Abstract
Long non-coding RNA (lncRNA) AGAP2-AS1 has been reported to be a potential biomarker for a variety of cancer types, while its function in clear cell renal carcinoma (ccRCC) has not yet been fully determined. The current study aimed to determine the value of lncRNA AGAP2-AS1 in ccRCC based on The Cancer Genome Atlas (TCGA) database. The association between AGAP2-AS1 expression and associated clinical characters were analyzed using the Wilcoxon signed-rank test and logistic regression. The diagnostic value of AGAP2-AS1 expression in ccRCC tissue was assessed using receiver operating characteristic (ROC) curve analysis. Clinicopathological characteristics associated with overall survival in patients with TCGA were analyzed using Cox regression and the Kaplan-Meier method. Gene set enrichment analysis (GSEA) was also performed to assess the biological function of AGAP2-AS1. The results demonstrated that increased expression of AGAP2-AS1 in ccRCC was significantly associated with male, T3/T4, lymph node metastasis, distant metastasis and high tumor stage (III/IV; all, P<0.05). The area under the ROC curve (normal vs. all tumors) was revealed to be 0.891. Kaplan-Meier survival analysis indicated that ccRCC with high lncRNA AGAP2-AS1 exhibited a worse prognosis compared with low AGAP2-AS1 (P<0.001). The univariate analysis revealed that high expression of AGAP2-AS1 was significantly associated with poor overall survival [hazard ratio (HR). 1.85; 95% confidence interval (CI), 1.48–2.33; P<0.001). Multivariate analysis revealed that AGAP2-AS1 remained independently associated with overall survival, with a HR of 1.57 (CI, 1.21–2.03; P<0.01). GSEA outcome demonstrated that stromal stimulation, angiogenesis, epithelial to mesenchymal transition, basal cell carcinoma, ECM receptor interaction and the Notch signaling pathway were differentially enriched in the AGAP2-AS1 high expression phenotype. Therefore, the high expression of AGAP2-AS1 may be an independent predictor of poor survival in patients with ccRCC.
Keywords: long non-coding RNA AGAP2-AS1, clear cell renal carcinoma, receiver operating characteristic, independent predictor, gene set enrichment analysis
Introduction
Renal cell carcinoma (RCC), is among the most common urological tumor, and accounts for 3.8% of adult malignancies in the United States (1). Clear cell renal cell carcinoma (ccRCC), which is the most common RCC subtype, associates with early distant metastasis (2). Accumulating knowledge and scientific research have demonstrated that localized ccRCC (even partly advanced stages) may be curable, however, up to 17% of patients with ccRCCs exhibit distant metastases at time of diagnosis (3,4). For patients with distant metastasis, chemotherapy and radiotherapy are common treatments. However, not all patients respond to these treatments, and the long-term survival rate for patients with ccRCC and distant metastasis remains poor (5). The poor long-term survival rate has been associated with late detection of advanced stage (6–8). Therefore, the determination of novel diagnostic and prognostic biomarkers for this disease is required.
LncRNA AGAP2-AS1 (AGAP2 Antisense 1), which is also known as PUNISHER ENSG00000255737, is located at 12q14.1 and has been identified to be associated with a variety of cancer types, including non-small cell lung cancer (9,10) and malignant glioma (11). Recent research has demonstrated that the overexpression of AGAP2-AS1 occurs in breast cancer compared with paired adjacent noncancerous tissues, and promotes cell growth and trastuzumab resistance (12). In pancreatic cancer, AGAP2-AS1 was indicated to be associated with highly metastatic tumor characters by recruiting zeste homolog 2 (13). Additionally, further studies have revealed that the upregulated expression of AGAP2-AS1 markedly correlates with clinical features in hepatocellular carcinoma (HCC) and promotes the effects of hypoxia on metastasis and EMT (14). AGAP2-AS1 has also been detected in tissues samples of patients with gastric cancer and gastric cancer cell lines, which suggests that AGAP2-AS1 may be a potential prognostic biomarker (15). However, few studies have reported the expression of AGAP2-AS1 and the prognosis of ccRCC until recently.
The aim of the current study was to assess the correlation between the expression of AGAP2-AS1 and ccRCC based on data obtained from The Cancer Genome Atlas (TCGA). Transcriptomes and clinical documents of 539 patients with ccRCC were downloaded from TCGA and the expression of AGAP2-AS1 was analyzed with clinical characters of ccRCC. Furthermore, the biological pathways associated with ccRCC were determined using gene set enrichment analysis (GSEA).
The results of the current study demonstrated that the upregulated expression of AGAP-AS1 was associated with poor prognosis of ccRCC, and that epithelial mesenchymal transition (EMT), angiogenesis, notch pathway, ECM receptor interaction, stromal stimulation, basal cell carcinoma and the high recurrence of bladder cancer were associated with AGAP2-AS1 expression.
Materials and methods
RNA-sequencing patient data and bioinformatics analysis
The gene expression data (total of 611 samples; 539 ccRCC samples and 72 samples of normal adjacent noncancerous tissues; workflow type: HTSeq-FPKM) and corresponding clinical information were downloaded from TCGA. The expression of AGAP2-AS1 in ccRCC was analyzed and compared with adjacent healthy tissues. The characteristics of patients, including age, gender, grade, clinical stage and TNM stage, were recorded. Some data were not available, so these were considered to be missing values.
GSEA
In the present study, according to the expression of AGAP2-AS1, all cases were divided into the high-AGAP2-AS1 expression group and low-AGAP2-AS1 expression group, then GSEA was subsequently performed to assess the significant survival difference that was observed between high- and low-AGAP2-AS1 groups. Gene set permutations were performed 1,000 times for each analysis. The expression of AGAP2-AS1 was used as a phenotype label. The nominal P-value and normalized enrichment score (NES) were used to analyze the enriched pathways.
Statistical analysis
All data were conducted using R (v.3.5.3; http://cran.r-project.org/). The comparison of the expression of AGAP2-AS1 between ccRCC and normal groups was performed using Wilcoxon rank sum tests, ccRCC and adjacent groups were analyzed using Wilcoxon signed-rank tests. Subjects were divided into two groups: Gene expression above the median value vs. subjects with gene expression below the median value. The relationship between AGAP2-AS1 and age, gender, M classification, N classification were analyzed using the Wilcoxon rank sum test, AGAP2-AS1 and T classification, stage, grade was used Kruskal-Wallis test. Clinicopathological characteristics associated with OS in patients with AGAP2-AS1 were assessed using Cox regression and the Kaplan-Meier method. Multivariate Cox analysis was used to compare the influence of AGAP2-AS1 expression on survival and other clinical characteristics.
Results
Patient characteristics
The clinical characteristics of patients, including age, gender, grade, TNM stage and clinical stage, were collected and are presented in Table I. A total of 191 female patients and 346 male patients were included in the current study. A total of 173 patients were aged >55 year old (32.52%) and 359 patients were aged >=55 years old (67.48%). Clinical stage included stage I (269; 50.37%), stage II (57; 10.67%), stage III (125; 23.41%) and stage IV (83; 15.55%). The topography distribution included T1 (275; 51.21%), T2 (69; 12.85%), T3 (182; 33.89%) and T4 (11; 2.05%). A total of 240 patients (93.39%) exhibited no lymph node metastases. A total of 79 (15.64%) patients had distant metastases. A number of case files of lymph node metastases and distant metastases were not available, and these were treated as missing cases.
Table I.
Characteristics of patients with clear cell renal carcinoma based on The Cancer Genome Atlas database.
| Clinical characteristics | Nο. of cases | Percentage |
|---|---|---|
| Topography | ||
| T1 | 275 | 51.21 |
| T2 | 69 | 12.85 |
| T3 | 182 | 33.89 |
| T4 | 11 | 2.05 |
| Lymph node | ||
| N0 | 240 | 93.39 |
| N1 | 17 | 6.61 |
| Metastasis | ||
| M0 | 426 | 84.36 |
| M1 | 79 | 15.64 |
| Stage | ||
| I | 269 | 50.37 |
| II | 57 | 10.67 |
| III | 125 | 23.41 |
| IV | 83 | 15.55 |
| Age, years | ||
| <55 | 173 | 32.52 |
| ≥55 | 359 | 67.48 |
| Sex | ||
| Female | 191 | 35.57 |
| Male | 346 | 64.43 |
AGAP2-AS1 were highly expressed in renal tissues
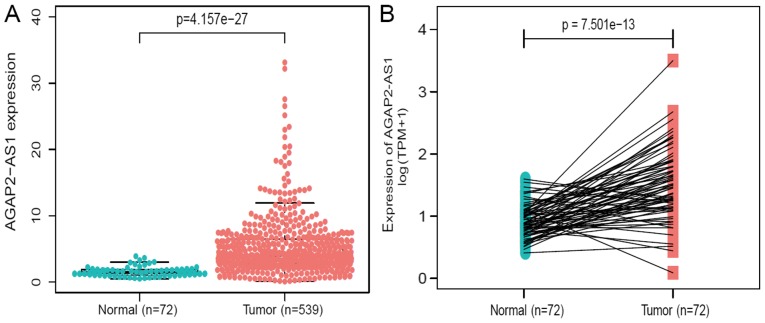
The expression of AGAP2-AS1 was detected in 539 ccRCC tissues and 72 adjacent healthy tissues using Wilcoxon rank sum test. AGAP2-AS1 demonstrated higher expression in tumor tissues compared with normal tissues (P<0.001; Fig. 1A). Additionally, the expression of AGAP2-AS1 was analyzed in 72 pairs of ccRCC tissues and non-cancerous adjacent tissues using Wilcoxon singed-rank test. The results indicated that AGAP2-AS1 was significantly overexpressed in ccRCC tumors (P<0.001; Fig. 1B), indicating AGAP2-AS1 may be associated with ccRCC carcinogenesis.
Figure 1.
lncRNA AGAP2-AS1 is significantly upregulated in clear cell renal carcinoma compared with in normal or adjacent normal tissues. (A) AGAP2-AS1 exhibited significantly higher expression in cancer tissues than in normal tissues (P<0.001). (B) AGAP2-AS1 was prominently overexpressed in clear cell renal carcinoma (P<0.001) compared with in 72 pairs of non-cancerous adjacent tissues according to a Wilcoxon single rank test. lncRNA, long non-coding RNA.
Correlation between the expression of AGAP2-AS1 and clinical characteristics in patients with ccRCC
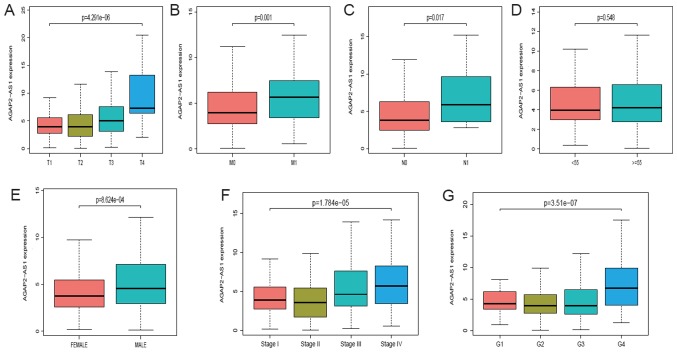
A total of 539 ccRCC samples with AGAP2-AS1 expression data were analyzed from TCGA. Upregulated expression of AGAP2-AS1 was significantly correlated with clinical stage (P<0.001; Fig. 2A), the grade of topography distribution (P<0.001; Fig. 2B), topography distribution (P<0.001; Fig. 2C), gender (P<0.001; Fig. 2D), distance metastasis (P<0.001; Fig. 2E) and lymph node metastasis (P<0.001; Fig. 2F). Univariate analysis revealed that increased AGAP2-AS1 expression (based on median value) was associated with poor prognostic clinicopathologic characteristics using logistic regression (Table II). Upregulated AGAP2-AS1 expression in ccRCC was significantly associated with TNM stage (OR, 1.75 for T3/T4 vs. T1/T2; P=0.002; OR, 3.23 for N1 vs. N0; P=0.047; OR, 1.98 for M1 vs. M0; P=0.007) and stage (OR, 1.77 for stage III/IV vs. stage I/II; P=0.001).
Figure 2.
Association between lncRNA AGAP2-AS1 expression and clinicopathologic characteristics. (A) Expression of AGAP2-AS1 grouped by T classification. (B) Expression of AGAP2-AS1 grouped by M classification. (C) Expression of AGAP2-AS1 grouped by N classification. (D) Expression of AGAP2-AS1 grouped by age. (E) Expression of AGAP2-AS1 grouped by sex. (F) Expression of AGAP2-AS1 grouped by clinical stage. (G) Expression of AGAP2-AS1 grouped by histologic grade. lncRNA, long non-coding RNA; T, topography distribution; N, lymph node metastasis; M, distant metastasis.
Table II.
lncRNA AGAP2-AS1 expression is associated with clinicopathological characteristics (logistic regression).
| Clinical characteristics | Total, n | Odds ratio (95% CI) | P-value |
|---|---|---|---|
| Age (≥55 vs. <55) | 530 | 1.16 (0.81–1.68) | 0.405 |
| Sex (male vs. female) | 530 | 1.83 (1.27–2.63) | 0.001 |
| T (T3/T4 vs. T1/T2) | 530 | 1.75 (1.23–2.52) | 0.002 |
| N (N1 vs. N0) | 255 | 3.23 (1.09–11.8) | 0.047 |
| M (M1 vs. M0) | 498 | 1.98 (1.21–3.31) | 0.007 |
| Grade (G3/G4 vs. G2/G1) | 521 | 1.33 (0.94–1.88) | 0.104 |
| Stage (III/IV vs. I/II) | 527 | 1.77 (1.24–2.52) | 0.001 |
Diagnostic value of AGAP2-AS1 expression in clear cell renal carcinoma
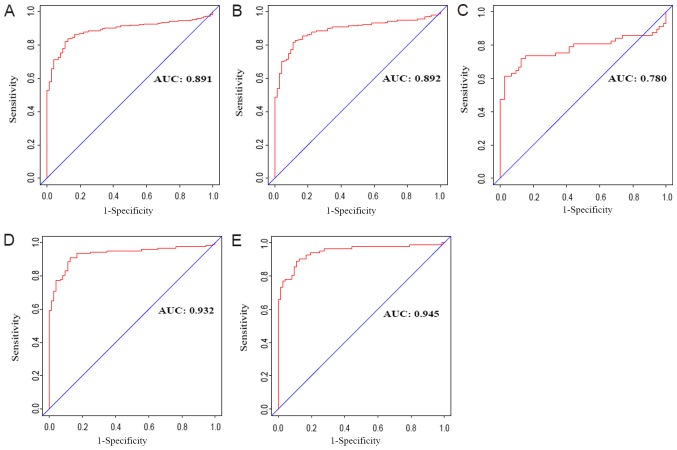
To assess the diagnostic value of AGAP2-AS1 in ccRCC, a ROC curve analysis was performed by testing the expression stage between patients with ccRCC and healthy, adjacent cases (Fig. 3A). The area under the ROC curve (AUC) was 0.891, which indicated an excellent diagnostic value. Subgroup analysis demonstrated the diagnostic value of AGAP2-AS1 expression in different stages of ccRCC, with AUC values of 0.892 for clinical stage I, 0.780 for clinical stage II, 0.932 for clinical stage III and 0.945 for clinical stage IV (Fig. 3B-3E).
Figure 3.
Diagnostic value of AGAP2-AS1expression in clear cell renal carcinoma. (A) Receiver operating characteristic curve for AGAP2-AS1 expression in normal renal tissue and clear cell carcinoma. (B) Normal vs. tumor in stage I. (C) Normal vs. tumor in stage II. (D) Normal vs. tumor in stage III. (E) Normal vs. tumor in stage IV. AUC, area under the curve.
Survival curve, univariate and multivariate analysis of AGAP2-AS1 in ccRCC
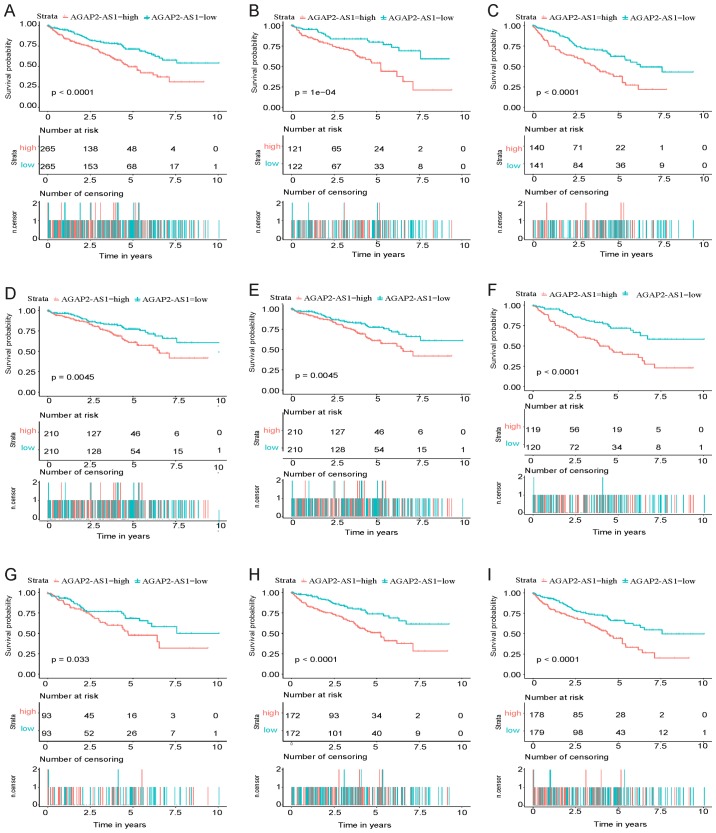
As presented in Fig. 4A, Kaplan-Meier survival analysis indicated that high-AGAP2-AS1 in ccRCC exhibited a worse prognosis compared with low-AGAP2-AS1 (P<0.001). The univariate analysis, which was performed using a Cox regression, revealed that high AGAP2-AS1 correlated significantly with poor OS [hazard ratio (HR), 1.85; 95% confidence interval (CI), 1.48–2.33; P<0.001]. Other clinicopathological variables were also indicated to be associated with poor survival included age, advanced stage and TNM stage (Table III).
Figure 4.
Kaplan-Meier curves for overall survival rate in clear cell renal carcinoma. (A) Kaplan-Meier curves for all cases. (B) Kaplan-Meier curves for stage G1/G2. (C) Kaplan-Meier curves for stage G3/G4. (D) Kaplan-Meier curves for clinical stage III/IV. (E) Kaplan-Meier curves for M0 and N0. (F) Kaplan-Meier curves for N0. (G) Kaplan-Meier curves for female. (H) Kaplan-Meier curves for male. (I) Kaplan-Meier curves for age.
Table III.
Univariate regression and multivariate survival model of prognostic covariates in patients with clear cell renal carcinoma (Cox regression).
| Clinicopathologic variables | Hazard ratio (95% CI) | P-value |
|---|---|---|
| Univariate analysis | ||
| Age (≥55 vs. <55) | 1.58 (0.98–2.56) | 0.060 |
| Sex (male vs. female) | 1.01 (0.66–1.54) | 0.951 |
| T (T3/T4 vs. T1/T2) | 1.82 (1.47–2.25) | <0.001 |
| N (N1 vs. N0) | 2.93 (1.52–5.67) | 0.001 |
| M (M1 vs. M0) | 4.07 (2.63–6.30) | <0.001 |
| Grade (G3/G4 vs. G1/G2) | 1.62 (1.29–2.04) | <0.001 |
| Stage (III/IV vs. I/II) | 1.92 (1.54–2.34) | <0.001 |
| AGAP2-AS1 | 1.85 (1.48–2.33) | <0.001 |
| Multivariate analysis | ||
| T (T3/T4 vs. T1/T2) | 1.41 (0.62–3.25) | 0.412 |
| N (N1 vs. N0) | 1.24 (0.62–2.49) | 0.548 |
| M (M1 vs. M0) | 2.22 (1.32–3.73) | 0.003 |
| Grade (G3/G4 vs. G1/G2) | 1.67 (1.01–2.75) | 0.045 |
| Stage (III/IV vs. I/II) | 1.48 (0.58–3.75) | 0.409 |
| AGAP2-AS1 | 1.57 (1.21–2.03) | 0.001 |
These results indicated ccRCC with increased AGAP2-AS1 expression correlated with the development into a more advanced stage (grade 3/4), lymph node metastasis and distance. Multivariate analysis was subsequently performed (Table III). The results demonstrated that high expression of AGAP2-AS1 was associated with poor OS in patients with ccRCC and a high HR.
AGAP2-AS1-associated biological pathways identified by GSEA
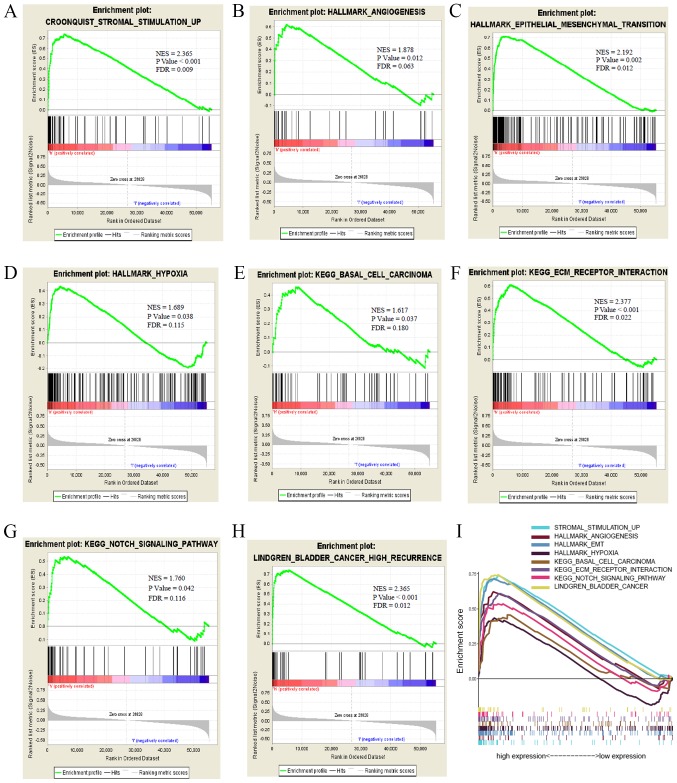
GSEA was performed to identify biological pathways in ccRCC by comparing the aberrant AGAP2-AS1 expression data sets. The significant differences (FDR <0.25; NOM P<0.05) in enrichment of MSigDB Collection (including h.all.v6.2.symbols.gmt and c2.cp.kegg.v6.2.symbols.gmt). A total of 7 critical biological pathways, including stromal stimulation, angiogenesis, epithelial to mesenchymal transition, basal cell carcinoma, ECM receptor interaction and the Notch signaling pathway indicated significantly differential enrichment in AGAP2-AS1 high expression phenotype based on NES, NOM P-value and FDR value (Figs. 5A–5I; Table IV), indicating the potential role of AGAP2-AS1 in the development of ccRCC.
Figure 5.
Enrichment plots from GSEA. GSEA results showing (A) stromal stimulation, (B) angiogenesis, (C) epithelial to mesenchymal transition, (D) hypoxia, (E) basal cell carcinoma, (F) ECM receptor interaction, (G) Notch signaling pathway, (H) bladder cancer high recurrence and (I) summary of GSEA. KEGG, Kyoto Encyclopedia of Genes and Genomes; NES, normalized enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis.
Table IV.
Gene sets enriched in phenotype high vs. low of AGAP2-AS1.
| MSigDB collection | Gene xet name | NES | NOM P-value | FDR q-value |
|---|---|---|---|---|
| h.all.v6.2.symbols.gmt | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION_6 | 2.192 | 0.002 | 0.011 |
| HALLMARK_HYPOXIA | 1.688 | 0.038 | 0.115 | |
| HALLMARK_ANGIOGENESIS | 1.877 | 0.012 | 0.063 | |
| c2.cp.kegg.v6.2.symbols.gmt | KEGG_ECM_RECEPTOR_INTERACTION | 2.077 | <0.001 | 0.021 |
| KEGG_NOTCH_SIGNALING_PATHWAY | 1.760 | 0.043 | 0.116 | |
| KEGG_BASAL_CELL_CARCINOMA | 1.617 | 0.037 | 0.180 | |
| c2.cgp.v6.2.symbols.gmt | LINDGREN_BLADDER_CANCER_HIGH_RECURRENCE | 2.367 | <0.001 | 0.012 |
| JECHLINGER_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_UP | 2.215 | <0.001 | 0.020 |
Gene sets with NOM P-value <0.05 and FDR q-value <0.25 were considered as significantly enriched. FDR, false discovery rate; NES, normalized enrichment score; NOM, normalized.
Discussion
ccRCC is a common subtype of kidney cancer, however, up to 17% of patients with ccRCCs present with primary metastatic disease (3). Therefore, the prognosis for patients with ccRCC and distance metastasis remains difficult to predict (16). lncRNAs form several groups of molecules that influence the gene expression of protein-coding genes in different ways. Currently, a large number of lncRNAs are increasingly considered to be associated with the recurrence and prognosis of cancer (17,18). lncRNAs exhibit the potential to be used as therapeutic targets and prognostic biomarkers. For example, lncRNA SNHG6-003 may function as a sponge for miR-26a/b to promote the progression of hepatocellular carcinoma (19). lncRNA LUCAT1 has been indicated to promote proliferation and invasion in ccRCC cells via the AKT/GSK-3β signaling pathway (20). lncRNA AGAP2-AS1 ectopic expression has been indicated in numerous types of carcinoma, including in hepatocellular carcinoma (14), non-small cell lung cancer (9,10), gastric cancer (15) and glioma (11). Furthermore, AGAP2-AS1 has been indicated to be co-expressed with HDGF (21) and ANGPTL4 (13), which are associated with tumor angiogenesis. However, few studies have reported the expression of AGAP2-AS1 and the effect of its expression on ccRCC prognosis. Therefore, in the current study, to determine the clinicopathological and prognostic value of AGAP2-AS1 in ccRCC, the mRNA expression of AGAP2-AS1 in ccRCC was assessed through bioinformatics analysis of data from TCGA database. Receiver operating characteristic (ROC) curve analysis, GSEA and other methods were also used to assess the diagnostic and prognostic value of AGAP2-AS1.
In the present study, high throughput RNA sequencing data of ccRCC were downloaded from the TCGA database, and the outcomes demonstrated that high lncRNA AGAP2-AS1 was significantly associated with worse survival status. Kaplan-Meier curves for OS also indicated that higher expression of lncRNA AGAP2-AS1 was associated with different genders, levels of ages, high clinical stage, and advanced TNM stage in patients with ccRCC. Logistic analyses, univariate and multivariate Cox analyses indicated that AGAP2-AS1 expression may be a potential indicator for ccRCC prognosis, and ROC analysis affirmed the diagnostic value of AGAP2-AS1 expression in ccRCC.
AGAP2-AS1, as a non-coding RNA, has been demonstrated to promote anaplastic glioma cells proliferation, migration and invasion, and the knockdown of AGAP2-AS1 has been indicated to increase apoptosis cell rates (11). Similar outcomes have been revealed in the human metastatic pancreatic cancer cell line AsPC-1 (13), and in non-small cell lung cancer cell lines A549 and SK-MES-1 (10). Consistent with previous studies, the results of the current study demonstrated that increased AGAP2-AS1 expression was associated with poor overall survival and the potential mechanisms governing this may be the connection with stromal simulation (22,23), angiogenesis (24,25), epithelial-mesenchymal transition (26), hypoxia (27) or the notch signaling pathway (28,29). Angiogenesis, epithelial-mesenchymal transition and hypoxia are well-known hallmarks of cancers, and angiogenesis serves an important role in the progression of ccRCC via VEGF (30), FGF-2, PDGF, angiopoietins, ephrins, apelin (APLN) and chemokines (31,32). In the present study, the aberrant expression of AGAP2-AS1 was enriched in the process of angiogenesis, which may be activated by the PI3K/Akt pathway (33). These previous studies indicated that lncRNA AGAP2-AS1 may function via the VEGF and Akt pathway, which could be used as a drug target for ccRCC. Furthermore, the prognostic value of AGAP2-AS1 expression was examined in different subgroups of ccRCC and it was indicated that high lncRNA AGAP2-AS1 expression was significantly associated with G1/G2, stage I/II and M0 cases, highlighting the potential value of AGAP2-AS1 in the development of ccRCC.
Currently, surgery is the most common treatment for ccRCC. However, the possibility of recurrence, which adversely impacts patient outcomes, is an important factor in the choice of treatment (34). The current study also assessed the correlation between AGAP2-AS1 expression and a number of clinicopathological characters, and the results revealed that this potential biomarker may help to guide treatment selection in patients with ccRCC. High expression of AGAP2-AS1 also negatively affected OS among patients with histological grade G1/G2, grade G3/G4 and clinical stage III/IV; but not patients with histological clinical stage I/II, which further demonstrated the specific prognostic role of AGAP2-AS1 expression in subgroup analysis and its potential contribution to precision therapy for ccRCC.
Although the results in the present study provided information regarding the relationship between AGAP2-AS1 and ccRCC, there were some limitations to the present study. All clinical factors should have been considered, including BMI, the details on treatments received by patients involved, smoking status and other biomarkers' levels. However, this information is often missing, or inconsistent treatments are stated in public databases. Only a total of 72 healthy samples and 539 ccRCC samples were evaluated in the current study which may limit the present work. Therefore, the sample size should be increased in future study.
Overall, the current study demonstrated the diagnostic and prognostic value of AGAP2-AS1 expression in patients with ccRCC. However, the current study was performed using RNA-seq of TCGA database, which lacks protein level files and direct mechanisms information. Additionally, the number of healthy subjects and information on later stages are limited. Therefore, further identification of effective biomarkers in ccRCC cases of advanced stage is required in the future.
In conclusion, we observed that lncRNA AGAP2-AS1 is up-regulated in ccRCC, which also correlates with clinical progression and serves as an independent risk factor for OS in ccRCC. Our findings partily demonstrated that lncRNA AGAP2-AS1 may be a potential biomarker in the diagnosis and prognosis of ccRCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Program of Graduate students innovation fund of Hebei Province (grant no. CXZZBS2019117).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
LG conceived the study and was the major contributor in writing the manuscript. AZ participated in its design, analyzed and interpreted the data. XW participated in design, visualization and critically revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387:894–906. doi: 10.1016/S0140-6736(15)00046-X. [DOI] [PubMed] [Google Scholar]
- 4.Gawlik-Jakubczak T, Matuszewski M, Biernat W. The metastasis of renal cell carcinoma to the urethra and local tumor recurrence. Urol Int. doi: 10.1159/000501699. doi: 10.1159/000501699. [DOI] [PubMed] [Google Scholar]
- 5.Yu L, Xiang L, Feng J, Li B, Zhou Z, Li J, Lin Y, Lv Y, Zou D, Lei Z, et al. miRNA-21 and miRNA-223 expression signature as a predictor for lymph node metastasis, distant metastasis and survival in kidney renal clear cell carcinoma. J Cancer. 2018;9:3651–3659. doi: 10.7150/jca.27117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blandin Knight S, Crosbie PA, Balata H, Chudziak J, Hussell T, Dive C. Progress and prospects of early detection in lung cancer. Open Biol. 2017;7:170070. doi: 10.1098/rsob.170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivera-Franco MM, Leon-Rodriguez E. Delays in breast cancer detection and treatment in developing countries. Breast Cancer (Auckl) 2018;12:1178223417752677. doi: 10.1177/1178223417752677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li P, Wong YN, Armstrong K, Haas N, Subedi P, Davis-Cerone M, Doshi JA. Survival among patients with advanced renal cell carcinoma in the pretargeted versus targeted therapy eras. Cancer Med. 2016;5:169–181. doi: 10.1002/cam4.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan KJ, Liu Y, Yang B, Tian XD, Li CR, Wang B. Prognostic and diagnostic significance of long non-coding RNA AGAP2-AS1 levels in patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2017;21:2392–2396. [PubMed] [Google Scholar]
- 10.Li W, Sun M, Zang C, Ma P, He J, Zhang M, Huang Z, Ding Y, Shu Y. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 2016;7:e2225. doi: 10.1038/cddis.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Yang F, Zhang L, Chen J, Zhao Z, Wang H, Wu F, Liang T, Yan X, Li J, et al. LncRNA profile study reveals four-lncRNA signature associated with the prognosis of patients with anaplastic gliomas. Oncotarget. 2016;7:77225–77236. doi: 10.18632/oncotarget.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong H, Wang W, Mo S, Chen R, Zou K, Han J, Zhang F, Hu J. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J Exp Clin Cancer Res. 2018;37:202. doi: 10.1186/s13046-018-0875-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Hui B, Ji H, Xu Y, Wang J, Ma Z, Zhang C, Wang K, Zhou Y. RREB1-induced upregulation of the lncRNA AGAP2-AS1 regulates the proliferation and migration of pancreatic cancer partly through suppressing ANKRD1 and ANGPTL4. Cell Death Dis. 2019;10:207. doi: 10.1038/s41419-019-1384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Wang Y, Wang L, Yao B, Sun L, Liu R, Chen T, Niu Y, Tu K, Liu Q. Long non-coding RNA AGAP2-AS1, functioning as a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16-5p and promotes proliferation and metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:194. doi: 10.1186/s13046-019-1188-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Qi F, Liu X, Wu H, Yu X, Wei C, Huang X, Ji G, Nie F, Wang K. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J Hematol Oncol. 2017;10:48. doi: 10.1186/s13045-017-0420-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motzer RJ, Molina AM. Targeting renal cell carcinoma. J Clin Oncol. 2009;27:3274–3276. doi: 10.1200/JCO.2009.21.8461. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Wu Z, Fu X, Han W. Long noncoding RNAs: Insights from biological features and functions to diseases. Med Res Rev. 2013;33:517–553. doi: 10.1002/med.21254. [DOI] [PubMed] [Google Scholar]
- 18.Seles M, Hutterer GC, Kiesslich T, Pummer K, Berindan-Neagoe I, Perakis S, Schwarzenbacher D, Stotz M, Gerger A, Pichler M. Current Insights into Long Non-Coding RNAs in Renal Cell Carcinoma. Int J Mol Sci. 2016;17:573. doi: 10.3390/ijms17040573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao C, Zhang T, Zhang D, Xie L, Zou X, Lei L, Wu D, Liu L. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene. 2017;36:1112–1122. doi: 10.1038/onc.2016.278. [DOI] [PubMed] [Google Scholar]
- 20.Zheng Z, Zhao F, Zhu D, Han J, Chen H, Cai Y, Chen Z, Xie W. Long non-coding RNA LUCAT1 promotes proliferation and invasion in clear cell renal cell carcinoma through AKT/GSK-3β signaling pathway. Cell Physiol Biochem. 2018;48:891–904. doi: 10.1159/000491957. [DOI] [PubMed] [Google Scholar]
- 21.Zheng Y, Lu S, Xu Y, Zheng J. Long non-coding RNA AGAP2-AS1 promotes the proliferation of glioma cells by sponging miR-15a/b-5p to upregulate the expression of HDGF and activating Wnt/β-catenin signaling pathway. Int J Biol Macromol. 2019;128:521–530. doi: 10.1016/j.ijbiomac.2019.01.121. [DOI] [PubMed] [Google Scholar]
- 22.Chang WK, Carmona-Fontaine C, Xavier JB. Tumour-stromal interactions generate emergent persistence in collective cancer cell migration. Interface Focus. 2013;3:20130017. doi: 10.1098/rsfs.2013.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong M, Cheng H, Song L, Wang W, Wang Q, Xu D, Xing W. Wogonin suppresses the activity of matrix metalloproteinase-9 and inhibits migration and invasion in human hepatocellular carcinoma. Molecules. 2018;23:384. doi: 10.3390/molecules23020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramjiawan RR, Griffioen AW, Duda DG. Anti-angiogenesis for cancer revisited: Is there a role for combinations with immunotherapy? Angiogenesis. 2017;20:185–204. doi: 10.1007/s10456-017-9552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: Alternative therapeutic targets. Angiogenesis. 2017;20:409–426. doi: 10.1007/s10456-017-9562-9. [DOI] [PubMed] [Google Scholar]
- 26.Chen T, You Y, Jiang H, Wang ZZ. Epithelial-mesenchymal transition (EMT): A biological process in the development, stem cell differentiation, and tumorigenesis. J Cell Physiol. 2017;232:3261–3272. doi: 10.1002/jcp.25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manoochehri Khoshinani H, Afshar S, Najafi R. Hypoxia: A double-edged sword in cancer therapy. Cancer Invest. 2016;34:536–545. doi: 10.1080/07357907.2016.1245317. [DOI] [PubMed] [Google Scholar]
- 28.Braune EB, Lendahl U. Notch -- a goldilocks signaling pathway in disease and cancer therapy. Discov Med. 2016;21:189–196. [PubMed] [Google Scholar]
- 29.Li L, Tang P, Li S, Qin X, Yang H, Wu C, Liu Y. Notch signaling pathway networks in cancer metastasis: A new target for cancer therapy. Med Oncol. 2017;34:180. doi: 10.1007/s12032-017-1039-6. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y, Zhou J, Zou D, Hou D, Zhang H, Zhao J, Li L, Hu J, Zhang Y, Jing Z. Overexpression of Limb-Bud and Heart (LBH) promotes angiogenesis in human glioma via VEGFA-mediated ERK signalling under hypoxia. EBioMedicine. 2019;48:36–48. doi: 10.1016/j.ebiom.2019.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lugano R, Ramachandran M, Dimberg A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. https://doi.org/10.1007/s00018-019-03351-7. Cell Mol Life Sci. 2019 doi: 10.1007/s00018-019-03351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chappell JC, Payne LB, Rathmell WK. Hypoxia, angiogenesis, and metabolism in the hereditary kidney cancers. J Clin Invest. 2019;129:442–451. doi: 10.1172/JCI120855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duan MX, Zhou H, Wu QQ, Liu C, Xiao Y, Deng W, Tang QZ. Andrographolide protects against HG-induced inflammation, apoptosis, migration, and impairment of angiogenesis via PI3K/AKT-eNOS signalling in HUVECs. Mediators Inflamm. 2019;2019:6168340. doi: 10.1155/2019/6168340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallardo E, Méndez-Vidal MJ, Pérez-Gracia JL, Sepúlveda-Sánchez JM, Campayo M, Chirivella-González I, García-Del-Muro X, González-Del-Alba A, Grande E, Suárez C. SEOM clinical guideline for treatment of kidney cancer (2017) Clin Transl Oncol. 2018;20:47–56. doi: 10.1007/s12094-017-1765-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.