Introduction
Research assessing the feasibility and value of incorporating genomic information into patient care is increasingly being reported. Published advances in the implementation of genomic medicine are reviewed on a monthly basis by the Genomic Medicine Working Group of the National Advisory Council for Human Genome Research of the National Human Genome Research Institute (NHGRI). Publications to be considered are identified by review of the contents of leading journals and by PubMed search for the terms “genomic medicine,” “genomic implementation,” and “evidence.” Emphasis is placed on papers focusing on health and/or cost-effectiveness outcomes. Publications viewed as having robust findings and broad implications for using genomics in clinical care are listed on NHGRI’s “Accomplishments in Genomic Medicine” site (see Web Resources).
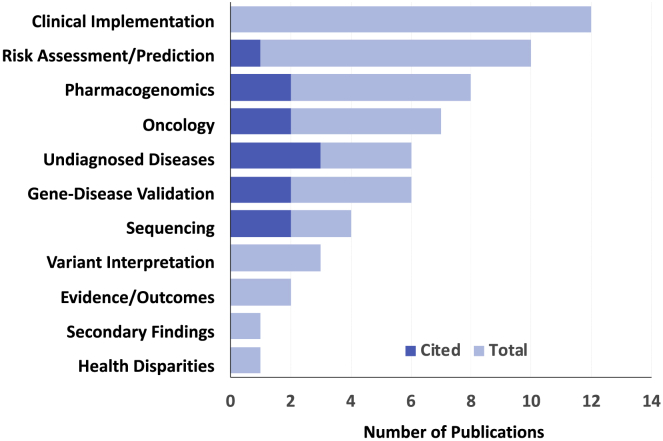
The Working Group defines genomic medicine as “an emerging medical discipline that involves using genomic information about an individual as part of their clinical care and the health outcomes and policy implications of that clinical use” (see Web Resources). In September 2019, the Working Group attempted to identify in its view the ten most significant advances among 48 recognized “accomplishments” published during the 12 months ending August 31, 2019. These papers clustered into 11 broad categories (Figure 1); 12 papers spanned two categories. Six admittedly subjective criteria were used (Box 1) in the selection of the ten papers cited for this 2019 review. Brief summaries, ordered by the papers’ dates of publication, are provided below.
Figure 1.
Topics Addressed by 48 “Accomplishments in Genomic Medicine”
Papers published in the 12 months ending August 30, 2019; 12 papers spanned two categories (most often oncology plus one other). Of ten papers cited in this review (dark blue), two spanned two categories.
Box 1. Criteria for Inclusion of Papers in Genomic Medicine Year in Review 2019.
-
•
Involve use of patients’ individual genomic variant information in clinical decision making
-
•
Demonstrate impact of direct clinical implementation
-
•
Are likely to be generalizable beyond original setting
-
•
Are likely to have implications for healthcare systems or practice guidelines
-
•
Are of sufficient size to be robust to sampling error
-
•
Are broadly representative of the field beyond NHGRI-sponsored or US-funded programs
Scaling up Variant Classification with Saturation Editing
Findlay, G.M., et al. (2018), Nature, 562, 217–22
Uncertainty about the functional consequences and disease relevance of gene variants remains a key challenge for the interpretation of genetic testing data and its translation to patient care. Findlay et al. applied saturation genome editing (SGE) to BRCA1 to measure the functional impact of nearly 4,000 variants. The team used CRISPR-Cas9 to generate all possible single-nucleotide variants in the 13 exons encoding the RING and BRCT domains in a haploid cell line and measured the functional effect of each variant by assessing cell survival. The resulting functional classifications were more than 95% accurate when compared to known pathogenic and benign variants annotated in NCBI’s ClinVar database, providing strong functional evidence that will help to improve interpretations for variants of uncertain significance and novel variants. SGE represents an innovative and scalable in vitro framework for characterizing risk associated with variants in many genes.
Narrowing the List of Genes Associated with Sudden Death
Hosseini, S.M., et al., Circulation. (2018) 138, 1195–1205
Brugada syndrome is a potentially lethal, heritable disorder characterized by susceptibility to sudden cardiac death, currently preventable only by implantation of an internal cardiac defibrillator. Multiple genes have been associated with this disorder and are included in the commercial testing panels often used as indications for defibrillator implantation, but levels of evidence for these associations varies. In this study, three independent curation groups—each comprising a medical geneticist and individuals with graduate degrees in genetics—used a standardized, ClinGen-derived semiquantitative scoring system to evaluate the strength of these associations with sudden arrhythmic death. Of 21 genes evaluated, biocurators classified only one (SCN5A) as having definitive evidence for pathogenicity; all other genes had only limited evidence, demonstrating that the strength of gene-disease associations should be critically evaluated before genes are incorporated into clinical testing.
What We’re Missing: Most BRCA1 and BRCA2 Variant Carriers Are Undetected
Manickam, K., et al. (2018). JAMA Netw Open. 1, e182140
BRCA1 and BRCA2, genes associated with hereditary breast/ovarian cancer (HBOC) syndrome, have a high degree of sequence variation, but the population prevalence of pathogenic or likely pathogenic (P/LP) variants is not known. Manickam and colleagues studied this in 50,000+ participants with exome sequences from the MyCode Community Health Initiative. The frequency of P/LP variants in this predominantly European ancestry population was 1 in 190, significantly higher than prior estimates. Participants carrying a P/LP variant had 5.95-fold increased odds of a personal history of breast cancer and 18.3-fold increased odds of ovarian cancer in comoparison to matched participants without a variant. Most importantly, almost half of all variant carriers did not meet current criteria for clinical testing, and of those meeting testing criteria, nearly half had not undergone clinical testing. Thus, three quarters of at-risk women were not identified as such and are not benefiting from evidence-based interventions; this is a significant care gap with implications for population health.
Cascade Screening for Familial Hypercholesterolemia
Alver, M., et al., (2019). Genet. Med. 21, 1173–1180
Health outcomes and the impact of leveraging population-based biobanks with integrated electronic health records to recall probands and undertake cascade screening for familial hypercholesterolemia (FH)-associated genomic variants are poorly understood. In a large biobank of more than 52,000 Estonians, of whom 4,776 had exome or whole-genome sequences, Alver and colleagues identified and recalled 27 individuals carrying FH-associated variants in LDLR, APOB, or PCSK9. Cascade screening of 64 family members identified an additional 20 carriers of FH-associated variants. Of the 41 FH variant carriers identified and further examined, 51% were reclassified from nonspecific hypercholesterolemia to definite FH; another 32% had been unrecognized by the medical system as being at high risk. Imaging-based risk stratification identified subclinical disease, and thus an indication for statin treatment, in 86% of the variant carriers. Genotype-guided recall of probands and subsequent cascade screening for FH is feasible within a population-based biobank and could facilitate more appropriate clinical management.
Positive Clinical Impact of Systematic Genomic Assessments in Patients Undergoing a Diagnostic Odyssey
Splinter, K., et al. (2018). N. Engl. J. Med. 379, 2131–2139
The clinical and practical impact of reaching a diagnosis in previously undiagnosed patients is poorly understood. The Undiagnosed Diseases Network (UDN), a multidisciplinary collaboration evaluating patients who have with complex presentations and have remained undiagnosed despite extensive clinical investigation, performed in-depth clinical evaluations along with exome and whole-genome sequencing, metabolomics testing, and studies in model organisms. From 2015–2017, the UDN accepted 601 of 1519 patients (321 female; 350 < 18 years) referred for evaluation. Of the first 382 patients with a completed evaluation, 132 (35%) received a diagnosis; these included 31 (11%) with new syndromes. Among diagnosed patients, the majority (58%) had medical-care changes, such as changes in therapy or shortening of the diagnostic odyssey. UDN features such as access to genome sequencing, systematic phenotyping, and secure data sharing could be implemented more broadly, but the scalability and sustainability of these methods remains to be demonstrated.
DNA Methylation Testing Improves Diagnosis Rate
Aref-Eshghi, E., et al. (2019). Am. J. Hum. Genet. 104, 685–700
The potential value of genome-wide DNA methylation in improving the current 42%–62% diagnostic yield of genomic sequencing and gene panels for neurodevelopmental presentations and congenital abnormalities (ND/CA) has not been explored. Aref-Eshghi and colleagues used genomic DNA methylation analysis to generate a computational model for diagnosing 14 ND/CA syndromes. When they applied this model to 67 individuals with suspected ND/CA, 31% showed a methylation profile consistent with the modeled syndromes; many of these individuals had genomic variants of unknown significance, whereas provisional diagnoses were ruled out in others, and still others received diagnoses not previously considered. When the model was applied to 965 ND/CA patients without a prior diagnosis, 27 were shown to have Mendelian or trinucleotide repeat disorders, and 106 had rare epi-variants, several on genes linked to the individual’s clinical disorder. Genomic DNA methylation analysis can facilitate molecular diagnosis of unresolved clinical cases and could be useful in clinical assessment of ND/CAs.
Rapid Genetic Diagnosis in Critically Ill Infants
Clark, M.M., et al. (2019). Sci. Transl. Med. 11. Published online April 24, 2019. 10.1126/scitranslmed.aat6177
Critically ill newborns suffering from serious undiagnosed conditions are increasingly being diagnosed by whole-genome sequencing, but this method can be too time consuming for the intensive-care unit (ICU) setting. Clark and colleagues described a rapid genome-sequencing approach that is linked to automated phenotyping interpretation and goes from blood draw to diagnosis in just over 20 h. They accomplished rapid automated phenotyping by using a mapping of Human Phenotype Ontology (HPO) terms to SnoMed-CT terms, enabling extraction from the health record of more than 25 times more HPO terms than manual extraction and providing greater information content and greater overlap with Online Mendelian Inheritance in Man (OMIM). Phenotypes ranked on their occurrence in OMIM were crossed with a ranked list of genomic variants to suggest a probability-ranked list of conditions. The automated system agreed with expert manual interpretation in 101 retrospective patients with 99% precision and correctly diagnosed 3 of 7 ICU patients prospectively, shaving 22 h off the manual system. Rapid genome sequencing with automated phenotyping expedites genomic diagnoses among critically ill infants, supporting broader adoption of this approach.
Cost-Effectiveness of DPYD Genotyping in Cancer Chemotherapy
Fragoulakis, V., et al. (2019). Am. J. Hum. Genet. 104, 1158–68
Dihydropyrimidine dehydrogenase (DPYD) is an important elimination enzyme for the fluoropyrimidine class of anticancer agents, variants in which can lead to altered drug efficacy and adverse drug reactions. The effectiveness and cost of assessing known variants producing low-activity products of DPYD were examined in 571 patients receiving a fluoropyrimidine agent. Outcomes included overall survival, occurrence of adverse drug reactions, and cost of resource utilization. DPYD normal metabolizers (528 individuals) had greater effectiveness (mean survival 7.13 versus 4.86 years) and lesser cost (mean €1,150 versus €3,712), representing a cost-saving option over DPYD intermediate and poor metabolizers (43 individuals) and mean quality-adjusted life years (QALYs) of 4.18 versus 3.02, respectively. Excess cost in the intermediate and poor metabolizers was due primarily to treatment of febrile neutropenia, mucositis, and diarrhea. DPYD-guided fluoropyrimidine treatment might represent a cost-saving choice for individuals undergoing fluoropyrimidine chemotherapy.
An Analyst’s Work Is Never Done
Liu. P., et al. (2019). N. Engl. J. Med. 380, 2478–2480
The rapid accrual of genomic knowledge can lead to changing interpretation of genomic variants, but the impact of reanalysis on diagnostic rates is not known. Liu and colleagues used a publicly available, semi-automated pipeline for reanalysis of exome sequence data to evaluate 250 cases from 2011–2012 (cohort 1) and 2,000 from 2012–2013 (cohort 2). Diagnostic rates increased from 24.8% to 46.8% in cohort 1 when manual clinical sign-out was used and from 25.2% to 36.7% in cohort 2 when the semi-automated approach conditioned on clinical phenotyping was used. Newly discovered gene-disease associations accounted for 75% of new findings in cohort 1 and 64% in cohort 2; new clinical information, variant reclassifications, copy-number variant (CNV) analyses, and family studies also contributed. Additionally, six molecular diagnoses were reversed, and dual genetic locus results increased. Benefits of periodic reanalyses of existing data are substantial, but integrating such analyses into healthcare will require reimbursement models that cover these additional costs.
CYP2C19 Genotyping in Antiplatelet Therapy
Martin, J., et al. (2019). Genet. Med. Published online July 18, 2019. 10.1038/s41436-019-0611-1
Availability of the CYP2C19 genotype could help patients to make decisions about antiplatelet therapy after percutaneous coronary intervention (PCI), but genotypes are often unknown at the time of the procedure. In the absence of pre-PCI genotyping, clinicians are increasingly prescribing prasugrel or ticagrelor, which could be switched to clopidogrel if CYP2C19 genotyping after PCI indicates normal metabolizer status. Martin and colleagues tested the impact of genotype-guided prescription advice on antiplatelet therapy switching in 1,063 individuals who had undergone PCI and who subsequently underwent CYP2C19 genotyping. Individuals who had high-risk CYP2C19 genotypes and were prescribed clopidogrel had a 2.89-fold-increased hazard of adverse events in comparison to those who were switched to prasugrel or ticagrelor. Those who did not have high-risk genotypes and switched to clopidogrel had outcomes similar to those of individuals who continued on prasugrel or ticagrelor. This study suggests that switching normal metabolizers to clopidogrel after PCI is safe, and it joins several others showing the benefits of pharmacogenetically based antiplatelet drug prescription.
Conclusion
Research into genomic medicine implementation is moving forward briskly; key themes this year focused on re-appraisal and re-analysis of genomic interpretations (Hosseini et al. and Liu et al.), functional interpretation of variants (Findlay et al.), advancing diagnoses in undiagnosed patients (Aref-Eshghi et al., Clark et al., and Splinter et al.), population screening for monogenic disorders (Alveret al. and Manickam et al.), and the impact of pharmacogenetic variants on treatment response (Fragoulakis et al. and Martin et al.). Of these, the approaches with the greatest potential for population health impact are most likely those involving common variants affecting common, serious diseases (Alver et al., Fragoulakis et al., Manickam et al., and Martin et al.), and the remainder provide new approaches for assessing rarer conditions or variants. Several (Splinter et al., Fragoulakis et al., Liu et al., and Martin et al.) assessed the impact of clinical testing on healthcare costs, another critical need for payer and policy-maker decision making.
Efforts to apply these approaches to other conditions and other settings to assess their generalizability will be needed. Replication and confirmation will be greatly facilitated by the public availability of the standardized gene-interpretation framework used by Hosseini et al., the mapping of Human Phenotype Ontology (HPO) to the Systematized Nomenclature of Medicine Clinical Terms (SNOMED-CT) provided by Clark et al., and the semi-automated pipeline for reanalysis of exome-sequence data produced by Liu et al. Additional populations should be screened for monogenic disease variants via the approaches of Alver et al. and Manickam et al., and additional pharmacogenes can be examined with the methods of Fragoulakis et al. and Martin et al. The groups represented by Aref-Eshghi and Splinter will most likely extend their studies to larger numbers of patients while others attempt to replicate their findings in other settings. Gene-specific functional assays based on saturation gene editing (Findlay et al.) are being developed and applied to other gene-disease entities, but demonstration of their clinical value might be a bit further off. Although all of these papers represent significant advances in the application of genomic medicine, they all will need expansion and dissemination if we are to realize the promise of genomics for improving patient care.
Web Resources
Genomic Medicine Working Group Accomplishments in Genomic Medicine, https://www.genome.gov/health/Genomics-and-Medicine/accomplishments
NHGRI definition of genomic medicine, https://www.genome.gov/health/Genomics-and-Medicine
ClinGen-derived semiquantitative scoring system, https://www.clinicalgenome.org/docs/summary-of-updates-to-the-clingen-gene-clinical-validity-curation-sop-version-7/
MyCode Community Health Initiative, https://www.geisinger.org/mycode