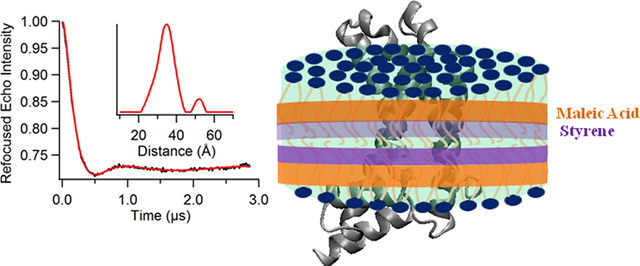
Abstract
Membrane proteins are responsible for conducting essential biological functions that are necessary for the survival of living organisms. In spite of their physiological importance, limited structural information is currently available as a result of challenges in applying biophysical techniques for studying these protein systems. Electron paramagnetic resonance (EPR) spectroscopy is a very powerful technique to study the structural and dynamic properties of membrane proteins. However, the application of EPR spectroscopy to membrane proteins in a native membrane-bound state is extremely challenging due to the complexity observed in inhomogeneity sample preparation and the dynamic motion of the spin label. Detergent micelles are very popular membrane mimetics for membrane proteins due to their smaller size and homogeneity, providing high-resolution structure analysis by solution NMR spectroscopy. However, it is important to test whether the protein structure in a micelle environment is the same as that of its membrane-bound state. Lipodisq nanoparticles or styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) have been introduced as a potentially good membrane-mimetic system for structural studies of membrane proteins. Recently, we reported on the EPR characterization of the KCNE1 membrane protein having a single transmembrane incorporated into lipodisq nanoparticles. In this work, lipodisq nanoparticles were used as a membrane mimic system for probing the structural and dynamic properties of the more complicated membrane protein system human KCNQ1 voltage sensing domain (Q1-VSD) having four transmembrane helices using site-directed spin-labeling EPR spectroscopy. Characterization of spin-labeled Q1-VSD incorporated into lipodisq nanoparticles was carried out using CW-EPR spectral line shape analysis and pulsed EPR double-electron electron resonance (DEER) measurements. The CW-EPR spectra indicate an increase in spectral line broadening with the addition of the styrene- maleic acid (SMA) polymer which approaches close to the rigid limit providing a homogeneous stabilization of the protein-lipid complex. Similarly, EPR DEER measurements indicated a superior quality of distance measurement with an increase in the phase memory time (Tm) values upon incorporation of the sample into lipodisq nanoparticles when compared to proteoliposomes. These results are consistent with the solution NMR structural studies on the Q1-VSD. This study will be beneficial for researchers working on investigating the structural and dynamic properties of more complicated membrane protein systems using lipodisq nanoparticles.
Graphical Abstract
INTRODUCTION
Membrane proteins are involved in essential biological functions that ensure the survival of living organisms. They are pharmacological targets of approximately 50% of all modern medical drugs.1,2 Despite its importance, structural and dynamic information on membrane-bound proteins is lacking when compared to water-soluble proteins. This is due to difficulties in studying membrane proteins. Electron paramagnetic resonance (EPR) spectroscopy is a rapidly growing powerful biophysical technique for studying the structural and dynamic properties of membrane proteins.3–5 The application of EPR spectroscopy is usually challenging due to diffculties in homogeneous sample preparation of membrane proteins in a functionally relevant membrane- bound state. Detergent micelles are the most commonly used membrane mimetics for membrane protein studies. It can form smaller size micelle complexes suitable for high-resolution solution NMR structural studies. However, it is often challenging to examine whether the structure of the proteins in a micelle environment is the same as that of its membrane- bound state. Additionally, the size and shape of detergent micelles may influence the native structural and functional activities of the protein.6–8
Several membrane mimetics have been used to solubilize membrane proteins for biophysical studies which can maintain the structural and functional properties of the protein. Examples of these membrane mimetics are liposomes, bicelles, nanodiscs, and lipodisq nanoparticles.9–14 Each of these membrane mimics have their own advantages and disadvantages. Liposomes or lipid bilayered vesicles are the most relevant membrane mimic of the lipid bilayer. However, it is inhomogeneous in size, having inaccessible interiors, making cytoplasmic domain studies difficult.15 Additionally, incorporation of high concentrations of proteins into liposomes is challenging, making spectroscopic EPR studies more difficult.16,17 Bicelles are another lipid bilayer mimic system forming discs that are composed of a mixture of both long-chain and short-chain phospholipids such as 1,2-dimyristoyl-sn-glycero-3- phosphocholine (DMPC) and 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC).18–21 Bicelles are beneficial because they can provide a lipid bilayer environment and accessibility for the interaction of both extracellular and cytoplasmic domains of membrane proteins.15,18–21 The drawback of using bicelles is that they requires specific types of lipids for bicelle formation which more often challenges its applications as the lipid compositions in the membrane can influence the function of membrane proteins.15,22,23 Membrane scaffold protein (MSP)-based nanodiscs are a very popular method to form homogeneous protein samples in a lipid bilayer environment. This helps to minimize the effect of pockets of high local electron spin concentrations, enhancing the transverse relaxation within a sample.24 The disadvantage of this method is that it requires detergents for protein incorporation which must then be completely removed for the formation of a protein-nanodisc complex.25 Furthermore, the absorbance properties of the membrane scaffold protein may interfere with the target protein. There may be specific scaffold protein-lipid interactions introducing challenges in preparation of nanodiscs.
Recently, lipodisq nanoparticles or styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) have been introduced as a relatively new membrane mimic system that can provide a native-like membrane environment for studying the structural and dynamic properties of membrane proteins. Lipodisq nanoparticles are formed from lipids solubilized by polymers instead of membrane scaffold proteins without interfering with the absorbance properties of the membrane proteins.25–29 The polymer used to solubilize lipids consists of styrene and maleic acid (SMA) at a molar ratio of 3:1. The SMA copolymer can be applied to any kind of lipids and can maintain the native lipid composition in solubilized nanoparticles.30–32 The lipodisq nanoparticles can be formed in a detergent-free environment. It can provide accessibility of the incorporated proteins to both sides of the membrane, which is also suitable for functional studies.33 It can help to understand the mechanistic process by maintaining the oligomerization states of proteins.34 Although lipodisq nanoparticles show a high potential to serve as a good membrane-mimetic system for biophysical studies of membrane proteins, characterization of lipodisq nanoparticles in the presence of membrane proteins is still lacking.13,26
In this study, lipodisq nanoparticles are characterized in the presence of a more complicated membrane protein, the human KCNQ1 voltage sensing domain (Q1-VSD). Human KCNQ1 is a voltage-gated potassium channel modulated by members of the KCNE protein family. KCNQ1 is involved in the cardiac repolarization phase of the heart beat and K+ homeostasis in the inner ear.35–37 Dysfunction of KCNQ1 causes several diseases including several cardiac arrhythmias, congenital deafness, and type II diabetes mellitus.38–40 Q1-VSD is a four-transmembrane protein consisting of 149 amino acids representing the first four helices (S1−S4) KCNQ1.41–43 Q1-VSD is an independent structural and functional unit maintaining the structural conformation and functional properties in a similar manner to the full-length channel (VSD + pore domain).44–48 The voltage-gated ion channels react to electrical fields via reorientations of the S4 helix carrying positive charges within the VSD.49 CW-EPR and pulsed EPR (double-electron electron resonance (DEER)) spectroscopic measurements were conducted on Q1-VSD mutants in 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-racglycerol) (sodium salt) (POPG) lipid with 3:1 SMA polymer for formation of the lipodisq nanoparticles system. The incorporation of Q1-VSD into POPC/POPG lipodisq nanoparticles indicated differences in the side-chain mobility pattern for the spin-labeled residues located in the aqueous phase when compared to the Q1-VSD residues located within the membrane bilayer. The pulse EPR measurements indicated a substantial improvement in the phase memory time (Tm) and DEER distance distribution for the sample in lipodisq nanoparticles when compared to the sample in lipid bilayered vesicles or proteoliposomes. These results are consistent with the structural behavior of Q1-VSD. This study provides useful information for researchers using lipodisq nanoparticles as a membrane mimetic system.
MATERIALS AND METHODS
Site-Directed Mutagenesis.
The His-tag expression vectors (pET-16b) containing a cysteine-less mutant of Q1-VSD were transformed into XL10-Gold Escherichia coli cells (Stratagene). Plasmid extracts from these cells were obtained using the QIAprep Spin Miniprep Kit (Qiagen). Site-directed cysteine mutants were introduced into the cysteine-less Q1-VSD gene using the QuickChange Lightning Site-Directed Mutagenesis kit (Stratagene). The Q1-VSD mutations were confirmed by DNA sequencing from XL10-Gold E. coli (Stratagene) transformants using the T7 primer (Integrated DNA Technologies). Successfully mutated vectors were transformed into Rosetta/C43(DE3) E. coli cells for protein overexpression. Single spin label mutants (Phe130, Gln147, and Val165) were generated by introducing Cys residues at positions 130, 147, and 165. These mutants were chosen to cover the Q1-VSD transmembrane domain (TMD) as well as one in the extracellular region of Q1-VSD. Double-spin label mutants (Phe123/Ser143) were generated by introducing a pair of Cys residues at positions 123 and 143.
Overexpression and Purification.
The overexpression and purification of Rosetta/C43(DE3) E. coli cells carrying mutated Q1-VSD genes were carried out using a previously described protocol.50 Rosetta/C43(DE3) E. coli cells carrying mutants of choice were grown in an M9 minimal medium with 100 μg/mL ampicillin and 50 μg/mL chloramphenicol. The cell culture was incubated at 25 °C and 240 rpm supplemented with MEM vitamin (Mediatech) and ZnCl2 (50 μM) until the OD600 reached 0.8, at which point protein expression was induced using 1 mM IPTG (isopropyl-1-thio-D-galactopyrano-side), followed by continued rotary shaking at 25 °C for 24 h. Cells were harvested by centrifugation of the cultures at 6500 rpm for 20 min at 4 °C. The cell pellets were resuspended in 20 mL of lysis buffer (75 mM Tris-HCl, pH 7.8, 300 mM NaCl, 0.2 mM EDTA) per gram of wet pellet with 2 mM TCEP, lysozyme (0.2 mg/mL), DNase (0.02 mg/mL), RNase(0.02 mg/mL), PMSF (0.2 mg/mL), and magnesium acetate (5 mM) and tumbled at room temperature for 1 h. The cell suspension was then sonicated for 10 min (5 s on/off cycles, Fisher Scientific Sonic Dismembrator model 500, amplitude 40%) on ice. The lysate was centrifuged at 18 000 rpm for 40 min at 4 °C, and the pellet containing the inclusion body was washed with lysis buffer, homogenized with Buffer A (40 Mm HEPES, pH 7.5, 300 mM NaCl) containing 2 mM TCEP and 0.5% dodecylphosphocholine (DPC), and rotated overnight at 4 °C to solubilize inclusion bodies. After detergent solubilization, the insoluble debris was removed by centrifugation at 18 000 rpm for 40 min at 4 °C. The supernatant was incubated with Ni(II)−NTA superflow resin (Qiagen) for 1 h at 4 °C. The protein was then purified using the gravity flow column by washing with 8–10 bed volumes of rinse buffer (Buffer A with 2 mM TCEP) containing 0.05% DPC. Nonspecific proteins were removed with wash buffer (Buffer A with 0.05% DPC, 50 mM imidazole, and 2 mM TCEP). Q1-VSD was eluted with elution buffer (buffer A, 500 mM imidazole with 2 mM TCEP) containing 0.05% 1-myristoyl-2-hydroxy-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (LMPG). Protein samples were concentrated using a Microcon YM-3 (molecular weight cutoff 3000) filter (Amicon). The protein concentration was determined from the A280 using an extinction coefficient of 1.2 mg/mL protein per OD280 on a NanoDrop 200c (Thermo Scientific). The protein purity from overexpression was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Spin Labeling and Reconstitution into Proteoliposomes.
Spin labeling and proteoliposomes reconstitution were carried out following the protocol previously described.51 After purification, each cysteine mutant was concentrated to 0.5 mM. Samples were then reduced with 2.5 mM DTT with gentle agitation at room temperature for 24 h to ensure complete conversion to Cys-SH. 1-Oxyl-2,2,5,5-tetramethylpyrro line-3-methylmethanethiosulfonate (MTSL) spin label was added to 10 mM from a 250 mM solution in methanol into a 0.5 mM Q1-VSD solution, which was then equilibrated at room temperature for 30 min, followed by incubation at 37 °C for 3 h, and further incubated overnight at room temperature. Samples were then buffer exchanged into a 50 mM phosphate, 0.05% LMPG, pH 7.8. Following buffer exchange, samples were bound to nickel resin in a column, which was then washed with 200 mL of 50 mM phosphate, 0.05% DPC, pH 7.8 to remove excess MTSL. The spin-labeled Q1-VSD was eluted using elusion buffer (buffer A, 500 mM imidazole with 2 mM TCEP) containing 0.5% DPC or 1% LMPG detergent. The spin-labeling efficiency (~75%) was determined by comparing the nanodrop UV A280 protein concentration with spin concentration obtained from CW-EPR spectroscopy.
The reconstitution of spin-labeled Q1-VSD protein into POPC/POPG (3:1) proteoliposomes was carried out via dialysis methods following a similar protocol in the literature.13,51,52 The concentrated spin-labeled Q1-VSD protein was mixed with a stock lipid slurry (400 mM SDS, 75 mM POPC, 25 mM POPG, 0.1 mM EDTA, 100 mM IMD, pH 7.5). The lipid slurry was pre-equilibrated to clear mixed micelles via several freeze-thaw cycles. The final protein:lipid molar ratio was set to 1:250. The Q1-VSD-lipid mixture was then subjected to extensive dialysis to remove all detergent present, during which process Q1-VSD/POPC/POPG vesicles spontaneously formed. The 4 L of dialysis buffer (10 mM imidazole and 0.1 mM EDTA at pH 7.5) was changed twice daily. Completion of detergent removal was determined when the Q1-VSD-lipid solution became cloudy and the surface tension of the dialysate indicated complete removal of detergent.
Reconstitution into Bicelles.
The reconstitution of spin-labeled protein into DMPC/DPC bicelles (3.2:1) was carried out following a similar protocol described previously.53,54 DMPC powder was added directly to the Q1-VSD solubilized in elusion buffer (500 mM IMD, pH 7) containing 0.5% DPC. The bicelles were formed by incubating on ice and 42 °C alternatively with gentle vortexing until the solution becomes clear. The final protein:lipid molar ratio was set to 1:500.
Reconstitution into Lipodisq Nanoparticles.
Lipodisq nanoparticles (prehydrolyzed styrene-maleic anhydride copolymer 3:1 ratio) were purchased from Sigma-Aldrich. The protein-lipid complex was incorporated into SMA-lipodisq nanoparticles following the published protocols.13,25,26 A 500 μL aliquot of proteoliposome-reconstituted protein sample (~30 mM POPC/POPG lipid) was added with an equal amount of 2.5% of lipodisq solution prepared in the same dialysis buffer (10 mM IMD, 0.1 mM EDTA at pH 7.5) dropwise over 3–4 min at a weight ratio of 1:1. The protein- lipodisq solution was allowed to equilibrate overnight at 4 °C. The resulting solution was centrifuged at 40 000g for 30 min to remove the nonsolubilized protein. The supernatant was further concentrated to the desired volume and concentration for EPR measurements.
EPR Spectroscopic Measurements.
EPR experiments were conducted at the Ohio Advanced EPR Laboratory. CWEPR spectra were collected at the X-band on a Bruker EMX CW-EPR spectrometer using an ER041xG microwave bridge and ER4119-HS cavity coupled with a BVT 3000 nitrogen gas temperature controller. Each spin-labeled CW-EPR spectrum was acquired by signal averaging 10 42 s field scans with a central field of 3315 G and sweep width of 100 G, modulation frequency of 100 kHz, modulation amplitude of 1 G, and microwave power of 10 mW at different temperatures (297– 325 K). The side-chain mobility was determined by calculating the inverse central line width from each CW-EPR spectrum.
Four pulse DEER experiments were performed using a Bruker ELEXSYS E580 spectrometer equipped with a SuperQFT pulse Q-band system with a 10 W amplifier and EN5107D2 resonator. All DEER samples were prepared at a spin concentration of 100–120 μM; 30% (w/w) deuterated glycerol was used as a cryoprotectant. The sample was loaded into a 1.1 mm inner diameter quartz capillary (Wilmad LabGlass, Buena, NJ) and mounted into the sample holder (plastic rod) inserted into the resonator. DEER data were collected using the standard four-pulse sequence55 [(π/2)ν1 − τ1 − (π)ν1 − t − (π)ν2 − (τ1 + τ2 − t) − (π)ν1 − τ2 − echo] at the Q-band with a probe pulse width of 8/16 ns, pump pulse width of 24 ns, 120 MHz frequency difference between the probe and the pump pulse, shot repetition time determined by the spin-lattice relaxation time (T1), 100 echoes/point, and 16-step phase cycling at 80 K collected out to ~2.0−3.0 μs for overnight data acquisition time (12 h).56 DEER data were analyzed using DEER Analysis 2015.57 The distance distributions P(r) were obtained by Tikhonov regularization58 in the distance domain, incorporating the constraint P(r) > 0. A homogeneous three-dimensional model for micelle samples and a homogeneous two-dimensional model for bicelles, liposomes, and lipodisq nanoparticles samples were used for background correction. The regularization parameter in the L curve was optimized by examining the fit of the time domain. The electron spin relaxation in the x−y plane is the spin echo dephasing time or phase memory time (Tm) of spin labels in lipid vesicles. It determines the feasibility of doing pulse experiments that depend upon echo detection. The electron spin relaxation arises from dipolar interaction among electron spins and the interaction of electron spins with the nuclear spins of the matrix. Transverse relaxation data were collected using the standard Hahn echo pulse sequence [(π/2) − τ1 − (π) − τ1 − echo] at the Q-band with 10/20 ns pulse widths, an initial τ1 of 200 ns, an increment of 16 ns, 100 echoes/point, and 2-step phase cycling at 80 K. The transverse relaxation time (T2) or phase memory time (Tm) was determined by fitting the data with a single-exponential decay.
EPR Spectral Simulations.
EPR spectra were simulated using the nonlinear least-squares (NLSL) program including the macroscopic order, microscopic disorder (MOMD) model developed by the Freed group.59,60 A previously published fitting procedure was used to simulate the CW-EPR spectra.61 The principle components of the hyperfine interaction tensor A = [5.5 ± 0.5, 5.5 ± 0.5, 34.8 ± 0.8] G and g tensors g =[2.0088 ± 0.0002, 2.0073 ± 0.0002, 2.0023 ± 0.0001] were obtained from a least-squares fit to the spectrum of F130C-Q1-VSD in a frozen state at 165 K. During the simulation process, the A and g tensors were held constant and the rotational diffusion tensors were varied. A two-site fit was used to account for both the rigid/slower and the higher/faster motional components of the EPR spectrum. The best-fit rotational correlation times and the relative population of both components were determined using the Brownian diffusion model.
RESULTS AND DISCUSSION
Recently, we characterized the structure of lipodisq nanoparticles in the absence and presence of a single transmembrane protein KCNE1 using dynamic light scattering (DLS), solid state nuclear magnetic resonance (SSNMR) spectroscopy, transmission electron microscopy (TEM), and EPR spectroscopy.62–66 In this study, the lipodisq nanoparticles were characterized using a more complicated and larger spin-labeled membrane protein system (Q1-VSD). The Q1-VSD consisting of four transmembrane domains was reconstituted into POPC/POPG lipid bilayers. The spin-labeled membrane protein was studied using CW-EPR line shape analysis, pulsed EPR phase memory time (Tm) measurements, DEER distance measurements, and side-chain mobility analysis for spin-labeled sites inside and outside of the membrane bilayer. A POPC:POPG (3:1) lipid bilayer was used to mimic phospholipids typically found in mammalian membranes.13,43,51,67 Figure 1 shows the schematic representation of the MTSL spin-label probe and the predicted topology of Q1-VSD in lipid bilayers based on a previous NMR study.50
Figure 1.
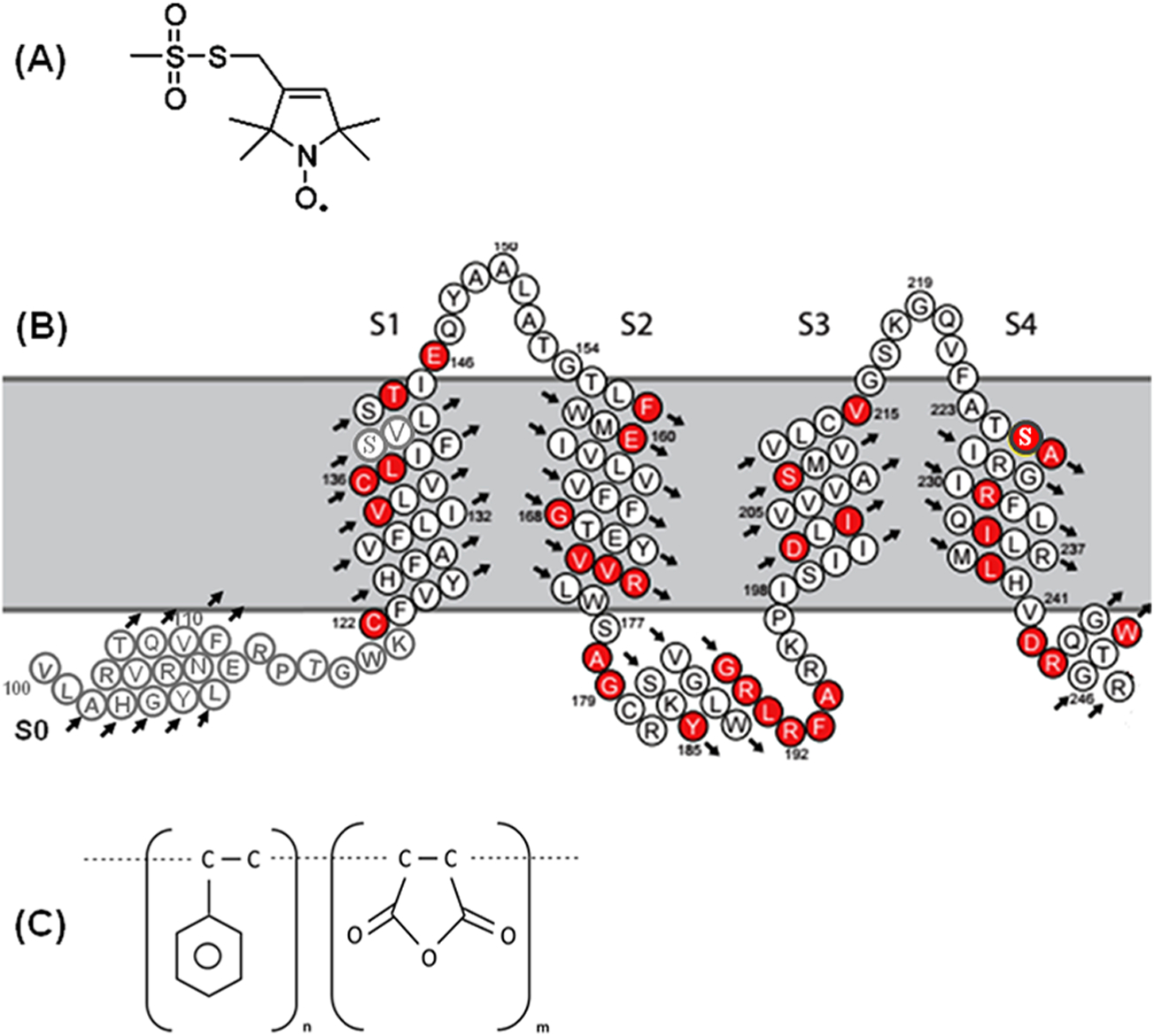
(A) Chemical structure of the MTSL spin label probe, (B) predicted topology of Q1-VSD (100–249) in lipid bilayers based on previous solution NMR studies,50 and (C) chemical structure of 3:1 SMA polymer. Red circles represent the mutants linked to long QT syndrome.
CW-EPR Spectral Measurements.
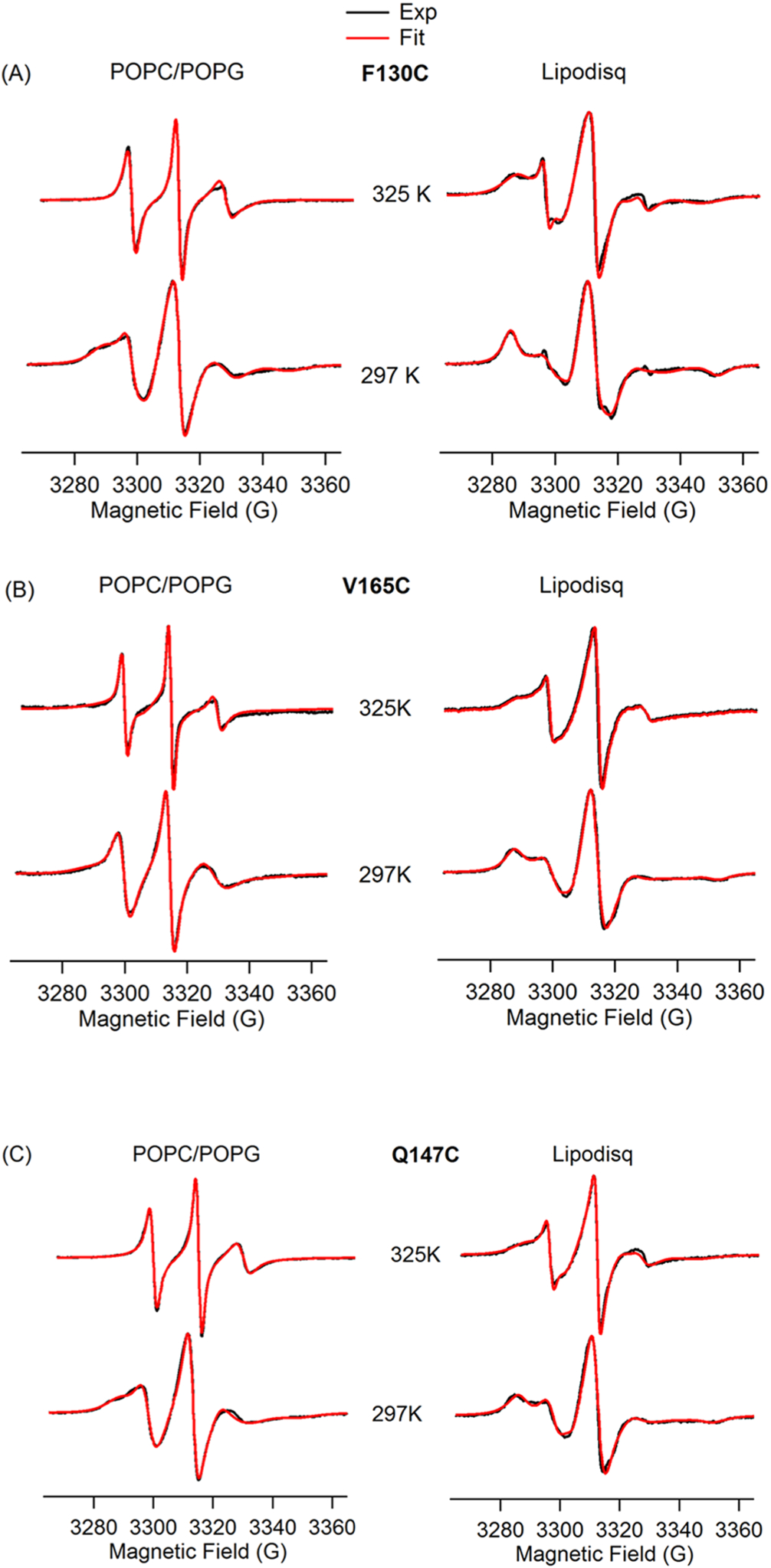
EPR spectroscopy in combination with site-directed spin labeling is a rapidly growing structural biology tool to study membrane proteins in different membrane environments.4,5,68 Important structural and dynamic information on membrane proteins can be obtained from line shape analysis of CW-EPR spectra of site-directed spin labels in different membrane mimetics.4,5,61,65,69–72 CW-EPR experiments were performed on three individual positions (F130C and V165C on the TMDs and Q147C on the outside of the membrane) on Q1-VSD at different temperatures from 297 to 325 K. CW-EPR spectra of spin-labeled Q1-VSD are shown in Figure 2A–C for the mutant F130C on TMD of helix S1, V165C on TMD of helix S2, and Q147C on the outside of the membrane in POPC/POPG lipid bilayers (left) and POPC/POPG lipodisq nanoparticles (right) at temperatures from 297 to 325 K. Inspection of the CW-EPR spectra for the mutant F130C indicates that the spectral line broadening decreases with increasing temperature from 297 to 325 K in POPC/POPG lipid bilayers. The CW-EPR spectrum of the sample in lipodisq nanoparticles shows broader line widths having a nearly immobilized shape at 297 K when compared to the EPR spectral line shape of the sample in POPC/POPG lipid bilayers. However, the line broadenings decrease with increasing temperature up to 325 K. Similarly, inspection of the CW-EPR spectra of the mutant V165C indicates that the spectral line broadening decreases with an increase in temperature from 297 to 325 K in POPC/POPG lipid bilayers. The CW-EPR spectrum of the sample in lipodisq nanoparticles shows a broader line shape having a nearly immobilized shape at 297 K when compared to the EPR spectral line shape of the sample in POPC/POPG lipid bilayers. However, the spectral line widths decrease as the temperature is increased (up to 325 K). The CW-EPR spectra of the mutant Q147C indicate that the spectral line broadening decreases with increasing temperature from 297 to 325 K in POPC/POPG lipid bilayers. The CW-EPR spectrum of the sample in lipodisq nanoparticles also shows broader linewidths having a nearly immobilized shape at 297 K when compared to the EPR spectral line shape of the sample in POPC/POPG lipid bilayers. However, the EPR spectral line widths decrease as the temperature increases (up to 325 K).
Figure 2.
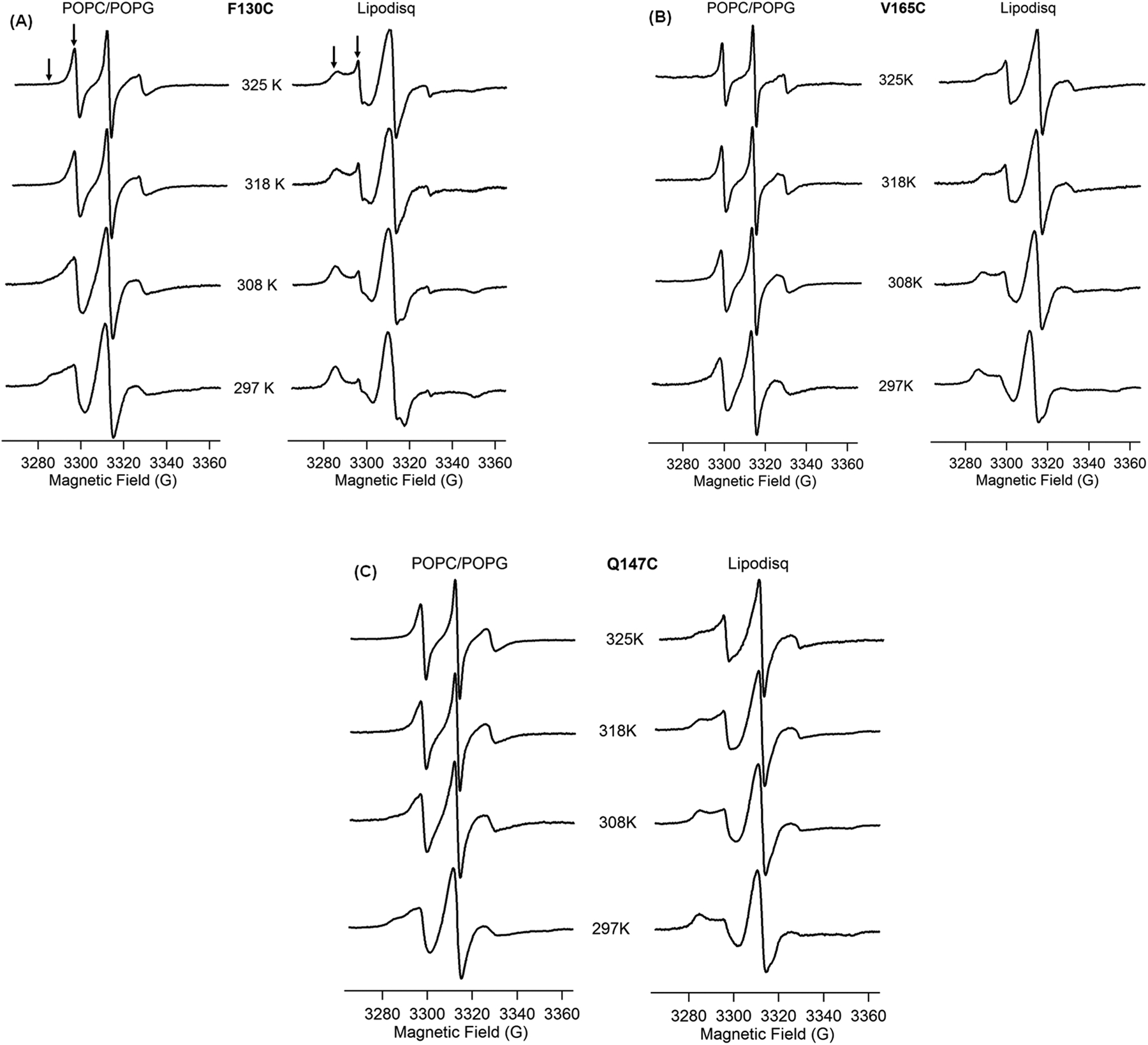
CW-EPR spectra of Q1-VSD bearing MTSL at F130C (A), V165C (B), and Q147C (C) in POPC/POPG liposomes (left) and POPC/POPG lipodisq nanoparticles (right) as a function of temperature. Arrows represent the two motional components in the spectra.
The EPR spectral line widths of spin-labeled Q1-VSD are broader in lipodisq nanoparticles samples when compared to proteoliposome samples, indicating that the motion of the spin-labeled sites is slower in lipodisq nanoparticle samples. The decrease in line broadening pattern with increasing temperature for lipodisq nanoparticles samples is consistent with the liposome samples. The majority of the CW-EPR spectra of all three mutants contain two motional components with a slower/rigid component (left arrow) and a more motional/less rigid component (right arrow). The CW-EPR spectra obtained for these spin-labeled samples are consistent with previously reported CW-EPR spectra for membrane proteins.61,65,67 A decrease in the line broadening in the spectra with increasing temperature is also consistent with the previously reported temperature-dependent SDSL CW-EPR spectra.65,73 The slower component is not so prominent on the spectra of all three sites in POPC/POPG lipid bilayers at higher temperatures from 318 to 325 K. A single-component EPR spectral line shape observed at 318 and 325 K for these sites might be due to the averaging of the two motional components at higher temperature when compared to the spectra collected at 297 K.65 The other possible reason may be the POPC/POPG lipid adopts a liquid phase at higher temperature, so that the lipid acyl chains have high fluidity causing less restrictions to the spin-label sites located inside the membrane, averaging the overall motion of the spin label resulting a single spectral component.74 The spin label at the Q147C site outside of the membrane is more dynamic at higher temperatures. This dynamic motion causes an averaging of the motion of the two components, resulting in a single component in the EPR spectrum.65
In order to describe the spin-label side-chain motion of Q1-VSD more quantitatively, the inverse central line widths were plotted against the temperatures for all three spin-labeled Q1-VSD mutants as shown in Figure 3. Figure 3 shows that the inverse central line width of the F130C site increases from 0.26 to 0.50 G−1 with increasing temperature from 297 to 325 K in liposomes and from 0.13 to 0.37 G−1 with increasing temperature from 297 to 325 K in lipodisq nanoparticles. Similarly, the inverse central line width of the site V165C increases from 0.35 to 0.55 G−1 with increasing temperature from 297 to 325 K in liposomes and from 0.22 to 0.39 G−1 with increasing temperature from 297 to 325 K in lipodisq nanoparticles. The inverse central line width pattern of the site Q147C increases from 0.30 to 0.47 G−1 with increasing temperature from 297 to 325 K in liposome and from 0.27 to0.46 G−1 with increasing temperature from 297 to 325 K in lipodisq nanoparticles. The overall increase in the inverse central line width pattern for all three sites is similar to the increase in temperature from 297 to 325 K. However, the inverse central line width pattern between liposome and lipodisq nanoparticle samples for site Q147C is narrower when compared to that of sites F130C and V165C between liposome and lipodisq nanoparticle samples. This is expected as the lipodisq nanoparticles do not have a significant effect or have a very minor effect on the motion of the spin label for the probe outside the membrane.65,66 The inverse central line width data indicate that the side-chain motion of the spin-labeled residues located inside the membrane bilayer is slower than that of the spin-labeled residues located outside the membrane. This motional behavior is consistent with the previously published membrane protein side-chain dynamics in lipid bilayers.61,65
Figure 3.
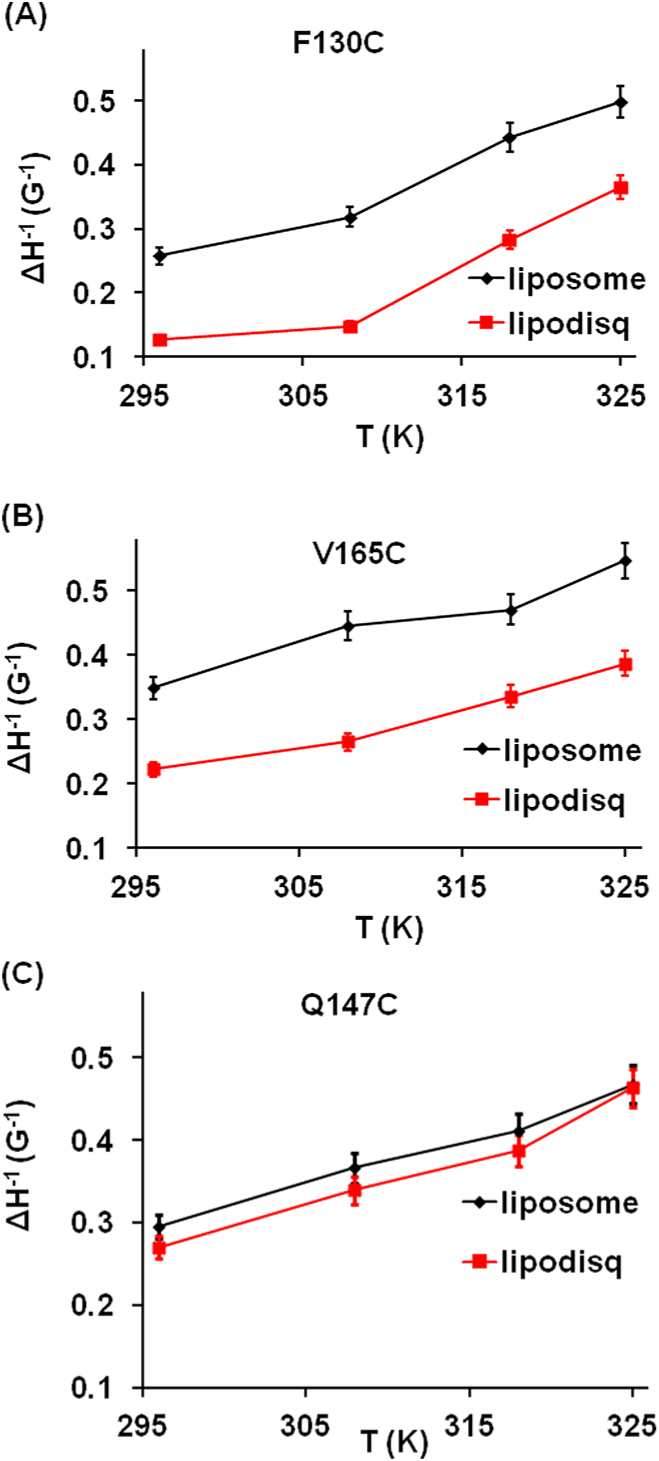
Inverse central line width as a function of temperature for Q1-VSD bearing MTSLs at sites F130C (A), V165C (B), and Q147C(C) calculated from EPR spectra in Figure 2.
In order to further quantify the spin-label side-chain motion of Q1-VSD, nonlinear least-squares (NLSL) MOMD EPR spectral simulations were carried out on the representative EPR spectra of spin-labeled sites F130C and V165C from the Q1-VSD TMD and Q147C from the extracellular domain of Q1-VSD at 297 and 325 K to determine the rotational correlation times and the relative population of the two motional components (see Table 1). The previously described method of simulation under the NLSL program was used for this study.59–61,65,66,75–77 The Zeeman interaction tensors (gxx, gyy, gzz) and hyperfine interaction tensors (Axx, Ayy, Azz) were held constant during the fitting process, and the rotational correlation times (τ) and the relative population of the two components were determined from the best-fit EPR spectra (see Figure 4, red lines). Table 1 shows rotational correlation times, the corresponding population of the slower/rigid (site1), and the more motional/less rigid component (site 2) components of EPR spectra. The EPR spectra were simulated using a two-site fit for the data at both temperatures 297 and 325 K to obtain the best-fit simulations. The simulation results indicated that the side-chain spin-label motion of the site F130C in the transmembrane helix S1 at 297 K in POPC/POPG liposome has correlation times of 23 ns for the slower/rigid component with a relative population of 63% and 1.0 ns for the more motional/less rigid component with a relative population of 37% and 17 ns for the slower/rigid component with a relative population of 64% and 1.1 ns for the more motional/less rigid component with a relative population of 34% for site V165C, while those of the extracellular mutant are 19 ns for the slower/rigid component with a relative population of 58% and 1.2 ns for the more motional/less rigid component with a relative population of 42% for site Q147C. Similarly, the side-chain spin-label motion of Q1-VSD mutants at 297 K in POPC/POPG lipodisq nanoparticles is closely immobilized with longer correlation times of 57 ns for the slower/rigid component with a relative population of 73% and 1.8 ns for the more motional/less rigid component with a relative population of 23% for site F130C and 58 ns for the slower/rigid component with a relative population of 63% and 2.3 ns for the more motional/less rigid component with a relative population of 27% for site V165C, while those of the extracellular mutant are 54 ns for the slower/rigid component with a relative population of 72% and 1.9 ns for the more motional/less rigid component with a relative population of 28% for site Q147C. The simulation results indicated that the rotational correlation times decreased to 1.1 ns for the slower/rigid component with a relative population of 33% and 1.0 ns for the more motional/less rigid component with a relative population of 67% for site F130C and 1.8 ns for the slower/rigid component with a relative population of 77% and 0.8 ns for the more motional/less rigid component with a relative population of 24% for site V165C, while those of the extracellular mutant decreased to 1.6 ns for the slower/rigid component with a relative population of 13% and 1.0 ns for the more motional/less rigid component with a relative population of 87% for the Q147C with increasing temperature to 325 K in POPC/POPG lipid bilayers. Similarly, the rotational correlation times decreased to 34 ns for the slower/rigid component with a relative population of 92% and 0.8 ns for the more motional/less rigid component with a relative population of 8% for site F130C and 18 ns for the slower/rigid component with a relative population of 82% and 1.0 ns for the more motional/less rigid component with a relative population of 18% for site V165C, while those of the extracellular mutant decreased to 15 ns for slower/rigid component with a relative population of 82% and 1.0 ns for the more motional/less rigid component with a relative population of 18% for site Q147C with increasing temperature to 325 K in lipodisq nanoparticles. The rotational correlation times (τ) for Q1-VSD samples in liposomes at 325 K are very close for both spectral components. These spectra could be also fit using a single-component fit, suggesting that both motional components are averaged at a higher temperature of 325 K. The decreasing pattern of rotational correlation times for all of the lipodisq nanoparticle samples is similar to that of the liposome samples as the temperature increases from 297 to 325 K. However, the values of the rotational correlation times are longer for lipodisq nanoparticle samples when compared to that of the Q1-VSD liposome samples. The decrease in the motional behavior of spin-labeled Q1-VSD lipodisq nanoparticle samples is consistent with the literature.65,66
Table 1.
Rotational Correlation Times (τ) and Relative Population (P) of Two Spectral Components of the EPR Spectra (Figure 4) Obtained from the Best-Fit NLSL MOMD Spectral Simulationa
| liposomes | lipodisq | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| sites | temperature (K) | τ, ns (site 1) | τ, ns (site 2) | P, % (site 1) | P, % (site 2) | τ, ns (site 1) | τ, ns (site 2) | P, % (site 1) | P, % (site 2) |
| F130C | 297 | 23 | 1 | 63 | 37 | 57 | 1.8 | 73 | 27 |
| 325 | 1.1 | 1.0 | 33 | 67 | 34 | 0.8 | 92 | 8 | |
| V165C | 297 | 17 | 1.1 | 64 | 36 | 58 | 2.3 | 63 | 37 |
| 325 | 1.8 | 0.8 | 77 | 23 | 18 | 1.0 | 82 | 18 | |
| Q147C | 297 | 19 | 1.5 | 58 | 42 | 54 | 1.9 | 72 | 28 |
| 325 | 1.6 | 1.0 | 13 | 87 | 15 | 1.0 | 82 | 18 | |
The uncertainty in the correlation time (τ) value is ±0.5−2 ns for liposome samples and ±0.5−5 ns for lipodisq samples, respectively. Similarly, the uncertainty in the relative population (P) value is ±3−6% for liposome samples and ±2−4% for lipodisq samples. These uncertainties were estimated based on the multiple batches of sample preparation and multiple simulations runs.
Figure 4.
CW-EPR spectral simulations of Q1-VSD mutants bearing MTSLs at 297 and 325 K at the sites F130C (A), V165C (B), and Q147C (C) using the NLSL MOMD program developed by Freed and co-workers.59,60
EPR spectroscopic studies require homogeneously prepared liposome samples to obtain better quality experimental data for the quantitative information about the structure and dynamic behavior of membrane proteins. A significant CW-EPR spectral line broadening is observed for lipodisq nanoparticle samples when compared to the spectral line shape of the liposome samples. This indicates a decrease in spin-label motion of lipodisq nanoparticle samples in comparison to the liposome samples. The increase in spectral line broadening is consistent with previously published EPR studies of membrane proteins.25,65,66,78,79 The lipodisq nanoparticle system is formed due to the lateral pressure generated during the interaction of the SMA polymer with the lipid acyl chain in the process of isolation of the individual complex with a specific smaller size.25 Recent biophysical studies have suggested that a bracelet of SMA polymer is formed around the lipid membrane where the styrene moieties oriented parallel to the membrane normal interacting directly with the lipid acyl chain and maleic acid groups are oriented in the same direction as the styrene moieties interacting with the lipid head groups.79 The driving force toward the burial of the styrene moieties into the hydrophobic core of the membrane may cause a decrease in the motion of the lipid acyl chain during formation of the lipodisq nanoparticles. Previous studies have also suggested that the SMA polymers may extract the patches of membrane containing well-incorporated membrane protein and stabilizes the specific small sizes in the process of formation of the lipodisq nanoparticles.32 Furthermore, formation of the lipodisq nanoparticles system induces an overall increase in the viscosity of the solution, slowing down the outside spin-label motion (see Figure 2).25,26 Our results also revealed a significant difference in the inverse central line width pattern between the spin-label probe inside the membrane and the spin-label probe outside the membrane (see Figure 3).
DEER Measurements on Q1-VSD in Various Membrane Mimetics.
Double-electron-electron resonance (DEER) spectroscopy coupled with SDSL is a rapidly growing powerful biophysical technique used to measure long-range distances of 18–80 Å for structural studies.13,55,68,80–85 However, application of DEER spectroscopy to spin-labeled membrane proteins is very challenging due to the much shorter phase memory times (Tm) of spin labels in lipid bilayered vesicles when compared to detergent micelles.85–89 The short phase memory times are due to inhomogeneous packing of the spin-labeled protein within the liposomes. This creates local inhomogeneous pockets of high spin concentrations.55 In order to characterize lipodisq nanoparticles for pulsed EPR studies, DEER experiments were performed on the inside probe of Q1-VSD (F123C–S143C) incorporated into various membrane mimetics such as DPC micelles, LMPG micelles, DMPC/DPC bicelles, POPC/POPG lipid bilayers, and POPC/POPG lipodisq nanoparticles for DEER distance measurements. Figure 5 shows DEER data for MTSL spin-labeled Q1-VSD (Phe123/Ser143) samples for 1% LMPG micelles (Figure 5A), 0.5% DPC micelles (Figure 5B), POPC/POPG liposomes (Figure 5C), bicelles (DMPC/DPC) (Figure 5D), and POPC/POPG lipodisq nanoparticles (Figure 5E). The left panel represents the time domain traces, and the right panel shows the distance distributions for Figure 5. All DEER distances derived from the maximum peak intensity and the approximate full width of the distribution at half maxima (fwhm) from DEER distance measurements on the Q1-VSD membrane protein are shown in Table 2. These distances on Q1-VSD are closely matching for each membrane environment within the experimental error and are also in agreement with the previous studies of Q1-VSD.50,90 These results also suggested that the secondary structural conformation of Q1-VSD is closely matching in all of these membrane environments. The fwhm for the lipodisq nanopaticles sample (~10Å) is lower than that of the liposome sample (~16 Å) and comparable to the micells (~9 Å for LMPG samples and ~11 Å for DPC sample) and bicelles (~12 Å) samples. Inspection of DEER data indicated that the signal-to-noise ratio (S/N) of DEER time domain data is significantly improved for lipodisq nanoparticles having well-defined oscillations with longer data acquisition out to 3 μs when compared to the POPC/POPG lipid bilayers (2 μs). This improvement in the S/N of the lipodisq nanoparticle sample is due to the improvement in the value of the phase memory time (Tm) for the lipodisq nanoparticle sample when compared to the liposome sample. Figure 6 shows the representative Tm curves for the dual-spin-labeled Q1-VSD (F123C/S143C) for POPC/POPG lipid bilayers and lipodisq nanoparticles samples. The phase memory time (Tm) data were analyzed in a similar manner to that previously reported in the literature.13,65 Figure 6 clearly indicates the signal intensity at a particular decay time of 3 μs is higher by ~2 fold for the lipodisq nanoparticle sample when compared to the liposome sample. The Tm data sets could not be adequately fit with a single-exponential decay to directly compare Tm values for different samples. However, qualitatively the Tm values of the lipodisq nanoparticles Q1- VSD samples have increased by a factor of ~2 when compared to Q1-VSD in liposomes. The data indicated the Q1-VSD Tm values can be enhanced by ~2-fold in lipodisq nanoparticles when compared to the same spin-labeled Q1-VSD protein in liposomes. DEER measurements can be extended for a longer range of distances with a higher signal-to-noise ratio (S/N) of the time domain DEER data with an improvement in the phase memory time.
Figure 5.
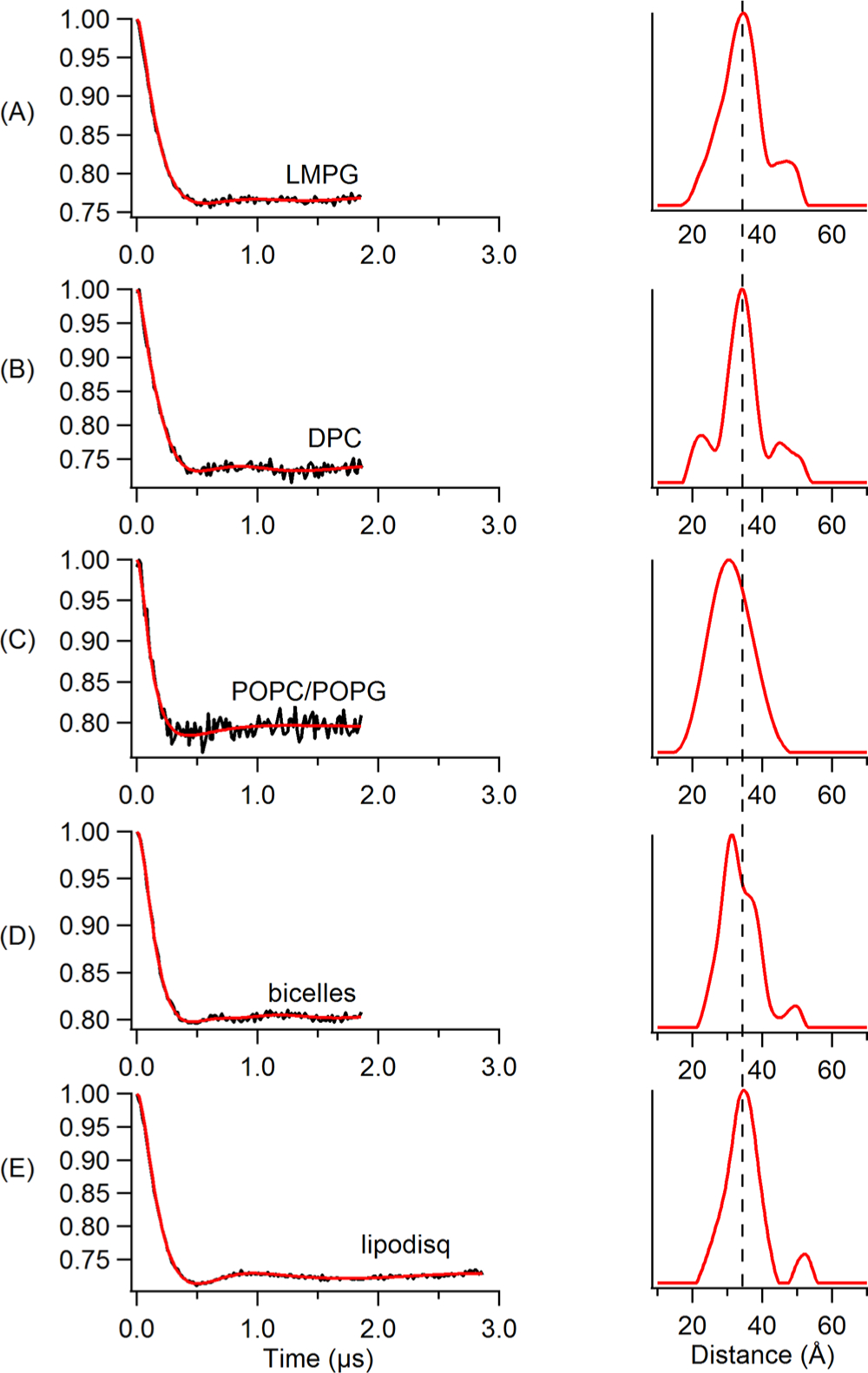
Q-band DEER data of Q1-VSD mutants (Phe123/Ser143) bearing two MTSL spin labels. Background-subtracted dipolar evolutions of the indicated mutants (left) and their corresponding distance probability distributions from Tikhonov regularization (right) for 1% LMPG micelles (A), 0.5% DPC micelles (B), proteoliposomes (POPC/POPG = 3:1) (C), bicelles (DMPC/DPC= 3.2:1) (D), and lipodisq nanoparticles (E).
Table 2.
Four Pulse Major Peak Distance and Approximate Full Width of the Distribution at Half Maxima (fwhm) from DEER Distance Measurements on the Q1-VSD Membrane Proteina
| Q1-VSD double mutant (Phe123/Ser143) | ||
|---|---|---|
| membrane mimetic | distance (Å) | fwhm (Å) |
| LMPG micelles | 34 | −11 |
| DPC micelles | 34 | −9 |
| POPC/POPG lipid bilayers | 31 | −16 |
| DMPC/DPC bicelles | 32 | −12 |
| POPC/POPG lipodisq | 34 | −10 |
The uncertainties for these distances are ±2−4 Å.
Figure 6.
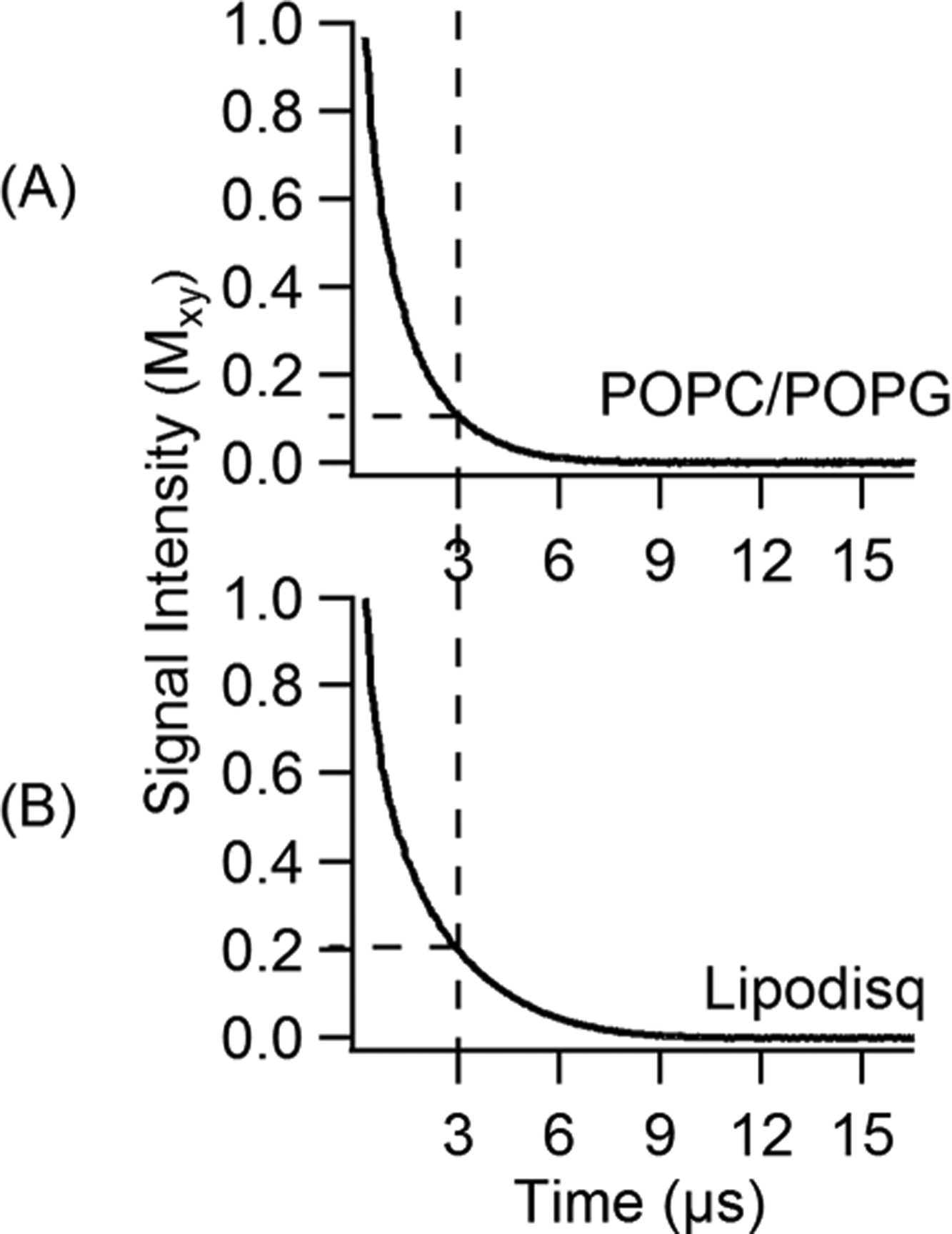
Experimental phase memory curves for dual-spin-labeled Q1-VSD mutants (Phe123/Ser143) bearing two MTSL spin labels for liposomes (POPC/POPG = 3:1) (A) and POPC/POPG lipodisq nanoparticles (B). Tm values are 1.2 ± 0.2 μs for proteoliposomes and 2.0 ± 0.2 μs for lipodisq nanoparticles.
In previous studies, we showed that the lipodisq nanoparticles-based sample preparation of the single-transmembrane KCNE1 protein and the influenza A M2 protein revealed significant improvement in the signal-to-noise ratio (S/N) for DEER time domain data and enhanced the phase memory time by ~2-fold.13,52,91 In this study, EPR measurements on the more complicated membrane protein Q1-VSD with a four-transmembrane domain also showed significant improvement in the signal-to-noise ratio (S/N) for DEER time domain data with the phase memory time increased by ~2-fold for lipodisq nanoparticle samples. A comparison of the most probable DEER distances obtained between double mutants of Q1-VSD (Phe123/Ser143) in various membrane environments including micelles (LMPG and DPC), POPC/POPG liposomes, bicelles, and lipodisq nanoparticles suggested that there is no significant structural perturbation of the protein due to formation of lipodisq nanoparticles.13,52,65,66 There is no significant change in the functional activities of the protein due to formation of lipodisq nanoparticles.26,32,92 The native membrane environment of the protein is preserved during biophysical measurements as there is no significant restriction applied by the SMA polymer to the protein.32 The overall reduction in the motion of the lipid-protein complex also helps to determine the local dynamics of the spin-label probe.76 The optimized lipodisq nanoparticles-based sample preparations reported in this study for the EPR spectroscopic studies are consistent with our previously optimized lipodisq nanoparticles for the SSNMR and TEM and EPR spectroscopic studies.62,64,65 This indicates that the protein molecules are well isolated from each other due to smaller and homogeneous sizes of lipodisq nanoparticles leading to even distribution of the spin labels causing a decrease in the local spin concentration and hence increasing the phase memory time for DEER measurements.13,26,52,64 The increase in the phase memory time improves the S/N of the time domain DEER data and allows longer DEER distance measurements more precisely for structural studies of challenging membrane proteins. The enhancement in phase memory time is consistent with the previously published phase memory time of membrane protein in lipodisq nanoparticles.13,52,65 The increase in EPR spectral line broadening and phase memory time enhancement in lipodisq nanoparticles samples observed in this study may vary depending upon the membrane protein systems and the length and choice of phospholipids used for the study.93 On the basis of our current and previous studies,13,52,62,65,66,91,94 we suggest that the lipodisq nanoparticles system is potentially a good membrane mimetic and useful for EPR studies of membrane proteins. However, further protein systems and lipid systems need to be investigated to generalize the use of lipodisq nanoparticles.
CONCLUSION
A more complicated membrane protein Q1-VSD embedded into lipodisq nanoparticles was characterized using site-directed spin-labeling EPR spectroscopy. The CW-EPR data revealed an increase in spectral line broadening for the spin-labeled Q1-VSD mutations in lipodisq nanoparticles when compared to that in liposomes. Similarly, DEER data revealed a substantial improvement in the signal-to-noise ratio (S/N) for DEER time domain data with the phase memory time increased by ~2-fold for lipodisq nanoparticle samples. The enhancement in the phase memory time allows longer DEER distance measurements more precisely for structural studies of challenging membrane proteins. This lipodisq nanoparticles solubilization method can also be used for a variety of different lipid compositions. The results of this study along with our previous studies provide a library of reference data for the use of lipodisq nanoparicles to advance EPR studies of challenging membrane proteins for solving structure- and dynamics-related problems.
ACKNOWLEDGMENTS
This work was generously supported by the NIGMS/NIH Maximizing Investigator’s Research Award (MIRA), R35 GM126935 award, and a NSF CHE-1807131 award. The pulsed EPR spectrometer was purchased through funding provided by the NSF (MRI-1725502), the Ohio Board of Regents, and Miami University. G.A.L. would also like to acknowledge support from the John W. Steube Professorship.
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jpcb.9b11506
The authors declare no competing financial interest.
Contributor Information
Indra D. Sahu, Natural Science Division, Campbellsville University, Campbellsville, Kentucky 42718, United States; Department of Chemistry and Biochemistry, Miami University, Oxford, Ohio 45056, United States
Gunjan Dixit, Department of Chemistry and Biochemistry, Miami University, Oxford, Ohio 45056, United States.
Warren D. Reynolds, Department of Chemistry and Biochemistry, Miami University, Oxford, Ohio 45056, United States
Ryan Kaplevatsky, Department of Chemistry and Biochemistry, Miami University, Oxford, Ohio 45056, United States.
Benjamin D. Harding, Department of Chemistry and Biochemistry, Miami University, Oxford, Ohio 45056, United States
Colleen K. Jaycox, Department of Chemistry and Biochemistry, Miami University, Oxford, Ohio 45056, United States
Robert M. McCarrick, Department of Chemistry and Biochemistry, Miami University, Oxford, Ohio 45056, United States
Gary A. Lorigan, Department of Chemistry and Biochemistry, Miami University, Oxford, Ohio 45056, United States;.
REFERENCES
- (1).Overington JP; Al-Lazikani B; Hopkins AL How many drug targets are there? Nat. Rev. Drug Discovery 2006, 5, 993–996. [DOI] [PubMed] [Google Scholar]
- (2).Sanders CR; Myers JK Disease-related misassembly of membrane proteins. Annu. Rev. Biophys. Biomol. Struct 2004, 33, 25–51. [DOI] [PubMed] [Google Scholar]
- (3).Hubbell WL; McHaourab HS; Altenbach C; Lietzow MA Watching proteins move using site-directed spin labeling. Structure 1996, 4, 779–783. [DOI] [PubMed] [Google Scholar]
- (4).Klug CS; Feix JB Methods and applications of site-directed spin labeling EPR spectroscopy. Methods Cell Biol 2008, 84, 617–658. [DOI] [PubMed] [Google Scholar]
- (5).Sahu ID; McCarrick RM; Lorigan GA Use of electron paramagnetic resonance to solve biochemical problems. Biochemistry 2013, 52, 5967–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Sachs JN; Engelman DM Introduction to the membrane protein reviews: The interplay of structure, dynamics, and environment in membrane protein function. Annu. Rev. Biochem 2006, 75, 707–712. [DOI] [PubMed] [Google Scholar]
- (7).Prive GG Detergents for the stabilization and crystallization of membrane proteins. Methods 2007, 41, 388–397. [DOI] [PubMed] [Google Scholar]
- (8).Garavito RM; Ferguson-Miller S Detergents as tools in membrane biochemistry. J. Biol. Chem 2001, 276, 32403–32406. [DOI] [PubMed] [Google Scholar]
- (9).Seddon AM; Curnow P; Booth PJ Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta, Biomembr 2004, 1666, 105–117. [DOI] [PubMed] [Google Scholar]
- (10).Park SH; Opella SJ Triton X-100 as the ″short-chain lipid″ improves the magnetic alignment and stability of membrane proteins in phosphatidylcholine bilayers for oriented-sample solid-state NMR spectroscopy. J. Am. Chem. Soc 2010, 132, 12552–12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).De Angelis AA; Opella SJ Bicelle samples for solid-state NMR of membrane proteins. Nat. Protoc 2007, 2, 2332–2338. [DOI] [PubMed] [Google Scholar]
- (12).Bayburt TH; Sligar SG Membrane protein assembly into Nanodiscs. FEBS Lett 2010, 584, 1721–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Sahu ID; McCarrick RM; Troxel KR; Zhang R; Smith HJ; Dunagan MM; Swartz MS; Rajan PV; Kroncke BM; Sanders CR; Lorigan GA DEER EPR measurements for membrane protein structures via bifunctional spin labels and lipodisq nanoparticles. Biochemistry 2013, 52, 6627–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Bayburt TH; Sligar SG Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci 2003, 12, 2476–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Raschle T; Hiller S; Etzkorn M; Wagner G Nonmicellar systems for solution NMR spectroscopy of membrane proteins. Curr. Opin. Struct. Biol 2010, 20, 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Geertsma ER; Nik Mahmood NAB; Schuurman-Wolters GK; Poolman B Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc 2008, 3, 256–266. [DOI] [PubMed] [Google Scholar]
- (17).Fang G; Friesen R; Lanfermeijer F; Hagting A; Poolman B; Konings WN Manipulation of activity and orientation of membrane-reconstituted di-tripeptide transport protein DtpT of Lactococcus lactis. Mol. Membr. Biol 1999, 16, 297–304. [DOI] [PubMed] [Google Scholar]
- (18).Lau T-L; Kim C; Ginsberg MH; Ulmer TS The structure of the integrin alpha IIb beta 3 transmembrane complex explains integrin transmembrane signalling. EMBO J 2009, 28, 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lee D; Walter KFA; Brueckner A-K; Hilty C; Becker S; Griesinger C Bilayer in small bicelles revealed by lipid-protein interactions using NMR spectroscopy. J. Am. Chem. Soc 2008, 130, 13822–13823. [DOI] [PubMed] [Google Scholar]
- (20).Sanders CR; Prosser RS Bicelles: a model membrane system for all seasons? Structure 1998, 6, 1227–1234. [DOI] [PubMed] [Google Scholar]
- (21).Lu Z; Van Horn WD; Chen J; Mathew S; Zent R; Sanders CR Bicelles at Low Concentrations. Mol. Pharmaceutics 2012, 9, 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Vold RR; Prosser RS Magnetically oriented phospholipid bilayered micelles for structural studies of polypeptides. Does the ideal bicelle exist? J. Magn. Reson., Ser. B 1996, 113, 267–271. [Google Scholar]
- (23).Duerr UHN; Gildenberg M; Ramamoorthy A The magic of bicelles lights up membrane protein structure. Chem. Rev 2012, 112, 6054–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zou P; McHaourab HS Increased sensitivity and extended range of distance measurements in spin-labeled membrane proteins: Q-band double electron-electron resonance and nanoscale bilayers. Biophys. J 2010, 98, L18–L20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Orwick MC; Judge PJ; Procek J; Lindholm L; Graziadei A; Engel A; Grobner G; Watts A Detergent-free formation and physicochemical characterization of nanosized lipid-polymer complexes: lipodisq. Angew. Chem., Int. Ed 2012, 51, 4653–4657. [DOI] [PubMed] [Google Scholar]
- (26).Orwick-Rydmark M; Lovett JE; Graziadei A; Lindholm L; Hicks MR; Watts A Detergent-free incorporation of a seven-transmembrane receptor protein into nanosized bilayer lipodisq particles for functional and biophysical studies. Nano Lett 2012, 12, 4687–4692. [DOI] [PubMed] [Google Scholar]
- (27).Knowles TJ; Finka R; Smith C; Lin Y-P; Dafforn T; Overduin M Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc 2009, 131, 7484–7485. [DOI] [PubMed] [Google Scholar]
- (28).Jamshad M; Lin Y-P; Knowles TJ; Parslow RA; Harris C; Wheatley M; Poyner DR; Bill RM; Thomas ORT; Overduin M; Dafforn TR Surfactant-free purification of membrane proteins with intact native membrane environment. Biochem. Soc. Trans 2011, 39, 813–818. [DOI] [PubMed] [Google Scholar]
- (29).Dorr JM; Scheidelaar S; Koorengevel MC; Dominguez JJ; Schafer M; van Walree CA; Killian JA The styrene-maleic acid copolymer: a versatile tool in membrane research. Eur. Biophys. J 2016, 45, 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Scheidelaar S; Koorengevel MC; Pardo JD; Meeldijk JD; Breukink E; Killian JA Molecular model for the solubilization of membranes into nanodisks by styrene maleic acid copolymers. Biophys. J 2015, 108, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Long AR; O’Brien CC; Malhotra K; Schwall CT; Albert AD; Watts A; Alder NN A detergent-free strategy for the reconstitution of active enzyme complexes from native biological membranes into nanoscale discs. BMC Biotechnol 2013, 13, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Logez C; Damian M; Legros C; Dupre C; Guery M; Mary S; Wagner R; M’Kadmi C; Nosjean O; Fould B; Marie J; Fehrentz J-A; Martinez J; Ferry G; Boutin JA; Baneres J-L Detergent-free isolation of functional G protein-coupled receptors into nanometric lipid particles. Biochemistry 2016, 55, 38–48. [DOI] [PubMed] [Google Scholar]
- (33).Gulati S; Jamshad M; Knowles TJ; Morrison KA; Downing R; Cant N; Collins R; Koenderink JB; Ford RC; Overduin M; Kerr ID; Dafforn TR; Rothnie AJ Detergent-free purification of ABC (ATP-binding-cassette) transporters. Biochem. J 2014, 461, 269–278. [DOI] [PubMed] [Google Scholar]
- (34).Shi L; Shen Q-T; Kiel A; Wang J; Wang H-W; Melia TJ; Rothman JE; Pincet F SNARE proteins: one to fuse and three to keep the nascent fusion pore open. Science 2012, 335, 1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Jespersen T; Grunnet M; Olesen SP The KCNQ1 potassium channel: From gene to physiological function. Physiology 2005, 20, 408–416. [DOI] [PubMed] [Google Scholar]
- (36).Robbins J KCNQ potassium channels: physiology, patho-physiology, and pharmacology. Pharmacol. Ther 2001, 90, 1–19. [DOI] [PubMed] [Google Scholar]
- (37).Peroz D; Rodriguez N; Choveau F; Baro I; Merot J; Loussouarn G Kv7.1 (KCNQ1) properties and channelopathies. J. Physiol 2008, 586, 1785–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Sun QM; Song K; Shen XZ; Cai Y The Association between KCNQ1 Gene Polymorphism and Type 2 Diabetes Risk: A Meta-Analysis. PLoS One 2012, 7, e48578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Unoki H; Takahashi A; Kawaguchi T; Hara K; Horikoshi M; Andersen G; Ng DPK; Holmkvist J; Borch-Johnsen K; Jorgensen T; et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in east asian and european populations. Nat. Genet 2008, 40, 1098–1102. [DOI] [PubMed] [Google Scholar]
- (40).Hedley PL; Jorgensen P; Schlamowitz S; Wangari R; Moolman-Smook J; Brink PA; Kanters JK; Corfield VA; Christiansen M The genetic basis of long QT and short QT syndromes: a mutation update. Hum. Mutat 2009, 30, 1486–1511. [DOI] [PubMed] [Google Scholar]
- (41).Kang C; Tian C; Sonnichsen FD; Smith JA; Meiler J; George ALJ; Vanoye CG; Kim HJ; Sanders CR Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel. Biochemistry 2008, 47, 7999–8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Tian C; Vanoye CG; Kang C; Welch RC; Kim HJ; George AL; Sanders CR Preparation, functional characterization, and NMR studies of human KCNE1, a voltage-gated potassium channel accessory subunit associated with deafness and long QT syndrome. Biochemistry 2007, 46, 11459–11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Coey AT; Sahu ID; Gunasekera TS; Troxel KR; Hawn JM; Swartz MS; Wickenheiser MR; Reid RJ; Welch RC; Vanoye CG; Kang CB; Sanders CR; Lorigan GA Reconstitution of KCNE1 into lipid bilayers: comparing the structural, dynamic, and activity differences in micelle and vesicle environments. Biochemistry 2011, 50, 10851–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zhao J; Blunck R The isolated voltage sensing domain of the Shaker potassium channel forms a voltage-gated cation channel. eLife 2016, 5, e18130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Chakrapani S; Cuello LG; Cortes DM; Perozo E Structural dynamics of an isolated voltage-sensor domain in a lipid bilayer. Structure 2008, 16, 398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Mishina Y; Mutoh M; Knopfel T Transfer of Kv3.1 voltage sensor features to the isolated Ci-VSP voltage-sensing domain. Biophys. J 2012, 103, 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Borjesson SI; Elinder F Structure, function, and modification of the voltage sensor in voltage-gated ion channels. Cell Biochem. Biophys 2008, 52, 149–174. [DOI] [PubMed] [Google Scholar]
- (48).Lau CHY; King GF; Mobli M Molecular basis of the interaction between gating modifier spider toxins and the voltage sensor of voltage-gated ion channels. Sci. Rep 2016, 6, 34333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Li Q; Wanderling S; Sompornpisut P; Perozo E Structural basis of lipid-driven conformational transitions in the KvAP voltage-sensing domain. Nat. Struct. Mol. Biol 2014, 21, 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Peng DG; Kim JH; Kroncke BM; Law CL; Xia Y; Droege KD; Van Horn WD; Vanoye CG; Sanders CR Purification and structural study of the voltage-sensor domain of the human KCNQ1 potassium ion channel. Biochemistry 2014, 53, 2032–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Barrett PJ; Song Y; Van Horn WD; Hustedt EJ; Schafer JM; Hadziselimovic A; Beel AJ; Sanders CR The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science 2012, 336, 1168–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Sahu ID; Kroncke BM; Zhang R; Dunagan MM; Smith HJ; Craig A; McCarrick RM; Sanders CR; Lorigan GA Structural investigation of the transmembrane domain of KCNE1 in proteoliposomes. Biochemistry 2014, 53, 6392–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Zhang R; Sahu ID; Gibson KR; Muhammad N; Bali AP; Comer RG; Liu L; Craig AF; McCarrick RM; Dabney-Smith C; Sanders CR; Lorigan GA Development of electron spin echo envelope modulation (ESEEM) spectroscopy to probe the secondary structure of recomninant membrane proteins in lipid bilayer. Protein Sci 2015, 24, 1707–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Nolandt OV; Walther TH; Grage SL; Ulrich AS Magnetically oriented dodecylphosphocholine bicelles for solid-state NMR structure analysis. Biochim. Biophys. Acta, Biomembr 2012, 1818, 1142–1147. [DOI] [PubMed] [Google Scholar]
- (55).Jeschke G DEER distance measurements on proteins. Annu. Rev. Phys. Chem 2012, 63, 419–446. [DOI] [PubMed] [Google Scholar]
- (56).Feldmann EA; Ni S; Sahu ID; Mishler CH; Risser DD; Murakami JL; Tom SK; McCarrick RM; Lorigan GA; Tolbert BS; Callahan SM; Kennedy MA Evidence for direct binding between HetR from anabaena sp PCC 7120 and PatS-5. Biochemistry 2011, 50, 9212–9224. [DOI] [PubMed] [Google Scholar]
- (57).Jeschke G; Chechik V; Ionita P; Godt A; Zimmermann H; Banham J; Timmel CR; Hilger D; Jung H DeerAnalysis2006 - a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson 2006, 30, 473–498. [Google Scholar]
- (58).Chiang YW; Borbat PP; Freed JH The determination of pair distance distributions by pulsed ESR using tikhonov regularization. J. Magn. Reson 2005, 172, 279–295. [DOI] [PubMed] [Google Scholar]
- (59).Schneider DJ; Freed JH Calculating slow motional magnetic resonance spectra: a user’s guide In Biological Magnetic Resonance; Berlinger LJ, Ed.; Plenum Publishing: New York, 1989. [Google Scholar]
- (60).Budil DE; Lee S; Saxena S; Freed JH Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified levenberg-marquardt algorithm. J. Magn. Reson., Ser. A 1996, 120, 155–189. [Google Scholar]
- (61).Sahu ID; Craig AF; Dunagan MM; Troxel KR; Zhang R; Meiberg AG; Harmon CN; McCarrick RM; Kroncke BM; Sanders CR; Lorigan GA Probing structural dynamics and topology of the KCNE1 membrane protein in lipid bilayers via site-directed spin labeling and electron paramagnetic resonance spectroscopy. Biochemistry 2015, 54, 6402–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Zhang R; Sahu ID; Liu L; Osatuke A; Comer RG; Dabney-Smith C; Lorigan GA Characterizing the structure of lipodisq nanoparticles for membrane protein spectroscopic studies. Biochim. Biophys. Acta, Biomembr 2015, 1848, 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Craig AF; Clark EE; Sahu ID; Zhang R; Frantz ND; Al-Abdul-Wahid S; Dabney-Smith C; Konkolewicz D; Lorigan GA Tuning the size of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) using RAFT polymerization for biophysical studies. Biochim. Biophys. Acta, Biomembr 2016, 1858, 2931–2939. [DOI] [PubMed] [Google Scholar]
- (64).Zhang R; Sahu ID; Bali AP; Dabney-Smith C; Lorigan GA Characterization of the structure of lipodisq nanoparticles in the presence of KCNE1 by dynamic light scattering and transmission electron microscopy. Chem. Phys. Lipids 2017, 203, 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Sahu ID; Zhang R; Dunagan MM; Craig AF; Lorigan GA Characterization of KCNE1 inside lipodisq nanoparticles for EPR spectroscopic studies of membrane proteins. J. Phys. Chem. B 2017, 121, 5312–5321. [DOI] [PubMed] [Google Scholar]
- (66).Sahu ID; Craig AF; Dunagum MM; McCarrick RM; Lorigan GA Characterization of bifunctional spin labels for investigating the structural and dynamic properties of membrane proteins using EPR spectroscopy. J. Phys. Chem. B 2017, 121, 9185–9195. [DOI] [PubMed] [Google Scholar]
- (67).Song Y; Hustedt EJ; Brandon S; Sanders CR Competition Between Homodimerization and Cholesterol Binding to the C99 Domain of the Amyloid Precursor Protein. Biochemistry 2013, 52, 5051–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Sahu ID; Lorigan GA Site-Directed Spin Labeling EPR for Studying Membrane Proteins. BioMed Res. Int 2018, 2018, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Jeschke G; Bender A; Schweikardt T; Panek G; Decker H; Paulsen H Localization of the N-terminal domain in light-harvesting chlorophyll a/b protein by EPR measurements. J. Biol. Chem 2005, 280, 18623–18630. [DOI] [PubMed] [Google Scholar]
- (70).Mchaourab HS; Perozo E; Berliner L; Eaton G; Eaton S Determination of protein folds and conformational dynamics using spin-labeling EPR spectroscopy Biological Magnetic Resonance; Springer US: Boston, MA, 2002; Vol. 19, pp 185–247. [Google Scholar]
- (71).Perozo E; Cortes DM; Cuello LG Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat. Struct. Biol 1998, 5, 459–469. [DOI] [PubMed] [Google Scholar]
- (72).Vasquez V; Sotomayor M; Marien Cortes D; Roux B; Schulten K; Perozo E Three-dimensional architecture of membrane-embedded MscS in the closed conformation. J. Mol. Biol 2008, 378, 55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Tkachev Y; Timofeev V Uniform EPR spectra analysis of spin-labeled macromolecules by temperature and viscosity dependences. Nitroxides-Theory, Experiment and Applications; InTech, 2012; pp 285–314. [Google Scholar]
- (74).Fan W; Evans RM Turning up the heat on membrane fluidity. Cell 2015, 161, 962–963. [DOI] [PubMed] [Google Scholar]
- (75).Toledo Warshaviak D; Khramtsov VV; Cascio D; Altenbach C; Hubbell WL Structure and dynamics of an imidazoline nitroxide side chain with strongly hindered internal motion in proteins. J. Magn. Reson 2013, 232, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Fleissner MR; Bridges MD; Brooks EK; Cascio D; Kalai T; Hideg K; Hubbell WL Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 16241–16246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).McCaffrey JE; James ZM; Svensson B; Binder BP; Thomas DD A bifunctional spin label reports the structural topology of phospholamban in magnetically-aligned bicelles. J. Magn. Reson 2016, 262, 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Stepien P; Polit A; Wisniewska-Becker A Comparative EPR studies on lipid bilayer properties in nanodiscs and liposomes. Biochim. Biophys. Acta, Biomembr 2015, 1848, 60–66. [DOI] [PubMed] [Google Scholar]
- (79).Jamshad M; Grimard V; Idini I; Knowles TJ; Dowle MR; Schofield N; Sridhar P; Lin Y; Finka R; Wheatley M; Thomas ORT; Palmer RE; Overduin M; Govaerts C; Ruysschaert J-M; Edler KJ; Dafforn TR Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Res 2015, 8, 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Borbat PP; McHaourab HS; Freed JH Protein structure determination using long-distance constraints from double-quantum coherence ESR: Study of T4 lysozyme. J. Am. Chem. Soc 2002, 124, 5304–5314. [DOI] [PubMed] [Google Scholar]
- (81).Jeschke G; Polyhach Y Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys. Chem. Chem. Phys 2007, 9, 1895–1910. [DOI] [PubMed] [Google Scholar]
- (82).Jao CC; Hegde BG; Chen J; Haworth IS; Langen R Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 19666–19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Mokdad A; Herrick DZ; Kahn AK; Andrews E; Kim M; Cafiso DS Ligand-induced structural changes in the escherichia coli ferric citrate transporter reveal modes for regulating protein-protein interactions. J. Mol. Biol 2012, 423, 818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Schiemann O; Cekan P; Margraf D; Prisner TF; Sigurdsson ST Relative orientation of rigid nitroxides by PELDOR: beyond distance measurements in nucleic acids. Angew. Chem., Int. Ed 2009, 48, 3292–3295. [DOI] [PubMed] [Google Scholar]
- (85).Endeward B; Butterwick JA; MacKinnon R; Prisner TF Pulsed electron-electron double-resonance determination of spin-label distances and orientations on the tetrameric potassium ion channel KcsA. J. Am. Chem. Soc 2009, 131, 15246–15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Jeschke G; Wegener C; Nietschke M; Jung H; Steinhoff HJ Interresidual distance determination by four-pulse double electron-electron resonance in an integral membrane protein: the Na+/proline transporter PutP of Escherichia coli. Biophys. J 2004, 86, 2551–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Hilger D; Jung H; Padan E; Wegener C; Vogel KP; Steinhoff HJ; Jeschke G Assessing oligomerization of membrane proteins by four-pulse DEER: pH-dependent dimerization of NhaA Na+/H+ antiporter of E-coli. Biophys. J 2005, 89, 1328–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Hilger D; Polyhach Y; Padan E; Jung H; Jeschke G High-resolution structure of a Na+/H+ antiporter dimer obtained by pulsed election paramagnetic resonance distance measurements. Biophys. J 2007, 93, 3675–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Xu Q; Ellena JF; Kim M; Cafiso DS Substrate-dependent unfolding of the energy coupling motif of a membrane transport protein determined by double electron-electron resonance. Biochemistry 2006, 45, 10847–10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Huang H; Kuenze G; Smith JA; Taylor KC; Duran AM; Hadziselimovic A; Meiler J; Vanoye CG; George AL; Sanders CR Mechanisms of KCNQ1 channel dysfunction in long QT syndrome involving voltage sensor domain mutations. Science Advances 2018, 4, eaar2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Kim SS; Upshur MA; Saotome K; Sahu ID; McCarrick RM; Feix JB; Lorigan GA; Howard KP Cholesterol-dependent conformational exchange of the C-terminal domain of the influenza A M2 protein. Biochemistry 2015, 54, 7157–7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Morrison KA; Akram A; Mathews A; Khan ZA; Patel JH; Zhou CM; Hardy DJ; Moore-Kelly C; Patel R; Odiba V; Knowles TJ; Javed MUH; Chmel NP; Dafforn TR; Rothnie AJ Membrane protein extraction and purification using styrene-maleic acid (SMA) copolymer: effect of variations in polymer structure. Biochem. J 2016, 473, 4349–4360. [DOI] [PubMed] [Google Scholar]
- (93).Kandasamy SK; Larson RG Molecular dynamics simulations of model trans-membrane peptides in lipid bilayers: A systematic investigation of hydrophobic mismatch. Biophys. J 2006, 90, 2326–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Zhang RF; Sahu ID; Bali AP; Dabney-Smith C; Lorigan GA Characterization of the structure of lipodisq nanoparticles in the presence of KCNE1 by dynamic light scattering and transmission electron microscopy. Chem. Phys. Lipids 2017, 203, 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]


