Abstract
miRNAs are endogenous noncoding single-stranded small RNAs of ~22 nucleotides in length that post-transcriptionally repress the expression of their various target genes. They contribute to the regulation of a variety of physiologic processes including embryonic development, differentiation and proliferation, apoptosis, metabolism, hemostasis and inflammation. In addition, aberrant miRNA expression is implicated in the pathogenesis of numerous diseases including cancer, hepatitis, cardiovascular diseases and metabolic diseases. Steroid hormones regulate virtually every aspect of metabolism, and acute and chronic steroid hormone biosynthesis is primarily regulated by tissue-specific trophic hormones involving transcriptional and translational events. In addition, it is becoming increasingly clear that steroidogenic pathways are also subject to post-transcriptional and post-translational regulations including processes such as phosphorylation/dephosphorylation, protein‒protein interactions and regulation by specific miRNAs, although the latter is in its infancy state. Here, we summarize the recent advances in miRNA-mediated regulation of steroidogenesis with emphasis on adrenal and gonadal steroidogenesis.
Keywords: ACTH, gonadotropins, steroid synthesis, StAR, cholesterol
MicroRNAs
miRNAs are endogenous single-stranded small RNAs of ~22 nucleotides in length and generated from endogenous hairpin-shaped transcripts (Bartel 2018). miRNAs post-transcriptionally downregulate gene expression by base-pairing to partially complementary sites in one or more target mRNAs, usually in the 3′ UTR, triggering their repression; mechanisms for miRNA-mediated downregulation of gene expression involve some combination of translational repression, mRNA deadenylation, decapping, 5′-to-3′ mRNA degradation and alteration of mRNA stability (Fabian et al. 2010, Jonas & Izaurralde 2015). miRNAs belong to a group of heterogeneous noncoding RNAs (ncRNAs) including long noncoding RNAs (lncRNAs) such as long intergenic ncRNAs, natural antisense transcripts, pseudogenes and circular RNAs (circRNAs), ribozymes, ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), small nucleolar RNAs (snRNAs), Piwi-associated RNAs (piRNAs), small nuclear RNAs (snoRNAs), telomere-associated RNAs (TERC, TERRA), miRNAs and miRNAs that participate in RNA interference (RNAi or siRNA) (Esteller 2011). Among these ncRNAs, miRNAs are one of the most extensively studied ncRNA species and have emerged as important regulators of gene expression (Bartel 2018). Using computational and experimental approaches, it has been demonstrated that a single miRNA can bind to and regulate on an average, more than 100 mRNAs (Di Leva et al. 2014). In contrast, multiple types of miRNAs can bind to the 3′-UTR of a single mRNA target and collectively control its expression (Kim 2005). Furthermore, single miRNAs can have multiple target sites in the 3′-UTRs, thereby increasing repression efficiency (Rottiers & Näär 2012). In addition, some miRNAs have also been shown to affect multiple targets in linear pathways or interconnected in regulatory networks, thereby causing a large additive effect (Rottiers & Näär 2012). miRNAs are predicted to regulate more than 60% of human protein-coding genes (Friedman et al. 2009).
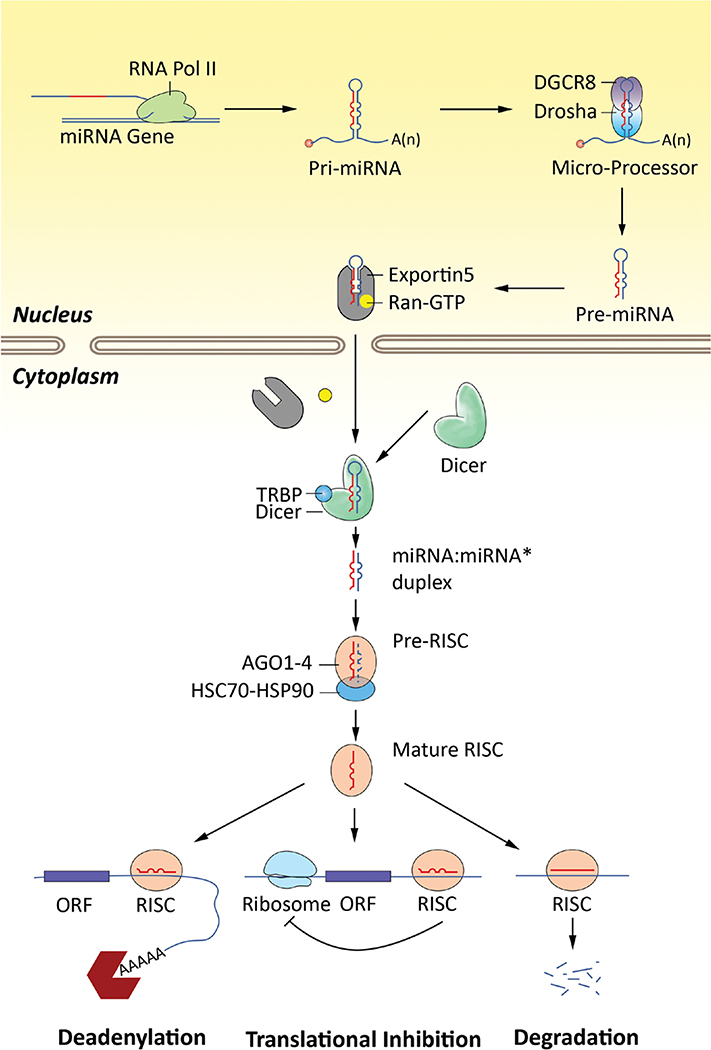
The biogenesis of miRNAs is a complex process involving multiple and distinct pathways (Ha & Kim 2014, Bartel 2018, Gebert & MacRae 2019). miRNA sequences are usually located in intergenic or intronic or polycistronic regions of the genome. As illustrated in Fig. 1, transcription of such sequences by RNA polymerase II generates a single miRNA hairpin precursor or clusters of multiple precursors, called primary miRNA (Pri-miRNAs) and consist of a 5′-cap (7MGpppG) and polyadenylated (AAAAA) tail. Specific promoters facilitate the transcription of intergenic miRNA genes; intragenic miRNA is processed by their own promoters if antisense oriented or by their host-gene promoters if sense oriented (Lima et al. 2017). In some cases, miRNAs are transcribed as a single long transcript called clusters, which may have analogous seed regions, and in this context, they are classified as a family. The biogenesis of miRNA is classified into canonical and non-canonical pathways (O’Brien et al. 2018). The canonical pathway of miRNA biogenesis is the principal pathway by which miRNAs are processed. In this pathway, primary-miRNA (pri-miRNA, ~1–3 kb in length) are transcribed from their genes by RNA polymerase II in the nucleus, and they are typically spliced, capped and polyadenylated. Pri-miRNAs fold into hairpin structures and these pri-miRNAs are cleaved in the nucleus by the microprocessor complex containing a dsRNA-binding protein, DiGeorge Syndrome Critical Region 8 Protein (DGCR8) and an RNase III enzyme, Drosha, into ~70–100 nucleotide-long stem-loop structures, termed precursor-miRNAs (pre-miRNA). The pre-miRNAs are then transported from the nucleus to the cytoplasm by RAN-GTP complex and karyopherin exportin 5 (Exp5).
Figure 1.
The miRNA biogenesis pathway. The canonical miRNA biogenesis pathway is shown here. MiRNAs are transcribed as pri-miRNA from intergenic, intronic or polycistronic genomic loci by RNA polymerase II. The nascent pri-miRNA transcripts are first processed into ~70-nucleotide pre-miRNAs by the Drosha and DGCR8 RNase III complex in the nucleus. Pre-miRNAs are transported from nucleus to cytoplasm by Exportin 5 and processed into mature ~22-nucleotide miRNA:miRNA* complexes by Dicer and TRBP RNase III enzyme complex, which are then loaded on RISC. Specific miRNA-RISC complexes translational repression of mRNA targets. AGO1–4, Argonaute proteins 1–4; DGCR8, microprocessor complex subunit DGCR8; HSC70, heat shock cognate 71 kDa protein, also known as HDPA8; HSP90, heat shock protein 90; Ran, RAS-related small GTPase nuclear protein; RISC, RNA-induced silencing complex; TRBP, HIV-1 TAR RNA-binding protein.
Once in the cytoplasm, the pre-miRNAs undergo an additional processing step by the RNase III enzyme Dicer and transactivation-response RNA-binding protein (TRBP) RNase III enzyme complex to generate ~22 bp miRNA:miRNA* duplex (Ha & Kim 2014, Bartel 2018, Gebert & MacRae 2019). This duplex is then loaded into the mRNA-associated multiprotein miRNA-induced silencing complex (miRISC), which includes Argonaute (AGO) 1–4 family of proteins. The mature single-stranded miRNA, termed guide strand, is preferentially retained in the complex. The mature miRNA/RISC complex then binds to complementary sites in the mRNA target to negatively regulate gene expression. The remaining strand, known as the passenger strand, is degraded as a RISC complex substrate. Although relatively less common, multiple non-canonical miRNA biogenesis pathways have been characterized (O’Brien et al. 2018). These pathways have been shown to utilize various combinations of the key proteins involved in the canonical pathway, including Drosha, Dicer, Exp5, and AGO (O’Brien et al. 2018).
Recent advances have led to clear understanding that miRNAs contribute to the regulation of a variety of physiologic processes including embryonic development, differentiation and proliferation, apoptosis, metabolism, hemostasis and inflammation (Bushati & Cohen 2007, O’Connell et al. 2012, Rottiers & Näär 2012, Dong et al. 2013, Alberti & Cochella 2017, Bartel 2018, Gebert & MacRae 2019). There is also ample evidence available that miRNAs play a key role in the pathogenesis of numerous diseases including cancer, hepatitis, cardiovascular diseases and metabolic diseases (Esteller 2011, Sayed & Abdellatif 2011, Rottiers & Näär 2012, Di Leva et al. 2014, Price et al. 2014, Adams et al. 2017, Lin & He 2017, Rupaimoole & Slack 2017). Moreover, because of their ability to contribute to the pathogenesis of various diseases, miRNAs have emerged as potential therapeutic targets to combat these diseases (Rupaimoole & Slack 2017).
Steroid hormones are involved in virtually every cellular process and are essential to maintain metabolic homeostasis (Table 1). In steroidogenic cells of the adrenal gland (zona glomerulosa cells, zona fasciculata cells and zona reticularis cells), ovary (granulosa cells, theca cells and luteal cells) and testis (Leydig cells), the overall rate of steroid hormone production is controlled by trophic hormones (ACTH, LH, FSH, or angiotensin II (Ang II)/potassium (K+)) at two levels: (a) at the level of cellular cholesterol (substrate) mobilization and its transport to and within the mitochondria for side chain cleavage by CYP11A1 (P450scc) occurring within minutes, reflecting the acute regulation of steroidogenesis (Miller & Bose 2011, Hattangady et al. 2012, Shen et al. 2016a, Selvaraj et al. 2018) and (b) at the level of gene transcription occurring between hours and days, reflecting chronic regulation of steroid hormone biosynthesis (Simpson & Waterman 1988, Simpson et al. 1992, Payne & Youngblood 1995, Romero et al. 2010, Hattangady et al. 2012, Shen et al. 2016a). In addition, in recent years, it has been increasingly clear that steroidogenic pathways are also subject to post-transcriptional and post-translational regulations including processes such as phosphorylation/dephosphorylation and protein‒protein interactions, as well as the involvement of specific miRNAs (Shen et al. 2016b). This review presents a detailed description of miRNA regulation of steroid hormone biosynthesis, with emphasis on adrenal and gonadal steroidogenesis.
Table 1.
Metabolic and physiological functions of key adrenal and gonadal steroids.
| Steroidogenic tissues and cells | Trophic hormone(s) | Steroid(s) | Metabolic and physiological functions |
|---|---|---|---|
| Adrenal Gland | |||
| Zona glomerulosa cells Zona fasciculata cells Zona reticularis cells |
Ang II, K+ and ACTH ACTH ACTH |
Aldosterone Cortisola DHEA, DHEHS, A4, Androstenediol, 11OHA |
Aldosterone is the principal mineralocorticoid synthesized by zona glomerulosa of the adrenal cortex. It helps maintain blood pressure and water and salt balance in the body by helping the kidneys retain sodium and excrete potassium or hydrogen. In humans, excessive secretion of this steroid (aldosteronism) results in hypertension, and induces end-organ damage, including in the kidneys and heart. Cortisol is one of the major glucocorticoids synthesized mostly in the zona fasciculata (but some is also synthesized in zona reticularis) of the adrenal cortex. It helps to control the metabolism of fats, proteins and carbohydrates; gluconeogenesis; suppresses inflammation; immune function; regulates blood pressure; increases blood sugar; controls gluconeogenesis and lipolysis of adipose tissue; and it affects water and electrolyte balance, blood pressure, body temperature and mineralization of bones. Decrease of cortisol induces Addison’s disease, while overproduction of cortisol is related to Cushing’s syndrome. A group of autosomal recessive genetic defects in cortisol biosynthesis result in congenital adrenal hyperplasia (CAH). The zona reticularis layer of human adrenal produces androgens, such as DHEA, DHEA sulfate (DHEAS), androstenedione (A4), androstenediol and 11β-hydroxyandrostenesione (11OHA), although DHEA and DHEAS are secreted in greater quantities than other adrenal androgens. These steroids have little androgenic activity by themselves, but they are precursor hormones for peripheral and gonadal conversion into female sex steroids (estrogens) and male sex hormones (androgens). |
| Ovary | |||
| Granulosa cells Luteinized granulosa cells/luteal cells Theca-interstitial cells |
FSH LH LH |
Estrogens (β-estradiol, estrone and estriol) Progesterone Androgens (Androstenedione, Testosterone) |
Estrogens are a group of steroid hormones and function as the primary female sex hormones. Three naturally occurring estrogens are β-estradiol (the most potent naturally occurring estrogen), estrone and estriol. Estrogens are produced primarily by granulosa cells of developing follicles in the ovaries, but also by the corpus luteum (CL), and placenta. Estrogens regulate development and maintenance of female sex characteristics. Estrogens regulate many physiologic functions including menstrual cycle, and reproduction, bone integrity, cognition, behavior and cholesterol mobilization. Estrogens are also implicated in the development and progression of numerous diseases such as various types of cancers (breast, ovarian, colorectal, prostate and endometrial), osteoporosis, neurodegenerative diseases, cardiovascular disease, insulin resistance, lupus erythematosus, endometriosis and obesity. Progesterone (P4) is an intermediate of sex steroids produced by all steroidogenic tissues/cells. It is secreted in large quantities by luteinized granulosa cells and luteal cells and by placenta during pregnancy. The most important function of progesterone is to support implantation of the ovum and preserve pregnancy through its action on the uterine endothelium. It plays an important role in postovulatory regulation of the menstrual cycle, inhibits the release of gonadotropins, and regulates many reproductive functions including sexual behavior in both females and males. In addition, progesterone contributes to the regulation of diverse neural cell functions throughout the nervous system. Theca-interstitial cells of the follicle as well as theca-luteal cells of the corpus luteum are the major site of ovarian androgen production. The androgen substrates are required for ovarian estrogen biosynthesis by granulosa cells. In humans, normal (physiological) production of androgens maintains follicular growth via promotion of early stage folliculogenesis and prevention of follicular atresia. In contrast, excessive androgen production results in abnormal follicular growth and reproductive dysfunction. In women, hyperandrogenemia associated with clinical conditions such as congenital adrenal hyperplasia (CAH), Cushing’s syndrome, and polycystic ovary syndrome (PCOS) also leads to infertility. |
| Testis | |||
| Leydig Cells | LH | Testosterone | Testosterone is synthesized in men by the testicular Leydig cells and in women either directly by the adrenals and ovaries, or by peripheral conversion of androstenedione. Testosterone is then converted to more potent dihydrotestosterone (DHT) by 5α reductase, or by aromatase (CYP19A1) to more potent estrogen (the estradiol). Testosterone is the hormone responsible for male sexual characteristics. In men, testosterone has been implicated in the regulation of sperm production, bone mass, fat distribution, and red blood cell production. Testosterone plays a critical role in carbohydrate, fat and protein metabolism. It exerts major influence on body fat composition and muscle mass in males and normal physiologic levels of testosterone exert favorable actions on male cardiovascular system. Testosterone deficiency is associated with an increased fat mass, reduced insulin sensitivity, impaired glucose tolerance and dyslipidemia. |
Cortisol is the major representative glucocorticoid in most mammals including humans; in rodents and other lower animals, corticosterone is the principal glucocorticoid.
Steroidogenesis: an overview
Although this review focuses on miRNA regulation of adrenal and gonadal steroid hormone biosynthesis, to familiarize the readers, a brief overview regarding critical events involved in steroidogenesis is warranted. The steroid hormones are primarily synthesized by endocrine glands, adrenal cortex, and gonads (ovary and testis) and by placenta during pregnancy. There are five major classes of steroid hormones: (i) cortisol is the major glucocorticoid in humans and most mammals (a similar analog of this hormone, designated as corticosterone, is synthesized by rodents, birds and reptiles); (ii) aldosterone is the major mineralocorticoid in humans; (iii) androgens, such as testosterone and dihydrotestosterone; (iv) estrogens, mainly estradiol and estrone; and (v) progestins, such as progesterone (Miller & Auchus 2011). The outermost zona glomerulosa layer of the adrenal cortex produces aldosterone; there are two major regulators of aldosterone biosynthesis: angiotensin II (AngII) and high extracellular potassium levels and a secondary regulator, adrenocorticotrophic hormone (ACTH) (Williams 2005, Hattangady et al. 2012, Bollag 2014). The zona fasciculata, the middle and largest layer of the adrenal cortex is the major site of cortisol (glucocorticoid) production (Turcu et al. 2014, Stewart & Newell-Price 2016). The innermost zone of the adrenal cortex, zona reticularis, predominantly secretes DHEA and DHEAS and to a lesser extent androstenedione (Turcu et al. 2014, Stewart & Newell-Price 2016). These zones are primarily regulated by ACTH. Follicle-stimulating hormone (FSH) controls progesterone and estrogen synthesis in ovarian granulosa cells, whereas luteinizing hormone (LH) regulates progesterone synthesis in luteinized granulosa cells and luteal cells, androgen production in ovarian theca-interstitial cells (Hoffman et al. 2012), and testosterone synthesis in testicular Leydig cells (Matsumoto & Bremmer 2016).
Cholesterol is the principal precursor for the biosynthesis of all steroid hormones in the adrenal gland, ovaries and testis (Azhar & Reaven 2002, Miller & Bose 2011). In these glands, there are multiple sources of cholesterol, which could potentially supply cholesterol substrate needed for steroidogenesis; the cholesterol can be synthesized de novo from acetate, directly obtained from circulating lipoproteins (low-density lipoprotein (LDL) and high-density lipoprotein (HDL)) or acquired via the hydrolysis of stored cytoplasmic cholesteryl ester lipid droplets (Azhar & Reaven 2002). In addition, cholesterol for steroidogenesis can also be derived from the plasma membrane (Deng et al. 2019). The current evidence suggests that plasma lipoproteins are the major source of cholesterol for steroidogenesis in adrenal gland, ovary and under certain conditions in testicular Leydig cells (Azhar & Reaven 2002, Miller & Bose 2011, Shen et al. 2018). Human steroidogenic cells obtain cholesterol from circulating LDL via the LDL-receptor/endocytic pathway (Miller & Bose 2011), whereas HDL particles supply cholesterol to mostly rodent steroidogenic cells via the SR-B1/selective pathway (Shen et al. 2018). The first common step in the production of all steroid hormones is the enzymatic conversion of cholesterol (a carbon27 (C27) steroid) to pregnenolone (the primary C21 product) catalyzed by cholesterol side-chain cleavage (P450scc) enzyme, encoded by the CYP11A1 gene and which is under the control of trophic hormones (ACTH, LH, FSH or AngII/high K+ depending on the tissue) using cAMP or calcium as second messenger. The P450scc is localized in the inner mitochondrial membrane (IMM) along with its cofactor protein ferredoxin reductase. The key steps in this reaction are the mobilization and transport of lipid droplet-associated cholesterol ester (cholesterol is esterified by acyl-coenzyme A:cholesteryl acyltransferase (ACAT), stored in lipid droplets, and released by trophic hormone stimulation of hormone-sensitive lipase (HSL)) to the outer mitochondrial membrane (OMM) mediated by HSL, cytoskeleton, SNAREs and possibly other factors (Shen et al. 2003, 2012, 2016b, Kraemer et al. 2004, 2017, Lin et al. 2016), and subsequent cholesterol transport from the OMM to the IMM and eventual loading of the precursor into the active site of P450scc. The movement of cholesterol from OMM to IMM is considered the rate-limiting step in all steroid hormone production and is primarily mediated by the StAR protein (encoded by the STARD1 gene) (Miller 2017, Selvaraj et al. 2018). Trophic hormones rapidly induce the expression of StAR protein, which then facilitates inter mitochondrial membrane cholesterol transfer and initiate steroidogenesis. Although a role for translocator protein (TSPO)/peripheral benzodiazepine receptor (PBR) has also been suggested in the movement of cholesterol from OMM to IMM (Papadopoulos et al. 2018), however, based on the current evidence, there is no consensus whether or not TSPO/PBR is critically involved in intermembrane cholesterol transfer (Costa et al. 2018, Selvaraj et al. 2018). Once P450scc-catalyzed pregnenolone is formed, it can then be transformed to progesterone, androgens, estrogens, and corticosteroids in a tissue-specific manner.
Biosynthesis of adrenal aldosterone
As noted above, the zona glomerulosa (ZG) layer of the human adrenal cortex is the site of synthesis of mineralocorticoid, aldosterone, and requires sequential action of three enzymes (Turcu & Auchus 2015): (1) 3β-hydroxysteroid dehydrogenase type 2 (HSD3B2), which catalyzes the irreversible conversion of the hydroxyl group to a keto group on carbon 3 and concurrent isomerization of the double bond from the position between C5/6 (’Δ5’) to C4/5 (’Δ4’) position; (2) the microsomal, type 2 cytochrome P450 enzyme 21-hydroxylase (P450c21, CYP21A2), which converts progesterone via hydroxylation on carbon 21 to the aldosterone precursor, 11-deoxycorticosterone (DOC); and (3) finally aldosterone biosynthesis is completed in the mitochondria, where DOC undergoes 11β- and 18-hydroxylation followed by 18-oxidation to form the final product, aldosterone, which in humans is catalyzed by a single enzyme, aldosterone synthase (P450c11AS, CYP11B2) (Fig. 2). AngII and high extracellular potassium ions are the potent primary stimulators of aldosterone, and ACTH serves as a secondary regulator.
Figure 2.
The major steroid biosynthetic pathways of the human adrenal gland. 3β HSD2, 3β-hydroxysteroid dehydrogenase/Δ5−4 isomerase, type, also called hydroxy-delta-5-steroid dehydrogenase, 3β- and steroid delta isomerase 2 (HSD3B2 gene); DHEA, dehydroepiandrosterone; DHEA-S, dehydroepiandrosterone-sulfate; P450c11, cytochrome P450c11 (steroid11 β-hydroxylase), also called Cytochrome P450 Family 11 Subfamily B Member 1 (CYP11B1 gene); P450c11AS, cytochrome P45011AS (aldosterone synthase), also called Cytochrome P450 Family 11 Subfamily B member 2 (CYP11B2 gene); P450c17, cytochrome P450c17 is the single enzyme mediating both 17α-hydroxylase/17,20 lyase activities, also called Cytochrome P450 Family 17 Subfamily A Member 1 (CYP17A1 gene); P450c21, cytochrome P450c21 (steroid 21-hydroxylase), also called Cytochrome P450 Family 21 Subfamily A Member 2 (CYP21A2 gene); P450scc, cytochrome P450scc side chain cleavage enzyme, also called Cytochrome P450 Family 11 Subfamily A Member 1 (CYP11A1 gene); StAR, steroidogenic acute regulatory protein (STAR gene); ST2A1, bile salt sulfotransferase 2A1/dehydroepiandrosterone sulfotransferase also called Sulfotransferase Family 2A Member 1 (SULT2A1 gene).
Biosynthesis of adrenal cortisol
The glucocorticoid cortisol is mainly synthesized in the zona fasciculata layer under the regulation of ACTH/cAMP/PKA signaling cascade. 17α-hydroxylase (P450c17, CYP17A1) introduces a hydroxyl group on carbon position 17 of pregnenolone or progesterone with roughly equal efficiency to form 17α-hydroxypregnenolone (17OHPreg) and 17α-hydroxyprogesterone (17OHP), respectively. With the sequential actions of 3βHSD2 (HSD3B2) and 21-hydroxylase, P450c21 (CYP21A2), 17-hydroxysteroids are converted to 11-deoxycortisol. In the final step, 11β-hydroxylase, P450c11β (CYP11B1) catalyzes the conversion of 11-deoxycortisol to cortisol (Turcu & Auchus 2015) (Fig. 2). In rodents the zona fasciculata lacks P450c17 (CYP17A1) and consequently nascent progesterone is 21-hydroxylated and 11β-hydroxylated to produce corticosterone, which is a dominant glucocorticoid in these species, but in humans it accounts for a minor product.
Biosynthesis of adrenal androgen
The adrenal androgens are mostly synthesized in the zona reticularis. 17α-Hydroxylase (P450c17, CYP17A1)/17,20 lyase is a dual function enzyme, which acts both as a hydroxylase hydroxylating pregnenolone, and as a lyase catalyzing the conversion of 17-hydroxypregnenolone to DHEA. Even though the affinity of the human 17α-hydroxylase (P450c17, CYP17A1) is similar either for Δ5 steroid substrate (pregnenolone) or Δ4 steroid substrate (progesterone), the predominant pathway for the C17,20 lyase is via the 17-hydroxypregnenolone (Δ5 substrate) (Fig. 2). DHEA is then converted to DHEA sulfate (DHEAS) by an adrenal sulfokinase (encoded by the SULT2A1 gene), which is the most abundant adrenal steroid (Turcu et al. 2014, Turcu & Auchus 2015). DHEA is also converted to androstenedione by the enzyme 3β-hydroxysteroid dehydrogenase (3βHSD). The adrenal also synthesizes small amounts of testosterone by action of 17β-hydroxysteroid dehydrogenase type 5 (17βHSD5, AKR1C3) on androstenedione.
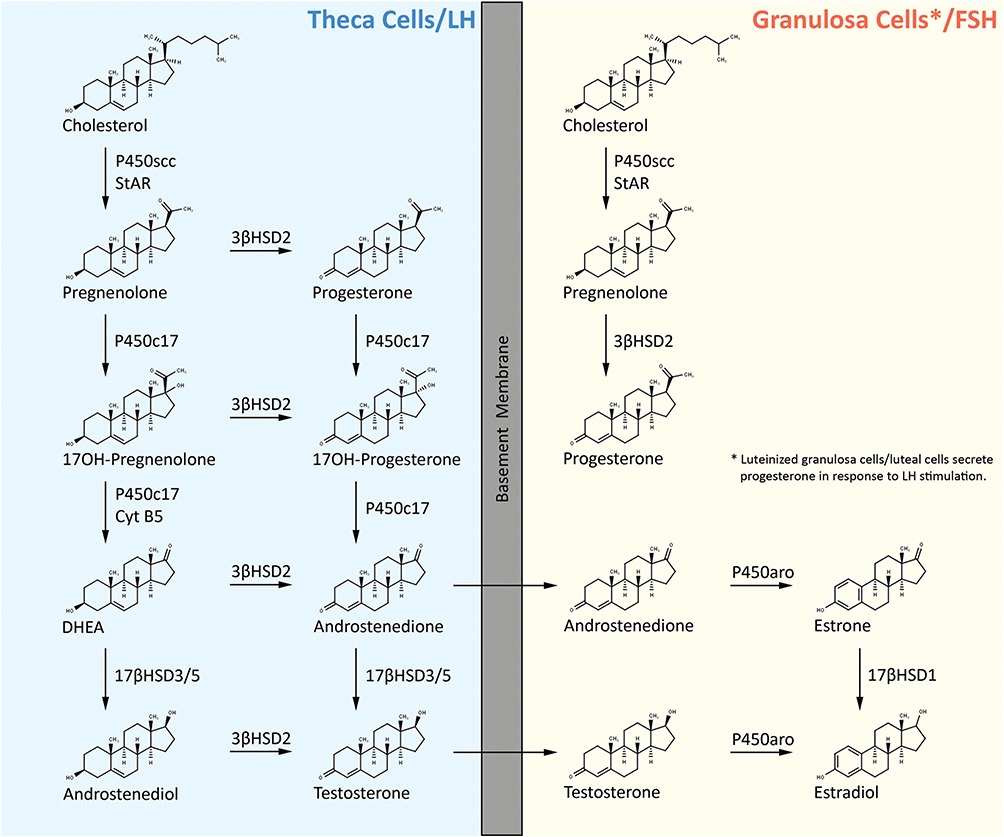
Biosynthesis of ovarian estrogen and progestin
Both the antral follicle and the corpus luteum synthesize ovarian sex steroids, estrogen and progestin. In most mammals (including mice and humans), antral follicle steroidogenesis has been described by the two-cell gonadotropin theory (Hoffman et al. 2012, Andersen & Ezcurra 2014). According to this theory, theca cells of the antral follicle respond to LH receptor signaling by inducing the transcription of genes encoding the enzymes necessary for the conversion of cholesterol-derived pregnenolone to androgens such as androstenedione and testosterone. Granulosa cells of the antral follicle respond to FSH signaling by upregulating the transcription of genes encoding the enzymes necessary for conversion of theca cell-derived androgens into estrogens. Following initial cleavage of cholesterol, the resulting pregnenolone in theca cells is metabolized in one of the two pathways often referred to as the Δ5 and the Δ4 pathway (Hoffman et al. 2012, Andersen & Ezcurra 2014). The Δ5 pathway begins by 17α-hydroxylase complex (P450c17, CYP17A1) catalyzed generation of 17α-hydroxypregnenolone from pregnenolone. Subsequently, 17α-hydroxypregnenolone is converted to DHEA by the C17,20 lyase activity of 17α-hydroxylase (both 17α-hydroxylase and C17,20 lyase activities reside on a single protein molecule and is located exclusively in theca cells). DHEA is next converted to androstenedione by 3β-hydroxysteroid dehydrogenase/Δ5−4 isomerase (3βHSD2). Androstenedione can either be converted to testosterone by 17β-hydroxysteroid dehydrogenase 3/17β-hydroxysteroid dehydrogenase 5 (17βHSD3/5) within the theca cells or diffuse into the granulosa cells for its conversion into estrogen by aromatase (P450arom, CYP19A1). DHEA can also be converted to androstenediol by 17βHSD3/5 and subsequently to testosterone by 3βHSD2 (Fig. 3). The Δ4 pathway begins with the conversion of pregnenolone to progesterone through a bifunctional enzyme, namely 3β-hydroxysteroid dehydrogenase (HSD3B)/Δ5,4-isomerase. Once progesterone is generated, the subsequent reactions in the Δ4 pathway proceed in identical fashion as that of Δ5 pathway employing the same enzymes and genes, that is, progesterone is first converted to 17α-hydroxyprogesterone, which is further metabolized to androstenedione. Androstenedione is then transformed to the final product of the Δ4 pathway, which is testosterone (Fig. 3). However, in humans, further metabolism of 17α-hydroxyprogesterone does not occur and, thus, the only route for estrogen synthesis in human ovary is via the Δ5 pathway. Also, it should be noted that both theca and granulosa cells express StAR, P450scc (CYP11A1) and 3βHSD2. Both cell types are therefore capable of producing pregnenolone/progesterone from cholesterol substrate. However, in the follicular phase of the menstrual cycle, the relatively avascular granulosa cells are exposed to little oxygen or cholesterol, and, thus, produce only low amounts of these steroids. In contrast, after exposure to gonadotropins during the luteal phase, the granulosa cells become ‘luteinized’ and synthesize robust amounts of pregnenolone and progesterone from cholesterol (Hoffman et al. 2012).
Figure 3.
Human ovarian progestin and estrogen biosynthetic pathways. 17β-HSD1, 17β-hydroxysteroid dehydrogenase type 1, also called Hydroxysteroid 17-Beta Dehydrogenase 1 (HSD17B1); 17β-HSD3/5, 17β-hydroxysteroid dehydrogenase type 3/17β-hydroxysteroid dehydrogenase type 5, also called Hydroxysteroid 17-Beta Dehydrogenase 3/Hydroxysteroid 17-Beta Dehydrogenase 5 (HSD17B3/HSD17B5); CYT B5, cytochrome B5 protein, also called Cytochrome B5 Type B (CYB5B gene); P450arom, cytochrome P450 aromatase, also called Cytochrome P450 Family 19 Subfamily A Member 1 (CYP19A1 gene); P450c17, cytochrome P450c17 (is a single enzyme mediating both 17α-hydroxylase/17,20 lyase activities), also called Cytochrome P450 Family 17 Subfamily A Member 1 (CYP17A1 gene); P450scc, cytochrome P450scc side chain cleavage enzyme, also called cytochrome P450 family 11 Subfamily A Member 1 (CYP11A1 gene); StAR, steroidogenic acute regulatory protein (STAR gene).
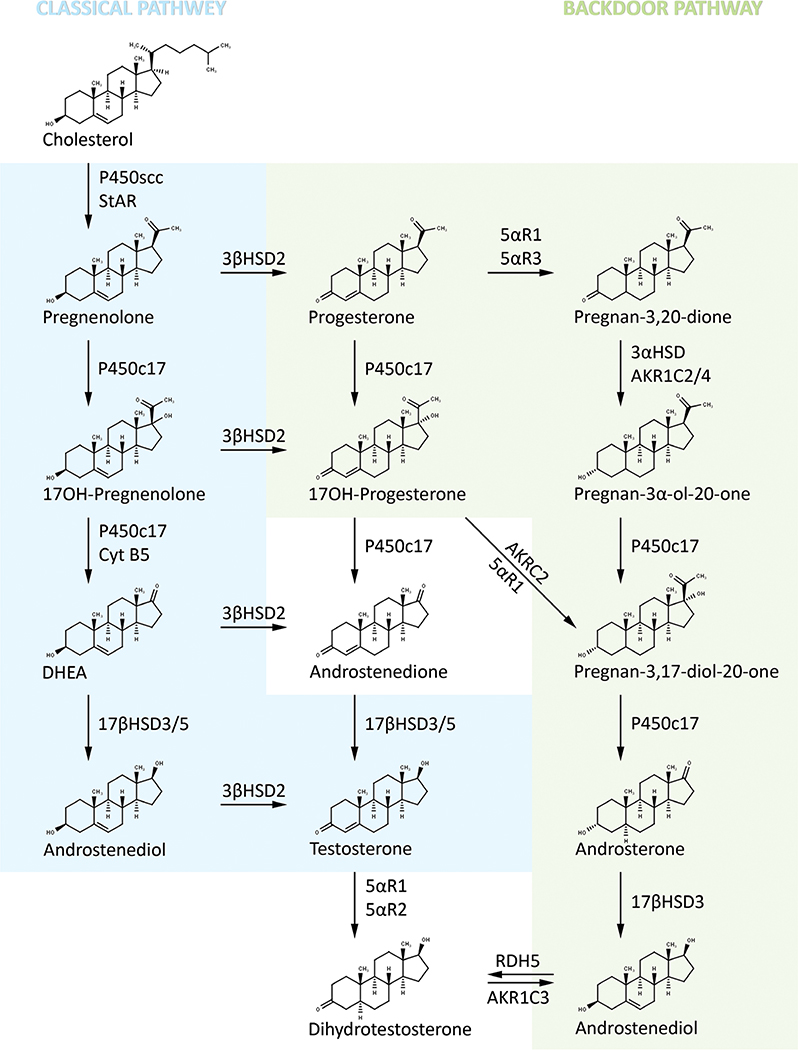
Biosynthesis of androgen via classic and backdoor pathways
Testicular Leydig cells are the principal source of testosterone; in fact, over 95% of the testosterone is secreted by the Leydig cells. In the ‘classic’ pathway, LH binds to its receptor on the Leydig cell surface, which is also coupled to the cAMP/PKA signaling cascade and initiates testosterone production (Matsumoto & Bremmer 2016). In this classic pathway, pregnenolone is converted through the Δ5 pathway by P450c17 to 17α-hydroxypregnenolone and then to DHEA with the first reaction catalyzed by its 17α-hydroxylase activity using P450 oxidoreductase (POR) as a coenzyme and the second reaction by the 17,20 lyase activity in the presence of POR and cytochrome b5 (CYB5). DHEA is then converted to testosterone through androstenedione or androstenediol catalyzed by 3βHSD2 and 17βHSD3/5, respectively (Fig. 4). In the Δ4 pathway, pregnenolone is converted sequentially to 17α-hydroxyprogesterone, androstenedione, and testosterone. It should be noted that biosynthesis of testosterone in human testis proceeds predominantly through the Δ5 pathway and very little testosterone is generated via the Δ4 pathway (Flück & Pandey 2014, Matsumoto & Bremmer 2016). The most potent endogenous androgen 5α-dihydrotestosterone (DHT) is formed from testosterone catalyzed by steroid 5α-reductase type 2 (SRD5A2). DHT formation is low in testis, but high levels are produced in genital skin and prostate.
Figure 4.
Biosynthesis of testicular androgen via classic front door and alternative backdoor pathways. AKR1C3, Aldo-Keto Reductase Family 1 Member C3 (AKR1C3 gene); 5α-R1, 5α-reductase type 1 or steroid 5α-reductase type 1, also called Steroid 5 alpha-Reductase 1 (SRD5A1 gene); 5α-R2, 5α-reductase type 2 or steroid 5α-reductase type 2, also called Steroid 5 alpha-Reductase 2 (SRD5A2 gene); 5α-R2, 5α-reductase type 3 or steroid 5α-reductase type 3, also called Steroid 5 alpha-Reductase 3 (SRD5A3 gene); 17β-HSD2, 17β-hydroxysteroid dehydrogenase type 2, also called Hydroxysteroid 17-Beta Dehydrogenase 2 (HSD17B2); 17β-HSD3, 17β-hydroxysteroid dehydrogenase type 3, also called Hydroxysteroid 17-Beta Dehydrogenase 3 (HSD17B3); 17β-HSD3/5, 17β-hydroxysteroid dehydrogenase type 3/17β-hydroxysteroid dehydrogenase type 5, also called Hydroxysteroid 17-Beta Dehydrogenase 3/Hydroxysteroid 17-Beta Dehydrogenase 5 (HSD17B3/HSD17B5); CYT B5, cytochrome B5 protein, also called Cytochrome B5 Type B (CYB5B gene); P450c17, cytochrome P450c17 (is a single enzyme mediating both 17α-hydroxylase/17,20 lyase activities), also called Cytochrome P450 Family 17 Subfamily A Member 1 (CYP17A1 gene); P450scc, cytochrome P450scc side chain cleavage enzyme, also called Cytochrome P450 Family 11 Subfamily A Member 1 (CYP11A1 gene); RDH5, retinol dehydrogenase 5 (11-cis and 9-cis) also called Retinol Dehydrogenase 5 (RDH5 gene); StAR, steroidogenic acute regulatory protein (STAR gene).
There is an alternative pathway or ‘backdoor’ pathway that was first described for androgen production in the tammar wallaby pouch young testis and subsequently in the immature mouse testis (Auchus 2004, Miller & Auchus 2019). In this pathway, androgen synthesis (DHT) occurs through conversion of 17α-hydroxyprogesterone to DHT without going through androstenedione or testosterone as intermediates (Fig. 4). Evidence is accumulating that this pathway plays an important role in hyperandrogenic disorders (Miller & Auchus 2019) such as 21-hydroxylase deficiency-linked congenital adrenal hyperplasia (Kamrath et al. 2012), polycystic ovary syndrome (PCOS) (Martin et al. 2017), some virilized female newborns with P450 oxidoreductase deficiency (Shackleton et al. 2004, Homma et al. 2006, Krone et al. 2012) and physiologic ‘minipuberty of infancy’ (Dhayat et al. 2017). Furthermore, AKR1C2 and AKR1C4 gene mutation studies (Flück & Pandey 2014) led to the conclusion that both ‘classic’ pathways of androgen synthesis and ‘backdoor’ are required for normal male genital development (Biason-Lauber et al. 2013). A very recent study demonstrated that the major human ‘backdoor’ androgen is androsterone and is primarily derived from placental progesterone (O’Shaughnessy et al. 2019).
miRNA regulation of adrenal steroidogenesis
To date most of the miRNA studies in adrenals have been directed toward understanding the role of miRNAs in aldosterone signaling (Butterworth & Alvarez de la Rosa 2019) and glucocorticoid action (Clayton et al. 2018) and in the pathogenesis of adrenal tumors (Igaz et al. 2015, Robertson et al. 2017). Only a limited number of studies have examined regulation of adrenal steroidogenesis by miRNAs. Previously, Robertson et al. (2013) demonstrated that knockdown of Dicer 1 disrupted CYP11B1 (11β-hydroxylase) and CYP11B2 (aldosterone synthase) mRNA levels in adrenocortical cells in vitro, pointing to a potential involvement of miRNAs in the regulation of adrenal steroidogenesis. The authors identified one such miRNA, miRNA-24, which was shown to modulate CYP11B1 and CYP11B2 expression along with alterations in aldosterone and corticosterone secretory rates. In a related study, it was shown that knockdown of miRNAs using Dicer 1 siRNA in H295R adrenocortical cells increased mRNA levels of CYP11A1, CYP21A1, and CYP17A1 and the secretion of cortisol, corticosterone, 11-deoxycorticosterone, 18-hydroxycorticosterone and aldosterone (Robertson et al. 2017). Loss- and gain-of-function studies demonstrated a direct effect of miRNA-125a-5p and miRNA-125b-5p on CYP11B2 and of miRNA-320a-3p levels on CYP11A1 and CYP17A1 mRNA levels. The miRNA let-7b was found to express in a negative correlated manner with cortisol levels of primary pigmented nodular adrenocortical disease (PPNAD) patients (Iliopoulos et al. 2009). miRNA-21 stimulated aldosterone secretion and promoted H295R cell proliferation (Romero et al. 2008). Furthermore, we showed that miRNA-132 inhibits steroidogenesis in Y1 adrenocortical cells by a dual mechanism: under basal conditions, miRNA-132 attenuates steroidogenesis by reducing StAR protein levels (Hu et al. 2017). In response to hormonal stimulation, miRNA-132 induces the expression of 3β-HSD and 20α-HSD through inhibition of transcription factor MeCP2 (Hu et al. 2017). This, in turn, causes sequential conversion of the CYP11A1 reaction product pregnenolone to progesterone and then into the biologically inactive 20α-hydroxyprogesterone. In another study aimed at understanding the molecular mechanism underlying hypoxia-induced dysregulation of steroidogenesis, H295R cells were used to identify miRNA(s) targeting steroidogenic gene expression under hypoxic conditions. All major steroidogenic genes, including HMGR, StAR, CYP17A1, 3β-HSD2, 17β-HSD1, and CYP19A1, except for CYP11A1, showed 3–4-fold reduction in their expression in the hypoxic cells (Yu et al. 2015). Use of gain- and loss-of-function strategies demonstrated that hypoxia-induced increased expression of miRNA-98 was associated with the downregulation of CYP19A1 mRNA and protein levels. The studies by Nusrin et al. (2014) further demonstrated that mRNA expression of CYP21A2 and CYP11B2 was significantly upregulated in hypoxic H295R cells, although CYP11B2 protein levels were downregulated under identical experimental conditions. No significant effect of hypoxia was observed on the expression of CYP11B1. Expression of some key transcription factors involved in the regulation of CYP21A2, CYP11B1 and CYP11B2, such as NURR-77, CITED2, and NOR-1, was upregulated, whereas expression of SF-1 and DAX-1 was significantly downregulated in H295 cells in response to hypoxia. Finally, evidence was presented showing that CYP11B1 and CYP11B2 mRNAs are direct targets of hypoxia-inducible miRNA-10b (Nusrin et al. 2014).
Several adrenal miRNAs are subject to cell-specific regulation by hormones. miRNA-21 was found to be upregulated in an angiotensin II-stimulated screen of greater than 200 miRNAs in human adrenocortical H295R cells (Romero et al. 2008). Riester et al. (2012) examined the miRNA expression profiles of adrenal glands from control (0 time) and mice treated with ACTH for 10, 30 and 60 min. Sixteen miRNAs showed significantly increased expression and one miRNA (miRNA-433) showed decreased expression when data were compared between 10 min vs 0 min (Supplementary Table 2, see section on supplementary materials given at the end of this article). Interestingly, a comparison between 30 min vs 10 min demonstrated that the expression of 16 miRNAs was downregulated, whereas expression of miRNA-433 was upregulated. Similar results were obtained when comparisons were made between 60 min vs 10 min. qRT-PCR measurements further confirmed the upregulation of miRNA-101a, miRNA-142–3p, and miRNA-96 with the 10-min group demonstrating the highest upregulation as compared to 30 or 60 min. In contrast, miRNA-433 showed significant upregulation only when comparison was made between baseline vs 10 min after ACTH treatment. Our previous studies demonstrated that adrenals from ACTH, 17α-ethinyl estradiol (17α-E2) and dexamethasone (DEX)-treated rats exhibited miRNA profiles distinct from control animals (Hu et al. 2013). (It should be noted that (a) chronic ACTH treatment leads to upregulation of steroidogenic machinery and causes enhanced expression of adrenal low-density lipoprotein receptor (LDL-R) and scavenger receptor class B, type 1 (SR-B1); (b) treatment of rats with 17α-E2, a potent hypocholesterolemic agent, also leads to induction of both adrenal LDL-R and SR-B1; and (c) DEX treatment inhibits the adrenal HPA axis, and pituitary ACTH secretion, and as a result, downregulates the adrenal LDL-R, SR-B1 and plasma corticosterone levels.) ACTH treatment upregulated the expression of five miRNAs and downregulated the levels of three miRNAs (Supplementary Table 2). Likewise, the levels of seven miRNAs were upregulated, whereas levels of eight miRNAs were downregulated in response to 17α-E2 treatment (Supplementary Table 2). On the other hand, DEX treatment decreased the levels of five miRNAs, but increased the level of one miRNA (Supplementary Table 2). Several adrenal mRNAs are subject to regulation by more than one hormone. In another study, we demonstrated that miRNA-132 expression is increased in intact adrenals and Y1 adrenocortical cells by ACTH and its second messenger cAMP, respectively (Hu et al. 2017). We also demonstrated that in vivo treatment of rat adrenals with ACTH decreased the expression of miRNA-125a, miRNA-125b, and miRNA-455 and reciprocally increased SR-B1 expression (Hu et al. 2012).
miRNA regulation of ovarian steroidogenesis
A large number of studies have been reported mainly describing the regulatory role of miRNAs in mammalian follicular, oocyte and luteal development (Donadeu et al. 2012, Tesfaye et al. 2018), ovarian function (Imbar & Eisenberg 2014, McGinnis et al. 2015, Maalouf et al. 2016), growth, proliferation, and apoptosis of ovarian granulosa and theca cells (Reza et al. 2019), female fertility and reproduction (Hasuwa et al. 2013, Reza et al. 2019), pituitary gonadotropin (FSH and LH) secretion (Hasuwa et al. 2013, Wang et al. 2015, Lannes et al. 2016, Ahmed et al. 2017, Messina & Prevot 2017), estrogen action (Klinge 2012) and gynecological reproductive diseases (Santamaria & Taylor 2014, Sørensen et al. 2014, Panir et al. 2018). In addition, a number of publications have directly addressed the molecular actions of miRNAs on ovarian steroidogenesis, particularly granulosa cells, which are the main source for steroid hormone synthesis (e.g., estradiol and progesterone) and secretion (for summary see Supplementary Table 1). Genome-wide screening of miRNAs revealed the potential involvement of miRNAs in the regulation of sex steroid hormone (i.e., estradiol, progesterone, and testosterone) production in human ovarian granulosa cells (Sirotkin et al. 2009). Transfection of cultured human granulosa cells with 80 miRNA constructs demonstrated that 51 miRNA constructs reduced estradiol production. Thirty-six caused significant inhibition in progesterone secretion, none enhanced estradiol secretion, whereas ten miRNA constructs significantly upregulated progesterone production (Supplementary Table 1) (Sirotkin et al. 2009). Under identical experimental conditions, 57 miRNAs inhibited testosterone production and only one miRNA enhanced testosterone secretion (Supplementary Table 1). Yao et al. (2010a), using expression profiling, reported that expression of 3 miRNAs were upregulated and 13 miRNAs were downregulated in TGF-β1-treated mouse preantral granulosa cells. Furthermore, it was shown that one of the upregulated miRNAs, miRNA-224, plays a regulatory role in TGF-β1-induced granulosa cell proliferation and estradiol release (at least in part via increased gene transcription of Cyp19a1) by targeting Smad4. A follow-up study by the same group demonstrated a transcriptional cooperation between p53 and NF-κB p65 in the regulation of miRNA 224 gene transcription in mouse granulosa cells (Liang et al. 2013). Xu et al. (2011), using loss-of-function and gain-of-function strategies, demonstrated that miRNA-378 negatively regulates estradiol production in porcine granulosa cells by targeting the estradiol synthesizing enzyme aromatase. miRNA-378 has also been shown to regulate oocyte maturation by downregulating estradiol production via the suppression of aromatase (Cyp19a1) activity in porcine cumulus granulosa cells (Pan et al. 2015).
A study by Yin et al. (2012) demonstrated that miRNA-383 stimulates estradiol production by targeting (inhibiting) RNA-binding motif, single-stranded-interacting protein 1 (RBMS1), at least in part through inactivation of its downstream target c-Myc. Furthermore, SF-1 serves as a positive regulator of miRNA-383 processing and function and mediates the positive effects of miRNA-383/RBMS1/c-Myc on granulosa cell estradiol secretion. Another study provided evidence that miRNA-133b downregulates forkhead L2 (Foxl2) transcription factor in mouse and human granulosa cells by directly targeting its 3′UTR, thus attenuating the Foxl2-mediated transcriptional repression of StAR and Cyp19A1 and subsequently increased release of estradiol (Dai et al. 2013). Studies by Yin et al. (2014) demonstrated that PMSG/eCG treatment of female mice (48 h) resulted in a time-dependent downregulation of miRNA-320 expression. In contrast, treatment of mice with hCG 24 h after PMSG/eCG treatment caused a rapid induction of miRNA expression. Administration of lentiviral miRNA-320 mimic into the ovarian bursa of mice suppressed cell proliferation and inhibited estradiol production. Likewise, overexpression of miRNA-320 in granulosa cells caused significant inhibition of estradiol synthesis and Cyp19a1 mRNA and protein expression and granulosa cell proliferation by directly targeting E2F1 and SF-1. In contrast, silencing of miRNA-320 was accompanied by increased biosynthesis of estradiol (Yin et al. 2014). It was also shown that overexpression of miRNA-132 in isolated mouse granulosa cells promoted increased secretion of estradiol. Further studies demonstrated that miRNA-132 promotes estradiol biosynthesis in granulosa cells via translational repression of Nurr1 and induction of Cyp19a1 gene expression. Using an overexpression strategy, Wang et al. (2016b) first showed that miRNA-764–3p downregulated the expression of Cyp19a1 by directly targeting SF-1, and as a result decreased synthesis of estradiol in mouse granulosa cells. In contrast, knockdown of miRNA-764–3p induced expression levels of SF-1 and Cyp19a1 along with the stimulation of estradiol synthesis. These alterations induced by knockdown of miRNA-764–3p, however, were prevented by simultaneous knockdown of SF-1.
Yu et al. (2016) investigated the relationship between miRNA-375 and corticotropin-releasing hormone (CRH) signaling cascade in the porcine ovary. Using PCR and in situ hybridization techniques, the authors showed that miRNA-375 was exclusively localized in granulosa cells, whereas CRH receptor 1 (CRHR1) expression was detected in both granulosa cells and oocytes. Overexpression of miRNA-375 in cultured granulosa cells attenuated estradiol production, whereas knockdown of miRNA-375 enhanced estradiol synthesis. These studies led the authors to conclude that miRNA-375 employs the CRH signaling pathway to inhibit estradiol synthesis in porcine ovary. Studies by Zhang et al. (2017b) provided evidence showing that miRNA-143 is highly expressed in granulosa cells of primary, secondary and antral follicles and functions as a negative regulator of FSH-induced estradiol production by targeting KRAS. In addition, miRNA-143 is also implicated in FSH-mediated granulosa cell proliferation and that FSH negatively regulates miRNA-143. It has recently been demonstrated that miRNA-1275 promotes apoptosis, initiates follicular atresia and inhibits CYP19A1 expression and estradiol production in cultured porcine granulosa cells. Using bioinformatics and reporter assay techniques, the authors further demonstrated that liver receptor homolog-1 (LRH-1) and not CYP19A1 is a direct target of miRNA-1275. Sang et al. (2013) examined the role of four miRNAs (miRNA-24, miRNA-132, miRNA-320, and miRNA-520c-3p) in the regulation of estradiol biosynthesis in a human granulosa cell line, KGN. Treatment of KGN cells with a miRNA-24 mimic decreased estradiol synthesis, whereas its inhibitor increased estradiol production. On the other hand, use of miRNA-132, miRNA-320, miRNA-520c-3p, and miRNA-222 mimics promoted estradiol production and their inhibitors caused significant reduction in estradiol synthesis (Supplementary Table 1). Transfection of primary human ovarian granulosa cells with anti-miRNA-15a resulted in a significant increase in estradiol secretion (Sirotkin et al. 2014). Transfection of cells with miRNA-15a mimic did not affect estradiol production. Robinson et al. (2018) investigated the role of miRNA-221 in bovine ovarian granulosa cell estradiol production. They reported that transfection of granulosa cells with a miRNA-221 mimic reduced granulosa cell estradiol production in response to FSH or IGF, although the use of miRNA-221 inhibitor showed little or no effect on steroid production. Zhang et al. (2019a), using bovine granulosa cells, demonstrated that overexpression of miRNA-143 led to decreased secretion of estradiol, and silencing of miRNA-143 was associated with increased estradiol production. This inhibitory action of miRNA-143 on estradiol synthesis is mediated via downregulation of functional expression of its target gene, FSH receptor (FSHR).
Additional miRNAs have been identified (serum, follicular fluid or granulosa cells) that either directly regulate progesterone or testosterone production (Sang et al. 2013, Mohammed et al. 2017, Robinson et al. 2018), or whose expression is altered in polycystic ovary syndrome (Sang et al. 2013, Sørensen et al. 2016a,b and references therein, Zhang et al. 2017a) or premature ovarian insufficiency (Imbar & Eisenberg 2014, Virant-Klun et al. 2016, Guo et al. 2017, Tu et al. 2019) with relevance to steroidogenesis. Sang et al. (2013) provided evidence that specific miRNAs regulate estradiol and progesterone in a model human granulosa cell line, KGN. Transfection of KGN cells with mimics of miRNA-222, miRNA-320, miRNA-520c-3p, and miRNA-132 suppressed estradiol production, while miRNA-24 mimic promoted estradiol production. Use of respective inhibitors of these miRNAs indicated that miRNA-222, miRNA-320, miRNA-520c-3p and miRNA-132 inhibitors increased, and miRNA-24 inhibitor decreased estradiol production. Likewise, transfection of KGN cells with mimics of miRNA-24, miRNA-193b and miRNA-483–5p suppressed progesterone synthesis, while use of their inhibitors stimulated progesterone production (Sang et al. 2013). Sirotkin et al. (2014) demonstrated that transfection of primary human ovarian granulosa cells with anti-miRNA15a caused a significant reduction in progesterone and testosterone production, whereas transfection of cells with pre-miRNA15a stimulated the secretion of both progesterone and testosterone.
Another study demonstrated that administration of miRNA-320 into mouse ovary in vivo significantly upregulated the production of progesterone and testosterone (Yin et al. 2014). Likewise, transfection of mouse ovarian granulosa cells with miRNA-320 mimic resulted in increased secretion of progesterone. In contrast, transfection of granulosa cells with miRNA-320 inhibitor decreased progesterone production (Yin et al. 2014). A recent work from our laboratory provided evidence that miRNA-132 promotes the production of 20α-hydroxyprogesterone, a biologically inactive analog of progesterone in rat ovarian granulosa cells (Hu et al. 2017). In cultured luteinized granulosa cells, inhibition of miRNA-96 was accompanied by decreased progesterone production via modulation of FOXO1 transcription factor (Mohammed et al. 2017). The studies carried out by Robinson et al. (2018) suggest that overexpression of miRNA-221 leads to inhibition of FSH/IGF-stimulated progesterone production in bovine granulosa cells, but surprisingly use of miRNA-221 inhibitor had little or no effect. Xu et al. (2018) reported that miRNA-29b promotes progesterone production in bovine corpus luteal cells via the modulation of oxytocin receptor expression. Recent studies by Zhang et al. (2019a) provide evidence that miRNA-31 and miRNA-143 both serve as a negative regulator of progesterone synthesis in bovine granulosa cells by inhibiting the expression of their common target, FSHR (follicle-stimulating hormone receptor).
Several recent studies have identified granulosa cell miRNAs whose expression has been shown to be regulated by gonadotropin (luteinizing hormone/human chorionic gonadotropin (LH/hCG), follicle-stimulating hormone (FSH)), cAMP (second messenger of gonadotropins), growth factor (TGF), androgens, or transcription factors (e.g., SF-1, NF-κB and p53) and as a consequence may also be involved in the regulation of steroidogenesis (summarized in Supplementary Table 2). Microarray analysis of miRNA expression in mouse mural granulosa cells showed that 212 mature miRNA transcripts were detected and 13 miRNAs as differentially expressed in mural granulosa cells collected at 0 and after 4-h treatment of mice with hCG (Fiedler et al. 2008). Additionally, the authors reported that that expression of two miRNAs, miRNA-132 and miRNA-212, was greatly upregulated following hCG treatment. Quantitative RT-PCR measurements further confirmed upregulation of miRNA-132 and miRNA-212 in a time-dependent manner in response to hCG treatment of mice in vivo and 8-Br-cAMP treatment of isolated granulosa cells in vitro. In another study, Carletti et al. (2010) demonstrated that mural granulosa cells isolated from preovulatory follicles exhibited ~6.4-fold increased expression or miRNA-21 transcript 12 h after equine gonadotropin (eCG) treatment and returned to basal levels at 36 h. A study by Yao et al. (2010a) using expression profiling reported that expression of three miRNAs was upregulated and thirteen miRNAs downregulated in TGF-β1-treated mouse preantral granulosa cells. In another study, it was demonstrated that treatment of cultured primary rat granulosa cells with FSH for 12 h upregulated the expression of 17 miRNAs and downregulated the expression of 14 miRNAs (Yao et al. 2010b). It should be noted that although expression of miRNA-29a and miRNA-30d was significantly downregulated 12 h after FSH treatment, their expression, however, was significantly increased 48 h after hormone treatment. A previous study from our laboratory had shown that exposure of cultured rat granulosa cell to Bt2cAMP for 24 h inhibited the expression of 6 miRNAs, while enhancing the expression of 4 miRNAs (Hu et al. 2013). It was demonstrated that expression of ovarian miRNA-320 was significantly downregulated in response to treatment of immature females with eCG for 24, 48 or 72 h. In contrast, hCG injection (24 h) after eCG treatment (48 h) caused a rapid increase in miRNA-320 expression levels. In addition, qRT-PCR measurement data demonstrated that FSH treatment of mouse granulosa cells leads to time- and dose-dependent inhibition of miRNA-320 expression (Supplementary Table 2). Studies by Sen et al. (2014) reported that miRNA-125b is expressed at relatively high levels in mouse granulosa cells, KGN granulosa tumor cells and primary human granulosa cells. Androgen (dihydrotestosterone (DHT) or testosterone) treatment of cultured mouse granulosa cells or KGN cells led to increased expression miRNA-125b, but not its homolog, miRNA-125a. Interestingly, estrogen (estradiol) or a synthetic steroid, R5020 (a progesterone receptor agonist), had no significant effect on miRNA-125b expression. Expression of some miRNAs is also regulated by transcription factors (Yin et al. 2012, 2014, Liang et al. 2013). For example, SF-1 functions as a positive regulator of miRNA-383 processing and function in granulosa cells (Yin et al. 2012). Moreover, it has been demonstrated that miRNA-320 regulates granulosa cell proliferation and estradiol secretion by targeting E2F1 and SF-1 during follicular development (Yin et al. 2014). Finally, recent evidence suggests that transcription factors p53 and NF-κB work in concert to regulate miRNA-224 transcription in mouse ovarian granulosa cells (Liang et al. 2013). A study of miRNA expression in mouse pre-antral granulosa cells after TGF-β1 treatment demonstrated that expression of 16 miRNAs was significantly altered, of which three miRNAs were upregulated, whereas expression of 13 miRNAs was downregulated (Yao et al. 2010a).
Polycystic ovary syndrome (PCOS), also known as polycystic ovarian syndrome, is a condition characterized by imbalance of sex hormones, ovulatory dysfunction, and polycystic ovarian morphologic features (Azziz et al. 2016, McCartney & Marshall 2016, Rosenfield & Ehrmann 2016). Two earlier studies were the first to report altered miRNA expression in the follicular fluid of women with PCOS (Sang et al. 2013, Roth et al. 2014). In one study, Sang et al. (2013) provided evidence that miRNA-132 and miRNA-320 are expressed at significantly reduced levels in the follicular fluid (both in micro-vesicles and the supernatant of follicular fluid) of PCOS patients compared to control subjects. Using specific miRNA inhibitors and mimics and a human granulosa cell line (KGN), the authors also reported that miRNA-132, miRNA-320, miRNA-520–3p, miRNA-24 and miRNA-222 regulate estradiol concentrations, whereas miRNA-24, miRNA-193b, and miRNA-483–5p regulate progesterone levels. In a second study, Roth et al. (2014), using microarray analysis, identified 235 miRNAs in human follicular fluid, of which 29 were differentially expressed between control and PCOS groups. A subset of the ten miRNAs (miRNA-32, miRNA-34c-5p, miRNA135a, miRNA-138, miRNA-142–5p, miRNA-18b, miRNA-489, miRNA-627, miRNA-888 and miRNA-9) displaying the largest differentially expressed miRNAs were selected for validation by qRT-PCR. Only five (50%) of these miRNAs, miRNA-9, miRNA-18b, miRNA-32, miRNA-34c, and miRNA-135a, were confirmed to be significantly increased in the PCOS group compared to the control group. Studies conducted by Xu et al. (2015) reported that compared to healthy women (control group), a total of 59 miRNAs were identified that were differentially expressed in cumulus granulosa cells from patients with PCOS; 21 miRNAs were upregulated, and 38 miRNAs were downregulated (Supplementary Table 3). Hossain et al. (2013) examined changes in miRNA expression in a chronically androgenized (5α-dihydrotestosterone (DHT)-treated rat model of PCOS, which duplicates many clinical characteristics of human PCOS, using miRNA PCR array. The authors reported significant changes within the ovary; expression of 17 miRNAs were upregulated and 72 were downregulated (Supplementary Table 3). Interestingly, most of the differentially expressed miRNAs were found to be mainly localized in the theca cells of the ovary. Mi-RNA-9, miRNA-32, and miRNA-489 were the only miRNAs that were found to be increased in both PCOS patients and DHT-induced PCOS rats.
Studies reported by Huang et al. (2016) demonstrated that expression of 16 miRNAs was increased and one miRNA decreased in cumulus cells derived from ovaries of patients with PCOS; among the miRNAs, miRNA-509–3p improves estradiol secretion by attenuating the expression of MAP3K8. Zhang et al. (2017a) provided evidence that miRNA-320 expression is downregulated in human PCOS cumulus granulosa cells (hCGCs) as compared to hCGCs cells from normal subjects. MiRNA-320 knockdown (miRNA-320 inhibitors) and overexpression (miRNA-320 mimics) experiments demonstrated that inhibition of miRNA-320 significantly attenuated IGF1-stimulated progesterone and estradiol production in normal granulosa cells, whereas increased miRNA-320a expression caused by miRNA-320 mimics considerably improved IGF-stimulated progesterone and estradiol production in PCOS cumulus cells. In contrast, miRNA-320 overexpression and its inhibition had no effect on FSH- or LH-stimulated progesterone and estrogen production in both control and PCOS cumulus cells. The results of this study further demonstrated that dysregulation of miRNA/RUNX2/CYP11A1/CYP19A1 plays a regulatory role in the development and progression of estrogen deficiency in CGCs from patients with PCOS (Zhang et al. 2017a). These studies, however, are at variance with a recent study by Yin et al. (2014) showing that miRNA-320 inhibits estradiol synthesis in and proliferation of mouse ovarian granulosa cells by targeting E2FA and SF-1 transcription factors. There are several additional reports showing PCOS-induced alterations in miRNAs expression as summarized in Supplementary Table 3 and in recent reviews (Sørensen et al. 2014, 2016b, Chen et al. 2019).
Premature ovarian failure (POF) – also known as premature ovarian insufficiency (POI) – is defined as the cessation of ovarian function before the age of 40 years. It is accompanied by menstrual irregularities, elevated gonadotropins, estrogen deficiency, loss of oocytes and lack of folliculogenesis (Nelson 2009, Tucker et al. 2016, Torrealday et al. 2017). Until now, only limited studies have been carried out to delineate the potential role of miRNAs in the pathophysiology of POF. One microarray study identified 29 miRNAs based on significance (P value), of which 12 were differentially expressed between patients with POF and normal women (Yang et al. 2012). Among these differentially expressed miRNAs, ten miRNAs were upregulated and two were downregulated. Quantitative RT-PCR validation studies using a larger cohort of 39 POF patients and 20 normal women confirmed high expression of miRNA-146a, miRNA-27a, miRNA-23a and miRNA-126 in serum samples from POF patients as compared to the controls. A further investigation in the potential role of miRNA-23a in POF demonstrated that it promotes apoptosis of granulosa cells by downregulating the X-linked inhibitor of apoptosis (XIAP) protein with a subsequent increase in caspase-3 cleavage (Yang et al. 2012, McGinnis et al. 2015, Guo et al. 2017). Furthermore, miRNA-27b, miRNA-190, miRNA-151, miRNA-672, miRNA-29a, and miRNA-144 have been implicated in the development of POF (Imbar & Eisenberg 2014). Dang et al. (2015) identified 51 differentially expressed miRNAs by carrying out miRNA expression profiling of serum samples from control subjects and POF patients; 22 miRNAs were significantly upregulated and 29 miRNAs significantly downregulated in POF patients compared to control subjects. Seven upregulated miRNAs were further validated by qRT-PCR. The expression of only one miRNA, miRNA-22–3p, was markedly downregulated in POF patients compared to normal subjects (Dang et al. 2015). In contrast, the other eight miRNAs showed no statistically significant differences between the two groups. MiRNA-379–5p is also known to be associated with premature ovarian insufficiency (Dang et al. 2018).
A total of 62 miRNAs were upregulated and 20 miRNAs were downregulated in a rat POF model (4-vinylcyclohexane diepoxide (VCD)-treated rats) (Kuang et al. 2014). qRT-PCR analysis of six miRNAs selected from the microarray data confirmed that expression of miRNA-29a and miRNA-144 was downregulated, whereas expression of miRNA-27b, miRNA-190, miRNA-151, and miRNA-672 was upregulated in POF rat ovarian tissues as compared to normal ovarian tissues. Another study by Li et al. (2014) demonstrated that 16 miRNAs were upregulated and 9 were downregulated in the ovaries of VCD-treated rats.
In addition, many miRNAs can modulate steroidogenesis by altering growth and proliferation and apoptosis of ovarian cells (Baley & Li 2012, Donadeu et al. 2012, Maalouf et al. 2016, Reza et al. 2019, Zhang et al. 2019b) (Supplementary Table 4). Sirotkin et al. (2010) examined the effect of transfection of cultured primary human ovarian granulosa cells with 80 different constructs encoding human pre-miRNAs on the expression of the proliferation marker, proliferating cell nuclear antigen (PCNA), a nuclear nonhistone protein, and the apoptosis marker, Bax, evaluated using immunocytochemical techniques. Eleven out of 80 tested miRNAs constructs stimulated, and 53 miRNAs inhibited expression of the proliferation marker PCNA (Supplementary Table 4). In addition, 11 of the 80 miRNAs tested promoted enhanced accumulation of Bax content, whereas 46 mRNAs caused attenuation of Bax levels in granulosa cells (Supplementary Table 4). In mouse granulosa cells, Carletti et al. (2010) provided evidence that miRNA-21 inhibits apoptosis both in vitro and in vivo. MiRNA-224 was shown to exert a stimulatory effect on TGF-β1-induced granulosa cell proliferation and estradiol secretion through modulation of SMAD4 activity and increased mRNA expression of Cyp19a1 (Yao et al. 2010a). Studies by Yin et al. (2012) suggest that miRNA-383 functions to promote estradiol secretion in mouse granulosa cells by targeting RBMS1 and through partial inactivation of c-Myc. Moreover, SF-1 functions as a positive regulator of miRNA-383 expression and function. Lin et al. (2012) reported that miRNA-26b, which is upregulated during follicular atresia, was associated with increased number of DNA breaks and promoted granulosa cell apoptosis by targeting the ataxia telangiectasia mutated (ATM) gene. In cultured human granulosa cells, miRNA-23a was shown to induce apoptosis via downregulation of XIAP along with increased caspase-3 cleavage (Yang et al. 2012). A report by Yan et al. (2012) indicates that miRNA-145 inhibits mouse granulosa cell proliferation by targeting activin receptor IB (ACVRIB) (Yan et al. 2012). It was demonstrated that transfection of primary human granulosa cells with anti-miRNA-15a led to significant increased accumulation of both proliferation and apoptosis markers and an inhibition in the secretion of progesterone and testosterone and stimulation of estradiol release (Sirotkin et al. 2014). In contrast, transfection of granulosa cells with pre-miRNA-15a resulted in decreased accumulation of proliferation and apoptosis-related proteins, MAPK/ERK1,2 and caspase 3, and enhanced secretion of progesterone and testosterone, but had no effect on estradiol production (Sirotkin et al. 2014). Using gain- and loss-of function approaches, Chen et al. (2015) demonstrated that overexpression of miRNA-146a in human granulosa cells resulted in accelerated apoptosis, whereas downregulation of miRNA-146a was associated with reduced apoptosis. Moreover, evidence was presented showing that miRNA-146a modulates ovarian granulosa cell apoptosis by simultaneously targeting interleukin-1 receptor-associated kinase (IRAK1) and tumor necrosis factor receptor-associated factor 6 (TRAF6). Yao et al. (2018) demonstrated that miRNA-181b attenuates porcine granulosa cell apoptosis by inhibiting the expression of SMAD7, activating the TGF-β signaling pathway and increasing expression with the transforming growth factor beta receptor (TGFBR1) promoter (Yao et al. 2018). Very recently, Zhou et al. (2019) provided evidence that miRNA-150 promotes apoptosis of ovine ovarian granulosa cells by targeting the StAR protein gene (STAR gene). Several additional miRNAs have been identified as summarized in Supplementary Table 2 that either modulate ovarian cell proliferation or apoptosis.
miRNA regulation of testicular steroidogenesis
Currently very little information is available about the miRNA regulation of steroidogenesis in testicular Leydig cells. Studies mainly from our laboratories have shown that some miRNAs may be involved in the regulation of testicular steroidogenesis either directly or indirectly using model Leydig cell lines, MLTC-1 and R2C cells (Hu et al. 2012, 2013). It was reported by us that miRNA-125a is expressed at much higher levels than miRNA-125b in MLTC-1 cells (Hu et al. 2012). In contrast, very little miRNA-455–5p or miRNA-145 expression was noted under basal conditions. Treatment of MLTC-1 cells with a cAMP analog, Bt2cAMP, significantly downregulated the expression of all four miRNAs (i.e., miRNA-125a, miRNA-125b, miRNA-455–5p and miRNA-145). We further demonstrated that miRNA-125a and miRNA-455 act as potent negative regulators of SR-BI and SR-BI-mediated selective HDL-CE transport function (Hu et al. 2012). Overexpression of pre-miRNA-125a and pre-miRNA-455 significantly decreased the levels of progesterone produced in both R2C and MLTC-1 cells. Additional follow-up experiments demonstrated that treatment of MLTC-1 cells with Bt2cAMP for 6 h increased the expression of miRNA-212, miRNA-183, miRNA-132, miRNA-182 and miRNA-96 and inhibited the expression of miRNA-138 and miRNA-19a (Hu et al. 2013). In mouse Leydig tumor MLTC-1 and murine Y1 adrenocortical cells, it has been demonstrated that inhibition of miRNA-200c is associated with increased vimentin expression and steroid production (Hu et al. 2017). A recent study by Geng et al. (2017) demonstrated that miRNA-150 by targeting StAR protein negatively regulates testosterone production in mouse Leydig cells both in vivo and in vitro. Furthermore, Men et al. (2017) demonstrated that overexpression of H19 in MLTC-1 cells stimulated StAR expression by antagonizing let-7, which inhibits StAR protein expression at the posttranscriptional levels. Gao et al. (2018) provided evidence that dysregulation of mouse testicular Leydig cell miRNA-146a-5p/Mta3 signaling cascade contributes to the pathogenesis of impaired steroidogenesis, at least in part, in response to exposure of testis to the environmental toxin, bisphenol A (BPA). A similar type of study demonstrated that miRNA-6321/Map3k1-regulated the JNK/c-Jun/Nur77 signaling cascade, mediating the Triclosan (TCS)-induced inhibition of steroidogenesis in primary rat testicular Leydig cells and mouse Leydig tumor TM3 cells (Ha et al. 2018).
Steroidogenic enzymes and protein factors as potential targets for miRNAs
Few studies have directly attempted to identify steroidogenic enzymes, protein factors that mediate cholesterol delivery to cells and its transport to and within the mitochondria for steroidogenesis and gonadotropin receptors, as potential targets for miRNAs. Using human adrenal tissue samples and human H295R adrenocortical cells, CYP11A1, CYP11B1, CYP17A1 and CYP19A1, have been identified as potential targets for various miRNAs (Table 2). Likewise, in mouse Y1 adrenocortical tumor cells, miRNA-132 was shown to directly target cholesterol transport protein, StAR and methyl CpG-binding protein with a direct relevance to steroidogenesis (Hu et al. 2017). Relatively more information is currently available about the ovarian steroidogenic targets for several miRNAs (Table 2). These include gonadotropin receptors (FSHR, LHCGR), steroidogenic enzymes (CYP19A1), transcription factors (Creb1, Foxl2, LRH-1, SF-1, E2F1, RUNX2, SREBP-1a, SREBP-2), cholesterol transport proteins (StAR) and signaling proteins (HRas, EFNA3, IGF-1, MAP3K8) (Table 2). Likewise, in testicular Leydig cells, miRNA-150 and let-7b target cholesterol transport protein, StAR (Geng et al. 2017, Men et al. 2017), whereas miRNA-200c targets vimentin, a protein involved in intracellular cholesterol transport (Hu et al. 2017). Two miRNAs, miRNA-125a and miRNA-455, directly target testicular Leydig cell SR-B1 (Hu et al. 2012), and miRNA-6321 targets Map3k1 (Ha et al. 2018) (Table 2).
Table 2.
Potential targets of miRNAs in steroidogenic pathways.
| Species/steroidogenic tissues/cells | miRNA | Potential targets of steroidogenic enzymes or protein factors | Reference |
|---|---|---|---|
| Adrenal steroidogenic pathways | |||
| Human tissue samples and human H295R adrenocortical cells Human H295R adrenocortical cells Human H295 adrenocortical cells Mouse Y1 adrenocortical tumor cells Human adrenal tissue, and human H295R adrenocortical cells |
miRNA-24 miRNA-10b miRNA-98 miRNA-132 DICER1 miRNA-125a-5p miRNA-125b-5p miRNA-24 miRNA-320a-3p miRNA-10b miRNA-579, miRNA-561 |
CYP11B1, CYP11B2 CYP11B1, CYP11B2 CYP19A1 StAR, Mecp2 CYP11A1, CYP11B1, CYP21A, CYP17A1, CYP11B2 CYP11B2 CYP11B2 CYP11B1, CYP11B2 CYP11A1, CYP17A1 CYP11B1, CYP11B2 HSD11B1 |
Robertson et al. 2013, Nusrin et al. 2014, Yu et al. 2015, Hu et al. 2017, Robertson et al. 2013, 2017, Clayton et al. 2018 |
| Ovarian steroidogenic pathways | |||
| Porcine ovarian cumulus granulosa cells Mouse ovarian granulosa cells Primary mouse ovarian granulosa cells, and human granulosa-like tumor cell line, KGN Rat ovaries Primary human ovarian granulosa cells Mouse ovarian granulosa cells Human ovarian granulosa-like tumor cell line Primary mouse ovarian granulosa cells Porcine ovarian granulosa cells Human ovarian cumulus granulosa cells and human granulosa-like tumor cell line, KGN Mouse ovarian granulosa cells Human ovarian granulosa like tumor cell line, KGN Mouse ovarian granulosa cells, and mouse granulosa-like tumor cell line, KK-1 Human ovarian granulosa-like tumor cell line, KGN Human ovarian cumulus granulosa cells Porcine ovarian granulosa cells Rat ovary Buffalo ovaries and buffalo ovarian granulosa cells. Human ovarian cumulus granulosa cells and human granulosa-like tumor cell line, KGN Human ovarian cumulus granulosa cells Ovine ovarian granulosa cells |
miRNA-378 miRNA-383 miRNA-133b miRNA-5131-3p miRNA-513a-3p miRNA-320, miRNA-383 miRNA-592 miRNA-132 miRNA-126* miRNA-509-3p miRNA-764-3p let-7b miRNA-27a-3p miRNA-375 miRNA-320a miRNA-1275 miRNA-122 miRNA-210 miRNA-323-3p miRNA-320a miRNA-150 |
CYP19A1 RBMS1 Foxl2 LHCGR LHCGR E2F1, SF-1 LHCGR Nurr1 FSHR MAP3K8 SF-1 StAR Creb1 SP1 RUNX2 LRH-1 LRBP, SREBP-1a/SREBP-2, LHCGR HRas, EFNA3 IGF-1 RUNX2 StAR |
Xu et al. 2011, Pan et al. 2015, Yin et al. 2012, Dai et al. 2013, Kitahara et al. 2013, Troppmann et al. 2014, Yin et al. 2014, Song et al. 2015, Wu et al. 2015, Du et al. 2016b, Huang et al. 2016, Wang et al. 2016a, Men et al. 2017, Wang et al. 2017, Tu et al. 2019, Zhang et al. 2017a, Liu et al. 2018, Menon et al. 2018, Shukla et al. 2018, Wang et al. 2019, Zhang et al. 2019a, Zhou et al. 2019 |
| Testicular steroidogenic pathways | |||
| Mouse testicular Leydig tumor MLTC-1 cells Mouse testicular Leydig tumor MLTC-1 cells Mouse testicular Leydig cells Mouse testicular Leydig tumor MLTC-1 cells Mouse testicular Leydig tumor MA-10 cells Primary rat testicular Leydig cells and Mouse testicular Leydig tumor TM3 cells |
miRNA-125a, miRNA-455 miRNA-200c miRNA-150 let-7b MiRNA-146a-5p miRNA-6321 |
SR-B1 Vimentin STAR Star Mta-3 Map3k1 |
Hu et al. 2012, Hu et al. 2017, Geng et al. 2017, Men et al. 2017, Gao et al. 2018, Ha et al. 2018 |
Creb1, cAMP-responsive element-binding protein1; E2F1, E2F transcription factor 1, FSHR, follicle-stimulating hormone receptor; Foxl2, forkhead Box L2; IGF-1, insulin-like growth factor-1; LHCGR, luteinizing hormone/chorionic gonadotropin receptor; LRBP, luteinizing hormone receptor (LHR) mRNA-binding protein; LRH-1, liver receptor homolog-1; MAP3K1, mitogen-activated protein kinase, kinase, kinase1; MAP3K8, mitogen-activated protein kinase kinase kinase 8; Mecp2, methyl CpG-binding protein; Mta-3, metastasis-associated 3; Nur77, orphan nuclear receptor Nur77 (also known as NR4A1, NGF1B, TR3TIS1, NAK or N10); Nurr1, Nurr1 transcription factor, also called nuclear receptor subfamily 4 group A member 2 (Nr4a2); RBMS1, RNA-binding motif single-stranded interacting protein 1; RUNX2, Runt-related transcription factor also known as core binding factor subunit alpha-1 (CBP-alpha-1); SR-B1, scavenger receptor class B, type 1; SREBP-1a, sterol regulatory element-binding protein1a; SREBP-2, sterol regulatory element-binding protein2; STA R/StAR/Star, steroidogenic acute regulatory protein; SF-1, steroidogenic factor 1.
Summary and conclusions
This review summarizes the current understanding of the role of miRNAs in the regulation of adrenal and gonadal steroidogenesis and related processes. MiRNAs are endogenous single-stranded small RNAs of ~22 nucleotides in length that post-transcriptionally downregulate gene expression. A single RNA could suppress hundreds of genes, and a single gene could be modulated by multiple miRNAs. Although miRNA regulation of steroidogenesis is an emerging field, extensive literature exists about the important role played by miRNAs in the regulation of many other cellular processes such as inflammatory and immune responses, cell-cycle progression and proliferation, differentiation, tissue remodeling, apoptosis and disease pathology. The limited information that is currently available for three steroidogenic tissues, adrenal, ovary and testis, already suggests that miRNAs are putatively involved in virtually every aspect of the steroidogenic process, including receptor-mediated lipoprotein cholesterol delivery to the cell interior, intracellular cholesterol processing and transport to and within the mitochondria, other steroidogenesis-related accessory protein and transcription factors, and steroidogenic enzymes themselves. Table 2 depicts the current understanding of the roles of specific miRNAs in steroidogenesis. The large amount of data generated through genome-wide analyses of steroid producing adrenal and gonadal tissues, particularly ovarian granulosa cells, should provide ample opportunity for the identification and functional characterization of miRNAs that will likely lead to a much greater mechanistic understanding of the posttranscriptional regulation of steroidogenesis. Besides, with the introduction and availability of high-throughput, next-generation RNA sequencing tools, as well as technical advances in sequencing methodologies and newer bioinformatics, future studies should allow investigators to precisely identify novel, or low abundance miRNAs in steroidogenic tissues and their pathophysiological relevance to steroidogenesis. Furthermore, studies to determine the expression and role of miRNAs in pathological settings should lead to identification of miRNAs as biomarkers or validated targets and functions that are likely to greatly aid in the clinical management of steroid hormone-related diseases and/or reproductive diseases.
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Institutes of Health grants 2R01HL33881 (S A) and P30DK116074 (F B K), and by Merit Review Awards # I01BX001923 (S A) and # I01BX000398 (F B K), and Senior Research Career Scientist Award # IK6B004200 (S A) from the United States Department of Veterans Affairs, Biomedical Laboratory Research Development Program.
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this review.
Supplementary materials
This is linked to the online version of the paper at https://doi.org/10.1530/JME-19-0105.
References
- Adams BD, Parsons C, Walker L, Zhang WC & Slack FJ 2017. Targeting noncoding RNAs in disease. Journal of Clinical Investigation 127 761–771. ( 10.1172/JCI84424) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed K, LaPierre MP, Gasser E, Denzier R, Yang Y, Rülicke T, Kero J, Latreille M & Stoffel M 2017. Loss of microRNA-7a2 induces hypogonadotropic hypogonadism and infertility. Journal of Clinical Investigation 127 1061–1074. ( 10.1172/JCI90031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti C & Cochella L 2017. A framework for understanding the roles of miRNAs in animal development. Development 144 2548–2559. ( 10.1242/dev.146613) [DOI] [PubMed] [Google Scholar]
- Andersen CY & Ezcurra D 2014. Human steroidogenesis: implications for controlled ovarian stimulation with exogenous gonadotropins. Reproductive Biology and Endocrinology 12 128 ( 10.1186/1477-7827-12-128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchus RJ 2004. The backdoor pathway to dihydrotestosterone. Trends in Endocrinology and Metabolism 15 432–438. ( 10.1016/j.tem.2004.09.004) [DOI] [PubMed] [Google Scholar]
- Azhar S & Reaven E 2002. Scavenger receptor class BI and selective cholesteryl ester uptake: partners in the regulation of steroidogenesis. Molecular and Cellular Endocrinology 195 1–26. ( 10.1016/s0303-7207(02)00222-8) [DOI] [PubMed] [Google Scholar]
- Azziz R, Carmina E, Chen Z, Dunaif A, Laven JSE, Legro RS, Lizneva D, Natterson-Horowitz B, Teede HJ & Yildiz BO 2016. Polycystic ovary syndrome. Nature Reviews: Disease Primers 2 16057 ( 10.1038/nrdp.2016.57) [DOI] [PubMed] [Google Scholar]
- Baley J & Li J 2012. Micro RNAs and ovarian function. Journal of Ovarian Research 5 8 ( 10.1186/1757-2215-5-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2018. Metazoan microRNAs. Cell 173 20–51. ( 10.1016/j.cell.2018.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biason-Lauber A, Miller WL, Pandey AV & Flück CE 2013. Of marsupials and men: ‘Backdoor’ dihydrotestosterone synthesis in male sexual differentiation. Molecular and Cellular Endocrinology 371 124–132. ( 10.1016/j.mce.2013.01.017) [DOI] [PubMed] [Google Scholar]
- Bollag WB 2014. Regulation of aldosterone synthesis and secreti and secretion. Comprehensive Physiology 4 1017–1055. ( 10.1002/cphy.c130037) [DOI] [PubMed] [Google Scholar]
- Bushati N & Cohen SM 2007. MicroRNA functions. Annual Review of Cell and Developmental Biology 23 175–205. ( 10.1146/annurev.cellbio.23.090506.123406) [DOI] [PubMed] [Google Scholar]
- Butterworth MB & Alvarez de la Rosa D 2019. Regulation of aldosterone signaling by nicroRNAs. Vitamins and Hormones 109 69–103. ( 10.1016/bs.vh.2018.09.002) [DOI] [PubMed] [Google Scholar]
- Carletti MZ, Fiedler SD & Christenson LK 2010. MicroRNA 21 blocks apoptosis in mouse periovulatory grenulosa cells. Biology of Reproduction 83 286–295. ( 10.1095/biolreprod.109.081448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xie M, Liu D & Shi K 2015. Downregulation of microRNA-146a inhibits ovarian granulosa cell apoptosis by simultaneously targeting interleukin-1 receptor-associated kinase and tumor necrosis factor receptor-associated factor 6. Molecular Medicine Reports 12 5155–5162. ( 10.3892/mmr.2015.4036) [DOI] [PubMed] [Google Scholar]
- Chen B, Xu P, Wang J & Zhang C 2019. The role of miRNA in polycystic ovary syndrome (PCOS). Gene 706 91–96. ( 10.1016/j.gene.2019.04.082) [DOI] [PubMed] [Google Scholar]
- Clayton SA, Jones SW, Kurowska-Stolarska M & Clark AR 2018. The role of microRNAs in glucocorticoid action. Journal of Biological Chemistry 293 1865–1874. ( 10.1074/jbc.R117.000366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa B, Da Pozzo E & Martini C 2018. Translocator protein and steroidogenesis. Biochemical Journal 475 901–904. ( 10.1042/BCJ20170766) [DOI] [PubMed] [Google Scholar]
- Dai A, Sun H, Fang T, Zhang Q, Wu S, Jiang Y, Ding L, Yan G & Hu Y 2013. MicroRNA-133b stimulates ovarian estradiol synthesis by targeting Foxl2. FEBS Letters 587 2474–2482. ( 10.1016/j.febslet.2013.06.023) [DOI] [PubMed] [Google Scholar]
- Dang Y, Zhao S, Qin Y, Han T, Li W & Chen ZJ 2015. MicroRNA-22–3p is down-regulated in the plasma of Han Chinese patients with premature ovarian failure. Fertility and Sterility 103 802.e1–807.e1. ( 10.1016/j.fertnstert.2014.12.106) [DOI] [PubMed] [Google Scholar]
- Dang Y, Wang X, Hao Y, Zhang X, Zhao S, Ma J, Qin Y & Chen ZJ 2018. MicroRNA-379–5p is associated with biochemical premature ovarian insufficiency through PARP1 and XRCC6. Cell Death and Disease 9 106 ( 10.1038/s41419-017-0163-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng B, Shen WJ, Dong D, Azhar S & Kraemer FB 2019. Plasma membrane cholesterol trafficking in steroidogenesis. FASEB Journal 33 1389–1400. ( 10.1096/fj.201800697RRR) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayat NA, Dick B, Frey BM, d’Uscio CH, Vogt B & Flück CE 2017. Androgen biosynthesis during minipuberty favors the backdoor pathway over the classic pathway: insights into the enzyme activities and steroid fluxes in healthy infants during the first year of life from the urinary steroid metabolome. Journal of Steroid Biochemistry and Molecular Biology 165 312–322. [DOI] [PubMed] [Google Scholar]
- Di Leva G, Garofalo M & Croce CM 2014. MicroRNAs in cancer. Annual Review of Pathology: Mechanisms of Disease 9 287–314. ( 10.1146/annurev-pathol-012513-104715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donadeu FX, Schauer SN & Sontakke SD 2012. Involvement of miRNAs in ovarian follicular and luteal development. Journal of Endocrinology 215 323–334. ( 10.1530/JOE-12-0252) [DOI] [PubMed] [Google Scholar]
- Dong H, Lei J, Ding L, Wen Y, Ju H & Zhang X 2013. MicroRNA: function, detection, and bioanalysis. Chemical Reviews 113 6207–6233. ( 10.1021/cr300362f) [DOI] [PubMed] [Google Scholar]
- Du X, Li Q, Pan Z & Li Q 2016b. Androgen receptor and miRNA-126* axis controls follicle-stimulating hormone receptor expression in porcine granulosa cells. Reproduction 152 161–169. ( 10.1530/REP-15-0517) [DOI] [PubMed] [Google Scholar]
- Esteller M 2011. Non-coding RNAs in human disease. Nature Reviews: Genetics 12 861–874. ( 10.1038/nrg3074) [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N & Filipowicz W 2010. Regulation of mRNA translation and stability by microRNAs. Annual Review of Biochemistry 79 351–379. ( 10.1146/annurev-biochem-060308-103103) [DOI] [PubMed] [Google Scholar]
- Fiedler SD, Carletti MZ, Hong X & Christenson LK 2008. Hormonal regulation of microRNA expression in periovulatory mouse mural granulosa cells. Biology of Reproduction 79 1030–1037. ( 10.1095/biolreprod.108.069690) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flück CE & Pandey AV 2014. Steroidogenesis of the testis – new genes and pathways. Annales d’Endocrinologie 75 40–47. ( 10.1016/j.ando.2014.03.002) [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK-H, Burge CB & Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research 19 92–105. ( 10.1101/gr.082701.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao GZ, Zhao Y, Li HX & Li W 2018. Bisphenol A-elicited miR-146a-5p impairs murine testicular steroidogenesis through negative regulation of Mta3 signaling. Biochemical and Biophysical Research Communications 501 478–485. ( 10.1016/j.bbrc.2018.05.017) [DOI] [PubMed] [Google Scholar]
- Gebert LFR & MacRae IJ 2019. Regulation of microRNA function in animals. Nature Reviews: Molecular Cell Biology 20 21–37. ( 10.1038/s41580-018-0045-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng XJ, Zhao DM, Mao GH & Tan L 2017. MicroRNA-150 regulates steroidogenesis of mouse testicular Leydig cells by targeting STAR. Reproduction 154 229–236. ( 10.1530/REP-17-0234) [DOI] [PubMed] [Google Scholar]
- Guo Y, Sun J & Lai D 2017. Role of microRNAs in premature ovarian insufficiency. Reproductive Biology and Endocrinology 15 38 ( 10.1186/s12958-017-0256-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M & Kim VN 2014. Regulation of microRNA biogenesis. Nature Reviews: Molecular Cell Biology 15 509–524. ( 10.1038/nrm3838) [DOI] [PubMed] [Google Scholar]
- Ha M, Zhang P, Li L & Liu C 2018. Triclosan suppresses testicular steroidogenesis via the miR-6321/JNK/Nur77 cascade. Cellular Physiology and Biochemistry 50 2029–2045. ( 10.1159/000495049) [DOI] [PubMed] [Google Scholar]
- Hasuwa H, Ueda J, Ikawa M & Okabe M 2013. MiR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science 341 71–73. ( 10.1126/science.1237999) [DOI] [PubMed] [Google Scholar]
- Hattangady NG, Olala LO, Bollag WB & Rainey WE 2012. Acute and chronic regulation of aldosterone production. Molecular and Cellular Endocrinology 350 151–162. ( 10.1016/j.mce.2011.07.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2012. Chapter 15: Reproductive endocrinology In Williams Gynecology, 2nd ed. Eds Hoffman BL, Schorge JO, Schaffer JI, Halvorson LM, Bradshaw KD, Cunningham E & Calver LE. New York, NY: McGraw-Hill; (available at: http://accessmedicine.mhmedical.com/content.aspx?); bookid 399 and sectioned = 41722304 Accessed on 3 March 2019. [Google Scholar]
- Homma K, Hasegawa T, Nagai T, Adachi M, Horikawa R, Fujiwara I, Tajima T, Takeda R, Fukami M & Ogata T 2006. Urine steroid hormone profile analysis in cytochrome P450 oxidoreductase deficiency: implication for the backdoor pathway to dihydrotestosterone. Journal of Clinical Endocrinology and Metabolism 91 2643–2649. ( 10.1210/jc.2005-2460) [DOI] [PubMed] [Google Scholar]
- Hossain MM, Cao M, Wang Q, Kim JY, Schellander K, Tesfaye D & Tsang BK 2013. Altered expression of miRNAs in a dihydrotestosterone-induced rat PCOS model. Journal of Ovarian Research 6 36 ( 10.1186/1757-2215-6-36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Shen WJ, Kraemer FB & Azhar S 2012. MicroRNAs 125a and 455 represses lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type 1 in steroidogenic cells. Molecular and Cellular Biology 32 5035–5045. ( 10.1128/MCB.01002-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Shen WJ, Cortez Y, Tang X, Liu LF, Kraemer FB & Azhar S 2013. Hormone regulation of microRNA expression in steroid producing cells of the ovary, testis and adrenal gland. PLoS ONE 8 e78040 ( 10.1371/journal.pone.0078040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Shen WJ, Kraemer FB & Azhar S 2017. Regulation of adrenal and ovarian steroidogenesis by miR-132. Journal of Molecular Endocrinology 59 269–283. ( 10.1530/JME-17-0011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Liu C, Hao C, Tang Q, Liu R, Lin S, Zhang L & Yan W 2016. Identification of altered microRNAs and mRNAs in the cumulus cells of PCOS patients: miRNA-509–3p promotes oestradiol secretion by targeting MAP3K8. Reproduction 151 643–655. ( 10.1530/REP-16-0071) [DOI] [PubMed] [Google Scholar]
- Igaz P, Igaz I, Nagy Z, Nyírȍ G, Szabó PM, Falus A, Patócs A & Rácz K 2015. MicroRNAs in adrenal tumors: relevance for pathogenesis, diagnosis, and therapy. Cellular and Molecular Life Sciences 72 417–428. ( 10.1007/s00018-014-1752-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Bimpaki EI, Nesterova M & Stratakis CA 2009. MicroRNA signature of primary pigmented nodular adrenocortical disease: clinical correlations and regulation of Wnt signaling. Cancer Research 69 3278–3282. ( 10.1158/0008-5472.CAN-09-0155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbar T & Eisenberg I 2014. Regulatory role of microRNAs in ovarian function. Fertility and Sterility 101 1524–1530. ( 10.1016/j.fertnstert.2014.04.024) [DOI] [PubMed] [Google Scholar]
- Jonas S & Izaurralde E 2015. Towards a molecular understanding of microRNA-mediated gene silencing. Nature Reviews: Genetics 16 421–433. ( 10.1038/nrg3965) [DOI] [PubMed] [Google Scholar]
- Kamrath C, Hochberg Z, Hartmann MF, Remer T & Wudy SA 2012. Increased activation of alternative ‘backdoor’ pathway in patients with 21-hydroxylase deficiency: evidence from urinary steroid hormone analysis. Journal of Clinical Endocrinology and Metabolism 93 E367–E375. [DOI] [PubMed] [Google Scholar]
- Kim VN 2005. MicroRNA biogenesis: coordinated cropping and dicing. Nature Reviews: Molecular Cell Biology 6 376–385. ( 10.1038/nrm1644) [DOI] [PubMed] [Google Scholar]
- Kitahara Y, Nakamura K, Kogure K & Minegishi T 2013. Role of microRNA-136–3p on the expression of luteinizing hormone-human chorionic gonadotropin receptor mRNA in rat ovaries. Biology of Reproduction 89 1–10. [DOI] [PubMed] [Google Scholar]
- Klinge CM 2012. miRNAs and estrogen action. Trends in Endocrinology and Metabolism 23 223–233. ( 10.1016/j.tem.2012.03.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer FB, Shen WJ, Harada K, Patel S, Osuga J, Ishibashi S & Azhar S 2004. Hormone-sensitive lipase is required for high-density lipoprotein cholesteryl ester-supported adrenal steroidogenesis. Molecular Endocrinology 18 549–557. ( 10.1210/me.2003-0179) [DOI] [PubMed] [Google Scholar]
- Kraemer FB, Shen WJ & Azhar S 2017. SNAREs and cholesterol movement for steroidogenesis. Molecular and Cellular Endocrinology 441 17–21. ( 10.1016/j.mce.2016.07.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krone N, Reisch N, Idkowiak J, Dhir V, Ivison HE, Hughes BA, Rose IT, O’Neil DM, Vijelaar R, Smith MJ, et al. 2012. Genotype-phenotype analysis in congenital hyperplasia due to P450 oxidoreductase due to 450 oxidoreductase deficiency. Journal of Clinical Endocrinology and Metabolism 97 E257–E267. ( 10.1210/jc.2011-0640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang H, Han D, Xie J, Yan Y, Li J & Ge P 2014. Profiling of differentially expressed microRNAs in premature ovarian failure in an animal model. Gynecological Endocrinology 30 57–61. ( 10.3109/09513590.2013.850659) [DOI] [PubMed] [Google Scholar]
- Lannes J, L’hôte D, Fernandez-Vega A, Garrel G, Laverrière JN, CohenTannoudji J & Quérat B 2016. A regulatory loop between miR-132 and miR-125b involved in gonadotrope cell desensitization to GnRH. Scientific Reports 6 31563 ( 10.1038/srep31563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Fan S, Han D, Xie J, Kuang H & Ge P 2014. Microarray gene expression profiling and bioinformatics analysis of premature ovarian failure in a rat model. Experimental and Molecular Pathology 97 535–541. ( 10.1016/j.yexmp.2014.10.015) [DOI] [PubMed] [Google Scholar]
- Li Y, Xiang Y, Song Y, Wan L, Yu G & Tan L 2019. Dysregulated miRNA142, −33b, and −423 in granulosa cells target TGFBR1 and SMAD7: a possible role is polycystic ovary syndrome. Molecular Human Reproduction 25 638–646. ( 10.1093/molehr/gaz014) [DOI] [PubMed] [Google Scholar]
- Liang M, Yao G, Yin M, Lü M, Tian H, Liu L, Lian J, Huang X & Sun F 2013. Transcriptional cooperation between p53 and NF-κB p65 regulates microRNA-224 transcription in mouse ovarian granulosa cells. Molecular and Cellular Endocrinology 370 119–129. ( 10.1016/j.mce.2013.02.014) [DOI] [PubMed] [Google Scholar]
- Lima J, Batty JA, Sinclair H & Kunadian V 2017. MicroRNAs in ischemic heart disease: from pathophysiology to potential clinical applications. Cardiology in Review 25 117–125. ( 10.1097/CRD.0000000000000114) [DOI] [PubMed] [Google Scholar]
- Lin C-P & He L 2017. Noncoding RNAs in cancer development. Annual Review of Cancer Biology 1 163–184. ( 10.1146/annurev-cancerbio-050216-034443) [DOI] [Google Scholar]
- Lin F, Li R, Pan ZX, Zhou B, Yu DB, Wang XG, Ma XS, Han J, Shen M & Liu HL 2012. miRNA-26b promotes granulosa cell apoptosis by targeting ATM during follicular atresia in porcine ovary. PLoS ONE 7 e38640 ( 10.1371/journal.pone.0038640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Hou X, Shen WJ, Hanssen R, Khor VK, Cortez Y, Roseman AN, Azhar S & Kraemer FB 2016. SNARE-mediated cholesterol movement to mitochondria supports steroidogenesis in rodent cells. Molecular Endocrinology 30 234–247. ( 10.1210/me.2015-1281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li X, Yao Y, Li G, Pan Z & Li Q 2018. miRNA-1275 control granulosa cell apoptosis and estradiol synthesis by impairing LRH-1/CYP19A1 axis. Biochimica et Biophysica Acta (BBA): Gene Regulatory Mechanisms 1861 246–257. [DOI] [PubMed] [Google Scholar]
- Maalouf SW, Liu WS & Pate JL 2016. MicroRNA in ovarian function. Cell and Tissue Research 363 7–18. ( 10.1007/s00441-015-2307-4) [DOI] [PubMed] [Google Scholar]
- Martin N, Galván JA, Pandey AV, Trippel M, Tapia C, Müller M, Perren A & Flück CE 2017. Genes and proteins of the alternative steroid backdoor pathway for dihydrotestosterone synthesis are expressed in the human ovary and seem enhanced in the polycystic ovary syndrome. Molecular and Cellular Endocrinology 441 116–123. ( 10.1016/j.mce.2016.07.029) [DOI] [PubMed] [Google Scholar]
- Matsumoto AM & Bremmer WJ 2016. Chapter 19: Testicular disorder In Williams Textbook of Endocrinology, 13th ed., pp 694–784 Eds Melmed S, Polonsky KS, Larsen PR & Kronenberg HM. Philadelphia, PA: Elsevier; ISBN: 978–0-323–29738-7; OCLC Number: 1075067557. [Google Scholar]
- McCartney CR & Marshall JC 2016. CLINICAL PRACTICE. Polycystic ovary syndrome. New England Journal of Medicine 375 54–64. ( 10.1056/NEJMcp1514916) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis LK, Luense LJ & Christenson LK 2015. MicroRNA in ovarian biology and disease. Cold Spring Harbor Perspectives in Medicine 5 a022962 ( 10.1101/cshperspect.a022962) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men Y, Fan Y, Shen Y, Lu L & Kallen AN 2017. The steroidogenic acute regulatory protein (StAR) is regulated by the H19/let-7axis. Endocrinology 158 402–409. ( 10.1210/en.2016-1340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon KMJ, Menon B & Gulappa T 2018. Regulation of luteinizing hormone receptor mRNA expression in the ovary: the role of miR122. Vitamins and Hormones 107 67–87. ( 10.1016/bs.vh.2018.01.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina A & Prevot V 2017. Hypothalamic microRNAs flip the switch for fertility. Oncotarget 8 8993–8994. ( 10.18632/oncotarget.14646) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WL 2017. Steroidogenesis: unanswered questions. Trends in Endocrinology and Metabolism 28 771–793. ( 10.1016/j.tem.2017.09.002) [DOI] [PubMed] [Google Scholar]
- Miller WL & Auchus RJ 2011. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocrine Reviews 32 81–151. ( 10.1210/er.2010-0013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WL & Auchus RJ 2019. The ‘backdoor pathway’ of androgen synthesis in human male sexual development. PLoS Biology 17 e3000198 ( 10.1371/journal.pbio.3000198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WL & Bose HS 2011. Early steps in steroidogenesis: intracellular cholesterol trafficking. Journal of Lipid Research 52 2111–2135. ( 10.1194/jlr.R016675) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed BT, Sontakke SD, Loannidis J, Duncan WC & Donadeu FX 2017. The adequate corpus luteum: miRNA-94 promotes luteal cell survival and progesterone production. Journal of Clinical Endocrinology and Metabolism 102 2188–2198. ( 10.1210/jc.2017-00259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson LM 2009. Primary ovarian insufficiency. New England Journal of Medicine 360 606–614. ( 10.1056/NEJMcp0808697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusrin S, Tong SKH, Chaturvedi G, Wu RS, Giesy JP & Kong RYC 2014. Regulation of CYP11B1 and CYP11B2 steroidogenic genes by hypoxia-inducible miR-10b in H295R cells. Marine Pollution Bulletin 85 344–351. ( 10.1016/j.marpolbul.2014.04.002) [DOI] [PubMed] [Google Scholar]
- O’Brien J, Hayder H, Zayed Y & Peng C 2018. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Frontiers in Endocrinology 9 402 ( 10.3389/fendo.2018.00402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS & Baltimore D 2012. MicroRNA regulation of inflammatory responses. Annual Review of Immunology 30 295–312. ( 10.1146/annurev-immunol-020711-075013) [DOI] [PubMed] [Google Scholar]
- O’Shaughnessy PJ, Antignac JP, Le Bizec B, Morvan ML, Svenchnikov K, Söder O, Savchuk I, Monteiro A, Soffientini U, Johnston ZC, et al. 2019. Alternative (backdoor) androgen production and masculinization in the human fetus. PLoS Biology 17 e3000002 ( 10.1371/journal.pbio.3000002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Tomas D, Shen W & Li J 2015. MicroRNA-378 regulates oocyte maturation via the suppression of aromatase in porcine cumulus cells. American Journal of Physiology: Endocrinology and Metabolism 308 E525–E534. ( 10.1152/ajpendo.00480.2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panir K, Schjenken JE, Robertson SA & Hull ML 2018. Non-coding RNAs in endometriosis: a narrative review. Human Reproduction Update 24 497–515. ( 10.1093/humupd/dmy014) [DOI] [PubMed] [Google Scholar]
- Papadopoulos V, Fan J & Zirkin B 2018. Translocator protein (18 kDa): an update on its function in steroidogenesis. Journal of Neuroendocrinology 30 e12500 ( 10.1111/jne.12500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne AH & Youngblood GL 1995. Regulation of expression of steroidogenic enzymes in Leydig cells. Biology of Reproduction 52 217–225. ( 10.1095/biolreprod52.2.217) [DOI] [PubMed] [Google Scholar]
- Price NL, Ramirez CM & Fernández-Hernando C 2014. Relevance of microRNA in metabolic diseases. Critical Reviews in Clinical Laboratory Sciences 51 305–320. ( 10.3109/10408363.2014.937522) [DOI] [PubMed] [Google Scholar]
- Reza AMMT, Cho YJ, Han SG, Song H, Park C, Hong K & Kim JH 2019. Roles of microRNAs in mammalian reproduction: from the commitment of germ cells to pre-implantation embryos. Biological Reviews of the Cambridge Philosophical Society 94 415–438. ( 10.1111/brv.12459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riester A, Issler O, Spyroglou A, Rodring SH, Chen A & Beuschlein F 2012. ACTH-dependent regulation of miRNAs as endogenous modulators of glucocorticoid receptor expression in the adrenal gland. Endocrinology 153 212–222. ( 10.1210/en.2011-1285) [DOI] [PubMed] [Google Scholar]
- Robertson S, MacKenzie SM, Alvarez-Madrazo S, Diver LA, Lin J, Stewart PM, Frazer R, Connell JM & Davies E 2013. MicroRNA-24 is a novel regulator of aldosterone and cortisol in the human adrenal cortex. Hypertension 62 572–578. ( 10.1161/HYPERTENSIONAHA.113.01102) [DOI] [PubMed] [Google Scholar]
- Robertson S, Diver LA, Alvarez-Madrazo S, Livine C, Eijaz A, Fraser R, Connell JM, MacKenzie SM & Davies E 2017. Regulation of corticosteroidogenic genes by microRNAs. International Journal of Endocrinology 2017 2021903 ( 10.1155/2017/2021903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CL, Zhang L, Schütz LF, Totty ML & Spicer LJ 2018. MicroRNA 221 expression in theca and granulosa cells: hormonal regulation and function. Journal of Animal Science 96 641–652. ( 10.1093/jas/skx069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero DG, Plonczynski MW, Carvajal CA, Gomez-Sanchez EP & Gomez-Sanchez CE 2008. Microribonucleic acid-21 increases aldosterone secretion and proliferation in H295R human adrenocortical cells. Endocrinology 149 2477–2483. ( 10.1210/en.2007-1686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero DG, Gomez-Sanchez EP & Gomez-Sanchez CE 2010. Angiotensin II-regulated transcription regulatory genes in adrenal steroidogenesis. Physiological Genomics 42A 259–266. ( 10.1152/physiolgenomics.00098.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfield RL & Ehrmann DA 2016. The pathogenesis of polycystic ovary syndrome (PCOS): the hypothesis of PCOS as functional ovarian hyperandrogenism revisited. Endocrine Reviews 37 467–520. ( 10.1210/er.2015-1104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth LW, McCallie B, Alvero R, Schoolcraft WB, Minjarez D & KatzJaffe MG 2014. Altered microRNA and gene expression in the follicular fluid with polycystic ovary syndrome. Journal of Assisted Reproduction and Genetics 31 355–362. ( 10.1007/s10815-013-0161-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V & Näär AM 2012. MicroRNAs in metabolism and disorders. Nature Reviews: Molecular Cell Biology 13 239–250. ( 10.1038/nrm3313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupaimoole R & Slack FJ 2017. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nature Reviews: Drug Discovery 16 203–222. ( 10.1038/nrd.2016.246) [DOI] [PubMed] [Google Scholar]
- Sang Q, Yao Z, Wang H, Feng R, Wang H, Zhao X, Xing Q, Jin L, He L, Wu L, et al. 2013. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. Journal of Clinical Endocrinology and Metabolism 98 3068–3079. ( 10.1210/jc.2013-1715) [DOI] [PubMed] [Google Scholar]
- Santamaria X & Taylor H 2014. MicRNA and gynecological reproductive diseases. Fertility and Sterility 101 1545–1551. ( 10.1016/j.fertnstert.2014.04.044) [DOI] [PubMed] [Google Scholar]
- Sayed D & Abdellatif M 2011. MicoRNAs in development and disease. Physiological Reviews 91 827–887. ( 10.1152/physrev.00006.2010) [DOI] [PubMed] [Google Scholar]
- Selvaraj V, Stocco DM & Clark BJ 2018. Current knowledge on the acute regulation of steroidogenesis. Biology of Reproduction 99 13–26. ( 10.1093/biolre/ioy102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Prizant H, Light A, Biswas A, Hayes E, Lee H-J, Brad D, Gleicher N & Hames SR 2014. Androgens regulate ovarian follicular development by increasing follicle stimulating hormone receptor and microRNA-125b expression. PNAS 111 3008–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton C, Marcos J, Malunowicz EM, Szarras-Czapnik M, Jira P, Taylor NF, Murphy N, Crushell E, Gottschalk M, Hauffa B, et al. 2004. Biochemical diagnosis of Antley-Bixler syndrome by steroid analysis. American Journal of Medical Genetics: Part A 128A 223–231. ( 10.1002/ajmg.a.30104) [DOI] [PubMed] [Google Scholar]
- Shen WJ, Patel S, Natu V, Hong R, Wang J, Azhar S & Kraemer FB 2003. Interaction of hormone-sensitive lipase with steroidogenic acute regulatory protein: facilitation of cholesterol transfer in adrenal. Journal of Biological Chemistry 278 43870–43876. ( 10.1074/jbc.M303934200) [DOI] [PubMed] [Google Scholar]
- Shen WJ, Zaidi SK, Patel S, Cortez Y, Ueno M, Azhar R, Azhar S & Kraemer FB 2012. Ablation of vimentin results in defective steroidogenesis. Endocrinology 153 3249–3257. ( 10.1210/en.2012-1048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WJ, Azhar S & Kraemer FB 2016a. Lipid droplets and steroidogenic cells. Experimental Cell Research 340 209–214. ( 10.1016/j.yexcr.2015.11.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W-J, Hu Z, Hu J, Kraemer FB & Azhar S 2016b. Post-transcriptional and post-translational regulation of steroidogenesis In Post-Translational Mechanisms in Endocrine Regulation, pp 253–275. Eds Menon KMJ & Goldstrohm AC. New York: Springer International Publishing. [Google Scholar]
- Shen WJ, Asthana S, Kraemer FB & Azhar S 2018. Scavenger receptor B type 1: expression, molecular regulation, and cholesterol transport function. Journal of Lipid Research 59 1114–1131. ( 10.1194/jlr.R083121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Dahiya S, Onteru SK & Singh D 2018. Differentially expressed miRNA-210 during follicular-luteal transition regulates pre-ovulatory granulosa cell function targeting HRas and EFNA3. Journal of Cellular Biochemistry 119 7934–7943. ( 10.1002/jcb.26508) [DOI] [PubMed] [Google Scholar]
- Simpson ER & Waterman MR 1988. Regulation of the synthesis of steroidogenic enzymes in adrenal cortical cells by ACTH. Annual Review of Physiology 50 427–440. ( 10.1146/annurev.ph.50.030188.002235) [DOI] [PubMed] [Google Scholar]
- Simpson E, Lauber M, Demester M, Means G, Mahendroo M, Kilgore M, Mendelson C & Waterman M 1992. Regulation of expression of the genes encoding steroidogenic enzymes in the ovary. Journal of Steroid Biochemistry and Molecular Biology 41 409–413. ( 10.1016/0960-0760(92)90366-q) [DOI] [PubMed] [Google Scholar]
- Sirotkin AV, Ovcharenko D, Grossmann R, Lauková M & Mlyncek M 2009. Identification of miRNAs controlling human ovarian cell steroidogenesis via a genome-scale screen. Journal of Cellular Physiology 219 415–420. ( 10.1002/jcp.21689) [DOI] [PubMed] [Google Scholar]
- Sirotkin AV, Laukova M, Ovcharenko D, Brenault P & Mlynček M 2010. Identification of miRNAs controlling human ovarian cell proliferation and apoptosis. Journal of Cellular Physiology 223 49–56. ( 10.1002/jcp.21999) [DOI] [PubMed] [Google Scholar]
- Sirotkin AV, Kisová G, Brenaut P, Ovcharenko D, Grossmann R & Mlyncek M 2014. Involvement of MiRNA15a in control of human ovarian granulosa cell proliferation, apoptosis, steroidogenesis, and response to FSH. MicroRNA 3 29–36. [DOI] [PubMed] [Google Scholar]
- Song J, Luo S & Li SW 2015. miRNA-592 is downregulated and may target LHCGR in polycystic ovary syndrome patients. Reproductive Biology 15 229–237. ( 10.1016/j.repbio.2015.10.005) [DOI] [PubMed] [Google Scholar]
- Sørensen AJ, Wissing ML, Salö S, Englund ALM & Dalgaard LT 2014. MicroRNAs related to polycystic ovary syndrome. Genes 5 648–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen AE, Udesen PB, Wissing ML, Englund ALM & Dalgaard LT 2016a. MicroRNA specifies in follicular fluid associating with polycystic ovary syndrome and related intermediary phenotypes. Journal of Clinical Endocrinology and Metabolism 101 1579–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen AE, Udesen PB, Wissing ML, Englund ALM & Dalgaard LT 2016b. MicroRNAs related to androgen metabolism and polycystic ovary syndrome. Chemico-Biological Interactions 259 8–16. ( 10.1016/j.cbi.2016.06.008) [DOI] [PubMed] [Google Scholar]
- Stewart PM & Newell-Price JDC 2016. Chapter 15: The adrenal cortex In Williams Textbook of Endocrinology, 13th ed, pp 490–555. Eds Melmed S, Polonsky KS, Larsen PR & Kronenberg HM. Philadelphia, PA: Elsevier; ISBN: 978–0-323–29738-7; OCLC Number: 1075067557. [Google Scholar]
- Tesfaye D, Gebremedhn S, Salilew-Wondim D, Hailay T, Hoelker M, Grosse-Brinkhaus C & Schellander K 2018. MicroRNAs: tiny molecules with a significant role in mammalian follicular and oocyte development. Reproduction 155 R121–R35. ( 10.1530/REP-17-0428) [DOI] [PubMed] [Google Scholar]
- Torrealday S, Kodaman P & Pal L 2017. Premature ovarian insufficiency: an update on recent advances in understanding and management. F1000Research 6 2069 ( 10.12688/f1000research.11948.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troppmann B, Kossack V, Schüring AN & Gromoll J 2014. MicroRNA513a-3p acts as a co-regulator of luteinizing hormone/chrionic gonadotropin receptor gene expression in human granulosa cells. Molecular and Cellular Endocrinology 390 65–72. [DOI] [PubMed] [Google Scholar]
- Tu J, Cheung AH-H, Chan CL-K & Chan WY 2019. The role of miRNAs in ovarian granulosa cells in health and disease. Frontiers in Endocrinology 10 174 ( 10.3389/fendo.2019.00174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker EJ, Grover SR, Bachelot A, Touraine P & Sinclair AH 2016. Premature ovarian insufficiency: new perspectives on genetic cause and phenotypic spectrum. Endocrine Reviews 37 609–635. ( 10.1210/er.2016-1047) [DOI] [PubMed] [Google Scholar]
- Turcu AF & Auchus RJ 2015. Adrenal steroidogenesis and congenital adrenal hyperplasia. Endocrinology and Metabolism Clinics of North America 44 275–296. ( 10.1016/j.ecl.2015.02.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcu A, Smith JM, Auchus R & Rainey WE 2014. Adrenal androgens and androgen precursors: definition, synthesis, regulation and physiologic actions. Comprehensive Physiology 4 1369–1381. ( 10.1002/cphy.c140006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virant-Klun I, Ståhlberg A, Kubista M & Skutella T 2016. MicroRNAs: from female fertility, germ cells, and stem cells to cancer in humans. Stem Cells International 2016 3984937 ( 10.1155/2016/3984937) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Graham I, Hastings R, Gunewardena S, Brinkmeier ML, Conn PM, Camper SA & Kumar TR 2015. Gonadotropic-specific deletion of Dicer results in severely suppressed gonadotropins and fertility defects. Journal of Biological Chemistry 290 2699–2714. ( 10.1074/jbc.M114.621565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li D, Zhang S, Xing Y, Gao Y & Wu J 2016a. MicroRNA-125a-5p induces mouse granulosa cell apoptosis by targeting signal transducer and activator of transcription 3. Menopause 23 100–107. ( 10.1097/GME.0000000000000507) [DOI] [PubMed] [Google Scholar]
- Wang L, Li C, Li R, Deng Y, Tan Y, Tong C & Qi H 2016b. MicroRNA-764–3p regulates 17β-estradiol synthesis of mouse ovarian granulosa cells by targeting steroidogenic factor-1. In Vitro Cellular and Developmental Biology: Animal 52 365–373. ( 10.1007/s11626-015-9977-9) [DOI] [PubMed] [Google Scholar]
- Wang M, Liu M, Sun J, Jia L, Ma S, Gao J, Xu Y, Zhang H, Tsang SY & Li X 2017. MicroRNA-27a-3p affects estradiol and androgen imbalance by targeting Creb1 in the granulosa cells in mouse polycystic ovary syndrome model. Reproductive Biology 17 295–304. ( 10.1016/j.repbio.2017.09.005) [DOI] [PubMed] [Google Scholar]
- Wang T, Liu Y, LV M, Xing Q, Zhang Z, He X, Xu Y, Wei Z & Cao Y 2019. MiR-323–3p regulates the steroidogenesis and cell apoptosis in polycystic ovary syndrome (PCOS) by targeting IGF-1. Gene 683 87–100. ( 10.1016/j.gene.2018.10.006) [DOI] [PubMed] [Google Scholar]
- Williams GH 2005. Aldosterone biosynthesis, regulation and classical mechanism of action. Heart Failure Reviews 10 7–13. ( 10.1007/s10741-005-2343-3) [DOI] [PubMed] [Google Scholar]
- Wu S, Sun H, Zhang Q, Jiang Y, Fang T, Cui I, Yang G & Hu Y 2015. MicroRNA-132 promotes estradiol synthesis in ovarian granulosa cells via translational repression of Nurr1. Reproductive Biology and Endocrinology 13 94 ( 10.1186/s12958-015-0095-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Linher-Melville K, Yang BB, Wu D & Li J 2011. Micro-RNA 378 (miR-378) regulates ovarian estradiol production by targeting aromatase. Endocrinology 152 3941–3951. ( 10.1210/en.2011-1147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Zhang YW, Tong XH & Liu YS 2015. Characterization of microRNA profile in human cumulus granulosa cells: identification of microRNAs that regulate Notch signaling and are associated with PCOS. Molecular and Cellular Endocrinology 404 26–36. ( 10.1016/j.mce.2015.01.030) [DOI] [PubMed] [Google Scholar]
- Xu MQ, Jiang H, Zhang LQ, Sun XL, Luo D, Fu Y, Gao Y, Yuan B & Zhang JB 2018. MiR-29b affects the secretion of PROG and promotes the proliferation of bovine corpus luteum cells. PLoS ONE 13 e0195562 ( 10.1371/journal.pone.0195562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Zhang L, Fang T, Zhang Q, Wu S, Jiang Y, Sun H & hu Y 2012. MicroRNA-145 suppresses mouse granulosa cell by targeting activin receptor IB. FEBS Letters 586 3263–3270. ( 10.1016/j.febslet.2012.06.048) [DOI] [PubMed] [Google Scholar]
- Yang X, Zhou Y, Peng S, Wu L, Lin HY, Wang S & Wang H 2012. Differentially expressed plasma microRNAs in premature ovarian failure patients and the potent regulatory function of mir-23a in granulosa cell apoptosis. Reproduction 144 235–244. ( 10.1530/REP-11-0371) [DOI] [PubMed] [Google Scholar]
- Yao G, Yin M, Lian J, Tian H, Liu L, Li X & Sun F 2010a. MicroRNA-224 is involved in transforming growth factor-β-mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Molecular Endocrinology 24 540–551. ( 10.1210/me.2009-0432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N, Yang BQ, Liu Y, Tan XY, Lu CL, Yuan XH & Ma X 2010b. Follicle-stimulating hormone regulation of microRNA expression on progesterone production in cultured rat granulosa cells. Endocrine 38 158–166. ( 10.1007/s12020-010-9345-1) [DOI] [PubMed] [Google Scholar]
- Yao W, Pan Z, Du X, Zhang J & Li Q 2018. miR-181b-induced SMAD7 downregulation controls granulosa cell apoptosis through TGF-β signaling by interacting with the TGFBR1 promoter. Journal of Cellular Physiology 233 6807–6821. ( 10.1002/jcp.26431) [DOI] [PubMed] [Google Scholar]
- Yin M, Lü M, Yao G, Tian H, Lian J, Liu L, Liang M, Wang Y & Sun F 2012. Transactivation of microRNA-383 by steroidogenic factor-1 promotes estradiol release from mouse granulosa cells by targeting RBMS1. Molecular Endocrinology 26 1129–1143. ( 10.1210/me.2011-1341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M, Wang X, Yao G, Lü M, Liang M, Sun Y & Sun F 2014. Transactivation of microRNA-320 by microRNA-383 regulates granulosa cell functions by targeting E2F1 and SF-1 proteins. Journal of Biological Chemistry 289 18239–18257. ( 10.1074/jbc.M113.546044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RMK, Chaturvedi G, Tong SKH, Nusrin S, Giesy JP, Wu RSS & Kong RYC 2015. Evidence for microRNA-mediated regulation of steroidogenesis by hypoxia. Environmental Science and Technology 49 1138–1147. ( 10.1021/es504676s) [DOI] [PubMed] [Google Scholar]
- Yu C, Li M, Wang Y, Liu Y, Yan C, Pan J, Liu J & Cui S 2016. Mir-375 mediates CRH signaling pathway in inhibiting E2 synthesis in porcine ovary. Reproduction 153 63–73. ( 10.1530/REP-16-0323) [DOI] [PubMed] [Google Scholar]
- Zhang C, Wang H, Yan C, Gao X & Ling X 2017a. Deregulation of RNX2 by miRNA-320a deficiency impairs steroidogenesis in cumulus granulosa cells from polycystic ovary syndrome (PCOS) patients. Biochemical and Biophysical Research Communications 482 1469–1476. ( 10.1016/j.bbrc.2016.12.059) [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang X, Zhang X, Lu Y, Li L & Cui S 2017b. MiRNA-143 mediates the proliferative signaling pathway of FSH and regulates estradiol production. Journal of Endocrinology 234 1–14. ( 10.1530/JOE-16-0488) [DOI] [PubMed] [Google Scholar]
- Zhang Z, Chen CZ, Xu MQ, Zhang LQ, Liu JB, Gao Y, Jiang H, Yuan B & Zhang JB 2019a. MiR-31 and MiR-143 affect steroid hormone synthesis and inhibit cell apoptosis in bovine granulosa cells through FSHR. Theriogenology 123 45–53. ( 10.1016/j.theriogenology.2018.09.020) [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu Y, Liu H & Pan Z 2019b. MicroRNAs in ovarian follicular atresia and granulosa cell apoptosis. Reproductive Biology and Endocrinology 17 9 ( 10.1186/s12958-018-0450-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Miao Y, Li Y, Li X, Xi J & Zhang Z 2019. MicroRNA-150 promote apoptosis of ovine granulosa cells by targeting STAR gene. Theriogenology 127 66–71. ( 10.1016/j.theriogenology.2019.01.003) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.