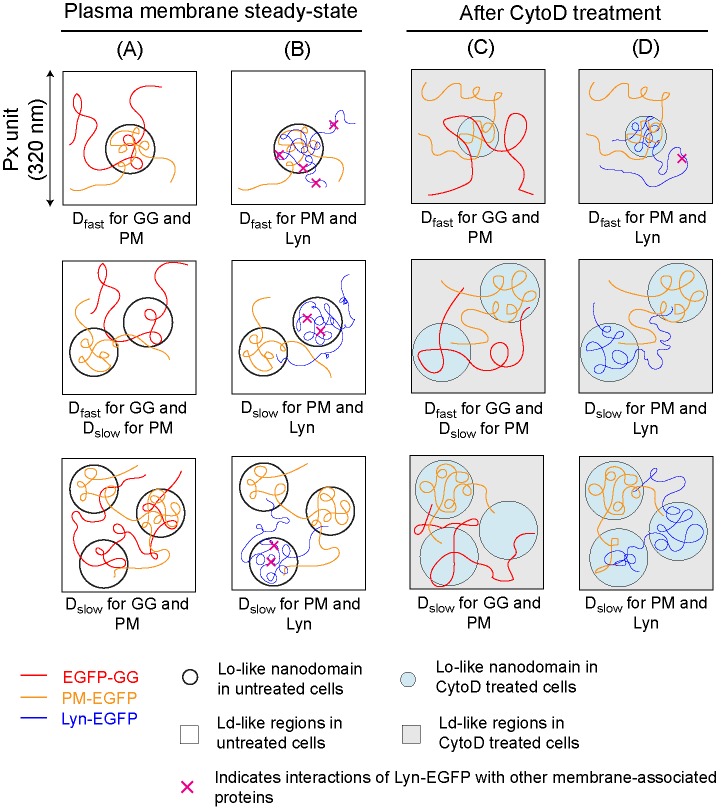
Diffusion of inner leaflet probes EGFP-GG, PM-EGFP, and Lyn-EGFP depends on lipid-based interactions inside and outside of Lo-like proteolipid nanodomains and is modulated by inhibition of actin polymerization. Representative Px units with low (top), moderate (middle), or high (bottom) levels of coverage by nanodomains (circles) are shown. EGFP-GG partitions less favorably into Lo-like environments so that this probe’s diffusion is less affected by the presence of the nanodomains and its Dfast corresponds to Px units with both low and moderate coverage; only Px with high levels of coverage manifest as Dslow for this probe. Dfast for PM-EGFP and Lyn-EGFP corresponds to Px units with low coverage by nanodomains, whereas Dslow for these probes corresponds to Px units with moderate and high levels of coverage. Diffusion of Lyn-EGFP is further retarded by interactions of its protein modules with other proteins in and out of nanodomains (indicated by × [pink]). Contrasting behavior of these probes is illustrated by their respective diffusion trajectories. PM-EGFP is compared with EGFP-GG (A) and to Lyn-EGFP (B) in resting steady state plasma membrane of RBL cells. Retardation of diffusion due to dynamic partitioning into nanodomains: Lyn-EGFP > PM-EGFP > EGFP-GG, as reflected by respective D CDFs and extracted D values (Figure 3), as well as relative τ0,av values determined for these probes (Table 1). (C, D) When resting cells are treated with 1 μM CytoD, the nanodomains become more ordered and probe interactions with membrane components change into and out of nanodomains, as reflected by their diffusion properties (Figure 4). Compared with untreated cells, EGFP-GG partitions less favorably into nanodomains, whereas PM-EGFP partitions more favorably, C. As reflected by τ0,av values, Lyn-EGFP loses interactions with proteins within nanodomains (absence of × inside nanodomains), behaving more like PM-EGFP in these regions, but gains interactions with proteins outside these regions (as reflected by Slopeav values), D. Differences in diffusion properties caused by CytoD treatment are also reflected in D CDFs (Supplemental Figure S4) and extracted D values (Table 1).
Differences between PM-EGFP and Lyn-EGFP, which have the same lipid anchor, suggest that nanodomains include proteins that interact with Lyn’s protein modules (Figure 5B). Lyn-EGFP has lower Dav and Dslow values and a somewhat higher Fslow value (Figure 3, B–E). Similarly, lower Dfast of Lyn-EGFP suggests that nanodomains in all Px units retard diffusion for Lyn-EGFP more than for PM-EGFP (e.g., Figure 5B, top). Lyn-EGFP may also interact with other proteins in the membrane or cytoplasm when this probe diffuses outside of the nanodomains. Interestingly, Slopeav for Lyn-EGFP is greater than for PM-EGFP and less than for EGFP-GG, and this measure of micrometer scale viscosity indicates that PM-EGFP has the lowest level of interaction outside of nanodomains. EGFP-GG’s apparent higher level of interaction may be due to the polybasic motif included in this construct to stabilize its membrane localization, such as electrostatic interactions with acidic phospholipids that localize to Ld-like regions.
Changes in the diffusion properties of inner leaflet probes after cells are treated with 1 μM CytoD (Figure 4 and Supplemental Figure S4; Table 1) are consistent with previous observations that lipid-based heterogeneity of the plasma membrane is modulated when actin polymerization is inhibited (Shelby et al., 2016). Our measurements after CytoD treatment point to a consistent picture that 1) the fraction of nanodomain-rich Px units increases, 2) the nanodomain coverage a) increases in nanodomain-rich Px units and b) decreases in nanodomain-poor Px units, and 3) overall the nanodomains become more ordered (Figure 5, C and D). Compared with their counterparts in untreated cells, EGFP-GG diffuses faster in both nanodomain-poor and nanodomain-rich Px units (Figure 5C), whereas PM-EGFP and Lyn-EGFP diffuse faster in nanodomain-poor Px units and slower in nanodomain-rich Px units (Figure 5D). These results provide new evidence that CytoD-treated live cells exhibit stronger phase-like segregation, so that the diffusion behavior of EGFP-GG and PM-EGFP becomes more distinguishable. Previous studies showed that the long chain actin meshwork serves to restrict phase separation (Ehrig et al., 2011; Machta et al., 2011; Gomez-Llobregat et al., 2013; Honigmann et al., 2014b; Vogel et al., 2017) and that CytoD increases the size of corrals formed by this meshwork (Fujiwara et al., 2016). Similarly, experiments and Ising-based simulations showed that inhibition of actin polymerization increases coclustering of Lo-preferring components (Shelby et al., 2016).
CytoD affects the diffusion of Lyn-EGFP differentially compared with PM-EGFP, reflecting the involvement of Lyn’s protein modules (Figure 5D). In contrast to PM-EGFP and EGFP-GG, large changes for Lyn-EGFP in τ0,av (decrease) and Slopeav (increase) after CytoD treatment (Figure 4 and Supplemental Figure S4) indicate that Lyn-EGFP’s protein-based interactions decrease within Lo-like nanodomains. Lyn may also interact more with partners outside nanodomains via its protein modules in an actin-dependent manner. Although our results with mild CytoD treatment are consistent with previous evidence that Lyn’s protein modules link indirectly with the actin cytoskeleton (Minoguchi et al., 1994), we cannot rule out other complicating effects of this reagent.
Diffusion of Lo-preferring lipid probes differs in outer vs inner leaflets of plasma membrane
The lipid probes PM-EGFP (inner leaflet) and YFP-GL-GPI (outer leaflet) both prefer Lo-like environments, and we found that the two leaflets exhibit a similar predominance of nanodomain-rich Px units as sensed by both of these probes, with Fslow values of 0.63 and 0.72, respectively (Figure 3E and Table 1). However, absolute values of Dslow, Dfast, and Dav of YFP-GL-GPI are substantially lower than for PM-EGFP, indicating greater sensitivity to confinement of the outer leaflet probe in both nanodomain-rich and nanodomain-poor populations of Px units (Figure 3, C and D). YFP-GL-GPI also exhibits substantially higher values of τ0,av and Slopeav than PM-EGFP (Figure 4, E and F). The higher level of nanoscale confinement (τ0,av) may be due to a higher level of order in the nanodomains in the outer leaflet, consistent with lower D values and as suggested by comparing lipid compositions in both leaflets (Gupta et al., 2020; Lorent et al., 2019). The higher apparent micrometer-scale viscosity (Slopeav) may reflect additional interaction partners of YFP-GL-GPI, such as the thick glycocalyx on the outer leaflet (Chapanian et al., 2014). CytoD treatment has no effect on τ0,av, while Slopeav increases markedly for YFP-GL-GPI (Table 1; Figure 4, C–E, and Supplemental Figure S4B), suggesting that the largest changes of YFP-GL-GPI interactions to increase the apparent viscosity occur outside of Lo-like nanodomains.
Notably, Lo-preferring probes in both leaflets, PM-EGFP, Lyn-EGFP, and YFP-GL-GPI, exhibit Fslow values in the range 0.63–0.72, independent of their Dslow and Dav values (Figure 3). This is consistent with previous measurements of predominant Lo-like character in the plasma membrane of RBL cells by electron spin resonance, fluorescence anisotropy, and fluorescence imaging (Gidwani et al., 2001; Swamy et al., 2006; Levental et al., 2009). In contrast, EGFP-GG exhibits Fslow = 0.41, consistent with this Ld-preferring probe being the least susceptible to confinement. Similar high coverage of Lo-like regions is also reported for other cell types (Owen et al., 2012; Honigmann et al., 2014a).
Diffusion of TM proteins is influenced primarily by protein-based interactions
AF488-IgE-FcεRI and other TM proteins evaluated in our study have different numbers of TM segments and different preferences for Lo-like environments, as evaluated by a variety of criteria (Kenworthy et al., 2004; Lorent et al., 2017). Not surprisingly, the Dav of these protein probes is substantially lower than for the lipid-anchored probes (EGFP-GG, PM-EGFP, Lyn-EGFP, YFP-GL-GPI), and those with a single TM segment (YFP-GL-GT46, EGFR-EGFP) diffuse somewhat faster than those with four (AcGFP-Orai1) or seven (AF488-IgE-FcεRI) TM segments. All TM probes show τ0,av values substantially higher than those for the lipid probes, indicating nanoscopic confinement involving protein interactions (Supplemental Table S2B). These confining interactions appear to be highly protein-dependent, as suggested by our results that the TM probes with one TM segment show the smallest τ0,av, but the TM probe with seven TM segments exhibits a lower value than the TM probe with four TM segments. The Slopeav value, which averages over all interactions at the micrometer scale, is highest for the probe with seven TM segments and lowest for the probe with four TM segments, and these two values also bracket the Slopeav values for all four of the lipid-based probes in inner and outer leaflets. Overall, our results indicate that diffusion of TM protein probes is not dramatically influenced by Lo-like nanodomains in the resting plasma membrane but retarded predominantly by interactions with other proteins that depend on the physical biochemistry of the specific TM probe monitored.
Concluding remarks
Collectives of weak lipid-based and protein-based interactions underlie the dynamic steady state of plasma membrane heterogeneity in live cells. We demonstrate here how unusually robust statistical analyses of D distributions measured with ImFCS for structurally distinctive probes can provide an integrated view of this heterogeneity. Valuable as a general approach for many cell types, with ImFCS we found that the membrane organization of resting RBL mast cells is predominantly Lo-like, as sensed by lipid probes, that protein modules in lipid-anchored probes modulate partitioning, and that diffusion of TM probes is selectively influenced by protein-based interactions. The simple analytical framework we describe to evaluate D distributions can be adapted readily to other spatially resolved fluorescence fluctuation methods (Ries et al., 2012; Wiseman, 2012; Digman et al., 2013; Bag and Wohland, 2014; Moens et al., 2015; Hendrix et al., 2016; Krmpot et al., 2019). In the context of mast cell activation, future studies will build on the foundation established here to examine stimulated signaling as mediated by the participating proteins within the environment of the responding membrane. A key step will be to quantitatively address the essential roles played by weak, lipid-based interactions in transmembrane signaling initiated by antigen-driven coupling of IgE-FcεRI with Lyn kinase anchored to the membrane inner leaflet (Holowka and Baird, 2016).
MATERIALS AND METHODS
Reagents
MEM, Opti-MEM, Trypsin-EDTA (0.01%), and gentamicin sulfate were purchased from Life Technologies (Carlsbad, CA). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Atlanta, CA). Alexafluor 488 (AF488) NHS ester (Invitrogen) was used to label monoclonal anti-DNP immunoglobulin E (IgE) as described previously (Larson et al., 2005). Cytochalasin D (CytoD) and phorbol 12,13-dibutyrate (PDB) were obtained from Sigma-Aldrich (St. Louis, MO). Stock solutions of PDB and CytoD were prepared in dimethylsulfoxide (DMSO) and stored at -80°C.
Cell culture, transfection, and labeling
RBL-2H3 mast cells (for brevity, RBL cells) were cultured in growth medium (80% MEM supplemented with 20% FBS and 10 mg/l gentamicin sulfate) at 37°C and 5% (vol/vol) CO2 environment. Cells in a confluent 75-cm2 flask were washed once with 2 ml Trypsin-EDTA, detached with 2 ml Trypsin-EDTA for 5 min at 37°C and 5% (vol/vol) CO2 environment, and harvested in growth medium. About 20,000 cells were homogeneously spread in a 35 mm MatTek dish (Ashland, MA) containing 2 ml growth medium and allowed to grow overnight. MatTek dishes containing the adhered cells were transfected using FuGENE HD transfection kit (Promega). Typically, 0.5–1 μg of plasmid DNA and 3 μl FuGENE/μg DNA were used per dish. For transfection, plasmid DNA and FuGENE were first mixed in 100 μl Opti-MEM and incubated at room temperature for 15 min. Next, MatTek dishes containing cells were washed once and then covered with 1 ml Opti-MEM. The DNA/FuGENE complex was spread evenly over the cells and incubated for 1 h, followed by incubation with prewarmed PDB (1 ml, 0.1 μg/ml) for 3 h at 37°C in 5% (vol/vol) CO2 environment. Finally, 2 ml of growth medium was added to each MatTek dish after Opti-MEMm was discarded. The transfected cells were allowed to grow for 18–22 h at 37°C in a 5% (vol/vol) CO2 environment before imaging. For imaging, the cells were washed twice with buffered salt solution (BSS: 135 mM NaCl, 5.0 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 5.6 mM glucose, and 20 mM HEPES; pH 7.4) and imaged in fresh BSS. The plasmids used in this study encode the following proteins: Lyn-EGFP (Hess et al., 2003), PM-EGFP (Pyenta et al., 2001), EGFP-GG (Pyenta et al., 2001), YFP-GL-GPI (Sengupta et al., 2008), AcGFP-Orai1 (Holowka et al., 2018), EGFR-EGFP (Bryant et al., 2013), and YFP-GL-GT46 (Sengupta et al., 2008).
For labeling FcεRI, the cells were washed twice with BSS, followed by addition of a mixture of AF488-labeled (0.5 μg/ml) and unlabeled (1.5 μg/ml) IgE for 40 min at room temperature. The cells were washed twice with BSS and directly imaged in fresh BSS.
For CytoD treatment, a fresh working solution of CytoD (1 μM) was prepared by diluting the stock solution in BSS before the experiment. The DMSO content in the final solution was <0.1%. The cells were preincubated with 1 ml of 1 μM CytoD for 15 min at room temperature before imaging.
ImFCS data acquisition
Fluorescently labeled ventral plasma membranes of RBL cells were imaged with a home-built total internal reflection fluorescence microscope (TIRFM; DMIRB, Leica Microsystems, Germany) equipped with an oil immersion objective (PlanApo, 100×, NA 1.47; Leica Microsystems, Germany), a 488-nm excitation laser (Coherent, Santa Clara, CA), and an electron-multiplying charge-coupled device (EMCCD) camera (black illuminated Andor iXON 897DU, pixel size 16 μm, Andor Technology, Belfast, UK). The excitation laser beam was introduced and focused on the back focal plane of the objective by a pair of tilting mirrors and a dichroic mirror (ZT405/488/561/640rpc, Chroma Technology). The same set of mirrors was used to adjust the TIRF angle of the excitation beam to illuminate the ventral membrane. The fluorescence signal from the sample was recorded by the EMCCD camera after it passed through the same objective and the dichroic mirror and reflected to the camera chip after being filtered by an emission filter (ZET488/561m, Chroma Technology). The laser power was 50 μW before the objective.
For ImFCS, a stack of 80,000 images from a region of interest (ROI) on the plasma membrane was recorded at an acquisition speed of 3.5 ms/frame. The ROI sizes were between 40 × 40 and 50 × 50 pixels (pixel size in the object plane = 160 nm), depending on the cell size. The images were acquired in “frame transfer” mode with readout speed 10 MHz, with an EM gain of 300 (scale 6–300). Image acquisition was performed with Andor Solis software. The acquisition conditions were optimized following the protocols reported previously (Sankaran et al., 2013; Krieger et al., 2015). All measurements were carried out at room temperature.
ImFCS data analysis: D values were determined at Px unit length scale
This raw image stack was further processed by a FIJI plug-in for ImFCS (Imaging_FCS 1.491, downloaded 1 October 2016 from the laboratory website of Thorsten Wohland, National University of Singapore). Raw autocorrelation functions (ACFs) were first computed after 2 × 2 pixel binning (Px unit) of an image stack. The ACF for each Px unit was fitted with a one-component, two-dimensional Brownian diffusion model (Eq. 1; Sankaran et al., 2013). This yields a map of lateral diffusion coefficient (D) with pixel dimension of 320 × 320 nm:
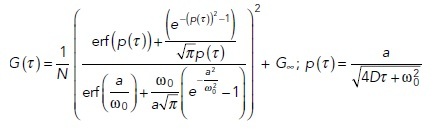 |
1 |
In the above equations, G(τ) is the ACF as a function of lag time (τ), N is the number of particles diffusing within the observation area, D is the lateral diffusion coefficient in the Px unit, a is the length of the Px unit in the object plane (320 nm), ω0 is the point spread function (PSF) of the microscope, Aeff is the effective observation area, which is determined by the convolution of measured area (a2) and point spread function, and G∞ is the convergence value of G(τ) at very large lag times. We used N, D, and G∞ as fit parameters, and ω0 was experimentally determined using the method described previously (Bag et al., 2012).
Fitting of cumulative distribution functions of D
The D values obtained in Px units (Eq. 1) from multiple cells for a given probe are grouped to create their respective distributions. First, cumulative frequencies for each D value were determined in ascending order, which were then plotted against corresponding D values to generate normalized CDFs of D using Igor Pro (Version 7 and 8; WaveMetrics, Portland, OR). This CDF was fitted with the following models (Eqs. 2 and 3 for one- and two-component models, respectively):
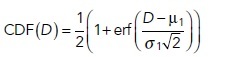 |
2 |
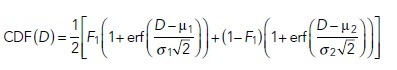 |
3 |
| 4 |
In the above equations, μ1 and σ1 are the mean and SD of the first component while μ2 and σ2 are the mean and SD of the second component, F1 is the fraction of the first component, and (1-F1) is the fraction of the second component of the D distribution. The best fitting model (Eq. 2 or Eq. 3) and goodness of fit were determined by reduced chi-squared values. We also compared the periodicity of the residual plots for the two models. If fitting with the one-component model yields a residual plot that strongly oscillates around zero and this periodicity largely disappears in the residual plot for fitting with two components, along with strong reduction of reduced chi-squared values, we accept a two-component model. A three-component model did not improve the quality of fitting in any case and therefore was not considered. The weighted average of D, <DCDF>, is given by Eq. 4.
The component with lower mean value, min {μ1, μ2} = Dslow, is defined as representing the population of Px units with more slowly diffusing probes, and the component with higher mean value, max {μ1, μ2} = Dfast, is defined as representing the population of Px units with faster diffusing probes. According to our working model for lipid-anchored probes (Supplemental Appendix, Schemes A1–A3), Dslow represents probes diffusing in nanodomain-rich Px units, and Dfast represents probes diffusing in nanodomain-poor Px units. The D components are influenced by protein-based interactions when probes possess protein modules and, correspondingly, Dslow and Dfast for those probes represent populations of interaction-rich and interaction-poor Px units, respectively.
The large number of D values (NPx) used here to create a given CDF allow us not only to detect subtle differences between Dfast and Dslow statistically for a given probe under a given condition (Supplemental Figure S2) but also to distinguish between two probes under a given condition. For example, Dfast of EGFP-GG and PM-EGFP is 0.66 ± 0.19 and 0.67 ± 0.24 μm2/s (mean ± SD; Table 1) in untreated cells, respectively. The Ffast (= 1 - Fslow) is 0.59 and 0.37, while the NPx is 10,527 and 9,375, respectively. This means that Dfast for EGFP-GG and PM-EGFP corresponds to respective diffusion coefficients averaged over n Px units: 6211 (= 0.59 × 10,527) and 3469 (= 0.37 × 9375), respectively. We performed an unpaired Student’s t test to compare the average values of Dfast of PM-EGFP and EGFP-GG using mean, SD, and n of 0.66, 0.19 and 6211 for EGFP-GG and 0.67, 0.27, and 3469 for PM-EGFP, which returned a statistically significant difference (p value = 0.02), even though the standard deviations overlap. Note that SD/√n = SEM of D components for both EGFP-GG and PM-EGFP is less than 1% of the mean values; the same is true for all of the probes in Table 1. We used this statistical analysis (Student’s t test) to make pairwise comparisons across experimental conditions for a given probe or across probes for a given condition (e.g., Figure 3).
Nanoscopic confinement (τ0) and micrometer-scale viscosity (Slope = 1/Deff) are determined in Sv units. As described previously (Bag et al., 2016), the τ0 map was created by analysis of spot variation FCS (svFCS) on small subregions (Sv units: 8 × 8 pixels or 4 × 4 Px units) within the same raw stack (Figure 2A, top). Diffusion times (τD) were first determined for four different observation area (Aeff) sizes generated by 2 × 2–, 3 × 3–, 4 × 4–, and 5 × 5–pixel binning within each Sv unit (Figure 2E). The Aeff thus obtained were 0.42, 0.57, 0.78, and 1.05 μm2, respectively. ImFCS for each set of Aeff yields a one-component ACF for each size and thereby a D value for each. The τD values for each were determined by dividing each Aeff by its corresponding D value. The plot of τD against Aeff for each Sv unit was fitted with a straight line (Eq. 5) to get the τ0 value as the intercept; the Slope of the fit is the inverse of the effective micrometer-scale diffusion coefficient (1/Deff). There are 36 Sv units in a 50 × 50–pixel raw stack, and the spatial scale of the τ0 and Slope maps is 1.28 × 1.28 μm2:
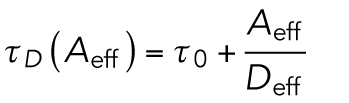 |
5 |
Limitations of current ImFCS data analysis
Some limitations of ImFCS measurements of diffusion coefficients of mobile probes are described in preceding sections, including their diffraction-limited resolution at the Px unit (320 × 320 nm in this study). The precise values we achieved are based on one- or two-component CDF analysis of D values, which combined data from multiple cells for adequate statistics to distinguish 10% differences between D components. More data points per cell are needed to achieve sensitive distinction between populations in individual cells. This may be possible using the same analytical framework with better cameras (and other array detectors) and/or brighter fluorophores such that good quality ACFs are determined at each pixel (e.g., 160 × 160 nm) without the need for binning. Evaluating probe diffusion to characterize the membrane’s spatial heterogeneity also depends on the stability of membrane features and the time resolution of the detector. Our study evaluated the steady state of the plasma membrane, either in resting cells or after CytoD treatment. Time-dependent processes, such as stimulated signaling, can also be addressed as long as good-quality raw ACFs are generated with sufficient time resolution (Bag et al., 2015). The camera used in this study provides good raw data only from 280-s measurement, which could be further assessed in 70-s segments. A combination of brighter fluorophores and higher quantum efficiency of the camera may yield high-quality data even with 10-s measurements (Bag et al., 2015; Sankaran et al., 2013). We envisage successful employment of ImFCS statistical analysis strategy for time-dependent processes (within minutes) at single-cell resolution in the future. Unlike STED-FCS and high-speed SPT, which both have substantial technical challenges, the fundamental diffraction limit of ImFCS does not allow direct estimates of the size and lifetime of nanoscopic heterogeneities in the plasma membrane. Emerging strategies such as spectral cross-correlation of polarity-sensitive probes and phasor analysis can be incorporated in the ImFCS platform as possible routes to mitigate this limitation (Ranjit et al., 2014; Nicovich et al., 2018).
Supplementary Material
Acknowledgments
We are grateful to Alice Wagenknecht-Wiesner for help in preparing the constructs. We thank Thorsten Wohland (National University of Singapore) for making the ImFCS software freely available and for helpful discussions. This work is supported by National Institute of General Medical Sciences (NIGMS) Grant R01GM117552. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIGMS or the National Institutes of Health.
Abbreviations used:
- CDF
cumulative distribution function
- EGFP
enhanced green fluorescent protein
- GG
geranyl–geranyl lipid anchor
- GPI
glycosylphosphatidylinositol anchor
- ImFCS
imaging fluorescence correlation spectroscopy
- Ld
liquid disordered
- Lo
liquid ordered
- PM
minimal palmitoyl–myristoyl lipid anchor of Lyn
- svFCS
spot-variation fluorescence correlation spectroscopy
- TM
transmembrane
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-10-0559) on January 2, 2020.
REFERENCES
- Akinlaja J, Sachs F. (1998). The breakdown of cell membranes by electrical and mechanical stress. Biophys J , 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht D, Winterflood CM, Ewers H. (2015). Dual color single particle tracking via nanobodies. Methods Appl Fluoresc , 024001. [DOI] [PubMed] [Google Scholar]
- Anderson RG, Jacobson K. (2002). A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science , 1821–1825. [DOI] [PubMed] [Google Scholar]
- Andrade DM, Clausen MP, Keller J, Mueller V, Wu C, Bear JE, Hell SW, Lagerholm BC, Eggeling C. (2015). Cortical actin networks induce spatio-temporal confinement of phospholipids in the plasma membrane—a minimally invasive investigation by STED-FCS. Sci Rep , 11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bag N, Huang S, Wohland T. (2015). Plasma membrane organization of epidermal growth factor receptor in resting and ligand-bound states. Biophys J , 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bag N, Ng XW, Sankaran J, Wohland T. (2016). Spatiotemporal mapping of diffusion dynamics and organization in plasma membranes. Methods Appl Fluoresc , 034003. [DOI] [PubMed] [Google Scholar]
- Bag N, Sankaran J, Paul A, Kraut RS, Wohland T. (2012). Calibration and limits of camera-based fluorescence correlation spectroscopy: a supported lipid bilayer study. Chemphyschem , 2784–2794. [DOI] [PubMed] [Google Scholar]
- Bag N, Wohland T. (2014). Imaging fluorescence fluctuation spectroscopy: new tools for quantitative bioimaging. Annu Rev Phys Chem , 225–248. [DOI] [PubMed] [Google Scholar]
- Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, Baird BA, Webb WW. (2007). Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci USA , 3165–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchaud JP, Georges A. (1990). Anomalous diffusion in disordered media—statistical mechanisms, models and physical applications. Phys Rep , 127–293. [Google Scholar]
- Brown DA. (2006). Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology (Bethesda) , 430–439. [DOI] [PubMed] [Google Scholar]
- Bryant KL, Antonyak MA, Cerione RA, Baird B, Holowka D. (2013). Mutations in the polybasic juxtamembrane sequence of both plasma membrane- and endoplasmic reticulum-localized epidermal growth factor receptors confer ligand-independent cell transformation. J Biol Chem , 34930–34942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapanian R, Kwan DH, Constantinescu I, Shaikh FA, Rossi NA, Withers SG, Kizhakkedathu JN. (2014). Enhancement of biological reactions on cell surfaces via macromolecular crowding. Nat Commun , 4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chein M, Perlson E, Roichman Y. (2019). Flow arrest in the plasma membrane. Biophys J , 810–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi V, Mendez-Villuendas E, Ingolfsson HI, Gu RX, Siuda I, Melo MN, Moussatova A, DeGagne LJ, Sejdiu BI, Singh G, et al (2018). Lipid–protein interactions are unique fingerprints for membrane proteins. ACS Cent Sci , 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Gratton E. (2011). Lessons in fluctuation correlation spectroscopy. Annu Rev Phys Chem , 645–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Stakic M, Gratton E. (2013). Raster image correlation spectroscopy and number and brightness analysis. Methods Enzymol , 121–144. [DOI] [PubMed] [Google Scholar]
- Dinic J, Ashrafzadeh P, Parmryd I. (2013). Actin filaments attachment at the plasma membrane in live cells cause the formation of ordered lipid domains. Biochim Biophys Acta , 1102–1111. [DOI] [PubMed] [Google Scholar]
- Douglass AD, Vale RD. (2005). Single-molecule microscopy reveals plasma membrane microdomains created by protein–protein networks that exclude or trap signaling molecules in T cells. Cell , 937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwald E, Stone MB, Gray EM, Wu J, Veatch SL. (2014). Oxygen depletion speeds and simplifies diffusion in HeLa cells. Biophys J , 1873–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schonle A, Hell SW. (2009). Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature , 1159–1162. [DOI] [PubMed] [Google Scholar]
- Ehrig J, Petrov EP, Schwille P. (2011). Near-critical fluctuations and cytoskeleton-assisted phase separation lead to subdiffusion in cell membranes. Biophys J , 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara TK, Iwasawa K, Kalay Z, Tsunoyama TA, Watanabe Y, Umemura YM, Murakoshi H, Suzuki KG, Nemoto YL, Morone N, Kusumi A. (2016). Confined diffusion of transmembrane proteins and lipids induced by the same actin meshwork lining the plasma membrane. Mol Biol Cell , 1101–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidwani A, Holowka D, Baird B. (2001). Fluorescence anisotropy measurements of lipid order in plasma membranes and lipid rafts from RBL-2H3 mast cells. Biochemistry-Us , 12422–12429. [DOI] [PubMed] [Google Scholar]
- Golebiewska U, Kay JG, Masters T, Grinstein S, Im W, Pastor RW, Scarlata S, McLaughlin S. (2011). Evidence for a fence that impedes the diffusion of phosphatidylinositol 4,5-bisphosphate out of the forming phagosomes of macrophages. Mol Biol Cell , 3498–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Llobregat J, Buceta J, Reigada R. (2013). Interplay of cytoskeletal activity and lipid phase stability in dynamic protein recruitment and clustering. Sci Rep , 2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goni FM. (2019). “Rafts”: a nickname for putative transient nanodomains. Chem Phys Lipids , 34–39. [DOI] [PubMed] [Google Scholar]
- Gupta A, Korte T, Herrmann A, Wohland T. (2020). Plasma membrane asymmetry of lipid organization: fluorescence lifetime microscopy and correlation spectroscopy analysis. J Lipid Res , 252–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Sim Y, Lagnado L, Irvine RF. (2009). Reversible binding and rapid diffusion of proteins in complex with inositol lipids serves to coordinate free movement with spatial information. J Cell Biol , 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HT, Marguet D. (2008). T-cell antigen receptor triggering and lipid rafts: a matter of space and time scales. Talking point on the involvement of lipid rafts in T-cell activation. EMBO Rep , 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HT, Marguet D. (2011). Detecting nanodomains in living cell membrane by fluorescence correlation spectroscopy. Annu Rev Phys Chem , 417–436. [DOI] [PubMed] [Google Scholar]
- Hendrix J, Dekens T, Schrimpf W, Lamb DC. (2016). Arbitrary-region raster image correlation spectroscopy. Biophys J , 1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess ST, Sheets ED, Wagenknecht-Wiesner A, Heikal AA. (2003). Quantitative analysis of the fluorescence properties of intrinsically fluorescent proteins in living cells. Biophys J , 2566–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowka D, Baird B. (2016). Roles for lipid heterogeneity in immunoreceptor signaling. Biochim Biophys Acta , 830–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowka D, Gosse JA, Hammond AT, Han X, Sengupta P, Smith NL, Wagenknecht-Wiesner A, Wu M, Young RM, Baird B. (2005). Lipid segregation and IgE receptor signaling: a decade of progress. Biochim Biophys Acta , 252–259. [DOI] [PubMed] [Google Scholar]
- Holowka D, Thanapuasuwan K, Baird B. (2018). Short chain ceramides disrupt immunoreceptor signaling by inhibiting segregation of Lo from Ld plasma membrane components. Biol Open , bio034702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigmann A, Mueller V, Ta H, Schoenle A, Sezgin E, Hell SW, Eggeling C. (2014a). Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nat Commun , 5412. [DOI] [PubMed] [Google Scholar]
- Honigmann A, Pralle A. (2016). Compartmentalization of the cell membrane. J Mol Biol , 4739–4748. [DOI] [PubMed] [Google Scholar]
- Honigmann A, Sadeghi S, Keller J, Hell SW, Eggeling C, Vink R. (2014b). A lipid bound actin meshwork organizes liquid phase separation in model membranes. eLife , e01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Simsek MF, Jin W, Pralle A. (2015). Effect of receptor dimerization on membrane lipid raft structure continuously quantified on single cells by camera based fluorescence correlation spectroscopy. PLoS One , e0121777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ike H, Kosugi A, Kato A, Iino R, Hirano H, Fujiwara T, Ritchie K, Kusumi A. (2003). Mechanism of Lck recruitment to the T-cell receptor cluster as studied by single-molecule-fluorescence video imaging. Chemphyschem , 620–626. [DOI] [PubMed] [Google Scholar]
- Ingley E. (2012). Functions of the Lyn tyrosine kinase in health and disease. Cell Commun Signal , 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K, Mouritsen OG, Anderson RGW. (2007). Lipid rafts: at a crossroad between cell biology and physics. Nat Cell Biol , 7–14. [DOI] [PubMed] [Google Scholar]
- Kannan B, Guo L, Sudhaharan T, Ahmed S, Maruyama I, Wohland T. (2007). Spatially resolved total internal reflection fluorescence correlation microscopy using an electron multiplying charge-coupled device camera. Anal Chem , 4463–4470. [DOI] [PubMed] [Google Scholar]
- Kenworthy AK, Nichols BJ, Remmert CL, Hendrix GM, Kumar M, Zimmerberg J, Lippincott-Schwartz J. (2004). Dynamics of putative raft-associated proteins at the cell surface. J Cell Biol , 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotzsch E, Schutz GJ. (2013). A critical survey of methods to detect plasma membrane rafts. Philos Trans R Soc Lond B Biol Sci , 20120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster DV, Mayor S. (2016). Cortical actin and the plasma membrane: inextricably intertwined. Curr Opin Cell Biol , 81–89. [DOI] [PubMed] [Google Scholar]
- Krieger JW, Singh AP, Bag N, Garbe CS, Saunders TE, Langowski J, Wohland T. (2015). Imaging fluorescence (cross-) correlation spectroscopy in live cells and organisms. Nat Protoc , 1948–1974. [DOI] [PubMed] [Google Scholar]
- Krmpot AJ, Nikolic SN, Oasa S, Papadopoulos DK, Vitali M, Oura M, Mikuni S, Thyberg P, Tisa S, Kinjo M, et al (2019). Functional fluorescence microscopy imaging: quantitative scanning-free confocal fluorescence microscopy for the characterization of fast dynamic processes in live cells. Anal Chem , 11129–11137. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki KG. (2012a). Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu Rev Cell Dev Biol , 215–250. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Fujiwara TK, Morone N, Yoshida KJ, Chadda R, Xie M, Kasai RS, Suzuki KG. (2012b) Membrane mechanisms for signal transduction: the coupling of the meso-scale raft domains to membrane-skeleton-induced compartments and dynamic protein complexes. Semin Cell Dev Biol , 126–144. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Fujiwara TK, Tsunoyama TA, Kasai RS, Liu AA, Hirosawa KM, Kinoshita M, Matsumori N, Komura N, Ando H, Suzuki KGN. (2019). Defining raft domains in the plasma membrane. Traffic , 106–137. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Shirai YM, Koyama-Honda I, Suzuki KG, Fujiwara TK. (2010). Hierarchical organization of the plasma membrane: investigations by single-molecule tracking vs. fluorescence correlation spectroscopy. FEBS Lett , 1814–1823. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Suzuki KG, Kasai RS, Ritchie K, Fujiwara TK. (2011). Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem Sci , 604–615. [DOI] [PubMed] [Google Scholar]
- Larson DR, Gosse JA, Holowka DA, Baird BA, Webb WW. (2005). Temporally resolved interactions between antigen-stimulated IgE receptors and Lyn kinase on living cells. J Cell Biol , 527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenne PF, Wawrezinieck L, Conchonaud F, Wurtz O, Boned A, Guo XJ, Rigneault H, He HT, Marguet D. (2006). Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J , 3245–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I, Byfield FJ, Chowdhury P, Gai F, Baumgart T, Janmey PA. (2009). Cholesterol-dependent phase separation in cell-derived giant plasma-membrane vesicles. Biochem J , 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I, Veatch S. (2016). The continuing mystery of lipid rafts. J Mol Biol , 4749–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidke DS, Andrews NL, Pfeiffer JR, Jones HDT, Sinclair MB, Haaland DM, Burns AR, Wilson BS, Oliver JM, Lidke KA. (2007). Exploring membrane protein dynamics by multicolor single quantum dot imaging using wide field, TIRF, and hyperspectral microscopy. Proc SPIE , 64480Y. [Google Scholar]
- Lorent JH, Diaz-Rohrer B, Lin X, Spring K, Gorfe AA, Levental KR, Levental I. (2017). Structural determinants and functional consequences of protein affinity for membrane rafts. Nat Commun , 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent JH, Levental KR, Ganesan L, Rivera-Longsworth G, Sezgin E, Doktorova MD, Lyman E, Levental I. (2019). The mammalian plasma membrane is defined by transmembrane asymmetries in lipid unsaturation, leaflet packing, and protein shape. BioRxiv 10.1101/698837. [Google Scholar]
- Lyman E, Hsieh CL, Eggeling C. (2018). From dynamics to membrane organization: experimental breakthroughs occasion a “modeling manifesto.” Biophys J , 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machta BB, Papanikolaou S, Sethna JP, Veatch SL. (2011). Minimal model of plasma membrane heterogeneity requires coupling cortical actin to criticality. Biophys J , 1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magde D, Elson E, Webb W.(1972). Thermodynamic fluctuations in a reacting system—measurement by fluorescence correlation spectroscopy. Phys Rev Lett , 705–708. [Google Scholar]
- Meder D, Moreno MJ, Verkade P, Vaz WL, Simons K. (2006). Phase coexistence and connectivity in the apical membrane of polarized epithelial cells. Proc Natl Acad Sci USA , 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon AK, Holowka D, Webb WW, Baird B. (1986). Clustering, mobility, and triggering activity of small oligomers of immunoglobulin E on rat basophilic leukemia cells. J Cell Biol , 534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoguchi K, Kihara H, Nishikata H, Hamawy MM, Siraganian RP. (1994). Src family tyrosine kinase Lyn binds several proteins including paxillin in rat basophilic leukemia-cells. Mol Immunol , 519–529. [DOI] [PubMed] [Google Scholar]
- Moens PD, Digman MA, Gratton E. (2015). Modes of diffusion of cholera toxin bound to GM1 on live cell membrane by image mean square displacement analysis. Biophys J , 1448–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng XW, Teh C, Korzh V, Wohland T. (2016). The secreted signaling protein Wnt3 is associated with plasma membrane lipid domains in vivo: a SPIM-FCS study. Biophys J , 418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson GL. (2014). The fluid-mosaic model of membrane structure: still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim Biophys Acta , 1451–1466. [DOI] [PubMed] [Google Scholar]
- Nicovich PR, Kwiatek JM, Ma Y, Benda A, Gaus K. (2018). FSCS reveals the complexity of lipid domain dynamics in the plasma membrane of live cells. Biophys J , 2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F, Salomon DS. (2006). Epidermal growth factor receptor (EGFR) signaling in cancer. Gene , 2–16. [DOI] [PubMed] [Google Scholar]
- Owen DM, Williamson DJ, Magenau A, Gaus K. (2012). Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nat Commun , 1256. [DOI] [PubMed] [Google Scholar]
- Prakriya M. (2013). Store-operated Orai channels: structure and function. Curr Top Membr , 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pralle A, Keller P, Florin E-L, H0rbor JKH. (2000). Sphingolipid–cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J Cell Biol , 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyenta PS, Holowka D, Baird B. (2001). Cross-correlation analysis of inner-leaflet-anchored green fluorescent protein co-redistributed with IgE receptors and outer leaflet lipid raft components. Biophys J , 2120–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyenta PS, Schwille P, Webb WW, Holowka D, Baird B. (2003). Lateral diffusion of membrane lipid-anchored probes before and after aggregation of cell surface IgE-receptors. J Phys Chem A , 8310–8318. [Google Scholar]
- Raghunathan K, Kenworthy AK. (2018). Dynamic pattern generation in cell membranes: current insights into membrane organization. Biochim Biophys Acta Biomembr , 2018–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghupathy R, Anilkumar AA, Polley A, Singh PP, Yadav M, Johnson C, Suryawanshi S, Saikam V, Sawant SD, Panda A, et al (2015). Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell , 581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjit S, Lanzano L, Gratton E. (2014). Mapping diffusion in a living cell via the phasor approach. Biophys J , 2775–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M, Mayor S. (2014). Active organization of membrane constituents in living cells. Curr Opin Cell Biol , 126–132. [DOI] [PubMed] [Google Scholar]
- Ries J, Weidemann T, Schwille P. (2012). Fluorescence correlation spectroscopy. In: Comprehensive Biophysics, ed. P Schwille, Cambridge, MA: Elsevier, 210–245. [Google Scholar]
- Sadegh S, Higgins JL, Mannion PC, Tamkun MM, Krapf D. (2017). Plasma membrane is compartmentalized by a self-similar cortical actin meshwork. Phys Rev X , 011031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl SJ, Leutenegger M, Hilbert M, Hell SW, Eggeling C. (2010). Fast molecular tracking maps nanoscale dynamics of plasma membrane lipids. Proc Natl Acad Sci USA , 6829–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran J, Bag N, Kraut RS, Wohland T. (2013). Accuracy and precision in camera-based fluorescence correlation spectroscopy measurements. Anal Chem , 3948–3954. [DOI] [PubMed] [Google Scholar]
- Sarkar P, Chattopadhyay A. (2019). Exploring membrane organization at varying spatiotemporal resolutions utilizing fluorescence-based approaches: implications in membrane biology. Phys Chem Chem Phys , 11554–11563. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Hammond A, Holowka D, Baird B. (2008). Structural determinants for partitioning of lipids and proteins between coexisting fluid phases in giant plasma membrane vesicles. Biochim Biophys Acta , 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin E, Levental I, Mayor S, Eggeling C. (2017). The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol , 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby SA, Holowka D, Baird B, Veatch SL. (2013). Distinct stages of stimulated FcepsilonRI receptor clustering and immobilization are identified through superresolution imaging. Biophys J , 2343–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby SA, Veatch SL, Holowka DA, Baird BA. (2016). Functional nanoscale coupling of Lyn kinase with IgE-FcepsilonRI is restricted by the actin cytoskeleton in early antigen-stimulated signaling. Mol Biol Cell , 3645–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvartsman DE, Donaldson JC, Diaz B, Gutman O, Martin GS, Henis YI. (2007). Src kinase activity and SH2 domain regulate the dynamics of Src association with lipid and protein targets. J Cell Biol , 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Gerl MJ. (2010). Revitalizing membrane rafts: new tools and insights. Nature reviews. Mol Cell Biol , 688–699. [DOI] [PubMed] [Google Scholar]
- Simons K, Sampaio JL. (2011). Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol , a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamy MJ, Ciani L, Ge M, Smith AK, Holowka D, Baird B, Freed JH. (2006). Coexisting domains in the plasma membranes of live cells characterized by spin-label ESR spectroscopy. Biophys J , 4452–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JL, Feder TJ, Webb WW. (1992). Effects of protein concentration on IgE receptor mobility in rat basophilic leukemia cell plasma membranes. Biophys J , 1402–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble WS, Grinstein S. (2015). Barriers to the free diffusion of proteins and lipids in the plasma membrane. J Cell Biol , 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkova NM, Horvath I, Torok Z, Wolkers WF, Balogi Z, Shigapova N, Crowe LM, Tablin F, Vierling E, Crowe JH, Vigh L. (2002). Small heat-shock proteins regulate membrane lipid polymorphism. Proc Natl Acad Sci USA , 13504–13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten TS, Mayor S. (2015). Current approaches to studying membrane organization. F1000Res , 1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten SV, Mackenzie GG, Oteiza PI. (2010). The plasma membrane plays a central role in cells response to mechanical stress. Biochim Biophys Acta , 1739–1749. [DOI] [PubMed] [Google Scholar]
- Viola A, Gupta N. (2007). Tether and trap: regulation of membrane-raft dynamics by actin-binding proteins. Nat Rev Immunol , 889–896. [DOI] [PubMed] [Google Scholar]
- Vogel SK, Greiss F, Khmelinskaia A, Schwille P. (2017). Control of lipid domain organization by a biomimetic contractile actomyosin cortex. eLife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrezinieck L, Rigneault H, Marguet D, Lenne P. (2005). Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys J , 4029–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser S, Moertelmaier M, Fuertbauer E, Stockinger H, Schutz GJ. (2007). (Un)confined diffusion of CD59 in the plasma membrane determined by high-resolution single molecule microscopy. Biophys J , 3719–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler PM, Regmi R, Flauraud V, Brugger J, Rigneault H, Wenger J, Garcia-Parajo MF. (2017). Transient nanoscopic phase separation in biological lipid membranes resolved by planar plasmonic antennas. Acs Nano , 7241–7250. [DOI] [PubMed] [Google Scholar]
- Wiseman PW. (2012). Image correlation spectroscopy. In: Comprehensive Biophysics, ed. P Schwille, Cambridge, MA: Elsevier, 246–259. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.