Abstract
Differential epidermal growth factor receptor (EGFR) phosphorylation is thought to couple receptor activation to distinct signaling pathways. However, the molecular mechanisms responsible for biased signaling are unresolved due to a lack of insight into the phosphorylation patterns of full-length EGFR. We extended a single-molecule pull-down technique previously used to study protein–protein interactions to allow for robust measurement of receptor phosphorylation. We found that EGFR is predominantly phosphorylated at multiple sites, yet phosphorylation at specific tyrosines is variable and only a subset of receptors share phosphorylation at the same site, even with saturating ligand concentrations. We found distinct populations of receptors as soon as 1 min after ligand stimulation, indicating early diversification of function. To understand this heterogeneity, we developed a mathematical model. The model predicted that variations in phosphorylation are dependent on the abundances of signaling partners, while phosphorylation levels are dependent on dimer lifetimes. The predictions were confirmed in studies of cell lines with different expression levels of signaling partners, and in experiments comparing low- and high-affinity ligands and oncogenic EGFR mutants. These results reveal how ligand-regulated receptor dimerization dynamics and adaptor protein concentrations play critical roles in EGFR signaling.
INTRODUCTION
The ability of a cell to respond rapidly and specifically to changes in the surrounding environment is controlled by protein–protein interactions at the plasma membrane and along the signaling cascade. While much is known about the biochemical events that govern signaling pathways, this information has mostly been derived from population-based measurements that average over millions of cells and/or proteins. However, there is growing evidence that system heterogeneity at both the cellular and molecular levels contribute to cellular information processing (Lahav et al., 2004; Feinerman et al., 2008; Coba et al., 2009; Spencer et al., 2009). The epidermal growth factor receptor (EGFR/ErbB1/HER1) has 20 cytoplasmic tyrosines, at least six of which (Y992, Y1045, Y1068, Y1086, Y1148, and Y1173) are capable of recruiting signaling proteins when phosphorylated (Jorissen et al., 2003; Schulze et al., 2005; Hause et al., 2012). The potential for multisite phosphorylation on individual receptors provides a mechanism to control signaling output that is dependent on complex factors such as steric hindrance, relative abundance of receptors and adaptors, cooperative interactions, and lifetimes of complexes (Gibson et al., 2000; Coba et al., 2009; Salazar and Höfer, 2009; Lau et al., 2011; Stites et al., 2015). Signal propagation is opposed by cellular phosphatases (Kleiman et al., 2011), whose activity is proposed to be blocked when phosphotyrosine substrates are engaged with binding partners (Rotin et al., 1992; Brunati et al., 1998; Jadwin et al., 2018). To better understand the contributions of these factors on the differential phosphorylation of EGFR, we have combined quantitative single-molecule measurements with rule-based modeling of EGFR signaling.
Limitations of existing techniques, such as Western blot analysis and those of quantitative mass spectrometry-based proteomics, preclude the detection or accurate quantification of receptor phosphorylation in intact, individual signaling proteins. Single-molecule pull down (SiMPull) is a powerful technique that allows for interrogation of macromolecular complexes at the individual protein level. This technique captures macromolecular complexes on glass coverslips, combining the power of immunoprecipitation with high-resolution imaging for single-molecule quantitative analysis (Jain et al., 2011). We made a number of critical improvements to SiMPull that allowed us to directly detect and rigorously quantify the phosphorylation state of thousands of individual membrane receptors. This represents a significant improvement over traditional, semiquantitative methods, which report only trends in phosphorylation state changes over the entire population. Our improvements to SiMPull protocols include pretreatment to reduce autofluorescence and corrections for receptor surface expression. We employed a simplified imaging chamber that accommodates up to 20 samples, each with sample volumes of only 10 μl. We also demonstrate the critical importance of optimizing antibody labeling and fixation conditions. To quantify receptor phosphorylation, we have used two- and three-color imaging to identify individual proteins and their corresponding phosphorylation status. These multiplex SiMPull measurements provide a new level of detail in the status of receptor phosphorylation that was previously inaccessible with traditional biochemical techniques.
The unique data provided by SiMPull were used to parameterize a mathematical model for site-specific phosphorylation kinetics of EGFR and to test model predictions. Particularly, we explored two traits of relevance for signaling processes: 1) the influence of adaptor protein abundances on the phosphorylation levels of the EGFR residues to which these proteins bind; and 2) the frequency of multisite phosphorylation across individual EGFRs in the population. We specifically explore how the phosphorylation status of individual receptors reflects the opposing contributions of adaptor binding versus phosphatase activity. For example, simulations predicted that overexpression of the adaptor protein Grb2 would lead to increased phosphorylation at the EGFR tyrosine residue where Grb2 binds (Y1068). This prediction was confirmed experimentally using SiMPull and compared with phosphorylation at the EGFR tyrosine residue where Shc1 binds (Y1173), which was found to be significantly less affected by Grb2 overexpression. Additionally, our model predicts that differences in adaptor protein abundances across cell lines should result in differences in phosphorylation patterns. We used our model to evaluate observed phosphorylation kinetics of mutant versus wild-type EGFR, as well as EGFR engaged by the low-affinity ligand epigen versus the high-affinity ligand EGF. Simulation results suggest that the ligand-specific differences we observed in phosphorylation can be explained by differences in receptor dimer lifetimes. These new insights into the extent of phosphorylation at individual tyrosines, along with the existence of multisite phosphorylation, have implications for how EGFR translates extracellular cues into downstream signaling outcomes.
RESULTS
Assessing receptor phosphorylation at the single-molecule level
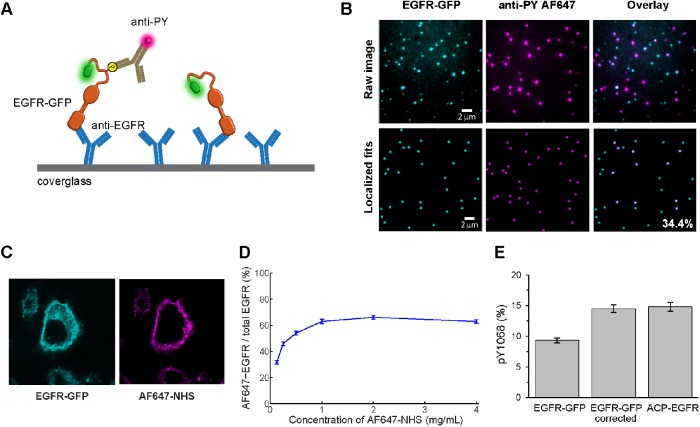
Our principal method to interrogate the phosphorylation status of individual EGFRs after different treatments is single-molecule immunoprecipitation via SiMPull. SiMPull samples are prepared in a manner similar to SDS–PAGE/Western blot protocols, but the sample is evaluated using single-molecule microscopy to dramatically improve quantification. Figure 1A illustrates the basic principles of our assay. Briefly, cells expressing GFP-tagged EGFR are lysed before or after treatments (as specified in the figure legends). Clarified lysates are diluted and dispensed onto coverslips precoated with anti-EGFR antibodies. Following incubation and washes, individual EGFR-GFPs are imaged by total internal reflection fluorescence (TIRF) microscopy. In the raw images, receptors appear as diffraction-limited fluorescent spots that may represent more than one molecule (Figure 1B, top left). Single molecules are selected by fitting each emission profile to a point spread function model and rejecting fluorescent spots that do not fit to a single-emitter model (Figure 1B, bottom left). To evaluate the overall phosphorylation state of individual receptors, coverslips are incubated with pan-reactive, anti-phosphotyrosine (anti-PY) antibodies bearing fluorescent tags with spectral emission distinct from GFP (Figure 1B, center). Colocalization between EGFR-GFP and anti-PY identifies phosphorylated receptors. In this example, the overlay image shows that 34.4% of EGFR-GFPs are phosphorylated on at least one tyrosine site (Figure 1B, right). Phosphorylation at specific tyrosines in the EGFR cytoplasmic tail is evaluated by substituting a site-specific phosphotyrosine antibody (anti-pY1068 or anti-pY1173) for the pan-PY antibodies.
FIGURE 1:
SiMPull to quantify protein phosphorylation. (A) Illustration depicting overall principle for assessing phosphorylation at the single-molecule level using GFP-tagged EGFR (EGFR-GFP) as an example. (B) Representative images showing raw data (top) and blob-reconstructed localized molecules (bottom). CHO-EGFR-GFP cells were stimulated for 5 min with 25 nM EGF at 37°C before lysis for SiMPull. Raw images are brightness and contrast enhanced for visualization. The EGFR-GFP fits were filtered based on their fit to the microscope point spread function and the GFP channel used as a mask to create the overlay. The number in the bottom right image represents the phosphorylation percentage estimated for this field of view. (C) Confocal images showing typical distribution of EGFR-GFP in CHO cells (left) and the labeling of surface proteins achieved with the AF647-NHS ester (right). (D) Cells were incubated with increasing concentrations of AF647-NHS and assayed by SiMPull to determine the percentage of EGFR-GFP molecules labeled with AF647. Number of receptors analyzed per data point, 850 < N < 1550. (E) Percentage of pY1068+ receptors estimated for EGFR-GFP before and after correcting for surface expression. The corrected phosphorylation percentage for EGFR-GFP corresponds to the value measured for ACP-EGFR, which includes only plasma membrane–localized receptors. N > 2400 for each EGFR type. Error bars are SE of measured phosphorylation percentages.
Over the course of this study, we implemented a series of improvements in the experimental process and image analysis to achieve efficient and accurate quantification. These are described briefly here and more detail is found in the Materials and Methods section and in legends to Supplemental Figures S1–S3. As has been previously noted (Jain et al., 2012), autofluorescence background was observed in our green spectral channel (503–548 nm) that was identified as single-GFP molecules in the absence of cell lysate. We found that incubating the PEG-coated coverglass with sodium borohydride (10 mg/ml NaBH4 for 4 min) significantly reduced the number of background fluorescent molecules (Supplemental Figure S1, A and B). To increase our throughput, we generated a simple sample chamber by using a hydrophobic barrier pen to create an array of up to 20 independent regions on a coverglass that requires as little as 10 µl of sample (Supplemental Figure S1C). Because antibodies are used to quantify protein phosphorylation, it is critical to optimize the antibody labeling conditions, including time and concentration (Supplemental Figure S1, D–F). We found that on the time scale of our image acquisition the antibody could dissociate from EGFR (Supplemental Figure S1E), which would lead to an underestimation of receptor phosphorylation. Post–antibody labeling fixation with 4% paraformaldehyde/0.1% glutaraldehyde (PFA/GA) for 10 min stabilized the antibody levels for at least 1 h (Supplemental Figure S1E, PFA/GA). Because binding affinity will vary for each antibody and fluorescent conjugation may also alter antibody affinity, it is necessary to perform a binding curve for each antibody (Supplemental Figure S1F) to ensure optimal labeling conditions. Therefore, in the SiMPull assays reported here, we used antibody doses yielding saturated labeling and chemical cross-linking to permanently couple antibodies to surface-captured receptors, removing potential artifacts from antibody affinities or dissociation. The phosphotyrosines probed in EGFR are located in an intrinsically disordered region of the C-terminal tail; therefore these sites are likely to be more accessible to antibodies than if they were located in folded regions. If a phosphorylation site of interest is found in a folded region of a protein, a denaturation step may be used (Kim et al., 2016).
Correcting for EGFR surface expression in SiMPull data analysis
Like all immunoprecipitation methods that rely on detergent permeabilization of cells as a first step, SiMPull does not distinguish between EGFR trafficking through different intracellular compartments. We observed intracellularly localized EGFR-GFP (Figure 1C, left) that would be inaccessible to ligand. To determine the fraction of EGFR at the plasma membrane, we labeled all surface proteins on the CHO-EGFR-GFP cells with membrane-impermeable AF647-NHS ester (Figure 1C, right) and used SiMPull to visualize the amount of EGFR-GFP colocalized with AF647. By increasing the concentration of AF647-NHS until saturation is achieved, we estimated that ∼65% of the receptors are located at the plasma membrane (Figure 1D). With this information, we corrected our measurements to account for only those receptors available to bind ligand (see Materials and Methods). For example, application of this correction shows that ∼14% of the receptors are phosphorylated at Y1068 after 1 min stimulation with 50 nM EGF (Figure 1E). We note that while surface labeling of receptors with AF647-NHS ester allows for identification of surface proteins, we found that prelabeling of EGFR in this way reduced EGF binding (unpublished data). Therefore, we did not use global NHS labeling of receptors for the study of EGFR activation. To validate our correction method, we analyzed the phosphorylation levels of receptors from CHO cells expressing ACP-tagged EGFR. We directly labeled the plasma membrane–localized EGFR using membrane-impermeable CoA-Atto488 as described previously (Ziomkiewicz et al., 2013; Valley et al., 2015). Cells were then exposed to EGF and probed for EGFR phosphorylation with SiMPull, this time using Atto488 as the marker for plasma membrane EGFR. The percentages of phosphorylated EGFR were similar when comparing the membrane-localized ACP-EGFR and the membrane-corrected EGFR-GFP samples (Figure 1E). Therefore, the effects of EGF binding to EGFR on the plasma membrane can be accurately determined from whole cell lysates and we apply this correction for the remainder of the results.
Extent of phosphorylation varies by tyrosine residue
Results of the use of SiMPull to characterize phosphorylation of EGFR in CHO-EGFR-GFP cells over a range of EGF doses are shown in Figure 2A. The multiwell hydrophobic array format made it possible to efficiently examine a full dose response of activation in a single imaging session. We quantified total EGFR tyrosine phosphorylation (PY) and compared it with the phosphorylation patterns for two specific tyrosine sites (Y1068 and Y1173). Cells stimulated for 5 min (Figure 2A) with increasing concentrations of EGF showed the expected increase in total phosphorylation with ligand dose (Figure 2A, PY, blue bars). This fraction reached 40.8 ± 1.3% with 50 nM EGF, a dose that is considered saturating. While the extent of phosphorylation on specific tyrosines is lower than total PY values across the dose response curve, the fraction of EGFR with phosphorylation at Y1173 was consistently higher than at Y1068 (Figure 2A).
FIGURE 2:
The extent of phosphorylation varies by tyrosine residue. (A) Dose response curve determined by SiMPull measurements for CHO-EGFR-GFP cells after 5 min of EGF addition at 37°C. Number of receptors analyzed per condition, N > 1500. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (B) Schematic of EGFR tyrosine site-specific model, represented as an extended contact map (Chylek et al., 2011). (C) The model is capable of generating dose-response behaviors similar to those observed experimentally (A). (D) Phosphorylation time course for CHO-EGFR-GFP cells stimulated with 25 nM EGF at 37°C. N > 1800. Symbols are the SiMPull data and error bars are SE of measured phosphorylation percentages. The series of experiments reported in A and D were each performed on the same day, but the two series of experiments were performed on different days. For this reason, results at the final time point in D are not combined with the results shown in A. Solid lines in D are derived from simulations using best-fit parameter values. As described in Materials and Methods, dose-response data in A and all times-series data in D were considered in curve fitting, that is, the parameter estimates are based on a global fit. Shaded areas in D indicate prediction uncertainties, determined by sampling parameter values from the multidimensional posterior established via Bayesian parameter estimation.
These results provide important observations. First, phosphorylation detection by SiMPull is sensitive, capable of detecting receptor phosphorylation at low ligand dose. Second, even under saturating ligand conditions, only a fraction of receptors is found to be phosphorylated, reaching a maximum of approximately 41% with 5 min stimulation. Third, the extent of phosphorylation varies by tyrosine residue. The detected phosphorylation levels are not restricted due to limitations in antibody labeling, because Western blot results show similar trends in tyrosine phosphorylation (Supplemental Figure S2, A–C) and cells stimulated in the presence of phosphatase inhibitors showed increased receptor phosphorylation (Supplemental Figure S2, D and E) for all three antibodies. The addition of high salt (500 mM NaCl) in the buffer for the immunoprecipitation protocol, which interferes with electrostatic protein–protein interactions, did not change the detected phosphorylation (Supplemental Figure S2F). This control suggests that, under our conditions, adaptor proteins that might interfere with antibody recognition in the SiMPull protocol are not coprecipitating.
Computational model suggests biased phosphorylation is explained by variations in adaptor protein abundances
To interpret the SiMPull data, we built a mathematical model based on our understanding of EGFR signaling mechanisms. We defined the model in terms of formal rules for interactions using the BioNetGen language (BNGL; Faeder et al., 2009). Supplemental Table S1 summarizes key references and measurements that provide estimates of model parameters, including our own estimates of receptor expression levels in the cell lines utilized in the study. Our model explicitly incorporates the asymmetric orientation of the EGFR kinase domains during dimerization, which assumes one monomer is active while the other monomer serves as an allosteric activator over the lifetime of that dimer pair (Zhang et al., 2006; Pryor et al., 2013). We also incorporated the observation that structural accessibility limits the efficiency of self-phosphorylation (i.e., within an active monomer’s own tail) by about 30% (Kovacs et al., 2015). An electronic version of the model is provided in the form of a BNGL file in the online Supplemental Material (Supplemental File S1). Included with the BNGL file are all of the files necessary to perform the fitting and Bayesian uncertainty quantification procedures (Supplemental File S2). These files are also available online at the RuleHub repository (https://github.com/RuleWorld/RuleHub/tree/2019Jun18/Published/Salazar-Cavazos2019).
Figure 2B illustrates the structure of our model. Our first goal for computational modeling was to explore the possible mechanisms giving rise to the observed biased phosphorylation in EGFR at Y1173 compared with Y1068. As depicted in Figure 2B, and described in detail in Materials and Methods, our model includes site-specific phosphorylation of Y1068 and Y1173, and the recruitment of adaptor proteins Grb2 and Shc1 to these sites, respectively. We considered a range of parameter values in the model that might be adjusted to reproduce the biased phosphorylation (Supplemental Table S2). First, we considered the possibility that the phosphorylation and/or dephosphorylation rates are not equivalent for each tyrosine. It is possible for the model to fit our SiMPull data by allowing phosphorylation and dephosphorylation rate constants for Y1068 and Y1173 to be different, but earlier work suggests that these rate constants have similar values for the two sites (Kleiman et al., 2011; Kim et al., 2012). Thus, our data do not allow site-specific differences in phosphorylation and dephosphorylation at Y1068 and Y1173 to be ruled out, but such differences are dissonant with results from earlier studies.
An alternative possibility is that the binding of adaptor proteins (i.e., Grb2 and Shc1) physically serves as a barrier to protect specific phosphorylated tyrosines from phosphatase activity. This hypothesis is supported by in vitro and cellular studies showing the ability of SH2 domains to protect phosphosites from dephosphorylation (Rotin et al., 1992; Brunati et al., 1998; Jadwin et al., 2018). To test whether variation in protein abundances alone could explain our observations, we allowed the concentration of Grb2 and Shc1 to independently vary during the fitting process, while all other parameters were held constant. As a simplification, we took binding parameters (forward and reverse rate constants) for Grb2 interaction with pY1068 (via Grb2’s SH2 domain) and Shc1 interaction with pY1173 (via Shc1’s phosphotyrosine binding [PTB] domain) to be identical. The equilibrium dissociation constants measured in vitro by fluorescence polarization for these two interactions are comparable (2.6 µM for Grb2 SH2-pY1068 and 1.4 µM for Shc1 PTB-pY1173; Hause et al., 2012). By allowing Grb2 and Shc1 to have different abundances, it became possible to parameterize the model (through curve fitting, as described in Materials and Methods) such that the model generates dose-response behaviors (Figure 2C) as well as dynamic behaviors at the saturating dose of 25 nM EGF (Figure 2D) similar to those observed with SiMPull. The results of uncertainty quantification, illustrated in Figure 2D, show that there is model discrepancy (i.e., variability in experiments not captured by the model). Nevertheless, we regard simulations to adequately reproduce the data, at least qualitatively. The similarity between simulations and observed behaviors suggests that different abundances of phosphotyrosine site-specific binding partners are a plausible explanation for the observed differences in phosphorylation at Y1068 and Y1173.
Predicted influence of Grb2 overexpression in phosphorylation levels is observed experimentally
Our next goal was to experimentally validate the model prediction that biased phosphorylation is best explained by having different expression levels of Grb2 and Shc1. To set up conditions for these experiments, we ran simulations over a range of Grb2 overexpression values (two-, four-, and eightfold). Results in Figure 3A predict that Grb2 overexpression will lead to increased phosphorylation at Y1068, where Grb2 binds. Notably, no change is predicted for phosphorylated Y1173, where Grb2 is not expected to bind (Figure 3B). To test these predictions, we created CHO-EGFR-GFP cells overexpressing Grb2-mCherry and evaluated the influence on EGFR phosphorylation using SiMPull (Figure 3, C and D). Consistent with trends in the simulation results, overexpression of Grb2 led to a significant (approximately threefold) increase in phosphorylation of Y1068 (Figure 3C) and a much less pronounced increase in phosphorylation of Y1173 (Figure 3D). The relatively small change in phosphorylation at Y1173 (Figure 3D) is consistent with reports that high expression of Grb2 can enhance phosphorylation at sites beyond the canonical Grb2 binding sites in EGFR (Jadwin et al., 2018).
FIGURE 3:
Predicted and observed phosphorylation kinetics in cells overexpressing Grb2. Predicted percentages of EGFR phosphorylation at tyrosines 1068 (A) and 1173 (B) after stimulation with 25 nM of EGF in cells with increasing overexpression (OE) of Grb2 (two-, four-, and eightfold). SiMPull quantification of EGFR phosphorylation at tyrosines 1068 (C) and 1173 (D) after stimulation with 25 nM of EGF in CHO-EGFR-GFP cells expressing endogenous levels of Grb2 (orange) or overexpressing Grb2-mCherry (blue). N > 1050. This CHO-EGFR-GFP Grb2-mCherry cell line overexpresses Grb2 by approximately threefold on average, as determined by Western blot analysis (unpublished data), compared with wild-type cells. Error bars represent mean ± SEM.
Model predicts cell-specific phosphorylation patterns based on differences in adaptor protein abundances
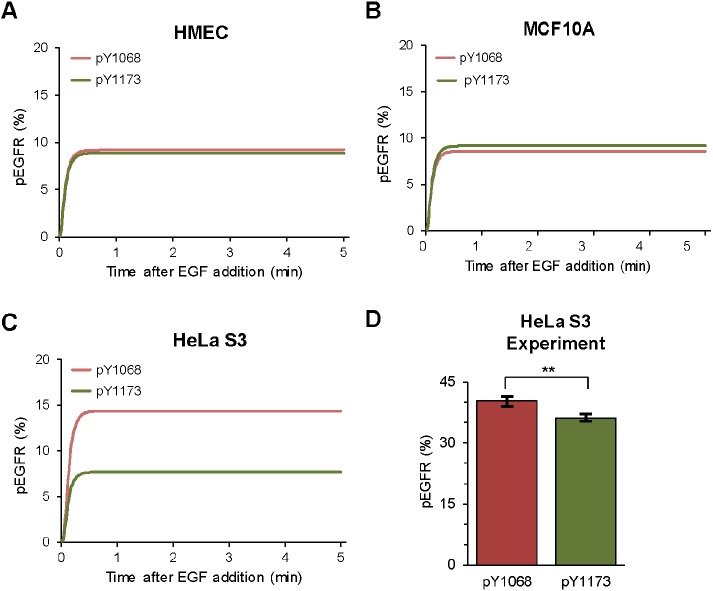
On the basis of these results, we hypothesized that cell types naturally expressing different levels of these adaptor proteins would display different phosphorylation patterns. Protein copy numbers have been assayed using both global and targeted mass spectrometry-based proteomics in various cell lines (Kulak et al., 2014; Shi et al., 2016). These estimates include the protein copy numbers (per cell) for EGFR, Grb2, and Shc1 in the nontumorigenic mammary epithelial HMEC and MCF10A cells, as well as in HeLa cervical cancer cells (see Supplemental Table S1). As described in Materials and Methods, we performed simulations using these measured cell-specific protein copy numbers (Supplemental Table S1) and obtained model predictions for the phosphorylation patterns and kinetics in each of the three cell lines (Figure 4, A–C). In HMEC cells, where the estimated abundances of both adaptor proteins are relatively low, the model predicts similar levels of phosphorylation at both tyrosine residues (Figure 4A). For MCF10A cells, the model predicts slightly higher phosphorylation at Y1173 given that its binding partner, Shc1, is expressed at a higher level than Grb2 (Figure 4B). The most striking difference in expression levels between the two adaptor proteins is found in HeLa S3 cells, where it is estimated that there are ∼600,000 copies of Grb2 per cell compared with ∼100,000 copies of Shc1 per cell (Supplemental Table S1). Accordingly, for these cells, the model predicts that phosphorylation at Y1068 should be higher than at Y1173 (Figure 4C), the reverse of what is predicted by the version of the model parameterized for consistency with our CHO-cell data. Experimental results using SiMPull are shown in Figure 4D. These results confirm that HeLa S3 cells exhibit the expected reversal in biased phosphorylation. The difference in phosphorylation is less pronounced than predicted, with phosphorylation at Y1068 only slightly higher than phosphorylation at Y1173, but there is qualitative consistency between the predicted and observed phosphorylation levels at the two sites.
FIGURE 4:
Predicted phosphorylation patterns in cell lines with varying adaptor protein expression. (A–C) Predictions of phosphorylation kinetics for HMEC (A), MCF10A (B), and HeLa S3 (C) cell lines. HMEC cells express low levels of both Grb2 and Shc1; MCF10A cells have slightly higher levels of Shc1 than Grb2; HeLa S3 cells express approximately seven times more Grb2 than Shc1 (Kulak et al., 2014; Shi et al., 2016; see Supplemental Table S1). (D) Phosphorylation pattern in HeLa S3 cells obtained by SiMPull measurements. N > 1550. **, P ≤ 0.01. Error bars represent mean ± SEM.
Three-color SiMPull reveals frequency of multisite phosphorylation on individual EGFR
Our results above suggest that EGF-treated cells bear subpopulations of receptors with differing phosphorylation patterns. SiMPull offers unique advantages over traditional methods, because individual molecules can be probed with more than one antibody providing each has a spectrally distinct fluorescent tag. We utilized the capability for simultaneous three-color SiMPull imaging to determine the frequency of multisite phosphorylation on individual EGFRs. Figure 5A illustrates the basic protocol used to evaluate the incidence of multisite phosphorylation at the single-receptor level. The image in Figure 5B shows a typical result, where each receptor is resolved based on GFP emission (dark green), and then overlaid with detection of anti–PY1068-CF555 (light green) and detection of anti–PY-AF647 (purple). Circled, white spots indicate the presence of EGFRs that are positive for both antibodies and, therefore, phosphorylated on PY1068 as well as at least one other tyrosine in the cytoplasmic tail.
FIGURE 5:
SiMPull reveals EGFR multisite phosphorylation. (A) Schematic of three-color labeling. (B) Representative three-color SiMPull image of EGFR-GFP (dark green), anti-pY1068 (light green), and anti–pan-PY (purple). White circles indicate receptors that contain all three labels and are therefore positive for both pY1068 and pan-PY. Cells were treated with 25 nM EGF for 5 min at 37°C. (C) Quantification of three-color SiMPull shows that multisite phosphorylation is readily observed over a range of EGF concentrations. Three-color images were analyzed for the fraction of EGFR demonstrating colabeling of pY1068+PY+ (orange) and for pY1173+PY+ (brown). N > 850. (D) Plot shows the fraction of pan-phosphorylation found in the total EGFR population (dark blue), the pY1068+ population (medium blue), and the pY1173+ population (light blue). N > 850. **, P ≤ 0.01. (E) Bar graph showing the fraction of pY1068+ receptors within the total EGFR population (red) as compared with within the phosphorylated receptor population (pink). N > 850. (F) Mathematical model parameterized by single-site phosphorylation data predicts the presence of dual-site phosphorylation (pYpY, orange line). (G) SiMPull experiments demonstrate dual phosphorylation at Y1068 and Y1173 that is qualitatively consistent with model predictions shown in F. N > 750. All error bars represent mean ± SEM.
When labeling a single protein with two or more antibodies, the potential for steric hindrance must be considered. We first tested whether colabeling of receptors with pairwise combinations of antibodies to PY1068, PY1173, and/or pan-PY was technically feasible. Results in Supplemental Figure S3 confirm the feasibility of a method that employs sequential, pairwise antibody labeling and also provides for correction of steric blocking during our labeling procedures (see also Materials and Methods). We also performed step-photobleaching analysis to assure that the doubly labeled receptors were associated with a single EGFR-GFP molecule and not two or more EGFRs in a diffraction-limited spot (Supplemental Figure S4A). Step photobleaching confirmed that at our pull-down density the majority of double-labeled receptors are indeed monomers (∼98%).
By combining anti–pan-PY labeling with each of the two specific sites (pY1173 or pY1068), we were able to quantify the frequency of multisite phosphorylation on individual receptors. First, we examined to what extent a receptor phosphorylated at Y1068 or Y1173 might also be phosphorylated at another tyrosine (pan-PY). Figure 5C reports the percentage of all receptors exhibiting multisite phosphorylation, found to reach 13.0 ± 1.8% as a function of EGF dose for each of the two tyrosines examined here. From this data, we can also interrogate the behavior of specific phosphorylation events with respect to a particular receptor pool. For example, Figure 5D shows the probability of a receptor being phosphorylated (PY) within three distinct subpopulations. As expected, the total percentage of phosphorylated EGFR increased with dose (Figure 5D, PY in total EGFR, dark blue). In the fraction of receptors phosphorylated at Y1068, there is a high probability of that same receptor being colabeled for pan-PY, reaching 80.3 ± 4.9% after 5 min of 50 nM EGF stimulation (Figure 5D, PY in pY1068+ EGFR, medium blue). Additionally, we found that pY1173+ receptors were frequently colabeled for pan-PY (Figure 5D, PY in pY1173+, light blue) at a high frequency, albeit lower than for pY1068+ EGFR. These data suggest that, at least in CHO cells, there is a slight increase in multisite phosphorylation in combination with pY1068 over pY1173. We also analyzed the data in a manner to determine the fraction of pY1068 within the total receptor (pY1068 in total EGFR) or phosphoreceptor (pY1068 in PY+ EGFR) populations. Figure 5E shows that the total fraction of EGFR phosphorylated at Y1068 increases with ligand dose (red), while the fraction of pY1068 within the PY+ EGFR population remains relatively constant (pink). Simulations corresponding to experiments in Figures 5, D and E, are qualitatively consistent with the observed results (Supplemental Figure S4, B and C), and predict that even at subnanomolar EGF concentrations the levels of pY1068 in PY+ EGFR remain at comparable levels as when EGF concentrations are high (Supplemental Figure S4C). This supports the idea that, at least for tyrosine sites with fast phosphorylation kinetics such as Y1068, the phosphorylation levels within a signaling unit (activated receptor) remain relatively constant and the influence of ligand dose is in the number of activated receptors. Together, these results directly demonstrate that it is common for individual EGFRs to acquire phosphorylation on multiple sites.
Dual phosphorylation at Y1068-Y1173 is predicted by the model and supported by SiMPull measurements
We next explored the mechanistic basis for multisite phosphorylation in our computational model, which was initially trained using only single-site phosphorylation data. Simulation results in Figure 5F predict that approximately 3% of receptors will become dually phosphorylated at Y1068 and Y1173 (pYpY) within 1 min of saturating ligand dose, reaching a steady-state value sustained for at least 5 min. To test this prediction experimentally, we performed three-color analysis by treating cells for 5 min with EGF, followed by lysis and SiMPull isolation. Samples were colabeled with anti-pY1068 and anti-pY1173 antibodies and corrected for reduced labeling efficiency due to steric blocking (see Supplemental Figure S3, B–D, and Materials and Methods). As shown in Figure 5G, we found that 6.9 ± 1.3% of total receptors have dual phosphorylation at Y1068 and Y1173 (pYpY). As a control, we treated cells with EGF in the presence of phosphatase inhibitors and found a corresponding increase in the percentage of dually phosphorylated EGFR (Supplemental Figure S3E). Therefore, both our computational model and quantitative SiMPull experiments indicate that a fraction of EGFR is dually phosphorylated at Y1068 and Y1173 in response to ligand stimulation.
Dimerization efficiency and lifetime are key factors in multisite phosphorylation
We performed a sensitivity analysis to determine how dual phosphorylation depends on parameter values (Supplemental Figure S5). Changes in phosphorylation and dephosphorylation rate constants had the greatest effect on the percentage of dual phosphorylation. Changes in parameters that govern adaptor protein recruitment had the next largest effects on dual phosphorylation.
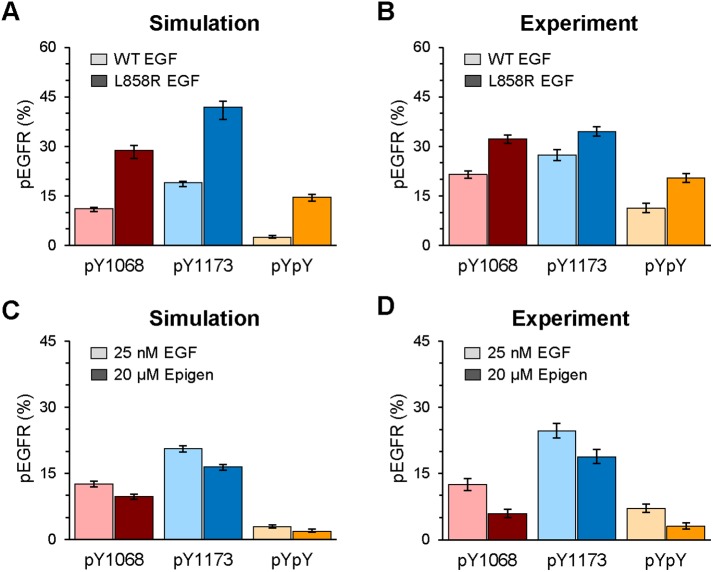
To validate the impact of changes in kinase activity, we turned to our prior work with activating EGFR mutations common in non–small cell lung cancer (Valley et al., 2015). The oncogenic properties of the L858R EGFR mutant have been attributed in part to increased kinase activity (Zhang et al., 2006), as well as structural changes that enhance dimerization potential in the absence of ligand (Valley et al., 2015). Using the mathematical model, we simulated the mono- and dual-site phosphorylation in response to increased kinase activity (reflected by the pseudo first-order rate constant used in the model to characterize autophosphorylation) for L858R EGFR dimers over wild-type (WT) EGFR dimers. The model predictions in Figure 6A show that enhanced kinase activity will lead to increases in phosphorylation at both Y1068 and Y1173, as well as in dual phosphorylation. Experimentally, we compared single and multisite phosphorylation in CHO cells expressing WT or L858R EGFR under saturating EGF conditions, where our prior work showed that ligand-bound WT and mutant EGFR have the same dimer lifetime (Valley et al., 2015). In qualitative agreement with modeling predictions, we found that L858R EGFR exhibits an increase in phosphorylation at each individual tyrosine as well as in dual phosphorylation at both Y1068 and Y1173 (Figure 6B). To obtain a quantitative estimate of the increased kinase activity, we fitted our model to the data of Figure 6B. In the fitting procedure, we allowed the rate constant for autophosphorylation to vary, but held all other parameters fixed at their previously estimated values. The one-parameter fit indicates that the intrinsic kinase activity of L858R EGFR is 3.6-fold higher than WT EGFR. Simulation results based on the best fit are shown in Figure 6A. Our estimate is consistent with previous in vitro measurements, in which the kinase activity for the L858R kinase domain was determined to be approximately 3.5-fold higher (Zhang et al., 2006).
FIGURE 6:
Altered multisite phosphorylation resulting from EGFR mutation or stimulation by a low-affinity ligand. (A) Model predicts higher single-site and multisite phosphorylation in EGFR-L858R vs. WT, where the mutant has a higher kinase activity. (B) SiMPull measurements for WT and mutant EGFR expressed in CHO cells display qualitative agreement with model predictions in A. Cells were stimulated with 50 nM EGF for 5 min at 37°C. N > 950. Here, anti–PY-AF647 was used to identify phosphorylated surface EGFR since the receptors were not GFP-tagged. (C) Model predicts a lower phosphorylation with epigen stimulation as compared with EGF, where the mechanism is that epigen induces less stable dimers. (D) SiMPull measurements for EGF- and epigen-stimulated cells display qualitative agreement with model predictions in C. CHO-EGFR-GFP cells were stimulated for 5 min at 37°C with the indicated saturating ligand dose. N > 1200. Error bars for simulations report the prediction uncertainties determined via the Bayesian analysis described in Materials and Methods; error bars for experimental results represent mean ± SEM.
We were particularly interested in exploring the interdependent relationships between ligand affinity, dimerization off-rates, and multisite phosphorylation. To explore this experimentally, we focused on comparing multisite EGFR phosphorylation in cells stimulated with either (high affinity) EGF or another natural EGFR ligand, (low affinity) epigen. We chose this system because shorter dimer lifetimes were recently proposed to be a key feature that explains altered cellular outcomes of epigen-mediated signaling (Freed et al., 2017). We used the mathematical model to predict the outcome of shorter dimer lifetimes on EGFR phosphorylation and found that receptor phosphorylation at pY1068 is expected to be reduced (Figure 6C), in qualitative agreement with Western blot results reported by Freed et al. (2017). The model also predicted that phosphorylation at Y1173 and dual phosphorylation should be reduced. We used three-color SiMPull to evaluate the differential levels of multisite phosphorylation for EGFR after stimulation with either EGF or epigen, using saturating levels of both ligands to ensure equivalent occupancy (Figure 6D). We found that both tyrosines were phosphorylated to a lesser extent by epigen than in EGF-stimulated cells. The number of receptors achieving dual phosphorylation was also reduced with epigen activation. To determine what dimer lifetime best explains these results, we fitted our model to the data of Figure 6D. In the fitting procedure, we allowed the rate constant for EGFR dimer dissociation to vary, but held all other parameters fixed at their previously estimated values. The one-parameter fit indicates that epigen-induced dimers have 4.8-fold shorter lifetime than EGF-induced dimers. Simulation results based on the best fit are shown in Figure 6C. These results strengthen the argument that altered epigen signaling is a result of shorter-lived dimer interactions.
DISCUSSION
Mathematical modeling of cellular signaling, including EGFR signaling, has historically relied on population-based measurements (Kholodenko et al., 1999; Hendriks et al., 2003). Parameterization of mathematical models has improved over time, incorporating more accurate measurements such as quantitative mass spectrometry techniques to profile overall protein abundance patterns for the EGFR–MAPK pathway (Shi et al., 2015), as well as ligand-induced changes in the phosphoproteome (Yi et al., 2018). Concomitantly, the systems biology field has increasingly recognized the influence of heterogeneity at the single-cell scale in determining signaling output (Kolitz and Lauffenburger, 2012). Recent advances in quantitative imaging now provide powerful new methods to interrogate single-cell and single-molecule dynamics, which are providing new insights on how cells encode and interpret information (Grecco et al., 2011; Welch et al., 2011; Purvis and Lahav, 2013). In prior work, we have built spatial stochastic models of EGFR/ErbB signaling (Pryor et al., 2013, 2015) that incorporate single-molecule measurements, including experimentally derived values for receptor diffusion and ligand-dependent changes in dimer off-rates (Low-Nam et al., 2011; Steinkamp et al., 2014). These properties of EGFR continue to be key features of our current phosphorylation site-specific, rule-based model.
Wollman and colleagues have proposed that a deeper understanding of signaling networks requires richer data sets collected at the single-cell level, coupled closely with computational modeling in an iterative process of prediction, experimental confirmation, and manipulation (Handly et al., 2016). In this work, we set out to purposely pair simulations with novel single-molecule quantitative data, capturing the dose-dependent heterogeneity in phosphorylation of thousands of single receptors. Improvements in the single-molecule technique, SiMPull, allowed us to obtain robust, quantitative information about EGFR multisite phosphorylation patterns and to identify the frequency of subpopulations. We found that there are distinct populations of receptors with varying phosphorylation patterns, indicating an early diversification of downstream signaling. These data were used to train a rule-based model of site-specific EGFR phosphorylation and adaptor recruitment (i.e., to estimate unknown model parameter values via a curve-fitting procedure).
Our rule-based model for EGFR signaling, illustrated in Figure 2B as an extended contact map (Chylek et al., 2011), has a structure consistent, to a first-order approximation, with known and well-established mechanisms of ligand–receptor binding, ligand-dependent receptor-mediated receptor dimerization, autophosphorylation, and phosphotyrosine-dependent recruitment of adaptor proteins. However, the model should be viewed as an idealization, because as a simplification, we purposely excluded a number of potentially relevant complicating factors, such as inside-out effects of signaling on ligand binding (Macdonald-Obermann and Pike, 2009), competition among the cellular milieu of SH2/PTB domain-containing signaling proteins for phosphotyrosine docking sites in EGFR (Stites et al., 2015), and cooperative recruitment of adaptor proteins (Sigismund et al., 2013). These and other aspects of EGFR signaling are not yet fully understood. Another reason to view the model as an idealization is the outcome of our Bayesian analysis of prediction uncertainty, described in Materials and Methods. Results from this analysis, which are illustrated in Figure 2D, indicate that the parameterized model is not able to fully capture variability in the training data. Despite this model discrepancy issue, we deemed the parameterized model to adequately reproduce the training data and to be useful for guiding further experiments, for the following reasons. Simulations are similar to and qualitatively consistent with observed dose-response behaviors (cf. Figure 2, A and C) and observed dynamic behaviors (cf. the curves and data points in Figure 2D). It should be noted that SiMPull assays are destructive. Thus, any training data spanning multiple conditions would necessarily need to be generated from multiple independent experiments performed in parallel. This requirement very likely introduces uncontrolled variability, which is not accounted for in the model.
We directly tested the widely held assumption that SH2 domain binding transiently protects specific phosphotyrosines from cellular phosphatases, as suggested by prior work using semiquantitative, population-based Western blot techniques (Rotin et al., 1992; Brunati et al., 1998; Sigismund et al., 2013; Jadwin et al., 2018). As our model predicted, overexpression of Grb2 led to a pronounced increase in phosphorylation at Y1068 in EGFR, the principal site where Grb2 binds, but largely left phosphorylation at Y1173, where Shc1 binds, unchanged. This behavior may be important for understanding EGFR signaling in cancer cells, where expression levels of signaling proteins are commonly altered (Verbeek et al., 1997; Santarius et al., 2010). In particular, we predicted and observed biased phosphorylation of Y1068 and Y1173 that shifts across cell lines having different adaptor protein abundances (cf. Figures 2 and 4).
It should be acknowledged that the observed reversal in biased phosphorylation of Y1068 and Y1173 for HeLa S3 cells (vs. CHO cells) was less pronounced than predicted (cf. Figure 3, C and D). There are at least two potential explanations for this discrepancy of degree. First, there could be errors in the copy number measurements that were used to obtain the cell-specific models. In a recent study, concerns were raised about the accuracy of available data sets (Erickson et al., 2019), including the data set that we used to set abundances of EGFR, Grb2, and Shc1 for the HeLa cell–specific model. Second, as a simplification, we held rate constants for receptor phosphorylation and dephosphorylation fixed across the different cell-specific versions of the model. However, these rate constants are potentially cell specific. Phosphatase abundances are known to vary across cell type (Shi et al., 2016), giving rise to the possibility that effective dephosphorylation rates may be cell specific. Furthermore, membrane properties are known to vary across cell type (Lajoie et al., 2007), with potential impacts on EGFR localization and dimerization, which could affect autophosphorylation. Finally, it should be recalled that our model is an idealization, and as such, it cannot be expected to match observations with fine-grained resolution. The model provides a plausible explanation for our findings about the interplay between adaptor protein abundances and site-specific EGFR phosphorylation patterns, but there may be other explanations, such as cell-specific kinase and phosphatase activities, which cannot be ruled out. Although we focused on two prototypical examples of binding partners, Grb2 and Shc1 (Kholodenko et al., 2000), the principles we describe likely apply to the broader interactome map of ∼89 SH2-bearing proteins that interact with ErbB receptors with varying degrees in affinity, abundance, and tissue-specific expression (Hause et al., 2012). Of course, some SH2-bearing proteins, such as Grb2, may be somewhat promiscuous and bind with weaker affinity to more than one phosphosite in the receptor cytoplasmic tail (Jadwin et al., 2018). Binding of adaptors at one or more sites may partially block the accessibility of other SH2 proteins or phosphatases at nearby sites or even serve as scaffolds to build larger macromolecular complexes (Hsieh et al., 2010; Telesco et al., 2011). Addressing the combinatorial complexity of these signaling interactions is a strength of rule-based approaches (Chylek et al., 2014), particularly when combined with single-molecule quantification.
We demonstrate the advantages of SiMPull for determining the fraction of receptors that simultaneously bear more than one posttranslational modification. This ability required considerable optimization of SiMPull protocols, representing a step forward over previous protocols that failed to detect an EGF-dependent increase in multisite EGFR phosphorylation (Kim et al., 2016, 2018). Notably, our modeling and experimental results indicating that individual copies of EGFR are phosphorylated at multiple sites are consistent with previous work indicating that multisite phosphorylation is important for recruitment of certain combinations of signaling proteins to activated EGFR (Sigismund et al., 2013; Fortian and Sorkin, 2014). Interestingly, our results demonstrate that not all receptors are phosphorylated identically. Differential phosphorylation patterns may allow individual receptors to connect to distinct pathways for biased signaling. It is particularly important to understand biased signaling in the context of different ligand types and doses, as well as cell variables such as the expression levels of receptors and their signaling partners (Wolf-Yadlin et al., 2006; Chen et al., 2009; Wilson et al., 2012; Freed et al., 2017).
Our combination of modeling and experiments has provided a connection between receptor dimer lifetimes and phosphorylation outcomes. Recent work from the Lemmon group has explored differential phosphorylation patterns and signaling outcomes after EGFR stimulation with high- and low-affinity ligands (Freed et al., 2017). Our contributions to that study supported the conclusion that low-affinity ligands, such as epigen, induce distinct dimer structures that are less stable than dimers induced by EGF. By allowing the dissociation rate constant for ligand-induced EGFR dimers to vary in a one-parameter fit, our model became capable of closely reproducing the SiMPull data from experiments with epigen-treated cells. The adjusted value for this rate constant corresponds to a reduced dimer lifetime, suggesting that shorter-lived epigen-induced EGFR dimers is a sufficient explanation for the observed differences in epigen- versus EGF-induced EGFR phosphorylation kinetics. Taken together with our results from EGFR mutants, the data indicate that dimer lifetime and kinase efficiency act together to dictate rates of multisite phosphorylation. We showed previously that EGF-bound WT and L858R EGFR dimers have similar lifetimes (Valley et al., 2015), while our new SiMPull results show that EGF treatment leads to significant increases in single- and dual-site phosphorylation for the mutant. Simulations reported in Figure 6 indicate that this can be explained by the 3.6-fold increase in intrinsic kinase activity, a result that is reasonably consistent with biochemical measurements using recombinant kinase domains (Zhang et al., 2006). On the other hand, L858R receptors also favor ligand-independent signaling, attributed to “inside-out” signaling that promotes extension of the extracellular domain and exposure of the dimer arm in the absence of ligand (Valley et al., 2015) and/or more favorable orientation of cytoplasmic kinase domains (Shan et al., 2012; Red Brewer et al., 2013). So, the oncogenic properties of the L858R mutation can be attributed to both the increase in catalytic activity and enhanced dimerization in the absence of ligand.
In summary, we have integrated the unique data sets provided by SiMPull with mathematical modeling to better understand the combinatorial aspects of EGFR signaling. We have shown that there are distinct populations of receptors with varying degrees of phosphorylation that can connect individual receptors to distinct pathways for biased signaling. Our results provide novel insight into the regulation of EGFR phosphorylation, demonstrating critical roles for adaptor protein abundance and receptor dimerization lifetimes. The abundances of adaptor proteins vary across cell types (Shi et al., 2016) and it has been proposed that these variations explain cell-specific functions of EGFR and other receptor tyrosine kinases (RTKs). Here, we have seen that there is an interplay between single-molecule patterns of phosphorylation and adaptor protein abundances. Thus, SiMPull assays complement measurements of these abundances. We have also shown that these types of data can be analyzed in an integrated manner using a mechanistic model. Finally, we have demonstrated that multisite phosphorylation is quite prevalent and needs to be considered in modeling of EGFR signaling. Further work along these lines will help us to better understand the context-dependent functions of RTKs in the future.
MATERIALS AND METHODS
Cell lines and reagents
CHO cells expressing GFP-tagged (Brock et al., 1999; Lidke et al., 2004) or ACP-tagged EGFR (provided by Donna Arndt-Jovin, Max Planck Institute for Biophysical Chemistry) were cultured in DMEM supplemented with 10% fetal bovine serum, penicillin–streptomycin, and 2 mM l-glutamine (Thermo Fisher Scientific). ACP-tagged EGFR was as described in Ziomkiewicz et al. (2013) and Valley et al. (2015) with the exception that a shortened 16 aa sequence was introduced at the EGFR N-terminus (George, 2006). EGF, protease and phosphatase inhibitor cocktail, Alexa Fluor 647 NHS ester, and NeutrAvidin were purchased from Thermo Fisher Scientific. CoA 488 and ACP synthase were purchased from New England Biolabs. N-(2-aminoethyl)-3-aminopropyltrimethoxysilane was purchased from United Chemical Technologies (#A0700). Sodium bicarbonate and sodium borohydride were purchased from EMD Millipore (#SX0320-1, #SX0380-3). mPEG-succinimidyl valerate (MPEG-SVA-5000-5g) and biotin-PEG-succinimidyl valerate (Biotin-PEG-SVA-5000-500 mg) were from Laysan Bio. Biotinylated anti-EGFR antibody (E101) was obtained from Leinco Technologies. Antibodies in carrier-free buffer were purchased from Cell Signaling Technologies: EGFR pY1068 (clone 1H12, 2236BF) and EGFR pY1173 (clone 53A5, 4407BF). Monoclonal antibody prelabeled with AF647 to detect pan-tyrosine phosphorylation (PY99 antibody, sc-7020 AF647) was purchased from Santa Cruz Biotechnology. Mix-n-Stain CF555 and CF640R antibody labeling kits were purchased from Biotium. PFA and GA were purchased from Electron Microscopy Sciences. Each of the three commercial antibodies used to detect phosphotyrosines in EGFR (1H12, 53A5, and PY99) is vendor-recommended for Western blotting. Recognition of epitopes is likely insensitive to whether surface-captured receptor is in a denatured or folded state, because the epitopes of PY99 lie mostly within the intrinsically disordered C-terminal region of the receptor and the epitopes of 1H12 and 53A5 (peptides encompassing pY1068 and pY1173) lie entirely within this region. Kim et al. (2016) evaluated and confirmed the site specificity of 1H12 and 53A5 using immunoblotting and mutant forms of EGFR (Y1068F and Y1173F).
Labeling of antibodies
Carrier-free antibodies (50 μg at 0.5–1 mg/ml per reaction) were labeled using Mix-n-Stain antibody labeling kits following the manufacturer’s instructions. Briefly, the labeling reaction was carried out for 30 min at room temperature and antibodies were centrifuged using the ultrafiltration vial provided to remove the unconjugated dye. Antibodies were resuspended in phosphate-buffered saline (PBS) and stored at 4°C. The labeling efficiency achieved was between 2.7 and 4.4 dyes/antibody.
Cell treatment and lysate preparation
CHO-EGFR-GFP cells were plated overnight in 60-mm tissue culture dishes at 800,000 cells/dish and CHO-ACP-EGFR cells in 24-well plates at 50,000 cells/well. For ACP labeling, CHO-ACP-EGFR cells were washed with serum-free DMEM medium (SFM), incubated with ACP labeling solution (SFM; 10 mM MgCl2, 4 μM CoA 488, and 1 μM ACP) for 20 min at 37°C and washed three times with SFM before stimulation. Cells were washed in Tyrode’s solution (135 mM NaCl, 10 mM KCl, 0.4 mM MgCl2, 1 mM CaCl2, 10 mM HEPES, 20 mM glucose, 0.1% bovine serum albumin [BSA], pH 7.2) and treated with ligand or Tyrode’s solution alone (resting cells) at 37°C. At the indicated time points, cells were placed on ice, washed once with cold PBS followed by the addition of lysis buffer (1% IGEPAL CA-630, 150 mM NaCl, 50 mM Tris, pH 7.2) containing protease and phosphatase inhibitors. Cell lysates were collected using cell scrapers (Greiner Bio-One North America; #541070), transferred to fresh tubes on ice, and gently vortexed every 5 min for a total of 20 min. Lysates were centrifuged at 16,000 × g for 20 min at 4°C and the supernatant was transferred to a new tube and stored at −80°C. For experiments involving treatment of cells with phosphatase inhibitors a stock solution of 30 mM pervanadate (PV) was prepared before each experiment by mixing equimolar concentrations of hydrogen peroxide and activated sodium orthovanadate incubated in the dark for at least 15 min before use. Cells were pre-treated for 15 min with a Tyrode’s solution containing 1 mM PV followed by incubation for 5 min in a solution with 50 nM EGF and 1 mM PV.
Fabrication of hydrophobic arrays and surface functionalization
Coverglasses (24 × 60 mm, #1.5; Electron Microscopy Sciences; #63793) were Piranha-cleaned (Labit et al., 2008) and placed in a coverglass holder (Fisher Scientific; #08-817). Coverglasses were sequentially sonicated in methanol and acetone for 10 min each, and in 1 M KOH for 20 min using a bath sonicator (Branson Ultrasonics; B1200R-1). These solutions were stored in polypropylene 50 ml tubes (VWR; #89401-564) and reused up to five times. Coverglasses were rinsed with Milli-Q water two times, dried by quickly passing them multiple times over the flame of a Bunsen burner using metal tweezers, and placed in a dry coverglass holder. A solution containing 95% methanol, 5% acetic acid, and 1% aminosilane (N-(2-aminoethyl)-3-aminopropyltrimethoxysilane) was prepared in an Erlenmeyer flask, immediately poured into the coverglass holder, and incubated at room temperature for 10 min in the dark, followed by 2 min sonication, and another 10 min incubation in the dark. Coverglasses were next washed with methanol for 2 min, rinsed and washed for 2 min with water, and dried in the dark. Treated coverglasses were placed on top of a parafilm-covered coverglass containing a guide pattern, which was used as reference to draw the sample array (Supplemental Figure S1C) with a hydrophobic barrier pen (Vector Laboratories; #H-4000). Ink was allowed to dry for at least 5 min before coverglasses were placed in a humidified chamber. For surface functionalization, 50 mg of mPEG-succinimidyl valerate, 1.3 mg of biotin-PEG-succinimidyl valerate, and 200 µl of freshly prepared 10 mM sodium bicarbonate were mixed thoroughly by pipetting up and down, centrifuged for 1 min at 10,000 × g at room temperature, and immediately applied to the SiMPull array (10–13 µl per region). After incubating for 3–4 h in the dark inside the humidified boxes, arrays were washed by sequential 30 s submersions into three water-filled 250-ml glass beakers. Coverglasses were dried with nitrogen gas, and stored in pairs (back to back) inside 50-ml tubes, which were filled with nitrogen gas before being closed and sealed with parafilm. Coverglasses were stored in the dark at −20°C for up to a week before use.
Labeling and quantification of surface receptors
CHO-EGFR-GFP cells grown in 24-well plates were placed on ice and washed three times with cold PBS. AF647-NHS ester was dissolved at the indicated concentrations in PBS. Cells were incubated with this solution for 30 min at 4°C with gentle agitation, washed three times with cold PBS, and subjected to cell lysis. The percentage of receptors labeled with AF647 across different dye concentrations was assessed with SiMPull (see also Figure 1D). To estimate the percentage of receptors at the cell surface the AF647-labeling curve was fitted to a biexponential decay curve in its increasing form using the “fit” function in MATLAB: y = C1 (1-e-ax) + C2 (1-e-bx), where y is the percentage of AF647-labeled receptors, x is the concentration of reactive AF647-NHS ester used, and a > 0, b > 0, C1, and C2 are coefficients to be fitted. The sum of the coefficients C1 and C2 represent the asymptote of the curve and an approximation of the fraction of receptors at the cell surface.
Single-molecule pull down and phosphosite labeling
T50 (10 mM Tris, pH 8.0, 50 mM NaCl) and T50-BSA (T50 with 0.1 mg/ml BSA) solutions were prepared and stored for up to a month at room temperature. SiMPull arrays were equilibrated at room temperature and placed on a TC100 plate lined with parafilm. Each region of the SiMPull array was treated with 10–15 µl of a 10 mg/ml sodium borohydride (NaBH4)/PBS solution for 4 min at room temperature to reduce background fluorescence (Supplemental Figure S1, A and B) and washed three times with PBS. SiMPull regions were then incubated with a 0.2 mg/ml NeutrAvidin/T50 solution for 5 min and washed three times with T50, followed by incubation with a 2 µg/ml biotinylated anti-EGFR/T50-BSA solution for 10 min and washed three times with T50-BSA.
The plate containing the SiMPull array(s) was kept on ice during sample preparation. Lysates were diluted in cold T50-BSA with protease and phosphatase inhibitors (T50-BSA/PPI), gently vortexed, and added to the SiMPull array. After 10 min incubation, the lysates were removed and the SiMPull regions washed four times with cold T50-BSA/PPI. To determine the appropriate dilution factor, the density of pull-down receptors as a function of lysate concentration was first assessed to achieve a pull-down density 0.04–0.08/μm2 (see also Supplemental Figure S4). Antibodies were diluted in cold T50-BSA/PPI, incubated for 1 h, washed six times with cold T50-BSA, and washed twice with cold PBS. Immediately after, antibodies were fixed for 10 min with a 4% PFA/0.1% GA solution and washed twice with 10 mM Tris (pH 7.4)/PBS for a total of 10 min to inactivate fixatives. See also Supplemental Figure S1 for discussion on the need for incubation with saturating antibody concentration and subsequent fixation. For three-color SiMPull experiments the same antibody incubation and fixation procedure was performed for the second antibody. Tris solution was replaced by T50-BSA and the SiMPull array was equilibrated to room temperature before proceeding to imaging.
SiMPull imaging
Imaging of SiMPull samples was performed using an inverted microscope (Olympus America; model IX71) equipped with a 150×/1.45 NA oil-immersion objective for TIRF microscopy (Olympus America; UAPON 150XOTIRF) and a three-dimensional piezo stage (Mad City Labs; Nano-LPS100). Excitation of CF640R- or AF647-labeled antibodies was done using a 642-nm laser (Thorlabs; HL63133DG), CF555-labeled antibodies using a 561-nm laser (Coherent; sapphire 561-100 CW CDRH), and of GFP- and CoA 488–tagged receptors using a 488-nm laser (Spectra Physics; cyan 100 mW). All lasers were set in total internal reflection configuration, and laser powers were adjusted to prevent photobleaching of the sample at the time scale of the image exposure time (300 ms). Sample illumination and emission were filtered using a quad-band dichroic and emission filter set (Semrock; LF405/488/561/635-A-000). Emission light was separated into four channels using a quad-view multichannel imaging system (Photometrics; model QV2) equipped with the appropriate dichroics (Chroma; 495 DCLP, 565 DCLP, 660 DCLP) and emission filters (Semrock; 685/40 nm, 600/37 nm, 525/45 nm). Emission light was collected with an electron-multiplying charge-coupled device camera (Andor Technology; DU-897E-C50-#BV) with EM gain set to 200. Each channel was 256 × 256 pixels, with a pixel size of 106.7 nm. Photobleaching and bleed through were prevented by controlling the laser shutters and microscope stage through a MATLAB script to sequentially excite and acquire the different fluorophores (642-nm laser first, 488-nm laser last). A minimum of 20 regions of interest were acquired per condition. For quantification of step photobleaching of EGFR-GFP molecules, a 100-frame time series (300 ms exposure time) was acquired after imaging of the other two channels.
Quantification of receptor phosphorylation
All image processing was performed using MATLAB together with the MATLAB toolbox for image-processing DIPImage (Delft University of Technology; Hendriks et al., 1999) and all software is available upon request. The location of emitters in each channel was calculated using graphics processor unit computing as previously described (Smith et al., 2010). Fits in the GFP channel were filtered based on the quality of the fit to the point spread function to reduce the chances of detecting multiple receptors in close proximity as a single molecule. Image registration was performed as previously described (Schwartz et al., 2017). In our studies, the root mean square error for image registration was <10 nm. For visualization purposes, Gaussian blob representations of the fluorophore localizations were generated. A receptor was considered to be phosphorylated when the localization centers of the receptor and labeled antibody were at a distance <106.7 nm (within 1 pixel).
Phosphorylation percentages were calculated as 100*(NPhos)/(NGFP), where NPhos is the number of receptors identified as phosphorylated and NGFP is the corrected number of single molecules in the GFP channel. The number of GFP localizations was calculated by subtracting background spots and accounting only for surface receptors as follows: NGFP = (NLOC-NBG)*SR, where NLOC is the total number of emitters localized, NBG is the expected number of background emitters in the area imaged, and SR (surface ratio) is the fraction of receptors located at the cell surface. The density of background emitters was quantified for each SiMPull array and used for background correction of samples in that array. For three-color SiMPull experiments where steric hindrance between sequentially incubated antibodies was observed (i.e., pY1068-pY1173 detection), estimations of dual phosphorylation were corrected to account for this hindrance as explained in Supplemental Figure S3. Note that use of a three-color imaging scheme to correlate phosphoantibody labeling directly with GFP-tagged receptors was critical, due to the relatively high nonspecific binding of the antibodies (Supplemental Figure S6). In the absence of the GFP channel to remove the nonspecific binding, the values for dual labeling are underestimated (Supplemental Figure S6).
Statistical analysis
Based on the consideration that the phosphorylation state of each receptor analyzed has the properties of a Bernoulli trial, SEs of phosphorylation measurements were calculated as for sample proportions in a binomial distribution: SE = p(1−p)/n, where p is the fraction of receptors phosphorylated and n is the total number of receptors. The condition np > 10 (with the exception of Figure 6E, np > 5) and np(1−p) > 10 was ensured to be met to allow this approximation to be adequate. Two-sample Z test (two-tailed) was used to estimate p values (LeBlanc, 2004).
Step-photobleaching analysis
For step-photobleaching analysis of multiphosphorylated receptors, the average fluorescence intensity of the area (200 × 200 nm) surrounding each of these EGFR-GFP molecules was quantified and plotted for the duration of the time series. Intensity plots were manually analyzed and the number of photobleaching steps was quantified.
Mathematical modeling
The model was formulated using BNGL (Faeder et al., 2009). The BioNetGen input file that defines the model is available as Supplemental File S1. This file is a plain-text file. The model accounts for the proteins, sites, and interactions depicted in Figure 2B. The model implicitly accounts for three compartments, which are each taken to be well mixed: the extracellular fluid, the plasma membrane, and the cytosol. In the model, EGFR dimer-dependent autophosphorylation is taken to be asymmetric, as discussed in the main text. In addition to Y1068 and Y1173, the model accounts for other tyrosines in EGFR, which are lumped together and labeled YN in the BNGL-formatted model-definition file. The model structure was optimized using model restructuration (reviewed in the supplementary tutorial of Erickson et al., 2019).
In the one-parameter fits carried out to characterize the increased kinase activity of L858R EGFR and the lifetime of epigen-induced EGFR dimers, we used simple grid search as the fitting method, meaning that we systematically varied the free parameter over a specified range, calculating the quality of fit at each parameter value. When the free parameter was the rate constant for autophosphorylation, we considered a range 0.1× below and 100× above the estimated value for WT EGFR. We considered a step size of 0.1 in the grid search. When the free parameter was the rate constant for EGFR dimer dissociation, we similarly considered a range 0.1× below and 100× above the estimated value for EGF-induced dimers. We again considered a step size of 0.1 in the grid search.
Simulations were performed as follows. In a preprocessing step, rules were interpreted by BioNetGen to obtain the rule-implied reaction network and the corresponding ordinary differential equations (ODEs) for mass-action chemical kinetics. Simulations of the dynamics of responses to ligand stimulation were performed by numerically integrating the BioNetGen-derived ODEs using CVODE with BioNetGen’s default CVODE settings. CVODE (https://computation.llnl.gov/projects/sundials; Hindmarsh et al., 2005) is a dependency of BioNetGen.
Model parameters were estimated via curve fitting, which was performed using the differential evolution (DE) algorithm implemented in PyBioNetFit (Mitra et al., 2019), a software package for parameterization of biological models that supersedes BioNetFit (Thomas et al., 2016). An important parameter of the DE algorithm is population size: this algorithmic parameter was set to 200. Fitting runs were allowed to continue until apparent convergence, that is, until the value of the objective function in the optimization problem stopped decreasing. PyBioNetFit takes as input three file types: a BioNetGen input file (i.e., a BNGL-formatted model definition), a configuration file, and one or more EXP files with experimental data. These files are packaged together in a ZIP archive as Supplemental File S2. The parameters listed in Supplemental Table S1 were held fixed in fitting. These parameters were estimated on the basis of prior knowledge, referenced in the table, or measured/determined in this study as indicated. The parameters listed in Supplemental Table S2 were taken to be the free parameters to be adjusted by PyBioNetFit during fitting within the indicated intervals. The data used in fitting included the dose-response data in Figure 2A (for EGF doses eliciting a significant change in phosphorylation, 0.5, 5, and 50 nM) and the time-series data in Figure 2D. Parameter estimates are based on a global fit to all of these data.
All parameter estimates were originally made for CHO cells. Additional cell-specific models were obtained by modifying CHO-cell protein copy numbers, as follows. As indicated in Supplemental Table S1, the EGFR abundance that we estimated for CHO cells via direct measurement (flow cytometry) and the Grb2 and Shc1 abundances that we estimated for CHO cells via fitting to data in Figure 2, A and D, were replaced with direct measurements of EGFR, Grb2, and Shc1 abundances made in earlier studies for HMEC, MCF10A, and HeLa S3 cells (Kulak et al., 2014; Shi et al., 2016). As a simplification, other parameter values were taken to be the same across cell types.
Confidence intervals for best-fit parameter values were estimated using PyBioNetFit’s bootstrapping procedure. A total of 1000 bootstrap replicates of fitting were performed, and the reported 90% confidence intervals give the range from the 5th percentile to the 95th percentile of the replicate results (Supplemental Table S3). The bootstrapping procedure used the same BioNetGen and EXP files as before and the configuration file fit_bootstrap.conf provided in Supplemental File S2.
Bayesian uncertainty quantification (UQ) was performed using the parallel tempering (PT) algorithm implemented in PyBioNetFit, which is a parallelized Markov chain Monte Carlo method. We used PT to find 90% credible intervals for each parameter listed in Supplemental Table S2. The prior distribution considered for each parameter was a log-uniform distribution over the range indicated for the parameter in Supplemental Table S2. Ten independent runs were performed, each consisting of 200,000 iterations (36 simulations per iteration), with samples recorded after a burn-in period of 50,000 iterations. Reported 90% credible intervals (Supplemental Table S3) for each parameter give the range from the 5th to 95th percentile of the sampled parameter sets. (Note that credible intervals are not identical to bootstrap confidence intervals.) Prediction uncertainties (Figure 2D, shaded areas; Figure 6, A and C, error bars) were calculated by running simulations with sampled parameter sets and finding the 2.5th and 97.5th percentile pEGFR percentage at each time point. UQ jobs used the same BioNetGen and EXP files as before and the configuration file fit_pt.conf provided in Supplemental File S2. Distributions of parameter values in sampled parameter sets and the correlation between each pair of parameters (Supplemental Figure S7) were plotted using the function plotmatrix from MATLAB.
Fitting was performed on a laptop. Bootstrapping was performed using 7000 core hours on a multicore workstation. PT was performed using 40,000 core hours on a computer cluster.
Supplementary Material
Acknowledgments
We thank Donna Arndt-Jovin and Mark Lemmon for helpful discussions. We also thank Donna Arndt-Jovin for the CHO-ACP-EGFR cell line and suggesting the ACP-EGFR comparison experiment. We thank Richard Pepermans for generating the Grb2-mCherry construct, Hannah Johnson for generation of the CHO-EGFR-GFP Grb2-mCherry cell line, Shayna Lucero for assistance with cell culture, and Aubrey Gibson for assistance with Western blots. This work was supported by the New Mexico Spatiotemporal Modeling Center (National Institutes of Health [NIH] P50GM-085273), Mexican Secretariat of Public Education SEP (E.S.-C.), National Science Foundation CAREER MCB-0845062 (D.S.L.), NIH R35GM-126934 (D.S.L.), and the UNM Comprehensive Cancer Center (NIH P30CA118100). We gratefully acknowledge use of the University of New Mexico Comprehensive Cancer Center fluorescence microscopy and flow cytometry facilities, as well as the NIH P30CA118100 support for these cores. We acknowledge use of computing resources provided by the Los Alamos National Laboratory Institutional Computing program, which is operated for the National Nuclear Security Administration of the Department of Energy under contract.
Abbreviations used:
- ACP
acyl carrier protein
- BNGL
BioNetGen language
- EGFR
epidermal growth factor receptor
- GA
glutaraldehyde
- GFP
green fluorescent protein
- NHS
N-hydroxysuccinimide
- PEG
polyethylene glycol
- PFA
paraformaldehyde
- PTB
phosphotyrosine binding
- PV
pervanadate
- PY
phosphotyrosine
- RTK
receptor tyrosine kinase
- SH2
Src homology 2
- SiMPull
single-molecule pull down
- TIRF
total internal reflection fluorescence
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-09-0548) on January 8, 2020.
REFERENCES
- Brock R, Hamelers IHL, Jovin TM. (1999). Comparison of fixation protocols for adherent cultured cells applied to a GFP fusion protein of the epidermal growth factor receptor. Cytometry , 353–362. [DOI] [PubMed] [Google Scholar]
- Brunati AM, Pinna LA, Bergantino E, Ruzzene M, Cirri P, Ramponi G, Donella-Deana A. (1998). Src homology-2 domains protect phosphotyrosyl residues against enzymatic dephosphorylation. Biochem Biophys Res Commun , 700–705. [DOI] [PubMed] [Google Scholar]
- Chen WW, Schoeberl B, Jasper PJ, Niepel M, Nielsen UB, Lauffenburger DA, Sorger PK. (2009). Input–output behavior of ErbB signaling pathways as revealed by a mass action model trained against dynamic data. Mol Syst Biol , 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylek LA, Harris LA, Tung C-S, Faeder JR, Lopez CF, Hlavacek WS. (2014). Rule-based modeling: a computational approach for studying biomolecular site dynamics in cell signaling systems. Wiley Interdiscip Rev Syst Biol Med , 13–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylek LA, Hu B, Blinov ML, Emonet T, Faeder JR, Goldstein B, Gutenkunst RN, Haugh JM, Lipniacki T, Posner RG, et al (2011). Guidelines for visualizing and annotating rule-based models. Mol Biosyst , 2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coba MP, Pocklington AJ, Collins MO, Kopanitsa MV, Uren RT, Swamy S, Croning MDR, Choudhary JS, Grant SGN. (2009). Neurotransmitters drive combinatorial multistate postsynaptic density networks. Sci Signal , ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson KE, Rukhlenko OS, Shahinuzzaman M, Slavkova KP, Lin YT, Suderman R, Stites EC, Anghel M, Posner RG, Barua D, et al (2019). Modeling cell line-specific recruitment of signaling proteins to the insulin-like growth factor 1 receptor. PLoS Comput Biol , e1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faeder JR, Blinov ML, Hlavacek WS. (2009). Rule-based modeling of biochemical systems with BioNetGen. Methods Mol Biol , 113–167. [DOI] [PubMed] [Google Scholar]
- Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. (2008). Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science , 1081–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortian A, Sorkin A. (2014). Live-cell fluorescence imaging reveals high stoichiometry of Grb2 binding to the EGF receptor sustained during endocytosis. J Cell Sci , 432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed DM, Bessman NJ, Kiyatkin A, Salazar-Cavazos E, Byrne PO, Moore JO, Valley CC, Ferguson KM, Leahy DJ, Lidke DS, et al (2017). EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell , 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George N. (2006). A New Method for Protein Labeling with Small Molecules Based on Acyl Carrier Protein. PhD Thesis Lausanne, Switzerland: EPFL. [Google Scholar]
- Gibson SK, Parkes JH, Liebman PA. (2000). Phosphorylation modulates the affinity of light-activated rhodopsin for G protein and arrestin. Biochemistry , 5738–5749. [DOI] [PubMed] [Google Scholar]
- Grecco HE, Schmick M, Bastiaens PIH. (2011). Signaling from the living plasma membrane. Cell , 897–909. [DOI] [PubMed] [Google Scholar]
- Handly LN, Yao J, Wollman R. (2016). Signal transduction at the single-cell level: approaches to study the dynamic nature of signaling networks. J Mol Biol , 3669–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause RJ, Leung KK, Barkinge JL, Ciaccio MF, Chuu CP, Jones RB. (2012). Comprehensive binary interaction mapping of SH2 domains via fluorescence polarization reveals novel functional diversification of ErbB receptors. PLoS One , e44471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks BS, Opresko LK, Wiley HS, Lauffenburger D. (2003). Quantitative analysis of HER2-mediated effects on HER2 and epidermal growth factor receptor endocytosis. Distribution of homo- and heterodimers depends on relative HER2 levels. J Biol Chem , 23343–23351. [DOI] [PubMed] [Google Scholar]
- Hendriks CLL, van Vliet LJ, Rieger B, van Kempen GMP, van Ginkel M. (1999). DIPimage: a scientific image processing toolbox for MATLAB. Quant Imaging Group, Faculty of Applied Sciences, Delft University of Technology, Delft, Netherlands.
- Hindmarsh AC, Brown PN, Grant KE, Lee SL, Serban R, Shumaker DE, Woodward CS. (2005). SUNDIALS: suite of nonlinear and differential/algebraic equation solvers. ACM Trans Math Softw , 363–396. [Google Scholar]
- Hsieh M, Yang S, Raymond-Stinz M, Edwards JS, Wilson BS. (2010). Spatio-temporal modeling of signaling protein recruitment to EGFR. BMC Syst Biol , 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadwin JA, Curran TG, Lafontaine AT, White FM, Mayer BJ. (2018). Src homology 2 domains enhance tyrosine phosphorylation in vivo by protecting binding sites in their target proteins from dephosphorylation. J Biol Chem , 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK, Ha T. (2011). Probing cellular protein complexes using single-molecule pull-down. Nature , 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Liu R, Xiang YK, Ha T. (2012). Single-molecule pull-down for studying protein interactions. Nat Protoc , 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. (2003). Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res , 31–53. [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Demin OV, Moehren G, Hoek JB. (1999). Quantification of short term signaling by the epidermal growth factor receptor. J Biol Chem , 30169–30181. [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Hoek JB, Westerhoff HV. (2000). Why cytoplasmic signalling proteins should be recruited to cell membranes. Trends Cell Biol , 173–178. [DOI] [PubMed] [Google Scholar]
- Kim KL, Kim D, Lee S, Kim S-J, Noh JE, Kim J-H, Chae YC, Lee J-B, Ryu SH. (2016). Pairwise detection of site-specific receptor phosphorylations using single-molecule blotting. Nat Commun , 11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Li Z, Apetri M, Luo B, Settleman E, Anderson KS. (2012). Temporal resolution of autophosphorylation for normal and oncogenic forms of EGFR and differential effects of gefitinib. Biochemistry , 5212–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KL, Park KM, Murray J, Kim K, Ryu SH. (2018). Direct profiling the post-translational modification codes of a single protein immobilized on a surface using Cu-free click chemistry. ACS Cent Sci , 614–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman LB, Maiwald T, Conzelmann H, Lauffenburger DA, Sorger PK. (2011). Rapid phospho-turnover by receptor tyrosine kinases impacts downstream signaling and drug binding. Mol Cell , 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz SE, Lauffenburger DA. (2012). Measurement and modeling of signaling at the single-cell level. Biochemistry , 7433–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs E, Das R, Wang Q, Collier TS, Cantor A, Huang Y, Wong K, Mirza A, Barros T, Grob P, et al (2015). Analysis of the role of the C-terminal tail in the regulation of the epidermal growth factor receptor. Mol Cell Biol , 3083–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. (2014). Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods , 319–324. [DOI] [PubMed] [Google Scholar]
- Labit H, Goldar A, Guilbaud G, Douarche C, Hyrien O, Marheineke K. (2008). A simple and optimized method of producing silanized surfaces for FISH and replication mapping on combed DNA fibers. Biotechniques , 649–658. [DOI] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. (2004). Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet , 147–150. [DOI] [PubMed] [Google Scholar]
- Lajoie P, Partridge EA, Guay G, Goetz JG, Pawling J, Lagana A, Bharat J, Dennis JW, Nabi IR. (2007). Plasma membrane domain organization regulates EGFR signaling in tumor cells. J Cell Biol , 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau EK, Trester-Zedlitz M, Trinidad JC, Kotowski SJ, Krutchinsky AN, Burlingame AL, von Zastrow M. (2011). Quantitative encoding of the effect of a partial agonist on individual opioid receptors by multisite phosphorylation and threshold detection. Sci Signal , ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc DC. (2004). Statistics: Concepts and Applications for Science, Burlington, MA: Jones & Bartlett Learning. [Google Scholar]
- Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM. (2004). Quantum dot ligands provide new insights into erbB/HER receptor–mediated signal transduction. Nat Biotechnol , 198–203. [DOI] [PubMed] [Google Scholar]
- Low-Nam ST, Lidke KA, Cutler PJ, Roovers RC, van Bergen en Henegouwen PMP, Wilson BS, Lidke DS. (2011). ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nat Struct Mol Biol , 1244–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald-Obermann JL, Pike LJ. (2009). The intracellular juxtamembrane domain of the epidermal growth factor (EGF) receptor is responsible for the allosteric regulation of EGF binding. J Biol Chem , 13570–13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra ED, Suderman R, Colvin J, Ionkov A, Hu A, Sauro HM, Posner RG, Hlavacek WS. (2019). PyBioNetFit and the biological property specification language. IScience , 1012–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor MM, Low-Nam ST, Halász ÁM, Lidke DS, Wilson BS, Edwards JS. (2013). Dynamic transition states of ErbB1 phosphorylation predicted by spatial stochastic modeling. Biophys J , 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor MM, Steinkamp MP, Halasz AM, Chen Y, Yang S, Smith MS, Zahoransky-Kohalmi G, Swift M, Xu X-P, Hanein D, et al (2015). Orchestration of ErbB3 signaling through heterointeractions and homointeractions. Mol Biol Cell , 4109–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis JE, Lahav G. (2013). Encoding and decoding cellular information through signaling dynamics. Cell , 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red Brewer M, Yun C-H, Lai D, Lemmon MA, Eck MJ, Pao W. (2013). Mechanism for activation of mutated epidermal growth factor receptors in lung cancer. Proc Natl Acad Sci USA , E3595–E3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Margolis B, Mohammadi M, Daly RJ, Daum G, Li N, Fischer EH, Burgess WH, Ullrich A, Schlessinger J. (1992). SH2 domains prevent tyrosine dephosphorylation of the EGF receptor: identification of Tyr992 as the high-affinity binding site for SH2 domains of phospholipase C gamma. EMBO J , 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar C, Höfer T. (2009). Multisite protein phosphorylation—from molecular mechanisms to kinetic models. FEBS J , 3177–3198. [DOI] [PubMed] [Google Scholar]
- Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. (2010). A census of amplified and overexpressed human cancer genes. Nat Rev Cancer , 59–64. [DOI] [PubMed] [Google Scholar]
- Schulze WX, Deng L, Mann M. (2005). Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol , 2005.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SL, Cleyrat C, Olah MJ, Relich PK, Phillips GK, Hlavacek WS, Lidke KA, Wilson BS, Lidke DS. (2017). Differential mast cell outcomes are sensitive to FcεRI-Syk binding kinetics. Mol Biol Cell , 3397–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y, Eastwood MP, Zhang X, Kim ET, Arkhipov A, Dror RO, Jumper J, Kuriyan J, Shaw DE. (2012). Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell , 860–870. [DOI] [PubMed] [Google Scholar]
- Shi T, Gao Y, Gaffrey MJ, Nicora CD, Fillmore TL, Chrisler WB, Gritsenko MA, Wu C, He J, Bloodsworth KJ, et al (2015). Sensitive targeted quantification of ERK phosphorylation dynamics and stoichiometry in human cells without affinity enrichment. Anal Chem , 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Niepel M, McDermott JE, Gao Y, Nicora CD, Chrisler WB, Markillie LM, Petyuk VA, Smith RD, Rodland KD, et al (2016). Conservation of protein abundance patterns reveals the regulatory architecture of the EGFR-MAPK pathway. Sci Signal , rs6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S, Algisi V, Nappo G, Conte A, Pascolutti R, Cuomo A, Bonaldi T, Argenzio E, Verhoef LGGC, Maspero E, et al (2013). Threshold-controlled ubiquitination of the EGFR directs receptor fate. EMBO J , 2140–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CS, Joseph N, Rieger B, Lidke KA. (2010). Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat Methods , 373–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. (2009). Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature , 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkamp MP, Low-Nam ST, Yang S, Lidke KA, Lidke DS, Wilson BS. (2014). erbB3 is an active tyrosine kinase capable of homo- and heterointeractions. Mol Cell Biol , 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stites EC, Aziz M, Creamer MS, Von Hoff DD, Posner RG, Hlavacek WS. (2015). Use of mechanistic models to integrate and analyze multiple proteomic datasets. Biophys J , 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesco SE, Shih AJ, Jia F, Radhakrishnan R. (2011). A multiscale modeling approach to investigate molecular mechanisms of pseudokinase activation and drug resistance in the HER3/ErbB3 receptor tyrosine kinase signaling network. Mol Biosyst , 2066–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BR, Chylek LA, Colvin J, Sirimulla S, Clayton AHA, Hlavacek WS, Posner RG. (2016). BioNetFit: A fitting tool compatible with BioNetGen, NFsim and distributed computing environments. Bioinformatics , 798–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valley CC, Arndt-Jovin DJ, Karedla N, Steinkamp MP, Chizhik AI, Hlavacek WS, Wilson BS, Lidke KA, Lidke DS. (2015). Enhanced dimerization drives ligand-independent activity of mutant epidermal growth factor receptor in lung cancer. Mol Biol Cell , 4087–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeek BS, Adriaansen-slot SS, Rijksen G, Vroom TM. (1997). Grb2 overexpression in nuclei and cytoplasm of human breast cells: a histochemical and biochemical study of normal and neoplastic mammary tissue specimens. J Pathol , 195–203. [DOI] [PubMed] [Google Scholar]
- Welch CM, Elliott H, Danuser G, Hahn KM. (2011). Imaging the coordination of multiple signalling activities in living cells. Nat Rev Mol Cell Biol , 749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KJ, Mill C, Lambert S, Buchman J, Wilson TR, Hernandez-Gordillo V, Gallo RM, Ades LMC, Settleman J, Riese DJ. (2012). EGFR ligands exhibit functional differences in models of paracrine and autocrine signaling. Growth Factors , 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf-Yadlin A, Kumar N, Zhang Y, Hautaniemi S, Zaman M, Kim H-D, Grantcharova V, Lauffenburger DA, White FM. (2006). Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol Syst Biol , 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Shi T, Gritsenko MA, X’avia Chan C-Y, Fillmore TL, Hess BM, Swensen AC, Liu T, Smith RD, Wiley HS, et al (2018). Targeted quantification of phosphorylation dynamics in the context of EGFR-MAPK pathway. Anal Chem , 5256–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. (2006). An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell , 1137–1149. [DOI] [PubMed] [Google Scholar]
- Ziomkiewicz I, Loman A, Klement R, Fritsch C, Klymchenko AS, Bunt G, Jovin TM, Arndt-Jovin DJ. (2013). Dynamic conformational transitions of the EGF receptor in living mammalian cells determined by FRET and fluorescence lifetime imaging microscopy. Cytom Part A , 794–805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.