Abstract
Progenitor cells at the basal layer of skin epidermis play an essential role in maintaining tissue homeostasis and enhancing wound repair in skin. The proliferation, differentiation, and cell death of epidermal progenitor cells have to be delicately regulated, as deregulation of this process can lead to many skin diseases, including skin cancers. However, the underlying molecular mechanisms involved in skin homeostasis remain poorly defined. In this study, with quantitative proteomics approach, we identified an important interaction between KDF1 (keratinocyte differentiation factor 1) and IKKα (IκB kinase α) in differentiating skin keratinocytes. Ablation of either KDF1 or IKKα in mice leads to similar but striking abnormalities in skin development, particularly in skin epidermal differentiation. With biochemical and mouse genetics approach, we further demonstrate that the interaction of IKKα and KDF1 is essential for epidermal differentiation. To probe deeper into the mechanisms, we find that KDF1 associates with a deubiquitinating protease USP7 (ubiquitin‐specific peptidase 7), and KDF1 can regulate skin differentiation through deubiquitination and stabilization of IKKα. Taken together, our study unravels an important molecular mechanism underlying epidermal differentiation and skin tissue homeostasis.
Keywords: deubiquitination, epidermal differentiation, IKKα, KDF1, skin
Subject Categories: Development & Differentiation; Post-translational Modifications, Proteolysis & Proteomics; Skin
KDF1 (Keratinocyte Differentiation Factor 1) promotes the deubiquitination and stability of IKKα by recruiting USP7 (Ubiquitin Specific Peptidase 7). This interaction is essential for skin epidermal differentiation.

Introduction
Mammalian skin serves as an essential water‐impermeable barrier that protects us from various environmental damages 1. Epidermis of skin is a stratified epithelial tissue that consists of four major tissue layers: basal layer, spinous layer, granular layer, and stratum corneum. Tissue homeostasis of skin epidermis is sustained by the potential epidermal progenitor/stem cells that localize at the basal layer. In adult skin, these cells periodically move upward from their niche at the basement membrane and undergo terminal differentiation to replenish lost skin cells during normal tissue homeostasis or upon skin injury 1, 2, 3. Deregulation of epidermal differentiation in skin can lead to the development of various skin diseases including skin cancers, such as SCC (squamous cell carcinoma).
Although the morphogenetic changes during epidermal differentiation have been studied 1, 2, 3, much remains unknown about the molecular mechanisms underlying this process. The role of KDF1 (keratinocyte differentiation factor 1) in epidermal differentiation was discovered before via an ENU‐induced mutagenesis screen in mice 4. It has been shown that the KDF1 mutant fetuses developed a thick, taut, and hyperplastic epidermis with diminished barrier ability. Loss of KDF1 also leads to aberrant cell proliferation, misexpression of basal and spinous differentiation markers, and an absence of terminal differentiation, suggesting that KDF1 is a novel player that is critically involved in epidermal differentiation. To determine the underlying molecular mechanism and dissect the signaling network involved in skin stratification, we examined KDF1's interactome by quantitative proteomics approach. Our MS (mass spectrometry) analysis identified an intriguing protein, IKKα (IκB kinase α), as a specific binding partner of KDF1 in differentiating keratinocytes.
IKKα is a well‐established component within the NF‐κB signaling pathway 5. However, loss of function of IKKα in mice leads to striking abnormalities in skin development. The IKKα KO (knockout) mice developed abnormally thick and shiny skin with diminished terminal differentiation in epidermis, resembling the phenotypes in KDF1 KO animals 2, 6, 7, 8. Interestingly, it has also been demonstrated that its function in skin differentiation is independent upon its kinase activity and its role in NF‐κB‐signal transduction 6, 7, 9. Emerging evidence suggests that IKKα regulates skin tissue homeostasis via its own transcriptional regulatory role. A dramatic increase of IKKα level in keratinocyte nucleus precedes epidermal differentiation, which could modulate the autocrine signaling of EGF and the expression of a potential secreted keratinocyte differentiation‐inducible factor (kDIF) 10. Consistent with its role in epidermal differentiation, deregulated IKKα in skin could lead to tumorigenesis, such as cutaneous SCCs 11, 12, 13, 14, 15.
To probe deeper into the molecular mechanisms, we found that KDF1 deletion will enhance IKKα ubiquitination and abolish IKKα accumulation in differentiating keratinocytes. Protein ubiquitination is a universal mechanism for protein degradation that controls a wide variety of cellular processes 16, 17, 18, 19. Deubiquitination is a reverse process of ubiquitination, which is carried out by deubiquitinating enzymes (DUBs) 20, 21. Interestingly, our proteomics analysis revealed that KDF1 associates with a key DUB, USP7/HAUSP (ubiquitin‐specific peptidase 7/herpesvirus‐associated ubiquitin‐specific protease), in keratinocytes. As a deubiquitination molecule, USP7 has been implicated in various signaling cascades, including NF‐κB pathway 22, 23, p53/MDM2 (mouse double minute 2 homolog), PTEN (phosphatase and tensin homolog), and FOXO4 regulation 24, 25, 26, 27. However, the role of USP7 in skin development and epidermal stratification remains unclear. In this study, we present compelling evidence that KDF1 regulates IKKα ubiquitination and stability through its interaction with USP7. Deletion of USP7 in epidermal keratinocytes leads to decreased IKKα and aberrant differentiation. Taken together, our results illuminate an important molecular mechanism whereby differentiation of epidermal progenitor cells is regulated by KDF1 and IKKα proteins.
Results
KDF1 interacts with IKKα
KDF1 deficiency leads to profound abnormalities in skin development. To dissect the underlying mechanisms, we engineered expression vectors encoding HA‐ and His6‐tagged KDF1. We used tandem affinity purification to isolate the KDF1 complex from transfected cells upon calcium‐induced keratinocyte differentiation and then employed SILAC (stable isotope labeling by amino acids in cell culture) coupled with LC‐MS/MS (liquid chromatography and tandem mass spectrometry) to determine the KDF1 interacting proteins (Fig 1A and Dataset EV1). Our analysis identified IKKα as a KDF1 binding partner in differentiating keratinocytes. To confirm the interaction, we first conducted co‐immunoprecipitation assay with ectopically expressed KDF1 and IKKα in HEK293 cells (Fig 1B and C). To examine the interaction of endogenous proteins, we immunoprecipitated KDF1 in WT (wild‐type) skin keratinocytes before or after high calcium‐induced epidermal differentiation. Interestingly, endogenous co‐immunoprecipitation can only be detected in differentiated keratinocytes, suggesting that endogenous KDF1 associates with IKKα specifically in differentiated cells (Fig 1D, quantification in Fig EV1A, and source data in Fig EV1B). The kinase activity of IKKα is dispensable for keratinocyte differentiation 6, 7, 9. As expected, the kinase‐dead mutant of IKKα retains strong binding affinity to KDF1 in vitro (Fig 1E).
Figure 1. KDF1 associates with IKKα.

-
ATandem affinity purification was used to isolate KDF1‐associated proteins from SILAC‐labeled keratinocytes. Precipitated proteins were resolved by SDS–PAGE and subjected to identification with LC‐MS/MS. Arrows denote IgG heavy and light chains. Star denotes KDF1. Ctrl: control. Kd: kilodalton for molecular weight markers.
-
B, CHEK293 cells were transfected with empty vector or plasmids encoding HA‐tagged KDF1 and/or GST‐tagged IKKα. Immunoprecipitation (α‐HA or α‐GST) was carried out to determine their interaction. Immunoprecipitates (IP) and whole cell lysate (WCL) were immunoblotted (IB) with different antibodies as indicated.
-
DImmunoprecipitation was carried out with α‐KDF1 antibody in keratinocyte cultured in medium with low or high concentration of calcium (Ca). IPs were blotted with different antibodies as indicated. Star denotes KDF1. The same IP was used for USP7 immunoblot as shown in Fig 5B.
-
EHEK293 cells were transfected to co‐express HA‐tagged KDF1 with GST‐tagged IKKα or its kinase‐dead (KD) mutant. IP and WCL were blotted with different antibodies as indicated.
-
FHEK293 cells were transfected to co‐express HA‐tagged KDF1 with GST‐tagged IKKα or its different truncation mutants (Fig EV1B). IP and WCL were blotted with different antibodies as indicated. Arrow denotes antibody heavy chain.
-
GHEK293 cells were transfected to co‐express GST‐tagged IKKα with HA‐tagged KDF1 or its different truncation mutants (Fig EV1C). IP and WCL were blotted with different antibodies as indicated.
Figure EV1. Association between IKKα and KDF1 .

-
ALevel of IKKα in α‐KDF1 immunoprecipitate was quantified by densitometry and presented as bar grafts. Statistical analysis is conducted using unpaired Student's t‐test. Error bar represents SD (standard deviation). N = 3. **P < 0.01.
-
BUnmodified blots for Fig 1D.
-
C, DDiagrams indicating the domain structure of IKKα (C) and KDF1 (D), as well as different truncation mutants generated in this study. LZ: leucine zipper; HLH: helix‐loop‐helix domain; aa: amino acid residues.
In order to map the potential binding regions within KDF1 and IKKα, we generated various truncation mutants of KDF1 and IKKα, based upon the known functional domains or protein characteristics (Fig EV1C and D). IKKα is a conserved helix‐loop‐helix kinase, which contains a serine–threonine kinase domain, a leucine zipper motif (LZ), a helix‐loop‐helix domain (HLH), and a NEMO‐binding domain (Fig EV1C) 28. Co‐immunoprecipitation analysis suggests that the kinase domain of IKKα (mutant 1) does not bind KDF1. The IKKα truncation mutant containing LZ and HLH domain (mutant 2 and 3) can associate with KDF1, whereas the truncation mutant harboring the HLH and NEMO‐binding domain (mutant 4) cannot. It strongly suggests that the LZ domain may be responsible for the interaction with KDF1. Consistent with this hypothesis, an IKKα truncation mutant containing LZ domain alone (mutant 5) can interact with KDF1, whereas a LZ‐deletion mutant of IKKα (mutant 6) fails to bind KDF1 (Fig 1F).
KDF1 contains a proline‐ and cysteine‐rich region near the N‐terminus 4 (Fig EV1D), which shows no binding affinity toward IKKα (mutant 1) (Fig 1G). Therefore, we further truncated the C‐terminal portion of KDF1 (mutant 2) into two halves (mutants 3 and 4). The co‐immunoprecipitation analysis indicates that the binding motif of KDF1 lies within central domain of KDF1 (mutant 3). Consistently, an in‐frame deletion of this region (mutant 5) abolished IKKα binding (Fig 1G). Taken together, our data show that the interaction is mediated by the LZ motif of IKKα and the central domain of KDF1.
Interaction of IKKα and KDF1 is essential for epidermal stratification
Loss of KDF1 or IKKα has been shown to affect skin development in vivo 4, 6, 7, 8, 9. To assess the relevance of IKKα and KDF1 interaction in skin differentiation, we first systematically examined epidermal stratification in embryonic E18.5 skin of mutant animals. Histological analysis coupled with immunofluorescence staining revealed striking similarities of KDF1 KO and IKKα KO in epidermal differentiation, including thickened epidermis, loss of stratum corneum (Fig 2A), expansion of basal cell marker, hyperplastic spinous layer, and loss of expression of late differentiation markers (Fig 2B, and quantification in Fig EV2A and B). To further dissect the molecular mechanisms, we isolated primary basal progenitor cells from KDF1 KO and IKKα KO animals. When induced to differentiate with calcium shift, KDF1 or IKKα KO cells fail to express loricrin or filaggrin, both are well‐established biochemical markers for epidermal differentiation (Fig 2C). The aberrant skin stratification upon loss of KDF1 is not due to potential changes in skin inflammation, as staining of macrophage, T‐cell, and dendritic cell markers shows no significant increase of immunocytes in KDF1 KO skin (Fig EV2C).
Figure 2. Loss of KDF1 or IKKα leads to aberrant epidermal differentiation.

-
AH/E staining of E18.5 skin sections from WT, IKKα KO, and KDF1 KO mice. Dotted lines denote dermal–epidermal boundaries. Epi: epidermis, Der: dermis. Scale bar = 50 μm.
-
BE18.5 skin sections from WT, IKKα KO, and KDF1 KO mice were immunostained with different antibodies as indicated (Krt14: keratin 14, Krt10: keratin 10, Lor: loricrin, β4: β4‐integrin, CD104). Dotted lines denote dermal–epidermal boundaries. Epi: epidermis, Der: dermis. Scale bar = 100 μm.
-
CImmunoblots of WCL collected at 0, 1, and 2 days after calcium shift with different antibodies as indicated.
Figure EV2. Skin thickening upon loss of IKKα or KDF1 .

-
A, BQuantification of the thickness of Krt14‐positive (A) or Krt10‐positive (B) cell layers in WT vs. IKKα‐ or KDF1‐deficient skin epidermis. At least 3 biological replicates (independent skin samples) and at least 3 sections for each skin sample were analyzed for each strain. Note significantly increased thickness upon deletion of IKKα or KDF1.
-
CE18.5 skin sections from WT and KDF1 KO mice were immunostained with different antibodies for skin inflammation as indicated (Krt14: keratin 14, β4: β4‐integrin, CD104, F4/80: macrophage marker, CD3: T‐cell marker, CD11c: dendritic cell marker). Adult WT mouse skin treated with imiquimod (IMQ) was stained as positive controls for skin inflammation. Dotted lines denote dermal–epidermal boundaries. Epi: epidermis, Der: dermis. Scale bar = 50 μm.
The phenotypic resemblance between KDF1 and IKKα null cells suggests that these two proteins may act in the same pathway to regulate epidermal tissue homeostasis. To test this hypothesis, we used PiggyBac transposon system to restore expression of WT IKKα or IKKα mutant deficient in KDF1 binding (mutant 6) in IKKα null cells (Fig 3A). However, constitutive expression of KDF1 in KDF1 KO cells may lead to cell cycle arrest. To resolve this issue, we used a tetracycline‐inducible system to restore expression of WT KDF1 or KDF1 mutant defective in IKKα interaction (mutant 5) in KDF1 KO keratinocytes. Induction with doxycycline can lead to a dose‐dependent expression of KDF1 or its mutant in the cells (Fig 3B).
Figure 3. IKKα and KDF1 interaction is essential for skin stratification.

-
AImmunoblots of WCL from IKKα KO cells with rescued expression of HA‐tagged IKKα or its mutant using different antibodies as indicated.
-
BImmunoblots of WCL from KDF1 KO cells with rescued expression of KDF1 or its mutant using different antibodies as indicated. Expression of exogenous KDF1 was induced by doxycycline (DOX) at different concentration.
-
C, DExpression of loricrin is determined by immunoblots and quantified by densitometry. The relative expression level of loricrin upon calcium shift in different cell types was calculated and presented as bar graphs. Statistical analysis is conducted using unpaired Student's t‐test. Error bar represents SD (standard deviation). N = 3 (biological replicates). ***P < 0.001; **P < 0.01; *P < 0.05
-
ESections of regenerated skin developed from engrafted IKKα KO, KDF1 KO, and their rescued cells were immunostained with different antibodies as indicated. Dotted lines denote dermal–epidermal boundaries. Epi: epidermis, Der: dermis. Scale bar = 50 μm.
With establishment of these cell lines, we first examined the ability of these constructs to rescue epidermal differentiation upon calcium shift in vitro. Quantitative analysis of the expression level of loricrin shows that expression of WT IKKα or WT KDF1 is able to significantly increase the expression of loricrin. By contrast, expression of IKKα or KDF1 mutant failed to rescue the differentiation defect in vitro (Figs 3C and D, and EV3A).
Figure EV3. IKKα and KDF1 interaction is essential for epidermal differentiation.

-
AExpression level of loricrin is determined by immunoblots.
-
BQuantification of epidermal thickness in skin grafts generated from WT or IKKα‐ or KDF1‐deficient cells, or KO cells rescued with IKKα, KDF1, or their corresponding mutants. The plot indicates first quartile (bottom line of the box), median (middle line of the box), third quartile (top line of the box), and minimum and maximum measurements (whiskers). Each cell type had at least 3 biological replicates (independent skin grafts). At least 3 sections were taken from each graft for analysis and quantification. Statistical analysis is conducted comparing each cell type to WT, using unpaired Student's t‐test. N > 3. ****P < 0.0001.
To investigate the relevance of KDF1 and IKKα interaction in an in vivo setting, we took advantage of the mouse epidermal organotypic culture system that has been recently developed by our laboratory. KDF1 or IKKα KO cells and different rescued cells were cultured on top of acellularized dermis. Exposure to the air/liquid interphase can induce stratification of cultured cells to generate a skin‐like organoid in vitro 29. Transplantation of this cultured skin organoid to nude host leads to efficient skin engraftments, which are stable and can readily express exogenous genes that have been transduced to the epidermal progenitor cells. Regenerated skin from KDF1 or IKKα KO cells displays striking epidermal abnormalities, including thickened epidermis, loss of cornified cells, and expansion of basal cell markers, resembling the phenotypes of KDF1 or IKKα KO in vivo (Fig 3E). Interestingly, when WT but not mutant KDF1 or IKKα was re‐expressed in the regenerated skin, epidermal differentiation was largely restored, including decreased epidermal thickness and restrictive expression of Krt14 (keratin 14, a basal cell marker) in the basal layer (Fig 3E and quantification in Fig EV3B). Taken together, our results strongly suggest that epidermal differentiation requires interaction between KDF1 and IKKα.
KDF1 regulates keratinocyte differentiation via controlling the protein stability of IKKα
It has been shown that IKKα governs epidermal differentiation through its own transcriptional regulatory role in cell nucleus 10, 30. Through protein fractionation, we found that both IKKα and KDF1 show significant localization in cell nuclear fractions (Fig EV4A). Protein level of IKKα exhibits a strong increase upon keratinocyte differentiation (Fig 4A). Interestingly, although IKKα level is comparable in undifferentiated WT or KDF1 KO epidermal progenitor cells, the level of IKKα is dramatically reduced in KDF1 KO cells upon calcium switch‐induced differentiation (Fig 4A). To investigate the potential mechanism, we found that IKKα mRNA level is not significantly changed upon loss of KDF1 (Fig EV4B). Instead, stability of IKKα exhibits a significant decrease in KDF1 KO cells (Fig 4B and quantification in 4C). Consistent with this notion, the ubiquitination level of endogenous or exogenously expressed IKKα 31 is significantly increased in KDF1 KO cells upon differentiation (Fig 4D and quantification in 4E). Additionally, expression of WT KDF1 but not its mutant can restore the IKKα level in KDF1 null cells upon calcium shift (Fig 4F, and quantification in Fig EV4C). Together, these results suggest that KDF1 can regulate epidermal differentiation through controlling the protein stability of IKKα.
Figure EV4. KDF1 regulates IKKα protein stability.

-
ANuclear and cytoplasmic protein fractions were obtained from epidermal keratinocytes cultured in low calcium or high calcium medium. Protein lysates were subjected to immunoblots with different antibodies as indicated. Mr: molecular weight marker.
-
BQuantification of IKKα mRNA level in WT or KDF1 null cells with qPCR. N = 3. AU: arbitrary unit.
-
CLevel of IKKα (main Fig 4F) was quantified by densitometry and presented as bar graph. Statistical analysis is conducted using unpaired Student's t‐test. Error bar represents SD (standard deviation). N = 3. *P < 0.05. Statistically significant increase of IKKα was detected in cells re‐expressing WT KDF1 but not mutant KDF1.
-
D, EQuantification of whole epidermal (D) or Krt14‐positive cell layer (E) thickness in skin grafts generated from WT or KDF1‐deficient cells, or KO cells rescued with IKKα expression. Each cell type had at least 3 biological replicates (independent skin grafts). At least 3 sections were taken from each graft for analysis and quantification.
Figure 4. IKKα protein level is crucial during keratinocyte differentiation, and KDF1 regulates the ubiquitination and the protein level of IKKα.

-
AWCL from WT and KDF1 KO cells before and after calcium shift were immunoblotted with different antibodies as indicated. Lo: low calcium; Hi: high calcium.
-
B, CWT and KDF1 KO keratinocytes were treated with 20 nM cycloheximide (CHX). WCL was collected at 0‐, 30‐, 60‐, 90‐, and 120‐min post‐CHX treatment and subjected to immunoblotting with IKKα antibody (B). Band intensity is determined by densitometry, and the amount of IKKα is calculated and quantified (C). Statistical analysis is conducted using two‐way ANOVA. N = 3 (biological replicates). *P < 0.05. Error bar represents SD (standard deviation).
-
D, EWT and KDF1 KO keratinocyte (right panels) or cells transfected with plasmid encoding exogenous IKKα (left panels) were treated with MG132 at 10 μM for 6 h and then were subjected to immunoprecipitation using anti‐ubiquitin (Ub) antibody. IP and WCL were analyzed by immunoblots with α‐IKKα antibody (D). Overall intensity of all ubiquitinated IKKα bands was determined by densitometry. Ratio of ubiquitinated IKKα was quantified and presented as bar graphs (E). Statistical analysis is conducted using unpaired Student's t‐test. Error bar represents SD (standard deviation). N = 3 (biological replicates). *P < 0.05. Kd: kilodalton for molecular weight markers.
-
FIKKα protein level in KDF1 KO keratinocyte expressing WT KDF1 or KDF1 mutant, under both low and high calcium conditions, was examined and quantified by Western blotting (quantification shown in Fig EV4C).
-
GPiggyBac transposon was used to ectopically express HA‐tagged IKKα in KDF1‐deficient cells. WCL was collected and analyzed by immunoblots with different antibodies as indicated.
-
HOverexpression of IKKα in KDF1 KO keratinocyte can restore normal skin stratification. Skin sections from grafted tissue were immunostained with different antibodies as indicated. Dotted lines denote dermal–epidermal boundaries. Epi: epidermis, Der: dermis. Scale bars = 50 μm.
To further test this hypothesis, we try to rescue KDF1 KO cells with exogenous expression of IKKα (Fig 4G). Interestingly, upon skin engraftment, we find that ectopic expression of IKKα can significantly restore skin stratification, including decreased epidermal thickness and normal expression of Krt14 in the basal cell layer (Fig 4H, and quantification in Fig EV4D and E). Although exogenous expression of IKKα can reduce overall epidermal thickness, it alone cannot fully restore the normal epidermal structure, suggesting that additional factors/pathways may be involved in KDF1‐regulation of epidermal differentiation as well.
KDF1 controls the ubiquitination level of IKKα via USP7
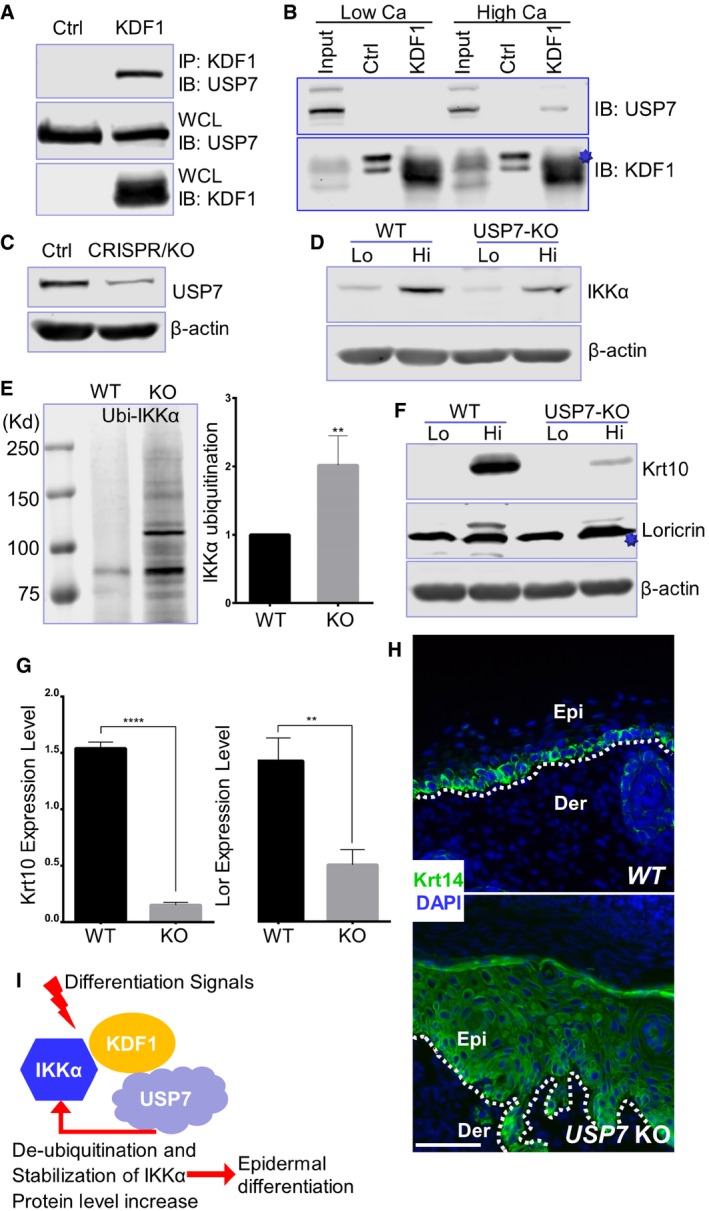
DUBs are special proteases that can recognize and specifically cleave ubiquitin or ubiquitin‐like proteins from target molecules. Different DUBs have been shown to be involved in various cellular processes including protein stability, cell cycle regulation, and chromatin remodeling 20, 21. Our proteomics analysis of KDF1 interactome (Fig 1A) demonstrates another potential binding partner of KDF1, USP7, which is a DUB and can reverse both poly‐ and mono‐ubiquitination of protein targets. The interaction between KDF1 and USP7 was confirmed in vitro by co‐immunoprecipitation assay (Fig 5A). Interestingly, as KDF1 interaction with IKKα, endogenous KDF1 association with USP7 can only be detected in differentiated primary skin keratinocytes (Fig 5B, and source data in Fig EV5A). Consistent with previous report 24, USP7 shows strong nuclear localization in cultured epidermal keratinocytes (Fig EV4A).
Figure 5. USP7 regulates IKKα ubiquitination and skin differentiation.
-
AHEK293 cells were transfected to co‐express KDF1 with USP7. IP and WCL were blotted with different antibodies as indicated.
-
BImmunoprecipitation was carried out with α‐KDF1 antibody in keratinocyte cultured in medium with low or high concentration of calcium (Ca), same as in Fig 1D. IPs were blotted with different antibodies as indicated. The star denotes KDF1.
-
CWCL from WT and USP7 CRISPR KO cells was subjected to immunoblotting with different antibodies as indicated.
-
DIKKα protein level in WT and USP7 KO keratinocyte was examined by immunoblotting before and after calcium shift.
-
EWT and USP7 KO keratinocytes were treated with MG132 at 10 μM for 6 h and then subjected to immunoprecipitation using anti‐ubiquitin antibody. Precipitated product was analyzed by immunoblotting with IKKα antibody. Band intensity was determined by densitometry and shown as bar graphs. Statistical analysis is conducted using unpaired Student's t‐test. Error bar represents SD (standard deviation). N = 3 (biological replicates). **P < 0.01. Kd: kilodalton for molecular weight markers.
-
F, GImmunoblot of Krt10 and loricrin (Lor) with WT and USP7 KO cells before and after calcium shift (F). Star denotes an unspecific band in α‐Loricrin blots. Band intensity was determined by densitometry and shown as bar graphs (G). Statistical analysis is conducted using unpaired Student's t‐test. Error bar represents SD (standard deviation). N = 3 (biological replicates). ****P < 0.0001; **P < 0.01.
-
HSections of engrafted skin developed from USP7 KO or control WT cells were immunostained with different antibodies as indicated. Dotted lines denote dermal–epidermal boundaries. Epi: epidermis, Der: dermis. Scale bar = 100 μm.
-
IA working model of epidermal differentiation regulated by KDF1. Upon differentiation signals, KDF1 associates with IKKα and recruits deubiquitination enzyme USP7 to the protein complex. USP7 can deubiquitinate IKKα and promote its protein stability, which will in turn promote epidermal differentiation.
Figure EV5. USP7 regulates epidermal differentiation.

-
AUnmodified blots for Fig 5B.
-
BWCL from WT and USP7 CRISPR KO cells after calcium shift were subjected to immunoblotting with different antibodies as indicated.
-
CLevel of IKKα (main Fig 5D) was quantified by densitometry and presented as bar graph. Statistical analysis is conducted using unpaired Student's t‐test. Error bar represents SD (standard deviation). N = 3. *P < 0.05. Level of IKKα in WT cells was significantly higher than that in USP7 KO cells upon calcium shift.
-
D, EThickness of Krt14‐positive cell layers (D) and the whole epidermis (E) in skin grafts generated from WT and USP7 KO cells were quantified and showed as box and whisker plots. Each cell type had at least 3 biological replicates (independent skin grafts). At least 3 sections were taken from each graft for analysis and quantification. Statistical analysis is conducted using unpaired Student's t‐test. N > 3. ****P < 0.0001.
The potential role of USP7 in skin development and keratinocyte differentiation has not been addressed before, although it has been shown to regulate the turnover of many signaling molecules, such as p53 and PTEN 24, 25. To this end, we first employed CRISPR (clustered regularly interspaced short palindromic repeats) technology and generated USP7 KO keratinocyte (Figs 5C and EV5B). Deletion of USP7 in skin keratinocytes leads to decreased IKKα protein level upon differentiation (Fig 5D, and quantification in Fig EV5C). Additionally, loss of USP7 results in enhanced IKKα ubiquitination (Fig 5E). When induced to differentiation by calcium shift, the USP7 KO cells exhibit significantly reduced expression of Krt10 and loricrin (Fig 5F and quantification in 5G). Regenerated skin from transplanted USP7 KO cells displays striking epidermal abnormalities similar to KDF1 and IKKα KO cells, including thickened epidermis and expansion of basal cells (Fig 5H and quantification in Fig EV5D and E). Together, our studies suggest that KDF1 regulates IKKα ubiquitination and protein stability by recruiting USP7, a deubiquitinating enzyme, which is essential for epidermal differentiation (Fig 5I).
Discussion
Adult tissue homeostasis and wound repair are mediated by the delicate balance between cell proliferation, cell death, and differentiation. Skin provides an essential barrier protecting us from various environmental damages. Aberrant tissue homeostasis or wound repair can lead to dire consequence for our survival. In skin, both processes are driven by the epidermal stem/progenitor cells that localize at the basal layer of the skin epidermis 1, 32, 33. Differentiation of epidermal progenitor cells is a complex but fascinating process, involving the permanent withdrawal of cells from the cell cycle, the synthesis and modification of various protein and lipid components of the cornified envelop, and the controlled dissolution of cellular organelles and the nucleus 1, 34. Aberrant skin differentiation contributes to the development of various skin diseases, including psoriasis, inflammatory skin diseases, and SCC. Cutaneous SCC is the second most common human cancer, afflicting more than 250,000 patients in the United States every year 35, 36. Cutaneous SCC can be highly invasive and metastatic (3–10% rate of metastasis), and a significant number of patients with a primary SCCs develop secondary lesions within 5 years of diagnosis, leading to severe morbidity and mortality 37. Thus, understanding epidermal differentiation and its underlying molecular mechanisms is critical for devising effective therapeutic strategies for the treatment of various skin diseases. In this study, by employing a combinatory approach encompassing mouse genetics with molecular and cell biology studies, we provided compelling evidence that KDF1 regulates epidermal differentiation by forming a regulatory complex with IKKα, and controlling IKKα protein stability via recruiting a deubiquitinating enzyme, USP7.
KDF1 was initially identified by a forward genetics screen as a key regulator of epidermal differentiation in mouse skin 4. Recent studies suggest that KDF1 is also involved in tooth agenesis, and mutation of KDF1 has been identified in patients with ectodermal dysplasia, a heterogeneous group of diseases that affects the ectoderm derivatives, such as skin, hair follicles, teeth, and nails 38. Despite its potentially important role in tissue development and homeostasis, little is known about the molecular role of KDF1. Previous study suggests that KDF1 can affect expression of p63 in skin keratinocytes, and genetically interacts with stratifin (Sfn, 14‐3‐3σ). Our current study shows that KDF1 association with IKKα is essential for epidermal differentiation, and ectopic expression of IKKα can rescue skin stratification in KDF1 null cells, strongly suggesting that IKKα acts as an important downstream partner of KDF1 in epidermal differentiation.
It has been demonstrated in multiple independent studies that loss of IKKα leads to striking skin abnormalities, including defective epidermal differentiation 6, 7, 39. Studies by Hu, et al 9 and by Gareus et al 40 also indicate that ablation of IKKα in cultured skin keratinocytes can directly inhibit epidermal differentiation, strongly suggesting that IKKα regulates skin differentiation via a keratinocyte cell‐autonomous manner. Conditional KO (knockout) of IKKα with a Krt14 promoter‐driven Cre transgenic line leads to perinatal lethality. Mutant mice have rather normal epidermal stratification but aberrant skin barrier function, suggesting impaired terminal differentiation capability of IKKα‐deficient keratinocytes 40. By contrast, mice with conditional deletion of IKKα by a Krt5 promoter‐driven Cre can survive to adulthood but exhibit epidermal hyperproliferation and skin carcinogenesis phenotypes 10. It is possible that this discrepancy results from potential difference from different Cre transgenic lines used 41. Additionally, it is noteworthy that IKKα may act in a paracrine or autocrine manner by controlling the secretion of a hitherto unidentified kDIF (keratinocyte differentiation‐inducing factor) to regulate epidermal differentiation 9. Thus, an incomplete deletion of IKKα may lead to mild phenotypes in vivo.
It remains incompletely understood how IKKα regulates epidermal stratification. IKKα has been shown to be able to translocate to the cell nucleus and directly regulate target gene expressions. In 2004, Sil et al 42 detected increased levels of nuclear IKKα induced by keratinocyte differentiation and identified the putative NLS (nuclear localization sequence) within the kinase domain of IKKα. The transcriptional activity of IKKα was further illustrated by Liu et al, 11 that IKKα negatively regulates VEGF‐A expression via binding to the distal VEGF‐A (vascular endothelial growth factor‐A) promoter. Besides VEGF‐A, a number of other transcriptional targets of IKKα have been identified, including EGF (epidermal growth factor), HB‐EGF (heparin‐binding EGF‐like growth factor), and amphiregulin 10. Interestingly, it is noteworthy that IKKα can also regulate transcription of Sfn by preventing its hypermethylation and silencing 43. Sfn has been shown to genetically interact with KDF1 in epidermal development pathways 4. In this study, we illustrated a critical interaction of KDF1 with IKKα, which is mediated by the LZ domain of IKKα. Deletion of LZ domain in IKKα will abolish this interaction and suppress epidermal differentiation. However, LZ domain is also involved in heterodimerization or homodimerization of IKKα and IKKβ 44, and deletion of LZ domain can block kinase activity of IKKα 45, 46. Future analysis will be required to delineate the precise molecular pathways whereby LZ domain of IKKα is involved in skin differentiation.
Protein ubiquitination and deubiquitination are highly dynamic but tightly controlled processes, regulating not only proteostasis but also function of the target proteins. The DUBs are special proteases that can reverse the modification of target proteins by single ubiquitin and polyubiquitin. The human genome encodes nearly one hundred DUBs, and their substrate specificity can be modulated by different mechanisms 20. Ubiquitin‐specific DUBs usually contain multiple domains with insertions and/or extensions that can control their substrate specificity. Additionally, the substrate specificity and subcellular localization of DUBs can be regulated by their protein binding partners. USP7 was first discovered as a binding protein for a herpes virus regulatory protein 47, 48. However, accumulating evidence suggests that USP7 plays critical but diverse roles in many different cellular processes including host–virus interaction, DNA repair, transcription, epigenetic regulation, and tumorigenesis, potentially through its many identified downstream targets, including p53, PTEN, FOXO4, and NFκB pathway proteins 22, 23, 24, 25, 26, 27. Null mutation of USP7 in mice leads to early embryonic lethality, suggesting its important role in development 27. Our data showed that it is also an essential gene involved in differentiation of skin epidermal cells. Although our results demonstrated regulation of IKKα deubiquitination by USP7, we cannot rule out the possibility that loss of USP7 in skin keratinocytes may lead to changes of other signaling proteins. Characterization of potential IKKα mutant deficient for ubiquitination together with proteomics analysis of USP7 KO epidermal cells and functional studies in vitro and in vivo will be essential to resolve this issue in the future.
In closing, our findings reveal an important molecular mechanism underlying epidermal differentiation and provide an important basis for the development of rationally based, molecularly targeted drugs for the treatment of various skin diseases, including skin cancers.
Materials and Methods
Antibodies, reagents, and plasmid DNA constructions
Loricrin and filaggrin antibodies were generous gifts from Dr. Elaine Fuchs at the Rockefeller University. Chicken Krt14, and rabbit Krt5 and Krt10 antibodies were obtained from Covance (Princeton, NJ). Rat monoclonal β4‐integrin (CD104) was obtained from BD Pharmingen (Franklin lakes, NJ). KDF1 antibody (HPA028639), α‐Flag antibody, and EZview™ Red anti‐HA affinity beads were obtained from Sigma (St. Louis, MO). IKKα antibody (#2682) and normal rabbit IgG were obtained from Cell Signaling Technology (Danvers, MA). Loricrin (55439‐1‐AP), α/β‐tubulin, and β‐actin were obtained from Proteintech® (Rosemont, IL). Rabbit polyclonal antibodies against HA and GST, and protein A/G beads were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Mono‐ and polyubiquitinylated conjugates monoclonal antibody (FK2) was obtained from Enzo (Farmingdale, NY). Other chemicals or reagents were obtained from Sigma (St. Louis, MO), unless otherwise indicated.
Primers used to generate IKKα and KDF1 mutants are listed as follows (core sequence only): IKKα‐mut‐1: forward‐ATG GAG CGG CCC, reverse‐ACG CTC AAT ACG AGA CTG TAG TGA ATG A; IKKα‐mut‐2: forward‐AGT CTT CAT TCA CTA CAG TCT CGT AT, reverse‐TCA TTC TGT TAA CCA ACT CCA ATC A; IKKα‐mut‐3: forward‐GTG CAC TAT GTG TCT GGA CTA A, reverse‐TTC TAG ACT GGA TCC TAC AAG GG; IKKα‐mut‐4: forward‐AGA CGT CAG GGA GAC TTG AT, reverse‐TCA TTC TGT TAA CCA ACT CCA ATC A; IKKα‐mut‐5: forward‐GTG CAC TAT GTG TCT GGA CTA A, reverse‐AGA TTC CAT CAA GTC TCC CTG AC; IKKα‐mut‐6 (bridge gap): forward‐ACA AAG GGC AGC AAT TCA GCT TGA CT, reverse‐AGT CAA GCT GAA TTG CTG CCC TTT GT; KDF1‐mut‐1: forward‐ATG CCC AGG CCG GGA CAG CCC CG, reverse‐GCC CAT GCT TGT CTT GAG CCT C; KDF1‐mut‐2: forward‐CAG AGG CTC AAG ACA AGC AT, reverse‐GCA GTA CAC CTG CAG CAG GGG TG; KDF1‐mut‐3: forward‐CAG AGG CTC AAG ACA AGC AT, reverse‐TGA GAT CTT GCT GGT CTT CTC; KDF1‐mut‐4: forward‐GAG AAG ACC AGC AAG ATC TCA G, reverse‐GCA GTA CAC CTG CAG CAG GGG TG; and KDF1‐mut‐5 (bridge gap): forward‐AGG CTC AAG ACA AGC GAG AAG ACC AGC AAG, reverse‐CTT GCT GGT CTT CTC GCT TGT CTT GAG CCT. The mRNA level of IKKα was examined via RT–qPCR using primers: GAC TGT ATA TGA AGG ACC ATT TGC; GTC TTC CTT TAG CCC AGA TAC G.
SILAC‐MS and proteomics analysis
Undifferentiated WT keratinocytes were subjected to SILAC label. l‐lysine‐2HCl (4, 4, 5, 5‐D4) and l‐arginine‐HCl (μ‐13C6) (Cambridge Isotope Laboratories Inc, Andover, MA) were used to replace the regular lysine and arginine in the medium for heavy isotope labeling. Cells with light isotope labeling were used as a control. Heavy isotope‐labeled cells were transfected with construct expressing HA‐ and His6‐tagged KDF1. The cells were subjected to calcium shift for 24 h before lysis with RIPA (radioimmunoprecipiation assay) buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 10% Glycerol, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X‐100, 1% sodium deoxycholate, 0.1% SDS) containing protease inhibitors. We mixed the same amount of heavy labeled proteins and light labeled proteins, and conducted sequential purification with Ni‐NTA column and immunoprecipitation with anti‐HA affinity agarose (Sigma, St. Louis, MO). The product from tandem affinity purification was resolved in SDS–PAGE and subjected to identification with LC‐MS/MS. Fractionation was conducted using trypsin digestion.
Cell culture
Primary mouse keratinocytes were isolated using previously reported methods 49. Epidermis of newborn mice or E18.5 was separated from dermis by an overnight treatment with dispase. Then, the primary keratinocytes were dispersed from the epidermis using trypsin. Keratinocytes were co‐cultured with mitomycin C‐treated 3T3 fibroblast feeder cells until the third passage. Cells were maintained in E‐media supplemented with 15% FBS. The final concentration of Ca2+ is 0.05 mM. High calcium shift was performed using E‐media supplemented with 15% FBS, with Ca2+ at a final concentration of 1.5 mM. HEK293 cells were cultured in DMEM supplemented with 10% FBS.
Cell transfection was carried out with Lipofectamine™ 3000 reagent (Invitrogen, CA), following manufacturer suggested protocol. A Tet‐On 3G tetracycline‐inducible expression system (Takara/Clontech) was used for inducible transgene expression. 50 ng/ml of doxycycline was used to induce gene expression in vitro.
Animals
KDF1 mutant strain was a generous gift from Dr. Scott D. Weatherbee (Yale University) 4. The IKKα KO strain was a generous gift from Dr. Anning Lin (the University of Chicago) 6. Nude mice for skin transplantation were purchased from the Jackson Laboratory (002019‐NU/J). Around 8‐week‐old female nude mice were used for skin grafting. All mice used in this study were bred and maintained at the ARC (animal resource center) of the University of Chicago in accordance with institutional guidelines. For skin inflammation analysis, adult CD1 WT mice (8–11 weeks old) were treated topically with commercial Aldara cream (5% imiquimod cream) for 7 days.
Protein biochemical analysis
Western blot was conducted as previously described 50. Cell lysates were prepared with RIPA buffer containing protease inhibitors. After the concentration of total protein is assessed, equal amounts of the cell lysates were resolved in sodium dodecyl sulfate–polyacrylamide gel and electroblotted onto a nitrocellulose membrane. The membrane was incubated with Odyssey blocking buffer (Li‐COR biosciences, Lincoln, NE) for 1 h at room temperature, followed by an overnight incubation with desired primary antibody at 4°C. Immunoblots were washed three times with 1× Tween 20/phosphate‐buffered saline (PBST) and incubated with secondary antibody (1:10,000 dilution) at room temperature for 1 h. Blot was washed with 1× PBST for another three times. LI‐COR Odyssey scanner was used to visualize the blotting signals, and LI‐COR Biosciences Software was used to conduct the quantification.
For immunoprecipitation, cell lysates were prepared with RIPA buffer containing protease inhibitors. After the concentration of total protein is assessed, equal amounts of the cell lysates (200–400 μg) were pre‐cleaned using 30 μl protein A/G beads (Santa Cruz, CA). 5–10% samples were kept as input, and the remaining samples were incubated with desired antibody overnight at 4°C. 30 μl protein A/G beads (Santa Cruz, CA) were added on the following day and kept rotating for another 2 h at 4°C. The antibody and associated proteins were precipitated with beads via centrifuge, followed by five times washing step using lysis buffer. The precipitated proteins were analyzed by Western blot as described above.
Skin organotypic culture and grafting
Skin organotypic culture and grafting were performed as previously described 49. Decellularized dermis (1 × 1 cm square shape) was prepared from newborn CD1 mice skin via EDTA treatment 29. 2 × 106 cultured keratinocytes with desired genomic modifications were seeded onto the dermis in cell culture insert. Then, the skin culture was exposed to air/liquid interphase after an overnight attachment to form skin organoids. For grafting with skin organoids, nude mice aged ~ 8 weeks were anaesthetized. Two 1 × 1 cm square shape wounds were introduced to the back skin of the nude mice. After transplantation of skin organoids to the fresh wounds, the wound edge was sealed with surgical glue. The animals with skin graft were housed separately, and the bandages over the wound could be removed 1 week after surgery 49, 51, 52. To induce exogenous gene expression in Tet regulated system, doxycycline food (TD. 120658, Envigo, Huntington, UK) was given after bandages removed until the end of the study. All the experiments were repeated more than three times (three biological replicates). For phenotypic analysis using immunostaining, at least three sections were taken from each graft for quantifications.
Histology and immunofluorescence
Skin samples were embedded in optimal cutting temperature (OTC) compound, sectioned, and fixed in 4% paraformaldehyde. Hematoxylin and eosin (HE) staining or immunofluorescence staining of desired sections was conducted as previously described 53. Antibodies were diluted following the manufacturer's instructions unless indicated. Images were taken using EVOS FL imaging system. Evaluation of epidermal differentiation markers and measurement of epidermal thickness were carried out using ImageJ.
Statistical analysis
Statistical analysis was performed using Excel or GraphPad Prism software. Box plots were used to represent the entire population without assumptions on the statistical distribution. In most experiments, Student's t‐test was used to evaluate the statistical significance of the difference (P value). For results in Fig 4C, two‐way ANOVA (analysis of variance) was used to assess the statistical significance.
Author contributions
YL, LT, AL, SDW, and XW designed the experiments. YL, JY, and XG performed the experiments. YL and XW analyzed the data. XW wrote the manuscript. All authors edited the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Dataset EV1
Review Process File
Acknowledgements
We are very grateful to Dr. Elaine Fuchs at the Rockefeller University for sharing reagents and technical assistance. We thank Linda Degenstein at the transgenic core facility and Dr. Don Wolfgeher at the proteomics core facility at the University of Chicago for excellent technical assistance. The animal studies were carried out in the ALAAC‐accredited animal research facility at the University of Chicago. This work was supported by grants NIH R01AR063630 and R01OD023700; the Research Scholar Grant (RSG‐13‐198‐01) from the American Cancer Society; and the V scholar award from V foundation to XW. YL is supported by the University of Chicago Cancer Biology Training Grant (T32, CA009594).
EMBO Reports (2020) 21: e48566
Data availability
The datasets produced in this study are available in the following databases: Proteomics data: PRIDE (accession: PXD015673; http://www.ebi.ac.uk/pride/archive/projects/PXD015673).
References
- 1. Fuchs E (2008) Skin stem cells: rising to the surface. J Cell Biol 180: 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lopez‐Pajares V, Yan K, Zarnegar BJ, Jameson KL, Khavari PA (2013) Genetic pathways in disorders of epidermal differentiation. Trends Genet 29: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Perdigoto CN, Valdes VJ, Bardot ES, Ezhkova E (2014) Epigenetic regulation of epidermal differentiation. Cold Spring Harb Perspect Med 4: a015263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee S, Kong Y, Weatherbee SD (2013) Forward genetics identifies Kdf1/1810019J16Rik as an essential regulator of the proliferation‐differentiation decision in epidermal progenitor cells. Dev Biol 383: 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pasparakis M (2009) Regulation of tissue homeostasis by NF‐kappaB signalling: implications for inflammatory diseases. Nat Rev Immunol 9: 778–788 [DOI] [PubMed] [Google Scholar]
- 6. Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M (1999) Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science 284: 316–320 [DOI] [PubMed] [Google Scholar]
- 7. Takeda K, Takeuchi O, Tsujimura T, Itami S, Adachi O, Kawai T, Sanjo H, Yoshikawa K, Terada N, Akira S (1999) Limb and skin abnormalities in mice lacking IKKalpha. Science 284: 313–316 [DOI] [PubMed] [Google Scholar]
- 8. Li ZW, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, Johnson R, Karin M (1999) The IKKbeta subunit of IkappaB kinase (IKK) is essential for nuclear factor kappaB activation and prevention of apoptosis. J Exp Med 189: 1839–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu Y, Baud V, Oga T, Kim KI, Yoshida K, Karin M (2001) IKKalpha controls formation of the epidermis independently of NF‐kappaB. Nature 410: 710–714 [DOI] [PubMed] [Google Scholar]
- 10. Liu B, Xia X, Zhu F, Park E, Carbajal S, Kiguchi K, DiGiovanni J, Fischer SM, Hu Y (2008) IKKalpha is required to maintain skin homeostasis and prevent skin cancer. Cancer Cell 14: 212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu B, Park E, Zhu F, Bustos T, Liu J, Shen J, Fischer SM, Hu Y (2006) A critical role for I kappaB kinase alpha in the development of human and mouse squamous cell carcinomas. Proc Natl Acad Sci USA 103: 17202–17207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maeda G, Chiba T, Kawashiri S, Satoh T, Imai K (2007) Epigenetic inactivation of IkappaB Kinase‐alpha in oral carcinomas and tumor progression. Clin Cancer Res 13: 5041–5047 [DOI] [PubMed] [Google Scholar]
- 13. Park E, Zhu F, Liu B, Xia X, Shen J, Bustos T, Fischer SM, Hu Y (2007) Reduction in IkappaB kinase alpha expression promotes the development of skin papillomas and carcinomas. Can Res 67: 9158–9168 [DOI] [PubMed] [Google Scholar]
- 14. Deng L, Li Y, Ai P, Xie Y, Zhu H, Chen N (2015) Increase in IkappaB kinase alpha expression suppresses the tumor progression and improves the prognosis for nasopharyngeal carcinoma. Mol Carcinog 54: 156–165 [DOI] [PubMed] [Google Scholar]
- 15. Park E, Liu B, Xia X, Zhu F, Jami WB, Hu Y (2011) Role of IKKalpha in skin squamous cell carcinomas. Future Oncol 7: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Swatek KN, Komander D (2016) Ubiquitin modifications. Cell Res 26: 399–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81: 203–229 [DOI] [PubMed] [Google Scholar]
- 18. Ikeda F, Crosetto N, Dikic I (2010) What determines the specificity and outcomes of ubiquitin signaling? Cell 143: 677–681 [DOI] [PubMed] [Google Scholar]
- 19. Hu H, Sun SC (2016) Ubiquitin signaling in immune responses. Cell Res 26: 457–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbe S (2013) Deubiquitylases from genes to organism. Physiol Rev 93: 1289–1315 [DOI] [PubMed] [Google Scholar]
- 21. Komander D, Clague MJ, Urbe S (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10: 550–563 [DOI] [PubMed] [Google Scholar]
- 22. Colleran A, Collins PE, O'Carroll C, Ahmed A, Mao X, McManus B, Kiely PA, Burstein E, Carmody RJ (2013) Deubiquitination of NF‐kappaB by ubiquitin‐specific protease‐7 promotes transcription. Proc Natl Acad Sci USA 110: 618–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li T, Guan J, Li S, Zhang X, Zheng X (2014) HSCARG downregulates NF‐kappaB signaling by interacting with USP7 and inhibiting NEMO ubiquitination. Cell Death Dis 5: e1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song MS, Salmena L, Carracedo A, Egia A, Lo‐Coco F, Teruya‐Feldstein J, Pandolfi PP (2008) The deubiquitinylation and localization of PTEN are regulated by a HAUSP‐PML network. Nature 455: 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brooks CL, Li M, Hu M, Shi Y, Gu W (2007) The p53–Mdm2–HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene 26: 7262–7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van der Horst A, de Vries‐Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM (2006) FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol 8: 1064–1073 [DOI] [PubMed] [Google Scholar]
- 27. Kon N, Kobayashi Y, Li M, Brooks CL, Ludwig T, Gu W (2010) Inactivation of HAUSP in vivo modulates p53 function. Oncogene 29: 1270–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Israel A (2010) The IKK complex, a central regulator of NF‐kappaB activation. Cold Spring Harb Perspect Biol 2: a000158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prunieras M, Regnier M, Woodley D (1983) Methods for cultivation of keratinocytes with an air‐liquid interface. J Invest Dermatol 81: 28s–33s [DOI] [PubMed] [Google Scholar]
- 30. Descargues P, Sil AK, Sano Y, Korchynskyi O, Han G, Owens P, Wang XJ, Karin M (2008) IKKalpha is a critical coregulator of a Smad4‐independent TGFbeta‐Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc Natl Acad Sci USA 105: 2487–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamaguchi T, Kimura J, Miki Y, Yoshida K (2007) The deubiquitinating enzyme USP11 controls an IkappaB kinase alpha (IKKalpha)‐p53 signaling pathway in response to tumor necrosis factor alpha (TNFalpha). J Biol Chem 282: 33943–33948 [DOI] [PubMed] [Google Scholar]
- 32. Blanpain C, Fuchs E (2006) Epidermal stem cells of the skin. Annu Rev Cell Dev Biol 22: 339–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Candi E, Schmidt R, Melino G (2005) The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol 6: 328–340 [DOI] [PubMed] [Google Scholar]
- 34. Koster MI, Roop DR (2007) Mechanisms regulating epithelial stratification. Annu Rev Cell Dev Biol 23: 93–113 [DOI] [PubMed] [Google Scholar]
- 35. Preston DS, Stern RS (1992) Nonmelanoma cancers of the skin. N Engl J Med 327: 1649–1662 [DOI] [PubMed] [Google Scholar]
- 36. Alam M, Ratner D (2001) Cutaneous squamous‐cell carcinoma. N Engl J Med 344: 975–983 [DOI] [PubMed] [Google Scholar]
- 37. Burton KA, Ashack KA, Khachemoune A (2016) Cutaneous squamous cell carcinoma: a review of high‐risk and metastatic disease. Am J Clin Dermatol 17: 491–508 [DOI] [PubMed] [Google Scholar]
- 38. Shamseldin HE, Khalifa O, Binamer YM, Almutawa A, Arold ST, Zaidan H, Alkuraya FS (2017) KDF1, encoding keratinocyte differentiation factor 1, is mutated in a multigenerational family with ectodermal dysplasia. Hum Genet 136: 99–105 [DOI] [PubMed] [Google Scholar]
- 39. Li Q, Lu Q, Hwang JY, Buscher D, Lee KF, Izpisua‐Belmonte JC, Verma IM (1999) IKK1‐deficient mice exhibit abnormal development of skin and skeleton. Genes Dev 13: 1322–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gareus R, Huth M, Breiden B, Nenci A, Rosch N, Haase I, Bloch W, Sandhoff K, Pasparakis M (2007) Normal epidermal differentiation but impaired skin‐barrier formation upon keratinocyte‐restricted IKK1 ablation. Nat Cell Biol 9: 461–469 [DOI] [PubMed] [Google Scholar]
- 41. Heffner CS, Herbert Pratt C, Babiuk RP, Sharma Y, Rockwood SF, Donahue LR, Eppig JT, Murray SA (2012) Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat Commun 3: 1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sil AK, Maeda S, Sano Y, Roop DR, Karin M (2004) IkappaB kinase‐alpha acts in the epidermis to control skeletal and craniofacial morphogenesis. Nature 428: 660–664 [DOI] [PubMed] [Google Scholar]
- 43. Zhu F, Xia X, Liu B, Shen J, Hu Y, Person M, Hu Y (2007) IKKalpha shields 14‐3‐3sigma, a G(2)/M cell cycle checkpoint gene, from hypermethylation, preventing its silencing. Mol Cell 27: 214–227 [DOI] [PubMed] [Google Scholar]
- 44. Kwak YT, Guo J, Shen J, Gaynor RB (2000) Analysis of domains in the IKKalpha and IKKbeta proteins that regulate their kinase activity. J Biol Chem 275: 14752–14759 [DOI] [PubMed] [Google Scholar]
- 45. Delhase M, Hayakawa M, Chen Y, Karin M (1999) Positive and negative regulation of IkappaB kinase activity through IKKbeta subunit phosphorylation. Science 284: 309–313 [DOI] [PubMed] [Google Scholar]
- 46. Zandi E, Chen Y, Karin M (1998) Direct phosphorylation of IkappaB by IKKalpha and IKKbeta: discrimination between free and NF‐kappaB‐bound substrate. Science 281: 1360–1363 [DOI] [PubMed] [Google Scholar]
- 47. Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J (1997) A novel ubiquitin‐specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J 16: 1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meredith M, Orr A, Everett R (1994) Herpes simplex virus type 1 immediate‐early protein Vmw110 binds strongly and specifically to a 135‐kDa cellular protein. Virology 200: 457–469 [DOI] [PubMed] [Google Scholar]
- 49. Lee P, Jiang S, Li Y, Yue J, Gou X, Chen SY, Zhao Y, Schober M, Tan M, Wu X (2017) Phosphorylation of Pkp1 by RIPK4 regulates epidermal differentiation and skin tumorigenesis. EMBO J 36: 1963–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu X, Suetsugu S, Cooper LA, Takenawa T, Guan JL (2004) Focal adhesion kinase regulation of N‐WASP subcellular localization and function. J Biol Chem 279: 9565–9576 [DOI] [PubMed] [Google Scholar]
- 51. Liu H, Yue J, Huang H, Gou X, Chen SY, Zhao Y, Wu X (2015) Regulation of focal adhesion dynamics and cell motility by the EB2 and Hax1 protein complex. J Biol Chem 290: 30771–30782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yue J, Zhang Y, Liang WG, Gou X, Lee P, Liu H, Lyu W, Tang WJ, Chen SY, Yang F et al (2016) In vivo epidermal migration requires focal adhesion targeting of ACF7. Nat Commun 7: 11692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, Fuchs E (2007) Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell 12: 313–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Dataset EV1
Review Process File
Data Availability Statement
The datasets produced in this study are available in the following databases: Proteomics data: PRIDE (accession: PXD015673; http://www.ebi.ac.uk/pride/archive/projects/PXD015673).


