Abstract
Deoxyhypusine synthase (DHPS) utilizes the polyamine spermidine to catalyze the hypusine modification of the mRNA translation factor eIF5A and promotes oncogenesis through poorly-defined mechanisms. Because germline deletion of Dhps is embryonically lethal, its role in normal postnatal cellular function in vivo remains unknown. We generated a mouse model that enabled the inducible, postnatal deletion of Dhps specifically in postnatal islet β cells, which function to maintain glucose homeostasis. Removal of Dhps did not have an effect under normal physiologic conditions. However, upon development of insulin resistance, which induces β-cell proliferation, Dhps deletion caused alterations in proteins required for mRNA translation and protein secretion, reduced production of the cell cycle molecule cyclin D2, impaired β-cell proliferation, and induced overt diabetes. We found that hypusine biosynthesis was downstream of protein kinase C-ζ and was required for c-Myc-induced proliferation. Our studies reveal a requirement for DHPS in β cells to link polyamines to mRNA translation to effect facultative cellular proliferation and glucose homeostasis.
The natural polyamines (putrescine, spermidine, and spermine), which are generated endogenously through the action of the enzyme ornithine decarboxylase (ODC), promote cellular proliferation and maintenance (1). Among the polyamines, only spermidine is a substrate for the enzyme deoxyhypusine synthase (DHPS), which transfers a butylamine moiety to the ε-amine of Lys50 of eukaryotic translation initiation factor 5A (eIF5A) to form the rare amino acid hypusine (2). The polyamines, DHPS and eIF5A have been studied largely in the context of cancer, where they promote oncogenesis by enabling the production of proteins involved in cellular migration and growth (2–5). These factors are also important in normal cells during development, because germline deletion of the genes encoding ornithine decarboxylase (ODC; the enzyme that regulates polyamine production), DHPS and eIF5A in mice induce lethality during early embryonic stages (6–8). Whereas conditional knockouts of ODC have been described in the context of macrophage polarization (9, 10), the absence of animal models to study either DHPS or eIF5A in the postnatal state have precluded analysis of the potential role of these factors in the maintenance of normal cellular function.
The growth and function of islet β cells is necessary for fuel homeostasis and viability in mammals. β cells produce insulin, a hormone necessary for glucose disposal and the suppression of ketogenesis and gluconeogenesis. β cells are considered to have facultative proliferative activity, because they are largely replication-quiescent postnatally (11, 12), but can be induced to proliferate under pathophysiological conditions. For example, during obesity, generalized insulin resistance increases the demand for insulin secretion from β cells to maintain glucose homeostasis (13). In rodents, this increase in insulin secretion is partially met though expansion of β-cell mass (14). Although no longitudinal data are available in humans, cross-sectional studies similarly suggest that obese humans have greater β-cell mass compared to lean controls (15). Cellular replication is believed to be the dominant mechanism for gains in β-cell mass in mice (16), and replicating β cells has been detected in humans, especially in younger individuals (11, 17). The molecular mechanisms governing proliferation in β cells remains incompletely characterized. Given a role for polyamines in oncogenesis, we surmised that DHPS may be critical for the function of polyamines in enabling proliferation of β cells. We previously showed that the hypusine modification is important in the production of stress-responsive proteins during cytokine signaling and endoplasmic reticulum (ER) stress in the pancreatic islet (18–20), but a role in facultative proliferation remained speculative.
In this study, we hypothesized that facultative gains in β-cell mass during obesity were mediated by the action of DHPS through its downstream hypusine modification of eIF5A. To test this hypothesis, we developed a mouse model to permit inducible, tissue-specific deletion to investigate the role of DHPS during cellular replication of β cells and glucose metabolism in vivo. Collectively, our studies reveal a requirement for hypusine to link specific mRNAs with the translational machinery to effect facultative cellular proliferation.
RESULTS
High fat diet feeding results in diabetes with attendant loss of β-cell mass in DhpsΔβ animals
To achieve inducible, tissue-specific deletion of Dhps in mice, we first generated mice in which exons 2 to 7 of the Dhps allele were flanked by Cre recombinase recognition sequences (loxP) (Supplemental fig. S1A). These mice were backcrossed for 10 generations onto the C57BL6/J background, then crossed to C57BL6/J mice harboring a transgene encoding the Cre recombinase-modified estrogen receptor fusion protein under control of the mouse Ins1 promoter (MIP1-CreERT) (21). Some of the mice were also crossed to Rosa26R-Tomato mice to monitor Cre-mediated recombination (22). The crosses resulted in the generation of multiple different mouse genotypes, which were confirmed by PCR genotyping (Supplemental fig. S1B). Administration of tamoxifen to DhpsloxP/loxP;MIP1-CreERT mice at 8 weeks of age resulted in the generation of β-cell-specific knockout mice (henceforth referred to as DhpsΔβ mice), as shown by overlapping immunostaining of Tomato fluorescent protein and insulin, indicating β-cell-specific recombination at the Rosa26R-Tomato locus (Supplemental fig. S1C) and by immunoblotting showing reduced DHPS protein in pancreatic islets but not in the hypothalamus or liver (Supplemental fig. S1D). One week following the final tamoxifen injection, no alterations in glucose tolerance (by intraperitoneal glucose tolerance test, GTT) were observed in DhpsΔβ mice compared to littermate controls (which included both Cre-positive and loxP/loxP mice) (Supplemental fig. S1E and S1F), a finding suggesting that the acute loss of DHPS in β cells does not immediately impact glucose homeostasis.
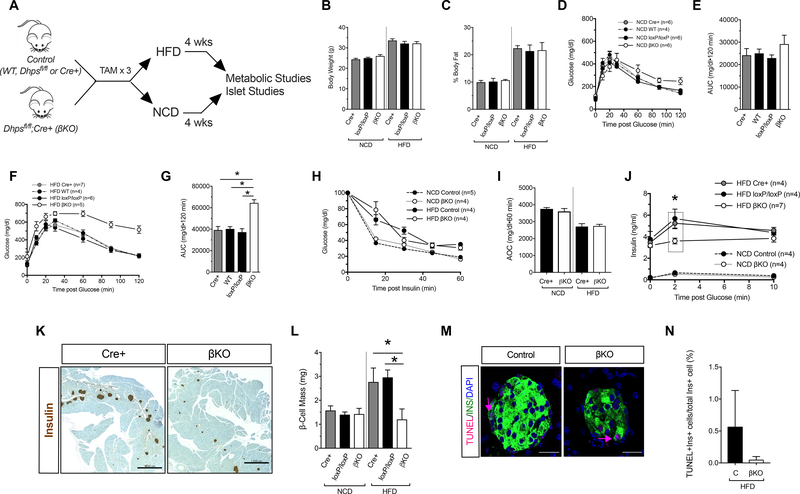
To interrogate the role of DHPS in facultative cellular proliferation, we next subjected DhpsΔβ and control littermates (including Cre-positive, loxP/loxP, and wild-type controls) to either a high fat diet (HFD; 60% calories from fat), which induces β-cell proliferation (23), or to a normal chow diet (NCD, 16% calories from fat). Following 4 weeks of feeding (Fig. 1A), both control and DhpsΔβ animals on a HFD gained equivalent weight and body fat, which was greater than their NCD-fed counterparts (Fig. 1B and 1C). Glucose tolerance as assessed by GTT did not differ between NCD-fed DhpsΔβ mice or control mice (Fig. 1D and 1E). By contrast, HFD-fed DhpsΔβ mice exhibited frank diabetes with significantly worse glucose tolerance compared to HFD-fed control animals (Fig. 1F and 1G). Notably, although insulin sensitivity (based on insulin tolerance testing) was reduced in HFD-fed animals, DhpsΔβ mice did not differ from control mice (Fig. 1H and 1I), suggesting that worsened glucose tolerance in DhpsΔβ mice was not due to inherent alterations in insulin sensitivity.
Figure 1: β-cell-specific knockout of Dhps impairs glucose tolerance after HFD-feeding.
Control (WT, Cre+ or DhpsloxP/loxP) and DhpsΔβ (βKO) mice were fed for 4 weeks with a normal chow diet (NCD) or high fat diet (HFD) and metabolic parameters were assessed. (A) Schematic timeline showing the period of tamoxifen (TAM) injections and feeding. (B) Body weight at the conclusion of the feeding regimen. Data from N=5 mice per group. (C) Percent body fat at the conclusion of the feeding regimen. Data from N=4 mice per group. (D) Glucose tolerance test in NCD-fed mice. (E) Area under the curve (AUC) analysis of glucose tolerance tests in (D). (F) Glucose tolerance test in HFD-fed mice. (G) AUC analysis of glucose tolerance tests in (F). (H) Insulin tolerance test in NCD- and HFD-fed mice. (I) Area over the curve (AOC) of insulin tolerance test in (H). (J) Serum insulin levels during a glucose tolerance test. (K) Images of whole pancreatic sections from representative HFD-fed control and DhpsΔβ mice immunostained for insulin (brown) and counterstained with hematoxylin. Scale bar, 1000 μm. (L) Quantitation of pancreatic β-cell mass. Data from N=3–6 mice per group. (M) Representative images of pancreata stained for TUNEL (red), insulin (green) and nuclei (DAPI, blue). Scale bar, 50 μm. (N) Quantification of TUNEL immunostaining (N=3 mice per group). Data presented as mean ± SEM; *p-value < 0.05 for the comparisons shown by one-way ANOVA.
To evaluate β-cell function in HFD-fed DhpsΔβ mice, we performed glucose-stimulated insulin secretion studies in vivo. Whereas insulin levels were similar between DhpsΔβ and controls on a NCD, HFD-fed DhpsΔβ mice showed significantly lower absolute insulin levels 2 min after an intraperitoneal glucose load, and lower incremental changes in insulin levels compared to HFD-fed controls, consistent with an impairment in insulin secretion (Fig. 1J). HFD-fed DhpsΔβ mice failed to gain β-cell mass in contrast to HFD-fed controls (whose mass was greater than that in animals placed on a NCD) (Fig. 1K and 1L). This failure to gain β-cell mass was likely caused by lack of cellular accrual (proliferation) rather than an increase in cellular death, because we did not observe increases in TUNEL staining of β cells in pancreas sections from DhpsΔβ mice compared to controls (Fig. 1M and 1N).
Gene expression and proteomics analyses suggest a potential defect in mRNA translation
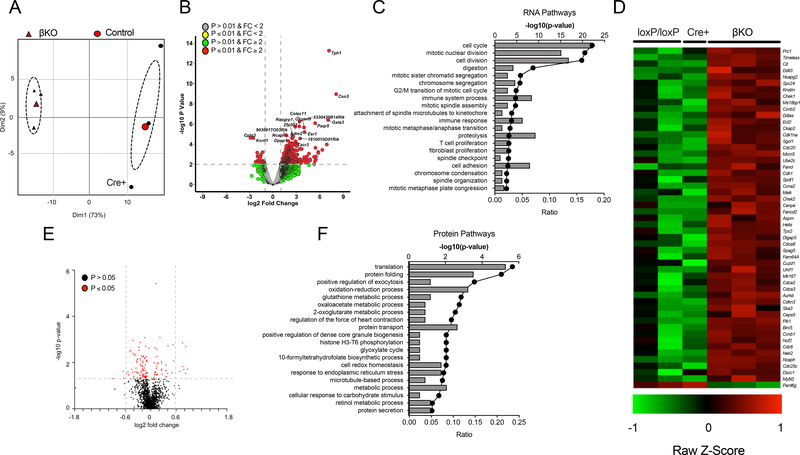
To interrogate the molecular effects of DHPS deficiency, we next performed RNA deep sequencing and global proteomics analyses on isolated islets from DhpsΔβ and control animals from the HFD-fed group. In RNA deep sequencing studies, islets from HFD-fed controls (one Cre-positive and two loxP/loxP animals) and DhpsΔβ mice showed distinct clustering in principal component analysis, with greater variance seen among control than DhpsΔβ islets (Fig. 2A). Deletion of Dhps resulted in a total of 747 genes that were significantly differentially expressed in islets from 4-week HFD-fed mice, when using the criteria of p-value<0.05 and fold-change>2.0 (Fig. 2B). By Gene Ontology Enrichment Analysis, the greatest enrichment of genes in DhpsΔβ islets was observed in pathways affecting cell cycle, mitotic nuclear division, and cell division (Fig. 2C). Virtually all of the genes involved in cellular replication were significantly increased in DhpsΔβ islets compared to control islets (Fig. 2D). Because the primary role of DHPS is the posttranslational activation of the mRNA translation factor eIF5A, we also performed tandem mass spectrometry (MS/MS)-based proteomics analysis on islets from HFD-fed mice. A total of 130 proteins (out of a total of 1991 proteins identified) were significantly altered in DhpsΔβ islets compared to controls (using a p-value <0.05; Fig. 2E). Application of Gene Ontology Enrichment Analysis showed that these proteins clustered in pathways affecting primarily mRNA translation, protein folding, exocytosis, and oxidation-reduction processes (Fig. 2F)—all pathways consistent with effects of eIF5A on translation and protein trafficking through the ER. Notably, because of their low abundance relative to others in the β cell, cell cycle proteins were either not detected or were not significantly changed in this analysis.
Figure 2: RNA-sequencing and proteomics of control and β-cell-specific Dhps knockout islets.
Control (Cre+ and DhpsloxP/loxP) and DhpsΔβ (βKO) mice were fed a NCD or HFD for 4 weeks. Data from N=3 mice per group. (A) Principal component analysis displaying the variance of RNA-sequencing data obtained from control and βKO islets. (B) Volcano plot of differentially-expressed mRNAs in comparison between control and DhpsΔβ islets. (C) Gene Ontology Enrichment Analysis for mRNA sequencing data. (D) Heatmap of differentially-expressed genes involved in cellular proliferation. (E) Volcano plot of differentially-expressed proteins in comparison between control and DhpsΔβ mice. Data from N=3–6 mice per group. (F) Gene Ontology Enrichment Analysis from proteomics data.
β-cell proliferation following HFD feeding is impaired in DhpsΔβ mice
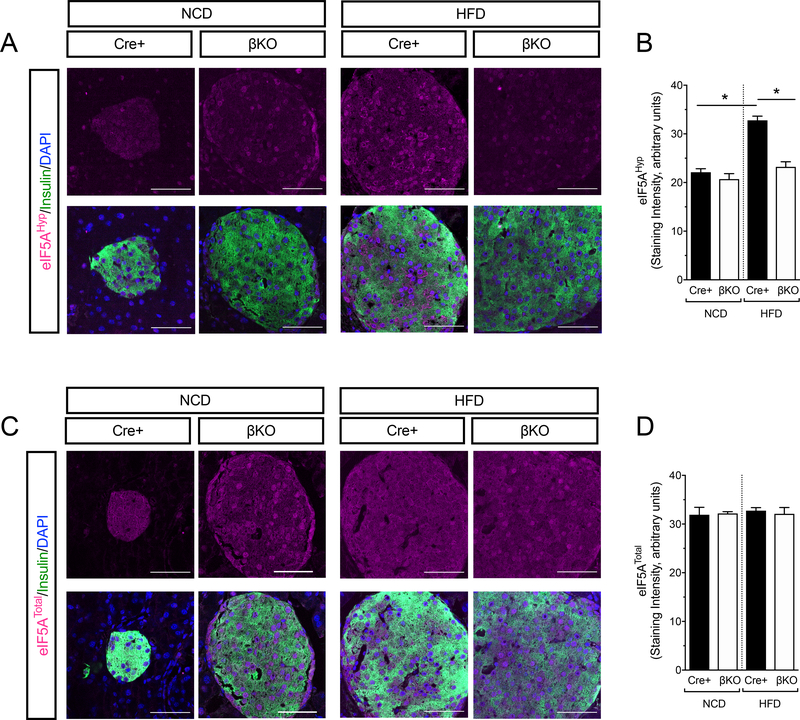
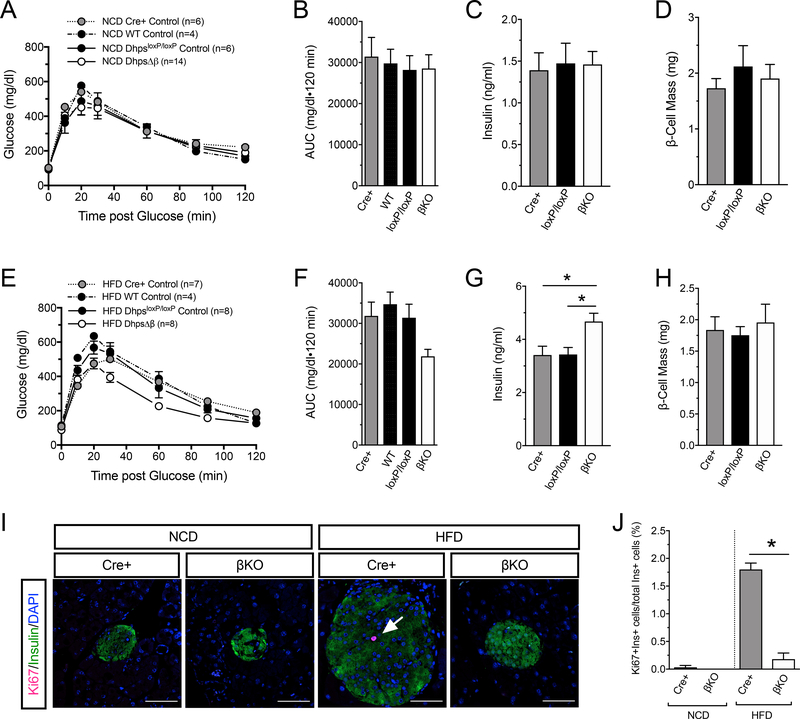
Because DhpsΔβ mice did not accrue β-cell mass, the increases in mRNAs encoding pro-proliferative proteins suggested to us a possible transcriptional compensatory response. β-cell proliferation is observed as early as 1 week following initiation of HFD feeding (23–26). We therefore examined phenotypic and molecular characteristics in control and DhpsΔβ mice after 1 week of NCD or HFD feeding. Immunoblots and immunostaining of islets revealed that the levels of hypusinated eIF5A (eIF5AHyp) increased in control animals upon HFD feeding (Fig. 3A and 3B and Supplemental fig. S1G), whereas in islets from DhpsΔβ animals, eIF5AHyp levels by immunostaining were significantly reduced and not detectably changed upon HFD feeding (Fig. 3A and 3B). The intensity of immunostaining for eIF5ATotal did not change with diet or genotype (Fig. 3C and 3D), suggesting that the changes in eIF5AHyp levels are a result of posttranslational modification. Glucose tolerance, circulating insulin, and β-cell mass did not differ between DhpsΔβ mice and littermate controls on a NCD (Fig. 4A–D), whereas HFD-fed DhpsΔβ mice exhibited a small, but statistically insignificant, improvement in glucose tolerance compared to control mice following 1 week of feeding (Fig. 4E and 4F). Consistent with this finding, circulating insulin levels in HFD-fed DhpsΔβ mice were slightly but significantly higher than in the control animals (Fig. 4G). At this timepoint, β-cell mass was similar between DhpsΔβ and control mice fed a HFD (Fig. 4H). To interrogate alterations in β-cell proliferation, we performed immunofluorescence staining of pancreas sections from NCD- and HFD-fed animals for Ki67, a marker of cellular proliferation. One week of HFD feeding increased the frequency of Ki67-positive β cells compared to NCD in control littermates, an increase that did not occur in DhpsΔβ mice (Fig. 4I and 4J). Moreover, Ki67-positivity remained significantly lower in DhpsΔβ compared to control littermates when fed either a NCD or HFD (Fig. 4J). Together, these results suggest that loss of Dhps impairs the early β-cell proliferative response to a HFD.
Figure 3: Hypusination of eIF5A is increased after 1 week of high fat diet feeding.
Control Cre+ and DhpsΔβ (βKO) mice were fed for 1 week with NCD or HFD and eIF5AHyp and eIF5ATotal levels were assessed by immunostaining of pancreas sections. (A) Representative images of pancreata stained for eIF5AHyp (magenta), insulin (green), and nuclei (DAPI, blue). Scale bar, 50 μm. (B) Quantification of eIF5AHyp immunostaining in (A) from N=3 animals per group. (C) Representative images of pancreata stained for eIF5ATotal (magenta), insulin (green), and nuclei (DAPI, blue). Scale bar, 50 μm. (D) Quantification of eIF5ATotal immunostaining in (C). Data from N=3 animals per group. Data presented as mean +/− SEM; *p-value < 0.05 for the comparisons shown by one-way ANOVA.
Figure 4: β-cell-specific Dhps knockout mice exhibit normal glucose tolerance after 1 week of high fat diet feeding.
Control (Cre+, WT, and DhpsloxP/loxP) and DhpsΔβ (βKO) mice were fed for 1 week with NCD or HFD and metabolic parameters were assessed. (A) Glucose tolerance test in NCD-fed mice. (B) Area under the curve (AUC) analysis for glucose tolerance tests from NCD-fed mice in (A). (C) Serum insulin levels in NCD-fed mice in (A). Data from N=3 Cre+ and DhpsloxP/loxP mice, N=5 DhpsΔβ mice. (D) β-cell mass in NCD-fed mice in (A). N=3 mice per group. (E) Glucose tolerance test in HFD-fed mice. (F) AUC analysis for glucose tolerance tests in (E). (G) Serum insulin levels in HFD-fed mice in (E). Data from N=3 Cre+ and DhpsloxP/loxP mice, N=4 DhpsΔβ mice. (H) β-cell mass in HFD-fed mice in (E). Data from N=3 Cre+ and DhpsloxP/loxP mice, N=5 DhpsΔβ mice. (I) Representative images of pancreata from the indicated mice stained for Ki67 (magenta, arrow), insulin (green), and nuclei (DAPI, blue). Scale bar, 50 μm. (J) Quantification of Ki67 immunostaining in (I), data from N=3–5 animals per group. Data presented as mean ± SEM; *p-value < 0.05 by one-way ANOVA.
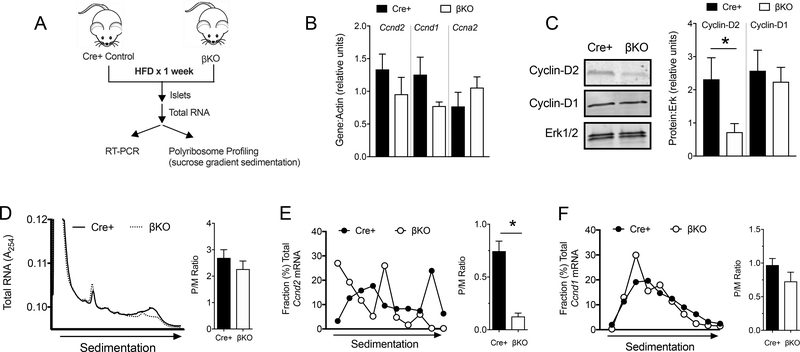
Dhps deficiency in β cells impairs Ccnd2 mRNA translation during HFD feeding
To clarify the molecular alterations that contribute to the defective β-cell proliferative phenotype in DhpsΔβ mice, we evaluated in isolated islets the expression of specific genes involved in cell cycle progression at the G1/S boundary (Fig. 5A). The levels of several key mRNAs crucial to β-cell proliferation, including Ccnd2 (encoding Cyclin-D2), Ccnd1 (encoding Cyclin-D1), and Ccna2 (encoding Cyclin-A2) did not differ between HFD-fed control and DhpsΔβ islets (Fig. 5B). However, Cyclin-D2 protein levels in islets were significantly reduced in DhpsΔβ islets compared to controls following 1 week of HFD feeding (Fig. 5C). By contrast, no differences were seen in the protein levels of Cyclin-D1 (Fig. 5C).
Figure 5: Decreased mRNA translation of Cyclin-D2 in DhpsΔβ mice.
(A) Schematic showing experimental design. Control (Cre+) and DhpsΔβ (βKO) mice were fed a HFD for 1 week and islets were isolated and subjected to RT-PCR and polyribosome profiling (PRP) analysis. (B) Quantitative RT-PCR from islets for the genes indicated. Data from N=5–6 animals per group. (C) Representative immunoblot analysis from islets for Cyclin-D2, Cyclin-D1, and Erk½ (left) and quantification of immunoblots (right). Data from N=3–5 animals per group. (D) PRP of isolated islets (left) and quantification of polyribosome:monoribosome ratio (right). Data from N=3 mice per group. (E) Quantitative RT-PCR of Cycd2 in polyribosomal fractions. Data from N=3–4 mice per group. (F) Quantitative RT-PCR of Cycd1 in polyribosomal fractions. Data from N=3–4 mice per group. Data presented as mean ± SEM; *p-value < 0.05 by T test.
Given the dissociation between mRNA levels and protein levels for Cyclin-D2, we next asked if loss of DHPS impaired the translation of Ccnd2 mRNA. We performed polyribosome profiling (PRP) studies using lysates from islets of 1-week HFD-fed animals. PRP involves sucrose gradient sedimentation of cellular extracts to fractionate total RNAs based on their association with ribosomes (27). RNAs associated with monoribosomes sediment higher in the gradient compared with those that are associated with multiple ribosomes (polyribosomes)—and hence are actively translated. DhpsΔβ islets were similar to Cre-positive control islets in terms of the polyribosome:monoribosome ratio (or P/M ratio, which reflects the relative association of RNAs with polyribosomes compared to monoribosomes) (Fig. 5D). This result suggested that global translation of RNAs was unaffected by deletion of Dhps in β cells. To interrogate the translation of specific mRNAs, we performed RT-PCR analysis for Ccnd2 and Ccnd1 on sedimentation gradient fractions. Ccnd2 mRNA exhibited a shift in occupancy toward monoribosomes in DhpsΔβ islets compared to Cre-positive control islets, indicating its absence of engagement with polyribosomes (Fig. 5E). In contrast, the polyribosome engagement of Ccnd1 mRNA was similar between DhpsΔβ islets and Cre-positive controls (Fig. 5F). Collectively, these results suggest reduced translational initiation of Ccnd2 in DhpsΔβ mouse islets.
Ornithine decarboxylase and c-Myc lie upstream of DHPS activity
DHPS utilizes the polyamine spermidine as a substrate to generate hypusine on eIF5A. We asked if depletion of polyamines in mice impaired β-cell proliferation similarly to loss of DHPS. The rate-limiting enzyme for the production of polyamines is ornithine decarboxylase (ODC), which is irreversibly inhibited by difluoromethylornithine (DFMO) (28). Male C57BL/6J mice were given DFMO (or vehicle) in their drinking water for 5 days prior to and concurrently with 1 week of HFD feeding. DFMO-treated mice had similar body weight, glucose tolerance, and β-cell mass as vehicle-treated mice (Supplemental fig. S2A–D). Notably, similar to DhpsΔβ mice, DFMO-treated mice exhibited reduced β-cell proliferation compared to controls (Supplemental fig. S2E and S2F), suggesting that ODC activity and polyamines are required for the β-cell proliferative response to HFD-feeding.
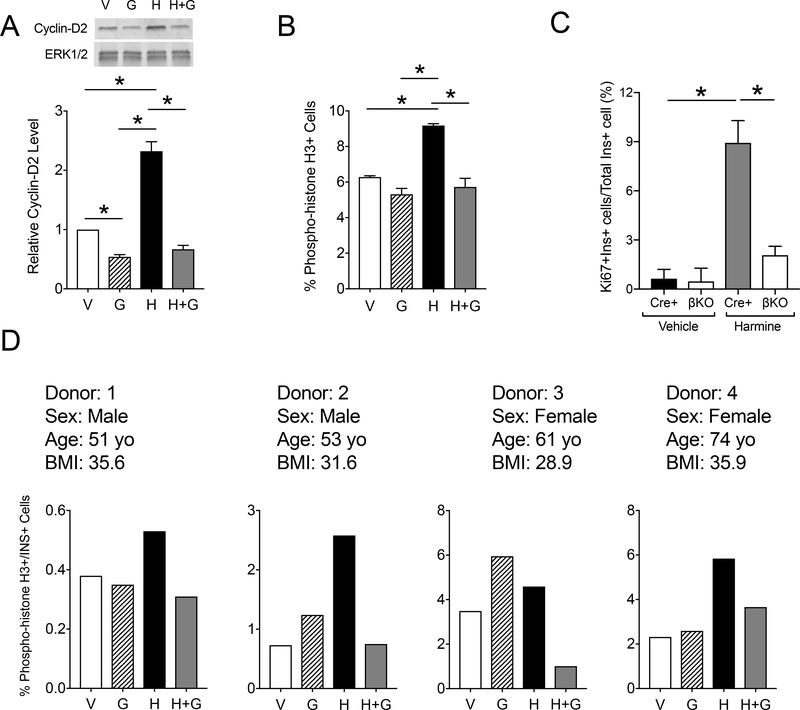
The transcriptional activity of the gene encoding ODC (Odc1) is regulated in part by the oncogenic transcription factor c-Myc (29). To test the possibility that the proliferation axis involving c-Myc and ODC depends upon the downstream activity of DHPS, we utilized a small molecule activator of c-Myc (harmine) (30) and a specific inhibitor of DHPS (Gc7) (31, 32) in mouse islets in vitro. Mouse islets treated with harmine exhibited an increase in levels of Cyclin-D2 by immunoblot, but concurrent treatment with both harmine and Gc7 abolished this effect (Fig. 6A). Flow cytometry analysis (Fig. 6B and Supplemental fig. S3A) of islet cells demonstrated that a marker of cellular mitosis (phosphorylated Histone H3) was increased upon 72 h of harmine treatment and was reduced to baseline upon concurrent treatment with Gc7. To confirm that the effects seen with Gc7 are likely mediated through inhibition of DHPS, we next tested the effects of harmine on islets from DhpsΔβ mice. After harmine treatment, islets from Cre-positive control mice exhibited a significant increase in proliferation as measured by Ki67 staining compared to vehicle treatment; by contrast, treatment of DhpsΔβ mouse islets did not result in a significant increase in Ki67 immunostaining after harmine treatment (Fig. 6C and S3B).
Figure 6: DHPS inhibition attenuates harmine-induced β cell proliferation in mouse and human islets.,
(A) Islets were isolated from male 8–9 week old CD1 mice, then treated in vitro with vehicle (V), harmine (H), and Gc7 (G). Representative immunoblot (top) and quantification of protein levels (bottom) for Cyclin-D2. Data are from N=6 mice per group. (B) Islets were isolated from male 8–9 week old CD1 mice, then treated in vitro with vehicle (V), harmine (H), and Gc7, followed by flow cytometry analysis of dispersed islet cells immunostained for phospho-histone H3. Data are from N=4–6 mice per group. (C). Islets were isolated from control (Cre+) and DhpsΔβ (βKO) mice, then treated in vitro with vehicle (V) or harmine (H), then percent of β cells that immunostained for Ki67 was calculated. Data are from N=3 animals per group. (D) Human islets from 4 donors were treated in vitro with vehicle, harmine, and/or Gc7 (G), dispersed, immunostained for phospho-histone H3, then subjected to flow cytometry analysis. Graphs show a single technical replicate for each donor. Data in A, B, and C are presented as mean ± SEM; *p-value < 0.05 by one-way ANOVA.
To interrogate the c-Myc/ODC/DHPS axis in human islets, we performed flow cytometry for phosphorylated Histone H3 in human islets incubated with Gc7 and/or harmine. Harmine treatment resulted in an increase in phosphorylated histone H3-positive cells in each of the four islet donors, with inhibition of this effect by concurrent Gc7 treatment (Fig. 6D and Supplemental fig. S4). Collectively, these data in mouse and human islets suggest that c-Myc induction of human islet cell proliferation requires DHPS activity.
The activity of DHPS depends on PKC-ζ
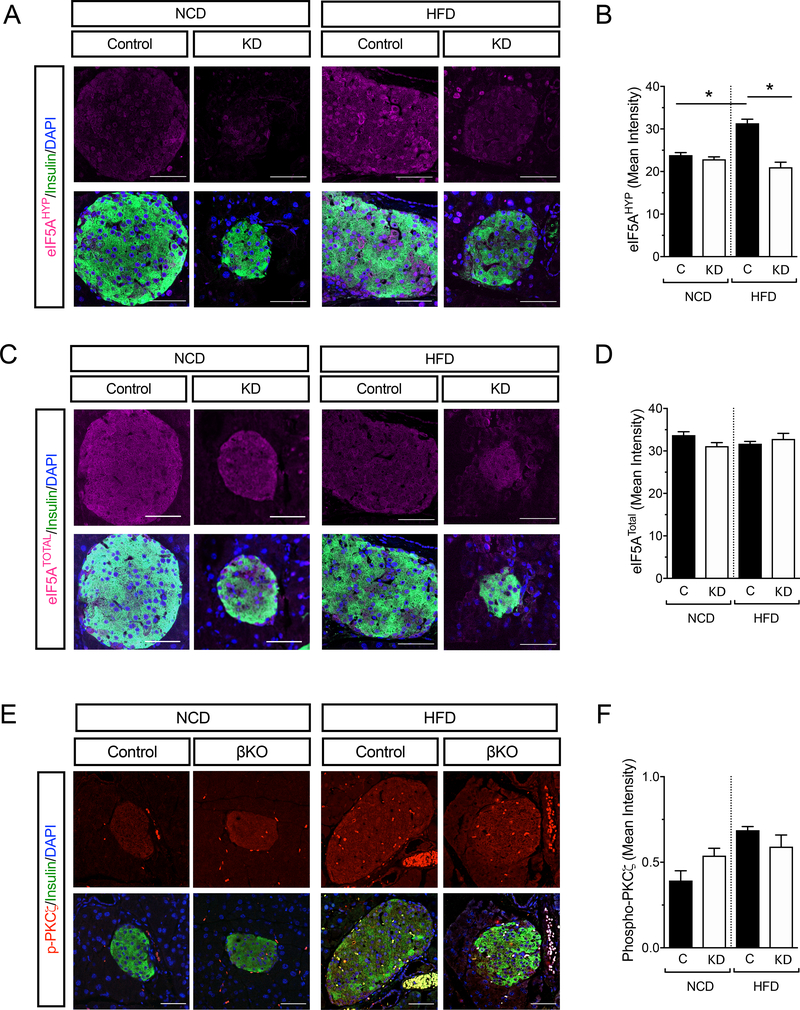
A key pathway that links extracellular mitogenic signals to mRNA translation and β-cell proliferation is the growth factor/mTORC1 pathway (23, 33–37). Protein kinase C-ζ (PKC-ζ) is proximal to mTORC1 (38, 39) and in mice harboring kinase-dead-PKC-ζ protein in β cells (β-KD-PKC-ζ), the proliferative response to obesity and insulin resistance is impaired (23)—a phenotype similar to DhpsΔβ mice. To determine how the mTORC1 pathway might be linked to the ODC/DHPS/eIF5A pathway, we examined the levels of eIF5AHyp in the islets of HFD-fed β-KD-PKC-ζ mice by immunostaining. The staining intensity for eIF5AHyp was increased in the islets of control animals, but not in those of β-KD-PKC-ζ mice upon HFD feeding (Fig. 7A and 7B). Notably, staining intensity of eIF5ATotal did not change with diet or genotype (Fig. 7C and 7D). These findings suggest that the phenotype of β-KD-PKC-ζ mice may be due to the loss of eIF5AHyp formation. To confirm that DHPS is downstream of PKC-ζ, we next examined if activation of PKC-ζ (through its phosphorylation at residue Thr410) occurred in DhpsΔβ mice. Immunostaining showed that the phosphorylation of PKC-ζ did not differ between control and DhpsΔβ mice fed either a NCD or HFD (Fig. 7E and 7F). Collectively, these data suggest that DHPS lies downstream of PKC-ζ.
Figure 7: Hypusine generation requires PKCζ.
Wildtype (WT) and kinase-dead-PKCζ (KD) mice were fed a NCD or HFD for 1 week. (A) Representative images of pancreata stained for eIF5AHyp (magenta), insulin (green), and nuclei (DAPI, blue). (B) Quantification of eIF5AHyp levels from (A). Data are from N=3 mice per group. (C) Representative images of pancreata stained for eIF5ATotal (magenta), insulin (green), and nuclei (DAPI, blue). (D) Quantification of eIF5ATotal levels from (B). Data are from N=3 mice per group. (E) Representative images of NCD- and HFD-fed control and DhpsΔβ (βKO) mouse pancreata immunostained for phospho-PKCζ (red), insulin (green), and nuclei (DAPI, blue). Scale bar, 50 μm. (F) Quantification of phospho-PKCζ immunostaining in insulin positive cells shown in (E). Data are from N=3 mice per group. Data presented as mean ± SEM; *p-value < 0.05 for the comparisons shown by one-way ANOVA.
DISCUSSION
The polyamines putrescine, spermidine, and spermine participate in cellular replication, and their depletion by inhibition of ODC has been a focus of antiproliferative therapies for several cancers (40, 41). A specific role for spermidine was suggested in our prior zebrafish study (42) showing that inhibition of ODC, which results in reduced pancreatic cell proliferation, can be rescued by supplementation with spermidine. However, the mechanism of this rescue remained elusive. A specific role of DHPS as a downstream effector of spermidine has not been rigorously tested in mammals in vivo, owing in part to the embryonic lethality of mice harboring germline homozygous mutations in Dhps (6, 7). Our studies utilizing a conditional knockout mouse model of Dhps allowed the demonstration of a role of Dhps in β-cell proliferation.
Two concepts regarding the growth and replication of postnatal β cells have emerged. First, although β cells were originally considered to be post-mitotic cells in the adult mammal, it is now recognized that they exhibit a slow rate of replication that decreases with age (11, 12, 17, 43). Second, the mass of β cells can be altered to compensate for insulin resistance that occurs during physiologic or pathologic processes, such as growth, gestation, and obesity and insulin resistance (44, 45). The mechanisms that underlie the compensatory gains in β-cell mass are thought to involve both increases in cell size and number, and with respect to the latter, the molecular pathways triggering cell cycle entry remain incompletely understood. In this study, we focused on the molecular mechanisms at the onset of compensatory β-cell mass expansion during obesity/insulin resistance. Using an inducible β-cell-specific knockout mouse model, we found that DHPS activity was increased in islets in response to HFD feeding, that DHPS was necessary for the induction of β-cell replication and for the maintenance of normal glucose homeostasis following HFD feeding, that DHPS promoted the mRNA translation of the critical cell cycle regulator Ccnd2, and that DHPS activity was downstream of c-Myc and PKCζ. Our studies do not entirely rule out a role for DHPS in the maintenance of β-cell function (in addition to mass); in many cases, mass and function are closely intertwined, and the defects we observed in our proteomics data (with alterations in pathways of protein translation and secretion) may indeed suggest some loss in β-cell function.
DHPS catalyzes the first and rate-limiting step of the hypusine modification of eIF5A (2). eIF5AHyp facilitates the elongation and termination phases of mRNA translation in yeast (46). In mammals, it is unclear if eIF5AHyp functions as a general translation factor or a factor specific for certain mRNAs. A proteomics study of eIF5A-depleted HeLa cells has revealed alterations in protein folding and mRNA translation pathways, suggestive of a role for the factor in ER stress-related processes (47). In our study, we demonstrated that loss of DHPS resulted in the expected reduction in eIF5AHyp levels and acute reduction in the translation of a key cell cycle gene whose product, Cyclin-D2, was necessary for adaptive β-cell replication. Notably, the loss of Dhps in β cells did not acutely affect translation globally, because there were no changes in either the islet PRP analysis or in the specific translation of another cell cycle gene (Ccnd1). Moreover, the loss of Dhps appeared to cause an mRNA translation initiation block, rather than a block in translation elongation or termination. With respect to the latter point, our findings suggest that Dhps had an effect that was independent of eIF5AHyp, a possibility that we think is unlikely, or that resulted from the accumulation of the unhypusinated form of eIF5A. Resolution of these possibilities will require the conditional deletion of the gene encoding eIF5A. We should note that our findings are distinct but mechanistically consistent with our prior observations that DHPS promotes the translation of key proteins involved in the apoptotic phase of ER stress in pancreatic islets (20).
Little is known about the regulation of eIF5AHyp formation. We linked two regulators of cellular proliferation, PKCζ and c-Myc, to hypusination. PKCζ is an atypical Ser/Thr protein kinase that lies proximal to mTOR activation (23, 39). The phenotype of β-cell PKCζ kinase-dead mice are similar to that of DhpsΔβ mice described here, in which defects in HFD-induced Cyclin-D2 activation and β-cell proliferation result in a diabetic phenotype (23). This observation led us to explore a possible link between the PKCζ/mTOR and DHPS/eIF5AHyp pathways, and we observed that eIF5A hypusination in β cells in response to HFD feeding required intact PKCζ activity. Conversely, activation of PKCζ (as assessed by phospho-PKCζ immunostaining) did not appear to be dependent upon DHPS. These findings therefore place PKCζ upstream of DHPS/eIF5AHyp in a pathway that links nutritional signals from HFD feeding to β-cell replication. c-Myc is an oncogenic transcription factor that can drive rodent and human β-cell replication (30, 48, 49). Because c-Myc is a direct transcriptional activator of the gene encoding ODC (Odc1) (29), we surmised that the effect of c-Myc in inducing proliferation may be mediated at least partially through the ODC/DHPS/eIF5AHyp axis. Our studies using the DYRK1A inhibitor harmine (which indirectly activates c-Myc) (30) suggest that the pro-proliferative effects of c-Myc are abrogated when DHPS is inhibited in mouse and human islets.
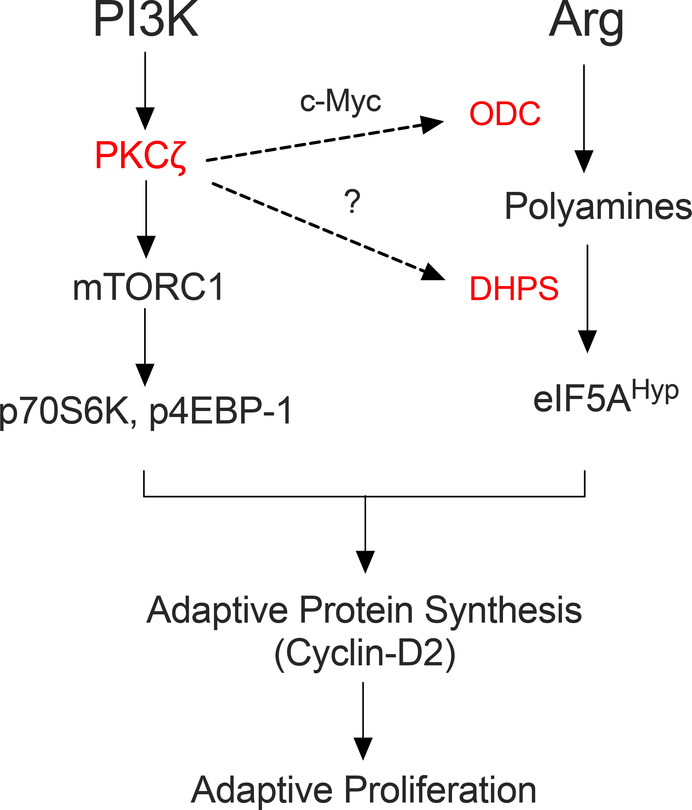
In conclusion, we demonstrated a role for DHPS in β-cell replication. Our studies involving the PKCζ/mTOR and c-Myc/ODC pathways implicate DHPS as a key node through which these pathways affect replication in the β cell (Fig. 8). Our work opens several future avenues of investigation, including the mechanism by which PKCζ influences eIF5A hypusination (through ODC compared to DHPS), the role of exogenous and endogenously-derived spermidine as a substrate for DHPS during facultative proliferation, and the potential independent effect of accumulating unhypusinated eIF5A on the proliferative response. The resolution of these and other related issues must await the analysis of animal models with conditional knockout of Odc1 and Eif5a1.
Figure 8. Proposed pathway linking PKCζ, c-Myc, and polyamines to adaptive proliferation in β cells.
The schematic diagram shows the proposed positioning of the PI3K- PKCζ-mTOR pathway (left arm of the diagram) and the polyamine-eIF5A pathway (right arm of the diagram) relative to the adaptive translational and proliferative responses. The hierarchy of the factors depicted in red (PKCζ, c-Myc, ODC, and DHPS) have been shown in this study or others to serve as key nodes through which proliferative responses are mediated – deletion or inhibition of these factors severely restricts the ability of the β cell to produce Cyclin-D2 and undergo replication. This study demonstrated that DHPS is downstream of PKCζ, c-Myc, and ODC functions. This study does not explicitly rule out the possibility that PKCζ might directly or indirectly regulate the function of DHPS (shown as a dashed line with a question mark).
MATERIALS AND METHODS
Animal Studies
Mice containing the floxed Dhps alleles were crossed to MIP1-CreERT (21) mice to generate DhpsΔβ mice on the C57BL/6J background. Animals were maintained under protocols approved by the Indiana University School of Medicine Institutional Animal Care and Use Committee. The following primers were used for genotyping DhpsΔβ mice: 5’-GTAAACTAGAGTTCTGCGATGGGTGG-3’ (forward) and 5’-TCAATCTGGTCATAAGGGCACAGG-3’ (reverse), and were expected to generate a 319 bp band for the wild-type allele and 396 bp for the floxed allele. Mice were crossed to B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J and pancreas tissue was harvested to visualize Tomato expression. Male mice with the desired genotype were then weaned and utilized for all the experiments in this study. Mice were administered 3 daily intraperitoneal injections of 2.5 mg of tamoxifen dissolved in peanut oil at 8 weeks of age. Mice were then allowed to acclimate for 1 week prior to being placed on either a NCD (16% kcal from fat; Research Diets; D12492) or HFD (60% kcal from fat; Research Diets; D12492). Glucose and insulin tolerance tests were performed as described (18). Mice harboring kinase-dead-PKC-ζ protein in β-cells (β-KD-PKC-ζ) (23) were placed on either a NCD or HFD at 8 weeks of age as previously described (23) and euthanized for pancreas tissue after 1 week.
Cell Isolation and Culture
Mouse islets were cultured in 11 mM glucose as previously described (50). Human islets were obtained from the Integrated Islet Distribution Program or from the University of Alberta and maintained in complete Standard Islet Medium (Prodo Labs) and supplemented with ciprofloxacin (Fisher Scientific). Mouse and human islets were pretreated with 100 μM Gc7 or vehicle (10 mM acetic acid) for 16 hours prior to concurrent treatment with 10 μM harmine (Sigma Aldrich) or vehicle (dimethylsulfoxide) for 48 or 72 hours prior to collection for western blot or flow cytometry analysis.
Immunostaining and morphometric assessment of β-cell mass and β-cell death
β-cell mass was calculated as previously detailed using at least five mice per group (51). Pancreata and isolated islets were stained using the following antibodies: anti-insulin (1:350, #A0564, Dako), anti-Ki67(1:200, #Ab66155–200, Abcam), anti-eIF5AHyp (1:200, IU-44), and anti-eIF5ATotal (1:200, #611977, BD Pharmingen), anti-phospho-PKC-ζ (1:100, SAB4503773, Sigma-Aldrich and DAPI (Thermo Fisher). Apoptosis was detected using the ApopTag Red In Situ Apoptosis Detection Kit (Millipore Sigma). Images were acquired using a Zeiss LSM800 microscope (Carl Zeiss).
Polyribosome Profiling (PRP) and RT-PCR
PRP experiments using isolated islets proceeded as described previously (52). Total RNA from the PRP fractions was reverse transcribed and subjected to SYBR Green-I-based quantitative real-time PCR. Polyribosome-to-monoribosome (P/M) ratios were quantitated by calculating the area under the curve corresponding to the polyribosome peaks (more than two ribosomes) divided by the area under the curve for the monoribosome (80S) peak. Primers for Ccnd2, Ccna2, and Ccnd1 were described previously (24, 26).
Immunoblot Analysis
Whole-cell extracts from mouse islets were prepared as described previously and subjected to a 4–20% gradient SDS-PAGE (53). For immunoblots, antibodies were as follows: anti-eIF5AHyp (1:3000) (54); anti-eIF5ATotal (1:3000, Clone 26, BD Biosciences); anti-ERK1/2 (1:1,000, sc-94, Santa Cruz); anti-Cyclin-D2 (1:1,000, AB-2, Thermo Fisher Scientific); anti-Cyclin-D1 (1:1000, AB-1, Thermo Fisher Scientific); anti-β-tubulin (1:3000, 9F3, Cell Signaling); anti-DHPS (1:1000, sc-365077, Santa Cruz). Immunoblots were visualized using fluorescently-labeled secondary antibodies (LI-COR Biosciences) and were quantified using LI-COR software.
RNA-sequencing library preparation and sequencing
Purified total RNA was first evaluated for its quantity and quality using and Agilent Bioanalyzer 2100. All RNA samples had a RIN (RNA Integrity Number) of 8 or higher. Ten ng of total RNA per sample were used for library preparation. cDNA was first synthesized through reverse transcription with poly-dT priming, using SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara Clontech Laboratories, Inc.), followed by cDNA shearing with Covaris AFA sonicator (Covaris) and size selection with AMPure beads (Beckman Coulter). Barcoded cDNA library was then prepared using Ion Plus Fragment Library Kit (Thermo Fisher Scientific). Each library was quantified and its quality assessed by Agilent Bioanalyzer, and multiple libraries were pooled in equal molarity. Average size of library insert was approximately 150 bp. Eight microliters of 100 pM pooled libraries were applied to Ion Sphere Particles (ISP) template preparation and amplification using Ion OneTouch 2, followed by ISP loading onto a PI® Chip and sequencing on Ion Proton semiconductor (Thermo Fisher Scientific). Each PI® Chip allows loading of about 140 million ISP templates, generating approximately 100 million reads up to 10–15 Gbp. A Phred quality score (Q score) was used to measure the quality of sequencing. More than 90% of the sequencing reads reached Q30 (99.9% base call accuracy).
RNA-Sequencing alignment and analysis
All sequenced libraries were mapped to the mouse genome (UCSC mm10) using STAR RNA-seq aligner (55). The reads distribution across the genome was assessed using bamutils (from ngsutils) (56). Uniquely mapped sequencing reads were assigned to mm10 refGene genes using featureCounts (57). Differential expression (DE) analyses were performed using edgeR v3.22.3 implemented in the Bioconductor package (58) to identify differentially-expressed mRNAs between control and DhpsΔβ samples. Biological coefficients of variation between the samples were estimated using an empirical Bayes approach under the assumption that the data follows a negative binomial distribution. We filtered out low expression transcripts based on percentage of samples (less than 50%) and CPM cutoff of 1. Statistical significance was defined as p-value ≤ 0.05 and a fold change (FC) ≥ 2 of expression level between comparison of knockout mice and controls. The heat map and locus-by-locus volcano plot were performed using R package. DAVID was used for the functional enrichment analysis (59), based on annotation files from GO (60), to identify biological pathways that were significantly enriched.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Isolated islet samples were treated with 8 M urea in 50 mM Tris-HCl and sonicated for protein extraction. For each sample set, an equal amount of protein starting material was reduced and alkylated with tris(2-carboxyethyl)phosphine (TCEP) and chloroacetamide (CAM) and digested with Trypsin Gold (Promega) over night. Samples were labeled with 0.2 mg of TMT mass tagging reagents (Proteome Sciences via Thermo Scientific). Approximately 100 μg of the mixed sample was fractionated using Pierce High pH Reversed-Phase Peptide Fractionation Kit (Thermo Scientific) and subject to LC-MS/MS analysis.
Column chromatography was performed in-line with a Fusion Lumos Orbitrap mass spectrometer with advanced precursor determination and Easy-IC using a Nanoflex Easy nanospray ion source. Data were acquired with the full MS acquisition performed at a resolution of 60,000 and MS/MS analysis performed at a resolution of 50,000. MS/MS fragmentation was performed by higher-energy collision dissociation (HCD) with a collision energy of 36.
MS/MS database search was performed with the relevant Uniprot FASTA database using SEQUEST HT within Proteome Discoverer 2.2 (PD 2.2, Thermo). The reporter ion quantitation node within PD 2.2 was used for quantitation following sample normalization based on the total amount of protein detected for each reporter group. For significance analysis, p-values were calculated using an ANOVA and a Tukey HSD post-hoc test. Locus-by-locus volcano plot were performed using R package.
Flow cytometry
Islets were dispersed using 1 ml of Accumax (#07921, StemCell Technologies) containing 2 U/mL of DNAse I and PluronicTM F-127 for 15 min at 37 °C. Single cells were stained with 25 μM Newport Green PDX for 90 min at 37 °C. Cells were washed, resuspended in 1 mL of PBS and incubated at 37 °C for 30 min. Cells were fixed with 2 % paraformaldehyde for 10 min and subsequently permeabilized with Transcription Factor Staining Buffer Set (Invitrogen). Intracellular staining was carried out in perm/wash buffer containing Ki67-PE (Clone 16A8, Biolegend) and Histone H3 (pS28)-Alexa Fluor 647 (Clone HTA28, BD), overnight 4 °C. After staining, the cells were washed, filtered and acquired on a FACS Canto II cytometer (BD). Data were analyzed using FlowJo software (Tree Star).
Statistical Analysis
All data are presented as mean ±SEM. For comparisons of two groups, a two-tailed unpaired Student’s t test was used. For comparisons of more than two groups, a one-way ANOVA was used followed by a Tukey post-test.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank Ms. K. Orr, Ms. K. Randhave, and Ms. J. Nelson from the Indiana University Diabetes Research Center Islet and Translation Cores for technical assistance with islet isolations and PCRs. The authors also wish to thank the Integrated Islet Distribution Program and the University of Alberta for provision of human islets, and Dr. L. Philipson (University of Chicago) for provision of the MIP-CreERT mice.
FUNDING
This work was supported by an American Physiological Society Porter Fellowship (to EML), a JDRF Career Development Award (to TLM), a JDRF Postdoctoral Fellowship (to FS), and NIH grants R01 DK060581 and R01 DK105588 (to RGM); R01 DK113079, R01 DK105015, R01 DK077096 (to AG-O). This work utilized core services supported by NIH grant P30 DK097512 (to Indiana University), and core services supported by NIH grant P30 DK020595 (to University of Chicago) were used for the creation of the MIP-CreERT mice. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
COMPETING INTERESTS
The authors declare that they have no competing interests.
DATA AND MATERIALS AVAILABILITY
The RNA sequencing data have been deposited to NCBI GEO with the dataset identifier GSE136581. The mass spectrometry proteomics data have been deposited to ProteomeXchange with the dataset identifier PXD015414. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. The DhpsloxP/loxP mice presented in this study were generated by the authors and require a materials transfer agreement for use outside of Indiana University.
REFERENCES AND NOTES
- 1.Pegg AE, Functions of Polyamines in Mammals, J. Biol. Chem 291, 14904–14912 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park MH, Nishimura K, Zanelli CF, Valentini SR, Functional significance of eIF5A and its hypusine modification in eukaryotes., Amino Acids 38, 491–500 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujimura K, Choi S, Wyse M, Strnadel J, Wright T, Klemke R, Eukaryotic Translation Initiation Factor 5A (EIF5A) Regulates Pancreatic Cancer Metastasis by Modulating RhoA and Rho-associated Kinase (ROCK) Protein Expression Levels, J. Biol. Chem 290, 29907–29919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishimura K, Murozumi K, Shirahata A, Park MH, Kashiwagi K, Igarashi K, Independent roles of eIF5A and polyamines in cell proliferation, Biochem. J 385, 779–785 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strnadel J, Choi S, Fujimura K, Wang H, Zhang W, Wyse M, Wright T, Gross E, Peinado C, Park HW, Bui J, Kelber J, Bouvet M, Guan K-L, Klemke RL, eIF5A-PEAK1 Signaling Regulates YAP1/TAZ Protein Expression and Pancreatic Cancer Cell Growth, Cancer Res. 77, 1997–2007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishimura K, Lee SB, Park JH, Park MH, Essential role of eIF5A-1 and deoxyhypusine synthase in mouse embryonic development., Amino Acids 42, 703–710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Templin AT, Maier B, Nishiki Y, Tersey SA, Mirmira RG, Deoxyhypusine synthase haploinsufficiency attenuates acute cytokine signaling., Cell Cycle 10, 1–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pendeville H, Carpino N, Marine JC, Takahashi Y, Muller M, Martial JA, Cleveland JL, The ornithine decarboxylase gene is essential for cell survival during early murine development, Mol. Cell. Biol 21, 6549–6558 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardbower DM, Asim M, Luis PB, Singh K, Barry DP, Yang C, Steeves MA, Cleveland JL, Schneider C, Piazuelo MB, Gobert AP, Wilson KT, Ornithine decarboxylase regulates M1 macrophage activation and mucosal inflammation via histone modifications, Proc. Natl. Acad. Sci. U. S. A 114, E751–E760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh K, Coburn LA, Asim M, Barry DP, Allaman MM, Shi C, Washington MK, Luis PB, Schneider C, Delgado AG, Piazuelo MB, Cleveland JL, Gobert AP, Wilson KT, Ornithine Decarboxylase in Macrophages Exacerbates Colitis and Promotes Colitis-Associated Colon Carcinogenesis by Impairing M1 Immune Responses, Cancer Res. (2018), doi: 10.1158/0008-5472.CAN-18-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregg BE, Moore PC, Demozay D, Hall BA, Li M, Husain A, Wright AJ, Atkinson MA, Rhodes CJ, Formation of a human β-cell population within pancreatic islets is set early in life., J Clin Endocrinol Metab 97, 3197–3206 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teta M, Long SY, Wartschow LM, Rankin MM, Kushner JA, Very slow turnover of β-cells in aged adult mice, Diabetes 54, 2557–2567 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Kahn SE, Prigeon RL, McCulloch DK, Boyko EJ, Bergman RN, Schwartz MW, Neifing JL, Ward WK, Beard JC, Palmer JP, Quantification of the Relationship Between Insulin Sensitivity and β-Cell Function in Human Subjects: Evidence for a Hyperbolic Function, Diabetes 42, 1663–1672 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Gupta D, Jetton TL, LaRock K, Monga N, Satish B, Lausier J, Peshavaria M, Leahy JL, Temporal characterization of β cell-adaptive and -maladaptive mechanisms during chronic high-fat feeding in C57BL/6NTac mice, J. Biol. Chem 292, 12449–12459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saisho Y, Butler AE, Manesso E, Elashoff D, Rizza RA, Butler PC, β-Cell Mass and Turnover in Humans: Effects of obesity and aging, Diabetes Care 36, 111–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dor Y, Brown J, Martinez OI, Melton DA, Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation, Nature 429, 41–46 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Meier JJ, Butler AE, Saisho Y, Monchamp T, Galasso R, Bhushan A, Rizza RA, Butler PC, Beta-cell replication is the primary mechanism subserving the postnatal expansion of beta-cell mass in humans., Diabetes 57, 1584–1594 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maier B, Ogihara T, Trace AP, Tersey SA, Robbins RD, Chakrabarti SK, Nunemaker CS, Stull ND, Taylor CA, Thompson JE, Dondero RS, Lewis EC, Dinarello CA, Nadler JL, Mirmira RG, The unique hypusine modification of eIF5A promotes islet beta cell inflammation and dysfunction in mice, J. Clin. Invest 120, 2156–2170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishiki Y, Adewola A, Hatanaka M, Templin AT, Maier B, Mirmira RG, Translational Control of Inducible Nitric Oxide Synthase by p38 MAPK in Islet beta-Cells, Mol Endocrinol 27, 336–349 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robbins RD, Tersey SA, Ogihara T, Gupta D, Farb TB, Ficorilli J, Bokvist K, Maier B, Mirmira RG, Inhibition of deoxyhypusine synthase enhances islet {beta} cell function and survival in the setting of endoplasmic reticulum stress and type 2 diabetes, J. Biol. Chem 285, 39943–39952 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamarina NA, Roe MW, Philipson LH, Characterization of mice expressing Ins1 gene promoter driven CreERT recombinase for conditional gene deletion in pancreatic β-cells, Islets 6, e27685 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H, A robust and high-throughput Cre reporting and characterization system for the whole mouse brain, Nat. Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lakshmipathi J, Alvarez-Perez JC, Rosselot C, Casinelli GP, Stamateris RE, Rausell-Palamos F, O’Donnell CP, Vasavada RC, Scott DK, Alonso LC, Garcia-Ocaña A, PKCζ Is Essential for Pancreatic β-Cell Replication During Insulin Resistance by Regulating mTOR and Cyclin-D2, Diabetes 65, 1283–1296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosser RE, Maulis MF, Moullé VS, Dunn JC, Carboneau BA, Arasi K, Pappan K, Poitout V, Gannon M, High-fat diet-induced β-cell proliferation occurs prior to insulin resistance in C57Bl/6J male mice, Am. J. Physiol. Endocrinol. Metab 308, E573–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma RB, O’Donnell AC, Stamateris RE, Ha B, McCloskey KM, Reynolds PR, Arvan P, Alonso LC, Insulin demand regulates β cell number via the unfolded protein response, J. Clin. Invest 125, 3831–3846 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamateris RE, Sharma RB, Hollern DA, Alonso LC, Adaptive β-cell proliferation increases early in high-fat feeding in mice, concurrent with metabolic changes, with induction of islet cyclin D2 expression., Am J Physiol Endocrinol Metab 305, E149–59 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans-Molina C, Hatanaka M, Mirmira RG, Lost in translation: endoplasmic reticulum stress and the decline of β-cell health in diabetes mellitus., Diabetes Obes Metab 15 Suppl 3, 159–169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poulin R, Lu L, Ackermann B, Bey P, Pegg AE, Mechanism of the irreversible inactivation of mouse ornithine decarboxylase by alpha-difluoromethylornithine. Characterization of sequences at the inhibitor and coenzyme binding sites, J. Biol. Chem 267, 150–158 (1992). [PubMed] [Google Scholar]
- 29.Bello-Fernandez C, Packham G, Cleveland JL, The ornithine decarboxylase gene is a transcriptional target of c-Myc, Proc. Natl. Acad. Sci. U. S. A 90, 7804–7808 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang P, Alvarez-Perez J-C, Felsenfeld DP, Liu H, Sivendran S, Bender A, Kumar A, Sanchez R, Scott DK, Garcia-Ocaña A, Stewart AF, A high-throughput chemical screen reveals that harmine-mediated inhibition of DYRK1A increases human pancreatic beta cell replication, Nat. Med 21, 383–388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park MH, Wolff EC, Lee YB, Folk JE, Antiproliferative effects of inhibitors of deoxyhypusine synthase. Inhibition of growth of Chinese hamster ovary cells by guanyl diamines., J Biol Chem 269, 27827–27832 (1994). [PubMed] [Google Scholar]
- 32.Umland TC, Wolff EC, Park MH, Davies DR, A new crystal structure of deoxyhypusine synthase reveals the configuration of the active enzyme and of an enzyme.NAD.inhibitor ternary complex., J Biol Chem 279, 28697–28705 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Balcazar N, Sathyamurthy A, Elghazi L, Gould A, Weiss A, Shiojima I, Walsh K, Bernal-Mizrachi E, mTORC1 activation regulates beta-cell mass and proliferation by modulation of cyclin D2 synthesis and stability., J Biol Chem 284, 7832–7842 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alejandro EU, Gregg B, Wallen T, Kumusoglu D, Meister D, Chen A, Merrins MJ, Satin LS, Liu M, Arvan P, Bernal-Mizrachi E, Maternal diet-induced microRNAs and mTOR underlie β cell dysfunction in offspring, J. Clin. Invest 124, 4395–4410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartolome A, Guillén C, Role of the mammalian target of rapamycin (mTOR) complexes in pancreatic β-cell mass regulation, Vitam. Horm 95, 425–469 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Blandino-Rosano M, Chen AY, Scheys JO, Alejandro EU, Gould AP, Taranukha T, Elghazi L, Cras-Méneur C, Bernal-Mizrachi E, mTORC1 signaling and regulation of pancreatic β-cell mass., Cell Cycle 11, 1892–1902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu Y, Lindner J, Kumar A, Yuan W, Magnuson MA, Rictor/mTORC2 is essential for maintaining a balance between β-cell proliferation and cell size., Diabetes 60, 827–837 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vasavada RC, Wang L, Fujinaka Y, Takane KK, Rosa TC, Mellado-Gil JMD, Friedman PA, Garcia-Ocaña A, Protein kinase C-zeta activation markedly enhances beta-cell proliferation: an essential role in growth factor mediated beta-cell mitogenesis, Diabetes 56, 2732–2743 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Velazquez-Garcia S, Valle S, Rosa TC, Takane KK, Demirci C, Alvarez-Perez JC, Mellado-Gil JM, Ernst S, Scott DK, Vasavada RC, Alonso LC, Garcia-Ocaña A, Activation of protein kinase C-ζ in pancreatic β-cells in vivo improves glucose tolerance and induces β-cell expansion via mTOR activation, Diabetes 60, 2546–2559 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexiou GA, Lianos GD, Ragos V, Galani V, Kyritsis AP, Difluoromethylornithine in cancer: new advances, Future Oncol. 13, 809–819 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M, Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell, J. Mol. Biol 427, 3389–3406 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Mastracci TL, Robertson MA, Mirmira RG, Anderson RM, Polyamine biosynthesis is critical for growth and differentiation of the pancreas, Sci. Rep 5, 13269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teta M, Rankin MM, Long SY, Stein GM, Kushner JA, Growth and regeneration of adult beta cells does not involve specialized progenitors., Dev Cell 12, 817–826 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Nolan CJ, Prentki M, The islet β-cell: fuel responsive and vulnerable., Trends Endocrinol Metab 19, 285–291 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Prentki M, Nolan CJ, Islet β cell failure in type 2 diabetes., J Clin Invest 116, 1802–1812 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuller AP, Wu CC-C, Dever TE, Buskirk AR, Green R, eIF5A Functions Globally in Translation Elongation and Termination, Mol. Cell 66, 194–205.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mandal A, Mandal S, Park MH, Global quantitative proteomics reveal up-regulation of endoplasmic reticulum stress response proteins upon depletion of eIF5A in HeLa cells, Sci. Rep 6, 25795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karslioglu E, Kleinberger JW, Salim FG, Cox AE, Takane KK, Scott DK, Stewart AF, cMyc Is a Principal Upstream Driver of β-Cell Proliferation in Rat Insulinoma Cell Lines and Is an Effective Mediator of Human β-Cell Replication, Mol. Endocrinol 25, 1760–1772 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laybutt DR, Weir GC, Kaneto H, Lebet J, Palmiter RD, Sharma A, Bonner-Weir S, Overexpression of c-Myc in β-Cells of Transgenic Mice Causes Proliferation and Apoptosis, Downregulation of Insulin Gene Expression, and Diabetes, Diabetes 51, 1793–1804 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Evans-Molina C, Robbins RD, Kono T, Tersey SA, Vestermark GL, Nunemaker CS, Garmey JC, Deering TG, Keller SR, Maier B, Mirmira RG, PPAR-{gamma} Activation Restores Islet Function in Diabetic Mice Through Reduction of ER Stress and Maintenance of Euchromatin Structure., Mol Cell Biol 29, 2053–2067 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tersey SA, Levasseur EM, Syed F, Farb TB, Orr KS, Nelson JB, Shaw JL, Bokvist K, Mather KJ, Mirmira RG, Episodic β-cell death and dedifferentiation during diet-induced obesity and dysglycemia in male mice, FASEB J. 32, 6150–6158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hatanaka M, Maier B, Sims EK, Templin AT, Kulkarni RN, Evans-Molina C, Mirmira RG, Palmitate Induces mRNA Translation and Increases ER Protein Load in Islet β-Cells via Activation of the Mammalian Target of Rapamycin Pathway, Diabetes 63, 3404–3415 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iype T, Francis J, Garmey JC, Schisler JC, Nesher R, Weir GC, Becker TC, Newgard CB, Griffen SC, Mirmira RG, Mechanism of insulin gene regulation by the pancreatic transcription factor Pdx-1: application of pre-mRNA analysis and chromatin immunoprecipitation to assess formation of functional transcriptional complexes, J Biol Chem 280, 16798–16807 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Nishiki Y, Farb TB, Friedrich J, Bokvist K, Mirmira RG, Maier B, Characterization of a novel polyclonal anti-hypusine antibody, SpringerPlus 2, 421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner, Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Breese MR, Liu Y, NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets, Bioinformatics 29, 494–496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features, Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 58.McCarthy DJ, Chen Y, Smyth GK, Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation, Nucleic Acids Res. 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang DW, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat. Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Gene ontology: tool for the unification of biology. The Gene Ontology Consortium, Nat. Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.