Abstract
Previous studies have suggested that a variety of tumor driver genetic alterations affected the treatment efficacy of chemotherapy and epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs) in advanced non-small cell lung cancer (NSCLC). The present study aimed to investigate the association between the tumor genetic alteration landscape and the treatment outcome of first-line chemotherapy and EGFR-TKIs in advanced NSCLC. A total of 94 patients with advanced NSCLC were recruited. All patients received first-line chemotherapy and/or EGFR-TKIs (either first- or second-generation EGFR-TKI, or third-generation EGFR-TKI) alone or sequentially. Prior to chemotherapy and/or EGFR-TKI treatment, plasma, effusion and/or tumor tissues from the included patients were subjected to next-generation sequencing, targeting 59 genes. The results indicated that the positive genetic alteration status prior to first-line chemotherapy was associated with prolonged progression-free survival (PFS) time compared with the negative status [9.1 vs. 4.0 months; hazard ratio (HR)=6.68; 95% CI, 2.25–19.82; P=0.001). Furthermore, patients with EGFR activating mutation harboring concomitant alterations exhibited a shorter PFS (11.1 vs. 7.4 months; HR=2.14; 95% CI, 1.03–4.44; P=0.04) and overall survival (OS) time [not reached (NR) vs. 32.8 months; HR=4.30; 95% CI, 1.41–13.16; P=0.01] than those without concomitant alterations, with first- and second-generation EGFR-TKI treatment. Similarly, patients with T79M mutation harboring concomitant alterations exhibited a shorter PFS (15.6 vs. 3.6 months; HR=9.48; 95% CI, 2.29–39.28; P=0.002) and OS time (NR vs. 32.8 months; HR=4.85; 95% CI, 1.16–20.29; P=0.03) with osimertinib treatment. Taken together, the results demonstrated that positive genetic alteration status predicted greater efficacy of first-line chemotherapy, while concomitant genetic alterations were associated with poor treatment outcome for first- or second-generation EGFR-TKI and third-generation EGFR-TKI treatment.
Keywords: NSCLC, tumor genetic alteration profiling, NGS, chemotherapy, EGFR-TKI
Introduction
In advanced non-small cell lung cancer (NSCLC), cytotoxic chemotherapy and epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs) account for a high proportion of the treatment regimens (1). Chemotherapy is one of the most essential treatment strategies for patients without targetable driver gene mutations and is commonly used following targeted drug resistance among patients with targetable driver genes (1). Previously, platinum-based doublet chemotherapy regimens served as the standard first-line treatment; however, these are considered insufficient, with a response rate of 30–40% and a short progression-free survival (PFS) time ranging between 5 and 7 months (2–6). Thus, it is crucial to identify and develop novel markers to predict the efficacy of chemotherapy.
First- and second-generation EGFR-TKIs serve a vital role in the treatment of advanced NSCLC with EGFR-activating mutations, demonstrating superior treatment efficacy in this population compared with traditional chemotherapy (1,7). However, previous clinical trials have reported that the response rate of first- or second-generation EGFR-TKIs is ~70%, and PFS time ranges between 9 and 14 months (7–13). Thus, numerous patients with EGFR-activating mutations gain limited treatment benefits from EGFR-TKIs (7–13). In addition to EGFR-activating mutations, the existing studies suggested that concomitant mutations may affect the therapeutic efficacy of first-generation EGFR-TKIs (14).
It was reported that during progression on first- or second-generation EGFR-TKI treatment, ~60% of patients with initial EGFR-activating mutations will develop T790M resistance mutations (15), while the third-generation EGFR-TKI osimertinib is selective for the T790M mutation. However, the response rate with osimertinib is 60–70%, and PFS time is ~10 months, while some patients with T790M mutations fail to benefit from osimertinib treatment (16–18). Whether concomitant alterations beyond EGFR affect the therapeutic efficacy of the third-generation EGFR-TKI warrants further investigation.
Extensive application of next-generation sequencing (NGS) genotyping in clinical practice and comprehensive genetic analysis have allowed for the simultaneous discovery of diverse genetic alterations from only one sample in NSCLC. However, in addition to the driver genes with targeted agents, the predictive and prognostic values of other mutated or amplified genes still remain contradictory. Thus, utilizing genetic alteration profiling based on NGS, in order to further optimize the selection or combination of targeted therapy and chemotherapy requires further research.
Therefore, in addition to sensitive driver gene status, it is essential to comprehensively take the entire genetic landscape into account in order to optimize therapeutic strategy. The present study aimed to retrospectively analyze the association between the genetic alteration landscape of the tumor and the treatment outcome of different treatment modalities, including chemotherapy, and first-, second- and third-generation EGFR-TKIs, in advanced NSCLC.
Materials and methods
Study design
The present study was approved by The Institutional Review Board of the Affiliated Tumor Hospital of Guangxi Medical University (Nanning, China) and conducted in accordance with The Declaration of Helsinki. Written informed consent was provided by all patients. The inclusion criteria were as follows: i) IIIB/IV NSCLC patients; ii) patients received first-line chemotherapy and/or EGFR-TKIs (either first-, second- or third-generation EGFR-TKIs) alone or sequentially; and iii) specimens from all patients were subjected to NGS genotyping prior to specified treatment. Exclusion criteria were: i) No complete documentation; ii) specified treatment was adjuvant or neoadjuvant therapy. The patients' medical records were retrospectively reviewed to identify eligible patients. A total of 94 patients with advanced NSCLC, diagnosed between June 2007 and May 2018 at Affiliated Tumor Hospital of Guangxi Medical University (Nanning, China) were identified. Fifty-seven (60.6%) were male and 37 (39.4%) were females, with median age of 58 years (range, 27–80 years). Patients were grouped into three partially overlapping analysis cohorts based on treatment regimens, as follows: 64 patients were classified into the first-line chemotherapy cohort (regardless of whether targeted therapy was given prior to chemotherapy); 41 patients were classified into the first- or second-generation EGFR-TKIs cohort (exhibiting EGFR-sensitive mutations prior to first- or second-generation EGFR-TKIs) and 21 patients were classified into the third-generation EGFR-TKIs cohort (exhibiting T790M-positive mutations prior to treatment with osimertinib). A total of 23 patients were included in two analysis cohorts, while five patients were simultaneously included in all three analysis cohorts.
NGS detection
Prior to chemotherapy and/or EGFR-TKI treatment, baseline peripheral blood, effusion and/or tumor tissues from included patients were subjected to NGS, targeting 59 genes (ALK, AKT, APC, AR, ATM, BIM, BRAF, BRCA1, BRCA2, CCND1, CDK4, CDK6, CDKN2A, CTNNB1, DDR2, EGFR, ERBB2, ESR1, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, HRAS, IDH1, IDH2, JAK2, KIT, KRAS, MAP2K1, MAP2K2, MET, MLH1, MSH2, MSH6, MTOR, MYC NF1, NRAS, NTRK1, PDGFRA, PIK3CA, PMS2, PTCH1, PTEN, PTPN11, RAF1, RB1, RET, ROS1, SMARCA4, SMO, SRC, STK11, THADA, TP53, TSC1, TSC2, VHL). A median raw average coverage depth of 600X was achieved for all formalin-fixed paraffin-free tumor samples and 2,000X was achieved for circulating free DNA samples. Regarding patients that underwent matched samples NGS detection at baseline, the genetic alteration status between matched samples was consistent, with a different variant allele frequency, of which the higher value was assessed.
Sample collection
For formalin-fixed paraffin-embedded tumor samples, only samples harbored tumor cell content above 20% were considered qualified and included. 5–10 ml peripheral blood was collected in EDTA-coated tubes (BD Biosciences). Plasma was isolated by fractionating the whole blood by centrifuging at 1,500-2,000 g for 10–15 min at room temperature within 2 h of blood collection and shipped to Nanjing Geneseeq Technology, Inc., within the next 48 h (19). Matched malignant pleural effusion were collected through catheter drainage from the patient. Samples were also sent to Nanjing Geneseeq Technology Inc. (Nanjing, Jiangsu, China) for DNA extraction and genetic testing.
DNA extraction and library preparation
DNA was extracted from tumor tissue, whole blood or effusion using the DNeasy Blood & Tissue kit (Qiagen GmbH), according to the manufacturer's protocol. Purified DNA was quantified by the Qubit dsDNA HS (High Sensitivity) Assay kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. Sequencing libraries were prepared using the KAPA Hyper Prep kit (KAPA Biosystems; Roche Diagnostics).
Hybridization capture and sequencing
Different libraries with unique indices were pooled together in desirable ratios for up to 2 µg total library input. Human cot-1 DNA (Thermo Fisher Scientific, Inc.) and xGen Universal Blocking Oligos (Integrated DNA Technologies, Inc.) were added as blocking reagents. Customized xGen Lockdown Probes (Integrated DNA Technologies, Inc.) targeting 59 cancer-associated genes (exons and selected introns) were used for hybridization enrichment. The capture reaction was performed with the NimbleGen SeqCap EZ Hybridization and Wash kit (Roche Applied Science) and Dynabeads M-270 (Thermo Fisher Scientific, Inc.), according to the manufacturers' protocols. Captured libraries were on-beads amplified with Illumina p5 (5′-AATGATACGGCGACCACCGA-3′) and p7 primers (5′-CAAGCAGAAGACGGCATACGAGAT-3′) in KAPA HiFi HotStart ReadyMix (KAPA Biosystems; Roche Diagnostics). The post-capture amplified library was purified using Agencourt AMPure XP beads and quantified via SYBR Green-based qPCR using a KAPA Library Quantification kit (KAPA Biosystems; Roche Diagnostics). Sequences of forward and reverse primers were as follows: Forward, 5′-AATGATACGGCGACCACCGA-3′ and reverse, 5′-CAAGCAGAAGACGGCATACGA-3′. Reference gene size of the library should not exceed 1,000 base pairs (1 kb). The thermocycling conditions were as follows: Initiation at 95°C for 5 min, denaturation at 95°C for 30 sec and extension at 60°C for 45 sec repeated for 34 cycles. A total of 0.1X IDTE buffer was used to dilute the library to 50,000-fold, and this was used as template for qPCR absolute quantitative detection.
Library fragment size was determined using the Agilent Technologies 2100 Bioanalyzer (Agilent Technologies, Inc.). The target-enriched library was then sequenced on the HiSeq4000 NGS platform (Illumina, Inc.), according to the manufacturer's protocol.
Sequencing data processing
Trimmomatic was used for FASTQ file quality control (below 15 or N bases were removed) (20). Reads were then mapped to the reference Human Genome (hg19) using Burrows-Wheeler Aligner (BWA-mem, version 0.7.12) (github.com/lh3/bwa/tree/master/bwakit). Local realignment around the indels and base quality score recalibration was applied with the Genome Analysis Toolkit version 3.4.0 (software.broadinstitute.org/gatk/), which was also applied to detect germline mutations. VarScan2 was used for somatic mutation detection (21). Common SNPs were filtered out using dbSNP version 137 software (22) and the 1,000 Genomes database, followed by annotation using ANNOVAR version 2016Apr25 (23). Genomic fusions were identified using FACTERA version 1.4 with default parameters (24). Copy number variations were detected using ADTEx version 2.0 (adtex.sourceforge.net) with default parameters (19).
Study endpoint
The assessed clinical endpoints were as follows: Objective response rate (ORR), PFS and overall survival (OS). ORR was defined as the proportion of patients who achieved complete or partial response, according to the Response Evaluation Criteria in Solid Tumors (RECIST) guideline (version 1.1) (25). PFS was calculated from the time of treatment initiation to the progression of the disease (as determined by means of the RECIST guidelines) or mortality for any reason. OS was assessed from the date of diagnosis to mortality for any reason. The date of last follow-up was 1st July 2019.
Statistical analysis
In this study, continuous variables are presented as median (range) and binary variables were presented as frequency. Fisher's exact test was used to compare categorical characteristics between molecular groups, and age was analyzed using the Wilcoxon rank-sum test. The association between predictive factors and ORR was assessed using logistic regression. The Kaplan-Meier method and multivariate Cox proportional hazards regression analysis were performed to detect predictive factors in PFS and OS. All statistical analyses were two-sided. P<0.05 was considered to indicate a statistically significant difference. All analyses were performed using SPSS version 17.0 (SPSS Inc.).
Results
Association between genetic alteration status and treatment outcome of first-line chemotherapy
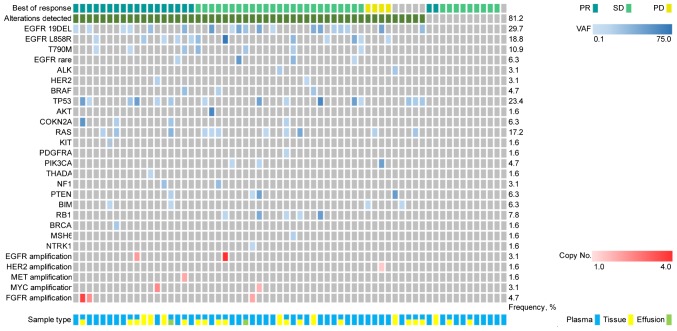
The first-line chemotherapy cohort included 64 patients with advanced NSCLC receiving platinum-based doublet first-line chemotherapy. The median follow-up on first-line chemotherapy was 20.3 months (range, 3.0–135.6 months). A total of 52 patients (81.3%) exhibited genetic alterations, whereas 12 patients (18.8%) did not present with any genetic alterations (Fig. 1). The baseline characteristics for patients with advanced NSCLC are presented in Table SI.
Figure 1.
Genetic alterations prior to first-line chemotherapy based on next-generation sequencing, targeting 59 genes from 64 patients with advanced non-small cell lung cancer. PR, partial response; SD, stable disease; PD, progressive disease; VAF, variant allele frequency; EGFR, epidermal growth factor receptor.
The following treatment variables were assessed: Genetic alterations status (detected or not detected), sex, age (≤65 or >65 years), smoking status, Eastern Cooperative Oncology Group (ECOG) performance status (0–1 or 2) (26), histology (squamous or non-squamous cell carcinoma), clinical stage, number of previous anticancer regimens for advanced disease (0 or ≥1) and central nervous system (CNS) metastases (Table SI). The ORR of first-line chemotherapy was 29.7% (19/64), while no significant difference was observed between patients with or without genetic alterations (32.7 vs. 16.7%; P=0.29; Table I).
Table I.
Treatment outcome for patients with advanced non-small cell lung cancer receiving first-line chemotherapy.
| PFS | OS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ORR | Univariate | Multivariate | Univariate | Multivariate | |||||||
| Variable | No., % | No., % | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Gene alternation status | 0.29 | 0.001a | 0.0003b | 0.87 | 0.51 | ||||||
| Not detected | 12 (18.8) | 2 (16.7) | 6.68 (2.25–19.82) | 4.43 (1.97–10.00) | 1.12 (0.31–4.07) | 1.64 (0.38–7.17) | |||||
| Detected | 52 (81.3) | 17 (32.7) | 1 (Reference) | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||||
| ECOG performance status | 0.36 | 0.02a | 0.21 | 0.02a | 0.03a | ||||||
| 0-1 | 57 (89.1) | 18 (31.6) | 3.06 (0.57–16.6) | 2.32 (0.62–8.70) | 2.94 (0.69–12.56) | 3.05 (1.10–8.47) | |||||
| 2 | 7 (10.9) | 1 (14.3) | 1 (Reference) | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||||
| CNS metastasis | 0.22 | 0.68 | 0.84 | 0.04a | 0.25 | ||||||
| No | 51 (79.7) | 17 (33.3) | 1.15 (0.55- 2.38 | 0.90 (0.33–2.44) | 2.22 (0.68–7.30) | 2.03 (0.60–6.82) | |||||
| Yes | 13 (20.3) | 2 (15.4) | 1 (Reference) | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||||
| Previous anticancer regimens | 0.70 | 0.07 | 0.02a | − | − | ||||||
| 0 | 45 (70.3) | 14 (31.1) | 1.80 (0.80–4.06) | 2.47 (1.17–5.20) | − | − | |||||
| ≥1 | 19 (29.7) | 5 (26.3) | 1 (Reference) | 1 (Reference) | – | – | |||||
P<0.05
P<0.001. ORR, objective response rate; PFS, progression-free survival; OS, overall survival; HR, hazard ratio; ECOG, Eastern Cooperative Oncology Group; CNS, central nervous system.
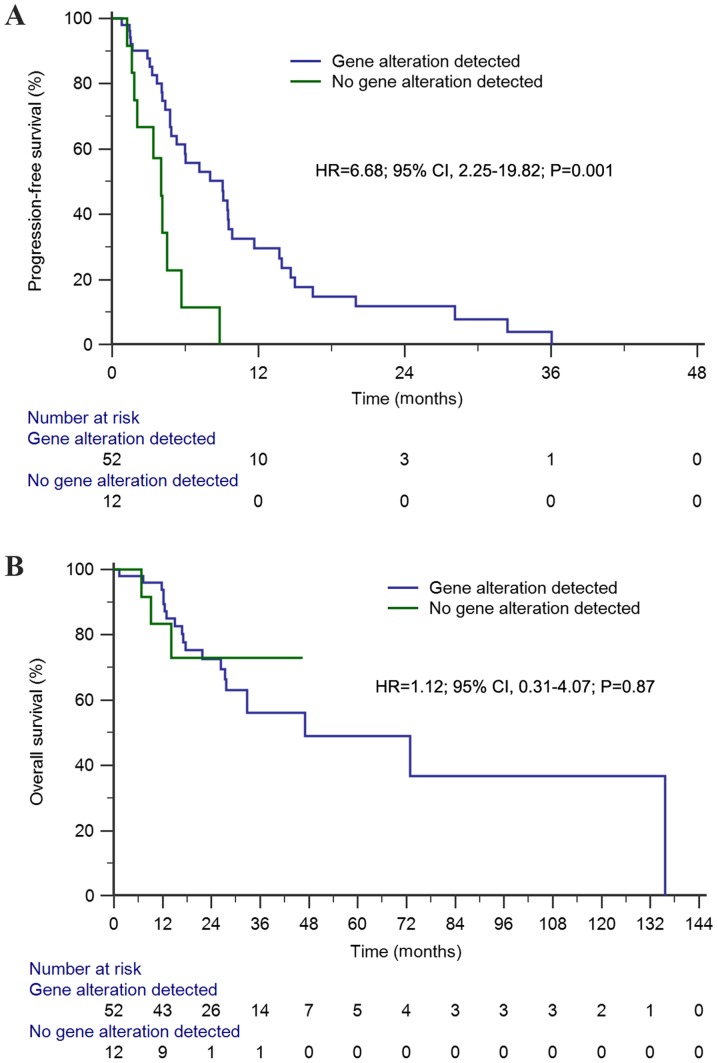
The median PFS time of this cohort was 6.0 months (95% CI, 3.1–8.9 months). Univariate analysis demonstrated that the median PFS time of first-line chemotherapy among patients with genetic alterations was significantly longer than that of patients without genetic alterations [9.1 vs. 4.0 months; hazard ratio (HR)=6.68; 95% CI, 2.25–19.82; P=0.001; Fig. 2A). Conversely, genetic alteration status (HR=4.43; 95% CI, 1.97–10.00; P=0.0003) and receiving previous anticancer regimens (HR=2.47; 95% CI, 1.17–5.20; P=0.02) were predictive factors of PFS time in the multivariate analysis (Table I).
Figure 2.
Kaplan-Meier survival analyses of first-line chemotherapy among patients with advanced non-small cell lung cancer. (A) Kaplan-Meier estimates of progression-free survival (9.1 months vs. 4.0 months) between patients with and without genetic alterations; (B) Kaplan-Meier estimates of overall survival (47.0 months vs. 47.0 months) between patients with and without genetic alterations. HR, hazard ratio.
The median OS time of the first-line chemotherapy cohort was 47.0 months (95% CI, 8.2–85.7 months). Univariate analysis demonstrated that OS for patients with genetic alterations was similar to that of patients without genetic alterations (47.0 vs. 47.0 months; HR=1.12; 95% CI, 0.31–4.07; P=0.87; Fig. 2B). Similarly, multivariate analysis demonstrated that there was no significant difference observed in OS time (HR=1.64; 95% CI, 0.38–7.17; P=0.51; Table I).
Association between concomitant alterations and treatment outcome of the first- or second-generation EGFR-TKIs cohort
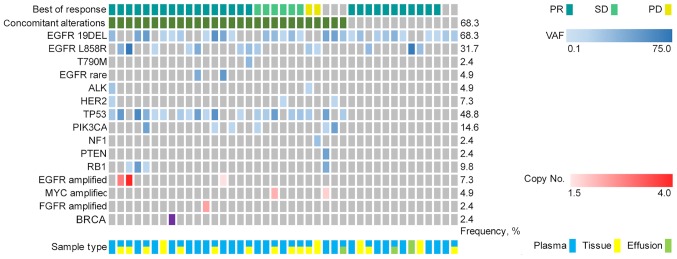
The first- or second-generation EGFR-TKI cohort included 41 patients with advanced NSCLC exhibiting a positive EGFR-activating mutation based on NGS and received first- or second-generation EGFR-TKI targeted therapy. The median follow-up for the first- or second-generation EGFR-TKIs cohort was 20.4 months (range, 6.0–121.5 months). A total of 28 patients (68.3%) exhibited concomitant alterations (Fig. 3). The baseline characteristics for patients with EGFR-mutant advanced NSCLC are presented in Table SII.
Figure 3.
Genetic alterations prior to first- or second-generation EGFR-tyrosine kinase inhibitors treatment based on next-generation sequencing targeting 59 genes from 41 patients with advanced non-small cell lung cancer harboring EGFR mutation. EGFR, epidermal growth factor receptor; PR, partial response; SD, stable disease; PD, progressive disease; VAF, variant allele frequency.
The following treatment variables were assessed: Genetic alterations, sex, age, smoking status, clinical stage, number of previous anticancer regimens for advanced disease, EGFR mutation site (exon 19 deletion or exon 21 mutation) and CNS metastases (Table SII). The ORR of the first- or second-generation EGFR-TKIs cohort was 73.2% (30/41). Although the ORR among patients with concomitant alterations was lower than that in patients without concomitant alterations, no significant difference was observed between the two groups (64.3 vs. 92.3%; P=0.07; Table II).
Table II.
Treatment outcome for patients with EGFR-mutant advanced non-small cell lung cancer receiving EGFR tyrosine kinase inhibitors.
| PFS | OS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ORR | Univariate | Multivariate | Univariate | Multivariate | |||||||
| Variable | No., % | No., % | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Concomitant alternation status | 0.07 | 0.04a | 0.047a | 0.01a | 0.03a | ||||||
| Not detected | 13 (31.7) | 12 (92.3) | 2.14 (95% CI, 1.03–4.44) | 2.30 (1.01–5.23) | 4.30 (1.41–13.16) | 9.20 (1.18–71.50) | |||||
| Detected | 28 (68.3) | 18 (64.3) | 1 (Reference) | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||||
| Smoking status | 0.48 | 0.04a | 0.10 | 0.04a | 0.13 | ||||||
| No | 26 (63.4) | 20 (76.9) | 2.02 (0.86–4.76) | 2.71 (0.83–8.87) | 2.87 (0.87–9.53) | 3.95 (0.74–21.10) | |||||
| Yes | 15 (36.6) | 10 (66.7) | 1 (Reference) | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||||
P<0.05. EGFR, epidermal growth factor receptor; ORR, objective response rate; PFS, progression-free survival; OS, overall survival; HR, hazard ratio; NR, not reached.
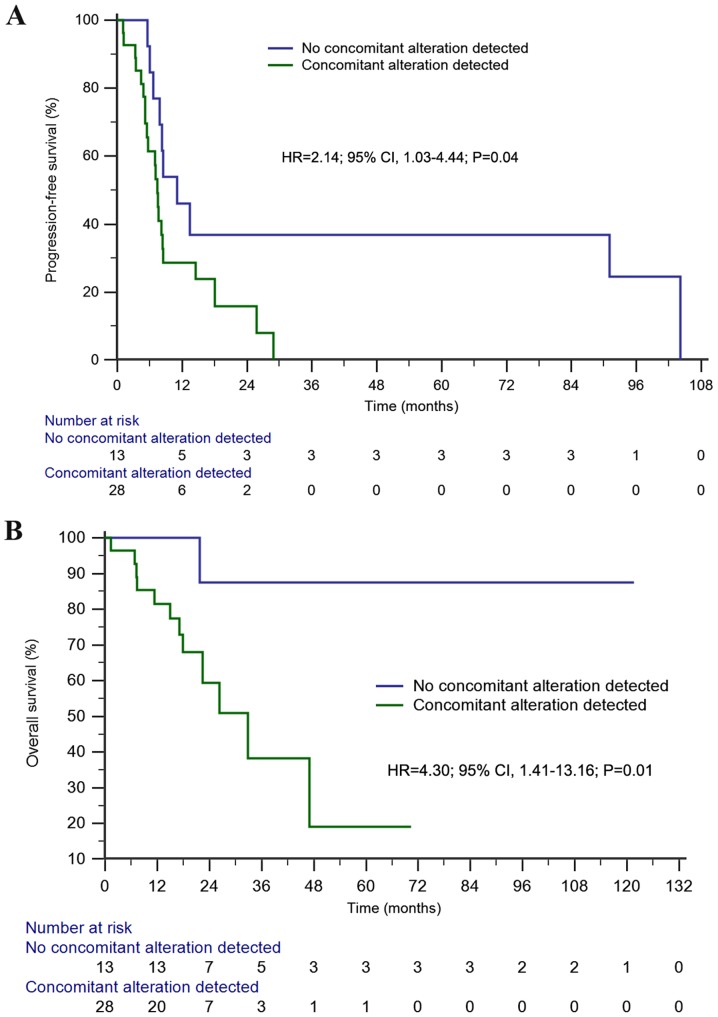
The median PFS time in the first- or second-generation EGFR-TKIs cohort was 8.1 months (95% CI, 7.1–9.2 months). Univariate analysis demonstrated a significantly longer PFS time in patients without concomitant alterations than in those with concomitant alterations (11.1 vs. 7.4 months; HR=2.14; 95% CI, 1.03–4.44; P=0.04; Fig. 4A). Furthermore, concomitant alterations were the only prognostic factor of PFS in the multivariate analysis (HR=2.30; 95% CI, 1.01–5.23; P=0.047; Table II).
Figure 4.
Kaplan-Meier survival analyses among advanced EGFR-mutant non-small cell lung cancer. (A) Kaplan-Meier estimates of progression-free survival (11.1 months vs. 7.4 months) of first- or second-generation EGFR-tyrosine kinase inhibitors treatment between patients with and without genetic alterations; (B) Kaplan-Meier estimates of overall survival (NR vs. 32.8 months) of first- or second-generation EGFR-tyrosine kinase inhibitors treatment between patients with and without genetic alterations . NR, not reached; EGFR, epidermal growth factor receptor; HR, hazard ratio.
The median OS time of the first- or second-generation EGFR-TKIs cohort was not reached. However, patients without concomitant alterations exhibited a significantly longer OS time than those with concomitant alterations in both univariate [not reached (NR) vs. 32.8 months; HR=4.30; 95% CI, 1.41–13.16; P=0.01; Fig. 4B] and multivariate (HR=9.20; 95% CI, 1.18–71.50; P=0.03) analyses.
Subgroup analysis of the 28 patients with exon 19 deletions indicated a significantly longer PFS (11.1 vs. 7.5 months; HR=2.75; 95% CI, 1.12–6.74; P=0.027; Fig. S1A) and OS time (NR vs. 32.8 months; HR=6.62; 95% CI, 1.69–26.01; P=0.007; Fig. S1B) in patients without concomitant alterations compared with those with concomitant alterations. Multivariate analyses demonstrated that concomitant alterations were the only prognostic factor of PFS (HR=2.86; 95% CI, 1.08–7.60; P=0.034) and OS time (HR=11.09; 95% CI, 1.34–91.98; P=0.026) in patients with exon 19 deletions. Univariate and multivariate analyses in patients with exon 21 mutations demonstrated that concomitant alterations had no significant effect on PFS time, while median OS was not reached (Fig. S2).
Association between concomitant alterations and treatment outcome of osimertinib
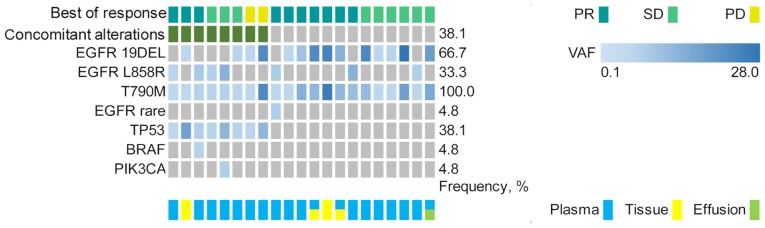
The third-generation EGFR-TKIs cohort included 21 patients with T790M-positive advanced NSCLC who had progressed disease following exposure to first- or second-generation EGFR-TKIs. All patients received third-generation EGFR-TKI osimertinib treatment, with a median follow-up of 32.8 months (range, 10.7–121.5 months). A total of 8 patients (38.1%) presented with concomitant alterations beyond EGFR (Fig. 5). Co-occurring EGFR mutation detection was identified in 14 patients (66.7%) with exon 19 deletions and 7 patients (33.3%) with exon 21 mutations. The baseline characteristics for patients with T790M-mutant advanced NSCLC are presented in Table SIII.
Figure 5.
Genetic alterations prior to treatment with osimertinib based on next-generation sequencing, targeting 59 genes from 21 patients with T790M-positive cases resistant to first- or second-generation EGFR-tyrosine kinase inhibitors. EGFR, epidermal growth factor receptor; PR, partial response; SD, stable disease; PD, progressive disease; VAF, variant allele frequency.
The following treatment variables were assessed: Concomitant genetic alterations, sex, age, smoking status, co-occurring EGFR mutation sites and CNS metastases (Table SIII). The ORR of the third-generation EGFR-TKI osimertinib treatment cohort was 47.6% (10/21), while the ORRs among patients with and without concomitant alterations were 37.5% (3/8) and 53.8% (7/13), respectively (Table III).
Table III.
Treatment outcome for patients with T790M-positive advanced non-small cell lung cancer receiving osimertinib.
| PFS | OS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ORR | Univariate | Multivariate | Univariate | Multivariate | |||||||
| Variable | No., % | No., % | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Concomitant alternation status | 0.47 | 0.002a | 0.001a | 0.03a | 0.05 | ||||||
| Not detected | 13 (61.9) | 7 (53.8) | 9.48 (2.29–39.28) | 24.4 (3.42–174.6) | 4.85 (1.16–20.29) | 4.93 (0.99–24.52) | |||||
| Detected | 8 | 3 | 1 (Reference) | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||||
| Co-occurring EGFR mutation sites | 0.54 | 0.83 | 0.04a | 0.66 | 0.66 | ||||||
| Exon 21 mutation | 7 (33.3) | 4 (57.1) | 0.89 (0.30–2.62) | 0.17 (0.03–0.96) | 0.71 (0.16–3.11) | 0.69 (0.13–3.56) | |||||
| Exon 19 deletion | 14 (66.7) | 6 (42.9) | 1 (Reference) | 1 (Reference) | 1 (Reference) | 1 (Reference) | |||||
P<0.05. ORR, objective response rate; PFS, progression-free survival; OS, overall survival; HR, hazard ratio; NR, not reached; EGFR, epidermal growth factor receptor.
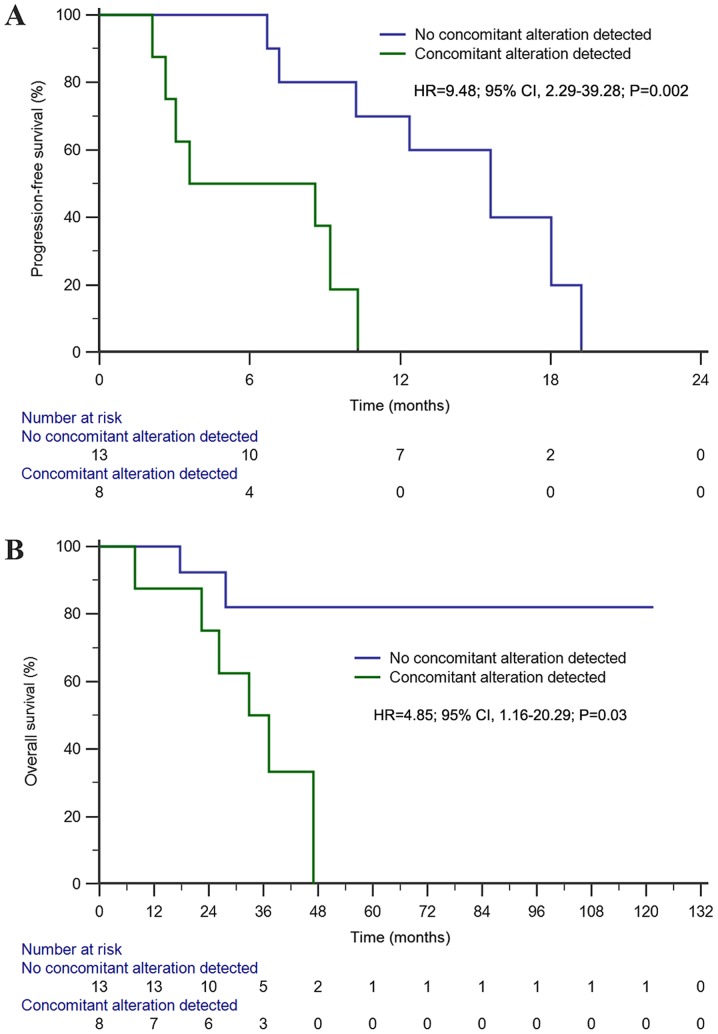
The median PFS time of this cohort was 10.3 months (95% CI, 8.2–12.4 months). Univariate analysis indicated that patients without concomitant alterations had a significantly longer PFS time than those with concomitant alterations (15.6 vs. 3.6 months; HR=9.48; 95% CI, 2.29–39.28; P=0.002; Fig. 6A). Furthermore, concomitant alterations (HR=24.4; 95% CI, 3.42–174.6; P=0.001) and co-occurring EGFR mutation sites (HR=0.17; 95% CI, 0.03–0.96; P=0.04) were both prognostic factors affecting PFS in the multivariate analysis.
Figure 6.
Kaplan-Meier survival analyses among T790M-positive NSCLC. (A) Kaplan-Meier estimates of progression-free survival (15.6 months vs. 3.6 months) of osimertinib treatment between patients with or without concomitant alterations beyond EGFR; (B) Kaplan-Meier estimates of overall survival (NR vs. 32.8 months) of osimertinib treatment between patients with or without concomitant alterations beyond EGFR. NR, not reached; EGFR, epidermal growth factor receptor; HR, hazard ratio.
The median OS time of the third-generation EGFR-TKI osimertinib treatment cohort was 47.0 months (95% CI, 29.3–64.6 months). Univariate analysis demonstrated that patients without concomitant alterations exhibited a significantly longer OS time than those with concomitant alterations (NR vs. 32.8 months; HR=4.85; 95% CI, 1.16–20.29; P=0.03; Fig. 6B). Furthermore, concomitant alterations were the only prognostic factor of OS in the multivariate analysis (HR=4.93; 95% CI, 0.99–24.52; P=0.05).
Discussion
Analysis of the NGS genotyping results prior to chemotherapy and EGFR-TKI treatment in the present study demonstrated that tumor genetic alteration profiling serves vital roles in both targeted therapy and cytotoxic chemotherapy. A positive genetic alteration status may significantly prolong PFS time in patients undergoing first-line chemotherapy. Furthermore, the presence of concomitant alterations was negatively associated with therapeutic outcomes of first- and second-generation EGFR-TKIs treatment. Similarly, concomitant alterations were demonstrated to affect PFS and OS time for third-generation EGFR-TKIs treatment. Understanding tumor genetic alteration profiling may improve the facilitation and development of effective and personalized therapeutic strategies, and will aid in predicting the therapeutic outcome either for cytotoxic chemotherapy or EGFR-TKIs in advanced NSCLC. Thus, genetic sequencing is essential in advanced NSCLC as it may be insufficient to develop treatment strategies based on limited genetic information.
Previous studies have reported conflicting results regarding the association between single gene mutations and chemotherapy efficacy. It has been demonstrated that patients with EGFR mutations tend to exhibit longer PFS time than patients without mutations for first-line chemotherapy (7,27,28). Regarding KRAS, BRAF and other genetic mutations, the predictive effect of these genetic alterations on the efficacy of chemotherapy is also controversial (29–31). However, to the best of our knowledge, existing evidence has only considered the alteration of a single gene and came to inconsistent conclusions. To the best of our knowledge, no study has taken comprehensive genetic profiling into account to investigate the effect on chemotherapy efficacy in advanced NSCLC. However, various prognostic models based on genetic profiling were demonstrated to successfully predict responders to adjuvant chemotherapy (32,33). With controversy concerning the test method, study design or sample selection and inconvenience as multiple specified genes were requested for testing in each model, the reported genetic models were not widely utilized in clinical practice. Nevertheless, as cytotoxic agents, chemotherapeutic drugs exhibit global regulation activities in more than one signaling pathway (34,35). Notably, in the present study, categorizing the population receiving first-line chemotherapy based on the alteration status of the detected genes demonstrated that patients with genetic alterations exhibited a significantly longer PFS time following first-line chemotherapy. Currently, the molecular mechanism by which genetic alterations affect the treatment outcome of chemotherapy remains unclear, thus further research is required.
The predictive value of concomitant mutations for targeted therapy in advanced NSCLC is gaining recognition. Hong et al (14) reported that co-occurring mutations beyond EGFR affect the therapeutic efficacy of first-generation EGFR-TKIs, with lower ORR and shorter PFS and OS time (14). Furthermore, Wang et al (36) demonstrated that accompanied gene aberrations may predict resistance to treatment with TKIs. Recently, Chen et al (37) confirmed the negative predictive role of co-occurring driver gene mutations in TKI therapy. Consistent with these findings, the results of the present study confirmed that PFS and OS time were significantly shorter in patients with concomitant alterations than those without concomitant alterations.
Notably, stratified analysis among patients with exon 19 deletions indicated that PFS and OS time were significantly longer among patients without concomitant alterations. However, analysis among patients with exon 21 mutations failed to demonstrate a significant effect of concomitant alterations on the efficacy of EGFR-TKI therapy. However, it is difficult to reach a definitive conclusion as the small number of cases with exon 21 mutations in the present study may have limited the ability to detect the impact of concomitant alterations on the efficacy of EGFR-TKI treatment. It was hypothesized that patients with exon 21 mutations may be more likely to develop other genetic alterations and activate signaling pathways responsible for TKI resistance, which is not associated with concomitant alterations. Regarding the population with exon 19 deletions, the results of the present study support the notion that the bypass signaling pathway activated by concomitant alterations may affect the efficacy of TKIs, ultimately affecting the survival of advanced EGFR-mutant NSCLC (14).
To the best of our knowledge, the present study was the first to reveal the predictive role of concomitant alterations on third-generation EGFR-TKI therapy. Among patients with T790M mutation receiving osimertinib treatment, concomitant alterations predicted significantly shortened PFS and OS time as compared with those without concomitant alterations. It was hypothesized that the bypass pathway activated by concomitant alterations may initiate rapid drug resistance as even the EGFR pathway is inhibited by osimertinib, ultimately resulting in poor treatment efficacy and prognosis among patients with T790M mutation receiving osimertinib. Patients with T790M-resistant mutations and co-occurring exon 19 deletions exhibited a longer PFS time with osimertinib than patients with co-occurring exon 21 mutations. Furthermore, co-occurring EGFR-activating genetic mutation type was one of the independent prognostic factors for PFS. Similarly, pooled analysis of two osimertinib studies in T790M-positive NSCLC revealed a trend toward an increased response rate (70% vs. 57%) and a prolonged PFS time (11.1 vs. 9.5 months) among patients with co-occurring exon 19 deletions (17,38,39). The underlying molecular mechanisms of poorer efficacy among patients with co-occurring exon 21 mutations compared with exon 19 deletions may be due to a higher number of concomitant genetic alterations (57.1 vs. 28.6%), which may contribute to the bypass pathway activation responsible for resistance as we speculation
Several limitations exist within the present study. First, as it was a retrospective study, patient selection bias may have attributed to somewhat longer PFS time in the first-line chemotherapy cohort, while patients receiving different treatments were included in the three analysis cohorts. Secondly, relatively small sample sizes of each analysis cohort were used, thus affecting the reliability of the results. Furthermore, NGS results based on the targeted gene panel have the potential to miss information of non-target genes in non-preselected regions, as well as the possible inability of plasma NGS to detect genetic alterations, and of tissue NGS to detect genetic heterogeneity in some cases, which may have affected the accuracy of the results (40,41).
In conclusion, diverse genetic alteration profiling in patients with advanced NSCLC identified by NGS resulted in the discovery of different therapeutic efficacies for different treatment modes. Positive genetic alterations statuses prior to first-line chemotherapy were predictive of favorable treatment outcomes, whereas concomitant alterations indicated poor therapeutic effect and survival with either first- or second-generation EGFR-TKI, or third-generation EGFR-TKI treatment. How genetic alteration profiling of tumors using NGS could guide the selection and combination of treatment regimens for advanced NSCLC remains an area for further investigation. Dynamic monitoring of genetic alteration evolution patterns caused by different therapeutic drugs, such as chemotherapy or EGFR-TKIs, may help understand the underlying molecular mechanisms affecting the efficacy of chemotherapy or EGFR-TKIs, in order to predict and improve treatment efficacy, and ultimately overcome drug resistance.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Dongqing Zhu, Dr Qiuxiang Ou, Mr. Yong Wu and Miss Lijuan Liu (Nanjing Geneseeq Technology Inc.) for their technical support.
Glossary
Abbreviations
- NGS
next-generation sequencing
- PFS
progression-free survival
- OS
overall survival
- NSCLC
non-small cell lung cancer
- EGFR
epidermal growth factor receptor
- TKI
tyrosine kinase inhibitor
- ORR
objective response rate
- RECIST
Response Evaluation Criteria in Solid Tumors
- CNS
central nervous system
- ECOG
Eastern Cooperative Oncology Group
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WJ, QY, AZ and RN conceived and designed the present study. WJ, QY, AZ, WZ and HW collected the data, and HW, WZ, CS and SZ analyzed the data. RN and QY supervised the present study. WJ drafted the initial manuscript, while QY, AZ, CS and RN subsequently reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by The Institutional Review Board of the Affiliated Tumor Hospital of Guangxi Medical University (Nanning, China), and conducted in accordance with The Declaration of Helsinki. Written informed consent was provided by all patients prior to the study start.
Patient consent for publication
Written informed consent was provided by all patients for their data to be published in the present study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Doroshow DB, Herbst RS. Treatment of advanced non-small cell lung Cancer in 2018. JAMA Oncol. 2018;4:569–570. doi: 10.1001/jamaoncol.2017.5190. [DOI] [PubMed] [Google Scholar]
- 2.Ohe Y, Ohashi Y, Kubota K, Tamura T, Nakagawa K, Negoro S, Nishiwaki Y, Saijo N, Ariyoshi Y, Fukuoka M. Randomized phase III study of cisplatin plus irinotecan versus carboplatin plus paclitaxel, cisplatin plus gemcitabine, and cisplatin plus vinorelbine for advanced non-small-cell lung cancer: Four-Arm cooperative study in Japan. Ann Oncol. 2007;18:317–323. doi: 10.1093/annonc/mdl377. [DOI] [PubMed] [Google Scholar]
- 3.Fossella F, Pereira JR, von Pawel J, Pluzanska A, Gorbounova V, Kaukel E, Mattson KV, Ramlau R, Szczesna A, Fidias P, et al. Randomized, multinational, phase III study of docetaxel plus platinum combinations versus vinorelbine plus cisplatin for advanced non-small-cell lung cancer: The TAX 326 study group. J Clin Oncol. 2003;21:3016–3024. doi: 10.1200/JCO.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 4.Scagliotti GV, Parikh P, von Pawel J, Biesma B, Vansteenkiste J, Manegold C, Serwatowski P, Gatzemeier U, Digumarti R, Zukin M, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-I patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:3543–3551. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 5.Rodrigues-Pereira J, Kim JH, Magallanes M, Lee DH, Wang J, Ganju V, Martinez-Barrera L, Barraclough H, van Kooten M, Orlando M. A randomized phase 3 trial comparing pemetrexed/carboplatin and docetaxel/carboplatin as first-line treatment for advanced, nonsquamous non-small cell lung cancer. J Thorac Oncol. 2011;6:1907–1914. doi: 10.1097/JTO.0b013e318226b5fa. [DOI] [PubMed] [Google Scholar]
- 6.Scagliotti GV, Gridelli C, de Marinis F, Thomas M, Dediu M, Pujol JL, Manegold C, San Antonio B, Peterson PM, John W, et al. Efficacy and safety of maintenance pemetrexed in patients with advanced nonsquamous non-small cell lung cancer following pemetrexed plus cisplatin induction treatment: A cross-trial comparison of two phase III trials. Lung Cancer. 2014;85:408–414. doi: 10.1016/j.lungcan.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 8.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 9.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harboring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 10.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, Zhang S, Wang J, Zhou S, Ren S, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicenter, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 11.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicenter, open-label, randomised multicenter phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 12.Sequist LV, Yang JC, Yamamoto N, O'Byrne K, Hirsh V, Mok T, Geater SL, Orlov S, Tsai CM, Boyer M, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 13.Wu YL, Cheng Y, Zhou X, Lee KH, Nakagawa K, Niho S, Tsuji F, Linke R, Rosell R, Corral J, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): A, randomised open-label, phase 3 trial. Lancet Oncol. 2017;18:1454–1466. doi: 10.1016/S1470-2045(17)30608-3. [DOI] [PubMed] [Google Scholar]
- 14.Hong S, Gao F, Fu S, Wang Y, Fang W, Huang Y, Zhang L. Concomitant genetic alterations with response to treatment and epidermal growth factor receptor tyrosine kinase inhibitors in patients with EGFR-mutant advanced non-small CELL lung cancer. JAMA Oncol. 2018;4:739–742. doi: 10.1001/jamaoncol.2018.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M, Riely GJ, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, Ahn MJ, Kim SW, Su WC, Horn L, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 17.Ahn MJ, Tsai CM, Shepherd FA, Bazhenova L, Sequist LV, Hida T, Yang JCH, Ramalingam SS, Mitsudomi T, Jänne PA, et al. Osimertinib in patients with T790M mutation-positive, advanced non-small cell lung cancer: Long-Term follow-up from a pooled analysis of 2 phase 2 studies. Cancer. 2018;15:892–901. doi: 10.1002/cncr.31891. [DOI] [PubMed] [Google Scholar]
- 18.Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, Shepherd FA, He Y, Akamatsu H, Theelen WS, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z, Yang N, Ou Q, Xiang Y, Jiang T, Wu X, Bao H, Tong X, Wang X, Shao YW, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res. 2018;24:3097–3107. doi: 10.1158/1078-0432.CCR-17-2310. [DOI] [PubMed] [Google Scholar]
- 20.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. Dbsnp: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M, Alizadeh AA. FACTERA: A practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30:3390–3393. doi: 10.1093/bioinformatics/btu549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Moone M, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 26.Zubrod CG, Schleiderman M, Frei S, Brindley C, Gold GL, Shnider B, Oviedo R, Gorman J, Jones Jr R, Jonsson U, et al. Appraisal of methods for the study of chemotherapy of cancer in man: Comparative therapeutic trial of nitrogen mustard and triethylene thiophosphoramide. J Chronic Dis. 1960;11:7–33. doi: 10.1016/0021-9681(60)90137-5. [DOI] [Google Scholar]
- 27.Fukuoka M, Wu YL, Thongprasert S, Sunpaweravong P, Leong SS, Sriuranpong V, Chao TY, Nakagawa K, Chu DT, Saijo N, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) J Clin Oncol. 2011;29:2866–2874. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 28.Wu M, Zhao J, Song SW, Zhuo M, Wang X, Bai H, Wang S, Yang L, An T, Zhang Y, et al. EGFR mutations are associated with prognosis but not with the response to front-line chemotherapy in the Chinese patients with advanced non-small cell lung cancer. Lung Cancer. 2010;67:343–347. doi: 10.1016/j.lungcan.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Marabese M, Ganzinelli M, Garassino MC, Shepherd FA, Piva S, Caiola E, Macerelli M, Bettini A, Lauricella C, Floriani I, et al. KRAS mutations affect prognosis of non-small-cell lung cancer patients treated with first-line platinum containing chemotherap. Oncotarget. 2015;6:34014–34022. doi: 10.18632/oncotarget.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macerelli M, Caramella C, Faivre L, Besse B, Planchard D, Polo V, Ngo Camus M, Celebic A, Koubi-Pick V, Lacroix L, et al. Does KRAS mutational status predict chemoresistance in advanced non-small cell lung cancer (NSCLC)? Lung Cancer. 2014;83:383–388. doi: 10.1016/j.lungcan.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 31.Cardarella S, Ogino A, Nishino M, Butaney M, Shen J, Lydon C, Yeap BY, Sholl LM, Johnson BE, Jänne PA. Clinical, pathologic, and biologic features associated with BRAF mutations in non-small cell lung cancer. Clin Cancer Res. 2013;19:4532–4540. doi: 10.1158/1078-0432.CCR-13-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Laar RK. Genomic signatures for predicting survival and adjuvant chemotherapy benefit in patients with non-small-cell lung cancer. BMC Med Genomics. 2012;5:30. doi: 10.1186/1755-8794-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, Thomas RK, Naoki K, Ladd-Acosta C, Liu N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Booth L, Poklepovic A, Dent P. Not the comfy chair! Cancer drugs that act against multiple active sites. Expert Opin Ther Targets. 2019;23:893–901. doi: 10.1080/14728222.2019.1691526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mun H, Lee SH, Lee CH, Jo SY, Oh JH, Lee A, Lee B, Jang SJ, Suh YA. Taxotere induces dephosphorylation of MET in patient-derived tumor models. Anticancer Res. 2020;40:109–119. doi: 10.21873/anticanres.13932. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Cheng Y, An T, Gao H, Wang K, Zhou Q, Hu Y, Song Y, Ding C, Peng F, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): A phase 2, single-arm, multicenter clinical trial. Lancet Respir Med. 2018;6:681–690. doi: 10.1016/S2213-2600(18)30264-9. [DOI] [PubMed] [Google Scholar]
- 37.Chen M, Xu Y, Zhao J, Zhong W, Zhang L, Bi Y, Wang M. Concurrent driver gene mutations as negative predictive factors in epidermal growth factor receptor-positive non-small cell lung cancer. Ebiomedicine. 2019;42:304–310. doi: 10.1016/j.ebiom.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang JCH, Ahn MJ, Kim DW, Ramalingam SS, Sequist LV, Su WC, Kim SW, Kim JH, Planchard D, Felip E, et al. Osimertinib in pretreated T790M-positive advanced non-small-cell lung cancer: AURA study phase II extension component. J Clin Oncol. 2017;35:1288–1296. doi: 10.1200/JCO.2016.70.3223. [DOI] [PubMed] [Google Scholar]
- 39.Goss G, Tsai CM, Shepherd FA, Bazhenova L, Lee JS, Chang GC, Crino L, Satouchi M, Chu Q, Hida T, et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): A multicenter, open-label, single-arm, phase 2 study. Lancet Oncol. 2016;17:1643–1652. doi: 10.1016/S1470-2045(16)30508-3. [DOI] [PubMed] [Google Scholar]
- 40.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 41.Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Ann Rev Pathol. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.