Abstract
Emerging evidence demonstrates that microRNAs (miRNAs) could serve as reliable biomarkers of inflammation and oncogenesis. The aim of this study was to determine whether miR-23a and miR-181b were suitable as biomarkers of irritable bowel syndrome (IBS) and colorectal cancer (CRC). Forty patients with IBS (29 females, 11 males), 33 with CRC (14 females, 19 males), and 33 healthy controls (17 females, 16 males) were prospectively included. Serum levels of miRNAs were evaluated by quantitative real-time PCR. The serum levels of miR-23a and miR-181b were significantly higher in the IBS group (p = 0.0009 and 0.004, respectively) and CRC group (p = 0.002 and 0.029, respectively) than in the control group. Serum levels of miR-23a and miR-181b were upregulated in CRC vs. IBS, but the differences did not reach statistical significance (p = 0.169 and 0.179, respectively). The miRNet and Reactome databases identified phosphatase and tensin homolog as a major common pathway, indicating inflammation as a central hallmark. Although miRNAs could serve as reliable biomarkers in clinical practice, future studies are needed to establish appropriate cut-off limits.
Keywords: Biomarker, colorectal cancer, irritable bowel syndrome, microRNA, miRNA, miR-23a, miR-181b, inflammation
INTRODUCTION
Irritable bowel syndrome (IBS) is a common functional gastrointestinal disorder [1] that is diagnosed based on the Rome criteria [2]. Although the Rome IV criteria were released in 2016 [3], the Rome III criteria are still frequently used for the diagnosis of IBS [2]. According to the Rome III criteria, IBS is characterized by recurrent abdominal pain or discomfort, 3 days per month over the last 3 months (12 weeks), associated with two or more of the following criteria with symptom onset for at least 6 months: 1) improved defecation, 2) onset associated with a change in stool frequency, and/or 3) onset associated with a change in stool form (appearance) [2-4]. IBS is still considered a multifactorial disease, although some pathogenic mechanisms have been proposed, including low-grade inflammation, which might be immune activation, rather than a true inflammatory state [5].
The involvement of inflammation in colorectal cancer (CRC) and inflammatory bowel disease (IBD) is well known [6,7]. Systemic inflammation has been suggested to be predictive of the survival of patients with various solid tumors [6]. Therefore, efforts have been made in order to develop scores with dual purposes: to measure systemic inflammation and to predict prognosis [8].
Although moderate and severe forms of IBD are easier to diagnose, the differential diagnosis of milder forms of IBD is often difficult, requiring multiple investigations [9]. Overlapping symptoms attributable to IBS, which have also been reported for milder forms of IBD in remission [7], are key factors that generate anxiety and the “defensive” attitude of the physician toward excluding organic diseases. Hence, a countable non-invasive biomarker is needed to differentiate the two entities because of the differences in disease onset, management, and prognosis. In search for a reliable biomarker, researchers have shifted from biochemical assessment toward more sophisticated approaches, such as genomics, for the assessment of coding and non-coding genes.
Because of intensive research, there has been much progress in understanding the functions of small, non-coding RNAs [10], such as microRNAs (miRNAs), which have been found to mediate mRNA cleavage, translational repression, and mRNA destabilization [11-13]. The dysregulation of miRNAs involved in crucial cellular processes has been described in many cell types in different diseases [14,15]. Various miRNAs can also have a tumor suppressor or oncogenic role [13]. Emerging evidence demonstrates that miRNAs could serve as reliable biomarkers [16,17] because of the high stability of these molecules in biological fluids, including serum/plasma [18]. The identification of circulating miRNA species as non-invasive biomarkers has led to significant progress in the treatment of a wide range of pathologies. The main advantage of miRNAs is the relatively high stability of the molecular structures in a wide range of biological specimens, including serum [13].
Although the utility of miRNAs as biomarkers has been investigated in a large spectrum of conditions, relatively few studies have reported miRNA expression patterns in IBS [19-22]. Hence, besides cytokines and gene polymorphisms, further studies of miRNA expression patterns in intestinal biopsy samples are needed to identify a reliable biomarker to easily discriminate between IBS and non-IBS diseases, such as IBD.
The miRNAs miR-23a and miR-181b have been investigated as potential biomarkers of carcinogenesis and inflammatory pathways but not IBS [23,24]. Moreover, recent data suggest that diet could induce changes to the gut microbiome, leading to the upregulation of specific miRNAs, such as miR-23a [25]. Intensive research concerning the role of miR-23a and miR-181b as therapeutic targets or early prognostic biomarkers of CRC has been ongoing, but few studies have investigated possible biomarkers of IBS.
Therefore, the aim of the present study was to assess the serum expression patterns of miR-23a and miR-181b as potential biomarkers in IBS and CRC and to evaluate potential correlations.
MATERIALS AND METHODS
Study approval
The study protocol was approved by the Ethical Committee of the “Iuliu Hatieganu” University of Medicine and Pharmacy (Cluj-Napoca, Romania; approval no. 24/2015) and conducted in accordance with the tenets of the Declaration of Helsinki and the Convention for the protection of Human Rights and Dignity of the Human Being with regard to the Application of Biology and Medicine: Convention on Human Rights and Biomedicine. All participants were informed about the study protocol and signed a consent form prior to inclusion.
Study design
This prospective, observational, pilot study was conducted in two teaching tertiary care units in Cluj-Napoca, Romania, between 2014 and 2015. The sample size was estimated based on previously published studies and adjusted in order to be considered appropriate for preliminary results and subsequent validation of data obtained from a larger group [26].
Study group
The study group included a total of 73 patients: 33 with CRC diagnosed by endoscopy plus biopsy and 40 with IBS diagnosed according to Rome III criteria. Three IBS subtypes were classified according to Rome III criteria: IBS with diarrhea (IBS-D), IBS with constipation (IBS-C), or mixed IBS (IBS-M). Patients with other confounding conditions, such as inflammatory status, severe organic diseases, and the use of anti-inflammatory drugs 1 month prior to inclusion, were excluded. Patients with IBS were also subdivided into post-infectious (PI-IBS) and non-post-infectious (non-PI-IBS) groups, according to previous methodology [27], based on the onset of IBS symptoms. Patients with symptoms occurring after a triggering event, such as an acute episode of gastroenteritis (nausea, vomiting, and diarrhea), were assigned to PI-IBS group.
The CRC group consisted of 33 patients diagnosed with CRC according to the tumor-node-metastasis (TNM) staging system of the Sixth Edition of the American Joint Commission. The subjects were diagnosed by lower endoscopy and histopathological assessment of biopsy samples and were included prior to any chemotherapy, immunomodulatory treatment, or radiotherapy. Information regarding vital status was obtained from death records from the National Population Register. The epidemiological and clinical variables recorded were age at diagnosis, sex, tumor stage, presence of lymph node invasion, presence of metastases, and grade of histopathological differentiation.
Thirty-three consecutive healthy controls were recruited by a similar approach, including local advertisements. Exclusion criteria for the control group were alcohol and substance abuse or dependence, presence of severe organic disorders, and presence of digestive symptoms or conditions.
After fasting for a minimum of 8 h, a venous blood sample (0.5 mL) was collected from patients and controls between 07:00 and 11:00 h and stored at −80°C within 30 min in the same facility until miRNA quantification. Freeze–thaw cycles were avoided until the assessment of all samples.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of serum miRNAs
Total RNA from 400 µL of serum was extracted using TRIzol™ reagent (Invitrogen Corporation, Carlsbad, CA, USA) based on TRI Reagentâ protocol, eluted in 10 µL of RNase-free water, and stored at −80°C until analysis. The total RNA concentration was determined using a NanoDrop ND-200 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and the quality of RNA was assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Serum levels of miR-23a (cat. no. 4427975; Applied Biosystems, Carlsbad, CA, USA) and miR-181b (cat. no. 440886; Applied Biosystems) were evaluated by qRT-PCR with the use of the TaqMan™ Advanced miRNA cDNA Synthesis Kit (cat. no. A28007; Applied Biosystems), TaqMan™ Fast Advanced Master Mix (cat. no. 4444557; Applied Biosystems), and specific TaqMan Primers. U6 (RNU6B; cat. no. 4427975; Applied Biosystems) was used for normalization of the data based on 2−ΔΔCT method as described previously [28].
Statistical analysis
Statistical analysis was conducted using GraphPad Prism software (version 6.0; GraphPad Software, Inc., San Diego, CA, USA). Both parametric and nonparametric data were compared. The Pearson’s correlation coefficient was determined. Statistical significance was determined with the Student’s t-test. A probability (p) value of <0.05 was considered statistically significant. The miRNet (https://www.mirnet.ca) database was used for identification of miRNAs as potential biomarkers [29], and the Reactome database (https://reactome.org/) was used for the classification of molecular pathways.
RESULTS
Baseline data
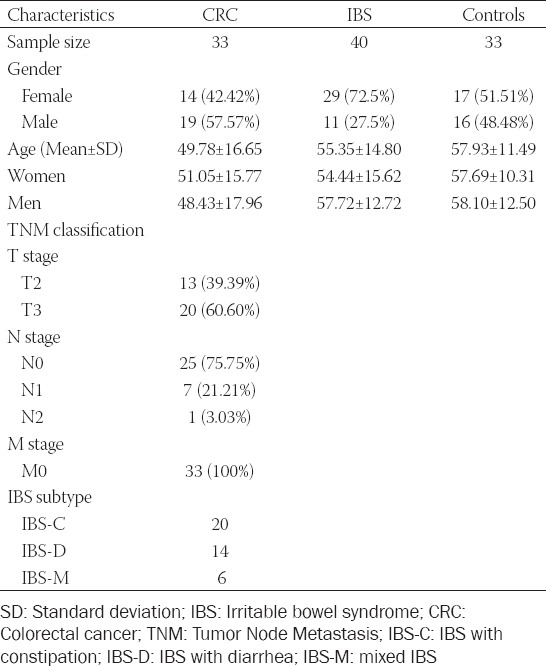
Thirty-three patients with CRC (14 females, 19 males; mean age, 57.93 ± 11.49 years), 40 with IBS (29 females, 11 males; mean age, 55.35 ± 14.80 years), and 33 consecutive healthy controls (17 females, 16 males; mean age, 49.78 ± 16.65 years) were included for analysis. The baseline data of the groups are presented as mean values in Table 1. The male: female ratio in the IBS group was 2.63. Twenty patients were diagnosed with IBS-C, 14 with IBS-D, and six with IBS-M. In the IBS group, eight (20%) patients were diagnosed with PI-IBS, of which five (62.5%) were female. The TNM classifications of patients in the CRC group are also presented in Table 1.
TABLE 1.
Main characteristics of the groups and TNM indicators for the CRC group
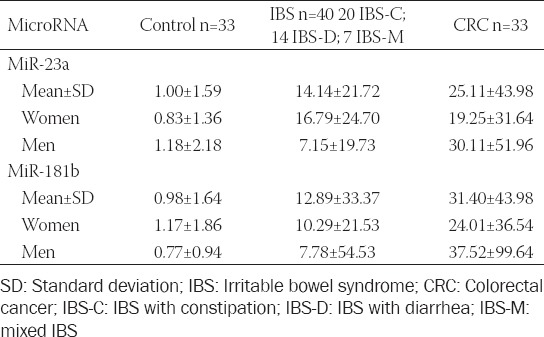
The mean serum miRNA levels in the IBS, CRC, and healthy control groups are shown in Table 2.
TABLE 2.
Serum levels for miR‑23a and miR‑181b in patients and a healthy control group
As shown in Figure 1, serum levels of miR-23a and miR-181b were significantly greater in the IBS group as compared with the control groups (p = 0.0009 and 0.004, respectively) and significantly greater in the CRC group than in the control group (p = 0.002 and 0.029, respectively).
FIGURE 1.
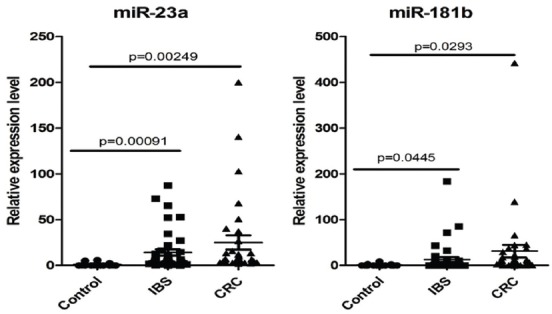
TaqMan relative expression level in serum for miR-23a and miR-181b, data normalized to U6 and expressed as fold change for controls, IBS and CRC. IBS: Irritable bowel syndrome; CRC: Colorectal cancer.
Regarding serum levels of the miRNAs in patients with CRC vs. IBS, the data suggest that miR-23a and miR-181b levels were comparatively upregulated in patients with CRC, but these differences did not reach statistical significance (p = 0.169 and 0.179, respectively). Serum levels of miR-23a and miR-181b tended to be higher in patients with stage T3 CRC, as compared with stage T2 (p = 0.223 and 0.334, respectively).
Of the CRC patients, 72.75% were classified as stage N0. There were no patients with metastatic disease (M0, 100%).
Regarding the IBS subtypes, miR-181b expression was significantly higher in IBS-D than in IBS-C and IBS-M (p = 0.033; Figure 2).
FIGURE 2.
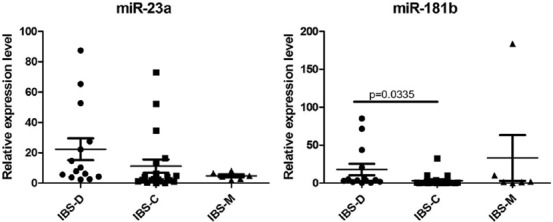
TaqMan relative expression level in serum for miR-23a and miR-181b, data normalized to U6 and expressed as fold change in IBS subtypes. IBS: Irritable bowel syndrome; IBS-C: IBS with constipation; IBS-D: IBS with diarrhea; IBS-M: mixed IBS.
There were no significant differences in any of the other comparisons (IBS-D vs. IBS-M, IBS-M vs. IBS-D, and PI-IBS vs. non-PI-IBS; Figure 3 and Table 3).
FIGURE 3.
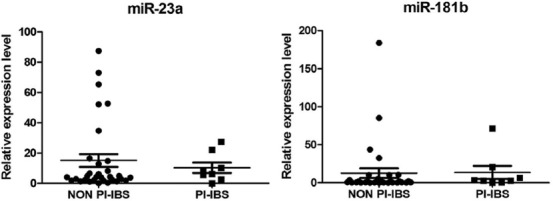
TaqMan relative expression level in serum for miR-23a and miR-181b, data normalized to U6 and expressed as fold change in PI-IBS and non PI-IBS. IBS: Irritable bowel syndrome; PI: Postinfectious.
TABLE 3.
Comparison of IBS subtypes and correlation established for miR‑23a and miR‑181b
The expression levels of miR-23a and miR-181b were positively correlated in the control group (p = 0.001) but not in the IBS and CRC groups (p = 0.208 and 0.156, respectively).
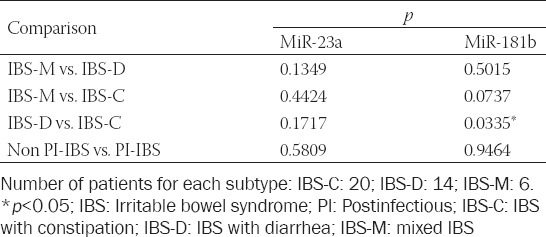
To analyze the link between serum levels of miR-23a and miR-181b with IBS and with CRC, correlation and receiver operating characteristic (ROC) curve analyses were performed. ROC curves of miR-23a and miR-181b in IBS and CRC are presented in Figure 4. ROC analysis was performed to assess the diagnostic value of both miRNAs. The areas under the ROC curves for miR-23a in IBS and CRC were 0.886 (95% confidence interval [CI] = 0.808–0.964, p < 0.001) and 0.961 (95% CI = 0.9185–1.004, p < 0.001), respectively, indicating that miR-23a could be used as a reliable biomarker for the differential diagnosis of these entities.
FIGURE 4.
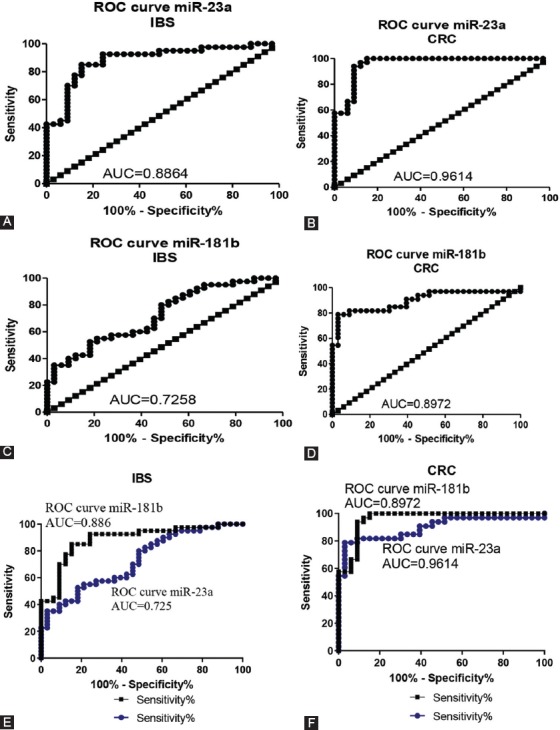
ROC for miR-23a and miR -181b in IBS (A-B) and in CRC (C-D) patients. ROC analysis and ROC curves for both miR in IBS (E) and CRC (F) patients. IBS: Irritable bowel syndrome; CRC: Colorectal cancer; ROC: Receiver operating characteristic; AUC: Area under the curve.
The integration-involvement of the most relevant target genes in biological processes based on the miRNet and Reactome databases is shown in Figure 5. The interrelations of miR-23a and miR-181b with other miRNAs and targeted genes and their products are integrated in a complex network of biological processes.
FIGURE 5.
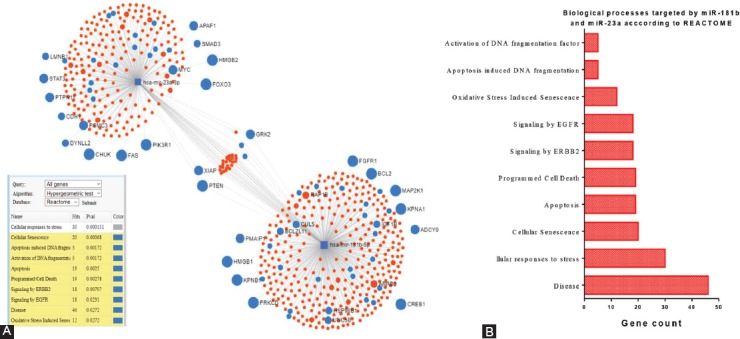
The miR-23a-miR-181b-mRNA network generated using miRNet. (A) These transcripts are presented as a representative node, (B) targeting important genes representative for important biological process presented in the panel.
DISCUSSION
IBS is a common functional gastrointestinal disorder worldwide [1]. Diagnosis of IBS is established based on clinical criteria, rather than exclusion [2,4]. In clinical practice, differential diagnosis implies multiple investigations; some of which are invasive. The correlation between inflammation and IBD is well known, as is the correlation between inflammation and IBS and oncogenesis. However, at present, there is no quantitative trait or biomarker to accurately quantify inflammation or a cut-off value for the risk of CRC.
Numerous investigations have supported the use of miRNAs as potential diagnostic and prognostic biomarkers of CRC and more recently for IBS. However, few studies have evaluated the use of miRNAs as biomarkers of IBS.
The stability of miRNAs in serum was demonstrated under various conditions, such as boiling, pH variations, extended storage, and freeze–thaw cycles. Notably, there were no significant differences in the expression levels of miRNAs under these conditions as compared with control (non-treated) serum [30]. A meta-analysis published in 2015 concluded that serum could be better for miRNA analysis than plasma for screening of CRC and that the use of multiple miRNAs, rather than just one, has higher predictive accuracy [31].
Although U6 is often used as an internal control, some authors report that this gene might be unsuitable since expression levels can vary with freeze–thaw cycles, but still is one of the most used internal control [32].
A recent study addressed the involvement of miRNAs in pain associated with IBS [21], as well as potential roles in other chronic pain conditions, in which miRNAs might modulate several genes associated with both the nociceptive and analgesic systems, which influence changes to neurons involved in the development or maintenance of pain.
The expression of miR-23a is reportedly significantly higher in CRC patients than in healthy controls (p < 0.0001) [33], which is in agreement with another study that also found higher expression levels in patients with CRC with microsatellite instability [34]. In addition, Yong et al. [16] reported that serum levels of miR-23b and miR-23a were greater in CRC patients than in healthy controls (p = 0.045) and proposed a combination of three biomarkers, including miR-23a, for screening of CRC [16]. They also found that miR-23b expression was upregulated in tissue and blood samples of CCR patients [16]. Recent data indicate that miR-23a might be related to inflammation and that miR-23a in combination with other three miRNAs was correlated with ulcerative colitis disease activity with higher sensitivity and specificity than C-reactive protein [35]. These data suggest that miR-23a could be used to discriminate between IBD and IBS, but a cut-off value must be determined.
To the best of our knowledge, this study is the first to investigate the use of serum miR-23a as a biomarker of IBS. The results of this study show that serum levels of miR-23a and miR-181b were significantly higher in patients with IBS than in healthy controls, emphasizing the point that although structural changes cannot be detected by current diagnostic tools and techniques in clinical practice (i.e., lower endoscopy and C-reactive protein), various mechanisms or epigenetic phenomena can lead to different clinical phenotypes.
Unfortunately, few studies have assessed serum levels of miR-181b, as most have focused on tissue samples either recent or archived. The use of miR-181b as a biomarker remains controversial because of its dual role in the pathology of various diseases [23,36,37].
There is evidence that miR-181b regulates nuclear factor-κB–mediated vascular inflammation in response to proinflammatory triggers [24]. Expression of miR-181b was about 12-fold higher than that of miR-181a and 274-fold higher than that of miR-181c by qRT-PCR [24]. The role of miR-181a in inflammation has already been suggested [38]. Dysregulation of the miR-181–phosphatase and tensin homolog (PTEN) axis has been suggested in the development of cancer and metabolic syndrome [39].
Differences in miRNA expression profiles between tissue and serum samples have been reported, which might explain our results. Our data indicate that miR-23a and miR-181b were overexpressed in stage T3 vs. T2 CRC, but this difference did not reach statistical significance (p = 0.223 and 0.334, respectively). It is possible that these miRNAs could have significant differences in stage T4 vs. T1.
Although the number of patients was limited, the aim of this pilot study was to assess serum levels of miR-23a and miR-181b in IBS patients as compared with CRC patients and healthy controls, as few studies have compared the expression profiles of potential biomarkers in IBS vs. IBD. The results of the present study revealed that although IBS is considered a functional disorder, there were differences in miRNA expression levels between the IBS and healthy control groups. These miRNAs have been proposed as biomarkers of inflammatory responses and tumorigenesis. The results of miRNet analysis revealed the complexity and interrelation of various pathways based on currently available information. Also, the miR-23a/miR-181b-mRNA network generated using the miRNet database identified PTEN as a major common pathway, which also noted inflammation as a central hallmark. These findings enhance prior findings that signal transducer and activator of transcription 3 activate miR-181b via PTEN and the cylindromatosis gene, as part of an epigenetic switch that links inflammation to cancer [23,40]. Our data indicate that miR-23a and miR-181b were significantly upregulated in IBS vs. healthy controls, supporting further investigation of these miRNAs in IBS and IBD patients. Our results also confirm those of previous studies that miR-23a and miR-181b were upregulated in patients with CRC vs. healthy controls [33].
CONCLUSION
The results of this study show that miRNA expression could provide a basis for future research of miRNAs as biomarkers for IBS and confirmed the findings of previous studies that miR-23a and miR-181b are upregulated in CRC, suggesting potential roles as biomarkers of CRC, although cut-off limits must be established.
ACKNOWLEDGMENTS
This study was funded by the European Social Fund, Human Resources Development Operational Programme 2007-2013, project no. POSDRU/159/1.5/S/138776 TRANSCENT [Institutional collaborative model of biomedical scientific research transposed in clinical practice – TRANSCENT]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The results of this study were firstly presented at Poster Section of The XXXVIth National Congress of Gastroenterology, Hepatology and Digestive Endoscopy.
Footnotes
Conflict of interest statement: The authors declare no conflict of interests
REFERENCES
- 1.Choung RS, Locke GR., 3rd Epidemiology of IBS. Gastroenterol Clin North Am. 2011;40:1–10. doi: 10.1016/j.gtc.2010.12.006. https://doi.org/10.1016/j.gtc.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480–91. doi: 10.1053/j.gastro.2005.11.061. https://doi.org/10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 3.Drossman DA, Hasler WL. Rome IV-Functional GI Disorders:Disorders of gut-brain interaction. Gastroenterology. 2016;150:1257–61. doi: 10.1053/j.gastro.2016.03.035. https://doi.org/10.1053/j.gastro.2016.03.035. [DOI] [PubMed] [Google Scholar]
- 4.Dumitrascu DL. Making a positive diagnosis of irritable bowel syndrome. J Clin Gastroenterol. 2011;45(Suppl):S82–5. doi: 10.1097/MCG.0b013e31821fbd5a. https://doi.org/10.1097/MCG.0b013e31821fbd5a. [DOI] [PubMed] [Google Scholar]
- 5.Bercik P, Verdu EF, Collins SM. Is irritable bowel syndrome a low-grade inflammatory bowel disease?Gastroenterol Clin North Am. 2005;34:235–45. doi: 10.1016/j.gtc.2005.02.007. vi-vii. https://doi.org/10.1016/j.gtc.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Roxburgh CS, McMillan DC. Role of systemic inflammatory response in predicting survival in patients with primary operable cancer. Future Oncol. 2010;6:149–63. doi: 10.2217/fon.09.136. https://doi.org/10.2217/fon.09.136. [DOI] [PubMed] [Google Scholar]
- 7.Keohane J, O'Mahony C, O'Mahony L, O'Mahony S, Quigley EM, Shanahan F. Irritable bowel syndrome-type symptoms in patients with inflammatory bowel disease:a real association or reflection of occult inflammation?Am J Gastroenterol. 2010;105:1788, 89-94;quiz 95. doi: 10.1038/ajg.2010.156. https://doi.org/10.1038/ajg.2010.156. [DOI] [PubMed] [Google Scholar]
- 8.Laird BJ, Kaasa S, McMillan DC, Fallon MT, Hjermstad MJ, Fayers P, et al. Prognostic factors in patients with advanced cancer:a comparison of clinicopathological factors and the development of an inflammation-based prognostic system. Clin Cancer Res. 2013;19:5456–64. doi: 10.1158/1078-0432.CCR-13-1066. https://doi.org/10.1158/1078-0432.CCR-13-1066. [DOI] [PubMed] [Google Scholar]
- 9.Schoepfer AM, Trummler M, Seeholzer P, Seibold-Schmid B, Seibold F. Discriminating IBD from IBS:comparison of the test performance of fecal markers, blood leukocytes, CRP, and IBD antibodies. Inflamm Bowel Dis. 2008;14:32–9. doi: 10.1002/ibd.20275. https://doi.org/10.1002/ibd.20275. [DOI] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V. The Celegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. https://doi.org/10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 11.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. https://doi.org/10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. https://doi.org/10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 13.Braicu C, Calin GA, Berindan-Neagoe I. MicroRNAs and cancer therapy - from bystanders to major players. Curr Med Chem. 2013;20:3561–73. doi: 10.2174/0929867311320290002. https://doi.org/10.2174/0929867311320290002. [DOI] [PubMed] [Google Scholar]
- 14.He L, Hannon GJ. MicroRNAs:small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. https://doi.org/10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 15.Berindan-Neagoe I, Calin GA. Molecular pathways:microRNAs, cancer cells, and microenvironment. Clin Cancer Res. 2014;20:6247–53. doi: 10.1158/1078-0432.CCR-13-2500. https://doi.org/10.1158/1078-0432.CCR-13-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yong FL, Law CW, Wang CW. Potentiality of a triple microRNA classifier:miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer. 2013;13:280. doi: 10.1186/1471-2407-13-280. https://doi.org/10.1186/1471-2407-13-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Q, Yang L, Larson S, Basra S, Merwat S, Tan A, et al. Decreased miR-199 augments visceral pain in patients with IBS through translational upregulation of TRPV1. Gut. 2015;65(5):797–805. doi: 10.1136/gutjnl-2013-306464. https://doi.org/10.1136/gutjnl-2013-306464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braicu C, Tomuleasa C, Monroig P, Cucuianu A, Berindan-Neagoe I, Calin GA. Exosomes as divine messengers:are they the Hermes of modern molecular oncology? Cell Death Differ. 2015;22:34–45. doi: 10.1038/cdd.2014.130. https://doi.org/10.1038/cdd.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fourie NH, Peace RM, Abey SK, Sherwin LB, Rahim-Williams B, Smyser PA, et al. Elevated circulating miR-150 and miR-342-3p in patients with irritable bowel syndrome. Exp Mol Pathol. 2014;96:422–5. doi: 10.1016/j.yexmp.2014.04.009. https://doi.org/10.1016/j.yexmp.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapeller J, Houghton LA, Monnikes H, Walstab J, Moller D, Bonisch H, et al. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17:2967–77. doi: 10.1093/hmg/ddn195. https://doi.org/10.1093/hmg/ddn195. [DOI] [PubMed] [Google Scholar]
- 21.Ciszek BP, Khan AA, Dang H, Slade GD, Smith S, Bair E, et al. MicroRNA expression profiles differentiate chronic pain condition subtypes. Transl Res. 2015;166(6):706–720. e11. doi: 10.1016/j.trsl.2015.06.008. https://doi.org/10.1016/j.trsl.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez C, Rodino-Janeiro BK, Lobo B, Stanifer ML, Klaus B, Granzow M, et al. miR-16 and miR-125b are involved in barrier function dysregulation through the modulation of claudin-2 and cingulin expression in the jejunum in IBS with diarrhoea. Gut. 2017;66:1537–8. doi: 10.1136/gutjnl-2016-311477. https://doi.org/10.1136/gutjnl-2016-311477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Huang SK, Zhao M, Yang M, Zhong JL, Gu YY, et al. Identification of a circulating microRNA signature for colorectal cancer detection. PLoS One. 2014;9:e87451. doi: 10.1371/journal.pone.0087451. https://doi.org/10.1371/journal.pone.0087451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun X, Icli B, Wara AK, Belkin N, He S, Kobzik L, et al. MicroRNA-181b regulates NF-kappaB-mediated vascular inflammation. J Clin Invest. 2012;122:1973–90. doi: 10.1172/JCI61495. https://doi.org/10.1172/JCI61495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohan M, Chow CT, Ryan CN, Chan LS, Dufour J, Aye PP, et al. Dietary gluten-induced gut dysbiosis is accompanied by selective upregulation of microRNAs with intestinal tight junction and bacteria-binding motifs in rhesus macaque model of celiac disease. Nutrients. 2016;8(11):pii:E684. doi: 10.3390/nu8110684. https://doi.org/10.3390/nu8110684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Q, Souba WW, Croce CM, Verne GN. MicroRNA-29a regulates intestinal membrane permeability in patients with irritable bowel syndrome. Gut. 2010;59:775–84. doi: 10.1136/gut.2009.181834. https://doi.org/10.1136/gut.2009.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.David LE, Surdea-Blaga T, Dumitrascu DL. Semiquantitative fecal calprotectin test in postinfectious and non-postinfectious irritable bowel syndrome:cross-sectional study. Sao Paulo Med J. 2014;133(4):343–9. doi: 10.1590/1516-3180.2014.8000815. https://doi.org/10.1590/1516-3180.2014.8000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ionescu C, Braicu C, Chiorean R, Cojocneanu Petric R, Neagoe E, Pop L, et al. TIMP-1 expression in human colorectal cancer is associated with SMAD3 gene expression levels:a pilot study. J Gastrointestin Liver Dis. 2014;23:413–8. doi: 10.15403/jgld.2014.1121.234.smad. https://doi.org/10.15403/jgld.2014.1121.234.smad. [DOI] [PubMed] [Google Scholar]
- 29.Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, Xia J. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44(W1):W135–41. doi: 10.1093/nar/gkw288. https://doi.org/10.1093/nar/gkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rome S. Use of miRNAs in biofluids as biomarkers in dietary and lifestyle intervention studies. Genes Nutr. 2015;10:483. doi: 10.1007/s12263-015-0483-1. https://doi.org/10.1007/s12263-015-0483-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng W, Tu Y, Zhu Y, Wang Z, Li C, Lao L, et al. Predictive power of circulating miRNAs in detecting colorectal cancer. Tumour Biol. 2015;36:2559–67. doi: 10.1007/s13277-014-2872-2. https://doi.org/10.1007/s13277-014-2872-2. [DOI] [PubMed] [Google Scholar]
- 32.Xiang M, Zeng Y, Yang R, Xu H, Chen Z, Zhong J, et al. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun. 2014;454:210–4. doi: 10.1016/j.bbrc.2014.10.064. https://doi.org/10.1016/j.bbrc.2014.10.064. [DOI] [PubMed] [Google Scholar]
- 33.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. https://doi.org/10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Liao D, Wang X, Wu Z, Nie J, Bai M, et al. Elevated microRNA-23a expression enhances the chemoresistance of colorectal cancer cells with microsatellite instability to 5-fluorouracil by directly targeting ABCF1. Curr Protein Pept Sci. 2015;16:301–9. doi: 10.2174/138920371604150429153309. https://doi.org/10.2174/138920371604150429153309. [DOI] [PubMed] [Google Scholar]
- 35.Polytarchou C, Oikonomopoulos A, Mahurkar S, Touroutoglou A, Koukos G, Hommes DW, et al. Assessment of circulating microRNAs for the diagnosis and disease activity evaluation in patients with ulcerative colitis by using the nanostring technology. Inflamm Bowel Dis. 2015;21(11):2533–9. doi: 10.1097/MIB.0000000000000547. https://doi.org/10.1097/MIB.0000000000000547. [DOI] [PubMed] [Google Scholar]
- 36.Seoudi AM, Lashine YA, Abdelaziz AI. MicroRNA-181a - a tale of discrepancies. Expert Rev Mol Med. 2012;14:e5. doi: 10.1017/S1462399411002122. https://doi.org/10.1017/s1462399411002122. [DOI] [PubMed] [Google Scholar]
- 37.Braicu C, Gulei D, Cojocneanu R, Raduly L, Jurj A, Knutsen E, et al. I miR-181a/b therapy in lung cancer:reality or myth? Mol Oncol. 2019;13:9–25. doi: 10.1002/1878-0261.12420. https://doi.org/10.1002/1878-0261.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–61. doi: 10.1016/j.cell.2007.03.008. https://doi.org/10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Henao-Mejia J, Williams A, Goff LA, Staron M, Licona-Limon P, Kaech SM, et al. The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity. 2013;38:984–97. doi: 10.1016/j.immuni.2013.02.021. https://doi.org/10.1016/j.immuni.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Molecular cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. https://doi.org/10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]


