Abstract
Hepatocellular carcinoma (HCC) is a malignant tumor with extensive metastasis. Changes in the tumor microenvironment provide favorable conditions for tumor metastasis. However, the role of changes to the tumor microenvironment in HCC metastasis is yet to be elucidated. The Gene Expression Omnibus expression profile GSE5093 consists of 20 noncancerous tissues surrounding HCC tissues, including 9 metastasis-inclined microenvironment samples with detectable metastases and 11 metastasis-averse microenvironment samples without detectable metastases. The present study assessed 35 HCC samples to verify the results of chip analysis. In total, 712 upregulated and 459 downregulated genes were identified, with 1,033 nodes, 7,589 edges and 10 hub genes. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis revealed that the differentially expressed genes were significantly enriched in ‘cell-cell adhesion’, ‘cell proliferation’ and ‘protein binding’. The top 10 hub genes were identified via a protein-protein interaction analysis. The 3 most significant modules were identified from the protein-protein network. Moreover, an association between hub genes and patient prognosis was identified. In conclusion, these candidate genes and pathways may help elucidate the mechanisms underlying HCC metastasis and identify more options for targeted therapy.
Keywords: hepatocellular carcinoma, tumor microenvironment, differentially expressed genes, enrichment analysis, protein-protein interaction, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the major histological subtype of primary liver cancer accounting for 85% of all liver cancer cases worldwide, and it was reported to be the most common liver malignancy in 2016 (1). HCC is one of the most aggressive types of cancer with a high mortality rate, whereby 326,000 people died of HCC in 2015 (2). There are several definite risk factors for HCC, such as chronic hepatitis B/C virus infection, nonalcoholic fatty liver diseases, aflatoxin consumption and smoking (2). In addition, there have been a number of identified prognostic markers of HCC, such as alpha fetoprotein, vascular endothelial growth factor and transforming growth factor β, in both large-scale clinical trials and research projects. However, the early diagnosis and effective treatment of HCC remains problematic (3).
To date, surgical resection remains the gold standard treatment for HCC; however, postoperative recurrence and metastasis is common (4). In addition, a large number of patients with HCC are diagnosed at the advanced stages, in which the tumor has already metastasized to other organs prior to surgery (4). Therefore, metastasis is a key challenge in the treatment of HCC. Tumor metastasis is a malignant biological process involving multiple factors and complex signaling pathways that depend not only on the genetic changes of malignant tumor cells, but also on the changes in the tumor microenvironment, such as the stroma, blood vessels and infiltrating inflammatory cells (5). Dysfunction of gene expression in the microenvironment surrounding a tumor serves an important role in the metastatic behavior of tumor cells, such as adhesion of tumor cells, degradation of the extracellular matrix, invasion of basal tissues, homing ability to enter specific tissues, movement and migration in the circulatory system and the promotion of the angiogenesis (6). Therefore, the identification of key genes which function in the tumor microenvironment of HCC may be helpful to identify new targeted therapeutic strategies for HCC metastasis.
In the present study, an original dataset (GSE5093) was obtained from the NCBI-Gene Expression Omnibus database (ncbi.nlm.nih.gov/geo/) containing 20 samples of noncancerous tissues surrounding HCC tissues from two distinct groups of patients with HCC, including 9 metastasis-inclined microenvironment (MIM) samples, with detectable metastases and 11 metastasis-averse microenvironment (MAM) samples, without detectable metastases (7). Differentially expressed genes (DEGs) were filtered in using the Morpheus Website with a data processing standard. Gene-Spring software (version 13.1.1; Agilent Technologies Inc.) was employed to screen the DEGs, followed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG; kegg.jp) pathway enrichment analysis. Furthermore, a protein-protein interaction (PPI) network was established and three significant modules were analyzed. A total of 35 HCC tissue samples were assessed to verify the results of chip analysis. The present study aimed to investigate the genetic molecular mechanisms underlying the metastatic phenotype of HCC and improve the diagnosis and treatment of metastases of primary hepatic carcinoma patients.
Materials and methods
Data collection
The gene expression profile GSE5093 was obtained from Gene Expression Omnibus (GEO) (7). The GSE5093 dataset was based on the GPL1262 dataset [National Cancer Institute (NCI)/Advanced Technology Corner (ATC) Hs-UniGEM2, Advanced Technology Center Microarray Facility (National Institutes of Health (NIH)/NCI/Cancer Research Center (CCR)/ATC)] and contained 20 samples, including 9 MIM samples and 11 MAM samples.
Patients and specimens
The present study was approved by The Institutional Review Board of the Second Affiliated Hospital of Chongqing Medical University (Chongqing, China) and written informed consent was provided by all patients according to The Declaration of Helsinki. A total of 35 HCC specimens (from 21 men and 14 women; age range, 22–73 years; mean age, 53 years) were collected during resection of HCC tumor at The Second Affiliated Hospital of Chongqing Medical University (Chongqing, China) between May 2015 and November 2016. Tissue samples included 16 noncancerous surrounding hepatic tissue samples with MIM and 19 noncancerous surrounding hepatic tissue samples with MAM (<5 cm away from HCC tissue). Specimens were stored in liquid nitrogen for subsequent experimentation. The clinicopathological features of the HCC specimens are listed in Table SI.
Identification and analysis of DEGs
MIM and MAM samples were divided into two groups and GEO2R (ncbi.nlm.nih.gov/geo/geo2r/) was used to analyze the DEGs between the two groups. The raw expression data files, which included TXT files (Agilent platform), were used for analysis by processing using the Morpheus Website (software.broadinstitute.org). A unpaired t-test was used to identify the DEGs and |log fold change (FC)|≥1 and P<0.05 were used as the cut-off criteria for statistical significance.
Functional and pathway enrichment of DEGs
After identifying the DEGs, GO enrichment and KEGG pathway analyses were performed for gene annotation and functional enrichment analysis using the online tool Database for Annotation, Visualization and Integrated Discovery (DAVID; david.abcc.ncifcrf.gov/). The resulting GO terms and KEGG pathways with P<0.05 were considered significantly enriched for the obtained DEGs.
Survival analysis of the hub genes in HCC
Kaplan-Meier survival analysis and the log-rank test were employed to determine the association between hub genes and HCC, using the Gene Expression Profiling Interactive Analysis (GEPIA) database (8) and prognostic data from The Cancer Genome Atlas (cancer.gov/tcga) database. The percentage of low and high expression groups was set at 50%.
Constructing the PPI network of DEGs
The Search Tool for the Retrieval of Interacting Genes (STRING) database offers both experimental and predicted interactive information (9). In the present study, the STRING database was used to explore the enrichment analysis results according to the biological process, molecular function and cell components determined for the DEGs. Finally, the interactions were selected, of which combined scores of >0.7 were used to construct the PPI network using the Molecular Complex Detection (MCODE) plugin within Cytoscape software [version 3.7.2; (10,11)].
RNA preparation and reverse transcription-quantitative (RT-q)PCR
Total RNA from tissue samples was extracted using TRIzol® reagent (Takara Bio, Inc.), according to the manufacturer's protocol. RNA was reverse transcribed into cDNA using the PrimeScript RT reagent (Takara Bio, Inc.). The temperature protocol for RT was as follows: 37°C for 5 min, followed by 45°C for 42 min and 75°C for 5 min. qPCR was subsequently performed using SYBR-Green Real-time PCR Master mix (Beijing Transgen Biotech Co., Ltd.). The primer sequences used for qPCR are listed in Table SII. The following thermocycling conditions were used for qPCR: Initial denaturation at 92°C for 15 min; 37 cycles of 95°C for 15 sec, 55°C for 30 sec; and a final extension at 72°C for 30 sec. Relative mRNA levels were measured using the 2−ΔΔCq method (12) and normalized to the internal reference gene GAPDH.
Statistical analysis
All data are presented as the mean ± standard deviation and analyzed using SPSS 20.0 software (IBM Corp.) and GraphPad Prism 7 (GraphPad Software, Inc.). Differences between groups were analyzed using paired Student's t-test and one-way analysis of variance, followed by Newman-Keuls post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of DEGs between MIM and MAM samples
Through the analysis and processing of data in GSE5093, a total of 1,171 DEGs were identified, which included 712 upregulated and 459 downregulated genes. The heat map of DEG expression levels (top 50 upregulated and 50 downregulated genes) is exhibited in (Fig. 1).
Figure 1.
Heat map of the top 100 differentially expressed genes in metastasis-inclined microenvironment and metastasis-averse microenvironment samples (50 upregulated and 50 downregulated genes). Red, upregulation; blue, downregulation.
GO and pathway enrichment analyses of DEGs
The 1,171 DEGs were selected for functional analysis, performed using the DAVID database. GO analysis of DEGs was carried out from three aspects, which covered molecular function (MF), cellular component (CC) and biological process (BP). BP analysis revealed that the upregulated DEGs were enriched in ‘cell-cell adhesion’, ‘mRNA splicing via spliceosome’ and ‘platelet degranulation’ (Table I), whereas the downregulated DEGs were enriched in ‘cell-cell adhesion’, ‘cell proliferation’ and ‘positive regulation to tyrosine phosphorylation of STAT5 protein’ (Table II). For CC, the upregulated DEGs were enriched in the ‘extracellular exosome and cytosol’, as well as the ‘membrane’ (Table I) and the downregulated DEGs were enriched in ‘receptor complex’, ‘integral component of plasma membrane’ and ‘postsynaptic density’ (Table II). In addition, the MF of upregulated DEGs were enriched in ‘protein binding’, ‘poly(A) RNA binding’ and ‘cadherin binding involved in cell-cell adhesion’ (Table I), whereas the downregulated DEGs were enriched in ‘protein binding’, ‘epidermal growth factor receptor binding’ and ‘amino acid transmembrane transfer activity’ (Table II). These results suggest that the DEGs were associated with the biological processes of cell migration and metastasis.
Table I.
Gene ontology analysis of upregulated differentially expressed genes associated with metastasis-inclined microenvironment and metastasis-averse microenvironment.
| Category | Term | Gene function | Gene count | P-value |
|---|---|---|---|---|
| BP | GO:0098609 | Cell-cell adhesion | 37 | 2.9×10−10 |
| BP | GO:0000398 | mRNA splicing, via spliceosome | 28 | 3.1×10−7 |
| BP | GO:0002576 | Platelet degranulation | 18 | 7.1×10−7 |
| BP | GO:0006457 | Protein folding | 22 | 1.2×10−5 |
| BP | GO:0048013 | Ephrin receptor signaling pathway | 14 | 3.8×10−5 |
| BP | GO:0043066 | Movement of cell or subcellular component | 14 | 3.8×10−5 |
| BP | GO:0043066 | Negative regulation of apoptotic process | 38 | 4.2×10−5 |
| BP | GO:0007165 | Signal transduction | 75 | 5.4×10−5 |
| BP | GO:0019886 | Antigen processing and presentation of exogenous peptide antigen via MHC class II | 14 | 7.8×10−5 |
| BP | GO:0050900 | Leukocyte migration | 16 | 1.1×10−4 |
| CC | GO:0070062 | Extracellular exosome | 220 | 1.1×10−28 |
| CC | GO:0016020 | Membrane | 170 | 7.6×10−21 |
| CC | GO:0005829 | Cytosol | 218 | 1.4×10−18 |
| CC | GO:0005654 | Nucleoplasm | 177 | 4.2×10−13 |
| CC | GO:0005913 | Cell-cell adherens junction | 44 | 5.2×10−13 |
| CC | GO:0031012 | Extracellular matrix | 37 | 5.5×10−10 |
| CC | GO:0042470 | Melanosome | 21 | 9.2×10−10 |
| CC | GO:0043209 | Myelin sheath | 25 | 2.5×10−9 |
| CC | GO:0005615 | Extracellular space | 90 | 1.1×10−7 |
| CC | GO:0030529 | Intracellular ribonucleoprotein complex | 21 | 1.8×10−7 |
| MF | GO:0005515 | Protein binding | 448 | 2.8×10−14 |
| MF | GO:0044822 | Poly(A) RNA binding | 100 | 7.6×10−14 |
| MF | GO:0098641 | Cadherin binding involved in cell-cell adhesion | 42 | 1.8×10−12 |
| MF | GO:0032403 | Protein complex binding | 23 | 2.8×10−5 |
| MF | GO:0003723 | RNA binding | 43 | 4.3×10−5 |
| MF | GO:0051287 | NAD binding | 9 | 1.1×10−4 |
| MF | GO:0051082 | Unfolded protein binding | 15 | 1.3×10−4 |
| MF | GO:0008134 | Transcription factor binding | 25 | 4.8×10−4 |
| MF | GO:0005524 | ATP binding | 86 | 5.9×10−4 |
| MF | GO:0019899 | Enzyme binding | 27 | 9.6×10−4 |
P<0.001. GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function.
Table II.
Gene ontology analysis of downregulated differentially expressed genes associated with metastasis-inclined microenvironment and metastasis-averse microenvironment.
| Category | Term | Gene function | Gene count | P-value |
|---|---|---|---|---|
| BP | GO:0007155 | Cell adhesion | 28 | c2.8×10−5 |
| BP | GO:0008283 | Cell proliferation | 22 | c3.0×10−4 |
| BP | GO:0042523 | Positive regulation of tyrosine phosphorylation of Stat5 protein | 5 | c6.7×10−4 |
| BP | GO:0007165 | Signal transduction | 47 | c9.4×10−4 |
| BP | GO:0031532 | Actin cytoskeleton reorganization | 6 | b5.8×10−3 |
| BP | GO:0009267 | Cellular response to starvation | 6 | b5.8×10−3 |
| BP | GO:0060749 | Mammary gland alveolus development | 4 | b7.8×10−3 |
| BP | GO:0007169 | Transmembrane receptor protein tyrosine kinase signaling pathway | 8 | b9.5×10−3 |
| BP | GO:0006865 | Amino acid transport | 5 | b1.0×10−2 |
| BP | GO:0045944 | Positive regulation of transcription from RNA polymerase II promoter | 37 | a1.1×10−2 |
| CC | GO:0043235 | Receptor complex | 90 | c1.9×10−4 |
| CC | GO:0005887 | Integral component of plasma membrane | 47 | c4.1×10−4 |
| CC | GO:0014069 | Postsynaptic density | 78 | b1.3×10−3 |
| CC | GO:0045211 | Postsynaptic membrane | 64 | b1.4×10−3 |
| CC | GO:0005829 | Cytosol | 52 | b2.1×10−3 |
| CC | GO:0005886 | Plasma membrane | 52 | b3.6×10−3 |
| CC | GO:0009986 | Cell surface | 52 | b4.5×10−3 |
| CC | GO:0030054 | Cell junction | 15 | b5.8×10−3 |
| CC | GO:0005794 | GOlgi apparatus | 8 | b6.7×10−3 |
| CC | GO:0005737 | Cytoplasm | 8 | b7.5×10−3 |
| MF | GO:0005515 | Protein binding | 258 | c1.5×10−5 |
| MF | GO:0005154 | Epidermal growth factor receptor binding | 6 | c8.6×10−4 |
| MF | GO:0015171 | Amino acid transmembrane transporter activity | 7 | c8.6×10−4 |
| MF | GO:0005524 | ATP binding | 56 | c9.5×10−4 |
| MF | GO:0042803 | Protein homodimerization activity | 32 | b1.5×10−3 |
| MF | GO:0004714 | Transmembrane receptor protein tyrosine kinase activity | 6 | b2.1×10−3 |
| MF | GO:0008179 | Adenylate cyclase binding | 3 | a1.9×10−2 |
| MF | GO:0001078 | Transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding | 8 | a1.9×10−2 |
| MF | GO:0005096 | Gtpase activator activity | 14 | a2.0×10−2 |
| MF | GO:0043236 | Laminin binding promoter | 4 | a2.2×10−2 |
P<0.05
P<0.01
P<0.001. GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function.
KEGG pathway analysis of MIM and MAM DEGs
To further analysis the functions of DEGs, KEGG pathway analysis was conducted to determine the most significantly enriched pathways of the upregulated DEGs and downregulated DEGs. The upregulated DEGs were enriched in ‘protein processing in the endoplasmic reticulum’, ‘antigen processing and presentation’ and ‘phagosome’ pathways (Table III). The downregulated DEGs were enriched in the ‘arrhythmogenic right ventricular cardiomyopathy’, ‘phosphatidylinositol signaling system’ and ‘inositol phosphate metabolism’ pathways (Table III).
Table III.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of DEGs associated with metastasis-inclined microenvironment and metastasis-averse microenvironment.
| A, Upregulated DEGs | ||||
|---|---|---|---|---|
| Pathway | Name | Gene count | P-value | Genes |
| hsa04141 | Protein processing in endoplasmic reticulum | 26 | c7.50×10−6 | HSP90AB1, SEC31A, GANAB, PDIA3, PDIA4, PRKCSH, CALR, SEC62, DERL3, CANX, SSR1, OS9, HSPH1,xBP1, RPN1, DNAJA1, HSPA6, HSPA5, HSPA8, SEC23A, P4HB, BCAP31, CAPN1, SEC23B, SEL1L, UBE2E1 |
| hsa04612 | Antigen processing and presentation | 15 | c7.23×10−5 | HSP90AB1, CIITA, PDIA3, CD8B, LGMN, CTSS, CALR, CANX, TAPBP, HSPA6, HLA-DRB5, CTSB, HLA-DPB1, HSPA8, HLA-DRA |
| hsa04145 | Phagosome | 22 | c9.24×10−5 | ACTB, C3, ATP6AP1, CTSS, CALR, ATP6V1B1, ITGB1, CANX, ITGAM, ACTG1, ATP6V1A, LAMP2, HLA-DRB5, MPO, VAMP3, HLA-DPB1, THBS1, FCGR3A, DYNC1H1, THBS2, CD14, HLA-DRA |
| hsa03040 | Spliceosome | 20 | c1.51×10−4 | SRSF1, TRA2A, PRPF3, DDX5, SF3A1, SF3B4, RBMX, HNRNPA1, HNRNPU, SRSF4, HNRNPK, PLRG1, DHX38, PCBP1, USP39, HSPA6, DHX15, ACIN1, HSPA8, THOC1 |
| hsa04918 | Thyroid hormone synthesis | 13 | c4.73×10−4 | TG, GPX2, ADCY7, GNAQ, ATP1B2, PAX8, TPO, CREB3L1, PRKCG, PRKACB, HSPA5, PDIA4, CANX |
| hsa04610 | Complement and coagulation cascades | 12 | b1.50×10−3 | KNG1, C8B, C7, A2M, FGA, C3, CFB, CD46, C6, SERPING1, C1S, C2 |
| hsa04520 | Adherens junction | 12 | b1.91×10−3 | ACTB, ACTG1, PTPN6, TGFBR2, CTNND1, CDH1, SMAD2, PTPN1, WASL, TCF7L2, IQGAP1, VCL |
| hsa05142 | Chagas disease (American trypanosomiasis) | 15 | b1.98×10−3 | CFLAR, GNAI3, GNAI2, C3, RELA, TGFBR2, MAP2K4, NFKB1, SMAD2, CALR, GNAQ, PPP2CB, IL12A, PPP2R2B, PPP2R2A |
| hsa05110 | Vibrio cholerae infection | 10 | b2.23×10−3 | ACTB, ACTG1, ATP6V1A, KDELR2, ATP6AP1, PRKCG, PRKACB, PDIA4, ATP6V1B1, TJP2 |
| hsa05146 | Amoebiasis | 15 | b2.37×10−3 | IL1R1, RELA, PRKCG, NFKB1, ITGAM, VCL, ARG1, C8B, SERPINB9, GNAQ, IL12A, PRKACB, COL1A1, CD14, FN1 |
| B, Downregulated DEGs | ||||
| hsa05412 | Arrhythmogenic right ventricular cardiomyopathy | 8 | b2.35×10−3 | ITGA8, SGCD, DSC2, GJA1, ITGA2, CACNB4, ITGA4, CACNA1C (ARVC) |
| hsa04070 | Phosphatidylinositol signaling system | 9 | b5.51×10−3 | PIK3CG, MTM1, DGKB, PIK3C2G, IMPA1, PIP5K1B, ITPKB, PTEN, ITPR2 |
| hsa00562 | Inositol phosphate metabolism | 7 | a1.33×10−2 | PIK3CG, MTM1, PIK3C2G, IMPA1, PIP5K1B, ITPKB, PTEN |
| hsa04020 | Calcium signaling pathway | 11 | a2.61×10−2 | P2RX7, SLC8A1, CCKBR, ERBB4, PHKB, GRPR, ITPKB, PPP3CA, PTGFR, CACNA1C, ITPR2 |
| hsa05202 | Transcriptional misregulation in cancer | 10 | a4.13×10−2 | MAF, CCNT2, CSF2, HHEX, LMO2, FLT3, GZMB, SMAD1, JMJD1C, HMGA2 |
| hsa04151 | PI3K-Akt signaling pathway | 16 | a5.39×10−2 | PHLPP1, PIK3CG, COL4A3, PPP2R5A, TCL1A, ITGA2, FGF13, ITGA4, KIT, PTEN, COL5A2, PRLR, ITGA8, PDGFD, GHR, IL2 |
| hsa05410 | Hypertrophic cardiomyopathy (HCM) | 6 | a6.39×10−2 | ITGA8, SGCD, ITGA2, CACNB4, ITGA4, CACNA1C |
| hsa04810 | Regulation of actin cytoskeleton | 11 | a6.54×10−2 | PIK3CG, DOCK1, ARHGEF7, DIAPH2, ITGA8, PIP5K1B, ITGA2, IQGAP2, FGF13, PDGFD, ITGA4 |
| hsa04080 | Neuroactive ligand-receptor interaction | 13 | a8.12×10−2 | CCKBR, NPY2R, GABRA5, PTGFR, P2RX7, AGTR2, P2RY10, PRLR, GRPR, CHRNA5, CHRNB3, TSHR, GHR |
| hsa05414 | Dilated cardiomyopathy | 6 | a8.21×10−2 | ITGA8, SGCD, ITGA2, CACNB4, ITGA4, CACNA1C |
P<0.05
P<0.01
P<0.001. DEGs, differentially expressed genes.
Analysis of the PPI network with DEGs in MIM and MAM
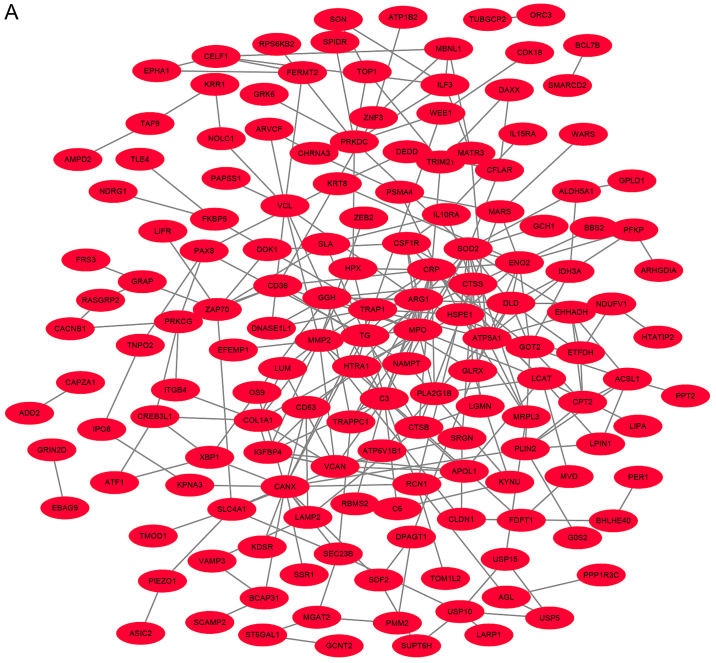
According to the information provided by the STRING, the interaction of the DEGs and acquired hub genes of potential diagnosis and treatment associated DEGs were analyzed. As depicted in Figs. 2A and S1, there were 955 nodes and 7,589 edges, of which 638 nodes represented upregulated DEGs and nodes represented downregulated DEGs (Fig. 2B). The top 10 hub genes with the highest degrees included: Heat shock protein family A member 8 (HSPA8); PH domain and leucine rich repeat protein phosphatase 1 (PHLPP1); phosphoribosylglycinamide formyltransferase (GART); carbamoyl-phosphate synthetase 2 (CAD); actin beta (ACTB); cadherin 1 (CDH1); phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit γ (PIK3CG); nuclear factor κB subunit 1 (NFκB1); signal transducer and activator of transcription 3 (STAT3); and heat shock protein family A (Hsp70) member 5 (HSPA5). Then, the expression of the top 10 hub genes were verified using RT-qPCR. There were 4 hub upregulated genes and 6 downregulated genes in MAM, compared with MIM samples (Fig. 3). Moreover, the top 3 significant modules were selected from the DEG PPI network using MCODE (Fig. 4A-C). Module 1 included 31 nodes and 458 edges, module 2 included 37 nodes and 267 edges and module 3 included 45 nodes. Furthermore, GO and KEGG pathway analysis results were used to analyze the functional and signal pathway enrichment of the three modules. The results showed that module 1 was primarily associated with ‘RNA splicing’, ‘catalytic step 2 spliceosome’, ‘nucleotide binding’ and ‘spliceosome’ pathways (Table IV). Module 2 was primarily enriched in ‘protein modification by small protein conjugation’, ‘catalytic complex’, ‘ubiquitin-protein transferase activity’ and ‘chemokine signaling’ pathways (Table V). Module 3 was most enriched in ‘protein folding’, ‘extracellular exosomes’, ‘protein disulfide isomerase activity’ and ‘chemokine signaling’ pathways (Table VI). These results indicated that the 10 hub genes may function in the biological behavior processes of cell migration and metastasis.
Figure 2.
DEG protein-protein interaction network in metastasis-inclined microenvironment and metastasis-averse microenvironment. (A) The most significant interaction network of the upregulated DEGs interaction network. (B) Interaction network of the 317 downregulated DEGs interaction network. DEG, differentially expressed gene.
Figure 3.
Top 10 hub genes were detected by reverse transcription-quantitative PCR. Expression of (A) CAD, (B) GART, (C) HSPA5, (D) NFKB1, (E) ACTB, (F) CDH1, (G) HSPA8, (H) PHLPP1, (I) PIK3GC and (J) STAT3 The data are presented as the mean ± standard deviation of 3 independent experiments. **P<0.01 MAM vs. MIM. CAD, carbamoyl-phosphate synthetase 2; GART, phosphoribosylglycinamide formyltransferase; HSPA5, heat shock protein family A member 5; NFκB1, nuclear factor κB subunit 1; ACTB, actin beta; CDH1, Cadherin 1; HSPA8, heat shock protein family A member 8; PHLPP1, PH domain and leucine rich repeat protein phosphatase 1; PIK3CG, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit γ; STAT3, signal transducer and activator of transcription 3.
Figure 4.
Top 3 modules from the protein-protein interaction network determined by the molecular complex detection score. (A) Module 1 and (B) Module 2. Top 3 modules from the protein-protein interaction network determined by the molecular complex detection score. (C) Module 3.
Table IV.
Functional and pathway enrichment analysis of the genes in module 1.
| Category | Term | Gene function | Gene count | P-value |
|---|---|---|---|---|
| BP | GO:0008380 | RNA splicing | 6 | c2.27×10−9 |
| BP | GO: 0000398 | mRNA splicing, via spliceosome | 6 | c8.01×10−7 |
| BP | GO:0006397 | mRNA processing | 5 | c2.06×10−6 |
| BP | GO:0048025 | Negative regulation of mRNA splicing, via spliceosome | 4 | c3.81×10−6 |
| BP | GO:0000381 | Regulation of alternative mRNA splicing, via spliceosome | 4 | c4.06×10−5 |
| CC | GO:0071013 | Catalytic step 2 spliceosome | 3 | c6.60×10−19 |
| CC | GO:0005681 | Spliceosomal complex | 2 | c1.58×10−10 |
| CC | GO:0030529 | Intracellular ribonucleoprotein complex | 2 | c8.66×10−9 |
| CC | GO:0019013 | Viral nucleocapsid | 2 | c7.85×10−8 |
| CC | GO:0005654 | Nucleoplasm | 12 | c8.50×10−6 |
| MF | GO:0000166 | Nucleotide binding | 7 | c3.01×10−10 |
| MF | GO:0044822 | Poly(A) RNA binding | 7 | c1.66×10−9 |
| MF | GO:0003723 | RNA binding | 6 | c1.95×10−5 |
| MF | GO:0003729 | mRNA binding | 12 | c4.65×10−5 |
| MF | GO:0003676 | Nucleic acid binding | 7 | c8.67×10−4 |
| KEGG_PATHWAY | has:03040 | Spliceosome | 20 | c1.54×10−28 |
| KEGG_PATHWAY | has: 03015 | mRNA surveillance pathway | 4 | b4.32×10−3 |
| KEGG_PATHWAY | has: 05168 | Herpes simplex infection | 4 | a3.18×10−2 |
P<0.05
P<0.01
P<0.001. GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table V.
Functional and pathway enrichment analysis of the genes in module 2.
| Category | Term | Gene function | Gene count | P-value |
|---|---|---|---|---|
| BP | GO:0032446 | Protein modification by small protein conjugation | 12 | b6.57×10−9 |
| BP | GO:0070647 | Protein modification by small protein conjugation or removal | 12 | b5.38×10−8 |
| BP | GO:0007186 | G-protein coupled receptor signaling pathway | 11 | b2.93×10−7 |
| BP | GO:0016567 | Protein ubiquitination | 10 | b5.90×10−7 |
| BP | GO:0007193 | Adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway | 5 | b6.02×10−6 |
| CC | GO:1902494 | Catalytic complex | 11 | b9.45×10−6 |
| CC | GO:0000151 | Ubiquitin ligase complex | 7 | b1.07×10−5 |
| CC | GO:0098552 | Side of membrane | 5 | b2.94×10−4 |
| CC | GO:1990234 | Transferase complex | 6 | b3.96×10−4 |
| CC | GO:0000152 | Nuclear ubiquitin ligase complex | 3 | a1.53×10−3 |
| MF | GO:0004842 | Ubiquitin-protein transferase activity | 8 | b3.79×10−6 |
| MF | GO:0001664 | G-protein coupled receptor binding | 6 | b4.47×10−6 |
| MF | GO:0019787 | Ubiquitin-like protein transferase activity | 8 | b5.20×10−6 |
| MF | GO:0008528 | G-protein coupled peptide receptor activity | 5 | b1.42×10−4 |
| MF | GO:0001653 | Peptide receptor activity | 5 | b1.52×10−4 |
| KEGG_PATHWAY | has: 04062 | Chemokine signaling pathway | 11 | b4.43×10−10 |
| KEGG_PATHWAY | has: 04120 | Ubiquitin mediated proteolysis | 6 | b1.26×10−4 |
| KEGG_PATHWAY | has: 04914 | Progesterone-mediated oocyte maturation | 5 | b2.19×10−4 |
P<0.01
P<0.001. GO, gene ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table VI.
Functional and pathway enrichment analysis of the genes in module 3.
| Category | Term | Gene function | Gene count | P-value |
|---|---|---|---|---|
| BP | GO:0006457 | Protein folding | 8 | b1.45×10−7 |
| BP | GO:0045454 | Cell redox homeostasis | 5 | b3.19×10−5 |
| BP | GO:0044710 | Single-organism metabolic process | 19 | b4.36×10−5 |
| BP | GO:0006091 | Generation of precursor metabolites and energy | 6 | b3.46×10−4 |
| BP | GO:0090066 | Regulation of anatomical structure size | 7 | b3.49×10−4 |
| CC | GO:0070062 | Extracellular exosome | 23 | b1.72×10−8 |
| CC | GO:0072562 | Blood microparticle | 5 | b7.56×10−5 |
| CC | GO:0005577 | Fibrinogen complex | 3 | b1.64×10−4 |
| CC | GO:0043209 | Myelin sheath | 5 | b5.65×10−4 |
| CC | GO:0005829 | Cytosol | 9 | a2.90×10−3 |
| MF | GO:0003756 | Protein disulfide isomerase activity | 3 | a1.07×10−3 |
| MF | GO:0005524 | ATP binding | 11 | a1.31×10−3 |
| MF | GO:0004070 | Aspartate carbamoyltransferase activity | 2 | a5.77×10−3 |
| MF | GO:0004088 | Carbamoyl-phosphatesynthase (glutamine-hydrolyzing) activity | 2 | a5.77×10−3 |
| MF | GO:0004087 | Carbon metabolism | 2 | a5.77×10−3 |
| KEGG_PATHWAY | has: 01200 | Chemokine signaling pathway | 5 | a1.23×10−3 |
| KEGG_PATHWAY | has: 04144 | Endocytosis | 6 | a2.89×10−3 |
| KEGG_PATHWAY | has: 05100 | Bacterial invasion of epithelial cells | 4 | a4.33×10−3 |
P<0.01
P<0.001. GO, gene ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
The Kaplan-Meier plot of hub genes in MIM and MAM
Through GEPIA prediction of the association between the 10 hub genes and HCC patient prognosis, it was observed that expression of CAD [Hazard Ratio (HR), 1.9; P<0.001], GART (HR, 1.8; P=0.018), HSPA5 (HR, 1.5; P=0.016), ACTB (HR, 1.6; P=0.0074), CDH1 (HR, 0.66; P=0.018) and HSPA8 (HR, 1.6; P=0.011) were associated with poor overall survival in patients with HCC (Fig. 5).
Figure 5.
Prognostic value of hub genes in patients with HCC. Kaplan-Meier analysis of OS time un patients with HCC with high and low expression levels of: (A) CAD, (B) GART, (C) HSPA5, (D) NFκB1, (E) ACTB, (F) CDH1, (G) HSPA8, (H) PHLPP1, (I) PIK3CG and (J) STAT3. The red dotted line above the red curve and the red dotted line below represent the high (50%) and low cutoff (50%) on the survival curve. The blue dotted line above the blue curve and the blue dotted line below represent high (50%) and low cutoff (50%) on the survival curve. HCC, hepatocellular carcinoma; OS, overall survival; CAD, carbamoyl-phosphate synthetase 2; GART, phosphoribosylglycinamide formyltransferase; HSPA5, heat shock protein family A member 5; NFκB1, nuclear factor κB subunit 1; ACTB, actin beta; CDH1, cadherin 1; HSPA8, heat shock protein family A member 8; PHLPP1, PH domain and leucine rich repeat protein phosphatase 1; PIK3CG, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit γ; STAT3, signal transducer and activator of transcription 3; HR, hazard ratio; TPM, transcripts per million.
Discussion
HCC is the most common malignant tumor with high morbidity and mortality, and was reported to be one of the major causes of cancer-associated mortalities worldwide, in 2002 (13). Patients with HCC are often diagnosed with extrahepatic metastasis, which poses a great challenge to the diagnosis and treatment of HCC (14). Currently, high-throughput gene chip technology can be used to explore the occurrence and development of diseases from the whole genome or transcriptome level, which has been widely used in gene expression analysis and biomarker discovery for a variety of illnesses (14). In order to further understand the molecular mechanisms underlying the recurrence and metastasis of HCC, the GSE5093 Biochip dataset on the metastatic microenvironment of HCC was obtained from the GEO database. This was used to perform a systematic and bioinformatics analysis, including enrichment analysis of differences in gene expression and protein-protein interaction.
In the present study, a total of 1,171 DEGs were identified, of which 712 were upregulated and 459 were downregulated. GO term functional analysis demonstrated that the upregulated genes were primarily involved in ‘cell-cell adhesion’, ‘extracellular exosome’ and ‘cadherin binding’, while downregulated DEGs were involved in ‘cell-cell adhesion’, ‘integral component of plasma membrane’ and ‘epidermal growth factor receptor binding’. The tumor cells leave the primary site, invade the extracellular matrix, adhere to macromolecular protein components in the basement membrane, degrade the basement membrane and the extracellular matrix, and move through the extracellular matrix to invade the surrounding tissues (15). Once in the circulation system, tumor cells evade the surveillance of the immune system. When passing through the vessel wall to the secondary site, tumor cells adhere to the secondary site, proliferate and eventually metastasize (16,17). Cell adhesion, receptor complex, integral component of plasma membrane, postsynaptic density and epidermal growth factor receptor binding are key auxiliary components (18–20). Cell adhesion molecules suppress the adhesive function of tumor cells and glycoproteins located on the cell surface (21). Moreover, KEGG pathway analysis indicated that the upregulated DEGs influenced ‘protein processing in endoplasmic reticulum’, ‘antigen processing and presentation’ and ‘phagosome’; the downregulated DEGs were associated with ‘arrhythmogenic right ventricular cardiomyopathy’, ‘phosphatidylinositol signaling system’ and ‘inositol phosphate metabolism’. Tumor metastasis involves detachment from the primary site of the tumor, entrance into the surrounding stroma and the circulation or lymphatic system, adhesion to endothelial cell walls, extravasation and invasion through vascular proliferation for the formation a novel metastatic lesion (22,23). GO and KEGG analyses suggested that the aforementioned DEGs may influence signaling pathways associated with tumor cell metastasis. The identified DEGs may provide novel research directions and targets for the treatment of HCC metastasis.
Based on the established PPI network with DEGs, the following top 10 hub genes were identified: HSPA8, PHLPP1, GART, CAD, ACTB, CDH1, PIK3CG, NFκB1, STAT3 and HSPA5. HSPA8 is a member of the heat shock proteins family, which influences molecular signal transduction, apoptosis and protein regulation (24). HSPA8 is located in the cytoplasm and lysosomes and is involved in mediating cell autophagy by binding to the substrate protein and transporting it into the lysosomal cavity (25,26). Novel research has shown that the Hsp70 protein may inhibit apoptosis by regulating the caspase-dependent pathway (27). Moreover, tumor vaccines against HSP70S have been successfully completed in animal models and clinical trials are underway (27).
PHLPP1 is an important regulator of Akt serine-threonine kinases and conventional/novel protein kinase C isoforms (28). PHLPP1 can inhibit growth factor-induced signal transduction pathways in cancer cells, so it may be used as a growth inhibitor in several types of cancer (28). In the human breast cancer cell line 21T, AK294 phosphorylation was inhibited by LY294002, and the expression of PHLPP1 in two metastatic cell lines (MT1 and MT2) was much lower compared with early breast cancer cells (29). PHLPP1 can negatively regulate the activity of AKT and its downstream kinase by phosphorylating the hydrophobic group of AKT at the Serine473 site. This resulted in inhibition of the PI3K/AKT signaling pathway, and the activation of various cancer promoting signaling pathways, including amplification or gain-of-function mutations in upstream receptor protein tyrosine kinases (RPTKs), activating mutations in PI3K and Akt and loss-of-function mutations in the regulatory phosphatase PTEN, which affect differentiation and proliferation of cancer cells and apoptosis in breast cancer cells (29). Therefore, it is hypothesized that PHLPP1 is associated with tumor metastasis.
Trifunctional purine biosynthetic protein adenosine-3 is an enzyme that is encoded by the GART gene (30). GART participates in several aspects of energy metabolism, including as a ATP binding source, metal ion binding source and phosphoribosylamine-glycine ligase activity source (30,31). CAD is a fusion gene that encodes three enzymes involved in pyrimidine biosynthesis; carbamoyl-phosphate synthetase 2 and aspartate transcarbamylase fused with dihydroorotase (31). CAD is a key enzyme the regulates multiple biological processes in the first three steps of pyrimidine biosynthesis (32). These three genes serve a role in the synthesis and metabolism of nucleic acids and proteins in cells, and their synthesized products provide essential substances and energy for cell proliferation (32).
Actin is a major constituent of the muscular contractile apparatus and ACTB is one of six different actin isoforms that have been identified in humans (33). ACTB binds RNA-binding protein Sam68 and participates in the regulation of the synaptic formation of dendritic spines (34). ACTB supports the muscular cytoskeleton during the formation of novel tumor cells following tumor cell proliferation and metastasis. Thus, ACTB is considered to be associated with cell proliferation and metastasis (34).
CDH1, also known as epithelial cadherin (E-cadherin), is a protein encoded by the CDH1 gene (35). Mutations in CDH1 are closely associated with liver, colorectal and gastric cancer. Moreover, a functional deficit of CDH1 promotes the proliferation and invasiveness of tumor cells, and promotes the malignant development of tumors (35). E-cadherin is a member of the cadherin family and downregulation of E-cadherin reduces cell-cell adhesion, which results in cells that are more susceptible to migration (36). This change can easily result in cancer cells passing through the cell basement membrane and invading surrounding tissues (36). A previous study demonstrated that E-cadherin is an important protein that regulates cell-cell adhesion. On the cell membrane, E-cadherin binds to β-catenin via its cytoplasmic tail, allowing epithelial cells to bind tightly together. Knockdown of E-cadherin expression results in β-catenin release into the cytoplasm and translocation into the nucleus, and this process can result in the expression of epithelial mesenchymal transition (EMT)-inducible transcription factors. Therefore, E-cadherin is an important gene that regulates EMT (37).
The PIK3CG subunit λ isoform is an enzyme encoded by the PIK3CG gene (38). Several studies have demonstrated that the function of PIK3CG is to regulate the transmission of extracellular signals, including E-cadherin-mediated cell-cell adhesion, which serves an important role in maintaining the structural and functional integrity of the epithelium (39,40).
The NFκB1 gene is a cellular transcription factor and encodes the NF-κB p105 subunit protein. The expression of NF-κB is activated via various stimuli, such as cytokines, oxidative free radicals and bacterial or viral products (41). Activated NF-κB is transported to the nucleus to participate in the transcription and synthesis of various proteins and signaling molecules, regulating various cell behaviors (41). It has been revealed that osteopontin can regulate the expression of MTI-MMP via the NF-κB signaling pathway and promote the migration of cancer cells and invasion into the extracellular matrix (42). A previous study confirmed that several adhesion molecules, such as ICAM-1, VCAM-1 and ELAM-1, are regulated by NF-κB, which serves an important role in tumor metastasis (43).
STAT3, which contains SH2 and SH3 domains, binds to specific phosphotyrosine-containing peptides. When STAT3 is activated by phosphorylation, it serves as a transcriptional activator, in either its homodimeric or heterodimeric form (44). Phosphorylated STAT3 binds a specific site in the target gene promoter sequence and promotes transcription of the gene (44). Dysregulation of this pathway in tumors leads to increased angiogenesis surrounding the tumor, promotes blood supply to the tumor, increases tumor survival and promotes malignant development. It has been demonstrated that tissue-specific inactivation of STAT3 in the lungs of mice is associated with the occurrence and progression of lung adenocarcinoma, which reduced the survival rate. Notably, decreased expression of STAT3 promotes the expression of NF-κB in the cytoplasm, thus regulating NF-κB-induced IL-8 expression (45). This process promotes IL-8-mediated tumor invasion, angiogenesis and progression (45).
Binding immunoglobulin protein, also known as HSPA5, is a protein that is encoded by the HSPA5 gene (46). A large number of studies have revealed the high expression of HSPA5 in all malignant tumors (47,48). Therefore, inhibiting HSPA5 gene expression may be used as an adjuvant therapy for cancer. In addition, reducing the large amount of HSPA5 produced during the stress response can increase the apoptosis of tumor cells and inhibit tumor growth (49).
In addition, tumor metastasis is a complex multi-step process, whereby tumor cells not only interact with each other and host cells, but also with extracellular matrix components (50). Certain characteristics, such as adhesion, metastasis, migration, skeleton assembly, signal transduction and proliferation are associated with tumor metastasis (50,51). Migration is considered a key factor in the process of tumor cell metastasis (51). Previous studies have demonstrated that RNA splicing, protein modification by small protein conjugation, catalytic complex, ubiquitin-protein transferase activity, chemokine signaling pathways, protein folding, extracellular exosomes, protein disulfide isomerase activity and chemokine signaling pathways are also involved in the process of tumor cell migration and metastasis, such as the expansion of the cell front, the formation of new adhesion sites and the release of the original adhesion sites in the tail of the cell (52–56).
A study reported that miR-26b-5p exerts metastatic properties and maintains epithelial cell adhesion molecule + cancer stem cells via HSPA8 in HCC (57). Moreover, a study investigating the role of PHLPP1 in HCC reported that miR-190 influences EMT by regulating the expression of PHLPP1, thus affecting the malignant biological behavior of HCC cells (58). Isobaric tags for relative and absolute quantification analysis of clinical samples of HCC indicated that ACTB serves an important role in the initiation stage of HCC (59). Long non-coding RNA ATRERNA1 promotes the metastasis and invasiveness of HCC cells by recruiting EHMT2 and/or ehmt2/snai1 complexes to inhibit CDH1 (60). Loss of NFκB1 promotes the occurrence of age-related chronic liver disease (CLD) which is characterized by steatosis, neutrophil proliferation, fibrosis, telomere damage of hepatocytes and HCC (61). The results of the present study demonstrated that STAT3 is one of the hub genes. Phosphorylated RPN2 activates the signal transducer and activator of STAT3 and is also responsible for the RPN2-stimulated elevated expression of MMP-9 and for invading HCC cells (62).
In the current study, biological function and signal pathway enrichment analysis revealed that the genes in module 1, 2 and 3 were primarily enriched in ‘RNA splicing’, ‘G-protein coupled receptor signaling pathway’ and ‘protein ubiquitination’. Studies have demonstrated that these signaling pathways are required for cell survival and metabolism and serve an important role in tumor recurrence and metastasis (63–65). Therefore, it is hypothesized that these molecular pathways may provide novel insight and potential targets for the diagnosis and treatment of HCC.
In conclusion, 1,171 DEGs were identified (including 712 upregulated and 459 downregulated genes, 1,033 nodes, 7,589 edges and 10 hub genes) via gene profile dataset and integrated bioinformatics analysis in noncancerous surrounding hepatic tissues. The 10 hub genes were significantly enriched in several signaling pathways which serve an important role in tumor metastasis. According to the present research and analysis, CAD, GART, HSPA5, ACTB, CDH1 and HSPA8 may have potential as targets for the diagnosis and treatment of metastatic HCC. The present study may provide a scientific basis for the investigation of these genes in HCC in the future.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by The Foundation of Scientific Research of Sichuan Medical Association (grant. no. S16007), The Sichuan Health Planning Commission Key Project Foundation (grant. no. 17ZD008), The Sichuan Science and Technology Project (grant. no. 2018JY0276) and The Chengdu Science and Technology Bureau Technology Innovation Project (grant. no. 2018-YF05-01228-SN).
Availability of data and materials
The datasets generated and/or analyzed during the present study are available in the [NCBI] repository, [https://www.ncbi.nlm.nih.gov/geo].
Authors' contributions
YL and HW performed the experiments, analyzed the data and drafted the manuscript. YW, HW, MD and CL conceptualized the study design and critically revised the manuscript. YL, MD, CL and HW wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by The Institutional Review Board of the Second Affiliated Hospital of Chongqing Medical University (Chongqing, China) and written informed consent was provided by all patients according to the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chen WQ, Zheng RS, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Greten TF, Wang XW, Korangy F. Current concepts of immune based treatments for patients with HCC: From basic science to novel treatment approaches. Gut. 2015;64:842–848. doi: 10.1136/gutjnl-2014-307990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pang RW, Joh JW, Johnson PJ, Monden M, Pawlik TM, Poon RT. Biology of hepatocellular carcinoma. Ann Surg Oncol. 2008;15:962–971. doi: 10.1245/s10434-007-9730-z. [DOI] [PubMed] [Google Scholar]
- 5.Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, Simon R, Li Y, Robles AI, Chen Y, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–423. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 6.Iizuka N, Oka M, Yamada-Okabe H, Nishida M, Maeda Y, Mori N, Takao T, Tamesa T, Tangoku A, Tabuchi H, et al. Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet. 2003;361:923–929. doi: 10.1016/S0140-6736(03)12775-4. [DOI] [PubMed] [Google Scholar]
- 7.Budhu A, Forgues M, Ye QH, Jia HL, He P, Zanetti KA, Kammula US, Chen Y, Qin LX, Tang ZY, Wang XW. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45((W1)):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 14.Fan W, Ye G. Microarray analysis for the identification of specific proteins and functional modules involved in the process of hepatocellular carcinoma originating from cirrhotic liver. Mol Med Rep. 2018;17:5619–5626. doi: 10.3892/mmr.2018.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulubova MV. Expression of cell adhesion molecules, their ligands and tumour necrosis factor alpha in the liver of patients with metastatic gastrointestinal carcinomas. Histochem J. 2002;34:67–77. doi: 10.1023/A:1021304227369. [DOI] [PubMed] [Google Scholar]
- 16.Ploverini P. Cellular adhesion molecules; newly identified mediators of angiogenesis. Am J Pathol. 1996;148:1023–1029. [PMC free article] [PubMed] [Google Scholar]
- 17.McGrogan D, Bookstein R. Tumor suppressor genes in prostate cancer. Semin Cancer Biol. 1997;8:11–19. doi: 10.1006/scbi.1997.0048. [DOI] [PubMed] [Google Scholar]
- 18.Folkman J. Tumor angiogenesis: Therapeutic implications. N Engl J Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 19.Folkman J. What is the evidence that tumors are angiogenesis dependent? J Natl Cancer Inst. 1990;82:4–6. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]
- 20.Akiyama SK, Larjava H, Yamada KM. Differences in the biosynthesis and localization of the fibronectin receptor in normal and transformed cultured human cells. Cancer Res. 1990;50:1601–1607. [PubMed] [Google Scholar]
- 21.Koop S, Schmidt EE, MacDonald IC, Morris VL, Khokha R, Grattan M, Leone J, Chambers AF, Groom AC. Independence of metastatic ability and extravasation: Metastatic ras-transformed and control fibroblasts extravasate equally well. Proc Natl Acad Sci USA. 1996;93:11080–11084. doi: 10.1073/pnas.93.20.11080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein CA. Cancer. The metastasis cascade. Science. 2008;321:1785–1787. doi: 10.1126/science.1164853. [DOI] [PubMed] [Google Scholar]
- 23.Chiang AC, Massagué J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Donnell JP, Marsh HM, Sondermann H, Sevier CS. Disrupted hydrogen-bond network and impaired ATPase activity in an Hsc70 cysteine mutant. Biochemistry. 2018;57:1073–1086. doi: 10.1021/acs.biochem.7b01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie W, Zhang L, Jiao H, Guan L, Zha J, Li X, Wu M, Wang Z, Han J, You H. Chaperone-mediated autophagy prevents apoptosis by degrading BBC3/PUMA. Autophagy. 2015;11:1623–1635. doi: 10.1080/15548627.2015.1075688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Wang Q, Lin H, Li S, Sun L, Yang Y. HSP72 and gp96 in gastroenterological cancers. Clin Chim Acta. 2013;417:73–79. doi: 10.1016/j.cca.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 29.Brognard J, Newton AC. PHLiPPing the switch on Akt and protein kinase C signaling. Trends Endocrinol Metab. 2008;19:223–230. doi: 10.1016/j.tem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiao M, Iglehart JD, Pardee AB. Metastatic potential of 21T human breast cancer cells depends on Akt/protein kinase B activation. Cancer Res. 2007;67:5293–5299. doi: 10.1158/0008-5472.CAN-07-0877. [DOI] [PubMed] [Google Scholar]
- 31.Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, Totoki Y, Choi DK, et al. The DNA sequence of human chromosome 21. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- 32.Banerjee D, Nandagopal K. Potential interaction between the GARS-AIRS-GART Gene and CP2/LBP-1c/LSF transcription factor in down syndrome-related Alzheimer disease. Cell Mol Neurobiol. 2007;27:1117–1126. doi: 10.1007/s10571-007-9217-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng BG, Wolfe LA, Ichikawa M, Markello T, He M, Tifft CJ, Gahl WA, Freeze HH. Biallelic mutations in CAD, impair de novo pyrimidine biosynthesis and decrease glycosylation precursors. Hum Mol Genet. 2015;24:3050–3057. doi: 10.1093/hmg/ddv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC. The evolution of compositionally and functionally distinct actin filaments. J Cell Sci. 2015;128:2009–2019. doi: 10.1242/jcs.165563. [DOI] [PubMed] [Google Scholar]
- 35.Guo C, Liu S, Wang J, Sun MZ, Greenaway FT. ACTB in cancer. Clin Chim Acta. 2013;417:39–44. doi: 10.1016/j.cca.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 36.Cappell SD, Chung M, Jaimovich A, Spencer SL, Meyer T. Irreversible APC (Cdh1) inactivation underlies the point of no return for cell-cycle entry. Cell. 2016;166:167–180. doi: 10.1016/j.cell.2016.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beavon IR. The E-cadherin-catenin complex in tumour metastasis: Structure, function and regulation. Eur J Cancer. 2000;36:1607–1620. doi: 10.1016/S0959-8049(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 38.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 39.Alloatti G, Montrucchio G, Lembo G, Hirsch E. Phosphoinositide 3-kinase gamma: Kinase-dependent and -independent activities in cardiovascular function and disease. Biochem Soc Trans. 2004;32:383–386. doi: 10.1042/bst0320383. [DOI] [PubMed] [Google Scholar]
- 40.Rubio I, Wittig U, Meyer C, Heinze R, Kadereit D, Waldmann H, Downward J, Wetzker R. Farnesylation of Ras is important for the interaction with phosphoinositide 3-kinase gamma. Eur J Biochem. 1999;266:70–82. doi: 10.1046/j.1432-1327.1999.00815.x. [DOI] [PubMed] [Google Scholar]
- 41.Uehara M, McGrath MM, Ohori S, Solhjou Z, Banouni N, Routray S, Evans C, DiNitto JP, Elkhal A, Turka LA, et al. Regulation of T cell alloimmunity by PI3Kγ and PI3Kδ. Nat Commun. 2017;8:951. doi: 10.1038/s41467-017-00982-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Concetti J, Wilson CL. NFKB1 and cancer: Friend or foe? Cells. 2018;7:E133. doi: 10.3390/cells7090133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Philip S, Bulbule A, Kundu GC. Matrix metalloproteinase-2: Mechanism and regulation of NF-κB-mediated activation and its role in cell motility and ECM-invasion. Glycoconj J. 2004;21:429–441. doi: 10.1007/s10719-004-5533-7. [DOI] [PubMed] [Google Scholar]
- 44.Bharti AC, Aggarwal BB. Nuclear factor-kappa B and cancer: Its role in prevention and therapy. Biochem Pharmacol. 2002;64:883–888. doi: 10.1016/S0006-2952(02)01154-1. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Shen Y, Wang S, Shen Q, Zhou X. The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett. 2018;415:117–128. doi: 10.1016/j.canlet.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caetano MS, Hassane M, Van HT, Bugarin E, Cumpian AM, McDowell CL, Cavazos CG, Zhang H, Deng S, Diao L, et al. Sex specific function of epithelial STAT3 signaling in pathogenesis of K-ras mutant lung cancer. Nat Commun. 2018;9:4589. doi: 10.1038/s41467-018-07042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Lee J, Liem D, Ping P. HSPA5 gene encoding Hsp70 chaperone BiP in the endoplasmic reticulum. Gene. 2017;618:14–23. doi: 10.1016/j.gene.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arap MA, Lahdenranta J, Mintz PJ, Hajitou A, Sarkis AS, Arap W, Pasqualini R. Cell surface expression of the stress response chaperone GRP78 enables tumor targeting by circulating ligands. Cancer Cell. 2004;6:275–284. doi: 10.1016/j.ccr.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 49.Shuda M, Kondoh N, Imazeki N, Tanaka K, Okada T, Mori K, Hada A, Arai M, Wakatsuki T, Matsubara O, et al. Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: A possible involvement of the ER stress pathway in hepatocarcinogenesis. J Hepatol. 2003;38:605–614. doi: 10.1016/S0168-8278(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 50.Um E, Oh JM, Granick S, Cho YK. Cell migration in microengineered tumor environments. Lab Chip. 2017;17:4171–4185. doi: 10.1039/C7LC00555E. [DOI] [PubMed] [Google Scholar]
- 51.Condeelis J, Pollard JW. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y, Zheng H, Zhan Y, Fan S. An emerging tumor invasion mechanism about the collective cell migration. Am J Transl Res. 2019;11:5301–5312. [PMC free article] [PubMed] [Google Scholar]
- 53.Denisenko TV, Gorbunova AS, Zhivotovsky B. Mitochondrial involvement in migration, invasion and metastasis. Front Cell Dev Biol. 2019;7:355. doi: 10.3389/fcell.2019.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li X, Miao Y, Pal DS, Devreotes PN. Excitable networks controlling cell migration during development and disease. Semin Cell Dev Biol. 2019 Dec 10; doi: 10.1016/j.semcdb.2019.11.001. (Epub ahead of print). doi: 10.1016/j.semcdb.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mishra AK, Campanale JP, Mondo JA, Montell DJ. Cell interactions in collective cell migration. Development. 2019;146:dev172056. doi: 10.1242/dev.172056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greenlee JD, King MR. Engineered fluidic systems to understand lymphatic cancer metastasis. Biomicrofluidics. 2020;14:011502. doi: 10.1063/1.5133970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Misra UK, Payne S, Pizzo SV. The monomeric receptor binding domain of tetrameric; α2-macroglobulin binds to cell surface GRP78 triggering equivalent; activation of signaling cascades. Biochemistry. 2013;52:4014–4025. doi: 10.1021/bi400376s. [DOI] [PubMed] [Google Scholar]
- 58.Khosla R, Hemati H, Rastogi A, Ramakrishna G, Sarin SK, Trehanpati N. MiR-26b-5p helps in EpCAM+cancer stem cells maintenance via HSC71/HSPA8 and augments malignant features in HCC. Liver Int. 2019;39:1692–1703. doi: 10.1111/liv.14188. [DOI] [PubMed] [Google Scholar]
- 59.Xiong Y, Wu S, Yu H, Wu J, Wang Y, Li H, Huang H, Zhang H. MiR-190 promotes HCC proliferation and metastasis by targeting PHLPP1. Exp Cell Res. 2018;371:185–195. doi: 10.1016/j.yexcr.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 60.Goh WW, Lee YH, Ramdzan ZM, Chung MC, Wong L, Sergot MJ. A network-based maximum link approach towards MS identifies potentially important roles for undetected ARRB1/2 and ACTB in liver cancer progression. Int J Bioinform Res Appl. 2012;8:155–170. doi: 10.1504/IJBRA.2012.048967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song W, Gu Y, Lu S, Wu H, Cheng Z, Hu J, Qian Y, Zheng Y, Fan H. LncRNA TRERNA1 facilitates hepatocellular carcinoma metastasis by dimethylating H3K9 in the CDH1 promoter region via the recruitment of the EHMT2/SNAI1 complex. Cell Prolif. 2019;52:e12621. doi: 10.1111/cpr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson CL, Jurk D, Fullard N, Banks P, Page A, Luli S, Elsharkawy AM, Gieling RG, Chakraborty JB, Fox C, et al. NFκB1 is a suppressor of neutrophil-driven hepatocellular carcinoma. Nat Commun. 2015;6:6818. doi: 10.1038/ncomms7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 64.King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- 65.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the present study are available in the [NCBI] repository, [https://www.ncbi.nlm.nih.gov/geo].