Abstract
Hepatocellular carcinoma (HCC) remains one of the most lethal malignant tumors worldwide; however, the etiology of HCC still remains poorly understood. In the present study, cancer-omics databases, including The Cancer Genome Atlas, GTEx and Gene Expression Omnibus, were systematically analyzed in order to investigate the role of the long non-coding RNA (lncRNA) zinc finger protein, FOG family member 2-antisense 1 (ZFPM2-AS1) and the zinc finger protein, FOG family member 2 (ZFPM2) gene in the occurrence and progression of HCC. It was identified that the expression levels of lncRNA ZFPM2-AS1 were significantly increased in HCC tissues, whereas expression levels of the ZFPM2 gene were significantly decreased in HCC tissues compared with normal liver tissues. Higher expression levels of ZFPM2-AS1 were significantly associated with a less favorable prognosis of HCC, whereas higher expression levels of the ZFPM2 gene were associated with a more favorable prognosis of HCC. Genetic alterations in the ZFPM2 gene may contribute to a worse prognosis of HCC. Validation of the GSE14520 dataset also demon stared that ZFPM2 gene expression levels were significantly decreased in HCC tissues (P<0.001). The receiver operating characteristic (ROC) analysis of the ZFPM2 gene indicated high accuracy of this gene in distinguishing between HCC tissues and non-tumor tissues. The areas under the ROC curves were >0.8. Using integrated strategies, the present study demonstrated that lncRNA ZFPM2-AS1 and the ZFPM2 gene may contribute to the occurrence and prognosis of HCC. These findings may provide a novel understanding of the molecular mechanisms underlying the occurrence and prognosis of HCC.
Keywords: hepatocellular carcinoma, ZFPM2-AS1, ZFPM2, bioinformatics analysis, prognosis
Introduction
Hepatocellular carcinoma (HCC) is one of the most lethal malignant tumors worldwide, with an incidence rate of 40.0% in men and 15.3% in women per 100,000 population in China (1,2). According to the Global Burden of Disease Study 2017, ~820,000 individuals succumbed to HCC worldwide (3). Among them, the number of HCC-associated mortalities in China (~422,000) accounted for 51.5% of global HCC-associated mortalities (4).
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs >200 nucleotides in length (5). Previous studies have shown that lncRNAs serve a regulatory role in tumor development and prognosis and can be potential tumor biomarkers and therapeutic targets (6,7). Meanwhile, lncRNAs have been found to serve a role in chromatin modification, transcription and post-transcriptional processing in HCC (8–11). Notably, overexpression of lncRNA HOTAIR, which was previously reported in breast cancer, was first identified to predict tumor recurrence in patients with HCC following liver transplantation (12,13). Subsequent studies have reported that lncRNAs: MALAT1, HULC, GAS5, NEAT1, PCNA-AS1, PVT1, TUG1 and HOTTIP are associated with the development of HCC (11,14–17). lncRNA zinc finger protein, FOG family member 2-antisense 1 (ZFPM2-AS1), located on the 8q23 chromosome and next to the zinc finger protein, FOG family member 2 (ZFPM2) gene, serves a role in carcinogenesis and tumor progression in HCC and gastric cancer (18,19). The ZFPM2 gene modulates the activity of GATA family proteins and serves a role in heart morphogenesis and development of coronary vessels (20,21). Previous studies also revealed that ZFPM2 could cooperate with GATA factors and contribute to the occurrence of ovarian tumors, neuroblastoma, testicular carcinoma, germ cell tumors, Wilms' tumor, gliomas, glioblastoma, lung cancer, breast cancer and osteosarcoma (22–31). The chromosome 8q23 region is a high susceptibility locus for several types of cancer and genome-wide association studies (GWAS) have identified a number of cancer-associated single nucleotide polymorphisms that are adjacent to the ZFPM2-AS1 and ZFPM2 gene in this region (32–37).
Considering the promising role of the lncRNA ZFPM2-AS1 and the ZFPM2 gene in carcinogenesis and prognosis of several types of cancer, it was hypothesized that lncRNA ZFPM2-AS1 and the ZFPM2 gene also contribute to the development and prognosis of HCC. In present study, a series of bioinformatic and clinical analyses were performed to investigate the potential functions of lncRNA ZFPM2-AS1 and the ZFPM2 gene in the process of carcinogenesis and progression of HCC.
Materials and methods
Expression of ZFPM2-AS1 and ZFPM2 gene in the cancer genome atlas (TCGA) and GTEx tissues
The comparison of the expression levels of ZFPM2-AS1 and ZFPM2 genes in HCC and non-tumor tissues was performed using GEPIA version 2.0 (37), during which TCGA (https://portal.gdc.cancer.gov) HCC samples were compared with GETx (https://www.gtexportal.org/home) samples, which were used as controls. The associations of expression levels of ZFPM2-AS1 and the ZFPM2 gene with the prognosis of HCC and other digestive system tumors were evaluated using the Kaplan Meier plotter (http://kmplot.com/analysis), which presents overall survival, disease free survival, relapse free and progression free survival (38), and GEPIA.
Validation of expression of ZFPM2-AS1 and ZFPM2 gene in clinical tissues
The present study was approved by the Ethics Committee of the Army Military Medical University (Chongqing, China) and written informed consent was provided by all participants prior to the study start. A total of 53 HCC and paired adjacent normal tissues (>2 cm from tumor tissues) 45 men and 8 women; age range, 30–74 years; median age, 53 years) were collected from the Department of Hepatobiliary Surgery (Chongqing, China) between November 2017 and May 2019, following surgical resection. All diagnoses were blindly confirmed by at least two pathologists at The First Affiliated Hospital of Army Military Medical University, and patients who received radiofrequency ablation, chemoradiotherapy or other treatments prior to surgery were excluded from the present study. Samples were subsequently stored at −80°C, prior to subsequent experimentation.
Reverse transcription-quantative (RT-q)PCR
Total RNA was extracted from HCC tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized using the PrimeScript RT reagent kit with gDNA Eraser (Takara Bio, Inc.), and qPCR was performed using TB Green Premix Ex Taq II (Takara Bio, Inc.). The following primer sequences were used for qPCR: ZFPM2-AS1 forward, 5′-GCTTCTATGCCTTCCTTCCCTT-3′, and reverse, 5′-CTCCATACTCTCCCTGGGTT-3′; ZFPM2 forward, 5′-GCTACCCTCCCGTCATTT-3′, and reverse, 5′-TTAGCCATCTGCTGCCAT-3′; and β-actin forward, 5′-CCACGAAACTACCTTCAACTCC-3; and reverse, 5′-GTGATCTCCTTCTGCATCCTGT-3′. The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 30 sec; 40 cycles of denaturation at 95°C for 5 sec, annealing and elongation at 60°C for 30 sec; and a final extension at 72°C for 30 sec. Relative ZFPM2-AS1 and ZFPM2 mRNA levels were measured using the 2−ΔΔCq method (39) and normalized to the internal reference gene β-actin.
Interaction network and functional enrichment analyses
To investigate the biological functions and pathways of ZFPM2-AS1 and the ZFPM2 gene, gene-gene and protein-protein interaction (PPI) network analysis of the ZFPM2 gene was conducted using the GeneMANIA (http://genemania.org) and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database version 11.0 (40). Genes associated with ZFPM2 and ZFPM2-AS1 were initially identified using the COXPRESdb database (version 7.3; http://coxpresdb.jp). Subsequently, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) analyses of ZFPM2-AS1 and ZFPM2-associated genes were performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.8 (david.ncifcrf.gov/home.jsp).
Determination of genetic mutation status of the ZFPM2-AS1 lncRNA and ZFPM2 gene
To investigate the underlying mechanisms relevant to mutation status of ZFPM2-AS1 and ZFPM2 gene, the cBioPortal database (cbioportal.org/) was utilized. Kaplan-Meier survival estimates for overall survival of HCC patients, with or without mutations of the ZFPM2 gene was also analyzed, using the log-rank test.
Validation of the GEO dataset
The validation of the expression levels of the ZFPM2 gene in HCC tissues and adjacent normal tissues was further conducted with the GEO dataset GSE14520 (41). The receiver operating curve (ROC) with the area under the curve (AUC) value for assessing the predictive accuracy and discriminative ability of ROC was drawn to identify the diagnostic significance of expression level of the ZFPM2 gene.
Statistical analysis
SPSS version 22.0 (IMB Corp.) and GraphPad Prism version 7.0 (GraphPad Software, Inc.) were used for statistical analyses. P<0.05 was considered to indicate a statistically significant difference. All results are presented as mean ± standard deviation (unless otherwise shown). One-way ANOVA tests were used to evaluate the differences in ZFPM2-AS1 and ZFPM2 expression in clinical stages of HCC, while Wilcoxon's test was used for paired continuous variables. The χ2 test was used to evaluate differences in categorical variables. All expression data were log transformed for differential analysis.
Results
Associations between expression levels of lncRNA ZFPM2-AS1 and the ZFPM2 gene with clinical significance of HCC
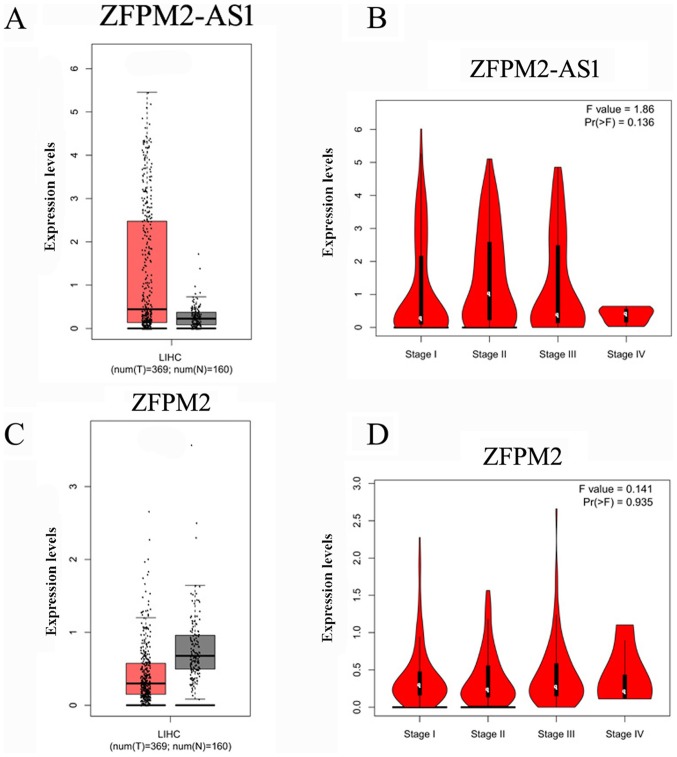
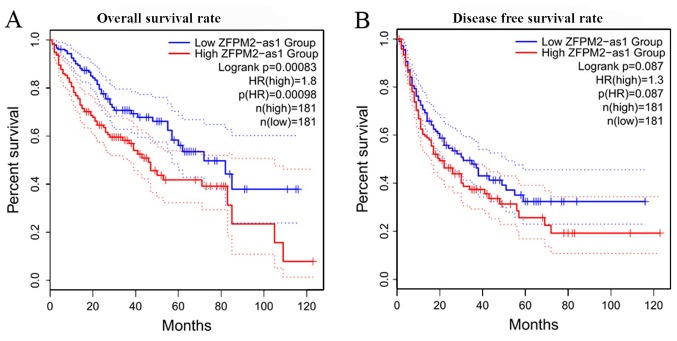
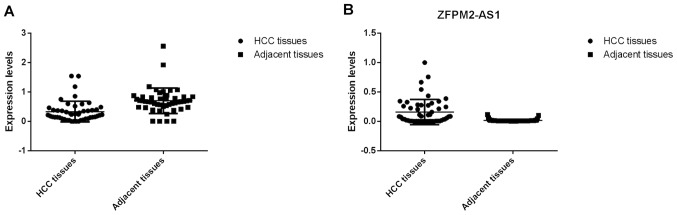
First the associations between the expression levels of ZFPM2-AS1 and the ZFPM2 gene and the clinical characteristics of HCC in TCGA and GTEx samples were analyzed. Table I presents the clinical characteristics of the patients from TCGA and GTEx databases (only sex available for GTEx), including sex, age at diagnosis, Child-Pugh score (42), creatinine value, HCC risk factor, family cancer history (data for 326 samples available), neoplasm histological grade (43) (data for 372 samples available) and metastasis status. The expression levels of lncRNA ZFPM2-AS1 were higher in HCC tissues compared with normal liver tissues (Fig. 1A), whereas the expression levels of the ZFPM2 gene were significantly lower in HCC tissues compared with normal liver tissues (Fig. 1C). No significant difference between the clinical stages of HCC and ZFPM2-AS1 (Fig. 1B; P=0.136) and ZFPM2 (Fig. 1D; P=0.935) expression levels were observed. For the survival of patients with HCC it was observed that higher expression levels of ZFPM2-AS1 were significantly associated with a less favorable prognosis (Fig. 2A), whereas higher expression levels of the ZFPM2 gene were significantly associated with better prognosis of HCC (Fig. 3). These bioinformatic results were also verified using clinical samples. The expression levels of lncRNA ZFPM2-AS1 were significantly higher in HCC tissues compared with adjacent normal tissues (Fig. 4B; P<0.001), whereas the expression levels of the ZFPM2 gene were significantly lower in HCC tissues compared with adjacent normal tissues (Fig. 4A; P<0.001).
Table I.
Characteristics of patients with HCC from TCGA and GTEx datasets.
| Variables | HCC cases (TCGA), n | Controls (GTEx), n | P-value |
|---|---|---|---|
| Sex | 0.799 | ||
| Male | 255 | 123 | |
| Female | 122 | 56 | |
| Age at diagnosis, years (mean ± standard deviation) | 59.5±13.5 | ||
| Child-Pugh score | |||
| A | 223 | ||
| B | 21 | ||
| C | 1 | ||
| Unknown | 132 | ||
| Creatinine value, mg/dl | 2.76±11.7 | ||
| HCC risk factor | |||
| Alcoholism | 76 | ||
| Hepatitis B infection | 98 | ||
| Hepatitis C infection | 52 | ||
| Family cancer history | |||
| Yes | 114 | ||
| No | 212 | ||
| Neoplasm histological grade | |||
| G1 | 55 | ||
| G2 | 180 | ||
| G3 | 124 | ||
| G4 | 13 | ||
| Metastasis | |||
| No | 272 | ||
| Yes | 105 |
The publicly available GTEX data only provided data on sex. HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas.
Figure 1.
Comparisons of expression levels of lncRNA ZFPM2-AS1 and the ZFPM2 gene. Red represents HCC tissues, grey represents normal liver tissues and black dots represent individual cases. (A) Expression level of lncRNA ZFPM2-AS1 in HCC (369) and normal liver (160) tissues. (B) Expression level of lncRNA ZFPM2-AS1 in tissues of patients with HCC of different clinical stages. (C) Expression level of the ZFPM2 gene in HCC (n=369) and normal liver (n=160) tissues. (D) Expression levels of the ZFPM2 gene in patients with HCC of different clinical stages. lncRNA, long non-coding RNA; AS1, antisense RNA 1; ZFPM2, zinc finger protein, FOG family member 2; HCC, hepatocellular carcinoma; LIHC, liver hepatocellular carcinoma.
Figure 2.
Kaplan-Meier survival curves of patients with HCC based on lncRNA ZFPM2-AS1 expression levels. (A) Overall survival rate of patients with HCC based on lncRNA ZFPM2-AS1 expression levels. (B) Disease-free survival rate of patients with HCC based on lncRNA ZFPM2-AS1 expression levels. lncRNA, long non-coding RNA; AS1, antisense RNA 1; ZFPM2, zinc finger protein, FOG family member 2; HCC, hepatocellular carcinoma; HR, hazard ratio.
Figure 3.
Kaplan-Meier survival curves of patients with HCC based on ZFPM2 gene expression levels. (A) Overall survival rate of patients with HCC based on ZFPM2 gene expression levels. (B) Relapse free survival rate of patients with HCC based on ZFPM2 gene expression levels. (C) Disease specific survival rate of patients with HCC based on ZFPM2 gene expression levels. (D) Progress free survival rate of patients with HCC based on ZFPM2 gene expression levels. lncRNA, long non-coding RNA; AS1, antisense RNA 1; ZFPM2, zinc finger protein, FOG family member 2; HCC, hepatocellular carcinoma; HR, hazard ratio.
Figure 4.
Expression levels of lncRNA ZFPM2-AS1 and the ZFPM2 gene. (A) Expression levels of ZFPM2 in HCC and adjacent tissues. (B) Expression levels of ZFPM2-AS1 in HCC and adjacent tissues. lncRNA, long non-coding RNA; AS1, antisense RNA 1; ZFPM2, zinc finger protein, FOG family member 2; HCC, hepatocellular carcinoma.
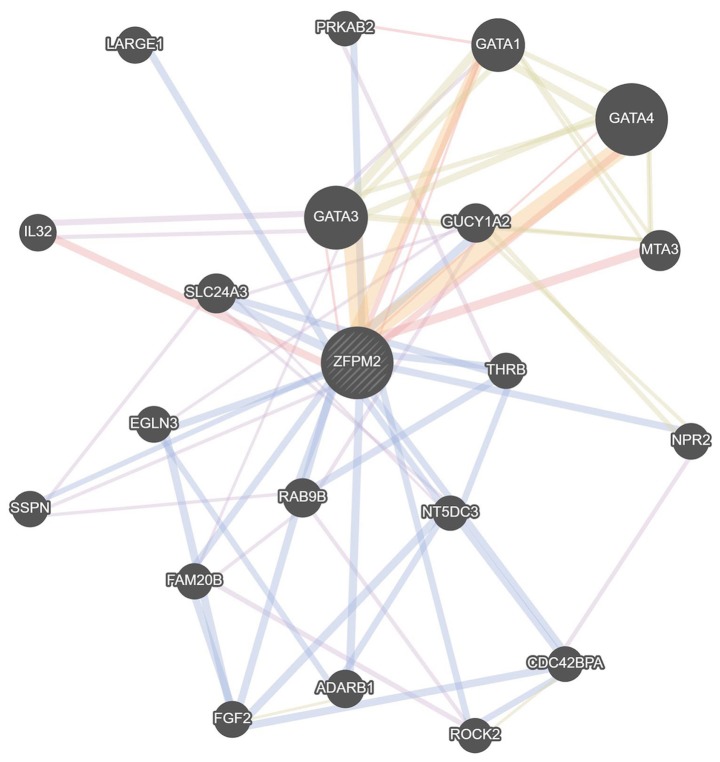
Gene-gene and PPI network of the lncRNA ZFPM2-AS1 and ZFPM2 gene
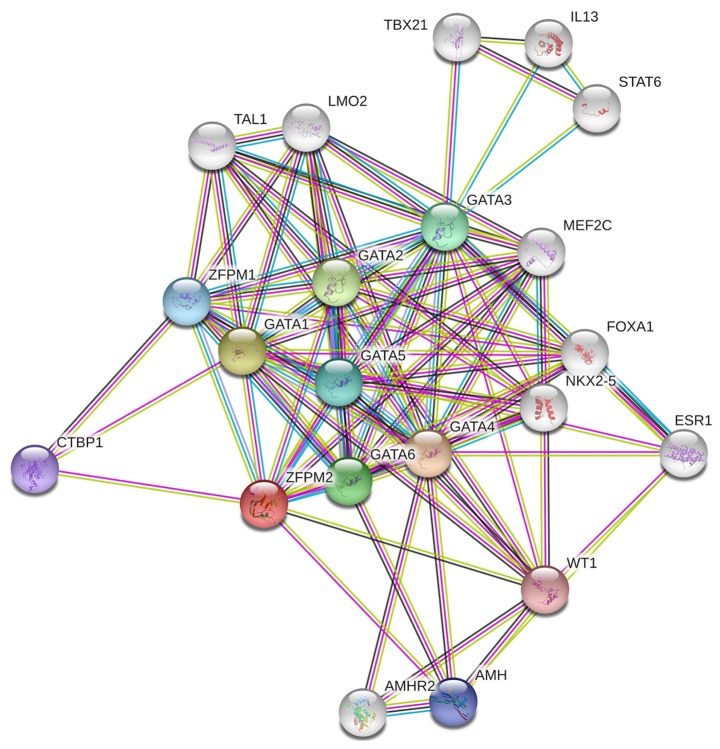
According to the results obtained from COXPRESdb, lncRNA ZFPM2-AS1 was associated with the ZFPM2 gene. Thus, gene-gene and PPI network analysis of the ZFPM2 gene were conducted using the GeneMANIA and STRING tools and it was demonstrated that ZFPM2 primarily associated with GATA factors, including GATA1, GATA3 and GATA4 (Figs. 5 and 6).
Figure 5.
Gene-gene association network of the ZFPM2 gene drawn using GeneMANIA. The circles indicate the function of the gene. ZFPM2, zinc finger protein, FOG family member 2.
Figure 6.
Protein-protein interaction network of the ZFPM2 gene. A network diagram of interactions between proteins associated with the ZFPM2 protein drawn using STRING. ZFPM2, zinc finger protein, FOG family member 2.
Clinical significance of genetic alterations of the lncRNA ZFPM2-AS1 and ZFPM2 gene
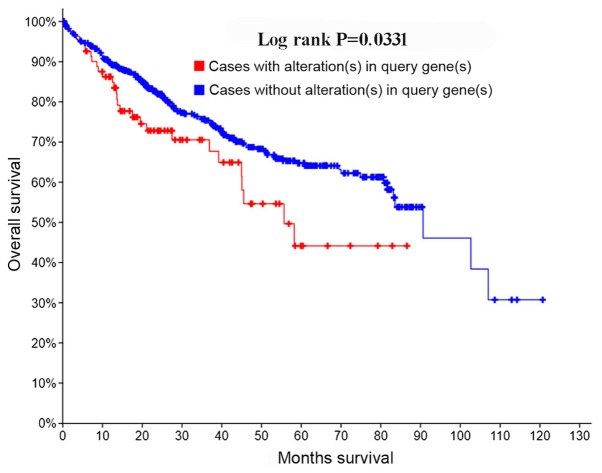
Using the cBioPortal database, 9% (93/1,052) of samples were identified as harboring a mutated ZFPM2 gene. From Kaplan-Meier survival analysis, the overall survival rate demonstrated statistical differences, which means patients with HCC with ZFPM2 mutations had a less favorable prognosis compared with those without ZFPM2 mutations (P=0.0331; Fig. 7).
Figure 7.
Survival curves for genetic alternations of the zinc finger protein, FOG family member 2 gene.
KEGG pathway and GO term analyses
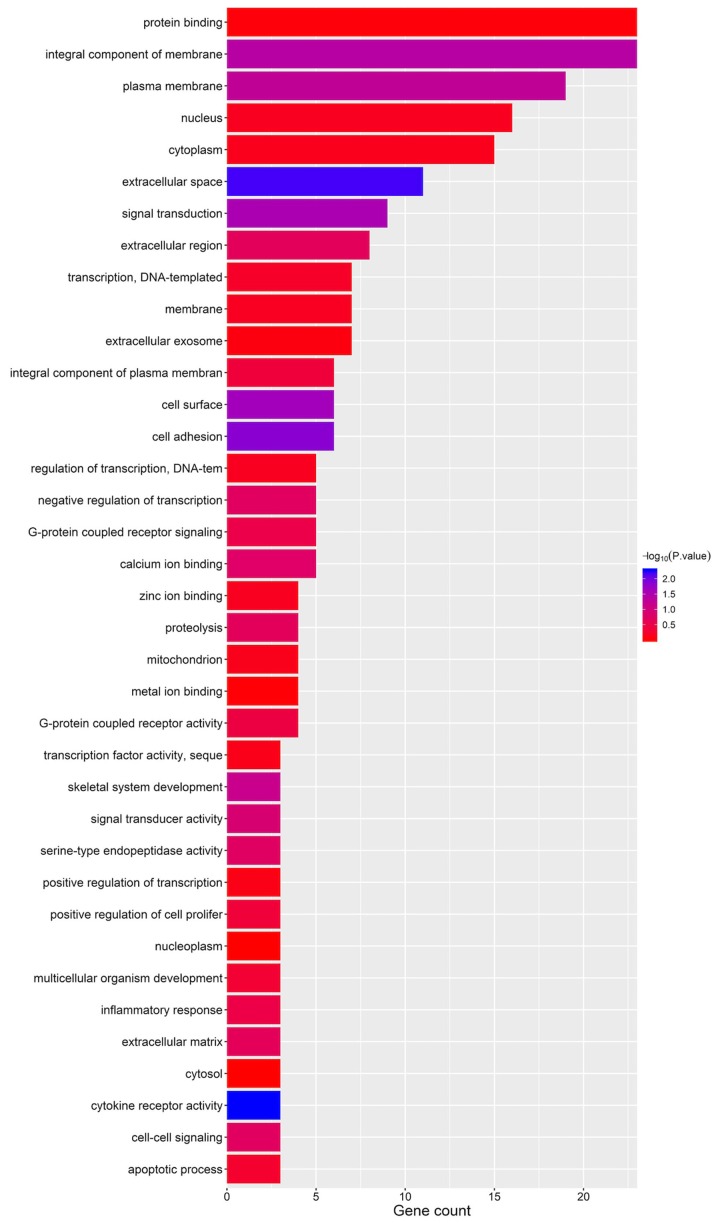
The top 200 ZFPM2 and ZFPM2-AS1 associated genes are presented in Table SI, which were identified using the COXPRESdb database. KEGG pathway and GO term analysis of the ZFPM2 associated genes were performed using DAVID. The GO term results demonstrated that these genes may be involved in the ‘integral component of plasma membrane’, ‘protein binding’ and ‘plasma membrane’ (Fig. 8).
Figure 8.
Gene Ontology term enrichment plots of the zinc finger protein, FOG family member 2 associated genes.
Validation of the ZFPM2 expression profiling in the GSE14520 dataset
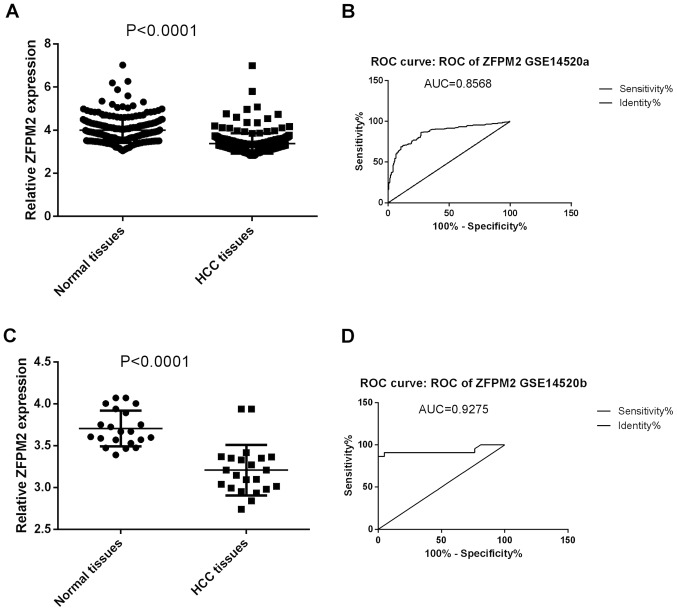
As shown in Fig. 9, the expression levels of the ZFPM2 gene in HCC and non-tumor tissues were consistent in the GSE14520 dataset, in stages I and II. Consistent with TCGA data, ZFPM2 gene expression were significantly decreased in HCC tissues compared with the non-tumor tissues in both stage I and II (P<0.001; Fig. 9A and C). The ROC analysis of the ZFPM2 gene demonstrated a high accuracy of ZFPM2 in distinguishing between HCC tissues and non-tumor tissues (AUCs, >0.8; Fig. 9B and D)
Figure 9.
Expression and ROC analysis of ZFPM2 in the GSE14520 dataset of hepatocellular carcinoma and non-tumor tissues. (A) Comparison of ZFPM2 gene expression in tumor and non-tumor tissues in stage I. (B) ROC curve of ZFPM2 between HCC and non-tumor tissues stage I. (C) Comparison of ZFPM2 gene expression in tumor and non-tumor tissues in stage II. (D) ROC curve of ZFPM2 between HCC and non-tumor tissues stage II. ZFPM2, zinc finger protein, FOG family member 2; ROC, receiver operating characteristic; HCC, hepatocellular carcinoma.
Discussion
At present, the etiology of HCC remains poorly understood. In the present study, datasets from the cancer-omics databases TCGA, GTEX and GEO were analyzed in order to confirm the role of lncRNA ZFPM2-AS1 and the ZFPM2 gene in HCC, which are located at the cancer susceptibility locus 8q23 implicated in the carcinogenesis and prognosis of HCC (44). In the present study, it was observed that the expression levels of lncRNA ZFPM2-AS1 and the ZFPM2 gene were significantly different between HCC tissues and normal liver tissues and that these expression levels were also associated with prognosis of HCC. Patients with HCC with ZFPM2 gene alterations had a less favorable prognosis compared with those without ZFPM2 gene alterations. Functional enrichment analysis demonstrated that the ZFPM2 associated genes were primarily involved in the formation of integral component of membrane, protein binding and plasma membrane. To the best of our knowledge, the present study is the first report that aimed to investigate the association between lncRNA ZFPM2-AS1, the ZFPM2 gene and the occurrence and progression of HCC.
Both lncRNA ZFPM2-AS1 and the ZFPM2 gene are located at 8q23 region, an aggregate of cancer susceptible loci (44–49). Tomlinson et al (48) first identified rs16892766 on chromosome 8q23.3 as a colorectal cancer susceptibility locus. A previous study identified 41 variants that are associated with venous thromboembolism, and mapped to the ZFPM2-AS1 and ZFPM2 gene region using GWAS catalog (50). In the present study, expression levels of lncRNA ZFPM2-AS1 and the ZFPM2 gene were associated with both the occurrence and prognosis of HCC and mutations of the ZFPM2 gene were associated with a less favorable prognosis of HCC. These results further confirmed the role of lncRNA ZFPM2-AS1 and the ZFPM2 gene in HCC carcinogenesis.
In the present study, gene-gene and PPI analyses revealed that ZFPM2-AS1 and ZFPM2 were primarily co-expressed and interacted with the GATA factors, including GATA1, GATA3 and GATA4. The GATA family, which controls the development of diverse tissues by activating or repressing transcription, widely participants in carcinogenesis, differentiation of several types of cancer (51,52). Furthermore, studies have shown that aberrant GATA-3 expression contributes to the occurrence of breast, prostate and pancreatic cancer (53–58). GATA1, GATA4 and GATA6 are also associated with different types of cancer, including colorectal and breast cancer (59,60). The results of the present study demonstrated that ZFPM2 was significantly associated with GATA factors, suggesting its potential role in the development of different types of cancer.
Conclusively, the present study demonstrated that lncRNA ZFPM2-AS1 and the ZFPM2 gene may contribute to the occurrence and progression of HCC. These findings may provide a novel perspective on the underlying molecular mechanisms of HCC and suggest valuable biomarkers and therapeutic targets for patients with HCC. However, further validations with experimental evidence and clinical research are needed to confirm the functions of lncRNA ZFPM2-AS1 and ZFPM2 gene in HCC carcinogenesis.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by the Science Foundation for Outstanding Young People of the Army Military Medical University (Chongqing, China; grant. no. 20170113).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XM and YL designed the study. YL, XW, LM, SL, ZM and XF performed the statistical analyses. YL, XW and LM drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Army Military Medical University (Chongqing, China) and written informed consent was provided by all participants prior to the study start (approval no. 20170307).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Forner A, Gilabert M, Bruix J, Raoul JL. Treatment of intermediate-stage hepatocellular carcinoma. Nat Rev Clin Oncol. 2014;11:525–535. doi: 10.1038/nrclinonc.2014.122. [DOI] [PubMed] [Google Scholar]
- 2.Zhu RX, Seto WK, Lai CL, Yuen MF. Epidemiology of hepatocellular carcinoma in the Asia-pacific region. Gut Liver. 2016;10:332–339. doi: 10.5009/gnl15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.GBD 2017 Causes of Death Collaborators, corp-author. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980-2017: A systematic analysis for the global burden of disease study 2017. Lancet. 2018;392:1736–1788. doi: 10.1016/S0140-6736(18)32203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 5.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 6.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mai H, Zhou B, Liu L, Yang F, Conran C, Ji Y, Hou J, Jiang D. Molecular pattern of lncRNAs in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:198. doi: 10.1186/s13046-019-1213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao J, Wu L, Meng X, Yang H, Ni S, Wang Q, Zhou J, Zhang Q, Su K, Shao L, et al. Profiling, clinicopathological correlation and functional validation of specific long non-coding RNAs for hepatocellular carcinoma. Mol Cancer. 2017;16:164. doi: 10.1186/s12943-017-0733-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Chen L, Gu J, Zhang H, Yuan J, Lian Q, Lv G, Wang S, Wu Y, Yang YT, et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun. 2017;8:14421. doi: 10.1038/ncomms14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu FJ, Zheng JJ, Dong PH, Fan XM. Long non-coding RNAs and hepatocellular carcinoma. Mol Clin Oncol. 2015;3:13–17. doi: 10.3892/mco.2014.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 13.Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16:129. doi: 10.1186/s12943-017-0696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J, Zhang Y, Chen J, Shen H, Hu Z. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. 2012;7:e35145. doi: 10.1371/journal.pone.0035145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 16.Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong M, Dang Y, Feng Z, Chen G. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol. 2015;8:5395–5402. [PMC free article] [PubMed] [Google Scholar]
- 17.Tu ZQ, Li RJ, Mei JZ, Li XH. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:4303–4309. [PMC free article] [PubMed] [Google Scholar]
- 18.Yan J, Zhou C, Guo K, Li Q, Wang Z. A novel seven-lncRNA signature for prognosis prediction in hepatocellular carcinoma. J Cell Biochem. 2019;120:213–223. doi: 10.1002/jcb.27321. [DOI] [PubMed] [Google Scholar]
- 19.Kong F, Deng X, Kong X, Du Y, Li L, Zhu H, Wang Y, Xie D, Guha S, Li Z, et al. ZFPM2-AS1, a novel lncRNA, attenuates the p53 pathway and promotes gastric carcinogenesis by stabilizing MIF. Oncogene. 2018;37:5982–5996. doi: 10.1038/s41388-018-0387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svensson EC, Tufts RL, Polk CE, Leiden JM. Molecular cloning of FOG-2: A modulator of transcription factor GATA-4 in cardiomyocytes. Proc Natl Acad Sci USA. 1999;96:956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, Fujiwara Y, Orkin SH. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/S0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- 22.Guo L, Wang J, Yang P, Lu Q, Zhang T, Yang Y. MicroRNA- 200 promotes lung cancer cell growth through FOG2-independent AKT activation. IUBMB Life. 2015;67:720–725. doi: 10.1002/iub.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aumsuwan P, Khan SI, Khan IA, Ali Z, Avula B, Walker LA, Shariat-Madar Z, Helferich WG, Katzenellenbogen BS, Dasmahapatra AK. The anticancer potential of steroidal saponin, dioscin, isolated from wild yam (Dioscorea villosa) root extract in invasive human breast cancer cell line MDA-MB-231 in vitro. Arch Biochem Biophys. 2016;591:98–110. doi: 10.1016/j.abb.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Tsang SY, Mei L, Wan W, Li J, Li Y, Zhao C, Ding X, Pun FW, Hu X, Wang J, et al. Glioma association and balancing selection of ZFPM2. PLoS One. 2015;10:e0133003. doi: 10.1371/journal.pone.0133003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoene V, Fischer M, Ivanova A, Wallach T, Berthold F, Dame C. GATA factors in human neuroblastoma: Distinctive expression patterns in clinical subtypes. Br J Cancer. 2009;101:1481–1489. doi: 10.1038/sj.bjc.6605276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laitinen MP, Anttonen M, Ketola I, Wilson DB, Ritvos O, Butzow R, Heikinheimo M. Transcription factors GATA-4 and GATA-6 and a GATA family cofactor, FOG-2, are expressed in human ovary and sex cord-derived ovarian tumors. J Clin Endocrinol Metab. 2000;85:3476–3483. doi: 10.1210/jcem.85.9.6828. [DOI] [PubMed] [Google Scholar]
- 27.Salonen J, Rajpert-De Meyts E, Mannisto S, Nielsen JE, Graem N, Toppari J, Heikinheimo M. Differential developmental expression of transcription factors GATA-4 and GATA-6, their cofactor FOG-2 and downstream target genes in testicular carcinoma in situ and germ cell tumors. Eur J Endocrinol. 2010;162:625–631. doi: 10.1530/EJE-09-0734. [DOI] [PubMed] [Google Scholar]
- 28.Karlsson J, Holmquist Mengelbier L, Elfving P, Gisselsson Nord D. High-resolution genomic profiling of an adult Wilms' tumor: Evidence for a pathogenesis distinct from corresponding pediatric tumors. Virchows Archiv. 2011;459:547–553. doi: 10.1007/s00428-011-1148-0. [DOI] [PubMed] [Google Scholar]
- 29.Virgone C, Cecchetto G, Ferrari A, Bisogno G, Donofrio V, Boldrini R, Collini P, Dall'Igna P, Alaggio R. GATA-4 and FOG-2 expression in pediatric ovarian sex cord-stromal tumors replicates embryonal gonadal phenotype: Results from the TREP project. PLoS One. 2012;7:e45914. doi: 10.1371/journal.pone.0045914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan D, Tian H. Integrated network analysis to explore the key genes regulated by parathyroid hormone receptor 1 in osteosarcoma. World J Surg Oncol. 2017;15:177. doi: 10.1186/s12957-017-1242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vastrad B, Vastrad C, Godavarthi A, Chandrashekar R. Molecular mechanisms underlying gliomas and glioblastoma pathogenesis revealed by bioinformatics analysis of microarray data. Med Oncol. 2017;34:182. doi: 10.1007/s12032-017-1043-x. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Lv L, Liang Y, Shen X, Zhou S, Zhu J, Ma R. Association of 8q23-24 region (8q23.3 loci and 8q24.21 loci) with susceptibility to colorectal cancer: A systematic and updated meta-analysis. Int J Clin Exp Med. 2015;8:21001–21013. [PMC free article] [PubMed] [Google Scholar]
- 33.Selenti N, Tzetis M, Braoudaki M, Gianikou K, Kitsiou-Tzeli S, Fryssira H. An interstitial deletion at 8q23.1-q24.12 associated with langer-giedion syndrome/trichorhinophalangeal syndrome (TRPS) type II and Cornelia de Lange syndrome 4. Mol Cytogenet. 2015;8:64. doi: 10.1186/s13039-015-0169-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plaza-Benhumea L, Valdes-Miranda JM, Toral-Lopez J, Perez- Cabrera A, Cuevas-Covarrubias S. Trichorhinophalangeal syndrome type II due to a novel 8q23.3-q24.12 deletion associated with imperforate hymen and vaginal stenosis. Br J Dermatol. 2014;171:1581–1583. doi: 10.1111/bjd.13177. [DOI] [PubMed] [Google Scholar]
- 35.Win AK, Jenkins MA. Is the reported modifying effect of 8q23.3 and 11q23.1 on colorectal cancer risk for MLH1 mutation carriers valid? Int J Cancer. 2013;133:1762–1763. doi: 10.1002/ijc.28179. [DOI] [PubMed] [Google Scholar]
- 36.Pereza N, Severinski S, Ostojic S, Volk M, Maver A, Dekanić KB, Kapović M, Peterlin B. Third case of 8q23.3-q24.13 deletion in a patient with langer-giedion syndrome phenotype without TRPS1 gene deletion. Am J Med Genet A. 2012;158A:659–663. doi: 10.1002/ajmg.a.35201. [DOI] [PubMed] [Google Scholar]
- 37.Carvajal-Carmona LG, Cazier JB, Jones AM, Howarth K, Broderick P, Pittman A, Dobbins S, Tenesa A, Farrington S, Prendergast J, et al. Fine-mapping of colorectal cancer susceptibility loci at 8q23.3, 16q22.1 and 19q13.11: Refinement of association signals and use of in silico analysis to suggest functional variation and unexpected candidate target genes. Hum Mol Genet. 2011;20:2879–2888. doi: 10.1093/hmg/ddr190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menyhart O, Nagy A, Gyorffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5:181006. doi: 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, Wang XW. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seimiya M, Ohno S, Yamamoto H, Fujiwara K, Yoshida T, Sawabe Y, Sogawa K, Matsushita K, Yokosuka O, Nomura F. Child-Pugh score is altered by the albumin measurement method. Hepatology. 2013;57:2093–2094. doi: 10.1002/hep.25972. [DOI] [PubMed] [Google Scholar]
- 43.Taniai M, Tomimatsu M, Okuda H, Saito A, Obata H. Immunohistochemical detection of proliferating cell nuclear antigen in hepatocellular carcinoma: Relationship to histological grade. J Gastroenterol Hepatol. 1993;8:420–425. doi: 10.1111/j.1440-1746.1993.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 44.Ghorbanoghli Z, Nieuwenhuis MH, Houwing-Duistermaat JJ, Jagmohan-Changur S, Hes FJ, Tops CM, Wagner A, Aalfs CM, Verhoef S, Gómez García EB, et al. Colorectal cancer risk variants at 8q23.3 and 11q23.1 are associated with disease phenotype in APC mutation carriers. Fam Cancer. 2016;15:563–570. doi: 10.1007/s10689-016-9877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, Gu Y. Quantitative assessment of the influence of common variation rs16892766 at 8q23.3 with colorectal adenoma and cancer susceptibility. Mol Genet Genomics. 2015;290:461–469. doi: 10.1007/s00438-014-0928-z. [DOI] [PubMed] [Google Scholar]
- 46.Talseth-Palmer BA, Wijnen JT, Brenne IS, Jagmohan-Changur S, Barker D, Ashton KA, Tops CM, Evans TJ, McPhillips M, Groombridge C, et al. Combined analysis of three Lynch syndrome cohorts confirms the modifying effects of 8q23.3 and 11q23.1 in MLH1 mutation carriers. Int J Cancer. 2013;132:1556–1564. doi: 10.1002/ijc.27843. [DOI] [PubMed] [Google Scholar]
- 47.Wijnen JT, Brohet RM, van Eijk R, Jagmohan-Changur S, Middeldorp A, Tops CM, van Puijenbroek M, Ausems MG, Gómez García E, Hes FJ, et al. Chromosome 8q23.3 and 11q23.1 variants modify colorectal cancer risk in Lynch syndrome. Gastroenterology. 2009;136:131–137. doi: 10.1053/j.gastro.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 48.Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A, Sullivan K, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 49.Okamoto H, Yasui K, Zhao C, Arii S, Inazawa J. PTK2 and EIF3S3 genes may be amplification targets at 8q23-q24 and are associated with large hepatocellular carcinomas. Hepatology. 2003;38:1242–1249. doi: 10.1053/jhep.2003.50457. [DOI] [PubMed] [Google Scholar]
- 50.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng R, Blobel GA. GATA transcription factors and cancer. Genes Cancer. 2010;1:1178–1188. doi: 10.1177/1947601911404223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chou J, Provot S, Werb Z. GATA3 in development and cancer differentiation: Cells GATA have it! J Cell Physiol. 2010;222:42–49. doi: 10.1002/jcp.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albergaria A, Paredes J, Sousa B, Milanezi F, Carneiro V, Bastos J, Costa S, Vieira D, Lopes N, Lam EW, et al. Expression of FOXA1 and GATA-3 in breast cancer: The prognostic significance in hormone receptor-negative tumours. Breast Cancer Res. 2009;11:R40. doi: 10.1186/bcr2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voduc D, Cheang M, Nielsen T. GATA-3 expression in breast cancer has a strong association with estrogen receptor but lacks independent prognostic value. Cancer Epidemiol Biomarkers Prev. 2008;17:365–373. doi: 10.1158/1055-9965.EPI-06-1090. [DOI] [PubMed] [Google Scholar]
- 55.Kouros-Mehr H, Bechis SK, Slorach EM, Littlepage LE, Egeblad M, Ewald AJ, Pai SY, Ho IC, Werb Z. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–152. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–6483. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- 57.Gulbinas A, Berberat PO, Dambrauskas Z, Giese T, Giese N, Autschbach F, Kleeff J, Meuer S, Büchler MW, Friess H. Aberrant gata-3 expression in human pancreatic cancer. J Histochem Cytochem. 2006;54:161–169. doi: 10.1369/jhc.5A6626.2005. [DOI] [PubMed] [Google Scholar]
- 58.Parikh P, Palazzo JP, Rose LJ, Daskalakis C, Weigel RJ. GATA-3 expression as a predictor of hormone response in breast cancer. J Am Coll Surg. 2005;200:705–710. doi: 10.1016/j.jamcollsurg.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 59.Gong X, Liu W, Wu L, Ma Z, Wang Y, Yu S, Zhang J, Xie H, Wei G, Ma F, et al. Transcriptional repressor GATA binding 1-mediated repression of SRY-box 2 expression suppresses cancer stem cell functions and tumor initiation. J Biol Chem. 2018;293:18646–18654. doi: 10.1074/jbc.RA118.003983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu K, Wang J, Gao J, Di J, Jiang B, Chen L, Wang Z, Wang A, Wu F, Wu W, et al. GATA binding protein 2 overexpression is associated with poor prognosis in KRAS mutant colorectal cancer. Oncol Rep. 2016;36:1672–1678. doi: 10.3892/or.2016.4961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.