Abstract
Purpose
Although the combination of anti-PD1/PD-L1 with platinum chemotherapy is a standard of care for NSCLC, clinical responses vary. While predictive biomarkers are validated for immunotherapy, including PD-L1 expression, tumor mutational burden, and inflamed immune microenvironment, their relevance to chemo-immunotherapy combinations is less clear. We have recently shown that activation of the STING innate immune pathway enhances immunotherapy response in SCLC. We hypothesize that STING pathway activation may predict and underlie predictive correlates of anti-tumor immunity in NSCLC.
Experimental Design
We analyzed transcriptomic and proteomic profiles in two NSCLC cohorts from our institution (treatment-naïve-PROSPECT; relapsed-BATTLE-2) and The Cancer Genome Atlas (total n=1320). Tumors were stratified by STING activation based on protein and/or mRNA expression of cGAS, phospho-STING, and STING-mediated-chemokines CCL5 and CXCL10. STING activation in patient tumors and in platinum-treated preclinical NSCLC models was correlated with biomarkers of immunotherapy response.
Results
STING activation is associated with higher levels of intrinsic DNA damage, targetable immune checkpoints, and chemokines in treatment-naïve and relapsed lung adenocarcinoma. We observed that tumors with lower STING and immune gene expression show higher frequency of STK11 mutations; however, we identified a subset of these tumors that are TP53 co-mutated and display high immune- and STING- related gene expression. Treatment with cisplatin increases STING pathway activation and PD-L1 expression in multiple NSCLC preclinical models, including adeno- and squamous cell carcinoma.
Conclusions
STING pathway activation in NSCLC predicts features of immunotherapy response and is enhanced by cisplatin treatment, suggesting a possible predictive biomarker, and mechanism, for improved response to chemo-immunotherapy combinations.
Keywords: lung cancer, immunotherapy, innate immunity, STING, immune checkpoints
Introduction
Recently, immune checkpoint blockade targeting the programmed-cell-death-1 (PD1)/programmed-cell-death-ligand-1 (PD-L1) axis, either alone or in combination with conventional chemotherapy, has become a new standard-of-care in non-small cell lung cancer (NSCLC) leading to improvements in survival (1). Analyses of clinical datasets have identified high tumor mutation burden (TMB) (2) and PD-L1 expression (3) as predictors of favorable T-cell responses, while somatic mutations (like EGFR and KRAS/STK11) are associated with lack of T-cell response (4, 5). Ayers et al. developed an immune score that includes genes related to immune-response, like IFN-γ–signaling, antigen presentation, chemokines, T-cells cytotoxic activity, and adaptive immunity, (6). This score has been validated as an independent positive biomarker for clinical responses to anti-PD1 monotherapy in pan-cancer clinical trials including lung cancer; tumors with positive score can be defined as “inflamed” and this is an independent biomarker of immunotherapy response (7, 8). However, there is still a need for more extensive characterization of the biology of immune responsive subsets in NSCLC as the underlying mechanism mediating the enhancement of immune checkpoint inhibitor response by chemotherapy and the biomarkers for this combination remain largely unexplored.
DNA damage, intrinsic due to tumor genomic instability (9) or increased under the pressure of DNA damaging treatments (radiotherapy, chemotherapy, or inhibitors of DNA damage response (DDR), may stimulate the immune system and enhance tumor response to immunotherapy (10–13). Cytosolic DNA fragments (e.g. from DNA damage) are detected by Cyclic GMP-AMP synthase (cGAS) which, in turn, activates the Stimulator of Interferon Genes (STING) pathway and, subsequently, a type-I interferon (IFN) response (14). Our group has demonstrated that the induction of DNA damage by treatment with DDR-inhibitors, such as PARP- or CHK1-inhibitors, synergize with anti-PD-L1 in a syngeneic model of small cell lung cancer by activating a STING mediated anti-tumor immune response (12). Therefore, we hypothesize that STING pathway activation markers may identify NSCLC tumors with increased expression of targetable immune checkpoints and, thus, sensitivity to immune checkpoint blockade (ICB). Furthermore, we hypothesize that concurrent treatment with DNA damaging agents such as platinum increase STING pathway activation creating a more immune responsive phenotype.
In this study, we explored the landscape of immune related genes that are associated with the activation of the STING pathway in NSCLC patient tumors. Using transcriptomic and proteomic profiling of five large, independent NSCLC patient cohorts (total of 1320 tumors), stratified according to STING activation, we correlated the mRNA and protein expression of immune-related genes to STING pathway activation. We then studied the effect of platinum treatment, with or without PD-L1 blockade, in vitro and in vivo in NSCLC models to determine if activation of the STING pathway by cisplatin contributes to anti-tumor immune response.
Materials and Methods
Reverse-phase protein array (RPPA)
Protein lysates were collected in a buffer containing 1% Triton X-100, 50mM HEPES (pH 7.4), 150mM NaCl, 1.5mM MgCl2, 1mM EGTA, 100mM NaF, 10mM NaPPi, 10% glycerol, 1mM PMSF, 1mM Na3VO4, and 10 μg/mL aprotinin. Samples were quantified and protein arrays were printed from lysates and stained as previously described (15, 16). Briefly, the slide images were quantified by using MicroVigene 4.0 (Vigene- Tech, Carlisle, MA). The spot level raw data were processed by using the R package SuperCurve, which returns the estimated protein concentration (raw concentration) and a quality control (QC) score for each slide. Only slides with a QC score >0.8 were used for downstream analysis. The raw concentration data were normalized by median-centering each sample across all the proteins to correct loading bias.
Clinical cohorts (Table 1)
Table1.
NSCLC cohorts analyzed in the present work.
| Clinical cohort | Histology | Treatment | n | Genomic profile | STING/Immune Transcriptomic profile | STING/Immune Proteomic profile |
|---|---|---|---|---|---|---|
| PROSPECT | LUSC | Naïve | 57 | n=57 | n=36 | |
| PROSPECT | LUAD | Naïve | 152 | n=152 | n=120 | |
| TCGA | LUSC | Naïve | 501 | n=501 | n=501 | |
| TCGA | LUAD | Naïve | 515 | n=511 | n=511 | |
| BATTLE-2 | LUAD | Relapsed | 255 | n=95 |
PROSPECT
The MD Anderson cohort obtained from the Profiling of Resistance patterns and Oncogenic Signaling Pathways in Evaluation of Cancers of the Thorax (PROSPECT) study (n=209) (17) includes surgically resected NSCLC tumors, collected between 2006 and 2010, and sub-classified in lung adenocarcinoma (LUAD) (n=152) and lung squamous (LUSC) (n=57) cohorts. Gene expression and protein data were available from 209 and 156 patients, respectively.TCGA. The Cancer Genome Atlas (TCGA) (18) included LUAD (n=515) and LUSC (n=501) cohorts, containing untreated samples from NSCLC (total n=1016), for which gene expression data (TPM) are available.
BATTLE-2
The MD Anderson Biomarker-integrated Approaches of Targeted Therapy for Lung Cancer Elimination (BATTLE-2) trial represents a biopsy-mandated study in 255 heavily pretreated lung cancer patients; with mandatory pre-randomization tumor biopsy profiling (19). Gene expression data was available from 95 patients.All gene expression data are represented as log2 trasformed.
For cell lines, western blot analysis, qPCR, immunohistochemistry, animal models, statistical analysis: see supplemental materials and methods
Results
STING pathway protein expression is associated with immune activation in lung adenocarcinoma cancer
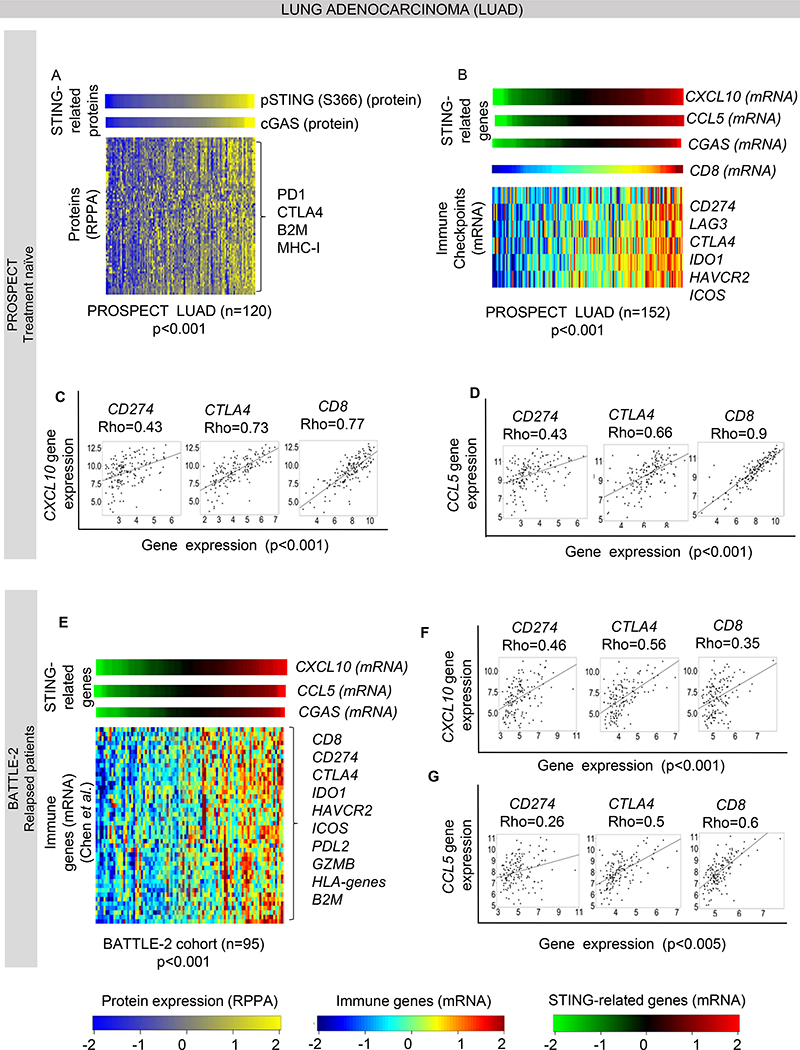
cGAS protein, in the presence of cytosolic DNA fragments, catalyzes the synthesis of cyclic GMP-AMP (cGAMP), which binds to the adaptor protein, STING and induces its phosphorylation at the activation site (S366) (12). Here, we investigated the relationship between cGAS and phospho-STING (S366) protein expression levels in treatment-naïve LUAD tumors from the MD Anderson Cancer Center (MDACC) PROSPECT trial (n=120).
As expected, phospho-STING (S366) protein levels were significantly correlated with cGAS protein levels (p=0.010) (Figure 1A). To explore if STING activation is associated with a distinct proteomic profile in lung cancer, we then stratified patient tumors by expression of phospho-STING and cGAS and identified proteins measured by RPPA that were correlated with cGAS or phospho-STING expression (p-value ≤0.05, Spearman rho (rho)≥0.3) (Figure 1A). As shown in Figure 1A, we found that high protein levels of phospho-STING were associated with high expression of 98 proteins, including the immune checkpoint proteins PD1 and CTLA-4 (both rho>0.4, p<0.001), antigen presentation markers beta2-microglobulin (B2M) and MHC class I (both rho>0.35, p<0.001). DNA damage response proteins (p53, BRCA2, P21, ATR, RAD51) and mesenchymal markers (AXL, Phospho-Axl (Y702), N-Cadherin) were strongly associated with phospho-STING protein levels (all rho>0.35, p<0.001), suggesting that intrinsic DNA damage and mesenchymal markers may be features of NSCLC tumors with higher basal levels of STING activation (20, 21).
Figure 1. STING pathway proteins expression is associated with immune activation in lung adenocarcinoma.
(A) Heatmap shows correlation of phospho-STING (Ser_366) protein levels with CGAS and other immune related proteins in the MD Anderson PROSPECT cohort, including treatment naïve adenocarcinoma (n=120, Spearman’s rho > 0.3, p<0.001).
(B) Expression of the STING downstream chemokine CXCL10 is correlated with the other STING related genes CCL5 and CGAS (p<0.001), with expression of selected targetable immune genes: CD274 (PD-L1), LAG3, CTLA4, IDO1, HAVCR2 and ICOS (Spearman’s rho > 0.5, p<0.001) and with CD8 mRNA levels, indicative of immune infiltration in PROSPECT LUAD cohort (n=120). (C-D) Individual dot-plots showing correlation of CXCL10 (C) and CCL5 (D) gene expression with selected targetable genes CD274 and CTLA4, and CD8 in PROSPECT LUAD cohort (n=120). (E) Correlation of CXCL10 with CCL5 and CGAS gene expression, with selected targetable immune genes and other immune markers from the list of Chen et al. in the treatment-refractory and relapsed BATTLE-2 cohort (n=95, Spearman’s rho > 0.3, p<0.001). (F-G) Individual dot-plots showing correlation of CXCL10 (F) and CCL5 (G) gene expression with selected targetable genes CD274 and CTLA4, and CD8 in BATTLE-2 cohort.
As pSTING cannot be assayed directly by gene expression, we used expression of three STING-related genes (CGAS and two downstream chemokines, CXCL10 and CCL5) as a surrogate and compared expression of these surrogates to expression levels of a curated list of immune markers (22) in the PROSPECT LUAD cohort (n=120). The list includes immune checkpoints, markers of immune cell infiltration, and chemokines, all known to be associated with an “inflamed” tumor phenotype. As expected, expression of the three surrogates was highly correlated (all rho>0.45, p<0.001). Within the curated list of markers, a number of genes were correlated with CGAS including CD8 (rho= 0.77, p<0.001), that indicates CD8+ T-cell infiltration, and with a list of targetable immune checkpoints, CD274 (PD-L1), CTLA4, LAG3, IDO1, HAVCR2, ICOS (all rho >0.4, p<0.001) (Figure 1B–D). These observations were then validated in the larger independent TCGA LUAD cohort (n=511, rho>0.6, p<0.001) (Supplemental Figure 1A). Thus, in two independent clinical cohorts that represent the entire heterogeneous genomic landscape of LUAD (including known driver mutation cohorts-e.g., KRAS, TP53, EGFR, ALK), STING pathway activation is highly associated with expression of PD-L1 and other immune markers that could be therapeutically targeted.
Since immunotherapy is currently more frequently used in the treatment of locally advanced or metastatic disease (1), we extended our analysis to the gene expression data from biopsies performed as part of the MDACC BATTLE-2 clinical trial (19). In this study, tumor biopsies were obtained from metastatic LUAD NSCLC patients who were refractory to at least one prior platinum-based chemotherapy prior to subsequent treatment. As shown in Figure 1E–G, we detected a strong correlation between CXCL10 gene expression and the other STING-related genes, CCL5 and CGAS, along with immune targetable genes (CD274, IDO1, CTLA4, HAVCR2, ICOS, PDCD1LG2) and markers of immune cell infiltration CD8, GZMB, HLA-genes, B2M (rho>0.35, p<0.001). These results suggest that the interplay between anti-tumor innate immune response, through the STING pathway, and the expression of other immune markers is maintained in previously treated, relapsed NSCLC.
STK11/LKB1 mutation is associated with different immune activation phenotypes
In recent studies from our group and others, LKB1 has emerged as an important biomarker of primary resistance to immune checkpoints blockade (ICB) in NSCLC. Skoulidis et al. (4, 23) demonstrated that, among KRAS mutated LUAD, tumors with STK11(LKB1) mutations are immunologically “cold” and are intrinsically resistant to anti-PD1/PD-L1 immunotherapy, and they have low expression of immune markers (including PD1, CTLA4, PD-L1, CD8) compared to other LUAD. Furthermore, Barbie et al. recently demonstrated that, mechanistically, LKB1 loss inducs a silencing of the STING pathway and impaired response to cytosolic DNA in KRAS/STK11(LKB1) mutant NSCLC models and is associated with downregulation of a type I IFN signature (24, 25).
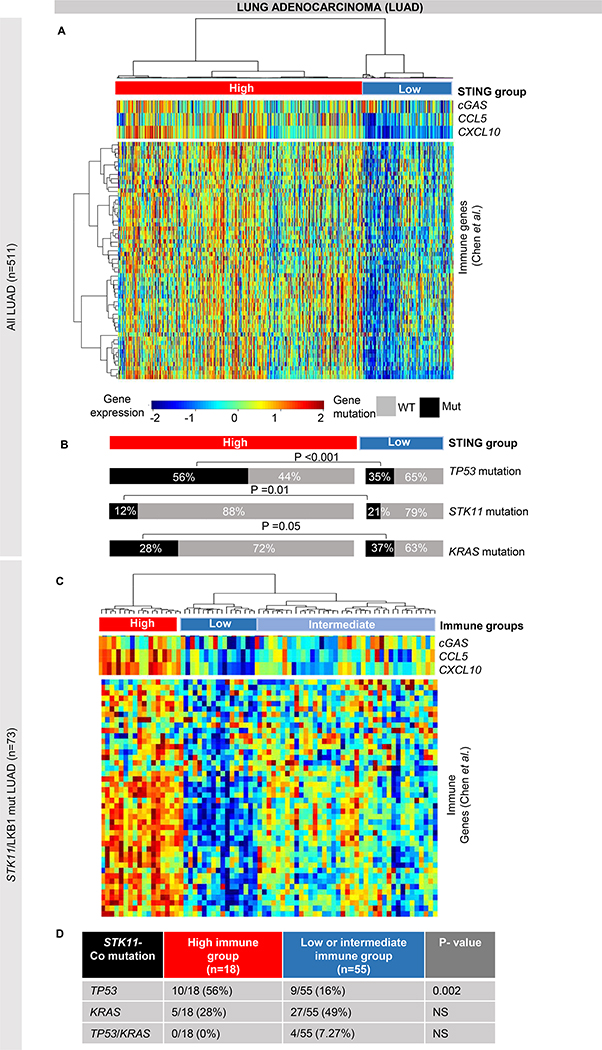
Using hierarchical clustering in the TCGA LUAD cohort based on gene expression of CGAS, CCL5 and CXCL10 we identified two main groups – STING-high (n=373) and STING-low (n=138). We then compared expression of immune related genes (22) and the frequency of mutations between these two groups. As expected from our observations, the STING-low group had lower expression of the immune related genes (including immune checkpoints and markers of immune infiltrations) (Figure 2A). Consistent with prior findings in KRAS-mutant lung cancers (4, 23), we also observed that STK11 (LKB1) loss of function mutations were significantly enriched in the STING-low immune group (21% versus 12%, p=0.01) (Supplemental Figure 2A, Figure 2B); similar results were obtained for KRAS mutation (37% versus 28%, p=0.05) (Figure 2B). Conversely, TP53 mutations were enriched in the high STING-high immune LUAD group (56% versus 35%, p<0.001) (Figure 2B).
Figure 2. STK11/LKB1 mutation is associated with different phenotypes of immune activation.
(A-B) Identification of STING/immune signatures genes (from Chen et al.) shows two main groups of LUAD carcinomas in TCGA LUAD cohort (n=515), a low STING/low immune group and a high STING/high immune group (A). We found an enrichment of STK11 (LKB1) (p=0.01) and of KRAS (p=0.05) mutations in low STING/low immune group and of TP53 mutations (p<0.001) in high STING/high immune tumors (B). (C-D) Hierarchical clustering of immune signatures genes (from Chen et al.) in STK11- mutant tumors in the TCGA LUAD cohort identifies three subgroups with low, intermediate and high expression of immune and STING genes (C). Comparison of the three subgroups detected a significant enrichment in the frequency of TP53 mutations (p=0.002, by Fisher’s exact test) in the “immune-high” tumors as compared to the other two subgroups (D). NS= not significant.
As we observed STK11 mutant LUAD in both the STING-high and –low groups, despite these tumors being universally considered to be immunologically cold, we next analyzed only the STK11/LKB1 mutant LUAD tumors (n=73) from the TCGA cohort. Using hierarchical clustering based on expression of CGAS, CCL5 and CXCL10 we identified three distinct groups that we named STING-low, -intermediate, and –high. We then performed a supervised analysis to compare expression of the immune related genes between these three groups. While the majority of STK11/LKB1 mutant tumors were in the STING-low/-intermediate group (n=55) and had low expression of immune related genes, there is a subset in the STING-high group (n=18) with higher levels of targetable immune genes, like CD274, HAVCR2, PDCD1LG2, T-cell infiltration markers, and chemokines (Figure 2C). These STING-high STK11/LKB1 mutant tumors showed an “inflamed” immune phenotype, enriched for immune checkpoints and cytokines expression, which is more similar to a LKB1 wild-type tumor (Figure 2C). Interestingly, when we compared the mutation profiles of the STING-high STK11/LKB1 mutant group with the other two groups, including low or intermediate expression of these genes (immune “cold” groups), we found that the STING-high group is enriched for TP53 mutations (56% versus 16%, p=0.002) (Figure 2D). Moreover, in the STING-high group, KRAS and TP53 mutations are mutually exclusive, while in the STING-low/-intermediate group they can overlap (7% of samples are co-mutant). While several groups have identified STK11 mutations as predictive of poor response to immunotherapy and that STK11/KRAS mutant lung adenocarcinoma have low PD-L1 expression and STING gene silencing (26, 27), our data suggest that among the STK11 mutant subgroup, there are a range of inflammatory phenotypes defined by the presence of co-mutations. Specifically, 25% of STK11 mutant tumors have an active STING pathway, immune checkpoint expression, and inflammatory characteristics consistent with other immune “hot” NSCLC subtypes and, among these, 56% are TP53 mutant and KRAS wild-type. We investigated the difference in cGAS protein expression by RPPA in our panel of LUAD cell lines, comparing STK11/KRAS mutant cells with STK11/TP53 mutant cells. Cell lines in the STK11/TP53 mutant group are heterogeneous in their expression of cGAS and include subsets with medium and high cGAS protein expression. In contrast (but consistent with the patient data), the STK11/KRAS mutant cell lines were all cGAS low (26, 27) (Supplemental Figure 2B).
Cisplatin treatment enhances the protein expression of STING pathway markers in pre-clinical models of NSCLC
Recent reports have described that DDR-inhibitors and radiotherapy can increase PD-L1 expression and immunogenicity of tumors in various cancer models (10–13). Phase III clinical trials in NSCLC have demonstrated that patients who receive standard chemotherapy (platinum-based doublet) plus immunotherapy in first line of treatment do better in terms of overall survival than patients treated either with chemotherapy or chemotherapy followed by immunotherapy (28, 29). We hypothesized that the improvement in clinical response/survival observed with the platinum plus immunotherapy combination (vs platinum alone) may be mediated by platinum induced DNA damage leading to activation of the innate immune (STING) pathway.
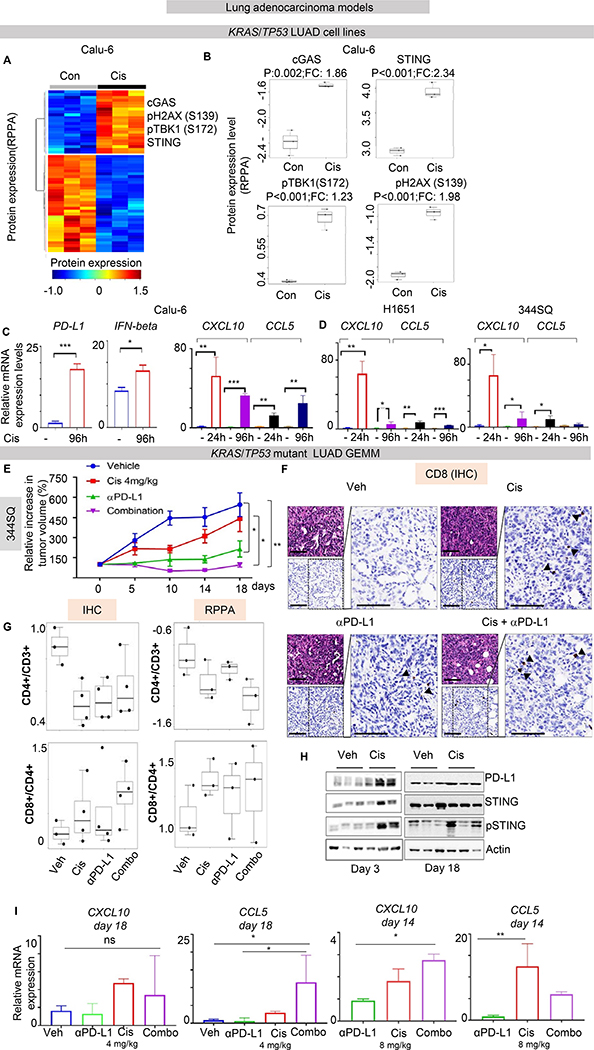
Platinum chemotherapyis the chemotherapeutic backbone for the frontline treatment of NSCLC and is included in all chemotherapy plus immunotherapy combinations, thus we investigated the influence of cisplatin treatment chemotherapy on the expression of immune-related proteins in NSCLC cell lines. We treated a panel of LUAD cell lines (n=3) with cisplatin for 96 hours and analyzed changes in protein expression and pathway activation between control and treated samples by RPPA. We selected three LUAD NSCLC cell lines representative of three biological profiles: Calu-6 (KRAS/TP53 mutant), HCC827 (EGFR mutant), and H1944 (KRAS/STK11 mutant). The KRAS/TP53 mutant represents immune sensitive disease, whereas the other two cell lines represent two immune-resistant phenotypes (4, 5, 30). In the KRAS/TP53 mutant LUAD cell line, Calu6, we observed a significant upregulation of multiple STING pathway proteins following cisplatin treatment, including total STING (fold change, FC=2.34, p<0.001), phospho-TBK1 (S172) (FC=1.23, p<0.001) and cGAS (FC=1.86, p=0.002), along with a moderate increase in PD-L1 (FC=1.15, p=0.028) and phospho-H2AX (S139) (FC=1.98, p<0.001), that is a marker of DNA damage, as compared to untreated controls (Figure 3A, B). We also investigated changes in gene expression of CD274 (PD-L1), IFNβ, and the inflammatory cytokines, CXCL10 and CCL5 after cisplatin treatment in Calu6 cells. We found that expression of all four genes was significantly increased following cisplatin treatment (Figure 3C). Similarly, CXCL10 and CCL5 expression was also increased after cisplatin treatment in two other KRAS/TP53 mutant LUAD cell lines, H1651 (human) and 344SQ (derived from KrasLA1/+p53R172HΔG/+ mice) (Figure 3D).
Figure 3. Cisplatin treatment activates STING pathway markers in NSCLC in vitro and in vivo.
(A-B) Supervised hierarchical clustering of protein expression profiles in untreated and cisplatin-treated LUAD cells Calu-6 (KRAS/TP53 mutant) shows upregulation of proteins in the STING pathway (STING, phospho-TBK1 (Ser_172), CGAS), PD-L1 and phospho-H2AX(Ser_139) following cisplatin treatment (p<0.01, by t-test). (C-D) Quantitative PCR (qPCR) measurement of PD-L1, CCL5, CXCL10 and IFNβ mRNA expression in three KRAS/TP53 mutant NSCLC cell lines treated with cisplatin for 24 and 96 hours. Data presented as mean ±SD and p values by t-test ***p<0.001, **p<0.01, *p<0.05.. (E) 344SQ tumor bearing mice were treated with vehicle, cisplatin (4 mg/kg, i.p., 1/7), anti-PD-L1 antibody (300μg, i.p., 1/7), or the combination. Relative tumor volumes (mean ± S.E.M.) are shown. The combination of cisplatin and anti-PD-L1 antibody significantly potentiated the anti-tumor response to either single agent (P values were calculated by ANOVA, followed by Tukey’s test. (F) Histological analysis of 344SQ tumors demonstrate the presence of CD8+ immune cells. Scale bar = 200 μM. (G) Results from IHC and RPPA analysis for T-cells infiltration: CD8+/CD4+ and CD4+/CD3+ ratio are presented. Ratio were calculated from number of positive cells for any marker/ mm2. CD4+/CD3+ ratio was decreased by IHC (p=0.038 calculated by ANOVA, followed by Dunnett’s test versus vehicle, p=0.02 for cisplatin, p=0.03 for anti-PDL1, p=0.098 for combination) and by RPPA (p=0.1 by ANOVA). Conversely, CD8+/CD4+ ratio was increased by IHC in 2/4 tumors from cisplatin and 3/4 tumors from combination arm (p>0.1 by ANOVA) and the same result were confirmed by RPPA (p>0.1 by ANOVA). (H) Images from western blot analysis on lysates from tumors harvested from the 344SQ tumor bearing mice after three days or at the end of treatment showed an increase in PD-L1 and phospho-STING levels in cisplatin-treated tumors compared to the vehicle group. (I) Quantitative PCR (qPCR) measurement of CCL5 and CXCL10 mRNA expression in tumors after indicated treatments. Data presented as mean ±SD and p values were calculated by ANOVA, followed by Tukey’s test. ***p<0.001, **p<0.01, *p<0.05. ns=not significant.
Retrospective analysis of immunotherapy clinical trials have shown that EGFR mutant patients do not benefit from immunotherapy (4, 30). This is thought to be due to their distinct biology, resulting from having a single dominant molecular driver with fewer potential neoantigens, low levels of PD-L1 expression (only 11% of EGFR mutant tumors have PD-L1>50%) and little CD8+ T-cell infiltration (36). Interestingly, in the EGFR mutant-cell line HCC827, we did observe that PD-L1 was modestly increased (similar to other NSCLC models), but cGAS was downregulated post-cisplatin treatment (FC-1.2, p<0.01) without any significant change in other STING markers (Supplemental Figure 3A). Similarly, since previous studies have characterized the KRAS/STK11 tumors as immune-resistant and less responsive to cytosolic DNA signal, with downregulated TMEM173 (STING) levels and decreased STING activation (32), we expected that KRAS/STK11 (LKB1) mutated NSCLC cells would not show DNA sensing pathway activation in response to cisplatin. Consistent with this, in the KRAS/STK11 (LKB1) mutant-cell line H1944, we detected a modest increase in PD-L1 protein levels after cisplatin without any change in cGAS levels (Supplemental Figure 3B). Having observed increased expression of STING-related genes in STING-high STK11/TP53 mutant LUAD tumors (Figure 2B) and increased expression of cGAS protein in STK11/TP53 mutant cell lines (Supplemental Figure 2A), we were interested to understand how STK11/TP53 cells respond to cisplatin in terms of production of IFNβ and inflammatory cytokines, CXCL10 and CCL5. We observed increase of CXCL10 and CCL5 after 96 hours of cisplatin treatment in two STK11/TP53 cell lines: both were significantly increased in H1573 cells (p= 0.02 for CCL5 and p=0.006 for CXCL10) and CXCL10 (p=0.01) in HCC2302, Supplemental Figure 4). These results demonstrate that while cisplatin treatment leads to increased PD-L1 expression across various NSCLC in vitro models, the presence of STING activation is genotype-dependent in LUAD.
Based on these results, we asked if chemotherapy treatment augments the anti-tumor immune response and the efficacy of anti-PD-L1 treatment in models that are partially sensitive to single agent anti-PD-L1 treatment, but display adaptive resistance (31, 32). We selected the lung adenocarcinoma cell line, 344SQ, derived from KrasLA1/+p53R172HΔG/+ mice, that produces highly metastatic, mesenchymal tumors and transplanted it into immunocompetent syngeneic mice. Tumor-bearing mice (n= 8 per group) were treated once a week with vehicle, cisplatin (4 mg/kg per week) and/or anti-PD-L1 (300μg per week) for 18 days. As expected, all vehicle-treated mice (n = 8) experienced rapid tumor progression (Figure 3E). At day 18, cisplatin alone (4 mg/kg per week) also did not cause any significant tumor regression in these models as compared to vehicle group (Figure 3E). Mice treated with anti-PD-L1 had inhibition of tumor growth as compared to vehicle (p=0.026, by ANOVA, followed by Tukey’s test) (Figure 3E). However, combination of anti-PD-L1 with cisplatin, even with submaximal dosing of cisplatin, significantly potentiated the anti-tumor response with tumor growth inhibition as compared to vehicle (p=0.003) and to cisplatin (p=0.026) (Figure 3E).
Histological analyses revealed the presence of infiltrating immune cells within the tumors, collected at day 18, as demonstrated by representative images for CD8 staining in Figure 3F. To identify the T-cells present within the tumors, we performed immunohistochemistry (IHC) for CD3, CD4, and CD8. We observed a significant reduction in CD4+/CD3+ T-cell ratio (p=0.038 by ANOVA, followed by Dunnett’s test, Figure 3G, left panel) in cisplatin and immunotherapy arms, and a trend in increase in CD8+/CD4+ T-cell ratio in cisplatin (2/4 tumors) and even more in combination arm (3/4 tumors) (Figure 3G, right panel). These changes suggest that cisplatin induces changes in T-cell infiltration similar to immunotherapy at a time point where immunotherapy treatment alone inhibits tumor growth. Similar trends were confirmed evaluating CD4 and CD8 protein expression by RPPA in the same samples (Figure 3G). To determine the effect of treatment with cisplatin on STING pathway markers in this in vivo model, we performed western blot analyses of tumors from vehicle and cisplatin treated animals following 3 days of treatment and at the end of treatment, at day 18 (Figure 3H). Tumors from cisplatin-treated animals showed higher levels of PD-L1 and phospho-STING (in 2/3 samples), compared to tumors treated with vehicle, as shown by western blot images and relative densitometric quantification (Supplementary Figure 5). In contrast, we did not detect an increase of these proteins in tumors treated with anti-PD-L1 alone or cisplatin/anti-PD-L1 combination at either time point. However, in the case of the combination, protein analysis may have been technically limited by the extent of necrotic tissue, as described above. To better explore the effect of cisplatin on intra-cellular signaling, we repeated the experiment using a higher dose of cisplatin (8mg/kg) that would be comparable to clinical dosing in patients. Unfortunately the use of this dose was limited by toxicity, with occurrence of loss in body weights more than 15%, and cisplatin treatment had to be terminated after 2 weeks. However, tumors were collected after two weeks of treatment. We assayed expression of the STING downstream inflammatory chemokines CXCL10 and CCL5 in treated tumors and we found that they are increased in tumors from both the cisplatin and combination arms (Figure 3I). In particular, CXCL10 was increased significantly in combination arm with cisplatin 8 mg/kg (p=0.03 by ANOVA, followed by Tukey’s test) and CCL5 was significantly increased by cisplatin 8mg/kg (p=0.01 by ANOVA, followed by Tukey’s test), as compared to single agent immunotherapy and in combination arm with cisplatin 4mg/kg (p=0.03 by ANOVA, followed by Tukey’s test) as compared to vehicle and single agent immunotherapy. Tumors with increase in CD8+/CD4+ ratio were the ones with high STING activation, as demonstrated by phospho-STING protein levels and increase of STING downstream chemokines. Together, these findings indicate that cisplatin treatment activates the STING pathway and induces the expression of PD-L1 in an aggressive mesenchymal NSCLC murine model and suggest that the addition of platinum-chemotherapy may augment the anti-tumor immune response of anti-PD-L1 through STING activation
These data support future exploration of the interplay between STING and PD-L1 pathway in response to chemo-immunotherapy treatment.
STING pathway expression is associated with immune activation in squamous lung cancer
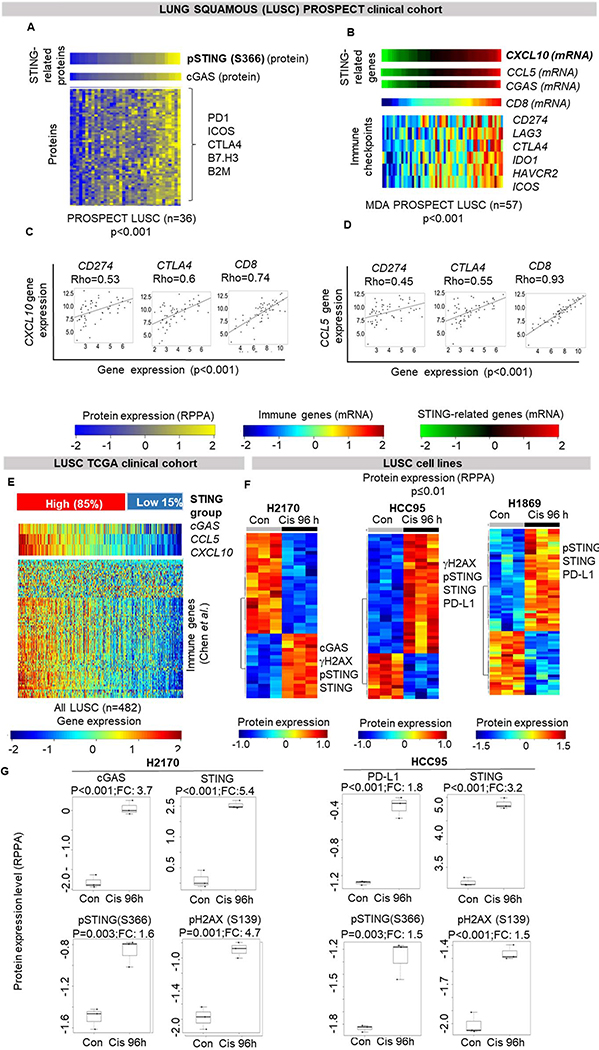
Since combination of chemotherapy and immunotherapy is currently approved for both lung adenocarcinoma and lung squamous carcinomas (LUSC) in the absence of driver mutations, we investigated the association between STING pathway activation and immune activation using RPPA data from a cohort of treatment-naïve squamous lung tumors from our institution (MDACC PROSPECT LUSC, n=36). As expected, phospho-STING protein levels were highly correlated with cGAS protein levels (Spearman’s rho (rho) = 0.58, p=0.0003) and targetable immune markers (PD1, ICOS, CTLA4, B7.H3, B2M) (all rho > 0.5, p<0.001) (Figure 4A). In addition, among the top markers associated with phospho-STING, we also detected the EMT marker N-cadherin and DNA damage proteins (DNAPKcs, ATR, CHK1) (rho > 0.4, p<0.01). LUSC tumors with high levels of STING proteins also have an “inflamed phenotype”, and EMT and high DNA damage may identify such tumors.
Figure 4. STING pathway activation is associated with expression of immune markers in squamous cell lung carcinoma.
(A) Heatmap shows correlation of phospho-STING with cGAS protein levels and other immune and inflammatory proteins in the MDACC PROSPECT lung squamous cohort (Spearman’s rho > 0.5, p<0.001). (B) Expression of CXCL10 is correlated with other STING genes, CCL5 and CGAS, CD8, and selected targetable immune genes in PROSPECT LUSC: CD274 (PD-L1) (Spearman’s rho = 0.4, p<0.001), LAG3, CTLA4, IDO1, HAVCR2 and ICOS (Spearman’s rho > 0.6, p<0.0001; respectively). (C-D) Individual dot-plots showing correlation of CXCL10 and CCL5 expression with selected targetable genes shown in 4B. (E) Hierchical clustering, as shown for LUAD in Figure 2A, was applied to TCGA LUSC cohort. (F-G) Supervised hierarchical clustering of protein expression profiles in three untreated and cisplatin-treated (3μM for 96hours) LUSC cells shows upregulation of proteins in the STING pathway, PD-L1 and γH2AX following cisplatin treatment (p<0.01, by t-test).
We further investigated, in the PROSPECT LUSC cohort for which we have gene expression data (n=57), the association between STING-related genes (CGAS, CXCL10 and CCL5), and expression of the curated list of immune markers by Chen et al. (29). Similar to lung adenocarcinoma, CXCL10 mRNA expression was positively correlated with other STING genes, CGAS and CCL5, and with the immune markers including targetable immune checkpoints genes, CD274 (PD-L1), LAG3, CTLA4, IDO1, HAVCR2, ICOS (rho >0.4, p<0.001) (Figure 4B–D). We further validated these findings in the TCGA LUSC cohort (n=501) (Supplemental Figure 6) where STING related genes were again correlated with immune markers (rho>0.3, p<0.001). In the same LUSC cohort, we also applied the hierarchical clustering used in the TCGA LUAD cohort, based on gene expression levels of CGAS, CCL5 and CXCL10 (Figure 2A) and identified two groups – STING-high (85%) and STING-low (15%). Unlike in LUAD, there were no significant difference in the frequency of TP53 and STK11 mutations between these two groups, as expected considering high frequency of TP53 mutation and almost null frequency of STK11 mutation (81% and 3% respectively) (33). Similar to LUAD results, the STING-high LUSC group had higher expression of the immune related genes (Figure 4E).
Finally, we treated three squamous lung carcinoma cell lines (H2170, HCC95 and H1869) with cisplatin to explore the effect on STING and PD-L1: phospho-STING (S366), STING and PD-L1 were among the top markers upregulated post-cisplatin treatment in all three cell lines (p<0.01; Figure 4F, G). cGAS, and phosph-H2AX (S139), marker of DNA damage, were also significantly upregulated post-cisplatin treatment in H2170 and HCC95 cells (p<0.01; Figure 4F, G).
Discussion
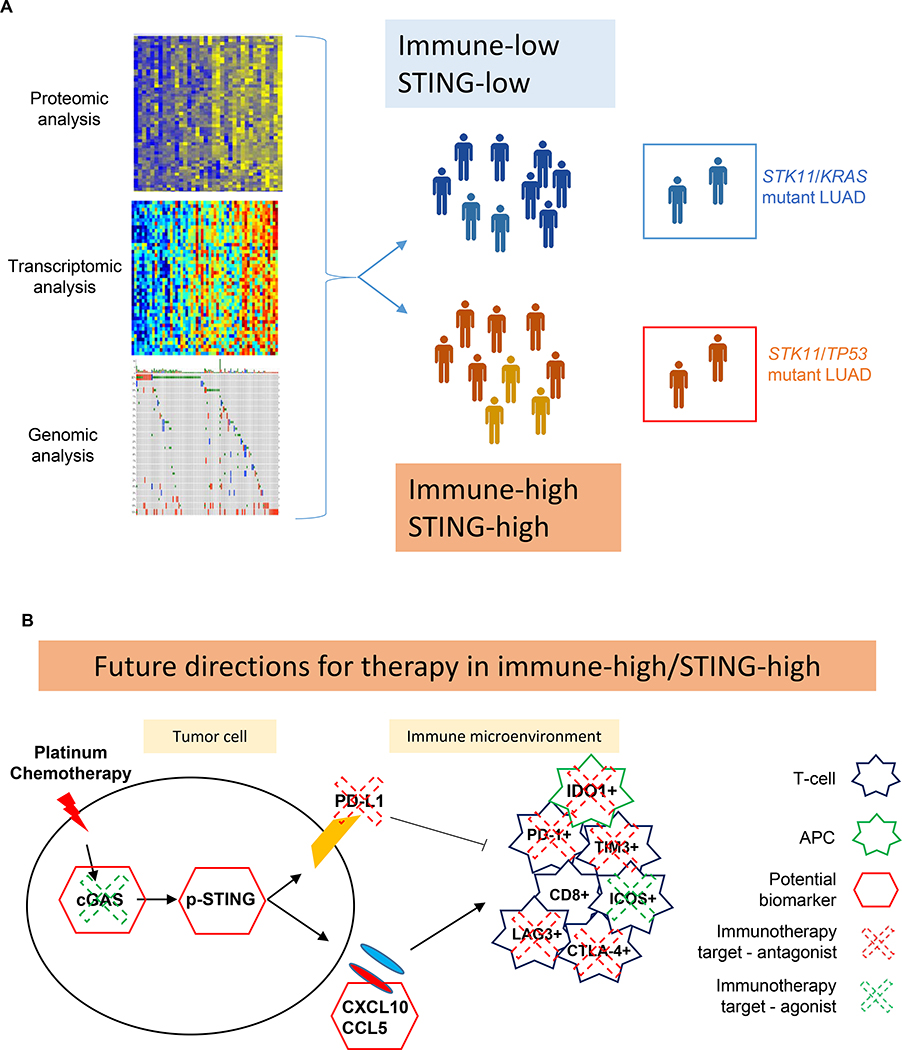
In this work, we explored for the first time the landscape of STING pathway activation in NSCLC tumors, including adenocarcinomas and squamous lung cancers. We found that NSCLCs with high STING pathway activation (detected by STING related protein or gene expression) had higher levels of targetable immune checkpoints and markers of an active immune microenvironment that are associated with clinical responses to immunotherapy (6, 34) (Figure 5A). In prior studies by our group and others, high levels of basal or post-treatment T-cell infiltration markers and immune checkpoints molecules were associated with greater immunogenicity and response to ICB (12). Thus, we show that STING activation correlates with each of these known immune-responsive features in NSCLC, as represented by expression of targetable immune genes by tumor or stromal cells (Figure 5B), and could be a potential biomarker for novel immunotherapy- and immunotherapy-based combination in NSCLC. When we correlated genomic features of LUAD tumors with STING activation, we found that STK11 (LKB1) mutant tumors have lower expression of immune genes, as compared to other LUAD tumors, as known from previous studies in KRAS/STK11 mutant tumors (4, 23). However, we identified a novel subset of the STK11 patients, characterized by co-mutation in TP53, that show high STING activation and immune genes expression, thus suggesting a subset of STK11 mutant lung adenocarcinoma that may potentially benefit from immunotherapy. Our findings suggest that in addition to STK11 (LKB1) mutation itself, specific co-mutations such as KRAS or TP53 may play important roles in regulating activation of the STING pathway and overall modulation of the immune microenvironment. These findings warrant further investigation in patient samples to investigate the predictive role of each genotype (KRAS/STK11 versus TP53/STK11) for response to chemo-immunotherapy combination and to better understand the molecular features of this subset of STING activated, STK11 (LKB1) mutant NSCLC. The intrinsic DNA damage associated with loss of p53 function (12) may counter-balance the epigenetic silencing of TMEM173 (STING) by LKB1 in STK11/TP53 mutant tumors (4, 23); whereas, the dual activation of mTOR pathway by both KRAS mutation and LKB1 loss may act cooperatively to suppress activation of the innate immune system in KRAS/STK11 mutant tumors.In LUSC tumors, we did not detect any significant difference in mutations between the STING-high and STING-low subgroup.
Figure 5. Working model.
(A) Schematic representation of the resources used in this work: genomic, transcriptomic and proteomic analysis led to identification of two main group of STING/immune expression in NSCLC, with distinct molecular subgroups in LUAD. (B) Schematic representation of the novel potential therapeutic implication for high STING/high immune NSCLC.
As with TP53 mutations, genetic alterations of DDR machinery genes may explain why many cancer cells have intrinsic levels of DNA damage (35). DNA damage can also be further increased secondary to anti-cancer therapies, including chemotherapy, radiotherapy, or therapies targeting DDR pathway proteins (e.g. PARP inhibitors) (10–13). For example, previous studies have shown that cisplatin treatment activates STING and PD-L1 expression in models of breast cancer (11) and SCLC (12), supporting the idea of a connection between innate and adaptive anti-cancer immunity. In this work, we detected a positive correlation between STING-, immune- and DNA damage-related proteins expression in PROSPECT cohorts. Recently, two clinical trials, Keynote-189 (LUAD) and Keynote-407 (LUSC) (28, 29), demonstrated that the addition of immunotherapy to standard platinum doublet chemotherapy was superior to chemotherapy, in terms of PFS and OS, also in PD-L1 low NSCLC. Therefore, there is a critical need to investigate the landscape of immune profiles in NSCLC in this context to optimize patients’ selection. A limit of the clinical cohorts used in this study is lack of paired pre- and post-treatment samples from patients treated with chemotherapy or chemo-immunotherapy. First, we tested the effect of cisplatin in pre-clinical models of NSCLC. We observed that chemotherapy induces STING and PD-L1 protein expression in KRAS/TP53 mutated human adenocarcinoma cells and murine tumors (32), and in human lung squamous cell lines; conversely, EGFR and KRAS/STK11 adenocarcinoma cells did not show increase in cGAS levels after cisplatin, even if there is an increase in PD-L1 protein levels, thus suggesting another reason for their known immune-resistance. The increase in inflammatory chemokines CXCL10 and CCL5 post cisplatin treatment in multiple models in vitro and in vivo, accompanied by the increase in CD8+ cytotoxic T-cells and decrease in CD4+ helper T-cells in murine tumors, suggest that chemotherapy contributes to an “inflamed” microenvironment, similarly to what we have previously demonstrated with DDR-inhibitors in SCLC (12, 36). We speculate that cisplatin-induced STING activation may explain the enhanced activity of platinum-based chemotherapy-immunotherapy combination. However, STING protein can be subjected to multiple post-translational modifications that affect its transient response and may also cause different downstream responses (37). Recently, STING activation has also been associated with poor prognosis and low immune infiltration (38). Crosstalk among immune-checkpoints could explain this apparent dual role of STING, since upregulation of immune checkpoints leads to immunosuppression. Consideration of multiple and, as yet, uncharacterized roles of the STING pathway may also explain the heterogeneous response in terms of STING activation and associated CD8+ T-cells infiltration that we observed in our animal models. Further studies should address this gap in knowledge, exploring changes in STING pathway and immune cell infiltration at multiple time points during treatment.
Together, our data support an interplay between innate immune pathways and immune-suppressive checkpoints in NSCLC and that intrinsic STING pathway activation may be an upstream mediator of immune checkpoint expression and, ultimately, immunotherapy response. However, inducing the activation of STING pathway by DNA damage, as we demonstrated with cisplatin, or directly, with STING agonists or other novel pharmacological modulators of STING (39–41), could potentially increase the immune responsive phenotype also in some otherwise resistant tumors and encourage further evaluation of novel combination strategies based on this rationale.
Supplementary Material
Acknowledgment
We acknowledge members of Translational Molecular Pathology Immunoprofiling Laboratory (TMP-IL), for their technical expertise provided on histology processing and immunohistochemistry staining: Wei Lu, Mei Jiang, Jianling Zhou, Jocelyn Coronel, Lakshmi Kakarala.
Financial support: This work was supported by: The NIH/NCI CCSG P30-CA016672 (Bioinformatics Shared Resource); Lung Cancer Research Foundation LCRF (TS); NIH/NCI T32 CA009666 (CMG); ASCO Young Investigator Award (CMG); Institutional research support from Eli Lilly, Novartis, Merck, Nektar,Astra Zeneca, F Hoffman -La Roche, Janssen, Checkmate,Incyte, Bristol Myers Squibb (VAP); The University of Texas-Southwestern and MD Anderson Cancer Center Lung SPORE (5 P50 CA070907); NCI R37 CA21460 (DLG); NIH/NCI R01-CA207295 (LAB); NIH/NCI R01-CA205150 (JVH); NIH/NCI U01-CA213273 (LAB, JVH); research Support from AstraZeneca, Bayer, GlaxoSmithKline, Spectrum (JVH); The Department of Defense LC170171 (LAB); Through generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shot Program (JVH, JW, LAB, DLG); CPRIT-MIRA RP160652 (JVH & DLG); The MD Anderson Cancer Center Small Cell Lung Cancer Working Group and Abell Hangar Foundation Distinguished Professor Endowment (LAB), MD Anderson Cancer Center Physician Scientist Award (LAB & DLG), The Rexanna Foundation for Fighting Lung Cancer (JVH, LAB, DLG), and Sabin fellowship (LAB).
Conflict of Interest
V.A.P. has served on advisory boards for: Nektar Therapeutics, Bristol Myers Squibb, Astra Zeneca, Novartis, Merck, Arrys, Zara Ed Pharma, LOXO oncology,F Hoffman-La-Roche, Janssen Research Foundation,Clovis Oncology,Eli Lilly,Takeda,Abbie,Tesaro, Exelixis,Gritstone and has received speaker fees from F Hoffman-La-Roche. She is currently affiliated with Pfizer. I. I. W. serves on advisory board forGenentech/Roche, Bayer, Bristol-Myers Squibb, Astra Zeneca/Medimmune, Pfizer, HTG Molecular, Asuragen, Merck, GlaxoSmithKline, Guardant Health and MSD and has received speaker fees from: Medscape, MSD, Genentech/Roche, Pfizer; he received research funding: Genentech, Oncoplex, HTG Molecular, DepArray, Merck, Bristol-Myers Squibb, Medimmune, Adaptive, Adaptimmune, EMD Serono, Pfizer, Takeda, Amgen, Karus, Johnson & Johnson, Bayer, Iovance, 4D, Novartis, and Akoya. F.S. received honoraria from Bristol-Myers Squibb. J.V.H. serves on advisory committees for AstraZeneca, Boehringer Ingelheim, Exelixis, Genentech, GSK, Guardant Health, Hengrui, Lilly, Novartis, Spectrum, EMD Serono, and Synta. D.L.G. serves on advisory committees for AstraZeneca, GlaxoSmithKline, Sanofi, Ribon Therapeutics Alethia Biotherapeutics, Inc., and Janssen; he receives research funding from AstraZeneca, Janssen R&D, Takeda and Ribon Therapeutics. L.A.B. provides consulting for AstraZeneca, AbbVie, GenMab, BergenBio, Pharma Mar, SA, Sierra Oncology, Bristol Myers Squibb, Alethia Biotherapeutics Inc. and research funding from AstraZeneca, AbbVie, GenMab and Sierra Oncology.
References
- 1.Ettinger DS, Aisner DL, Wood DE, Akerley W, Bauman J, Chang JY, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 5.2018. J Natl Compr Canc Netw 2018;16:807–21. [DOI] [PubMed] [Google Scholar]
- 2.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823–33. [DOI] [PubMed] [Google Scholar]
- 4.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 Mutations and PD1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov 2018; 8:822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CK, Man J, Lord S, Links M, Gebski V, Mok T, et al. Checkpoint Inhibitors in Metastatic EGFR-Mutated Non-Small Cell Lung Cancer-A Meta-Analysis. J Thorac Oncol 2017;12:403–7. [DOI] [PubMed] [Google Scholar]
- 6.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017; 127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ott PA, Bang YJ, Piha-Paul SA, Razak ARA, Bennouna J, Soria JC, et al. T-Cell-Inflamed Gene-Expression Profile, Programmed Death Ligand 1 Expression, and Tumor Mutational Burden Predict Efficacy in Patients Treated With Pembrolizumab Across 20 Cancers: KEYNOTE-028. J Clin Oncol 2019;37:318–27. [DOI] [PubMed] [Google Scholar]
- 8.Trujillo JA, Sweis RF, Bao R, Luke JJ. T Cell-Inflamed versus Non-T Cell-Inflamed Tumors: A Conceptual Framework for Cancer Immunotherapy Drug Development and Combination Therapy Selection. Cancer Immunol Res 2018;6:990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teo MY, Bambury RM, Zabor EC, Jordan E, Al-Ahmadie H, Boyd ME, et al. DNA Damage Response and Repair Gene Alterations Are Associated with Improved Survival in Patients with Platinum-Treated Advanced Urothelial Carcinoma. Clin Cancer Res 2017;23:3610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding L, Kim H, Wang Q, Kearns M, Jiang T, Ohlson CE, et al. PARP Inhibition Elicits STING-Dependent Antitumor Immunity in Brca1-Deficient Ovarian Cancer. Cell Rep 2018;25:2972–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkes EE, Walker SM, Taggart LE, McCabe N, Knight LA, Wilkinson R, et al. Activation of STING-Dependent Innate Immune Signaling By S-Phase-Specific DNA Damage in Breast Cancer. J Natl Cancer Inst 2016; 109(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, et al. Targeting DNA Damage Response Promotes Antitumor Immunity through STING-Mediated T-cell Activation in Small Cell Lung Cancer. Cancer Discov 2019. [epub}. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato H, Niimi A, Yasuhara T, Permata TBM, Hagiwara Y, Isono M, et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat Commun 2017;8:1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barber GN. STING: infection, inflammation and cancer. Nat Rev Immunol 2015;15:760–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardnell RJ, Li L, Sen T, Bara R, Tong P, Fujimoto J, et al. Protein expression of TTF1 and cMYC define distinct molecular subgroups of small cell lung cancer with unique vulnerabilities to aurora kinase inhibition, DLL3 targeting, and other targeted therapies. Oncotarget 2017;8:73419–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byers LA, Wang J, Nilsson MB, Fujimoto J, Saintigny P, Yordy J, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov 2012;2:798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardnell RJ, Behrens C, Diao L, Fan Y, Tang X, Tong P, et al. An Integrated Molecular Analysis of Lung Adenocarcinomas Identifies Potential Therapeutic Targets among TTF1-Negative Tumors, Including DNA Repair Proteins and Nrf2. Clin Cancer Res 2015;21:3480–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Cancer Genome Atlas Research N. Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim ES, Herbst RS, Wistuba II, Lee JJ, Blumenschein GR Jr, Tsao A, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov 2011;1:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, et al. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin Cancer Res 2016;22:609–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lou Y, Diao L, Cuentas ER, Denning WL, Chen L, Fan YH, et al. Epithelial-Mesenchymal Transition Is Associated with a Distinct Tumor Microenvironment Including Elevation of Inflammatory Signals and Multiple Immune Checkpoints in Lung Adenocarcinoma. Clin Cancer Res 2016;22:3630–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Diao L, Yang Y, Yi X, Rodriguez BL, Li Y, Villalobos PA, et al. CD38-Mediated Immunosuppression as a Mechanism of Tumor Cell Escape from PD1/PD-L1 Blockade. Cancer Discov 2018;8:1156–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015; 5:860–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbie DA, Kitajima S, Ivanova E, Guo S, Yoshida R, Campisi M, et al. Suppression of STING Associated with LKB1 Loss in KRAS-Driven Lung Cancer. Cancer Discov 2019; 9:34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Della Corte CM, Byers LA. Evading the STING: LKB1 Loss Leads to STING Silencing and Immune Escape in KRAS-Mutant Lung Cancers. Cancer Discov 2019; 9:16–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wargo JA, Reddy SM, Reuben A, Sharma P. Monitoring immune responses in the tumor microenvironment. Curr Opin Immunol 2016;41:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:2078–92. [DOI] [PubMed] [Google Scholar]
- 29.Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gümüş M, Mazières J, et al. Pembrolizumab plus Chemotherapy for Squamous Non-Small-Cell Lung Cancer. N Engl J Med 2018;379:2040–51. [DOI] [PubMed] [Google Scholar]
- 30.Gainor JF, Shaw AT, Sequist LV, Fu X, Azzoli CG, Piotrowska Z, et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin Cancer Res 2016;22:4585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev 2009;23:2140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 20145:5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J1, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. doi: 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whiteside TL, Demaria S, Rodriguez-Ruiz ME, Zarour HM, Melero I. Emerging Opportunities and Challenges in Cancer Immunotherapy. Clin Cancer Res 2016;22:1845–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sen T, Gay CM, Byers LA. Targeting DNA damage repair in small cell lung cancer and the biomarker landscape. Transl Lung Cancer Res 2018;7:50–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen T, Della Corte CM, Milutinovic S, Cardnell RJ, Diao L, Ramkumar K,et al. Combination Treatment of the Oral CHK1 Inhibitor, SRA737, and Low-Dose Gemcitabine Enhances the Effect of Programmed Death Ligand 1 Blockade by Modulating the Immune Microenvironment in SCLC. J Thorac Oncol 2019: S1556–0864(19)30692–6. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jønsson KL, et al. Non-canonical Activation of the DNA Sensing Adaptor STING by ATM and IFI16 Mediates NF-κB Signaling after Nuclear DNA Damage. Mol Cell 2018; 71:745–60.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An X, Zhu Y, Zheng T, Wang G, Zhang M, Li J, et al. An Analysis of the Expression and Association with Immune Cell Infiltration of the cGAS/STING Pathway in Pan-Cancer. Mol Ther Nucleic Acids 2018;14:80–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berger G, Lawler SE. Novel non-nucleotidic STING agonists for cancer immunotherapy. Future Med Chem 2018;10:2767–9. [DOI] [PubMed] [Google Scholar]
- 40.Ramanjulu JM, Pesiridis GS, Yang J, Concha N, Singhaus R, Zhang SY, et al. Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature 2018;564:439–43. [DOI] [PubMed] [Google Scholar]
- 41.Cheng N, Watkins-Schulz R, Junkins RD, David CN, Johnson BM, Montgomery SA, et al. A nanoparticle-incorporated STING activator enhances antitumor immunity in PD-L1-insensitive models of triple-negative breast cancer. JCI Insight 2018;3(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.