Abstract
Inbreeding is typically detrimental to fitness. However, some animal populations are reported to inbreed without incurring inbreeding depression, ostensibly due to past “purging” of deleterious alleles. Challenging this is the position that purging can, at best, only adapt a population to a particular environment; novel selective regimes will always uncover additional inbreeding load. We consider this in a prominent test case: the eusocial naked mole‐rat (Heterocephalus glaber), one of the most inbred of all free‐living mammals. We investigated factors affecting mortality in a population of naked mole‐rats struck by a spontaneous, lethal coronavirus outbreak. In a multivariate model, inbreeding coefficient strongly predicted mortality, with closely inbred mole‐rats (F≥ 0.25) over 300% more likely to die than their outbred counterparts. We demonstrate that, contrary to common assertions, strong inbreeding depression is evident in this species. Our results suggest that loss of genetic diversity through inbreeding may render populations vulnerable to local extinction from emerging infectious diseases even when other inbreeding depression symptoms are absent.
Keywords: Coronavirus, disease susceptibility, Heterocephalus glaber, inbreeding depression, purging
Inbreeding depression, the negative fitness effects of mating among relatives, has been well documented in laboratory, agricultural, and natural settings (e.g., Charlesworth and Charlesworth 1987; Crnokrak and Roff 1999; Hedrick and Kalinowski 2000; Keller and Waller 2002). Nonetheless, great variance exists in the magnitude of inbreeding depression within and among populations (e.g., Pray and Goodnight 1995; Lacy and Ballou 1998; Haag et al. 2003) and some populations, despite high levels of inbreeding, show little or no evidence of inbreeding depression (e.g., Keane et al. 1986; Doums et al. 1996; Kalinowski et al. 1999; Visscher et al. 2001; Duarte et al. 2003; Bilde et al. 2005; Peer and Taborsky 2005; Milot et al. 2007). Such a lack of inbreeding depression is often attributed to “purging,” the removal of deleterious alleles by strong selection, during historical periods of extended inbreeding (Lande and Schemske 1985; Barrett and Charlesworth 1991). Although there is empirical support for reduced inbreeding depression with continued inbreeding (e.g., Barrett and Charlesworth 1991; McCall et al. 1994; Crnokrak and Barrett 2002), recent studies, both theoretical (Wang et al. 1999; Wang 2000; Whitlock 2002; Glémin 2003) and empirical (Barrett and Charlesworth 1991; Ballou 1997; Lacy and Ballou 1998; Byers and Waller 1999; Crnokrak and Barrett 2002; Boakes et al. 2007), have demonstrated that purging is not always effective. Alleles that are only deleterious under certain environmental conditions, for example, will only be disfavored by natural selection when those particular conditions arise (Bijlsma et al. 1999). Even habitual inbreeders would thus be expected to still harbor alleles that would be deleterious under some different, novel selection regime.
The eusocial naked mole‐rat, Heterocephalus glaber, is a classic example of a habitual inbreeder seemingly impervious to inbreeding depression; “the only mammal species that has been shown to undergo continuous close inbreeding with no obvious effects of inbreeding depression” (Bromham and Harvey 1996). An early microsatellite study of wild colonies suggested that over 80% of all mating occurs between first‐degree relatives (Reeve et al. 1990)—an unprecedented level of inbreeding among vertebrates. The lack of inbreeding avoidance and the naked mole‐rats' intense xenophobia (Lacey and Sherman 1991) have been described as “mechanisms that apparently minimize the chance of ever outbreeding” (Jarvis et al. 1994). More recently, however, some circumstantial evidence has emerged of some latent selection for outbreeding: O'Riain et al. (1996) and Braude (2000) demonstrated the existence of rare dispersal morphs, and Clarke and Faulkes (1999) and Ciszek (2000) demonstrated a weak mating preference for nonkin. However, in 25 years of intensive study no overt signs of inbreeding depression have been reported. This makes the naked mole‐rat a particularly interesting case to test the hypothesis that novel selection regimes may uncover inbreeding depression.
In this study, we performed a retrospective investigation of the effects of inbreeding on naked mole‐rat mortality through a spontaneous outbreak of a novel viral pathogen. In doing so, we uncovered the first evidence of a substantial cost to continuous close inbreeding in this species.
Methods
STUDY POPULATION
Our population, initiated with wild‐caught founders from various localities in Kenya, has been maintained since 1981 in custom‐built facilities at the University of Cape Town. Husbandry details have been described previously by Jarvis (1991). Colonies typically comprised a single breeding pair and up to 55 nonbreeding relatives of both sexes. In total, our sample comprised 365 animals in 10 complete colonies: 209 males and 159 females, including 11 and 10 breeders of either sex, respectively. Age and body mass were known for all animals (age: 5 to 273 months, mean 89.4 ± 60.4 SD; mass: 11.9 to 83.2 g, mean 35.7 ± 12.7 SD). Furthermore, for each individual, a single dam and sire could be identified with certainty, on the basis of their distinctive morphologies and behavioral profiles (Clarke and Faulkes 1998). Based on these records, we constructed a pedigree for each individual (maximum length: five generations) and used it to estimate a coefficient of inbreeding (F) as per Wright (1922). Our study violates a key assumption of Wright's approach, however: that pedigree founders are unrelated. Population genetic studies (Faulkes et al. 1990; Reeve et al. 1990) suggest that intracolony relatedness is typically very high in wild naked mole‐rats. Thus, here we explicitly assumed that pedigree founders captured from the same exact locality (i.e., same colony) were siblings. Parallel analyses in which founders were held to be unrelated yielded qualitatively concordant analyses (results not shown). Inbreeding coefficients ranged from F= 0 (outbred) through to F= 0.5 (highly inbred) with mean F= 0.163 ± 0.158 SD.
DISEASE OUTBREAK
In September 2002, an emergent strain of virulent enteric coronavirus swept unchecked through our captive naked mole‐rat study population, causing acute diarrhea, dehydration, and severe enteric hemorrhaging. The day of first exposure in each colony was determined from the first appearance of symptoms (dry, unused toilet chamber), and in each case, the first deaths were recorded within two days. Coronaviruses spread easily by faeco‐oral or aerosol transmission (Weiss and Navas‐Martin 2005), so we assumed that all individuals in a colony were exposed to the virus simultaneously. No attempts were made to medicate infected animals. Mortality was monitored daily and dead animals were removed immediately for identification and confirmation of the presence of disease symptoms.
STATISTICAL ANALYSIS
To identify factors affecting survival, we used proportional hazards regressions (Therneau and Grambsch 2002), implemented with the PHREG procedure (with the TIES = exact option), in SAS (SAS Institute, Cary, NC). Our data conformed to the assumption of proportional hazards; no evidence of nonproportionality was apparent in the smoothed, scaled Schoenfeld residual plots (Therneau and Grambsch 2002) and the inclusion of a time‐dependent covariate had a nonsignificant effect (P= 0.09). Model selection followed the “main‐effects first” model‐building strategy of Hosmer and Lemeshow (2000). Only the effects shown in Table 1 were entered into the final model; we excluded body mass as an explanatory variable after preliminary runs showed that its presence did not significantly improve the model (likelihood ratio test: χ2= 1.27; P= 0.26). All two‐ and three‐way interaction terms not shown in Table 1 were excluded in a similar fashion. Because the 10 colonies were housed in three separate constant environment rooms, we accounted for possible location effects by stratifying the final model by “room number,” and nested within that, “colony.” An unstratified model, however, yielded virtually identical results. To assess the relative effect of different levels of inbreeding on survival, we calculated hazard ratios (Therneau and Grambsch 2002) for F at discrete values. Finally, inbreeding depression was estimated in terms of the number of lethal equivalents per gamete, calculated as per Kalinowski and Hedrick (1998).
1.
Factors affecting the probability of death during the coronavirus epizootic.
| Variable | df | Parameter estimate | SE | χ2 | P | Hazard ratio | 95% confidence limits for hazard ratio | |
| Inbreeding F | 1 | 4.422 | 1.444 | 9.379 | 0.0022 | 83.295 | 4.914 | 1411.805 |
| Age (months) | 1 | 0.006 | 0.002 | 8.008 | 0.0047 | 1.006 | 1.002 | 1.011 |
| Sex | 1 | 0.362 | 0.179 | 4.078 | 0.0434 | 1.436 | 1.011 | 2.041 |
| Breeding status | 1 | 1.012 | 1.383 | 0.536 | 0.4642 | 2.752 | 0.183 | 41.399 |
| Age × Breeding status | 1 | −0.017 | 0.006 | 7.677 | 0.0056 | 0.983 | 0.971 | 0.995 |
| Sex × Breeding status | 1 | 2.821 | 1.096 | 6.624 | 0.0101 | 16.789 | 1.959 | 143.863 |
Results
In just eight weeks, acute diarrhea, dehydration, and severe enteric haemorrhaging associated with the coronavirus killed 161 of 365 animals (44.1%). Survival was significantly lower among more inbred animals (Fig. 1). Statistically controlling for the effects of other explanatory variables (Table 1), we found that offspring produced by half‐sibling (F= 0.125) and full‐sibling (F= 0.250) parent pairs were, respectively, 174% and 302% more likely to die than the offspring of unrelated parents (F= 0). Inbreeding depression in survival, measured as lethal equivalents, was calculated as B= 1.13 (95% CI: 0.50–1.81).
1.
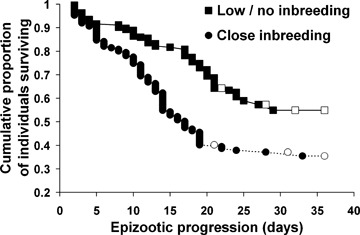
Kaplan–Meier plots showing the proportional survival of highly inbred (F≥ 0.25) versus outbred and mildly inbred naked mole‐rats (0 ≤F≤ 0.125) through the course of the coronavirus outbreak. Open markers denote censored datapoints. Although represented categorically here, inbreeding was treated as a continuous variable for all regression analyses.
Age was also highly significantly correlated with susceptibility, with 10 years of age increasing mortality risk by 212.5% among nonbreeders (Fig. 2). Sex too was a significant factor, with females (143.6%) more likely to survive than males. Breeding status interacted significantly with sex. Queens had high survival despite their advanced age (eight of 10 survived), whereas only two of 11 male breeders survived.
2.
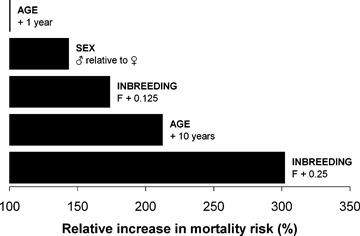
Comparison of selected variables' effects on the risk of death during the coronavirus epizootic. These values are derived from hazard ratio estimates generated by the proportional hazards model and reflect the change in likelihood of death following a given change of a predictor variable from its base condition (i.e., 0 for continuous variables, females in the case of sex). Age and sex effect estimates shown here apply to nonbreeders only.
Discussion
This study provides the first clear evidence of a substantial cost to inbreeding in this habitual inbreeder. Inbred naked mole‐rats were significantly more likely to die than their outbred counterparts, displaying inbreeding depression of a magnitude similar to that observed in other captive rodents (naked mole‐rats: B = 1.13; other rodents: mean B = 1.15; calculated for n= 7 captive taxa; Ralls et al. 1988). Note, however, that this estimate of lethal equivalents does not account for variation in other variables (sex, age, breeding status) that were found to affect mortality (Table 1), and may thus be an underestimate. Moreover, this figure does not account for Allee effects associated with the obligate social lifestyle of naked mole‐rats. In the wild, naked mole‐rat colonies persist under harsh environmental conditions because the cooperative efforts of multiple foragers enable them to exploit patchy food resources. The loss of 87% of the workforce (as we observed in one colony) would almost certainly lead to local extinction of a wild colony. Thus, the true costs of inbreeding in the wild may well be higher than the estimates presented here.
Mechanistically, inbreeding increases homozygosity, leaving inbred individuals with lower genetic diversity than their outbred counterparts. There is much interest in the relationship between host genetic diversity (at key loci such as the major histocompatibility complex, as well as more generally in the genome) and resistance to parasites and disease (O'Brien and Evermann 1988; Hamilton et al. 1990; Hedrick 1994). The results of this study augment mounting evidence from a broad range of taxa that reduced genetic diversity, as can arise through inbreeding, is associated with increased susceptibility to infection (O'Brien et al. 1985; Black 1992; Carrington et al. 1999; Coltman et al. 1999; Meagher 1999; Schmid‐Hempel and Crozier 1999; Cassinello et al. 2001; Hedrick et al. 2001; Messaoudi et al. 2002; Acevedo‐Whitehouse et al. 2003; McClelland et al. 2003; Reid et al. 2003; Spielman et al. 2004; Wegner et al. 2004; Hawley et al. 2005; Calleri et al. 2006; Whiteman et al. 2006; Luong et al. 2007; Reid et al. 2007; Seeley and Tarpy 2007).
Our secondary findings, that susceptibility increased with age and was higher in males, both conform to established theory. Immune responsiveness is known to decline with age in various taxa (Adamo et al. 2001; Linton and Dorshkind 2004), with a range of symptoms including a marked decline in T‐cell production. Similarly, a male bias in susceptibility is a well‐documented phenomenon (Klein 2000), and in vertebrates is hypothesized to result at least in part from the immunosuppressive burden of testosterone (Folstad and Karter 1992). Our data provide circumstantial support for this hypothesis, in that breeding males, which typically show highly elevated testosterone levels (Clarke and Faulkes 1998), were especially vulnerable to the virus. The high survival among queens was unexpected, but may also have been partially hormone driven: glucocorticoid stress hormones, also known to be immunosuppressive (Sapolsky 1992), are typically lower in naked mole‐rat queens than in other colony members, except during pregnancy (Faulkes and Abbott 1997). At such times stress hormone levels are elevated, most likely in response to acute energetic stress (Råberg et al. 1998), and immune function is typically impaired (e.g., Nordling et al. 1998) Accordingly, of four mole‐rat queens pregnant at the onset of the infection, two aborted and survived, whereas the two that carried their pups to full term succumbed to the virus. Other potential sources of energetic stress, and hence immunosuppression, such as differences in nutritional status (Gershwin et al. 1985), workload (Deerenberg et al. 1997), or thermoregulatory burden (Nelson and Demas 1996), are unlikely to have contributed substantially to these results because all animals had lifelong access to ad libitum food and lived in a finely regulated physical environment.
In this study, we have demonstrated that inbreeding is associated with increased susceptibility to a viral epidemic in a habitual close inbreeder, ostensibly free from other inbreeding depression effects. These results have important implications for our understanding of animal populations with reduced genetic variability. Inbred individuals may remain more vulnerable to emergent infectious diseases than their outbred counterparts, despite extensive purging of the background genetic load. Thus, assessment of inbreeding depression in the absence of severe disease outbreaks may dramatically underestimate the true fitness costs of inbreeding.
More generally, our results emphasize that purging cannot remove deleterious alleles unless these alleles are exposed to selection (Bijlsma et al. 1999). Alleles temporarily “hidden” from selection, such as those conferring susceptibility to novel or rare diseases, may accumulate over time without negatively affecting fitness. However, when an appropriate pathogen is ultimately encountered, these alleles may start to strongly affect fitness and thus will be exposed to selection. Our finding of increased mortality among inbred mole‐rats during a coronavirus outbreak reflects such purging in action.
Associate Editor: H. Kokko
ACKNOWLEDGMENTS
We thank J. U. M. Jarvis for pedigree records and H. Kokko and two anonymous reviewers for helpful discussions. Financial support was provided to JOR by the South African National Research Foundation. This research adhered to the Animal Behavior Society Guidelines for the Use of Animals in Research, the legal requirements of the country in which the work was carried out, and all institutional guidelines.
Contributor Information
Adin Ross‐Gillespie, Department of Zoology, University of Cape Town, Rondebosch 7700, South Africa; E‐mail: A.Ross‐Gillespie@ed.ac.uk.
M. Justin O'Riain, Department of Zoology, University of Cape Town, Rondebosch 7700, South Africa.
Lukas F. Keller, Zoologisches Museum, Universität Zürich, Winterthurerstrasse 190, CH‐8057 Zürich, Switzerland
LITERATURE CITED
- Acevedo‐Whitehouse,K., Gulland F., Greig D., and Amos W.. 2003. Disease susceptibility in California sea lions. Nature 422:35. [DOI] [PubMed] [Google Scholar]
- Adamo,S. A., Jensen M., and Younger M.. 2001. Changes in lifetime immunocompetence in male and female Gryllus texensis (formerly G. integer): trade‐offs between immunity and reproduction. Anim. Behav. 62:417–425. [Google Scholar]
- Ballou,J. D. 1997. Ancestral inbreeding only minimally affects inbreeding depression in mammalian populations. J. Hered. 88:169–178. [DOI] [PubMed] [Google Scholar]
- Barrett,S. C. H., and Charlesworth D.. 1991. Effects of a change in the level of inbreeding on the genetic load. Nature 352:522–524. [DOI] [PubMed] [Google Scholar]
- Bijlsma,R., Bundgaard J., and Van Putten W. F.. 1999. Environmental dependence of inbreeding depression and purging in Drosophila melanogaster. J. Evol. Biol. 12;1125–1137. [Google Scholar]
- Bilde,T., Lubin Y., Smith D., Schneider J. M., and Maklakov A. A.. 2005. The transition to social inbred mating systems in spiders: role of inbreeding tolerance in a subsocial predecessor. Evolution 59:160–174. [PubMed] [Google Scholar]
- Black,F. L. 1992. Why did they die? Science 258:1739–1740. [DOI] [PubMed] [Google Scholar]
- Boakes,E. H., Wang J., and Amos W.. 2007. An investigation of inbreeding depression and purging in captive pedigreed populations. Heredity 98:172–182. [DOI] [PubMed] [Google Scholar]
- Braude,S. 2000. Dispersal and new colony formation in wild naked‐mole‐rats: evidence against inbreeding as the system of mating. Behav. Ecol. 11:7–12. [Google Scholar]
- Bromham,L., and Harvey P. H.. 1996. Naked mole‐rats on the move. Curr. Biol. 6:1082–1083. [DOI] [PubMed] [Google Scholar]
- Byers,D., and Waller D. M.. 1999. Do plant populations purge their genetic load? Effects of population size and mating history on inbreeding depression. Annu. Rev. Ecol. Syst. 30:479–513. [Google Scholar]
- Calleri,D. V. II, Reid E. M., Rosengaus R. B., Vargo E. L., and Traniello J. F. A.. 2006. Inbreeding and disease resistance in a social insect: effects of heterozygosity on immunocompetence in the termite Zootermopsis angusticollis. Proc. R. Soc. Lond. B 273:2633–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington,M., Nelson G. W., Martin M. P., Kissner T., Vlahov D., Goedert J. J., Kaslow R., Buchbinder S., Hoots K., and O'Brien S.. 1999. HLA and HIV‐1: heterozygote advantage and B*35‐Cw*04 disadvantage. Science 283:1748–1752. [DOI] [PubMed] [Google Scholar]
- Cassinello,J., Gomendio M., and Roldan E. R. S.. 2001. Relationship between coefficient of inbreeding and parasite burden in endangered gazelles. Conserv. Biol. 15:1171–1174. [Google Scholar]
- Charlesworth,D., and Charlesworth B.. 1987. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 18:237–268. [Google Scholar]
- Ciszek,D. 2000. New colony formation in the “highly inbred” eusocial naked mole‐rat: outbreeding is preferred. Behav. Ecol. 11:1–6. [Google Scholar]
- Clarke,F. M., and Faulkes C. G.. 1998. Hormonal and behavioural correlates of male dominance and reproductive status in captive colonies of the naked mole‐rat, Heterocephalus glaber. Proc. R. Soc. Lond. B 265:1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke,F. M., and Faulkes C. G.. 1999. Kin discrimination and female mate choice in the naked mole‐rat Heterocephalus glaber. Proc. R. Soc. Lond. B 266:1995–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltman,D. W., Pilkington J. G., Smith J. A., and Pemberton J. M.. 1999. Parasite‐mediated selection against inbred soay sheep in a free‐living, island population. Evolution 53:1259–1267. [DOI] [PubMed] [Google Scholar]
- Crnokrak,P., and Barrett S. C. H.. 2002. Purging the genetic load: a review of the experimental evidence. Evolution 56:2347–2358. [DOI] [PubMed] [Google Scholar]
- Crnokrak,P., and Roff D. A.. 1999. Inbreeding depression in the wild. Heredity 83:260–270. [DOI] [PubMed] [Google Scholar]
- Deerenberg,C., Apanius V., Daan S., and Bos N.. 1997. Reproductive effort decreases antibody responsiveness. Proc. R. Soc. Lond. B 264:1021–1029. [Google Scholar]
- Doums,C., Viard F., Pernot A. F., Delay B., and Jarne P.. 1996. Inbreeding depression, neutral polymorphism, and copulatory behavior in freshwater snails: a self‐fertilization syndrome. Evolution 50:1908–1918. [DOI] [PubMed] [Google Scholar]
- Duarte,L. C., Bouteiller C., Fontanillas P., Petit E., and Perrin N.. 2003. Inbreeding in the greater white‐toothed shrew, Crocidura russula. Evolution 57:638–645. [DOI] [PubMed] [Google Scholar]
- Faulkes,C. G., and Abbott D. H.. 1997. The physiology of a reproductive dictatorship: regulation of male and female reproduction by a single breeding female in colonies of naked mole‐rats. Pp. 302–334 in Solomon N. G. and French J. A., eds. Cooperative breeding in mammals. Cambridge Univ. Press, Cambridge, UK. [Google Scholar]
- Faulkes,C. G., Abbott D. H., and Mellor A. L.. 1990. Investigation of genetic diversity in wild colonies of naked mole‐rats (Heterocephalus glaber) by DNA fingerprinting. J. Zool. Lond. 221:87–97. [Google Scholar]
- Folstad,I., and Karter A. J.. 1992. Parasites, bright males, and the immunocompetence handicap. Am. Nat. 139:603–622. [Google Scholar]
- Gershwin,M. E., Beach R. S., and Hurley L. S.. 1985. Nutrition and immunity. Academic Press, Orlando, FL. [Google Scholar]
- Glémin,S. 2003. How are deleterious mutations purged? Drift versus nonrandom mating. Evolution. 57:2678–2687. [DOI] [PubMed] [Google Scholar]
- Haag,C. R., Sakwinska O., and Ebert D.. 2003. Test of synergistic interaction between infection and inbreeding in Daphnia magna. Evolution 57:777–783. [DOI] [PubMed] [Google Scholar]
- Hamilton,W. D., Axelrod R., and Tanese R.. 1990. Sexual reproduction as an adaptation to resist parasites. Proc. Natl. Acad. Sci. 87:3566–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley,D. M., Sydenstricker K. V., Kollias G. V., and Dhondt A. A.. 2005. Genetic diversity predicts pathogen resistance and cell‐mediated immunocompetence in house finches. Biol. Lett. 1:326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick,P. W. 1994. Evolutionary genetics of the major histocompatibility complex. Am. Nat. 143:945–964. [Google Scholar]
- Hedrick,P. W., and Kalinowski S. T.. 2000. Inbreeding depression in conservation biology. Annu. Rev. Ecol. Syst. 31:139–162. [Google Scholar]
- Hedrick,P. W., Kim T. J., and Parker K. M.. 2001. Parasite resistance and genetic variation in the endangered Gila topminnow. Anim. Conserv. 4:103–109. [Google Scholar]
- Hosmer,D. W., and Lemeshow S.. 2000. Applied logistic regression. 2nd. ed. John Wiley & Sons, New York. [Google Scholar]
- Jarvis,J. U. M. 1991. Methods for capturing, transporting and maintaining naked mole‐rats in captivity. Pp. 467–483 in Sherman P. W., Jarvis J. U. M., and Alexander R. D., eds. The biology of the naked mole‐rat. Princeton Univ. Press, Princeton, NJ. [Google Scholar]
- Jarvis,J. U. M., O'Riain M. J., Bennett N. C., and Sherman W. C.. 1994. Eusociality: a family affair. Trends Ecol. Evol. 9:47–51. [DOI] [PubMed] [Google Scholar]
- Kalinowski,S. T., and Hedrick P. W.. 1998. An improved method for estimating inbreeding depression in pedigrees. Zoo. Biol. 17:481–497. [Google Scholar]
- Kalinowski,S. T., Hedrick P. W., and Miller P. S.. 1999. No inbreeding depression observed in Mexican red wolf captive breeding programs. Conserv. Biol. 13:1371–1377. [Google Scholar]
- Keane,B., Creel S. R., and Waser P. M.. 1996. No evidence of inbreeding avoidance or inbreeding depression in a social carnivore. Behav. Ecol. 7:480–489. [Google Scholar]
- Keller,L. F., and Waller D. M.. 2002. Inbreeding effects in wild populations. Trends Ecol. Evol. 17:230–241. [Google Scholar]
- Klein,S. L. 2000. Hormones and mating system affect sex and species differences in immune function among vertebrates. Behav. Processes 51:149–166. [DOI] [PubMed] [Google Scholar]
- Lacey,E. A., and Sherman P. W.. 1991. Social organization of naked mole‐rat colonies: evidence for divisions of labour. Pp. 275–336 in Sherman P. W., Jarvis J. U. M., and Alexander R. D., eds. The biology of the naked mole‐rat. Princeton Univ. Press, Princeton, NJ. [Google Scholar]
- Lacy R. C., and Ballou J. D.. 1998. Effectiveness of selection in reducing the genetic load in populations of Peromyscus polionotus during generations of inbreeding. Evolution 52:900–909. [DOI] [PubMed] [Google Scholar]
- Lande,R., and Schemske D. W.. 1985. The evolution of self‐fertilization and inbreeding depression in plants. I. genetic models. Evolution 39:24–40. [DOI] [PubMed] [Google Scholar]
- Linton,P. J., and Dorshkind K.. 2004. Age‐related changes in lymphocyte development and function. Nat. Immunol. 5:133–139. [DOI] [PubMed] [Google Scholar]
- Luong,L. T., Heath B. D., and Polak M.. 2007. Host inbreeding increases susceptibility to ectoparasitism. J. Evol. Biol. 20:79–86. [DOI] [PubMed] [Google Scholar]
- McCall,C., Waller D. M., and Mitchell‐Olds T.. 1994. Effects of serial inbreeding on fitness components in Impatiens capensis. Evolution 48:818–827. [DOI] [PubMed] [Google Scholar]
- McClelland,E. E., Penn D. J., and Potts W. K.. 2003. Major histocompatibility complex heterozygote superiority during coinfection. Infect. Immun. 71:2079–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher,S. 1999. Genetic diversity and Capillaria hepatica (Nematoda) prevalence in Michigan deer mouse populations. Evolution 53:1318–1324. [DOI] [PubMed] [Google Scholar]
- Messaoudi,I., Guevara Patiño J. A., Dyall R., LeMaoult J., and Nikolich‐Žugich J.. 2002. Direct link between MHC polymorphism, T cell avidity, and diversity in immune defense. Science 298:1797–1800. [DOI] [PubMed] [Google Scholar]
- Milot,E., Weimerskirsch H., Duchesne P., and Bernatchez L.. 2007. Surviving with low genetic diversity: the case of albatrosses. Proc. R. Soc. Lond. B 274:779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson,R. J., and Demas G. E.. 1996. Seasonal changes in immune function. Q. Rev. Biol. 71:511–548. [DOI] [PubMed] [Google Scholar]
- Nordling,D., Andersson M., Zohari S., and Gustafsson L.. 1998. Reproductive effort reduces specific immune response and parasite resistance. Proc. R. Soc. Lond. B 265:1291–1298. [Google Scholar]
- O'Brien,S. J., and Evermann J. F.. 1988. Interactive influences of infectious diseases and genetic diversity in natural populations. Trends Ecol. Evol. 3:254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien,S. J., Roelke M. E., Marker L., Newman A., Winkler C. A., Meltzer D., Colly L., Evermann J. F., Bush M., and Wildt D. E.. 1985. Genetic basis for species vulnerability in the cheetah. Science 227:1428–1434. [DOI] [PubMed] [Google Scholar]
- O'Riain,M. J., Jarvis J. U. M., and Faulkes C. G.. 1996. A dispersive morph in the naked mole‐rat. Nature 380:619–621. [DOI] [PubMed] [Google Scholar]
- Peer,K., and Taborsky M.. 2005. Outbreeding depression, but no inbreeding depression in haplodiploid ambrosia beetles with regular sibling mating. Evolution 59:317–323. [PubMed] [Google Scholar]
- Pray,L. A., and Goodnight C. J.. 1995. Genetic variation in inbreeding depression in the red flour beetle Tribolium castaneum. Evolution 49:176–188. [DOI] [PubMed] [Google Scholar]
- Råberg,L., Grahn M., Hasselquist D., and Svensson E.. 1998. On the adaptive significance of stress induced immunosuppression. Proc. R. Soc. Lond. B 265:1637–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralls,K., Ballou J. D., and Templeton A.. 1988. Estimates of lethal equivalents and the cost of inbreeding depression in mammals. Conserv. Biol. 2:185–193. [Google Scholar]
- Reeve,H. K., Westneat D. F., Noon W. A., Sherman P. W., and Aquadro C. F.. 1990. DNA “fingerprinting” reveals high levels of inbreeding in colonies of the eusocial naked mole‐rat. Proc. Natl. Acad. Sci. USA 87:2496–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid,J. M., Arcese P., and Keller L. F.. 2003. Inbreeding depresses immune response in song sparrows (Melospiza melodia): direct and inter‐generational effects. Proc. R. Soc. Lond. B 270:2151–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid,J. M., Arcese P., Keller L. F., Elliott K. H., Sampson L., and Hasselquist D.. 2007. Inbreeding effects on immune response in free‐living song sparrows (Melospiza melodia). Proc. R. Soc. Lond. B 274:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapolsky,R. M. 1992. Neuroendocrinology of the stress response. Pp. 287–324 in Becker J. B., ed. Behavioral endocrinology. MIT Press, Boston, MA. [Google Scholar]
- Schmid‐Hempel,P., and Crozier R. H.. 1999. Polyandry versus polygyny versus parasites. Philos. Trans. R. Soc. B 354:507–515. [Google Scholar]
- Seeley,T. D., and Tarpy D. R.. 2007. Queen promiscuity lowers disease within honeybee colonies. Proc. R. Soc. Lond. B 274:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman,D., Brook B. W., Briscoe D. A., and Frankham R.. 2004. Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv. Genet. 5:439–448. [Google Scholar]
- Therneau,T. M., and Grambsch P. M.. 2002. Modeling survival data: extending the Cox model. Springer, New York. [Google Scholar]
- Visscher,P. M., Smith D., Hall S. J. G., and Williams J. L.. 2001. A viable herd of genetically uniform cattle. Nature 409:303. [DOI] [PubMed] [Google Scholar]
- Wang,J. L. 2000. Effects of population structures and selection strategies on the purging of inbreeding depression due to deleterious mutations. Genet. Res. 76:75–86. [DOI] [PubMed] [Google Scholar]
- Wang,J. L., Hill W. G., Charlesworth D., and Charlesworth B.. 1999. Dynamics of inbreeding depression due to deleterious mutations in small populations: mutation parameters and inbreeding rate. Genet. Res. 74:165–178. [DOI] [PubMed] [Google Scholar]
- Wegner,K. M., Kalbe M., Schaschl H., and Reusch T. B. H.. 2004. Parasites and individual major histocompatibility complex diversity—an optimal choice? Microb. Infect. 6:1110–1116. [DOI] [PubMed] [Google Scholar]
- Weiss,S. R. and Navas‐Martin S.. 2005. Coronavirus pathogenicity and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman,N. K., Matson K. D., Bollmer J. L., and Parker P. G.. 2006. Disease ecology in the Galapagos Hawk (Buteo galapagoensis): host genetic diversity, parasite load and natural antibodies. Proc. R. Soc. Lond. B 273:797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock,M. C. 2002. Selection, load and inbreeding depression in a large metapopulation. Genetics 160;1191–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright,S. 1922. Coefficients of inbreeding and relationship. Am. Nat. 56:330–338. [Google Scholar]


