Abstract
Regulation of body growth remains a fascinating and unresolved biological mystery. One key component of body growth is skeletal and longitudinal bone growth. Children grow taller because their bones grew longer, and the predominant driver of longitudinal bone growth is a cartilaginous structure found near the ends of long bone named the growth plate. Numerous recent studies have started to unveil the importance of microRNAs in regulation of growth plate functions, therefore contributing to regulation of linear growth. In addition to longitudinal growth, other organs in our body need to increase in size and cell number as we grow, and the regulation of organ growth involves both systemic factors like hormones; and other intrinsic mechanisms, which we are just beginning to understand. This review aims to summarize some recent important findings on how microRNAs are involved in both of these processes: the regulation of longitudinal bone growth, and the regulation of organs and overall body growth.
Keywords: miRNA, body growth, body size, growth plate, cartilage
Introduction
microRNAs (miRNAs) are small (18–24 nucleotides long) non-coding RNAs that play an important role in the regulation of gene expression. Intergenic or exonic miRNAs are transcribed by RNA polymerase II as primary miRNAs (pri-miRNAs), which are long hairpin-shaped transcripts each containing one or more miRNA sequences. (Lee et al. 2004). pri-miRNAs are then processed in the nucleus by the microprocessor complex into hairpin-shaped transcripts of 80 nucleotides called pre-miRNA. On the other hand, intronic miRNA sequences are transcribed as part of pre-mRNA and excised by sliceosome generating pri-miRNAs or pre-miRNAs that are processed like intergenic or exonic miRNAs. These pre-miRNAs are then actively exported to the cytoplasm and processed by endoribonuclease Dicer into a small double-stranded RNA structure typically with 22 nucleotides (Lund et al. 2004). This miRNA duplex is unwound into the mature single-stranded RNA and incorporated into the RNA-induced silencing complex (RISC), guiding this complex to bind a complementary sequence within the 3’-untranslated region (UTR) of target mRNAs (Fig.1). In animal cells, this 3’complementary sequence is typically 6 to 8 nucleotides long, and also known as the seed region (Bartel 2009). Binding of the miRNAs to the seed region of its target genes suppresses gene expression, either through mRNA cleavage or translational repression, depending on the extent of the miRNA-mRNA pairing. In general, short and imperfect base pairing favors translational repression; while perfect base pairing favors mRNA cleavage (Mathonnet et al. 2007; Petersen et al. 2006).
Fig 1.
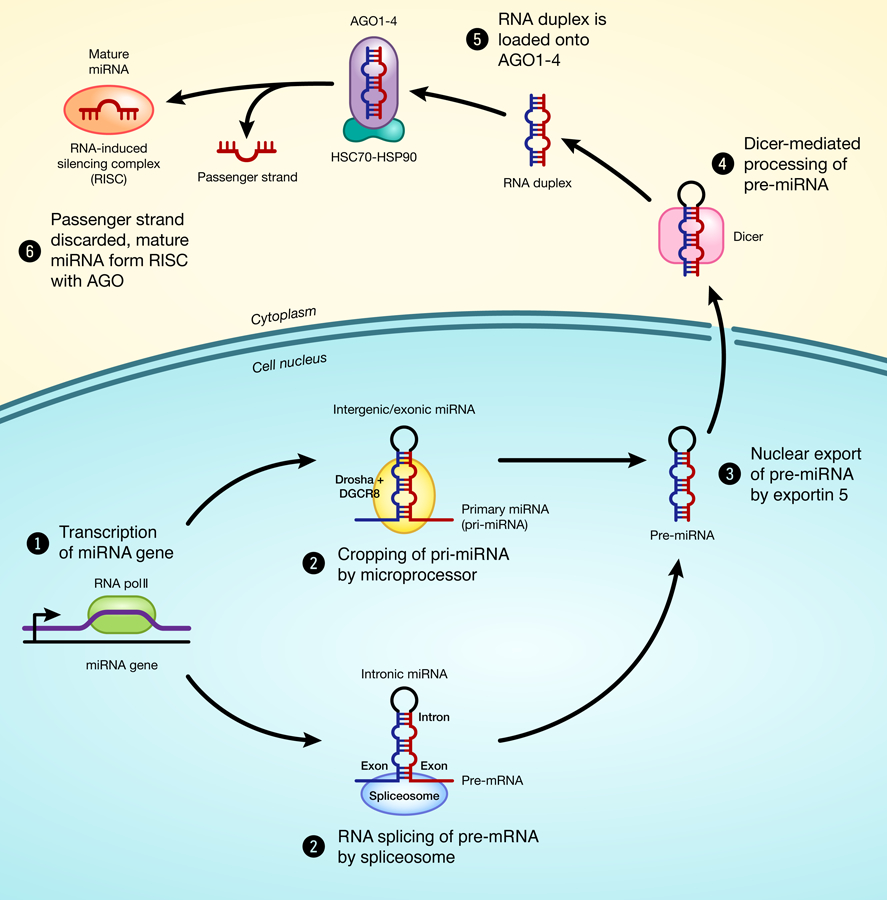
Schematic model of miRNA biogenesis
In fact, most genes in the human genome are thought to be regulated by one or more miRNAs (Lewis et al. 2005), and each miRNA can regulate a larger number of target mRNAs (Fabian et al. 2010). Because of their ability to regulate gene expression, miRNAs has been shown to play important roles in many physiological and developmental processes, as well as human diseases (for review, see (Sayed & Abdellatif 2011)). In this review, I will discuss our current understanding on the regulation of growth by miRNAs, primarily focusing on two areas: the regulation by miRNAs of longitudinal bone growth and of overall body growth.
Longitudinal Bone Growth: Endochondral Ossification
Longitudinal bone growth represents an important component of overall body growth in mammals. During embryonic development, bone formation begins with the condensation of mesenchymal stem cells. In a few places of our body, such as in the skull, mesenchymal condensations differentiate directly into bone-forming osteoblasts. These osteoblasts form bones by laying down bone matrix, mostly composed of type I collagen, and this process is called intramembranous bone formation. However, in most other condensations, bones are formed by a different process known as endochondral ossification (for review, see (Kronenberg 2003)). In this process, mesenchymal stem cells first differentiate into type II collagen-producing chondrocytes, forming a cartilage mold. This cartilage mold gradually expands due to chondrocyte proliferation. Then in the center of the cartilage, cells stop dividing and start enlarging to become type X collagen-producing hypertrophic chondrocytes. These hypertrophic chondrocytes drive cartilage matrix mineralization and eventually undergo apoptosis, leaving a scaffold of cartilage matrix for invasion of osteoblasts to lay down bone matrix in the center of the cartilage, known as the primary ossification center. This process of chondrocyte proliferation, followed by hypertrophy and osteoblast invasion lead to the formation and lengthening of long bones, which significantly contribute to the overall increase in body size and length.
As bones continue to increase in length into early postnatal life, this process of endochondral ossification persist, but become restricted to an area near the ends of long bones known as the epiphyseal growth plate. Within the growth plate, chondrocytes are now arranged histologically into three distinct zones called resting, proliferative, and hypertrophic zones (Fig.2). Resting zone near the top of the growth plate consists of round and slowly-proliferating chondrocytes that serve as precursors (Abad et al. 2002) for the proliferating columnar chondrocytes right underneath, which then undergo the same progression toward hypertrophy and apoptosis as they travel toward the center of the bone. In all mammalian species, the growth plate gradually becomes narrower while the overall growth rate declines with age, until growth essentially ceases as adult body size is attained (Lui et al. 2011).
Fig 2.
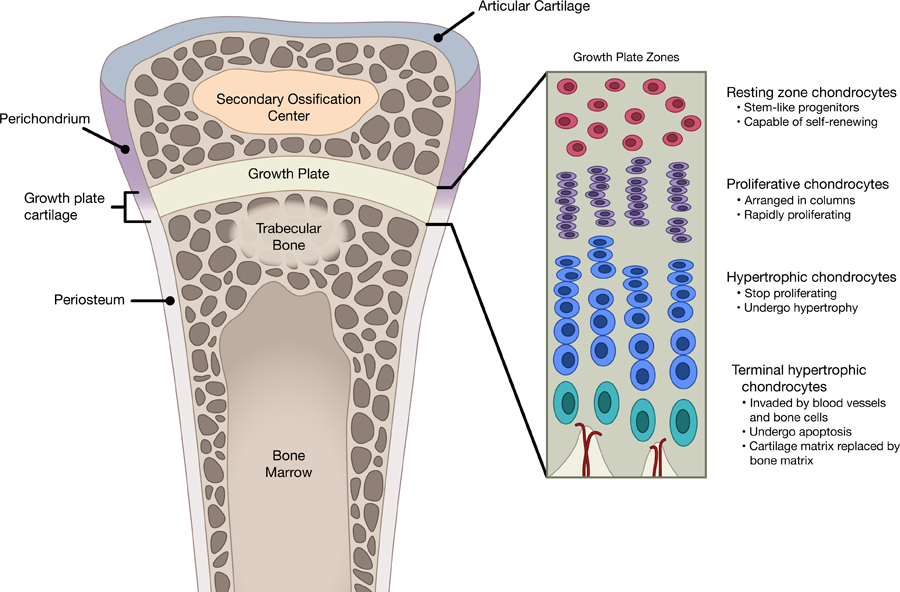
Histological and functional differences between growth plate zones
Regulation of Local Growth Plate Function by miRNA
Endochondral ossification requires careful spatial and temporal interplay between different transcription factors and multiple signaling pathways at the growth plate (Lui et al. 2010a), and recent evidence suggested that miRNAs may play an important role in regulating this process. Mice with cartilage-specific knockout of Dicer (driven by cartilage specific Col2a1 promoter), which is critical for biogenesis of miRNAs, showed severe skeletal growth defects, with decreased chondrocyte proliferation and accelerated hypertrophy (Kobayashi et al. 2008), strongly indicating that miRNAs is essential for normal function of the growth plate.
High-throughput screening of miRNAs expression in growth plate identified miR-140 as one of the most highly expressed and specific miRNAs in chondrocytes (Kobayashi et al. 2008; Tuddenham et al. 2006; Wienholds et al. 2005). Two different groups have since elucidated the role of miR-140 in the growth plate using different mouse models. First, Miyaki et al. showed that skeletal ablation of miR-140 (cre-recombination driven by somitic mesoderm specific Meox promoter) resulted in a mild skeletal defect with shortening of limbs (Miyaki et al. 2010). Growth plates were examined histologically at 1-week of age postnatally, and the authors found decreased proliferative columns height (and reduced proliferation), but hypertrophic zone height were not affected, therefore suggesting miR-140 contribute to regulation of chondrocyte proliferation. Subsequently, Nakamura et al. used a mouse model with complete knockout of miR-140, and found similarly mild skeletal defect and decreased proliferative column height at different ages (Nakamura et al. 2011). However, careful histological examination of these mice suggested that the shortening of proliferative zone may instead be due to inhibition of resting chondrocyte recruitment into the proliferative columns and accelerated hypertrophic differentiation (Nakamura et al. 2011). These authors therefore conclude that miR-140 may instead regulate resting chondrocytes differentiation and hypertrophic differentiation.
Recent studies have provided insights into the molecular mechanisms by which miR-140 is regulated in the growth plate and how miR-140 in turn regulates chondrocyte functions. miR-140 expression was found to be positively regulated by Sox9 (Nakamura et al. 2012), which is an important transcription factors for cartilage formation and subsequent chondrocyte differentiation. Earlier efforts have identified several downstream targets of miR-140 in chondrocytes, including Aspartyl aminopeptidase (Dnpep) (Nakamura et al. 2011), which modulates bone morphogenetic protein (BMP) signaling and therefore affects hypertrophy; and A disintegrin and metalloproteinase with thrombospondin motifs 5 (Adamts5) (Miyaki et al. 2010), an enzyme involved in cartilage-matrix turnover. More recently, genome-wide screening has identified and experimentally validated many bona fide targets of miR-140 in chondrocytes, including a list of genes involved in Wnt signaling (Barter et al. 2015). The same study went on to show that miR-140 promoted activation of the non-canonical Wnt signaling pathway (Barter et al. 2015).
In addition to miR-140, another group of miRNAs recently demonstrated to have important functions in the growth plate is the let-7 family of miRNAs. The murine let-7 family is composed of 12 members expressed from eight genomic loci (let-7a-1, let-7a-2, let-7b, let-7c-1, let-7c-2, let-7d, let-7e, let-7f-1, let-7f-2, let-7g, let-7i, miR-98), and multiple members were found to be among the most abundantly expressed miRNAs in the growth plate (Kobayashi et al. 2008). Due to the large number of members and their genomic locations, it is impractical to genetically knockout the whole let-7 family of miRNAs. However, one study showed that RNA binding protein Lin28a and lin28b both inhibit biogenesis of let-7 (Piskounova et al. 2011), therefore suggesting that suppression of pan-let-7 expression could be achieved through transgenic overexpression of Lin28a or b. Interestingly, transgenic mice ubiquitously overexpressing Lin28a demonstrated overgrowth (Zhu et al. 2010), while ubiquitous let-7 overexpression caused a slight decrease in body size (Frost & Olson 2011). Nevertheless, it was not clear at the time whether these growth phenotypes were attributed to the function of Lin28-let-7 directly on skeletal development. More recently, transgenic mice with cartilage specific overexpression of Lin28a have been generated (Papaioannou et al. 2013). Lin28a transgenic mice demonstrated significant suppression of all let-7 miRNA family in the growth plate, and surprisingly, led to growth retardation rather than overgrowth (as in the ubiquitous Lin28a transgenic). Mice with cartilage-specific Lin28a expression showed decreased chondrocyte proliferation and increased apoptosis in the proliferative zone (Papaioannou et al. 2013), leading to a mild reduction in bone length. Interestingly, when these transgenic mice were crossed with the miR-140 knockout mice (which showed a similarly mild reduction in bone length), the simultaneous loss of let-7 and miR-140 resulted in a substantially greater reduction (~50%) of bone length and body size, suggesting a synergistic (rather than additive) effect of miR-140 and let-7 on bone growth. Because the combined Lin28a transgenic/miR-140 knockout produced a growth phenotype reminiscent of mice with chondrocytes missing Dicer, it is likely that miR-140 and the let-7 family represents two major miRNA species important for skeletal development and chondrocyte functions, with let-7 regulating proliferation, and miR-140 regulating resting chondrocyte recruitment and hypertrophic differentiation (Fig.3).
Fig 3.
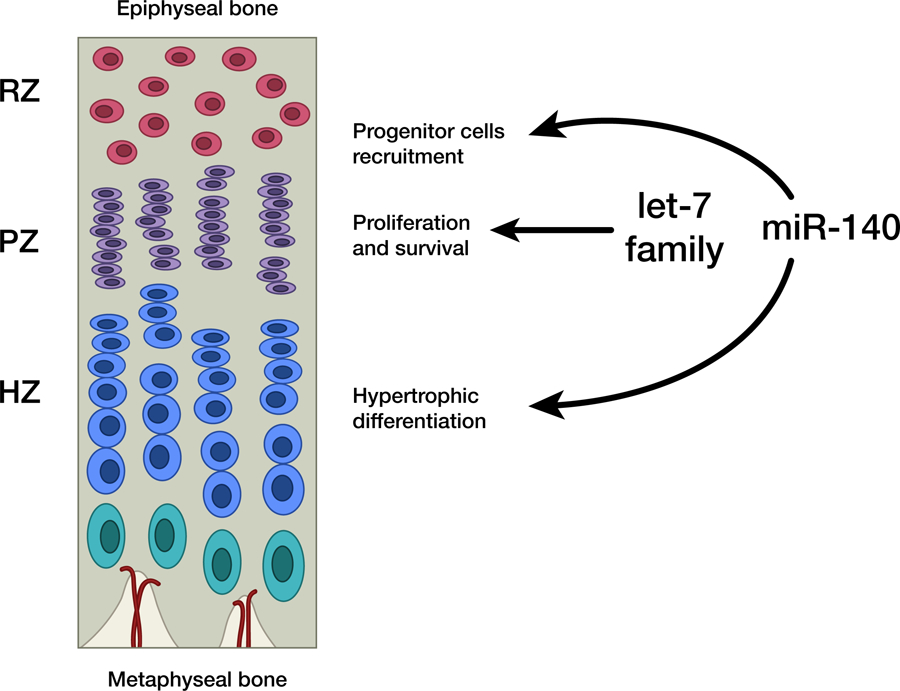
Proposed model for regulation of growth plate by miRNAs.
Numerous recent studies have identified other miRNAs that may have an effect on chondrocyte differentiation. For example, in a micromass culture model of mesenchymal stem cell line C3H10T1/2, BMP2 strongly suppresses expression of miR-199a, which negatively regulate downstream signaling by Smad1 and 4 (Lin et al. 2009). Similarly, transforming growth factor- beta (TGF-β) induced chondrogenic differentiation of mesenchymal stem cells causes downregulation of another miRNA miR-145, which directly target SRY-related high mobility group-Box 9 (Sox9), which is a key transcription factor for chondrogenesis (Yang et al. 2011). In the articular cartilage, a study showed that expression of type II collagen is regulated by miR-675, whose production itself is positively regulated by Sox9 (Dudek et al. 2010). More in vivo evidence will be needed to address whether these miRNAs indeed have important physiological functions in endochondral ossification and bone growth.
miRNAs and GH-IGF-I-axis
So far I have been discussing local regulation of miRNAs on growth plate functions. However, longitudinal bone growth, as well as somatic growth of other organs, are also regulated systemically by different hormonal factors, and particularly, by growth hormone (GH) and insulin-like growth factor-I (IGF-I). Both GH and IGF-I are potent stimulators of body growth. The anterior pituitary in the brain produces GH, which increases production of IGF-I in the liver, muscles, and other organs with growth hormone receptors (GHR). IGF-I, both in the circulation (from the liver) and local, in turn acts on the growth plate and other organs to stimulate growth (Fig.4). Children with GH deficiency or GH insensitivity therefore results in poor growth (for review, see (Lui et al. 2015)). Emerging evidence suggested that miRNAs may play important roles in regulating IGF-I signaling. For example, IGF-I signaling is frequently elevated in breast cancer, and is found to be due in part to overexpression of the IGF-I receptor (IGF1R) (Tamimi et al. 2011). In these breast cancer cells, two miRNAs that suppresses IGF1R expression, miR-148a and miR-152, were found to be downregulated (Xu et al. 2013). In addition, a long list of miRNAs have been identified to negatively regulate IGF-I signaling but were downregulated in different cancers, strongly linking miRNAs to tumorigenesis through modulation of IGF-I signaling (for review, see (Jung & Suh 2014)).
Fig 4.
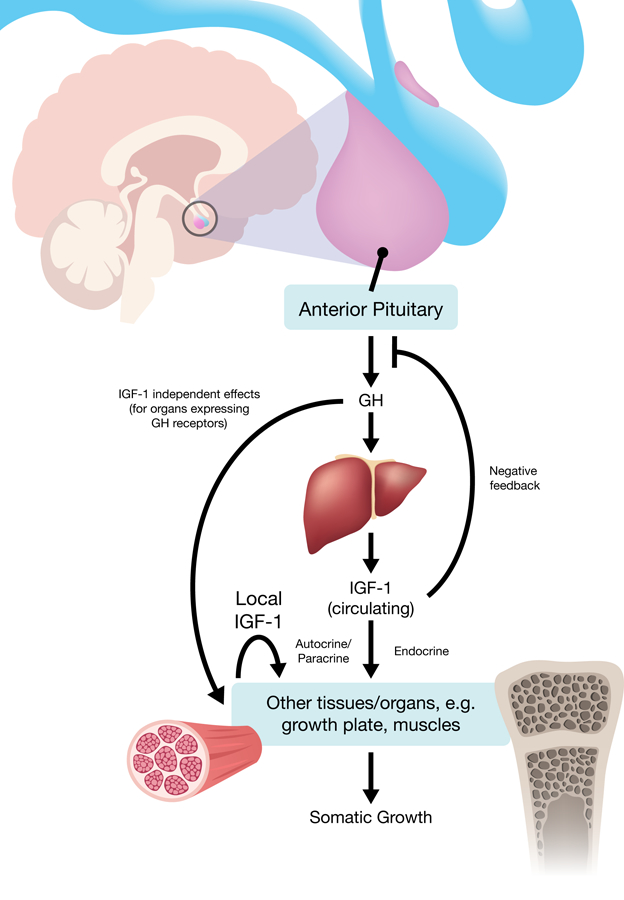
Stimulation of Growth by GH-IGF-I axis.
In contrast to the extensive literature available on in vitro functions of miRNAs on IGF-I signaling, much is left to be explored on the in vivo role of miRNAs on IGF-I signaling and organs and body growth. In 2009, Hyun et al. found in drosophila that deletion of miR-8 (the sole Drosophila ortholog of the mammalian miR-200 family) resulted in defective insulin/IGF signaling and decreased body size (Hyun et al. 2009). Interestingly, in mammalian cells, miR-200 targets FOG2 (Friend of GATA2), which inhibits PI3K activity. Therefore, ablation of miR-8 led to increased expression of ush (the drosophila homolog of FOG2), hence suppressing PI3K activity and cell growth (Hyun et al. 2009). In mammals, the miR-200 family consists of multiple members (miR-200a, miR-200b, miR-200c, miR-141, and miR-429), making it difficult to completely knockout the miR-200 family. One mouse model with deletion of miR-200b and miR-429 has been generated but was not found to affect body size (Hasuwa et al. 2013). It remains unclear whether deletion of all miR-200 family members would have an effect on body size in mammals. In zebrafish, injection of exogenous miR-141 or miR-429a (two members of the miR-200 family) in embryos led to suppression of GH and GH receptor expressions, therefore causing decreased IGF1 expression and decreased somatic growth in embryos (Jing et al. 2015). These findings are not consistent with that in drosophila, where deletion, rather than overexpression of miR-8/miR-200, resulted in decreased IGF signaling and growth. In addition, in both mice (Lupu et al. 2001) and humans (Mehta et al. 2005), GH does not have an effect on prenatal growth and birth size. Therefore it remains unclear whether these findings in zebrafish embryos could be replicated in mammals. Taken together, recent in vivo findings on miRNAs and GH/IGF-I signaling have shown early promising signs but much is yet to be elucidated.
Regulation of Growth-Limiting Genetic Program by miRNA
Body growth is regulated both systemically (through different hormones) and locally, as indicated by transplantation experiments showing juvenile organs continue to grow at a fast pace when placed into adult recipients (Pape et al. 2006; Metcalf 1964; Montgomery et al. 1981), suggesting intrinsic mechanisms exist within organs to regulate somatic growth rate. Our group has previously identified a growth-limiting genetic program that may help explain the local regulation of organ growth. Because organ growth is rapid in early life but gradually slows with age, and this deceleration of growth happens in a coordinated fashion to help maintain body proportions, we hypothesized the existence of a genetic program common between different organs to regulate growth in this period. To test our hypothesis, we performed microarray and specifically looked for changes of gene expression that occurs during early postnatal life, and sought to identify genes that were commonly up- or downregulated with age in multiple organs. Our study uncovered in mice and rats, an evolutionarily conserved growth-limiting genetic program that occurs simultaneously in multiple organs during juvenile life (Finkielstain et al. 2009; Lui et al. 2010b). This program is common among liver, kidney, lung, and heart, and involves the downregulation with age of a large number of growth-promoting genes (including Ezh2, Igf2, Mest, Mycn), therefore driving somatic growth to gradually slow with age in multiple organs. More recently, we have identified the same genetic program in juvenile organs in sheep (Delaney et al. 2014), and interestingly, the downregulation of these growth-promoting genes appears to occur over a longer period of time in sheep, suggesting that the pace of this program may be evolutionarily modulated to achieve different body sizes among different mammalian species (Delaney et al. 2014).
Because a single miRNA may regulate a large number of target mRNAs, we reasoned that downregulation of growth-promoting genes in the program might be driven by upregulation of a common miRNA with age. In support of our hypothesis, we performed miRNA microarray in juvenile mouse organs and found four miRNAs, 3 of which belongs to the miR-29 family (miR-29a, miR-29b, and miR-29c) to be coordinately upregulated with age in multiple organs (Kamran et al. 2015). Bioinformatic analysis showed that predicted targets of miR-29 were overrepresented in genes that were downregulated with age in multiple organs, and we went on to experimentally validated three target genes implicated in body growth, namely Igf1, Mesoderm-specific transcript (Mest), and Igf2 binding protein 1 (Igf2bp1), to be bona fide targets of miR-29 (Kamran et al. 2015). Our findings suggested that the miR-29 family of miRNAs negatively regulates organ growth, and the upregulation of miR-29 during juvenile life may help drive the downregulation of growth promoting genes, thus contributing to the gradual slowing of organ growth with age. Our hypothesis would also predict that knockout of miR-29 family will result in increase in body growth and body size. However, ubiquitous miR-29 knockout mice have been generated and these mice did not have overgrowth, but instead showed growth retardation and die around 4 weeks of age (Cushing et al. 2015). Examination of these mice showed significant defects in lung vascular smooth muscle cells differentiation, leading to respiratory problems and early postnatal fatality. Therefore, it remains unclear whether miR-29, in addition to its essential role in lung development, may serve as a major negative regulator of postnatal body growth.
Concluding remarks
Numerous recent studies have uncovered the importance of miRNAs in the regulation of growth, particularly in chondrocytes, and how that affects the overall function of the growth plate to drive longitudinal bone growth. However, due to our limited knowledge on the mechanisms of overall body growth regulation, we are just beginning to understand how miRNAs may be involved in this process (a summary is provided in Table 1). It would be important to consolidate findings between different model organisms to try to improve the applicability of the information gathered in drosophila and zebrafish into mammalian species. Additionally, considering the clear connection between miRNAs and unrestrained growth in tumor cells, one may be tempted to explore if any of these exciting discovery in cancer may help yield important insights into the regulation of miRNAs on the normal physiology of cellular, organ, and overall body growth.
Table 1.
miRNA involved in overall organs/body growth regulation
| miRNA | Species | Proposed effect on growth | Molecular target(s) | Reference |
|---|---|---|---|---|
| miR-8/ miR200 | Drosophila | Promotes growth by increasing PI3K activation | USH/FOG2 | (Hyun et al. 2009) |
| miR-200 | Zebrafish | Suppresses growth by targeting GH-IGF-I signaling | GH, GHR, IGF2a | (Jing et al. 2015) |
| miR-9a | Drosophila | Suppresses growth by targeting insulin signaling | sNPFR1/NPY2R | (Suh et al. 2015) |
| miR-1/ miR-133 | Mouse | Suppresses cardiac hypertrophy | RohA, Cdc42, Nelf-A/WHSC2 | (Care et al. 2007) |
| let-7 | Mouse | Suppresses growth by regulating glucose metabolism and insulin sensitivity | Myc, Kras, Igf2bp1, Hmga2 | (Zhu et al. 2010) (Frost & Olson 2011) |
| miR-29 | Mouse | Suppresses organs growth by targeting a genetic program | Igf1, Mest, Igfbp1 | (Kamran et al. 2015) |
Acknowledgements
I would like to thank Jeremy Swan and Nichole C. Swan from the Computer Support Services Core of Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, for recreating the figures in this manuscript.
Funding Source
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH.
Abbreviations
- miRNAs
microRNAs
- Pri-miRNAs
primary miRNAs
- GH
growth hormone
- GHR
growth hormone receptor
- IGF-I
insulin-like growth factor-I
- IGF1R
insulin-like growth factor-I receptor
- RISC
RNA-induced silencing complex
- mRNA
Messenger RNA
- UTR
untranslated region
- BMP
bone morphogenetic protein
- TGF
transforming growth factor
Reference List
- Abad V, Meyers JL, Weise M, Gafni RI, Barnes KM, Nilsson O, Bacher JD & Baron J 2002. The role of the resting zone in growth plate chondrogenesis. Endocrinology 143 1851–1857. [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: target recognition and regulatory functions. Cell 136 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barter MJ, Tselepi M, Gomez R, Woods S, Hui W, Smith GR, Shanley DP, Clark IM & Young DA 2015. Genome-Wide MicroRNA and Gene Analysis of Mesenchymal Stem Cell Chondrogenesis Identifies an Essential Role and Multiple Targets for miR-140–5p. Stem Cells 33 3266–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushing L, Costinean S, Xu W, Jiang Z, Madden L, Kuang P, Huang J, Weisman A, Hata A, Croce CM & Lu J 2015. Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature. PLoS Genet. 11 e1005238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney A, Padmanabhan V, Rezvani G, Chen W, Forcinito P, Cheung CS, Baron J & Lui JC 2014. Evolutionary conservation and modulation of a juvenile growth-regulating genetic program. J Mol.Endocrinol. 52 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek KA, Lafont JE, Martinez-Sanchez A & Murphy CL 2010. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol.Chem. 285 24381–24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N & Filipowicz W 2010. Regulation of mRNA translation and stability by microRNAs. Annu.Rev.Biochem. 79 351–379. [DOI] [PubMed] [Google Scholar]
- Finkielstain GP, Forcinito P, Lui JC, Barnes KM, Marino R, Makaroun S, Nguyen V, Lazarus JE, Nilsson O & Baron J 2009. An extensive genetic program occurring during postnatal growth in multiple tissues. Endocrinology 150 1791–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost RJ & Olson EN 2011. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc.Natl.Acad.Sci.U.S.A 108 21075–21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasuwa H, Ueda J, Ikawa M & Okabe M 2013. miR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science 341 71–73. [DOI] [PubMed] [Google Scholar]
- Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J & Kim VN 2009. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 139 1096–1108. [DOI] [PubMed] [Google Scholar]
- Jing J, Xiong S, Li Z, Wu J, Zhou L, Gui JF & Mei J 2015. A feedback regulatory loop involving p53/miR-200 and growth hormone endocrine axis controls embryo size of zebrafish. Sci.Rep. 5 15906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HJ & Suh Y 2014. Regulation of IGF −1 signaling by microRNAs. Front Genet. 5 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamran F, Andrade AC, Nella AA, Clokie SJ, Rezvani G, Nilsson O, Baron J & Lui JC 2015. Evidence That Up-Regulation of MicroRNA-29 Contributes to Postnatal Body Growth Deceleration. Mol.Endocrinol. 29 921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, Merkenschlager M & Kronenberg HM 2008. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc.Natl.Acad.Sci.U.S.A 105 1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg HM 2003. Developmental regulation of the growth plate. Nature 423 332–336. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH & Kim VN 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB & Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120 15–20. [DOI] [PubMed] [Google Scholar]
- Lin EA, Kong L, Bai XH, Luan Y & Liu CJ 2009. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol.Chem. 284 11326–11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC, Andrade AC, Forcinito P, Hegde A, Chen W, Baron J & Nilsson O 2010a. Spatial and temporal regulation of gene expression in the mammalian growth plate. Bone 46 1380–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC, Forcinito P, Chang M, Chen W, Barnes KM & Baron J 2010b. Coordinated postnatal down-regulation of multiple growth-promoting genes: evidence for a genetic program limiting organ growth. FASEB J 24 3083–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC, Garrison P & Baron J 2015. Regulation of body growth. Curr.Opin.Pediatr 27 502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC, Nilsson O & Baron J 2011. Growth plate senescence and catch-up growth. Endocr.Dev. 21 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE & Kutay U 2004. Nuclear export of microRNA precursors. Science 303 95–98. [DOI] [PubMed] [Google Scholar]
- Lupu F, Terwilliger JD, Lee K, Segre GV & Efstratiadis A 2001. Roles of growth hormone and insulin-like growth factor 1 in mouse postnatal growth. Dev.Biol. 229 141–162. [DOI] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, Filipowicz W, Duchaine TF & Sonenberg N 2007. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 317 1764–1767. [DOI] [PubMed] [Google Scholar]
- Mehta A, Hindmarsh PC, Stanhope RG, Turton JP, Cole TJ, Preece MA & Dattani MT 2005. The role of growth hormone in determining birth size and early postnatal growth, using congenital growth hormone deficiency (GHD) as a model. Clin.Endocrinol.(Oxf) 63 223–231. [DOI] [PubMed] [Google Scholar]
- Metcalf D 1964. Restricted Growth Capacity of Multiple Spleen Grafts. Transplantation 2 387–392. [DOI] [PubMed] [Google Scholar]
- Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, Takada S, Lotz MK, Ueno-Kudo H & Asahara H 2010. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 24 1173–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RK, Sybicki MA & Grand RJ 1981. Autonomous biochemical and morphological differentiation in fetal rat intestine transplanted at 17 and 20 days of gestation. Dev.Biol. 87 76–84. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, He X, Kato H, Wakitani S, Kobayashi T, Watanabe S, Iida A, Tahara H, Warman ML, Watanapokasin R & Postlethwait JH 2012. Sox9 is upstream of microRNA-140 in cartilage. Appl.Biochem.Biotechnol. 166 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Inloes JB, Katagiri T & Kobayashi T 2011. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol.Cell Biol. 31 3019–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou G, Inloes JB, Nakamura Y, Paltrinieri E & Kobayashi T 2013. let-7 and miR-140 microRNAs coordinately regulate skeletal development. Proc.Natl.Acad.Sci.U.S.A 110 E3291–E3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape L, Hoppe J, Becker T, Ehrich JH, Neipp M, Ahlenstiel T & Offner G 2006. Superior long-term graft function and better growth of grafts in children receiving kidneys from paediatric compared with adult donors. Nephrol.Dial.Transplant. 21 2596–2600. [DOI] [PubMed] [Google Scholar]
- Petersen CP, Bordeleau ME, Pelletier J & Sharp PA 2006. Short RNAs repress translation after initiation in mammalian cells. Mol.Cell 21 533–542. [DOI] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D & Gregory RI 2011. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 147 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D & Abdellatif M 2011. MicroRNAs in development and disease. Physiol Rev. 91 827–887. [DOI] [PubMed] [Google Scholar]
- Tamimi RM, Colditz GA, Wang Y, Collins LC, Hu R, Rosner B, Irie HY, Connolly JL & Schnitt SJ 2011. Expression of IGF1R in normal breast tissue and subsequent risk of breast cancer. Breast Cancer Res Treat. 128 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I & Dalmay T 2006. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 580 4214–4217. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de BE, Horvitz HR, Kauppinen S & Plasterk RH 2005. MicroRNA expression in zebrafish embryonic development. Science 309 310–311. [DOI] [PubMed] [Google Scholar]
- Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y, Qi YT, Xu Q, Li W, Lu B, Peiper SS, Jiang BH & Liu LZ 2013. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol.Cell Biol. 5 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Guo H, Zhang Y, Chen L, Ying D & Dong S 2011. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS One 6 e21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, Hirschhorn JN, Palmert MR & Daley GQ 2010. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat.Genet. 42 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]


