Abstract
MicroRNAs (miRNAs) have been proven to regulate the development and progression of cancer through various mechanisms. The aim of the present study was to compare miRNA expression between primary melanomas from different sites. We analyzed the expression of 84 miRNAs in 27 primary melanoma and 5 nevus formalin-fixed paraffin-embedded (FFPE) samples using the Human Cancer PathwayFinder miScript miRNA PCR Array. The FFPE samples were obtained from the archives of the Municipal Clinical Emergency Hospital of Timisoara and included 10 cutaneous melanomas, 10 uveal melanomas, 7 mucosal melanomas, and 5 cutaneous nevi. Out of 84 miRNAs, 11 miRNAs showed altered expression in all types of melanoma compared with the nevi. Among these, miR-155-5p, miR-9-5p, miR-142-5p, miR-19a-3p, miR-134-5p, and miR-301a-3p were upregulated, while miR-205-5p, miR-203a-3p, miR-27b-3p, miR-218-5p, and miR-23b-3p were downregulated. The highest similarity in miRNA expression pattern was found between uveal and mucosal melanoma groups, i.e., 15 miRNAs had altered expression in both groups. Overall, we identified several miRNAs with significantly altered expression in primary melanomas, including those reported for the first time in this type of cancer. Among them, mir-9-5p, mir-203a-3p, mir-19a-3p, mir-27b-3p, and mir-218-5p showed altered expression in all three melanoma types vs. nevi. Further research should explore the potential of these miRNAs in melanoma.
Keywords: microRNA, primary melanoma, miScript miRNA PCR Array
INTRODUCTION
Primary melanoma, a malignant neoplasm of melanocytes, can be highly aggressive and has an increasing incidence worldwide [1]. In 2016, 76,380 new cases of melanoma were estimated in the United States (US) from which approximately 10,130 cases would be fatal [2]. Romania and other countries from Central and Eastern Europe show a higher incidence of 111 primary melanomas at advanced stage compared with Western Europe, which may be due to the lack of proper health education in these countries, among other reasons [3]. Based on this information, we consider melanoma to be one of the most important research areas in our country.
Melanoma can occur in any tissue that contains melanocytes. These cells are predominantly present in the skin, but they can also be found in organs such as the eyes, inner ear, and brain (the substantia nigra and locus coeruleus) as well as in the mucosal lining of the leptomeninges, oral cavity, esophagus, rectum, anal canal, nasal cavity, paranasal sinuses, larynx, vagina, and cervix [4-13]. In each of these tissues melanocytes have different functions and are influenced by different local factors [13].
Primary melanomas originating from different sites show characteristic tumor mutational burden [14-20]. According to the site of involvement, primary melanomas are classified as: cutaneous melanoma (CM), uveal melanoma (UM), mucosal melanoma (MM), and melanoma of the internal organs.
Numerous genomic studies showed different mutational patterns in melanoma, which was followed by the investigation of epigenetic factors involved in melanoma development. MicroRNAs (miRNAs) are small noncoding RNAs (∼22 nt in length) that can regulate the development and progression of cancer through various mechanisms. Different miRNAs have been shown to be upregulated or downregulated in melanoma, which suggests their use as diagnostic and prognostic biomarkers as well as therapeutic targets [21]. Furthermore, circulating miRNAs can be used for non-invasive diagnosis and prognosis of early metastatic disease, representing a more cost-effective method for monitoring patients and deciding about the treatment. These biomarkers have a higher sensitivity for detection in early stages of disease than the current imaging techniques (e.g., computed tomography [CT], positron emission tomography [PET] scan, etc.), and they are less invasive compared with tumor excision and sentinel lymph node biopsy, used for staging of melanoma [22].
Several studies investigated miRNA expression changes in melanomas, most notably in cutaneous and uveal types, however, there is little data on miRNA expression in primary mucosal melanoma. To the best of our knowledge, only two studies have investigated changes in miRNA expression in conjunctival melanomas [23,24] and none in mucosal melanomas involving other sites. Therefore, the current study is the first to analyze miRNA expression in primary mucosal melanomas involving mucosal surfaces other than the conjunctiva. Here, we compared miRNA expression among three primary melanoma types (CM, UM, and MM) and control cutaneous nevi. We identified several miRNAs with significantly altered expression in primary melanomas, including those reported for the first time in this type of cancer.
MATERIALS AND METHODS
Tissue samples
We obtained 27 primary melanoma and 5 nevus formalin-fixed paraffin-embedded (FFPE) samples from the archives of the Pathology Department at the Municipal Clinical Emergency Hospital of Timisoara. The FFPE samples included 10 cutaneous melanomas (stage III-IV), 10 uveal melanomas, 7 mucosal melanomas (2 intestinal mucosa, 2 genital mucosa, 1 nasal mucosa, and 2 oral mucosa) and 5 cutaneous nevi (Table 1). To reduce genomic and transcriptomic changes that occur because of environmental factors, we collected the control nevus samples from younger individuals.
TABLE 1.
Characteristics of patients with primary melanoma (n = 27) and controls (n = 5)

All participants signed informed consent to participate in the study, and the study was approved by the Institutional Ethics Committee (approval number 1-015922/2019). Patients did not receive any treatment prior to tumor excision.
Real-time polymerase chain reaction (real-time PCR)
MiRNAs were purified from FFPE samples using a miRNeasy FFPE Kit (Qiagen, MD, US), according to the manufacturer’s instructions. We analyzed the expression of 84 miRNAs using the Human Cancer PathwayFinder miScript miRNA PCR Array (Qiagen, MD, US), on an ABI 7900HT real-time PCR instrument (Thermo Fisher Scientific, MA, US).
Real-time PCR data were analyzed using the online QIAGEN GeneGlobe Data Analysis Center, with a threshold of 2 for fold changes of miRNA expression in melanoma samples. Afterwards, we compared these results with miRNA expression in control nevus samples. We corrected the results using the Benjamini-Hochberg procedure to account for any possible false discovery rate. Moreover, we used the Qiagen Ingenuity pathway analysis (IPA) tool to identify pathways and diseases associated with miRNAs with altered expression.
RESULTS
The distribution of upregulated and downregulated miRNAs in the three primary melanoma types compared with control nevi is presented in Figure 1-3. MiRNAs with most significant changes in their expression in the primary melanoma samples compared with nevus group are presented in Table 2. We used these results to generate a cluster dendrogram that highlights the segregation of miRNA expression according to the 4 studied groups (Figure 4). The samples clustered together for each melanoma type and for nevi, showing similar RNA expression patterns in all three repetitions of miRNA expression analysis.
FIGURE 1.
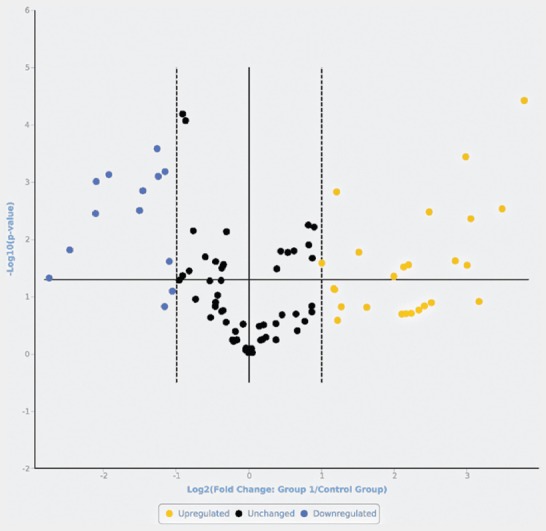
Volcano plot representing the microRNAs (miRNAs) in cutaneous melanomas compared with cutaneous nevi. Upregulated (yellow) and downregulated (blue) miRNAs with a fold change >2 vs. nevi, and miRNAs that were not significantly differently expressed (black, fold change <2 vs. nevi).
FIGURE 3.
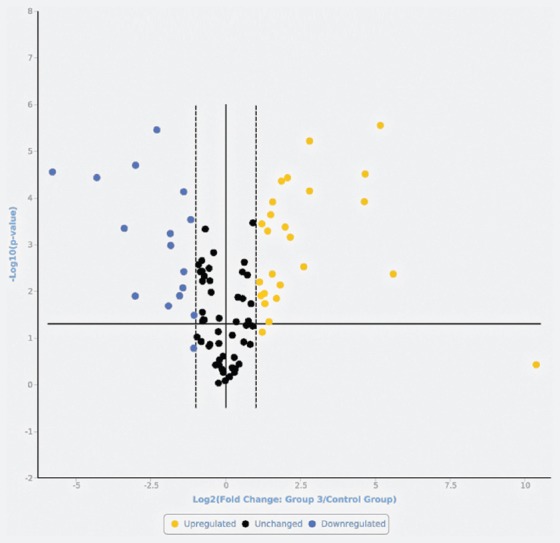
Volcano plot representing the microRNAs (miRNAs) in mucosal melanomas compared with cutaneous nevi. Upregulated (yellow) and downregulated (blue) miRNAs with a fold change >2 vs. nevi, and miRNAs that were not significantly differently expressed (black, fold change <2 vs. nevi).
TABLE 2.
MicroRNAs (miRNAs) with most significant changes in their expression in primary melanoma samples (n = 27) compared with cutaneous nevi (n = 5)
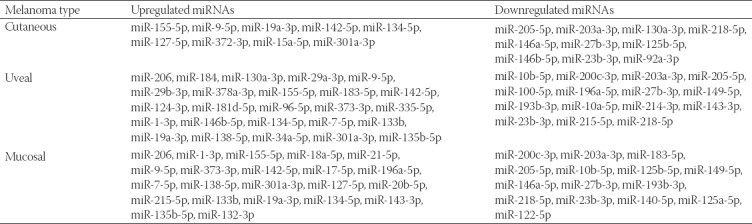
FIGURE 4.
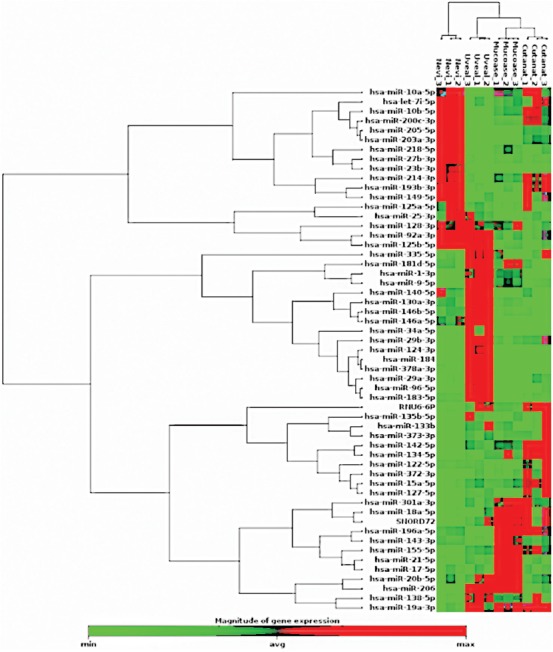
Cluster analysis of the changes in microRNA (miRNA) expression in 4 analyzed groups (cutaneous nevi, cutaneous melanoma, uveal melanoma, and mucosal melanoma). The samples (the top of the figure) clustered together for each melanoma type and for nevi, showing similar RNA expression patterns (the right of the figure) in all three repetitions of miRNA expression analysis.
FIGURE 2.
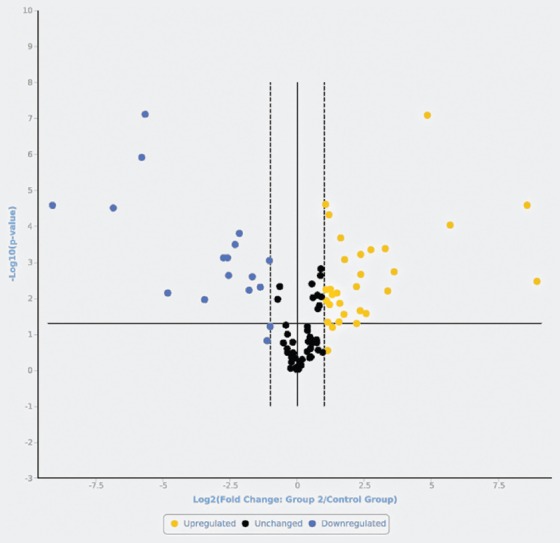
Volcano plot representing the microRNAs (miRNAs) in uveal melanomas compared with cutaneous nevi. Upregulated (yellow) and downregulated (blue) miRNAs with a fold change >2 vs. nevi, and miRNAs that were not significantly differently expressed (black, fold change <2 vs. nevi).
Out of 84 miRNAs, 11 miRNAs showed altered expression in all types of melanoma compared with the nevi. Among these, miR-155-5p, miR-9-5p, miR-142-5p, miR-19a-3p, miR-134-5p, and miR-301a-3p were upregulated, while miR-205-5p, miR-203a-3p, miR-27b-3p, miR-218-5p, and miR-23b-3p were downregulated. The highest similarity in miRNA expression pattern was found between uveal and mucosal melanoma groups, i.e., 15 miRNAs had altered expression in both groups.
Using the IPA, we determined the diseases, pathways, and biological functions associated with the miRNAs with altered expression. The IPA revealed that the dysregulated miRNAs are involved in many physiological and pathological processes. Table S1 shows tumor stages, tumor sites, and diseases associated with the dysregulated miRNAs for each melanoma type. It is worth noting that both primary and metastatic disease are common to all three melanoma types. We used IPA for each data set, identifying the targets of the miRNAs that were significantly overexpressed or underexpressed in our study (p < 0.05). Matching our results against the IPA data sets we generated pathway networks for each data set and, by identifying shared molecules, we created merged networks for each type of melanoma (Figure S1-S3). The targets common to all three types of melanoma were Smad2/3, insulin, sirtuin 1 (SIRT1), and tumor protein p53. The other identified targets were either unique to each type of melanoma (35 targets in MM, 29 targets in CM, and 38 targets in UM) or common to at least 2 of the analyzed melanoma types (MM and CM: Dickkopf WNT signaling pathway inhibitor 1 [DKK1], G protein signaling modulator 2 [GPSM2], one cut homeobox 2 [ONECUT2], protein regulator of cytokinesis 1 [PRC1], reversion-inducing-cysteine-rich protein with kazal motifs [RECK], and secretory carrier membrane protein 1 [SCAMP1]; CM and UM: protein kinase B [PKB or Akt], and insulin receptor [INSR]; UM and MM: interferon alpha [IFN-α], pro-inflammatory cytokines, parathyroid hormone like hormone [PTHLH], retinoblastoma [Rb] protein, and telomerase reverse transcriptase [TERT]).
TABLE S1.
Tumor stages, tumor sites, and diseases associated with dysregulated microRNAs (miRNAs) for each melanoma type, according to the Qiagen Ingenuity pathway analysis (IPA)
FIGURE S1.
The Qiagen Ingenuity pathway analysis (IPA) in cutaneous melanoma (n = 10)
FIGURE S3.
The Qiagen Ingenuity pathway analysis (IPA) in uveal melanoma (n = 10)
FIGURE S2.
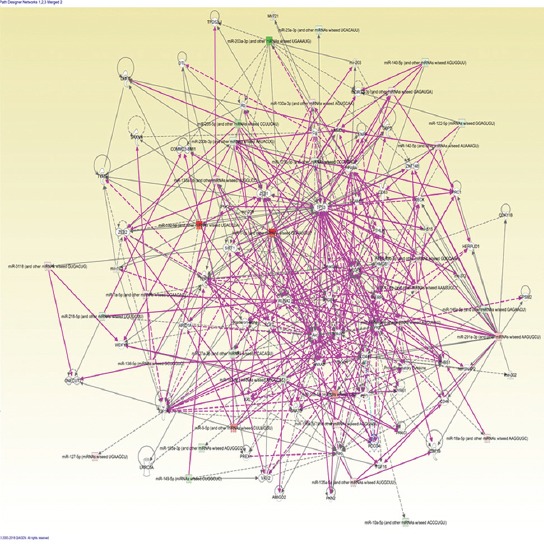
The Qiagen Ingenuity pathway analysis (IPA) in mucosal melanoma (n = 7)
DISCUSSION
In this study, we analyzed the expression of 84 miRNAs in cutaneous, uveal, and mucosal melanomas compared with miRNA expression in control cutaneous nevi. Out of 84 miRNAs, 11 miRNAs showed altered expression in all types of melanoma compared with the nevi. Among these miRNAs, only miR-155-5p, miR-142-5p, miR-205-5p, miR-23b-3p, miR-134-5p, and miR-301a-3p were previously studied in melanoma. Thus, for the first time, here we reported altered expression of miR-9-5p, miR-203a-3p, miR-19a-3p, miR-27b-3p, and miR-218-5p in the three melanoma types vs. nevi.
We found that miR-155-5p was upregulated in all three types of melanoma. Similarly, a previous study reported miR-150 and miR-155 to be upregulated in primary and metastatic melanoma compared with nevi [25]. In addition, miR-155-5p was suggested to play a role in the development of other solid and hematopoietic cancers [26-29]. Nevertheless, in melanoma cell lines, ectopic expression of miR-155-5p had an anti-proliferative and pro-apoptotic effect [30], and higher miR-155-5p expression in metastatic melanoma could predict longer post-recurrence survival [25,31]. The expression of miR-155-5p increases during inflammatory response, especially during lymphocyte proliferation, and some authors suggest this to support the role of miR-155 in melanoma progression [32,33].
In this study, miR-205-5p was downregulated in melanoma vs. nevus samples, which is consistent with previous studies [34,35]. MiR-205 was shown to act as a tumor suppressor, inhibiting melanoma cell proliferation and inducing apoptosis by targeting vascular endothelial growth factor (VEGF) and transcription factor E2F1 in vitro as well as in vivo [35-37].
MiR-142-5p was upregulated in our melanoma samples compared with control nevi, and previous research indicated that miR-142 is one of five miRNAs with important clinical implications and a high prognostic value in metastatic melanoma [38].
MiR-23b-3p was downregulated in our melanoma samples vs. control nevi. This miRNA was shown to have altered expression in different malignant tumors, including melanoma, with clinical, therapeutic, and prognostic significance [34,39].
We showed for the first time that miR-9-5p is upregulated in melanoma compared with nevi. MiR-9-5p promotes cell proliferation and metastasis in non-small cell lung cancer (NSCLC) and colorectal cancer [40,41]. It is involved in the differentiation of B lymphocytes into plasma cells by the negative regulation of the transcription factor PR domain zinc finger protein 1 (PRDM1/BLIMP-1), showing lower levels as the lymphocytes differentiate. This explains high levels of miR-9-5p in primary large-B cell lymphomas [42]. On the other hand, miR-9 is downregulated in human ovarian cancer compared with normal ovary, and the overexpression of miR-9 inhibits cell growth in ovarian cancer in vitro through the negative regulation of nuclear factor NF-kappa-B p105 (NFκB1) [42,43]. MiR-9-5p may represent a new prognostic marker in melanoma and possibly a new therapeutic target.
We found that miR-19a-3p is upregulated in melanoma vs. nevi, which is another novel finding in this cancer type. In gastric cancer, miR-19a-3p had a negative prognostic impact and promoted cell malignancy [44].
MiR-134-5p was upregulated in our melanoma samples compared with nevi. Previous studies showed that miR-134-5p is downregulated in NSCLC cells [45] and nasopharyngeal carcinoma cells [46] and that it has a role in inhibiting tumor progression. Interestingly, another study on melanoma showed that miR-134-5p is downregulated in melanoma patients compared with healthy controls [47], and this discrepancy with our results should be further investigated.
We reported in this study upregulation of miR-301a-3p in melanoma vs. nevi. MiR-301a was previously reported to be upregulated in melanoma samples compared to benign melanocytic lesions [48]. In addition, in hepatocellular carcinoma cell lines, miR-301a-3p overexpression was shown to stimulate cell proliferation, invasion, and chemoresistance [49].
For the first time, we showed that miR-203a-3p was downregulated in melanoma compared with nevi. The overexpression of miR-203a-3p in colorectal cancer cell lines inhibited cell proliferation and reduced chemoresistance [50] and similarly, in nasopharyngeal carcinoma, overexpressed miR-203a-3p inhibited cell proliferation, migration, and invasion in vitro as well as xenograft tumor growth and lung metastasis in vivo [51]. On the other hand, in hepatocellular carcinoma cells, miR-203a-3p.1 overexpression was reported to be oncogenic [52].
MiR-27b-3p and miR-218-5p were another two downregulated miRNAs in our case study that are reported for the first time in melanoma. MiR-27b-3p was also downregulated in lung cancer tissues [53] and in breast cancer tissues and cell lines, where it was associated with chemoresistance [54]. However, miR-27b-3p expression was increased in Dox-resistant anaplastic thyroid cancer (ATC) cells [55]. MiR-218-5p expression was reduced in gastric cancer [56], NSCLC [57], and gallbladder cancer (GBC) tissue [58]; moreover, miR-218-5p overexpression reversed the resistance of GBC cells to gemcitabine [58].
Among miRNAs with altered expression in at least one type of melanoma (Table 3), we focused on those previously studied in melanoma. Although some studies reported that miR-127-5p and miR-15a-5p were not significantly differently expressed in melanoma compared to melanocytic nevi [59,60], our results showed these miRNAs to be overexpressed in both cutaneous and mucosal melanomas vs. nevi. In addition, miR-335-5p was reported to be significantly differentially expressed in the melanoma cell line MML-1 compared to benign nevi [33], but our results showed a significant overexpression of miR-335-5p only in uveal melanoma group compared to control nevi.
TABLE 3.
MicroRNAa (miRNAs) with altered expression in at least one group of the studied melanoma types (n = 27) that were or were not investigated before
The main limitation to our study is the small sample size. Thus, although we consider some of our findings to be groundbreaking in melanoma research, they still need to be confirmed in large-scale studies. To this end, we propose analyzing miRNAs in each melanoma sample separately and in combination with follow-up data, so to determine changes in miRNA expression specific to each cell type and site of origin, the impact of local factors, and the prognostic potential of miRNAs in melanoma. To achieve this, we plan to conduct a prospective study in the future.
CONCLUSION
Overall, we identified several miRNAs with significantly altered expression in primary melanomas, including those reported for the first time in this type of cancer. Mir-9-5p, miR-203a-3p, miR-19a-3p, miR-134-5p, miR-301a-3p, miR-155-5p, miR-142-5p, miR-205-5p, miR-23b-3p, miR-27b-3p, and miR-218-5p had altered expression in all three melanoma types. Further research should explore the potential of these miRNAs in melanoma. The primary contribution of this study is to demonstrate that despite originating from the same cell type, the three melanoma types are still separate entities characterized by different miRNA expression patterns.
ACKNOWLEDGMENTS
This project was financed by the Victor Babes University of Medicine and Pharmacy Timisoara, through the project for young researchers PII-C5-TC-2017-02 C.I.
Footnotes
Conflict of interest statement: The authors declare no conflict of interests
REFERENCES
- 1.Ocanha-Xavier JP, Xavier-Junior JCC, Marques MEA. Melanoma:clinical, evolutive and histopathological characteristics of a series of 136 cases. An Bras Dermatol. 2018;93(3):373–6. doi: 10.1590/abd1806-4841.20186690. https://doi.org/10.1590/abd1806-4841.201⇝0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. https://doi.org/10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Fechete O, Ungureanu L, Şenilă S, Vornicescu D, Dănescu S, Vasilovici A, et al. Risk factors for melanoma and skin health behaviour:an analysis on Romanian melanoma patients. Oncol Lett. 2019;17(5):4139–44. doi: 10.3892/ol.2018.9737. https://doi.org/10.3892/ol.2018.9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett AW, Raja AM. The immunohistochemical identification of human oral mucosal melanocytes. Arch Oral Biol. 1997;42(1):77–81. doi: 10.1016/s0003-9969(96)00113-6. https://doi.org/10.1016/S0003-9969(96)00113-6. [DOI] [PubMed] [Google Scholar]
- 5.Tolleson WH. Human melanocyte biology, toxicology, and pathology. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2005;23(2):105–61. doi: 10.1080/10590500500234970. https://doi.org/10.1080/10590500500234970. [DOI] [PubMed] [Google Scholar]
- 6.De La Pava S, Nigogosyan G, Pickren JW, Cabrera A. Melanosis of the esophagus. Cancer. 1963;16:48–50. doi: 10.1002/1097-0142(196301)16:1<48::aid-cncr2820160107>3.0.co;2-m. https:// doi.org/ 10.1002/1097-0142 (196301)16:1<48:AID- CN CR 282 01 60 107>3.0. CO ;2- M. [DOI] [PubMed] [Google Scholar]
- 7.Werdin C, Limas C, Knodell RG. Primary malignant melanoma of the rectum. Evidence for origination from rectal mucosal melanocytes. Cancer. 1988;61(7):1364–70. doi: 10.1002/1097-0142(19880401)61:7<1364::aid-cncr2820610715>3.0.co;2-b. https:// doi.org/10.10 02/109 7-0142 (1988 0401)61:7 <13 64:AID - CNCR2 820 6 10715 >3.0 .CO ;2-B. [DOI] [PubMed] [Google Scholar]
- 8.Zak FG, Lawson W. The presence of melanocytes in the nasal cavity. Ann Otol Rhinol Laryngol. 1974;83(4):515–9. doi: 10.1177/000348947408300414. https://doi.org/10.1177/000348947408300414. [DOI] [PubMed] [Google Scholar]
- 9.Uehara T, Matsubara O, Kasuga T. Melanocytes in the nasal cavity and paranasal sinus. Incidence and distribution in Japan. Acta Pathol Jpn. 1987;37(7):1105–14. doi: 10.1111/j.1440-1827.1987.tb00427.x. https://doi.org/10.1111/j.1440-1827.1987.tb00427.x. [DOI] [PubMed] [Google Scholar]
- 10.Goldman JL, Lawson W, Zak FG, Roffman JD. The presence of melanocytes in the human larynx. Laryngoscope. 1972;82(5):824–35. doi: 10.1288/00005537-197205000-00009. https://doi.org/10.1288/00005537-197205000-00009. [DOI] [PubMed] [Google Scholar]
- 11.Nigogosyan G, De La Pava S, Pickren JW. Melanoblasts in vaginal mucosa. Origin for primary malignant melanoma. Cancer. 1964;17:912–3. doi: 10.1002/1097-0142(196407)17:7<912::aid-cncr2820170711>3.0.co;2-f. https://doi.org/ 10.1002/ 1097 - 0142 (196407) 17:7 <912: AID-CNCR2820170711 >3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 12.Uehara T, Takayama S, Takemura T, Kasuga T. Foci of stromal melanocytes (so-called blue naevus) of the uterine cervix in Japanese women. Virchows Arch A Pathol Anat Histopathol. 1991;418(4):327–31. doi: 10.1007/BF01600162. https://doi.org/10.1007/BF01600162. [DOI] [PubMed] [Google Scholar]
- 13.Brenner M, Hearing VJ. What are melanocytes really doing all day long…?:from the ViewPoint of a keratinocyte:Melanocytes - cells with a secret identity and incomparable abilities. Exp Dermatol. 2009;18(9):799–819. doi: 10.1111/j.1600-0625.2009.00912.x. https://doi.org/10.1111/j.1600-0625.2009.00912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beadling C, Jacobson-Dunlop E, Hodi FS, Le C, Warrick A, Patterson J, et al. KIT gene mutations and copy number in melanoma subtypes. Clin Cancer Res. 2008;14(21):6821–8. doi: 10.1158/1078-0432.CCR-08-0575. https://doi.org/10.1158/1078-0432.CCR-08-0575. [DOI] [PubMed] [Google Scholar]
- 15.Griewank K, Westekemper H, Murali R, Mach M, Schilling B, Wiesner T, et al. Conjunctival melanomas harbor BRAF and NRAS mutations and copy number changes similar to cutaneous and mucosal melanomas. Clin Cancer Res. 2013;19(12):3143–52. doi: 10.1158/1078-0432.CCR-13-0163. https://doi.org/10.1158/1078-0432.CCR-13-0163. [DOI] [PubMed] [Google Scholar]
- 16.Bastian BC, Olshen AB, LeBoit PE, Pinkel D. Classifying melanocytic tumors based on DNA copy number changes. Am J Pathol. 2003;163(5):1765–70. doi: 10.1016/S0002-9440(10)63536-5. https://doi.org/10.1016/S0002-9440(10)63536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abysheva SN, Iyevleva AG, Efimova NV. KIT mutations in Russian patients with mucosal melanoma. Melanoma Res. 2011;21(6):555–9. doi: 10.1097/CMR.0b013e32834bf398. https://doi.org/10.1097/CMR.0b013e32834bf398. [DOI] [PubMed] [Google Scholar]
- 18.Antonescu CR, Busam KJ, Francone TD, Wong GC, Guo T, Agaram NP, et al. KIT mutation in anal melanomas correlates with KIT protein expression and is sensitive to specific kinase inhibition. Int J Cancer. 2007;121(2):257–64. doi: 10.1002/ijc.22681. https://doi.org/10.1002/ijc.22681. [DOI] [PubMed] [Google Scholar]
- 19.Aulmann S, Sinn HP, Penzel R, Gilks CB, Schott S, Hassel JC, et al. Comparison of molecular abnormalities in vulvar and vaginal melanomas. Mod Pathol. 2014;27(10):1386–93. doi: 10.1038/modpathol.2013.211. https://doi.org/10.1038/modpathol.2013.211. [DOI] [PubMed] [Google Scholar]
- 20.Furney SJ, Turajlic S, Stamp G, Nohadani M, Carlisle A, Thomas JM, et al. Genome sequencing of mucosal melanomas reveals that they are driven by distinct mechanisms from cutaneous melanoma. J Pathol. 2013;230(3):261–9. doi: 10.1002/path.4204. https://doi.org/10.1002/path.4204. [DOI] [PubMed] [Google Scholar]
- 21.Mirzaei H, Gholamin S, Shahidsales S, Sahebkar A, Jaafari MR, Mirzaei HR, et al. MicroRNAs as potential diagnostic and prognostic biomarkers in melanoma. Eur J Cancer. 2016;53:25–32. doi: 10.1016/j.ejca.2015.10.009. https://doi.org/10.1016/j.ejca.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Ross CL, Kaushik S, Valdes-Rodriguez R, Anvekar R. MicroRNAs in cutaneous melanoma:role as diagnostic and prognostic biomarkers. J Cell Physiol. 2018;233(7):5133–41. doi: 10.1002/jcp.26395. https://doi.org/10.1002/jcp.26395. [DOI] [PubMed] [Google Scholar]
- 23.Stark MS, Tyagi S, Nancarrow DJ, Boyle GM, Cook AL, Whiteman DC, et al. Characterization of the melanoma miRNAome by deep sequencing. PLoS One. 2010;5(3):e9685. doi: 10.1371/journal.pone.0009685. https://doi.org/10.1371/journal.pone.0009685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen AC, Mikkelsen LH, Borup R, Kiss K, Toft PB, von Buchwald C, et al. MicroRNA expression profile in conjunctival melanoma. Invest Ophthalmol Vis Sci. 2016;57(10):4205–12. doi: 10.1167/iovs.16-19862. https://doi.org/10.1167/iovs.16-19862. [DOI] [PubMed] [Google Scholar]
- 25.Segura MF, Belitskaya-Lévy I, Rose AE, Zakrzewski J, Gaziel A, Hanniford D, et al. Melanoma MicroRNA signature predicts post-recurrence survival. Clin Cancer Res. 2010;16(5):1577–86. doi: 10.1158/1078-0432.CCR-09-2721. https://doi.org/10.1158/1078-0432.CCR-09-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–70. doi: 10.1158/0008-5472.CAN-05-1783. https://doi.org/10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 27.Jung M, Mollenkopf HJ, Grimm C, Wagner I, Albrecht M, Waller T, et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med. 2009;13(9B):3918–28. doi: 10.1111/j.1582-4934.2009.00705.x. https://doi.org/10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. https://doi.org/10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106(17):7113–8. doi: 10.1073/pnas.0902636106. https://doi.org/10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levati L, Alvino E, Pagani E, Arcelli D, Caporaso P, Bondanza S, et al. Altered expression of selected microRNAs in melanoma:antiproliferative and proapoptotic activity of miRNA-155. Int J Oncol. 2009;35(2):393–400. [PubMed] [Google Scholar]
- 31.Segura MF, Greenwald HS, Hanniford D, Osman I, Hernando E. MicroRNA and cutaneous melanoma:from discovery to prognosis and therapy. Carcinogenesis. 2012;33(10):1823–32. doi: 10.1093/carcin/bgs205. https://doi.org/10.1093/carcin/bgs205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tili E, Croce CM, Michaille JJ. MiR-155:on the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28(5):264–84. doi: 10.1080/08830180903093796. https://doi.org/10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 33.Lunavat TR, Cheng L, Kim DK, Bhadury J, Jang SC, Lässer C, et al. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells - Evidence of unique microRNA cargos. RNA Biology. 2015;12(8):810–23. doi: 10.1080/15476286.2015.1056975. https://doi.org/10.1080/15476286.2015.1056975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozubek J, Ma Z, Fleming E, Duggan T, Wu R, Shin DG, et al. In-depth characterization of microRNA transcriptome in melanoma. PLoS One. 2013;8(9):e72699. doi: 10.1371/journal.pone.0072699. https://doi.org/10.1371/journal.pone.0072699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y, Brenn T, Brown ER, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression:miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer. 2012;106(3):553–61. doi: 10.1038/bjc.2011.568. https://doi.org/10.1038/bjc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011;286(19):16606–14. doi: 10.1074/jbc.M111.227611. https://doi.org/10.1074/jbc.M111.227611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noguchi S, Iwasaki J, Kumazaki M, Mori T, Maruo K, Sakai H, et al. Chemically modified synthetic microRNA-205 inhibits the growth of melanoma cells in vitro and in vivo. Mol Ther. 2013;21(6):1204–11. doi: 10.1038/mt.2013.70. https://doi.org/10.1038/mt.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jayawardana K, Schramm SJ, Tembe V, Mueller S, Thompson JF, Scolyer RA, et al. Identification, review, and systematic cross-validation of microRNA prognostic signatures in metastatic melanoma. J Invest Dermatol. 2016;136(1):245–54. doi: 10.1038/JID.2015.355. https://doi.org/10.1038/JID.2015.355. [DOI] [PubMed] [Google Scholar]
- 39.Grossi I, Salvi A, Baiocchi G, Portolani N, De Petro G. Functional role of microRNA-23b-3p in cancer biology. Microrna. 2018;7(3):156–66. doi: 10.2174/2211536607666180629155025. https://doi.org/10.2174/2211536607666180629155025. [DOI] [PubMed] [Google Scholar]
- 40.Li G, Wu F, Yang H, Deng X, Yuan Y. MiR-9-5p promotes cell growth and metastasis in non-small cell lung cancer through the repression of TGFBR2. Biomed Pharmacother. 2017;96:1170–8. doi: 10.1016/j.biopha.2017.11.105. https://doi.org/10.1016/j.biopha.2017.11.105. [DOI] [PubMed] [Google Scholar]
- 41.Snezhkina AV, Krasnov GS, Zhikrivetskaya SO, Karpova IY, Fedorova MS, Nyushko KM. Overexpression of microRNAs miR-9, -98, and -199 correlates with the downregulation of HK2 expression in colorectal cancer. Mol Biol (Mosk) 2018;52(2):220–30. doi: 10.7868/S0026898418020052. https://doi.org/10.1134/S0026893318020140. [DOI] [PubMed] [Google Scholar]
- 42.Koens L, Qin Y, Leung WY, Corver WE, Jansen PM, Willemze R, et al. MicroRNA profiling of primary cutaneous large B-cell lymphomas. PLoS One. 2013;8(12):e82471. doi: 10.1371/journal.pone.0082471. https://doi.org/10.1371/journal.pone.0082471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo LM, Pu Y, Han Z, Liu T, Li YX, Liu M, et al. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009;276(19):5537–46. doi: 10.1111/j.1742-4658.2009.07237.x. https://doi.org/10.1111/j.1742-4658.2009.07237.x. [DOI] [PubMed] [Google Scholar]
- 44.Qiao F, Gong P, Song Y, Shen X, Su X, Li Y, et al. Downregulated PITX1 modulated by miR-19a-3p promotes cell malignancy and predicts a poor prognosis of gastric cancer by affecting transcriptionally activated PDCD5. Cell Physiol Biochem. 2018;46(6):2215–31. doi: 10.1159/000489590. https://doi.org/10.1159/000489590. [DOI] [PubMed] [Google Scholar]
- 45.Liu Q, Wu Y, Xiao J, Zou J. Long non-coding RNA prostate cancer-associated transcript 7 (PCAT7) induces poor prognosis and promotes tumorigenesis by inhibiting mir-134-5p in non-small-cell lung (NSCLC) Med Sci Monit. 2017;23:6089–98. doi: 10.12659/MSM.907904. https://doi.org/10.12659/MSM.907904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Tao Z, Qu J, Zhou X, Zhang C. Long non-coding RNA PCAT7 regulates ELF2 signaling through inhibition of miR-134-5p in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2017;491(2):374–81. doi: 10.1016/j.bbrc.2017.07.093. https://doi.org/10.1016/j.bbrc.2017.07.093. [DOI] [PubMed] [Google Scholar]
- 47.Solé C, Tramonti D, Schramm M, Goicoechea I, Armesto M, Hernandez LI, et al. The circulating transcriptome as a source of biomarkers for melanoma. Cancers (Basel) 2019;11(1):70. doi: 10.3390/cancers11010070. https://doi.org/10.3390/cancers11010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sand M, Skrygan M, Sand D, Georgas D, Gambichler T, Hahn SA, et al. Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell Tissue Res. 2013;351(1):85–98. doi: 10.1007/s00441-012-1514-5. https://doi.org/10.1007/s00441-012-1514-5. [DOI] [PubMed] [Google Scholar]
- 49.Hu J, Ruan J, Liu X, Xiao C, Xiong J. MicroRNA-301a-3p suppressed the progression of hepatocellular carcinoma via targeting VGLL4. Pathol Res Pract. 2018;214(12):2039–45. doi: 10.1016/j.prp.2018.09.008. https://doi.org/10.1016/j.prp.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 50.Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K, Huang Z, et al. LncRNA HOTAIR is a prognostic biomarker for the proliferation and chemoresistance of colorectal cancer via miR-203a-3p-mediated Wnt/ß-catenin signaling pathway. Cell Physiol Biochem. 2018;46(3):1275–85. doi: 10.1159/000489110. https://doi.org/10.1159/000489110. [DOI] [PubMed] [Google Scholar]
- 51.Jiang N, Jiang X, Chen Z, Song X, Wu L, Zong D, et al. MiR-203a-3p suppresses cell proliferation and metastasis through inhibiting LASP1 in nasopharyngeal carcinoma. J Exp Clin Cancer Res . 2017;36:138. doi: 10.1186/s13046-017-0604-3. https://doi.org/10.1186/s13046-017-0604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huo W, Du M, Pan X, Zhu X, Gao Y, Li Z. miR-203a-3p.1 targets IL-24 to modulate hepatocellular carcinoma cell growth and metastasis. FEBS Open Bio. 2017;7(8):1085–91. doi: 10.1002/2211-5463.12248. https://doi.org/10.1002/2211-5463.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Y, Xu T, Cao YW, Ding XQ. Antitumor effect of miR-27b-3p on lung cancer cells via targeting Fzd7. Eur Rev Med Pharmacol Sci. 2017;21(18):4113–23. [PubMed] [Google Scholar]
- 54.Chen D, Si W, Shen J, Du C, Lou W, Bao C, et al. miR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis. 2018;9(2):188. doi: 10.1038/s41419-017-0211-4. https://doi.org/10.1038/s41419-017-0211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y, Han YF, Ye B, Zhang YL, Dong JD, Zhu SJ, et al. miR-27b-3p is involved in doxorubicin resistance of human anaplastic thyroid cancer cells via targeting peroxisome proliferator-activated receptor gamma. Basic Clin Pharmacol Toxicol. 2018;123(6):670–7. doi: 10.1111/bcpt.13076. https://doi.org/10.1111/bcpt.13076. [DOI] [PubMed] [Google Scholar]
- 56.Deng M, Zeng C, Lu X, He X, Zhang R, Qiu Q, et al. miR-218 suppresses gastric cancer cell cycle progression through the CDK6/Cyclin D1/E2F1 axis in a feedback loop. Cancer Lett. 2017;403:175–85. doi: 10.1016/j.canlet.2017.06.006. https://doi.org/10.1016/j.canlet.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 57.Zhu K, Ding H, Wang W, Liao Z, Fu Z, Hong Y, et al. Tumor-suppressive miR-218-5p inhibits cancer cell proliferation and migration via EGFR in non-small cell lung cancer. Oncotarget. 2016;7(19):28075–85. doi: 10.18632/oncotarget.8576. https://doi.org/10.18632/oncotarget.ↀ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H, Zhan M, Xu SW, Chen W, Long MM, Shi YH, et al. miR-218-5p restores sensitivity to gemcitabine through PRKCE/MDR1 axis in gallbladder cancer. Cell Death Dis. 2017;8(5):e2770. doi: 10.1038/cddis.2017.178. https://doi.org/10.1038/cddis.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zehavi L, Avraham R, Barzilai A, Bar-Ilan D, Navon R, Sidi Y, et al. Silencing of a large microRNA cluster on human chromosome 14q32 in melanoma:biological effects of mir-376a and mir-376c on insulin growth factor 1 receptor. Mol Cancer. 2012;11:44. doi: 10.1186/1476-4598-11-44. https://doi.org/10.1186/1476-4598-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chai L, Kang XJ, Sun ZZ, Zeng MF, Yu SR, Ding Y, et al. MiR-497-5p, miR-195-5p and miR-455-3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manag Res. 2018;10:989–1003. doi: 10.2147/CMAR.S163335. https://doi.org/10.2147/CMAR.S163335. [DOI] [PMC free article] [PubMed] [Google Scholar]


