Abstract
Pruning that selectively eliminates inappropriate projections is crucial for sculpting neural circuits during development. During Drosophila metamorphosis, ddaC sensory neurons undergo dendrite‐specific pruning in response to the steroid hormone ecdysone. However, the understanding of the molecular mechanisms underlying dendrite pruning remains incomplete. Here, we show that protein phosphatase 2A (PP2A) is required for dendrite pruning. The catalytic (Microtubule star/Mts), scaffolding (PP2A‐29B), and two regulatory subunits (Widerborst/Wdb and Twins/Tws) play important roles in dendrite pruning. Functional analyses indicate that PP2A, via Wdb, facilitates the expression of Sox14 and Mical prior to dendrite pruning. Furthermore, PP2A, via Tws, governs the minus‐end‐out orientation of microtubules (MTs) in the dendrites. Moreover, the levels of Klp10A, a MT depolymerase, increase when PP2A is compromised. Attenuation of Klp10A fully rescues the MT orientation defects in mts or pp2a‐29b RNAi ddaC neurons, suggesting that PP2A governs dendritic MT orientation by suppressing Klp10A levels and/or function. Taken together, this study sheds light on a novel function of PP2A in regulating dendrite pruning and dendritic MT polarity in sensory neurons.
Keywords: dendrite pruning, Klp10A, microtubule orientation, neuron, protein phosphatase
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Neuroscience; Post-translational Modifications, Proteolysis & Proteomics
Protein phosphatase PP2A plays a crucial role in dendrite pruning in Drosophila sensory neurons. PP2A acts via two regulatory subunits Widerborst and Twins to mediate ecdysone signaling and to control microtubule orientation in dendrites.

Introduction
Neuronal pruning that precisely removes exuberant neuronal connections and excessive neurites is essential for the maturation of the nervous system during animal development 1. Neuronal pruning is a highly conserved process widely occurring in both vertebrates and invertebrates during the development of the nervous systems 2, 3. In both central and peripheral nervous systems of mammals, neurons selectively eliminate the superfluous and unneeded neurites to establish functional circuits 4. Drosophila has emerged as an attractive model to elucidate the underlying mechanisms of neuronal pruning over the last decade 5, 6. In Drosophila, the nervous system undergoes dramatic remodeling during metamorphosis in order to build up adult‐specific neuronal networks 3, 7. In the central nervous system (CNS), mushroom body γ neurons remove their axonal branches from the dorsal and medial lobes as well as their entire dendrites and subsequently regrow to form adult‐specific circuits 8. Likewise, in the peripheral nervous system (PNS), some sensory neurons, including dorsal dendritic arborization (da) neurons, undergo either apoptosis or pruning during early metamorphosis. Class I (ddaD/E) and class IV (ddaC, also known as C4da) neurons selectively remove their larval dendrite branches but keep their axons intact 9, 10, while class II (ddaB) and class III (ddaA/F) neurons are apoptotic 10. Defects in neuronal pruning have been shown to be associated with brain disorders, such as autism spectrum disorder (ASD) and schizophrenia 11, 12. Thus, studying the molecular and cellular mechanisms of developmental pruning would potentially contribute to our understanding of the pathogenesis of human brain disorders.
Drosophila ddaC sensory neuron is an excellent model to study developmental dendrite pruning due to its highly branched dendrites and the ease of live‐cell imaging. In response to a late‐larval pulse of the steroid molting hormone 20‐hydroxyecdysone (20E), ddaC neurons undergo a stereotyped pruning process in which their larval dendrites are first severed at the proximal regions, subsequently fragmented and removed by phagocyte‐mediated debris clearance (Fig 1A) 9, 10, 13. Dendrite pruning of ddaC neurons is initiated by an ecdysone‐induced signaling cascade. The neuron‐specific ecdysone receptor EcR‐B1, which is upregulated in response to the late‐larval ecdysone pulse, is required to induce the expression of the transcription factor Sox14. Sox14 in turn upregulates the expression of the cytoskeletal regulator Mical 14. Other molecules and signal pathways including ubiquitin–proteasome system 15, 16, Ik2/Kat60L 17, 18, caspases 15, 19, headcase 20, microtubule (MT) regulators/motors 21, 22, 23, calcium signaling 24, insulin pathway 16, endocytosis 25, 26, 27, and the JNK signaling pathway 28 have been documented to regulate dendrite pruning of ddaC neurons. It has been recently reported that PAR‐1 and Patronin play important roles in regulating MT stability and orientation during dendrite pruning 22, 23, 29. However, our understanding of the molecular and cellular mechanisms underlying dendrite pruning is still far from complete.
Figure 1. Mts is required for dendrite pruning in ddaC neurons.
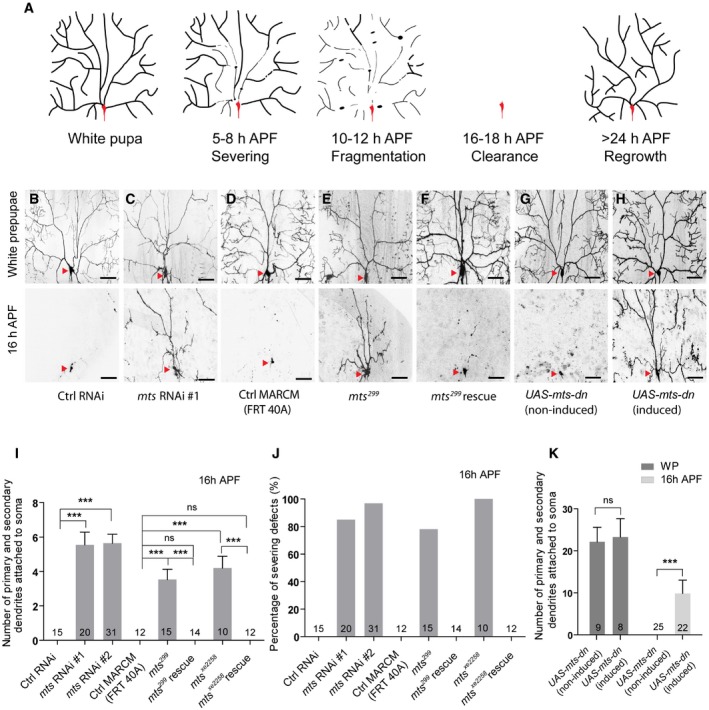
-
AA Schematic representation of dendrite pruning in ddaC neurons.
-
B–FLive confocal images of ddaC neurons expressing UAS‐mCD8‐GFP driven by ppk‐Gal4 at WP and 16 h APF stages. Dendrites of ctrl RNAi (B), mts RNAi #1 (C), ctrl MARCM (D), mts 299 MARCM (E), and mts 299 rescue (F) ddaC neurons at WP and 16 h APF stages. Red arrowheads point to the ddaC somas.
-
G, HUAS‐mts‐dn ddaC neurons from animals in RU486‐induced condition (H) driven by GeneSwitch‐Gal4‐2295 exhibited normal arbors at WP stage and severe dendrite pruning defects at 16 h APF, compared to those in a non‐induced condition (G). Red arrowheads point to the ddaC somas.
-
I–KQuantification of number of primary and secondary dendrites attached to soma and percentage of severing defects at 16 h APF.
In a genome‐wide RNA interference (RNAi) screen, we isolated Microtubule star (Mts), a catalytic subunit of protein phosphatase 2A (PP2A), which is required for dendrite pruning of ddaC neurons. PP2A is a highly conserved and ubiquitous serine–threonine phosphatase 30, 31. In mammals, the core enzyme exists as a dimer (PP2AD) consisting of a catalytic C subunit (PP2AC) and a scaffolding A subunit of 65 kDa (PR65). A regulatory B subunit is associated with this core architecture to form the holoenzyme 30. Several isoforms of A (α and β), C (α and β), and B (B, B′, B″, and B‴) subunits have been found to be encoded by distinct genes or alternative splicing of a single gene 30 (Appendix Fig S1A). PP2A is involved in many essential cellular functions including cell cycle progression, cellular metabolism, migration, and apoptosis 32, 33, 34. In Drosophila, heterotrimeric PP2A holoenzyme comprises the A subunit PP2A‐29B, the C subunit Mts, and one of the following regulatory B subunits, including Twins (Tws, B subunit), Well‐rounded (Wrd, B′ subunit), Widerborst (Wdb, B″ subunit), and PR72 (B‴ subunit) 31. PP2A regulates various developmental processes, including tissue pattern 35, planar cell polarity 36, asymmetric cell division 37, 38, and synapse formation 39. PP2A has been reported to regulate MT cytoskeletal organization in both mitotic and postmitotic cells 39, 40.
In this study, we report an important role of PP2A in dendrite pruning of sensory neurons. We show that PP2A, via its regulatory subunit Wdb, regulates dendrite pruning by activating the expression of Sox14 and Mical. Interestingly, PP2A, via another regulatory subunit Tws, is responsible for the uniform MT orientation in dendrites. Thus, our study reveals, for the first time, that the protein phosphatase PP2A regulates dendrite pruning and dendritic MT polarity via ecdysone signaling and a MT‐depolymerizing kinesin, respectively.
Results
The catalytic subunit of PP2A Mts is required for ddaC dendrite pruning
To identify novel players of dendrite pruning, we previously conducted an unbiased genome‐wide RNAi screen in ddaC neurons 14. In this screen, we isolated mts which is required for dendrite pruning. The neurons expressing the control RNAi construct completely eliminated larval dendrites at 16 h APF (Fig 1B, I and J). In contrast, knockdown of mts, via two independent RNAi transgenes (#1, BSC27723 and #2, v41924), led to consistent dendrite pruning defects in the vast majority of ddaC neurons at the same time point (85 and 97%, respectively) (Fig 1C, I and J, Appendix Fig S1B). mts encodes the catalytic subunit of PP2A which was shown to localize as star‐like structures on mitotic spindles in dividing cells 40. To further verify the role of mts in ddaC dendrite pruning, we next generated mutant clones using the strong hypomorphic allele mts 299 38 and the null allele mts xe2258 41. Consistently, in contrast to no pruning defects in the control clones (Fig 1D, I and J), we observed similar severing defects in 78 and 100% of mts 299 and mts xe2258 ddaC clones, respectively (Figs 1E, I and J, and EV1A). The pruning phenotypes associated with mts 299 and mts xe2258 mutants were fully rescued by the expression of the wild‐type Mts protein (Figs 1F, I and J, and EV1A). In addition, Mts is required for initial dendrite arborization, as the number of primary and secondary dendrites was significantly reduced in mts RNAi or mutant neurons (Appendix Fig S1C). We also observed the simplified dendrite morphology in mts xe2258 mutant neurons from the wandering 3rd instar larvae, compared to that in the wild‐type ones (Fig EV1B and C). Taken together, Mts is required for both dendrite pruning and initial dendrite arborization in ddaC neurons. To examine whether PP2A is required at the timing of pruning, and the dendrite pruning defect in mts mutant neurons is not caused by the initial morphology defect, we made use of the RU486‐inducible GeneSwitch system 42 to induce a dominant‐negative mutant form of Mts (Mts‐dn) 36, which lacks the N‐terminal region of its phosphatase domain. Mts‐dn expression was induced from the 2nd instar stage upon RU486 feeding. While RU486 treatment did not alter the number of primary and secondary dendrites of Mts‐dn‐expressing ddaC neurons at the white prepupal (WP) stage (Fig 1G, H and K), severe dendrite pruning defects were observed at 16 h APF with 9.9 primary and secondary dendrites attached to the soma (Fig 1G, H and K).
Figure EV1. Mts is essential for dendrite pruning and larval dendrite morphology in ddaC neurons.
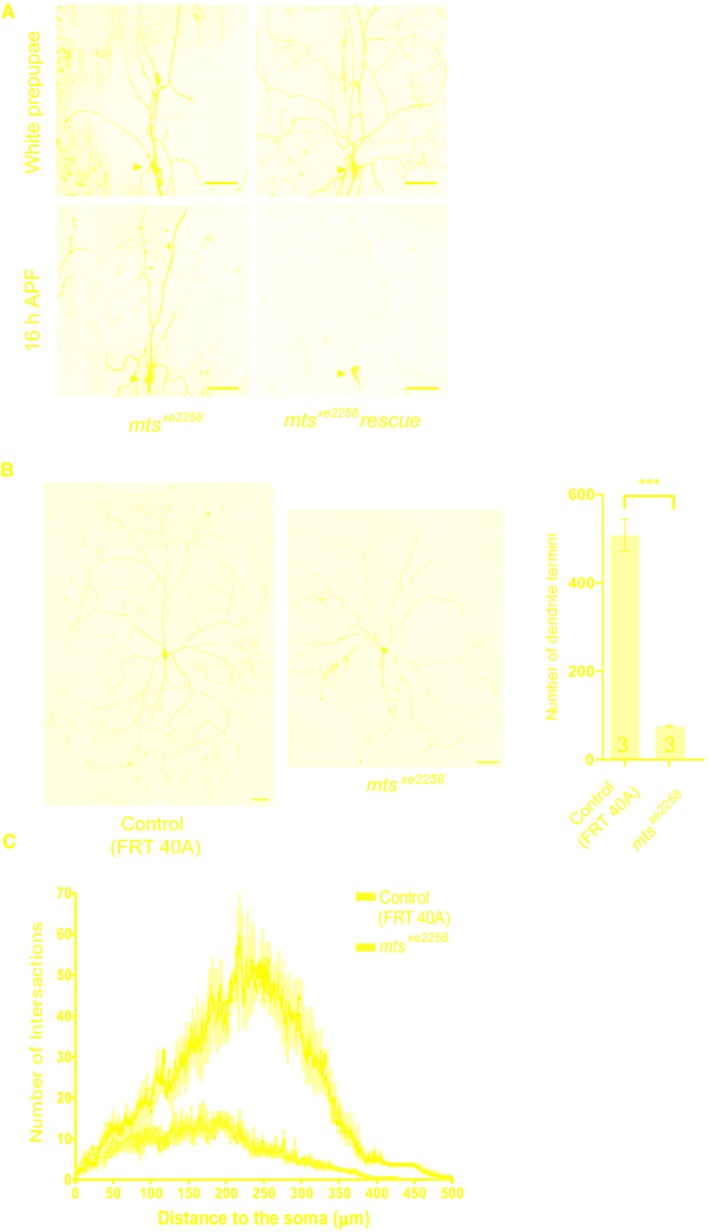
-
ALive confocal images of ddaC neurons expressing mCD8‐GFP driven by ppk‐Gal4 at WP and 16 h APF stages. Dendrites of mts xe2258 MARCM and mts xe2258 rescue ddaC neurons at WP and 16 h APF stages. Red arrowheads point to the ddaC somas.
-
B, CLive confocal images of ddaC neurons visualized by ppk‐Gal4‐driven mCD8::GFP expression at wL3 stage. ddaC neurons of mts xe2258 MARCM clone exhibited simplified dendrite arbors at WP stage. The rightest panel in B is the quantification of number of dendrite terminal and sholl analysis (C) for control and mts xe2258 MARCM ddaC clones.
Collectively, Mts acts as a new regulator that mediates dendrite pruning in a cell‐autonomous manner during early metamorphosis.
The scaffolding subunit PP2A‐29B is essential for ddaC dendrite pruning
The catalytic subunit of PP2A associates with the scaffolding subunit to form a core enzyme, which subsequently binds to the regulatory subunit to form the holoenzyme 43. Given that PP2A‐29B is the only known scaffolding subunit in Drosophila, we next examined a potential requirement of PP2A‐29B for dendrite pruning. In contrast to the controls (Fig 2A), attenuation of pp2a‐29b by three RNAi transgenes, v49671 (#1), v49672 (#2), and BSC29384 (#3), resulted in consistent dendrite pruning defects in ddaC neurons (Fig 2B, F and G, Appendix Fig S1B), similar to mts knockdown. Compared to the control clones (Fig 2C, F and G), ddaC MARCM clones derived from a lethal P‐element insertion allele, pp2a‐29b rs, exhibited similar severing defects with full penetrance; 6.1 primary and secondary dendrites were attached to the soma at 16 h APF (Fig 2D, F and G). The expression of wild‐type PP2A‐29B protein in pp2a‐29b rs mutant ddaC neurons completely rescued the dendrite pruning defects (Fig 2E–G). Thus, the scaffolding subunit PP2A‐29B also plays an important role in dendrite pruning; the core PP2A enzyme consisting of Mts and PP2A‐29B is critical for regulating dendrite pruning in ddaC sensory neurons.
Figure 2. PP2A‐29B is required for dendrite pruning in ddaC neurons.
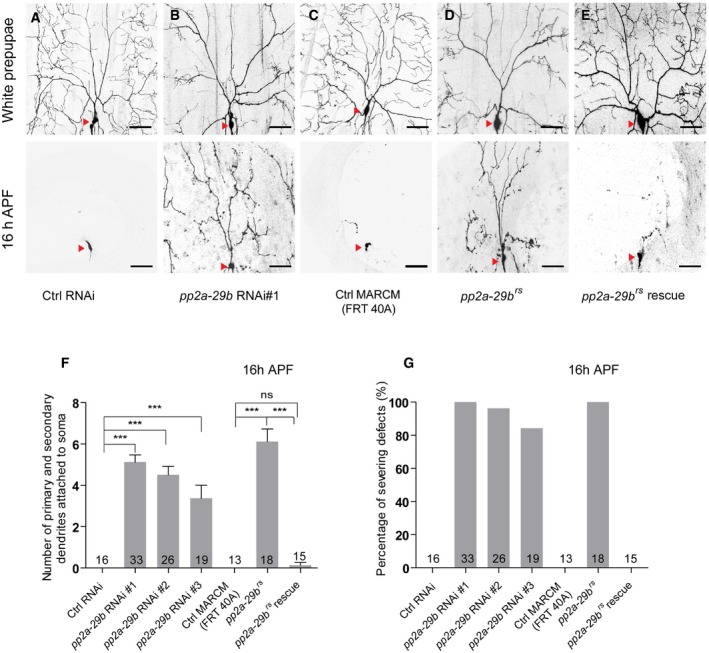
-
A–ELive confocal images of ddaC neurons expressing UAS‐mCD8‐GFP driven by ppk‐Gal4 at WP and 16 h APF stages. Dendrites of ctrl RNAi (A), pp2a‐29b RNAi #1 (B), ctrl MARCM (C), pp2a‐29b rs MARCM (D), and pp2a‐29b rs rescue (E) ddaC neurons at WP and 16 h APF stages. Red arrowheads point to the ddaC somas.
-
F, GQuantification of number of primary and secondary dendrites attached to soma and percentage of severing defects at 16 h APF.
Two regulatory subunits of PP2A, Wdb and Tws, are required for ddaC dendrite pruning
We next attempted to identify possible regulatory subunits of PP2A, which may act together with the core enzyme to regulate dendrite pruning. In Drosophila, there are four regulatory subunits for PP2A, including Tws, Wrd, Wdb, and PR72. We therefore knocked down all these regulatory subunits via a variety of available RNAi lines. Compared to the controls (Fig 3A), knockdown of either wdb or tws caused dendrite pruning defects (Figs 3B, C, I and J, and EV2A). Double knockdown of wdb (RNAi #1) and tws (RNAi #1) exacerbated the dendrite pruning phenotype, compared to either wdb or tws knockdown (Fig EV2B). To further confirm the requirement of wdb and tws in dendrite pruning, we conducted MARCM analyses using wdb or tws mutants. Compared to no pruning defect in the wild‐type clones (Fig 3D, I and J), wdb dw and wdb 14 mutants exhibited the pruning defects in 91 and 100% of their ddaC mutant clones, which retained approximately 7.4 and 4.6 major dendrites, respectively (Fig 3E, F, I and J). Likewise, MARCM analysis of tws 60, a previously published allele 35, revealed consistent dendrite pruning defects in 41% of the mutant clones (Fig 3G, I and J), which appear to be weaker than those in wdb mutants. In a parallel clonal screen, we also isolated another new tws allele, tws 603 , which showed similar dendrite pruning defects in 50% of its mutant clones (Fig 3H–J). Thus, these results show that two PP2A regulatory subunits, Wdb and Tws, are required to regulate ddaC dendrite pruning during metamorphosis.
Figure 3. Two regulatory subunits of the PP2A, Wdb and Tws, are required for ddaC dendrite pruning.
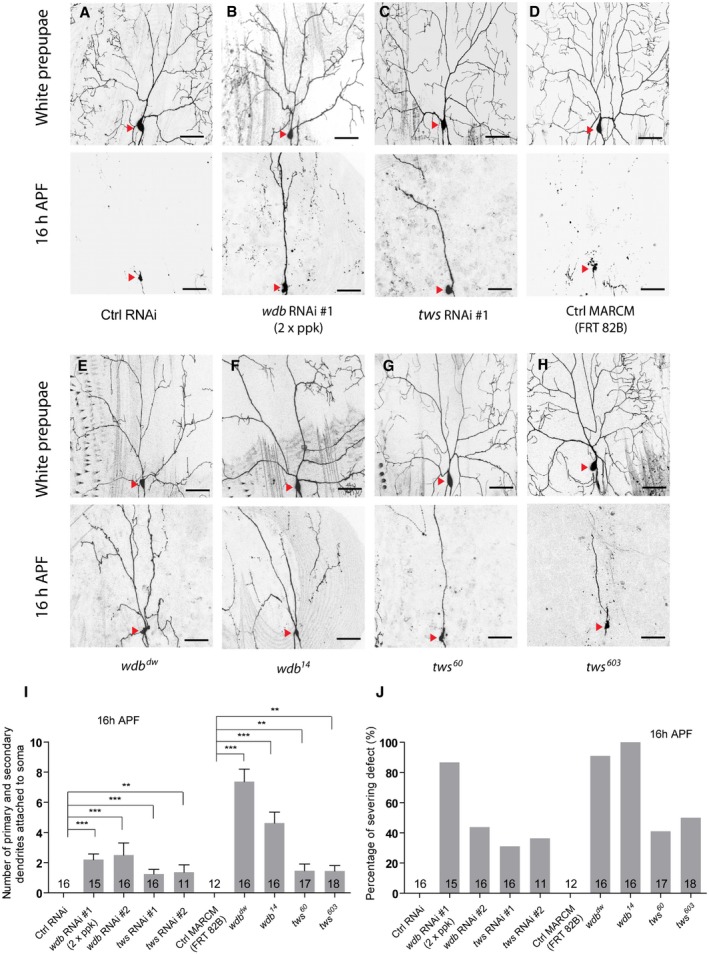
-
A–HLive confocal images of ddaC neurons expressing UAS‐mCD8‐GFP driven by ppk‐Gal4 at WP and 16 h APF stages. Dendrites of ctrl RNAi (A), wdb RNAi #1 driven by two copies of ppk‐Gal4 (B), tws RNAi #1 (C), ctrl MARCM (D), wdb dw MARCM (E), wdb 14 MARCM (F), tws 60 MARCM (G), and tws 603 MARCM (H) ddaC neurons at WP and 16 h APF stages. Red arrowheads point to the ddaC somas.
-
I, JQuantification of number of primary and secondary dendrites attached to soma and percentage of severing defects at 16 h APF.
Figure EV2. Tws knockdown enhanced the pruning defects in wdb RNAi ddaC neurons.
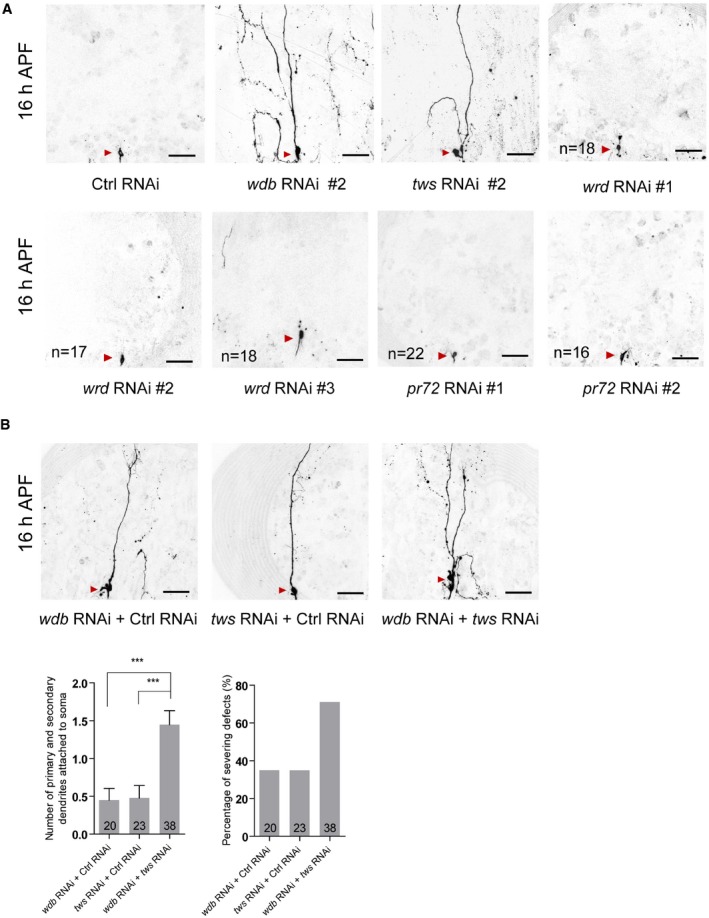
- Live confocal images of ddaC neurons expressing mCD8‐GFP driven by ppk‐Gal4 at 16 h APF stage. Dendrites of ctrl RNAi, wdb RNAi #2, tws RNAi #2, wrd RNAi #1, wrd RNAi #2, wrd RNAi #3, pr72 RNAi #1, and pr72 RNAi #2 ddaC neurons at 16 h APF stage. Red arrowheads point to the ddaC somas. The numbers of neurons (n) examined for wrd and pr72 are shown on the panels.
- Live confocal images of ddaC neurons expressing mCD8‐GFP driven by ppk‐Gal4 at 16 h APF stage. Dendrites of wdb RNAi + ctrl RNAi, tws RNAi + ctrl RNAi, and wdb RNAi + tws RNAi ddaC neurons at 16 h APF stage. Red arrowheads point to the ddaC somas. Quantification of number of primary and secondary dendrites attached to soma and percentage of severing defects in these genotypes at 16 h APF (bottom panels).
We next investigated whether PP2A also regulates dendrite pruning of other da neurons during metamorphosis. Class I da neurons, ddaD and ddaE, undergo stereotyped dendrite pruning, similar to ddaC neurons. By 19 h APF, the dendrites of class I neurons were completely pruned in the wild‐type animals (Appendix Fig S2A). In contrast, some dendritic branches of class I neurons remained attached to the soma in most of mts xe2258, pp2a‐29b rs, or wdb dw mutant neurons at the same time point (Appendix Fig S2B–D). Taken together, these findings indicate that PP2A is also required for dendrite pruning of class I da neurons.
PP2A is required for the expression of Sox14 and Mical, two downstream targets of ecdysone signaling during dendrite pruning
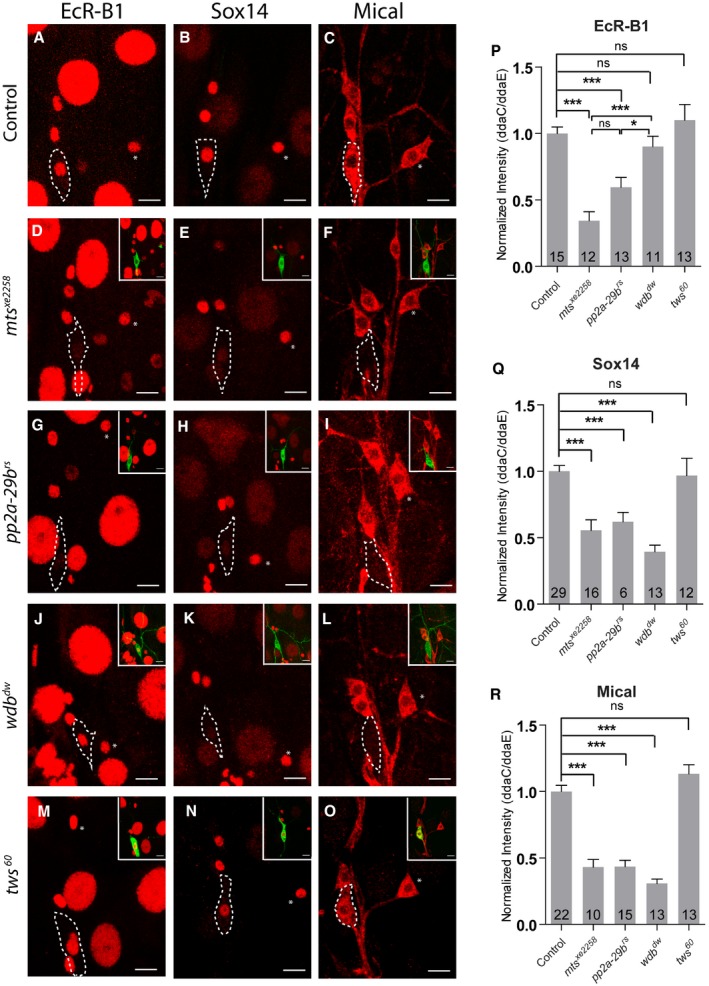
Our previous studies have demonstrated that dendrite pruning of ddaC neurons requires the activation of ecdysone signaling which leads to upregulation of two downstream targets, the key transcription factor Sox14 and the actin regulators Mical 14. We next investigated whether PP2A modulates the ecdysone pathway during dendrite pruning. To this end, we first examined the protein levels of EcR‐B1, a neuron‐specific ecdysone receptor, in various PP2A mutant ddaC neurons. Compared to the wild‐type controls (Fig 4A and P), the levels of EcR‐B1 protein were reduced in MARCM ddaC clones from mts xe2258 (Fig 4D and P) or pp2a‐29b rs mutant (Fig 4G and P) and Mts‐dn‐overexpressing ddaC neurons (Appendix Fig S3A) at WP stage. It has been documented that Sox14 and Mical are dramatically upregulated in protein level during the larval–pupal transition 14. Interestingly, compared to that in the control neurons (Fig 4B and Q), Sox14 protein levels were reduced at WP stage in the ddaC clones derived from mts xe2258 (Fig 4E and Q), pp2a‐29b rs (Fig 4H and Q), or mts‐dn neurons (Appendix Fig S3A), suggesting that Mts and PP2A‐29B are required for the upregulation of Sox14 protein expression prior to dendrite pruning. Given that Sox14 is critical for upregulation of the actin regulator Mical, we next investigated Mical levels in mts or pp2a‐29b mutant neurons at WP stage. In the wild‐type neurons, Mical protein was abundantly expressed at WP stage (Fig 4C and R). Similar to Sox14, Mical protein levels were reduced in ddaC neurons from mts xe2258 (Fig 4F and R), pp2a‐29b rs (Fig 4I and R), or mts‐dn (Appendix Fig S3A) mutant neurons at WP stage. Using a mical‐β‐gal reporter that drives the LacZ expression in ddaC neurons under the control of a mical enhancer 16, the LacZ expressions were absent or strongly reduced in mts or pp2a‐29b RNAi neurons (Appendix Fig S3B). These data suggest that Mts and PP2A‐29B likely regulate the transcription of mical in ddaC neurons. Interestingly, EcR‐B1 protein levels appeared to be unchanged in wdb dw (Fig 4J and P) or wdb 14 neurons (Appendix Fig S3A). In contrast, either Sox14 or Mical expression levels were significantly reduced in wdb dw (Fig 4K, L Q and R) or wdb 14 neurons (Appendix Fig S3A). Moreover, tws 60 MARCM ddaC clones did not exhibit a significant alteration in the levels of EcR‐B1 (Fig 4M and P), Sox14 (Fig 4N and Q), and Mical (Fig 4O and R) at WP stage. Taken together, these results suggest that PP2A, via Wdb, regulates the expression of Sox14 and Mical to facilitate ecdysone signaling in ddaC neurons prior to dendrite pruning.
Figure 4. PP2A is required to regulate ecdysone signaling during dendrite pruning.
-
A–OConfocal images of control (A–C), mts xe2258 (D–F), pp2a‐29b rs (G–I), wdb dw (J–L), and tws 60 (M–O), MARCM ddaC clones that were immunostained for EcR‐B1 (A, D, G, J, M) and Sox14 (B, E, H, K, N), and Mical (C, F, I, L, O) at WP stage. ddaC somas are labeled by dashed lines, ddaE by asterisks. ddaC neurons were identified by ppk‐Gal4‐driven mCD8::GFP (green channel) expression, as shown at the top right corner.
-
P–RQuantitative analyses of normalized EcR‐B1, Sox14, and Mical fluorescence intensities in ddaC neurons.
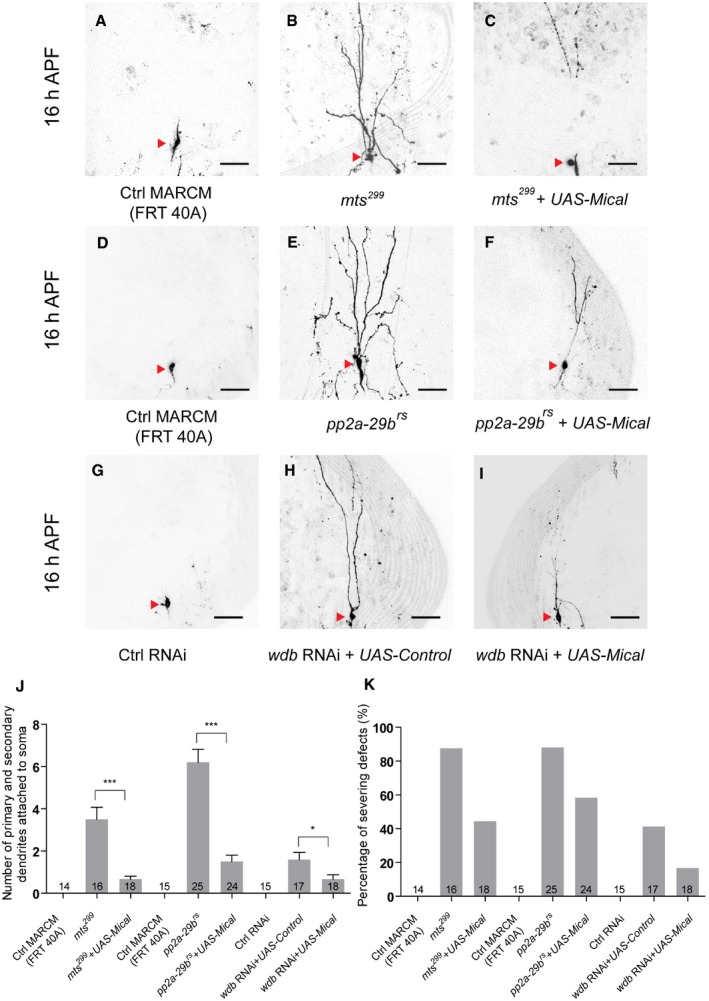
We further tested whether PP2A regulates dendrite pruning through upregulation of Mical protein. We overexpressed Mical protein via a UAS‐mical transgene in mts 299 , pp2a‐29b rs, or wdb RNAi ddaC neurons to assess if overexpressed Mical is able to suppress their dendrite pruning defects. The wild‐type clones or control RNAi neurons completely removed their larval dendrites (Fig 5A, D and G). Interestingly, the pruning defects associated with mts 299 (Fig 5B, J and K), pp2a‐29b rs (Fig 5E, J and K), or wdb RNAi (Fig 5H, J and K) mutant neurons were significantly suppressed by the overexpression of Mical at 16 h APF (Fig 5C, F and I–K). Thus, these data suggest that PP2A controls dendrite pruning of ddaC neurons at least partially via Mical upregulation.
Figure 5. Mical overexpression significantly suppressed the dendrite pruning defects in PP2A mutant neurons.
-
A–ILive confocal images of ddaC neurons expressing UAS‐mCD8‐GFP driven by ppk‐Gal4 at WP and 16 h APF stages. Dendrites of ctrl MARCM (A), mts 299 MARCM (B), UAS‐Mical + mts 299 MARCM (C), ctrl MARCM (D), pp2a‐29b rs MARCM (E), UAS‐Mical + pp2a‐29b rs MARCM (F), ctrl RNAi (G), wdb RNAi + UAS‐Control (H), and wdb RNAi + UAS‐Mical (I) ddaC neurons at 16 h APF stage. Red arrowheads point to the ddaC somas.
-
J, KQuantification of number of primary and secondary dendrites attached to soma and percentage of severing defects at 16 h APF.
PP2A is required for proper distribution of dendritic and axonal MT markers
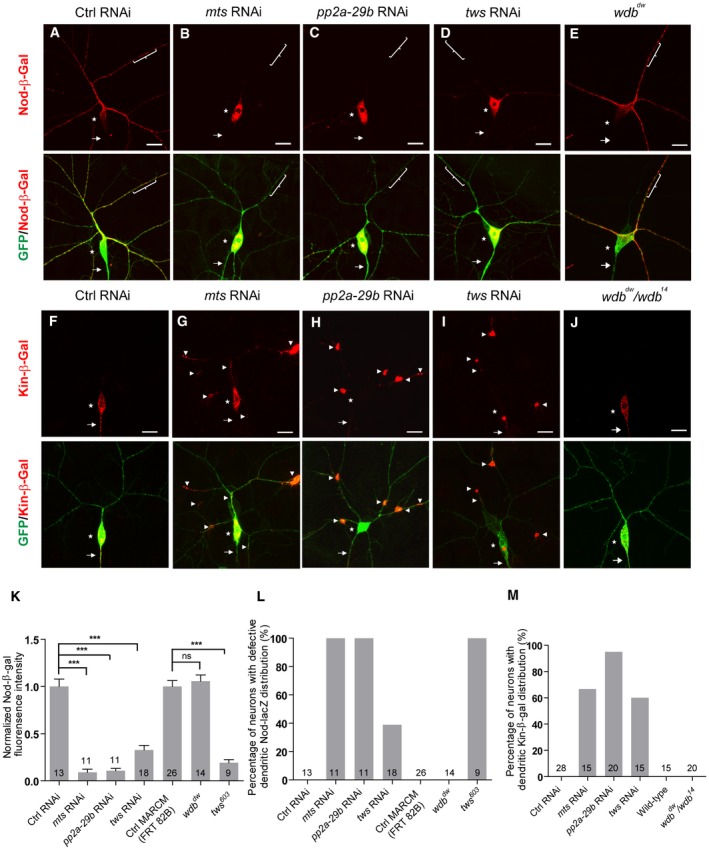
It has been shown that Mical functions as an actin disassembly factor to disassemble F‐actin in vitro and in bristles 44. However, we did not observe any obvious defect in the phalloidin staining in the proximal dendrites and soma of mts and pp2a‐29b RNAi ddaC neurons from the 3rd instar larvae (Fig EV3A and B), suggesting that PP2A does not regulate overall F‐actin level. However, PP2A is shown to modulate F‐actin dynamics in a separate study 45. Recent studies have also documented that MT dynamics, stability, and orientation play important roles in dendrite pruning 17, 21, 22, 23. Since PP2A can regulate the structure of MT cytoskeleton in motor neurons 39, we then proceeded to investigate its potential role in regulating MTs in ddaC sensory neurons. We examined the distribution/level of MT‐associated proteins (Patronin, TACC and Mini spindles/Msps) as well as two MT markers (Nod‐β‐gal and Kin‐β‐gal; also known as Khc::Nod::LacZ and Khc::LacZ, respectively) when PP2A subunits were depleted. While the levels/distributions of Patronin, TACC, and Msps appeared to be normal (Appendix Fig S4A and B), the distributions of Nod‐β‐gal and Kin‐β‐gal were drastically affected in various PP2A mutant or RNAi neurons (Fig 6). The chimera Nod‐β‐gal, which consists of the Nod motor domain (residues 1–320), the kinesin‐1 coiled‐coil domain (residues 334–606), and the β‐galactosidase (β‐gal) tag, was previously used as a marker to label the sites enriched with the MT minus ends in multiple Drosophila cell types 46, 47. In sensory neurons, Nod‐β‐gal was specifically enriched in dendrites but absent in axons and thus used as a marker for potentially detecting the presence of minus‐end‐out MTs in dendrites 23, 47, 48, 49. Nod‐β‐gal was localized in the dendrites, but not in the axons, of the control ddaC neurons (Fig 6A, Appendix Fig S5A and B). Strikingly, Nod‐β‐gal signals were significantly reduced in the dendrites but highly enriched in the soma when Mts (Fig 6B, K and L), PP2A‐29B (Fig 6C, K and L), or Tws (Fig 6D, K and L) were knocked down in ddaC neurons. Consistent with the RNAi knockdown, ddaC clones from tws 603 mutant also exhibited a strong reduction in the dendrites but an accumulation in the soma in terms of Nod‐β‐gal levels (Appendix Fig S5A and Fig 6K and L). To quantify the alterations of dendritic Nod‐β‐gal distribution, we measured the intensity of β‐gal fluorescence in the dendrites that are 30 μm away from the soma. In mts RNAi, pp2a‐29b RNAi, and tws 603 mutant neurons, dendritic Nod‐β‐gal signals were significantly reduced to approximately 10–20% of the original level (Fig 6K), suggesting drastic decreases in dendritic minus‐end‐out MTs. Interestingly, the Nod‐β‐gal distribution was not affected in wdb dw mutant clones (Fig 6E, K and L). Overexpression of Mical did not rescue the defective Nod‐β‐gal distribution in tws RNAi neurons (Appendix Fig S5B). Thus, these data indicate that PP2A might regulate dendritic MT orientation through the regulatory subunit Tws but not Wdb.
Figure EV3. The distribution of F‐actin appeared to be largely normal between wild‐type and mts/pp2a‐29b RNAi ddaC neurons at wL3.
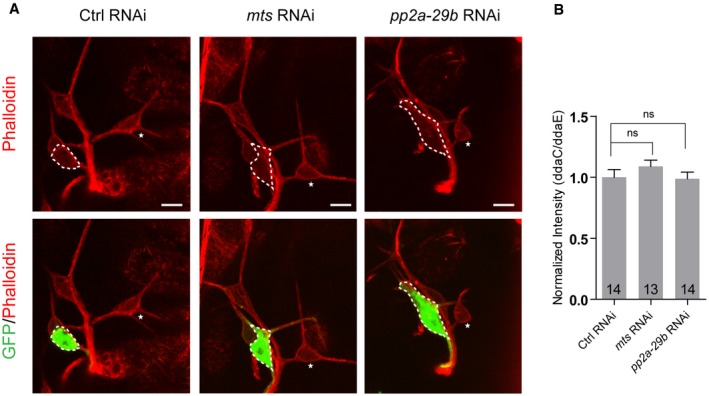
- Confocal images of ctrl RNAi, mts RNAi, and pp2a‐29b RNAi ddaC neurons immunostained for phalloidin at wL3 stage. ddaC somas are labeled by dashed lines. ddaC neurons were identified by ppk‐Gal4‐driven mCD8::GFP (green channel) expression. ddaC somas are labeled by dashed lines, and ddaE somas are marked by asterisks.
- Quantitative analysis of normalized phalloidin fluorescence intensities of ddaC somas.
Figure 6. PP2A is essential for proper distribution of dendritic and axonal MT markers.
-
A–EConfocal images of ddaC neurons at the wandering 3rd instar (wL3) stage immunostained for anti‐β‐galactosidase. Nod‐β‐gal signals were localized in the dendrites of the ctrl RNAi (A) and wdb dw (E) mutant ddaC neurons; however, Nod‐β‐gal levels were strongly reduced in the dendrites and accumulated in the somas in mts RNAi (B), pp2a‐29b RNAi (C), and tws RNAi (D) ddaC neurons. ddaC somas are marked by asterisks, axons by arrows, and dendrites used for analyzing by curly brackets.
-
F–JKin‐β‐gal was mislocalized to the dendrites in mts RNAi (G), pp2a‐29b RNAi (H), tws RNAi (I), and wdb dw/ wdb 14 (J) mutant ddaC neurons, compared to the ctrl RNAi neurons (F), while the localization of Kin‐β‐gal in wdb dw/ wdb 14 mutant (J) is normal. ddaC somas are marked by asterisks, axons by arrows, and white arrowheads point to dendritic Kin‐β‐gal signals.
-
KQuantification of normalized Nod‐β‐gal fluorescence intensity in dendrites ofddaC neurons.
-
L, MQuantification of the percentage of neurons with defective Nod‐β‐gal and Kin‐β‐gal distribution in ddaC neurons.
We next detected the localization of the axon‐specific marker Kin‐β‐gal, in which the motor and coiled‐coil domains of kinesin‐1 (residues 1–606) are fused with the β‐gal tag. Kin‐β‐gal localizes at the sites enriched with MT plus ends 46. In the wild‐type ddaC neurons, the Kin‐β‐gal signals were detectable in the axons but not in the dendrites (Fig 6F) 23, 47, suggesting the presence of plus‐end‐out MTs predominantly in axons. Notably, punctate Kin‐β‐gal signals were observed in the dendrites of mts (Fig 6G and M), pp2a‐29b (Fig 6H and M), or tws (Fig 6I and M) RNAi ddaC neurons, suggesting the presence of plus‐end‐out MTs in the mutant dendrites. However, Kin‐β‐gal was properly localized in the axons of wdb dw/wdb 14 mutant neurons (Fig 6J and M). Thus, Wdb is dispensable for the distribution of Nod‐β‐gal and Kin‐β‐gal in dendrites and axons, respectively.
Taken together, Mts, PP2A‐29B, and Tws govern proper distribution of dendrite and axon‐specific MT markers in ddaC neurons.
PP2A is required for uniform minus‐end‐out MT orientation in the dendrites
Given that Nod‐β‐gal and Kin‐β‐gal distributions were impaired in the dendrites of PP2A mutant neurons, we next made use of the MT plus‐end marker EB1‐GFP to precisely determine dendritic MT orientation via live imaging. EB1‐GFP marks growing plus ends of MTs; the direction of EB1‐GFP movement is defined as MT orientation in neurons 49, 50, 51. In Drosophila da neurons, > 95% of dendritic EB1‐GFP comets move toward the soma (retrograde), whereas almost all axonal EB1‐GFP comets migrate away from the soma (anterograde), suggesting a nearly uniform minus‐end‐out MT orientation in dendrites and a plus‐end‐out MT orientation in axons 52, 53. To determine whether PP2A regulates dendritic MT orientation, we set out to observe sequential MT growth by detecting the moving EB1‐GFP comets in the dendrites of ddaC neurons. In the control ddaC neurons, almost all of dendritic EB1‐GFP comets migrated toward the soma (retrograde), whereas approximately 1% of the dendritic comets moved away from the soma (anterograde) (Fig 7A and E, Appendix Fig S6A and B), indicating a uniform minus‐end‐out MT orientation in the major dendrites. In contrast, RNAi knockdown of mts or pp2a‐29b led to approximately 16‐fold and 20‐fold increases, respectively, in the percentage of anterogradely moving comets (Fig 7B, C and E). Likewise, we also observed a similar defect in dendritic MT orientation in Mts‐dn‐overexpressing ddaC neurons (Appendix Fig S6A). Moreover, knockdown of Tws led to mixed MT orientations in the dendrites of ddaC neurons (Fig 7D and E). We observed an 8.5‐fold increase in the percentage of anterograde EB1‐GFP comets in the dendrites of tws RNAi neurons (Fig 7E). Similar to that in the wild‐type neurons (Fig 7I and K), the minus‐end‐out MT orientation was not disturbed in the dendrites of wdb dw /wdb 14 mutant neurons (Fig 7J and K), confirming that Wdb is not important for dendritic MT orientation in ddaC neurons. In addition, knockdown of wdb in the tws RNAi background did not significantly enhance the dendritic MT misorientation phenotype (Appendix Fig S6B), suggesting that Tws and Wdb unlikely act redundantly to regulate dendritic MT orientation. We also measured the number, track length, and speed of EB1‐GFP comets, which reflect overall MT dynamics and nucleation. In mts, pp2a‐29b, tws RNAi, or wdb mutant neurons, we observed no or neglectable alterations on the number, track length, and speed of the EB1‐GFP comets (Fig 7F–H and L–N).
Figure 7. PP2A is required for uniform minus‐end‐out orientation of dendritic MTs in ddaC neurons.
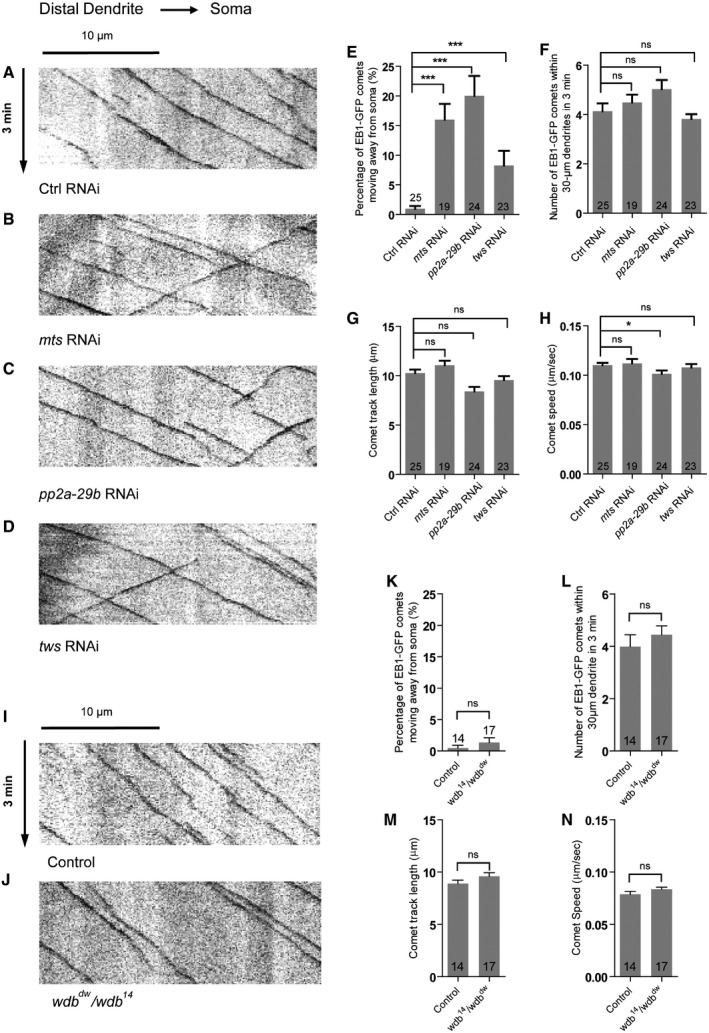
-
A–DRepresentative kymographs depicting the movement patterns of EB1 comets in the proximal dendrites of ddaC neurons at 96 h AEL. In control RNAi (A) ddaC dendrites, EB1‐GFP comets predominantly moved toward the somas (retrograde). However, in mts RNAi (B), pp2a‐29b RNAi (C), and tws RNAi (D) ddaC dendrite branches, some EB1‐GFP comets moved away from the somas (anterograde).
-
E–HQuantitative analyses of the percentages of anterograde EB1 comets in each neuron imaged (E), the average numbers of EB1‐GFP comets within 30 μm dendrite in 3 min (F), the average comet track length of each neuron (G), and the average comet speed of each neuron (H).
-
I, JRepresentative kymographs depicting the movement patterns of EB1 comets in the proximal dendrites of ddaC neurons at 96 h AEL. In both control (I) and wdb dw /wdb 14 mutant (J) ddaC dendrites, EB1‐GFP comets predominantly moved toward the somas (retrograde).
-
K–NQuantitative analyses of the percentages of anterograde EB1 comets in each neuron imaged (K), the average numbers of EB1‐GFP comets within 30 μm dendrite in 3 min (L), the average comet track length of each neuron (M), and the average comet speed of each neuron (N).
Collectively, these data suggest that PP2A, via the regulatory subunit Tws but not Wdb, regulates the minus‐end‐out MT orientation in ddaC dendrites.
PP2A regulates dendritic MT orientation through suppressing Klp10A level
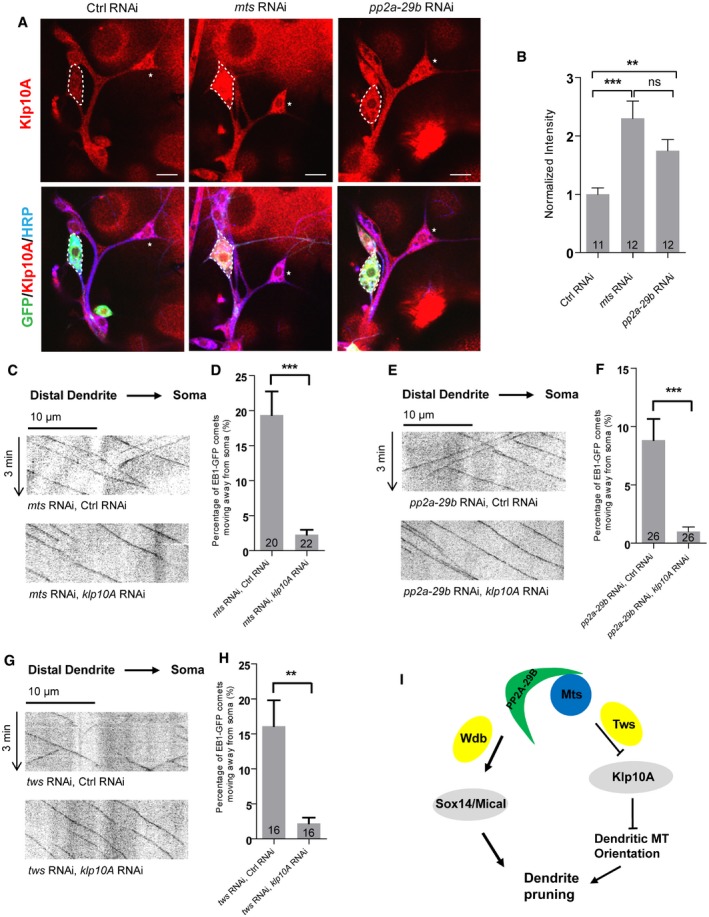
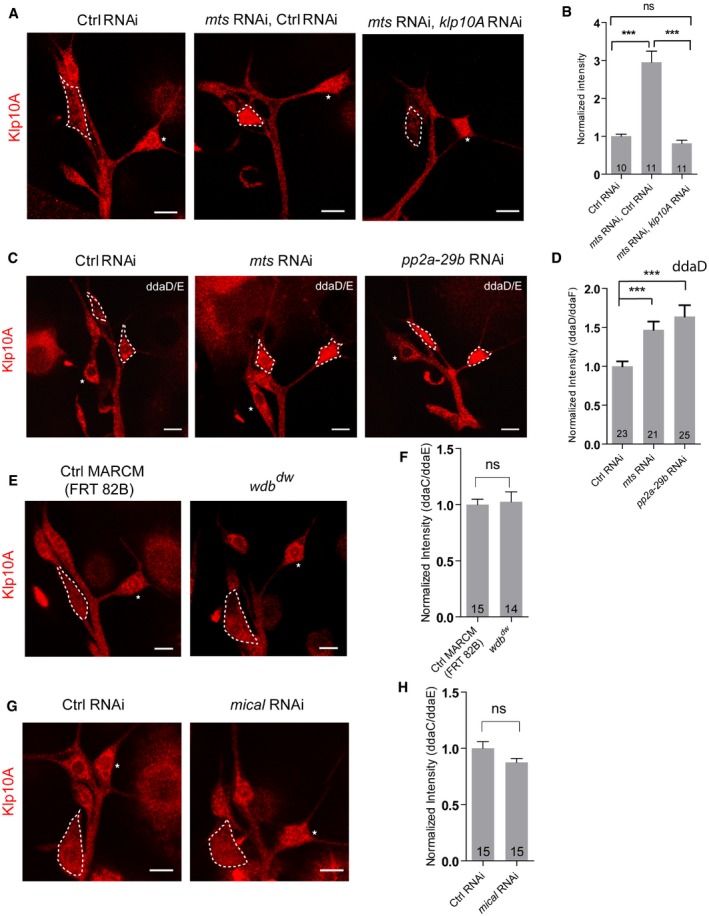
To gain insight into the molecular mechanism of PP2A in the regulation of dendritic MT orientation, we analyzed the level and localization of several MT regulators including Patronin, Klp10A, TACC, and Msps. Klp10A levels were significantly elevated upon mts or pp2a‐29b knockdown (Fig 8A and B), whereas the rest of MT regulators remained unchanged (Appendix Fig S4A and B). We observed a 2.3‐fold increase in Klp10A levels in the soma of mts RNAi ddaC neurons (Fig 8A and B), when compared to the controls. Klp10A levels were also significantly increased in the soma of pp2a‐29b RNAi ddaC neurons (Fig 8A and B). Klp10A is a kinesin‐13‐family MT depolymerase that has been reported to regulate MT orientation in the dendrites of ddaC neurons 23. Thus, PP2A appears to negatively regulate Klp10A level. To confirm the specificity of Klp10A signals, we further knocked down Klp10A in mts RNAi ddaC neurons. Knockdown of Klp10A strongly eliminated its signals in mts RNAi neurons (Fig EV4A and B), confirming that the Klp10A levels are elevated upon Mts knockdown. Likewise, knockdown of either Mts or PP2A‐29B in class I ddaD/E neurons also exhibited a significant increase in Klp10A levels (Fig EV4C and D), suggesting a general role of PP2A in suppressing Klp10A level in sensory neurons. Klp10A level was not affected in wdb dw ddaC clones (Fig EV4E and F). Similar to Wdb, Mical is not required to regulate Klp10A level and dendritic MT orientation, as knockdown of Mical did not affect the Klp10A level (Fig EV4G and H), Nod‐β‐gal distribution (Appendix Fig S7A), or EB1‐GFP comet directionality (Appendix Fig S7B).
Figure 8. PP2A regulates dendritic MT orientation through suppressing Klp10A levels.
-
AConfocal images of ctrl RNAi, mts RNAi, and pp2a‐29b RNAi ddaC neurons immunostained for Klp10A at wL3 stages. ddaC somas are labeled by dashed lines; ddaE somas are marked by asterisks.
-
BQuantitative analysis of normalized Klp10A fluorescence intensities of ddaC somas.
-
C–HRepresentative kymographs depicting the movement patterns of EB1 comets in the proximal dendrites of ddaC neurons at 96 h AEL in (C) mts RNAi + ctrl RNAi and mts RNAi + klp10a RNAi; (E) pp2a‐29b RNAi + ctrl RNAi and pp2a‐29b RNAi + klp10a RNAi; and (G) tws RNAi + control RNAi and tws RNAi + klp10a RNAi. (D, F, H) Quantitative analyses of the percentages of anterograde EB1 comets in ddaC neurons.
-
IA possible model. PP2A, via Wdb, regulates ddaC dendritepruning through the expression of Sox14 and Mical, whereas PP2A, via Tws, regulates dendritic MT polarity via suppressing the Klp10A levels.
Figure EV4. PP2A is required to suppress the levels of Klp10A in class I ddaD/E neurons.
-
A–HConfocal images of ddaC neurons of (A) ctrl RNAi, mts RNAi + control RNAi and mts RNAi + klp10a RNAi; (C) ctrl RNAi, mts RNAi and pp2a‐29b RNAi; (E) ctrl MARCM and wdb dw MARCM; and (G) ctrl RNAi and mical RNAi. All were immunostained for Klp10A at wL3 stage. ddaC (A, E, G) and ddaD/E (C) somas are labeled by dashed lines, and ddaE somas (A, E, G) are marked by asterisks. ddaC (A, E, G) and ddaD/E (C) neurons were identified by the mCD8::GFP expression driven by ppk‐Gal4 and Gal4 2‐21‐driven, respectively. (B, D, F, H) Quantitative analysis of normalized Klp10A fluorescence intensities of ddaC or ddaD somas.
To examine whether the MT orientation defects in mts RNAi or pp2a‐29b RNAi neurons are caused by elevated Klp10A level, we further attenuated Klp10A in these RNAi neurons and examined dendritic MT orientation. Klp10A knockdown almost completely restored the retrograde movement of dendritic EB1‐GFP comets in mts RNAi ddaC neurons (Fig 8C and D). Likewise, Klp10A knockdown led to a significant reduction in the percentage of anterograde EB1‐GFP comets in the pp2a‐29b RNAi ddaC dendrites (Fig 8E and F). Consistently, knocking down klp10A in tws RNAi ddaC neurons almost fully rescued the dendritic MT orientation defects (Fig 8G and H). Thus, mixed MT orientations in the dendrites of PP2A RNAi neurons are likely caused by elevated Klp10A level or activity. Moreover, Klp10A knockdown slightly suppressed the dendrite pruning defects in mts or pp2a‐29b RNAi neurons (Fig EV5A), however, did not restore Mical expression (Fig EV5B). The partial suppression suggests that PP2A regulates dendrite pruning partly via Klp10‐mediated MT orientation.
Figure EV5. PP2A regulates dendrite pruning partially through suppressing the Klp10A level.
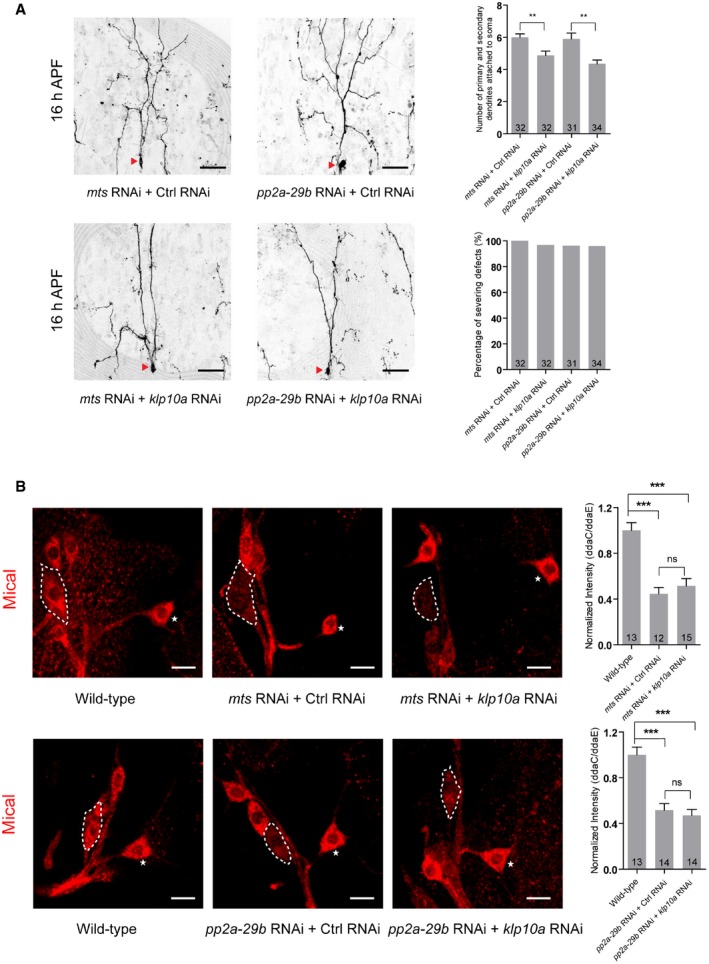
- Live confocal images of ddaC neurons expressing mCD8‐GFP driven by ppk‐Gal4 at 16 h APF stage. Dendrites of mts RNAi + Ctrl RNAi, mts RNAi + klp10a RNAi, pp2a‐29b RNAi + Ctrl RNAi, and pp2a‐29b RNAi + klp10a RNAi ddaC neurons at 16 h APF stage. Red arrowheads point to the ddaC somas. Quantification of number of primary and secondary dendrites attached to soma and percentage of severing defects at 16 h APF (rightest panels).
- Confocal images of ddaC neurons of wild‐type, mts RNAi + Ctrl RNAi, mts RNAi + klp10a RNAi, wild‐type, pp2a‐29b RNAi + Ctrl RNAi, and pp2a‐29b RNAi + klp10a RNAi that were immunostained for Mical at WP stage. Quantitative analyses of normalized Mical fluorescence (rightest panels). ddaC somas are labeled by dashed lines, and ddaE somas are marked by asterisks.
Discussion
Pruning is essential for the development and refinement of neuronal circuits 54. During Drosophila metamorphosis, ddaC neurons selectively eliminate their dendrites while keeping their axons intact. Recent studies report that PP2A dysfunction is associated with a series of neurodegenerative diseases in humans, such as Parkinson's disease and Alzheimer's disease 55, 56. In this study, we identified Drosophila PP2A as an important regulator of dendrite pruning during development. We first isolated the catalytic subunit Mts, the scaffolding subunit PP2A‐29B, and two regulatory subunits, namely Wdb and Tws. Our data suggest that PP2A, via Wdb, facilitates the expression of Sox14 and Mical, two downstream targets of ecdysone signaling, whereas PP2A, via Tws, regulates dendritic minus‐end‐out MT orientation which is required for dendrite pruning 23 (see the model in Fig 8I).
PP2A regulates ecdysone signaling during dendrite pruning
In this study, we found that the dendrite pruning phenotypes in various pp2a mutants are reminiscent of those of EcR, sox14, or mical mutants 9, 10, 14. Indeed, both Mts and PP2A‐29B are required to upregulate the expression of EcR‐B1, Sox14, and Mical in ddaC neurons. Furthermore, PP2A promotes dendrite pruning via the activation of Mical expression, as Mical overexpression dramatically suppressed the dendrite pruning defects in mts and pp2a‐29b mutant neurons. Different from the catalytic (Mts) and scaffolding subunits (PP2A‐29B), the regulatory subunit Wdb is important for Sox14 and Mical expression, but appears dispensable for EcR‐B1 expression. Wdb might regulate Sox14 and Mical expression in parallel to or downstream of EcR‐B1. Alternatively, multiple regulatory subunits of PP2A might act redundantly with Wdb to facilitate EcR‐B1 expression. How PP2A regulates EcR‐B1 expression remains unclear. EcR‐B1 expression is induced by the TGF‐β signaling, the cohesion complex, and the nuclear receptors Ftz‐f1/Hr39 before the onset of axon pruning in mushroom body γ neurons 57, 58, 59, which are likely conserved in ddaC neurons. We speculate that PP2A may regulate the expression of EcR‐B1 via the modulation of its upstream regulators/pathways, for example, TGF‐β pathway and the cohesin complex. It has been reported that phosphorylation of TGF‐β receptors and cohesin is important for their functions 60, 61. PP2A might modulate their phosphorylation levels to elicit their downstream pathways including ecdysone signaling.
PP2A regulates dendritic MT polarity by inhibiting the MT‐depolymerizing kinesin Klp10A
During dendrite pruning, MT breakdown appears to precede the scission of dendritic membrane in ddaC neurons, leading to severing of proximal dendrites from the soma 10, 17. Growing evidence indicates that regulators of MT cytoskeletons are involved in dendrite pruning. Par‐1 kinase promotes MT breakdown probably via Tau phosphorylation and thereby facilitates dendrite pruning of ddaC neurons 22. Moreover, recent studies from us and others have demonstrated a high correlation between dendritic minus‐end‐out orientation and dendrite pruning. Kinesin‐1/2 motors and a MT minus‐end‐binding protein Patronin regulate minus‐end‐out MT orientation in dendrites and are required for dendrite pruning in ddaC neurons 21, 23, 29, 62. The kinesin‐13 MT depolymerase Klp10A antagonizes the function of Patronin and thereby negatively regulates dendritic MT orientation during dendrite pruning 23. In addition, other regulators of dendritic MT polarity, for example, CNN and APC1/2 52, 63, appear to be required for dendrite pruning in ddaC neuron 23.
In this study, we report that PP2A also regulates both dendritic MT orientation and dendrite pruning in ddaC neurons. We observed Nod‐β‐gal/Kin‐β‐gal mislocalization as well as mixed EB1‐GFP comets in the dendrites of mts, pp2a‐29b, and tws RNAi neurons, suggesting increased plus‐end‐out MTs in mutant dendrites. These results raise the possibility that PP2A may regulate polarized MT nucleation and/or proper distribution of the MT nucleator γ‐tubulin in the dendrites of ddaC neurons. Golgi outposts have been reported to locally nucleate MTs in the dendrites of sensory neurons 64, but a recent study suggested otherwise 53. γ‐tubulin might be associated with intracellular membrane of different organelles, for example, Golgi and the endoplasmic reticulum, to nucleate minus‐end‐out MTs in dendrites. PP2A might be involved in the proper distribution of γ‐tubulin in dendrites and thereby cause polarized MT polymerization. Moreover, we found that PP2A regulates dendritic MT orientation and dendrite pruning at least partially via suppressing the levels or functions of Klp10A. Klp10A protein levels were significantly elevated in mts or pp2a‐29b RNAi neurons. Further knockdown of klp10a suppressed the defects in both dendritic MT orientation and dendrite pruning in these RNAi backgrounds. Therefore, loss of PP2A function phenocopies Klp10A overexpression in terms of dendrite pruning and dendritic MT orientation 23. It is conceivable that elevated Klp10A might be able to attack the MT ends and depolymerize long MTs to short filaments. Short MTs were proposed to re‐orient randomly and serve as seeds for polymerization 65, potentially resulting in a mixed MT polarity in dendrites.
Consistently, an accompanying paper by the Rumpf laboratory also reports the roles of PP2A in regulating Mical expression and dendrite pruning in the same neurons 45. In addition to its important role in dendritic MT orientation in our study, their study, however, highlights another critical role of PP2A in regulating the actin dynamics in the dendrites 45. F‐actin undergoes disassembly at the proximal dendrites of ddaC neurons at the onset of dendrite pruning 17. Since Mical can directly disassemble F‐actin via its N‐terminal flavoprotein monooxygenase (FM) domain in vitro and in bristles 44, it is an excellent candidate for promoting F‐actin disassembly during dendrite pruning. Taken together, these two studies provide complementary insights into PP2A functions in regulating both microtubule orientation and actin dynamics during dendrite pruning.
In summary, we have demonstrated a vital role of PP2A in governing dendrite pruning. Mechanistically, PP2A, via two regulatory subunits, Wdb and Tws, regulates the activation of ecdysone signaling as well as the minus‐end‐out MT orientation in dendrites.
Materials and Methods
Fly strains
UAS‐mical and UAS‐mical NT 66, SOP‐flp (#42) 67, ppk‐Gal4 on II and III chromosome 68, UAS‐Kin‐β‐gal 46 , UAS‐EB1‐GFP 69 , UAS‐mts‐dn 36, mts 299 38, pp2a‐29b rs 38, tws 60 35, tws 603 (Yu lab), mical‐β‐gal 16, wdb dw, and wdb 14 36.
The following stocks were obtained from Bloomington Stock Centre (BSC): UAS‐mCD8::GFP, FRT40A, FRT82B, UAS‐Dicer2, Gal4 4‐77 (BL#8737), UAS‐Nod‐β‐gal (BL#9912), mts RNAi #1 (BL27723), pp2a‐29b RNAi #3 (BL#29384), wdb RNAi #2 (BL#28939), wrd RNAi #1 (BL#38900), mts xe2258 (BL5684), klp10a RNAi # 2 (BL#33963), tubP‐Gal80, Gal4 109(2)80 , elav‐Gal4 C155 (BL#458), and UAS‐LifeAct‐RFP (BL#58362).
The following stocks were obtained from the Vienna Drosophila RNAi Centre (VDRC): mts RNAi #2 (v41924), pp2a‐29b RNAi #1 (v49671), pp2a‐29b RNAi #2 (v49672), wdb RNAi #1 (v101406), tws RNAi #1 (v34340), tws RNAi #2 (v104167), wrd RNAi #2 (V22614), wrd RNAi #3 (V107057), pr72 RNAi #1 (V34894), pr72 RNAi #2 (V107621), klp10a RNAi #1 (v41534), mical RNAi (V46096), and control RNAi (v36355, v37288).
Immunohistochemistry and antibodies
The following primary antibodies were used for immunohistochemistry at the indicated dilution: mouse anti‐β‐galactosidase (Promega, 1:1,000), rabbit anti‐Klp10A (1:1,000, a gift from D. J. Sharp), mouse anti‐EcR‐B1 (1:50; AD4.4, DSHB), mouse polyclonal anti‐Sox14 (1:200; F. Yu), rabbit anti‐Mical (1:200; a gift from A. L. Kolodkin), Guinea pig anti‐Msps (1:500; a gift from H. Wang), rabbit anti‐TACC (1:500; a gift from J.W.Raff and H.Wang), mouse anti‐Patronin (1:250; F. Yu), Phalloidin (Thermo Fisher; 1:100), Cy3 or Cy5‐conjugated secondary antibodies (1:500; Jackson Laboratories, Cat#: 111‐165‐003, 112‐095‐003). For immunostaining, pupae or larvae were dissected in cold PBS and fixed with 4% formaldehyde for 15 min. Mounting was performed in VECTASHIELD mounting medium, and the samples were directly visualized by the confocal microscopy Leica TSC SP2.
Live imaging of EB1‐GFP comet
Larvae at 96 h AEL were immersed with a drop of halocarbon oil (Santa Cruz, Cat#sc‐250077) and mounted to slides for confocal imaging. Time‐lapse imaging for EB1‐GFP comets was recorded with Olympus FV3000 using 60× oil lens with 3× zoom. Eighty‐three frames were acquired at 2.25‐s intervals with 6 Z‐steps. Kymographs were generated for Z‐projected time‐lapse images using KymographBuilder plugin in Image J.
Live imaging analysis
To image Drosophila da neurons, larvae at the wandering 3rd instar (wL3) or pupae at WP stage were first washed in PBS buffer briefly and followed by immersion with 90% glycerol. For imaging da neurons, pupal cases at 16 h APF or 19 h APF were carefully removed before mounted with 90% glycerol. Dendrite images were recorded with Leica TSC SP2.
MARCM analysis of da sensory neurons
MARCM analysis, dendrite imaging, and quantification were carried out as previously described 14. ddaC clones were chosen and imaged according to their location and morphology at the WP stage. ddaC neurons were examined for dendrite pruning defects at 16 h APF.
RU486/mifepristone treatment for the GeneSwitch system
RU486/mifepristone treatment for the GeneSwitch system was performed as previously described 70. Embryos were collected at 6 h intervals and reared on standard food to the early 3rd instar larva stage. Subsequently, the larvae grew in the standard culture medium with 240 μg/ml mifepristone (Sigma Aldrich M8046). White prepupae were picked and subjected to phenotypic analysis at WP and/or 16 h APF.
Quantification of ddaC dendrites
Live confocal images of ddaC neurons expressing UAS‐mCD8‐GFP driven by ppk‐Gal4 were performed at WP, 16 h APF stage. Dorsal is up in all images. The severing defect was defined by the presence of dendrites that remain attached to the soma at 16 h APF. The severing defect in wild‐type or mutant ddaC neurons was quantified in a 275 × 275 μm region of the dorsal dendritic field, originating from the abdominal segments 2–5. The percentage of severing defects is the percentage of ddaC neurons with larval dendrites attached to the soma at 16 h APF. The average number of primary and secondary dendrites attached to soma was computed from wild‐type or mutant ddaC neurons. The number of neurons (n) examined in each group is shown on the bars. Sholl analyses of dendrite morphogenesis were conducted using ImageJ. Plots of average length, number of intersections, and SEM were generated using GraphPad Prism software.
Quantification of immunostaining
Images were acquired from projected z‐stacks (at 1.5 μm intervals) to cover the entire volume of ddaC/D/E sensory neurons using the confocal microscopy Leica TSC SP2. To quantify the fluorescence intensities, cell nuclei (EcR‐B1/Sox14 immunostaining) or whole soma (Mical/Klp10A/Patronin/Msps/TACC immunostaining) contours were drawn on the appropriate fluorescent channel based on the GFP channel or relative cellular position in ImageJ software. After subtracting the background (Rolling Ball Radius = 30) on the entire image of that channel, we measured the mean gray value in the marked area in ddaC and ddaE on the same images and calculated their ratios. The ratios were normalized to the corresponding average control values and subjected to statistical analysis for comparison between different conditions. Graphs display the average values of the ddaC/ddaE ratios and the standard error of the mean (SEM) normalized to controls. The number of ddaC neurons (n) examined in each group is shown on the bars. Insets show the ddaC neurons labeled by ppk‐Gal4‐driven UAS‐mCD8‐GFP expression.
To quantify the alterations of dendritic Nod‐β‐gal distribution, we measured its intensity in the 20 μm of major dorsal dendrites which were 30 μm away from the soma. The number of ddaC neurons (n) examined in each group is shown on the bars. Dorsal is up in all images.
Statistics
For pairwise comparison, two‐tailed Student's t‐test was applied to determine statistical significance. One‐way ANOVA with Bonferroni test was applied to determine significance when multiple groups were present. Error bars in all graphs represent SEM. Statistical significance was defined as ***P < 0.001, **P < 0.01, *P < 0.05, n.s., not significant. The number of neurons (n) in each group is shown on the bars.
Author contributions
FY, KSN, and MR conceived and designed the study. KSN and MR performed most of the experiments. QT conducted some Nod‐lacZ and Kin‐lacZ experiments. SB repeated some EB1‐GFP experiments. KSN, MR, and FY analyzed the data. FY and MR wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Acknowledgements
We thank S. Eaton, Y. Jan, A. L. Kolodkin, J.W. Raff, S. Rogers, M.M. Rolls, D.J. Sharp, T. Uemura, H. Wang, the Bloomington Stock Center (BSC), DSHB (University of Iowa), Kyoto Stock Center (Japan), and VDRC (Austria) for generously providing antibodies and fly stocks. We would like to thank S. Rumpf for communicating the results prior to publication. We thank the Yu lab members for helpful discussion. This work was supported by Temasek Life Sciences Laboratory (TLL) Singapore (TLL‐2040) and National Research Foundation Singapore (NRF) (SBP‐P3 and SBP‐P8) (F.Y.).
EMBO Reports (2020) 21: e48843
References
- 1. Luo L, O'Leary DD (2005) Axon retraction and degeneration in development and disease. Annu Rev Neurosci 28: 127–156 [DOI] [PubMed] [Google Scholar]
- 2. O'Leary DD, Koester SE (1993) Development of projection neuron types, axon pathways, and patterned connections of the mammalian cortex. Neuron 10: 991–1006 [DOI] [PubMed] [Google Scholar]
- 3. Truman JW (1990) Metamorphosis of the central nervous system of Drosophila . J Neurobiol 21: 1072–1084 [DOI] [PubMed] [Google Scholar]
- 4. Lichtman JW, Colman H (2000) Synapse elimination and indelible memory. Neuron 25: 269–278 [DOI] [PubMed] [Google Scholar]
- 5. Kanamori T, Togashi K, Koizumi H, Emoto K (2015) Dendritic remodeling: lessons from invertebrate model systems. Int Rev Cell Mol Biol 318: 1–25 [DOI] [PubMed] [Google Scholar]
- 6. Yu F, Schuldiner O (2014) Axon and dendrite pruning in Drosophila . Curr Opin Neurobiol 27: 192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schubiger M, Wade AA, Carney GE, Truman JW, Bender M (1998) Drosophila EcR‐B ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development 125: 2053–2062 [DOI] [PubMed] [Google Scholar]
- 8. Lee T, Marticke S, Sung C, Robinow S, Luo L (2000) Cell‐autonomous requirement of the USP/EcR‐B ecdysone receptor for mushroom body neuronal remodeling in Drosophila . Neuron 28: 807–818 [DOI] [PubMed] [Google Scholar]
- 9. Kuo CT, Jan LY, Jan YN (2005) Dendrite‐specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin‐proteasome, and ecdysone signaling. Proc Natl Acad Sci USA 102: 15230–15235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Williams DW, Truman JW (2005) Cellular mechanisms of dendrite pruning in Drosophila: insights from in vivo time‐lapse of remodeling dendritic arborizing sensory neurons. Development 132: 3631–3642 [DOI] [PubMed] [Google Scholar]
- 11. Selemon LD, Zecevic N (2015) Schizophrenia: a tale of two critical periods for prefrontal cortical development. Transl Psychiatry 5: e623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A et al (2014) Loss of mTOR‐dependent macroautophagy causes autistic‐like synaptic pruning deficits. Neuron 83: 1131–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han C, Song Y, Xiao H, Wang D, Franc NC, Jan LY, Jan YN (2014) Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila . Neuron 81: 544–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kirilly D, Gu Y, Huang Y, Wu Z, Bashirullah A, Low BC, Kolodkin AL, Wang H, Yu F (2009) A genetic pathway composed of Sox14 and Mical governs severing of dendrites during pruning. Nat Neurosci 12: 1497–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuo CT, Zhu S, Younger S, Jan LY, Jan YN (2006) Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron 51: 283–290 [DOI] [PubMed] [Google Scholar]
- 16. Wong JJ, Li S, Lim EK, Wang Y, Wang C, Zhang H, Kirilly D, Wu C, Liou YC, Wang H et al (2013) A Cullin1‐based SCF E3 ubiquitin ligase targets the InR/PI3K/TOR pathway to regulate neuronal pruning. PLoS Biol 11: e1001657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee HH, Jan LY, Jan YN (2009) Drosophila IKK‐related kinase Ik2 and Katanin p60‐like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc Natl Acad Sci USA 106: 6363–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin T, Pan PY, Lai YT, Chiang KW, Hsieh HL, Wu YP, Ke JM, Lee MC, Liao SS, Shih HT et al (2015) Spindle‐F is the central mediator of Ik2 kinase‐dependent dendrite pruning in Drosophila sensory neurons. PLoS Genet 11: e1005642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW (2006) Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci 9: 1234–1236 [DOI] [PubMed] [Google Scholar]
- 20. Loncle N, Williams DW (2012) An interaction screen identifies headcase as a regulator of large‐scale pruning. J Neurosci 32: 17086–17096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herzmann S, Gotzelmann I, Reekers LF, Rumpf S (2018) Spatial regulation of microtubule disruption during dendrite pruning in Drosophila . Development 145: dev156950 [DOI] [PubMed] [Google Scholar]
- 22. Herzmann S, Krumkamp R, Rode S, Kintrup C, Rumpf S (2017) PAR‐1 promotes microtubule breakdown during dendrite pruning in Drosophila . EMBO J 36: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y, Rui M, Tang Q, Bu S, Yu F (2019) Patronin governs minus‐end‐out orientation of dendritic microtubules to promote dendrite pruning in Drosophila . Elife 8: e39964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanamori T, Kanai MI, Dairyo Y, Yasunaga K, Morikawa RK, Emoto K (2013) Compartmentalized calcium transients trigger dendrite pruning in Drosophila sensory neurons. Science 340: 1475–1478 [DOI] [PubMed] [Google Scholar]
- 25. Kanamori T, Yoshino J, Yasunaga K, Dairyo Y, Emoto K (2015) Local endocytosis triggers dendritic thinning and pruning in Drosophila sensory neurons. Nat Commun 6: 6515 [DOI] [PubMed] [Google Scholar]
- 26. Zhang H, Wang Y, Wong JJ, Lim KL, Liou YC, Wang H, Yu F (2014) Endocytic pathways downregulate the L1‐type cell adhesion molecule neuroglian to promote dendrite pruning in Drosophila . Dev Cell 30: 463–478 [DOI] [PubMed] [Google Scholar]
- 27. Loncle N, Agromayor M, Martin‐Serrano J, Williams DW (2015) An ESCRT module is required for neuron pruning. Sci Rep 5: 8461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu S, Chen R, Soba P, Jan YN (2019) JNK signaling coordinates with ecdysone signaling to promote pruning of Drosophila sensory neuron dendrites. Development 146: dev163592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feng C, Thyagarajan P, Shorey M, Seebold DY, Weiner AT, Albertson RM, Rao KS, Sagasti A, Goetschius DJ, Rolls MM (2019) Patronin‐mediated minus end growth is required for dendritic microtubule polarity. J Cell Biol 218: 2309–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janssens V, Longin S, Goris J (2008) PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail). Trends Biochem Sci 33: 113–121 [DOI] [PubMed] [Google Scholar]
- 31. Janssens V, Goris J (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J 353: 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Santoro MF, Annand RR, Robertson MM, Peng YW, Brady MJ, Mankovich JA, Hackett MC, Ghayur T, Walter G, Wong WW et al (1998) Regulation of protein phosphatase 2A activity by caspase‐3 during apoptosis. J Biol Chem 273: 13119–13128 [DOI] [PubMed] [Google Scholar]
- 33. Turowski P, Fernandez A, Favre B, Lamb NJ, Hemmings BA (1995) Differential methylation and altered conformation of cytoplasmic and nuclear forms of protein phosphatase 2A during cell cycle progression. J Cell Biol 129: 397–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kiely PA, O'Gorman D, Luong K, Ron D, O'Connor R (2006) Insulin‐like growth factor I controls a mutually exclusive association of RACK1 with protein phosphatase 2A and beta1 integrin to promote cell migration. Mol Cell Biol 26: 4041–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uemura T, Shiomi K, Togashi S, Takeichi M (1993) Mutation of twins encoding a regulator of protein phosphatase 2A leads to pattern duplication in Drosophila imaginal discs. Genes Dev 7: 429–440 [DOI] [PubMed] [Google Scholar]
- 36. Hannus M, Feiguin F, Heisenberg CP, Eaton S (2002) Planar cell polarization requires Widerborst, a B’ regulatory subunit of protein phosphatase 2A. Development 129: 3493–3503 [DOI] [PubMed] [Google Scholar]
- 37. Ogawa H, Ohta N, Moon W, Matsuzaki F (2009) Protein phosphatase 2A negatively regulates aPKC signaling by modulating phosphorylation of Par‐6 in Drosophila neuroblast asymmetric divisions. J Cell Sci 122: 3242–3249 [DOI] [PubMed] [Google Scholar]
- 38. Wang C, Chang KC, Somers G, Virshup D, Ang BT, Tang C, Yu F, Wang H (2009) Protein phosphatase 2A regulates self‐renewal of Drosophila neural stem cells. Development 136: 2287–2296 [DOI] [PubMed] [Google Scholar]
- 39. Viquez NM, Li CR, Wairkar YP, DiAntonio A (2006) The B’ protein phosphatase 2A regulatory subunit well‐rounded regulates synaptic growth and cytoskeletal stability at the Drosophila neuromuscular junction. J Neurosci 26: 9293–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Snaith HA, Armstrong CG, Guo Y, Kaiser K, Cohen PT (1996) Deficiency of protein phosphatase 2A uncouples the nuclear and centrosome cycles and prevents attachment of microtubules to the kinetochore in Drosophila microtubule star (mts) embryos. J Cell Sci 109(Pt 13): 3001–3012 [DOI] [PubMed] [Google Scholar]
- 41. Wassarman DA, Solomon NM, Chang HC, Karim FD, Therrien M, Rubin GM (1996) Protein phosphatase 2A positively and negatively regulates Ras1‐mediated photoreceptor development in Drosophila . Genes Dev 10: 272–278 [DOI] [PubMed] [Google Scholar]
- 42. Osterwalder T, Yoon KS, White BH, Keshishian H (2001) A conditional tissue‐specific transgene expression system using inducible GAL4. Proc Natl Acad Sci USA 98: 12596–12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu Y, Xing Y, Chen Y, Chao Y, Lin Z, Fan E, Yu JW, Strack S, Jeffrey PD, Shi Y (2006) Structure of the protein phosphatase 2A holoenzyme. Cell 127: 1239–1251 [DOI] [PubMed] [Google Scholar]
- 44. Hung RJ, Yazdani U, Yoon J, Wu H, Yang T, Gupta N, Huang Z, van Berkel WJ, Terman JR (2010) Mical links semaphorins to F‐actin disassembly. Nature 463: 823–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Neele W, Ulrike G, Sebastian R (2020) PP2A phosphatase is required for dendrite pruning via actin regulation in Drosophila . EMBO Rep 21: e48870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clark IE, Jan LY, Jan YN (1997) Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development 124: 461–470 [DOI] [PubMed] [Google Scholar]
- 47. Zheng Y, Wildonger J, Ye B, Zhang Y, Kita A, Younger SH, Zimmerman S, Jan LY, Jan YN (2008) Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nat Cell Biol 10: 1172–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Satoh D, Sato D, Tsuyama T, Saito M, Ohkura H, Rolls MM, Ishikawa F, Uemura T (2008) Spatial control of branching within dendritic arbors by dynein‐dependent transport of Rab5‐endosomes. Nat Cell Biol 10: 1164–1171 [DOI] [PubMed] [Google Scholar]
- 49. Rolls MM, Satoh D, Clyne PJ, Henner AL, Uemura T, Doe CQ (2007) Polarity and intracellular compartmentalization of Drosophila neurons. Neural Dev 2: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, Galjart N (2003) Visualization of microtubule growth in cultured neurons via the use of EB3‐GFP (end‐binding protein 3‐green fluorescent protein). J Neurosci 23: 2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vaughan KT (2005) TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J Cell Biol 171: 197–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mattie FJ, Stackpole MM, Stone MC, Clippard JR, Rudnick DA, Qiu Y, Tao J, Allender DL, Parmar M, Rolls MM (2010) Directed microtubule growth, +TIPs, and kinesin‐2 are required for uniform microtubule polarity in dendrites. Curr Biol 20: 2169–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nguyen MM, McCracken CJ, Milner ES, Goetschius DJ, Weiner AT, Long MK, Michael NL, Munro S, Rolls MM (2014) Gamma‐tubulin controls neuronal microtubule polarity independently of Golgi outposts. Mol Biol Cell 25: 2039–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Riccomagno MM, Kolodkin AL (2015) Sculpting neural circuits by axon and dendrite pruning. Annu Rev Cell Dev Biol 31: 779–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martin L, Latypova X, Wilson CM, Magnaudeix A, Perrin ML, Terro F (2013) Tau protein phosphatases in Alzheimer's disease: the leading role of PP2A. Ageing Res Rev 12: 39–49 [DOI] [PubMed] [Google Scholar]
- 56. Du TT, Chen YC, Lu YQ, Meng FG, Yang H, Zhang JG (2018) Subthalamic nucleus deep brain stimulation protects neurons by activating autophagy via PP2A inactivation in a rat model of Parkinson's disease. Exp Neurol 306: 232–242 [DOI] [PubMed] [Google Scholar]
- 57. Boulanger A, Clouet‐Redt C, Farge M, Flandre A, Guignard T, Fernando C, Juge F, Dura JM (2011) ftz‐f1 and Hr39 opposing roles on EcR expression during Drosophila mushroom body neuron remodeling. Nat Neurosci 14: 37–44 [DOI] [PubMed] [Google Scholar]
- 58. Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L (2008) piggyBac‐based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell 14: 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zheng X, Wang J, Haerry TE, Wu AY, Martin J, O'Connor MB, Lee CH, Lee T (2003) TGF‐beta signaling activates steroid hormone receptor expression during neuronal remodeling in the Drosophila brain. Cell 112: 303–315 [DOI] [PubMed] [Google Scholar]
- 60. Liu T, Feng XH (2010) Regulation of TGF‐beta signalling by protein phosphatases. Biochem J 430: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y (2006) Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441: 46–52 [DOI] [PubMed] [Google Scholar]
- 62. Stone MC, Nguyen MM, Tao J, Allender DL, Rolls MM (2010) Global up‐regulation of microtubule dynamics and polarity reversal during regeneration of an axon from a dendrite. Mol Biol Cell 21: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yalgin C, Ebrahimi S, Delandre C, Yoong LF, Akimoto S, Tran H, Amikura R, Spokony R, Torben‐Nielsen B, White KP et al (2015) Centrosomin represses dendrite branching by orienting microtubule nucleation. Nat Neurosci 18: 1437–1445 [DOI] [PubMed] [Google Scholar]
- 64. Ori‐McKenney KM, Jan LY, Jan YN (2012) Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron 76: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. del Castillo U, Winding M, Lu W, Gelfand VI (2015) Interplay between kinesin‐1 and cortical dynein during axonal outgrowth and microtubule organization in Drosophila neurons. Elife 4: e10140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Terman JR, Mao T, Pasterkamp RJ, Yu HH, Kolodkin AL (2002) MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin‐mediated axonal repulsion. Cell 109: 887–900 [DOI] [PubMed] [Google Scholar]
- 67. Matsubara D, Horiuchi SY, Shimono K, Usui T, Uemura T (2011) The seven‐pass transmembrane cadherin Flamingo controls dendritic self‐avoidance via its binding to a LIM domain protein, Espinas, in Drosophila sensory neurons. Genes Dev 25: 1982–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Grueber WB, Ye B, Moore AW, Jan LY, Jan YN (2003) Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol 13: 618–626 [DOI] [PubMed] [Google Scholar]
- 69. Stone MC, Roegiers F, Rolls MM (2008) Microtubules have opposite orientation in axons and dendrites of Drosophila neurons. Mol Biol Cell 19: 4122–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang Y, Zhang H, Shi M, Liou YC, Lu L, Yu F (2017) Sec71 functions as a GEF for the small GTPase Arf1 to govern dendrite pruning of Drosophila sensory neurons. Development 144: 1851–1862 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8


